Abstract
Temporal ecological niche partitioning is an underappreciated driver of speciation. While insects have long been models for circadian biology, the genes and circuits that allow adaptive changes in diel-niches remain poorly understood. We compared gene expression in closely related day- and night-active non-model wild silk moths, with otherwise similar ecologies. Using an ortholog-based pipeline to compare RNA-Seq patterns across two moth species, we find over 25 pairs of gene orthologs showing differential expression. Notably, the gene disco, involved in circadian control, optic lobe and clock neuron development in Drosophila, shows robust adult circadian mRNA cycling in moth heads. Disco is highly conserved in moths and has additional zinc-finger domains with specific nocturnal and diurnal mutations. We propose disco as a candidate gene for the diversification of temporal diel-niche in moths.
Keywords: chronobiology, de novo transcriptome, Lepidoptera, light cycle, RNA-Seq, temporal
1. Introduction
Ecological niche, including the temporal partitioning of activities within a day (diel-niche), is an underappreciated driver of speciation and adaptive evolution [1]. Partitioning can help organisms exploit resources, avoid competition and reduce predation risk [2–7]. Differences in physiology and performance at various temperatures, light levels and humidity can also drive partitioning [8]. In insects, circadian rhythms are crucial for regulating daily locomotor, reproductive behaviour and feeding activity [5,9–11]. Over the past few decades, research in Drosophila has led to a better understanding of the molecular and neural mechanisms underlying insect circadian rhythms [12,13]. They are governed by transcriptional–translational feedback loops, self-sustained molecular oscillations that affect patterns of gene expression at the cellular level and, due to expression in the central nervous system, scale up to determine whole-organism patterns of activity (reviewed in D. melanogaster in [12,14]).
Four key elements have been identified in the circadian control system that may be relevant to temporal niche evolution: (i) genes and proteins that govern the central clock, (ii) neural circuits connecting the clock to downstream processes, (iii) neuropeptides, such as pigment-dispersing factor (PDF), released by these clock neurons to modulate the synchronization within the clock network, and (iv) light detection by photoreceptors and photosensitive molecules, which entrain the clock to match external light cycles. Behaviours with strong diel periodicity that also impact fitness could drive selection on genes and circuits of the molecular clock or on those involved in entrainment to external stimuli.
While the identity and function of core clock genes have been explored extensively in D. melanogaster [15–17], the clock proteins period (Per), timeless (Tim) and cryptochrome (Cry) are highly conserved across animals [18,19]. They participate in auto-regulatory feedback loops to produce changes in gene expression on an approximately 24 h cycle. Nearly one-third of cell types in D. melanogaster express clock genes [20], but daily circadian patterns only require expression of core molecular clock components in about 150 clock neurons [21–24]. These cluster into 3 dorsal groups (DNs) and 4 lateral groups (LNs) based on cell body location and cluster into 17 groups based on gene expression [13,25]. A subset of LNs and DNs release PDFs and other neuromodulators to keep clock cells synchronized or signal to other neuroendocrine centres [26]. These oscillators can be light entrained via light-sensitive proteins expressed in clock cells (Cry) and peripheral photoreceptors (rhodopsin Rh6) [27,28]. Flies with different activity periods have displayed species-specific expression patterns of Cry and PDF [29].
Comparisons among circadian genes and circuits across evolutionary time scales indicate a high degree of conservation between invertebrates [30–32] and vertebrates [33], and insects serve as useful genetic models of human circadian disorders [32,34,35]. However, since diel switches can occur over shorter evolutionary time scales [29], examining more divergent circadian genes can shed light on circadian evolution. Lepidoptera (the moths and butterflies) contain many extreme diel-niche switches, often between closely related species, and their evolutionary history is well known, making them ideal for studying diel-niche evolution [36,37]. They also have some of the best characterized circadian and sensory genes and circuits, outside of Drosophila [38–44]. For example, the wild silk moth family, Saturniidae is an important model for understanding chronobiology [45,46]. Moths are thus a useful system to explore the adaptive significance of circadian rhythms in the context of ecological niche partitioning [47,48].
We investigate two closely related saturniid moth species, the pink-striped oakworm moth (Anisota pellucida) and the rosy maple moth (Dryocampa rubicunda) that overlap in geographic distribution and habitat but differ in daily activity patterns, with Anisota being diurnal and Dryocampa being nocturnal. These species diverged approximately 3.8 Ma [49] and represent a relatively recent shift in diel-niche. We compare the day–night cycling genes to identify candidate drivers of temporal partitioning. We identify differentially expressed orthologs and focus our analysis on gene pairs that show day–night cycling patterns in both species. We consider genes that switch from upregulation to downregulation or vice versa across species, and those that are strongly differentially regulated in only one species, as suitable candidates for being involved in diel shifts. We explore the evolution of the gene disco, one of these candidates, with known circadian function in flies. We use modelling to predict and compare its protein structure, function and evolutionary conservation in moths and flies.
2. Methods
(a). Insect rearing and sampling design
Moths were reared under natural light–dark cycles at room temperature (25°C). In total, three–five adult males were sampled 2 days after eclosion at the two time points, midday (‘day’) and midnight (electronic supplementary material S1). Moths were decapitated and heads flash-frozen using liquid nitrogen and stored at −80°C.
(b). RNA extraction, library preparation and sequencing
Tissues were homogenized using a bead beater and extracted using a modified Trizol extraction protocol (electronic supplementary material, data15). RNA clean-up was done using a Thermo Scientific RapidOut DNA Removal Kit (Waltham, MA, USA). Samples were shipped overnight to the NERC-NBAF, Liverpool, UK after dehydrating in a biosafety chamber using GenTegraRNA tubes from (GenTegra, CA, USA) and assessed for DNA and RNA yields. We selected samples for library preparation based on RNA quality to ensure equal sampling across treatments. RNA libraries were prepared using the NEB polyA selection and NEBNext Ultra II directional stranded kit (New England Biolabs, MA, USA) suitable for low input yields. Twelve samples were run on one lane of an Illumina NovaSeq using SP chemistry (Paired-end, 2 × 150 bp sequencing).
(c). Read trimming and clean-up
For A. pellucida and D. rubicunda, read trimming was undertaken by the NERC-NBAF core, and the raw Fastq files were trimmed for the presence of Illumina adapter sequences using Cutadapt v. 1.2 [50]. The option ‘-O 3’ was used, so that the 3′ end of any read that matched the adapter sequence for greater than 3 bp was trimmed. Reads were further trimmed using Sickle v. 1.200 [51] with a minimum window quality score of 20. Reads less than 15 bp after trimming were removed.
(d). De novo transcriptomes, transcriptome library sizes, quality control and annotation
Quality control (QC) was conducted on trimmed reads; library size varied for each species (electronic supplementary material, figure S2). We examined expression data and removed genes with transcripts per million (TPM) less than 1. We combined reads from multiple samples and generated several reference de novo transcriptomes of these assemblers, combined them and measured duplication, completeness and redundancy for the different versions (electronic supplementary material, table S2). Further QC and filtering were conducted using BUSCO v. 5.2.0 [52], TransRate v. 1.0.3 [53], QUAST v. 5.02 [54], CD-HIT v. 4.6.8 [55,56] and cascaded clustering with MMseqs2 v. 12 [57]. We used the least duplicated and most complete assemblies; however, we repeated certain analyses with less conservative assemblies (electronic supplementary material, S1). Transcriptomes were annotated with eggNOG-mapper v. 2.1.9 [58] and OrthoFinder v. 2.5.2 [58,59].
(e). Differential gene expression enrichment, orthology and network analysis
Reads were mapped using Salmon v. 0.14.1 [60] and differential gene expression analysis was conducted using EdgeR v. 3.38.1 [61] and DESeq2 v. 1.36.0 [62]; the former was used to normalize and test for significantly expressed genes, and the latter was used to normalize and to generate principal component analyses (PCAs) and other summary statistics. Genes were assigned into orthogroups using OrthoFinder v. 2.5.2 [59,63], which used predicted genes from Bombyx mori, Antheraea pernyi and Antheraea yammai as templates [64–66]. These orthogroups were used to transfer annotations from Bombyx (described later) or to identify pairs of differentially expressed orthologs using orthologous gene pairs that showed significant fold change (FC) in both species pairs. We performed gene enrichment analysis using GO terms with the tools TopGO v. 2.48.0 [67], ReviGo (https://github.com/rajko-horvat) and ShinyGo v. 0.75c [68] (http://bioinformatics.sdstate.edu/goc/). Gene network analysis was undertaken with WGCNA v. 1.71 [69,70].
(f). Gene functional annotation
We functionally annotated genes by cross-referencing them with the B. mori annotations and adding EggNOG annotations. We divided these into functional groups of vision, smell, hearing, circadian, behaviour and brain using GO terms from amigo (http://amigo.geneontology.org/) (electronic supplementary material, table S8).
