Abstract
Arrhythmogenic cardiomyopathy is an inherited entity characterized by irregular cell–cell adhesion, cardiomyocyte death and fibro-fatty replacement of ventricular myocytes, leading to malignant ventricular arrythmias, contractile dysfunction and sudden cardiac death. Pathogenic variants in genes that encode desmosome are the predominant cause of arrhythmogenic cardiomyopathy. Moreover, signalling pathways such as Wnt/ß-catenin and transforming growth factor-β have been involved in the disease progression. However, still little is known about the molecular pathophysiological mechanisms that underlie arrhythmogenic cardiomyopathy pathogenesis. We used mRNA and small RNA sequencing to analyse the transcriptome of health and arrhythmogenic cardiomyopathy of autopsied human hearts. Our results showed 697 differentially expressed genes and eight differentially expressed miRNAs. Functional enrichment revealed mitochondrial respiratory-related pathways, impaired response to oxidative stress, apoptotic signalling pathways and inflammatory response-related and extracellular matrix response pathways. Furthermore, analysis of the miRNA–mRNA interactome identified eleven negatively correlated miRNA-target pairs for arrhythmogenic cardiomyopathy. Our finding revealed novel arrhythmogenic cardiomyopathy-related miRNAs with important regulatory function in disease pathogenesis, highlighting their value as potential key targets for therapeutic approaches.
Keywords: arrhythmogenic cardiomyopathy, sudden cardiac death, RNA sequencing, microRNA, miRNA–mRNA
1. Introduction
Arrhythmogenic cardiomyopathy (ACM) is a heterogeneous genetic entity characterized by ventricular arrhythmias, contractile dysfunctions and progressive fibro-adipose replacement of myocardium [1]. ACM encompasses arrhythmogenic right ventricular cardiomyopathy (ARVC), arrhythmogenic biventricular cardiomyopathy (ABVC) and arrhythmogenic left ventricular cardiomyopathy (ALVC), all predisposing the patient to ventricular electrical instability and sudden cardiac death (SCD) [2]. ACM is a rare condition with a prevalence of 1/5000–6.5/5000 (OMIM #107970; ORPHA247) that affects mainly young individuals and athletes. In 2020, the “Padua criteria” were proposed for the diagnosis of ACM but it has been recently improved [3]. Several features such as male sex, previous non-sustained ventricular tachyarrhythmias, extent of T-wave inversion, recurrent premature ectopic beats and lower biventricular ejection fraction are risk factors for subsequent malignant arrhythmogenic events [4].
Pathogenic variants in genes encoding desmosomal proteins, such as PKP2, DSG2, DSC2 and DSP, constitute the main cause of ACM, although non-desmosomal forms of ACM also exist [5]. The inheritance of the disease is generally autosomic dominant; however, recessive forms with similar phenotypes have also been reported (JUP and DSP leading to Naxos and Carvajal diseases, respectively) [3]. All forms of ACM are characterized by low penetrance and variable expressivity, even in carriers of the same rare causative variant [6]. Genetic screening is crucial in ACM as it is one of the criteria on which the diagnosis of ACM is currently based [3].
The early identification of patients and adoption of personalized therapeutic measures remain the main current challenges. Hence, unravelling the pathophysiological mechanisms involved in the onset, progression and outcome of ACM will help with clinical diagnosis as well as risk stratification. From a pathophysiological perspective, hearts affected by ACM show a progressive loss of cardiomyocytes and fibro-fatty tissue replacement [7]. Although the pathological hallmark of the disease is usually present in the epicardial surface, septal and left ventricle areas can also show fibro-fatty infiltrations [8]. The desmosomes’ impairment affects the cell–cell junction, signalling at the intercalated discs and gaps union that functionally leads to intraventricular delay and re-entry circuits that provoke ventricular arrythmias. Furthermore, cardiomyocytes cannot cope with the mechanical strain, thus contributing to cardiomyocyte apoptosis.
Previous research focused on the comprehension of molecular mechanisms driving the ACM phenotype has demonstrated alterations of the Wnt/β-catenin pathway, which activates adipogenesis, fibrosis and apoptosis [9,10,11]. Another dysregulated pathway in ACM is the Hippo pathway, which responds to mechanical stimuli or cell–cell interactions regulating cell proliferation, apoptosis and cell fate. Moreover, activation of the Hippo pathway might inhibit the canonical Wnt signalling, thus promoting adipogenesis [12,13]. MicroRNAs (miRNAs) are small non-coding RNAs with a crucial role in the physiological development of the heart and cardiovascular diseases [14]. More recently, miRNA have also been shown to play an important role in ACM [11,15]. In the present work, we used mRNA and miRNA sequencing to analyse the transcriptome in ACM and healthy hearts.
2. Materials and Methods
2.1. Study Population
We recruited four frozen right ventricle (RV) myocardial biopsies from ACM patients (A1.R, A2.R, A4.R and A6.R) and four RV myocardial samples from unrelated ACM subjects (B1.R, B2.R, B3.R and B5.R) who died from conditions other than cardiac diseases as control group (Table 1). In ACM samples, pathogenic variant in each case is showed in Table 1. In postmortem analysis, all samples showed less than 60% of residual myocytes, with fibrosis substitution in right ventricular myocardium free wall, some of them even with fat-ty replacement. In addition, family history (first-degree relative) of ACM was confirmed in all cases. The criteria to collect samples was a definite clinical diagnosis of ACM. The second step was to include samples carrying genetic variants with conclusive deleterious role, all in genes currently associated with ACM.
Table 1.
Genetic data of samples with disease confirmed and pathogenic variant identified. ACM: Arrhythmogenic Cardiomyopathy; LP: Likely Pathogenic; NA: Not Available; P: Pathogenic; RV: Right Ventricle; VUS: Variant of Unknown Significance.
| Sample | Phenotype | Gene | Protein | Nucleotide | dbSNP | gnomAD | ClinVar | ACMG |
|---|---|---|---|---|---|---|---|---|
| A1.R | ACM | FLNC | p.Arg1370Ter | c.4108C > T | rs1342121466 | 4/1450766 (0.0002%) | P | P |
| A2.R | ACM |
PKP2
DSG2 |
p.Lys678ArgfsTer12 - |
c.1881del c.523 + 2dup |
rs764817683 rs2073126642 |
NA 2/1451740 (0.0001%) |
P VUS |
P LP |
| A4.R | ACM | PKP2 | p.Arg79Ter | c.235C > T | rs121434420 | 22/1577370 (0.001%) | LP | LP |
| A6.R | ACM | TMEM43 | p.Ser358Leu | c.1073C > T | rs63750743 | 2/1461886 (0.0001%) | P | P |
| B1.R | Control | - | - | - | - | - | - | - |
| B2.R | Control | - | - | - | - | - | - | - |
| B3.R | Control | - | - | - | - | - | - | - |
| B5.R | Control | - | - | - | - | - | - | - |
2.2. Ethics
This study was approved by the appropriate ethics committees. The ethical research principles were fulfilled following the Helsinki Declaration, and further amendments, and the Belmont report. This study also adhered to two legal provisions governing human research and the Spanish Organic Law 15/1999 for the Regulation of Automated Processing of Personal Data. All procedures performed were in accordance with the ethical standards of the institutional research committee and was approved by the local Ethics Committee (CEIM: 2021.185, CEIC 1315-N-21, 2018/0417 and 2014/0306).
2.3. Myocardial Tissue Collection
This is a multicentre study that involved Research Biomedicine Institute Josep Trueta (IDIBGI), Girona; Health Research Institute La Fe, Hospital La Fe of Valencia, Valencia; and Research Institute of Biomedicine and Innovation (INIBICA), Cádiz. Heart samples from human autopsies were stored at −80 °C until they were analysed.
2.4. RNA Extraction
The TRI Reagent (Sigma-Aldrich, St Louis, MO, USA) was used to isolate and purify total RNA according to the manufacturer’s instructions. Then, DNase treatment was performed using RNA clean and concentrator-5 kit (Zymo Research, Irvine, CA, USA). RNA was quantified using a Qubit RNA High-Sensitivity Assay kit in the Qubit® 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). The quality and integrity of total RNA were controlled on the Agilent Technologies 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
2.5. RNA-Sequencing Analysis and Bioinformatics
Standard specific mRNA-sequencing (mRNA-seq) libraries were generated using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina using the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs, Ipswich, MA), and single-end libraries were sequenced on an Illumina SE100 Platform with an output of ~680M reads per sample. Standard miRNA libraries were generated using the NEXTFLEX small RNA-seq kit v3 (Perkin Elmer, Waltham, MA, USA), and single-end libraries were sequenced on an Illumina SE60 Platform with an output of ~2 M reads per sample.
For trimming and aligning raw data, fastq sequence reads were uploaded to the European version of the Galaxy platform [16]. Reads were trimmed with the Trim Galore software (Galaxy Version 0.6.7 + galaxy0) and aligned to the built-in human reference genome December 2013 (GRCh38/hg38) with the RNA STAR Gapped-read mapper (Galaxy Version 2.7.10b + galaxy3). For gene expression analyses, bam files were downloaded from the Galaxy server and further analysed with the different RStudio packages downloaded from the Bioconductor website (http://bioconductor.org, accessed on 8 July 2023). Reads were assigned to genes by means of the “featureCounts” function of the “Rsubread” package, version 2.10.5 [17], and annotation files human release 43 (GRCh38.p13) (https://www.gencodegenes.org/human/release_43.html, accessed on 18 April 2024) and Chromosomal coordinates of Homo sapiens microRNAs (https://www.mirbase.org, accessed on 18 April 2024) were used for mRNA and miRNA analysis, respectively. Only the mapped reads were used to calculate gene expressions. The library size of each experimental point ranged from 37,342,862 to 51,420,486 sequences and from 318,097 to 1,898,024 sequences for mRNA and miRNA analysis, respectively.
