Abstract
It is well known that modifiers play a role in ameliorating or exacerbating disease phenotypes in patients and carriers of recessively inherited disorders such as sickle cell disease and thalassemia. Here, we give an overview of the literature concerning a recently described association in carriers of SUPT5H Loss-of-Function variants with a beta-thalassemia-like phenotype including the characteristic elevated levels of HbA2. That SUPT5H acts as modifier in beta-thalassemia carriers became evident from three reported cases in whom combined heterozygosity of SUPT5H and HBB gene variants was observed to resemble a mild beta-thalassemia intermedia phenotype. The different SUPT5H variants and hematologic parameters reported are collected and reviewed to provide insight into the possible effects on hematologic expression, as well as potential disease mechanisms in carriers and patients.
Keywords: hemoglobin, SUPT5H, beta-thalassemia, hematology, modifying factor, molecular diagnosis, β-thalassemia intermedia
1. Introduction
Hemoglobinopathies are the most common monogenic disorders in the world, with an ever-increasing global disease burden each year [1,2]. The thalassemias are characterized by reduced synthesis of the globin chains of hemoglobin, specifically beta-globin chains in beta-thalassemia. As most hemoglobinopathies show recessive inheritance, carriers are usually clinically silent, although they can be identified based on hematological features including microcytic hypochromic erythrocytes and elevated levels of the minor adult hemoglobin known as HbA2. Regular hemoglobinopathy diagnostics as well as pre-marital, pre-conceptional and/or antenatal thalassemia screening programs occasionally identify individuals with microcytic hypochromic parameters and elevated HbA2 in whom no beta-thalassemia variants in the HBB gene can be identified [3,4]. In very rare cases, haplotype analysis may reveal a pattern of inheritance that does not segregate with the locus of the HBB gene located within the so-called beta-globin gene cluster (chromosome 11 p15), so-called unlinked beta-thalassemia [3]. This can be caused by other genes in the human genome involved in regulating beta-globin gene expression. Several erythroid-specific genes have been reported to carry variants causing elevation of the percentage of HbA2, such as GTF2E2, GATA1, ASH1L and KLF1 [5,6,7,8,9], although not necessarily with beta-thalassemia traits like red cell indices.
Recently, Whole Exome Sequencing (WES) identified Loss-of-Function (LoF) variants in SUPT5H (NM_001111020.3) in several members of two independent Dutch families showing microcytic hypochromia and elevated HbA2 but no beta-thalassemia variants in the HBB genes [3]. The discovery of splicing variants (including both donor- and acceptor variants) in SUPT5H in these two independent families and subsequent RNA sequencing confirmed an altered splice pattern leading to intron retention and a premature stop codon.
Additional LoF variants in SUPT5H were found in several other individuals of Dutch, French and Italian ancestry, all of whom presented hematological characteristics consistent with beta-thalassemia traits but were negative for variants in the HBB genes [3,10,11]. Three individuals from two independent Greek families expressing a non-transfusion-dependent beta-thalassemia intermedia phenotype were characterized to have co-inherited a known Mediterranean beta-thalassemia variant and an additional variant in SUPT5H. These findings indicated that SUPT5H may play a role in regulating beta-globin gene expression. Elevated HbA2 in the absence of a beta-thalassemia variant in the HBB gene is one of the key hematological findings that may suggest the presence of a variant in SUPT5H. More cases have been reported with unknown SUPT5H variants expressing beta-thalassemia traits after the first publication by Achour et al. in 2020 [3].
2. SUPT5H Gene and hSpt5 Protein: Structure and Function
Promoter proximal pausing is an important regulatory step in eukaryotic transcription catalyzed by the enzyme RNA polymerase II (RNA Pol II). Participation in the general transcription factor DSIF is required for normal RNA synthesis. Human DSIF is a heterodimer composed of two subunits, hSpt4 and hSpt5. From the N-terminus, the hSpt5 protein contains an acidic domain, a region that is homologous to the bacterial transcription factor NusG (NGN domain), which interacts with Spt4h, followed by five Kyrpides–Ouzounis–Woese (KOW) domains, two C-terminal repeat regions and another two C-terminal KOW domains (Figure 1). The two C-terminal repeat regions (CTR-1 and CTR-2) can be phosphorylated by the positive transcription elongation factor pTEFb activating RNA Pol II elongation [12]. The C-terminal region of hSpt5 includes the tandem KOW domain, designated KOW6 and 7 in Figure 1, which is near the exiting RNA and might play a role in recruiting factors for RNA capping and in 3′RNA processing [13,14]. Zuber et al. (2018) [12] demonstrated a tight domain interaction between KOW4, KOW6 and KOW7, a typical beta-barrel fold that possibly enlarges the structure of the basic KOW fold compared to other KOW domains of hSpt5. This extended structure might be of importance to present a larger binding surface for additional molecular interactions. The KOW6-7 domains were shown to interact exclusively with protein factors necessary for RNA elongation and/or processing during transcription termination.
