Abstract
Background
Leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5) is a cancer stem cell (CSC) marker of colorectal cancer and may be a CSC marker of other cancer types. Few studies have been conducted on LGR5 expression in extrahepatic cholangiocarcinoma (ECC).
Methods
We analyzed LGR5 expression using RNAscope, a highly sensitive RNA in situ hybridization technique. Fifty-three ECCs were selected from the medical archives at Shinshu University Hospital and analyzed using a tissue microarray. LGR5 expression levels were divided into expression and no expression groups. LGR5 expression and clinicopathological characteristics were analyzed.
Results
Among 25 cases, no LGR5-positive dots were identified. Among 28 cases, some LGR5-positive dots were observed in carcinoma cells, together with a wide range of LGR5-positive cells. LGR5 expression was conspicuous in glandular duct formations. Well- to moderately differentiated types showed significantly higher LGR5 expression than the poorly differentiated type (p = 0.0268). LGR5 expression was associated with good overall survival (p = 0.0219) and good disease-free survival (DFS) (p = 0.0228). High LGR5 expression was associated with well- to moderately-differentiated types, indicating a favorable prognosis. In terms of DFS, multivariate analysis showed that high LGR5 expression was an independent favorable prognostic factor (p = 0.0397).
Conclusions
These findings suggest that LGR5 is a promising, novel prognostic marker.
Keywords: Leucine-rich repeat-containing G protein-coupled receptor 5, Extrahepatic cholangiocarcinoma, RNA in situ hybridization
Background
Extrahepatic cholangiocarcinoma (ECC) is a rare subtype of cholangiocarcinoma that is increasing in frequency and lethality [1]. ECC progresses asymptomatically in its early stages, making it difficult to diagnose [2]. Surgery is the only curative option for ECC patients, but surgery is not possible for many cases [2]. Systemic chemotherapy is the main treatment for unresectable ECC. However, the effect is limited and does not improve overall survival (OS) [3].
Signaling pathways that regulate self-renewal and differentiation of cancer stem cells are under intense investigation to develop effective therapeutic strategies for cancer [4]. Surface markers of cancer stem cells are being assessed as potential therapeutic targets [4]. We have focused on leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5). LGR5 is structurally similar to members of the G protein-coupled receptor family, which consists of seven transmembrane domain proteins. G protein-coupled receptors function as receptors for various classes of ligands, including peptide hormones and chemokines [5]. Lgr5 is a stem cell marker of the colon [6]. LGR5 is also a stem cell marker of colorectal cancer [7]. LGR5 has recently been analyzed as a novel marker of various human cancers, including colorectal [7], gastric [8], liver [9], and breast [10] cancers. However, few studies have investigated LGR5 expression in the biliary system, including extrahepatic bile ducts. LGR5 is a potential prognostic marker of ECC and may be a therapeutic target. Most analyses of LGR5 expression in clinical specimens have been conducted by immunohistochemistry [11], but no reliable antibodies exist [12–14]. Therefore, we analyzed LGR5 expression in ECC by RNAscope, a highly sensitive RNA in situ hybridization method, for comparison with clinicopathological data.
Materials and methods
Patients and materials
We identified 61 ECC cases that underwent surgical resection between January 2015 and December 2021 at Shinshu University Hospital (Matsumoto, Japan). Two cases were excluded because of a lack of clinical data. Six cases were excluded because they were negative for the positive control (housekeeping gene). Therefore, 53 ECC cases remained as suitable candidates for the analysis. The 8th edition of the UICC TNM Classification [15], 5th edition of the World Health Organization classification [16], and General Rules for Clinical and Pathological Studies on Cancer of the Biliary Tree were used for pathological evaluation of ECC [17]. The TNM stage was divided into two groups, I-II and III-V, and the depth of infiltration was divided into T1-T2 and T3-T4, for subgroup analysis as reported previously [18–20]. Histological features of all specimens were confirmed by two pathologists (TU and MI). This study was approved by the ethics committee of Shinshu University, Japan (Approval No. 5836). All researchers were blinded to the patients’ data during experimental analysis.