The difference in the library size ranges between mRNA and miRNA is attributed to the inherent characteristics of the RNA types and their respective sequencing techniques. mRNA, being longer and more varied compared to miRNA, requires a greater number of reads to ensure adequate coverage and precise quantification of gene expression. In contrast, miRNA, which is significantly shorter, needs fewer reads to achieve comprehensive representation. These differing sequencing requirements account for the observed variations in library sizes between mRNA and miRNA.
The differential gene expression analyses were performed with package ‘DESeq2’ version 1.36.0 [17]. All the gene comparisons with a p-value <0.05 and a fold-change >2 were considered differentially expressed under the experimental conditions. Functional Over Representation Analysis (ORA) based on Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) were conducted with the package ‘clusterProfiler’ version 4.4.4. [18]. The gene sets with a p-value <0.05 were considered overrepresented under the experimental conditions. Finally, to identify miRNA–mRNA interaction, we constructed a pipeline using the DEMs and DEGs between ACM and control cohorts. We used Diana-microT, ElMMo, Microcosm, Miranda, Mirdb, Pictar, PITA and Targetscan prediction tools to select mRNA targets.
2.6. Statistical Analysis
Data are expressed as mean ± SEM, and n denotes the number of replicates for each experiment. Outliers were identified through the Rout method, using Q = 1%. The normal distribution of each variable was verified with the Shapiro–Wilk test. Statistical differences (p < 0.05) between the experimental groups were assessed using a two-tailed, unpaired Student’s t test for Gaussian distributions. For non-Gaussian distributions, a Mann–Whitney non-parametric test was used. All the statistical analyses were performed using GraphPad Prism 9.0 software (San Diego, CA, USA).
3. Results
3.1. ACM and Controls mRNA and miRNA Expression Profiles
We performed RNA-seq to investigate whether differences exist in the mRNA expression profiles of the myocardium of ACM and control patients. The genetic data of the 8 patients included in this study are shown in Table 1. The filtered RNA-seq read-set identified ≈ 29,568 genes. We used principal component analysis (PCA) to visualize the sample clustering for the most variably expressed genes (Figure 1). No outliers were observed in the PCA (Figure 2A).
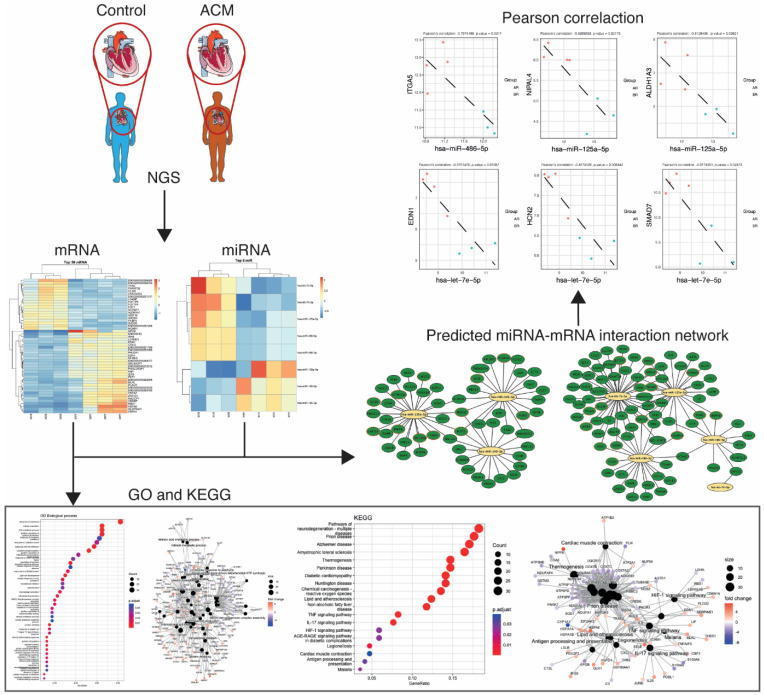
Figure 1.
A flowchart of the study design.
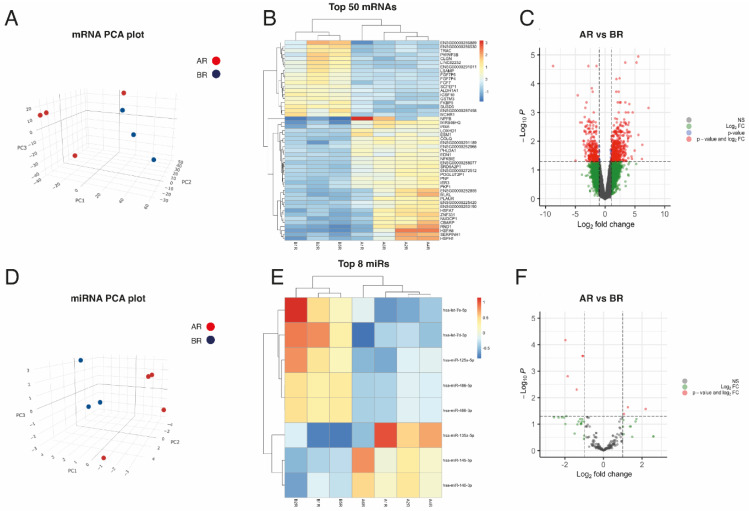
Figure 2.
Exploratory analysis of paired miRNA and mRNA expression in heart samples. (A) 3D-Principal Components Analysis plot, based on correlation matrix, for mRNA expression in ACM (n = 4) and control (n = 3) tissue samples. (B) Heatmap of the top 50 most DEMs sorted by absolute FC (all of them having FDR < 0.05). (C) Volcano plot of the mRNAs, highlighting in grey those not statistically significant with FDR > 0.05 and absolute FC < 2 (abslog2FC < 1); in blue, those with FDR < 0.05 but absolute FC < 2 (abslog2FC < 1); in green, those with absolute FC > 2 (abslog2FC > 1) but FDR > 0.05; and in red, those with FDR < 0.05 and absolute FC > 2 (abslog2FC > 1). (D) 3D-Principal Components Analysis plot, based on correlation matrix, for miRNA expression in ACM (n = 4) and control (n = 3) tissue samples. (E) Heatmap of the top 8 most DEMs sorted by absolute FC (all of them having FDR < 0.05). (F) Volcano plot of the miRNAs, highlighting in grey those not statistically significant with FDR > 0.05 and absolute FC < 2 (abslog2FC < 1); in green, those with absolute FC > 2 (abslog2FC > 1) but FDR > 0.05; and in red, those with FDR <0.05 and absolute FC > 2 (abslog2FC > 1).
In total, 689 genes were differentially expressed between ACM and controls, with a Log2 fold-change (FC) > 0.5 using a 5% false discovery rate (FDR). From those genes, 383 were upregulated and 306 were downregulated in the ACM samples (Figure 2B,C). We also analysed the miRNA expression profile by way of small RNA-seq to investigate the differential expression between ACM and the control groups. According to the PCA, the miRNA expression profile of the ACM cohort differed significantly from that of the control sample’s autopsies and no outliers were observed (Figure 2D). After RNA-seq analysis, 290 miRNAs were detected in all heart samples. The most highly expressed miRNA in all myocardial samples was miR-1-3p (Table S1). We identified eight DEMs between the ACM and control heart samples (Log2 FC > 0.5; FDR ≤ 0.05). Three of them (miR-135a-5p, miR-140-3p and miR-145-5p) were upregulated, and five (miR-486-5p, miR-486-3p, miR-125a-5p, let-7e-5p and let-7d-3p) were downregulated in ACM hearts as compared to the controls (Figure 2E,F).