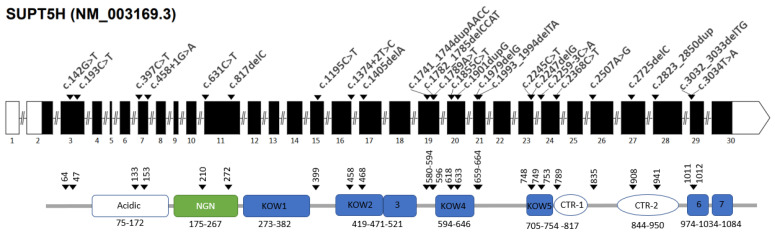
Figure 1.
Schematic presentation of the SUPT5H gene. The white boxes are the untranslated regions, and the black boxes are the coding exons. Arrows indicate the position and HGVS annotation of the variants. The protein structure is shown below from left (N-terminus) to right (C-terminus). The functional domains are the acidic region, the NGN domain which interacts with Spt4, the five KOW domains (Kyrpides–Ouzounis–Woese domains), two C-terminal repeat domains (CTR-1 and CTR-2) which can be phosphorylated and, finally, two KOW domains (KOW6 and 7). The numbers below the domains indicate the amino acid (a.a.) positions in the protein chain and the arrows correspond with the arrows in the upper graph, indicating the a.a. position of each variant.
If an SUPT5H LoF variant leads to a truncated Spt5 protein, parts of the C-terminal domains are predicted to be missing. Depending on the domains that are missing, important regions include the NGN region (critical for Spt4 dimerization), the CTR-1 and 2 regions (sensitive to phosphorylation and, therefore, activation of the DSIF complex) and the KOW6-7 domains (necessary for binding protein factors involved in RNA elongation, processing and termination). It is currently unclear whether a non-functional truncated protein product is formed or whether the transcript is degraded by Nonsense-Mediated Decay (NMD). In any case, there is no apparent correlation between the location of the premature stop position and the hematologic indices observed amongst carriers of the diverse SUPT5H variants. For example, heterozygotes for the nonsense variants c.193C>T and c.142G>T, both predicted to lead to a truncated protein missing all functional domains, present with hematologic parameters comparable to those in heterozygotes for the most terminal c.3032_3033delTG frameshift variant. The latter codes for an almost intact Spt5 protein in which only the last KOW domain and the C-terminus are missing [4,15]. These findings could support the hypothesis of NMD; however, more studies are necessary to shed light on how the reduction of Spt5 levels affects the beta-globin LCR recruitment of transcription factors to eventually reduce the expression of the HBB genes.
3. Molecular SUPT5H Variants Reported
A total of 28 SUPT5H variants have been reported to date. In the first report describing SUPT5H as a gene associated with a beta-thalassemia trait, eight different variants were observed [3]. Subsequent papers have reported an additional 20 variants. Heterozygotes may present either near-to-normal hematology or microcytic hypochromic parameters, but all were observed to have elevated HbA2 levels comparable to those in beta-thalassemia traits [4,10,11,15,16,17]. To date, there are no further reports of cases with double heterozygosity for variants in SUPT5H and HBB, and the only reason SUPT5H variants were suspected was due to elevated HbA2 levels with or without microcytic hypochromic indices in the absence of variants in the HBB gene.
The SUPT5H variants presently known are listed in Table 1 and shown in Figure 1. The majority are either nonsense or frameshift variants leading to a premature stop codon. Of four variants reported which alter the splice consensus sequence, one splice donor variant and one splice acceptor variant (c.458+1G>A and c.2259-3C>A, respectively) have been studied at the RNA level, demonstrating intron retention and an altered reading frame leading to a premature stop codon. Of the other splicing variants, two have not been examined at the RNA level, but in silico prediction software suggests an effect on splicing. The in silico prediction of two missense variants, SUPT5H:c. 2245C>T, p.(Arg749Trp) and c.2507A>G, p.(Tyr836Cys), also indicates alternative splicing, a subsequent altered reading frame and a premature stop codon. All variants, therefore, are consistent with the synthesis of a truncated protein, missing the 3′carboxy-terminus. The only exception is the variant SUPT5H: c.3034T>A, p.(Cys1012Ser), a missense variant in the penultimate exon of the gene without a clear in silico prediction in favor of alternative splicing.
Table 1.
