Abstract
Renal cell carcinoma (RCC) is characterized by high mortality and morbidity rates. Vav guanine nucleotide exchange factors (VAVs), crucial for signal transduction between cell membrane receptors and intracellular mediators, have been implicated in carcinogenesis. However, their potential prognostic value in RCC remains unclear. The impact of 150 common VAV polymorphisms on RCC risk and survival was investigated in a cohort of 630 individuals. Publicly available gene expression datasets were utilized to analyze VAV gene expression in relation to patient outcomes. The VAV3 rs17019888 polymorphism was significantly associated with RCC risk and overall survival after adjusting for false discovery rates. Expression quantitative trait loci analysis revealed that the risk allele of rs17019888 is linked to reduced VAV3 expression. Analysis of 19 kidney cancer gene expression datasets revealed lower VAV3 expression in RCC tissues compared to normal tissues, with higher expression correlating with better prognosis. Gene set enrichment analysis demonstrated that VAV3 negatively regulates the ubiquitin–proteasome system, extracellular matrix and membrane receptors, inflammatory responses, matrix metalloproteinases, and cell cycle pathways. Furthermore, elevated VAV3 expression was associated with increased infiltration of B cells, macrophages, and neutrophils into the RCC tumor microenvironment. Our findings suggest that VAV3 gene variants influence RCC risk and survival, contributing to a favorable prognosis in RCC.
Keywords: renal cell carcinoma, vav guanine nucleotide exchange factor, genetic variants, gene set enrichment analysis, prognosis
1. Introduction
Renal cell carcinoma (RCC) is among the most prevalent forms of genitourinary cancer, with global incidence rates rising by approximately 2–3% annually [1]. From 1990 to 2019, the number of RCC cases worldwide has increased by 154.78% [2]. The mortality rate of RCC ranges from 30 to 40%, with a higher prevalence in males and in developed countries. The age of onset has shifted to younger populations, likely due to widespread health screening [3]. Despite this, the 5-year survival rate remained significantly lower in metastatic cases (13%) than in regional cases (70%) [4]. Current treatments for advanced unresectable RCC are inadequate, underscoring the urgent need for novel therapeutic strategies. Genetic factors and single-nucleotide polymorphisms (SNPs) such as VHL rs7629500 and IRF5 rs3807306 play significant roles in RCC susceptibility [5]. Therefore, elucidating the genetic and molecular mechanisms underlying RCC may lead to the development of early diagnostic markers and effective treatments.
VAV genes function as guanine nucleotide exchange factors (GEFs) that activate Rho family GTPases [6]. In mammals, the VAV family comprises VAV1, VAV2, and VAV3, which have similar but not identical functions. VAV1 is primarily expressed in hematopoietic cells, whereas VAV2 and VAV3 exhibit more widespread expression patterns [7]. VAVs play a crucial role in the intracellular signaling pathways downstream of receptors with tyrosine kinase activity [8]. Upon receptor stimulation, VAV proteins undergo tyrosine phosphorylation, activating Rho GTPases such as RhoA, Rac1, and Cdc42 [6], which, in turn, regulate various cellular processes, including phagocytosis, vesicular transport, cell growth, chemotaxis, migration, and adhesion [9].
Recent studies suggest that VAV proteins are essential for maintaining the homeostasis of cardiovascular, central nervous, and immune systems [6]. They are also implicated in numerous aspects of cancer biology, such as epithelial–mesenchymal transition, tumorigenesis, chemosensitivity, and metastasis [10,11]. VAV1, initially identified for its oncogenic activity [12], has been associated with poor survival rates in pancreatic [13], ovarian [14], and medulloblastoma tumors [15]. Conversely, VAV1 may function as a tumor suppressor in immature T cells, with its loss leading to acute lymphoblastic leukemia in T cells [16,17]. Recent studies have investigated the relationship between genetic variants of VAV family genes and cancer susceptibility. For example, VAV3 rs7521681 and rs4915076 have been linked to thyroid cancer risk [18], whereas rs12410676 has been associated with prostate cancer in Chinese males [19]. Additionally, VAV2 rs12002767 may influence the overall survival of patients with non-small cell lung cancer by regulating VAV2 expression [20]. However, the clinical significance of the genetic variants of VAV family genes in RCC remains unclear.
Given the critical role of VAV GEF family genes in signal transduction and cellular behavior, we hypothesized that genetic variants in VAV family genes are associated with RCC risk and survival. To test this hypothesis, we examined the relationship between 150 SNPs in VAV family genes and RCC risk and survival outcomes in a cohort of 630 participants. Additionally, we explored the functional significance of the implicated gene, VAV3, through gene ontology (GO) enrichment and pathway analyses to elucidate the potential biological mechanisms influencing RCC development.
