Abstract
OBJECTIVE
To evaluate rabies virus (RABV) characterization data obtained from animal specimens submitted to the US public health rabies surveillance system and propose a standardized approach to sample selection for RABV characterization that could enhance early detection of important rabies epizootic events in the United States.
SAMPLE
United States public health rabies surveillance system data collected from January 1, 2010, through December 31, 2015.
PROCEDURES
Data were reviewed to identify RABV-positive specimens for which virus characterization would likely provide information regarding any of 4 overarching events (discovery of novel variants, translocation of RABV variants, host-shift events, and any unusual rabies-related event) that could substantially alter animal rabies epizootiology in the United States. These specimens were designated as specimens of epizootiological importance (SEIs). Estimates of the additional number of specimens that public health laboratories could expect to process each year if all SEIs underwent RABV characterization were calculated.
RESULTS
During the 6-year period, the mean annual number of SEIs was 855 (95% CI, 739 to 971); the mean number of SEIs that underwent virus characterization was 270 (95% CI, 187 to 353). Virus characterization of all SEIs would be expected to increase the public health laboratories’ test load by approximately 585 (95% CI, 543 to 625) specimens/y.
CONCLUSIONS AND CLINICAL RELEVANCE
Prioritization of RABV characterization of SEIs may improve early detection of rabies events associated with RABV host shifts, variant translocations, and importation. Characterization of SEIs may help refine wildlife rabies management practices. Each public health laboratory should evaluate testing of SEIs to ensure diagnostic laboratory capacity is not overstretched.
Rabies virus is a neurotropic virus of the genus Lyssavirus that causes fatal encephalitis in nearly 100% of infected mammals. There are 8 terrestrial RABV variants that circulate in the United States1 in 5 terrestrial wildlife reservoir species, namely striped skunks (Mephitis mephitis), raccoons (Procyon lotor), mongooses (Herpestes javanicus), arctic foxes (Vulpes lagopus), and gray foxes (Urocyon cinereoargenteus). Although rabid domestic animals are identified every year, > 90% of all rabid animals in the United States identified since 1980 have been wildlife species.2
Rabies in an animal is a notifiable event in the United States.3 Each year, state and territorial health departments and the National Rabies Management Program of the USDA APHIS Wildlife Services submit data to the US animal RSS, which is maintained by the CDC’s Poxvirus and Rabies Branch. The public health animal RSS in the United States is used to monitor rabies-related events in domestic and wildlife species. Virus characterization by molecular sequencing can determine the variant of the virus in a rabies-infected animal and is a valuable tool for identifying and understanding important events in the epizootiology of rabies in the United States. However, virus characterization is not routinely performed on all RABV-positive specimens. Although the public health animal RSS is robust, there is no clear protocol for identification of specimens that should undergo virus characterization. It would be ideal to characterize all RABV-positive specimens; however, this is neither financially nor logistically feasible. A strategy for selection of RABV-positive specimens for virus characterization that is based on existing animal RSS data would be useful.
Changes in the epizootiology of RABV have the potential to increase the risk of exposure of humans and domestic animals, impact animal control measures, and influence public health policy. Four types of events that have or could substantially alter animal rabies epizootiology in the United States include changes that occur in relation to host-shift events, translocation of RABV variants not previously documented within geographic areas, introduction of novel RABV variants (including importation events) over a given period, and any unusual rabies-related incidents.
Several examples of recent host-shift events highlight their wide-reaching impact. Of the 8 terrestrial wildlife RABV variants in the United States, 5 (California skunk, north-central skunk, mongoose, Texas gray fox, and Arizona gray fox) were the result of a host-shift event from dogs. For example, in the late 1980s, the gray fox (U cinereoargenteus) was recognized as an RABV reservoir species, with a novel RABV variant (Texas gray fox virus variant) consistent with a canine lineage.4,5 The raccoon RABV variant and south-central skunk RABV variant were the result of host-shift events from bats.6 Host-shift events from bats to carnivores can also have important public health implications, especially in areas where terrestrial animal rabies has been eliminated.6,7 In 2001, cross-species transmission of a bat RABV variant into the striped skunk population in Flagstaff, Ariz, occurred, causing a rabies outbreak in striped skunks.7 Bouricki et al8 determined that a 2009 rabies outbreak among gray foxes in California was likely caused by a host shift of the California skunk RABV variant into gray foxes.8 The frequency with which RABV has shifted between hosts6,8,9 emphasizes the importance of ongoing surveillance of RABV distribution among wildlife species and application of virus characterization to identify and understand changes in the epizootiology of rabies in the United States.
Translocation of rabid animals has led to large-scale epizootiological changes with regard to rabies. In the 1970s, a presumed human-mediated translocation of raccoons from their enzootic zone (Florida) to a rabies-free zone in the Mid-Atlantic region was responsible for arguably one of the most important public health events in the United States.10 The newly established raccoon RABV variant spread throughout the east coast of the United States, from Georgia to the border with Canada.10 The raccoon RABV variant now accounts for > 70% of all animal rabies cases in the United States and was the cause of 2 human deaths directly and 2 additional human deaths through transplantation of infected organs.2,11
Since 1995, the USDA has been working with local, state, and federal government partners to conduct ORV of wildlife in targeted areas to prevent the spread of specific terrestrial RABV variants. Enhanced rabies surveillance in these areas is critical, and the data are used to adapt the USDA’s rabies management practices and to monitor for RABV translocation events.12,13
The canine RABV variant was eliminated from the United States by the 1970s.5 However, in 1988, a novel domestic dog–coyote RABV variant was introduced from Mexico and became enzootic in coyotes (Canis latrans) along the border between the United States and Mexico.14 In addition to numerous domestic animal and wildlife cases of rabies, the outbreak resulted in 2 human deaths.15,16 In 1994, the dog-coyote RABV variant was detected in Alabama and Florida (likely a result of interstate transport of infected coyotes for hunting purposes) and caused local outbreaks in domestic dogs.4
The United States regained its canine RABV variant–free status in 2007,5 but multiple importations of infected dogs since that time highlight the importance of early identification of such dogs to prevent future outbreaks. Since 2007, the importation of dogs infected with the dog RABV variant has resulted in administration of postexposure prophylaxis to > 50 people, and > 90 animals (unpublished data) have had to be given booster vaccinations and undergo quarantine. Total public health response costs for each importation event have been > $250,000.17–19
The purpose of the evaluation reported here was to identify RABV-positive specimens submitted to public health laboratories that should be considered for routine virus characterization because of their potential to optimize detection of or provide information regarding the 4 key events of epizootiological importance (ie, SEIs). Characterization of specimens not classified as SEIs was also undertaken. In addition, the increased test load and associated costs that laboratories could expect if all those RABV-positive SEIs underwent virus characterization were estimated. We considered that identification and prioritized testing of SEIs would enhance rabies stakeholders’ abilities to better target rabies management efforts.