Variants in SUPT5H (NM_001111020.3). * Delta score [0–1.0] generated by SpliceAI # GnomAD v4.0.0.
| Nr. | HGVS Annotation | Molecular Effect | In Silico Prediction * | Description | Etnic Origin | GnomAD Allele Frequency # | Reference |
|---|---|---|---|---|---|---|---|
| 1 | c.142G>T | p.Glu48* | nonsense | Chinese | [4] | ||
| 2 | c.193C>T | p.Arg65* | nonsense | Chinese | 7.15 × 10−7 | [15] | |
| 3 | c.397C>T | p.Arg133* | nonsense | Chinese | 3.05 × 10−6 | [17] | |
| 4 | c.458+1G>A | 1.00 | Donor loss | Dutch | [3] | ||
| 5 | c.631C>T | p.Gln211* | nonsense | Chinese | [4] | ||
| 6 | c.817delC | p.Leu273* | nonsense | French | [3] | ||
| 7 | c.1195C>T | p.Gln399* | nonsense | Chinese | [16] | ||
| 8 | c.1374+2T>C | 0.71 | Donor loss | Greek | [3] | ||
| 9 | c.1405delA | p.Arg469Glufs*10 | Frameshift | Chinese | [4] | ||
| 10 | c.1741_1744dupAACC | p.Arg582Glnfs*21 | Frameshift | Greek | [3] | ||
| 11 | c.1782_1785delCCAT | p.Ile594Metfs*2 | Frameshift | Chinese? | [3] | ||
| 12 | c.1789A>T | p.Leu597* | nonsense | ? | [11] | ||
| 13 | c.1855C>T | p.Arg619* | nonsense | ? | [4] | ||
| 14 | c.1901dupG | p.Met635Hisfs*19 | Frameshift | Chinese | [4] | ||
| 15 | c.1979delG | p.Gly660Valfs*6 | Frameshift | Dutch | [3] | ||
| 16 | c.1993_1994delTA | p.Met665Glufs*19 | Frameshift | ? | [11] | ||
| 17 | c.2245C>T | p.Arg749Trp | 0.69 | Donor gain | ? | 2.40 × 10−6 | [11] |
| 18 | c.2247delG | p.Leu750Serfs*11 | Frameshift | ? | [11] | ||
| 19 | c.2259-3C>A | 0.09 | - | Dutch | [3] | ||
| 20 | c.2368C>T | p.Gln790* | nonsense | French | [10] | ||
| 21 | c.2507A>G | p.Tyr836Cys | Alternative splicing | ? | [11] | ||
| 22 | c.2725delC | p.Gln909Argfs*45 | Frameshift | Dutch | [3] | ||
| 23 | c.2823_2850dup | p.Ser951* | nonsense | Chinese | [4] | ||
| 24 | c.3032_3033delTG | p.Met1011Metfs*9 | Frameshift | Chinese | [17] | ||
| 25 | c.3034T>A | p.Cys1012Ser | 0.02 | - | ? | [11] | |
| 26 | EXON 21 -2 A>G | Splice acceptor | ? | [11] | |||
| 27 | p.Glu455Aspfs*23 | Frameshift | ? | [11] | |||
| 28 | del(chr19:39936531-40030719) | ? | Chinese | [4] |
As almost the entire spectrum of SUPT5H variants in individuals expressing an unlinked beta-thalassemia phenotype from different geographic areas lead to an LoF, this suggests that an intact C-terminus is essential for the correct function of the Spt5 protein. The haplo-insufficiency of Spt5, or a truncated Spt5 which misses the carboxy-terminus, is apparently the mechanism which gives rise to the typical phenotype of beta-thalassemia traits with the characteristically elevated levels of HbA2.
4. In Vitro Studies of SUPT5H LoF
The SUPT5H gene codes for human Spt5, a protein known to play a role in controlling the release of promoter-proximal pausing of RNA Pol II, which is crucial for gene regulation and cellular differentiation. That RNA pol II pausing plays a role in modulating hematopoietic stem cell emergence was already shown previously in zebrafish [18,19]. During erythropoiesis, cell cycle genes are largely paused as cells transition from progenitors to precursors. RNA Pol II pauses ~30 to 50 bp downstream of promoters and its release into productive elongation is a highly regulated process [20,21,22,23]. One of the factors involved in the convergence from paused to released RNA Pol II is the DRB (5,6,-dichloro-1-beta-D-ribofuranosylbenzimidazole) sensitivity-inducing factor (DSIF complex), composed of Spt5 and Spt4. As Spt5 is an essential and highly conserved elongation factor, depletion often leads to cell death and its role in cellular differentiation is therefore difficult to study [24,25,26,27,28]. The identification of LoF mutations in SUPT5H in patients with a beta-thalassemia trait-like phenotype in the absence of HBB mutations created a unique opportunity to study RNA Pol II pausing [11]. By perturbing SUPT5H in human hematopoietic stem and progenitor cells (HSPCs) using CRISPR-Cas9 to generate a heterozygous population of cells with a 50% reduced level of Spt5 protein levels, it was possible to study the effect on globin expression and cell differentiation.
