Abstract
During tumor development, promoter CpG islands that are normally silenced by Polycomb repressive complexes (PRCs) become DNA-hypermethylated. The molecular mechanism by which de novo DNA methyltransferase(s) [DNMT(s)] catalyze CpG methylation at PRC-regulated regions remains unclear. Here, we report a cryo–electron microscopy structure of the DNMT3A long isoform (DNMT3A1) amino-terminal region in complex with a nucleosome carrying PRC1-mediated histone H2A lysine-119 monoubiquitination (H2AK119Ub). We identify regions within the DNMT3A1 amino terminus that bind H2AK119Ub and the nucleosome acidic patch. This bidentate interaction is required for effective DNMT3A1 engagement with H2AK119Ub-modified chromatin in cells. Further, aberrant redistribution of DNMT3A1 to Polycomb target genes recapitulates the cancer-associated DNA hypermethylation signature and inhibits their transcriptional activation during cell differentiation. This effect is rescued by disruption of the DNMT3A1–acidic patch interaction. Together, our analyses reveal a binding interface critical for mediating promoter CpG island DNA hypermethylation, a major molecular hallmark of cancer.
A structural and genomic analysis of the DNMT3A1 interaction with H2AK119Ub nucleosomes explains its mistargeting in cancer.
INTRODUCTION
An aberrant DNA methylation [DNAme; primarily 5-methylcytosine (5mC)] landscape is a well-established pan-cancer molecular hallmark (1). As an example, megabase domains that are gene-poor, late-replicating, and of low CG density gradually lose DNAme in tumors. Recent evidence suggests that this progressive hypomethylation is coupled to cancer cell mitotic division and represents a byproduct of replicative aging (2). On the other hand, CpG islands (CGIs)—regions of high CG density found at >60% of human gene promoters (3)—are normally free of DNAme but become hypermethylated in cancer cells (4–6). This promoter CGI hypermethylation can repress downstream genes, including many tumor suppressors such as CDKN2A and BRCA1, thereby critically contributing to cancer initiation and development (7–10).
The molecular mechanisms underlying cancer-associated promoter CGI hypermethylation remain a major focus in the field. Notably, multiple independent analyses of large cohorts of patient tumor samples demonstrate that promoter CGIs that gain DNAme are marked by trimethylation of histone H3K27 (H3K27me3) in embryonic or tissue stem/progenitor cells (11–14). This histone posttranslational modification (PTM) is established by the Polycomb repressive complex 2 (PRC2), which cooperates with another Polycomb complex (PRC1) and its enzymatic product H2AK119 monoubiquitination (H2AK119Ub) to maintain the temporal silencing of genes involved in cell differentiation (15). Why Polycomb target genes are particularly vulnerable to DNA hypermethylation is not well understood. Previous studies have reported the genomic colocalization of Polycomb complex members and various DNA methyltransferases (DNMTs) (16, 17). However, the molecular and structural bases for their interactions, if any, are unclear.
DNMT3A is one of the two de novo DNMTs in mammalian cells (18). DNMT3A encodes two major isoforms (19), with short DNMT3A2 specifically expressed in early embryonic development and long DNMT3A1 universally expressed across somatic tissues. The catalytic activity of DNMT3A can be stimulated by catalytically inactive accessory proteins DNMT3L or DNMT3B3 (20, 21). The crystal structure of the C-terminal domains of DNMT3A and DNMT3L showed a heterotetramer complex of DNMT3L-DNMT3A-DNMT3A-DNMT3L (22). More recently, a cryo–electron microscopy (cryo-EM) structure of DNMT3A2-DNMT3B3 heterotetramer bound to a nucleosome revealed binding between the DNMT3B3 catalytic-like domain and the nucleosome acidic patch (23). Adding further complexity, DNMT3A can also function as homotetramer or oligomer without accessory DNMT3 proteins (24, 25). However, it remains unclear how DNMT3A oligomers interact with the nucleosome core and whether various DNMT3A-containing higher-order complexes operate at distinct genomic regions.
In addition to its methyltransferase domain, DNMT3A harbors several regulatory domains that can bind histone PTMs (26). In particular, its PWWP domain recognizes histone H3K36 di- and trimethylation (H3K36me2/3), which facilitates targeting to gene bodies and promoter-proximal intergenic regions of highly expressed genes (27–31). Missense mutations that disrupt the DNMT3A PWWP-H3K36me2/3 interaction have been reported in patients with Heyn-Sproul-Jackson syndrome or rare neuroendocrine tumors (32–34). Cells harboring DNMT3A PWWP mutations showed increased DNMT3A1 localization and DNA hypermethylation at promoter CGIs of Polycomb target genes (32, 35–37). In this regard, the N-terminal region specific to DNMT3A1 long isoform can bind the Polycomb PTM H2AK119Ub in vitro (37, 38). These studies suggest that imbalanced DNMT3A1 targeting, regulated by the competition between two histone reader activities, may contribute to aberrant DNA hypermethylation of Polycomb target genes.
In this study, we provide structural, biochemical, and cellular analyses to explore the molecular details, functional significance, and cancer relevance of the interaction between DNMT3A1 and H2AK119Ub nucleosomes. We identify critical regions within the DNMT3A1 N terminus that interact with ubiquitin (Ub) and the nucleosome acidic patch, which collectively contribute to the genome-wide localization of DNMT3A1 to H2AK119Ub+ Polycomb CGIs. We further show that this dual recognition of H2AK119Ub nucleosome by DNMT3A1 is essential for CGI hypermethylation during tumor progression and the stable silencing of Polycomb target genes. Together, these results provide insights to the cause, consequence, and therapeutic targeting of cancer-specific CGI hypermethylation.
RESULTS
Identifying minimal regions within N terminus of DNMT3A1 necessary for its localization to H2AK119Ub chromatin
We and others previously reported that disruptions of its PWWP domain led to DNMT3A1 mislocalization to H2AK119Ub-high genomic regions (36–38). Building on these findings, we further explored the nuclear distribution of epitope-tagged DNMT3A1 using immunofluorescence (IF) staining in C3H10T1/2 (hereafter 10T), a tetraploid female mouse mesenchymal stem cell line. In contrast to the uniform nuclear distribution of wild-type (WT) DNMT3A1 (DNMT3A1WT), a mutant lacking its PWWP domain (DNMT3A1ΔPWWP) localized to discrete foci (fig. S1A). These foci were also enriched for Xist—a marker for inactive X chromosome (Xi)—as assessed by RNA fluorescence in situ hybridization (FISH). Consistent with previous studies (39, 40), we found that Xi harbors high levels of histone H2AK119Ub readily visible by IF (fig. S1B). Double deletion of Ring1a/b, the catalytic subunits of PRC1, ablated H2AK119Ub and reverted DNMT3A1ΔPWWP distribution to that of DNMT3A1WT (fig. S1, B and C). The DNMT3A1ΔPWWP redistribution to Xi was recapitulated by point mutations (W330R and D333N) that compromise the PWWP-H3K36me2 interaction (fig. S1D). Notably, deleting the PWWP domain of the DNMT3A2 short isoform, which lacks the DNMT3A1 with ~220–amino acid N-terminal region, did not lead to accumulation at Xi (fig. S1D). Furthermore, swapping the DNMT3A1 N-terminal region into DNMT3B was sufficient to drive DNMT3BΔPWWP to Xi. These results agree with previous genome-wide studies showing a critical role for its N-terminal region in targeting DNMT3A1 to H2AK119Ub chromatin (37, 38). Therefore, we reasoned that IF could serve as a rapid and robust approach to investigating the interaction between DNMT3A1 and H2AK119Ub.
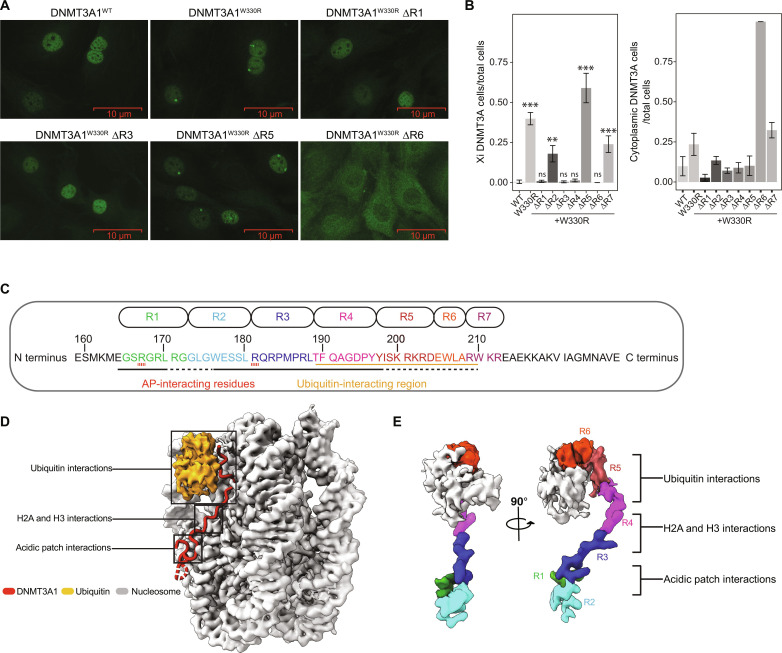
The DNMT3A1 N terminus contains a disordered region (amino acids 1 to 159) and a predicted α helix (amino acids 160 to 219) (fig. S1E). We found that amino acids160 to 219, but not amino acids 1 to 159, of DNMT3A1ΔPWWP were essential for the H2AK119Ub interaction (fig. S1E) and, thus, refer to amino acids 160 to 219 as the Ub-dependent recruitment (UDR) region. To further define the minimal element(s) within the UDR required for the DNMT3A1-H2AK119Ub interaction, we systematically removed four–to eight–amino acid blocks (denoted ΔR1 to ΔR7) across amino acids 165 to 212 of DNMT3A1W330R, followed by IF for DNMT3A1 (Fig. 1, A to C). Here, ΔR1 (amino acids 165 to 172), ΔR3 (amino acids 181 to 188), and ΔR4 (amino acids 189 to 196), but not ΔR2 (amino acids 173 to 180), ΔR5 (amino acids 197 to 204), or ΔR7 (amino acids 209 to 212), fully reversed the Xi accumulation of DNMT3A1W330R (Fig. 1, A and B). Notably, DNMT3A1W330R ΔR6 (amino acids 205 to 208) was primarily cytoplasmic, consistent with loss of a nuclear localization signal within this region (Fig. 1, A and B) (41). These genetic analyses suggest that multiple regions within the UDR contribute to the DNMT3A1 recognition of H2AK119Ub-modified nucleosome.
Fig. 1. Identification of critical regions within N terminus of DNMT3A1 for DNMT3A1-H2AK119Ub interaction.
(A) Representative IF staining of FLAG-DNMT3A1 in 10T cells expressing DNMT3A1WT, DNMT3A1W330R (compromising the PWWP domain), or DNMT3A1W330R with four–to eight–amino acid deletions within the UDR domain [∆R1, ∆R3, ∆R5, and ∆R6: see (C) for more detail]. (B) Quantification of FLAG-DNMT3A1 IF in 10T cells expressing DNMT3A1WT, DNMT3A1W330R, or DNMT3A1W330R with ∆R1 to ∆R7. Left: Ratio of 10T cells displaying DNMT3A1 Xi accumulation compared to all 10T cells. Right: Ratio of 10T cells displaying DNMT3A1 cytoplasmic localization compared to all 10T cells. Error bars represent SD of five replicates. Student’s t test, (ns) P > 0.05, **P < 0.01, and ***P < 0.001. (C) Amino acid sequence of the DNMT3A1 UDR domain (amino acids 159 to 228). Regions subject to deletion analysis in (A) (R1 to R7) are indicated at top. Interaction surfaces for the nucleosome acidic patch and Ub are indicated at bottom. (D) Cryo-EM map of DNMT3A1 N-terminal region (amino acids 159 to 228) bound to H2AK119Ub nucleosome. (E) Portion of the cryo-EM map highlighting various regions within DNMT3A1 UDR domain [colored as per (C)] and Ub, shown at two different views. AP, acidic patch.
A cryo-EM structure of DNMT3A1 UDR in complex with H2AK119Ub nucleosome
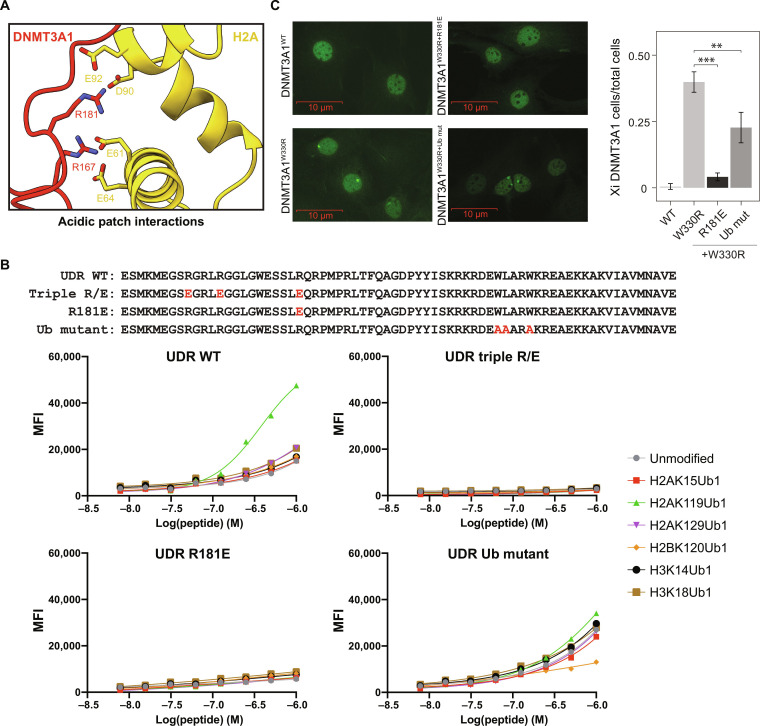
To gain a detailed understanding of their potential mode(s) of engagement, we determined the structure of the DNMT3A1 UDR (amino acids 159 to 228) bound to a H2AK119Ub nucleosome at an overall resolution of 2.8 Å (Fig. 1D). This structure shows well-resolved densities for the DNMT3A1 UDR peptide and nucleosome, as well as a density for Ub (figs. S2 and S3). From our cryo-EM map, we can unambiguously assign 33 residues of the DNMT3A1 UDR (amino acids 165 to 197) that span the nucleosome face and make histone contacts (fig. S4). Density for parts of the peptide interacting with Ub and the Ub density itself were more mobile, precluding our ability to define specific side-chain interactions (fig. S4). Further, the Ub interaction region contains several charged residues (DNMT3A1 with amino acids 200 to 203: KRKR) that could also potentially interact with nucleosomal DNA (Fig. 1, C and D). Notably, the structure suggested critical regions of interaction between the DNMT3A1 UDR and H2AK119Ub (R6), histones H2A and H3 (R4), and the nucleosome acidic patch (R1 and R3) (Fig. 1E). The structural importance of these blocks is consistent with our deletion analyses (Fig. 1, A to C). Our structure also identifies interactions between DNMT3A1 R181 and H2A E61, D90, and E92 of the nucleosome acidic patch (Fig. 2A and fig. S4). While it appears that R181 is its primary acidic patch interacting residue, the UDR peptide folds into a flexible loop, allowing DNMT3A1 R167 to also engage H2A E61 and E64 of the acidic patch. Last, DNMT3A1 R171 and W176, located in the flexible loop between the acidic patch binding residues, also engage the nucleosome, further contributing to complex formation (fig. S4).