(g). Gene mining and in silico evolution
We mined genes of interest from moths and insects using well-annotated genomes on Ensembl and NCBI, insectbase [71]. Bombyx mori and A. pernyi were taken from the source papers [64–66]. Two sets of analyses were conducted: (i) 18 Bombycoidea moths and their relatives and (ii) 38 insect genomes (electronic supplementary material, data 11). We assigned a reference protein sequence for each gene of interest from the B. mori-predicted proteome. We used OrthoFinder v. 2.5.2 to identify orthologs [59,63] and filtered orthogroups containing the reference sequence. To ensure a single sequence per species in each orthogroup, custom python scripts filtered the longest and most similar sequences to the reference.
We ran AlphaFold v. 2.1.2 [72] to predict the three-dimensional structures for each reference B. mori sequence. For alignments less than 1500 amino acids and with high conservation scores, calculated using Alistat v. 1.12 [73], we modelled conservation using the webserver of Consurf (https://consurf.tau.ac.il/consurf_index.php) [74–78]. Results were displayed using Jmol first glance viewer (http://firstglance.jmol.org/). Outputs for alpha fold runs and consurf are available as electronic supplementary material data. PyMol v. 2.5.5 [79] was used to align the three-dimensional structures, and Aliview was used to compare alignments. We used InterProScan webserver v. 5.66-98.0 [80,81] (https://www.ebi.ac.uk/interpro/about/interproscan/) and NetPhos v. 3.1 [82,83] for predicting the domains and phosphorylated sites.
3. Results
We compared gene expression across two wild silk moth species, A. pellucida and D. rubicunda (referred to as Anisota and Dryocampa hereon) whose males are diurnal and nocturnal, respectively (figure 1, electronic supplementary material, table S1). We generated head (eyes and brain) transcriptomes from moths collected and flash frozen at midday and midnight, referred to as ‘day’ and ‘night’ hereafter. Using multiple programs to assemble high-quality de novo assemblies (electronic supplementary material, table S2), we characterized the level of gene (mRNA) expression mapping reads to them.
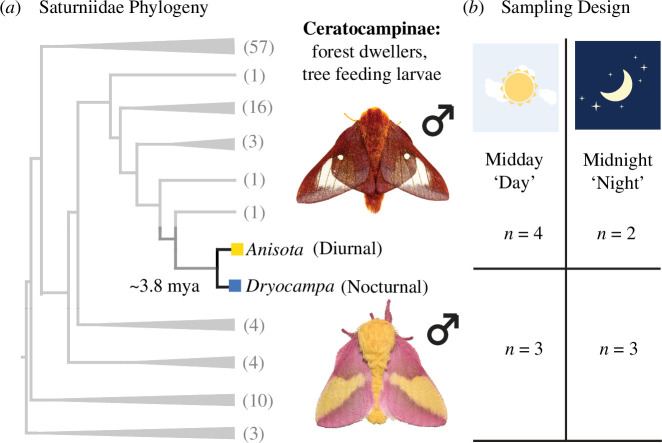
Figure 1.
Nocturnal and diurnal moths on a phylogeny with RNA-Seq sampling design. (a) Collapsed phylogeny of Saturniidae, adapted to show the placement of the two study species, diurnal A. pellucida (pink-striped oakworm moth) and nocturnal D. rubicunda (rosy maple moth). Grey numbers on tips represent the number of described genera in the tree before collapsing. Phylogeny adapted from Rougerie et al. [49]. (b) Sampling design showing the number of replicates sampled for each species and collection period (day/night). Collection of heads was done 2 days post eclosion at midday (sun) and midnight (moon). Photo credits A. pellucida © Mike Chapman; D. rubicunda (Creative Commons) Andy Reacgo and Chrissy Mclearan.
(a). Day–night gene expression patterns switch between nocturnal and diurnal species
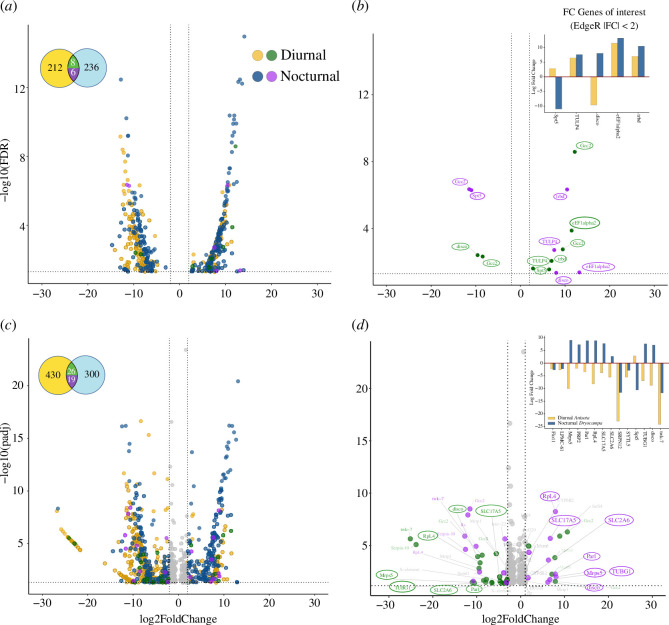
We found 350 and 393 significantly differentially expressed genes (DEGs) when comparing day and night treatments for each species (figure 2a ; electronic supplementary material, table S3,data 1). Anisota had more day-upregulated genes (56%), and Dryocampa was slightly more night-upregulated (53%). To compare DEG sets between species, we mapped our DEGs to B. mori (electronic supplementary material, data S1). Approximately 60% of DEGs from each species had identifiable orthologs in B. mori (electronic supplementary material, table S3), and only a small number of DEGs (6–8 genes) overlapped between both species (figure 2b ). We also replicated this analysis using DESeq2, another differential gene expression (DGE) tool, to ensure that our results were robust to different normalization methods [84]. With DESeq2, we found 498 and 697 DEGs (figure 2c ; electronic supplementary material, table S3, data S2), with similar B. mori annotation rates (61%), although the proportion of day upregulated genes increased considerably in Anisota (79%) compared with being more evenly split in Dryocampa (50%; electronic supplementary material, table S3). The total number of overlapping genes increased (19–26; figure 2d ) when using DESeq2. A comparison of the two methods revealed that 174 and 216 genes were shared between Anisota and Dryocampa, respectively.
Figure 2.
Nocturnal and diurnal species show divergent patterns of gene expression with different analytical approaches (a,c): Left panels display volcano plots illustrating fold change and adjusted p-values for the significant differentially expressed genes between samples collected at midday and midnight for EdgeR (a) and DESeq2 (c). Circles at the upper left corner of (a) and (c) represent the number of genes expressed in both species and colours correspond to FC values for those genes in volcano plots. Yellow and blue represent genes expressed only in the nocturnal or diurnal species, green/purple indicates DEGs present in both species. (b,d) Right panels display only genes expressed in both species for EdgeR (b) and DESeq2 (d). Genes that display opposite trends in expression are highlighted and displayed in the boxplot inset in the upper left corner. Positive fold change indicates night overexpression and negative fold change indicates day overexpression. Genes had false discovery rate (FDR) or adjusted p‐value < 0.05 and −2 < FC < 2. Gene names and annotations are from B. mori.
(b). Divergently expressed genes are linked to brain optic lobe, antennal and neural development
To identify important regulators involved in diel-niche evolution, we applied two filtering criteria to our gene expression data. First, we selected genes that exhibited highly significant differential expression in both species. Second, we focused on genes that displayed upregulation or downregulation patterns consistent with the natural diel activity of each species. Our rationale was that this subset of genes was more likely to contain key candidate regulatory genes. To compare DGE overlap between the two species, we grouped transcripts to their matching orthologs from B. mori; if two transcripts from different species mapped to the same ortholog, we treated them as being the same. This allowed us to examine overlapping genes between the species to see if any genes switched fold-change sign from positive to negative or vice versa (figure 2, electronic supplementary material, data S3). We found 51 overlapping DEG transcripts that mapped to 28 unique B. mori genes. Nine genes showed flipped patterns of expression between the two species, and eight coincided with known diel activity patterns (electronic supplementary material, table S7). Examining gene ontology (GO) annotations and comparing orthologs from flybase (https://flybase.org/), we found genes linked to optic lobe and antennal development (disco), locomotion and energy use (SLC2A6 and SLC17A5), brain and neural development (TUBG1) and other essential biological processes like transcription, ribosomal translation, protein processing, mitochondrial maintenance (RpS4, PARL and Mrps5) and wound response (PRP2) (figure 2b,d ; electronic supplementary material, table S7). Of these, only three, disco, Spt5 and Gcc2, were recovered using both methods.