2. Materials and Methods
2.1. Study Population and Data Collection
This study involved 630 participants recruited from Taipei Medical University Hospital, Taipei Municipal Wan Fang Hospital, and National Taiwan University Hospital. The cohort comprised 312 patients with pathologically confirmed RCC and 318 cancer-free controls matched for age (±1 year) and sex [21,22]. Demographic information was gathered through structured interviews, and clinical data were extracted from medical records. Our patient cohort consisted of 75.3% clear cell RCC (KIRC), 8.7% papillary RCC (KIRP), and 8% chromophobe RCC (KICH), closely mirroring the general cancer statistics where these subtypes represent approximately 75%, 10%, and 5% of all kidney cancers, respectively. As shown in Table S1, the control group and patients with RCC were similar regarding sex, age, body mass index (BMI), and smoking status, but showed significant differences in alcohol intake, hypertension, and diabetes prevalence (p < 0.001). Most RCC cases were at stages I–II (81.4%) or grade I–II (75.2%). During the median follow-up period of 90 months, 34 (10.9%) patients died. All participants provided written informed consent prior to the interviews and specimen collection. This study was conducted in compliance with the Declaration of Helsinki and received approval from the Research Ethics Committee of the National Taiwan University Hospital (approval no. 9100201527, 2 July 2012).
2.2. SNP Selection and Genotyping
We selected haplotype-tagged SNPs for the VAV genes (VAV1, VAV2, and VAV3) using Han Chinese data from the 1000 Genomes Project and Haploview version 4.2, applying the criteria of minor allele frequency >0.03 and pairwise linkage disequilibrium r2 > 0.8 [23,24]. A total of 150 SNPs was identified and genotyped. Genomic DNA was extracted from peripheral blood using the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA). Genotyping was performed using the Affymetrix Axiom Genotyping Arrays at the National Centre for Genome Medicine, Taiwan, in accordance with established protocols [25]. All SNPs were in Hardy–Weinberg equilibrium (p > 0.001), with genotyping rates between 97.9 and 100.0%.
2.3. Bioinformatic Analyses
Expression quantitative trait loci (eQTL) associations between SNPs and gene expression were evaluated using the Genotype-Tissue Expression (GTEx) portal [26]. Functional predictions of the SNPs were performed using HaploReg [27]. We analyzed 19 kidney cancer gene expression datasets from Gene Expression Omnibus [28,29,30,31,32,33,34,35,36,37,38,39,40,41], ArrayExpress [42,43], and The Cancer Genome Atlas (TCGA) [44] to explore correlations between gene expression and patient prognosis. For a deeper insight into the molecular mechanisms and pathways involving VAV3, we analyzed the genes correlated with VAV3 in TCGA-KIRC kidney renal clear cell carcinoma samples using Pearson’s correlation via LinkedOmics [45]. Enrichment analysis of GO terms and WikiPathways was conducted using gene set enrichment analysis (GSEA), with thresholds set at a false discovery rate (FDR) <0.05 and 1000 simulations. Tumor-infiltrating immune cell infiltration levels in TCGA-KIRC in relation to VAV3 somatic copy number alterations and expression were compared using the TIMER database [46].
2.4. Statistical Analyses
Logistic regression was used to assess SNP associations with RCC risk by calculating odds ratios (ORs) and 95% confidence intervals (CIs) using the Statistical Package for the Social Sciences (version 19.0.0; IBM, Armonk, NY, USA). Statistical significance was defined as p < 0.05, and multiple testing corrections were applied using the FDR (q-value) [47]. VAV3 expression levels in cancer and adjacent normal tissues were compared using standardized mean difference (SMD) and 95% CI with a random-effects model in Review Manager (RevMan version 5.4.1; Cochrane, London, UK). Associations between VAV3 mRNA expression and RCC patient survival were evaluated by pooling hazard ratios (HRs) and 95% CIs using a random-effects model with RevMan.
3. Results
Logistic regression analysis was conducted to examine the association between genetic variants in VAV genes and the risk of RCC (Table S2). Of the 150 SNPs analyzed, seven SNPs within the VAV genes demonstrated a significant association with the risk of RCC (p < 0.05). Notably, the most significant association was observed for SNP rs17019888 in the VAV3 gene, which had a q-value of 0.320, suggesting that 32.0% of these associations may be false positives (less than one false discovery). Specifically, each additional minor allele C of VAV3 rs17019888 was associated with a 33% reduction in RCC risk (OR = 0.67, 95% CI = 0.50–0.88, p = 0.005; Table 1). This association remained significant after adjusting for confounding factors including sex, age, BMI, smoking status, alcohol intake, and history of diabetes and hypertension (adjusted OR = 0.59, 95% CI = 0.43–0.81, p = 0.001; Table 1).
Table 1.
Association of VAV3 rs17019888 with the risk of renal cell carcinoma.
| Genotype | Controls, n (%) | Patients, n (%) | OR (95% CI) | p | q | OR (95% CI) a | p a |
|---|---|---|---|---|---|---|---|
| TT | 191 (60.8) | 214 (70.9) | 1.00 | 1.00 | |||
| TC | 104 (33.1) | 79 (26.2) | 0.68 (0.48–0.96) | 0.030 | 0.56 (0.38–0.82) | 0.003 | |
| CC | 19 (6.1) | 9 (3.0) | 0.42 (0.19–0.96) | 0.039 | 0.42 (0.17–1.05) | 0.064 | |
| TC/CC | 0.64 (0.46–0.89) | 0.009 | 0.54 (0.37–0.78) | 0.001 | |||
| Trend | 0.67 (0.50–0.88) | 0.005 | 0.320 | 0.59 (0.43–0.81) | 0.001 |
Abbreviations: OR, odds ratio; CI, confidence interval. a ORs were adjusted for sex, age, body mass index, smoking status, alcohol intake, and history of diabetes and hypertension.