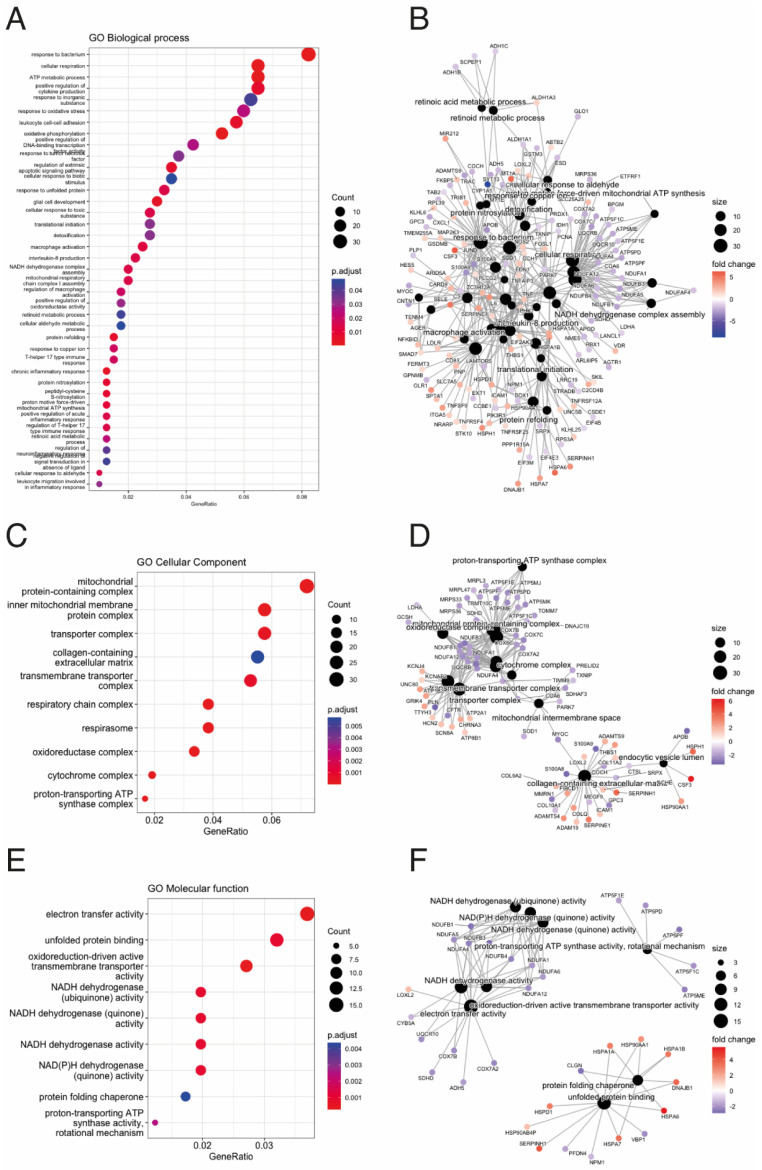
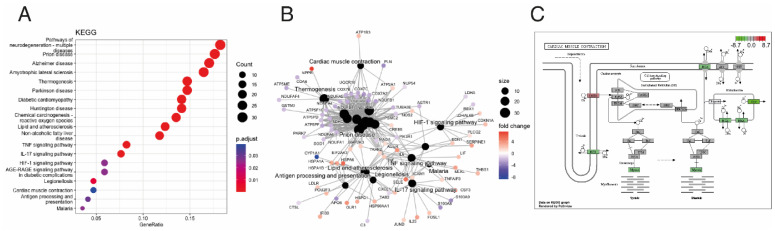
To identify the biological mechanisms related to ACM disease, we performed pathway enrichment on the DEGs in our study. According to the number of counts and categorized by the biological process (BP), we found mitochondrial respiratory-related pathways, response to oxidative stress, apoptotic signalling pathways, inflammatory response-related pathways and retinoic acid metabolic processes to be among the most enriched GO terms (Figure 3A,B). In the cellular component (CC), mitochondrial respiratory- and extracellular matrix-related pathways were among the most enriched pathways in ACM (Figure 3C,D). Similarly, in terms of molecular function (MF), mitochondrial respiratory- and ER stress-related pathways were among the most enriched pathways in ACM (Figure 3E,F). Finally, KEGG pathway analysis showed that these DEGs were mainly involved in diabetic cardiomyopathy, lipid and atherosclerosis, inflammation response-related pathways and cardiac muscle contraction (Figure 4A–C).
Figure 3.
Gene Ontology (GO) Over Representation Analysis (ORA) between ACM and control groups. (A) Dot plot for Biological Process (BP) analysis. (B) Gene-Concept Network plot showing the linkages of differential expressed genes and BP terms. (C) Dot plot for Cellular Component (CC) analysis. (D) Gene-Concept Network plot showing the linkages of differential expressed genes and CC terms. (E) Dot plot for Molecular Function (MF) analysis. (F) Gene-Concept Network plot showing the linkages of differential expressed genes and MF terms.
Figure 4.
Kyoto Encyclopedia of Genes and Genomes (KEGG) Over Representation Analysis (ORA) between ACM and control groups. (A) Dot plot for KEGG analysis. (B) Gene-Concept Network plot showing the linkages of differential expressed genes and KEGG pathways. (C) Down-regulated “cardiac muscle contraction” KEGG pathway; gene expression values are mapped to gradient.
3.2. Integrative Analysis Identified miRNA–mRNA Interaction for ACM
Considering that mRNAs are typically targeted by many miRNAs and each miRNA targets multiple mRNAs, we established an effective pipeline to identify the miRNA–mRNA interaction for ACM using the DEMs and DEGs between ACM and controls. We used Diana-microT, ElMMo, Microcosm, Miranda, Mirdb, Pictar, PITA and Targetscan prediction tools to select the mRNA targets. We only selected negatively correlated miRNA–mRNA pairs. This prediction and filtering approach resulted in 304 pairs comprising eight miRNAs and their 135 likely target mRNAs (Figure 5A and Tables S1–S3). The list of miRNA–gene pairs includes already validated pairs using the Mirtarbase and Tarbase databases (Figure 5A and Tables S1–S3).
Figure 5.
miRNA–mRNA interaction analysis. (A) Network of selected miRNA–mRNA interactions. (B) Negatively correlated miRNA–mRNA pairs predicted simultaneously, at least in three databases used in our pipeline.
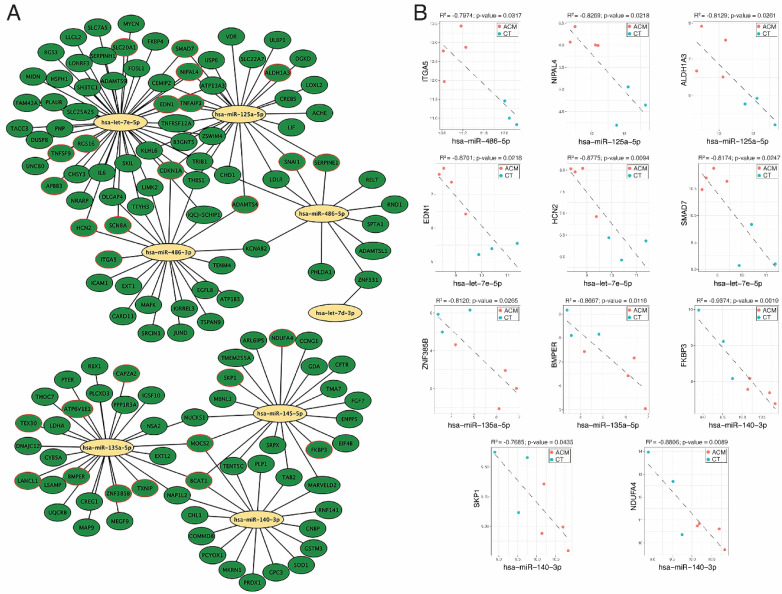
Among all miRNA–mRNA pairs, we selected those genes meeting the following criteria: (1) genes predicted as the target of a DEM by at least three miRNA target prediction tools, (2) target genes that were experimentally validated and stored in a database such as the Mirtarbase and/or Tarbase databases and (3) target genes shared by at least two DEMs (Table 2). Pearson correlation tests were performed to identify the miRNA-target pairs. Our miRNA–mRNA analysis identified 11 negatively correlated miRNA-target pairs for ACM: miR-486-5p/ITGA5, miR-125a-5p/NIPAL4, miR-125a-5p/ALDH1A3, let-7e-5p/EDN1, let-7e-5p/HCN2, let-7e-5p/SMAD7, miR-135a-5p/ZNF385B, miR-135a-5p/BMPER, miR-140-3p/FKBP3, miR-140-3p/SKP1 and miR-140-3p/NDUFA4 (R < −0.7, p-value < 0.05) (Figure 5B).
Table 2.
Main miRNA–gene interactions predicted.
| Counts | |||||||
|---|---|---|---|---|---|---|---|
| Sample A1R | Sample A2R | Sample A4R | Sample A6R | Sample B1R | Sample B2R | Sample B5R | Gene Symbol |
| 268,147 | 333,296 | 319,027 | 39,769 | 149,975 | 223,451 | 826,406 | hsa-miR-1-3p |
| 110,587 | 91,623 | 77,600 | 49,265 | 38,476 | 47,353 | 180,699 | hsa-miR-143-3p |
| 26,647 | 70,761 | 71,180 | 51,234 | 63,687 | 81,431 | 160,503 | hsa-let-7a-5p |
| 27,074 | 45,652 | 39,339 | 18,103 | 25,298 | 41,027 | 111,000 | hsa-let-7f-5p |
| 25,882 | 52,083 | 43,790 | 25,771 | 21,360 | 25,545 | 85,002 | hsa-miR-26a-5p |
| 28,171 | 45,954 | 34,588 | 8582 | 17,237 | 12,243 | 56,518 | hsa-miR-30d-5p |
| 13,406 | 53,514 | 23,570 | 6424 | 8590 | 6284 | 34,382 | hsa-miR-133a-3p |
| 17,567 | 26,356 | 25,662 | 9669 | 11,579 | 12,041 | 39,905 | hsa-miR-24-3p |
| 17,871 | 16,799 | 14,542 | 4624 | 5552 | 4409 | 25,811 | hsa-miR-30a-5p |
| 10,120 | 14,886 | 12,774 | 4398 | 5066 | 5808 | 20,783 | hsa-miR-126-3p |
| 8811 | 13,206 | 12,865 | 4841 | 5804 | 6031 | 20,016 | hsa-miR-3074-5p |
| 4907 | 18,329 | 10,663 | 3449 | 6000 | 6092 | 18,876 | hsa-miR-30c-5p |
| 4371 | 12,644 | 10,991 | 5905 | 5633 | 5314 | 18,177 | hsa-miR-125b-5p |
| 4524 | 11,010 | 10,114 | 5501 | 6172 | 6933 | 18,603 | hsa-let-7g-5p |
| 6351 | 9710 | 7213 | 3066 | 3287 | 5284 | 15,622 | hsa-miR-27b-3p |
| 2106 | 7043 | 6079 | 2008 | 5542 | 7547 | 16,297 | hsa-miR-125a-5p |
| 3642 | 10,930 | 6973 | 2556 | 3627 | 4263 | 12,207 | hsa-miR-23b-3p |
| 4042 | 10,743 | 6644 | 1492 | 2689 | 2598 | 10,319 | hsa-miR-378a-3p |
| 2029 | 6048 | 4382 | 2687 | 4514 | 3126 | 9480 | hsa-miR-92a-3p |
| 2838 | 7362 | 5436 | 3769 | 2454 | 2178 | 6331 | hsa-miR-23a-3p |
| 3568 | 4394 | 3894 | 2565 | 3678 | 2126 | 9118 | hsa-miR-16-5p |
| 5040 | 4232 | 4240 | 1244 | 2229 | 2111 | 8984 | hsa-miR-22-3p |
| 2482 | 5222 | 4175 | 2261 | 1938 | 3021 | 8180 | hsa-miR-26b-5p |
| 2953 | 4191 | 2876 | 2977 | 4368 | 1119 | 7761 | hsa-miR-451a |
| 1496 | 3889 | 2983 | 1273 | 2556 | 2826 | 10,222 | hsa-miR-486-5p |
| 1496 | 3889 | 2983 | 1273 | 2556 | 2826 | 10,222 | hsa-miR-486-3p |
| 1872 | 3873 | 2903 | 2441 | 2029 | 2386 | 8070 | hsa-let-7i-5p |
| 1796 | 4135 | 3928 | 3380 | 1426 | 1829 | 7013 | hsa-miR-199a-3p |