Martell et al. (2023) [11] observed that RNA Pol II pause release, which facilitates productive elongation, was hindered in human hematopoiesis with disrupted SUPT5H expression. As these cells began transitioning from progenitors to precursors, both cell cycle kinetics and the onset of erythroid gene expression programs were delayed, although cells still underwent terminal differentiation. This suggests that RNA Pol II pausing plays a role in the regulation of cell cycle progression during erythroid differentiation as cells begin transitioning from progenitors into more mature erythroid precursors. The authors also examined the level of non-coding enhancer-derived RNAs (eRNAs) in the regulatory regions of both the beta- and alpha-globin gene clusters, as eRNA transcription is considered a reliable marker of enhancer activity [29,30]. The reduced levels of eRNA within the beta-globin Locus Control Region (beta-LCR) observed in SUPT5H-edited cells suggests that this may contribute to the reduced HBB expression in carriers of SUPT5H LoF variants. HBB expression is more susceptible than HBA expression to transcription perturbations, particularly at a later stage of erythropoiesis, when there is a higher demand for globin expression. It is not clear why HBB expression is affected more than HBA expression, but as the modes of regulation of gene expression in the beta- and alpha-globin gene clusters are different, it is possible that the effect of perturbation of RNA Pol II pausing may also be different. As Martell et al. (2023) [11] clearly state, patients harboring these mutations are relatively healthy, exhibiting subtle but consistent phenotypes characterized by mild or no anemia but clear signs of beta-/alpha-globin chain imbalance. This was confirmed by the imbalance in globin chain-synthesis results in one of the SUPT5H carriers reported by Achour et al. (2020) [3] and supports a down-regulation of HBB expression but not of HBA1/2 expression, or at least not to the same extent. The fact that HBB expression is regulated through the recruitment of Spt5 via the beta-LCR suggests that this may be an explanation for the reduced HBB expression observed in carriers of SUPT5H variants. In contrast, HBA enhancer sites were largely unaffected in SUPT5H-edited cells, with the exception of one site which was significantly up-regulated, which suggests that LoF variants can have differential effects on enhancer transcription across these different co-expressed genes [31].
5. Hematologic Phenotype of Carriers of LoF SUPT5H Variants
How do carriers of SUPT5H variants compare to carriers of known beta-thalassemia variants at the hematological level? The mean and range of hemoglobin concentration (Hb g/L), Mean Cellular Volume (MCV fL), Mean Corpuscular Concentration of Hemoglobin (MCH pg) and HbA2 values were collected from 47 SUPT5H heterozygotes reported in the literature (Table S1) and compared to the mean values and ranges for Hb, MCV, MCH and HbA2 levels for the normal [32] and beta-thalassemia carriers [1] as known from the literature. For SUPT5H/HBB double heterozygotes, the numbers were too limited to determine a mean or range; they are depicted as values in the graph (Figure 2).
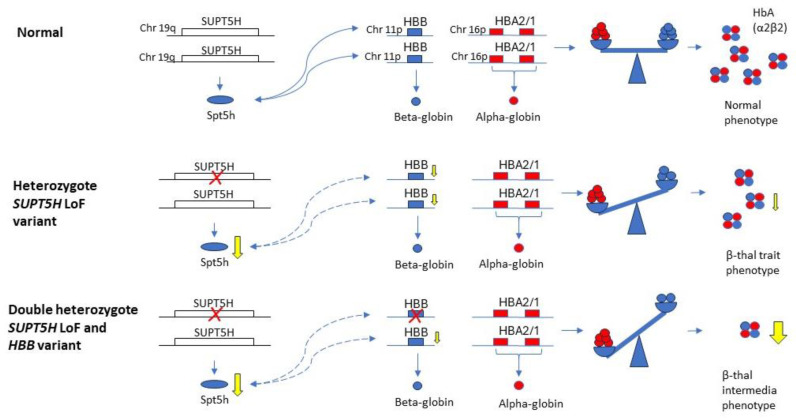
Figure 2.
Schematic presentation of the effect of reduced SUPT5H expression in carriers and in double heterozygous beta-thalassemia trait carriers. The SUPT5H gene is indicated as an open box, HBB and HBA1/2 genes as blue and red boxes respectively. The proteins are indicated below the genes as oval (Spt5h) and circles (alpha-/beta-globin). The tetramer represents hemoglobin HbA, the yellow arrows at the genes indicate the reduced levels of expression and reduction of protein synthesis, the curved lines indicate a suspected influence of reduced Spt5h synthesis on the HBB expression, for which the exact mechanism is unknown.