Fig. 2. DNMT3A1 UDR domain binds to H2AK119Ub nucleosome via dual recognition of acidic patch and Ub.
(A) Close-up view of the interactions between the N-terminal region of DNMT3A1 and acidic patch of the nucleosome. (B) Top: Amino acid sequence of the UDR peptide for WT, triple acidic patch mutants (R167E, R171E, and R181E), single acidic patch mutant (R181E), and Ub mutants (W207A, L208A, and W210A). Bottom: dCypher Luminex assay to measure interaction between 6×His-tagged UDR peptides (WT or mutant as noted: the Queries) and a multiplexed panel of fully defined nucleosomes (the Targets; unmodified or with diverse KUb: H2AK15Ub1, H2AK119Ub1, H2AK129Ub1, H2BK120Ub1, H3K14Ub1, or H2BK18Ub1). A four-parameter logistic equation where X is log(peptide concentration) was applied. (C) Left: Representative IF of FLAG-DNMT3A1 in 10T cells expressing DNMT3A1WT, DNMT3A1W330R (PWWP mutant) DNMT3A1W330R+R181E or DNMT3A1W330R+Ub mut. Right: Quantification of FLAG-DNMT3A1 IF in 10T cells expressing DNMT3A1WT, DNMT3A1W330R, DNMT3A1W330R+R181E or DNMT3A1W330R+Ub mut, shown as ratio of 10T cells displaying DNMT3A1 Xi accumulation compared to all 10T cells. Error bars represent SD of five replicates. Student’s t test, **P < 0.01, and ***P < 0.001.
DNMT3A1 UDR-H2AK119Ub nucleosome binding in vitro requires both acidic patch and Ub interactions
On the basis of structural analysis of the DNMT3A1 UDR-H2AK119Ub nucleosome interaction, we generated UDR peptides mutated for residues contacting the acidic patch (R167E/R171E within R1 and R181E within R3, also known as UDRtriple R/E) or Ub (W206A, L207A and W210A within R6, also known as UDRUb mut) (Figs. 1C and 2B). We then used multiplexed dCypher Luminex (see Methods) to test interactions between the various forms of DNMT3A1 UDR (the Queries) and a panel of fully defined nucleosomes (the Targets). We first confirmed enhanced interaction between WT UDR (UDRWT) and H2AK119Ub over other histone lysine monoubiquitinations (H2AK15Ub1, H2AK129Ub1, H2BK120Ub1, H3K14Ub1, and H3K18Ub1; Fig. 2B). In contrast, UDRR181E or UDRtriple R/E (acid patch binding mutants) completely lost nucleosome binding (Fig. 2B), while UDRUb mut largely lost the preference for H2AK119Ub (Fig. 2B). Consistently, IF of 10T cells expressing epitope-tagged DNMT3A1W330R showed that overlaid mutations R181E (i.e., DNMT3A1W330R+R181E), and to a lesser extent 206WLARW210/AAARA (i.e., DNMT3A1W330R+Ub mut), reversed Xi accumulation (Fig. 2C). Together, these results demonstrate that interaction between DNMT3A1 and H2AK119Ub requires distinct regions of the UDR to mediate bidentate recognition of the nucleosome acidic patch and Ub.
Genome-wide localization and methyltransferase activity of DNMT3A1 are regulated by its interface with H2AK119Ub and nucleosome acidic patch
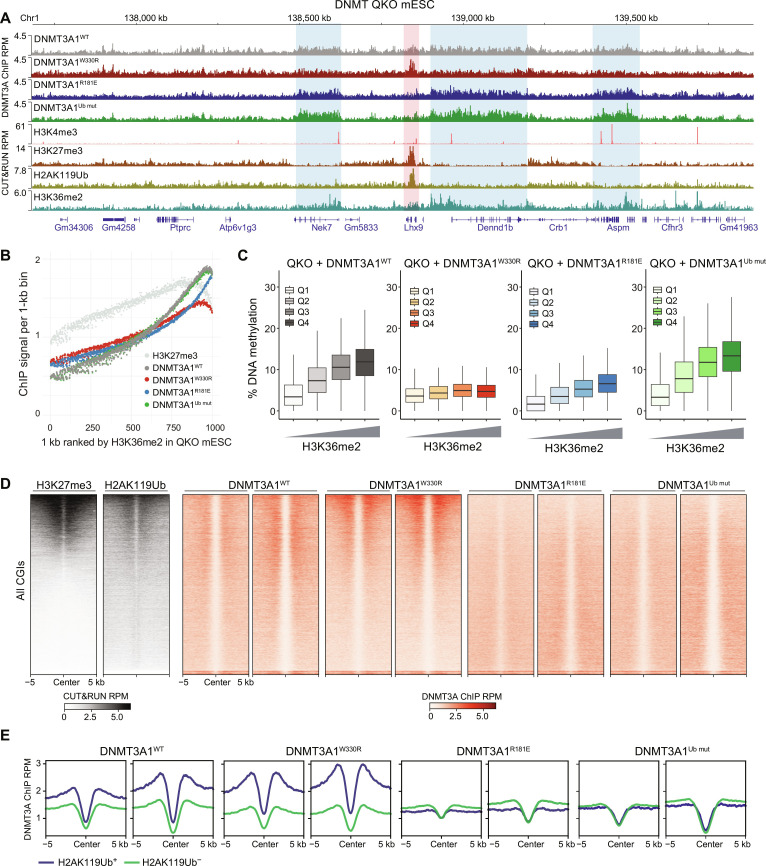
We next determined how the genome-wide targeting and de novo methyltransferase activity of DNMT3A1 were affected by mutations in its UDR or PWWP domains. To this end, we created DNMT quadruple knockout (QKO) mouse embryonic stem cells (mESCs) by targeted deletion of Dnmt3l in triple knockout (TKO) mESCs (42) already deficient for Dnmt3a, Dnmt3b, and Dnmt1 (fig. S5A). We then reexpressed either WT Dnmt3a1 (DNMT3A1WT) or alleles carrying mutations in the PWWP domain (DNMT3A1W330R), acidic patch interface (DNMT3A1R181E), or Ub-binding region (DNMT3A1Ub mut) (fig. S5B). This system enabled us to assess DNMT3A1 enrichment [by chromatin immunoprecipitation sequencing (ChIP-seq)] and de novo DNAme [5mC/5-hydroxymethylcytosine by enzymatic methyl sequencing (EM-seq)] in the absence of any additional DNMT machinery. As expected, while DNMT3A1WT, DNMT3A1R181E, and DNMT3A1Ub mut localization and methylation activity positively correlated with H3K36me2, this was lost with DNMT3A1W330R (Fig. 3, A to C, and fig. S5C). Despite the colocalization of DNMT3A1R181E and H3K36me2, we noticed a general decrease in DNMT3A1R181E methyltransferase binding and activity compared to DNMT3A1WT (Fig. 3, A to C, and fig. S5C). We also analyzed TKO mESCs expressing DNMT3A1 mutants (fig. S5D). Unlike QKO cells, the overall DNAme level (fig. S5E) and its positive correlation with H3K36me2 in TKO cells expressing DNMT3A1R181E were similar to that of DNMT3A1WT (compare Fig. 3C and fig. S5D). This suggested that in the presence of DNMT3L adaptor protein, diminished UDR interaction with the nucleosome acidic patch does not compromise DNMT3A1-mediated methylation globally or at H3K36me2 regions.
Fig. 3. Genomic targeting of DNMT3A1 is regulated by its interface with H2AK119Ub and nucleosome acidic patch.
(A) Genome browser view (at chromosome 1, 137.7 to 139.8 Mb, GRCm38) of DNMT3A1 ChIP-seq reads for DNMT3A1WT, DNMT3A1W330R, DNMT3A1R181E, and DNMT3A1Ub mut QKO mESCs. CUT&RUN reads for H3K4me3, H3K27me3, H2AK119Ub, and H3K36me2 in DNMT3A1WT QKO mESCs are also shown. Genes from RefSeq database are annotated at the bottom. (B) CUT&RUN reads per 1000 averaged 1-kb bin for H3K27me3 and ChIP-seq reads per 1000 averaged 1-kb bin for DNMT3A1 in DNMT3A1WT, DNMT3A1W330R, DNMT3A1R181E, and DNMT3A1Ub mut QKO mESCs. To generate 1000 rank-ordered bins, 1-kb genomic tiles were ranked by H3K36me2 enrichment in DNMT3A1WT QKO mESC and grouped. (C) Percent of DNAme (EM-seq) in DNMT3A1WT, DNMT3A1W330R, DNMT3A1R181E, and DNMT3A1Ub mut QKO mESCs per 10,000 bins grouped by H3K36me2 CUT&RUN enrichment [Q1 (lowest) to Q4 (highest)]. (D) Enrichment heat map of H3K27me3 and H3K36me2 CUT&RUN reads in DNMT3A1WT QKO mESCs and DNMT3A1 ChIP-seq reads in DNMT3A1WT, DNMT3A1W330R, DNMT3A1R181E, and DNMT3A1Ub mut QKO mESCs sorted by H3K27me3 centered at CGIs ± 5 kb. (E) Enrichment plot of ChIP-seq reads of DNMT3A1 in DNMT3A1WT, DNMT3A1W330R, DNMT3A1R181E, and DNMT3A1Ub mut QKO mESCs centered at CGIs ± 5 kb and grouped into the highest 20% H2AK119Ub-enriched CGIs (H2AK119Ub+) and the lowest 80% H2AK119Ub-enriched CGIs (H2AK119Ub−).
We next performed cleavage under targets and release using nuclease (CUT&RUN) for histone PTMs associated with transcriptionally active (H3K4me3) or Polycomb-repressed (H3K27me3 and H2AK119Ub) CGIs. DNMT3A1WT enrichment was negatively correlated with H3K4me3 but detectable at H3K27me3+ and H2AK119Ub+ CGIs (Fig. 3, D and E, and fig. S5F). DNMT3A1W330R, however, showed increased enrichment at Polycomb-repressed CGIs, consistent with the notion that H2AK119Ub competes with H3K36me2 for DNMT3A1 recruitment. In contrast, DNMT3A1R181E and DNMT3A1Ub mut showed reduced colocalization to H3K27me3+ and H2AK119Ub+ CGIs (Fig. 3, A, D, and E, and fig. S5F). These findings support that the DNMT3A1 PWWP promotes recruitment to nucleosomal H3K36me2, while the DNMT3A1 UDR is essential for DNMT3A1 localization at Polycomb-regulated CGIs via nucleosomal H2AK119Ub and the acidic patch.
Redistribution of DNMT3A1 leads to DNA hypermethylation at Polycomb CGIs
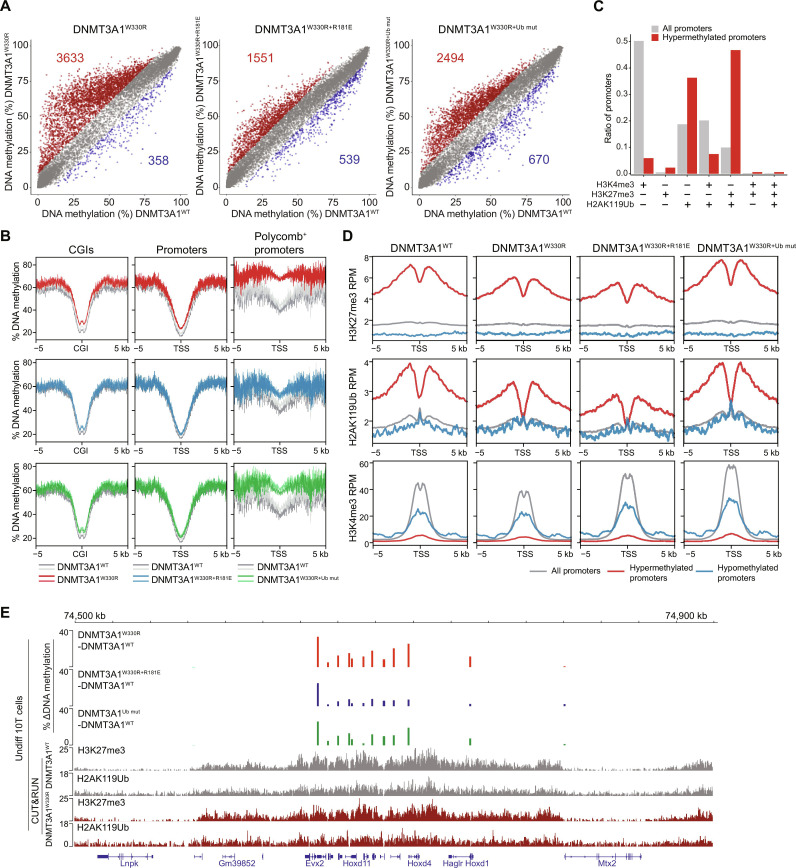
Despite WT or mutant DNMT3A1 targeting to H3K27me3+ and H2AK119Ub+ CGIs, we observed minimal DNAme increase at these regions in QKO mESCs (fig. S5G). This is unlikely due to the absence of DNMT1 in these cells since knock-in of DNMT3AW330R to WT mESCs also fails to induce CGI hypermethylation (32). We speculate that high expression of the DNA demethylating ten eleven translocation methylcytosine dioxygenases (TET) enzymes in mESCs may serve to protect CGIs from aberrant DNAme gain (43, 44). Supporting this idea, ectopic expression of DNMT3A1W330R in a more differentiated environment, such as 10T mouse mesenchymal stem cells (which also express intact DNMTs), was sufficient to drive promoter CGI DNA hypermethylation (Fig. 4, A and B, and fig. S6A). By integrating reduced representation bisulfite sequencing (RRBS) data with CUT&RUN histone PTM analyses, we found that promoters more likely to become hypermethylated in DNMT3AW330R-expressing cells were either H2AK119Ub+ or [H2AK119Ub+, H3K27me3+] in control cells (Fig. 4, B and C, and fig. S6B). This DNMT3A1W330R-induced hypermethylation of Polycomb CGIs can be reversed by overlaying a UDR mutation that compromises acid patch binding (DNMT3A1W330R+R181E) and, to a lesser extent, one that reduces Ub binding (DNMT3A1W330R+Ub mut) (Fig. 4, A and B).