| 817 | 2593 | 2470 | 3058 | 2414 | 3316 | 6456 | hsa-let-7c-5p |
| 3914 | 3111 | 3522 | 2173 | 1043 | 1457 | 5324 | hsa-miR-21-5p |
| 1840 | 3173 | 2807 | 1327 | 1658 | 1086 | 4991 | hsa-miR-103a-3p |
| 1840 | 3173 | 2807 | 1327 | 1658 | 1086 | 4991 | hsa-miR-103b |
| 2528 | 2397 | 2317 | 272 | 1096 | 1841 | 6253 | hsa-miR-499b-3p |
| 2528 | 2397 | 2317 | 272 | 1096 | 1840 | 6253 | hsa-miR-499a-5p |
| 1664 | 3486 | 2368 | 1272 | 1048 | 1185 | 5165 | hsa-miR-99b-5p |
| 1518 | 3079 | 2508 | 2021 | 848 | 946 | 4498 | hsa-miR-99a-5p |
| 2306 | 2956 | 2142 | 874 | 1256 | 1034 | 4838 | hsa-miR-181a-5p |
| 764 | 2217 | 1593 | 2737 | 1488 | 1875 | 4178 | hsa-let-7b-5p |
| 3068 | 2358 | 2571 | 523 | 976 | 664 | 4526 | hsa-miR-30e-5p |
| 1377 | 3945 | 2391 | 2752 | 671 | 508 | 2690 | hsa-miR-145-5p |
| 894 | 2055 | 1956 | 1684 | 712 | 909 | 3489 | hsa-miR-199b-3p |
| 1378 | 2869 | 1863 | 519 | 888 | 720 | 3044 | hsa-miR-30e-3p |
| 927 | 1861 | 1642 | 873 | 897 | 963 | 3769 | hsa-miR-423-3p |
| 927 | 1861 | 1642 | 873 | 897 | 963 | 3769 | hsa-miR-3184-5p |
| 912 | 1552 | 1243 | 2236 | 719 | 516 | 2638 | hsa-miR-100-5p |
| 951 | 2629 | 1462 | 1280 | 536 | 278 | 1718 | hsa-miR-140-3p |
| 1289 | 1841 | 1288 | 706 | 619 | 593 | 1990 | hsa-miR-27a-3p |
| 928 | 1654 | 1220 | 515 | 378 | 367 | 1917 | hsa-miR-151a-3p |
| 729 | 1841 | 1080 | 386 | 513 | 522 | 1785 | hsa-miR-30a-3p |
| 1095 | 1195 | 1101 | 748 | 560 | 487 | 1617 | hsa-miR-29a-3p |
| 913 | 1935 | 1215 | 466 | 387 | 249 | 1449 | hsa-miR-30b-5p |
| 603 | 1136 | 945 | 598 | 764 | 562 | 1959 | hsa-miR-191-5p |
| 267 | 588 | 593 | 469 | 840 | 1560 | 2217 | hsa-let-7e-5p |
| 543 | 1081 | 910 | 461 | 539 | 605 | 1712 | hsa-miR-151a-5p |
| 71 | 203 | 353 | 759 | 862 | 416 | 2886 | hsa-miR-10a-5p |
| 267 | 768 | 737 | 486 | 849 | 777 | 1490 | hsa-let-7d-5p |
| 431 | 821 | 735 | 377 | 481 | 704 | 1800 | hsa-miR-98-5p |
| 96 | 163 | 236 | 1239 | 872 | 287 | 1942 | hsa-miR-10b-5p |
| 495 | 983 | 831 | 383 | 366 | 320 | 1333 | hsa-miR-148a-3p |
4. Discussion
In the present study, we performed for the first time a wide-ranging characterization of the mRNA and miRNA transcriptome of myocardial tissue from ACM patients. We generated an ACM-specific miRNA–target transcript interaction network, thereby providing the first unbiased analysis of miRNAs and their targets in the context of ACM. Hereto, we identified 689 mRNAs and 53 miRNAs with significant differential expression in ACM. Subsequent pathway enrichment analysis of the DEGs showed significant enrichment for genes, mainly regarding mitochondrial respiration, extracellular matrix, oxidative stress, ER stress, apoptosis, cell–cell adhesion, inflammation, retinoic acid metabolic processes, diabetic cardiomyopathy, lipid and atherosclerosis and cardiac muscle contraction, among others. We created an ACM-associated miRNA interactome of eight miRNAs and their 135 likely target mRNAs.
ACM is characterized by the death of cardiomyocytes followed by inflammation and the progressive accumulation of fibro-fatty tissue. Alterations in Wnt/ß-catenin and the Hippo pathway due to abnormal cell–cell adhesion and intracellular signalling, caused by deleterious rare variants located in genes encoding desmosomal proteins, is considered the main cause [10,12]. This is in consonance with the deregulation of genes involved in cell–cell adhesion, extracellular matrix-related pathways, apoptosis and cardiac muscle contraction. Our results showed dysregulation of cardiac inflammation and lipid transport pathways consistent with another research. Rainer et al. analysed the coding and non-coding transcriptome of human cardiac stromal cells derived from endomyocardial biopsies of ACM patients. They found that, in addition to cell–cell adhesion, deregulated genes were also involved in cardiac inflammation and lipid transport [19]. Similarly, RNA-seq data from left ventricular tissue from deceased ACM patients showed dysregulation of cell–cell adhesion, extracellular matrix, and inflammation [20]. In alignment with these findings, Lin et al. (2023) profiled the region-resolved transcriptome and proteome of healthy and dilated cardiomyopathy (DCM) human myocardial tissue and obtained an extensive dataset. Based on the core proteome and transcriptome characteristics of healthy hearts, chamber-specific proteome alterations were further revealed in end-stage DCM, among which extracellular matrix (ECM), mitochondrial function, and muscle contraction were the most dysregulated biological processes. These results reinforce the relevance of ECM and mitochondrial dysfunction in cardiomyopathies and provide additional context for our findings in ACM, highlighting the broader implications of these dysregulated processes across different types of cardiomyopathies [21].
The mitochondrial impairment might constitute substrates for electrical and structural remodelling in ACM hearts, playing, therefore, a role in the electrical stability [22]. Lippi et al. recently demonstrated epigenetic and gene expression profiles of cardiac mesenchymal stromal cells and confirmed for the first time that mitochondrial dysfunction is present in ACM [23]. This group also proposed that oxidative stress represents a cofactor contributing to the pathogenesis of ACM [24], which might be a consequence of mitochondrial dysfunction. This fact was evidenced in patients carrying pathogenic PKP2 variants [25]. Consistently, our results revealed dysregulation of mitochondrial respiration pathways and oxidative stress-related pathways. Similarly, we found that DEGs in ACM are involved in ER stress pathways, which is in line with previous observations showing the overexpression of ER stress markers in the myocardial tissue of ACM murine models [26]. A comparative analysis of pathways identified in this study versus the reported literature is shown in Table 3.
Table 3.
Comparative analysis of pathways identified in this study versus the reported literature.