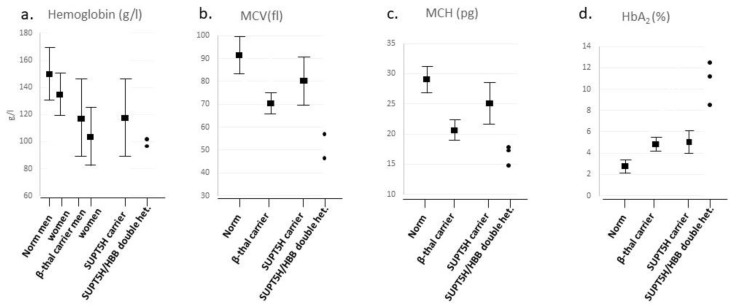
The comparison between the four groups, consisting of normal, beta-thalassemia carriers, SUPT5H carriers and double heterozygotes for SUPT5H and HBB variants, shows a decrease in Hb, MCV and MCH values between the normal and the beta-thalassemia carriers (Figure 3a–c). The comparison of hematologic indices (Hb, MCV and MCH) between the SUPT5H carriers, the normal range and the beta-thalassemia carrier range demonstrates that SUPT5H carriers show anemia due to a decrease in hemoglobin concentration equivalent to that seen in beta-thalassemia carriers. As far as MCV and MCH are concerned, the mean value for the SUPT5H carriers is between the normal and beta-thalassemia carriers, while the range overlaps between normal and beta-thalassemia carriers; this is a finding consistent with the relatively mild effect on the bi-allelic reduction in beta-gene expression of the SUPT5H carriers. Only three cases of double heterozygosity for an SUPT5H variant and a beta-thalassemia determinant in the HBB gene have been reported, showing Hb levels falling within the lower range of beta-thalassemia carriers. The MCV and MCH values seem to be below average compared to the beta-thalassemia group. Even though the number of double heterozygotes is not enough to calculate reliable statistical differences, these findings suggest that there is a modifying effect induced by SUPT5H haplo-insufficiency that down-regulates the beta-globin gene expression in beta-thalassemia heterozygotes. As the SUPT5H protein Spt5 acts transiently, it seems most likely that HBB expression on both alleles is down-regulated, enhancing the imbalance between alpha- and beta-globin polypeptide synthesis. Considering that carriers of SUPT5H variants have a hematological phenotype comparable to that of beta-thalassemia carriers, the expected phenotypic expression of double heterozygotes is more severe, which seems to be confirmed by the hematologic data.
Figure 3.
(a). Mean and 95% range of red cell indices for reference values from Dacie and Lewis, Practical Haematology 12th ed. [32], β-thalassemia carriers from Weatherall and Clegg, The Thalassemia Syndromes 4th ed. (1), SUPT5H carriers and double heterozygous carriers of SUPT5H and HBB variants (Table S1). (a). Hb (g/L) mean ± 2SD for reference group of men is 150 ± 20 g/L and that for women is 135 ± 15 g/L; Hb (g/L) mean ± 2SD for SUPT5H carriers is 119.5 g/L (91–149 g/L), which overlaps largely with the Hb of β-thalassemia carriers (men, 118 ± 15 g/L, and women, 108 ± 9.0 g/L). Mean Hb level of double heterozygote β-thalassemia and SUPT5H carriers is 98 g/L (SD = 2.89). (b). Mean MCV of reference men and women is 92 ± 9 femtoliter; the MCV of SUPT5H carriers is 79 femtoliter (69–91); the MCV of β-thalassemia carriers is 70.5 + 4.2 fl. The mean MCV of double heterozygote β-thalassemia and SUPT5H carriers is 53.3 femtoliter (SD = 6.35). (c). Mean MCH of reference is 29.5 ± 2.5 picogram, that of β-thalassemia carriers is 21.5 ± 1.3 picogram and that of SUPT5H carriers is 25.60 picogram (21.6–27.4). Mean MCH of double heterozygote β-thalassemia and SUPT5H carriers is 16.6 (SD = 1.42). (d). Mean HbA2 percentage for reference group is 2.2–3.5, mean HbA2% of SUPT5H carriers is 5.2% (3.6–6.4), and mean HbA2% of β carriers is 4.9 ± 0.5%. Mean of HbA2% for double heterozygote β-thalassemia and SUPT5H carriers is 10.67% (SD = 1.99).
