Abstract
Background
We have previously demonstrated the significant reliance of pancreatic Cancer Stem Cells (PaCSCs) on mitochondrial oxidative phosphorylation (OXPHOS), which enables versatile substrate utilization, including fatty acids (FAs). Notably, dysregulated lipid scavenging and aberrant FA metabolism are implicated in PDAC progression.
Methods & results
Our bioinformatics analyses revealed elevated expression of lipid metabolism-related genes in PDAC tissue samples compared to normal tissue samples, which correlated with a stemness signature. Additionally, PaCSCs exhibited heightened expression of diverse lipid metabolism genes and increased lipid droplet accumulation compared to differentiated progenies. Treatment with palmitic, oleic, and linolenic FAs notably augmented the self-renewal and chemotherapy resistance of CD133+ PaCSCs. Conversely, inhibitors of FA uptake, storage and metabolism reduced CSC populations both in vitro and in vivo. Mechanistically, inhibition of FA metabolism suppressed OXPHOS activity, inducing energy depletion and subsequent cell death in PaCSCs. Importantly, combining a FAO inhibitor and Gemcitabine treatment enhanced drug efficacy in vitro and in vivo, effectively diminishing the CSC content and functionality.
Conclusion
Targeting FAO inhibition represents a promising therapeutic strategy against this highly tumorigenic population.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-024-05598-6.
Keywords: Pancreatic cancer, Cancer stem cells, Lipid metabolism, Fatty acid, Oxidative phosphorylation, Chemoresistance
Introduction
Pancreatic ductal adenocarcinoma (PDAC), the most common form of pancreatic cancer, is a disease with an unfavorable prognosis due to its late diagnosis, as symptoms are nonspecific even at advanced disease stages, and its intrinsic resistance to established therapeutic options such as chemotherapy and radiotherapy [1]. Despite its relatively low incidence, PDAC is the seventh leading cause of cancer-related deaths worldwide and one of the most lethal solid tumors [2] with a poor long-term outcome: the estimated one-year overall survival rate of these patients is 24% [3].
The main malignant features of PDAC, i.e., intrinsic chemoresistance and elevated metastasis rate, can be partially attributed to specific subpopulations of cancer cells with tumor and metastasis-initiating properties, known as pancreatic cancer stem cells (PaCSCs) [4, 5]. CSCs are characterized by the capacity to undergo unlimited cell division while retaining their stem cell identity (self-renewal) and the ability to differentiate into diverse specialized cell types [4, 5]. Although migratory and invasive abilities are not restricted to CSCs, only metastatic stem-like cells would be able to initiate secondary lesions upon surviving in the bloodstream as circulating tumor cells (CTCs) [6]. Considering their intrinsic chemoresistance leading to tumor relapse, the design of combined treatments targeting both PaCSCs and non-CSC populations may represent a promising strategy for improving the long-term survival of PDAC patients.
In the recent years, our group reported that mitochondria are essential organelles for stemness maintenance and tumorigenicity, representing a key vulnerability for PaCSCs. Indeed, perturbations in various processes throughout the mitochondrial life cycle, ranging from biogenesis [7], fission [8] and recycling via mitophagy [9], to interference with mitochondrial activity via oxidative phosphorylation (OXPHOS) inhibition [7] and alteration of redox state [10], all of which significantly impair the tumorigenicity and chemoresistance of PaCSCs.
ATP production via OXPHOS requires a large amount of acetyl-CoA, which is commonly supplied by glycolysis, but can also be produced by β-oxidation of fatty acids (FAs). FAs from exogenous sources can be internalized via membrane transporters or lipid receptors such as CD36 or LRPs and then directly metabolized or stored in lipid droplets (LDs). Importantly, increased lipid uptake and aberrant FA metabolism have been linked to disease progression and poor prognosis in PDAC patients [11–13]. Additionally, studies in other cancers have suggested that lipid metabolism plays a critical role in CSC maintenance, thereby supporting cell membrane formation and energy production [14, 15]. Accordingly, we hypothesized that lipid metabolism, particularly FA metabolism, represents a pharmacologically targetable vulnerability for PaCSCs.
Indeed, we showed that several lipid metabolism genes are upregulated in PaCSCs and are correlated with stemness and poor survival in PDAC patients. PaCSCs show increased lipid storage in LDs and FA oxidation (FAO), and FAO inhibition markedly impaired OXPHOS activity, leading to an energy crisis and cell death. Finally, FAO inhibition improved the response to Gemcitabine both in vitro and in vivo, suggesting a new therapeutic strategy that may help improve the outcome of PDAC patients.
Materials and methods
Human data analysis
Expression data from human PDAC tissue and normal pancreatic tissue were analyzed using the webserver GEPIA2 (TCGA and the GTEx project databases; http://gepia2.cancer-pku.cn/) [16]. The Pearson correlation coefficient was calculated to study the association of the individual genes corresponding to lipid metabolism with a stemness signature defined by the combined expression of the pluripotency-related genes KLF4, OCT4, NANOG and SOX2. Additionally, we calculated the overall survival for pancreatic cancer patients from the respective upper and lower quartiles of the expression of these specific lipid metabolism genes; the hazard ratio (HR) was calculated from GEPIA2 using the Cox proportional hazards model.
Cell culture
PDAC patient-derived xenografts (PDXs): A6L, 185, 215, 253, 265 and 354 were obtained from the Biobank of the Spanish National Cancer Research Centre (CNIO), Madrid, Spain (MTAs #CNIO20-027, #CNIO21-253). PDAC PDX-derived cultures were established as previously described [17]. Pancreatic circulating tumor cells (CTCs): The metastatic model CTCA was established from circulating tumor cells and obtained through the Barts Pancreas Tissue Bank of the Barts Cancer Institute (https://www.bartspancreastissuebank.org.uk/; BCI, London, United Kingdom; 2019/02/IISA/PS/E/Cellcultures).
The cells were grown in RPMI 1640 medium (61870044) supplemented with 10% FBS and 50 U/mL penicillin/streptomycin (all from Gibco, Life Technologies, Carlsbad, CA, USA). The cells were cultured under standard conditions of 5% CO2, 95% humidity, and 37 °C, propagated by treatment with 1X trypsin with 0.2% EDTA (Corning, Oneonta, NY, USA) and subjected to a maximum of 15 passages. For experiments, the medium was changed to sphere medium [DMEM/F-12 (31331028) supplemented with 2% B27 (both from Gibco) and 20 ng/mL FGFbasic (Pan-Biotech, Aidenbach, Germany)], ensuring proper comparison of cells grown in adherent conditions with cells grown as spheres and minimizing any interference resulting from the different concentrations of glucose and other factors present in each media.
Cancer stem cell-enriching culture
For enrichment of CSCs, cells were grown as spheres as previously described [7]. Briefly, the cells were trypsinized, centrifuged at 1200 rpm for 5 min and resuspended in sphere medium [DMEM/F-12 supplemented with 2% B27 (both from Gibco) and 20 ng/mL FGFbasic (Pan-Biotech)]. The cells were then seeded at a density of 105 cells/mL in flasks covered with 10% poly-HEMA (2-hydroxyethyl methacrylate, Sigma–Aldrich, Saint Louis, MO, USA) in 96% ethanol. First generation spheres were grown for seven days. For serial passaging, spheres were harvested using a 40 μm cell strainer, dissociated with trypsin (Corning, Oneonta, NY, USA) and regrown at 105 cells/mL for five more days.
In vitro treatments
FA inhibitors included 200 µM Etomoxir (CPT1A inhibitor) [18] (E1905, Sigma–Aldrich), 100 µM Mildronate (carnitine synthesis inhibitor) [19] (15997, Cambridge Biosciences, Cayman, UK), 100 µM Ranolazine (3-ketoacyl-CoA thiolase inhibitor) [18] (15604, Cambridge Biosciences), all of which were dissolved in dH2O, and 1 µM Perhexiline (SML010, CPT1/CPT2 dual inhibitor) [18] (Sigma-Aldrich) which was dissolved in DMSO following the manufacturer’s instructions. The cells were treated for 24 to 72 h.
FA supplementation included 50 and 100 µM oleic acid (OA) (O3008, Sigma Aldrich), 50 µM sodium palmitate (P9767, Sigma Aldrich) or 200 µM linolenic acid (LNA) (L2376, Sigma Aldrich) conjugated with bovine serum albumin, Fraction V, Fatty Acid-Free, Nuclease-Free and Protease-Free (126609 Sigma-Aldrich). The cells were treated for 24 to 72 h.
Chemotherapy: Gemcitabine 0.9% sodium chloride (Eli Lilly and Company, IN, USA) was used at concentrations ranging from 10 to 5000 nM for 48 h.
Real time quantitative polymerase chain reaction (RTqPCR)
RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The RNA concentration and purity were determined by spectrophotometry (Nanodrop™ 2000, ThermoFisher Scientific). 1 µg of RNA was used for cDNA synthesis using Maxima H minus cDNA synthesis Master Mix with dsDNase kit (Thermo Fisher Scientific), followed by SYBR Green RTqPCR (PowerUp™ SYBR Green Master Mix, Applied Biosystems, Thermo Fisher Scientific) according to the manufacturer’s instructions. The primers used are detailed in Table 1.
Table 1.