Materials and Methods
Source of data
Beginning in 2013, results of RABV characterization of rabies-positive animal specimens, when available, were published in the CDC’s annual RSS summary,2 but the CDC has maintained RABV characterization data since 2008. State health departments do not typically report which testing method was used by each laboratory that submits data; therefore, the term RABV characterization in the present report refers to either antigenic or genetic typing methods. Rabies surveillance data for terrestrial nonvolant mammals (excluding humans) in the United States that were submitted to the CDC during the period of January 2010 through December 2015 were included in this evaluation. Bat data were not included but underwent separate analysis.20
Selection criteria for SEIs
Data were reviewed to determine which RABV-positive specimens were SEIs, namely specimens for which virus characterization would likely identify or provide relevant information regarding any 1 of 4 rabies-related epizootiological events. The 4 overarching events that have substantially altered or could substantially alter animal rabies epizootiology in the United States were introduction of novel RABV variants (including importation events), translocation of RABV variants not previously documented within geographic areas in the United States, changes that occur in relation to host-shift events, and any unusual rabies-related incidents.
Subcategorization of SEIs
The four overarching events were defined, and SEIs were then further refined into 12 subcategories. The SEIs were grouped into the 12 subcategories on the basis of each RABV-infected animal’s species or type, geographic location, travel history, or herd history or the epizootiological events the SEIs would identify or for which they would provide important information. With regard to introduction of novel variants (including importation events), the subcategories were species (specifically dog), domestic animal or livestock in a southern border state (ie, California, Arizona, New Mexico, Texas, or Florida), and mammals with a history of international travel in the preceding 12 months. With regard to translocation of RABV variants not previously documented within geographic areas, the subcategories were species (specifically, coyote, cougar, bobcat, wolf, deer, bison, and raccoon [ie, raccoons located west of the USDA’s ORV zone front {Figure 1} including enhanced rabies surveillance zones]); domestic and livestock mammals with a history of travel across RABV variant territory boundaries in the preceding 6 months; and terrestrial mammals located in a USDA enhanced rabies surveillance zone. With regard to host-shift events, the subcategories were species (specifically fox) and mammals in nonterrestrial mammal (ie, bat) reservoir areas. With regard to unusual rabies-related incidents, the subcategories were species (specifically rodents), cluster event (≥ 2 cases) in a livestock herd in a 60-day period, and other (eg, rabid cat in a location that had not had an RABV-positive cat in > 2 years). There was no prioritized ranking among the SEIs, and they were all deemed equally epizootiologically important.
Figure 1—
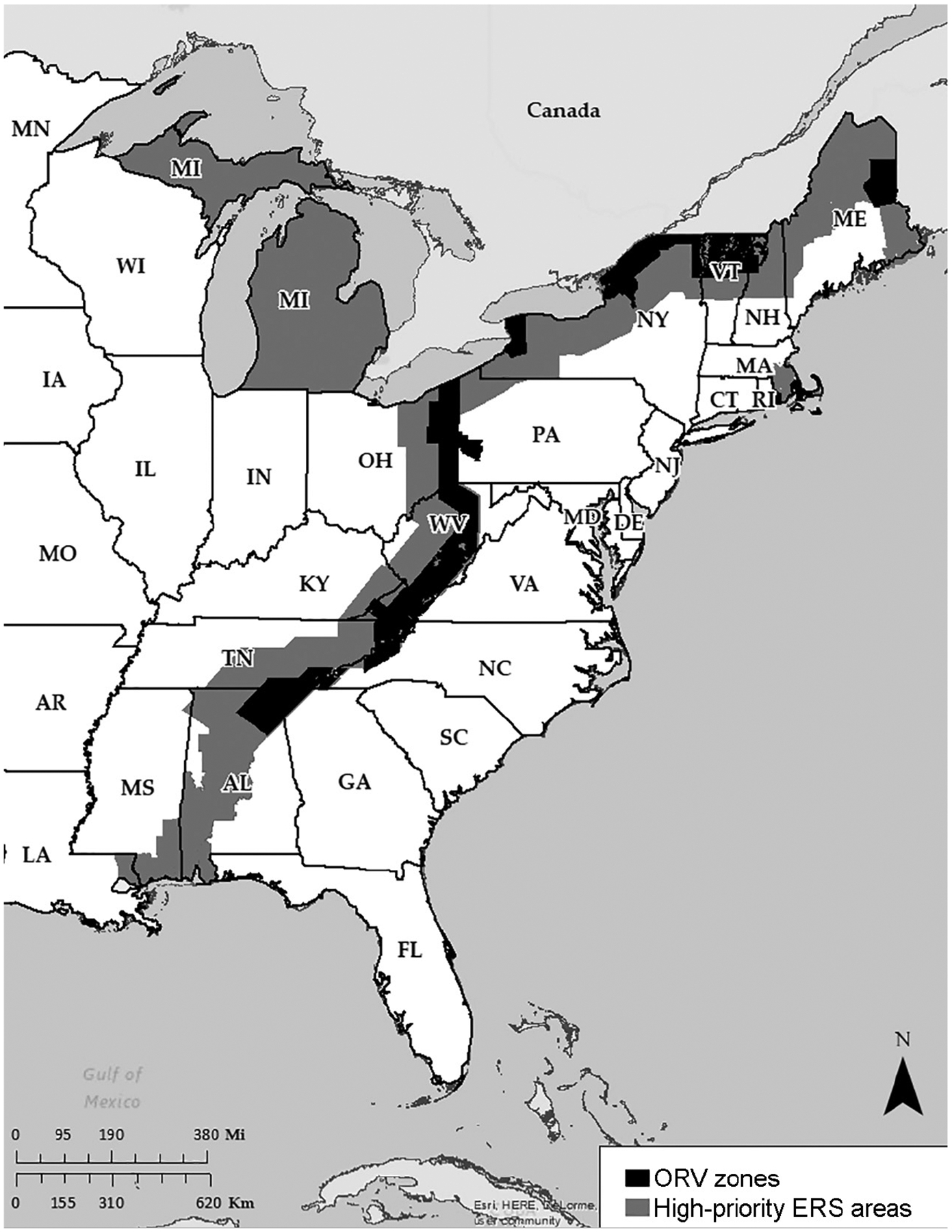
Geographic distribution of USDA enhanced rabies surveillance (ERS) areas (including ORV areas) in 2017.
