ABSTRACT
Although antibiotics remain a cornerstone of modern medicine, the issues of widespread antibiotic resistance and collateral damage to the microbiome from antibiotic use are driving a need for drug developers to consider more tailored, patient-directed products to avoid antibiotic-induced perturbations of the structure and function of the indigenous microbiota. This perspective summarizes a cascade of microbiome health effects that is initiated by antibiotic-mediated microbiome disruption at an individual level and ultimately leads to infection and transmission of multidrug-resistant pathogens across patient populations. The scientific evidence behind each of the key steps of this cascade is presented. The interruption of this cascade through the use of highly targeted, microbiome-sparing antibiotics aiming to improve health outcomes is discussed. Further, this perspective reflects on some key clinical trial design and reimbursement considerations to be addressed as part of the drug development path.
KEYWORDS: antibiotic-induced dysbiosis, multidrug-resistant pathogens, colonization, Clostridioides difficile infection, transmission
PERSPECTIVE
The introduction of antibiotics into clinical practice was a significant medical breakthrough that drastically reduced mortality rates related to infectious disease, increased the average human lifespan, and paved the way for modern procedures that would otherwise not be possible (1). Antimicrobial resistance (AR) among microorganisms has expanded over time through selective pressure, leading to more AR infections in humans, and is now a leading cause of mortality globally (2). An additional and underappreciated effect of antibiotic use is the collateral damage to our microbiota and resulting antibiotic-induced dysbiosis (see Table 1 for key terminology). In the ongoing search for clinical and public health solutions to the AR crisis, it will be key to develop new classes of antibiotics that combat infections without accelerating the development of resistance, with potential additional benefits of avoiding antibiotic-induced dysbiosis and thereby reducing secondary infections as well as transmission.
TABLE 1.
Microbiome-related terminology (3)
| Term | Definition |
|---|---|
| Microbiota | A community of microorganisms that occupy a particular site or habitat |
| Microbiome | A characteristic microbial community that occupies a reasonably well-defined habitat and has distinct physicochemical properties. The term not only refers to the microorganisms involved but also encompasses their theater of activity. Some people use the term microbiome to refer only to the organisms themselves (i.e., microbiota) or to refer to the collective genome of a microbial consortium or community (otherwise referred to as the metagenome). |
| Dysbiosis | Disruption of the composition, abundance, diversity, and functionality of a microbial community that leads to susceptibility to a given outcome, in this instance susceptibility to colonization by an AR organism |
| Colonization | The asymptomatic carriage of a microorganism, including opportunistic pathogens, in or on the body |
| Colonization resistance | The state of an intact or non-dysbiotic microbiome that is not conducive to the establishment of additional microbes, including pathogens, as stable members of the existing community |
| Pathogen abundance | The absolute or relative number of a given pathogen (species or genera) in a microbiota. In the case of relative pathogen abundance, this is the fraction of the total number of organisms in the microbiota |
The role of microbiome disruption in the pathogenesis of healthcare-associated infections
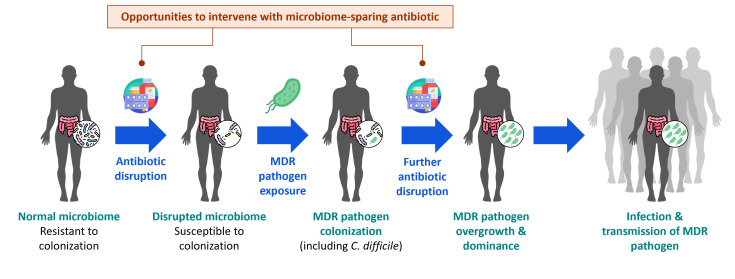
In homeostasis, the human microbiome is known to have a central role in overall health (4, 5), and in colonization resistance (6), through four main actions: (i) direct inhibition of pathogen growth through the production of bioactive small molecules (ii); barrier maintenance through preservation of the mucous layer and enterocyte health promotion, thereby preventing pathogen invasion or translocation (iii); cross-talk of the microbiome with the human host resulting in immune modulation (iv); nutrient utilization, thereby outcompeting pathogens and preventing their establishment. There is extensive evidence demonstrating the role of antibiotic-mediated microbiome disruption in the pathogenesis of Clostridioides difficile infection (CDI); the mechanisms by which antibiotic-mediated disruption of the microbiome increase susceptibility to CDI have been studied in detail (7). What is less understood is the role antibiotics have on disrupting colonization resistance against pathogens other than C. difficile and the risks for infection this may confer. Disruption of microbiome-conferred protective mechanisms can lead to gut colonization with potential pathogens, including multidrug-resistant (MDR) bacteria, which is the first step in a cascade of events resulting from exposure to antibiotics, with important consequences for healthcare-associated infections and public health (Fig. 1).