List of primers used for real time qPCR
| Gene | Forward primer (5’→3’) | Reverse primer (5’→3’) |
|---|---|---|
| ACTB | GGCACCACACCTTCTACAATG | GTGGTGGTGAAGCTGTAG |
| ALOX5 | TACATCGAGTTCCCCTGCTAC | GTTCTTTACGTCGGTGTTGCT |
| APOBR | AGACAGAGACAGACAGGATGGA | GGTGCCGAGGGAATCCAGTG |
| APOER2 | AGACGCTGATCTCCTCCACT | CAGGCCATTGAGCCGATTTG |
| ASAH1 | GCACAAGTTATGAAGGAAGCCAAG | TCCAATGATTCCTTTCTGTCTCG |
| BDH1 | TTTGGAACCACCGGGAGGA | CAATAGTGGGCGTCTTGCTC |
| CPT1A | CTGTGGAGAAAGCAGCGTTCT | TCGTAACATCGGCCGTGTAGT |
| DEGS2 | CAGCGACTTCGAGTGGGTCT | GGGTACTTGGCCAGTATCTCCTT |
| DGAT1 | TCGCCTGCAGGATTCTTTAT | GCATCACCACACACCAGTTC |
| HACD2 | TGCATGGACGATCACGGAAA | CCTGGCCCATTTGATGAGGT |
| HADHA | TTCAGCAACTTGACTGGGCA | CAGTGATCTGGAATCACCGCT |
| HPRT | TGACCTTGATTTATTTTGCATACC | CGAGCAAGACGTTCAGTCCT |
| KLF4 | AATTACCCATCCTTCCTGCCC | CCGGGACTGACCTTGGTAAT |
| LDLR | CACAACCAGGACGGCTACAG | AGATGTTCACGCCACGTCAT |
| LPCAT2 | TCTTCGGCTCAGTTTTGGCT | CATAGTTGAGCCGGGCGTAG |
| LPCAT4 | TGTGACCTGCCCAAAGTTGT | TGTGACCTGCCCAAAGTTGT |
| MGLL | CTCATTTCGCCTCTGGTTCTTG | GTGAGACGGCATTCAGCAGTTG |
| NANOG | AGAACTCTCCAACATCCTGAACCT | TGCCACCTCTTAGATTTCATTCTCT |
| OCT3/4 | CTTGCTGCAGAAGTGGGTGGAGGAA | CTGCAGTGTGGGTTTCGGGCA |
| PC | GCGTGTTTGACTACAGTGAG | TCTTGACCTCCTTGAACTTG |
| PEX13 | CGAGCAGCTACCTCAGCAAA | TGGTCATCCTCACCACTTGC |
| PLA2G4 | AGCACCAGTATTCCCACAAGTT | TCAAAGGTCTCATTCCACACAG |
| PPARD | CTCTATCGTCAACAAGGACG | GTCTTCTTGATCCGCTGCAT |
| PPARG | GACCTGAAACTTCAAGAGTACCAA | TGAGGCTTATTGTAGAGCTGAGT |
| PTGES3 | ACACTTTCCTCTTCTCCCCG | GGCGACTCCGCTTTTTCTCT |
| SCD | GCAAACACCCAGCTGTCAAA | AAGCCAGGTTTGTAGTACCTCC |
| SGPP2 | ACTCCTGCTCTGTGGTGAGA | GGCTAGCAAGACAAGGGTGT |
| SLC27A1 | CTCTCTGCTTCCCCAGGATG | CCCCAGCAGCCACAACAG |
| SLC27A4 | GGCTCAGGGGCCAATAAACT | ACAGATGAGGCGGGTCAATG |
| SOX2 | AGAACCCCAAGATGCACAAC | AGAACCCCAAGATGCACAAC |
| SPT1 | AGGAAGCGGCTAACTATGGC | TCGTAAAGCGCCTGTACCAT |
| SPTLC2 | ATGGCACCAGCCTTGGTAAA | TTTGGCAGGCATGTAGAGCA |
| TBXAS1 | CCTTCTCCTGGCTCATTTA | TCGTCTCGGTTCTTATTGG |
Droplet digital PCR (ddPCR)
Sample preparation
Blood and pancreatic tissue samples from mice bearing orthotopic tumors were harvested and processed for FACS sorting. Total blood samples were centrifuged and resuspended in ACK lysing buffer (Thermo Fisher Scientific) for 5 min. Pancreatic tumors were minced mechanically and enzymatically digested with collagenase P for 15 min at 37 °C followed by trypsin for 3 min at 37 °C. Next, cell suspensions were blocked in Flebogamma (1:10 dilution; Grifols) for 15 min at 4 °C and incubated with an hEpCAM-APC antibody (Miltenyi Biotec) or an appropriate isotype-matched control antibody (IgG2a-APC, BD Bioscience) for 30 min at 4 °C. Then, 96 single cells double positive for hEpCAM-APC and GFP and negative for DAPI (Sigma-Aldrich), were sorted in a BD FACSAria™ II into a 96-well plate containing 100 µl of GTC mix [4.19 M Guanidine thiocyanate; 25 mM Na Citrate pH = 7.3; 15 mM Sarcosyl; 11 mM 2-Mercaptoetanol; 18 µM Glycogen] and plates stored at -80 °C.
RNA extraction, cDNA synthesis and pre-amplification reaction
Individual cells were mixed with 10 µl of 2 M NaOAc and 100 µl 100% Phenol/Chloroform (both at pH 4) and then centrifuged for 13,000 rpm at 4 °C for 10 min. The upper aqueous phase was transferred to low binding tubes and mixed with 2 µl of 5 mg/mL linear acrylamide (Amresco®). After precipitation with isopropanol and ethanol washes, the RNA pellets were resuspended in RNasefree water and immediately reverse-transcribed with SuperScript® VILO cDNA Synthesis Kit (Thermo Fisher Scientific) following manufacturer´s instructions. Then, the cDNAs were mixed with the appropriate primers (CPT1A and HPRT) and a standard PCR using AmpliTaq Gold® 360 Master Mix (Thermo Fisher) was performed.
Droplet digital PCR (ddPCR)
The ddPCR was performed following the official ddPCR™ application guide performed in a QX100™ Droplet Digital™ PCR system (Bio-Rad). Briefly, mixes containing the pre-amplified cDNA, the specific primers used for each reaction, together with QX200™ ddPCR™ EvaGreen Supermix and QX200™ Droplet Generation Oil for EvaGreen (Bio-Rad), were plated into ddPCR™ cartridges (DG8™ Cartridges for QX200™/QX100™, Bio-Rad) and incubated in a QX100™ Droplet Generator (Bio-Rad). After droplet generation, 40 µl of the generated droplet emulsions were transferred into a new 96-well PCR plate (Eppendorf), foil sealed (PX1™ PCR Plate Sealer, Bio-Rad) and amplified into a C1000 Touch™ Thermal Cycler (Bio-Rad) following manufacturer´s instructions. Following PCR amplification, the plates were measured in a QX100™ Droplet Reader (Bio-Rad) and data analyzed using the software QuantaSoft 1.3.2.0 (Bio-Rad).
BODIPY staining
Cells grown on coverslips were fixed in 4% paraformaldehyde for 30 min at 4°C. Then, the cells were incubated with BODIPY® 493/503 (Thermo Fisher Scientific) at 2.5 µg/mL for 1 h at 4°C, inside a humid chamber. Coverslips were then washed with PBS and mounted with ProLong® with DAPI (1 µg/mL). Images were taken with a Zeiss LSM 710 confocal microscope.
Flow cytometry analysis
The cells were resuspended in blocking buffer (2% FBS, 0.5% BSA in PBS) for 15 min on ice under agitation. The cells were stained for 30 min at 4°C with APC-conjugated anti-CD133 or anti-Epcam antibodies (BioLegend, San Diego, USA) or the corresponding control immunoglobulin G1 antibody (IgG1, BioLegend). When indicated, the cells were also incubated for 15 min at 4°C (LD540, Nile red, LipidTOX™, Bodipy®) or 90 min at 37°C (FAO Blue) (Table 2). Annexin V-FITC staining was performed on attached and floating cells according to the manufacturer’s instructions (550474 & 556454, BD Biosciences, San Diego, CA, USA). Zombie Violet Dye (77477, Biolegend) or DAPI were used to exclude nonviable cells. A total of 50,000 cells per sample were analyzed using a FACSCanto II (BD, Franklin Lakes, NJ, USA) or ImageStream X Mark II (Amnis, Seattle, WA, USA) and analyzed with FlowJo 9.2 software (Ashland, OR, USA).
Table 2.
Flow cytometry probes
| Probe | Concentration | Manufacturer | Staining |
|---|---|---|---|
| LD540 | 25 µg/ml | Kindly donated by Luca Tirinato (King Abdullah University of Science and Technology) | Lipid droplets |
| Nile red | 0.5 µg/ml | Sigma Aldrich | Membrane and intracellular lipid droplets |
| LipidTOX™ Deep Red solution | 1:1000 | ThermoFisher Scientific | Neutral lipids |
| BODIPY® 493/503 | 0.5 µg/ml | ThermoFisher Scientific | Neutral lipids |
| FAO Blue | 20 µM | Funakoshi (Tokyo, Japan) | Fatty acid β-oxidation (FAO) activity |
XF extracellular flux analyzer experiments
The oxygen consumption rate (OCR) was determined by using the XF Mito Cell Stress Test, XF Long Chain Fatty Acid Oxidation Stress Test, or Palmitate Oxidation Stress Test (Agilent Technologies, Santa Clara, CA, USA). A total of 30,000 cells per well were cultured in an XF 96-well cell culture microplate (Agilent Technologies) previously coated with Cell-Tak (BD Biosciences) in growth medium for 24 h. Then, the cells were incubated for 1 h in base assay medium (D5030, Sigma Aldrich) supplemented with 2 mM glutamine, 10 mM glucose, and 1 mM pyruvate at 37 °C. The concentrations of Oligomycin and FCCP were adjusted for each primary cell type as follows: Oligomycin, 1.2 mM for 215 and 253 cells; and 0.8 µM for 354 cells; FCCP 1.2 µM for 215 and 253; and 0.4 µM for 354 cells. Oligomycin, FCCP, Rotenone (1 µM) and Antimycin A (1 µM) were dissolved in DMSO (all from Sigma-Aldrich). For the Long Chain Fatty Acid Oxidation Stress Test, Etomoxir (40 or 100 µM) or Ranolazine (50 µM) were injected into port A prior to Oligomycin, FCCP and Rotenone + Antimycin. For the Palmitate Oxidation Stress Test, cells were incubated for 1 h with FAO medium containing 1 mM glutamine, 2.5 mM glucose and 0.5 mM carnitine, and BSA-palmitate (100 µM) was injected in port A prior to Oligomycin, FCCP and Rotenone + Antimycin.
The percentage of complex I inhibition was calculated as the percentage of OCR inhibited upon compound injection with respect to the inhibition obtained with Rotenone, the latter used as 100%, as described previously [7]. Experiments were run in a XF96 analyzer (Seahorse Bioscience, Agilent Technologies), and the raw data were normalized to the protein content using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific).
IncuCyte live-cell assay
Cells were seeded at 20,000 cells/well in 96-well culture plates in 100 µL complete RPMI medium. After 24 h, the cells were subjected to different treatments together with LD540 (25 ng/ml) or Annexin V-FITC (1:100) and incubated for 15 min, after which the plate was inserted into an IncuCyte® Live-Cell Analysis System (Sartorius, Göttingen, Germany) for real-time imaging, with two fields imaged per well at 10x magnification every 2 h for a total of 4 days. The fluorescence and confluence data were analyzed using IncuCyte® Confluence version 2021 C software. The data shown represent fluorescence normalized to confluence at each time point.
For cytosolic ATP, PDX265 and 215 cells were infected with the Incucyte® CytoATP Lentivirus Reagent Kit following the manufacturer’s instructions and subjected to puromycin selection to generate a stably expressing population. PDX cells stably expressing CytoATP or the non-binding control were seeded at 10,000 cells/well in a 96-well microplate. After 24 h, the cells were exposed to different treatments and the ATP ratio was analyzed by luminescence using IncuCyte® Confluence version 2021 C software.
Sphere formation assay
A total of 104 cells were seeded in triplicate in sphere medium using polyHEMA-coated 24-well plates in the presence of different treatments. Seven days later, spheres were harvested for subsequent assays or counted with an inverted EVOS FL microscope (Thermo Fisher Scientific) using a 10x objective with phase contrast.
Colony formation assay
For the colony formation assay (CFA), 500 or 1000 cells were seeded per well in 2 mL of complete RPMI medium. After 24 h, the treatments were added to sphere medium. The media and treatments were refreshed every 7 days. After 21 days, the cells were stained with crystal violet (Acros Organics, Thermo Fisher Scientific) and the number of colonies was manually counted.
ATP
The cell pellets were washed with PBS, resuspended in ultrapure water (10977035, Invitrogen) and frozen for lysis. ATP was quantified using an ATP Determination Kit (A22066, Invitrogen) following the manufacturer’s instructions. Bioluminiscence was determined using a Synergy™ HT Multi-Mode Microplate Reader (BioTek, Winooski, Vermont, USA). The results were normalized to the protein concentration measured for the same samples with the Pierce™ BCA Protein Assay Kit.
