Abstract
Musculoskeletal sarcomas pose major challenges to researchers and clinicians due to their rarity and heterogeneity. Xenografting human cells or tumor fragments in rodents is a mainstay for the generation of cancer models and for the preclinical trial of novel drugs. Lately, though, technical, intrinsic and ethical concerns together with stricter regulations have significantly curbed the employment of murine patient-derived xenografts (mPDX). In alternatives to murine PDXs, researchers have focused on embryonal systems such as chorioallantoic membrane (CAM) and zebrafish embryos. These systems are time- and cost-effective hosts for tumor fragments and near-patient cells. The CAM of the chick embryo represents a unique vascularized environment to host xenografts with high engraftment rates, allowing for ease of visualization and molecular detection of metastatic cells. Thanks to the transparency of the larvae, zebrafish allow for the tracking of tumor development and metastatization, enabling high-throughput drug screening. This review will focus on xenograft models of musculoskeletal sarcomas to highlight the intrinsic and technically distinctive features of the different hosts, and how they can be exploited to elucidate biological mechanisms beneath the different phases of the tumor’s natural history and in drug development. Ultimately, the review suggests the combination of different models as an advantageous approach to boost basic and translational research.
Keywords: patient-derived xenografts (PDX), cell-derived xenografts (CDX), murine models, zebrafish, chorioallantoic membrane (CAM), musculoskeletal sarcomas
1. Introduction
Sarcomas account for less than 1% of adult malignancies and 12–15% of paediatric tumors [1]. Historically, sarcomas have been categorised according to the anatomical site of occurrence in Bone Sarcomas (BS) (15%), Soft Tissue Sarcomas (STS) (80%) and Gastro-Intestinal Stromal Tumors (GIST) (5%) [2]. Nowadays, the World Health Organization distinguishes over 100 histological subtypes with peculiar morphology and molecular traits [3,4]. Moreover, many histotypes are exceedingly rare (less than 1 case/1,000,000 persons) [5]. More than 90% of BS are classified as osteosarcomas (OS), Ewing sarcomas (EWS), or chondrosarcomas (CHS) [3]. Among STS, liposarcoma (LPS) and leiomyosarcoma (LMS) represent the most common histologic subtypes in adults, while rhabdomyosarcoma (RMS) is the most prevalent histotype in children [6]. The reader is referred to Table 1 for clinicopathological details. To date, the gold standard treatment for localised sarcoma is surgery, often in combination with radio- and/or polychemotherapy [1]. Unfortunately, around one third of sarcoma patients still have a dismal prognosis, and those with metastatic disease medially survive only 12 months [2]. Targeted drugs may overcome the limits of current therapies, however, the selection of proper actionable targets in sarcomas is challenging [1]. Based on omic analyses, a plethora of novel therapies has been proposed, however, their translation into the clinical praxis is discouraged by the unsatisfactory results obtained in clinical studies so far [7,8]. This reflects the lack of faithful research tools to model cancer phenotype at the molecular and organism levels [1,7]. Therefore, preclinical models that incorporate the biology and genetics of human cancers and preserve inter- and intra-tumor heterogeneity are compelling to obtain reliable and translatable results [7,8]. On the other hand, the drug-screening pipeline requires high-throughput and time-effective systems. Cell line-derived xenograft (CDXs) models have represented, up to now, the workhorse for basic and translational research, thanks to their consistency and cost-effectiveness, but patient-derived xenografts (PDXs) are currently recognized as the most effective preclinical model for phenocopying cancer biology and architecture, and studying drug response and resistance [8,9]. Both technical (low success rate and through-put, long experimental time, lack of tools to follow tumor growth) and intrinsic (lack of immune milieu, stroma substitution by host cells [10], different pharmacokinetics, and long-term model instability [11,12]) concerns profoundly burden this model [7]. The transposition of the EU Directive 2010/63/EU on the protection of animals used for scientific purposes under national laws has resulted in strict regulation, reflecting the growing ethical concerns around the use of adult animal experimentation [13]. Therefore, researchers are turning their attention to non-mammalian embryonal models, such as zebrafish larvae [14] and chick embryos [15]. Embryonal models fall under the aegis of less restrictive regulation [16] until they are capable of independent feeding (chick embryo: hatching; zebrafish: 5 days post-fertilization (dpf) [17]) and, most importantly, until they acquire the ability to feel pain (chick embryo: >13 egg development day (EDD) [18,19]; zebrafish: 5 dpf). Moreover, mice are kept under conditions that respect animal welfare, but do not represent life “as in the wild”—on the contrary, embryonal systems do not suffer environmental distress. Technically speaking, embryonal systems hold the promise of higher manageability without the need for animal facilities, higher throughput, and lower costs [14,15,20]. Zebrafish (ZF) has emerged as an innovative in vivo model due to its unique combination of genetic and physiological features. Its rapid development, embryo body transparency, high fecundity and easy genetic manipulation make ZF a suitable approach for preclinical studies [14]. Moreover, the ZF genome has been completely sequenced showing high homology to human DNA, and the larvae are therefore recognized as a suitable organism for genetic manipulation [21]. In this scenario, ZF xenografts represent a unique approach to investigate complex biological processes, to uncover molecular mechanisms, and to develop drug-screening platforms [14,22]. The chicken chorioallantoic membrane (CAM) model has been widely used for studies on angiogenesis and tumorigenesis on prostate cancer, glioblastoma, OS, and lung adenocarcinoma, thanks to its transparency and strongly vascularized structure [23]. Moreover, the CAM can host xenografts of human tumor cells and patient-derived fragments, given its immature immune system [24]. CAM has many advantages, such as ease of access, short experimental times, and cost-effectiveness. Xenografting human tumor cells or tissues on CAM induces angiogenesis, and this can be exploited for both basic research and the development of novel antiangiogenic therapies [25]. The choice of the host and of the procedure for a xenograft strictly depends on the experimental question of interest. The intrinsic and technical peculiarities of the different models can be exploited to represent and elucidate biological mechanisms which are specific to the different phases of the tumor’s natural history. This review of musculoskeletal sarcomas (MSKT) models aims to highlight the peculiar features and potentialities of the different xenograft hosts in the study of the different steps of tumor natural history and drug development, with the aim of generating awareness of the choice of the proper model and to prompt the integration of different models, boosting basic and translational research.
Table 1.
Sarcomas of the musculoskeletal system. The table reports the main molecular and clinical features of selected MSKT.
| Sarcoma | Type | Pathognomonic Feature | Incidence @ | 5-Year Overall Survival |
Primary Site | Main Metastasis Site | Therapy | Refs. |
|---|---|---|---|---|---|---|---|---|
| Osteosarcoma (OS) | BS | Multiple chromosomal aberrations; frequent TP53 and RB1 mutations; DNA helicase disorders |
3–4.5 * | 60% * | Knee, humerus | Lungs | Neo-adjuvant and adjuvant chemotherapy (Methotrexate, Adriamycin, Cisplatin (MAP)); surgery or radiotherapy where surgery is not possible | [26,27] |
| Ewing sarcoma (EWS) | BS | 85–90% EWS::FLI1 translocation |
<1 | 70% # | Long and flat bones | Lungs, bone, bone marrow | Induction chemotherapy (doxorubicin, etoposide, cyclophosphamide, vincristine, and ifosfamide); surgery; radiotherapy | [28] |
| Chondrosarcoma (CHS) | BS | IDH1/2, EXT1/2 mutations | <6 | >90% (low grade); 75% (II grade); 30% (III grade) | Proximal femur, humerus, tibia, pelvis and scapula | Lungs | Surgery | [29,30,31] |
| Liposarcoma (LPS) | STS | MDM2, CDK4 amplification | 10 | 93% (grade I §); 57% (grade II §§); 21% (grade III §§) |
Most commonly in thigh, retroperitoneum, inguinal region and popliteal fossa | Retroperitoneum, distant sites and soft tissue sites §§ | Surgery; chemoradiotherapy | [32,33] |
| Rhabdo-myosarcoma (RMS) | STS | PAX3::FOXOA1 and PAX7::FOXOA1 translocations; RAS-PI3K, RTK signaling, loss of PTEN, TP53 and CDKN2A in fusion negative RMS | 4.5 ^ | >70% | Head, neck, genitourinary tract, limbs | Lungs, bone and bone marrow | Surgery, ionizing radiation, chemotherapy (vincristine, actinomycin D, cyclophosphamide (VAC); Ifosfamide) | [34] |
| Myxofibrosarcoma (MFS) | STS | Highly complex karyotypes | 1 | 75% | Lower limbs, trunk, head and neck | Lungs, bone and lymph nodes | Surgery; radiotherapy; chemotherapy (doxorubicin, ifosfamide) | [35,36] |
| Synovial Sarcoma (SS) | STS | 95% SYT::SSX1/2/4 translocation | 1–2 | 76% # | Extremities | Lungs, bone and lymph nodes | Surgery; radiotherapy; chemotherapy (anthracycline plus ifosfamide, gemcitabine, docetaxel, trabectidin, VAC) | [37] |
| Leiomyosarcoma (LMS) | STS | Highly complex karyotypes with genomic instability | 6 | 50% | Commonly in peritoneum and extremities | Lungs, peritoneum, liver, and bone | Surgery; radiotherapy; chemotherapy (doxorubicin) |
[33] |
| Undifferentiated pleomorphic sarcoma (UPS) |
STS | Highly complex karyotypes | 8–10 | 48% | Long bones, preference for proximal tibia and distal femur | Lungs | Surgery; radiotherapy; chemotherapy (anthracycline and ifosfamide) |
[38,39] |
Notes: @ cases/106 population/year; * in children and adolescents; # local disease; § well-differentiated LPS; §§ de-differentiated LPS; ^ aged under 20.
2. Xenografting MSKT
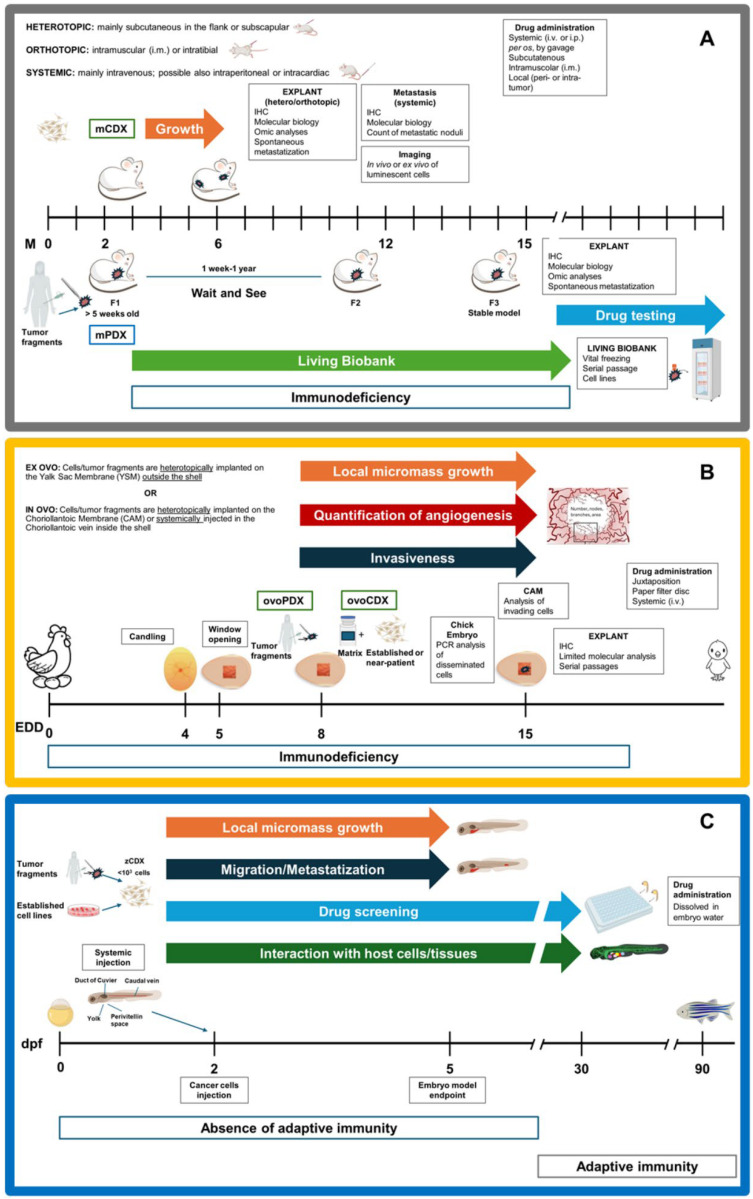
According to the National Cancer Institute Dictionary of Cancer Terms, the term “xenograft” refers to the transplant of an organ, tissue, or cells to an individual of another species [40]. While xenografts obtained by injection of established or near-patient cells (cell-derived xenografts, CDXs) are more reproducible and provide higher throughput, xenografting tumor fragments (patient-derived xenografts, PDXs) has the advantage of maintaining cellular heterogeneity and architectural features of the original tissue [8]. Xenografts of MSKT have been reported in rodents, especially in mice (m), embryonic and adult ZF (z), and CAM (ovo). Notably, xenografts in mice are regulated by the EU Directive on the protection of animals used for scientific purposes, while embryonal models are not considered animals until they are able to feed themselves, hence simplifying bureaucracy [16]. In the following paragraphs, technical details of the xenografting procedure in the different hosts are discussed for the sake of comparison (Figure 1 and Figure 2). The description of the detailed protocols for the obtainment of CDXs and PDXs from each host is beyond the scope of this review, therefore, the readers are referred to technical papers and reviews for further information [41,42,43,44,45,46,47,48].
Figure 1.
Methodologies and applications of CDXs and PDXs in the different hosts. Each figure reports a timeline of xenografting experiment, including technical details and downstream applications. (A) mice; (B) chick embryo; (C) zebrafish larvae. Timeline specifications: M: months; EDD: egg development days; dpf: days post-fertilization.
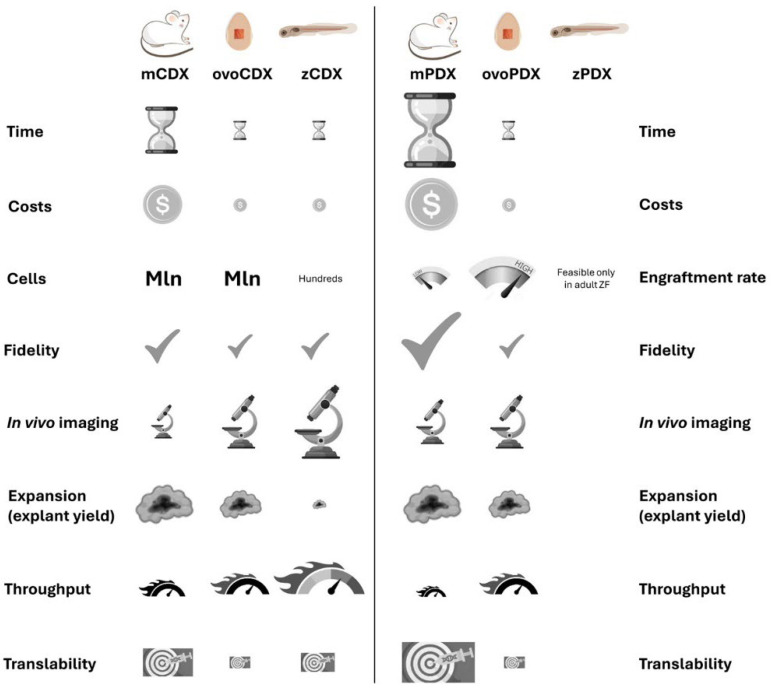
Figure 2.
Comparison of CDXs and PDXs in the different hosts. The table highlights the limits and potentialities of xenograft in mouse, chick embryo, and zebrafish larvae.
The earliest reports of xenotransplantation of MSKT into nude mice were published around the 1980s [49,50,51], but mPDX have re-gained momentum only in the last two decades. Obtaining an mPDX is a demanding endeavour, requiring an adequate tumor specimen (>1 mm3 and 10% viability [52]) and immune-compromised mice, which entail cumbersome sterile handling, significant costs and time (latency reaches 1 year [53]), imaging systems adapted to animal hosts, and experience in handling, given the low engraftment rate (Table 2). Moreover, an mPDX should be serially transplanted at least twice to obtain a stable model, and validated for its fidelity to the original tumor (Figure 1A) [7]. Indeed, preserving the tumor microenvironment from the donor tumor is the premise for the study of the tumor behaviour in vivo. However, passage after passage, the tumor stroma is increasingly replaced by the host stroma, and a decrease in tumor fidelity is generally observed. The replacement of the stroma is mostly considered an acceptable drawback of the model, since Arnaud Blomme et al. demonstrated that the metabolic profiles of both tumor cells and stromal cells remain stable for at least four passages, even though the replacement began at the second passage [10]. Ultimately, this large amount of work is rewarded by the creation of living biobanks of MSKT, which virtually eternalize a tumor specimen and render it available for future studies (Table 2). In respect to mPDX, establishing ovoPDXs is simpler to accomplish. Back in 1911, CAM represented the very first host for xenografts, while MSKT were transplanted only in 2012 [44,54]. Indeed, the high density of blood vessels creates an ideal milieu for tumor growth due to the ubiquitous supply of oxygen, nutrients, and growth factors [55]. In addition, ovoPDXs present unique advantages as grafts are visible to the naked eye, allowing for easy monitoring under a stereomicroscope [15]. Chick embryos are inexpensive and easy to manipulate as they are immunodeficient up to 18 EDD, experiments run in 10 days, and engraftment rates reach 80% in MSKT [44]. However, this ease comes at a price. Indeed, the amount of tumor tissue retrievable at the end of the experiment is similar to that at the start, limiting the possibility of down-stream molecular analysis (Figure 1B). For this reason, even if serial passages of ovoPDX are feasible up to 8 times in OS [56], these models cannot contribute to the creation of living biobanks. Notably, ovoPDXs allow for the mimicking of neoangiogenesis in vivo: indeed, after a first avascular phase of around 72–96 h after implantation, neovascularization occurs, in which the anastomosis between the CAM vasculature and the tumor vasculature is established, providing nutrients for a rapid growing phase [57]. Finally, one research group reported the feasibility of zPDX of gastrointestinal tumors in embryos [58,59] and in adult fish [60], however, the application of this technique is discouraged by the small embryo size.
CDXs can be obtained from established or near-patient cells, including primary and mPDX-derived cells. mCDX with established cell lines and their gene-edited or drug-resistant variants have been considered the workhorse for basic and translational research [9]. They usually require 1–5 × 106 cells/mouse and experiments run over a few months (Table 3 and Table 4; Figure 1A). mCDX of near-patient cells represent a win–win approach that couples the fidelity and heterogeneity of patient-derived samples with the manageability, time-effectiveness, and scalability of CDX [61]. Even though CAM is an established in vivo model to evaluate the progression of CDXs [15], there are only a few reports describing its use for BS and STS analysis, mostly focused on OS. In 2010, Balke et al. reported the ability of different OS cell lines (MNNG-HOS, U2OS and SaOS) to form vascularized solid tumors on CAM after four days of incubation [25]. Of note, the amount of xenografted cells for ovoCDX may be equal or even superior to mCDX, but experimental times are shorter (Figure 1B). On the other hand, zCDX can be considered miniature models as they only require <103 cells, rendering them particularly suitable for precious near-patient cells [62] (Figure 1C). zCDX also minimises experimental timelines to just a few days, as well as costs and the manipulation. Moreover, ZF embryos stand out for their optical transparency, facilitating in vivo imaging of engrafted cells [14].