Fig 1.
The microbiome health-effects cascade: from antibiotic-mediated microbiome disruption to infection and transmission. MDR, multidrug resistant.
It is now well-established in the literature that gut colonization with MDR pathogens carries a substantial risk of subsequent infection with the same or phenotypically similar MDR pathogen (8). In a large meta-analysis by Willems et al., the cumulative incidence of infection following gut colonization with MDR pathogens ranged from 7% to 19%, in most cases over a median period of 30 days, depending on the pathogen, which represents a sizable risk (8). Moreover, there is evidence that colonization with MDR pathogens is associated with an increase in the all-cause risk of infection (i.e., infection caused by any pathogen) (9). In a prospective cohort study of 3,600 patients who underwent colorectal surgery, patients who were colonized with extended-spectrum beta-lactamase (ESBL)-producing Enterobacterales had more than double the odds of all-cause subsequent surgical site infection compared with non-colonized patients (adjusted odds ratio, 2.36; 95% confidence interval [CI], 1.50–3.71), and these increased odds extended beyond merely increased infections caused by the ESBL-producing Enterobacterales itself (9). This extension suggests how dysbiosis that predisposes to colonization with ESBL-producing Enterobacterales has broader effects, including colonization with other antibiotic-resistant and antibiotic-susceptible pathogens.
Emerging data on fecal microbiota transplantation (FMT) bring this argument full circle by supporting the etiological role of the microbiome in the development of infection following MDR colonization. In a prospective cohort study by Ianiro et al., patients treated with FMT for recurrent C. difficile infection had 23% fewer bloodstream infections, a 14% reduction in length of hospitalization, and 32% greater 90-day survival when compared with matched patients who were treated with antibiotics (10).
Furthermore, several studies have shown that increased pathogen abundance can result from dysbiosis and may have an important role in the degree of increased infection risk (11–15). In a longitudinal study by Taur et al. of 94 patients undergoing allogeneic hematopoietic stem cell transplantation, increased abundance of Enterococcus species was associated with greater risk of subsequent vancomycin-resistant enterococcal bacteremia (hazard ratio, 9.35; 95% CI, 2.43–45.44), whereas increased abundance of Proteobacteria was associated with greater risk of subsequent Gram-negative bacteremia (hazard ratio, 5.46; 95% CI 1.03–19.91) (15). Studies of Klebsiella species in long-term acute care hospital patients, have shown that a relative abundance of 22%, as determined by 16S rRNA gene sequence analysis of rectal swab cultures, predicts subsequent Klebsiella infection (13, 14).
Beyond the impact on the individual patient, increased pathogen abundance has also been shown to increase the risk of transmission of MDR pathogens (16, 17), broadening the scope of the consequences of dysbiosis to the population. This is evident from a study of skin and environmental contamination among long-term care facility residents with asymptomatic C. difficile colonization: as the number of colonies recovered per perirectal swab increased, so did the percentage of positive cultures from both the skin of the patient and the patient care environment, such as bed rails or overbed tables. Such skin and environmental contamination is associated with contamination of the hands of healthcare personnel and transmission in healthcare settings (17). The same relationship between pathogen load, and patient skin and environmental contamination has been demonstrated with MDR Gram-negative bacilli (16). Meanwhile, evidence shows that treating patients for C. difficile infections with fidaxomicin, an antibiotic that is relatively microbiome-sparing with little or no activity against Gram-negative aerobic and anaerobic bacteria, reduces pathogen load and environmental contamination compared with patients who receive vancomycin/metronidazole (18, 19).