(c). Gene network analysis identifies diel activity and species-specific co-expressed clusters
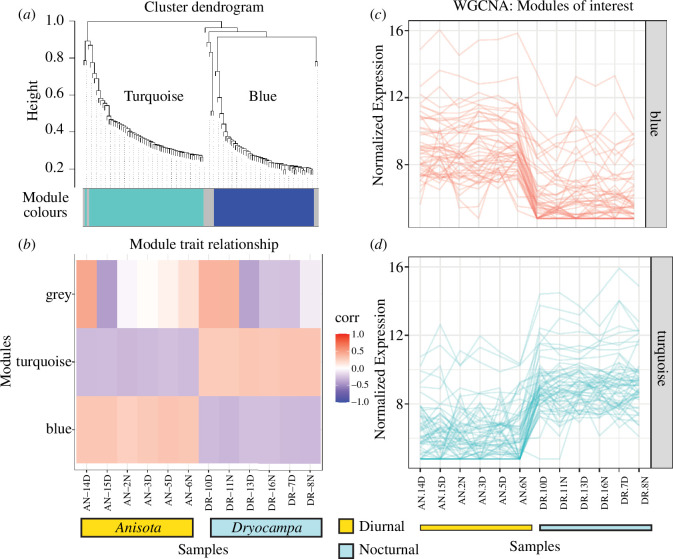
Identifying highly expressed genes helps understand which genes are activated during certain biological processes. However, determining only those that are highly expressed can often overlook genes with important biological functions [85]. We examined co-expressed genes that may be correlated with diel-niche or RNA collection time. We used WGCNA, a weighted correlation network analysis tool to cluster genes together using their normalized counts [70]. Examining co-expression patterns for each species separately, we found one module in each species that clustered with day–night treatment (cluster-grey60 and cluster-tan) (electronic supplementary material, figure S6 and data S4). Since we were interested in species-specific differences, we re-ran analyses and combined counts from Anisota and Dryocampa, using only normalized counts for genes that had valid B. mori annotations for both species resulting in a list of 2000 genes. Among these, we discovered two clusters (cluster-blue and cluster-turquoise) with 50 genes each that exhibited species-specific expression patterns (figure 3).
Figure 3.
Modules of clustered co-expressed genes grouped using normalized expression. (a) Dendrogram showing WGCNA clusters. (b) Patterns of gene expression correlate across samples and modules. (c) Normalized expression for all genes across samples. Two sets of 50 genes (blue) and (turquoise) have clear species-specific expression patterns. Normalization was done with DESeq2 and reads were mapped to the more stringently filtered transcriptome. A soft power analysis was conducted and a power of 9 was used for the WGCNA analysis. AN: Anisota, DR: Dryocampa. D and N represent different samples collected at day and night time points.
(d). Gene ontology enrichment of photoperiodism, circadian control, muscle and neural growth genes
We used a gene enrichment analysis to determine if GO terms were significantly over-represented in the DEG and WGCNA sets compared with the appropriate background of GO terms. Using TopGo, which allows custom gene sets, we found an over-representation of genes involved in several biological functions (electronic supplementary material, data S5). Neural retinal development, such as folic acid serine, glycine and retinoic acid metabolism had potential links to vision (electronic supplementary material, figure S15). We also used ShinyGo to examine WGCNA clusters (electronic supplementary material, data S5). The less duplicated, filtered transcriptomes were annotated using B. mori orthologs (electronic supplementary material, table S4). We examined the enrichment of both tan and grey60 modules, listing the non-redundant terms using ReviGo (electronic supplementary material, figure S7 and data S5). Gene clusters that co-expressed in the same direction together in the day and night treatments of both species included photoperiodism, circadian control, negative phototaxis and nervous system development. Next, we checked for the enrichment of modules that showed species-specific patterns (blue and turquoise; electronic supplementary material, figure S8 and data S5). These included genes involved in muscle proliferation and nerve growth, neural signalling, glycolysis, oxidative stress response and basic cellular functioning such as protein processing and transcriptional regulation.
(e). Day upregulation of vision genes in the diurnal moth
Since both EdgeR and DESeq2 analyses use different normalization methods and statistical model assumptions to calculate fold change between groups [84,86], we repeated enrichment analyses by examining genes that appeared in both analyses. For Anisota, we tested over-enrichment of a smaller subset (FC ≤ −5) of diurnally highly upregulated genes (figure S15, electronic supplementary material, table S5). We found gene enrichment for visual perception, excretion regulation, negative gravitaxis, synaptic plasticity, along with genes associated with other biological processes, such as RNA interference, endopeptidase activity and endocytosis. A reduction in stringency (FC ≤ −2) did not alter results considerably (electronic supplementary material, figure S4). Night-upregulated genes (FC ≥ 2) included ocellary pigment genes, eye-photoreceptor cell development, snRNA processing, post-embryonic development and neurotransmitter secretion, among a host of other processes that may be required for growth, development and metabolism (wnt signalling, tricarboxylic acid cycle and cellular response to insulin, glucose transport; electronic supplementary material, figure S4). We also report upregulation (FC ≤ 0) and downregulation (FC ≤ 0) enrichment analyses (electronic supplementary material, data S5).
(f). Night upregulation of antennal and olfactory brain regions mushroom development genes
We repeated the same analyses for Dryocampa and tested the nocturnally upregulated genes (FC ≥ 5). Our results show upregulation in genes associated with mushroom body development, locomotor rhythm, synaptic growth, energy utilization (sialin transport) and mitochondrial translation (figure S15, electronic supplementary material, table S6). A reduction in stringency (|FC| ≥ 2) showed entrainment of the clock cycle and antennal development genes. Genes associated with innate immune response, DNA repair, cell division, histone acetylation, circadian rhythm and retinoid cycle were upregulated during the day, possibly indicating a period of cellular repair during a time when these moths are inactive (electronic supplementary material, figure S5). We also report upregulation (FC ≤ 0) and downregulation (FC ≤ 0) enrichment analyses (electronic supplementary material, data S5).
(g). Sensory, circadian, eye development and behavioural genes show up in gene network analyses
We combined results from the DEG (EdgeR and DESeq2) and gene network analyses (WGCNA) to create a cumulative list of 1700 transcripts (electronic supplementary material, data S6). Focusing on genes that were recovered across Anisota and Dryocampa reduced the set to 274 transcripts (electronic supplementary material, data S7). Because many transcripts had poorly annotated B. mori hits, we improved annotations using the program eggNOG-mapper [58]. We tested if these genes had GO terms associated with sensory, circadian, brain and neural development or behavioural regulatory genes (electronic supplementary material, table S7 and data S8). We found that several genes in each category had GO terms associated with vision and brain development (electronic supplementary material, figure S13 and data S9).
(h). Expanded genes of interest set identified using a differentially expressed ortholog approach
Since B. mori annotations might miss certain genes, we used OrthoFinder to identify orthologs from the de novo transcriptomes of Anisota and Dryocampa. We used the differentially expressed transcript list from the DESeq2 analyses for each species to find if any transcript had a corresponding ortholog in the other species with differential expression. Thereby identifying pairs of differentially expressed transcripts and their respective fold change values (electronic supplementary material, figure S14). Many of these shared orthologs from the B. mori sets; but they also identified 10 additional orthologs (electronic supplementary material, data S6).
(i). Predicted functional regions and homology patterns identified for genes of interest
We examined protein and gene evolution for a set of genes which we found were of interest based on results from DGE, WGCNA and GO annotations (figure 4; electronic supplementary material, data S10). We obtained orthologs from publicly available Bombycoidea moth genomes, choosing representative species with the highest identity sequence relative to the B. mori reference. We modelled the three-dimensional structure of B. mori proteins and mapped the evolutionary conservation onto the three-dimensional predicted structure for proteins above a certain conservation threshold (electronic supplementary material, figure S13 and data S12). These analyses predict structurally and functionally conserved regions of proteins (electronic supplementary material, figure S9 and data 13). We repeated this analysis with 38 insect genomes (electronic supplementary material, data 11) and mapped evolutionary conservation onto three-dimensional protein structure (electronic supplementary material, data S12 and S13). We include results of evolutionary conservation analyses for two regulatory candidates (disco and tk) that showed varying levels of sequence and protein evolution between insects and moths (electronic supplementary material, figure S10).