Histopathology
Paraffin-embedded tissue blocks containing sufficient representative tumor areas from all cases were selected for tissue microarray (TMA) preparation. TMAs included invasive fronts. Tissue cores were punched out from each donor tumor block using thin-walled 3 mm stainless steel needles (Azumaya Medical Instruments Inc., Tokyo, Japan). The cores were arrayed in a recipient paraffin block. Serial Sect. 4 μm in thickness were cut from the blocks and stained with hematoxylin-eosin (HE).
LGR5 RNA in situ hybridization
LGR5 mRNA in TMAs was analyzed using the newly developed RNAscope kit (Advanced Cell Diagnostics, Hayward, CA, USA) in accordance with the manufacturer’s instructions using unstained sample tissue sections. The detailed procedure has been described in a previous study [21]. Mm-PPIB (ACD-313902) was used as a positive control. DapB (ACD-310043) was used as a negative control. Normal human colon was used as a positive control tissue. LGR5 is expressed at the crypt base in the normal human colon. Positive staining is indicated by brown punctate dots in the nucleus and/or cytoplasm. LGR5 expression levels were quantified by to a five-grade scoring system as described previously [22]: 0 = no staining; 1 = one to three dots per cell; 2 = four to 10 dots per cell and no or very few dot clusters; 3 = > 10 dots per cell and < 10% positive cells overall; 4 = > 10 dots per cell and > 10% positive cells with dot clusters. The overall score for each patient was evaluated in a high-power field (×400 magnification). Furthermore, LGR5 mRNA expression was categorized into no expression (score 0) and expression (scores 1–4). We then analyzed the relationship between LGR5 expression and clinicopathological data involving ECC patient prognosis.
Statistical analysis
Statistical analysis was performed using JMP version 13 (SAS Institute Japan, Tokyo, Japan). Categorical variables were compared using Fisher’s exact test. A p value < 0.05 was considered statistically significant. OS and disease-free survival (DFS) of ECC patients was calculated using the Kaplan–Meier method. Differences were compared using the log-rank test. Univariate and multivariate analyses of prognostic factors were performed using the Cox proportional hazard regression model.
Results
LGR5 expression in ECC
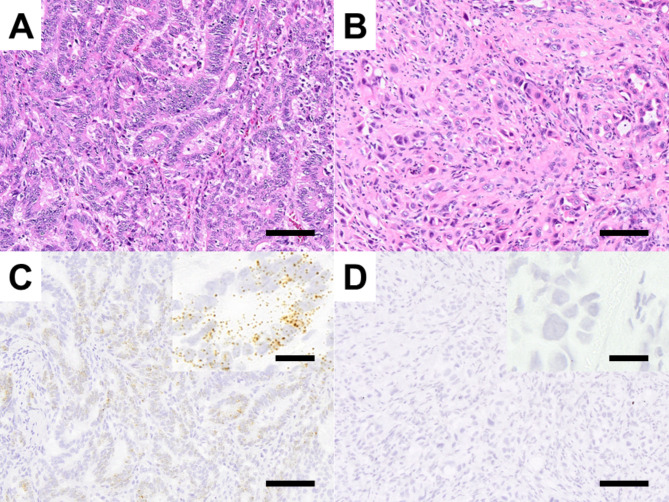
Among 28 cases, some LGR5-positive dots were observed in carcinoma cells (Fig. 1A, C), revealing a wide range of LGR5-positive cells. Conversely, among 25 cases, no positive dots were observed in cancer cells (Fig. 1B, D).
Fig. 1.