Data and statistical analyses
Data regarding SEIs from the US public health RSS were analyzed with commercially available softwarea and statisticalb,c programs. Data included information reported to the US public health RSS and did not include individual state data published on the web. Descriptive statistics, frequencies, and proportions are reported. The total number of RABV-positive specimens obtained for each SEI subcategory during the 6-year period of interest was used to calculate the estimated number (with corresponding CIs) of specimens that public health laboratories could expect to process if the disease rate remained relatively constant and all SEIs underwent RABV characterization. Some SEIs could be placed in multiple subcategories; however, duplicate samples from multiple SEI subcategories were removed to calculate the total sample increase expected by public health laboratories. Sample size and proportional frequencies with 95% CIs were calculated to assess the number of samples that would need to undergo virus characterization to detect a difference in RABV variants. A cost analysis was performed to estimate the financial burden on public health laboratories that perform rabies testing if all submitted SEIs underwent RABV characterization.
Results
RABV-positive terrestrial mammals and identification of SEIs
During 2010 through 2015, 453,674 terrestrial mammals were tested for RABV, of which 26,230 (5.8%) were RABV positive. Of those RABV-positive specimens, 7,878 (30.0%) underwent RABV characterization. Annually, the mean number of terrestrial mammal specimens submitted for rabies testing was 75,612 (95% CI, 71,768 to 79,456); 4,371 (95% CI, 4,022 to 4,720) specimens were RABV positive, of which 1,315 (95% CI, 1,089 to 1,542) underwent RABV characterization (Table 1). Volant terrestrial mammal (ie, bat) surveillance data are described elsewhere.20
Table 1—
Number of terrestrial mammal specimens submitted to public health laboratories for RABV testing, number of RABV-positive specimens, and number of specimens that underwent RABV characterization in the United States by year during the period of January 2010 through December 2015.
| Year | |||||||
|---|---|---|---|---|---|---|---|
| Variable | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | Mean (95% CI) |
| No. of specimens tested | 81,278 | 77,644 | 76,379 | 71,041 | 74,615 | 72,717 | 75,612 (71,768–79,456) |
| No. of RABV-positive specimens (%) | 4,723 (5.8) | 4,650 (6.0) | 4,488 (5.9) | 4,288 (6.0) | 4,277 (5.7) | 3,804 (5.2) | 4,371 (4,022–4,720) |
| No. of RABV-positive specimens that underwent virus characterization (%) | 1,341 (28.4) | 1,728 (37.2) | 1,257 (28.0) | 1,227 (28.6) | 1,209 (28.3) | 1,116 (29.3) | 1,315 (1,089–1,542) |
Rabies surveillance data for terrestrial nonvolant mammals (excluding humans) in the United States that were submitted to the CDC during the period of January 2010 through December 2015 were included in this evaluation. Bat data were not included but underwent separate analysis.20
On review of the data, the mean annual number of SEIs among the RABV-positive specimens was 855 (95% CI, 739 to 971); the mean annual number of SEIs that underwent virus characterization was 270 (31.6%; 95% CI, 187 to 353). Virus characterization of all SEIs in the United States would be expected to increase public health laboratories’ test load by approximately 585 (95% CI, 543 to 625) specimens/y.
Subcategories of SEIs that would identify or provide information regarding the introduction of novel RABV variants
Three subcategories of SEIs were found to provide the most meaningful information regarding the introduction of novel RABV variants. Among the SEIs, those in the subcategory of domestic animal or livestock (ie, cattle, horses, donkeys, sheep, goats, pigs, alpacas, and llamas) in a southern border state (ie, California, Arizona, New Mexico, Texas, or Florida) were considered prime candidates for RABV characterization. Livestock serve as sentinel species to detect the introduction of vampire bat (Desmodus rotundus) RABV variants linked to the potential incursion of vampire bat populations in the tropical or subtropical southern United States.21 Livestock can also serve as sentinel species for circulating wildlife RABV variants (both bat and terrestrial mammal variants) within a region. Domestic animals should also be monitored for the introduction of novel variants (eg, the dog-coyote variant identified in Texas).
During 2010 through 2015, 70,973 specimens from domestic animals or livestock (cattle, horses, donkeys, sheep, goats, pigs, alpacas, and llamas) in southern border states were tested; 440 (0.6%) were RABV positive, of which 360 (81.8%) underwent virus characterization. In the 360 SEIs that were characterized, variants detected included south-central skunk (n = 339), raccoon (11), US-enzootic bat variants (6), California skunk (2), Texas gray fox (1), and vampire bat (1). The RABV in 1 bovine SEI was characterized as a vampire bat variant (from D rotundus) in Texas in 2010, and the affected animal had a history of importation from Mexico. In the southern border states, the mean number of RABV-positive domestic and livestock animals identified annually, the mean number of SEI that underwent virus characterization, and the estimated laboratory test load increase were summarized (Table 2).
Table 2—
Public health laboratory RABV testing data related to RABV-positive specimens assigned to 11 SEI subcategories* on the basis that they would identify or provide important information regarding 4 overarching events of interest and the estimated annual increase in laboratory test load if all SEIs in a subcategory were to undergo RABV characterization.
| Event of interest | SEI subcategory† | Mean annual No. (95% CI) of RABV-positive specimens | Mean annual No. (95% CI) of RABV-positive specimens that underwent virus characterization | Estimate of mean (95% CI) annual No. of additional specimens processed if all SEIs were to undergo RABV characterization |
|---|---|---|---|---|
| Novel variant introduction | Domestic animal or livestock (ie, cattle, horses, donkeys, sheep, goats, pigs, alpacas, and llamas) in a southern border state (ie, California, Arizona, New Mexico, Texas, or Florida) | 78 (66–89) | 60 (49–71) | 24 (10–24) |
| Dog | 73 (61–84) | 37 (29–46) | 17 (22–48) | |
| Mammal with a history of international travel in the preceding 12 mo | Unknown | Unknown | < 5 | |
| Translocation events | Terrestrial mammal in the USDA ORV zone front (including enhanced rabies surveillance zones) | 301 (245–356) | 65 (12–118) | 235 (200–271) |
| Raccoon in an area west of the USDA ORV zone front (including enhanced rabies surveillance zones) | 36 (29–43) | 31 (24–37) | 5 (2–9) | |
| Mammal with large or migratory home range (specifically coyotes, cougars, bobcats, wolves, deer, and bison) | 33 (27–42) | 8 (4–12) | 24 (18–31) | |
| Domestic mammals with recent travel history (< 6 mo) across RABV variant territory boundaries‡ | Unknown | Unknown | < 10 | |
| Host shift | Fox | 362 (308–471) | 86 (61–110) | 276 (237–315) |
| Mammal in nonterrestrial mammal (ie, bat) reservoir areas | 12 (6–17) | 5 (1–9) | 6 (1–11) | |
| Unusual rabies-related incident | All rodents and lagomorphs (eg, squirrel, beaver, muskrat, groundhog, and rabbit) | 40 (33–47) | 7 (4–9) | 33 (28–39) |
| Cluster event (≥ 2 cases) in a livestock herd in a 60-d period | Unknown | Unknown | < 25 | |
| Total | 855 (739–971) | 270 (187–353) | 585 (543–625) |
A 12th SEI subcategory related to unusual rabies-related incidents was other; there were no data available for this subcategory.