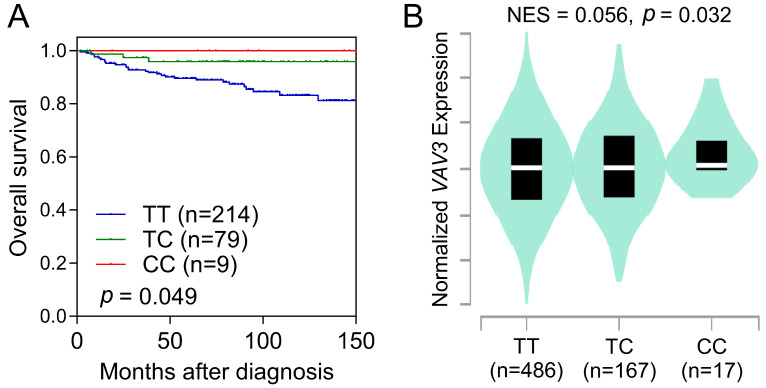
The minor C allele of VAV3 rs17019888 was also linked to overall survival among patients with RCC. Individuals carrying the minor allele C had a 68% reduction in the risk of all-cause mortality compared to those with the major allele homozygous genotype (TT) (HR = 0.32, 95% CI = 0.11–0.90, p = 0.032; Table 2 and Figure 1A). This association persisted after adjusting for sex, age, BMI, smoking status, alcohol intake, history of diabetes and hypertension, and disease stage and grade (adjusted HR = 0.12, 95% CI = 0.02–0.89, p = 0.038; Table 2).
Table 2.
Association of VAV3 rs17019888 with overall survival in patients with renal cell carcinoma.
| Genotype | n of Patients | n of Events | 5-y Survival Rate (%) | HR (95% CI) | p | HR (95% CI) a | p a |
|---|---|---|---|---|---|---|---|
| TT | 214 | 30 | 89.6 | 1.00 | 1.00 | ||
| TC | 79 | 3 | 95.9 | 0.27 (0.08–0.88) | 0.029 | ||
| CC | 9 | 1 | 100.0 | 0.74 (0.10–5.43) | 0.767 | ||
| TC/CC | 88 | 4 | 96.3 | 0.32 (0.11–0.90) | 0.032 | 0.12 (0.02–0.89) | 0.038 |
| Trend | 0.42 (0.17–1.03) | 0.057 | 0.28 (0.07–1.08) | 0.064 |
Abbreviations: HR, hazard ratio; CI, confidence interval. a HRs were adjusted for sex, age, body mass index, smoking status, alcohol intake, history of diabetes and hypertension, and disease stage and grade.
Figure 1.
Survival and expression quantitative trait loci analyses for VAV3 rs17019888. (A) Kaplan–Meier estimation displays overall survival of patients with renal cell carcinoma (RCC) categorized based on the rs17019888 genotype of VAV3. (B) Association between the genotypes of rs17019888 and the expression of VAV3 in whole blood based on the Genotype-Tissue Expression dataset. NES, normalized effect size.
Subsequent eQTL analyses of 670 whole-blood samples from the GTEx project revealed that the minor C allele of rs17019888 was associated with increased VAV3 mRNA expression (p = 0.032; Figure 1B). Despite rs17019888 being located within an intron of VAV3, functional prediction using HaploReg indicated that several SNPs in high linkage disequilibrium with rs17019888 coincided with potential promoter or enhancer regions (Table S3).
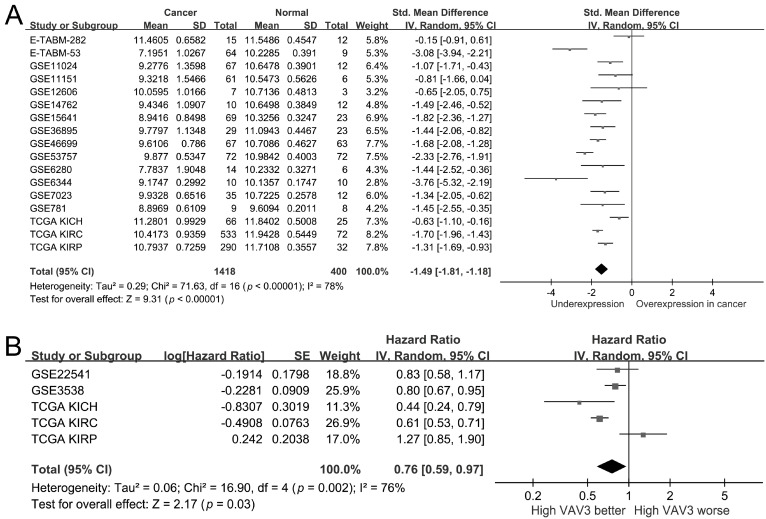
To better understand VAV3’s role in RCC, public kidney cancer datasets were analyzed. A pooled analysis of 1418 kidney cancer and 400 adjacent normal tissues across 17 studies revealed significant downregulation of VAV3 in kidney cancer (SMD = −1.49, 95% CI = −1.81 to −1.18, p < 0.001; Figure 2A). Moreover, higher VAV3 expression was significantly linked to improved patient survival (HR = 0.76, 95% CI = 0.59–0.97, p = 0.03; Figure 2B). Interestingly, VAV3 exhibited downregulation (SMD from −0.63 to −1.70; Figure 2A) across all three TCGA datasets for the main RCC subtypes: KIRC, KIRP, and KICH. High VAV3 expression was significantly associated with improved survival in the TCGA KIRC and KICH datasets, while it tended to be linked to poor survival in the KIRP dataset (Figure 2B).