Fig. 4. Polycomb promoter hypermethylation following DNMT3A1 redistribution requires DNMT3A1 interaction with the nucleosome acidic patch and H2AK119Ub.
(A) Scatter plots showing the percentage of DNAme (by RRBS) at promoters [TSS ± 500 base pairs (bp)] in DNMT3A1W330R, DNMT3AW330R+181E, and DNMT3A1W330R+Ub mut versus DNMT3A1WT-expressing 10T cells. Each dot represents a single promoter. Significantly hypermethylated (>10% and q < 0.01) and hypomethylated (<−10% and q < 0.01) promoters are colored in red and blue, respectively (two biological replicates). (B) Metagene plot showing the percentage of DNAme over CGIs ± 5 kb (left), promoters (TSS ± 5 kb) (middle), and Polycomb-regulated promoters (H3K27me3+ and H2AK119Ub+) (right) in DNMT3A1W330R, DNMT3AW330R+R181E, and DNMT3A1W330R+Ub mut compared to DNMT3A1WT 10T cells. (C) Bar plot showing representation among all promoters, or 10T DNMT3AW330R–hypermethylated promoters, for each group of promoters defined by histone PTM status in control cells as in fig. S6B. (D) Metagene plot showing enrichment of H2AK119Ub, H3K27me3, and H3K4me3 CUT&RUN reads from DNMT3A1WT, DNMT3A1W330R, DNMT3AW330R+R181E, and DNMT3A1W330R+Ub mut 10T cells, centered at TSS ± 5 kb, and grouped by all, hypermethylated, and hypomethylated promoters. (E) Genome browser view (chromosome 2, 74.5 to 75.0 Mb), showing differences in the percentage of DNAme (by RRBS) between DNMT3A1WT and various mutant 10T cells. Bottom: H2AK119Ub, H3K27me3, and H3K4me3 CUT&RUN reads from DNMT3A1WT and DNMT3A1W330R 10T cells. Genes from RefSeq database are annotated at the bottom.
We next performed CUT&RUN for H3K4me3, H3K27me3, and H2AK119Ub in various DNMT3A1 backgrounds to further characterize the chromatin state at CGIs that gained DNAme. In DNMT3A1W330R cells, while both genome-wide analysis and inspection of representative regions (e.g., HoxD genes) showed modest (~22%) reductions in H3K27me3 and H2AK119Ub at DNA-hypermethylated CGIs, these regions still retained >2-fold higher enrichment of these Polycomb marks relative to all promoters (Fig. 4, D and E). In addition, there was no correlation between the change in H3K27me3 and degree of DNAme increase (fig. S6C). Therefore, it appears that PWWP mutant–induced DNMT3A1 redistribution leads to aberrant DNAme at Polycomb-regulated CGIs in a UDR-dependent manner. This results in a hybrid chromatin state defined by the coexistence of three repressive epigenetic marks: H3K27me3, H2AK119Ub, and DNAme, which is rarely observed in normal somatic tissues but more common in cancer cells.
DNA hypermethylation at Polycomb CGIs impairs differentiation-induced transcriptional activation of target genes
We next performed RNA sequencing (RNA-seq) to determine whether the colocalization of Polycomb PTMs and DNAme affects gene expression in DNMT3A1 mutant cells. Only a few genes were significantly differentially expressed between 10T cells expressing DNMT3A1WT or DNMT3A1W330R (Fig. 5A), and the up-regulated and down-regulated genes showed minimal and comparable changes in DNAme (fig. S6D). In addition, genes that were hypermethylated in DNMT3A1W330R cells already belonged to the lowest expressed percentiles in DNMT3A1WT cells (Fig. 5B), suggesting that they are effectively repressed by Polycomb complexes and the introduction of DNAme has no further silencing effect at the steady-state level.
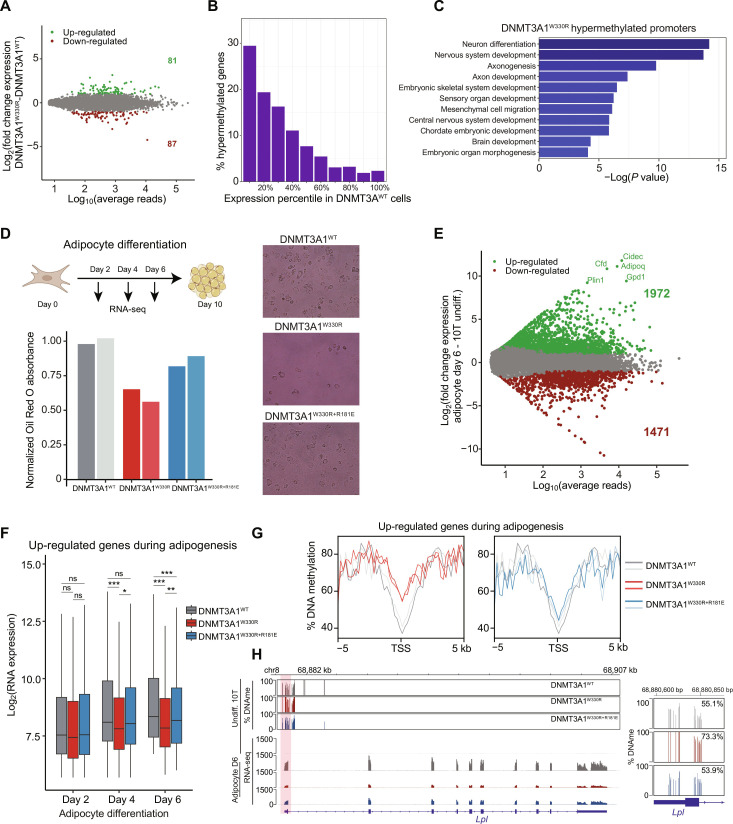
Fig. 5. Increased DNAme at Polycomb CGIs impairs differentiation-induced transcriptional activation of target genes.
(A) MA plot comparing log10 of average normalized reads per gene versus log2 fold change (LFC) in gene expression between DNMT3A1W330R and DNMT3A1WT. Significantly differentially expressed genes are highlighted in green (LFC > 2 and q < 0.01) or red (LFC < −2 and q < 0.01) (two biological replicates). (B) Bar plot showing distribution of 10T DNMT3A1W330R–hypermethylated genes into gene expression deciles in DNMT3A1WT 10T cells. (C) Bar plot of log10 P values of select GO terms of 10T DNMT3A1W330R–hypermethylated genes. (D) Top: Schematic of 10T differentiation to adipocytes. Bottom: Bar plot of normalized Red Oil O absorption in day 10 adipocytes. Representative images of Red Oil O staining are shown. (E) MA plot comparing log10 of average normalized reads per gene in DNMT3A1WT undifferentiated and day 6 adipocyte-differentiated 10T cells versus LFC in gene expression. Significantly differently expressed genes are highlighted in green [LFC > 2 and false discovery rate (FDR) < 0.01] or red (LFC < −2 and FDR < 0.01) (two biological replicates). (F) Gene expression (log2 RPM) box plot of top 500 up-regulated genes in adipocytes from days 2 to 6 (two biological replicates). Student’s t test, (ns) P > 0.05, *P < 0.05, **P < 0.01, and ***P < 0.001. (G) Enrichment plot showing the distribution of the percentage of DNAme over the promoters of up-regulated genes during adipogenesis for DNMT3A1WT, DNMT3A1W330R, and DNMT3AW330R+181E undifferentiated 10T cells. (H) Genome browser view of the percentage of DNAme and gene expression of undifferentiated and day 6 adipocyte-differentiated DNMT3A1WT, DNMT3A1W330R, and DNMT3AW330R+181E 10T cells over Lpl. Right: Average DNAme at Lpl promoter.
Gene ontology (GO) analysis of the hypermethylated genes in DNMT3A1W330R cells revealed enrichment for developmental gene clusters such as “neuron differentiation,” “embryonic skeletal system development,” and “embryonic organ morphogenesis” (Fig. 5C). Therefore, we examined the dynamics of gene expression during cell differentiation. We induced adipocyte differentiation of DNMT3A1 WT or mutant 10T cells for 10 days as previously (45), with total RNA collected for RNA-seq on days 2, 4, and 6 after initiation (Fig. 5D). Red Oil O staining of day 10 adipocytes found visibly reduced differentiation in DNMT3A1W330R cells (Fig. 5D, 61% of DNMT3A1WT) and a partial rescue of this effect in double mutant DNMT3A1W330R+R181E cells (85% of DNMT3A1WT). In DNMT3A1WT cells, 3443 genes were differentially expressed comparing days 6 versus 0 of adipocyte differentiation (1972 up-regulated/1471 down-regulated) (Fig. 5E). As expected, GO analysis of the up-regulated genes showed enrichment for adipocyte-specific groups (e.g., “fatty acid catabolic process” or “fatty acid metabolic process”) (fig. S6E). Consistent with Red Oil O staining, many adipogenesis-associated genes (46, 47), such as Cfd, Cidec, and Adipoq, failed to be activated by day 6 in DNMT3A1W330R, but not DNMT3A1W330R+R181E, cells (fig. S6F). Focusing on the top 500 genes up-regulated during adipogenesis, we profiled their expression kinetics through differentiation (days 2 to 6) and observed a significantly delayed increase in DNMT3AW330R cells with a significant rescue observed for DNMT3AW330R+R181E cells (Fig. 5F).
Next, we examined the promoter DNAme state of genes up-regulated during adipogenesis, using the RRBS data of undifferentiated 10T cells. Compared to all gene promoters, the up-regulated set gained a higher degree of DNAme in DNMT3A1W330R cells, with this partially reversed in DNMT3AW330R+R181E cells (Fig. 5G and fig. S6G). This is especially evident for adipogenesis regulators known to be sensitive to DNAme regulation, such as Slc27a1 (Fatp1), Klf15, and Lpl (Fig. 5H and fig. S6H) (46). Therefore, the incorporation of DNAme into promoter CGIs harboring Polycomb marks did not affect basal silencing but limited their potential to become activated upon induction. In the context of adipocyte differentiation, impaired expression of hypermethylated master regulators could have a downstream effect on the less DNAme-sensitive (e.g., low promoter CG density) adipogenesis factors (such as Cfd, Cidec, and Adipoq), causing a systematic delay to adipocyte commitment.
Cancer-associated Polycomb CGI hypermethylation requires DNMT3A1–acidic patch interaction
We next sought to explore the cancer relevance of our findings. Analysis of patient tumor samples suggests that CGIs that gain aberrant DNAme are more likely to be enriched for H3K27me3 in human embryonic or adult stem cells (11–14). However, this correlation has not been mechanistically dissected in a controlled experimental setting, nor has the potential involvement of DNMT3A been explored. To this end, we used a well-established carcinogen-induced esophagus squamous cell carcinoma (ESCC) mouse model (48). We generated organoid cultures from esophagi harvested from normal mice [normal esophagus (EN)] or ESCC developed in mice after 16 weeks treatment with the carcinogen 4-nitroquinoline 1-oxide (4NQO) (Fig. 6A). These organoid cultures have been extensively characterized and recapitulate the genomic and histological features of human EN and ESCC (48, 49). DNAme profiling by RRBS, as well as cleavage Under targets and tagmentation (CUT&Tag) for H3K4me3, H3K27me3, and H2AK119Ub, was performed in six independently generated organoid lines (3× EN and 3× ESCC). We also performed RRBS in EN organoids after introducing DNMT3A1WT, DNMT3A1W330R, and DNMT3A1W330R+R181E (Fig. 6A). Using these datasets, we examined (i) whether H2AK119Ub/H3K27me3 enrichment in esophageal progenitors predicts CGI DNA hypermethylation in tumors arising from these cells of origin and (ii) whether redistribution of DNMT3A1 to Polycomb CGIs through impaired PWWP reader function, in a UDR-dependent manner, is sufficient to recapitulate the cancer-specific DNA hypermethylation signature.
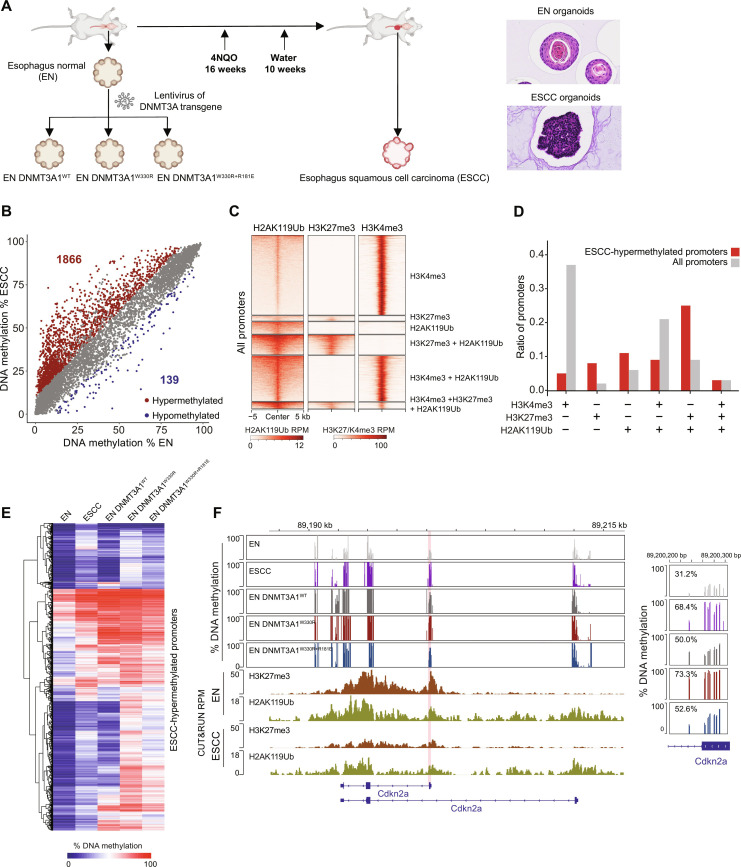
Fig. 6. Cancer-associated Polycomb CGI hypermethylation requires DNMT3A1–acidic patch interaction.