Some SEIs could be placed in multiple subcategories; however, duplicate samples from multiple SEI subcategories were removed to calculate the total sample increase expected by public health laboratories.
An example would be an animal that travels from Maryland (raccoon RABV enzootic territory) to Indiana (nonraccoon RABV territory).
See Table 1 for remainder of key.
Another SEI subcategory for which RABV characterization was considered to yield meaningful information about the introduction of novel RABV variants was dog. Characterization of all SEIs obtained from RABV-positive dogs is recommended to meet World Organisation for Animal Health international standards22 that allow a country to be classified as canine RABV variant–free.
During 2010 through 2015, 139,073 dog specimens were submitted for RABV testing in the United States. Results of testing indicated that 437 (0.3%) dogs were positive for RABV; 223 (50.7%) of those SEIs underwent RABV characterization. In the 223 characterized SEIs, variants detected included south-central skunk (n = 111), raccoon (60), north-central skunk (38), arctic fox (7), US-enzootic bat (4), California skunk (2), and canine (1). The single canine case of rabies was imported into Virginia from Egypt. The mean number of RABV-positive dog specimens identified annually, the mean number of SEIs that underwent virus characterization, and the estimated laboratory test load increase were summarized (Table 2).
A third subcategory of SEIs for which RABV characterization was considered to yield important information regarding the introduction of novel RABV variants was mammals with a history of international travel in the preceding 12 months. International travel included importation from a country or territory in which terrestrial mammal rabies is enzootic.
For the period of 2010 through 2015, the number of animals in this subcategory was difficult to estimate because this information is not routinely collected in national surveillance data sets. There were documented cases of rabies among animals imported into the United States, but fewer than 1 case was reported each year. We estimated the number of SEIs submitted that could be included in this subcategory would not exceed 5 specimens/y.
Subcategories of SEIs that would identify or provide information regarding the translocation of RABV variants not previously documented within geographic areas in the United States
Four subcategories of SEIs were found to provide the most meaningful information regarding translocation of RABV variants not previously documented within geographic areas in the United States. Among the SEIs, those from terrestrial mammals in the USDA ORV zone front (including enhanced rabies surveillance zones) were considered prime candidates for RABV characterization. Virus characterization of rabies-positive terrestrial mammals in areas such as the USDA enhanced rabies surveillance regions (which include ORV bait zones) provides information that is critical to landscape-level wildlife rabies management practices and monitoring for vaccine-induced RABV infections.
During 2010 through 2015, there were 1,870 RABV-positive terrestrial mammals identified in USDA enhanced rabies surveillance zones (Figure 1); of those SEIs, 560 (29.9%) underwent RABV characterization. The RABV variants detected included raccoon (n = 399), north-central skunk (153), and US-enzootic bat (8). The mean annual number of SEIs in USDA enhanced rabies surveillance zones, the mean number of SEIs that underwent virus characterization, and the estimated laboratory test load increase were summarized (Table 2).
The SEIs obtained from raccoons in areas west of the USDA ORV zone front (including enhanced rabies surveillance zones) were also considered to provide important information regarding translocation of RABV variants not previously documented within geographic areas in the Unites States. Within enhanced rabies surveillance zones, accurate and timely reporting of RABV characterization data is critical for adaptation of raccoon rabies management practices, response to challenges of raccoon RABV spillover into sympatric skunk populations, and detection of translocation of RABV variants.1
During 2010 through 2015, 15,317 raccoon specimens from states west of the ORV zone front were submitted for testing; 180 (1.2%) were RABV positive, and 160 (88.9%) of those SEIs underwent virus characterization. The RABV variants detected included south-central skunk (n = 158), north-central skunk (1), and Arizona gray fox (1). The mean annual number of RABV-positive raccoon specimens from states west of the ORV zone front, the mean number of SEIs that underwent virus characterization, and the estimated laboratory test load increase were summarized (Table 2).
Another SEI subcategory species for which RABV characterization was considered to yield important information regarding translocation of RABV variants not previously documented within geographic areas in the Unites States was mammals with large or migratory home ranges (specifically coyotes, cougars, bobcats, wolves, deer, and bison). It is known that routine RABV characterization is a useful tool in states located on the edge of an epizootic rabies zone.23 However, virus characterization of RABV-positive specimens submitted from states along boundaries between areas in which different RABV variants are found should be undertaken to monitor geographic translocation of variants.
In 2010 through 2015, 6,866 coyotes, cougars, bobcats, wolves, deer, and bison were tested for RABV. Of those animals, 200 (2.9%) were RABV positive. Fifty-one of the 200 (25.5%) SEIs underwent virus characterization. The RABV variants detected included raccoon (n = 27), south-central skunk (14), Arizona gray fox (6), arctic fox (2), US-enzootic bat (1), and California skunk (1). The mean number of SEIs identified annually, the mean number of SEIs that underwent virus characterization, and the estimated laboratory test load increase were summarized (Table 2). However, captivity status of wildlife species is not routinely reported; therefore, this may be an over-estimation of the true increase in numbers of tested specimens because captive wildlife would not be considered to have a large or migratory home range and any associated SEI would not be assigned to this subcategory.
The fourth SEI subcategory for which RABV characterization was considered to yield meaningful information about translocation of RABV variants not previously documented within geographic areas in the Unites States was domestic mammals with a recent travel history (< 6 months) across RABV variant territory boundaries. Accurate data regarding the number of reported rabid animals with a history of recent travel (< 6 months) across RABV variant territory boundaries were not available. However, movement of rabid animals between terrestrial RABV variant regions has been documented. Thus, it was estimated that the number of RABV-positive specimens submitted that could be assigned to this SEI subcategory for virus characterization would not exceed 10 specimens/y.