No significant differences are seen in the percentage of HbA2 in beta and SUPT5H carriers (Figure 3d). However, HbA2 levels were significantly elevated in double heterozygous SUPT5H/beta-thalassemia carriers. As there are only three reported cases of combined beta-thalassemia and SUPT5H LoF variants, it is difficult to determine if this is due to the specific feature of the beta-thalassemia variant or the effect of the interaction of the two variant alleles. Two of the cases described to date are double heterozygotes for HBB:c.118C>T and SUPT5H:c.1374+2T>C (with HbA2 levels of 8.5% and 11.1%, respectively). Heterozygotes for HBB:c.118C>T usually present with relatively lower HbA2 levels of 4.89 ± 0.84%. The third case is a double heterozygote for HBB:c.92+1G>A and SUPT5H:c.1741_1744dup (HbA2 12.4%). The heterozygote HBB:c.92+1G>A presents with a higher HbA2 of approximately 9–10%. The extra-ordinary elevation in HbA2 percentage seen in two double heterozygotes, HBB:c.118C>T and SUPT5H:c.1374+2T>C, cannot be attributed to the HBB variant. In beta-thalassemia carriers expressing a more severe clinical phenotype than expected from beta-thalassemia traits, such elevated HbA2 percentages might be an indication for a co-inherited SUPT5H variant.
6. Conclusions
LoF variants in SUPT5H are associated with a beta-thalassemia-like phenotype in so-called unlinked beta-thalassemia cases with typically elevated HbA2 and, in most cases, microcytic hypochromic erythrocyte parameters. SUPT5H is a modifying factor which may play a role in beta-thalassemia carriers who express a more severe phenotype than is seen in simple beta-thalassemia heterozygotes. However, examples from the literature are apparently scarce, with only three reported cases to date.
Recent studies have demonstrated that diminished SUPT5H expression levels can induce a stage-specific delay in erythroid differentiation [11]. In spite of extensive analysis of the Spt5 mutations in human HSPCs, we do not fully understand the mechanism with which Spt5 acts on beta-globin gene expression. It is also not clear why Spt5, which seems to play such an important role in RNase II polymerase pausing and elongation, has no detectable effect on alpha-globin gene expression in these cell lines, nor gives rise to a more severe pathological phenotype in carriers with abnormal development in other tissues.
The comparison of red cell indices amongst different groups of reference samples, beta-thalassemia carriers and carriers of SUPT5H variants and compound heterozygotes provided more insight into the clinical phenotype of reduced SUPT5H expression. The elevated levels of HbA2 in the absence of a beta-thalassemia trait causing variants in the HBB gene is one of the most suggestive elements to identify variants in the SUPT5H gene. Carriers of SUPT5H LoF variants express a mild beta-thalassemia trait, while double heterozygotes for SUPT5H and a beta-thalassemia variant present with a mild beta-thalassemia intermedia phenotype. For the latter, elevated HbA2 levels of 10.7% (SD = 2.0%) potentially implicate a diagnosis of combined SUPT5H/HBB in beta-thalassemia carriers expressing beta-thalassemia intermedia. However, little is known about the clinical impact of double heterozygosity as detailed information about increased transfusion need, splenomegaly, manifestations of extramedullary hematopoiesis, etc., is missing, with only three cases reported in the literature. The clinical severity may also impact genetic counseling when couples are identified as being carriers of an SUPT5H LoF variant and a regular beta-thalassemia trait variant in HBB, respectively. As only a few cases are known from the literature, it remains difficult to predict the severity of the beta-thalassemia intermedia phenotype in an affected double heterozygous child. If both parents are carriers of SUPT5H LoF variants, 25% of pregnancies will not lead to a viable embryo as the total lack of SUPT5H expression is likely incompatible with life, with hSpt5 playing such an essential role in the biologically essential mechanism of RNA Pol II pausing and elongation.
As the modifying effect of SUPT5H LoF is relatively subtle, it might be underdiagnosed amongst beta-thalassemia carriers with a more severe clinical expression. The extraordinarily elevated HbA2 levels may be a biomarker for the presence of an additional SUPT5H variant in cases in which a single HBB variant does not explain the clinical phenotype. Next Generation Sequencing (NGS) to investigate the SUPT5H gene would be the next diagnostic step or, if not available, contacting local, more specialized clinical genetic laboratories for further testing.
More studies are necessary to investigate the mechanism underlying reduced beta-gene expression in carriers of SUPT5H variants. The hemoglobinopathy network INHERENT (URL https://www.inherentnetwork.org (accessed on 14 August 2024)) is instrumental in investigating this topic further. International collaboration between diagnostic laboratory scientists and hematologists is essential in identifying cases where the clinical severity is not fully explained by the genotype or the elevated HbA2 is not explained by variants in the HBB gene.
Acknowledgments
This article/publication is based upon work from COST Action HELIOS, CA22119, supported by COST (European Cooperation in Science and Technology).
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms25168928/s1.