Based on the collective evidence presented, and the cascade of events described (Fig. 1), it becomes clear that by preventing or reducing colonization and pathogen burden in an index patient, it is possible to protect both the patient from infection and the population from transmission of and infection by MDR pathogens.
Microbiome-sparing antibiotics: precision therapy as a tool for microbiome preservation
Antibiotics are lifesaving drugs, but when used to treat an infection, they impact not only the target pathogen but also the susceptible portion of the microbiome, leaving the host vulnerable to colonization and possible infection by MDR pathogens, such as C. difficile. These negative effects are typically more prominent with broad-spectrum than with narrow-spectrum antibiotics. Experts have previously called for new approaches in antibiotic development where this collateral damage to the microbiome is minimized (20, 21). In concept, antibiotics that are targeted and highly specific to pathogenic organisms would not impact the microbiome, thus called “microbiome-sparing” antibiotics. No longer should killing activity alone be the driver of drug development candidates but rather a balance of killing activity with microbiome-sparing effects. Although broad-spectrum antibiotics would remain critical for the empiric treatment of certain presentations (e.g., sepsis), the use of microbiome-sparing antibiotics to treat infections where the causative pathogen is known could significantly reduce the adverse effects associated with microbiome disruption (Fig. 1). There are currently numerous agents with potential microbiome-sparing profiles in development for the treatment of infections caused by various key pathogens (Table 2).
TABLE 2.
Examples of targeted pathogen-specific agents in development with potential microbiome-sparing profiles (22–25)a
| Name | Phase | Company | Target/mechanism | Pathogen |
|---|---|---|---|---|
| Ridinilazole | III | Summit Therapeutics Inc. | Minor groove binder | Clostridioides difficile |
| CRS3123 | II | Crestone, Inc. | Methionyl-tRNA synthetase | C. difficile |
| Afabicin | II | Debiopharm | FabI | Staphylococcus spp. |
| AR-101 (mAb) | II | Aridis Pharmaceuticals Inc. | LPS serotype 011 | Pseudomonas aeruginosa |
| Ibezapolstat | II | Acurx Pharmaceuticals Inc. | DNA polymerase IIIC | C. difficile |
| Ribaxamaseb | II | Theriva Biologics | Orally ingested beta-lactamase | Various |
| TXA709 | I | Taxis Pharmaceuticals | FtsZ | MRSA |
| FP-100 | Preclinical | Flightpath Biosciences | 23S rRNA, selectively taken up via spirochete-specific nucleoside transporter | Borrelia burgdorferi |
| SMT-738 | Preclinical | Summit Therapeutics | LolC/E complex | Enterobacteriaceae |
| Lolamicin | Preclinical | – | LolCDE complex |
Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae |
| Debio 1453 | Preclinical | Debiopharm | FabI | Neisseria gonorrhoeae |
| Antisense (various peptide conjugate–peptide nucleic acids) | Preclinical | Techulon Inc. | Specific inhibition of gene translation | MRSA, Acinetobacter baumannii, and P. aeruginosa |
DNA, deoxyribonucleic acid; FabI, enoyl-acyl carrier protein reductase enzyme; FtsZ, filamenting temperature-sensitive mutant Z; LPS, lipopolysaccharide; MRSA, methicillin-resistant Staphylococcus aureus; tRNA, transport ribonucleic acid.
Ribaxamase is intended to be used in conjunction with a parenteral beta-lactam antibiotic, breaking down the antibiotic in the gut, rendering it microbiome-sparing.
What is needed for the development of microbiome-sparing antibiotics?