The morphological and morphometric evaluation of newly formed tissues is feasible in all xenograft hosts, but it is more challenging in ZF samples due to their small size and the limited number of available antiZF antibodies [63]. Down-stream molecular analysis is theoretically feasible for all the models, yet the amount of the available material is limited in the case of ZF and chick embryo. Technological development, especially in the field of imaging, has opened up new possibilities to quantify primary tumor growth and the study of tumor cells’ migration in the embryonal models.
In the next sections, we will compare different xenograft hosts and modalities, and highlight their peculiarities with respect to the different steps of tumor natural history, i.e., tumorigenesis, angiogenesis, and metastatization, as well as their potentialities in translational research.
Table 2.
mPDX collections for MSKT. The table reports injection site, engraftment rate, latency of selected collections of MSKT mPDX, and their applications.
| Tumor-Related Features | PDX-Related Features | Applications | Refs. | |||
|---|---|---|---|---|---|---|
| Sarcoma Histotype | Inoculated Mass | Injection Site | Tumor Take (%) | Latency of 1° Passage | ||
| OS and EWS | nd | flank, s.c. | 25/49 (51%) | 16–280 days | modeling | [64] |
| LPS, LMS, MFS, SS, UPS, CHS and other $ | 25–75 mm3 | bilaterally s.c. | 32/188 (17%) | ns | drug testing; Xenosarc platform | [65,66] |
| CHS | 2 mm3 | s.c. | ns | ns | omic analysis | [67] |
| OS | 2–3 mm3 | flank, s.c. | 15/37 (40.5%) | 19–225 days | modeling; clinicopathological correlations | [52] |
| OS and EWS | 4 mm3 | transscapular brown fat, s.c. | OS:22/61 (36%) EW:7/29 (24%) | 1 week–1 year | modeling; target dependencies; drug testing | [53] |
| LPS | 4 × 4 mm | bilaterally s.c. | 7/10 (70%) | 2–9 months | drug testing | [68,69] |
| OS, EWS, RMS | 2 × 2 × 1 mm | s.c. | EW: 17/41 (41.55%); OS: 5/12 (41.7%); RMS: 8/15 (53.3%) | ns | modeling; clinicopathological correlations; drug testing | [70] |
| OS, RMS | 2 × 2 mm3 | flank, s.c. | OS: 51.4% RMS: 53.8% |
300 days | target dependencies | [71] |
| OS | 3 mm3 | flank, s.c. | 9/21 (42.9%) | 3–9 weeks | modeling | [72] |
| LPS | diameter of 3–5 mm | lower back | 25/56 (44.64%) | ns | drug testing | [73] |
| OS | 2 mm in diameter | flank, s.c. | 20/57 (35.1%) | ns | modeling; clinicopathological correlations | [74] |
| LMS | ns | s.c. | 17/49 (35%) | ns | target dependencis | [75] |
| OS, EWS, RMS, SS, other * | 2–5 mm3 | interscapular fat pad, or i.m. | 76/131 (58%) | 12–285 days | modeling; target dependencies; drug testing; MAPPYACTS | [76] |
| Sarcoma (NCI PDXNet consortium) |
ns | s.c. | ns | ns | modeling; clinicopathological correlations; target dependencies | [77] |
| RMS | 25–30 mm3 | flank, s.c. | 50% (6/12) | 195 days (median) | modeling | [78] |
| OS, LMS, LPS | 1–2 mm3 | flank, s.c. | ns | ns | modeling; EuroBoNeT | [79,80] |
| BS | 3 × 3 × 3 mm | flank, s.c. | 16 | 19–125 days | modeling | [63] |
| STS | 5-mm fragments | orthotopic, i.m. or s.c. | 32/107 (29.9%) | 9–184 days | modeling; clinicopathological correlations | [81] |
| OS, EWS, RMS, other § | single-cell suspension | orthotopic | OS 15/31 (48%); RMS 13/20 (65%); EWS 2/7 (29%); other 5/6 (83%) | 1–11 months | modeling; drug screening on primary colture; drug testing | [82] |
| OS, EWS, LPS, UPS, SS other | 2 mm3 | s.c. | 7/12 (58%) | ns | target dependencies and drug identification | [83] |
Notes: ns: not specified; * also includes solid tumors, CNS, lymphoma, and leukemia; $ atypical lipomatous tumor, undifferentiated sarcoma not otherwise specified, solitary fibrous tumor, undifferentiated spindle cell sarcoma, extra-skeletal myxoid chondrosarcoma plus ultra-rarer histotypes, BCOR/CIC; § desmoid small round cell tumor; i.m.: intramuscular; s.c.: subcutaneous.
Table 3.
mCDX for the evaluation of oncogenes and oncosuppressors. For each paper, the table describes the main features of xenografted cells and experimental details of selected mCDX determining the role of genes in tumorigenesis.
| Sarcoma | Cells | Mice | Gene | Refs. | |||||
|---|---|---|---|---|---|---|---|---|---|
| MSKT | ID | Type | Quantity (×106) | Injection Site | Mice/Experimental Group | Experimental Time (Days) | ID | Role | |
| OS | 143.98.2 OS | Est. | 1 | Heterotopic | 6 | 25 | STAT3 | ↑ | [84] |
| OS | 143B PML BK TK | Est. | 10 | Heterotopic | 5 | nd | CHEK1 | ↑ | [85] |
| OS | MG63 | Est. | 1 | Heterotopic | nd | 30 | GFRA1 | ↑ | [86] |
| OS | MNNG-HOS | Est. | 1.5 | Heterotopic | 5 | 20 | S1PR3 | ↑ | [87] |
| OS | MNNG-HOS | Est. | 5 | Heterotopic | 20 | 21 | FGD1 | ↑ | [88] |
| OS | MNNG-HOS | Est. | 2 | Heterotopic/Orthotopic | ≤6 | 25 * | KIF18B | ↑ | [89] |
| OS | U2OS | Est. | 1 | Heterotopic | 10 | 28 | NG6X | ↓ | [90] |
| OS | 143B | Est. | 0.5 | Orthotopic | 5 | 30 | FOXP1 | ↑ | [91] |
| OS | pOS-1 | Near-pt. | 5 | Heterotopic | 6 | 42 | Gαi3 | ↑ | [92] |
| OS | pOS-1 | Near-pt. | 4 | Orthotopic | na | 28 | TIMM13 | ↑ | [93] |
| OS | K-HOS | Est. | 1 | Heterotopic | 7 | ≥35 | RIPK1 | ↑ | [94] |
| EWS | RDES, TC-32 | Est. | 3/0.2 | Heterotopic | ≤6 | nd | SOX6 | ↑ | [95] |
| EWS | A673 | Est. | 2 | Heterotopic | 6 | 25 | KLF15 or TCF4 | ↑ | [96] |
| EWS | A4573 | Est. | 4 | Heterotopic | 10 | 20 | CAV1 | ↑ | [97] |
| EWS | TC-71 | Est. | 1 | Heterotopic | 7 | 50 | RIPK1 | ↑ | [94] |
| EWS | A673 | Est. | 2 | Heterotopic | 5 | >60 | miR34a | ↓ | [98] |
| RMS | RD | Est. | 1.25/1.5 | Heterotopic | ≤7 | 45 | HES1-YAP1-CDKN1C | ↑ | [99] |
| RMS | Rh30, Rh41 | Est. | 0.75–1 | Heterotopic | 40 | 40/75 | miR-486-5p | ↓ | [100] |
| RMS | Rh18, Rh30, C265S-Rh30 mutant | Est. | 2 | Orthotopic | ≤10 | ≤45 | PANX1 | ↓ | [101] |
| LPS | DDLPS 246 | Near-pt. | 1–2 | Heterotopic | 6 | 45 | miR-133a | ↓ | [102] |
Notes: Est: established; near-pt: near-patient; nd: not disclosed; na: not available ↑ oncogene; ↓ oncosuppressor; * time to amputation.
Table 4.
mCDX for the evaluation of metastatic potential. For each paper, the table describes the main features of xenografted cells and experimental details of selected mCDX determining the role of genes in metastatic potential.
| Sarcoma | Cells | Mice | Gene | Refs. | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MSKT | ID | Type | Quantity (×106) | Injection Route | Metastasis Site | Mice/Experimental Group | Experiment Time (Days) | ID | Role | |
| OS | 143B, MG63.2 | Est. | 1.5 | Orthotopic | Lung | 5 | 28, 56 | IGFBP5 | ↓ | [103] |
| OS | 143B | Est. | 0.5 | Orthotopic | Lung | ≥6 | 20 | CXCR4 | ↑ | [104] |
| OS | SaOS-LM7 | Est. * | 1 | Systemic | Lung | 10 | 49/56 | BMP-2 | nd | [105] |
| OS | 143B | Est., drug-resistant | 0.5 | Orthotopic | Lung | 8 | 40 | MIG-7 | ↑ | [106] |
| OS | U2OS | Est. | 10 | Heterotopic | Lung | 6 | 70 | ROCK2 | ↑ | [107] |
| OS | MG-63/Akaluc | Est. | 1 | Systemic | Lung | ≤8 | nd | LPAR1 | ↑ | [108] |
| OS | MG63, SaOS2 | Est. | 1 | Systemic | Lung | 11 | 21 | TRIM7 | ↑ | [109] |
| EWS | TC71 | Est. | 0.5 | Metastatic site (chestwall) | Lung | 9 | nd | PDGFR-β | ↑ | [110] |
| EWS | A673 | Est. | 2 | Systemic | Lung and lymph nodes | 6 | 98 | IGF2BP3 | ↑ | [111] |
| EWS | TC71, EWS4 | Est., near-pt | 0.01 | Systemic | Lung, kidneys, retroperitoneum | 5, 10 | 150, 250 | PORCN | ↑ | [112] |
| EWS | A673 Luc |
Est. | 0.25 | Orthotopic | Lung and bone | ≤15 | 28 | ZYX/ITGA5 | ↓ | [113] |
Notes: Est: established; near-pt: near-patient; nd: not disclosed; ↑ oncogene; ↓ oncosuppressor; * established by 7 cycling of parental cell line through the lungs of nude mice.
3. Tumorigenesis
A better understanding of the initial molecular events that lead to an irreversibly transformed cellular state might revolutionise early detection strategies as well as therapeutic success rates [114]. The models of election for the study of the formation of tumors are represented by carcinogen exposure-induced tumors, genetically engineered mouse models (GEMMs) [115,116,117], and gene-edited ZF [118,119,120]. However, these models are out of the scope of this review and the reader is referred to the cited literature for MSKT de novo models. Regardless of the host, PDX are not ideal models to study tumorigenesis as they intrinsically consist of the injection of a preformed tumor in a living organism. However, the transplanted tumors can be gene-edited in vivo to determine the contribution of a specific oncogene to tumor growth: for example, OS mPDXs were administered adenovirus-associated virus (AAV) carrying shFOXP1 to confirm its oncogenic role in OS [91]. Similarly, the onco-suppressive role of miR193b was verified in vivo, transfecting established mPDX of dedifferentiated LPS with the miRNA itself or a small molecule inhibiting its target [121]. Other than this approach, mPDXs are correlatively exploited if they express and/or lack the gene of interest. mPDXs with high basal expression of TAp73 or activated Hedgehog pathways were used to confirm the pro-tumorigenic role of PLK2 [122] and Gli/Smo [123] and the efficacy of their pharmacological inhibition in OS. Additionally, the pro-tumorigenic role of SOX6, demonstrated via gene-editing approaches, was eventually validated in a collection of mPDX of EWS, where SOX6 levels correlated with a higher Ki67 index [95]. Gene-edited cells have been the workhorses for studying the impact of genes on tumorigenesis for years, both in vitro and in vivo. This approach, together with the use of specific inhibitors, has elucidated the contribution of oncogenes, oncosuppressors, miRNAs [98,100,124], and related pathways in sarcomagenesis (Table 3). Notably, Slemmons and colleagues generated and xenografted YHR and YHV cells harbouring multiple transgenes to elucidate the role of YAP in RMS [125]. Other researchers have, instead, exploited inducible models to study the interaction of other oncogenes with EWS::FLI1 [96]. To circumvent the limitations in the gene-editing of near-patient cells, mice bearing OS primary-CDX were infected with AAV carrying shGαi3 [92], with a similar approach to that used on mPDX. Finally, the oncogenic role of Gαi3 [92] and TIMM13 [93] in OS was corroborated via mCDX of CrispR/Cas9-edited near-patient cells. mCDXs of gene-edited cells require the obtainment of stable-modified models, which is time-consuming and may introduce variability due to sub-cloning and inconsistencies with in vitro experiments. The use of fast-developing embryonal models allows us to employ the same transient strategies (siRNA, antisense, miRNA antagonists, soluble molecules, and transfections) used in vitro in the in vivo setting, combining time-effectiveness and consistency in methodology and robustness of results. This approach was exploited to evaluate the role of the activation of Akt in RMS, using the same plasmid for myrAkt, both in vitro and in ZF, completely avoiding the use of mice [126]. A similar approach in ovoCDX permit the uncovering of the reliance of SS on YAP/TAZ-TEAD-mediated transcriptional activity and confirmed the efficacy of its inhibition via verteporfin [127].
4. Angiogenesis
Angiogenesis is essential for tumors’ growth beyond the size of 1 cm3, as this is an empirical limit beyond which nutrient deficiency and hypoxia occur [128]. Understanding how cancer cells hijack the natural process of angiogenesis to their favour could highlight novel therapeutic targets and help restrain tumor progression ab initio [129,130]. Angiogenesis is a very complex, spatially dynamic, and temporarily determined phenomenon, which is very difficult to dissect and visualize in an adult organism. Indeed, the qualitative evaluation and quantification of angiogenesis in vivo requires sophisticated techniques, such as injection of fluorescent and non-fluorescent markers [131], Magnetic Resonance Imaging (MRI), Positron Emission Tomography (PET), or Optical Doppler Tomography [128,132]. An alternative is represented by the in vivo angiogenesis assay (DIVAA), consisting of subcutaneous implantation of semi-closed silicone cylinders (angioreactors) into nude mice. This system was used, in combination with mCDXs, to highlight the role of microvesicular cargo in OS angiogenesis [133]. The role of EWS::FLI1 in hijacking angiogenesis and the recruitment of bone marrow-derived cells was described in mCDX of EWS cells [134,135]. mPDX are employed to evaluate the efficacy of anti-angiogenic drugs on tumor growth, rather than to study the angiogenetic mechanism per se. As one of the few examples, EYA3-targeting monoclonal antibody-drug conjugates and benzarone were tested in EWS [136].
Embryonal models, on the other hand, offer the advantage of incomparable ease of visualization and imaging. The CAM is the system of election for the study of angiogenesis, thanks to its highly vascularized bed. Xenografted cells, for example SaOS2, can induce sprouting angiogenesis, irrespective of the number of seeded cells, although the degree of the angiogenic response significantly correlates with cell number [25]. Analogously, xenografted tumor fragments also induce angiogenesis around 72–96 h after implantation and anastomose to the CAM vessels [57]. The CAM assay enables a straightforward observation of angiogenesis to the naked eye, which is not possible with other animal in vivo models. Moreover, tumor angiogenesis and vasculogenesis can further be visualized using in vivo microscopy in the CAM adjacent to the tumor [137]. Additionally, acquired images can be manually and/or semi-automatically analysed through specific software (ImageJ, AngioQuant, AngioTool) to gather quantitative information, which is of utmost interest for the screening of antiangiogenic drugs [137,138]. The study of angiogenesis in the CAM often proceeds stepwise, including chick aortic ring sprouting assays in vitro, CAM and/or yalk sac membrane (YSM) assay in ovo or ex ovo, and disease-related evaluation based on ovoCDX: such an approach has highlighted the anti-angiogenic properties of reversine in OS [139] and the various peptides in fibrosarcoma [140]. The angiogenic properties of cytokines, chemokines, growth factors, chemotherapeutics, and targeted drugs can be assessed by the direct placing of the substance of interest on the naked membranes of the chick embryo. Vimalraj and colleagues proposed a variant of YSM assay, which is classified as an in vitro model, where cells were not actually xenografted but grown on coverslips, which were simply juxtaposed on YSM. This approach highlighted the role of miR-432-5p and miR-424-5p in regulating angiogenesis in OS [141,142]. Interestingly, the data on the inhibitory effect of miR-432-5p on angiogenesis obtained in ovo were confirmed in the zCDX of OS cells previously transfected with the miRNA mimic [141].
ZF embryos represent an ideal model to study the interplay between tumor and endothelial cells [143]. Their transparency allows the monitoring of blood vessel development to evaluate the effects induced by the presence of cancer cells. The sub-intestinal vein (SIV) is a major vessel plexus in proximity to the yolk sack, commonly used to quantify the angiogenic potential through the measure of vessels spreading in surrounding tissues [144]. This model can be applied to study the angiogenic alterations induced by cancer cells or the preclinical effect of anti-angiogenic drugs. The generation of ZF strains with fluorescent vessels, together with the ease of high-content imaging, also represent great opportunities offered by this host (reviewed in [145]), which are quite underexploited in MSKT.
5. Metastatization
Metastatization is a multistep process, including local invasion, intravasation, and dissemination via hematogenous or lymphatic vessels, extravasation, homing, dormancy and final outgrowth of a new tumor [146]. As metastases represent the main cause of cancer-related death [147], representative metastasis models are pivotal to dissecting the molecular mechanism underlying each step and highlighting the related target dependencies [148].
Spontaneous metastasis from heterotopically implanted sarcoma mPDX represent rare events (Table 1) [53]. On the other hand, orthotopic mPDXs or mCDXs do lead to the formation of distant metastases in a wide range of target organs, other than lung and lymph nodes [149,150,151], probably due to the interaction of tumor cells with the native tumor-niche. However, metastatization takes such a long time that orthotopic primary tumors, especially in the limbs, often need to be excised and mice followed-up until metastases appear, however, this surgery represents a major ethical concern. Spontaneous metastasis models from orthotopic mPDX or mCDX were reported for EWS [152], OS [150,153,154], and UPS [155]. Interestingly, this technique represents the best setting for the preclinical evaluation of anti-metastatic drugs and was exploited in the mPDX of OS treated with a monoclonal antibody against the Wnt-signalling inhibitor dickkopf-1 [156], a β-catenin/transducin β-like protein 1 (TBL1) inhibitor alone and in combination with doxorubicin [157] and in mPDX of the Giant Cell Tumor of Bone (GCTB) treated with a monoclonal antibody targeting CD44 [158]. Established or rarely near-patient [159] mCDXs via systemic injection, which is performed mainly intravenously, lead to the formation of experimental metastasis. This route of administration eliminates the first steps of metastatization and directly disseminates cells into the bloodstream, enhancing replicability and reducing experimental times. The main metastatic site in these models is represented by the lungs, being the first organs reached by venous circulation. Systemic, orthotopic, and rarely heterotopic, mCDX served to elucidate the contribution of several genes and pathways to metastasization (Table 4). Notably, multiple mCDX with gene-edited cells were used to elucidate the pro-metastatic role of Rab22a-NeoF1 in OS and to dissect its molecular mechanism [160]. Moreover, orthotopic spontaneous and systemic experimental metastasis models have also been employed to assess the anti-metastatic efficacy of CSF1R [161], HDAC [100], PDPN [162], CDK [159], and LPAR1 [108] inhibitors in OS.