LGR5 expression in ECC. Representative features of HE-stained tissues (A) and RNAscope analysis of the LGR5 expression group (B). Representative features of HE-stained tissues (C) and RNAscope analysis of the no LGR5 expression group (D). Bar indicates 100 μm (magnified panel = 20 μm)
Relationships between LGR5 expression and clinicopathological characteristics
The clinicopathological characteristics of ECC patients are shown in Table 1. Well- to moderately differentiated adenocarcinoma showed significantly higher LGR5 expression than poorly differentiated adenocarcinoma (p = 0.0268). No other significant differences were found in age, sex, vascular invasion, depth of infiltration, lymph node metastasis, or TNM stage between the no LGR5 expression and LGR5 expression groups.
Table 1.
LGR5 expression and clinicopathological characteristics of ECC patients
| LGR5 | ||||
|---|---|---|---|---|
| Factors | n | No expression (n = 25) | Expression (n = 28) | p-value |
| Age | 0.4145 | |||
| =<71 years | 29 | 12 | 17 | |
| >71 years | 24 | 13 | 11 | |
| Sex | 1 | |||
| Female | 20 | 9 | 11 | |
| Male | 33 | 16 | 17 | |
| Vascular invasion | 0.5857 | |||
| Absent | 26 | 11 | 15 | |
| Present | 27 | 14 | 13 | |
| Differentiation | 0.0268 | |||
| Wel-Mod | 24 | 7 | 17 | |
| Por | 29 | 18 | 11 | |
| Depth of infiltration | 0.7688 | |||
| T1-T2 | 36 | 16 | 20 | |
| T3-T4 | 17 | 9 | 8 | |
| Lymph node metastasis | 1 | |||
| Absent | 29 | 14 | 15 | |
| Present | 24 | 11 | 13 | |
| TNM stage | 0.2725 | |||
| I-II | 32 | 13 | 19 | |
| III-IV | 21 | 12 | 9 | |
Wel: well-differentiated type, Mod: moderately differentiated type, Por: poorly differentiated type
Prognostic value of LGR5 in ECC
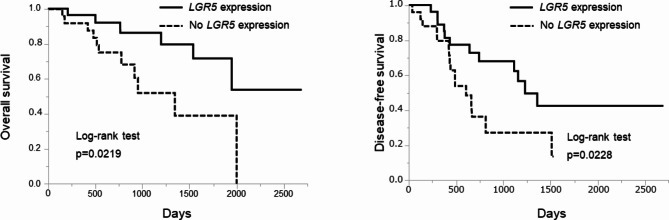
The prognostic value of LGR5 expression in ECC was determined using the Kaplan–Meier method and log-rank test (Fig. 2). High LGR5 expression was associated with good OS (log-rank test, p = 0.0219) and good DFS (log-rank test, p = 0.0228).
Fig. 2.
Prognostic value of LGR5 expression in ECC determined by Kaplan–Meier analysis. LGR5 expression was associated with good OS (log-rank test, p = 0.0219) and good DFS (log-rank test, p = 0.0228)
We evaluated the relationship of clinicopathological factors and LGR5 expression with OS using a Cox proportional hazard regression model (Table 2). In the univariate analysis, the presence of lymph node metastases (p = 0.0313), an advanced TNM stage (p = 0.0046), and no LGR5 expression (p = 0.0286) were significantly associated with worse OS. Variables that were statistically significant in the univariate analysis were entered into the multivariate analysis. The multivariate analysis showed that there was no significant independent prognostic factor, including LGR5 expression.
Table 2.