The current clinical trial model for antibiotics is one of non-inferiority versus standard of care. Beyond this, two key questions in the development of microbiome-sparing antibiotics are (i) how to design clinical trials that not only accomplish this demonstration of non-inferiority but also demonstrate an advantage in terms of impact on the microbiome, and (ii) what criteria would be considered acceptable evidence of clinical, likely clinical, and public health advantages of sparing the microbiome. Reflecting on the microbiome health-effects cascade (Fig. 1), these criteria could range from measuring a lack of microbiome disruption on the far-left end of the cascade through various indices to reducing infection and transmission of MDR pathogens on the far-right end of the cascade, potentially by measuring secondary infection rate or skin and environmental contamination. In between are studies that measure the rate of new instances of colonization with an MDR pathogen and, once colonized, the development of MDR pathogen dominance. Depending upon the effect size of the intervention and incidence of the outcome in controls, different endpoints across the spectrum may require vastly different scales and levels of resources, with only about 10 to just over 100 patients needed to assess microbiome indices (e.g., 50%–80% effect sizes, 50%–80% incidence; depending on relative degree of microbiome-sparing, indices used, and their thresholds) (26) compared with 1,000 to over 10,000 (e.g., 20%–35% effect sizes, 5%–15% incidence; depending on the underlying infection risk in the patient population and infection type) (8, 10) needed to assess secondary infection rates (https://clincalc.com/stats/samplesize.aspx).
Drug developers might also start to consider whether combining clinical and microbiome measures could provide evidence of superiority over comparator antibiotics in confirmatory phase 3 trials using a hierarchical nested design (27) (https://www.fda.gov/regulatory-information/search-fda-guidance-documents/multiple-endpoints-clinical-trials). The key precondition for regulatory approval of the new antibiotic remains unchanged: the primary efficacy endpoint (clinical response to the target infection) needs to be compared in a non-inferiority design. If clinical non-inferiority is confirmed, predetermined additional endpoints (i.e., microbiome-related endpoints) can be tested. Additional innovation in compositing outcomes in a manner that can better reflect the totality of clinical outcomes and potential patient preferences is the desirability of outcome ranking (DOOR) strategy in which the overall outcome of each patient is ranked in three domains of clinical response, infectious complications, and serious adverse events (28). Each of these domains includes pre-determined criteria, and one could envision the future addition of such criteria as persistent microbiome disruption, colonization, and infections occurring during a pre-determined follow-up period resulting from colonization as additional criteria in one or more of these domains. Approaches such as these reduce the level of risk for drug developers and have the potential to increase investment in antibiotic development, paving the way for a new approach to drug discovery that strikes a balance between target pathogen coverage and impact on the microbiome, as called for by experts (21).
To increase the plausibility of clinical trials for microbiome-sparing antibiotics, there is a need to generate robust data and grow the evidence base connecting surrogate study endpoints, such as microbiome indices, to hard clinical outcomes. Encouragingly, microbiome indices are already playing a role in drug development. For example, species engraftment and concentration of secondary bile salts were assessed as prespecified exploratory endpoints in the phase 3 clinical trial assessing SER-109, now FDA-approved as VOWST (Seres Therapeutics, Inc.), for the treatment of recurrent CDI (29). An additional example is the recent United Kingdom (UK) National Institute for Health and Care Excellence (NICE) assessment process for applicants to a subscription model payment system, which includes a microbiome effects criterion (https://www.engage.england.nhs.uk/survey/the-antimicrobial-products-subscription-model/).
In further contrast to the typical antibiotic model of last-line use to reduce the emergence of AR, microbiome-sparing antibiotics provide evolutionary favor in the prevention of AR (i.e., reduction of selection pressure on indigenous microbiota and containment of the emergence of resistance) and will need to be used widely for their benefits to be realized (30, 31). This represents a new value paradigm for antibiotics, and innovative models of reimbursement will be vital to support access to and use of microbiome-sparing antibiotics. One such example of innovation could be the “population health agreement” between NICE, NHS England, and Novartis for access to inclisiran, a medication indicated to treat familial hypercholesterolemia or clinical atherosclerotic cardiovascular disease. To support such a model for antibiotics, it will be necessary to demonstrate the population benefits of preserving the microbiome using criteria relevant to specific payers (e.g., Medicare, Veterans Affairs, NHS England), possibly through real-world evidence via risk-sharing agreements. It should be possible to identify settings with high rates of colonization and infection, such as nursing homes and inpatient rehabilitation facilities, hematology–oncology care settings, or organ transplant centers, where an approved microbiome-sparing agent for routine treatment of a target infection could be used on a trial basis. This might be first evaluated in a cluster-randomized study, introduced in a stepped-wedge design, or simply evaluated after wholesale introduction in quasi-experimental fashion, looking for impacts on population health and healthcare costs. The real-world population data generated using this strategy could be used by payors to inform longer-term formulary decisions.