In contrast, embryonal models can be effective systems for studying the first steps of metastases (i.e., local invasion, intra- and extravasation, and dissemination). In this case, the formation of full-blown metastasis is not observable, due to the short-time experiments and to the lack of target organs. Nevertheless, it was previously demonstrated that cells systemically injected into the chick embryo do lead to tumor formation in the hatched chicks [163]. CAM assays allow the easy monitoring of the intravasation of tumor cells into the microvasculature and the determination of the invasive phenotype of ovoCDX and ovoPDX, heterotopically deposited on the CAM surface or of the experimental metastatic potential after intravenous injection in the chorioallantoic vein [41,138,164]. Metastatic cells can be distinguished from the chicken ones and quantified by IHC or qPCR of ALU sequences from the DNA of isolated chicken embryo organs [165]. Imaging of metastatic cells is feasible via CT, MRI for magnetically labeled cells, and video-microscopy or fluorescent imaging under ex ovo conditions. In the context of sarcomas, Snail, a master gene of epithelial-to-mesenchymal transition (EMT), was modulated in OS cells, which were heterotopically injected on the CAM: cells overexpressing Snail showed a higher ability to invade the stroma, and enter and migrate along the vasculature of the CAM [166]. Analogously, the pro-migratory role of IL6 was demonstrated ex ovo upon injection of EWS cells and paracrine administration of the soluble interleukin [167]. Authors have demonstrated the higher invasiveness of treated cells by visualising migrated cells under a fluorescent microscope and categorising them according to the distance from the tumor edge [167]. Moreover, the analysis of ALU sequences has revealed a greater invasion ability to the lung and liver of FUS-CHOP-transfected LPS cells [168]. No less important, the CAM system may represent a complementary assay for anti-metastatic drug testing [138,164], but no examples are available for sarcomas.
Moving attention to the ZF model, the most attractive feature is the absence of pigmentation, which makes it particularly suitable for the imaging analysis of inner organs and tissues [169]. Moreover, researchers have generated several transgenic strains expressing fluorescent proteins in specific cell populations and organs, or upon pathway activations [170]. This, together with the injection of cancer cells marked with fluorescent trackers, allows the monitoring of single-cell behaviour and the dynamic interactions with surrounding and distant tissues. High-resolution imaging technologies, such as light sheet and confocal microscopy, represent the best approach to visualize the cross-talk between endothelial and cancer cells at a single-cell level—all outputs that are difficult to reach with other animal models [9]. Through this model, Van der Ent and colleagues first demonstrated the ability of EWS cells to proliferate, migrate, and disseminate via the hematogenous route to fins, head, and body. Afterwards they exploited an Albino Casper, TG(fli1:EGFP) strain, characterised by fluorescent vessels, to track the amount, size, and migratory routes of tumor cell foci for each embryo upon treatment with nutlin-3a and/or YK-4-279 [171]. Using a similar approach, fluorescent RON-deficient and proficient cells were injected into a ZF line with red fluorescently tracked endothelium: RON deficiency reduced local tumor burden at injection sites and perivascular areas with metastatic cells [172]. Lastly, mCherry-labelled EWS cells were injected into the yolk sac of fli1:EGFP ZF embryos to verify that the inhibition of SIRT1/2 via tenovin-6 halted the invasiveness of tumor cells by tracking the distance and direction of cell clusters inside the embryo in vivo [173].
6. Drug Development
Modeling sarcomas is essential for designing and validating novel drugs based on the identification of specific target dependencies. mPDXs allow the amplification of patient-derived tumor samples and the faithful preservation of histologic, genomic, epigenetic, genetic, metabolic, proteomic, and clonal traits [7,9] (Table 2). Several collections are available at local laboratories (Table 2), distributors, and within European programmes (listed in [174]), providing a virtually unlimited repository of clinically annotated and molecularly characterized human samples. The integration of different omic data via algorithms, mathematical modeling, or, lately, AI-systems, can provide even more informative and comprehensive results. RNA sequencing combined with drug prediction algorithms has been employed to rank therapies in RMS tumors and related PDX: notably, in this study, all the selected drugs were evaluated in mPDX [175]. Pandyia and colleagues set up a multi-OMICS analytical pipeline to prioritize drugs based on biomarkers: their approach indicated CDK4/6 hyperactivation and BETs as druggable targets in OS, which were validated in the same mPDX cohort in vivo [71]. While mPDXs constitute living biobanks, embryonal models represent instead short-time hosts from which a limited amount of material is retrievable at the end of xenografting experiments. However, given their fidelity [176,177,178], embryonal models have peculiar advantages in drug-screening applications. Indeed, they enable middle-to-high throughput studies that are feasible in the context of an intact living system recapitulating human physiology and pharmacokinetics in a reasonable timeframe [165,170]. Moreover, drug administration can be performed by juxtaposition in ovo or by dissolution into ZF embryo water, simplifying handling. A streamlined experimental procedure employing only one drug has been employed to calculate the IC50 of vincristine in ovoCDX of established RMS cells [179], providing a proof-of-principle for the employment of the CAM model as a platform for drug screening. However, a drawback of the use of ovoCDX in drug screening is the current lack of automated high-throughput procedures, which are feasible in ZF. By analogy with ZeOncoTest [180], Grissenberger and colleagues exploited high-content images of xenografted fluorescent EWS cells to follow tumor growth in ZF. This approach permitted the screening of 16 compounds and their combinations in groups of 30–95 larvae over a 3-day period of time in two cell lines and related models with inducible knock-down of EWS::FLI1 [181]. The obtained results were validated in a mPDX, with a significant reduction of mammalian animal models, costs, and experimental time. Understandably, mPDXs are difficult to employ for large high-throughput drug screening due to high costs and cumbersome handling [174]. Once the drug has been selected, mCDXs and, lately, mPDXs, represent the most effective preclinical models for the evaluation of drug efficacy. mCDXs have represented the principal modality of preclinical drug testing for decades—their employment is still pivotal for near-patient [182], drug-resistant [183], and gene-edited [184] cells, rarer disease entities (SS [185,186], Epithelioid sarcoma [187], MFS [188]), and they often represent a preliminary step to mPDXs. In recent decades, preclinical drug evaluation has moved to mPDXs as the most trustworthy model. Accordingly, all the most promising molecules currently in clinical trials for sarcomas have been tested on mPDX (Table 5). Moreover, the efficacy of the inhibitors of (i) proteasome (VLX1570) [189], H3K27ac (GSK-J4), CDK7-mediated transcriptional process (THZ1) [190], mevalonate pathway (atorvastatin) [191] in EWS, (ii) ATR [192] and the combination of Aurora Kinase A plus navitoclax [193] in RMS, (iii) irinotecan and axitinib in LPS [194], and (iv) ATR in SS [195] was validated in mPDX after drug screening. Most importantly, the creation of mPDX models of ultra-rare sarcomas permitted the evaluation of trabectedin, eribulin, and various combinations of chemotherapeutics in solitary fibrous tumors [196], doxorubicin, eribulin, temozolomide plus irinotecan, and the targeted drugs palbociclib and linsitinib in an orthotopic mPDX of FUS-ERG EWS [197,198,199], as well as the combination of c-MET, EZH2, mTOR inhibitors, and ADM plus trametinib in epithelioid sarcoma [200,201]. However, preclinical testing on mPDX does not warrant success in clinical evaluation [9]. Indeed, the robustness of results is restrained by the limited number of experimental groups (<10 mice/group) and disease entities of origin. Some notable examples of drugs which failed to enter clinical praxis after exceptional preclinical results are represented by eribulin-, olaratumab-, and Akt-directed therapies. In some cases, after an unsuccessful clinical trial, researchers returned to their preclinical models to identify possible biomarkers of response. Low Bcl-2 expression was identified as a possible biomarker of response to nab-paclitaxel, and its inhibition via venetoclax was found to be synergistic [202], while low SPARC expression was associated with low drug retention and inefficacy [203]. Several papers go through the entire path of drug discovery, especially in the context of ultra-rare tumors. This approach permitted the identification, screening, and validation of anti-cancer drugs in the context of (i) CIC-DUX4 Ewing-like sarcomas [204], (ii) desmoplastic round cell sarcoma [205,206,207,208], (iii) mesenchymal chondrosarcoma [209], (iv) GCTB [210], and (v) clear cell sarcoma [211].
A particular mention should be paid to the extremely reduced number of cells needed for zCDX (<103), which makes it an attractive and suitable system to xenograft near-patient cells. In this context, the efficacy of bone-targeted therapy (denosumab), TK inhibitors (lenvatinib), and their combination was evaluated on the zCDX of primary cells of desmoplastic fibroma [212], while trabectedin was evaluated on patient-derived primary cultures of UPS [213].
Table 5.
Open clinical trials with matching preclinical research on MSKT. The table reports details of recent clinical trials in MSKT on the left, while on the right the details of the matched preclinical research on mPDX are reported. n.d., not disclosed.
| Clinical Trial | Preclinical Research | ||||
|---|---|---|---|---|---|
| NCT Number | Study Title | Conditions | Mice/Group | N° mPDX/ Trial |
Refs. |
| NCT03838744 | Randomized Trial in Advanced, Metastatic, or Unresectable Soft Tissue Sarcoma After Failure of Standard Treatments | Advanced Soft Tissue Sarcoma | 5 | 2 | [214] |
| NCT04076579 | Trabectedin in Combination With Olaparib in Advanced Unresectable or Metastatic Sarcoma | Sarcoma; Sarcoma Metastatic | [214] | ||
| NCT03936465 | Study of the Bromodomain (BRD) and Extra-Terminal Domain (BET) Inhibitors BMS-986158 and BMS-986378 in Pediatric Cancer | Pediatric tumors and Lymphoma | 7 | 1 | [215] |
| NCT04095221 | A Study of the Drugs Prexasertib, Irinotecan, and Temozolomide in People With Desmoplastic Small Round Cell Tumor and Rhabdomyosarcoma | Rhabdomyosarcoma | 6 | 6 | [216] |
| NCT03709680 | Study Of Palbociclib Combined With Chemotherapy In Pediatric Patients With Recurrent/Refractory Solid Tumors | Pediatric solid tumors | 6 | 6 | [199] |
| NCT04238819 | A Study of Abemaciclib (LY2835219) in Combination With Other Anti-Cancer Treatments in Children and Young Adult Participants With Solid Tumors, Including Neuroblastoma | Relapsed Solid Tumor; Refractory Solid Tumor | 3 | 1 | [217] |
| NCT05071209 | Elimusertib for the Treatment of Relapsed or Refractory Solid Tumors | Recurrent/Refractory Alveolar Rhabdomyosarcoma; Recurrent/Refactory Ewing Sarcoma; | 5–6 | 2 | [218] |
| NCT04067115 | SARC037: A Phase I/II Study to Evaluate the Safety of Trabectedin in Combination With Irinotecan in Ewing Sarcoma Patients | Ewing Sarcoma | 6 | 1 | [208] |
| NCT04537715 | Study to Describe the Interaction Between Tazemetostat and Itraconazole and Between Tazemetostat and Rifampicin in Participants With Advanced Cancer | Epitelioid Sarcoma; Synovial Sarcoma | n.d. | 6 | [219] |
| NCT03793361 | Phase II Study of Regorafenib as Maintenance Therapy | Metastatic Soft Tissue Sarcoma | 10 | 7 | [220] |
| NCT05515575 | A Study of Niraparib in People With Soft Tissue Sarcoma Who Have Changes in Their Tumor DNA | Sarcoma, Soft Tissue | 7 | 1 | [221] |
| NCT05218499 | Brightline-1: A Study to Compare Brigimadlin (BI 907828) With Doxorubicin in People With a Type of Cancer Called Dedifferentiated Liposarcoma | Dedifferentiated Liposarcoma | 6 | 2 | [222] |
| NCT04742959 | Study of TT-00420 (Tinengotinib) Tablet as Monotherapy and Combination Therapy in Patients With Advanced Solid Tumors | Advanced solid tumors | 7–8 | 3 | [203] |
| NCT02095132 | Adavosertib and Irinotecan Hydrochloride in Treating Younger Patients With Relapsed or Refractory Solid Tumors | Relapsed or Refractory Solid Tumors | 10 | 4 | [223] |
| NCT04950075 | Study of INBRX-109 in Conventional Chondrosarcoma (ChonDRAgon) | Conventional Chondrosarcoma | 8 | 2 | [224] |
| NCT03718091 | M6620 (VX-970) in Selected Solid Tumors | Advanced solid tumor | 6 | 1 | [225] |
| NCT03810976 | A Study of Eribulin With Gemcitabine in Patients With Advanced Liposarcoma or Leiomyosarcoma | Liposarcoma or Leiomyosarcoma | 3–4 | 3 | [226] |
| NCT04794127 | Study of Trabectedin in Combination With Pioglitazone in Patients Myxoid Liposarcomas With Stable Disease After T Alone (TRABEPIO) | Myxoid Liposarcomas | 10 | 3 | [227] |
7. Combinatorial Approaches
The combination of different living hosts for xenografts represents a win-win approach which exploits the peculiar traits of each model for specific scientific questions, providing on the one hand the possibility to dissect disease mechanisms with higher robustness, and on the other a more comprehensive insight at the organismal level [228]. Below, some examples of this virtuous approach are reported.
Pignochino et al. demonstrated the efficacy of sorafenib in OS stepwise: the anti-angiogenic action was assessed on naked CAM exposed to the drug itself or to the conditioned medium of cells treated with the drug. As a second instance, the efficacy of sorafenib in restraining the tumorigenic and metastatic ability of OS cells was evaluated via heterotopic mCDX [229]. Analogously, in both ovoCDX and mCDX, and in a mPDX model, the treatment with verteporfin significantly slowed tumor growth. In this case, models were used stepwise from the most time-effective and scalable (ovoCDX) to the most complex, time-consuming, yet near-patient (mPDX), yielding consistent results [127]. Tome and colleagues seeded 143B-LM-RFP OS cells in ovo—the derived tumor masses were then transplanted into several other eggs and treated with echistatin. This protocol allowed the drug to be tested in an established tumor. Subsequently, the efficacy of echistatin diminishing the tumorigenic and metastatic potential of the same cells was demonstrated in mCDXs, in combination with doxorubicin [230]. Analogously, the pro-angiogenic role of soluble BDGF in CHS was firstly assessed in the absence of cells both in ovo and in mice, thereafter, an mCDX of BDGF knock-down cells confirmed the finding at the organismal level [231]. Additionally, the role of IGFB6 on angiogenesis was firstly elucidated injecting its mRNA in the one-stage flk1-GFP embryo in a physiological context—the anti-angiogenic effect was later confirmed in a disease-related context with the injection of RMS cells in mice [232]. Concerning studies in the metastatic setting, Adane and colleagues, after intensive molecular investigations, found confirmation of the anti-tumorigenic role of STAG2 firstly in ZF, where its loss was associated with a higher percentage of embryos with migrated cells, and later in mice, where the injection of the same STAG2-KO cells yielded more lung metastasis with respect to control cells [233].
Generally, drug screening studies proceed stepwise. For example, after identifying active compounds targeting FAK and Aurora Kinase B in EWS via a high-throughput chemical and genetic screening, Wang et al. preliminarily assessed the pharmacokinetic properties and efficacy of zCDX prior to mCDX and mPDX [234]. Remarkably, since all the hosts were characterized by immunodeficiency, the interspecific transfer of cells or tissues from one to another was feasible. Indeed, Yan and colleagues performed high-throughput drug screening on near-patient material in vitro—the most promising drugs were selected for further evaluation and performed on zCDX of dissociated mPDX tumors of two rhabdoid sarcomas [235].
8. Discussion
Survival rates for most MSKT have not improved since the 1980s, when chemotherapy was introduced. This stagnation reflects, on the one hand, a lack of understanding of the biological mechanisms underlying the sarcomagenesis and metastasis processes, and on the other a lack of reliable models to identify effective tumor targets and to testing new drugs preclinically [7,9].
Historically, most of the preclinical research has involved xenografts of cells or tumor fragments in adult rodents, especially mice. Recently, researchers have turned their attention to embryonal models such as chick embryo and zebrafish, which not only represent time- and cost-effective alternative hosts, but most importantly minimize pain, environmental distress and, hence, ethical concerns related to animal experimentation.
With regard to basic research, mCDX involving established gene-edited or drug-resistant cells have permitted the elucidation of the role of tens of genes modifying their tumorigenic or metastatic power (Table 3 and Table 4). In this experimental setting, the integration of fast-developing embryonal models has allowed, on the one hand, the employment of the same transient strategies used in vitro in the in vivo setting and, on the other hand, has raised the possibility of evaluating the phenotype of gene-edited near-patient cells. Thus, embryonal models contribute to time-effectiveness, consistency in methodology, and translatability of results. Murine models offer the unique possibility of studying all the different phases of the natural history of tumors with incomparable reliability, due to the close homology with humans at many levels [7,9,115], albeit blindly. Conversely, embryonal models permit the dissection and visualization of the dynamic interactions between cancer cells, vessels, and the host’s organs, even at single-cell level. Particularly, embryonal models allow the visualization and quantification of sprouting vessels, and the dissemination of tumor cells upon xenografting tumor fragments or cells [144,164,170,177,236]. Notwithstanding these features, researchers must keep in mind that embryos are miniaturized immunodeficient organisms under development, with high levels of specific growth factors, and thus lack target organs and time to model a full-blown metastasis.
Concerning translational research, preclinical trials have successfully relied on mCDX until mPDX took root at the beginning of the century. This practice allowed the expansion and biobanking of patient-derived material, which is particularly relevant in the case of rare and heterogeneous tumors, such as bone and soft tissue sarcomas [9]. mPDXs are acknowledged as the most effective preclinical model for recapitulating intrinsic cancer biology and architecture, and studying drug response and resistance [9]. Preclinical trials taking advantage of mPDXs are relevant throughout the whole course of drug development, including the phases in clinical experimentation. This is important in the context of rare tumors, like sarcomas, for which most clinical studies include patients of all sarcoma histotypes and in small case series. For these reasons, the use of PDX models represents a precious method in preclinical cancer research. Trabectedin is a paradigm of the usefulness of mPDX in the drug development process. This drug was approved in Europe in 2007 for the treatment of advanced STS after the failure of anthracycline and ifosfamide [237]. In 2015, the FDA endorsed its use for the treatment of previously treated advanced metastatic liposarcomas and leyomiosarcomas [238]. mPDXs were used to test trabectedin’s efficacy on sarcomas [239], and recently they were leveraged for the investigation of the molecular mechanisms of actions and of resistance [69]. These findings may give rise to innovative therapeutic combinatorial strategies enhancing the trabectedin effects. Another example of mPDXs’ utility in the management of trabectedin is represented by a recent paper by Zuco et al. studying new therapeutic approaches for desmoplastic small round cell tumor (DSRCT). The authors found that the combination of trabectedin with irinotecan resulted in complete responses in mice trials. These findings strongly support further investigations of this combination in clinical studies on relapsing DSRCT, moreover, these results can also be translated for the development of Lurbinectidin, a novel synthetic agent derived from trabectedin with a similar mechanism of action and that is currently under investigation in combination with irinotecan in sarcomas (NCT02611024) [208]. Nevertheless, preclinical testing on mPDX does not warrant success in clinical evaluation. In this scenario, the introduction of embryonal models in the screening phase of drug development, upon their validation as New Approach Methodologies (NAM), could provide mid-to-high throughput systems in the context of an intact living organism recapitulating human physiology and pharmacokinetics in a reasonable timeframe [165,170]. Finally, the little material required for zCDX and the short experimental time in both embryonal systems makes them appealing as “avatar models” or “mirror models” in a time-frame congruent with clinical decision making [212,213].