Univariate and multivariate analyses of prognostic factors associated with OS of ECC patients
| Factors | Univariate analysis (p-value) | Multivariate analysis (p-value) |
|---|---|---|
| Age (≤ 71 vs. >71 years) | 0.2812 | |
| Sex (Female vs. Male) | 0.2525 | |
| Differentiation (Wel–Mod vs. Por) | 0.1887 | |
| Vascular invasion (Absent vs. Present) | 0.1471 | |
| Depth of infiltration (T1-T2 vs. T3-T4) | 0.0903 | |
| Lymph node metastasis (absent vs. present) | 0.0313 | 0.7407 |
| TNM stage (I–II vs. III–V) | 0.0046 | 0.0839 |
| LGR5 (Expression vs. No expression) | 0.0286 | 0.0897 |
Wel: well-differentiated type, Mod: moderately differentiated type, Por: poorly differentiated type
We also evaluated the relationship of clinicopathological factors and LGR5 expression with DFS using a Cox proportional hazard regression model (Table 3). In the univariate analysis, the presence of lymph node metastases (p < 0.0001), an advanced TNM stage (p = 0.0020), and low LGR5 expression (p = 0.0267) were significantly associated with worse DFS. Variables that were statistically significant in the univariate analysis were entered into the multivariate analysis. The multivariate analysis showed that the presence of lymph node metastases and LGR5 expression were independent prognostic factors (p = 0.0013 and p = 0.0101, respectively).
Table 3.
Univariate and multivariate analyses of prognostic factors associated with DFS of ECC patients
| Factors | Univariate analysis (p-value) | Multivariate analysis (p-value) |
|---|---|---|
| Age (≤ 71 vs. >71 years) | 0.6793 | |
| Sex (Female vs. Male) | 0.0575 | |
| Differentiation (Wel–Mod vs. Por) | 0.0979 | |
| Vascular invasion (Absent vs. Present) | 0.0837 | |
| Depth of infiltration (T1-T2 vs. T3-T4) | 0.2441 | |
| Lymph node metastasis (absent vs. present) | < 0.0001 | 0.0013 |
| TNM stage (I–II vs. III–V) | 0.002 | 0.6054 |
| LGR5 (Expression vs. No expression) | 0.0267 | 0.0101 |
Wel: well-differentiated type, Mod: moderately differentiated type, Por: poorly differentiated type
Discussion
In extrahepatic bile ducts, high LGR5 expression was significantly associated with well- to moderately-differentiated adenocarcinoma. High LGR5 expression also indicated a favorable prognosis. Interestingly, LGR5 was not associated with high expression in poorly differentiated cancers or poor prognostic factors, which are generally considered to be characteristics of cancer stem cell markers [23]. However, LGR5 may be a novel prognostic marker of ECC.
Few studies have reported Lgr5 expression in normal bile ducts. Bile duct reactive cells in the vicinity of peribiliary glands, which differentiate into bile duct cells, have been suggested to be a type of proliferative intermediate stem cell present at the time of bile duct injury in mice [23, 24]. In a study by Yoshizawa et al. [25], LGR5 expression was significantly higher in highly differentiated adenocarcinoma than in intrahepatic cholangiocarcinoma, and high LGR5 expression was associated with a favorable prognosis. Among specific subtypes, LGR5 expression was significantly higher in the large duct type subtype. In intrahepatic cholangiocarcinoma, KRAS mutations are often found in large duct types [26]. ECCs also often have KRAS mutations [27], and there may be similarities to the large duct type of intrahepatic cholangiocarcinoma. In colorectal cancer, although well-differentiated adenocarcinomas have been reported to strongly express LGR5 as determined by RNA in situ hybridization [28], studies have also reported high LGR5 expression in poorly differentiated adenocarcinomas. Similarly, conflicting results regarding LGR5 expression and prognosis have been reported, and no unified view has been reached [28]. The most common method used to analyze LGR5 expression in previous reports is immunostaining [11]. The discrepancy in LGR5 expression analysis results in colorectal cancer may be due to differences in the analysis methods. Because recent studies have identified a tumor-suppressive role of LGR5 signaling in colorectal cancer [29], [30], RNA in situ hybridization may provide more accurate results.