Subcategories of SEIs that would identify or provide information regarding host-shift events
Two subcategories of SEIs were found to provide the most meaningful information regarding host-shift events. Among the SEIs, those in the subcategory of fox were considered prime candidates for RABV characterization. Foxes have been associated with rabies host-shift events5 and should be monitored closely.
During 2010 through 2015, there were 11,129 foxes tested for RABV, of which 2,176 (19.6%) were RABV positive. Of those SEIs, 519 (23.9%) underwent virus characterization. The RABV variants detected included raccoon (n = 363), south-central skunk (105), US-enzootic bat (20), arctic fox (19), Arizona gray fox (7), and north-central skunk (5). The mean number of RABV-positive foxes identified annually, the mean number of SEIs that underwent virus characterization, and the estimated laboratory test load increase were summarized (Table 2).
The other SEI subcategory for which RABV characterization was considered to yield meaningful information regarding host-shift events was mammals in nonterrestrial mammal (ie, bat) reservoir areas. Specimens from terrestrial mammals in rabies-free regions that are suspected of being rabid should be tested for RABV and, if positive, undergo RABV characterization to monitor for host-shift events as well as translocation events that may lead to the introduction of newly established variants within a reservoir species (eg, translocation of the raccoon variant from North Carolina to Washington state, where the variant becomes established in the resident raccoon population).
Washington, Oregon, Idaho, Illinois, Indiana, Nevada, Ohio, Mississippi, and Utah are considered terrestrial animal rabies-free regions.2 During 2010 through 2015, there were 47,901 mammals submitted for RABV testing from terrestrial animal rabies-free regions, and 73 (0.15%) of those mammals were RABV positive. Of those SEIs, 29 (39.7%) underwent virus characterization. The RABV variants detected included raccoon (n = 24) and US-enzootic bat (5). The mean annual number of RABV-positive mammals, the mean number of SEI that underwent virus characterization, and the estimated laboratory test load increase were summarized (Table 2).
Subcategories of SEIs that would identify or provide information regarding unusual rabies-related incidents
Three subcategories of SEIs were found to provide the most meaningful information regarding unusual rabies-related incidents. Among the SEIs, those in the subcategory of all rodents were considered prime candidates for RABV characterization. Currently, the Advisory Committee of Immunization Practices and National Association of State Public Health Veterinarians recognize that rabies in rodents is rare and that rodents do not have a role as RABV reservoir hosts. Thus, exposure to these animals in general do not result in recommendations to treat with rabies postexposure prophylaxis unless there is an unprovoked exposure, the animal is unavailable for testing, the animal is RABV positive, or postexposure prophylaxis is otherwise recommended by public health authorities. If an ecological shift, host shift, or virus shift results in an increased number of rodents with rabies, these recommendations may need to be reviewed.
Rodents and lagomorphs (squirrels, beavers, muskrats, groundhogs, and rabbits) submitted for testing are not often positive for rabies, given that most small rodents and lagomorphs do not survive an attack by a larger rabid animal and often develop paralytic rabies; however, every year, RABV-positive rodents (most commonly groundhogs) are identified.24,25 In RABV-positive rodents, virus characterization is important because the animals’ habitats often overlap with areas enzootic for raccoon rabies and other rabies reservoir hosts among terrestrial animals. Thus, information gathered from characterization of RABV in infected rodents can provide information about the species affected and the RABV variants that most frequently spill over in rodent populations, and allow for early detection of potential host shifts.
During 2010 through 2015, there were 13,368 rodents tested for RABV, of which 245 (1.8%) were RABV positive. Of those SEIs, 43 (17.6%) underwent virus characterization. The RABV variants detected included raccoon (n = 41), south-central skunk (1), and north-central skunk (1). The mean number of RABV-positive rodents identified annually, the mean number of SEIs that underwent virus characterization, and the estimated laboratory test load increase were summarized (Table 2).
Another SEI subcategory for which RABV characterization was considered to yield meaningful information about unusual rabies-related incidents was cluster events (≥ 2 cases) in a livestock herd in a 60-day period. Accurate information for this subcategory is not currently available among national surveillance data; however, it was likely that the number of rabies cluster events was underestimated because multiple cattle from a given herd may not be tested, despite clinical signs.26
The National Association of State Public Health Veterinarians acknowledges that herbivore-to-herbivore transmission of RABV is rare; therefore, herd quarantines are rarely recommended.27 However, there are anecdotal reports of animal-to-animal transmission of vampire bat rabies among cattle, among kudus in Southern Africa, and among deer located within raccoon rabies–epizootic areas.28,29 Alternatively, clusters of cases in livestock may also signal an incursion of vampire bat rabies21 in the United States. Early detection of case clusters could serve as a signal event. If changes in management practices or RABV variant epidemiology result in increases in livestock case clusters, this recommendation may need review. Therefore, RABV characterization of specimens collected during such events is important for guiding future recommendations. Thus, it was estimated that the number of RABV-positive specimens submitted that could be assigned to this SEI subcategory for virus characterization would not exceed 25 specimens/y.
A third SEI subcategory for which RABV characterization was considered to yield meaningful information about unusual rabies-related incidents was other. State and local health departments that identify unique rabies cases (eg, an RABV-positive cat in a region that has not reported an RABV-positive cat in the preceding 2 or more years) may also wish to pursue RABV characterization of collected specimens. Circumstances in which RABV characterization of collected specimens would be prudent include rabies in a species that is not commonly affected, novel locations of rabies outbreaks as determined from the most recent reservoir map published in the CDC’s annual rabies surveillance report,2 or reported animal rabies cases in an area where there have been no cases among terrestrial mammals documented in the preceding 2 to 5 years.
Characterization of specimens not classified as SEIs
In areas in which the raccoon RABV variant is enzootic (Maine, New Hampshire, Vermont, Pennsylvania, New York, Massachusetts, Connecticut, District of Columbia, Rhode Island, New Jersey, Delaware, Maryland, West Virginia, Virginia, North Carolina, South Carolina, Georgia, Florida, and Alabama), there were 11,359 RABV-positive raccoons identified during 2010 through 2015; 2,014 (17.7%) raccoon specimens underwent virus characterization, of which 2,007 (99.6%) were raccoon variant (Table 3). The nonraccoon RABV variants detected included north-central skunk (n = 5) and US-enzootic bat (2).