Currently, thanks to their predictive value, xenografts are used in co-clinical trials in which the xenograft is treated in parallel with the corresponding patient. This approach facilitates the prioritization of optimal treatments, simplifies rapid classification of responders, identifies biomarkers, and detects mechanisms of resistance [240]. This format of trial has also been conducted for sarcomas [241], with excellent results of concordance between drug response in mice and in corresponding patients. Moreover, not only are mPDXs used in co-clinical trials, but zPDXs are also, although not for sarcomas and instead for other solid tumors [51].
Consisting of multiple cell types and an extracellular matrix, tumors engage in complex interactions with surrounding tissues and the entire body. As a result, they are increasingly perceived as organs [242], the complexity of which cannot be modeled by a single system, neither in vitro nor in vivo. Therefore, while the choice of the xenograft host should align with the experimental question, the combination of different xenograft models based on cells and patient-derived material and the integration of omic, in vitro, and in vivo data could provide robustness to preclinical results [228]. Moreover, the adoption of stepwise approaches envisioning embryonal systems as a bridge between in vitro and in vivo murine experiments could corroborate the first, while reducing the number of more complex animals needed, in compliance with the 3R principles.
9. Conclusions and Future Perspectives
In the present work, we reviewed the literature reporting xenografts of human musculoskeletal sarcomas in mouse, zebrafish larvae, and chick embryos. The categorization of papers based on the underlying biological question allowed us to highlight the intrinsic and technical peculiarities of patient-derived and cell-derived xenografts across the phases of tumor natural history and drug development. Embryonal models emerged as potential tools to corroborate results obtained in gene-edited systems in vitro, using the same transient approaches, thereby increasing robustness and reducing experimental times. Embryonal models are promising systems for the study and visualization of stepwise, dynamic, and interactive processes such as angiogenesis and metastatization, thanks to the ease of imaging and reduced experimental windows. On the other hand, murine xenografts allow the study of overt metastatization and the creation of living biobanks of sarcomas. Thanks to their fidelity, mPDXs represent the gold standard for drug development, which could benefit from the introduction of high-throughput embryonal systems in the screening phase.
Overall, the application of embryonal systems in sarcomas is still limited. Yet, the combination of different hosts for xenografts in stepwise approaches involving near-patient material promises to provide reliable high-throughput and highly informative models, boosting both basic and translational research in respect of the 3R principles
Acronyms
| Adm | Adrenomedullin |
| Akt | AKT Serine/Threonine Kinase 1 |
| AI | Artificial intelligence |
| ALU | stretch of DNA characterized by the action of endonuclease from Arthrobacter luteus (Alu) |
| Atr | ATR Serine/Threonine Kinase |
| Bcl-2 | B-Cell Leukemia/Lymphoma 2 Protein. |
| Bdgf | Brain-Derived Nerve Growth Factor |
| Bets | Bromodomain Extraterminal Domain Proteins |
| Bmp-2 | Bone Morphogenetic Protein 2 |
| Cav1 | Caveolin 1 |
| Cdk | Cyclin-Dependent Kinase |
| Cdkn1c/2a | Cyclin Dependent Kinase Inhibitor 1c/2a |
| Chek1 | Checkpoint Kinase 1 |
| Cic-Dux4 | Fusion; Capicua Transcriptional Repressor- Double Homeobox 4 |
| CMET | Hepatocyte Growth Factor Receptor |
| Csf1r | Colony-Stimulating Factor-1 |
| Cxcr4 | C-X-C Chemokine Receptor Type 4 |
| Ews::Fli1 | Fusion; EWS RNA Binding Protein 1:: Fli-1 Proto-Oncogene, ETS Transcription Factor |
| Ext1/2 | Exostosin Glycosyltransferase 1 |
| Eya3 | Eya Transcriptional Coactivator and Phosphatase 3 |
| Ezh2 | Enhancer of Zeste 2 Polycomb Repressive Complex 2 Subunit |
| Fak | Focal Adhesion Kinase |
| FDA | Food and Drug Administration |
| Fgd1 | Fyve, Rhogef And Ph Domain Containing 1 |
| Fli1 | Friend Leukemia Integration 1 Transcription Factor |
| Foxp1 | Forkhead Box Protein P1 |
| Fus-Chop | Fusion Protein; Fus RNA Binding Protein- DNA Damage Inducible Transcript 3 |
| Fus-Erg | Fusion; FUS RNA Binding Protein- ETS Transcription Factor ERG |
| Gfra1 | Gdnf Family Receptor Alpha-1 |
| Gli | GLI Family Zinc Finger 1 |
| Gαi3 | Guanine Nucleotide-Binding Protein G(I) Subunit Alpha-3 |
| H3k27ac | Acetylation of Histon3 in lysine (k) 27 |
| Hdac | Histone Deacetylases |
| Hes1 | Hes Family Bhlh Transcription Factor 1 |
| IC50 | Half Maximal Inhibitory Concentration |
| IDH1/2 | Isocitrate Dehydrogenase (NADP(+)) 1/2 |
| Igf2bp3 | Insulin Like Growth Factor 2 mRNA Binding Protein 3 |
| Igfb6 | Insulin Like Growth Factor Binding Protein 6 |
| Igfbp5 | Insulin-Like Growth Factor-Binding Protein 5 |
| IHC | immunohistochemistry |
| Itga5 | Integrin Subunit Alpha 5 |
| Kif18b | Kinesin Family Member 18b |
| Klf15 | Klf Transcription Factor 15 |
| Lpar1 | Lysophosphatidic Acid Receptor 1 |
| Mdm2 | MDM2 Proto-Oncogene |
| Mig-7 | Migration Inducting Gene 7 |
| mTOR | Mammalian Target of Rapamycin |
| Ng6x | Nasopharyngeal Carcinoma-Associated Gene 6 |
| Panx1 | Pannexin 1 |
| Pax(n)::FoxoA1 | Fusion; Paired Box (n):: Forkhead Box O1 |
| PDGRF-Β | Platelet-Derived Growth Factor Receptor Beta |
| Pdpn | Podoplanin |
| Plk2 | Polo Like Kinase 2 |
| Porcn | Porcupine O-Acyltransferase |
| Pten | Phosphatase And Tensin Homolog |
| qPCR | Quantitative Polyerase Chain Reaction |
| Rab22a::Neof1 | Fusion Protein; Ras-Related Protein Rab-22A |
| Ras-PI3K | RAS Proto-Oncogene, GTPase-Phosphatidylinositol-4,5-Bisphosphate 3-Kinase |
| Rb1 | RB Transcriptional Corepressor 1 |
| Ripk1 | Receptor Interacting Serine/Threonine Kinase 1 |
| Rock2 | Rho Associated Coiled-Coil Containing Protein Kinase 2 |
| Ron | Macrophage Stimulating 1 Receptor |
| S1pr3 | Sphingosine-1-Phosphate Receptor 3 |
| Sh Gαi3 | Short Hairpin Guanine Nucleotide-Binding Protein G(I) Subunit Alpha-3 |
| Sh Foxp1 | Short Hairpin Forkhead Box P1 |
| Sirt1/2 | Sirtuin 1/2 |
| Smo:: | Smoothened, Frizzled Class Receptor |
| Sox6 | Sry-Box Transcription Factor 6 |
| Syt::Ssx1/2/4 | SS18 Subunit Of BAF Chromatin Remodeling Complex:: SSX Family Member 1/2/4 |
| Sparc | Secreted Protein Acidic and Cysteine Rich |
| Stag2 | Stag2 Cohesin Complex Component |
| Stat3 | Signal Transducer and Activator of Transcription 3 |
| Tap73 | Tumor Protein P73 |
| Taz | Transcriptional Coactivator with Pdz-Binding Motif |
| Tcf4 | Transcription Factor 4 |
| Tead | Tea Domain Transcription Factor |
| Timm13 | Translocase Of Inner Mitochondrial Membrane 13 |
| Tp53 | Tumor Protein P53 |
| Tk | Tyrosine Kinases |
| Trim7 | Tripartite Motif Containing 7 |
| Wnt | Wingless/Integrated |
| Yap1 | Yes-Associated Protein 1 |
| Yhr | Yaps127a-Htert-Hrasg12v |
| Yhv | Yaps127a-Htert-Vector |
| Zyx | Zyxin |
Author Contributions
Conceptualization, V.G., M.T., L.M., E.L., G.M. and A.D.V.; writing—original draft preparation, V.G., M.T., G.M. and M.P.; writing—review and editing, L.M., E.L., C.M.H., M.C., G.S., L.F., A.N.G., C.B., C.S., D.M.D. and T.I. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This work was supported by the European Union—Next Generation EU—NRRP M6C2—Investment 2.1 Enhancement and strengthening of biomedical research in the NHS—project PNRR-POC-2022-12376579 (CUP: D33C22001940006).
![]()
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Grünewald T.G., Alonso M., Avnet S., Banito A., Burdach S., Cidre-Aranaz F., Di Pompo G., Distel M., Dorado-Garcia H., Garcia-Castro J., et al. Sarcoma Treatment in the Era of Molecular Medicine. EMBO Mol. Med. 2020;12:e11131. doi: 10.15252/emmm.201911131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Damerell V., Pepper M.S., Prince S. Molecular Mechanisms Underpinning Sarcomas and Implications for Current and Future Therapy. Signal Transduct. Target. Ther. 2021;6:246. doi: 10.1038/s41392-021-00647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi J.H., Ro J.Y. The 2020 WHO Classification of Tumors of Bone: An Updated Review. Adv. Anat. Pathol. 2021;28:119–138. doi: 10.1097/PAP.0000000000000293. [DOI] [PubMed] [Google Scholar]
- 4.Sbaraglia M., Bellan E., Dei Tos A.P. The 2020 WHO Classification of Soft Tissue Tumours: News and Perspectives. Pathologica. 2021;113:70–84. doi: 10.32074/1591-951X-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stacchiotti S., Frezza A.M., Blay J., Baldini E.H., Bonvalot S., Bovée J.V.M.G., Callegaro D., Casali P.G., Chiang R.C., Demetri G.D., et al. Ultra-rare Sarcomas: A Consensus Paper from the Connective Tissue Oncology Society Community of Experts on the Incidence Threshold and the List of Entities. Cancer. 2021;127:2934–2942. doi: 10.1002/cncr.33618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoang N.T., Acevedo L.A., Mann M.J., Tolani B. A Review of Soft-Tissue Sarcomas: Translation of Biological Advances into Treatment Measures. Cancer Manag. Res. 2018;10:1089–1114. doi: 10.2147/CMAR.S159641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu W., Chao T., Ruiqi C., Juan S., Zhihong L. Patient-Derived Xenograft Models in Musculoskeletal Malignancies. J. Transl. Med. 2018;16:107. doi: 10.1186/s12967-018-1487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Bono J.S., Ashworth A. Translating Cancer Research into Targeted Therapeutics. Nature. 2010;467:543–549. doi: 10.1038/nature09339. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y., Wu W., Cai C., Zhang H., Shen H., Han Y. Patient-Derived Xenograft Models in Cancer Therapy: Technologies and Applications. Signal Transduct. Target. Ther. 2023;8:160. doi: 10.1038/s41392-023-01419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blomme A., Van Simaeys G., Doumont G., Costanza B., Bellier J., Otaka Y., Sherer F., Lovinfosse P., Boutry S., Palacios A.P., et al. Murine Stroma Adopts a Human-like Metabolic Phenotype in the PDX Model of Colorectal Cancer and Liver Metastases. Oncogene. 2018;37:1237–1250. doi: 10.1038/s41388-017-0018-x. [DOI] [PubMed] [Google Scholar]
- 11.Landuzzi L., Palladini A., Ceccarelli C., Asioli S., Nicoletti G., Giusti V., Ruzzi F., Ianzano M.L., Scalambra L., Laranga R., et al. Early Stability and Late Random Tumor Progression of a HER2-Positive Primary Breast Cancer Patient-Derived Xenograft. Sci. Rep. 2021;11:1563. doi: 10.1038/s41598-021-81085-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sunil H.S., O’Donnell K.A. Capturing Heterogeneity in PDX Models: Representation Matters. Nat. Commun. 2024;15:4652. doi: 10.1038/s41467-024-47607-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillén J., Robinson S., Decelle T., Exner C., van Vlissingen M.F. Approaches to Animal Research Project Evaluation in Europe after Implementation of Directive 2010/63/EU. Lab Anim. 2015;44:23–31. doi: 10.1038/laban.604. [DOI] [PubMed] [Google Scholar]
- 14.Brown H.K., Schiavone K., Tazzyman S., Heymann D., Chico T.J. Zebrafish Xenograft Models of Cancer and Metastasis for Drug Discovery. Expert Opin. Drug Discov. 2017;12:379–389. doi: 10.1080/17460441.2017.1297416. [DOI] [PubMed] [Google Scholar]
- 15.Miebach L., Berner J., Bekeschus S. In Ovo Model in Cancer Research and Tumor Immunology. Front. Immunol. 2022;13:1006064. doi: 10.3389/fimmu.2022.1006064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.European Union Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes. Off. J. Eur. Union. 2010;L276:33–80. [Google Scholar]
- 17.Strähle U., Scholz S., Geisler R., Greiner P., Hollert H., Rastegar S., Schumacher A., Selderslaghs I., Weiss C., Witters H., et al. Zebrafish Embryos as an Alternative to Animal Experiments—A Commentary on the Definition of the Onset of Protected Life Stages in Animal Welfare Regulations. Reprod. Toxicol. 2012;33:128–132. doi: 10.1016/j.reprotox.2011.06.121. [DOI] [PubMed] [Google Scholar]
- 18.Kollmansperger S., Anders M., Werner J., Saller A.M., Weiss L., Süß S.C., Reiser J., Schneider G., Schusser B., Baumgartner C., et al. Nociception in Chicken Embryos, Part II: Embryonal Development of Electroencephalic Neuronal Activity In Ovo as a Prerequisite for Nociception. Animals. 2023;13:2839. doi: 10.3390/ani13182839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss L., Saller A.M., Werner J., Süß S.C., Reiser J., Kollmansperger S., Anders M., Potschka H., Fenzl T., Schusser B., et al. Nociception in Chicken Embryos, Part I: Analysis of Cardiovascular Responses to a Mechanical Noxious Stimulus. Animals. 2023;13:2710. doi: 10.3390/ani13172710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zanella E.R., Grassi E., Trusolino L. Towards Precision Oncology with Patient-Derived Xenografts. Nat. Rev. Clin. Oncol. 2022;19:719–732. doi: 10.1038/s41571-022-00682-6. [DOI] [PubMed] [Google Scholar]
- 21.Howe K., Clark M.D., Torroja C.F., Torrance J., Berthelot C., Muffato M., Collins J.E., Humphray S., McLaren K., Matthews L., et al. The Zebrafish Reference Genome Sequence and Its Relationship to the Human Genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacRae C.A., Peterson R.T. Zebrafish as Tools for Drug Discovery. Nat. Rev. Drug Discov. 2015;14:721–731. doi: 10.1038/nrd4627. [DOI] [PubMed] [Google Scholar]
- 23.Mapanao A.K., Che P.P., Sarogni P., Sminia P., Giovannetti E., Voliani V. Tumor Grafted—Chick Chorioallantoic Membrane as an Alternative Model for Biological Cancer Research and Conventional/Nanomaterial-Based Theranostics Evaluation. Expert Opin. Drug Metab. Toxicol. 2021;17:947–968. doi: 10.1080/17425255.2021.1879047. [DOI] [PubMed] [Google Scholar]
- 24.Kue C.S., Tan K.Y., Lam M.L., Lee H.B. Chick Embryo Chorioallantoic Membrane (CAM): An Alternative Predictive Model in Acute Toxicological Studies for Anti-Cancer Drugs. Exp. Anim. 2015;64:129–138. doi: 10.1538/expanim.14-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manjunathan R., Ragunathan M. Chicken Chorioallantoic Membrane as a Reliable Model to Evaluate Osteosarcoma—An Experimental Approach Using SaOS2 Cell Line. Biol. Proced. Online. 2015;17:10. doi: 10.1186/s12575-015-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picci P., Manfrini M., Fabbri N., Gambarotti M., Vanel D., editors. Atlas of Musculoskeletal Tumors and Tumorlike Lesions: The Rizzoli Case Archive. Springer International Publishing; Cham, Switzerland: 2014. [DOI] [Google Scholar]
- 27.Beird H.C., Bielack S.S., Flanagan A.M., Gill J., Heymann D., Janeway K.A., Livingston J.A., Roberts R.D., Strauss S.J., Gorlick R. Osteosarcoma. Nat. Rev. Dis. Primers. 2022;8:7. doi: 10.1038/s41572-022-00409-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riggi N., Suvà M.L., Stamenkovic I. Ewing’s Sarcoma. N. Engl. J. Med. 2021;384:154–164. doi: 10.1056/NEJMra2028910. [DOI] [PubMed] [Google Scholar]
- 29.Weinschenk R.C., Wang W.-L., Lewis V.O. Chondrosarcoma. J. Am. Acad. Orthop. Surg. 2021;29:553–562. doi: 10.5435/JAAOS-D-20-01188. [DOI] [PubMed] [Google Scholar]
- 30.Gazendam A., Popovic S., Parasu N., Ghert M. Chondrosarcoma: A Clinical Review. J. Clin. Med. 2023;12:2506. doi: 10.3390/jcm12072506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorkildsen J., Myklebust T.Å. The National Incidence of Chondrosarcoma of Bone; a Review. Acta Oncol. 2023;62:110–117. doi: 10.1080/0284186X.2023.2177975. [DOI] [PubMed] [Google Scholar]
- 32.Thway K. Well-Differentiated Liposarcoma and Dedifferentiated Liposarcoma: An Updated Review. Semin. Diagn. Pathol. 2019;36:112–121. doi: 10.1053/j.semdp.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Gronchi A., Miah A.B., Dei Tos A.P., Abecassis N., Bajpai J., Bauer S., Biagini R., Bielack S., Blay J.Y., Bolle S., et al. Soft Tissue and Visceral Sarcomas: ESMO-EURACAN-GENTURIS Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up☆. Ann. Oncol. 2021;32:1348–1365. doi: 10.1016/j.annonc.2021.07.006. [DOI] [PubMed] [Google Scholar]
- 34.Skapek S.X., Ferrari A., Gupta A.A., Lupo P.J., Butler E., Shipley J., Barr F.G., Hawkins D.S. Rhabdomyosarcoma. Nat. Rev. Dis. Primer. 2019;5:1. doi: 10.1038/s41572-018-0051-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishio J., Nakayama S. Biology and Management of High-Grade Myxofibrosarcoma: State of the Art and Future Perspectives. Diagnostics. 2023;13:3022. doi: 10.3390/diagnostics13193022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vanni S., De Vita A., Gurrieri L., Fausti V., Miserocchi G., Spadazzi C., Liverani C., Cocchi C., Calabrese C., Bongiovanni A., et al. Myxofibrosarcoma Landscape: Diagnostic Pitfalls, Clinical Management and Future Perspectives. Ther. Adv. Med. Oncol. 2022;14:17588359221093973. doi: 10.1177/17588359221093973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blay J.-Y., von Mehren M., Jones R.L., Martin-Broto J., Stacchiotti S., Bauer S., Gelderblom H., Orbach D., Hindi N., Dei Tos A., et al. Synovial Sarcoma: Characteristics, Challenges, and Evolving Therapeutic Strategies. ESMO Open. 2023;8:101618. doi: 10.1016/j.esmoop.2023.101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun H., Liu J., Hu F., Xu M., Leng A., Jiang F., Chen K. Current Research and Management of Undifferentiated Pleomorphic Sarcoma/Myofibrosarcoma. Front. Genet. 2023;14:1109491. doi: 10.3389/fgene.2023.1109491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.PDQ Cancer Information Summaries [Internet]. Bethesda (MD): National Cancer Institute (US); 2002. Dictionary of Cancer Terms. [(accessed on 15th June 2024)]; Available online: https://www.ncbi.nlm.nih.gov/books/NBK65951/
- 40.Orphanet: Knowledge on Rare Diseases and Orphan Drugs. [(accessed on 4 July 2024)]. Available online: https://www.orpha.net.