| Pathway | This Study | Reported Literature |
|---|---|---|
| Adipogenesis | ✓ | [27,28] |
| Apoptosis | ✓ | [29] |
| Cardiac Electrophysiology | ✓ | [30,31] |
| Cardiac Muscle Contraction | ✓ | [10,12] |
| Cardiovascular System Development | [20] | |
| Cell–Cell Adhesion | ✓ | [19,20,22] |
| Chromatin Organization | [22] | |
| Circulatory System Development | [20] | |
| EMT Process | ✓ | [32] |
| ER Stress | ✓ | [25] |
| Extracellular Matrix | ✓ | [19,20] |
| Inflammation | ✓ | [19,20] |
| Lipid and Atherosclerosis | ✓ | [19] |
| Lipid Metabolism | ✓ | [33] |
| Mitochondrial Respiration | ✓ | [22] |
| Oxidative Stress | ✓ | [23] |
| Oxidized LDL-Dependent Pathway | [23] | |
| Platelet Degranulation | [19] | |
| Regulation of Protein Secretion | [22] | |
| Retinoic Acid Metabolic Process | ✓ | [34] |
| TGF-ß Signalling | ✓ | [35] |
| Tissue Development | [20] | |
We identified eleven negatively correlated miRNA-target transcript pairs: miR-486-5p/ITGA5, miR-125a-5p/NIPAL4, miR-125a-5p/ALDH1A3, let-7e-5p/EDN1, let-7e-5p/HCN2, let-7e-5p/SMAD7, miR-135a-5p/ZNF385B, miR-135a-5p/BMPER, miR-140-3p/FKBP3, miR-140-3p/SKP1 and miR-140-3p/NDUFA4. The epicardium is a source of multiple cardiac cell types, including fibroblasts, playing a key role during cardiac development and remodelling through the epithelial-to-mesenchymal (EMT) process [36,37,38,39]. Several studies have proposed the epicardium as the initial site of manifestation of classic ACM [40,41,42,43,44]. Integrin alpha 5 (ITGA5) is necessary for proper heart morphogenesis, and the binding of ITGA5 to the epicardial secreted fibronectin (FN1) is required for cardiomyocyte maturation [45,46,47,48]. In the adult heart, ITGA5 has been found to be upregulated in the atrial tissues of patients with atrial fibrillation, whereas epicardial secreted fibronectin has been identified as a source of fibroblasts through the EMT process [32]. Moreover, ITGA5 has been suggested as a promotor of adipocyte fibrosis-related gene expression [49]. Although in distinct biological contexts both ITGA5 and miR-486-5p have been reported to modulate the TGF-β-mediated EMT process, adipogenesis and fibrosis [35,49,50,51,52,53], the miR-486-5p/ITGA5 axis has been suggested as a potential target to modulate fibro-fatty infiltration of subepicardial layers into the myocardium. Interestingly, SMAD7, a negative regulator of TGF-ß signalling [54], was also deregulated in ACM samples, supporting the notion that TGF-ß signalling might be a key regulator of epicardial EMT processes in ACM. In colorectal cancer (CRC), TGF-β1 induces a partial EMT (pEMT) process and collective cell invasion without full mesenchymal transition, maintained by Smad transcription factors. This mechanism might similarly induce EMT processes in epicardial cells in ACM, leading to fibroblast accumulation [51]. miR-486-5p also plays a role in aging and adipogenesis in human adipose tissue-derived mesenchymal stem cells (hAT-MSCs) by targeting SIRT1, inhibiting proliferation and differentiation. This suggests miR-486-5p might similarly affect epicardial adipogenesis in ACM, contributing to its fibro-fatty phenotype [53]. ITGA5 plays a crucial role in adipocyte differentiation, where its reduction promotes adipogenesis and overexpression inhibits it. This regulatory role of ITGA5 could be disrupted in ACM, leading to pathological adipocyte accumulation [50]. Additionally, miR-486-5p inhibits adipogenesis in mesenchymal stem cells and prevents steroid-induced osteonecrosis by targeting TBX2 and upregulating p21. These anti-adipogenic effects could be leveraged to counteract ACM’s adipogenic component. Thus, the miR-486-5p/ITGA5 axis is a critical regulatory pathway in ACM, influencing both fibroblast and adipocyte behaviour. miR-486-5p has also been identified as a tumour suppressor in non-small cell lung cancer (NSCLC) progression, where it modulates TGF-β signalling and EMT processes. High expression of SMAD2, a downstream effector of TGF-β, correlates with poor prognosis in NSCLC and promotes EMT processes. miR-486-5p targets SMAD2, inhibiting TGF-β-induced EMT processes and metastasis in NSCLC cells [51]. Similarly, in ACM, miR-486-5p could inhibit TGF-β-mediated EMT processes by targeting ITGA5, thus preventing fibro-fatty infiltration. This highlights miR-486-5p as a potential modulator of epicardial EMT processes in ACM, which may be crucial for controlling disease progression. Understanding the miR-486-5p/ITGA5 axis offers potential therapeutic insights. Therapeutically, modulating this pathway could inhibit pathological EMT processes and adipogenesis, reducing fibro-fatty infiltration in ACM. Although little is known about the role of let-7e-5p in cardiac disease, several studies suggest an active role in the pathogenesis of heart failure [55,56]; however, its role in regulating TGF-ß signalling remains to be investigated.
Bone morphogenetic proteins (BMPs) are members of the TGF-ß superfamily. Several studies have described BMPER as a regulator of BMP signalling [57,58,59]. Besides its regulatory function in endothelial biology [57,60,61,62,63], BMPER has also been identified as an EMT process-related gene [64]. Furthermore, BMPER has been proposed as a potential regulator of epicardial adipose tissue (EAT) mesenchymal stem cells (MSCs). EAT is an ectopic visceral fat deposit enveloping the heart that acts as an active endocrine organ with effects on myocardial metabolism [65,66]. Curiously, the amount of right and left ventricular EAT has been associated with ACM and the disease’s severity in humans [27]. Hence, BMPER may play a role in ACM regulating EAT stem cells. Interestingly, miR-135a-5p has been reported to modulate adipogenesis in EAT MSCs through the Hippo signalling pathway [28,34]. This miRNA is downregulated during preadipocyte differentiation, and its overexpression impairs adipogenic marker gene expression, emphasizing its regulatory role in adipogenesis [27]. Furthermore, miR-135a-5p influences the canonical Wnt/β-catenin signalling pathway by targeting APC, thereby affecting cell differentiation processes [28]. In the context of ACM, both BMPER and miR-135a-5p are relevant due to their roles in regulating adipogenesis and fibrosis. BMPER has been shown to regulate BMP2-mediated signalling, influencing EMT processes and extracellular matrix deposition [63]. Additionally, miR-135a-5p has been reported to modulate adipogenesis in EAT MSCs through the Hippo signalling pathway, and its dysregulation may contribute to pathological changes in ACM [28]. The connection between BMPER and miR-135a-5p in regulating EAT MSCs presents a novel perspective on ACM treatment. The miR-135a-5p/BMPER axis may offer new therapeutic opportunities by targeting key pathways involved in fibrosis and adipogenesis. Given the association between EAT volume and ACM severity, modulating this axis could potentially mitigate fibro-fatty infiltration and improve disease outcomes.
ALDH1A3 is a member of the Aldehyde dehydrogenase 1 family (ALDH1) that participates in the retinoic acid (RA) biosynthesis [67], a vitamin A metabolite that plays a significant role in the contribution of the epicardium to heart development and function [68]. In the mouse heart, RA has been described to mediate dilated cardiomyopathy (DCM) and cardiomyocyte apoptosis after myocardial infarction, suggesting a role in the adult human heart as well [33,69]. Interestingly, RA has been implicated in adipogenesis through transcriptional regulation of PPARγ [70]. In this sense, the induction of PPARγ expression mediated by RA can convert both primary myogenic cells and the myoblast cell line to adipogenic cells in avian samples [71]. Our data showed miR-125a-5p as a potential target of ALDH1A3. Curiously, it has been shown that inhibition of this miRNA accelerates 3T3-L1 preadipocyte differentiation and upregulation of fatty acid metabolism-related genes [30]. Accordingly, we observed downregulation of miR-125a-5p and upregulation of ALDH1A3 in ACM samples. The ALDH1A3/miR-125a-5p axis therefore offers a novel perspective on the regulation of adipogenesis in ACM. ALDH1A3-mediated RA synthesis and subsequent RA-induced PPARγ expression could drive adipogenic differentiation in the epicardium, contributing to the fibro-fatty remodelling characteristic of ACM. The downregulation of miR-125a-5p, which typically inhibits adipogenesis, further supports this process, suggesting a coordinated regulatory mechanism involving both ALDH1A3 and miR-125a-5p. Therapeutically, targeting the ALDH1A3/miR-125a-5p axis could modulate adipogenesis and fibrosis in ACM, potentially mitigating disease progression. However, further research is needed to validate these findings and explore the clinical applications of manipulating the ALDH1A3/miR-125a-5p axis in ACM.
Altogether, our results reinforce the concept of the epicardial contribution to fibro-fatty remodelling in ACM and offer potential new therapeutic targets for the treatment of the disease. HCN2 is one of a family of four genes (HCN1-4) that encodes a hyperpolarization-activated, cyclic nucleotide-gated cation channel [31] whose altered function is associated with arrhythmogenic events [72]. Although alteration in the expression of HCN2 has not been related to arrhythmias in physiological conditions, under pathological conditions HCN2 overexpression induces arrhythmias [73,74], which is consistent with higher expression levels of HCN2, therefore suggesting that HCN2 might play a role in ACM. Dysregulation of Endothelin-1 (EDN1) has been linked to DCM [72]. Furthermore, EDN1 expression has been positively correlated with the levels of superoxide (SOD1 and SOD2) and the amount of collagen in the mouse heart [75]. Accordingly, we observed upregulation of EDN1 in ACM hearts, suggesting EDN1 as an important player in controlling fibrosis and oxidative stress in ACM.
Regarding the miRNA–mRNA pairs, miR-125a-5p/NIPAL4, miR-135a-5p/ZNF385B, miR-140-3p/FKBP3, miR-140-3p/SKP1 and miR-140-3p/NDUFA4, very little or nothing is known about the function of these genes in cardiac disease. Whereas Skp1 has been related to cardiac hypertrophy and the degradation of key sarcomeric proteins [29,76], NDUFA4 has been associated with cardiomyocyte apoptosis and mitochondrial dysfunction, two ACM-related processes [77]. Further research will be required to explore the role of NIPAL4, ZNF385B and FKBP3 in ACM. Finally, miR-135a-5p and miR-140-3p have been involved in a wide range of cardiac-related processes, such as cardiomyocyte apoptosis, cardiomyocyte hypoxia, oxidative stress, fibrosis, cardiac inflammation, cardiac hypertrophy and heart failure, making them good candidates for therapeutic targets for ACM.
Targeting miRNAs in preclinical and clinical settings shows promise for treating various diseases, including cancer. Preclinical studies demonstrate the potential of miRNA modulation in altering disease progression and overcoming resistance. Advanced techniques like CRISPR and next-generation sequencing enhance our understanding of miRNA dysregulation. However, challenges such as toxicity and off-target effects need to be addressed for successful clinical translation. Ongoing clinical trials are evaluating miRNA-based therapies, emphasizing the importance of ensuring safety and efficacy in clinical applications. Continued research is crucial to harness the therapeutic potential of targeting miRNAs and non-coding RNAs for improved patient outcomes [78].