(A) Schematic of EN- and 4NQO-induced ESCC organoid collection. Right: Representative images of EN and ESCC organoid histology. (B) Scatter plots showing the percentage of DNAme (by RRBS) at all promoters (TSS ± 500 bp) in ESCC versus EN organoids. Each dot represents a single promoter. Significantly hypermethylated (>10% and q < 0.01) and hypomethylated (<−10% and q < 0.01) promoters are respectively colored in red and blue (three biological replicates), respectively. (C) Heatmap showing enrichment of H3K27me3, H2AK119Ub, and H3K4me3 CUT&Tag reads at all promoters (TSS ± 5 kb) in EN organoids. Promoters are classified to six groups: H3K4me3+; H3K27me3+; H2AK119Ub+; H3K4me3+ and H2AK119Ub+; H3K27me3+ and H2AK119Ub+; and H3K4me3+, H3K27me3+, and H2AK119Ub+. (D) Bar plots showing the representation among all promoters, or ESCC-hypermethylated promoters, for each promoter group defined in (C). (E) Heatmap showing the percentage of promoter DNAme (by RRBS) in EN organoids, ESCC organoids, and EN organoids expressing DNMT3A1WT, DNMT3A1W330R, and DNMT3A1W330R+R181E for all ESCC-hypermethylated promoters. (F) Genome browser view of Cdkn2a (chromosome 4, 89,187 to 89,216 Mb, GRCm39), showing the percentage of DNAme (by RRBS) in EN organoids, ESCC organoids, and EN organoids expressing DNMT3A1WT, DNMT3A1W330R, and DNMT3A1W330R+R181E. Bottom: H3K27me3 and H2AK119Ub CUT&Tag reads in EN and ESCC organoids. Right: Average DNAme at Cdkn2a promoter (chromosome 4, 89,200,200 to 89,200,300 bp, GRCm39). Genes from RefSeq database are annotated at the bottom.
RRBS revealed widespread hypermethylation of >1800 promoters and limited hypomethylation of ~150 promoters in ESCC compared to EN organoids (Fig. 6B), despite comparable global DNAme levels (fig. S7A). We classified the promoters based on their levels of H2AK119Ub, H3K27me3, and H3K4me3 in EN organoids (Fig. 6C) and found that those with either H2AK119Ub and H3K27me3 or H2AK119Ub alone were more prone to gaining DNAme in ESCC organoids (Fig. 6D). In contrast, H3K4me3+ promoters were more resistant to cancer-associated DNA hypermethylation (Fig. 6D). These results provide strong support for the notion that Polycomb activity in normal tissue predisposes promoter CGIs to hypermethylation during oncogenesis. Similar to 10T cells, DNA-hypermethylated promoters in ESCC organoids showed reduced but higher-than-genome average levels of H3K27me3, possibly signifying H3K27me3 and DNAme co-occupancy at these regions (fig. S7B). However, we observed that compared to 10T cells, the ESCC-associated decreases in H3K27me3 at Polycomb CGIs were stronger and better correlated with the DNAme increase (fig. S7B).
Notably, expressing DNMT3A1W330R in EN organoids largely recapitulated the DNA hypermethylation signature of ESCC organoids (Fig. 6E and fig. S7, C and D). Over half of the ESCC-hypermethylated promoters were also hypermethylated in DNMT3A1W330R EN organoids, including classic tumor suppressor genes such as Cdkn2a (Fig. 6F) (50). Furthermore, using DNAme data from The Cancer Genome Atlas (51), we identified sets of hypermethylated promoters in human esophageal cancer and head and neck cancer (HNSC). Over 75% of the esophageal cancer–and HNSC-hypermethylated promoters were also hypermethylated in the DNMT3A1W330R EN organoids (fig. S7E). The DNMT3A1W330R-induced aberrant hypermethylation phenotype was markedly reversed in DNMT3A1W330R+R181E organoids (Fig. 6, E and F, and fig. S7, C and D), suggesting that interactions between DNMT3A1 UDR and nucleosomal H2AK119Ub/acidic patch play a critical role in establishing cancer-associated CGI hypermethylation.
DISCUSSION
In this study, we provide structural, biochemical, and genetic evidence that DNMT3A1 localization to Polycomb CGIs requires binding by its N-terminal UDR to both nucleosomal H2AK119Ub and the acidic patch. This targeting mechanism competes with the DNMT3A1 PWWP–H3K36me2/3 interaction to regulate genomic distribution of the methyltransferase. Furthermore, perturbation of the normal competition can lead to the CGI hypermethylation phenotype commonly found in cancer cells. These findings advance our mechanistic understanding of DNMT3A1 recruitment and regulation, reveal how mistargeting may contribute to tumor development, and represent a therapeutic opportunity.
DNMT3A1 does not have a classical Ub-binding domain and is not reported to bind free Ub but still shows preference for H2AK119Ub. This is not the first instance where H2AK119Ub is implicated in pathological mislocalization of chromatin regulators in cancer. The fusion oncoprotein SS18-SSX1 delocalizes BRG/Brahma-associated factors (BAF) chromatin–remodeling complex from promoters/enhancers toward Polycomb-repressed regions marked by H2AK119Ub (52). This (mis)targeting is facilitated by SSX1, which binds H2AK119Ub nucleosomes despite lacking a canonical Ub-binding domain and having no affinity for free Ub. Selective binding is accomplished by SSX1 docking to the nucleosome acidic patch and establishing interactions with Ub, much similar to the case of DNMT3A1. However, the interface with Ub differs between these two structures. In SS18-SSX1 interactions extend from the acidic patch through H2A α2 and α3 helices to a surface created by histone H3 and H2AK119Ub (53). In DNMT3A1, the interactions instead extend from the acidic patch through α2, a short loop between α3 and αC οf H2A, and ends between α2 οf H3 and Ub. Despite these distinctions, the general mode of interaction is preserved. These data raise important questions: Are H2AK119Ub nucleosomes uniquely capable in their aberrant delocalization of chromatin complexes (DNMT3A1 PWWP mutant, SS18-SSX1 fusion), and, if so, then are other complexes mistargeted to Polycomb loci using similar mechanisms?
Short isoform DNMT3A2 lacking the DNMT3A1 N terminus, including the UDR region, is predominantly expressed in early embryonic development (54). This coincides with the expression of noncatalytic DNMT3L (55), and the resulting DNMT3A2-DNMT3L heterotetramer establishes genome-wide DNAme (22, 56, 57). In this context, the catalytic-like domains of accessory DNMT3 proteins such as DNMT3B3 or DNMT3L can stabilize DNMT3A2 and/or provide interaction with the nucleosome acidic patch (23) in the absence of UDR as studied herein. DNMT3A1 becomes the dominant DNMT3A isoform around the same development window when DNMT3L expression decreases (58). We speculate that this switch allows DNMT3A1 targeting to Polycomb-regulated genes for de novo DNAme and long-term silencing (59). For example, a group of maternal genes are noncanonically imprinted by Polycomb activity before they acquire DNAme in extraembryonic tissues after implantation (60). In these regions, the ability of the DNMT3A1 UDR region to interact with the nucleosome acidic patch may allow the enzyme to act “solo” as a homotetramer or another type of homo-oligomer without DNMT3L or DNMT3B3. Notably, deletion of the mouse DNMT3A1 N terminus results in abnormal postnatal development (38). We postulate that the growth delay and behavior deficits in these mice are associated with a dysregulated switch from Polycomb to DNAme at certain developmental genes.
At the cellular level, perturbing interaction between the DNMT3A1 UDR and the nucleosome acidic patch (R181E) essentially abolished localization to H2AK119Ub+ CGIs but only reduced enzyme binding and activity toward H3K36me2 domains. Two nonmutually exclusive mechanisms could contribute to this differential impact. First, the UDR–acidic patch interaction may play a more critical role in stabilizing the DNMT3A1 engagement with H2AK119Ub over H3K36me2 nucleosomes. This could be due to the dual affinity of the PWWP domain for H3K36me2 and DNA, which may overcome the need for acidic patch binding (27). Second, as discussed above, of all DNMT proteins and isoforms, only DNMT3A1 has a UDR to mediate H2AK119Ub binding and is thus the most effective in vivo as a homotetramer or homo-oligomer to engage Polycomb-marked promoters (43, 61). In contrast, DNMT3A1 may be targeted to H3K36me2 regions in complex with DNMT3L and/or DNMT3B3, which provide additional contacts with the nucleosome acidic patch. This is observed in ESCs where DNMT3L colocalizes with and directs DNMT3A toward H3K36me2/3-marked gene bodies (62). We showed that in TKO mESCs (i.e., presence of DNMT3L), R181E mutation did not reduce DNMT3A1’s activity globally or at H3K36me2 domains (fig. S5D, compared to QKO data in Fig. 3C). Further structure-function analysis is warranted to test these hypotheses.
We and others have previously shown that mutations in TET DNA demethylases, or inhibition by the “oncometabolite” 2-hydroxyglutatrate produced by gain-of-function mutant isocitrate dehydrogenase (IDH), can establish CGI hypermethylation (63–67). These findings are consistent with the notion that TET enzymes act as guardians to protect CGIs from aberrant DNAme. However, how de novo DNA methylating activity is targeted to CGIs, particularly those enriched for Polycomb marks, remains elusive (14). An early model proposes that DNMT3 mislocalization in cancer may be facilitated by sequence-specific transcription factors (68, 69). However, the ectopic insertion of CGIs in cancer cells does not lead to their hypermethylation, arguing against a cis-element mechanism (70). Using organoids isolated from a well-established ESCC model, we could rigorously test and confirm the association between H2AK119Ub/H3K27me3 enrichment in tissue progenitor cells and CGI hypermethylation in tumors arising from these cells of origin. In addition, our results using normal organoids expressing various DNMT3A1 mutants suggest that perturbed reader function of the PWWP domain facilitates cancer-associated CGI hypermethylation in a UDR-dependent manner. These findings provide a unifying model for the establishment of Polycomb CGI DNA hypermethylation in cancer. In normal somatic tissues, a subpopulation of DNMT3A1 can localize to Polycomb CGIs via the UDR-H2AK119Ub interaction, but its methyltransferase activity is countered by TET enzymes. In cancer cells, DNA hypermethylation at these regions can result from either insufficient TET activity or increased binding of DNMT3A1 redistributed from H3K36me2/3 domains. Notably, missense mutations that impair the PWWP-H3K36me2 interaction are only found in rare types of neuroendocrine tumors (31, 34), suggesting that nongenetic mechanisms, such as PTMs within the DNMT3A1 PWWP domain, may contribute more generally to cancer-specific CGI hypermethylation.
Catalytic inhibitors of DNMT (e.g., decitabine) have been approved to treat myeloid malignancies. However, their use in solid tumors is limited in part by toxic side effects, as these inhibitors induce genome-wide demethylation that is also deleterious to normal cells. Our structure-function studies pave the way for developing inhibitors that target the bidentate interaction between DNMT3A1 and the H2AK119Ub nucleosome, which would be expected to specifically prevent or reverse CGI hypermethylation in cancer cells, while preserving the DNA methylome in normal tissues.
METHODS
Expressing DNMT3A1 mutants in 10T cells
cDNA of mouse Dnmt3A1WT and DnmtA1W326R (corresponding to human W330R) previously cloned into pCDH-EF1-MCS-Neo (System Biosciences) with an N-terminal FLAG–hemagglutinin (HA) epitope tag (37) was used with standard mutagenesis to generate deletions within UDR [ΔR1, amino acids 161 to 168; ΔR2, amino acids 169 to 176; ΔR3, amino acids 177 to 184; ΔR4, amino acids 185 to 192; ΔR5, amino acids 193 to 200; ΔR6, amino acids 201 to 204; ΔR7, amino acids 205 to 208; corresponding to human ΔR1 (amino acids 165 to 172), ΔR2 (amino acids 173 to 180), ΔR3 (amino acids 181 to 188), ΔR4 (amino acids 189 to 196), ΔR5 (amino acids 197 to 204), ΔR6 (amino acids 205 to 208), and ΔR7 (amino acids 209 to 212)], the acidic patch mutation R177E (corresponding to human R181E), and the Ub mutant (W202A, L203A, and W206A; corresponding to human W206A, L207A, and W210A). Primers (table S1) were designed using NEBbaseChanger tool and desired mutations confirmed by Sanger sequencing. To generate concentrated lentivirus, DNMT3A1 mutant vectors were cotransfected with helper plasmids psPAX2 and pVSVG into human embryonic kidney–293T. After 72 hours, supernatant was collected and concentrated using the PEG-it system as per the manufacturer’s instructions (System Biosciences).
C3H10T1/2 mouse fibroblast cells (10T; American Type Culture Collection, #CCL-226) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) with 10% fetal bovine serum (FBS; Gemini). To generate transgenic cell lines expressing DNMT3A1 mutants, 10T cells were transduced with concentrated lentivirus. Forty-eight hours after transduction, cells were selected under G418 (1 mg/ml) for 1 week. Methylation analysis was done after 21 days in culture.
CRISPR-KO to generate QKO mESC
Single guide RNAs (sgRNAs) directed against mouse Dnmt3L were cloned into px458. Dnmt3L KO clones (QKO) were generated by transfecting TKO J1 mESC (42) with sgRNA-containing px458 using Lipofectamine 3000 (Invitrogen) and sorting green fluorescent protein–positive (GFP+) cells after 72 hours. For single-clone isolation, transfected cells were seeded at low density (1000 cells per 9.6 cm2) for expansion, and successful KO lines were confirmed by immunoblot (to DNMT3L) and sequencing over the sgRNA target location (analysis with Tracking of Indels by Decomposition tool) (71).
Expressing DNMT3A1 mutants in TKO and QKO mESC
TKO and QKO mESC were cultured in serum/LIF medium [DMEM-KO, 15% FBS, murine recombinant LIF (1000 U/ml; MilliporeSigma, #ESG1106), 1× nonessential amino acids (100×; Gibco, #11140), 1× GlutaMAX (100×; Gibco, #35050-061), and 0.1 mM 2-mercaptoethanol (Gibco)].
The pCDH-EF1-MCS-Neo vectors carrying Dnmt3A1WT, Dnmt3A1W330R, Dnmt3A1R181E, or Dnmt3A1Ub mut were digested with Xba I and Bam HI and cloned to piggyBac backbone vector with GFP visualization marker (pPB-Dnmt3A1-GFP). These were cotransfected with piggybac transpose vector using Lipofectamine 3000 (Invitrogen) to the QKO mESC cell line. GFP+ cells were sorted after 7 days, and stable DNMT3A QKO mESC lines were confirmed by flow analysis and immunoblotting for DNMT3A and sequence confirmation of the appropriate allele. Methylation analysis and DNMT3A ChIP were done after 21 days in culture.