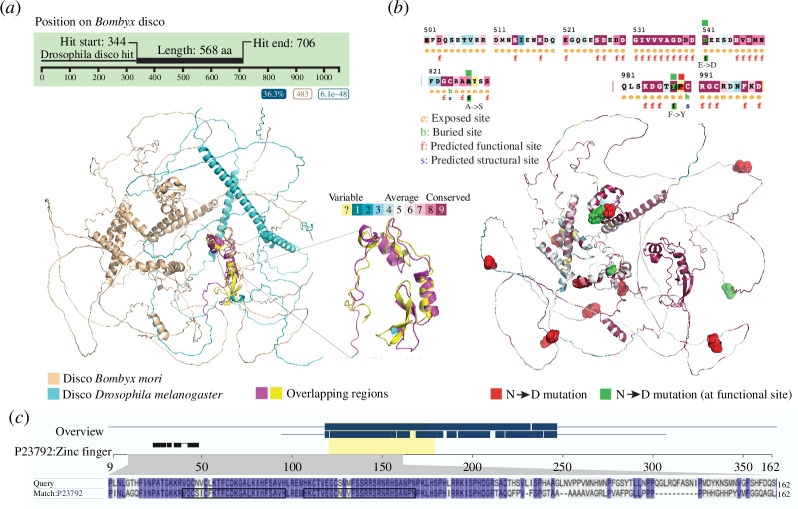
Figure 4.
Three-dimensional conservation and domain distribution of disco across Bombyx mori (1054 aa) and Drosophila melanogaster (568 aa). (a) Top: disco best Uniprot hit in Drosophila using default settings (blastp, e-threshold 10, Auto-Blosum62). Bottom: disco aligned and superimposed predicted structures for B. mori and D. melanogaster. Peach colour: B. mori alpha-fold predicted structure for disco, Cyan: D. melanogaster alpha-fold predicted structure for disco. (b) Top: partial views of the Consurf predicted residues for disco. Views encompassing the region where 3/14 mutated sites between nocturnal Dryocampa and diurnal Anisota overlap with predicted functional residues. Bottom: residues that were mutated between the nocturnal and diurnal species are highlighted on an overlay of the Consurf scores mapped onto the predicted alpha fold structure of disco from B. mori. Green residues indicate mutated and predicted functional sites, red indicates mutated sites that did not have a predicted residue. (c) Overlap of the highly conserved region of disco, this region includes the zinc-finger domain that is characteristic of the disco transcription factor.
(j). Modelling predicts additional functional zinc finger domains for disco in Lepidoptera
The gene disco was recovered across multiple analyses. In D. melanogaster, disco is known for its role in eye development, circadian maintenance, clock neuron development and appendage formation [87–91]. To determine if disco was conserved between moths and D. melanogaster, we compared the primary sequence and three-dimensional protein structure of B. mori and D. melanogaster. The sequence length of disco in B. mori was nearly double that of D. melanogaster (figure 4a ). However, a region spanning over 100 amino acids was highly conserved, contained the zinc-finger domain important for its function and showed strong three-dimensional structural conservation measured by various structural similarity metrics (whole protein alignment: RMSD: align = 37.833, super = 2.566, MatchAlign score: align = 540 (2234 atoms), super = 333.1 (554 atoms) versus alignment of conserved region: RMSD: align = 2.699, super = 2.566, MatchAlignScore: align = 443 (671 atoms), 351 (615) atoms). These results indicate that the DNA binding domain of disco has probably been conserved. However, an additional approximately 500 amino acid region absent in D. melanogaster is highly conserved across moths and includes several regions predicted to be functional (figure 4a , electronic supplementary material, figure S10). We hypothesize that disco has functions in moths that it may not play in flies. To further test this hypothesis, we compared disco sequences across Anisota and Dryocampa finding 23 mutations, 3 of which mapped to the predicted functional region (figure 4b , electronic supplementary material, data S14). InterProScan predicted four zinc-finger domains, three in this region, although the CATH-Gene3D databases prediction combined the two separate domains into a single predicted domain (electronic supplementary material, figure S11). We also found 53 sites with predicted phosphorylation potential, with roughly a third of these (18 sites) around the second zinc-finger domain. Reducing the stringency from 0.9 to 0.5 increased the total to 142 sites, which were still enriched around the second domain.
4. Discussion
We examined gene expression in the heads of two closely related moths with different activity times: the diurnal A. pellucida and the nocturnal D. rubicunda. We found 300–700 genes with different expression levels between day and night. Diurnal moths upregulated genes associated with vision while nocturnal moths upregulated genes linked to olfaction and locomotion. Notably, the gene disconnected (disco) showed differential expression in both species even accounting for the different models and parameters that were implicated in vision, circadian, hearing, locomotion and brain development in vinegar flies [88,91,92]. Interestingly, disco in moths has a larger size and potentially more functional domains compared with its counterpart in fruit flies, suggesting a broader role in moth biology. Several point mutations between Anisota and Dryocampa disco also mapped to these domains, highlighting the need to further study disco.
(a). Vision and olfaction
Visual systems often accompany diel and photic environment shifts [93], many of these changes are morphological, for example, nocturnal carpenter bees have much larger facets in their eyes than their diurnal counterparts [94], with poorly characterized single genes. Colour vision genes opsin are known to have diel-niche-linked evolution [42,95,96]. However, we found no diel-expression patterns in colour vision opsins, a result corroborated by a recent study [95]. However, we discovered a cerebral opsin (ceropsin), implicated in photoperiodism [97] upregulated during the day.
We also found several eye development genes (ANKRD17, EHD4, JAK2 and TENM2), phototransduction genes (PPAP2, RDH11), and retina homeostasis, eye-antennal disc development and photoreceptor cell maintenance genes (disco, glass) [98]. Surprisingly a few visual genes, such as garnet and rugose also appeared to have different isoforms present, showing both day and night upregulation. Garnet is an eye-colour mutant gene in flies [99], and rugose is implicated in retinal pattern formation [100].
There is evidence for increased investment in olfaction in dim light, with larger mushroom bodies in nocturnal species [93,101,102]. We also found several differently expressed olfactory genes, including those involved in odorant binding (Obp84a, Obp58b), pheromone response (tk), mushroom body development (DAAM2, DST) and antennal development (disco).
(b). Brain and neural rewiring
We found an upregulation of neural and brain development genes that are also implicated in adult brain plasticity in Lepidoptera and other insects [103–105]. These include genes linked to axon regeneration (APOD), central complex development (ALDH3A2, DST, OGT, Ten-a and TENM2), central nervous system development (disco, RpL4) and neuropeptide hormonal activity (tk). We speculate that plasticity could occur through neural wiring shaping sensory adaptation. Many Lepidoptera show seasonal plasticity in foraging preferences [106], and some override innate preferences for novel visual and olfactory cues after eclosion [107]. Diel-niche and circadian rhythms may also show plasticity, such as Hyles lineata showing relatively labile diel-niches possibly driven by temperature and resources [108–110].
(c). Circadian and behavioural regulators
We found differential expression of genes involved in locomotion (KCTD15, Tk, unc-22) and circadian or rhythmic behaviour (disco, JAK2, OGT and SREBF1/SREBF2) in both species. We also found several key clock genes such as per and tim, although they were downregulated only in Dryocampa. Clock-like (also called takeout), another gene under circadian control [111], was expressed in both species, although without any significant upregulation or downregulation. Clock-like was moderately conserved in the moths we examined but recovered fewer orthologs across the insects. In D. melanogaster, its closest homologs, Jhbp5 and takeout, had only 21–25% sequence identity (electronic supplementary material, data S10 and S14). Despite this, its three-dimensional structure was highly conserved (electronic supplementary material, data S14 and figure S12), indicating likely functional convergence.
(d). Disco as candidate diel-niche gene in adult moths
To identify candidates that might be key regulators of diel-niche, we searched for genes that were (i) expressed in both species, (ii) showed coincident expression patterns with respect to diel-niche, and (iii) played a role in sensory and circadian control. Only disconnected (disco) fit all criteria.
Disco was first described as a locus required for proper optic lobe formation in Drosophila melanogaster [112] but is implicated in the disruption of circadian rhythms [87,88,113], potentially through its role in the development of neurosecretory cells in the fly brain [114]. Wild-type flies show a bimodal, crepuscular activity pattern entrained to the external light–dark cycle, which becomes free-running in the absence of the entrainment cue (such as in constant darkness). Disco mutants show diurnal entrainment to a light–dark cycle but become arrhythmic in constant darkness [88,114], which appears to be owing to the loss of a set of PDF neurons, from disco mutant brains [115,116]. Pdf mutants show reduced morning activity bouts in light–dark conditions and, similar to disco mutants, become arrhythmic in constant darkness [117,118]. Importantly, these clusters of PDF-expressing cells appear conserved across many insect groups, having been identified in locusts, crickets, stick insects and cockroaches [119,120]. There are no previous studies on disco specifically in moths, but publicly available bulk RNA-Seq data from B. mori and Manduca sexta show disco expression in adult in heads and antennae with high larval but minimal pupal expression [65,121]. However, single cell-expression analyses, antibody staining and mRNA in situ hybridization are needed to determine if disco is expressed in or required for the development of clock neurons in Lepidoptera [122].