In our study, there are some limitations to mention. First is the number of samples. It is difficult to obtain human ventricular samples in proper condition to perform analyses such as those we performed in our study. However, we have a set of ACM samples which underwent clinical diagnostics and the gene variant was determined as the definite cause of disease; this is one of the main points nowadays as a large portion of rare variants remain with no definitive causality. Another limitation is the confirmation/validation of all these interactions, despite some of them having been previously validated. Moreover, it is important to take into account that the heart samples came from deceased individuals and RNA integrity could be affected by this condition. It is because of this that our control group shares similar postmortem intervals.
In summary, the evaluation of these miRNA–mRNA interactions highlight the potential for developing therapeutic agents that target these miRNAs as a promising approach in treating ACM. Therefore, this study successfully identified key miRNAs associated with ACM, offering new perspectives on the underlying mechanisms of ACM’s development.
Acknowledgments
We thank the Bosch i Aymerich Foundation for their support. IDIBGI and Institut de Recerca Sant Joan de Déu are a “CERCA Programme/Generalitat de Catalunya”. CIBERER and CIBERCV are an initiative of the ISCIII, Spanish Ministry of Economy and Competitiveness.
Abbreviations
| ALDH1 | Aldehyde dehydrogenase 1 family |
| ACM | Arrhythmogenic cardiomyopathy |
| BMPs | Bone morphogenetic proteins |
| FN1 | Fibronectin |
| ITGA5 | Integrin alpha 5 |
| LP | Likely pathogenic |
| miRNAs | MicroRNAs |
| P | Pathogenic |
| RV | Right ventricle |
| SCD | Sudden cardiac death |
| VUS | Variant of unknown significance |
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/biomedicines12081807/s1: Table S1: Raw counts of top 60 miRNAs; Table S2: list of miRNA (down)–gene (up) pairs; Table S3: list of miRNA (up)–gene (down) pairs.
Author Contributions
R.T. and A.M. conceived the experiments; O.C., M.A., G.S.-B. and A.B.-B. recruited the subjects. F.B. and R.T. conducted the experiments, and F.H.-T., J.C.-C., R.B., J.A.G.R., M.Q.-F. and M.R. analysed the results. F.B., J.A.G.R., O.C. and R.T. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was approved by the appropriate ethics committees. The ethical research principles were fulfilled following the Helsinki Declaration, and further amendments, and the Belmont report. This study also adhered to two legal provisions governing human research and the Spanish Organic Law 15/1999 for the Regulation of Automated Processing of Personal Data. All procedures performed were in accordance with the ethical standards of the institutional research committee and were approved by the local Ethics Committee (CEIM: 2021.185, CEIC 1315-N-21, 2018/0417 and 2014/0306).
Informed Consent Statement
Not applicable.
Data Availability Statement
The RNA-seq data generated in this manuscript have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession codes GSE273295 and GSE273298.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential competing interests.
Funding Statement
This work was funded by the European Regional Development Fund (ERDF) Integrated Territorial Initiative (ITI0017_2019) and Foundation Progreso y Salud PEER (2020-019). This study has been supported by the Spanish Ministry of Economy and Competitiveness with European Regional Development Fund PID2022-140047OB-C21. The EURAS project receives funding from the European Union’s Horizon Europe Research and Innovation Programme under Grant Agreement No. 101080580. (HORIZON-HLTH-2022-DISEASE-06). Funded by the European Union. This study has been also supported by Instituto de Salud Carlos III through the projects PI21/01282 and PI21/00094 (co-funded by European Regional Development Fund “A way to make Europe”), Sociedad Española de Cardiología (SEC/FEC-INV-BAS 20/003) and by Marató TV3 (736/C/2020) Health Research Institute La Fe (Memorial Nacho Barberá).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Arbelo E., Protonotarios A., Gimeno J.R., Arbustini E., Barriales-Villa R., Basso C., Bezzina C.R., Biagini E., Blom N.A., de Boer R.A., et al. 2023 ESC Guidelines for the management of cardiomyopathies. Eur. Heart J. 2023;44:3503–3626. doi: 10.1093/eurheartj/ehad194. [DOI] [PubMed] [Google Scholar]
- 2.Finocchiaro G., Papadakis M., Robertus J.L., Dhutia H., Steriotis A.K., Tome M., Mellor G., Merghani A., Malhotra A., Behr E., et al. Etiology of Sudden Death in Sports: Insights From a United Kingdom Regional Registry. J. Am. Coll. Cardiol. 2016;67:2108–2115. doi: 10.1016/j.jacc.2016.02.062. [DOI] [PubMed] [Google Scholar]
- 3.Corrado D., Anastasakis A., Basso C., Bauce B., Blomstrom-Lundqvist C., Bucciarelli-Ducci C., Cipriani A., De Asmundis C., Gandjbakhch E., Jimenez-Jaimez J., et al. Proposed diagnostic criteria for arrhythmogenic cardiomyopathy: European Task Force consensus report. Int. J. Cardiol. 2024;395:131447. doi: 10.1016/j.ijcard.2023.131447. [DOI] [PubMed] [Google Scholar]
- 4.Bosman L.P., Te Riele A. Arrhythmogenic right ventricular cardiomyopathy: A focused update on diagnosis and risk stratification. Heart. 2022;108:90–97. doi: 10.1136/heartjnl-2021-319113. [DOI] [PubMed] [Google Scholar]
- 5.Wilde A.A.M., Semsarian C., Marquez M.F., Sepehri Shamloo A., Ackerman M.J., Ashley E.A., Sternick E.B., Barajas-Martinez H., Behr E.R., Bezzina C.R., et al. European Heart Rhythm Association (EHRA)/Heart Rhythm Society (HRS)/Asia Pacific Heart Rhythm Society (APHRS)/Latin American Heart Rhythm Society (LAHRS) Expert Consensus Statement on the State of Genetic Testing for Cardiac Diseases. Heart Rhythm. 2022;19:e1–e60. doi: 10.1016/j.hrthm.2022.03.1225. [DOI] [PubMed] [Google Scholar]
- 6.Pinamonti B., Brun F., Mestroni L., Sinagra G. Arrhythmogenic right ventricular cardiomyopathy: From genetics to diagnostic and therapeutic challenges. World J. Cardiol. 2014;6:1234–1244. doi: 10.4330/wjc.v6.i12.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoorntje E.T., Te Rijdt W.P., James C.A., Pilichou K., Basso C., Judge D.P., Bezzina C.R., van Tintelen J.P. Arrhythmogenic cardiomyopathy: Pathology, genetics, and concepts in pathogenesis. Cardiovasc. Res. 2017;113:1521–1531. doi: 10.1093/cvr/cvx150. [DOI] [PubMed] [Google Scholar]
- 8.Boogerd C.J., Lacraz G.P.A., Vertesy A., van Kampen S.J., Perini I., de Ruiter H., Versteeg D., Brodehl A., van der Kraak P., Giacca M., et al. Spatial transcriptomics unveils ZBTB11 as a regulator of cardiomyocyte degeneration in arrhythmogenic cardiomyopathy. Cardiovasc. Res. 2023;119:477–491. doi: 10.1093/cvr/cvac072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sommariva E., Brambilla S., Carbucicchio C., Gambini E., Meraviglia V., Dello Russo A., Farina F.M., Casella M., Catto V., Pontone G., et al. Cardiac mesenchymal stromal cells are a source of adipocytes in arrhythmogenic cardiomyopathy. Eur. Heart J. 2016;37:1835–1846. doi: 10.1093/eurheartj/ehv579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Gras E., Lombardi R., Giocondo M.J., Willerson J.T., Schneider M.D., Khoury D.S., Marian A.J. Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J. Clin. Investig. 2006;116:2012–2021. doi: 10.1172/JCI27751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piquer-Gil M., Domenech-Dauder S., Sepulveda-Gomez M., Machi-Camacho C., Braza-Boils A., Zorio E. Non Coding RNAs as Regulators of Wnt/beta-Catenin and Hippo Pathways in Arrhythmogenic Cardiomyopathy. Biomedicines. 2022;10:2619. doi: 10.3390/biomedicines10102619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S.N., Gurha P., Lombardi R., Ruggiero A., Willerson J.T., Marian A.J. The hippo pathway is activated and is a causal mechanism for adipogenesis in arrhythmogenic cardiomyopathy. Circ. Res. 2014;114:454–468. doi: 10.1161/CIRCRESAHA.114.302810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rouhi L., Fan S., Cheedipudi S.M., Braza-Boils A., Molina M.S., Yao Y., Robertson M.J., Coarfa C., Gimeno J.R., Molina P., et al. The EP300/TP53 pathway, a suppressor of the Hippo and canonical WNT pathways, is activated in human hearts with arrhythmogenic cardiomyopathy in the absence of overt heart failure. Cardiovasc. Res. 2022;118:1466–1478. doi: 10.1093/cvr/cvab197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalayinia S., Arjmand F., Maleki M., Malakootian M., Singh C.P. MicroRNAs: Roles in cardiovascular development and disease. Cardiovasc. Pathol. 2021;50:107296. doi: 10.1016/j.carpath.2020.107296. [DOI] [PubMed] [Google Scholar]