Adipocyte differentiation
10T cells were seeded at 500,000 cells per 9.6 cm2 (in DMEM + 10% FBS) and, the next day, were changed to adipocyte differentiation medium [DMEM, 10% FBS, 0.5 mM isobutylmethylxanthine, 1 μM dexamethasone, insulin (5 μg/ml), and 5 μM troglitazone]. After 2 days, medium was changed to DMEM + 10% FBS + insulin (5 μg/ml) and changed every 2 days until day 10 of adipocyte differentiation. To quantify adipocytes with Oil Red O staining, cells were directly cross-linked (3.7% paraformaldehyde for 2 min) on a plate after medium removal and phosphate-buffered saline (PBS) wash. After a water wash, Oil Red O solution (1 ml per 9.6cm2; Sigma-Aldrich, #01391) was added and plates incubated at room temperature (RT) for 1 hour. Oil Red O solution was removed, the plate was dried, and 1 ml of 100% isopropanol was added for 20 min at RT. This isopropanol (200 μl) was used to measure an absorbance at 520 nm in triplicates on a 96-well plate reader.
Chromatin immunoprecipitation sequencing
Cross-linking ChIP in QKO was done using 2 × 107 cells per immunoprecipitation. Cells were directly cross-linked (1% paraformaldehyde for 10 min at RT with gentle shaking) on a plate after medium removal and PBS wash, and reactions were quenched with glycine (added to 125 mM for 5 min at RT). Cells were washed with cold PBS containing protease inhibitors (Roche, #5056489001), scraped off the plates, pelleted by centrifugation (1200 rpm at 4°C), and flash-frozen.
To prepare [antibody:bead] complexes, 75 μl of Protein A Dynabeads (Invitrogen, #10002D) per ChIP sample were washed three times with PBS + 0.01% Tween 20 (collecting on a magnet at each step), and 10 μl of anti-DNMT3A (Cell Signaling Technology, #3598) was added. Binding reactions were incubated overnight (4°C under rotation), washed three times with PBS + 0.01% Tween 20 (collecting on a magnet at each step), and resuspended in 100 μl of LB3.
To obtain a soluble chromatin, extract cell pellets were thawed, resuspended in 1 ml of LB1 [50 mM Hepes (pH 7.5), 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, and 1× protease inhibitor (Roche, #5056489001)] and placed on a rotator (10 min at 4°C). Samples were collected by centrifugation (1350g for 5 min), resuspended in 1 ml of LB2 [10 mM tris-HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, and protease inhibitor], and placed on a rotator (10 min at 4°C). Samples were collected by centrifugation (1350g for 5 min), resuspended in 1 ml of LB3 [10 mM tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, 1% Triton X-100, and protease inhibitor]. To prepare chromatin, extracts were sonicated for 20 min using a Covaris M220 focused ultrasonicator (peak power, 140; duty factor, 5; cycles/burst, 200) and centrifuged at 4°C for 20 min to remove cell debris.
Aliquots of sonicated chromatin (5% each) were removed as input DNA or to confirm effective sonication (by resolving on an agarose gel and staining for DNA). The remaining material was incubated (overnight at 4°C on a rotator) with 100 μl of anti-DNMT3A–bound magnetic beads. Beads were then sequentially collected on a magnet and washed with low-salt buffer [150 mM NaCl, 0.1% SDS, 1% Triton X-100, 1 mM EDTA, and 50 mM tris-HCl (pH 8.0)], high-salt buffer [500 mM NaCl, 0.1% SDS, 1% Triton X-100, 1 mM EDTA, and 50 mM tris-HCl (pH 8.0)], LiCl buffer [150 mM LiCl, 0.5% Na-deoxycholate, 0.1% SDS, 1% NP-40, 1 mM EDTA, and 50 mM tris-HCl (pH 8.0)], and TEN buffer [1 mM EDTA, 10 mM tris-HCl (pH 8.0), and 50 mM NaCl]. After the final wash and 3 min, centrifugation beads were resuspended in elution buffer [1% SDS, 50 mM tris-HCl (pH 8.0), 10 mM EDTA and 200 mM NaCl] and incubated for 30 min at 65°C. After brief centrifugation (20,000g), the supernatant was collected, and cross-links were reversed by incubating overnight at 65°C. The eluate was then sequentially treated with ribonuclease A (Thermo Fisher Scientific; 1 hour at 37°C) and proteinase K (Roche; 1 hour at 55°C), and DNA was recovered using polymerase chain reaction (PCR) purification kit (QIAGEN).
For ChIP-seq, libraries were prepared with NEBNext Ultra II Library Prep Kit reagents as per the manufacturer’s instructions. Libraries were sequenced on an Illumina NextSeq 550 to generate 75–base pair (bp) single-end reads.
CUT&Tag
A total of 2.5 × 105 cells were collected and washed with 1 ml of wash buffer [20 mM Hepes (pH 7.5), 150 mM NaCl, and 0.5 mM spermidine (Thermo Fisher Scientific, #13274)] before being resuspended in 1 ml of wash buffer (72). Concanavalin A–coated magnetic beads (Bangs Laboratories, #BP531) were washed twice with 1 ml of binding buffer [20 mM Hepes (pH 7.5), 10 mM KCl, 1 mM MnCl2, and 1 mM CaCl2]. Ten microliters of washed magnetic beads were added to each sample of 2.5 × 105 cells and incubated with rotation at RT for 15 min. Bead-bound cells were collected on a magnetic rack and resuspended in 100 μl of antibody buffer [wash buffer + 0.05% digitonin (Sigma-Aldrich, #D141) and 2 mM EDTA] and incubated with a primary antibody at 4°C overnight on nutator. After washing (with wash buffer + 0.05% digitonin), bead-bound cells were incubated with 1 μl of anti-rabbit immunoglobulin G (IgG) secondary (Antibodies-online, #ABIN101961) and 2 μl of pA-Tn5 adapter complex in 100 μl of Dig-300 buffer (wash buffer + 300 mM NaCl and 0.01% digitonin) at RT for 1 hour on nutator. Cells were washed three times with 1 ml Dig-300 buffer, resuspended in 250 μl Tagmentation buffer (Dig-300 buffer +10 mM MgCl2), and incubated at 37°C for 1 hour. To each sample, 10 μl of 0.5 M EDTA, 3 μl of 10% SDS, and 5 μl of proteinase K (10 mg/ml; Roche, #31158) were added and incubated at 50°C for 1 hour. DNA was purified using PCR purification kit (QIAGEN) and eluted with 25 μl of double-distilled H2O.
For library amplification, 2 μl of i5 unique index primer (10 μM), 2 μl of i7 unique index primer (10 μM), and 25 μl of NEBNext High-Fidelity 2× PCR Master Mix [New England Biolabs (NEB)] were added to 21 μl of purified CUT&Tag DNA, and the mix was subjected to PCR (72°C for 5 min; 98°C for 30 s; 12 cycles of 98°C for 10 s and 63°C for 10 s; 72°C for 1 min; and hold at 10°C). To purify PCR products, 1.1× volumes (55 μl) of Ampure XP beads (Beckman Coulter, #A63880) were added and incubated for 10 min at RT. Libraries were washed twice with 80% ethanol and eluted in 20 μl of 10 mM tris-HCl (pH 8). Libraries were sequenced on an Illumina NextSeq 550 to generate 75-bp paired-end reads.
CUT&RUN
CUTANA CUT&RUN was performed with 10T and QKO samples on an automated protocol (autoCUT&RUN) derived from those previously described (73–75). Briefly, for each CUT&RUN reaction, 500,000 cells (from 5 million cells/ml prepared in FBS with 10% dimethyl sulfoxide) were dispensed to individual wells of a 96-well plate, immobilized onto concanavalin A beads (EpiCypher, #21-1401), and incubated overnight (4°C) with 0.5 μg of antibody (IgG, H3K4me3, H3K27me3, H3K36me2, and H2AK119Ub1) [all antibodies validated to histone PTM-defined sample normalization and antibody profiling (SNAP)-ChIP nucleosome standards as previously (76)]. pAG-MNase (EpiCypher, #15-1016) was added and activated (2 hours at 4°C), and CUT&RUN-enriched DNA was purified using Serapure beads after mixing at 2:1 (bead:DNA) ratio. Recovered DNA was quantified using PicoGreen and reactions normalized to 5 ng of DNA (or entirety of the reaction if <5 ng of DNA was recovered) before preparing sequencing libraries (CUTANA CUT&RUN Library Prep kit; EpiCypher, #14-1001). All autoCUT&RUN steps were optimized and performed on Tecan Freedom EVO robotics platforms with gentle rocking for incubation steps and magnetic capture for medium exchange and washing steps. Sequencing was performed on an Illumina NextSeq2000 to generate 2× 50-bp paired-end reads.
IF staining
10T cells were seeded on eight-well chamber slides and, after a PBS wash, fixed with paraformaldehyde (4% for 15 min), and permeabilized with Triton X-100 (0.5% for 15 min). After a PBS wash, slides were blocked with 3% bovine serum albumin (BSA) on rotator for 30 min, washed with PBS, and incubated with anti-Flag (Sigma-Aldrich, #F1804) or anti-HA antibody (BioLegend, #901501) overnight at 4°C (1:200 dilution in 3% BSA). Samples were washed three times with PBS before incubating with Alexa Fluor anti-mouse IgG secondary (Thermo Fisher Scientific) for 1 hour (1:1000 in 3% BSA). Samples were then counterstained with 4′,6-diamidino-2-phenylindole (Thermo Fisher Scientific) solution in PBS for 15 min and washed three times with PBS. Slides were inverted onto gel mount on microscope slides, viewed, and photographed with an Olympus BX43 microscope.
Immunolabeling with FISH
Cells were grown on glass #1.5 coverslips coated with poly-d-lysine in 24-well plates. For imaging, cells were washed with PBS and fixed with 4% formaldehyde solution (10 min at RT). After washing twice with PBS, cells were permeabilized with 0.1% Triton X-100 in PBS (5 min at RT) and immunolabeled with anti-HA (BioLegend, #901501; 1:500 in PBS) for 1 hour at RT. After three washes with PBS, cells were incubated with secondary antibody (1 hour at RT) and again washed three times with PBS. Cells were refixed with 4% formaldehyde for 10 min and then hybridized with Stellaris RNA FISH probe for mouse Xist with Quasar 570 dye (BioSearch Tech, #SMF-3011-1). First, cells were washed with PBS and incubated with Stellaris wash buffer A (#SMF-WA1-60) for 5 min. Coverslips were then transferred to a dark, humidified a container, and incubated (37°C for 4 hours) with Stellaris FISH hybridization buffer (#SMF-HB1-10) containing the probe (1:100). Next, cells were washed with wash buffer A (30 min at 37°C), counterstained with 4′,6-diamidino-2-phenylindole in wash buffer A (30 min at 37°C), and then incubated with Stellaris wash buffer B (#SMF-WB1-20; 5 min at RT) before being mounted onto a slide. Images were taken and deconvoluted using a DeltaVision Restoration Microscopy system (GE Healthcare) with an Inverted Olympus IX-70 microscope using a 40× objective.
RNA sequencing
Total RNA was isolated using TRIzol and precipitated with isopropanol. Total RNA was quality-checked using Bioanalyzer [RNA integrity number (RIN) > 9], and libraries were generated using NEBNext Ultra II Library Prep Kit (NEB, #E7770) as per the manufacturer’s protocol. Libraries were sequenced on an Illumina NextSeq 550 to generate 75-bp single-end reads.
Organoid culture isolation and propagation
Murine organoids were generated as previously (49). Briefly, K5CreERT2 transgenic mice were crossed to R26tdTomatolsl/lsl and Trp53loxP/loxP (the Jackson Laboratory) to yield K5CreERT2;R26tdTomatolsl/lslt;Trp53loxP/loxP. Tamoxifen (Sigma-Aldrich; 0.25 mg/g of body weight) was administered via oral gavage to 8- to 12-week-old littermates to KO Trp53 and activate TdTomato expression. Two weeks after tamoxifen treatment, 4NQO (100 μg/ml; Sigma-Aldrich) in 2% propylene glycol (MP Biomedicals) was added to drinking water. Mice were exposed to 4NQO for 16 weeks and then followed up for 8 to 10 weeks to allow for tumorigenesis (48, 49). Mice were euthanized, and the esophagi were harvested. A portion of the esophagus was reserved for histological evaluation of disease progression. The remaining tissue was placed in dispase/PBS (5 U/μl; BD Biosciences) for 10 min. The muscle layer was dissected from the epithelial layer, and the latter was treated with 0.25% trypsin and passed through a 40-μm cell strainer (BD Biosciences) to generate a single-cell suspension. Using 24-well plates, 5000 cells were seeded per well in 50 μl of 3:1 Matrigel:organoid medium solution. After solidification, 500 μl of organoid medium (1:1 conditioned medium prepared as described (77) and advanced DMEM supplemented as in table S2) were added per well and replenished every other day. To create transgenics, EN organoids were transduced with concentrated lentivirus to introduce DNMT3A1WT, DNMT3A1W330R, or DNMT3A1W330R+R181E, placed under G418 (300 μg/ml) after 1 week, and selected for two additional weeks. Methylation analysis was done after at least 21 days in culture. All experiments were performed under approved protocols from the Columbia University Institutional Animal Care and Use Committee.
Western blot
Whole cell lysates were made in SDS lysis buffer (Thermo Fisher Scientific) and resolved on 3 to 8% or 4 to 12% gradient SDS–polyacrylamide gel electrophoresis (PAGE) gels (Invitrogen, NuPAGE). After transferring to a polyvinylidene difluoride membrane, same were blocked with 5% nonfat milk in PBST (PBS with 0.5% Tween 20) for 1 hour. Primary antibodies (1:1000) were added, incubated (overnight at 4°C), washed in PBST, and detected with horseradish peroxidase–conjugated anti-rabbit IgG (1:10,000 in PBST). The following antibodies were used: DNMT3A (Cell Signaling Technology, #3598), DNMT3L (Cell Signaling Technology#13451), β-actin (Abcam, #ab8227), and total H3 (Abcam, #ab1791).
Reduced representation bisulfite sequencing
Genomic DNA (1 μg) was collected from cells and sent to CD Genomics for RRBS sample preparation, library preparation, and sequencing (paired-end 150 bp).
Enzymatic methyl sequencing
EM-seq libraries were prepared from genomic DNA as previously (78) using the NEB EM-seq kit (NEB, #P7120L). Briefly, genomic DNA was purified using the NEB Monarch Genomic DNA Extraction kit (#T3010L), and 10 ng was sheared to an average fragment size of ~300 bp (Diagenode Bioruptor Pico, #B01060010) for enzymatic conversion or library preparation. Before sonication, fully 5mC-methylated pUC19 (0.01 ng) and fully unmethylated lambda DNA (0.02 ng) were spiked into each reaction to monitor conversion efficiency. After 9 cycles of PCR, library quality and quantification were assessed on the Agilent Tapestation 4200 using high-sensitivity D1000 reagents (#5067-5584 and #5067-5585). Libraries were sequenced (100 M reads per biological replicate) on an Illumina NovaSeq 6000 to generate 100-bp paired-end reads.