Table 3—
Number of RABV-positive raccoon and skunk specimens submitted to public health laboratories, number of specimens that underwent RABV characterization, and RABV variant results classified by variant territory in the United States and by year during the period of January 2010 through December 2015.
| Rabies reservoir host from enzootic host territory | Variable | Year | Mean (95% CI) annual percentage of species-specific reservoir variants detected | Mean (95% CI) annual No. of specimens | |||||
|---|---|---|---|---|---|---|---|---|---|
| 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | ||||
| Raccoons from raccoon variant territory | No. of RABV-positive specimens | 2,227 | 1,942 | 1,928 | 1,884 | 1,793 | 1,585 | — | 1,893 (1,672–2,113) |
| No. (%) of RABV-positive specimens that underwent virus characterization | 379 (17) | 374 (19.3) | 352 (18.3) | 356 (18.9) | 281 (15.7) | 272 (17.2) | — | 335 (286–385)* | |
| No. (%) of characterized specimens with raccoon variant | 377 (99.5) | 374 (100) | 351 (99.7) | 353 (99.2) | 280 (99.6) | 272 (100) | 99.6 (99.3–99.9) | 334 (285–383) | |
| Skunks from south-central skunk variant territory | No. of RABV-positive specimens | 638 | 786 | 653 | 786 | 894 | 729 | — | 747 (647–847) |
| No. (%) of RABV-positive specimens that underwent virus characterization | 395 (62) | 677 (86.1) | 373 (57.1) | 406 (51.7) | 501 (56) | 429 (58.8) | — | 463 (344–582)* | |
| No. (%) of characterized specimens with south-central skunk variant | 393 (99.5) | 672 (99.3) | 372 (99.7) | 405 (99.7) | 501 (100) | 429 (100) | 99.7 (99.4–100) | 462 (344–579) | |
| Skunks from north-central skunk variant† territory | No. of RABV-positive specimens | 167 | 144 | 177 | 101 | 120 | 64 | — | 128 (84–173) |
| No. (%) of RABV-positive specimens that underwent virus characterization | 73 (43.7) | 49 (34) | 50 (28.2) | 21 (20.7) | 12 (10) | 30 (46.9) | — | 39 (15–62) | |
| No. (%) of characterized specimens with north-central skunk variant | 70 (95.8) | 48 (97.9) | 50 (100) | 21 (100) | 11 (91.6) | 29 (96.6) | 96.9 (93.8–100) | 38 (15–60) | |
| Skunks from California skunk variant territory‡ | No. of RABV-positive specimens | 23 | 12 | 16 | 7 | 28 | 29 | — | 17 (6–27) |
| No. (%) of RABV-positive specimens that underwent virus characterization | Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | — | Unknown | |
| No. (%) of characterized specimens with California skunk variant | Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | |
Number of RABV-positive specimens that underwent virus characterization was sufficient to detect a difference in the population with 95% confidence.
Data for north-central skunk and western Canadian skunk variants were grouped together for analysis.
California did not report RABV variants to the US national RSS; however, there are some annual data for that state available online.
— = Not applicable.
See Table 1 for remainder of key.
In areas in which the south-central skunk RABV variant is enzootic (Texas, New Mexico, Arizona, Louisiana, Arkansas, Missouri, Nebraska, Kansas, Oklahoma, Colorado, and Wyoming), there were 4,486 RABV-positive skunks identified; 2,781 (62.0%) skunk specimens underwent virus characterization, of which 2,772 (99.7%) were south-central skunk variant. Other RABV variants detected included north-central skunk (n = 6) and US-enzootic bat (3). During the 2010–2015 period, the numbers of RABV-positive raccoon specimens obtained in areas in which the raccoon variant is enzootic and RABV-positive skunk specimens obtained in areas in which the south-central skunk variant is enzootic that underwent virus characterization were sufficient to detect differences in variants within the populations (Table 3).
In areas in which the north-central skunk RABV variant is enzootic (Montana, Michigan, Tennessee, Kentucky, Wyoming, North Dakota, South Dakota, Minnesota, Iowa, and Wisconsin), there were 773 RABV-positive skunks identified during 2010 through 2015; 235 (30.4%) skunk specimens underwent virus characterization, of which 229 (96.9%) were a skunk variant (north-central skunk, 178 [75.7%]; western Canada skunk, 51 [21.7%]), 5 (2.1%) were a raccoon variant, and 1 (0.4%) was a US-enzootic bat variant (Table 3). In California, where California skunk RABV variant is enzootic, there were 115 RABV-positive skunks identified; 0 (0%) skunk specimens underwent virus characterization.
Overall impact of testing all SEIs for public health laboratories
During 2010 through 2015, the mean annual number of animal specimens that underwent RABV characterization was 1,315 (95% CI, 1,089 to 1,542). Of those 1,315 specimens, 855 (95% CI, 739 to 971) were considered SEIs (Table 2). Virus characterization of all SEIs would be expected to increase the test load at public health laboratories by approximately 585 (95% CI, 543 to 625) specimens/y. If RABV characterization of specimens that were not considered SEIs was eliminated, the laboratory test load would be expected to decrease by 798 specimens/y (raccoon specimens, 335 [95% CI, 286 to 385]; skunk specimens, 463 [95% CI, 344 to 582]; Table 3). This would have resulted in a net reduction of 675 specimens undergoing RABV characterization in the United States annually, there-by saving both time and resources while maximizing detection of host-shift, translocation, and novel variant introduction events.
Discussion
Results of the analysis of RSS data for terrestrial nonvolant mammals (excluding humans) in the United States that were submitted to the CDC during the period of January 2010 through December 2015 revealed that a lack of RABV characterization of > 585 SEIs each year may have left the United States vulnerable to undetected RABV host-shift, translocation, or novel introduction events, which may have further precluded or delayed implementation of adequate control strategies leading to an increased risk of rabies to humans and animals. Rabies virus characterization of all SEIs identified in the present study could have improved the effectiveness of surveillance for epizootiological events of concern; although that would, at face value, have increased the number of specimens tested, there would have been minimal to no increase in the workload of public health laboratories during that period had no virus characterization of specimens that were not identified as SEIs been performed.