Although LGR5 is a robust CSC marker, there are other candidate markers. Melo et al. reported that, at least in colorectal cancer, more primitive and immature CSC marker expression, defined by SOX2, OCT4, and Nanog, as opposed to gut tissue-specific stem cell signatures such as LGR5, is associated with disease progression [31]. Furthermore, LGR5 expression and glandular duct formation have been reported to be high in the region of gland formation in colorectal cancer and colorectal adenomas [32]. It is unclear whether gland duct formation is regulated by LGR5 itself, but it may be related to the degree of cancer differentiation, which may have prognostic implications. If a similar phenomenon occurs in ECC, LGR5 expression may be associated with differentiation status and might play a role in prognosis.
LGR5 is a Wnt target gene enriched in intestinal stem cells and colorectal cancer [6]. In colorectal cancer, APC abnormalities activate Wnt/β-catenin signaling. KRAS abnormalities are well known in cholangiocarcinoma [26, 27]. KRAS abnormalities have also been reported to stimulate Wnt/β-catenin signaling [33, 34]. Therefore, LGR5 expression may be affected by genetic abnormalities in each cancer type. Additionally, LGR5 activates Wnt/β-catenin signaling by binding to R-spondin [35]. Binding of R-spongin 1 to LGR5, together with TGF-β type II receptors in colon cancer cells, directly activates TGFβ signaling and suppresses tumor growth [30]. The regulation of LGR5 expression is complex and may be elucidated in the future.
We previously reported that high LGR5 expression in poorly differentiated gastric carcinomas is associated with a poor prognosis [36]. Poorly differentiated gastric carcinoma is a diffuse type that may correspond to genomically stable in the molecular characterization of gastric adenocarcinoma [37]. Various mutations in this phenotype are known [37]. Genetic abnormalities other than truncation of APC or KRAS mutations may cause differences in the prognostic value of LGR5 expression.
A limitation of this study is that we observed LGR5 expression only in clinical specimens. However, RNA in situ hybridization is highly sensitive and accurately captures actual LGR5 expression. Using cholangiocarcinoma cell cultures, expression of LGR5 and cytokines such as TGF-β should be examined in coculture or by other methods.
Immunostaining for LGR5 is unreliable. Therefore, observing LGR5 expression using RNAscope may be more useful than immunostaining, although it is not yet commonly used. Additionally, there have been reports of serological LGR5 measurements [38], suggesting that comparing these findings with tissue expression should be performed in future studies. However, LGR5 is expressed throughout the body, with particularly high expression in the colon. Therefore, measuring its expression in the bile duct, where its expression is minimal, using serological analysis might not be meaningful. While methods such as sequencing and mass spectrometry are excellent for analyzing expression levels, RNAscope allows for direct observation of the expression site, enabling analysis in conjunction with histological features. This approach provides a more comprehensive understanding of the local expression patterns of LGR5 and its role within the tissue. Additionally, western blot analysis is challenging because of the lack of suitable antibodies for immunostaining of LGR5. This issue represents one of the technical limitations in the analysis of LGR5. Therefore, we decided to use RNAscope in this study to accurately determine the local expression of LGR5 in clinical specimens.
There are various regulatory mechanisms of LGR5 expression, and the significance of expression varies by organ and tissue type. By understanding these points, the use of LGR5 as a therapeutic target or CSC marker should be explored, and further studies are desirable.
Conclusion
In ECC, the relationship between LGR5 expression and a favorable prognosis may be a potential prognostic marker, and further exploration of the molecular mechanism of LGR5 expression is warranted.
Acknowledgements
We thank Masanobu Momose, Yasuyo Shimojo, Naoko Ogiwara, Chitose Arai, Marina Nuno, Kanade Wakabayashi, Naoko Yamaoka, Saki Mukai, Shotaro Komamura, Daiki Ogura, Daiki Gomyo, and Tsukane Seki at Shinshu University Hospital for their excellent technical assistance, and Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Author contributions
Hisashi Tamada: Writing – original draft. Takeshi Uehara: Conceptualization, Methodology, Formal analysis, Supervision, Writing – review & editing. Takahiro Yoshizawa: Investigation, Data curation. Mai Iwaya: Investigation. Shiho Asaka: Investigation. Tomoyuki Nakajima: Investigation. Masato Kamakura: Investigation, Data curation. Hiroyoshi Ota: Supervision, Writing – review & editing.
