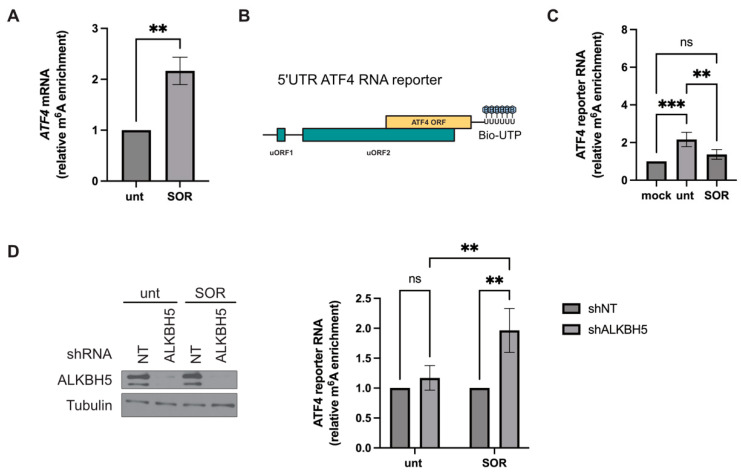
Figure 3.
Role of ALKBH5 in ATF4 mRNA methylation level. (A) Methylated RNA immunoprecipitation (MeRIP)-qPCR analysis. Hep3B were treated with sorafenib (SOR; 10 μM) for two hours or left untreated (unt), lysed, and their extracts were subjected to MeRIP using m6A antibodies. Immunoprecipitated m6A RNAs are then quantified by RT-qPCR using oligos specific to the main ORF of ATF4 mRNA. The amounts of m6A ATF4 mRNA were normalized against IgG precipitate and then expressed relative to untreated conditions. ** p ≤ 0.01. (B) Schematic representation of the 5’UTR ATF4 RNA reporter. (C) MeRIP-qPCR analysis of m6A level of the biotinylated 5′UTR ATF4 RNA reporter incubated with protein extracts prepared from either untreated (unt) or sorafenib (SOR)-treated Hep3B. RNA is then subjected to MeRIP using m6A antibodies. Immunoprecipitated m6A RNA is then incubated with streptavidin-agarose beads to purify the biotinylated reporter RNA, which is quantified by RT-qPCR using oligos specific to the 5′end of ATF4 mRNA. The amounts of m6A ATF4 reporter RNA were normalized against IgG precipitate and then expressed relative to the mock condition. Data are representative of 3 separate experiments, and values are given as mean ± SD. ** p ≤ 0.01, *** p ≤ 0.001, ns: not significant. (D) MeRIP-qPCR analysis of m6A level of 5′UTR ATF4 RNA reporter incubated with proteins extracts from Hep3B stably expressing either a control shRNA (shNT) or shALKBH5 and treated with SOR (10 µM, 2 h). Depletion of ALKBH5 is validated by western blot using specific antibodies as described in Figure 1 (left panels). m6A methylated ATF4 reporter RNAs were isolated and quantified by RT-qPCR (right graphs) as above. The amounts of m6A ATF4 reporter RNA were normalized against IgG precipitate and then expressed relative to the shNT condition. Data are representative of 3 separate experiments, and values are given as mean ± SD. ** p ≤ 0.01, ns: not significant. Original images can be found in Supplementary Materials.