Fully defined nucleosomes
Fully defined (by mutation or PTM status) nucleosomes (EpiCypher) were assembled by salt-dialysis of semisynthetic histones with 5′ biotinylated DNA (147 bp of 601-nucleosome positioning sequence) (79) as previously (73–80) and confirmed by mass spectrometry and SDS-PAGE or immunoblotting (if an antibody was available). All Kub1 histones contain a native gamma-lysine isopeptide linkage (81, 82).
Nucleosome binding assay: dCypher Luminex
Biotinylated nucleosomes (unmodified, Kub1, and acidic patch) were individually coupled to distinct magnetic avidin–coated xMAP bead regions (Luminex) at saturation and washed to remove excess. Individual [bead:Nuc] were adjusted to 1 million beads/ml, equimolar-multiplexed, transferred to storage buffer [10 mM cacodylate (pH 7.5), 0.01% BSA, 0.01% Tween 20, 1 mM EDTA, 10 mM β-mercaptoethanol, and 50% glycerol], and kept at −20°C. Panel balance and individual [Nuc:bead] identity were confirmed in Luminex assays with anti-dsDNA (EMD Millipore, #MAB030; 1:5, 1:50, and 1:500), anti-H3.1/2 (Active Motif, #61629; 1:250, 1:1000, and 1:4000), and anti-PTM (EMD Millipore, #04-263; Cell Signaling Technology, #8240S; and Cell Signaling Technology, #5546S; all 1:250, 1:1000, and 1:4000). For the dCypher Luminex assay, multiplexed nucleosomes (1000 beads per well: the Targets) were used to examine the binding of serially diluted (twofold: 1000 to 7.8 nM final) peptides [WT or mutant 6×His-DNMT3A1 UDR (Biomatik): the Queries]. Briefly, 50 μl of multiplexed dNuc panel was combined with 50 μl of query peptide in UDR buffer [final: 20 mM tris (pH 7.5), 150 mM NaCl, 0.01% BSA, 0.01% NP-40, salmon sperm DNA (0.04 μg/ml; Invitrogen, #15632-011), and 1 mM dithiothreitol (DTT)] in a black, flat-bottom 96-well plate (GreinerBio, #655900) and incubated in the dark for 1 hour with shaking (800 rpm). Beads were captured and washed twice (100 μl of UDR buffer) on a plate-based magnet, and 100 μl anti-6×His (Thermo Fisher Scientific #MA1-135; 1:1000) was added and incubated in the dark for 30 min with shaking (800 rpm). Beads were captured and washed once (100 μl of UDR buffer) on a plate-based magnet, and 100 μl of phycoerythrin goat anti-mouse IgG (BioLegend, #405307; 2 μg/ml) was added and incubated in the dark for 30 min with shaking (800 rpm). After a final wash, beads were resuspended in 100 μl of UDR buffer, and median fluorescence intensity (MFI) was read using a FlexMap3D instrument (PerkinElmer), counting a minimum of 50 events per target. Data were analyzed using GraphPad Prism V10 using a sigmoidal 4-parameter logistic equation where X is (log) concentration.
DNMT3A1 N-terminal peptide
DNMT3A1 UDR peptides (76 nucleotide oligomers: amino acids 159 to 228, N-terminal 6×His-tagged) were synthesized by Biomatik. Peptides were resuspended to 1 mg/ml in buffer containing 20 mM Hepes (pH 7.5), 0.5 mM EDTA, and 1 mM DTT.
Preparation and reconstitution of ubiquitinated nucleosomes
Plasmids containing WT histones were gifts from K. Luger, and mutant H2A-K119C was generated using the Q5 mutagenesis kit (NEB). Briefly, each histone was expressed in Escherichia coli BL21(DE3) pLysS cells (Novagen, catalog no. 69451), extracted from inclusion bodies, and sequentially purified by size exclusion and anion chromatography as previously described (83). Purified histones were freeze-dried using a Sentry lyophilizer (VirTis).
Ub plasmid (pET-His-Ub G76C) DNA was a gift from T. Yao. It was transformed into E. coli SoluBL21 (Amsbio, catalog no. C700200) competent cells and grown in 2xYT-Amp medium. Ub was expressed as soluble protein by inducing with 0.5 mM isopropyl-β-d-thiogalactopyranoside for 4 hours at 37°C upon the culture reaching optical density at 600 nm = 0.4 to 0.6. Bacteria cells were harvested and lysed (AvestinEmulsiflexC3). Ub was purified on Ni–nitrilotriacetic acid agarose beads (QIAGEN), lysis buffer [300 mM NaCl, 50 mM tris (pH 8.0), 10 mM imidazole, 5 mM beta-mercaptoethanol (BME), and 1× protease inhibitor], and elution buffer [300 mM NaCl, 50 mM tris (pH 8.0), 300 mM imidazole, and 5 mM BME], followed by HiTrap Q HP (GE Healthcare) liquid chromatography column with buffer A [50 mM NaCl, 20 mM tris (pH 8.0), 0.2 mM EDTA, and 10 mM BME] and buffer B [1 M NaCl, 20 mM tris (pH 8.0), 0.2 mM EDTA, and 10 mM BME]. Purified Ub was then dialyzed against water supplemented with 1 mM acetic acid, followed by flash-freezing in liquid nitrogen and lyophilized using a Sentry lyophilizer (VirTis).
Ubiquitinated H2A was generated following an established protocol (84). Briefly lyophilized Ub (G76C) and histone H2AK119C were resuspended in resuspension buffer (10 mM acetic acid and 7 M urea-deionized) and mixed in the ratio of 2:1. Sodium tetraborate, urea, and (tris(2-carboxyethyl)phosphine) (TCEP) were added to final concentrations of 50 mM, 6 M, and 5 mM, respectively. The mixture was incubated at RT for 30 min. Then, an amount of cross-linker (1,3-dichloroacetone, Sigma-Aldrich, catalog no. 167479) diluted in N,N′-dimethylformamide (Sigma-Aldrich, catalog no. D4551) equal to one-half molar ratio of total sulfhydryl groups was added to the solution and incubated on ice for 30 min. The reaction was stopped by the addition of BME to a final concentration of 5 mM. Then, the solution was diluted 10 times with denaturing binding buffer [50 mM sodium phosphate, 50 mM tris (pH 8.0), 300 mM NaCl, 6 M urea, 10 mM imidazole, and 5 mM BME] and purified through Ni–nitrilotriacetic acid agarose beads (QIAGEN) [denaturing elution buffer: 50 mM sodium phosphate, 50 mM tris (pH 8.0), 300 mM NaCl, 6 M urea, 250 mM imidazole, and 5 mM BME]. Purified ubiquitinated H2AK119C was dialyzed into water supplemented with 1 mM BME and lyophilized using Sentry lyophilizer (VirTis).
Nucleosomal DNA was generated from a plasmid containing eight copies of the Widom 601 positioning sequence, each flanked by an Eco RV site. This was transformed to E. coli DH5α (Thermo Fisher Scientific) competent cells and grown in 2xYT-Amp medium overnight. Cells were lysed, and plasmid DNA was purified using an established protocol (83). The 601 DNA fragment was excised using Eco RV (NEB), and fragments were further separated from the vector backbone using repeated steps of polyethylene glycol precipitation.
Nucleosome reconstitutions from individual histones were as described (83, 85). Briefly, equimolar amounts of H3, H4, H2B, and ubiquinated H2A were mixed and dialyzed into refolding buffer [2 M NaCl, 10 mM tris-HCl (pH 7.5), 1 mM EDTA, and 5 mM DTT]. The octamer in refolding buffer was purified by size exclusion chromatography on a Superdex 200 16/600 column (GE Healthcare). Nucleosomes were assembled by incubating purified Widom 601 DNA and histone octamers overnight with gradient salt dialysis using a peristaltic pump (Gilson Rapid Pump). Reconstitution was completed by dialyzing into Hepes buffer [20 mM Hepes (pH 7.5), 0.5 mM EDTA, and 2 mM DTT], and reconstitution efficiency was analyzed by 4.5% PAGE and quantified by DNA content.
Cryo-EM sample preparation
A 5:1 complex between DNMT3A1 N-terminal peptide (25 μM) and H2A-Ub nucleosome (5 μM) was assembled in cross-linking buffer [50 mM Hepes (pH 7.9), 20 mM NaCl, 1 mM EDTA, and 1 mM DTT]. The complex was incubated for 2 hours at 4°C. The cross-linking reaction was performed by adding an equal volume of cross-linking buffer containing 0.06% glutaraldehyde and incubated for 1 hour at 4°C. The cross-linking reaction was quenched with 100 mM tris (pH 7.9), dialyzed into buffer containing 20 mM tris (pH 7.9), 100 mM NaCl, and 2 mM DTT, and concentrated for grid freezing. Cryo-EM grids of the peptide-nucleosome complex were prepared following the established protocol (86). Briefly, 3.0 μl of the samples with a concentration of 0.6 mg/ml was applied to Quantifoil gold grids (300 mesh, 1.2-μm hole size) glow-discharged for 25 s. The grid was blotted for 3 s with a blot force of 3 using Vitrobot Mark IV (FEI Company) at 4°C and 100% humidity and plunge-frozen in liquid ethane.
Cryo-EM data acquisition and processing
DNMT3A1 N-terminal peptide and H2A-Ub nucleosome complex data were collected on an FEI Titan Krios 300 kV equipped with a Gatan K3 Summit camera at a nominal magnification of ×105,000 and a calibrated pixel size of 0.852 Å. Super-resolution images (0.426 Å per pixel) were collected as movie stacks for 2.4 s, fractionated into 48 subframes, and an accumulated dose of 51.32 e−/Å2. A total of 3078 images were collected at a defocus range of 1.3 to 2.0 mm. The movies were motion-corrected, dose-weighed, and binned to 0.852 Å per pixel using cryoSPARC’s “Patch Motion Correction” function (87). The contrast transfer function (CTF) was calculated using “Patch CTF Estimation (multi).” Particles were picked using cryoSPARC’s “Blob Picker” with a diameter between 80 and 120 Å, extracted from the micrographs in a 256 by 256 box, and Fourier-cropped to box 96 by 96 for the initial processing steps. Particles were subjected to two-dimensional (2D) classification into 50 classes, and the best particles were used to generate a model using the “Ab initio” function, classified using “Heterogeneous Refinement,” and subsequently classified locally in 3D using “3D Classification” with a wide mask focused on terminal DNA and Ub. The resultant map from these initial steps reflecting the best ubiquitinated-nucleosome features was refined, and its particles were re-extracted in a 256 by 256 box. These particles and the corresponding map were further locally classified with a focus on Ub and refined to obtain the final reported map resolved at 2.88 Å from 55,652 particles.
Model building
The cryo-EM map of the DNMT3A1 N-terminal peptide and H2A-Ub nucleosome complex allowed for the unambiguous fitting of DNMT3A1 peptide, histones proteins, and DNA. Available x-ray structure of nucleosome from Protein Data Bank (PDB): 3TU4 was used for rigid-body fit into cryo-EM reconstructions for the nucleosome in Chimera (88), while the peptide was built de novo manually using Coot (89). The PDB was manually fit into the density and then locally optimized using UCSF Chimera’s “Fit in map” function (88). The complete model was refined using PHENIX (phenix.real_space_refine), using first-only rigid-body setting and then secondary structure, atomic displacement parameters, rotamer, and Ramachandran restraints in 100 iterations (90). Ramachandran outliers and problematic regions were manually fixed in Coot. ChimeraX and PyMOL were used to prepare figures of the model and cryo-EM densities (89, 91, 92). Final model statistics are reported in table S3.
Bioinformatic analyses
Chromatin immunoprecipitation sequencing
Reads were aligned to mouse genome (GRCm38) using bowtie2 (v2.4.4) (93). Duplicated reads were removed by the “markdup” function from sambamba (v0.6.8) (94). DeepTools (v3.5.1) (95) was used for read depth normalization and visualization. Coverage tracks (bigwig) were generated by the bamCoverage function of DeepTools, normalizing to 1× depth (reads per genome coverage) with bin size as 50 bp and read extension as 200 bp. Heatmap and enrichment plot of DNMT3A reads over CGIs were made with DeepTools with the “computeMatrix” (reference-point, -a 5000 -b 5000) and “plotHeatmap” functions. Genomic enrichment of DNMT3A reads were visualized using IGV (Integrative Genomics Viewer).
RNA sequencing
Reads were trimmed using cutadapt (v3.6) (96) and pseudo-aligned to the mouse genome (GRCm39) using kallisto (v0.48.0, quant) (97). Transcript-level abundances were read with tximport (v.1.28.0) to get gene-level abundances. Raw counts per gene were used with DEseq2 (v.1.40) (98) to find significantly differently expressed genes between samples [|log2 fold change| (|LFC|) > 2 and q < 0.01]. For visualization with IGV, RNA-seq reads were aligned to GRCm38 using hisat2 (v2.2.1) and converted to bigwig files with DeepTools function “bamCoverage.”
CUT&Tag and CUT&RUN data analysis
CUT&Tag and CUT&RUN reads were trimmed using cutadapt (v3.6) and mapped to the mouse genome (GRCm39 for CUT&Tag and GRCm38 for CUT&RUN) using bowtie2 (parameters --local --very-sensitive-local --no-unal --no-mixed --no-discordant --phred33 -I 10 -X 700), and duplicated reads were removed by the markdup function from sambamba. Coverage tracks (bigwig) were generated by the bamCoverage function of DeepTools with bin size as 50 bp and normalizing to 1× depth (reads per genome coverage). Heatmap and enrichment plot of H3K27me3, H2AK119Ub, and H3K4me3 reads over promoters were made with DeepTools functions computeMatrix (reference-point, -a 5000 -b 5000) and plotHeatmap. Peaks of H3K27me3, H2AK119Ub, and H3K4me3 CUT&Tag data were called using SEACR (v1.3) (99) and bedtools (v2.26.0) (100). Intersect function was used to find promoters positive for each chromatin peak. Genomic enrichment of CUT&Tag and CUT&RUN signals were visualized using IGV.