Funding
This work was supported by a Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (KAKENHI) to Takeshi Uehara (20K07405). This study was partially supported by the Hokuto Foundation for Bioscience (grant awarded to TU). This study was also supported by a Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (KAKENHI) to Hiroyoshi Ota (22K07414). These funding bodies had no role in the study design, collection, analysis, or interpretation of data, or manuscript writing.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
This study was approved by the ethics committee of Shinshu University School of Medicine (Approval No: 5836). The requirement of informed consent was waived, and an opt-out method was used because of the retrospective design of the study. The investigation was conducted in compliance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vithayathil M, Khan SA. Current epidemiology of cholangiocarcinoma in western countries. J Hepatol. 2022;77(6):1690–8. 10.1016/j.jhep.2022.07.022 [DOI] [PubMed] [Google Scholar]
- 2.Tantau AI, Mandrutiu A, Pop A, Zaharie RD, Crisan D, Preda CM, et al. Extrahepatic cholangiocarcinoma: current status of endoscopic approach and additional therapies. World J Hepatol. 2021;13(2):166–86. 10.4254/wjh.v13.i2.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu L, Merath K, Farooq A, Hyer JM, Tsilimigras DI, Paredes AZ, et al. Photodynamic therapy may provide a benefit over systemic chemotherapy among non-surgically managed patients with extrahepatic cholangiocarcinoma. J Surg Oncol. 2020;121(2):286–93. 10.1002/jso.25773 [DOI] [PubMed] [Google Scholar]
- 4.Dragu DL, Necula LG, Bleotu C, Diaconu CC, Chivu-Economescu M. Therapies targeting cancer stem cells: current trends and future challenges. World J Stem Cells. 2015;7(9):1185–201. 10.4252/wjsc.v7.i9.1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoneberg T, Schultz G, Gudermann T. Structural basis of G protein-coupled receptor function. Mol Cell Endocrinol. 1999;151(1–2):181–93. 10.1016/S0303-7207(99)00017-9 [DOI] [PubMed] [Google Scholar]
- 6.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–7. 10.1038/nature06196 [DOI] [PubMed] [Google Scholar]
- 7.Han Y, Xue X, Jiang M, Guo X, Li P, Liu F, et al. LGR5, a relevant marker of cancer stem cells, indicates a poor prognosis in colorectal cancer patients: a meta-analysis. Clin Res Hepatol Gastroenterol. 2015;39(2):267–73. 10.1016/j.clinre.2014.07.008 [DOI] [PubMed] [Google Scholar]
- 8.Wang B, Chen Q, Cao Y, Ma X, Yin C, Jia Y, et al. LGR5 is a gastric Cancer stem cell marker Associated with Stemness and the EMT signature genes NANOG, NANOGP8, PRRX1, TWIST1, and BMI1. PLoS ONE. 2016;11(12):e0168904. 10.1371/journal.pone.0168904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao W, Li M, Liu J, Zhang S, Noordam L, Verstegen MMA, et al. LGR5 marks targetable tumor-initiating cells in mouse liver cancer. Nat Commun. 2020;11(1):1961. 10.1038/s41467-020-15846-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogasawara S, Uehara T, Nakajima T, Iwaya M, Maeno K, Tsuchiya S, et al. Correlation of clinicopathological features and LGR5 expression in triple-negative breast cancer. Ann Diagn Pathol. 2020;46:151491. 10.1016/j.anndiagpath.2020.151491 [DOI] [PubMed] [Google Scholar]
- 11.Chen Q, Zhang X, Li WM, Ji YQ, Cao HZ, Zheng P. Prognostic value of LGR5 in colorectal cancer: a meta-analysis. PLoS ONE. 2014;9(9):e107013. 10.1371/journal.pone.0107013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tempest N, Baker AM, Wright NA, Hapangama DK. Does human endometrial LGR5 gene expression suggest the existence of another hormonally regulated epithelial stem cell niche? Hum Reprod. 2018;33(6):1052–62. 10.1093/humrep/dey083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee HJ, Myung JK, Kim HS, Lee DH, Go HS, Choi JH, et al. Expression of LGR5 in mammary myoepithelial cells and in triple-negative breast cancers. Sci Rep. 2021;11(1):17750. 10.1038/s41598-021-97351-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirsch D, Barker N, McNeil N, Hu Y, Camps J, McKinnon K, et al. LGR5 positivity defines stem-like cells in colorectal cancer. Carcinogenesis. 2014;35(4):849–58. 10.1093/carcin/bgt377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brierley JD, Gospodarowicz MK, Wittekind C. TNM classification of malignant tumours. Wiley; 2017.