The rabies epizootics that occurred in the mid-Atlantic states in the 1970s and in Texas in the 1980s serve as examples of what happens when early detection and characterization of RABV variants do not occur and opportunities to slow or prevent geographic spread of those viruses are missed. Although the epizootic of rabid coyotes in Texas was eventually eliminated, the translocation of a rabid raccoon from Florida to RABV-naive raccoon populations in West Virginia resulted in the largest rabies epizootic in US history, which is still being combatted in 2019.10 The establishment of variants in novel locations resulted in a massive outpouring of public health resources to mitigate negative human and animal health consequences. Although these efforts have been largely successful, at least 6 human deaths were directly attributable to those rabies epidemiological changes.11,14 Early detection of rabies epizootiological changes has been successful several times in the United States. Host shifts of bat-variant RABV into skunks and foxes in Arizona and into foxes in Oregon were detected early, and mitigation efforts were rapidly undertaken. As a result, those host shifts were prevented from becoming established.
The concern that RABV characterization of all SEIs from rabid domestic mammals and terrestrial wildlife would increase the testing burden on public health laboratories may be offset, to some extent, by reductions in the numbers of specimens that are not SEIs but that undergo virus characterization, particularly in regions in which raccoon or south-central skunk RABV is enzootic. Given that > 99% of rabid raccoon specimens obtained in raccoon rabies–enzootic areas were characterized as raccoon RABV variant and > 99% of rabid skunk specimens obtained in south-central skunk rabies–enzootic areas were characterized as south-central skunk RABV variant in the present study, states with limited resources could focus characterization efforts on specimens that meet the SEI criteria developed for the evaluation of the present report. As the study data revealed, few north-central skunk, western Canada skunk, and California skunk specimens are reported to the CDC and characterized every year, and continued virus characterization of specimens is recommended. Limiting virus characterization of raccoon specimens obtained in raccoon RABV variant–enzootic areas (excluding USDA ORV zones and enhanced rabies surveillance areas) could decrease public health laboratories’ overall test load by as much as 335 (95% CI, 285 to 383) specimens/y. Limiting virus characterization of skunk specimens in south-central skunk RABV variant–enzootic areas could decrease the nationwide test load by as much as 463 (95% CI, 344 to 582) specimens/y. However, incorporating such limitations could preclude identification of the temporal or geographic origin of a translocation of the south-central skunk or raccoon RABV variants; therefore, storage of a selected subset of specimens from rabid skunks and raccoons within those enzootic areas for RABV characterization should be considered (Appendix).
One limitation of the present study was the unknown captivity status of animals with large home ranges (ie, coyotes, cougars, bobcats, wolves, deer, and bison) from which SEIs were obtained. Although the address of the location where a specimen was collected is sometimes provided, this information is not always available and captivity status is not recorded in the RSS. Captive wildlife submitted for rabies testing may not meet the definition of this SEI classification if their home ranges were limited. Additionally, the present study was a national-level analysis, and the number of samples tested by each state was not highlighted. It is likely that the impact of the recommendations derived from the analyses performed in the present evaluation would affect some states more than others. The CDC requests RABV characterization information be provided voluntarily by state health departments that report rabies-positive animals, but there is no requirement to do so, which may explain why some state health departments are not further characterizing the RABV variant in specimens or reporting those results to the CDC. Therefore, the SEIs that did not undergo variant characterization identified in the present evaluation may be attributable to a lack of data reporting rather than to a lack of testing. It is also possible that the absence of virus characterization data reflected a lack of awareness of the value and applicability of this type of information in the control and prevention of rabies. Furthermore, the lack of data reporting may also have reflected a deficiency in laboratory diagnostic capabilities owing to shortages in staff, training, or equipment. In addition to RABV detection in specimens by direct fluorescent antibody testing, antigenic typing of terrestrial mammal RABV variants can be performed. However, depending on the variety of monoclonal antibodies against viral nucleoproteins included in the test panel, antigenic typing may not have sufficient discriminatory power to differentiate certain terrestrial RABV variants (eg, some wildlife variants of dog origin), and state health departments may need improved access and increased capacity to perform PCR assays and molecular sequencing to obtain RABV characterization data. Currently, not all public health laboratories have the capacity to characterize RABV in specimens from animals with rabies, but access to appropriate test procedures is improving.
The Council of State and Territorial Epidemiologists and the CDC recommend, but do not require, RABV variant reporting for rabies-positive specimens. However, RABV characterization is a critical component necessary in longitudinal analyses to evaluate or reconstruct relevant epizootiological and evolutionary events. Furthermore, a robust archived set of historical specimens provides integral information to understand the spatial-temporal dynamics of rabies epizootics, which in turn allows for adequate monitoring of the progress of control and elimination strategies.
The success of RSS within and between states requires collaboration between epidemiologists and public health laboratory personnel. However, the infrastructure, resources, and surveillance system of each state differ, requiring each state to independently evaluate its current surveillance system and determine how to incorporate the proposed SEI selection criteria into current laboratory practices. The findings of the present evaluation have the potential to assist established laboratories target their current testing approach, help new laboratories develop a specimen-testing strategy, and encourage laboratories in which virus characterization is not performed to implement procedures for testing of selected specimens and reporting results to the CDC. Prioritized testing of SEIs could reduce laboratory testing costs for some states while simultaneously providing epizootiologically relevant data that allow further refinement of control and prevention measures and reduction of unnecessary testing in the future.
Acknowledgments
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the CDC or USDA.
The authors thank Xiaoyue Ma and Lauren Greenberg for technical assistance.
ABBREVIATIONS
- CI
Confidence interval
- ORV
Oral rabies vaccination
- RABV
Rabies virus
- RSS
Rabies surveillance system
- SEI
Specimen of epizootiological importance
Appendix
Recommendations for storage of specimens obtained from rabid animals.
Suggested duration of storage
Any RABV-positive specimen that undergoes virus characterization should be retained for long-term storage. Additionally, 10% of RABV-positive specimens from reservoir species (raccoon, skunk, arctic fox, and gray fox) should be similarly stored. Specimens submitted for long-term storage should be geographically and temporally representative throughout the year.
Suggested preservation methods
Fresh, well-preserved cross sections of brainstem that contain a 3+ or 4+ intensity of fluorescein isothiocyanate–labeled antibody against RABV should be stored long term. The ideal amount of tissue to freeze and store is the amount of cross-sectioned brainstem tissue or whole small rodent brain that can be fitted in half the capacity of a 2-mL cryotube (with an O-ring cap assembly). Specimens submitted in tin or large containers (volume, > 2 mL) should be aliquoted in 2-mL cryotubes after an adequate 3+ or 4+ section was selected by direct fluorescent antibody testing. For small rodents, if whole brains are not available, complete carcasses should be submitted in plastic biosafety bags.
Footnotes
Microsoft Access 2013, Microsoft Corp, Redmond, Wash.