Figure 2.
The mRNA expression of VAV3 in kidney cancer. (A) Forest plot of differential expression of VAV3 in kidney cancer and normal tissues. (B) Forest plot showing the association between VAV3 expression and kidney cancer prognosis. SD, standard deviation; SE, standard error; IV, inverse variance; CI, confidence interval; Std, standardized; TCGA, The Cancer Genome Atlas; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; df, degrees of freedom [28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44].
To understand the biological significance of VAV3 in RCC, we investigated VAV3-correlated genes in the TCGA-KIRC cohort. We identified 3298 positively correlated and 5606 negatively correlated genes based on FDR < 0.01 for Pearson’s correlation. GO term annotation indicated that VAV3-correlated genes were involved in cellular components such as the collagen trimer, condensed chromatin, and extracellular matrix (Figure 3A). These genes were primarily involved in biological processes such as collagen metabolic processes, chromosome segregation, and extracellular structure organization (Figure 3B). Molecular functions, such as extracellular matrix structural constituents, collagen binding, and nucleosome binding, were negatively regulated by VAV3 (Figure 3C). WikiPathway enrichment analysis suggested that VAV3 negatively regulates pathways related to the ubiquitin–proteasome system, extracellular matrix and membrane receptors, inflammatory response, matrix metalloproteinases, and cell cycle (Figure 3D), which indicate a protective role of VAV3 against RCC.
Figure 3.
Potential biological functions of VAV3 in kidney cancer. Gene ontology annotations of (A) cellular components, (B) biological processes, (C) molecular functions, and (D) WikiPathway enrichment analysis of VAV3 correlated genes.
Given the association between the inflammatory response and cancer development, we examined the correlation between VAV3 expression and immune cell infiltration in RCC. Arm-level deletion of VAV3 was associated with the infiltration of various immune cells, including B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells (Figure 4A). Furthermore, the infiltration levels of B cells, macrophages, and neutrophils positively correlated with VAV3 expression in RCC (Figure 4B). These findings suggest that VAV3 plays a crucial role in modulating immune cell infiltration, particularly in B cells, macrophages, and neutrophils, in RCC.
Figure 4.
Correlation between VAV3 expression and immune cell infiltration levels in the tumor microenvironment. (A) VAV3 copy number variations affect the infiltration levels of multiple immune cell types in The Cancer Genome Atlas kidney renal clear cell carcinoma dataset. ** p < 0.01; *** p < 0.001. (B) Correlation between VAV3 expression and the infiltrating levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells.
4. Discussion
In this study, we employed a combination of genetic and bioinformatics analyses to investigate the associations between genetic variants in VAV family genes and the risk and survival of RCC thoroughly. We identified a significant association between the VAV3 rs17019888 T > C polymorphism and the risk and overall survival of patients with RCC, which remained robust after multivariate analysis and multiple testing correction. Carriers of the rs17019888 C allele exhibited a reduced risk of developing RCC and lower rates of all-cause mortality. Additionally, the rs17019888 C allele was significantly correlated with increased VAV3 mRNA expression. Our pooled analysis of 19 studies indicated that VAV3 may have tumor-suppressive properties, as it is downregulated in kidney cancers, and that higher VAV3 mRNA expression levels were associated with better survival outcomes in patients with RCC. GSEA further suggested that VAV3 may negatively regulate pathways related to the ubiquitin–proteasome system, extracellular matrix and membrane receptors, inflammatory response, matrix metalloproteinases, and cell cycle. Higher VAV3 expression was also correlated with increased infiltration of anticancer immune cells, which is consistent with our genetic findings.
The VAV family of proteins, known as signal transducers, was initially identified for their protumorigenic activities. Various studies have reported that VAV3 enhances cell proliferation in cancers such as breast cancer [11], gastric cancer [48], endometrial cancer [49], osteosarcoma [50], and acute lymphoblastic leukemia [51]. In addition, VAV3 promotes cell migration and invasion in breast cancer [52], pancreatic cancer [53], gastric cancer [48], and osteosarcoma [50]. These findings highlight VAV3 as the pivotal player in cancer progression. However, despite its oncogenic effects and upregulation in several tumors, VAV3 expression is downregulated in certain cancers, including colon adenocarcinoma, head and neck squamous cell carcinoma, kidney cancer, lung adenocarcinoma, and prostate adenocarcinoma, as indicated by TCGA datasets. These data suggest that VAV3 expression levels and their prognostic significance are highly cancer-dependent, necessitating further investigations to confirm VAV3’s specific role in different cancer types.