Author Contributions
C.L.H., N.F.F.M.H. and A.A.: writing-original draft preparation. C.L.H., T.T.K., D.R.H., C.B. and J.T.-S.: writing-review and editing. J.L., J.t.H., S.J.G.A., H.e.I., M.V., S.B.-B., R.S., L.V., H.e.I. and C.V.: visualization. F.B.: supervision. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Weatherall D.J., Clegg J.B. The Thalassemia Syndromes. 4th ed. Wiley; Hoboken, NJ, USA: 2001. [Google Scholar]
- 2.Piel F.B. The Present and Future Global Burden of the Inherited Disorders of Hemoglobin. Hematol. Oncol. Clin. N. Am. 2016;30:327–341. doi: 10.1016/j.hoc.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Achour A., Koopmann T., Castel R., Santen G.W.E., den Hollander N., Knijnenburg J., Ruivenkamp C.A., Arkesteijn S.G., Ter Huurne J., Bisoen S., et al. A new gene associated with a beta-thalassemia phenotype: The observation of variants in, SUPT5H. Blood. 2020;136:1789–1793. doi: 10.1182/blood.2020005934. [DOI] [PubMed] [Google Scholar]
- 4.Lou J., Ye Y., Sun M., Zhao Y., Fu Y., Liu Y. A stepwise haematological screening and whole-exome sequencing reveal multiple mutations from SUPT5H causing an elevation of Hb A(2) from a cohort of 47336 individuals. Int. J. Lab. Hematol. 2023;45:90–95. doi: 10.1111/ijlh.13959. [DOI] [PubMed] [Google Scholar]
- 5.Kuschal C., Botta E., Orioli D., Digiovanna J.J., Seneca S., Keymolen K., Tamura D., Heller E., Khan S.G., Caligiuri G., et al. GTF2E2 Mutations Destabilize the General Transcription Factor Complex TFIIE in Individuals with DNA Repair-Proficient Trichothiodystrophy. Am. J. Hum. Genet. 2016;98:627–642. doi: 10.1016/j.ajhg.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ling T., Crispino J.D. GATA1 mutations in red cell disorders. IUBMB Life. 2020;72:106–118. doi: 10.1002/iub.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breton A., Theodorou A., Aktuna S., Sonzogni L., Darling D., Chan L., Menzel S., van der Spek P.J., Swagemakers S.M., Grosveld F., et al. ASH1L (a histone methyltransferase protein) is a novel candidate globin gene regulator revealed by genetic study of an English family with beta-thalassaemia unlinked to the beta-globin locus. Br. J. Haematol. 2016;175:525–530. doi: 10.1111/bjh.14256. [DOI] [PubMed] [Google Scholar]
- 8.Perseu L., Satta S., Moi P., Demartis F.R., Manunza L., Sollaino M.C., Barella S., Cao A., Galanello R. KLF1 gene mutations cause borderline HbA2. Blood. 2011;118:4454–4458. doi: 10.1182/blood-2011-04-345736. [DOI] [PubMed] [Google Scholar]
- 9.Viprakasit V., Gibbons R.J., Broughton B.C., Tolmie J.L., Brown D., Lunt P., Winter R.M., Marinoni S., Stefanini M., Brueton L., et al. Mutations in the general transcription factor TFIIH result in beta-thalassaemia in individuals with trichothiodystrophy. Hum. Mol. Genet. 2001;10:2797–2802. doi: 10.1093/hmg/10.24.2797. [DOI] [PubMed] [Google Scholar]
- 10.Charnay T., Cerino M., Gonnet K., Bonello-Palot N., Brechard M.P., Badens C. A novel SUPT5H variant associated with a beta-thalassaemia trait. Br. J. Haematol. 2022;196:e70–e71. doi: 10.1111/bjh.17985. [DOI] [PubMed] [Google Scholar]
- 11.Martell D.J., Merens H.E., Caulier A., Fiorini C., Ulirsch J.C., Ietswaart R., Choquet K., Graziadei G., Brancaleoni V., Cappellini M.D., et al. RNA polymerase II pausing temporally coordinates cell cycle progression and erythroid differentiation. Dev. Cell. 2023;58:2112–2127 e4. doi: 10.1016/j.devcel.2023.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuber P.K., Hahn L., Reinl A., Schweimer K., Knauer S.H., Gottesman M.E., Rösch P., Wöhrl B.M. Structure nucleic acid binding properties of KOW domains, 4.; 6-7 of human transcription elongation factor, DSIF. Sci. Rep. 2018;8:11660. doi: 10.1038/s41598-018-30042-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayer A., Schreieck A., Lidschreiber M., Leike K., Martin D.E., Cramer P. The spt5 C-terminal region recruits yeast 3′ RNA cleavage factor I. Mol. Cell. Biol. 2012;32:1321–1331. doi: 10.1128/MCB.06310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pei Y., Shuman S. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J. Biol. Chem. 2002;277:19639–19648. doi: 10.1074/jbc.M200015200. [DOI] [PubMed] [Google Scholar]