- 15.Alcalde M., Toro R., Bonet F., Cordoba-Caballero J., Martinez-Barrios E., Ranea J.A., Vallverdu-Prats M., Brugada R., Meraviglia V., Bellin M., et al. Role of microRNAs in arrhythmogenic cardiomyopathy: Translation as biomarkers into clinical practice. Transl. Res. J. Lab. Clin. Med. 2023;259:72–82. doi: 10.1016/j.trsl.2023.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Gruning B.A., et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rainer J., Meraviglia V., Blankenburg H., Piubelli C., Pramstaller P.P., Paolin A., Cogliati E., Pompilio G., Sommariva E., Domingues F.S., et al. The arrhythmogenic cardiomyopathy-specific coding and non-coding transcriptome in human cardiac stromal cells. BMC Genom. 2018;19:491. doi: 10.1186/s12864-018-4876-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall C.L., Gurha P., Sabater-Molina M., Asimaki A., Futema M., Lovering R.C., Suarez M.P., Aguilera B., Molina P., Zorio E., et al. RNA sequencing-based transcriptome profiling of cardiac tissue implicates novel putative disease mechanisms in FLNC-associated arrhythmogenic cardiomyopathy. Int. J. Cardiol. 2020;302:124–130. doi: 10.1016/j.ijcard.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin L., Liu S., Chen Z., Xie J., Fu M., Lu D., Wu Y., Shen H., Yang P., Qian J. Anatomically resolved transcriptome and proteomelandscapes reveal disease-relevant molecular signaturesand systematic changes in heart function of end-stagedilated cardiomyopathy. VIEW. 2023;4:1–18. doi: 10.1002/VIW.20220040. [DOI] [Google Scholar]
- 22.van Opbergen C.J.M., den Braven L., Delmar M., van Veen T.A.B. Mitochondrial Dysfunction as Substrate for Arrhythmogenic Cardiomyopathy: A Search for New Disease Mechanisms. Front. Physiol. 2019;10:1496. doi: 10.3389/fphys.2019.01496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lippi M., Maione A.S., Chiesa M., Perrucci G.L., Iengo L., Sattin T., Cencioni C., Savoia M., Zeiher A.M., Tundo F., et al. Omics Analyses of Stromal Cells from ACM Patients Reveal Alterations in Chromatin Organization and Mitochondrial Homeostasis. Int. J. Mol. Sci. 2023;24:17. doi: 10.3390/ijms241210017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sommariva E., Stadiotti I., Casella M., Catto V., Dello Russo A., Carbucicchio C., Arnaboldi L., De Metrio S., Milano G., Scopece A., et al. Oxidized LDL-dependent pathway as new pathogenic trigger in arrhythmogenic cardiomyopathy. EMBO Mol. Med. 2021;13:e14365. doi: 10.15252/emmm.202114365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Hernandez M., van Opbergen C.J.M., Bagwan N., Vissing C.R., Marron-Linares G.M., Zhang M., Torres Vega E., Sorrentino A., Drici L., Sulek K., et al. Loss of Nuclear Envelope Integrity and Increased Oxidant Production Cause DNA Damage in Adult Hearts Deficient in PKP2: A Molecular Substrate of ARVC. Circulation. 2022;146:851–867. doi: 10.1161/CIRCULATIONAHA.122.060454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pitsch M., Kant S., Mytzka C., Leube R.E., Krusche C.A. Autophagy and Endoplasmic Reticulum Stress during Onset and Progression of Arrhythmogenic Cardiomyopathy. Cells. 2021;11:96. doi: 10.3390/cells11010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aliyari Ghasabeh M., Te Riele A., James C.A., Chen H.S.V., Tichnell C., Murray B., Eng J., Kral B.G., Tandri H., Calkins H., et al. Epicardial Fat Distribution Assessed with Cardiac CT in Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy. Radiology. 2018;289:641–648. doi: 10.1148/radiol.2018180224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C., Peng Y., Peng Y., Peng J., Jiang S. miR-135a-5p inhibits 3T3-L1 adipogenesis through activation of canonical Wnt/beta-catenin signaling. J. Mol. Endocrinol. 2014;52:311–320. doi: 10.1530/JME-14-0013. [DOI] [PubMed] [Google Scholar]
- 29.Gao W., Guo N., Zhao S., Chen Z., Zhang W., Yan F., Liao H., Chi K. FBXW7 promotes pathological cardiac hypertrophy by targeting EZH2-SIX1 signaling. Exp. Cell Res. 2020;393:112059. doi: 10.1016/j.yexcr.2020.112059. [DOI] [PubMed] [Google Scholar]
- 30.Xu Y., Du J., Zhang P., Zhao X., Li Q., Jiang A., Jiang D., Tang G., Jiang Y., Wang J., et al. MicroRNA-125a-5p Mediates 3T3-L1 Preadipocyte Proliferation and Differentiation. Molecules. 2018;23:317. doi: 10.3390/molecules23020317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaccari T., Moroni A., Rocchi M., Gorza L., Bianchi M.E., Beltrame M., DiFrancesco D. The human gene coding for HCN2, a pacemaker channel of the heart. Biochim. Biophys. Acta. 1999;1446:419–425. doi: 10.1016/S0167-4781(99)00092-5. [DOI] [PubMed] [Google Scholar]
- 32.van den Berg N.W.E., Kawasaki M., Fabrizi B., Nariswari F.A., Verduijn A.C., Neefs J., Wesselink R., Al-Shama R.F.M., van der Wal A.C., de Boer O.J., et al. Epicardial and endothelial cell activation concurs with extracellular matrix remodeling in atrial fibrillation. Clin. Transl. Med. 2021;11:e558. doi: 10.1002/ctm2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Da Silva F., Jian Motamedi F., Weerasinghe Arachchige L.C., Tison A., Bradford S.T., Lefebvre J., Dolle P., Ghyselinck N.B., Wagner K.D., Schedl A. Retinoic acid signaling is directly activated in cardiomyocytes and protects mouse hearts from apoptosis after myocardial infarction. eLife. 2021;10:e68280. doi: 10.7554/eLife.68280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao S., Yang D., Huang W., Wang T., Li W. miR-135a-5p affects adipogenic differentiation of human adipose-derived mesenchymal stem cellsby promoting the Hippo signaling pathway. Int. J. Clin. Exp. Pathol. 2018;11:1347–1355. [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y., Tang B., Jiang W., Sun M., Zhang H., Tao Y., Wang H., Xiang D., Bai H., Guo M., et al. miR-486-5p Attenuates Steroid-Induced Adipogenesis and Osteonecrosis of the Femoral Head Via TBX2/P21 Axis. Stem Cells. 2023;41:711–723. doi: 10.1093/stmcls/sxad038. [DOI] [PubMed] [Google Scholar]
- 36.Smart N., Bollini S., Dube K.N., Vieira J.M., Zhou B., Riegler J., Price A.N., Lythgoe M.F., Davidson S., Yellon D., et al. Myocardial regeneration: Expanding the repertoire of thymosin beta4 in the ischemic heart. Ann. N. Y. Acad. Sci. 2012;1269:92–101. doi: 10.1111/j.1749-6632.2012.06708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei K., Serpooshan V., Hurtado C., Diez-Cunado M., Zhao M., Maruyama S., Zhu W., Fajardo G., Noseda M., Nakamura K., et al. Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature. 2015;525:479–485. doi: 10.1038/nature15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou B., von Gise A., Ma Q., Hu Y.W., Pu W.T. Genetic fate mapping demonstrates contribution of epicardium-derived cells to the annulus fibrosis of the mammalian heart. Dev. Biol. 2010;338:251–261. doi: 10.1016/j.ydbio.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kikuchi K., Gupta V., Wang J., Holdway J.E., Wills A.A., Fang Y., Poss K.D. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011;138:2895–2902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips M.D., Mukhopadhyay M., Poscablo C., Westphal H. Dkk1 and Dkk2 regulate epicardial specification during mouse heart development. Int. J. Cardiol. 2011;150:186–192. doi: 10.1016/j.ijcard.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Gise A., Zhou B., Honor L.B., Ma Q., Petryk A., Pu W.T. WT1 regulates epicardial epithelial to mesenchymal transition through beta-catenin and retinoic acid signaling pathways. Dev. Biol. 2011;356:421–431. doi: 10.1016/j.ydbio.2011.05.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corrado D., Basso C., Judge D.P. Arrhythmogenic Cardiomyopathy. Circ. Res. 2017;121:784–802. doi: 10.1161/CIRCRESAHA.117.309345. [DOI] [PubMed] [Google Scholar]
- 43.Cortes C., Francou A., De Bono C., Kelly R.G. Epithelial Properties of the Second Heart Field. Circ. Res. 2018;122:142–154. doi: 10.1161/CIRCRESAHA.117.310838. [DOI] [PubMed] [Google Scholar]
- 44.Yuan P., Cheedipudi S.M., Rouhi L., Fan S., Simon L., Zhao Z., Hong K., Gurha P., Marian A.J. Single-Cell RNA Sequencing Uncovers Paracrine Functions of the Epicardial-Derived Cells in Arrhythmogenic Cardiomyopathy. Circulation. 2021;143:2169–2187. doi: 10.1161/CIRCULATIONAHA.120.052928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J.T., Rayburn H., Hynes R.O. Embryonic mesodermal defects in alpha 5 integrin-deficient mice. Development. 1993;119:1093–1105. doi: 10.1242/dev.119.4.1093. [DOI] [PubMed] [Google Scholar]
- 46.Mittal A., Pulina M., Hou S.Y., Astrof S. Fibronectin and integrin alpha 5 play essential roles in the development of the cardiac neural crest. Mech. Dev. 2010;127:472–484. doi: 10.1016/j.mod.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mittal A., Pulina M., Hou S.Y., Astrof S. Fibronectin and integrin alpha 5 play requisite roles in cardiac morphogenesis. Dev. Biol. 2013;381:73–82. doi: 10.1016/j.ydbio.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ong L.P., Bargehr J., Knight-Schrijver V.R., Lee J., Colzani M., Bayraktar S., Bernard W.G., Marchiano S., Bertero A., Murry C.E., et al. Epicardially secreted fibronectin drives cardiomyocyte maturation in 3D-engineered heart tissues. Stem Cell Rep. 2023;18:936–951. doi: 10.1016/j.stemcr.2023.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y., Li Y., Liang J., Sun Z., Wu Q., Liu Y., Sun C. The Mechanism of Leptin on Inhibiting Fibrosis and Promoting Browning of White Fat by Reducing ITGA5 in Mice. Int. J. Mol. Sci. 2021;22:12353. doi: 10.3390/ijms222212353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim Y.J., Hwang S.H., Lee S.Y., Shin K.K., Cho H.H., Bae Y.C., Jung J.S. miR-486-5p induces replicative senescence of human adipose tissue-derived mesenchymal stem cells and its expression is controlled by high glucose. Stem Cells Dev. 2012;21:1749–1760. doi: 10.1089/scd.2011.0429. [DOI] [PubMed] [Google Scholar]