Furthermore, recent studies have shown that VAV1-deficient mice develop T-cell tumors at high frequencies upon aging or carcinogen treatment [17,54]. VAV1 silences NOTCH1 signaling and suppresses T cell acute lymphoblastic leukemia by promoting the degradation of the intracellular domain of NOTCH1 (ICN1) through ubiquitination and proteasomal degradation [17]. Given the structural similarity and overlapping functions of VAV proteins, it is plausible that VAV3 may also play tumor suppressor roles akin to VAV1. Studies have reported that VAV3-deficient mice develop certain tumors more frequently than their wild-type counterparts [55].
This study is the first to explore the clinical significance and role of VAV3 in RCC using bioinformatic and functional analyses. Our pooled analysis of multiple datasets demonstrated that VAV3 expression was reduced in RCC tumors and that patients with higher VAV3 expression had longer survival times. GSEA revealed that genes co-expressed with VAV3 were enriched in pathways related to the ubiquitin–proteasomal system, extracellular matrix and membrane receptors, inflammatory response, matrix metalloproteinases, and cell cycle. Additionally, high VAV3 expression was correlated with increased infiltration of anticancer immune cells. These findings suggested that VAV3 plays a suppressive role in RCC, possibly through ubiquitin-mediated proteasomal degradation, thereby influencing cancer-associated pathways. However, further research is required to elucidate the precise molecular mechanisms underlying VAV3’s involvement in RCC.
Despite these significant findings, this study has several limitations. First, the small sample size may have reduced the statistical power of detecting significant associations. Second, all study participants were Taiwanese, which may limit the generalizability of our findings to other ethnicities. Third, the lack of detailed clinical information from public datasets precluded adjustments to our analyses. Finally, the exact molecular mechanisms underlying the associations between the identified SNP and RCC survival remain unclear and require further investigation.
5. Conclusions
Our findings suggest that VAV3 rs17019888 T > C polymorphism may serve as a novel prognostic biomarker for RCC. The rs17019888 variant may influence RCC progression by modulating VAV3 expression and affecting cancer-related pathways and immune cell infiltration. However, it is essential to validate these results in larger studies and to conduct further mechanistic investigations to gain a comprehensive understanding of the underlying mechanisms.
Acknowledgments
The authors thank Chao-Shih Chen for data analysis, and the National Centre for Genome Medicine, Taiwan, for technical support. The results published here are based, in part, on data generated by the HaploReg, 1000 Genomes, GTEx, and TCGA projects.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/biomedicines12081694/s1, Table S1. Clinical and demographic characteristics of healthy controls and patients with renal cell carcinoma. Table S2. Association between vav guanine nucleotide exchange factor gene polymorphisms and the risk of renal cell carcinoma. Table S3. Regulatory annotation of VAV3 rs17019888.
Author Contributions
Conceptualization and methodology, C.-F.C., B.-Y.B., Y.-M.H., C.-Y.H. and S.-P.H.; resources and funding acquisition, B.-Y.B., C.-Y.H. and S.-P.H.; formal analysis and investigation, B.-Y.B., P.-L.C., L.-H.C. and C.-Y.L.; data curation and visualization, C.-F.C., J.-H.G. and T.-L.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Research Ethics Committee of the National Taiwan University Hospital (approval no. 9100201527, 2 July 2012).
Informed Consent Statement
Written informed consent was obtained from all participants prior to the interviews and specimen collection.