The disco gene encodes the acid zinc-finger transcription factor, 500 amino acids in length in D. melanogaster and 1000 in B. mori. Our modelling showed that an approximately 150 amino acid region is conserved structurally at the sequence level, and this region also contains zinc-finger motifs associated with DNA binding [123]. This indicates that disco probably has retained its DNA binding and pupal and appendage patterning function in moths. We predicted the functional regions of disco in B. mori based on evolutionary conservation modelling and found that an additional 500 amino acids that were absent in D. melanogaster are predicted to be functional (conserved and exposed). Domain- and family-level modelling predicted at least two additional zinc-finger domains in this region (electronic supplementary material, figures S10 and S11). We also found many phosphorylated sites surrounding the zinc-finger domains. Examining mutations between Anisota and Dryocampa disco revealed three mutations mapped to these predicted functional regions (figure 5b ). Given disco’s adult diel-specific expression in moths, its additional zinc-finger DNA binding domains and the high number of phosphorylated sites, we believe that it may be a candidate gene for diel-regulation in adult moths. we also identify other candidate genes for diel-niche evolution in Lepidoptera, which can be useful targets for further exploration in other diel-species pairs as well as functional validation using more direct experimental techniques.
Acknowledgements
We thank Jesse Breinholt, Martijn Timmermans and Andreas Zwick for help with project conceptualization. Kelly Dexter, Amanda Markee and members of the McGuire Center for Lepidoptera and Biodiversity at the University of Florida assisted with animal care and wet-lab troubleshooting. We thank Danielle DeLeo, Heather Bracken-Grissom, Zhou Lei, Jorge Perez-Moreno and Belinda Pinto for discussions, advice and assistance with bioinformatic pipelines and wet-lab protocols. Nicolas Alexandre, Sachit Daniel, Riddhi Deshmukh, David Plotkin, Nitin Ravikanthachari and Chelsea Skojec provided comments and feedback on the manuscript. Yu Fahong, Jessica Liberles, Janelle Nunez-Castilla, Raghavan Venket, Yi-Ming Weng and Xiaokang Zhang assisted with troubleshooting bioinformatic pipelines and provided analytical recommendations. The authors acknowledge the University of Florida Research Computing for providing computational resources and support that have contributed to the research results reported in this publication (http://researchcomputing.ufl.edu). We acknowledge NBAF and UF-ICBR for assistance with sequencing and generating libraries. This is publication #1716 from the Institute of Environment at Florida International University.
Contributor Information
Yash Sondhi, Email: yashsondhi@gmail.com.
Anthony J. Bellantuono, Email: anthjbell@gmail.com.
Caroline G. Storer, Email: cgstorer@gmail.com.
Scott D. Cinel, Email: cinel1@ufl.edu.
R. Keating Godfrey, Email: rkgodfrey@floridamuseum.ufl.edu.
Andrew J. Mongue, Email: andrew.mongue@ufl.edu.
Yi-Ming Weng, Email: weng76111@gmail.com.
Deborah Glass, Email: deborah_glass@hotmail.co.uk.
Ryan A. St Laurent, Email: thesaint913@gmail.com.
Chris A. Hamilton, Email: hamiltonlab@uidaho.edu.
Chandra Earl, Email: sunray1@ufl.edu.
Colin J. Brislawn, Email: cbrisl@gmail.com.
Ian J. Kitching, Email: i.kitching@nhm.ac.uk.
Seth M. Bybee, Email: seth.bybee@gmail.com.
Jamie C. Theobald, Email: theobald@fiu.edu.
Akito Y. Kawahara, Email: kawahara@flmnh.ufl.edu.
Ethics
This work did not require ethical approval from a human subject or animal welfare committee.
Data accessibility
The transcriptome libraries are also archived on GenBank under BioProject PRJNA1102514, and all associated sequence data are archived on the SRA database (SRR28778930-SRR28778945). Datasets S1 to S17: Available at: 10.6084/m9.figshare.23661603.
Supplementary material is available online [124].
Declaration of AI use
ChatGPT 4 and Github Co-pilot were used to assist with creating the python code for data visualization for electronic supplementary material, figure S14.
Authors’ contributions
Y.S.: conceptualization, data curation, formal analysis, investigation, methodology, project administration, software, visualization, writing—original draft, writing—review and editing; R.L.M.: data curation, investigation, methodology, project administration, writing—review and editing; A.J.B.: conceptualization, writing—review and editing; C.G.S.: conceptualization, data curation, methodology, project administration, supervision; S.D.C.: formal analysis, software; R.K.G.: software, visualization, writing—review and editing; A.J.M.: formal analysis, validation, writing—review and editing; Y.-M.W.: conceptualization, data curation, formal analysis, software; D.G.: data curation, investigation, project administration; R.A.S.L.: conceptualization, investigation, methodology, project administration, writing—original draft, writing—review and editing; C.A.H.: conceptualization, data curation, investigation, methodology, project administration, writing—review and editing; C.E.: data curation, formal analysis, software, visualization, writing—original draft, writing—review and editing; C.J.B.: software, validation, writing—review and editing; I.J.K.: conceptualization, funding acquisition, methodology, resources, writing—review and editing; S.M.B.: funding acquisition, methodology, supervision, validation, writing—review and editing; J.C.T.: conceptualization, funding acquisition, supervision, writing—review and editing; A.Y.K.: conceptualization, funding acquisition, methodology, resources, supervision, visualization, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
Funding for the project was through the National Science Foundation (NSF DEB-1557007, NSF PRFB-1612862, NSF IOS-1750833), the National Environment Research Council NERC (NE/P003915/1), (NSF IOS-1750833 to J.C.T.) and the U.S. Air Force Office of Scientific Research (AFOSR MURI award FA9550-22-1-0315). The Florida International University Presidential fellowship supported Y.S.
References
- 1. Kronfeld-Schor N, Dayan T. 2003. Partitioning of time as an ecological resource. Annu. Rev. Ecol. Evol. Syst. 34 , 153–181. ( 10.1146/annurev.ecolsys.34.011802.132435) [DOI] [Google Scholar]
- 2. Somanathan H, Krishna S, Jos EM, Gowda V, Kelber A, Borges RM. 2020. Nocturnal bees feed on diurnal leftovers and pay the price of day – night lifestyle transition. Front. Ecol. Evol. 8 . ( 10.3389/fevo.2020.566964) [DOI] [Google Scholar]
- 3. Bloch G, Bar-Shai N, Cytter Y, Green R. 2017. Time is honey: circadian clocks of bees and flowers and how their interactions may influence ecological communities. Phil. Trans. R. Soc. B 372 , 20160256. ( 10.1098/rstb.2016.0256) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Palmer MS, Gaynor KM, Becker JA, Abraham JO, Mumma MA, Pringle RM. 2022. Dynamic landscapes of fear: understanding spatiotemporal risk. Trends Ecol. Evol. 37 , 911–925. ( 10.1016/j.tree.2022.06.007) [DOI] [PubMed] [Google Scholar]
- 5. Fullard JH, Napoleone N. 2001. Diel flight periodicity and the evolution of auditory defences in the Macrolepidoptera. Anim. Behav. 62 , 349–368. ( 10.1006/anbe.2001.1753) [DOI] [Google Scholar]
- 6. van der Vinne V, Tachinardi P, Riede SJ, Akkerman J, Scheepe J, Daan S, Hut RA. 2019. Maximising survival by shifting the daily timing of activity. Ecol. Lett. 22 , 2097–2102. ( 10.1111/ele.13404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Székely D, Cogălniceanu D, Székely P, Denoël M. 2020. Adult-juvenile interactions and temporal niche partitioning between life-stages in a tropical amphibian. PLoS One 15 , e0238949. ( 10.1371/journal.pone.0238949) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Daan S. 1981. Adaptive daily strategies in behavior. In Biological rhythms (ed. Aschoff J), pp. 275–298. Boston, MA: Springer US. ( 10.1007/978-1-4615-6552-9_15) [DOI] [Google Scholar]
- 9. Wang D, Yang G, Chen W. 2021. Diel and circadian patterns of locomotor activity in the adults of diamondback moth (Plutella xylostella). Insects 12 , 727. ( 10.3390/insects12080727) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Niepoth N, Ke G, de Roode JC, Groot AT. 2018. Comparing behavior and clock gene expression between caterpillars, butterflies, and moths. J. Biol. Rhythms 33 , 52–64. ( 10.1177/0748730417746458) [DOI] [PubMed] [Google Scholar]
- 11. Groot AT. 2014. Circadian rhythms of sexual activities in moths: a review. Front. Ecol. Evol. 2 . ( 10.3389/fevo.2014.00043) [DOI] [Google Scholar]