Western blot
Cell pellets were lysed with RIPA buffer (Sigma Aldrich) supplemented with protease and phosphatase inhibitors (both from Alfa Aesar, Thermo Fisher Scientific). The protein concentration was determined with the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific). Proteins (30 µg protein/lane) were separated on Novex™ WedgeWell™ 10% Tris-Glycine precast gels using BenchMark™ pre-stained protein ladder (Invitrogen) and transferred to PVDF membranes (Thermo Fisher Scientific). The membranes were blocked in 5% BSA-1X PBS-0.1% Tween 20 (Thermo Fisher Scientific) for 1 h at room temperature and incubated overnight at 4°C with the following primary antibodies: pAMPK (Thr172), AMPK (both from Cell Signaling Technology, Inc, Danvers, MA, USA), NANOG, DGAT1, MGLL, PPARD (all from Santa Cruz Biotechnology, Inc, Dallas, Texas, USA) and β-actin as loading control (clone AC74, Sigma Aldrich). Afterwards, the membranes were subjected to peroxidase–conjugated secondary antibody (Invitrogen) and developed by chemiluminescence (Pierce™ ECL Western Blotting Substrate) using CL-XPosure™ (Thermo Fisher Scientific) films. The bands were analyzed using ImageJ software.
Metabolic activity
A total of 104 cells were seeded in triplicate in 96-well plates 24 h before treatment. 72 h after treatment, resazurin (Alfa Aesar) was added to the cells at a concentration of 10 µM in 1X PBS and the cells were incubated for 1 h at 37°C with 5% CO2. The fluorescence was assessed according to the manufacturer’s instructions by using a Synergy HT plate reader.
In vivo experiments
Mice were housed according to institutional guidelines and all experimental procedures were performed in compliance with the institutional guidelines for the welfare of experimental animals as approved by the University of Zaragoza Ethics Committee (CEICA PI22/17, PI35/19, PI41/20) and in accordance with the guidelines for Ethical Conduct in the Care and Use of Animals as stated in The International Guiding Principles for Biomedical Research involving Animals, developed by the Council for International Organizations of Medical Sciences (CIOMS).
For the ddPCR, a total of 5 × 104 PDX354-GFP cells were orthotopically injected into NU-Foxn1nu (Charles River, UK) nude mice (n = 8). Once the mice showed signs of disease, they were humanely sacrificed, and pancreatic tumors and total blood were harvested.
For the tumorigenicity assay (Extreme-Limiting Dilution Assay, ELDA), cells were treated in vitro for 48 h, trypsinized and resuspended in sphere medium with Matrigel™ (Corning) (50:50). Two cell densities (104 and 103) were subcutaneously injected into both the top and bottom flanks of six week-old Foxn1nu nude mice of both sexes (n = 4 mice per group, n = 8 injections per group). Tumor size was monitored once a week using a digital caliper and tumor volume was calculated using the formula (length*width2)/2. All the mice were sacrificed at the same time, when one of the tumors had reached the humane endpoint. ELDA calculations were performed at https://bioinf.wehi.edu.au/software/elda/.
For in vivo treatment, tumor pieces of approximately 15 mm3 were soaked in Matrigel™ prior to subcutaneous implantation in both flanks of six week-old Foxn1nu nude female mice (Envigo, IN, USA) (n = 5 mice per group, n = 10 implants per group) under isofluorane-induced anesthesia. When the tumor size was approximately 300 mm3, the mice were treated with 70 mg/kg Gemcitabine (i.p.) once a week for three weeks with or without 130 mg/kg Ranolazine or the corresponding dose of vehicle (PBS) (oral gavage) once a day until the endpoint. Tumor size was monitored twice a week using a caliper and tumor volume was calculated using the formula (length*width2)/2. After 13 weeks, when the control tumors had reached the humane endpoint criteria (maximum 1000 mm3), the mice were euthanized, the tumors were collected and weighed and pictures were taken. A small piece of tumor tissue was processed for RNA to assess lipid metabolism genes by RTqPCR as described above, or for protein extraction to assess p-AMPK/AMPK by Western blot. The remaining tumor tissues were dissociated as previously reported [7] and stained with EpCAM-FITC, CD133-PE and CD44-APC antibodies for FACS analysis as described above.
Statistical analysis
The data are expressed as the mean value of at least three experiments ± SEM. The statistical analysis was carried out with GraphPad Prism 8 (GraphPad Software Inc, USA). The significant differences were determined using, in general, analysis of variance (ANOVA, Chicago, IL, USA) and post hoc Bonferroni correction or Kruskal-Wallis tests, depending on the results of the Shapiro-Wilk normality test; p < 0.05 was considered to indicate statistical significant. Significant differences were classified as follows: ∗: p < 0.05; ∗∗: p < 0.01; ∗∗∗: p < 0.001.
Results
Lipid metabolism genes are linked to stemness in PDAC
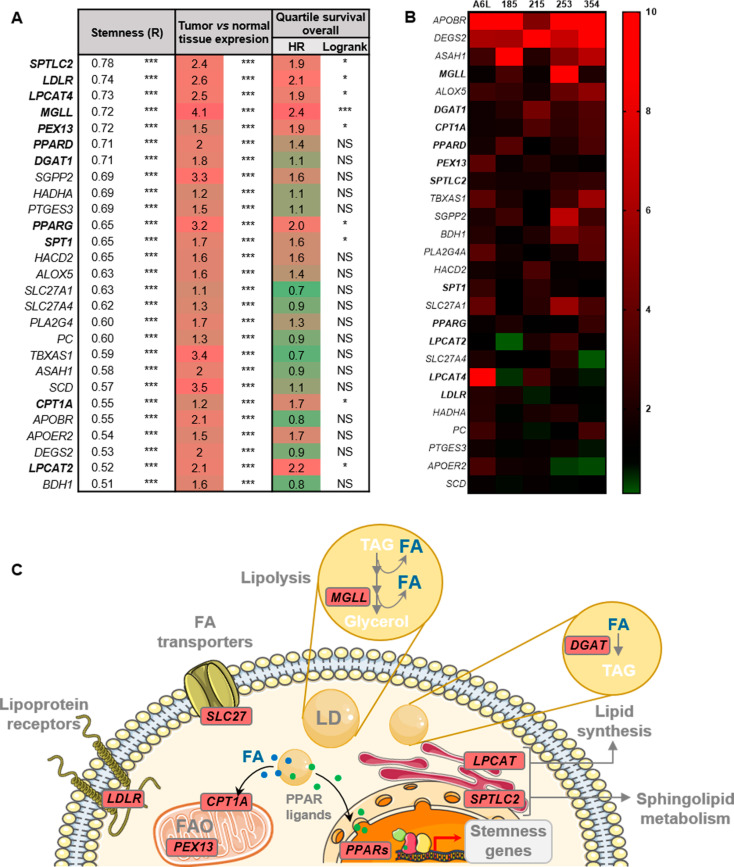
To explore the possible association of lipid metabolism with aggressiveness and stemness in PDAC, we first carried out correlation analyses of human gene expression data from the TCGA and GTEx datasets (normal pancreas/PDAC). For these analyses, we included all the genes related to lipid metabolism (GO pathways: sphingolipid acid metabolic process, fatty acid oxidation, fatty acid biosynthetic process, fatty acid metabolic process, medium/long-chain fatty acid catabolic process, positive/negative regulation of fatty acid biosynthetic process and fatty acid transporters) and our established stemness signature composed of the combination of NANOG, KLF4, OCT3/4 and SOX2 [7]. Among the genes with greater correlations with stemness, we identified lipoprotein receptors (LDLR, APOBR, APOER2), fatty acid transporters (SLC27A1, SLC27A4), genes related to LD maintenance and metabolism (LPCAT2, LPCAT4, MGLL, DGAT1), sphingolipid and arachidonic acid metabolism (SPTLC2, SGPP2, PTGES3, ALOX5, PLA2G4, TBXAS1), FAO (PEX13, HADHA, BDH2, ASAH1) and key general lipid metabolism regulators (PPARD, PPARG) (Fig. 1A). Importantly, all these genes were commonly overexpressed in PDAC tissue compared to normal tissue and we found that most of these overexpressed genes were predictive of poor overall survival in PDAC patients, which further suggests their important role in the aggressiveness of PDAC. Next, we further corroborated our finding that lipid metabolism genes with a greater correlation with stemness in PDAC patients were also significantly upregulated in PaCSC-enriched cultures (spheres, with increased expression of stemness genes and CD133 (Figure S1)) from five primary PDAC PDXs compared to their non-CSC counterparts (Fig. 1B). Figure 1C depicts some of the genes commonly upregulated in PDAC vs. normal tissue and CSCs vs. differentiated cells in their corresponding cellular pathways.
Fig. 1.
Lipid metabolism genes correlate with stemness in PDAC. (A) Bioinformatics analyses of lipid metabolism genes in human data included in the TCGA and GTEx project databases, were carried out with the webserver GEPIA2 (http://gepia2.cancer-pku.cn/). The genes were sorted by their correlation (R, first column) with a stemness signature composed of the genes NANOG, KLF4, OCT3/4 and SOX2. Data relative to expression in tumor vs. normal tissue and overall survival in the highest and lowest expression quartiles for each gene are shown in the second and third columns, respectively. (B) Heatmap of lipid metabolism genes overexpressed in PaCSC-enriched cultures (spheres) from five PDAC PDXs (A6L, 185, 215, 253, 354), evaluated by RTqPCR. In bold, genes determined in A with R > 0.7 or significantly correlated with poor survival. (C) Schematic representation of the main genes and pathways correlated with stemness in PDAC. Genes indicated in bold in A and B are highlighted in red in the drawing, indicating their corresponding pathways. FA: Fatty acid; LD: Lipid droplets; TAG: Triglycerides. The data are presented as the mean ± SEM of at least three independent experiments. * p < 0.05; *** p < 0.001
Increased lipid storage and metabolism in PaCSCs
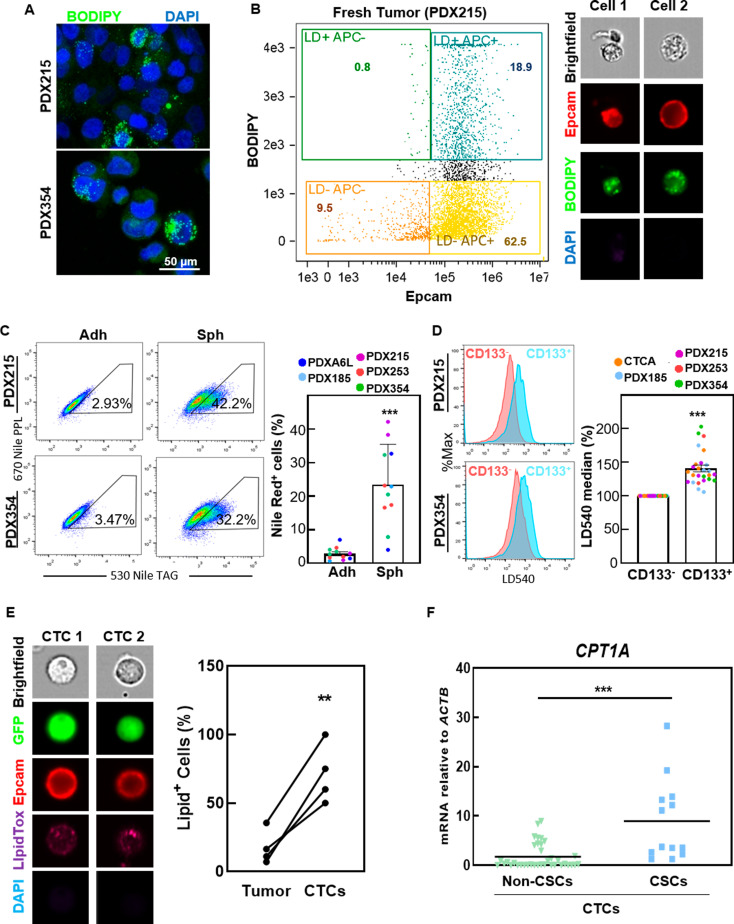
To confirm that PaCSC overexpress genes related with lipid storage in LDs, we first analyzed the LD content in our PDX models. The LD content was heterogeneous, with only a minority of cells showing significant enrichment of LDs, both in vitro (Fig. 2A) and in vivo (Fig. 2B). Interestingly, PaCSC-enriched conditions (spheres or CD133+ cells) showed a significantly greater LD content than non-CSCs (adherent cells or CD133−) (Fig. 2C and D), suggesting that the differential distribution of lipid content is dependent on stemness. We also confirmed this observation in a highly metastatic PDAC PDX model (CTCA), derived from circulating tumor cells of a stage IV PDAC patient (Fig. 2D). Interestingly, the percentage of cells with high lipid content was significantly greater in CTCs isolated from the blood of mice bearing orthotopic PDX tumors than in cells isolated from the primary tumor (Fig. 2E), implying a potential survival advantage in blood for cells with high lipid storage. Single-cell ddPCR analysis of CTCs demonstrated significant CPT1A overexpression, particularly in the CSC compartment of CTCs (Fig. 2F). Considering the important regulatory role of CPT1A in FAO via mitochondrial uptake of long-chain FAs, our results suggest that PaCSCs with active FA metabolism could be capable of initiating metastasis.