Data Availability Statement
Data will be available on reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This work was supported by the National Science and Technology Council of Taiwan (grant nos: 110-2314-B-002-113, 111-2314-B-002-240-MY3, 111-2320-B-039-021-MY3, 111-2218-E-037-001, and 112-2314-B-037-127), the Kaohsiung Medical University (grant no: KMUH109-9R63, KMUH109-9R64, KMUH112-2R59 and NHRIKMU-111-I002), and the China Medical University (grant nos: CMU110-MF-59, CMU111-MF-09, and CMU112-MF-10). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Capitanio U., Bensalah K., Bex A., Boorjian S.A., Bray F., Coleman J., Gore J.L., Sun M., Wood C., Russo P. Epidemiology of Renal Cell Carcinoma. Eur. Urol. 2019;75:74–84. doi: 10.1016/j.eururo.2018.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zi H., He S.H., Leng X.Y., Xu X.F., Huang Q., Weng H., Zhu C., Li L.Y., Gu J.M., Li X.H., et al. Global, regional, and national burden of kidney, bladder, and prostate cancers and their attributable risk factors, 1990–2019. Mil. Med. Res. 2021;8:60. doi: 10.1186/s40779-021-00354-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pang C., Guan Y., Li H., Chen W., Zhu G. Urologic cancer in China. Jpn. J. Clin. Oncol. 2016;46:497–501. doi: 10.1093/jjco/hyw034. [DOI] [PubMed] [Google Scholar]
- 4.Kase A.M., George D.J., Ramalingam S. Clear Cell Renal Cell Carcinoma: From Biology to Treatment. Cancers. 2023;15:665. doi: 10.3390/cancers15030665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Purdue M.P., Dutta D., Machiela M.J., Gorman B.R., Winter T., Okuhara D., Cleland S., Ferreiro-Iglesias A., Scheet P., Liu A., et al. Multi-ancestry genome-wide association study of kidney cancer identifies 63 susceptibility regions. Nat. Genet. 2024;56:809–818. doi: 10.1038/s41588-024-01725-7. [DOI] [PubMed] [Google Scholar]
- 6.Bustelo X.R. Vav family exchange factors: An integrated regulatory and functional view. Small GTPases. 2014;5:9. doi: 10.4161/21541248.2014.973757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Movilla N., Bustelo X.R. Biological and regulatory properties of Vav-3, a new member of the Vav family of oncoproteins. Mol. Cell Biol. 1999;19:7870–7885. doi: 10.1128/MCB.19.11.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez-Fdez S., Bustelo X.R. The Vav GEF Family: An Evolutionary and Functional Perspective. Cells. 2019;8:465. doi: 10.3390/cells8050465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katoh H., Hiramoto K., Negishi M. Activation of Rac1 by RhoG regulates cell migration. J. Cell Sci. 2006;119:56–65. doi: 10.1242/jcs.02720. [DOI] [PubMed] [Google Scholar]
- 10.Citterio C., Menacho-Marquez M., Garcia-Escudero R., Larive R.M., Barreiro O., Sanchez-Madrid F., Paramio J.M., Bustelo X.R. The rho exchange factors vav2 and vav3 control a lung metastasis-specific transcriptional program in breast cancer cells. Sci. Signal. 2012;5:ra71. doi: 10.1126/scisignal.2002962. [DOI] [PubMed] [Google Scholar]
- 11.Lee K., Liu Y., Mo J.Q., Zhang J., Dong Z., Lu S. Vav3 oncogene activates estrogen receptor and its overexpression may be involved in human breast cancer. BMC Cancer. 2008;8:158. doi: 10.1186/1471-2407-8-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornstein I., Pikarsky E., Groysman M., Amir G., Peylan-Ramu N., Katzav S. The haematopoietic specific signal transducer Vav1 is expressed in a subset of human neuroblastomas. J. Pathol. 2003;199:526–533. doi: 10.1002/path.1314. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez-Zapico M.E., Gonzalez-Paz N.C., Weiss E., Savoy D.N., Molina J.R., Fonseca R., Smyrk T.C., Chari S.T., Urrutia R., Billadeau D.D. Ectopic expression of VAV1 reveals an unexpected role in pancreatic cancer tumorigenesis. Cancer Cell. 2005;7:39–49. doi: 10.1016/j.ccr.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 14.Wakahashi S., Sudo T., Oka N., Ueno S., Yamaguchi S., Fujiwara K., Ohbayashi C., Nishimura R. VAV1 represses E-cadherin expression through the transactivation of Snail and Slug: A potential mechanism for aberrant epithelial to mesenchymal transition in human epithelial ovarian cancer. Transl. Res. 2013;162:181–190. doi: 10.1016/j.trsl.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Lindsey J.C., Kawauchi D., Schwalbe E.C., Solecki D.J., Selby M.P., McKinnon P.J., Olson J.M., Hayden J.T., Grundy R.G., Ellison D.W., et al. Cross-species epigenetics identifies a critical role for VAV1 in SHH subgroup medulloblastoma maintenance. Oncogene. 2015;34:4746–4757. doi: 10.1038/onc.2014.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abate F., da Silva-Almeida A.C., Zairis S., Robles-Valero J., Couronne L., Khiabanian H., Quinn S.A., Kim M.Y., Laginestra M.A., Kim C., et al. Activating mutations and translocations in the guanine exchange factor VAV1 in peripheral T-cell lymphomas. Proc. Natl. Acad. Sci. USA. 2017;114:764–769. doi: 10.1073/pnas.1608839114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robles-Valero J., Lorenzo-Martin L.F., Menacho-Marquez M., Fernandez-Pisonero I., Abad A., Camos M., Toribio M.L., Espinosa L., Bigas A., Bustelo X.R. A Paradoxical Tumor-Suppressor Role for the Rac1 Exchange Factor Vav1 in T Cell Acute Lymphoblastic Leukemia. Cancer Cell. 2017;32:608–623.E9. doi: 10.1016/j.ccell.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren W., Xu C., Wang S., Li H., Dai H., Yang F., Shao Y., Bai Y. The effect of VAV3 polymorphisms on thyroid cancer. Endocrine. 2022;75:178–184. doi: 10.1007/s12020-021-02827-6. [DOI] [PubMed] [Google Scholar]