- 41.Ribatti D. The Chick Embryo Chorioallantoic Membrane Patient-Derived Xenograft (PDX) Model. Pathol. Res. Pract. 2023;243:154367. doi: 10.1016/j.prp.2023.154367. [DOI] [PubMed] [Google Scholar]
- 42.Fazio M., Ablain J., Chuan Y., Langenau D.M., Zon L.I. Zebrafish Patient Avatars in Cancer Biology and Precision Cancer Therapy. Nat. Rev. Cancer. 2020;20:263–273. doi: 10.1038/s41568-020-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morton C.L., Houghton P.J. Establishment of Human Tumor Xenografts in Immunodeficient Mice. Nat. Protoc. 2007;2:247–250. doi: 10.1038/nprot.2007.25. [DOI] [PubMed] [Google Scholar]
- 44.Sys G.M.L., Lapeire L., Stevens N., Favoreel H., Forsyth R., Bracke M., De Wever O. The In Ovo CAM-Assay as a Xenograft Model for Sarcoma. J. Vis. Exp. 2013;77:50522. doi: 10.3791/50522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shoji C., Kikuchi K., Yoshida H., Miyachi M., Yagyu S., Tsuchiya K., Nakaya T., Hosoi H., Iehara T. In Ovo Chorioallantoic Membrane Assay as a Xenograft Model for Pediatric Rhabdomyosarcoma. Oncol. Rep. 2023;49:76. doi: 10.3892/or.2023.8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teng Y., Xie X., Walker S., White D.T., Mumm J.S., Cowell J.K. Evaluating Human Cancer Cell Metastasis in Zebrafish. BMC Cancer. 2013;13:453. doi: 10.1186/1471-2407-13-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paatero I., Alve S., Gramolelli S., Ivaska J., Ojala P. Zebrafish Embryo Xenograft and Metastasis Assay. Bio-Protocol. 2018;8:e3027. doi: 10.21769/BioProtoc.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Villanueva H., Sikora A.G. Methods in Molecular Biology. Volume 2471. Humana Press; New York, NY, USA: 2022. The Chicken Embryo Chorioallantoic Membrane (CAM): A Versatile Tool for the Study of Patient-Derived Xenografts; pp. 209–220. [DOI] [PubMed] [Google Scholar]
- 49.Giovanella B.C., Stehlin J.S., Williams L.J., Lee S.S., Shepard R.C. Heterotransplantation of Human Cancers into Nude Mice: A Model System for Human Cancer Chemotherapy. Cancer. 1978;42:2269–2281. doi: 10.1002/1097-0142(197811)42:5<2269::aid-cncr2820420527>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 50.Growth and Characterization of Childhood Rhabdomyosarcomas as Xenografts—PubMed. [(accessed on 27 June 2024)]; Available online: https://pubmed.ncbi.nlm.nih.gov/6950170/
- 51.Bauer H.C., Brosjö O., Broström L.A., Nilsson O.S., Reinholt F.P., Tribukait B. Growth and Ploidy of Human Osteosarcoma Xenografts in Serial Passage in Nude Mice. Eur. J. Cancer Clin. Oncol. 1986;22:821–830. doi: 10.1016/0277-5379(86)90369-x. [DOI] [PubMed] [Google Scholar]
- 52.Fortuna-Costa A., Alcantara Granato R., Meohas W., de Sá Lopes A.C., Cunha Caruso A., Castro e Silva Pinheiro R., da Gama d’Eça P., Braga Dias R., Perini J.A., Fernandes Barbosa A.P., et al. An Association between Successful Engraftment of Osteosarcoma Patient-Derived Xenografts and Clinicopathological Findings. Histol. Histopathol. 2020;35:1295–1307. doi: 10.14670/HH-18-256. [DOI] [PubMed] [Google Scholar]
- 53.Nanni P., Landuzzi L., Manara M.C., Righi A., Nicoletti G., Cristalli C., Pasello M., Parra A., Carrabotta M., Ferracin M., et al. Bone Sarcoma Patient-Derived Xenografts Are Faithful and Stable Preclinical Models for Molecular and Therapeutic Investigations. Sci. Rep. 2019;9:12174. doi: 10.1038/s41598-019-48634-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sys G., Van Bockstal M., Forsyth R., Balke M., Poffyn B., Uyttendaele D., Bracke M., De Wever O. Tumor Grafts Derived from Sarcoma Patients Retain Tumor Morphology, Viability, and Invasion Potential and Indicate Disease Outcomes in the Chick Chorioallantoic Membrane Model. Cancer Lett. 2012;326:69–78. doi: 10.1016/j.canlet.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 55.Eckrich J., Kugler P., Buhr C.R., Ernst B.P., Mendler S., Baumgart J., Brieger J., Wiesmann N. Monitoring of Tumor Growth and Vascularization with Repetitive Ultrasonography in the Chicken Chorioallantoic-Membrane-Assay. Sci. Rep. 2020;10:18585. doi: 10.1038/s41598-020-75660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feder A.-L., Pion E., Troebs J., Lenze U., Prantl L., Htwe M.M., Phyo A., Haerteis S., Aung T. Extended Analysis of Intratumoral Heterogeneity of Primary Osteosarcoma Tissue Using 3D-in-Vivo-Tumor-Model. Clin. Hemorheol. Microcirc. 2020;76:133–141. doi: 10.3233/CH-209204. [DOI] [PubMed] [Google Scholar]
- 57.Knighton D., Ausprunk D., Tapper D., Folkman J. Avascular and Vascular Phases of Tumour Growth in the Chick Embryo. Br. J. Cancer. 1977;35:347–356. doi: 10.1038/bjc.1977.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Di Franco G., Usai A., Funel N., Palmeri M., Montesanti I.E.R., Bianchini M., Gianardi D., Furbetta N., Guadagni S., Vasile E., et al. Use of Zebrafish Embryos as Avatar of Patients with Pancreatic Cancer: A New Xenotransplantation Model towards Personalized Medicine. World J. Gastroenterol. 2020;26:2792–2809. doi: 10.3748/wjg.v26.i21.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Usai A., Di Franco G., Piccardi M., Cateni P., Pollina L.E., Vivaldi C., Vasile E., Funel N., Palmeri M., Dente L., et al. Zebrafish Patient-Derived Xenografts Identify Chemo-Response in Pancreatic Ductal Adenocarcinoma Patients. Cancers. 2021;13:4131. doi: 10.3390/cancers13164131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan C., Brunson D.C., Tang Q., Do D., Iftimia N.A., Moore J.C., Hayes M.N., Welker A.M., Garcia E.G., Dubash T.D., et al. Visualizing Engrafted Human Cancer and Therapy Responses in Immunodeficient Zebrafish. Cell. 2019;177:1903–1914.e14. doi: 10.1016/j.cell.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Claridge S.E., Cavallo J.-A., Hopkins B.D. Patient-Derived In Vitro and In Vivo Models of Cancer. Adv. Exp. Med. Biol. 2022;1361:215–233. doi: 10.1007/978-3-030-91836-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liverani C., La Manna F., Groenewoud A., Mercatali L., Van Der Pluijm G., Pieri F., Cavaliere D., De Vita A., Spadazzi C., Miserocchi G., et al. Innovative Approaches to Establish and Characterize Primary Cultures: An Ex Vivo 3D System and the Zebrafish Model. Biol. Open. 2017;6:133–140. doi: 10.1242/bio.022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meohas W., Granato R.A., Guimarães J.A.M., Dias R.B., Fortuna-Costa A., Duarte M.E.L. Patient-derived xenografts as a preclinical model for bone sarcomas. Acta Ortop. Bras. 2018;26:98–102. doi: 10.1590/1413-785220182602186998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rainusso N., Cleveland H., Hernandez J.A., Quintanilla N.M., Hicks J., Vasudevan S., Marco R.A.W., Allen-Rhoades W., Wang L.L., Yustein J.T. Generation of Patient-Derived Tumor Xenografts from Percutaneous Tumor Biopsies in Children with Bone Sarcomas. Pediatr. Blood Cancer. 2019;66:e27579. doi: 10.1002/pbc.27579. [DOI] [PubMed] [Google Scholar]
- 65.Cornillie J., Wozniak A., Li H., Wang Y., Boeckx B., Gebreyohannes Y.K., Wellens J., Vanleeuw U., Hompes D., Stas M., et al. Establishment and Characterization of Histologically and Molecularly Stable Soft-Tissue Sarcoma Xenograft Models for Biological Studies and Preclinical Drug Testing. Mol. Cancer Ther. 2019;18:1168–1178. doi: 10.1158/1535-7163.MCT-18-1045. [DOI] [PubMed] [Google Scholar]
- 66.Schoffski P., Van Renterghem B., Cornillie J., Wang Y., Gebreyohannes Y.K., Lee C.-J., Wellens J., Vanleeuw U., Nysen M., Hompes D., et al. XenoSarc: A Comprehensive Platform of Patient-Derived Xenograft (PDX) Models of Soft Tissue Sarcoma (STS) for Early Drug Testing. J. Glob. Oncol. 2019;5:37. doi: 10.1200/JGO.2019.5.suppl.37. [DOI] [Google Scholar]
- 67.Pathmanapan S., Ilkayeva O., Martin J.T., Loe A.K.H., Zhang H., Zhang G.-F., Newgard C.B., Wunder J.S., Alman B.A. Mutant IDH and Non-Mutant Chondrosarcomas Display Distinct Cellular Metabolomes. Cancer Metab. 2021;9:13. doi: 10.1186/s40170-021-00247-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frapolli R., Tamborini E., Virdis E., Bello E., Tarantino E., Marchini S., Grosso F., Sanfilippo R., Gronchi A., Tercero J.C., et al. Novel Models of Myxoid Liposarcoma Xenografts Mimicking the Biological and Pharmacologic Features of Human Tumors. Clin. Cancer Res. 2010;16:4958–4967. doi: 10.1158/1078-0432.CCR-10-0317. [DOI] [PubMed] [Google Scholar]
- 69.Mannarino L., Craparotta I., Ballabio S., Frapolli R., Meroni M., Bello E., Panini N., Callari M., Sanfilippo R., Casali P.G., et al. Mechanisms of Responsiveness to and Resistance against Trabectedin in Murine Models of Human Myxoid Liposarcoma. Genomics. 2021;113:3439–3448. doi: 10.1016/j.ygeno.2021.07.028. [DOI] [PubMed] [Google Scholar]
- 70.Castillo-Ecija H., Pascual-Pasto G., Perez-Jaume S., Resa-Pares C., Vila-Ubach M., Monterrubio C., Jimenez-Cabaco A., Baulenas-Farres M., Muñoz-Aznar O., Salvador N., et al. Prognostic Value of Patient-Derived Xenograft Engraftment in Pediatric Sarcomas. J. Pathol. Clin. Res. 2021;7:338–349. doi: 10.1002/cjp2.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pandya P.H., Jannu A.J., Bijangi-Vishehsaraei K., Dobrota E., Bailey B.J., Barghi F., Shannon H.E., Riyahi N., Damayanti N.P., Young C., et al. Integrative Multi-OMICs Identifies Therapeutic Response Biomarkers and Confirms Fidelity of Clinically Annotated, Serially Passaged Patient-Derived Xenografts Established from Primary and Metastatic Pediatric and AYA Solid Tumors. Cancers. 2022;15:259. doi: 10.3390/cancers15010259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen F., Zhang Z., Shen R., Chen M., Li G., Zhu X. Generation and Characterization of Patient-Derived Xenografts from Patients with Osteosarcoma. Tissue Cell. 2022;79:101911. doi: 10.1016/j.tice.2022.101911. [DOI] [PubMed] [Google Scholar]
- 73.Xu C., Yan L., An Q., Zhang S., Guan X., Wang Z., Lv A., Liu D., Liu F., Dong B., et al. Establishment and Evaluation of Retroperitoneal Liposarcoma Patient-Derived Xenograft Models: An Ideal Model for Preclinical Study. Int. J. Med. Sci. 2022;19:1241–1253. doi: 10.7150/ijms.70706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun M., Wang Z., Sun W., Chen M., Ma X., Shen J., Fu Z., Zuo D., Wang G., Wang H., et al. Correlation between Patient-Derived Xenograft Modeling and Prognosis in Osteosarcoma. Orthop. Surg. 2022;14:1161–1166. doi: 10.1111/os.13211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hemming M.L., Bhola P., Loycano M.A., Anderson J.A., Taddei M.L., Doyle L.A., Lavrova E., Andersen J.L., Klega K.S., Benson M.R., et al. Preclinical Modeling of Leiomyosarcoma Identifies Susceptibility to Transcriptional CDK Inhibitors through Antagonism of E2F-Driven Oncogenic Gene Expression. Clin. Cancer Res. 2022;28:2397–2408. doi: 10.1158/1078-0432.CCR-21-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marques Da Costa M.E., Zaidi S., Scoazec J.-Y., Droit R., Lim W.C., Marchais A., Salmon J., Cherkaoui S., Morscher R.J., Laurent A., et al. A Biobank of Pediatric Patient-Derived-Xenograft Models in Cancer Precision Medicine Trial MAPPYACTS for Relapsed and Refractory Tumors. Commun. Biol. 2023;6:949. doi: 10.1038/s42003-023-05320-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun H., Cao S., Mashl R.J., Mo C.-K., Zaccaria S., Wendl M.C., Davies S.R., Bailey M.H., Primeau T.M., Hoog J., et al. Comprehensive Characterization of 536 Patient-Derived Xenograft Models Prioritizes Candidatesfor Targeted Treatment. Nat. Commun. 2021;12:5086. doi: 10.1038/s41467-021-25177-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gasparini P., Casanova M., Centonze G., Borzi C., Bergamaschi L., Collini P., Testi A., Chiaravalli S., Massimino M., Sozzi G., et al. Establishment of 6 Pediatric Rhabdomyosarcoma Patient’s Derived Xenograft Models Closely Recapitulating Patients’ Tumor Characteristics. Tumori. 2023;109:314–323. doi: 10.1177/03008916221110266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kresse S.H., Meza-Zepeda L.A., Machado I., Llombart-Bosch A., Myklebost O. Preclinical Xenograft Models of Human Sarcoma Show Nonrandom Loss of Aberrations. Cancer. 2012;118:558–570. doi: 10.1002/cncr.26276. [DOI] [PubMed] [Google Scholar]
- 80.Kuijjer M.L., Namløs H.M., Hauben E.I., Machado I., Kresse S.H., Serra M., Llombart-Bosch A., Hogendoorn P.C.W., Meza-Zepeda L.A., Myklebost O., et al. mRNA Expression Profiles of Primary High-Grade Central Osteosarcoma Are Preserved in Cell Lines and Xenografts. BMC Med. Genom. 2011;4:66. doi: 10.1186/1755-8794-4-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Russell T.A., Eckardt M.A., Murakami T., Elliott I.A., Kawaguchi K., Kiyuna T., Igarashi K., Li Y., Crompton J.G., Graham D.S., et al. Clinical Factors That Affect the Establishment of Soft Tissue Sarcoma Patient-Derived Orthotopic Xenografts: A University of California, Los Angeles, Sarcoma Program Prospective Clinical Trial. JCO Precis. Oncol. 2017;1:1–13. doi: 10.1200/PO.17.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stewart E., Federico S.M., Chen X., Shelat A.A., Bradley C., Gordon B., Karlstrom A., Twarog N.R., Clay M.R., Bahrami A., et al. Orthotopic Patient-Derived Xenografts of Paediatric Solid Tumours. Nature. 2017;549:96–100. doi: 10.1038/nature23647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Calvert N., Wu J., Sneddon S., Woodhouse J., Carey-Smith R., Wood D., Ingley E. The Use of Whole Exome Sequencing and Murine Patient Derived Xenografts as a Method of Chemosensitivity Testing in Sarcoma. Clin. Sarcoma Res. 2018;8:4. doi: 10.1186/s13569-018-0090-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yan J., Wang Q., Zou K., Wang L., Schwartz E.B., Fuchs J.R., Zheng Z., Wu J. Inhibition of the JAK2/STAT3 Signaling Pathway Exerts a Therapeutic Effect on Osteosarcoma. Mol. Med. Rep. 2015;12:498–502. doi: 10.3892/mmr.2015.3439. [DOI] [PubMed] [Google Scholar]
- 85.Lee H.-J., Cao Y., Pham V., Blackwood E., Wilson C., Evangelista M., Klijn C., Stokoe D., Settleman J. Ras-MEK Signaling Mediates a Critical Chk1-Dependent DNA Damage Response in Cancer Cells. Mol. Cancer Ther. 2017;16:694–704. doi: 10.1158/1535-7163.MCT-16-0504. [DOI] [PubMed] [Google Scholar]
- 86.Kim M., Jung J.-Y., Choi S., Lee H., Morales L.D., Koh J.-T., Kim S.H., Choi Y.-D., Choi C., Slaga T.J., et al. GFRA1 Promotes Cisplatin-Induced Chemoresistance in Osteosarcoma by Inducing Autophagy. Autophagy. 2017;13:149–168. doi: 10.1080/15548627.2016.1239676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shen Y., Zhao S., Wang S., Pan X., Zhang Y., Xu J., Jiang Y., Li H., Zhang Q., Gao J., et al. S1P/S1PR3 Axis Promotes Aerobic Glycolysis by YAP/c-MYC/PGAM1 Axis in Osteosarcoma. EBioMedicine. 2019;40:210–223. doi: 10.1016/j.ebiom.2018.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu W., Jing D., Meng Z., Hu B., Zhong B., Deng X., Jin X., Shao Z. FGD1 Promotes Tumor Progression and Regulates Tumor Immune Response in Osteosarcoma via Inhibiting PTEN Activity. Theranostics. 2020;10:2859–2871. doi: 10.7150/thno.41279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu W., Yu Z., Tang H., Wang X., Zhang B., Zhao J., Liu X., Zhang J., Wei M. Silencing KIF18B Enhances Radiosensitivity: Identification of a Promising Therapeutic Target in Sarcoma. EBioMedicine. 2020;61:103056. doi: 10.1016/j.ebiom.2020.103056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu L., Wang R., Zhang Z., Wang X. Nasopharyngeal Carcinoma-associated Gene 6 Inhibits Cell Viability, Migration, Invasion and Induces Apoptosis in Osteosarcoma Cells by Inactivating the Wnt/Β-catenin Signaling Pathway. Mol. Med. Rep. 2021;23:93. doi: 10.3892/mmr.2020.11732. [DOI] [PubMed] [Google Scholar]
- 91.Li H., Han X., Yang S., Wang Y., Dong Y., Tang T. FOXP1 Drives Osteosarcoma Development by Repressing P21 and RB Transcription Downstream of P53. Oncogene. 2021;40:2785–2802. doi: 10.1038/s41388-021-01742-4. [DOI] [PubMed] [Google Scholar]