- 51.Morandi E.M., Verstappen R., Zwierzina M.E., Geley S., Pierer G., Ploner C. ITGAV and ITGA5 diversely regulate proliferation and adipogenic differentiation of human adipose derived stem cells. Sci. Rep. 2016;6:28889. doi: 10.1038/srep28889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen T., Zhu J., Cai T., Du W., Zhang Y., Zhu Q., Liu Z., Huang J.A. Suppression of non-small cell lung cancer migration and invasion by hsa-miR-486-5p via the TGF-beta/SMAD2 signaling pathway. J. Cancer. 2019;10:6014–6024. doi: 10.7150/jca.35017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flum M., Dicks S., Teng Y.H., Schrempp M., Nystrom A., Boerries M., Hecht A. Canonical TGFbeta signaling induces collective invasion in colorectal carcinogenesis through a Snail1- and Zeb1-independent partial EMT. Oncogene. 2022;41:1492–1506. doi: 10.1038/s41388-022-02190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakao A., Afrakhte M., Moren A., Nakayama T., Christian J.L., Heuchel R., Itoh S., Kawabata M., Heldin N.E., Heldin C.H., et al. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 55.Marques F.Z., Vizi D., Khammy O., Mariani J.A., Kaye D.M. The transcardiac gradient of cardio-microRNAs in the failing heart. Eur. J. Heart Fail. 2016;18:1000–1008. doi: 10.1002/ejhf.517. [DOI] [PubMed] [Google Scholar]
- 56.Chouvarine P., Legchenko E., Geldner J., Riehle C., Hansmann G. Hypoxia drives cardiac miRNAs and inflammation in the right and left ventricle. J. Mol. Med. 2019;97:1427–1438. doi: 10.1007/s00109-019-01817-6. [DOI] [PubMed] [Google Scholar]
- 57.Heinke J., Wehofsits L., Zhou Q., Zoeller C., Baar K.M., Helbing T., Laib A., Augustin H., Bode C., Patterson C., et al. BMPER is an endothelial cell regulator and controls bone morphogenetic protein-4-dependent angiogenesis. Circ. Res. 2008;103:804–812. doi: 10.1161/CIRCRESAHA.108.178434. [DOI] [PubMed] [Google Scholar]
- 58.Serpe M., Umulis D., Ralston A., Chen J., Olson D.J., Avanesov A., Othmer H., O’Connor M.B., Blair S.S. The BMP-binding protein Crossveinless 2 is a short-range, concentration-dependent, biphasic modulator of BMP signaling in Drosophila. Dev. Cell. 2008;14:940–953. doi: 10.1016/j.devcel.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kelley R., Ren R., Pi X., Wu Y., Moreno I., Willis M., Moser M., Ross M., Podkowa M., Attisano L., et al. A concentration-dependent endocytic trap and sink mechanism converts Bmper from an activator to an inhibitor of Bmp signaling. J. Cell Biol. 2009;184:597–609. doi: 10.1083/jcb.200808064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dyer L., Wu Y., Moser M., Patterson C. BMPER-induced BMP signaling promotes coronary artery remodeling. Dev. Biol. 2014;386:385–394. doi: 10.1016/j.ydbio.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Helbing T., Rothweiler R., Heinke J., Goetz L., Diehl P., Zirlik A., Patterson C., Bode C., Moser M. BMPER is upregulated by statins and modulates endothelial inflammation by intercellular adhesion molecule-1. Arterioscler. Thromb. Vasc. Biol. 2010;30:554–560. doi: 10.1161/ATVBAHA.109.201087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pi X., Lockyer P., Dyer L.A., Schisler J.C., Russell B., Carey S., Sweet D.T., Chen Z., Tzima E., Willis M.S., et al. Bmper inhibits endothelial expression of inflammatory adhesion molecules and protects against atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2012;32:2214–2222. doi: 10.1161/ATVBAHA.112.252015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moreno-Miralles I., Ren R., Moser M., Hartnett M.E., Patterson C. Bone morphogenetic protein endothelial cell precursor-derived regulator regulates retinal angiogenesis in vivo in a mouse model of oxygen-induced retinopathy. Arterioscler. Thromb. Vasc. Biol. 2011;31:2216–2222. doi: 10.1161/ATVBAHA.111.230235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dyer L., Lockyer P., Wu Y., Saha A., Cyr C., Moser M., Pi X., Patterson C. BMPER Promotes Epithelial-Mesenchymal Transition in the Developing Cardiac Cushions. PLoS ONE. 2015;10:e0139209. doi: 10.1371/journal.pone.0139209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patel V.B., Shah S., Verma S., Oudit G.Y. Epicardial adipose tissue as a metabolic transducer: Role in heart failure and coronary artery disease. Heart Fail. Rev. 2017;22:889–902. doi: 10.1007/s10741-017-9644-1. [DOI] [PubMed] [Google Scholar]
- 66.Perez L.M., de Lucas B., Galvez B.G. BMPER is upregulated in obesity and seems to have a role in pericardial adipose stem cells. J. Cell. Physiol. 2021;236:132–145. doi: 10.1002/jcp.29829. [DOI] [PubMed] [Google Scholar]
- 67.Yao H., Hill S.F., Skidmore J.M., Sperry E.D., Swiderski D.L., Sanchez G.J., Bartels C.F., Raphael Y., Scacheri P.C., Iwase S., et al. CHD7 represses the retinoic acid synthesis enzyme ALDH1A3 during inner ear development. JCI Insight. 2018;3:e97440. doi: 10.1172/jci.insight.97440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Okumura K., Nakamachi K., Hosoe Y., Nakajima N. Identification of a novel retinoic acid-responsive element within the lamin A/C promoter. Biochem. Biophys. Res. Commun. 2000;269:197–202. doi: 10.1006/bbrc.2000.2242. [DOI] [PubMed] [Google Scholar]
- 69.Colbert M.C., Hall D.G., Kimball T.R., Witt S.A., Lorenz J.N., Kirby M.L., Hewett T.E., Klevitsky R., Robbins J. Cardiac compartment-specific overexpression of a modified retinoic acid receptor produces dilated cardiomyopathy and congestive heart failure in transgenic mice. J. Clin. Investig. 1997;100:1958–1968. doi: 10.1172/JCI119727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bost F., Caron L., Marchetti I., Dani C., Le Marchand-Brustel Y., Binetruy B. Retinoic acid activation of the ERK pathway is required for embryonic stem cell commitment into the adipocyte lineage. Biochem. J. 2002;361:621–627. doi: 10.1042/bj3610621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim D.H., Lee J., Suh Y., Ko J.K., Lee K. Transdifferentiation of Myoblasts Into Adipocytes by All-Trans-Retinoic Acid in Avian. Front. Cell Dev. Biol. 2022;10:856881. doi: 10.3389/fcell.2022.856881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rivolta I., Binda A., Masi A., DiFrancesco J.C. Cardiac and neuronal HCN channelopathies. Pflügers Arch. -Eur. J. Physiol. 2020;472:931–951. doi: 10.1007/s00424-020-02384-3. [DOI] [PubMed] [Google Scholar]
- 73.Oshita K., Itoh M., Hirashima S., Kuwabara Y., Ishihara K., Kuwahara K., Nakao K., Kimura T., Nakamura K., Ushijima K., et al. Ectopic automaticity induced in ventricular myocytes by transgenic overexpression of HCN2. J. Mol. Cell Cardiol. 2015;80:81–89. doi: 10.1016/j.yjmcc.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 74.Oshita K., Kozasa Y., Nakagawa Y., Kuwabara Y., Kuwahara K., Nakagawa T., Nakashima N., Hiraki T., Takano M. Overexpression of the HCN2 channel increases the arrhythmogenicity induced by hypokalemia. J. Physiol. Sci. 2019;69:653–660. doi: 10.1007/s12576-019-00684-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hathaway C.K., Grant R., Hagaman J.R., Hiller S., Li F., Xu L., Chang A.S., Madden V.J., Bagnell C.R., Rojas M., et al. Endothelin-1 critically influences cardiac function via superoxide-MMP9 cascade. Proc. Natl. Acad. Sci. USA. 2015;112:5141–5146. doi: 10.1073/pnas.1504557112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spaich S., Will R.D., Just S., Spaich S., Kuhn C., Frank D., Berger I.M., Wiemann S., Korn B., Koegl M., et al. F-box and leucine-rich repeat protein 22 is a cardiac-enriched F-box protein that regulates sarcomeric protein turnover and is essential for maintenance of contractile function in vivo. Circ. Res. 2012;111:1504–1516. doi: 10.1161/CIRCRESAHA.112.271007. [DOI] [PubMed] [Google Scholar]
- 77.Chen D., Hou Y., Cai X. MiR-210-3p Enhances Cardiomyocyte Apoptosis and Mitochondrial Dysfunction by Targeting the NDUFA4 Gene in Sepsis-Induced Myocardial Dysfunction. Int. Heart J. 2021;62:636–646. doi: 10.1536/ihj.20-512. [DOI] [PubMed] [Google Scholar]
- 78.Bayraktar E., Bayraktar R., Oztatlici H., Lopez-Berestein G., Amero P., Rodriguez-Aguayo C. Targeting miRNAs and Other Non-Coding RNAs as a Therapeutic Approach: An Update. Noncoding RNA. 2023;9:27. doi: 10.3390/ncrna9020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data generated in this manuscript have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession codes GSE273295 and GSE273298.