- 16.Nagtegaal ID, Odze RD, Klimstra D, Paradis V, Rugge M, Schirmacher P, et al. The 2019 WHO classification of tumours of the digestive system. Histopathology. 2020;76(2):182. 10.1111/his.13975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Surgery JSoH-B-P. General rules for clinical and pathological studies on cancer of the biliary tree (in Japanese), 7th eidtion edn. Tokyo: Kanehara Pub; 2021.
- 18.Ercolani G, Dazzi A, Giovinazzo F, Ruzzenente A, Bassi C, Guglielmi A, et al. Intrahepatic, peri-hilar and distal cholangiocarcinoma: three different locations of the same tumor or three different tumors? Eur J Surg Oncol. 2015;41(9):1162–9. 10.1016/j.ejso.2015.05.013 [DOI] [PubMed] [Google Scholar]
- 19.Jain A, Borad MJ, Kelley RK, Wang Y, Abdel-Wahab R, Meric-Bernstam F, et al. Cholangiocarcinoma with FGFR genetic aberrations: a unique clinical phenotype. JCO Precis Oncol. 2018;2:1–12. 10.1200/PO.17.00080 [DOI] [PubMed] [Google Scholar]
- 20.Otsuka S, Ebata T, Yokoyama Y, Mizuno T, Tsukahara T, Shimoyama Y, et al. Clinical value of additional resection of a margin-positive distal bile duct in perihilar cholangiocarcinoma. Br J Surg. 2019;106(6):774–82. 10.1002/bjs.11125 [DOI] [PubMed] [Google Scholar]
- 21.Ukpo OC, Flanagan JJ, Ma XJ, Luo Y, Thorstad WL, Lewis JS Jr. High-risk human papillomavirus E6/E7 mRNA detection by a novel in situ hybridization assay strongly correlates with p16 expression and patient outcomes in oropharyngeal squamous cell carcinoma. Am J Surg Pathol. 2011;35(9):1343–50. 10.1097/PAS.0b013e318220e59d [DOI] [PubMed] [Google Scholar]
- 22.Baker AM, Graham TA, Elia G, Wright NA, Rodriguez-Justo M. Characterization of LGR5 stem cells in colorectal adenomas and carcinomas. Sci Rep. 2015;5:8654. 10.1038/srep08654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu HJ, Chu PY. Role of Cancer Stem cells in Cholangiocarcinoma and therapeutic implications. Int J Mol Sci. 2019; 20(17). [DOI] [PMC free article] [PubMed]
- 24.Huch M, Dorrell C, Boj SF, van Es JH, Li VS, van de Wetering M, et al. In vitro expansion of single Lgr5 + liver stem cells induced by wnt-driven regeneration. Nature. 2013;494(7436):247–50. 10.1038/nature11826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshizawa T, Uehara T, Iwaya M, Asaka S, Kobayashi S, Nakajima T, et al. Correlation of LGR5 expression and clinicopathological features in intrahepatic cholangiocarcinoma. Pathol Res Pract. 2022;232:153832. 10.1016/j.prp.2022.153832 [DOI] [PubMed] [Google Scholar]
- 26.Zen Y. Intrahepatic cholangiocarcinoma: typical features, uncommon variants, and controversial related entities. Hum Pathol. 2023;132:197–207. 10.1016/j.humpath.2022.06.001 [DOI] [PubMed] [Google Scholar]