Fig. 2.
Increased lipid storage in PaCSCs. (A) Confocal microscopy of PDX215 and 354 stained with BODIPY (green) and DAPI (blue). (B) Imaging flow cytometry of a fresh PDX215 tumor stained with BODIPY (neutral lipids, green), EpCAM (human PDAC cells, red) and DAPI (live/dead cells). Left, representative flow cytometry plot for BODIPY and EpCAM expression in DAPI negative cells. Right, representative images of two individual cells. (C) Flow cytometry of Nile red staining (lipid droplets) in the indicated PDXs cultured in monolayers (adherent) or spheres. Left, representative plots. Right, pooled data. (D) Flow cytometry of LD540 staining (lipid droplets) in CD133- and CD133+ populations in the indicated PDXs. Left, representative histograms. Right, pooled data. (E) Imaging flow cytometry of cells isolated from the blood (CTCs) or primary tumors (Tumor) of mice bearing orthotopic PDX354-GFP tumors (n = 4). The cells were stained with LipidTox (neutral lipids, magenta), EpCAM (red) and DAPI (blue). Left, representative images of two individual CTCs. Right, quantification of the percentage of LipidTox+ cells in the blood and primary tumors of each mouse. (F) Single-cell mRNA expression by ddPCR of CPT1A in individual cells isolated from blood (CTCs), classified as non-CSCs and CSCs based on the expression of the pluripotency genes NANOG, KLF4, OCT3/4 and SOX2. The data are presented as mean ± SEM of at least three independent experiments. ** p < 0.01; *** p < 0.001
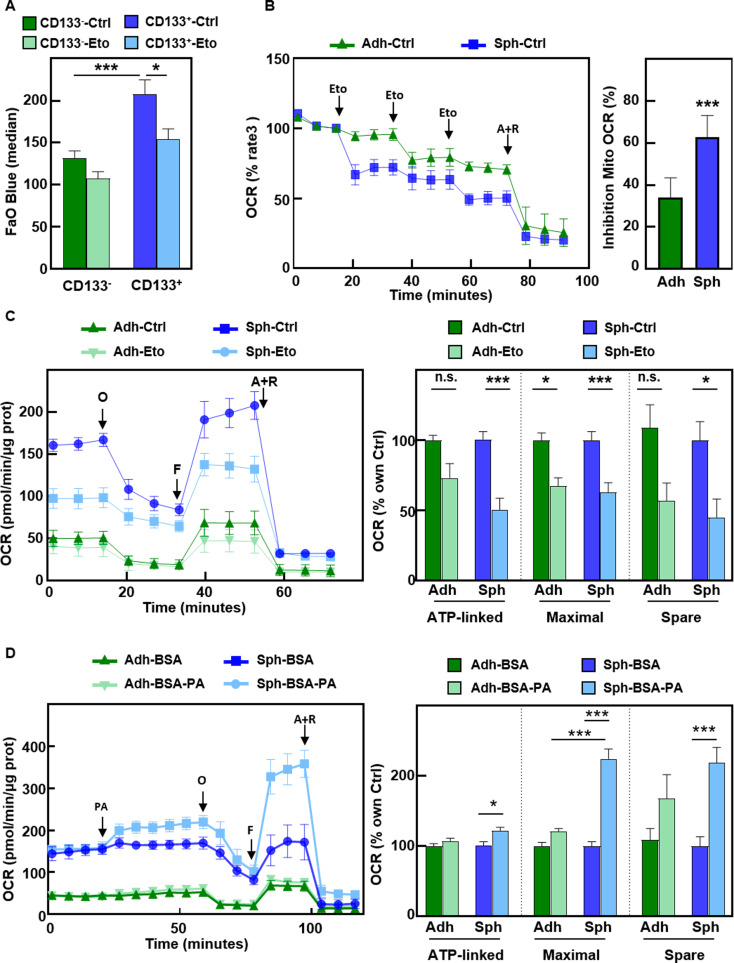
Next, we aimed to functionally validate our expression data, which suggested increased FAO activity in PaCSCs. FAO Blue staining assessed by flow cytometry confirmed that CD133+ cells exhibited increased FAO activity, which was abrogated by the CPT1A inhibitor Etomoxir (Fig. 3A). In addition, we measured the oxygen consumption rate (OCR) upon sequential injections of Etomoxir in cells previously grown either in adherent or in CSC-enriched sphere conditions. We found a greater inhibition of the OCR in CSC-enriched cultures (spheres) upon treatment with Etomoxir than in adherent cells (Fig. 3B). When cells were treated for Etomoxir for 30 min and subjected to the Long Chain FAO Stress Test, only sphere-derived cells exhibited a significant reduction in ATP-linked and maximal respiration, as well as spare respiratory capacity (Fig. 3C), suggesting increased FAO-dependent mitochondrial respiration in PaCSCs. Moreover, injection of the FA palmitate led to a significant increase in mitochondrial respiration parameters in sphere-derived cells only (Fig. 3D). These results suggest that PaCSCs exhibit enhanced FA metabolism compared with differentiated cells, both at the basal level and upon FA supplementation.
Fig. 3.
PaCSCs show enhanced mitochondrial FAO. (A) Median fluorescence intensity of FAO Blue staining as assessed by flow cytometry in cells gated for CD133 expression. Pooled data for PDX185 and 354 cells. (B) Oxygen consumption rate (OCR) by Seahorse analysis upon consecutive injections of 40 µM Etomoxir (Eto) and a final injection of the complex III inhibitor Antimycin A and the complex I inhibitor Rotenone (A + R, 1 µM). Left panel, kinetics in adherent vs. sphere-derived PDX215 cells. Right, percentage of mitochondrial OCR inhibition by Etomoxir, with respect to A + R, considered as 100% inhibition. (C) Long Chain FAO Stress Test in adherent and sphere-derived PDX215 cells pre-treated for 30 min with Etomoxir. Left, OCR kinetic profile. O, ATP synthase inhibitor Oligomycin; F, mitochondrial oxidative phosphorylation uncoupler FCCP. Right, ATP-linked and maximal respiration, and spare respiratory capacity, shown as percentages of the control for each condition. (D) Palmitate Oxidation Stress Test in adherent vs. sphere-derived PDX215 cells upon injection of palmitic acid conjugated with BSA (BSA-PA, 100 µM). Left panel, OCR kinetic profile. ATP-linked, maximal respiration and spare respiratory capacity are shown as percentages of the control for each condition. The data are presented as mean ± SEM of at least three independent experiments. * p < 0.05; *** p < 0.001
Fatty acid supplementation promotes CSC features in PDAC
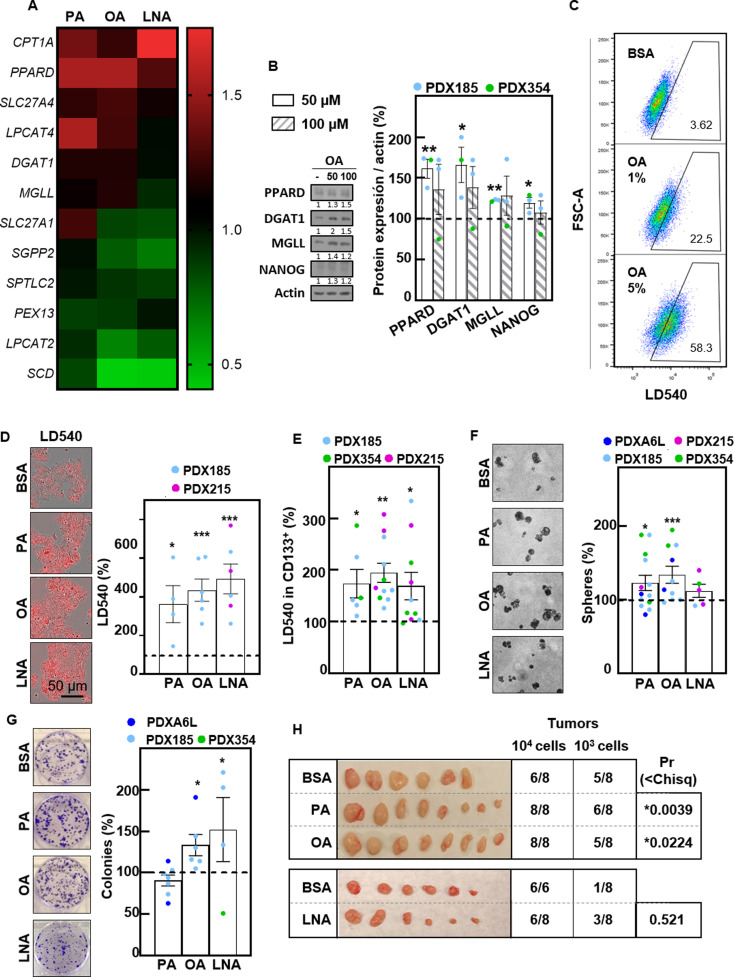
Free FAs are essential sources of energy within cells. Among them, the saturated nonesterified FA palmitic acid (PA), the monounsaturated FA oleic acid (OA) and the polyunsaturated FA linolenic acid (LNA) are the most common. Interestingly, treatment for 48 h with PA, OA and LNA in 2D cultures increased the expression of CPT1A, PPARD, LPCAT4, DGAT1 and MGLL (Fig. 4A), suggesting that exogenous FA supplementation may be able to induce the expression of genes that we previously found to be correlated with a high stemness signature (Fig. 1A). These changes were confirmed at the protein level, together with increased expression of the stemness marker NANOG (Fig. 4B). Considering these results together with the increased OXPHOS in PaCSCs upon incubation with PA (Fig. 3D), we next evaluated the effects of FA supplementation on CSC functionality. First, we confirmed by flow cytometry and live microscopy that treatment with exogenous FAs increased the LD content in PDAC cells (Fig. 4C and D). Interestingly, lipid storage in LDs was particularly relevant in the CD133+ population (Fig. 4E and data not shown). Treatment with exogenous FAs significantly increased the sphere and colony formation abilities of different PDX models (Fig. 4F and G), indicating enhanced self-renewal capacity. Crucially, in vitro pretreatment with FAs also enhanced in vivo tumorigenicity, as assessed by ELDA (Fig. 4H). Notably, the most consistent results were obtained with OA supplementation, while PA and LNA only showed significant results in some of the assays used. In summary, we concluded that supplementation with free FAs increases the accumulation of LDs and enhances self-renewal capacity in vitro and tumorigenicity in vivo.