Conclusion
Although it is encouraging that some progress has been made in controlling some forms of AR, many challenges remain (32, 33). Both antibiotic stewardship and infection prevention and control have been the main contributors to the progress to date, yet it is unknown how much further progress can be made utilizing these tools alone. The development of microbiome-sparing antibiotics is a key strategy for maintaining future progress and reducing the morbidity, mortality, and excess costs of AR. It will be critical that industry, academia, regulators, and public health band together. With the dawning of the age of microbiome-aware medical care and greater insights into the spread of AR, it is time to redesign our antibiotic therapies beginning with the end in mind.
ACKNOWLEDGMENTS
L.C.M. and A.L.H.: the findings and conclusions in this presentation are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the U.S. Department of Health and Human Services.
Writing support was provided by Laura Saunderson, with financial support from Debiopharm International SA.
Footnotes
Presented at: The content of this perspective was presented in part at the World Antimicrobial Resistance Congress, September 2023.
Contributor Information
L. Clifford McDonald, Email: ljm3@cdc.gov.
Aaron P. Mitchell, University of Georgia, Athens, Georgia, USA
REFERENCES
- 1. Hutchings MI, Truman AW, Wilkinson B. 2019. Antibiotics: past, present and future. Curr Opin Microbiol 51:72–80. doi: 10.1016/j.mib.2019.10.008 [DOI] [PubMed] [Google Scholar]
- 2. Antimicrobial Resistance Collaborators . 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399:629–655. doi: 10.1016/S0140-6736(21)02724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Young VB. 2017. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ 356:j831. doi: 10.1136/bmj.j831 [DOI] [PubMed] [Google Scholar]
- 4. Li J, Li Y, Ivey KL, Wang DD, Wilkinson JE, Franke A, Lee KH, Chan A, Huttenhower C, Hu FB, Rimm EB, Sun Q. 2022. Interplay between diet and gut microbiome, and circulating concentrations of trimethylamine N-oxide: findings from a longitudinal cohort of US men. Gut 71:724–733. doi: 10.1136/gutjnl-2020-322473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stiemsma LT, Michels KB. 2018. The role of the microbiome in the developmental origins of health and disease. Pediatrics 141:e20172437. doi: 10.1542/peds.2017-2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McKenney PT, Pamer EG. 2015. From hype to hope: the gut microbiota in enteric infectious disease. Cell 163:1326–1332. doi: 10.1016/j.cell.2015.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schnizlein MK, Young VB. 2022. Capturing the environment of the Clostridioides difficile infection cycle. Nat Rev Gastroenterol Hepatol 19:508–520. doi: 10.1038/s41575-022-00610-0 [DOI] [PubMed] [Google Scholar]
- 8. Willems RPJ, van Dijk K, Vehreschild MJGT, Biehl LM, Ket JCF, Remmelzwaal S, Vandenbroucke-Grauls CMJE. 2023. Incidence of infection with multidrug-resistant Gram-negative bacteria and vancomycin-resistant enterococci in carriers: a systematic review and meta-regression analysis. Lancet Infect Dis 23:719–731. doi: 10.1016/S1473-3099(22)00811-8 [DOI] [PubMed] [Google Scholar]
- 9. Dubinsky-Pertzov B, Temkin E, Harbarth S, Fankhauser-Rodriguez C, Carevic B, Radovanovic I, Ris F, Kariv Y, Buchs NC, Schiffer E, Cohen Percia S, Nutman A, Fallach N, Klausner J, Carmeli Y, R-GNOSIS WP4 Study Group . 2019. Carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae and the risk of surgical site infection after colorectal surgery: a prospective cohort study. Clin Infect Dis 68:1699–1704. doi: 10.1093/cid/ciy768 [DOI] [PubMed] [Google Scholar]