- 27.Montal R, Sia D, Montironi C, Leow WQ, Esteban-Fabro R, Pinyol R, et al. Molecular classification and therapeutic targets in extrahepatic cholangiocarcinoma. J Hepatol. 2020;73(2):315–27. 10.1016/j.jhep.2020.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang BG, Kim HS, Chang WY, Bae JM, Kim WH, Kang GH. Expression Profile of LGR5 and its prognostic significance in Colorectal Cancer Progression. Am J Pathol. 2018;188(10):2236–50. 10.1016/j.ajpath.2018.06.012 [DOI] [PubMed] [Google Scholar]
- 29.Wu C, Qiu S, Lu L, Zou J, Li WF, Wang O, et al. RSPO2-LGR5 signaling has tumour-suppressive activity in colorectal cancer. Nat Commun. 2014;5:3149. 10.1038/ncomms4149 [DOI] [PubMed] [Google Scholar]
- 30.Zhou X, Geng L, Wang D, Yi H, Talmon G, Wang J. R-Spondin1/LGR5 activates TGFbeta Signaling and suppresses Colon Cancer Metastasis. Cancer Res. 2017;77(23):6589–602. 10.1158/0008-5472.CAN-17-0219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Sousa EMF, Colak S, Buikhuisen J, Koster J, Cameron K, de Jong JH, et al. Methylation of cancer-stem-cell-associated wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell. 2011;9(5):476–85. 10.1016/j.stem.2011.10.008 [DOI] [PubMed] [Google Scholar]
- 32.Martin ML, Zeng Z, Adileh M, Jacobo A, Li C, Vakiani E, et al. Logarithmic expansion of LGR5(+) cells in human colorectal cancer. Cell Signal. 2018;42:97–105. 10.1016/j.cellsig.2017.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong CC, Xu J, Bian X, Wu JL, Kang W, Qian Y, et al. In colorectal Cancer cells with mutant KRAS, SLC25A22-Mediated glutaminolysis reduces DNA demethylation to increase WNT signaling, stemness, and Drug Resistance. Gastroenterology. 2020;159(6):2163–e21802166. 10.1053/j.gastro.2020.08.016 [DOI] [PubMed] [Google Scholar]
- 34.Lemieux E, Cagnol S, Beaudry K, Carrier J, Rivard N. Oncogenic KRAS signalling promotes the Wnt/beta-catenin pathway through LRP6 in colorectal cancer. Oncogene. 2015;34(38):4914–27. 10.1038/onc.2014.416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung C, Tan SH, Barker N. Recent advances in Lgr5(+) stem cell research. Trends Cell Biol. 2018;28(5):380–91. 10.1016/j.tcb.2018.01.010 [DOI] [PubMed] [Google Scholar]
- 36.Ehara T, Uehara T, Nakajima T, Kinugawa Y, Kobayashi S, Iwaya M, et al. LGR5 expression is associated with prognosis in poorly differentiated gastric adenocarcinoma. BMC Cancer. 2021;21(1):228. 10.1186/s12885-021-07913-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–9. 10.1038/nature13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ebeid SA, Abd El Moneim NA, El-Benhawy SA, Ramadan R, Ismail SE. Znhit1 and HIF-2alpha are correlated with cancer stem cell markers in breast cancer patients. Sci Rep. 2022;12(1):13918. 10.1038/s41598-022-18133-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.