STATA 13.1, StataCorp LLC, College Station, Tex.
OpenEpi: Open Source Epidemiologic Statistics for Public Health, version 3.01. Available at: www.OpenEpi.com. Accessed Sep 1, 2016.
References
- 1.Elmore SA, Chipman RB, Slate D, et al. Management and modeling approaches for controlling raccoon rabies: the road to elimination (Erratum published in PLoS Negl Trop Dis 2017;11:e0005579). PLoS Negl Trop Dis 2017;11:e0005249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma X, Monroe BP, Cleaton JM, et al. Rabies surveillance in the United States during 2016. J Am Vet Med Assoc 2018;252:945–957. [DOI] [PubMed] [Google Scholar]
- 3.Adams DA, Thomas KR, Jajosky RA, et al. Summary of notifiable diseases and conditions—United States, 2015. MMWR Morb Mortal Wkly Rep 2017;64:1–143. [DOI] [PubMed] [Google Scholar]
- 4.CDC. Translocation of coyote rabies—Florida, 1994. MMWR Morb Mortal Wkly Rep 1995;44:580–581, 587. [PubMed] [Google Scholar]
- 5.Velasco-Villa A, Reeder SA, Orciari LA, et al. Enzootic rabies elimination from dogs and reemergence in wild terrestrial carnivores, United States. Emerg Infect Dis 2008;14:1849–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuzmin IV, Shi M, Orciari LA, et al. Molecular inferences suggest multiple host shifts of rabies viruses from bats to mesocarnivores in Arizona during 2001–2009. PLoS Pathog 2012;8:e1002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leslie MJ, Messenger S, Rohde RE, et al. Bat-associated rabies virus in skunks. Emerg Infect Dis 2006;12:1274–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borucki MK, Chen-Harris H, Lao V, et al. Ultra-deep sequencing of intra-host rabies virus populations during cross-species transmission. PLoS Negl Trop Dis 2013;7:e2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding NZ, Xu DS, Sun YY, et al. A permanent host shift of rabies virus from Chiroptera to Carnivora associated with recombination. Sci Rep 2017;7:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nettles VF, Shaddock JH, Sikes RK, et al. Rabies in translocated raccoons. Am J Public Health 1979;69:601–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vora NM, Basavaraju SV, Feldman KA, et al. Raccoon rabies virus variant transmission through solid organ transplantation. JAMA 2013;310:398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slate D, Algeo TP, Nelson KM, et al. Oral rabies vaccination in North America: opportunities, complexities and challenges. PLoS Negl Trop Dis 2009;3:e549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirby JD, Chipman RB, Nelson KM, et al. Enhanced rabies surveillance to support effective oral rabies vaccination of raccoons in the Eastern United States. Trop Med Infect Dis 2017;2:E34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark KA, Wilson PJ. The coyote’s role in a rabies epizootic. In: Rollins D, Richardson C, Blankenship T, et al. , eds. Coyotes in the Southwest: a compendium of our knowledge. Symposium Proceedings. San Angelo, Tex: Texas Parks and Wildlife Department, 1995. [Google Scholar]
- 15.CDC. Human rabies—Texas, Arkansas and Georgia, 1991. MMWR Morb Mortal Wkly Rep 1991;40:765–769. [PubMed] [Google Scholar]
- 16.CDC. Human rabies—Alabama, Tennesse and Texas, 1994. MMWR Morb Mortal Wkly Rep 1995;44:269–272. [PubMed] [Google Scholar]
- 17.Sinclair JR, Wallace RM, Gruszynski K, et al. Rabies in a dog imported from Egypt with a falsified rabies vaccination certificate—Virginia, 2015. MMWR Morb Mortal Wkly Rep 2015;64:1359–1362. [DOI] [PubMed] [Google Scholar]
- 18.Castrodale L, Walker V, Baldwin J, et al. Rabies in a puppy imported from India to the USA, March 2007. Zoonoses Public Health 2008;55:427–430. [DOI] [PubMed] [Google Scholar]
- 19.CDC. Rabies in a dog imported from Iraq—New Jersey, June 2008. MMWR Morb Mortal Wkly Rep 2008;57:1076–1078. [PubMed] [Google Scholar]
- 20.Pieracci EG, Brown JA, Bergman DL, et al. Evaluation of species identification and rabies virus characterization among bat rabies cases in the United States. J Am Vet Med Assoc 2020;256:77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez-Burnes J, López A, Medellín J, et al. An outbreak of vampire bat-transmitted rabies in cattle in northeastern Mexico. Can Vet J 1997;38:175–177. [PMC free article] [PubMed] [Google Scholar]
- 22.World Organization for Animal Health. Terrestrial Animal Health Code (2016). Available at: www.oie.int/en/international-standard-setting/terrestrial-code/access-online/. Accessed Jul 12, 2017.
- 23.Blanton JD, Palmer D, Dyer J, et al. Rabies surveillance in the United States during 2010. J Am Vet Med Assoc 2011;239:773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDaniels AK. Rabid groundhog found at the Maryland zoo. Baltimore Sun 2017;Jul 13. Available at: www.baltimoresun.com/health/bs-hs-ground-hog-rabies-20170713-story.html. Accessed Nov 13, 2017. [Google Scholar]
- 25.WSPA 7 News. Rabid woodchuck found in Oconee Co. Available at: wspa.com/2017/07/06/rabid-woodchuck-found-in-oconee-co/. Accessed Nov 13, 2017.
- 26.Chipman RB, Cozzens TW, Shwiff SA, et al. Costs of raccoon rabies incidents in cattle herds in Hampshire County, West Virginia, and Guernsey County, Ohio. J Am Vet Med Assoc 2013;243:1561–1567. [DOI] [PubMed] [Google Scholar]
- 27.National Association of State Public Health Veterinarians Compendium of Animal Rabies Prevention and Control Committee. Compendium of Animal Rabies Prevention and Control, 2016. J Am Vet Med Assoc 2016;248:505–517. [DOI] [PubMed] [Google Scholar]
- 28.Scott TP, Fischer M, Khaiseb S, et al. Complete genome and molecular epidemiological data infer the maintenance of rabies among kudu (Tragelaphus strepsiceros) in Namibia. PLoS One 2013;8:e58739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen BW, Tack DM, Longenberger A, et al. Rabies in captive deer, Pennsylvania, USA, 2007–2010. Emerg Infect Dis 2012;18:138–141. [DOI] [PMC free article] [PubMed] [Google Scholar]