- 10. Ianiro G, Murri R, Sciumè GD, Impagnatiello M, Masucci L, Ford AC, Law GR, Tilg H, Sanguinetti M, Cauda R, Gasbarrini A, Fantoni M, Cammarota G. 2019. Incidence of bloodstream infections, length of hospital stay, and survival in patients with recurrent Clostridioides difficile infection treated with fecal microbiota transplantation or antibiotics: a prospective cohort study. Ann Intern Med 171:695–702. doi: 10.7326/M18-3635 [DOI] [PubMed] [Google Scholar]
- 11. Pérez-Nadales E, M Natera A, Recio-Rufián M, Guzmán-Puche J, Marín-Sanz JA, Martín-Pérez C, Cano Á, Castón JJ, Elías-López C, Machuca I, Gutiérrez-Gutiérrez B, Martínez-Martínez L, Torre-Cisneros J. 2022. Prognostic significance of the relative load of KPC-producing Klebsiella pneumoniae within the intestinal microbiota in a prospective cohort of colonized patients. Microbiol Spectr 10:e0272821. doi: 10.1128/spectrum.02728-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rao K, Seekatz A, Bassis C, Sun Y, Mantlo E, Bachman MA. 2020. Enterobacterales infection after intestinal dominance in hospitalized patients. mSphere 5:e00450-20. doi: 10.1128/mSphere.00450-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shimasaki T, Seekatz A, Bassis C, Rhee Y, Yelin RD, Fogg L, Dangana T, Cisneros EC, Weinstein RA, Okamoto K, Lolans K, Schoeny M, Lin MY, Moore NM, Young VB, Hayden MK, Centers for Disease Control and Prevention Epicenters Program . 2019. Increased relative abundance of Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae within the gut microbiota is associated with risk of bloodstream infection in long-term acute care hospital patients. Clin Infect Dis 68:2053–2059. doi: 10.1093/cid/ciy796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun Y, Patel A, SantaLucia J, Roberts E, Zhao L, Kaye K, Rao K, Bachman MA. 2021. Measurement of Klebsiella intestinal colonization density to assess infection risk. mSphere 6:e0050021. doi: 10.1128/mSphere.00500-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, Lee YJ, Dubin KA, Socci ND, Viale A, Perales M-A, Jenq RR, van den Brink MRM, Pamer EG. 2012. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 55:905–914. doi: 10.1093/cid/cis580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alhmidi H, Cadnum JL, Koganti S, Jencson AL, Bonomo RA, Wilson BM, Mayer J, Samore MH, Donskey CJ. 2020. Shedding of multidrug-resistant gram-negative bacilli by colonized patients during procedures and patient care activities. Am J Infect Control 48:1336–1340. doi: 10.1016/j.ajic.2020.06.004 [DOI] [PubMed] [Google Scholar]
- 17. Donskey CJ, Sunkesula VCK, Jencson AL, Stone ND, Gould CV, McDonald LC, Samore M, Mayer J, Pacheco S, Sambol S, Petrella L, Terry D, Gerding DN. 2014. Utility of a commercial PCR assay and a clinical prediction rule for detection of toxigenic Clostridium difficile in asymptomatic carriers. J Clin Microbiol 52:315–318. doi: 10.1128/JCM.01852-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Biswas JS, Patel A, Otter JA, Wade P, Newsholme W, van Kleef E, Goldenberg SD. 2015. Reduction in Clostridium difficile environmental contamination by hospitalized patients treated with fidaxomicin. J Hosp Infect 90:267–270. doi: 10.1016/j.jhin.2015.01.015 [DOI] [PubMed] [Google Scholar]
- 19. Davies K, Mawer D, Walker AS, Berry C, Planche T, Stanley P, Goldenberg S, Sandoe J, Wilcox MH. 2020. An analysis of Clostridium difficile environmental contamination during and after treatment for C difficile infection. Open Forum Infect Dis 7:faa362. doi: 10.1093/ofid/ofaa362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blaser MJ. 2016. Antibiotic use and its consequences for the normal microbiome. Science 352:544–545. doi: 10.1126/science.aad9358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rooney CM, Ahmed S, Wilcox MH. 2021. Protecting the microbiota. J Infect Dis 223:S290–S295. doi: 10.1093/infdis/jiab143 [DOI] [PubMed] [Google Scholar]