- 15.Xiao Z.Q., Jiang F., Li D.Z. beta-Thalassemia Trait Caused by SUPT5H Defects: Another Case Report. Hemoglobin. 2023;47:145–146. doi: 10.1080/03630269.2023.2265294. [DOI] [PubMed] [Google Scholar]
- 16.Huang X., Zhong L., Zhang R. Beta-thalassemia trait associated with a heterozygous loss-of-function variant of SUPT5H in a Southern chinese family. QJM. 2024 doi: 10.1093/qjmed/hcae112. Epub ahead of print . [DOI] [PubMed] [Google Scholar]
- 17.Lin Z., Liang X., Wei X., Liang G., Zhu D., Xie H., Yan T., Shang X. SUPT5H mutations associated with elevation of Hb A2 level: Identification of two novel variants and literature review. Gene. 2024 May 25;908:148294. doi: 10.1016/j.gene.2024.148294. [DOI] [PubMed] [Google Scholar]
- 18.Yang Q., Liu X., Zhou T., Cook J., Nguyen K., Bai X. RNA polymerase II pausing modulates hematopoietic stem cell emergence in zebrafish. Blood. 2016;128:1701–1710. doi: 10.1182/blood-2016-02-697847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taneda T., Zhu W., Cao Q., Watanabe H., Yamaguchi Y., Handa H., Wada T. Erythropoiesis is regulated by the transcription elongation factor Foggy/Spt5 through gata1 gene regulation. Genes Cells. 2011;16:231–242. doi: 10.1111/j.1365-2443.2010.01481.x. [DOI] [PubMed] [Google Scholar]
- 20.Chen F.X., Smith E.R., Shilatifard A. Born to run: Control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2018;19:464–478. doi: 10.1038/s41580-018-0010-5. [DOI] [PubMed] [Google Scholar]
- 21.Mayer A., Landry H.M., Churchman L.S. Pause & go: From the discovery of RNA polymerase pausing to its functional implications. Curr. Opin. Cell Biol. 2017;46:72–80. doi: 10.1016/j.ceb.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Core L.J., Lis J.T. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Core L., Adelman K. Promoter-proximal pausing of RNA polymerase II: A nexus of gene regulation. Genes Dev. 2019;33:960–982. doi: 10.1101/gad.325142.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tastemel M., Gogate A.A., Malladi V.S., Nguyen K., Mitchell C., Banaszynski L.A., Bai X. Transcription pausing regulates mouse embryonic stem cell differentiation. Stem Cell Res. 2017;25:250–255. doi: 10.1016/j.scr.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai X., Trowbridge J.J., Riley E., Lee J.A., DiBiase A., Kaartinen V.M., Orkin S., Zon L. TiF1-gamma plays an essential role in murine hematopoiesis and regulates transcriptional elongation of erythroid genes. Dev. Biol. 2013;373:422–430. doi: 10.1016/j.ydbio.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fitz J., Neumann T., Pavri R. Regulation of RNA polymerase II processivity by Spt5 is restricted to a narrow window during elongation. EMBO J. 2018;37:e97965. doi: 10.15252/embj.201797965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rahl P.B., Lin C.Y., Seila A.C., Flynn R.A., McCuine S., Burge C.B., Sharp P.A., Young R.A. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo S., Yamaguchi Y., Schilbach S., Wada T., Lee J., Goddard A., French D., Handa H., Rosenthal A. A regulator of transcriptional elongation controls vertebrate neuronal development. Nature. 2000;408:366–369. doi: 10.1038/35042590. [DOI] [PubMed] [Google Scholar]
- 29.Lam M.T., Li W., Rosenfeld M.G., Glass C.K. Enhancer RNAs and regulated transcriptional programs. Trends Biochem. Sci. 2014;39:170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kowalczyk M.S., Hughes J.R., Garrick D., Lynch M.D., Sharpe J.A., Sloane-Stanley J.A., McGowan S.J., De Gobbi M., Hosseini M., Vernimmen D., et al. Intragenic enhancers act as alternative promoters. Mol. Cell. 2012;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 31.Larke M.S.C., Schwessinger R., Nojima T., Telenius J., Beagrie R.A., Downes D.J., Oudelaar A.M., Truch J., Graham B., Bender M.A., et al. Enhancers predominantly regulate gene expression during differentiation via transcription initiation. Mol. Cell. 2021;81:983–997 e7. doi: 10.1016/j.molcel.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dacie J.V., Lewis S.M. Practical Haematology. 7th ed. Churchill Livingstone; Edinburgh, UK: 1991. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.