- 19.Liu M., Miao N., Zhu Y., Gu C.Y., Shi X.L., Cui W.L., Zhang W., Li Q.X. Association between polymorphism in Vav3 genes and risk of primary prostatic cancer in Chinese Han population. Chin. J. Pathol. 2016;45:451–456. doi: 10.3760/cma.j.issn.0529-5807.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Bai Y., Zheng J., Cheng L., Liu Q., Zhao G., Li J., Gu Y., Xu W., Wang M., Wei Q., et al. Potentially functional genetic variants of VAV2 and PSMA4 in the immune-activation pathway and non-small cell lung cancer survival. J. Gene Med. 2022;24:e3447. doi: 10.1002/jgm.3447. [DOI] [PubMed] [Google Scholar]
- 21.Huang C.Y., Hsueh Y.M., Chen L.C., Cheng W.C., Yu C.C., Chen W.J., Lu T.L., Lan K.J., Lee C.H., Huang S.P., et al. Clinical significance of glutamate metabotropic receptors in renal cell carcinoma risk and survival. Cancer Med. 2018;7:6104–6111. doi: 10.1002/cam4.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang C.Y., Su C.T., Chu J.S., Huang S.P., Pu Y.S., Yang H.Y., Chung C.J., Wu C.C., Hsueh Y.M. The polymorphisms of P53 codon 72 and MDM2 SNP309 and renal cell carcinoma risk in a low arsenic exposure area. Toxicol. Appl. Pharmacol. 2011;257:349–355. doi: 10.1016/j.taap.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 23.Genomes Project C., Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C.Y., Huang S.P., Chen Y.T., Wu H.E., Cheng W.C., Huang C.Y., Yu C.C., Lin V.C., Geng J.H., Lu T.L., et al. TNFRSF13B is a potential contributor to prostate cancer. Cancer Cell Int. 2022;22:180. doi: 10.1186/s12935-022-02590-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang H.H., Lee C.H., Chen Y.T., Huang C.Y., Yu C.C., Lin V.C., Geng J.H., Lu T.L., Huang S.P., Bao B.Y. Genetic Analysis Reveals the Prognostic Significance of the DNA Mismatch Repair Gene MSH2 in Advanced Prostate Cancer. Cancers. 2022;14:223. doi: 10.3390/cancers14010223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Consortium G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward L.D., Kellis M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44:D877–D881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eckel-Passow J.E., Serie D.J., Bot B.M., Joseph R.W., Cheville J.C., Parker A.S. ANKS1B is a smoking-related molecular alteration in clear cell renal cell carcinoma. BMC Urol. 2014;14:14. doi: 10.1186/1471-2490-14-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furge K.A., Chen J., Koeman J., Swiatek P., Dykema K., Lucin K., Kahnoski R., Yang X.J., Teh B.T. Detection of DNA copy number changes and oncogenic signaling abnormalities from gene expression data reveals MYC activation in high-grade papillary renal cell carcinoma. Cancer Res. 2007;67:3171–3176. doi: 10.1158/0008-5472.CAN-06-4571. [DOI] [PubMed] [Google Scholar]
- 30.Gumz M.L., Zou H., Kreinest P.A., Childs A.C., Belmonte L.S., LeGrand S.N., Wu K.J., Luxon B.A., Sinha M., Parker A.S., et al. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin. Cancer Res. 2007;13:4740–4749. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- 31.Jones J., Otu H., Spentzos D., Kolia S., Inan M., Beecken W.D., Fellbaum C., Gu X., Joseph M., Pantuck A.J., et al. Gene signatures of progression and metastasis in renal cell cancer. Clin. Cancer Res. 2005;11:5730–5739. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 32.Kort E.J., Farber L., Tretiakova M., Petillo D., Furge K.A., Yang X.J., Cornelius A., Teh B.T. The E2F3-Oncomir-1 axis is activated in Wilms’ tumor. Cancer Res. 2008;68:4034–4038. doi: 10.1158/0008-5472.CAN-08-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lenburg M.E., Liou L.S., Gerry N.P., Frampton G.M., Cohen H.T., Christman M.F. Previously unidentified changes in renal cell carcinoma gene expression identified by parametric analysis of microarray data. BMC Cancer. 2003;3:31. doi: 10.1186/1471-2407-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pena-Llopis S., Vega-Rubin-de-Celis S., Liao A., Leng N., Pavia-Jimenez A., Wang S., Yamasaki T., Zhrebker L., Sivanand S., Spence P., et al. BAP1 loss defines a new class of renal cell carcinoma. Nat. Genet. 2012;44:751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stickel J.S., Weinzierl A.O., Hillen N., Drews O., Schuler M.M., Hennenlotter J., Wernet D., Muller C.A., Stenzl A., Rammensee H.G., et al. HLA ligand profiles of primary renal cell carcinoma maintained in metastases. Cancer Immunol. Immunother. 2009;58:1407–1417. doi: 10.1007/s00262-008-0655-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Roemeling C.A., Radisky D.C., Marlow L.A., Cooper S.J., Grebe S.K., Anastasiadis P.Z., Tun H.W., Copland J.A. Neuronal pentraxin 2 supports clear cell renal cell carcinoma by activating the AMPA-selective glutamate receptor-4. Cancer Res. 2014;74:4796–4810. doi: 10.1158/0008-5472.CAN-14-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Roche O., Yan M.S., Finak G., Evans A.J., Metcalf J.L., Hast B.E., Hanna S.C., Wondergem B., Furge K.A., et al. Regulation of endocytosis via the oxygen-sensing pathway. Nat. Med. 2009;15:319–324. doi: 10.1038/nm.1922. [DOI] [PubMed] [Google Scholar]