Fig. 4.
Fatty acid supplementation enhances CSC functionality. (A) Gene expression determined by RTqPCR of the indicated genes after 48 h of treatment with palmitic acid (PA) (50 µM), oleic acid (OA 100 µM) or linolenic acid (LNA) (200 µM). (B) Western blot analysis of PPARD, DGAT1, MGLL and NANOG after 48 h of treatment with OA at 50 and 100 µM. Left, representative experiment in PDX185. Right, densitometric analyses of Western blots from PDX185 and PDX354. β-Αctin was used as loading control. (C) Representative flow cytometry plots of LD540 for PDX253 cells treated with increasing concentrations of OA (1 and 5%) for 24 h. (D) LD540 staining by IncuCyte imaging for PDX185 cells upon treatment with PA, OA or LNA as indicated in A. Left, representative images. Right, quantification, calculated as a percentage of the control conditions. (E) LD540 staining by flow cytometry in CD133+ cells from the indicated PDX models treated as described in A. (F) Sphere formation after seven days of treatment with PA, OA or LNA. Left, representative phase contrast images. Right, quantification, shown as a percentage of the control conditions. (G) Colony formation after 21 days of treatment with PA, OA or LNA. Left, representative images of colonies stained with crystal violet. Right, quantification of colony number, shown as compared percentage of the control conditions. (H) In vivo ELDA upon injection of the indicated number of PDX185 cells pretreated with PA, OA or LNA as indicated in A. Pictures of the tumors at the endpoint. ELDA calculations were performed on https://bioinf.wehi.edu.au/software/elda/. The data are presented as mean ± SEM of at least three independent experiments. * p < 0.05; ** p < 0.01; *** p < 0.001
Suppression of FAO activity in PDAC disrupts self-renewal and tumorigenic potential through the induction of energy stress
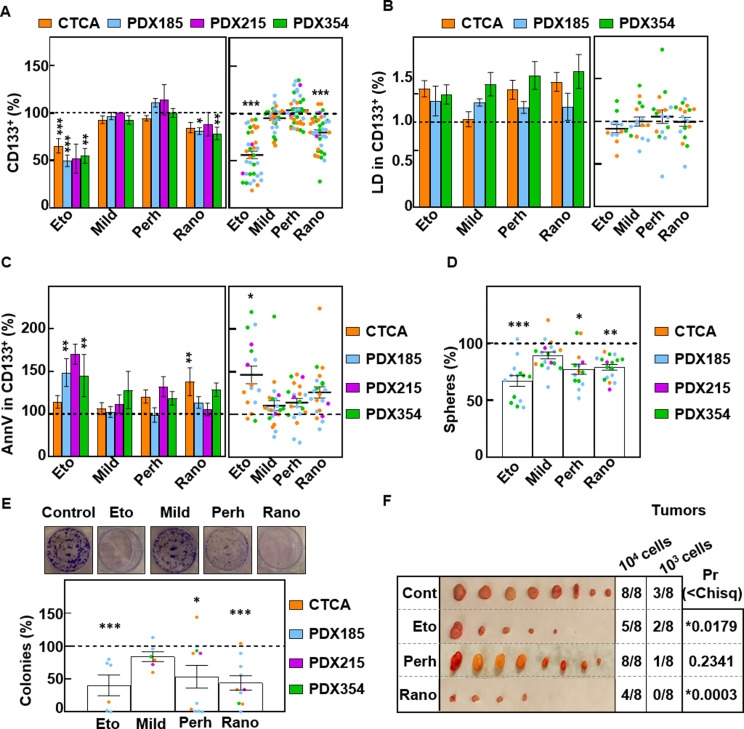
Considering that CSC-enriched cultures showed enhanced FAO activity and that FA supplementation increased stemness in PDAC cells, we next evaluated whether stemness is critically related to FAO and whether the respective inhibitors may be a potential new treatment strategy. For this purpose, we treated PDAC cells for 48 h with Etomoxir [18], Mildronate (carnitine synthesis inhibitor) [19], Perhexiline (CPT1/CPT2 dual inhibitor) [18] and Ranolazine (3-ketoacyl-CoA thiolase inhibitor) [18]. Interestingly, while Mildronate and Perhexiline were not effective, Etomoxir and Ranolazine both significantly reduced the percentage of CD133+ cells (Fig. 5A). However, this effect was not accompanied by a reduction in LD content, neither in the CSC population (Fig. 5B) nor in the total cell population (Figure S2A). Although only Etomoxir significantly increased apoptosis in the CSC population (Fig. 5C), but not in the total cell population (Figure S2B), treatment with either Etomoxir or Ranolazine consistently reduced sphere formation (Fig. 5D) and colony formation in vitro (Fig. 5E) and tumorigenicity in vivo (Fig. 5F). Therefore, Etomoxir and Ranolazine are able to target CSC functionality in PDAC.
Fig. 5.
FAO inhibition impairs PaCSC functionality. Unless otherwise specified, the cells were treated for 48 h with Etomoxir (Eto) (200 µM), Mildronate (Mild) (100 µM), Perhexiline (Perh) (1 µM) or Ranolazine (Rano) (100 µM). (A) Percentage of CD133+ cells by FACS. (B) LD540 staining of CD133+ cells. (C) Annexin V staining in CD133+ cells. Left panels, mean value of each cell type. Right panels, pooled data. (D) Sphere formation assay after seven days of treatment with the inhibitors. (E) Colony formation assay after 21 days of treatment with inhibitors. Upper panel representative images of crystal violet staining of PDX185 cells. Lower panel, colony number quantification. (F) Tumors at the endpoint after the inoculation of pretreated PDX185 cells. ELDA calculations were performed at https://bioinf.wehi.edu.au/software/elda/. The data are presented as mean ± SEM of at least three independent experiments. The dashed lines represent the control conditions. * p < 0.05; ** p < 0.01; *** p < 0.001
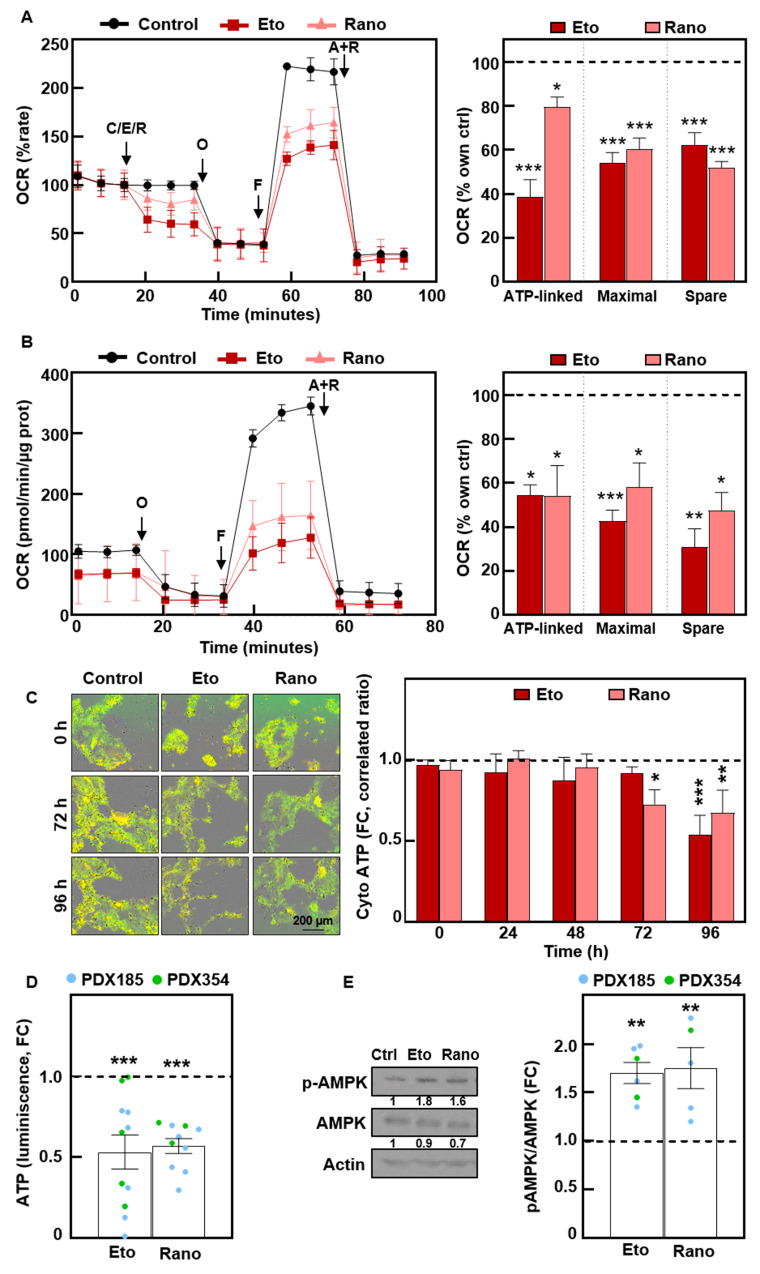
Interestingly, both compounds inhibited mitochondrial oxygen consumption rate when administered acutely, impacting mitochondrial respiration parameters (Fig. 6A). These effects were durable, since we detected mitochondrial OCR inhibition even after 72 h of treatment (Fig. 6B). As expected, the inhibition of mitochondrial activity resulted in a significant decrease in intracellular ATP (Fig. 6C and D) and a subsequent significant increase in AMPK stress kinase phosphorylation (Fig. 6E). In summary, FAO inhibition impairs CSC functionality by inhibiting mitochondrial respiration and consequently inducing energy stress.
Fig. 6.