- 12. Dubowy C, Sehgal A. 2017. Circadian rhythms and sleep in Drosophila melanogaster. Genetics 205 , 1373–1397. ( 10.1534/genetics.115.185157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma D, Przybylski D, Abruzzi KC, Schlichting M, Li Q, Long X, Rosbash M. 2021. A transcriptomic taxonomy of Drosophila circadian neurons around the clock. Elife 10 , e63056. ( 10.7554/eLife.63056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hardin PE. 2005. The circadian timekeeping system of Drosophila . Curr. Biol. 15 , R714–22. ( 10.1016/j.cub.2005.08.019) [DOI] [PubMed] [Google Scholar]
- 15. Konopka RJ, Benzer S. 1971. Clock mutants of Drosophila melanogaster. Proc. Natl Acad. Sci. USA 68 , 2112–2116. ( 10.1073/pnas.68.9.2112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hardin PE, Hall JC, Rosbash M. 1990. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343 , 536–540. ( 10.1038/343536a0) [DOI] [PubMed] [Google Scholar]
- 17. Bargiello TA, Jackson FR, Young MW. 1984. Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature 312 , 752–754. ( 10.1038/312752a0) [DOI] [PubMed] [Google Scholar]
- 18. Yu W, Hardin PE. 2006. Circadian oscillators of Drosophila and mammals. J. Cell. Sci. 119 , 4793–4795. ( 10.1242/jcs.03174) [DOI] [PubMed] [Google Scholar]
- 19. Engelen E, Janssens RC, Yagita K, Smits VAJ, van der Horst GTJ, Tamanini F. 2013. Mammalian TIMELESS is involved in period determination and DNA damage-dependent phase advancing of the circadian clock. PLoS One 8 , e56623. ( 10.1371/journal.pone.0056623) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patop IL, Anduaga AM, Bussi IL, Ceriani MF, Kadener S. 2023. Organismal landscape of clock cells and circadian gene expression in Drosophila. bioRxiv. 2023.05.23.542009. ( 10.1101/2023.05.23.542009) [DOI]
- 21. Zeng H, Qian Z, Myers MP, Rosbash M. 1996. A light-entrainment mechanism for the Drosophila circadian clock. Nature 380 , 129–135. ( 10.1038/380129a0) [DOI] [PubMed] [Google Scholar]
- 22. Glaser FT, Stanewsky R. 2005. Temperature synchronization of the Drosophila circadian clock. Curr. Biol. 15 , 1352–1363. ( 10.1016/j.cub.2005.06.056) [DOI] [PubMed] [Google Scholar]
- 23. Miyasako Y, Umezaki Y, Tomioka K. 2007. Separate sets of cerebral clock neurons are responsible for light and temperature entrainment of Drosophila circadian locomotor rhythms. J. Biol. Rhythms 22 , 115–126. ( 10.1177/0748730407299344) [DOI] [PubMed] [Google Scholar]
- 24. Miyazaki Y, Watari Y, Tanaka K, Goto SG. 2016. Temperature cycle amplitude alters the adult eclosion time and expression pattern of the circadian clock gene period in the onion fly. J. Insect Physiol. 86 , 54–59. ( 10.1016/j.jinsphys.2016.01.002) [DOI] [PubMed] [Google Scholar]
- 25. Ma D, Herndon N, Le JQ, Abruzzi KC, Zinn K, Rosbash M. 2023. Neural connectivity molecules best identify the heterogeneous clock and dopaminergic cell types in the Drosophila adult brain. Sci. Adv. 9 , eade8500. ( 10.1126/sciadv.ade8500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reinhard N, et al. 2023. Synaptic and peptidergic connectomes of the Drosophila circadian clock. bioRxiv. ( 10.1101/2023.09.11.557222) [DOI]
- 27. Helfrich-Förster C. 2020. Light input pathways to the circadian clock of insects with an emphasis on the fruit fly Drosophila melanogaster. J. Comp. Physiol. A Neuroethol. Sens. Neural. Behav. Physiol. 206 , 259–272. ( 10.1007/s00359-019-01379-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Senthilan PR, Grebler R, Reinhard N, Rieger D, Helfrich-Förster C. 2019. Role of rhodopsins as circadian photoreceptors in the Drosophila melanogaster. Biology 8 . ( 10.3390/biology8010006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hermann C, Saccon R, Senthilan PR, Domnik L, Dircksen H, Yoshii T, Helfrich-Förster C. 2013. The circadian clock network in the brain of different Drosophila species. J. Comp. Neurol. 521 , 367–388. ( 10.1002/cne.23178) [DOI] [PubMed] [Google Scholar]
- 30. Xiao Y, Yuan Y, Jimenez M, Soni N, Yadlapalli S. 2021. Clock proteins regulate spatiotemporal organization of clock genes to control circadian rhythms. Proc. Natl Acad. Sci. USA 118 . ( 10.1073/pnas.2019756118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Beer K, Helfrich-Förster C. 2020. Model and non-model insects in chronobiology. Front. Behav. Neurosci. 14 , 601676. ( 10.3389/fnbeh.2020.601676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sandrelli F, Costa R, Kyriacou CP, Rosato E. 2008. Comparative analysis of circadian clock genes in insects. Insect Mol. Biol. 17 , 447–463. ( 10.1111/j.1365-2583.2008.00832.x) [DOI] [PubMed] [Google Scholar]
- 33. Foster RG, Hughes S, Peirson SN. 2020. Circadian photoentrainment in mice and humans. Biology 9 , 180. ( 10.3390/biology9070180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Závodská R, Sauman I, Sehnal F. 2003. Distribution of PER protein, pigment-dispersing hormone, prothoracicotropic hormone, and eclosion hormone in the cephalic nervous system of insects. J. Biol. Rhythms 18 , 106–122. ( 10.1177/0748730403251711) [DOI] [PubMed] [Google Scholar]
- 35. Wager-Smith K, Kay SA. 2000. Circadian rhythm genetics: from flies to mice to humans. Nat. Genet. 26 , 23–27. ( 10.1038/79134) [DOI] [PubMed] [Google Scholar]
- 36. Kawahara AY, Plotkin D, Hamilton CA, Gough H, St Laurent R, Owens HL, Homziak NT, Barber JR. 2018. Diel behavior in moths and butterflies: a synthesis of data Illuminates the evolution of temporal activity. Org. Divers. Evol. 18 , 13–27. ( 10.1007/s13127-017-0350-6) [DOI] [Google Scholar]
- 37. Hamilton CA, St Laurent RA, Dexter K, Kitching IJ, Breinholt JW, Zwick A, Timmermans M, Barber JR, Kawahara AY. 2019. Phylogenomics resolves major relationships and reveals significant diversification rate shifts in the evolution of silk moths and relatives. BMC Evol. Biol. ( 10.1186/s12862-019-1505-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kobelková A, Závodská R, Sauman I, Bazalová O, Dolezel D. 2015. Expression of clock genes period and timeless in the central nervous system of the Mediterranean flour moth, Ephestia kuehniella. J. Biol. Rhythms 30 , 104–116. ( 10.1177/0748730414568430) [DOI] [PubMed] [Google Scholar]
- 39. Brady D, Saviane A, Cappellozza S, Sandrelli F. 2021. The circadian clock in Lepidoptera. Front. Physiol. 12 , 776826. ( 10.3389/fphys.2021.776826) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Iwai S, Fukui Y, Fujiwara Y, Takeda M. 2006. Structure and expressions of two circadian clock genes, period and timeless in the commercial silkmoth, Bombyx mori. J. Insect Physiol. 52 , 625–637. ( 10.1016/j.jinsphys.2006.03.001) [DOI] [PubMed] [Google Scholar]
- 41. Macias-Muñoz A, Rangel Olguin AG, Briscoe AD. 2019. Evolution of phototransduction genes in Lepidoptera. Genome Biol. Evol. 11 , 2107–2124. ( 10.1093/gbe/evz150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sondhi Y, Ellis EA, Bybee SM, Theobald JC, Kawahara AY. 2021. Light environment drives evolution of color vision genes in butterflies and moths. Commun. Biol. 4 , 177. ( 10.1038/s42003-021-01688-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jiang XC, et al. 2021. Identification of olfactory genes from the greater wax moth by antennal transcriptome analysis. Front. Physiol. 12 , 663040. ( 10.3389/fphys.2021.663040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. de Fouchier A, et al. 2017. Functional evolution of Lepidoptera olfactory receptors revealed by deorphanization of a moth repertoire. Nat. Commun. 8 , 15709. ( 10.1038/ncomms15709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Truman JW, Riddiford LM. 1970. Neuroendocrine control of ecdysis in silkmoths. Science 167 , 1624–1626. ( 10.1126/science.167.3925.1624) [DOI] [PubMed] [Google Scholar]
- 46. Sauman I, Reppert SM. 1996. Circadian clock neurons in the silkmoth Antheraea pernyi: novel mechanisms of period protein regulation. Neuron 17 , 889–900. ( 10.1016/s0896-6273(00)80220-2) [DOI] [PubMed] [Google Scholar]
- 47. Helm B, Visser ME, Schwartz W, Kronfeld-Schor N, Gerkema M, Piersma T, Bloch G. 2017. Two sides of a coin: ecological and chronobiological perspectives of timing in the wild. Phil. Trans. R. Soc. B 372 , 20160246. ( 10.1098/rstb.2016.0246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kyriacou CP, Peixoto AA, Sandrelli F, Costa R, Tauber E. 2008. Clines in clock genes: fine-tuning circadian rhythms to the environment. Trends Genet. 24 , 124–132. ( 10.1016/j.tig.2007.12.003) [DOI] [PubMed] [Google Scholar]
- 49. Rougerie R, et al. 2022. Phylogenomics Illuminates the evolutionary history of wild silkmoths in space and time (Lepidoptera: Saturniidae). bioRxiv. ( 10.1101/2022.03.29.486224) [DOI]
- 50. Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.j 17 , 10. ( 10.14806/ej.17.1.200) [DOI] [Google Scholar]
- 51. Joshi NA, Fass JN. 2011. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ Files. See https://github.com/najoshi/sickle.