- 38.Wuttig D., Zastrow S., Fussel S., Toma M.I., Meinhardt M., Kalman K., Junker K., Sanjmyatav J., Boll K., Hackermuller J., et al. CD31, EDNRB and TSPAN7 are promising prognostic markers in clear-cell renal cell carcinoma revealed by genome-wide expression analyses of primary tumors and metastases. Int. J. Cancer. 2012;131:E693–E704. doi: 10.1002/ijc.27419. [DOI] [PubMed] [Google Scholar]
- 39.Yusenko M.V., Kuiper R.P., Boethe T., Ljungberg B., van Kessel A.G., Kovacs G. High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinomas and renal oncocytomas. BMC Cancer. 2009;9:152. doi: 10.1186/1471-2407-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yusenko M.V., Zubakov D., Kovacs G. Gene expression profiling of chromophobe renal cell carcinomas and renal oncocytomas by Affymetrix GeneChip using pooled and individual tumours. Int. J. Biol. Sci. 2009;5:517–527. doi: 10.7150/ijbs.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao H., Ljungberg B., Grankvist K., Rasmuson T., Tibshirani R., Brooks J.D. Gene expression profiling predicts survival in conventional renal cell carcinoma. PLoS Med. 2006;3:e13. doi: 10.1371/journal.pmed.0030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cifola I., Spinelli R., Beltrame L., Peano C., Fasoli E., Ferrero S., Bosari S., Signorini S., Rocco F., Perego R., et al. Genome-wide screening of copy number alterations and LOH events in renal cell carcinomas and integration with gene expression profile. Mol. Cancer. 2008;7:6. doi: 10.1186/1476-4598-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corbin M., de Reynies A., Rickman D.S., Berrebi D., Boccon-Gibod L., Cohen-Gogo S., Fabre M., Jaubert F., Faussillon M., Yilmaz F., et al. WNT/beta-catenin pathway activation in Wilms tumors: A unifying mechanism with multiple entries? Genes Chromosomes Cancer. 2009;48:816–827. doi: 10.1002/gcc.20686. [DOI] [PubMed] [Google Scholar]
- 44.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vasaikar S.V., Straub P., Wang J., Zhang B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956–D963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li T., Fan J., Wang B., Traugh N., Chen Q., Liu J.S., Li B., Liu X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan B., Li Y., Zhao Q., Fan L., Wang D., Liu Y. Inhibition of gastric cancer cell growth and invasion through siRNA-mediated knockdown of guanine nucleotide exchange factor Vav3. Tumour Biol. 2014;35:1481–1488. doi: 10.1007/s13277-013-1204-2. [DOI] [PubMed] [Google Scholar]
- 49.Jing L., Hua X., Yuanna D., Rukun Z., Junjun M. Exosomal miR-499a-5p Inhibits Endometrial Cancer Growth and Metastasis via Targeting VAV3. Cancer Manag. Res. 2020;12:13541–13552. doi: 10.2147/CMAR.S283747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao Y., Li C., Wang H., Liu Y. LINC00265 targets miR-382-5p to regulate SAT1, VAV3 and angiogenesis in osteosarcoma. Aging. 2020;12:20212–20225. doi: 10.18632/aging.103762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nayak R.C., Chang K.H., Singh A.K., Kotliar M., Desai M., Wellendorf A.M., Wunderlich M., Bartram J., Mizukawa B., Cuadrado M., et al. Nuclear Vav3 is required for polycomb repression complex-1 activity in B-cell lymphoblastic leukemogenesis. Nat. Commun. 2022;13:3056. doi: 10.1038/s41467-022-30651-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ojala V.K., Knittle A.M., Kirjalainen P., Merilahti J.A.M., Kortesoja M., Tvorogov D., Vaparanta K., Lin S., Kast J., Pulliainen A.T., et al. The guanine nucleotide exchange factor VAV3 participates in ERBB4-mediated cancer cell migration. J. Biol. Chem. 2020;295:11559–11571. doi: 10.1074/jbc.RA119.010925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsuboi M., Taniuchi K., Furihata M., Naganuma S., Kimura M., Watanabe R., Shimizu T., Saito M., Dabanaka K., Hanazaki K., et al. Vav3 is linked to poor prognosis of pancreatic cancers and promotes the motility and invasiveness of pancreatic cancer cells. Pancreatology. 2016;16:905–916. doi: 10.1016/j.pan.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 54.Ruiz S., Santos E., Bustelo X.R. The use of knockout mice reveals a synergistic role of the Vav1 and Rasgrf2 gene deficiencies in lymphomagenesis and metastasis. PLoS ONE. 2009;4:e8229. doi: 10.1371/journal.pone.0008229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cuadrado M., Robles-Valero J. VAV Proteins as Double Agents in Cancer: Oncogenes with Tumor Suppressor Roles. Biology. 2021;10:888. doi: 10.3390/biology10090888. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be available on reasonable request.