- 92.Bian Z.-J., Shan H.-J., Zhu Y.-R., Shi C., Chen M.-B., Huang Y.-M., Wang X.-D., Zhou X.-Z., Cao C. Identification of Gαi3 as a Promising Target for Osteosarcoma Treatment. Int. J. Biol. Sci. 2022;18:1508–1520. doi: 10.7150/ijbs.68861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Han Q., Yan P., Song R., Liu F., Tian Q. HOXC13-Driven TIMM13 Overexpression Promotes Osteosarcoma Cell Growth. Cell Death Dis. 2023;14:398. doi: 10.1038/s41419-023-05910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brion R., Gantier M., Biteau K., Taurelle J., Brounais-Le Royer B., Verrecchia F., Rédini F., Guiho R. TRAIL-Based Therapies Efficacy in Pediatric Bone Tumors Models Is Modulated by TRAIL Non-Apoptotic Pathway Activation via RIPK1 Recruitment. Cancers. 2022;14:5627. doi: 10.3390/cancers14225627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marchetto A., Ohmura S., Orth M.F., Knott M.M.L., Colombo M.V., Arrigoni C., Bardinet V., Saucier D., Wehweck F.S., Li J., et al. Publisher Correction: Oncogenic Hijacking of a Developmental Transcription Factor Evokes Vulnerability toward Oxidative Stress in Ewing Sarcoma. Nat. Commun. 2020;11:6068. doi: 10.1038/s41467-020-20017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shi X., Zheng Y., Jiang L., Zhou B., Yang W., Li L., Ding L., Huang M., Gery S., Lin D.-C., et al. EWS-FLI1 Regulates and Cooperates with Core Regulatory Circuitry in Ewing Sarcoma. Nucleic Acids Res. 2020;48:11434–11451. doi: 10.1093/nar/gkaa901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tirado O.M., Mateo-Lozano S., Villar J., Dettin L.E., Llort A., Gallego S., Ban J., Kovar H., Notario V. Caveolin-1 (CAV1) Is a Target of EWS/FLI-1 and a Key Determinant of the Oncogenic Phenotype and Tumorigenicity of Ewing’s Sarcoma Cells. Cancer Res. 2006;66:9937–9947. doi: 10.1158/0008-5472.CAN-06-0927. [DOI] [PubMed] [Google Scholar]
- 98.Sciandra M., De Feo A., Parra A., Landuzzi L., Lollini P.-L., Manara M.C., Mattia G., Pontecorvi G., Baricordi C., Guerzoni C., et al. Circulating miR34a Levels as a Potential Biomarker in the Follow-up of Ewing Sarcoma. J. Cell Commun. Signal. 2020;14:335–347. doi: 10.1007/s12079-020-00567-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kovach A.R., Oristian K.M., Kirsch D.G., Bentley R.C., Cheng C., Chen X., Chen P.-H., Chi J.-T.A., Linardic C.M. Identification and Targeting of a HES1-YAP1-CDKN1C Functional Interaction in Fusion-Negative Rhabdomyosarcoma. Mol. Oncol. 2022;16:3587–3605. doi: 10.1002/1878-0261.13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bharathy N., Berlow N.E., Wang E., Abraham J., Settelmeyer T.P., Hooper J.E., Svalina M.N., Ishikawa Y., Zientek K., Bajwa Z., et al. The HDAC3-SMARCA4-miR-27a Axis Promotes Expression of the PAX3:FOXO1 Fusion Oncogene in Rhabdomyosarcoma. Sci. Signal. 2018;11:eaau7632. doi: 10.1126/scisignal.aau7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xiang X., Langlois S., St-Pierre M.-E., Barré J.F., Grynspan D., Purgina B., Cowan K.N. Pannexin 1 Inhibits Rhabdomyosarcoma Progression through a Mechanism Independent of Its Canonical Channel Function. Oncogenesis. 2018;7:89. doi: 10.1038/s41389-018-0100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yu P.Y., Lopez G., Braggio D., Koller D., Bill K.L.J., Prudner B.C., Zewdu A., Chen J.L., Iwenofu O.H., Lev D., et al. miR-133a Function in the Pathogenesis of Dedifferentiated Liposarcoma. Cancer Cell Int. 2018;18:89. doi: 10.1186/s12935-018-0583-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Su Y., Wagner E.R., Luo Q., Huang J., Chen L., He B.-C., Zuo G.-W., Shi Q., Zhang B.-Q., Zhu G., et al. Insulin-like Growth Factor Binding Protein 5 Suppresses Tumor Growth and Metastasis of Human Osteosarcoma. Oncogene. 2011;30:3907–3917. doi: 10.1038/onc.2011.97. [DOI] [PubMed] [Google Scholar]
- 104.Brennecke P., Arlt M.J.E., Campanile C., Husmann K., Gvozdenovic A., Apuzzo T., Thelen M., Born W., Fuchs B. CXCR4 Antibody Treatment Suppresses Metastatic Spread to the Lung of Intratibial Human Osteosarcoma Xenografts in Mice. Clin. Exp. Metastasis. 2014;31:339–349. doi: 10.1007/s10585-013-9632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gill J., Connolly P., Roth M., Chung S.H., Zhang W., Piperdi S., Hoang B., Yang R., Guzik H., Morris J., et al. The Effect of Bone Morphogenetic Protein-2 on Osteosarcoma Metastasis. PLoS ONE. 2017;12:e0173322. doi: 10.1371/journal.pone.0173322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ren K., Zhang J., Gu X., Wu S., Shi X., Ni Y., Chen Y., Lu J., Gao Z., Wang C., et al. Migration-Inducing Gene-7 Independently Predicts Poor Prognosis of Human Osteosarcoma and Is Associated with Vasculogenic Mimicry. Exp. Cell Res. 2018;369:80–89. doi: 10.1016/j.yexcr.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 107.Zucchini C., Manara M.C., Cristalli C., Carrabotta M., Greco S., Pinca R.S., Ferrari C., Landuzzi L., Pasello M., Lollini P.-L., et al. ROCK2 Deprivation Leads to the Inhibition of Tumor Growth and Metastatic Potential in Osteosarcoma Cells through the Modulation of YAP Activity. J. Exp. Clin. Cancer Res. 2019;38:503. doi: 10.1186/s13046-019-1506-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Takagi S., Sasaki Y., Koike S., Takemoto A., Seto Y., Haraguchi M., Ukaji T., Kawaguchi T., Sugawara M., Saito M., et al. Platelet-Derived Lysophosphatidic Acid Mediated LPAR1 Activation as a Therapeutic Target for Osteosarcoma Metastasis. Oncogene. 2021;40:5548–5558. doi: 10.1038/s41388-021-01956-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhou C., Zhang Z., Zhu X., Qian G., Zhou Y., Sun Y., Yu W., Wang J., Lu H., Lin F., et al. N6-Methyladenosine Modification of the TRIM7 Positively Regulates Tumorigenesis and Chemoresistance in Osteosarcoma through Ubiquitination of BRMS1. EBioMedicine. 2020;59:102955. doi: 10.1016/j.ebiom.2020.102955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Y.X., Mandal D., Wang S., Hughes D., Pollock R.E., Lev D., Kleinerman E., Hayes-Jordan A. Inhibiting Platelet-Derived Growth Factor Beta Reduces Ewing’s Sarcoma Growth and Metastasis in a Novel Orthotopic Human Xenograft Model. In Vivo. 2009;23:903–909. [PubMed] [Google Scholar]
- 111.Mancarella C., Pasello M., Ventura S., Grilli A., Calzolari L., Toracchio L., Lollini P.-L., Donati D.M., Picci P., Ferrari S., et al. Insulin-Like Growth Factor 2 mRNA-Binding Protein 3 Is a Novel Post-Transcriptional Regulator of Ewing Sarcoma Malignancy. Clin. Cancer Res. 2018;24:3704–3716. doi: 10.1158/1078-0432.CCR-17-2602. [DOI] [PubMed] [Google Scholar]
- 112.Hayashi M., Baker A., Goldstein S.D., Albert C.M., Jackson K.W., McCarty G., Kahlert U.D., Loeb D.M. Inhibition of Porcupine Prolongs Metastasis Free Survival in a Mouse Xenograft Model of Ewing Sarcoma. Oncotarget. 2017;8:78265–78276. doi: 10.18632/oncotarget.19432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chaturvedi A., Hoffman L.M., Jensen C.C., Lin Y.-C., Grossmann A.H., Randall R.L., Lessnick S.L., Welm A.L., Beckerle M.C. Molecular Dissection of the Mechanism by Which EWS/FLI Expression Compromises Actin Cytoskeletal Integrity and Cell Adhesion in Ewing Sarcoma. Mol. Biol. Cell. 2014;25:2695–2709. doi: 10.1091/mbc.E14-01-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Balani S., Nguyen L.V., Eaves C.J. Modeling the Process of Human Tumorigenesis. Nat. Commun. 2017;8:15422. doi: 10.1038/ncomms15422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jacques C., Renema N., Ory B., Walkley C.R., Grigoriadis A.E., Heymann D. Methods in Molecular Biology. Volume 1914. Humana Press; New York, NY, USA: 2019. Murine Models of Bone Sarcomas; pp. 331–342. [DOI] [PubMed] [Google Scholar]
- 116.Dodd R.D., Añó L., Blum J.M., Li Z., Van Mater D., Kirsch D.G. Methods in Molecular Biology. Volume 1267. Humana Press; New York, NY, USA: 2015. Methods to Generate Genetically Engineered Mouse Models of Soft Tissue Sarcoma; pp. 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dodd R.D., Mito J.K., Kirsch D.G. Animal Models of Soft-Tissue Sarcoma. Dis. Model. Mech. 2010;3:557–566. doi: 10.1242/dmm.005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jacques C., Renema N., Lezot F., Ory B., Walkley C.R., Grigoriadis A.E., Heymann D. Small Animal Models for the Study of Bone Sarcoma Pathogenesis:Characteristics, Therapeutic Interests and Limitations. J. Bone Oncol. 2018;12:7–13. doi: 10.1016/j.jbo.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Merlino G., Khanna C. Fishing for the Origins of Cancer. Genes Dev. 2007;21:1275–1279. doi: 10.1101/gad.1563707. [DOI] [PubMed] [Google Scholar]
- 120.Leacock S.W., Basse A.N., Chandler G.L., Kirk A.M., Rakheja D., Amatruda J.F. A Zebrafish Transgenic Model of Ewing’s Sarcoma Reveals Conserved Mediators of EWS-FLI1 Tumorigenesis. Dis. Model. Mech. 2012;5:95–106. doi: 10.1242/dmm.007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mazzu Y.Z., Hu Y., Soni R.K., Mojica K.M., Qin L.-X., Agius P., Waxman Z.M., Mihailovic A., Socci N.D., Hendrickson R.C., et al. miR-193b-Regulated Signaling Networks Serve as Tumor Suppressors in Liposarcoma and Promote Adipogenesis in Adipose-Derived Stem Cells. Cancer Res. 2017;77:5728–5740. doi: 10.1158/0008-5472.CAN-16-2253. [DOI] [PubMed] [Google Scholar]
- 122.Li W., Zhang X., Xi X., Li Y., Quan H., Liu S., Wu L., Wu P., Lan W., Shao Y., et al. PLK2 Modulation of Enriched TAp73 Affects Osteogenic Differentiation and Prognosis in Human Osteosarcoma. Cancer Med. 2020;9:4371–4385. doi: 10.1002/cam4.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lo W.W., Wunder J.S., Dickson B.C., Campbell V., McGovern K., Alman B.A., Andrulis I.L. Involvement and Targeted Intervention of Dysregulated Hedgehog Signaling in Osteosarcoma. Cancer. 2014;120:537–547. doi: 10.1002/cncr.28439. [DOI] [PubMed] [Google Scholar]
- 124.Hanna J.A., Garcia M.R., Lardennois A., Leavey P.J., Maglic D., Fagnan A., Go J.C., Roach J., Wang Y.-D., Finkelstein D., et al. PAX3-FOXO1 Drives miR-486-5p and Represses miR-221 Contributing to Pathogenesis of Alveolar Rhabdomyosarcoma. Oncogene. 2018;37:1991–2007. doi: 10.1038/s41388-017-0081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Slemmons K.K., Crose L.E.S., Rudzinski E., Bentley R.C., Linardic C.M. Role of the YAP Oncoprotein in Priming Ras-Driven Rhabdomyosarcoma. PLoS ONE. 2015;10:e0140781. doi: 10.1371/journal.pone.0140781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Codenotti S., Zizioli D., Mignani L., Rezzola S., Tabellini G., Parolini S., Giacomini A., Asperti M., Poli M., Mandracchia D., et al. Hyperactive Akt1 Signaling Increases Tumor Progression and DNA Repair in Embryonal Rhabdomyosarcoma RD Line and Confers Susceptibility to Glycolysis and Mevalonate Pathway Inhibitors. Cells. 2022;11:2859. doi: 10.3390/cells11182859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Isfort I., Cyra M., Elges S., Kailayangiri S., Altvater B., Rossig C., Steinestel K., Grünewald I., Huss S., Eßeling E., et al. SS18-SSX-Dependent YAP/TAZ Signaling in Synovial Sarcoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019;25:3718–3731. doi: 10.1158/1078-0432.CCR-17-3553. [DOI] [PubMed] [Google Scholar]
- 128.Eklund L., Bry M., Alitalo K. Mouse Models for Studying Angiogenesis and Lymphangiogenesis in Cancer. Mol. Oncol. 2013;7:259–282. doi: 10.1016/j.molonc.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Folkman J. Tumor Angiogenesis: Therapeutic Implications. N. Engl. J. Med. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 130.De Palma M., Biziato D., Petrova T.V. Microenvironmental Regulation of Tumour Angiogenesis. Nat. Rev. Cancer. 2017;17:457–474. doi: 10.1038/nrc.2017.51. [DOI] [PubMed] [Google Scholar]
- 131.Weiss A., van Beijnum J.R., Bonvin D., Jichlinski P., Dyson P.J., Griffioen A.W., Nowak-Sliwinska P. Low-Dose Angiostatic Tyrosine Kinase Inhibitors Improve Photodynamic Therapy for Cancer: Lack of Vascular Normalization. J. Cell. Mol. Med. 2014;18:480–491. doi: 10.1111/jcmm.12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mangir N., Dikici S., Claeyssens F., MacNeil S. Using Ex Ovo Chick Chorioallantoic Membrane (CAM) Assay To Evaluate the Biocompatibility and Angiogenic Response to Biomaterials. ACS Biomater. Sci. Eng. 2019;5:3190–3200. doi: 10.1021/acsbiomaterials.9b00172. [DOI] [PubMed] [Google Scholar]
- 133.Palinski W., Monti M., Camerlingo R., Iacobucci I., Bocella S., Pinto F., Iannuzzi C., Mansueto G., Pignatiello S., Fazioli F., et al. Lysosome Purinergic Receptor P2X4 Regulates Neoangiogenesis Induced by Microvesicles from Sarcoma Patients. Cell Death Dis. 2021;12:797. doi: 10.1038/s41419-021-04069-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Reddy K., Cao Y., Zhou Z., Yu L., Jia S.-F., Kleinerman E.S. VEGF165 Expression in the Tumor Microenvironment Influences the Differentiation of Bone Marrow-Derived Pericytes That Contribute to the Ewing’s Sarcoma Vasculature. Angiogenesis. 2008;11:257–267. doi: 10.1007/s10456-008-9109-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schadler K.L., Zweidler-McKay P.A., Guan H., Kleinerman E.S. Delta like Ligand 4 Plays a Critical Role in Pericyte/Vascular Smooth Muscle Cell Formation during Vasculogenesis and Tumor Vessel Expansion in Ewing’s Sarcoma. Clin. Cancer Res. 2010;16:848. doi: 10.1158/1078-0432.CCR-09-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang Y., Pandey R.N., Roychoudhury K., Milewski D., Kalin T.V., Szabo S., Pressey J.G., Hegde R.S. Targeting EYA3 in Ewing Sarcoma Retards Tumor Growth and Angiogenesis. Mol. Cancer Ther. 2021;20:803–815. doi: 10.1158/1535-7163.MCT-20-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ribatti D., Annese T., Tamma R. The Use of the Chick Embryo CAM Assay in the Study of Angiogenic Activiy of Biomaterials. Microvasc. Res. 2020;131:104026. doi: 10.1016/j.mvr.2020.104026. [DOI] [PubMed] [Google Scholar]
- 138.Chu P.-Y., Koh A.P.-F., Antony J., Huang R.Y.-J. Applications of the Chick Chorioallantoic Membrane as an Alternative Model for Cancer Studies. Cells Tissues Organs. 2022;211:222–237. doi: 10.1159/000513039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hu L., Li K., Lin L., Qian F., Li P., Zhu L., Cai H., You L., Song J., Kok S.H.L., et al. Reversine Suppresses Osteosarcoma Cell Growth through Targeting BMP-Smad1/5/8-Mediated Angiogenesis. Microvasc. Res. 2021;135:104136. doi: 10.1016/j.mvr.2021.104136. [DOI] [PubMed] [Google Scholar]
- 140.Colman R.W., Pixley R.A., Sainz I.M., Song J.S., Isordia-Salas I., Muhamed S.N., Powell J.A., Mousa S.A. Inhibition of Angiogenesis by Antibody Blocking the Action of Proangiogenic High-Molecular-Weight Kininogen. J. Thromb. Haemost. JTH. 2003;1:164–170. doi: 10.1046/j.1538-7836.2003.00025.x. [DOI] [PubMed] [Google Scholar]
- 141.Vimalraj S., Subramanian R., Saravanan S., Arumugam B., Anuradha D. MicroRNA-432-5p Regulates Sprouting and Intussusceptive Angiogenesis in Osteosarcoma Microenvironment by Targeting PDGFB. Lab. Investig. J. Tech. Methods Pathol. 2021;101:1011–1025. doi: 10.1038/s41374-021-00589-3. [DOI] [PubMed] [Google Scholar]
- 142.Vimalraj S., Saravanan S., Raghunandhakumar S., Anuradha D. Melatonin Regulates Tumor Angiogenesis via miR-424-5p/VEGFA Signaling Pathway in Osteosarcoma. Life Sci. 2020;256:118011. doi: 10.1016/j.lfs.2020.118011. [DOI] [PubMed] [Google Scholar]
- 143.Chávez M.N., Aedo G., Fierro F.A., Allende M.L., Egaña J.T. Zebrafish as an Emerging Model Organism to Study Angiogenesis in Development and Regeneration. Front. Physiol. 2016;7:56. doi: 10.3389/fphys.2016.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nicoli S., Presta M. The Zebrafish/Tumor Xenograft Angiogenesis Assay. Nat. Protoc. 2007;2:2918–2923. doi: 10.1038/nprot.2007.412. [DOI] [PubMed] [Google Scholar]
- 145.Tobia C., De Sena G., Presta M. Zebrafish Embryo, a Tool to Study Tumor Angiogenesis. Int. J. Dev. Biol. 2011;55:505–509. doi: 10.1387/ijdb.103238ct. [DOI] [PubMed] [Google Scholar]
- 146.Fan T., Kuang G., Long R., Han Y., Wang J. The Overall Process of Metastasis: From Initiation to a New Tumor. Biochim. Biophys. Acta Rev. Cancer. 2022;1877:188750. doi: 10.1016/j.bbcan.2022.188750. [DOI] [PubMed] [Google Scholar]