- 52. Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 38 , 4647–4654. ( 10.1093/molbev/msab199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Smith-Unna R, Boursnell C, Patro R, Hibberd JM, Kelly S. 2016. Transrate: reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 26 , 1134–1144. ( 10.1101/gr.196469.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mikheenko A, Prjibelski A, Saveliev V, Antipov D, Gurevich A. 2018. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 34 , i142–i150. ( 10.1093/bioinformatics/bty266) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22 , 1658–1659. ( 10.1093/bioinformatics/btl158) [DOI] [PubMed] [Google Scholar]
- 56. Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28 , 3150–3152. ( 10.1093/bioinformatics/bts565) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Steinegger M, Söding J. 2017. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data SETS. Nat. Biotechnol. 35 , 1026–1028. ( 10.1038/nbt.3988) [DOI] [PubMed] [Google Scholar]
- 58. Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. 2021. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38 , 5825–5829. ( 10.1093/molbev/msab293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20 , 238. ( 10.1186/s13059-019-1832-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14 , 417–419. ( 10.1038/nmeth.4197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 , 139–140. ( 10.1093/bioinformatics/btp616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-Seq data with DEseq2. Genome Biol. 15 , 550. ( 10.1186/s13059-014-0550-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16 , 157. ( 10.1186/s13059-015-0721-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Duan J, et al. 2020. A chromosome-scale genome assembly of Antheraea pernyi (Saturniidae, Lepidoptera). Mol. Ecol. Resour. 20 , 1372–1383. ( 10.1111/1755-0998.13199) [DOI] [PubMed] [Google Scholar]
- 65. Lu F, Wei Z, Luo Y, Guo H, Zhang G, Xia Q, Wang Y. 2020. SilkDB 3.0: visualizing and exploring multiple levels of data for silkworm. Nucleic Acids Res. 48 , D749–D755. ( 10.1093/nar/gkz919) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kim SR, et al. 2018. Genome sequence of the Japanese oak silk moth, Antheraea yamamai: the first draft genome in the family Saturniidae. Gigascience 7 , 1–11. ( 10.1093/gigascience/gix113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Alexa R, Rahnenfuhrer J. 2010. TopGO: enrichment analysis for gene ontology. R package version 2(0) See https://bioconductor.riken.jp/packages/3.0/bioc/manuals/topGO/man/topGO.pdf.
- 68. Ge SX, Jung D, Yao R. 2020. Shinygo: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36 , 2628–2629. ( 10.1093/bioinformatics/btz931) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Langfelder P, Horvath S. 2012. Fast R functions for robust correlations and hierarchical clustering. J. Stat. Softw. 46 . ( 10.18637/jss.v046.i11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 9 , 559. ( 10.1186/1471-2105-9-559) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mei Y, et al. 2022. Insectbase 2.0: a comprehensive gene resource for insects. Nucleic Acids Res. 50 , D1040–D1045. ( 10.1093/nar/gkab1090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jumper J, et al. 2021. Highly accurate protein structure prediction with alphafold. Nature 596 , 583–589. ( 10.1038/s41586-021-03819-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wong TKF, Kalyaanamoorthy S, Meusemann K, Yeates DK, Misof B, Jermiin LS. 2020. A minimum reporting standard for multiple sequence alignments. NAR Genom. Bioinform. 2 , lqaa024. ( 10.1093/nargab/lqaa024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yariv B, Yariv E, Kessel A, Masrati G, Chorin AB, Martz E, Mayrose I, Pupko T, Ben-Tal N. 2023. Using evolutionary data to make sense of macromolecules with a “face-lifted” Consurf. Protein Sci. 32 , e4582. ( 10.1002/pro.4582) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N. 2016. Consurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44 , W344–W350. ( 10.1093/nar/gkw408) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Celniker G, Nimrod G, Ashkenazy H, Glaser F, Martz E, Mayrose I, Pupko T, Ben‐Tal N. 2013. Consurf: using evolutionary data to raise testable hypotheses about protein function. Isr. J. Chem. 53 , 199–206. ( 10.1002/ijch.201200096) [DOI] [Google Scholar]
- 77. Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. 2010. Consurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38 , W529–W533. ( 10.1093/nar/gkq399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. 2005. Consurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33 , W299–302. ( 10.1093/nar/gki370) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schrödinger LLC. 2015. The PyMOL molecular graphics system . Version 1.8.
- 80. Paysan-Lafosse T, et al. 2023. Interpro in 2022. Nucleic Acids Res. 51 , D418–D427. ( 10.1093/nar/gkac993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jones P, et al. 2014. Interproscan 5: genome-scale protein function classification. Bioinformatics 30 , 1236–1240. ( 10.1093/bioinformatics/btu031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Blom N, Gammeltoft S, Brunak S. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294 , 1351–1362. ( 10.1006/jmbi.1999.3310) [DOI] [PubMed] [Google Scholar]
- 83. Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S. 2004. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4 , 1633–1649. ( 10.1002/pmic.200300771) [DOI] [PubMed] [Google Scholar]
- 84. Li D, Zand MS, Dye TD, Goniewicz ML, Rahman I, Xie Z. 2022. An evaluation of RNA-Seq differential analysis methods. PLoS One 17 , e0264246. ( 10.1371/journal.pone.0264246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sánchez-Baizán N, Ribas L, Piferrer F. 2022. Improved biomarker discovery through a plot twist in transcriptomic data analysis. BMC Biol. 20 , 208. ( 10.1186/s12915-022-01398-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Press LL. 2016. A survey of best practices for RNA-seq data analysis. CreateSpace Independent Publishing Platform. See https://genomebiology.biomedcentral.com/articles/10.1186/s13059-016-0881-8. [Google Scholar]
- 87. Hardin PE, Hall JC, Rosbash M. 1992. Behavioral and molecular analyses suggest that circadian output is disrupted by disconnected mutants in D. melanogaster. EMBO J. 11 , 1–6. ( 10.1002/j.1460-2075.1992.tb05020.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Dushay MS, Rosbash M, Hall JC. 1989. The disconnected visual system mutations in Drosophila melanogaster drastically disrupt circadian rhythms. J. Biol. Rhythms 4 , 1–27. ( 10.1177/074873048900400101) [DOI] [PubMed] [Google Scholar]
- 89. Lee KJ, Mukhopadhyay M, Pelka P, Campos AR, Steller H. 1999. Autoregulation of the Drosophila disconnected gene in the developing visual system. Dev. Biol. 214 , 385–398. ( 10.1006/dbio.1999.9420) [DOI] [PubMed] [Google Scholar]
- 90. Suri V, Qian Z, Hall JC, Rosbash M. 1998. Evidence that the TIM light response is relevant to light-induced phase shifts in Drosophila melanogaster. Neuron 21 , 225–234. ( 10.1016/s0896-6273(00)80529-2) [DOI] [PubMed] [Google Scholar]
- 91. Dey BK, Zhao XL, Popo-Ola E, Campos AR. 2009. Mutual regulation of the Drosophila disconnected (disco) and Distal-less (Dll) genes contributes to proximal-distal patterning of antenna and leg. Cell Tissue Res. 338 , 227–240. ( 10.1007/s00441-009-0865-z) [DOI] [PubMed] [Google Scholar]
- 92. Valentino P, Erclik T. 2022. Spalt and disco define the dorsal-ventral neuroepithelial compartments of the developing Drosophila medulla. Genetics 222 , iyac145. ( 10.1093/genetics/iyac145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tierney SM, Friedrich M, Humphreys WF, Jones TM, Warrant EJ, Wcislo WT. 2017. Consequences of evolutionary transitions in changing photic environments. Aust. Entomol. 56 , 23–46. ( 10.1111/aen.12264) [DOI] [Google Scholar]