- 22. Alshrari AS, Hudu SA, Elmigdadi F, Imran M. 2023. The urgent threat of Clostridioides difficile infection: a glimpse of the drugs of the future, with related patents and prospects. Biomedicines 11:426. doi: 10.3390/biomedicines11020426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Avis T, Wilson FX, Khan N, Mason CS, Powell DJ. 2021. Targeted microbiome-sparing antibiotics. Drug Discov Today 26:2198–2203. doi: 10.1016/j.drudis.2021.07.016 [DOI] [PubMed] [Google Scholar]
- 24. Ruggieri F, Compagne N, Antraygues K, Eveque M, Flipo M, Willand N. 2023. Antibiotics with novel mode of action as new weapons to fight antimicrobial resistance. Eur J Med Chem 256:115413. doi: 10.1016/j.ejmech.2023.115413 [DOI] [PubMed] [Google Scholar]
- 25. Muñoz KA, Ulrich RJ, Vasan AK, Sinclair M, Wen P-C, Holmes JR, Lee HY, Hung C-C, Fields CJ, Tajkhorshid E, Lau GW, Hergenrother PJ. 2024. A Gram-negative-selective antibiotic that spares the gut microbiome. Nature 630:429–436. doi: 10.1038/s41586-024-07502-0 [DOI] [PubMed] [Google Scholar]
- 26. Anthony WE, Wang B, Sukhum KV, D’Souza AW, Hink T, Cass C, Seiler S, Reske KA, Coon C, Dubberke ER, Burnham C-AD, Dantas G, Kwon JH. 2022. Acute and persistent effects of commonly used antibiotics on the gut microbiome and resistome in healthy adults. Cell Rep 39:110649. doi: 10.1016/j.celrep.2022.110649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Timsit J-F, de Kraker MEA, Sommer H, Weiss E, Bettiol E, Wolkewitz M, Nikolakopoulos S, Wilson D, Harbarth S, COMBACTE-NET consortium . 2017. Appropriate endpoints for evaluation of new antibiotic therapies for severe infections: a perspective from COMBACTE's STAT-Net. Intensive Care Med 43:1002–1012. doi: 10.1007/s00134-017-4802-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Howard-Anderson J, Hamasaki T, Dai W, Collyar D, Rubin D, Nambiar S, Kinamon T, Hill C, Gelone SP, Mariano D, Baba T, Holland TL, Doernberg SB, Chambers HF, Fowler VG, Evans SR, Boucher HW. 2023. Improving traditional registrational trial end points: development and application of a desirability of outcome ranking end point for complicated urinary tract infection clinical trials. Clin Infect Dis 76:e1157–e1165. doi: 10.1093/cid/ciac692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Feuerstadt P, Louie TJ, Lashner B, Wang EEL, Diao L, Bryant JA, Sims M, Kraft CS, Cohen SH, Berenson CS, Korman LY, Ford CB, Litcofsky KD, Lombardo M-J, Wortman JR, Wu H, Auniņš JG, McChalicher CWJ, Winkler JA, McGovern BH, Trucksis M, Henn MR, von Moltke L. 2022. SER-109, an oral microbiome therapy for recurrent Clostridioides difficile infection. N Engl J Med 386:220–229. doi: 10.1056/NEJMoa2106516 [DOI] [PubMed] [Google Scholar]
- 30. Diamantis S, Retur N, Bertrand B, Lieutier-Colas F, Carenco P, Mondain V, On Behalf Of Promise Professional Community Network On Antimicrobial Resistance. . 2022. The production of antibiotics must be reoriented: repositioning old narrow-spectrum antibioticsdeveloping new microbiome-sparing antibiotics. Antibiotics (Basel) 11:924. doi: 10.3390/antibiotics11070924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wollein Waldetoft K, Brown SP. 2017. Alternative therapeutics for self-limiting infections-An indirect approach to the antibiotic resistance challenge. PLoS Biol 15:e2003533. doi: 10.1371/journal.pbio.2003533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. CDC . 2019. Antibiotic resistance threats in the United States, 2019. U.S. Department of Health and Human Services, CDC, Atlanta, GA. [Google Scholar]
- 33. European Centre for Disease Prevention and Control . 2023. Antimicrobial resistance in the EU/EEA (EARS-Net) - annual epidemiological report 2022. Stockholm, ECDC. [Google Scholar]