DNAme analysis (RRBS and EM-seq)
Reduced representation bisulfite sequencing
Raw reads were filtered with trim galore (v0.6.6, --rrbs --paired for RRBS reads) and aligned to the mouse genome (GRCm39) using Bismark (v0.23.1) (101) with the functions “bismark” and “bismark_methylation_extractor” (--no_overlap --ignore 3 --ignore_r2 2 --ignore_3prime 2 --ignore_3prime_r2 2). The resulting bismark coverage files were analyzed using methylkit (v1.26) (102) to calculate overall DNAme over all promoters and to find significantly differently methylated promoters [>10% methylation change and false discovery rate (FDR) < 0.01, default chi-square test]. To compare methylation between RRBS samples, we restricted further analysis to CpGs that meet 10× coverage threshold across all samples and, furthermore, promoters [transcription start site (TSS) ± 500 bp] with at least five CpG meeting the 10× coverage threshold. DeepTools functions “computeMatrix” and “plotHeatmap” were used for visualization of methylation across TSS and CGIs (±5000 bp) using bismark generated bedgraph files. For visualization of promoter methylation on IGV tracks, methylKit was used to generate bedGraph files for promoters with at least five CpGs meeting the 10× coverage threshold.
Enzymatic methyl sequencing
Raw reads were filtered with trim galore (v0.6.6) and aligned to the mouse genome (GRCm39) using Bismark v0.23.1 with the functions bismark and bismark_methylation_extractor (--no_overlap --ignore 3 --ignore_r2 2 --ignore_3prime 2 --ignore_3prime_r2 2). The resulting bismark coverage files were analyzed using methylkit (v1.26) All CpGs were retained, and 10-kb bins or promoters (TSS ± 500 bp) with at least 10 CpGs were retained for analysis.
Acknowledgments
We thank colleagues for the supply of plasmids and materials (see Methods) and members of the Lu laboratory for critical reading of the manuscript. Schematic illustrations were created using BioRender.
Funding: This study was supported by National Institutes of Health (NIH) grants (R01CA266978 to C.L. and K.-J.A.; R44GM119893, R44DE029633, R44CA212733, and R44HG011875 to EpiCypher; R01DE031873 and R35GM138181 to C.L.; T32GM007739 and F30CA224971 to D.N.W; and P01CA098101 and R01AA026297 to H.N.). K.-J.A. was also supported by the Mark Foundation for Cancer Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Research in this publication used the resources of the Columbia HICCC Flow Cytometry Shared Resource, Confocal and Specialized Microscopy Shared Resource, and the Judith P. Sulzberger Columbia Genome Center, funded in part through the Office of the Director, NIH under awards S10OD020056 and NCI Cancer Center Support Grant P30CA013696.
Author contributions: Investigation: K.H.G., S.A.-A., S.L.G., V.S., I.K.P., R.L., D.N.W., J.L.M., X.X., V.U.S.K., and D.N.M. Methodology: K.H.G., S.A.-A., S.L.G., V.S., S.F., D.N.W., C.L.W., X.X., M.-C.K., K.-J.A., and C.L. Resources: S.A.-A., S.L.G., S.F., I.K.P., H.N., J.L.M., B.J.V., X.X., O.A.A., V.U.S.K., H.F.T., M.-C.K., and K.-J.A. Data curation: K.H.G., S.L.G., S.F., and B.J.V. Validation: K.H.G., S.A.-A., S.L.G., V.S., A.R.H., D.N.W., J.L.M., C.L.W., X.X., M.-C.K., and K.-J.A. Supervision: K.H.G., S.F., H.N., B.J.V., K.N., M.-C.K., K.-J.A., and C.L. Formal analysis: K.H.G., S.A.-A., S.L.G., A.R.H., I.K.P., N.L., J.L.M., V.U.S.K., D.N.M., and K.-J.A. Software: K.H.G., A.R.H., and B.J.V. Project administration: K.H.G., S.F., C.L.W., B.J.V., K.N., D.N.M., M.-C.K., K.-J.A., and C.L. Visualization: K.H.G., S.A.-A., D.N.W., S.L.G., V.S., K.-J.A., and C.L. Conceptualization: K.H.G., S.A.-A., D.N.W., M.-C.K., K.-J.A., and C.L. Writing—original draft: K.H.G., S.A.-A., S.L.G., M.-C.K., K.-J.A., and C.L. Writing—review and editing: K.H.G., S.A.-A., S.L.G., D.N.W., V.S., M.-C.K., K.-J.A., and C.L. Funding Acquisition: M.-C.K., K.-J.A., and C.L.
Competing interests: EpiCypher is a commercial developer and supplier of reagents (e.g., PTM-defined semisynthetic nucleosomes) and platforms (dCypher, CUTANA CUT&RUN) used in this study. S.L.G., J.L.M., V.U.S.K., A.R.H., O.A.A., I.K.P., H.F.T., K.N., C.L.W., D.N.M., B.J.V., and M.-C.K. are employed by (and own shares in) EpiCypher. M.-C.K. is a board member of EpiCypher. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RRBS, EM-seq, ChIP-seq, RNA-seq, CUT&RUN, and CUT &Tag data reported in this paper have been deposited at GEO. Accession numbers can be found at GSE247019. The cryo-EM density map of DNMT3A1 N-terminal peptide–H2AK119Ub nucleosome complex generated in this study has been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-42636. The resulting atomic coordinates have been deposited in the PDB with accession numbers PDB 8UW1. Any material generated in this study would be provided upon request to the corresponding authors.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 to S4
REFERENCES AND NOTES
- 1.Baylin S. B., Jones P. A., Epigenetic determinants of cancer. Cold Spring Harb. Perspect. Biol. 8, a019505 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou W., Dinh H. Q., Ramjan Z., Weisenberger D. J., Nicolet C. M., Shen H., Laird P. W., Berman B. P., DNA methylation loss in late-replicating domains is linked to mitotic cell division. Nat. Genet. 50, 591–602 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deaton A. M., Bird A., CpG islands and the regulation of transcription. Genes Dev. 25, 1010–1022 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feinberg A. P., Cui H., Ohlsson R., DNA methylation and genomic imprinting: Insights from cancer into epigenetic mechanisms. Semin. Cancer Biol. 12, 389–398 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Toyota M., Ahuja N., Ohe-Toyota M., Herman J. G., Baylin S. B., Issa J.-P. J., CpG island methylator phenotype in colorectal cancer. Proc. Natl. Acad. Sci. U.S.A. 96, 8681–8686 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Issa J.-P., CpG island methylator phenotype in cancer. Nat. Rev. Cancer 4, 988–993 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Tao Y., Kang B., Petkovich D. A., Bhandari Y. R., In J., Stein-O’Brien G., Kong X., Xie W., Zachos N., Maegawa S., Vaidya H., Brown S., Chiu Yen R.-W., Shao X., Thakor J., Lu Z., Cai Y., Zhang Y., Mallona I., Peinado M. A., Zahnow C. A., Ahuja N., Fertig E., Issa J.-P., Baylin S. B., Easwaran H., Aging-like spontaneous epigenetic silencing facilitates Wnt activation, stemness, and BrafV600E-induced tumorigenesis. Cancer Cell 35, 315–328.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esteller M., CpG island hypermethylation and tumor suppressor genes: A booming present, a brighter future. Oncogene 21, 5427–5440 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Merlo A., Herman J. G., Mao L., Lee D. J., Gabrielson E., Burger P. C., Baylin S. B., Sidransky D., 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat. Med. 1, 686–692 (1995). [DOI] [PubMed] [Google Scholar]
- 10.Dobrovic A., Simpfendorfer D., Methylation of the BRCA1 gene in sporadic breast cancer. Cancer Res. 57, 3347–3350 (1997). [PubMed] [Google Scholar]
- 11.Widschwendter M., Fiegl H., Egle D., Mueller-Holzner E., Spizzo G., Marth C., Weisenberger D. J., Campan M., Young J., Jacobs I., Laird P. W., Epigenetic stem cell signature in cancer. Nat. Genet. 39, 157–158 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Ohm J. E., McGarvey K. M., Yu X., Cheng L., Schuebel K. E., Cope L., Mohammad H. P., Chen W., Daniel V. C., Yu W., Berman D. M., Jenuwein T., Pruitt K., Sharkis S. J., Watkins D. N., Herman J. G., Baylin S. B., A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 39, 237–242 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Easwaran H., Johnstone S. E., Van Neste L., Ohm J., Mosbruger T., Wang Q., Aryee M. J., Joyce P., Ahuja N., Weisenberger D., Collisson E., Zhu J., Yegnasubramanian S., Matsui W., Baylin S. B., A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res. 22, 837–849 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlesinger Y., Straussman R., Keshet I., Farkash S., Hecht M., Zimmerman J., Eden E., Yakhini Z., Ben-Shushan E., Reubinoff B. E., Bergman Y., Simon I., Cedar H., Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 39, 232–236 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Blackledge N. P., Klose R. J., The molecular principles of gene regulation by Polycomb repressive complexes. Nat. Rev. Mol. Cell Biol. 22, 815–833 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viré E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.-M., Bollen M., Esteller M., Di Croce L., de Launoit Y., Fuks F., The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439, 871–874 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Rush M., Appanah R., Lee S., Lam L. L., Goyal P., Lorincz M. C., Targeting of EZH2 to a defined genomic site is sufficient for recruitment of Dnmt3a but not de novo DNA methylation. Epigenetics 4, 404–414 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Gujar H., Weisenberger D. J., Liang G., The roles of human DNA methyltransferases and their isoforms in shaping the epigenome. Genes 10, 172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okano M., Bell D. W., Haber D. A., Li E., DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Weisenberger D. J., Velicescu M., Cheng J. C., Gonzales F. A., Liang G., Jones P. A., Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Mol. Cancer Res. 2, 62–72 (2004). [PubMed] [Google Scholar]
- 21.Chedin F., Lieber M. R., Hsieh C.-L., The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl. Acad. Sci. U.S.A. 99, 16916–16921 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia D., Jurkowska R. Z., Zhang X., Jeltsch A., Cheng X., Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449, 248–251 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu T.-H., Liu M., Zhou X. E., Liang G., Zhao G., Xu H. E., Melcher K., Jones P. A., Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B. Nature 586, 151–155 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holz-Schietinger C., Matje D. M., Harrison M. F., Reich N. O., Oligomerization of DNMT3A controls the mechanism of de novo DNA methylation. J. Biol. Chem. 286, 41479–41488 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jurkowska R. Z., Rajavelu A., Anspach N., Urbanke C., Jankevicius G., Ragozin S., Nellen W., Jeltsch A., Oligomerization and binding of the Dnmt3a DNA methyltransferase to parallel DNA molecules: Heterochromatic localization and role of Dnmt3L. J. Biol. Chem. 286, 24200–24207 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y., Chen X., Lu C., The interplay between DNA and histone methylation: Molecular mechanisms and disease implications. EMBO Rep. 22, e51803 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinberg D. N., Papillon-Cavanagh S., Chen H., Yue Y., Chen X., Rajagopalan K. N., Horth C., McGuire J. T., Xu X., Nikbakht H., Lemiesz A. E., Marchione D. M., Marunde M. R., Meiners M. J., Cheek M. A., Keogh M.-C., Bareke E., Djedid A., Harutyunyan A. S., Jabado N., Garcia B. A., Li H., Allis C. D., Majewski J., Lu C., The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 573, 281–286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H., Hu B., Horth C., Bareke E., Rosenbaum P., Kwon S. Y., Sirois J., Weinberg D. N., Robison F. M., Garcia B. A., Lu C., Pastor W. A., Majewski J., H3K36 dimethylation shapes the epigenetic interaction landscape by directing repressive chromatin modifications in embryonic stem cells. Genome Res. 32, 825–837 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamagami N., Wu D. Y., Clemens A. W., Nettles S. A., Li A., Gabel H. W., NSD1 deposits histone H3 lysine 36 dimethylation to pattern non-CG DNA methylation in neurons. Mol. Cell 83, 1412–1428.e7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shirane K., Miura F., Ito T., Lorincz M. C., NSD1-deposited H3K36me2 directs de novo methylation in the mouse male germline and counteracts Polycomb-associated silencing. Nat. Genet. 52, 1088–1098 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Dukatz M., Holzer K., Choudalakis M., Emperle M., Lungu C., Bashtrykov P., Jeltsch A., H3K36me2/3 binding and DNA binding of the DNA methyltransferase DNMT3A PWWP domain both contribute to its chromatin interaction. J. Mol. Biol. 431, 5063–5074 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Heyn P., Logan C. V., Fluteau A., Challis R. C., Auchynnikava T., Martin C.-A., Marsh J. A., Taglini F., Kilanowski F., Parry D. A., Cormier-Daire V., Fong C.-T., Gibson K., Hwa V., Ibáñez L., Robertson S. P., Sebastiani G., Rappsilber J., Allshire R. C., Reijns M. A. M., Dauber A., Sproul D., Jackson A. P., Gain-of-function DNMT3A mutations cause microcephalic dwarfism and hypermethylation of Polycomb-regulated regions. Nat. Genet. 51, 96–105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mellid S., Coloma J., Calsina B., Monteagudo M., Roldán-Romero J. M., Santos M., Leandro-García L. J., Lanillos J., Martínez-Montes Á. M., Rodríguez-Antona C., Montero-Conde C., Martínez-López J., Ayala R., Matias-Guiu X., Robledo M., Cascón A., Novel DNMT3A germline variant in a patient with multiple paragangliomas and papillary thyroid carcinoma. Cancers 12, 3304 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Remacha L., Currás-Freixes M., Torres-Ruiz R., Schiavi F., Torres-Pérez R., Calsina B., Letón R., Comino-Méndez I., Roldán-Romero J. M., Montero-Conde C., Santos M., Pérez L. I., Pita G., Alonso M. R., Honrado E., Pedrinaci S., Crespo-Facorro B., Percesepe A., Falcioni M., Rodríguez-Perales S., Korpershoek E., Ramón-Maiques S., Opocher G., Rodríguez-Antona C., Robledo M., Cascón A., Gain-of-function mutations in DNMT3A in patients with paraganglioma. Genet. Med. 20, 1644–1651 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Kibe K., Shirane K., Ohishi H., Uemura S., Toh H., Sasaki H., The DNMT3A PWWP domain is essential for the normal DNA methylation landscape in mouse somatic cells and oocytes. PLOS Genet. 17, e1009570 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sendžikaitė G., Hanna C. W., Stewart-Morgan K. R., Ivanova E., Kelsey G., A DNMT3A PWWP mutation leads to methylation of bivalent chromatin and growth retardation in mice. Nat. Commun. 10, 1884 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinberg D. N., Rosenbaum P., Chen X., Barrows D., Horth C., Marunde M. R., Popova I. K., Gillespie Z. B., Keogh M.-C., Lu C., Majewski J., Allis C. D., Two competing mechanisms of DNMT3A recruitment regulate the dynamics of de novo DNA methylation at PRC1-targeted CpG islands. Nat. Genet. 53, 794–800 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu T., Hao D., Woo J., Huang T.-W., Guo L., Lin X., Guzman A. G., Tovy A., Rosas C., Jeong M., Zhou Y., Deneen B., Huang Y., Li W., Goodell M. A., The disordered N-terminal domain of DNMT3A recognizes H2AK119ub and is required for postnatal development. Nat. Genet. 