- 147.Fares J., Fares M.Y., Khachfe H.H., Salhab H.A., Fares Y. Molecular Principles of Metastasis: A Hallmark of Cancer Revisited. Signal Transduct. Target. Ther. 2020;5:28. doi: 10.1038/s41392-020-0134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gómez-Cuadrado L., Tracey N., Ma R., Qian B., Brunton V.G. Mouse Models of Metastasis: Progress and Prospects. Dis. Model. Mech. 2017;10:1061–1074. doi: 10.1242/dmm.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.VAN Noord R.A., Thomas T., Krook M., Chukkapalli S., Hoenerhoff M.J., Dillman J.R., Lawlor E.R., Opipari V.P., Newman E.A. Tissue-Directed Implantation Using Ultrasound Visualization for Development of Biologically Relevant Metastatic Tumor Xenografts. In Vivo. 2017;31:779–791. doi: 10.21873/invivo.11131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Marques Da Costa M.E., Daudigeos-Dubus E., Gomez-Brouchet A., Bawa O., Rouffiac V., Serra M., Scotlandi K., Santos C., Geoerger B., Gaspar N. Establishment and Characterization of in vivo Orthotopic Bioluminescent Xenograft Models from Human Osteosarcoma Cell Lines in Swiss Nude and NSG Mice. Cancer Med. 2018;7:665–676. doi: 10.1002/cam4.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Igarashi K., Kawaguchi K., Kiyuna T., Miyake K., Murakami T., Yamamoto N., Hayashi K., Kimura H., Miwa S., Tsuchiya H., et al. Effective Metabolic Targeting of Human Osteosarcoma Cells In Vitro and in Orthotopic Nude-Mouse Models with Recombinant Methioninase. Anticancer Res. 2017;37:4807–4812. doi: 10.21873/anticanres.11887. [DOI] [PubMed] [Google Scholar]
- 152.Goldstein S.D., Hayashi M., Albert C.M., Jackson K.W., Loeb D.M. An Orthotopic Xenograft Model with Survival Hindlimb Amputation Allows Investigation of the Effect of Tumor Microenvironment on Sarcoma Metastasis. Clin. Exp. Metastasis. 2015;32:703–715. doi: 10.1007/s10585-015-9738-x. [DOI] [PubMed] [Google Scholar]
- 153.Zhang T., Li J., Yin F., Lin B., Wang Z., Xu J., Wang H., Zuo D., Wang G., Hua Y., et al. Toosendanin Demonstrates Promising Antitumor Efficacy in Osteosarcoma by Targeting STAT3. Oncogene. 2017;36:6627–6639. doi: 10.1038/onc.2017.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang T., Li S., Li J., Yin F., Hua Y., Wang Z., Wang H., Zuo D., Xu J., Cai Z. Pectolinarigenin Acts as a Potential Anti-Osteosarcoma Agent via Mediating SHP-1/JAK2/STAT3 Signaling. Biomed. Pharmacother. 2022;153:113323. doi: 10.1016/j.biopha.2022.113323. [DOI] [PubMed] [Google Scholar]
- 155.Oshiro H., Kiyuna T., Tome Y., Miyake K., Kawaguchi K., Higuchi T., Miyake M., Zhang Z., Razmjooei S., Barangi M., et al. Detection of Metastasis in a Patient-Derived Orthotopic Xenograft (PDOX) Model of Undifferentiated Pleomorphic Sarcoma with Red Fluorescent Protein. Anticancer Res. 2019;39:81–85. doi: 10.21873/anticanres.13082. [DOI] [PubMed] [Google Scholar]
- 156.Goldstein S.D., Trucco M., Bautista Guzman W., Hayashi M., Loeb D.M. A Monoclonal Antibody against the Wnt Signaling Inhibitor Dickkopf-1 Inhibits Osteosarcoma Metastasis in a Preclinical Model. Oncotarget. 2016;7:21114–21123. doi: 10.18632/oncotarget.8522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nomura M., Rainusso N., Lee Y.-C., Dawson B., Coarfa C., Han R., Larson J.L., Shuck R., Kurenbekova L., Yustein J.T. Tegavivint and the β-Catenin/ALDH Axis in Chemotherapy-Resistant and Metastatic Osteosarcoma. J. Natl. Cancer Inst. 2019;111:1216–1227. doi: 10.1093/jnci/djz026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.He Y., Cheng D., Lian C., Liu Y., Luo W., Wang Y., Ma C., Wu Q., Tian P., He D., et al. Serglycin Induces Osteoclastogenesis and Promotes Tumor Growth in Giant Cell Tumor of Bone. Cell Death Dis. 2021;12:868. doi: 10.1038/s41419-021-04161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schott C.R., Koehne A.L., Sayles L.C., Young E.P., Luck C., Yu K., Lee A.G., Breese M.R., Leung S.G., Xu H., et al. Osteosarcoma PDX-Derived Cell Line Models for Preclinical Drug Evaluation Demonstrate Metastasis Inhibition by Dinaciclib through a Genome-Targeted Approach. Clin. Cancer Res. 2024;30:849–864. doi: 10.1158/1078-0432.CCR-23-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Liao D., Zhong L., Yin J., Zeng C., Wang X., Huang X., Chen J., Zhang H., Zhang R., Guan X.-Y., et al. Chromosomal Translocation-Derived Aberrant Rab22a Drives Metastasis of Osteosarcoma. Nat. Cell Biol. 2020;22:868–881. doi: 10.1038/s41556-020-0522-z. [DOI] [PubMed] [Google Scholar]
- 161.Smeester B.A., Slipek N.J., Pomeroy E.J., Laoharawee K., Osum S.H., Larsson A.T., Williams K.B., Stratton N., Yamamoto K., Peterson J.J., et al. PLX3397 Treatment Inhibits Constitutive CSF1R-Induced Oncogenic ERK Signaling, Reduces Tumor Growth, and Metastatic Burden in Osteosarcoma. Bone. 2020;136:115353. doi: 10.1016/j.bone.2020.115353. [DOI] [PubMed] [Google Scholar]
- 162.Takemoto A., Takagi S., Ukaji T., Gyobu N., Kakino M., Takami M., Kobayashi A., Lebel M., Kawaguchi T., Sugawara M., et al. Targeting Podoplanin for the Treatment of Osteosarcoma. Clin. Cancer Res. 2022;28:2633–2645. doi: 10.1158/1078-0432.CCR-21-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Endo Y., Sasaki T., Harada F., Noguchi M. Specific Detection of Metastasized Human Tumor Cells in Embryonic Chicks by the Polymerase Chain Reaction. Jpn. J. Cancer Res. 1990;81:723–726. doi: 10.1111/j.1349-7006.1990.tb02635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ribatti D. The CAM Assay in the Study of the Metastatic Process. Exp. Cell Res. 2021;400:112510. doi: 10.1016/j.yexcr.2021.112510. [DOI] [PubMed] [Google Scholar]
- 165.Pawlikowska P., Tayoun T., Oulhen M., Faugeroux V., Rouffiac V., Aberlenc A., Pommier A.L., Honore A., Marty V., Bawa O., et al. Exploitation of the Chick Embryo Chorioallantoic Membrane (CAM) as a Platform for Anti-Metastatic Drug Testing. Sci. Rep. 2020;10:16876. doi: 10.1038/s41598-020-73632-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sharili A.-S., Allen S., Smith K., Price J., McGonnell I.M. Snail2 Promotes Osteosarcoma Cell Motility through Remodelling of the Actin Cytoskeleton and Regulates Tumor Development. Cancer Lett. 2013;333:170–179. doi: 10.1016/j.canlet.2013.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lissat A., Joerschke M., Shinde D.A., Braunschweig T., Meier A., Makowska A., Bortnick R., Henneke P., Herget G., Gorr T.A., et al. IL6 Secreted by Ewing Sarcoma Tumor Microenvironment Confers Anti-Apoptotic and Cell-Disseminating Paracrine Responses in Ewing Sarcoma Cells. BMC Cancer. 2015;15:552. doi: 10.1186/s12885-015-1564-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Patil N., Ahmed Kabeer Rasheed S., Abba M., Hendrik Leupold J., Schwarzbach M., Allgayer H. A Mechanistic Study on the Metastasis Inducing Function of FUS-CHOP Fusion Protein in Liposarcoma. Int. J. Cancer. 2014;134:2808–2819. doi: 10.1002/ijc.28638. [DOI] [PubMed] [Google Scholar]
- 169.Choi T.-Y., Choi T.-I., Lee Y.-R., Choe S.-K., Kim C.-H. Zebrafish as an Animal Model for Biomedical Research. Exp. Mol. Med. 2021;53:310–317. doi: 10.1038/s12276-021-00571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Patton E.E., Zon L.I., Langenau D.M. Zebrafish Disease Models in Drug Discovery: From Preclinical Modelling to Clinical Trials. Nat. Rev. Drug Discov. 2021;20:611–628. doi: 10.1038/s41573-021-00210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Van der Ent W., Jochemsen A.G., Teunisse A.F.A.S., Krens S.F.G., Szuhai K., Spaink H.P., Hogendoorn P.C.W., Snaar-Jagalska B.E. Ewing Sarcoma Inhibition by Disruption of EWSR1-FLI1 Transcriptional Activity and Reactivation of P53. J. Pathol. 2014;233:415–424. doi: 10.1002/path.4378. [DOI] [PubMed] [Google Scholar]
- 172.Berning P., Hennemann C., Tulotta C., Schaefer C., Lechtape B., Hotfilder M., El Gourari Y., Jürgens H., Snaar-Jagalska E., Hempel G., et al. The Receptor Tyrosine Kinase RON and Its Isoforms as Therapeutic Targets in Ewing Sarcoma. Cancers. 2020;12:904. doi: 10.3390/cancers12040904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ban J., Aryee D.N.T., Fourtouna A., van der Ent W., Kauer M., Niedan S., Machado I., Rodriguez-Galindo C., Tirado O.M., Schwentner R., et al. Suppression of Deacetylase SIRT1 Mediates Tumor-Suppressive NOTCH Response and Offers a Novel Treatment Option in Metastatic Ewing Sarcoma. Cancer Res. 2014;74:6578–6588. doi: 10.1158/0008-5472.CAN-14-1736. [DOI] [PubMed] [Google Scholar]
- 174.Landuzzi L., Manara M.C., Lollini P.-L., Scotlandi K. Patient Derived Xenografts for Genome-Driven Therapy of Osteosarcoma. Cells. 2021;10:416. doi: 10.3390/cells10020416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Monsma D.J., Cherba D.M., Richardson P.J., Vance S., Rangarajan S., Dylewski D., Eugster E., Scott S.B., Beuschel N.L., Davidson P.J., et al. Using a Rhabdomyosarcoma Patient-Derived Xenograft to Examine Precision Medicine Approaches and Model Acquired Resistance. Pediatr. Blood Cancer. 2014;61:1570–1577. doi: 10.1002/pbc.25039. [DOI] [PubMed] [Google Scholar]
- 176.Komatsu A., Matsumoto K., Yoshimatsu Y., Sin Y., Kubota A., Saito T., Mizumoto A., Ohashi S., Muto M., Noguchi R., et al. The CAM Model for CIC-DUX4 Sarcoma and Its Potential Use for Precision Medicine. Cells. 2021;10:2613. doi: 10.3390/cells10102613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ali Z., Vildevall M., Rodriguez G.V., Tandiono D., Vamvakaris I., Evangelou G., Lolas G., Syrigos K.N., Villanueva A., Wick M., et al. Zebrafish Patient-Derived Xenograft Models Predict Lymph Node Involvement and Treatment Outcome in Non-Small Cell Lung Cancer. J. Exp. Clin. Cancer Res. 2022;41:58. doi: 10.1186/s13046-022-02280-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Almstedt E., Rosén E., Gloger M., Stockgard R., Hekmati N., Koltowska K., Krona C., Nelander S. Real-Time Evaluation of Glioblastoma Growth in Patient-Specific Zebrafish Xenografts. Neuro-Oncology. 2022;24:726–738. doi: 10.1093/neuonc/noab264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Guder W.K., Hartmann W., Trautmann M., Hardes J., Wardelmann E., Balke M., Streitbürger A. Analysis of Drug Sensitivity of Human High-Grade Osteosarcoma in a Chick Chorioallantoic Membrane (CAM) Model: A Proof of Principle Study. BMC Res. Notes. 2020;13:432. doi: 10.1186/s13104-020-05269-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cornet C., Dyballa S., Terriente J., Di Giacomo V. ZeOncoTest: Refining and Automating the Zebrafish Xenograft Model for Drug Discovery in Cancer. Pharmaceuticals. 2019;13:1. doi: 10.3390/ph13010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Grissenberger S., Sturtzel C., Wenninger-Weinzierl A., Radic-Sarikas B., Scheuringer E., Bierbaumer L., Etienne V., Némati F., Pascoal S., Tötzl M., et al. High-Content Drug Screening in Zebrafish Xenografts Reveals High Efficacy of Dual MCL-1/BCL-XL Inhibition against Ewing Sarcoma. Cancer Lett. 2023;554:216028. doi: 10.1016/j.canlet.2022.216028. [DOI] [PubMed] [Google Scholar]
- 182.Loganathan S.N., Tang N., Holler A.E., Wang N., Wang J. Targeting the IGF1R/PI3K/AKT Pathway Sensitizes Ewing Sarcoma to BET Bromodomain Inhibitors. Mol. Cancer Ther. 2019;18:929–936. doi: 10.1158/1535-7163.MCT-18-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hingorani P., Zhang W., Piperdi S., Pressman L., Lin J., Gorlick R., Kolb E.A. Preclinical Activity of Palifosfamide Lysine (ZIO-201) in Pediatric Sarcomas Including Oxazaphosphorine-Resistant Osteosarcoma. Cancer Chemother. Pharmacol. 2009;64:733–740. doi: 10.1007/s00280-008-0922-4. [DOI] [PubMed] [Google Scholar]
- 184.Vanhoefer U., Cao S., Harstrick A., Seeber S., Rustum Y.M. Comparative Antitumor Efficacy of Docetaxel and Paclitaxel in Nude Mice Bearing Human Tumor Xenografts That Overexpress the Multidrug Resistance Protein (MRP) Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 1997;8:1221–1228. doi: 10.1023/a:1008290406221. [DOI] [PubMed] [Google Scholar]
- 185.Cassinelli G., Dal Bo L., Favini E., Cominetti D., Pozzi S., Tortoreto M., De Cesare M., Lecis D., Scanziani E., Minoli L., et al. Supersulfated Low-Molecular Weight Heparin Synergizes with IGF1R/IR Inhibitor to Suppress Synovial Sarcoma Growth and Metastases. Cancer Lett. 2018;415:187–197. doi: 10.1016/j.canlet.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 186.Yamada S., Imura Y., Nakai T., Nakai S., Yasuda N., Kaneko K., Outani H., Takenaka S., Hamada K., Myoui A., et al. Therapeutic Potential of TAS-115 via c-MET and PDGFRα Signal Inhibition for Synovial Sarcoma. BMC Cancer. 2017;17:334. doi: 10.1186/s12885-017-3324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Harttrampf A.C., da Costa M.E.M., Renoult A., Daudigeos-Dubus E., Geoerger B. Histone Deacetylase Inhibitor Panobinostat Induces Antitumor Activity in Epithelioid Sarcoma and Rhabdoid Tumor by Growth Factor Receptor Modulation. BMC Cancer. 2021;21:833. doi: 10.1186/s12885-021-08579-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Okada T., Lee A.Y., Qin L.-X., Agaram N., Mimae T., Shen Y., O’Connor R., López-Lago M.A., Craig A., Miller M.L., et al. Integrin-A10 Dependency Identifies RAC and RICTOR as Therapeutic Targets in High-Grade Myxofibrosarcoma. Cancer Discov. 2016;6:1148–1165. doi: 10.1158/2159-8290.CD-15-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Shukla N., Somwar R., Smith R.S., Ambati S., Munoz S., Merchant M., D’Arcy P., Wang X., Kobos R., Antczak C., et al. Proteasome Addiction Defined in Ewing Sarcoma Is Effectively Targeted by a Novel Class of 19S Proteasome Inhibitors. Cancer Res. 2016;76:4525–4534. doi: 10.1158/0008-5472.CAN-16-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Heisey D.A.R., Jacob S., Lochmann T.L., Kurupi R., Ghotra M.S., Calbert M.L., Shende M., Maves Y.K., Koblinski J.E., Dozmorov M.G., et al. Pharmaceutical Interference of the EWS-FLI1-Driven Transcriptome By Cotargeting H3K27ac and RNA Polymerase Activity in Ewing Sarcoma. Mol. Cancer Ther. 2021;20:1868–1879. doi: 10.1158/1535-7163.MCT-20-0489. [DOI] [PubMed] [Google Scholar]
- 191.Buchou C., Laud-Duval K., van der Ent W., Grossetête S., Zaidi S., Gentric G., Corbé M., Müller K., Del Nery E., Surdez D., et al. Upregulation of the Mevalonate Pathway through EWSR1-FLI1/EGR2 Regulatory Axis Confers Ewing Cells Exquisite Sensitivity to Statins. Cancers. 2022;14:2327. doi: 10.3390/cancers14092327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Dorado García H., Pusch F., Bei Y., von Stebut J., Ibáñez G., Guillan K., Imami K., Gürgen D., Rolff J., Helmsauer K., et al. Therapeutic Targeting of ATR in Alveolar Rhabdomyosarcoma. Nat. Commun. 2022;13:4297. doi: 10.1038/s41467-022-32023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ommer J., Selfe J.L., Wachtel M., O’Brien E.M., Laubscher D., Roemmele M., Kasper S., Delattre O., Surdez D., Petts G., et al. Aurora A Kinase Inhibition Destabilizes PAX3-FOXO1 and MYCN and Synergizes with Navitoclax to Induce Rhabdomyosarcoma Cell Death. Cancer Res. 2020;80:832–842. doi: 10.1158/0008-5472.CAN-19-1479. [DOI] [PubMed] [Google Scholar]
- 194.Angeles C.V., Velez A., Rios J., Laxa B., Shum D., Ruiz P.D., Shen Y., Ostrovnaya I., Gularte-Mérida R., Nacev B.A., et al. A High-Content Screen for C/EBPα Expression Identifies Novel Therapeutic Agents in Dedifferentiated Liposarcoma. Clin. Cancer Res. 2022;28:175–186. doi: 10.1158/1078-0432.CCR-19-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Jones S.E., Fleuren E.D.G., Frankum J., Konde A., Williamson C.T., Krastev D.B., Pemberton H.N., Campbell J., Gulati A., Elliott R., et al. ATR Is a Therapeutic Target in Synovial Sarcoma. Cancer Res. 2017;77:7014–7026. doi: 10.1158/0008-5472.CAN-17-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Stacchiotti S., Saponara M., Frapolli R., Tortoreto M., Cominetti D., Provenzano S., Negri T., Dagrada G.P., Gronchi A., Colombo C., et al. Patient-Derived Solitary Fibrous Tumour Xenografts Predict High Sensitivity to Doxorubicin/Dacarbazine Combination Confirmed in the Clinic and Highlight the Potential Effectiveness of Trabectedin or Eribulin against This Tumour. Eur. J. Cancer. 2017;76:84–92. doi: 10.1016/j.ejca.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 197.Miyake K., Murakami T., Kiyuna T., Igarashi K., Kawaguchi K., Li Y., Singh A.S., Dry S.M., Eckardt M.A., Hiroshima Y., et al. Eribulin Regresses a Doxorubicin-Resistant Ewing’s Sarcoma with a FUS-ERG Fusion and CDKN2A-Deletion in a Patient-Derived Orthotopic Xenograft (PDOX) Nude Mouse Model. J. Cell. Biochem. 2018;119:967–972. doi: 10.1002/jcb.26263. [DOI] [PubMed] [Google Scholar]