Treatment with Etomoxir and Ranolazine impairs mitochondrial respiration, inducing energy stress. (A) Long Chain FAO Stress Test with acute injections of 100 µM Etomoxir (Eto) or 50 µM Ranolazine (Rano). Left, Oxygen Consumption Rate (OCR) kinetics. Right, ATP-linked, maximal and spare respiration. Pooled data for 354 and CTCA cells. O, Oligomycin; F, FCCP; A + R, Antimycin A + Rotenone. (B) Mito Stress after 72 h of treatment of PDX215 cells with Etomoxir or Ranolazine. Left, OCR kinetics. Right, ATP-linked, maximal and spare respiration. (C) Cytosolic ATP levels in PDX215 cells visualized by time-lapse fluorescence microscopy (0–96 h). Left, representative images at the indicated times. Right, quantification. (D) Total ATP levels quantified by bioluminescence at 48 h. (E) Western blot analysis of phospho-AMPK and total AMPK after 48 h of treatment. Left, representative experiment in PDX185. Right, densitometric analysis. β-Αctin was used as loading control. The data are presented as mean ± SEM of at least three independent experiments. * p < 0.05; ** p < 0.01; *** p < 0.001
Fatty acid metabolism and response to Gemcitabine
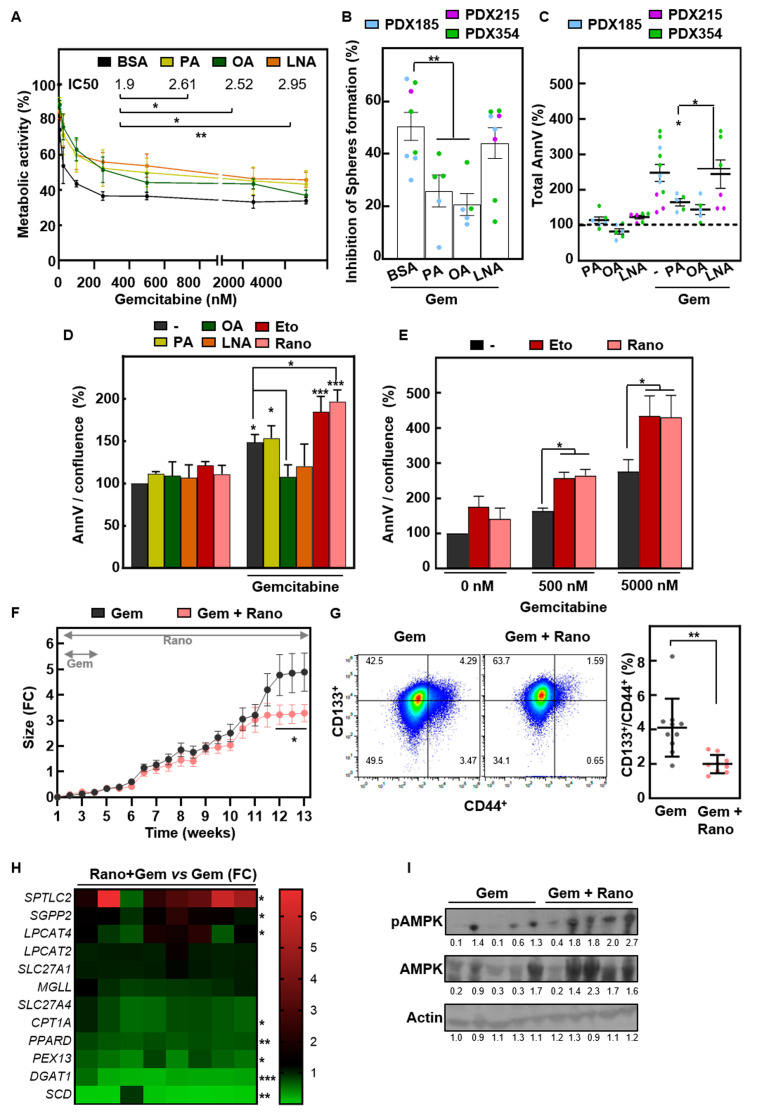
Finally, we explored whether modulating FAO activity also affects another key feature of CSCs, i.e., chemoresistance [4, 5]. Indeed, pretreatment with exogenous FAs for 24 h increased the IC50 of the chemotherapeutic agent Gemcitabine (Fig. 7A), thus attenuating the effects of the drug on self-renewal (Fig. 7B). Consistently, OA supplementation protected PDAC cells from Gemcitabine-induced apoptosis, as measured by FACS or IncuCyte (Fig. 7C and D, S3A). In contrast, cotreatment with either Etomoxir or Ranolazine significantly enhanced the response to Gemcitabine (Fig. 7D). Interestingly, the chemosensitizing effect of Etomoxir or Ranolazine was also observed when lipid inhibitors were applied after treatment with Gemcitabine for 48 h (Fig. 7E and Figure S3B). Importantly, in vivo treatment with Gemcitabine (3 weeks) combined with Ranolazine (until the endpoint) significantly delayed tumor growth rate, but only at late time points when the tumors reached approximately 2.5-3 times the initial tumor size (Fig. 7F). At the endpoint, the number of CD44+/CD133+ CSCs was significantly lower in the Ranolazine-treated tumors (Fig. 7G). Interestingly, the expression profiles of lipid metabolism genes significantly changed in the Ranolazine-treated tumors (Fig. 7H), and the p-AMPK/AMPK ratio increased (Fig. 7I), suggesting that tumors were suffering metabolic stress similar to what we observed in vitro (Fig. 6E). Therefore, combined treatment with FAO inhibitors and Gemcitabine improves the response to Gemcitabine alone in PDAC PDXs, providing a new perspective for a more effective treatment.
Fig. 7.
Fatty acid metabolism determines the response to Gemcitabine. (A) Percentage of metabolic activity determined by resazurin measurement and the IC50 of Gemcitabine after 72 h of treatment alone and in combination with palmitic acid (PA) (50 µM), oleic acid (OA) (100 µM) or linolenic acid (LNA) (200 µM) (pretreatment 24 h). (B) Sphere formation assay of cells pretreated with FAs for 24 h followed by treatment with Gemcitabine (1000 nM) under sphere forming conditions. (C) Annexin V levels were determined by flow cytometry after 48 h of treatment with PA, OA or LNA with or without 100 nM Gemcitabine. Pooled data of the indicated PDXs. (D) Annexin V levels were measured by IncuCyte imaging at 48 h in PDX185. (E) Annexin V measured by Incucyte imaging after 48 h treatment with Gemcitabine and 72 h additional hours in the presence of absence of FAO inhibitors in PDX185. In F-I, tumor pieces of PDX185 were subcutaneously implanted in nude mice, and when they reached around 300 mm3 they were treated for 3 weeks with Gemcitabine (70 mg/kg) with or without Ranolazine (130 mg/kg), which was administered daily for the whole duration of the experiment. (F) Tumor size is shown as the fold change vs. day 1 of treatment. (G) CSC content was measured as the percentage of CD44+/CD133+ double positive cells in tumors at the endpoint. (H) Lipid metabolism genes determined by RTqPCR. (I) Western blot analysis of phospho-AMPK and total AMPK in total lysates from tumors at the endpoint. β-Αctin was used as loading control. The data are presented as mean ± SEM of at least three independent experiments. * p < 0.05; ** p < 0.01; *** p < 0.001
Discussion
The concept of metabolic rewiring is now widely accepted as one of the hallmarks of cancer [20], but it has increasingly been recognized as a highly complex and intertwined process in recent years. Indeed, metabolic heterogeneity within tumors, related to fluctuating local microenvironments and distinct cellular populations, is becoming apparent. Specifically, we (and others) have described the unique metabolic features of the CSC population in PDAC compared to their more differentiated progenies: while CSCs acquire a more glycolytic metabolism in some cancer types [21, 22], most CSCs, including PaCSCs, rely on mitochondrial OXPHOS for maintenance of stemness [7, 23, 24]. Additionally, recent reports also highlight lipid metabolism as one of the main metabolic pathways regulating CSC functions in several tumor types [25]. Here, we describe the significance of mitochondrial FAO activity in regulating the self-renewal, tumorigenicity and, most importantly, chemoresistance of PaCSCs. Our findings are in line with the essential role of mitochondrial respiration in stemness in PDAC, as OXPHOS requires acetyl-CoA as a substrate for ATP production, which can be obtained not only via glucose metabolism but also through lipid catabolism via FAO.
Increased lipid uptake and aberrant FA metabolism have previously been associated with tumor progression and poor prognosis in PDAC patients [11–13]. Indeed, we found that several genes related to FA transport, storage and metabolism are correlated with a stemness signature in PDAC patients, some of which are significantly correlated with overall survival (Fig. 1). These genes were overexpressed under CSC-enriched culture conditions in several PDAC PDX models, strongly suggesting a causal relationship between active lipid metabolism and the malignant properties of CSCs. Among these genes, MGLL [26, 27], PPARD [28] and CPT1A [29] have been correlated with poor prognosis in various cancers. Indeed, increased CPT1A expression specifically in CSCs has been associated with worse outcomes in breast [30] and colon [31] cancer, suggesting a key role for mitochondrial FAO activity in CSC functionality. Our results demonstrate that this gene expression profile indeed translates into enhanced FAO activity in CD133+ and sphere-derived cells in PDAC, as previously demonstrated for CSCs in breast cancer [32] and hepatocellular carcinoma [33], where NANOG directly upregulates the expression of genes related to FAO.
Our results further indicated increased lipid accumulation in LDs of CD133+ PaCSCs (Fig. 2), corroborating our initial gene expression analyses. While similar results were previously reported in vitro for CSCs isolated from primary cultures of colorectal cancer [34] and breast and ovarian cancer cell lines [35, 36], we now also demonstrate differential LD storage for CSCs isolated from fresh tumors. Interestingly, we also detected an enrichment in LD content and CPT1A expression in CTCs isolated from an orthotopic PDX model. CPT1A overexpression was particularly prominent in circulating PaCSCs compared to primary tumors, suggesting a survival advantage for cells with increased lipid storage and metabolism in the blood. In fact, LD staining has been reported to significantly improve CTC detection in a prostate cancer model [37], suggesting that a similar approach might improve CTC detection in liquid biopsies from PDAC patients.
The addition of exogenous FAs upregulated the expression of several genes involved in lipid transport, storage and metabolism similar to their upregulation in sphere-derived cells (Fig. 4A and B). Interestingly, lipid droplet accumulation upon exposure to FAs was especially relevant for CD133+ PaCSCs (Fig. 4C and D). Moreover, PaCSCs also showed a significant increase in FAO activity coupled to mitochondrial respiration in response to PA (Fig. 3C). This preferential effect of exogenous FAs on CSC metabolism translated into significantly increased self-renewal (measured as sphere and colony formation; Fig. 4F and G) and in vivo tumorigenicity (Fig. 4H). Notably, incubation with OA most consistently enhanced self-renewal and chemoresistance, while the effects of incubation with PA and LNA were more variable (Figs. 4F-H and 7A-D) and incubation with linoleic acid did not have any significant effect (data not shown). In this sense, our data are in agreement with a previous report suggesting that a panel of FAs exerted differential effects on PDAC viability and growth in vitro and in vivo [38]. Nevertheless, our results strongly suggest that FA supplementation promoted stemness in PDAC, in agreement with previous reports in different cancer types. OA supplementation was previously shown to maintain self-renewal in breast CSCs treated with soraphen A, a de novo FA synthesis inhibitor [39]. A similar phenotype has been reported for metastasis initiation in oral carcinomas, melanoma and breast cancer where a high-fat diet or PA supplementation favored sphere formation and tumor initiation in metastatic sites [40–42]. However, epigenetic histone modifications and the activation of cellular signaling pathways independent of energy production seem to mediate the prometastatic effects of PA in these models.