- 94. Somanathan H, Kelber A, Borges RM, Wallén R, Warrant EJ. 2009. Visual ecology of Indian carpenter bees II: adaptations of eyes and ocelli to nocturnal and diurnal lifestyles. J. Comp. Physiol. A Neuroethol. Sens. Neural. Behav. Physiol. 195 , 571–583. ( 10.1007/s00359-009-0432-9) [DOI] [PubMed] [Google Scholar]
- 95. Akiyama T, Uchiyama H, Yajima S, Arikawa K, Terai Y. 2022. Parallel evolution of opsin visual pigments in hawkmoths by tuning of spectral sensitivities during transition from a nocturnal to a diurnal ecology. J. Exp. Biol. 225 , jeb244541. ( 10.1242/jeb.244541) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mulhair PO, Crowley L, Boyes DH, Lewis OT, Holland PWH. 2023. Opsin gene duplication in Lepidoptera: retrotransposition sex linkage, and gene expression. Mol. Biol. Evol. 40 , msad241. ( 10.1093/molbev/msad241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Shimizu I, Yamakawa Y, Shimazaki Y, Iwasa T. 2001. Molecular cloning of Bombyx cerebral opsin (Boceropsin) and cellular localization of its expression in the silkworm brain. Biochem. Biophys. Res. Commun. 287 , 27–34. ( 10.1006/bbrc.2001.5540) [DOI] [PubMed] [Google Scholar]
- 98. Moses K, Ellis MC, Rubin GM. 1989. The glass gene encodes a zinc-finger protein required by Drosophila photoreceptor cells. Nature 340 , 531–536. ( 10.1038/340531a0) [DOI] [PubMed] [Google Scholar]
- 99. Tearle R. 1991. Tissue specific effects of ommochrome pathway mutations in Drosophila melanogaster. Genet. Res. 57 , 257–266. ( 10.1017/S0016672300029402) [DOI] [PubMed] [Google Scholar]
- 100. Shamloula HK, Mbogho MP, Pimentel AC, Chrzanowska-Lightowlers ZMA, Hyatt V, Okano H, Venkatesh TR. 2002. Rugose (rg), a Drosophila A kinase anchor protein, is required for retinal pattern formation and interacts genetically with multiple signaling pathways. Genetics 161 , 693–710. ( 10.1093/genetics/161.2.693) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Stöckl A, Heinze S, Charalabidis A, El Jundi B, Warrant E, Kelber A. 2016. Differential investment in visual and olfactory brain areas reflects behavioural choices in hawk moths. Sci. Rep. 6 , 26041. ( 10.1038/srep26041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sourakov A, Chadd RW. 2022. The lives of moths: a natural history of our planet’s moth life. Princeton, NJ: Princeton University Press. [Google Scholar]
- 103. Dell’Aglio DD, McMillan WO, Montgomery SH. 2022. Shifting balances in the weighting of sensory modalities are predicted by divergence in brain morphology in incipient species of Heliconius butterflies. Anim. Behav. 185 , 83–90. ( 10.1016/j.anbehav.2022.01.003) [DOI] [Google Scholar]
- 104. Montgomery SH, Merrill RM, Ott SR. 2016. Brain composition in Heliconius butterflies, posteclosion growth and experience-dependent neuropil plasticity. J. Comp. Neurol. 524 , 1747–1769. ( 10.1002/cne.23993) [DOI] [PubMed] [Google Scholar]
- 105. Anton S, Rössler W. 2021. Plasticity and modulation of olfactory circuits in insects. Cell Tissue Res. 383 , 149–164. ( 10.1007/s00441-020-03329-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kuenzinger W, Kelber A, Weesner J, Travis J, Raguso RA, Goyret J. 2019. Innate colour preferences of a hawkmoth depend on visual context. Biol. Lett. 15 , 20180886. ( 10.1098/rsbl.2018.0886) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Kelber A. 2002. Pattern discrimination in a hawkmoth: innate preferences, learning performance and ecology. Proc. R. Soc. Lond. B 269 , 2573–2577. ( 10.1098/rspb.2002.2201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Broadhead GT, Basu T, von Arx M, Raguso RA. 2017. Diel rhythms and sex differences in the locomotor activity of hawkmoths. J. Exp. Biol. 220 , 1472–1480. ( 10.1242/jeb.143966) [DOI] [PubMed] [Google Scholar]
- 109. Bischoff M, Raguso RA, Jürgens A, Campbell DR. 2015. Context-dependent reproductive isolation mediated by floral scent and color. Evolution 69 , 1–13. ( 10.1111/evo.12558) [DOI] [PubMed] [Google Scholar]
- 110. Jaeger S, Girvin C, Demarest N, LoPresti E. 2023. Secondary pollinators contribute to reproductive success of a pink-flowered sand verbena population. Ecology 104 , e3977. ( 10.1002/ecy.3977) [DOI] [PubMed] [Google Scholar]
- 111. So WV, Sarov-Blat L, Kotarski CK, McDonald MJ, Allada R, Rosbash M. 2000. Takeout, a novel Drosophila gene under circadian clock transcriptional regulation. Mol. Cell. Biol. 20 , 6935–6944. ( 10.1128/MCB.20.18.6935-6944.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Steller H, Fischbach KF, Rubin GM. 1987. Disconnected: a locus required for neuronal pathway formation in the visual system of Drosophila. Cell 50 , 1139–1153. ( 10.1016/0092-8674(87)90180-2) [DOI] [PubMed] [Google Scholar]
- 113. Blanchardon E, Grima B, Klarsfeld A, Chélot E, Hardin PE, Préat T, Rouyer F. 2001. Defining the role of Drosophila lateral neurons in the control of circadian rhythms in motor activity and eclosion by targeted genetic ablation and period protein overexpression. Eur. J. Neurosci. 13 , 871–888. ( 10.1046/j.0953-816x.2000.01450.x) [DOI] [PubMed] [Google Scholar]
- 114. Helfrich-Förster C. 1998. Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: a brain-behavioral study of disconnected mutants. J. Comp. Physiol. A 182 , 435–453. ( 10.1007/s003590050192) [DOI] [PubMed] [Google Scholar]
- 115. Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. 1999. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99 , 791–802. ( 10.1016/s0092-8674(00)81676-1) [DOI] [PubMed] [Google Scholar]
- 116. Liang X, Holy TE, Taghert PH. 2016. Synchronous Drosophila circadian pacemakers display nonsynchronous Ca2+ rhythms in vivo. Science 351 , 976–981. ( 10.1126/science.aad3997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Helfrich-Förster C. 2005. Neurobiology of the fruit fly’s circadian clock. Genes Brain Behav. 4 , 65–76. ( 10.1111/j.1601-183X.2004.00092.x) [DOI] [PubMed] [Google Scholar]
- 118. Helfrich-Förster C. 2006. The neural basis of Drosophila’s circadian clock. Sleep Biol. Rhythms 4 , 224–234. ( 10.1111/j.1479-8425.2006.00223.x) [DOI] [Google Scholar]
- 119. Homberg U, Würden S, Dircksen H, Rao KR. 1991. Comparative anatomy of pigment-dispersing hormone-immunoreactive neurons in the brain of orthopteroid insects. Cell Tissue Res. 266 , 343–357. ( 10.1007/BF00318190) [DOI] [Google Scholar]
- 120. Shafer OT, Yao Z. 2014. Pigment-dispersing factor signaling and circadian rhythms in insect locomotor activity. Curr. Opin. Insect Sci. 1 , 73–80. ( 10.1016/j.cois.2014.05.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Mongue AJ, Kawahara AY. 2022. Population differentiation and structural variation in the Manduca sexta genome across the United States. G3 12 . ( 10.1093/g3journal/jkac047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Liu X, Zhang Z, Hu B, Chen K, Yu Y, Xiang H, Tan A. 2023. Single-cell transcriptomes provide insights into expansion of glial cells in Bombyx mori. Insect Sci. ( 10.1111/1744-7917.13294) [DOI] [PubMed] [Google Scholar]
- 123. Keller AD, Maniatis T. 1992. Only two of the five zinc fingers of the eukaryotic transcriptional repressor PRDI-BF1 are required for sequence-specific DNA binding. Mol. Cell. Biol. 12 , 1940–1949. ( 10.1128/mcb.12.5.1940-1949.1992) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Sondhi Y, Messcher RL, Bellantuono A, Storer CG, Cinel SD, Godfrey RKet al. 2024. Data from: Day-night gene expression reveals circadian gene disco as a candidate for diel-niche evolution in moths. Figshare. ( 10.6084/m9.figshare.c.7403413) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The transcriptome libraries are also archived on GenBank under BioProject PRJNA1102514, and all associated sequence data are archived on the SRA database (SRR28778930-SRR28778945). Datasets S1 to S17: Available at: 10.6084/m9.figshare.23661603.
Supplementary material is available online [124].