- 198.Miyake K., Murakami T., Kiyuna T., Igarashi K., Kawaguchi K., Miyake M., Li Y., Nelson S.D., Dry S.M., Bouvet M., et al. The Combination of Temozolomide-Irinotecan Regresses a Doxorubicin-Resistant Patient-Derived Orthotopic Xenograft (PDOX) Nude-Mouse Model of Recurrent Ewing’s Sarcoma with a FUS-ERG Fusion and CDKN2A Deletion: Direction for Third-Line Patient Therapy. Oncotarget. 2017;8:103129–103136. doi: 10.18632/oncotarget.20789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Murakami T., Singh A.S., Kiyuna T., Dry S.M., Li Y., James A.W., Igarashi K., Kawaguchi K., DeLong J.C., Zhang Y., et al. Effective Molecular Targeting of CDK4/6 and IGF-1R in a Rare FUS-ERG Fusion CDKN2A-Deletion Doxorubicin-Resistant Ewing’s Sarcoma Patient-Derived Orthotopic Xenograft (PDOX) Nude-Mouse Model. Oncotarget. 2016;7:47556–47564. doi: 10.18632/oncotarget.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Stacchiotti S., Zuco V., Tortoreto M., Cominetti D., Frezza A.M., Percio S., Indio V., Barisella M., Monti V., Brich S., et al. Comparative Assessment of Antitumor Effects and Autophagy Induction as a Resistance Mechanism by Cytotoxics and EZH2 Inhibition in INI1-Negative Epithelioid Sarcoma Patient-Derived Xenograft. Cancers. 2019;11:1015. doi: 10.3390/cancers11071015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Imura Y., Yasui H., Outani H., Wakamatsu T., Hamada K., Nakai T., Yamada S., Myoui A., Araki N., Ueda T., et al. Combined Targeting of mTOR and C-MET Signaling Pathways for Effective Management of Epithelioid Sarcoma. Mol. Cancer. 2014;13:185. doi: 10.1186/1476-4598-13-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Pascual-Pasto G., Resa-Pares C., Castillo-Ecija H., Aschero R., Baulenas-Farres M., Vila-Ubach M., Burgueño V., Balaguer-Lluna L., Cuadrado-Vilanova M., Olaciregui N.G., et al. Low Bcl-2 Is a Robust Biomarker of Sensitivity to Nab-Paclitaxel in Ewing Sarcoma. Biochem. Pharmacol. 2023;208:115408. doi: 10.1016/j.bcp.2022.115408. [DOI] [PubMed] [Google Scholar]
- 203.Pascual-Pasto G., Castillo-Ecija H., Unceta N., Aschero R., Resa-Pares C., Gómez-Caballero A., Vila-Ubach M., Muñoz-Aznar O., Suñol M., Burgueño V., et al. SPARC-Mediated Long-Term Retention of Nab-Paclitaxel in Pediatric Sarcomas. J. Control. Release. 2022;342:81–92. doi: 10.1016/j.jconrel.2021.12.035. [DOI] [PubMed] [Google Scholar]
- 204.Carrabotta M., Laginestra M.A., Durante G., Mancarella C., Landuzzi L., Parra A., Ruzzi F., Toracchio L., De Feo A., Giusti V., et al. Integrated Molecular Characterization of Patient-Derived Models Reveals Therapeutic Strategies for Treating CIC-DUX4 Sarcoma. Cancer Res. 2022;82:708–720. doi: 10.1158/0008-5472.CAN-21-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Gedminas J.M., Kaufman R., Boguslawski E.A., Gross A.C., Adams M., Beddows I., Kitchen-Goosen S.M., Roberts R.D., Grohar P.J. Lurbinectedin Inhibits the EWS-WT1 Transcription Factor in Desmoplastic Small Round Cell Tumor. Mol. Cancer Ther. 2022;21:1296–1305. doi: 10.1158/1535-7163.MCT-21-1003. [DOI] [PubMed] [Google Scholar]
- 206.Slotkin E.K., Bowman A.S., Levine M.F., Dela Cruz F., Coutinho D.F., Sanchez G.I., Rosales N., Modak S., Tap W.D., Gounder M.M., et al. Comprehensive Molecular Profiling of Desmoplastic Small Round Cell Tumor. Mol. Cancer Res. 2021;19:1146–1155. doi: 10.1158/1541-7786.MCR-20-0722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Smith R.S., Odintsov I., Liu Z., Lui A.J.-W., Hayashi T., Vojnic M., Suehara Y., Delasos L., Mattar M.S., Hmeljak J., et al. Novel Patient-Derived Models of Desmoplastic Small Round Cell Tumor Confirm a Targetable Dependency on ERBB Signaling. Dis. Model. Mech. 2022;15:dmm047621. doi: 10.1242/dmm.047621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zuco V., Pasquali S., Tortoreto M., Percio S., Doldi V., Barisella M., Collini P., Dagrada G.P., Brich S., Gasparini P., et al. Effectiveness of Irinotecan plus Trabectedin on a Desmoplastic Small Round Cell Tumor Patient-Derived Xenograft. Dis. Model. Mech. 2023;16:dmm049649. doi: 10.1242/dmm.049649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Safaric Tepes P., Segovia D., Jevtic S., Ramirez D., Lyons S.K., Sordella R. Patient-Derived Xenografts and in Vitro Model Show Rationale for Imatinib Mesylate Repurposing in HEY1-NCoA2-Driven Mesenchymal Chondrosarcoma. Lab. Investig. J. Tech. Methods Pathol. 2022;102:1038–1049. doi: 10.1038/s41374-021-00704-4. [DOI] [PubMed] [Google Scholar]
- 210.Suzuki R., Wakamatsu T., Yoshida K., Matsuoka Y., Takami H., Nakai S., Tamiya H., Kakunaga S., Yagi T., Yoshida K.-I., et al. Genetic Characterization of a Novel Organoid from Human Malignant Giant-Cell Tumor. J. Bone Oncol. 2023;41:100486. doi: 10.1016/j.jbo.2023.100486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Sakumoto M., Oyama R., Takahashi M., Takai Y., Kito F., Shiozawa K., Qiao Z., Endo M., Yoshida A., Kawai A., et al. Establishment and Proteomic Characterization of Patient-Derived Clear Cell Sarcoma Xenografts and Cell Lines. In Vitro Cell. Dev. Biol. Anim. 2018;54:163–176. doi: 10.1007/s11626-017-0207-5. [DOI] [PubMed] [Google Scholar]
- 212.De Vita A., Vanni S., Miserocchi G., Fausti V., Pieri F., Spadazzi C., Cocchi C., Liverani C., Calabrese C., Casadei R., et al. A Rationale for the Activity of Bone Target Therapy and Tyrosine Kinase Inhibitor Combination in Giant Cell Tumor of Bone and Desmoplastic Fibroma: Translational Evidences. Biomedicines. 2022;10:372. doi: 10.3390/biomedicines10020372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.De Vita A., Recine F., Miserocchi G., Pieri F., Spadazzi C., Cocchi C., Vanni S., Liverani C., Farnedi A., Fabbri F., et al. The Potential Role of the Extracellular Matrix in the Activity of Trabectedin in UPS and L-Sarcoma: Evidences from a Patient-Derived Primary Culture Case Series in Tridimensional and Zebrafish Models. J. Exp. Clin. Cancer Res. 2021;40:165. doi: 10.1186/s13046-021-01963-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ordóñez J.L., Amaral A.T., Carcaboso A.M., Herrero-Martín D., del Carmen García-Macías M., Sevillano V., Alonso D., Pascual-Pasto G., San-Segundo L., Vila-Ubach M., et al. The PARP Inhibitor Olaparib Enhances the Sensitivity of Ewing Sarcoma to Trabectedin. Oncotarget. 2015;6:18875–18890. doi: 10.18632/oncotarget.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Chin D.H., Osman I., Porch J., Kim H., Buck K.K., Rodriguez J., Carapia B., Yan D., Moura S.B., Sperry J., et al. BET Bromodomain Degradation Disrupts Function but Not 3D Formation of RNA Pol2 Clusters. Pharmaceuticals. 2023;16:199. doi: 10.3390/ph16020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Lowery C.D., Dowless M., Renschler M., Blosser W., VanWye A.B., Stephens J.R., Iversen P.W., Lin A.B., Beckmann R.P., Krytska K., et al. Broad Spectrum Activity of the Checkpoint Kinase 1 Inhibitor Prexasertib as a Single Agent or Chemopotentiator Across a Range of Preclinical Pediatric Tumor Models. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019;25:2278–2289. doi: 10.1158/1078-0432.CCR-18-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Dowless M., Lowery C.D., Shackleford T., Renschler M., Stephens J., Flack R., Blosser W., Gupta S., Stewart J., Webster Y., et al. Abemaciclib Is Active in Preclinical Models of Ewing Sarcoma via Multipronged Regulation of Cell Cycle, DNA Methylation, and Interferon Pathway Signaling. Clin. Cancer Res. 2018;24:6028–6039. doi: 10.1158/1078-0432.CCR-18-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wengner A.M., Siemeister G., Lücking U., Lefranc J., Wortmann L., Lienau P., Bader B., Bömer U., Moosmayer D., Eberspächer U., et al. The Novel ATR Inhibitor BAY 1895344 Is Efficacious as Monotherapy and Combined with DNA Damage-Inducing or Repair-Compromising Therapies in Preclinical Cancer Models. Mol. Cancer Ther. 2020;19:26–38. doi: 10.1158/1535-7163.MCT-19-0019. [DOI] [PubMed] [Google Scholar]
- 219.Kurmasheva R.T., Erickson S.W., Earley E., Smith M.A., Houghton P.J. In Vivo Evaluation of the EZH2 Inhibitor (EPZ011989) Alone or in Combination with Standard of Care Cytotoxic Agents against Pediatric Malignant Rhabdoid Tumor Preclinical Models-A Report from the Pediatric Preclinical Testing Consortium. Pediatr. Blood Cancer. 2021;68:e28772. doi: 10.1002/pbc.28772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Harrison D.J., Gill J.D., Roth M.E., Zhang W., Teicher B., Erickson S., Gatto G., Kurmasheva R.T., Houghton P.J., Smith M.A., et al. Initial in Vivo Testing of a Multitarget Kinase Inhibitor, Regorafenib, by the Pediatric Preclinical Testing Consortium. Pediatr. Blood Cancer. 2020;67:e28222. doi: 10.1002/pbc.28222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Li H., Tu J., Zhao Z., Chen L., Qu Y., Li H., Yao H., Wang X., Lee D.-F., Shen J., et al. Molecular Signatures of BRCAness Analysis Identifies PARP Inhibitor Niraparib as a Novel Targeted Therapeutic Strategy for Soft Tissue Sarcomas. Theranostics. 2020;10:9477–9494. doi: 10.7150/thno.45763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Cornillie J., Wozniak A., Li H., Gebreyohannes Y.K., Wellens J., Hompes D., Debiec-Rychter M., Sciot R., Schöffski P. Anti-Tumor Activity of the MDM2-TP53 Inhibitor BI-907828 in Dedifferentiated Liposarcoma Patient-Derived Xenograft Models Harboring MDM2 Amplification. Clin. Transl. Oncol. 2020;22:546–554. doi: 10.1007/s12094-019-02158-z. [DOI] [PubMed] [Google Scholar]
- 223.Kolb E.A., Houghton P.J., Kurmasheva R.T., Mosse Y.P., Maris J.M., Erickson S.W., Guo Y., Teicher B.A., Smith M.A., Gorlick R. Preclinical Evaluation of the Combination of AZD1775 and Irinotecan against Selected Pediatric Solid Tumors: A Pediatric Preclinical Testing Consortium Report. Pediatr. Blood Cancer. 2020;67:e28098. doi: 10.1002/pbc.28098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Subbiah V., Chawla S.P., Conley A.P., Wilky B.A., Tolcher A., Lakhani N.J., Berz D., Andrianov V., Crago W., Holcomb M., et al. Preclinical Characterization and Phase I Trial Results of INBRX-109, A Third-Generation, Recombinant, Humanized, Death Receptor 5 Agonist Antibody, in Chondrosarcoma. Clin. Cancer Res. 2023;29:2988–3003. doi: 10.1158/1078-0432.CCR-23-0974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Laroche-Clary A., Chaire V., Verbeke S., Algéo M.-P., Malykh A., Le Loarer F., Italiano A. ATR Inhibition Broadly Sensitizes Soft-Tissue Sarcoma Cells to Chemotherapy Independent of Alternative Lengthening Telomere (ALT) Status. Sci. Rep. 2020;10:7488. doi: 10.1038/s41598-020-63294-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.López-Álvarez M., González-Aguilera C., Moura D.S., Sánchez-Bustos P., Mondaza-Hernández J.L., Martín-Ruiz M., Renshaw M., Ramos R., Castilla C., Blanco-Alcaina E., et al. Efficacy of Eribulin Plus Gemcitabine Combination in L-Sarcomas. Int. J. Mol. Sci. 2022;24:680. doi: 10.3390/ijms24010680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Frapolli R., Bello E., Ponzo M., Craparotta I., Mannarino L., Ballabio S., Marchini S., Carrassa L., Ubezio P., Porcu L., et al. Combination of PPARγ Agonist Pioglitazone and Trabectedin Induce Adipocyte Differentiation to Overcome Trabectedin Resistance in Myxoid Liposarcomas. Clin. Cancer Res. 2019;25:7565–7575. doi: 10.1158/1078-0432.CCR-19-0976. [DOI] [PubMed] [Google Scholar]
- 228.Miserocchi G., Bocchini M., Cortesi M., Arienti C., De Vita A., Liverani C., Mercatali L., Bravaccini S., Ulivi P., Zanoni M. Combining Preclinical Tools and Models to Unravel Tumor Complexity: Jump into the next Dimension. Front. Immunol. 2023;14:1171141. doi: 10.3389/fimmu.2023.1171141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Pignochino Y., Dell’Aglio C., Basiricò M., Capozzi F., Soster M., Marchiò S., Bruno S., Gammaitoni L., Sangiolo D., Torchiaro E., et al. The Combination of Sorafenib and Everolimus Abrogates mTORC1 and mTORC2 Upregulation in Osteosarcoma Preclinical Models. Clin. Cancer Res. 2013;19:2117–2131. doi: 10.1158/1078-0432.CCR-12-2293. [DOI] [PubMed] [Google Scholar]
- 230.Tome Y., Kimura H., Sugimoto N., Tsuchiya H., Kanaya F., Bouvet M., Hoffman R.M. The Disintegrin Echistatin in Combination with Doxorubicin Targets High-Metastatic Human Osteosarcoma Overexpressing Aνβ3 Integrin in Chick Embryo and Nude Mouse Models. Oncotarget. 2016;7:87031–87036. doi: 10.18632/oncotarget.13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Lin C.-Y., Hung S.-Y., Chen H.-T., Tsou H.-K., Fong Y.-C., Wang S.-W., Tang C.-H. Brain-Derived Neurotrophic Factor Increases Vascular Endothelial Growth Factor Expression and Enhances Angiogenesis in Human Chondrosarcoma Cells. Biochem. Pharmacol. 2014;91:522–533. doi: 10.1016/j.bcp.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 232.Zhang C., Lu L., Li Y., Wang X., Zhou J., Liu Y., Fu P., Gallicchio M.A., Bach L.A., Duan C. IGF Binding Protein-6 Expression in Vascular Endothelial Cells Is Induced by Hypoxia and Plays a Negative Role in Tumor Angiogenesis. Int. J. Cancer. 2012;130:2003–2012. doi: 10.1002/ijc.26201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Adane B., Alexe G., Seong B.K.A., Lu D., Hwang E.E., Hnisz D., Lareau C.A., Ross L., Lin S., Dela Cruz F.S., et al. STAG2 Loss Rewires Oncogenic and Developmental Programs to Promote Metastasis in Ewing Sarcoma. Cancer Cell. 2021;39:827–844.e10. doi: 10.1016/j.ccell.2021.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wang S., Hwang E.E., Guha R., O’Neill A.F., Melong N., Veinotte C.J., Conway Saur A., Wuerthele K., Shen M., McKnight C., et al. High-Throughput Chemical Screening Identifies Focal Adhesion Kinase and Aurora Kinase B Inhibition as a Synergistic Treatment Combination in Ewing Sarcoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019;25:4552–4566. doi: 10.1158/1078-0432.CCR-17-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Gatzweiler C., Ridinger J., Herter S., Gerloff X.F., ElHarouni D., Berker Y., Imle R., Schmitt L., Kreth S., Stainczyk S., et al. Functional Therapeutic Target Validation Using Pediatric Zebrafish Xenograft Models. Cancers. 2022;14:849. doi: 10.3390/cancers14030849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ribatti D. Chick Embryo Chorioallantoic Membrane as a Useful Tool to Study Angiogenesis. Int. Rev. Cell Mol. Biol. 2008;270:181–224. doi: 10.1016/S1937-6448(08)01405-6. [DOI] [PubMed] [Google Scholar]
- 237.Demetri G.D., Chawla S.P., von Mehren M., Ritch P., Baker L.H., Blay J.Y., Hande K.R., Keohan M.L., Samuels B.L., Schuetze S., et al. Efficacy and Safety of Trabectedin in Patients with Advanced or Metastatic Liposarcoma or Leiomyosarcoma after Failure of Prior Anthracyclines and Ifosfamide: Results of a Randomized Phase II Study of Two Different Schedules. J. Clin. Oncol. 2009;27:4188–4196. doi: 10.1200/JCO.2008.21.0088. [DOI] [PubMed] [Google Scholar]
- 238.Demetri G.D., Von Mehren M., Jones R.L., Hensley M.L., Schuetze S.M., Staddon A., Milhem M., Elias A., Ganjoo K., Tawbi H., et al. Efficacy and Safety of Trabectedin or Dacarbazine for Metastatic Liposarcoma or Leiomyosarcoma after Failure of Conventional Chemotherapy: Results of a Phase III Randomized Multicenter Clinical Trial. J. Clin. Oncol. 2016;34:786–793. doi: 10.1200/JCO.2015.62.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Meco D., Colombo T., Ubezio P., Zucchetti M., Zaffaroni M., Riccardi A., Faircloth G., Jose J., D’Incalci M., Riccardi R. Effective Combination of ET-743 and Doxorubicin in Sarcoma: Preclinical Studies. Cancer Chemother. Pharmacol. 2003;52:131–138. doi: 10.1007/s00280-003-0636-6. [DOI] [PubMed] [Google Scholar]
- 240.Abdolahi S., Ghazvinian Z., Muhammadnejad S., Saleh M., Asadzadeh Aghdaei H., Baghaei K. Patient-Derived Xenograft (PDX) Models, Applications and Challenges in Cancer Research. J. Transl. Med. 2022;20:206. doi: 10.1186/s12967-022-03405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Yagishita S., Nishikawa T., Yoshida H., Shintani D., Sato S., Miwa M., Suzuki M., Yasuda M., Ogitani Y., Jikoh T., et al. Co-Clinical Study of [Fam-] Trastuzumab Deruxtecan (DS8201a) in Patient-Derived Xenograft Models of Uterine Carcinosarcoma and Its Association with Clinical Efficacy. Clin. Cancer Res. 2023;29:2239–2249. doi: 10.1158/1078-0432.CCR-22-3861. [DOI] [PubMed] [Google Scholar]
- 242.Egeblad M., Nakasone E.S., Werb Z. Tumors as Organs: Complex Tissues That Interface with the Entire Organism. Dev. Cell. 2010;18:884–901. doi: 10.1016/j.devcel.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.