In addition, FA supplementation, particularly OA, also protected PDAC cells from the cytotoxic effects of Gemcitabine treatment (Fig. 7A-D, Fig. S3A). Therefore, our data add to the growing body of evidence indicating that FAs support chemoresistance in different cancer types. Specifically, OA reduced the cytotoxic effects of docetaxel treatment in prostate cancer cells [43], while PA supplementation protected against cisplatin treatment in ovarian cancer [44], both via activation of ERK/AKT-mediated survival signals. On the other hand, saturated FAs modulate the response to 5-fluorouracil in colorectal cancer by regulating cell membrane fluidity [45].
Although we cannot formally exclude the relevance of other protective effects in our experimental settings, our results clearly indicate that FA-mediated chemoresistance in pancreatic PDX models depends on FA catabolism, as FAO inhibition restores response to Gemcitabine (Fig. 7E, S2B). This finding suggested that FAO inhibitors may be successful adjuvant drugs for improving PDAC chemotherapy efficacy. Considering that Etomoxir is not suitable for use in humans, we also tested alternative FAO inhibitors that are already approved in different countries for the treatment of angina pectoris: Mildronate (carnitine synthesis inhibitor) [46], Perhexiline (CPT1/CPT2 dual inhibitor) [18] and Ranolazine (3- ketoacyl -CoA thiolase inhibitor) [47]. Among these inhibitors, Ranolazine was the only compound that significantly reduced CD133+ expression, self-renewal and in vivo tumorigenicity, similar to Etomoxir (Fig. 5). We confirmed that the mechanism of action of these inhibitors was OXPHOS inhibition, which decreased oxygen consumption and mitochondrial spare respiratory capacity, resulting in a significant reduction in ATP production, and an increase in the pAMPK/AMPK ratio. Indeed, FAO has been shown to contribute to the mitochondrial spare respiratory capacity associated with survival under chemotherapeutic stress conditions [48, 49]. Interestingly, Ranolazine sensitized PDAC cells to Gemcitabine both in vitro and in vivo (Fig. 7), similar to previous results for human leukemia cells [50, 51], prostate cancer [47], and melanoma [52], where enhanced OXPHOS via FAO was also described as the main mechanism for resistance to apoptosis induced by chemotherapy and immunotherapy both in vitro and in vivo.
Interestingly, the fact that not only FA synthesis but also external supplementation of FA resulted in increased chemoresistance, suggests an interesting link between a high-fat diet/obesity and chemoresistance. Indeed, Incio et al. demonstrated that obesity reduced drug delivery and toxicity in PDAC [53]. Remarkably, a previous publication reported that cross-talk between adipose tissue and chronic myeloid leukemia cells results in lipolysis to fuel FAO, inducing chemoresistance in chronic myeloid leukemia cells [54]. Further research in this area may improve current treatment designs that also consider systemic metabolism, which has become a topic of particular interest in the recent years.
Conclusions
Our results demonstrated that PaCSCs accumulate lipids that serve as substrates for FAO to sustain mitochondrial respiration, which is necessary for maintaining stemness. CSC functionality such as self-renewal and tumorigenicity can be enhanced via FA supplementation or reduced by pharmacological inhibition of FAO. Importantly, we showed that existing FAO inhibitors approved for clinical use under other conditions (e.g. Ranolazine) could represent potential tools for overcoming pancreatic CSC-associated chemoresistance.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to acknowledge the use of the CIBA (Centro de Investigación Biomédica de Aragón) Flow Cytometry and Animal Facilities (Servicios Científico-Técnicos, IACS Universidad de Zaragoza). We also thank Laura Sancho Andrés for proofreading the manuscript.
Abbreviations
- CFA
Colony formation assay
- CSC
Cancer stem cell
- CTCs
Circulating tumor cells
- FA
Fatty acid
- HR
Hazard ratio
- LD
Lipid droplet
- LNA
Linolenic acid
- OA
Oleic acid
- OCR
Oxygen consumption rate
- OXPHOS
Oxidative phosphorylation
- PA
Palmitic acid
- PaCSCs
Pancreatic cancer stem cells
- PDAC
Pancreatic ductal adenocarcinoma
- PDX
Patient-derived xenografts
Author contributions
MM: investigation, formal analysis, writing-original draft, writing-review and editing. SC: conceptualization, investigation, formal analysis and writing-review. AR-G: investigation and writing-editing. DB: conceptualization and investigation. AMS, IV, PE, AB, ACG, PJ: investigation. CH: conceptualization, resources, writing-review and editing. PS: conceptualization, project administration, supervision, funding acquisition, investigation, writing-original draft, writing-review and editing.
Funding
The research was supported by the Instituto de Salud Carlos III through the Miguel Servet Program (CP16/00121 and CPII21/00005, to P.S.), the pFIS program (FI21/00031, to P. E-R) and Fondo de Investigaciones Sanitarias (PI17/00082 and PI20/00921, to P.S.) (all co-financed by European funds (FSE: “El FSE invierte en tu futuro” and FEDER: “Una manera de hacer Europa”, respectively), the Worldwide Cancer Research (WCR) Charity together with Asociación Española contra el Cáncer (AECC) (19–0250, to P.S.), and a LAB AECC grant (LABAE223389SANC, to P.S.). M.M. was recipient of a Margarita Salas fellowship from the Universidad Autónoma de Madrid (CA1/RSUE/202100646). A.R-G. was a recipient of a predoctoral contract from the Spanish AECC (PRDAR222458ROYO). I.V. was a recipient of a predoctoral contract from the Aragon Government.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethical approval
Human material: PDAC patient-derived xenografts (PDXs): A6L, 185, 215, 253, 265 and 354 were obtained from the Biobank of the Spanish National Cancer Research Centre (CNIO), Madrid, Spain (MTAs #CNIO20-027, #CNIO21-253). PDAC PDX-derived cultures were established as previously described [17]. Pancreatic circulating tumor cells (CTCs): The metastatic model CTCA was established from circulating tumor cells and obtained through the Barts Pancreas Tissue Bank of the Barts Cancer Institute (https://www.bartspancreastissuebank.org.uk/; BCI, London, United Kingdom; 2019/02/IISA/PS/E/Cellcultures). In vivo experiments: Mice were housed according to institutional guidelines and all experimental procedures were performed in compliance with the institutional guidelines for the welfare of experimental animals as approved by the University of Zaragoza Ethics Committee (CEICA PI22/17, PI35/19, PI41/20) and in accordance with the guidelines for Ethical Conduct in the Care and Use of Animals as stated in The International Guiding Principles for Biomedical Research involving Animals, developed by the Council for International Organizations of Medical Sciences (CIOMS).
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Marta Mascaraque and Sarah Courtois contributed equally to this work.
Contributor Information
Christopher Heeschen, Email: christopher.heeschen@ircc.it.
Patricia Sancho, Email: psancho@iisaragon.es.
References
- 1.Kleeff J, Korc M, Apte M, et al. Pancreatic cancer. Nat Rev Dis Primers. 2016;2(1):16022. 10.1038/nrdp.2016.22. 10.1038/nrdp.2016.22 [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. 10.3322/caac.21660. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 3.Rawla P, Sunkara T, Gaduputi V. Epidemiology of pancreatic Cancer: global trends, etiology and risk factors. World J Oncol. 2019;10(1):10–27. 10.14740/wjon1166. 10.14740/wjon1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishiwata T, Matsuda Y, Yoshimura H, et al. Pancreatic cancer stem cells: features and detection methods. Pathol Oncol Res. 2018;24(4):797–805. 10.1007/s12253-018-0420-x. 10.1007/s12253-018-0420-x [DOI] [PubMed] [Google Scholar]
- 5.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic Cancer stem cells. Cancer Res. 2007;67(3):1030–7. 10.1158/0008-5472.CAN-06-2030. 10.1158/0008-5472.CAN-06-2030 [DOI] [PubMed] [Google Scholar]
- 6.Amantini C, Morelli MB, Nabissi M, et al. Expression profiling of circulating Tumor cells in pancreatic ductal adenocarcinoma patients: biomarkers Predicting overall survival. Front Oncol. 2019;9. 10.3389/fonc.2019.00874. [DOI] [PMC free article] [PubMed]
- 7.Sancho P, Burgos-Ramos E, Tavera A, et al. MYC/PGC-1α balance determines the metabolic phenotype and plasticity of pancreatic Cancer stem cells. Cell Metab. 2015;22(4):590–605. 10.1016/j.cmet.2015.08.015. 10.1016/j.cmet.2015.08.015 [DOI] [PubMed] [Google Scholar]
- 8.Courtois S, de Luxán-Delgado B, Penin-Peyta L, et al. Inhibition of mitochondrial dynamics preferentially targets pancreatic Cancer cells with enhanced tumorigenic and invasive potential. Cancers (Basel). 2021;13(4):698. 10.3390/cancers13040698. 10.3390/cancers13040698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alcalá S, Sancho P, Martinelli P, et al. ISG15 and ISGylation is required for pancreatic cancer stem cell mitophagy and metabolic plasticity. Nat Commun. 2020;11(1):2682. 10.1038/s41467-020-16395-2. 10.1038/s41467-020-16395-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jagust P, Alcalá S Jr, Heeschen BS, Sancho C. Glutathione metabolism is essential for self-renewal and chemoresistance of pancreatic cancer stem cells. World J Stem Cells. 2020;12(11):1410–28. 10.4252/wjsc.v12.i11.1410. 10.4252/wjsc.v12.i11.1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tadros S, Shukla SK, King RJ, et al. De NovoLipid Synthesis facilitates Gemcitabine Resistance through endoplasmic reticulum stress in pancreatic Cancer. Cancer Res. 2017;77(20):5503–17. 10.1158/0008-5472.CAN-16-3062. 10.1158/0008-5472.CAN-16-3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swierczynski J. Role of abnormal lipid metabolism in development, progression, diagnosis and therapy of pancreatic cancer. World J Gastroenterol. 2014;20(9):2279. 10.3748/wjg.v20.i9.2279. 10.3748/wjg.v20.i9.2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillaumond F, Bidaut G, Ouaissi M, et al. Cholesterol uptake disruption, in association with chemotherapy, is a promising combined metabolic therapy for pancreatic adenocarcinoma. Proc Natl Acad Sci. 2015;112(8):2473–8. 10.1073/pnas.1421601112. 10.1073/pnas.1421601112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jagust P, de Luxán-Delgado B, Parejo-Alonso B, Sancho P. Metabolism-based therapeutic strategies targeting Cancer Stem cells. Front Pharmacol. 2019;10. 10.3389/fphar.2019.00203. [DOI] [PMC free article] [PubMed]