54, 625–636 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schoeftner S., Sengupta A. K., Kubicek S., Mechtler K., Spahn L., Koseki H., Jenuwein T., Wutz A., Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 25, 3110–3122 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Napoles M., Mermoud J. E., Wakao R., Tang Y. A., Endoh M., Appanah R., Nesterova T. B., Silva J., Otte A. P., Vidal M., Koseki H., Brockdorff N., Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 7, 663–676 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Zeng Y., Ren R., Kaur G., Hardikar S., Ying Z., Babcock L., Gupta E., Zhang X., Chen T., Cheng X., The inactive Dnmt3b3 isoform preferentially enhances Dnmt3b-mediated DNA methylation. Genes Dev. 34, 1546–1558 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsumura A., Hayakawa T., Kumaki Y., Takebayashi S., Sakaue M., Matsuoka C., Shimotohno K., Ishikawa F., Li E., Ueda H. R., Nakayama J., Okano M., Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11, 805–814 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Gu T., Lin X., Cullen S. M., Luo M., Jeong M., Estecio M., Shen J., Hardikar S., Sun D., Su J., Rux D., Guzman A., Lee M., Qi L. S., Chen J.-J., Kyba M., Huang Y., Chen T., Li W., Goodell M. A., DNMT3A and TET1 cooperate to regulate promoter epigenetic landscapes in mouse embryonic stem cells. Genome Biol. 19, 88 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koh K. P., Yabuuchi A., Rao S., Huang Y., Cunniff K., Nardone J., Laiho A., Tahiliani M., Sommer C. A., Mostoslavsky G., Lahesmaa R., Orkin S. H., Rodig S. J., Daley G. Q., Rao A., Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8, 200–213 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang Q.-Q., Otto T. C., Lane M. D., Commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc. Natl. Acad. Sci. U.S.A. 101, 9607–9611 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang S., Wang L., Li S., Zhang W., Ma X., Cheng G., Yang W., Zan L., Identification of potential key genes associated with adipogenesis through integrated analysis of five mouse transcriptome datasets. Int. J. Mol. Sci. 19, 3557 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tovy A., Reyes J. M., Zhang L., Huang Y.-H., Rosas C., Daquinag A. C., Guzman A., Ramabadran R., Chen C.-W., Gu T., Gupta S., Ortinau L., Park D., Cox A. R., Rau R. E., Hartig S. M., Kolonin M. G., Goodell M. A., Constitutive loss of DNMT3A causes morbid obesity through misregulation of adipogenesis. eLife 11, e72359 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Natsuizaka M., Whelan K. A., Kagawa S., Tanaka K., Giroux V., Chandramouleeswaran P. M., Long A., Sahu V., Darling D. S., Que J., Yang Y., Katz J. P., Wileyto E. P., Basu D., Kita Y., Natsugoe S., Naganuma S., Klein-Szanto A. J., Diehl J. A., Bass A. J., Wong K.-K., Rustgi A. K., Nakagawa H., Interplay between Notch1 and Notch3 promotes EMT and tumor initiation in squamous cell carcinoma. Nat. Commun. 8, 1758 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flashner S., Martin C., Matsuura N., Shimonosono M., Tomita Y., Morimoto M., Okolo O., Yu V. X., Parikh A. S., Klein-Szanto A. J. P., Yan K., Gabre J. T., Lu C., Momen-Heravi F., Rustgi A. K., Nakagawa H., Modeling oral-esophageal squamous cell carcinoma in 3D organoids. J. Vis. Exp. 10.3791/64676, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou C., Li J., Li Q., CDKN2A methylation in esophageal cancer: A meta-analysis. Oncotarget 8, 50071–50083 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y., Ge D., Lu C., The SMART App: An interactive web application for comprehensive DNA methylation analysis and visualization. Epigenetics Chromatin 12, 71 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McBride M. J., Mashtalir N., Winter E. B., Dao H. T., Filipovski M., D’Avino A. R., Seo H.-S., Umbreit N. T., St Pierre R., Valencia A. M., Qian K., Zullow H. J., Jaffe J. D., Dhe-Paganon S., Muir T. W., Kadoch C., The nucleosome acidic patch and H2A ubiquitination underlie mSWI/SNF recruitment in synovial sarcoma. Nat. Struct. Mol. Biol. 27, 836–845 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tong Z., Ai H., Xu Z., He K., Chu G.-C., Shi Q., Deng Z., Xue Q., Sun M., Du Y., Liang L., Li J.-B., Pan M., Liu L., Synovial sarcoma X breakpoint 1 protein uses a cryptic groove to selectively recognize H2AK119Ub nucleosomes. Nat. Struct. Mol. Biol. 31, 300–310 (2024). [DOI] [PubMed] [Google Scholar]
- 54.Chen T., Ueda Y., Xie S., Li E., A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with activede novo methylation. J. Biol. Chem. 277, 38746–38754 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Bourc’his D., Xu G.-L., Lin C.-S., Bollman B., Bestor T. H., Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Zhang Z.-M., Lu R., Wang P., Yu Y., Chen D., Gao L., Liu S., Ji D., Rothbart S. B., Wang Y., Wang G. G., Song J., Structural basis for DNMT3A-mediated de novo DNA methylation. Nature 554, 387–391 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ooi S. K. T., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.-P., Allis C. D., Cheng X., Bestor T. H., DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hata K., Okano M., Lei H., Li E., Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129, 1983–1993 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Mohn F., Weber M., Rebhan M., Roloff T. C., Richter J., Stadler M. B., Bibel M., Schübeler D., Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell 30, 755–766 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Inoue A., Jiang L., Lu F., Suzuki T., Zhang Y., Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manzo M., Wirz J., Ambrosi C., Villaseñor R., Roschitzki B., Baubec T., Isoform-specific localization of DNMT3A regulates DNA methylation fidelity at bivalent CpG islands. EMBO J. 36, 3421–3434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neri F., Krepelova A., Incarnato D., Maldotti M., Parlato C., Galvagni F., Matarese F., Stunnenberg H. G., Oliviero S., Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell 155, 121–134 (2013). [DOI] [PubMed] [Google Scholar]
- 63.Noushmehr H., Weisenberger D. J., Diefes K., Phillips H. S., Pujara K., Berman B. P., Pan F., Pelloski C. E., Sulman E. P., Bhat K. P., Verhaak R. G. W., Hoadley K. A., Hayes D. N., Perou C. M., Schmidt H. K., Ding L., Wilson R. K., Van Den Berg D., Shen H., Bengtsson H., Neuvial P., Cope L. M., Buckley J., Herman J. G., Baylin S. B., Laird P. W., Aldape K.; Cancer Genome Atlas Research Network , Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17, 510–522 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turcan S., Rohle D., Goenka A., Walsh L. A., Fang F., Yilmaz E., Campos C., Fabius A. W. M., Lu C., Ward P. S., Thompson C. B., Kaufman A., Guryanova O., Levine R., Heguy A., Viale A., Morris L. G. T., Huse J. T., Mellinghoff I. K., Chan T. A., IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483, 479–483 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu C., Venneti S., Akalin A., Fang F., Ward P. S., DeMatteo R. G., Intlekofer A. M., Chen C., Ye J., Hameed M., Nafa K., Agaram N. P., Cross J. R., Khanin R., Mason C. E., Healey J. H., Lowe S. W., Schwartz G. K., Melnick A., Thompson C. B., Induction of sarcomas by mutant IDH2. Genes Dev. 27, 1986–1998 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ko M., Huang Y., Jankowska A. M., Pape U. J., Tahiliani M., Bandukwala H. S., An J., Lamperti E. D., Koh K. P., Ganetzky R., Liu X. S., Aravind L., Agarwal S., Maciejewski J. P., Rao A., Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468, 839–843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Figueroa M. E., Abdel-Wahab O., Lu C., Ward P. S., Patel J., Shih A., Li Y., Bhagwat N., Vasanthakumar A., Fernandez H. F., Tallman M. S., Sun Z., Wolniak K., Peeters J. K., Liu W., Choe S. E., Fantin V. R., Paietta E., Löwenberg B., Licht J. D., Godley L. A., Delwel R., Valk P. J. M., Thompson C. B., Levine R. L., Melnick A., Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18, 553–567 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fang M., Ou J., Hutchinson L., Green M. R., The BRAF oncoprotein functions through the transcriptional repressor MAFG to mediate the CpG island methylator phenotype. Mol. Cell 55, 904–915 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Keshet I., Schlesinger Y., Farkash S., Rand E., Hecht M., Segal E., Pikarski E., Young R. A., Niveleau A., Cedar H., Simon I., Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat. Genet. 38, 149–153 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Masalmeh R. H. A., Taglini F., Rubio-Ramon C., Musialik K. I., Higham J., Davidson-Smith H., Kafetzopoulos I., Pawlicka K. P., Finan H. M., Clark R., Wills J., Finch A. J., Murphy L., Sproul D., De novo DNA methyltransferase activity in colorectal cancer is directed towards H3K36me3 marked CpG islands. Nat. Commun. 12, 694 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brinkman E. K., Chen T., Amendola M., van Steensel B., Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168–e168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaya-Okur H. S., Wu S. J., Codomo C. A., Pledger E. S., Bryson T. D., Henikoff J. G., Ahmad K., Henikoff S., CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marunde M. R., Fuchs H. A., Burg J. M., Popova I. K., Vaidya A., Hall N. W., Weinzapfel E. N., Meiners M. J., Watson R., Gillespie Z. B., Taylor H. F., Mukhsinova L., Onuoha U. C., Howard S. A., Novitzky K., McAnarney E. T., Krajewski K., Cowles M. W., Cheek M. A., Sun Z.-W., Venters B. J., Keogh M.-C., Musselman C. A., Nucleosome conformation dictates the histone code. eLife 13, e78866 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Skene P. J., Henikoff J. G., Henikoff S., Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 13, 1006–1019 (2018). [DOI] [PubMed] [Google Scholar]
- 75.Yusufova N., Kloetgen A., Teater M., Osunsade A., Camarillo J. M., Chin C. R., Doane A. S., Venters B. J., Portillo-Ledesma S., Conway J., Phillip J. M., Elemento O., Scott D. W., Béguelin W., Licht J. D., Kelleher N. L., Staudt L. M., Skoultchi A. I., Keogh M.-C., Apostolou E., Mason C. E., Imielinski M., Schlick T., David Y., Tsirigos A., Allis C. D., Soshnev A. A., Cesarman E., Melnick A. M., Histone H1 loss drives lymphoma by disrupting 3D chromatin architecture. Nature 589, 299–305 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shah R. N., Grzybowski A. T., Cornett E. M., Johnstone A. L., Dickson B. M., Boone B. A., Cheek M. A., Cowles M. W., Maryanski D., Meiners M. J., Tiedemann R. L., Vaughan R. M., Arora N., Sun Z.-W., Rothbart S. B., Keogh M.-C., Ruthenburg A. J., Examining the roles of H3K4 methylation states with systematically characterized antibodies. Mol. Cell 72, 162–177.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miyoshi H., Stappenbeck T. S., In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 8, 2471–2482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vaisvila R., Ponnaluri V. K. C., Sun Z., Langhorst B. W., Saleh L., Guan S., Dai N., Campbell M. A., Sexton B. S., Marks K., Samaranayake M., Samuelson J. C., Church H. E., Tamanaha E., Corrêa I. R., Pradhan S., Dimalanta E. T., Evans T. C., Williams L., Davis T. B., Enzymatic methyl sequencing detects DNA methylation at single-base resolution from picograms of DNA. Genome Res. 31, 1280–1289 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lowary P. T., Widom J., New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998). [DOI] [PubMed] [Google Scholar]
- 80.Schachner L. F., Jooß K., Morgan M. A., Piunti A., Meiners M. J., Kafader J. O., Lee A. S., Iwanaszko M., Cheek M. A., Burg J. M., Howard S. A., Keogh M.-C., Shilatifard A., Kelleher N. L., Decoding the protein composition of whole nucleosomes with Nuc-MS. Nat. Methods 18, 303–308 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thomas J. F., Valencia-Sánchez M. I., Tamburri S., Gloor S. L., Rustichelli S., Godínez-López V., De Ioannes P., Lee R., Abini-Agbomson S., Gretarsson K., Burg J. M., Hickman A. R., Sun L., Gopinath S., Taylor H. F., Sun Z.-W., Ezell R. J., Vaidya A., Meiners M. J., Cheek M. A., Rice W. J., Svetlov V., Nudler E., Lu C., Keogh M.-C., Pasini D., Armache K.-J., Structural basis of histone H2A lysine 119 deubiquitination by Polycomb repressive deubiquitinase BAP1/ASXL1. Sci. Adv. 9, eadg9832 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bi X., Yang R., Feng X., Rhodes D., Liu C.-F., Semisynthetic UbH2A reveals different activities of deubiquitinases and inhibitory effects of H2A K119 ubiquitination on H3K36 methylation in mononucleosomes. Org. Biomol. Chem. 14, 835–839 (2016). [DOI] [PubMed] [Google Scholar]
- 83.P. N. Dyer, R. S. Edayathumangalam, C. L. White, Y. Bao, S. Chakravarthy, U. M. Muthurajan, K. Luger, “Reconstitution of nucleosome core particles from recombinant histones and DNA” in Methods in Enzymology (Elsevier, 2003), vol. 375, pp. 23–44. [DOI] [PubMed] [Google Scholar]
- 84.Long L., Furgason M., Yao T., Generation of nonhydrolyzable ubiquitin–histone mimics. Methods 70, 134–138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Armache K.-J., Garlick J. D., Canzio D., Narlikar G. J., Kingston R. E., Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science 334, 977–982 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li X., Mooney P., Zheng S., Booth C. R., Braunfeld M. B., Gubbens S., Agard D. A., Cheng Y., Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 88.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 89.Emsley P., Cowtan K., Coot : Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 90.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX : A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Goddard T. D., Huang C. C., Meng E. C., Pettersen E. F., Couch G. S., Morris J. H., Ferrin T. E., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.L. L. C. Schrödinger, The PyMOL Molecular Graphics System, Version 2.0 (2015).
- 93.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tarasov A., Vilella A. J., Cuppen E., Nijman I. J., Prins P., Sambamba: Fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F., Manke T., deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10 (2011). [Google Scholar]
- 97.Bray N. L., Pimentel H., Melsted P., Pachter L., Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 98.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Meers M. P., Tenenbaum D., Henikoff S., Peak calling by sparse enrichment analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin 12, 42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Krueger F., Andrews S. R., Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Akalin A., Kormaksson M., Li S., Garrett-Bakelman F. E., Figueroa M. E., Melnick A., Mason C. E., methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S4