- 15.Liu Q, Luo Q, Halim A, Song G. Targeting lipid metabolism of cancer cells: a promising therapeutic strategy for cancer. Cancer Lett. 2017;401:39–45. 10.1016/j.canlet.2017.05.002. 10.1016/j.canlet.2017.05.002 [DOI] [PubMed] [Google Scholar]
- 16.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–60. 10.1093/nar/gkz430. 10.1093/nar/gkz430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mueller M, Hermann PC, Witthauer J, et al. Combined targeted treatment to Eliminate Tumorigenic Cancer Stem cells in human pancreatic Cancer. Gastroenterology. 2009;137(3):1102–13. 10.1053/j.gastro.2009.05.053. 10.1053/j.gastro.2009.05.053 [DOI] [PubMed] [Google Scholar]
- 18.Ma Y, Wang W, Devarakonda T, et al. Functional analysis of molecular and pharmacological modulators of mitochondrial fatty acid oxidation. Sci Rep. 2020;10(1):1450. 10.1038/s41598-020-58334-7. 10.1038/s41598-020-58334-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang JX, Rahimnejad S, Zhang YY, et al. Mildronate triggers growth suppression and lipid accumulation in largemouth bass (Micropterus salmoides) through disturbing lipid metabolism. Fish Physiol Biochem. 2022;48(1):145–59. 10.1007/s10695-021-01040-6. 10.1007/s10695-021-01040-6 [DOI] [PubMed] [Google Scholar]
- 20.Hanahan D. Hallmarks of Cancer: New dimensions. Cancer Discov. 2022;12(1):31–46. 10.1158/2159-8290.CD-21-1059. 10.1158/2159-8290.CD-21-1059 [DOI] [PubMed] [Google Scholar]
- 21.Mamouni K, Kim J, Lokeshwar BL, Kallifatidis G. ARRB1 regulates metabolic reprogramming to promote glycolysis in Stem Cell-Like bladder Cancer cells. Cancers (Basel). 2021;13(8):1809. 10.3390/cancers13081809. 10.3390/cancers13081809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Neill S, Porter RK, McNamee N, Martinez VG, O’Driscoll L. 2-Deoxy-D-Glucose inhibits aggressive triple-negative breast cancer cells by targeting glycolysis and the cancer stem cell phenotype. Sci Rep. 2019;9(1):3788. 10.1038/s41598-019-39789-9. 10.1038/s41598-019-39789-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raggi C, Taddei ML, Sacco E, et al. Mitochondrial oxidative metabolism contributes to a cancer stem cell phenotype in cholangiocarcinoma. J Hepatol. 2021;74(6):1373–85. 10.1016/j.jhep.2020.12.031. 10.1016/j.jhep.2020.12.031 [DOI] [PubMed] [Google Scholar]
- 24.Valle S, Alcalá S, Martin-Hijano L, et al. Exploiting oxidative phosphorylation to promote the stem and immunoevasive properties of pancreatic cancer stem cells. Nat Commun. 2020;11(1):5265. 10.1038/s41467-020-18954-z. 10.1038/s41467-020-18954-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Royo-García A, Courtois S, Parejo-Alonso B, Espiau-Romera P, Sancho P. Lipid droplets as metabolic determinants for stemness and chemoresistance in cancer. World J Stem Cells. 2021;13(9):1307–17. 10.4252/wjsc.v13.i9.1307. 10.4252/wjsc.v13.i9.1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye L, Zhang B, Seviour EG, et al. Monoacylglycerol lipase (MAGL) knockdown inhibits tumor cells growth in colorectal cancer. Cancer Lett. 2011;307(1):6–17. 10.1016/j.canlet.2011.03.007. 10.1016/j.canlet.2011.03.007 [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Liu Z, Lian Z, et al. Monoacylglycerol lipase: a novel potential therapeutic target and Prognostic Indicator for Hepatocellular Carcinoma. Sci Rep. 2016;6(1):35784. 10.1038/srep35784. 10.1038/srep35784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshinaga M, Taki K, Somada S, et al. The expression of both Peroxisome Proliferator-Activated Receptor Delta and Cyclooxygenase-2 in tissues is Associated with Poor Prognosis in Colorectal Cancer patients. Dig Dis Sci. 2011;56(4):1194–200. 10.1007/s10620-010-1389-9. 10.1007/s10620-010-1389-9 [DOI] [PubMed] [Google Scholar]
- 29.Das M, Giannoudis A, Sharma V. The role of CPT1A as a biomarker of breast cancer progression: a bioinformatic approach. Sci Rep. 2022;12(1):16441. 10.1038/s41598-022-20585-x. 10.1038/s41598-022-20585-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han S, Wei R, Zhang X, et al. CPT1A/2-Mediated FAO Enhancement—A metabolic target in radioresistant breast Cancer. Front Oncol. 2019;9. 10.3389/fonc.2019.01201. [DOI] [PMC free article] [PubMed]
- 31.Xiong X, Wen YA, Fairchild R, et al. Upregulation of CPT1A is essential for the tumor-promoting effect of adipocytes in colon cancer. Cell Death Dis. 2020;11(9):736. 10.1038/s41419-020-02936-6. 10.1038/s41419-020-02936-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang T, Fahrmann JF, Lee H, et al. JAK/STAT3-Regulated fatty acid β-Oxidation is critical for breast Cancer Stem Cell Self-Renewal and Chemoresistance. Cell Metab. 2018;27(1):136–e1505. 10.1016/j.cmet.2017.11.001. 10.1016/j.cmet.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen CL, Uthaya Kumar DB, Punj V, et al. NANOG metabolically reprograms Tumor-initiating stem-like cells through tumorigenic changes in oxidative phosphorylation and fatty acid metabolism. Cell Metab. 2016;23(1):206–19. 10.1016/j.cmet.2015.12.004. 10.1016/j.cmet.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tirinato L, Liberale C, Di Franco S, et al. Lipid droplets: a New Player in Colorectal Cancer Stem cells unveiled by Spectroscopic Imaging. Stem Cells. 2015;33(1):35–44. 10.1002/stem.1837. 10.1002/stem.1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hershey BJ, Vazzana R, Joppi DL, Havas KM. Lipid droplets define a sub-population of breast Cancer stem cells. J Clin Med. 2019;9(1):87. 10.3390/jcm9010087. 10.3390/jcm9010087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Condello S, Thomes-Pepin J, et al. Lipid desaturation is a metabolic marker and therapeutic target of ovarian Cancer stem cells. Cell Stem Cell. 2017;20(3):303–e3145. 10.1016/j.stem.2016.11.004. 10.1016/j.stem.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitra R, Goodman OB, Le TT. Enhanced detection of metastatic prostate cancer cells in human plasma with lipid bodies staining. BMC Cancer. 2014;14(1):91. 10.1186/1471-2407-14-91. 10.1186/1471-2407-14-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu M, Liu H, Duan Y, Zhang D, Li S, Wang F. Four types of fatty acids exert differential impact on pancreatic cancer growth. Cancer Lett. 2015;360(2):187–94. 10.1016/j.canlet.2015.02.002. 10.1016/j.canlet.2015.02.002 [DOI] [PubMed] [Google Scholar]
- 39.Corominas-Faja B, Cuyàs E, Gumuzio J, et al. Chemical inhibition of acetyl-CoA carboxylase suppresses self-renewal growth of cancer stem cells. Oncotarget. 2014;5(18):8306–16. 10.18632/oncotarget.2059. 10.18632/oncotarget.2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pascual G, Domínguez D, Elosúa-Bayes M, et al. Dietary palmitic acid promotes a prometastatic memory via Schwann cells. Nature. 2021;599(7885):485–90. 10.1038/s41586-021-04075-0. 10.1038/s41586-021-04075-0 [DOI] [PubMed] [Google Scholar]
- 41.Pascual G, Avgustinova A, Mejetta S, et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017;541(7635):41–5. 10.1038/nature20791. 10.1038/nature20791 [DOI] [PubMed] [Google Scholar]
- 42.Palmitate oxidation drives a pro-metastatic post-translational modification. Nat Cancer. Published Online Febr 15, 2023. 10.1038/s43018-023-00514-1 [DOI] [PubMed]
- 43.Liotti A, Cosimato V, Mirra P, et al. Oleic acid promotes prostate cancer malignant phenotype via the G protein-coupled receptor FFA1/GPR40. J Cell Physiol. 2018;233(9):7367–78. 10.1002/jcp.26572. 10.1002/jcp.26572 [DOI] [PubMed] [Google Scholar]
- 44.Bauerschlag DO, Maass N, Leonhardt P, et al. Fatty acid synthase overexpression: target for therapy and reversal of chemoresistance in ovarian cancer. J Transl Med. 2015;13(1):146. 10.1186/s12967-015-0511-3. 10.1186/s12967-015-0511-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MORITA HIRAIDET, HORIKAWA Y. Saturated fatty acids in cell membrane lipids induce resistance to 5-Fluorouracil in Colorectal Cancer cells. Anticancer Res. 2022;42(7):3313–24. 10.21873/anticanres.15819. 10.21873/anticanres.15819 [DOI] [PubMed] [Google Scholar]
- 46.Agency EM. SMH. Opinion of the Paediatric Committee on the Granting of a Product-Specific Waiver EMEA-002212-PIP01-17.; 2017.
- 47.Flaig TW, Salzmann-Sullivan M, Su LJ, et al. Lipid catabolism inhibition sensitizes prostate cancer cells to antiandrogen blockade. Oncotarget. 2017;8(34):56051–65. 10.18632/oncotarget.17359. 10.18632/oncotarget.17359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfleger J, He M, Abdellatif M. Mitochondrial complex II is a source of the reserve respiratory capacity that is regulated by metabolic sensors and promotes cell survival. Cell Death Dis. 2015;6(7):e1835–1835. 10.1038/cddis.2015.202. 10.1038/cddis.2015.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marchetti P, Fovez Q, Germain N, Khamari R, Kluza J. Mitochondrial spare respiratory capacity: mechanisms, regulation, and significance in non-transformed and cancer cells. FASEB J. 2020;34(10):13106–24. 10.1096/fj.202000767R. 10.1096/fj.202000767R [DOI] [PubMed] [Google Scholar]
- 50.Farge T, Saland E, de Toni F, et al. Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov. 2017;7(7):716–35. 10.1158/2159-8290.CD-16-0441. 10.1158/2159-8290.CD-16-0441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samudio I, Harmancey R, Fiegl M, et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clin Invest. 2010;120(1):142–56. 10.1172/JCI38942. 10.1172/JCI38942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Redondo-Muñoz M, Rodriguez-Baena FJ, Aldaz P, et al. Metabolic rewiring induced by ranolazine improves melanoma responses to targeted therapy and immunotherapy. Nat Metab. 2023;5(9):1544–62. 10.1038/s42255-023-00861-4. 10.1038/s42255-023-00861-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Incio J, Liu H, Suboj P, et al. Obesity-Induced inflammation and Desmoplasia promote pancreatic Cancer progression and resistance to Chemotherapy. Cancer Discov. 2016;6(8):852–69. 10.1158/2159-8290.CD-15-1177. 10.1158/2159-8290.CD-15-1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye H, Adane B, Khan N, et al. Leukemic stem cells evade chemotherapy by metabolic adaptation to an adipose tissue niche. Cell Stem Cell. 2016;19(1):23–37. 10.1016/j.stem.2016.06.001. 10.1016/j.stem.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.