Abstract
Phyllachora (Phyllachoraceae, Phyllachorales) species are parasitic fungi with a wide global distribution, causing tar spots on plants. In this study, we describe three newly discovered species: Phyllachora chongzhouensis, Phyllachora neidongensis, and Phyllachora huiliensis from Poaceae in China. These species were characterized using morphological traits and multi-locus phylogeny based on the internal transcribed spacer region (ITS) with the intervening 5.8S rRNA gene, the large subunit of the rRNA gene (LSU), and the 18S ribosomal RNA gene (SSU). Three known species of P. chloridis, P. graminis, and P. miscanthi have also been redescribed, because, in reviewing the original references of P. chloridis, P. graminis, and P. miscanthi, these were found to be relatively old and in Chinese or abbreviated. In addition, the illustrations were simple. In molecular identification, the ITS sequence is short, while the ITS, LSU, and SSU are incomplete. Therefore, this study provides new important references for the redescription of three known species and provides further evidence for the identification of new taxa.
Keywords: three new taxa, Poaceae, Phyllachora, multigene phylogeny, taxonomy
1. Introduction
The order Phyllachorales is a group of biotrophic, plant-parasitic fungi with high host specificity and a global distribution [1]. The species of Phyllachorales have a common name, ‘tar spot fungi’ [2], because they are usually leaf- or stem-inhabiting with shiny black stromata. Initially, the families Phyllachoraceae and Phaeochoraceae were classified within this order mostly based on their morphological characteristics and host preferences [3,4]. Then, a new family, Telimenaceae, typified with Telimena erythrinae Racib., was proposed to be separated from the family Phyllachoraceae with the aid of ancestral state reconstruction [5]. Subsequently, Guterres et al. [6] presented a monogeneric new family, Neopolystigmataceae, which appeared to be well supported within Phyllachorales in both maximum likelihood (ML) and Bayesian inference (BI) phylogenetic relationship analyses.
The family Phyllachoraceae, introduced by Theissen and Sydow [7], comprises approximately 54 genera [5]. Phyllachora is the largest genus; it houses 1084 species. (https://www.speciesfungorum.org/Names/Names.asp, accessed on 10 May 2024). The majority of these genera exhibit a pronounced affinity for plants, manifesting from sub-epidermal to extensive intracellular infection within the leaf tissues, thereby instigating plant pathogenesis. In Phyllachora, sexual ascomata are more commonly observed, whereas the asexual spermogonia are less conspicuous. They are often intermixed within the ascomata, which makes them difficult to observe. The sexual morphs are distinguished by conspicuous tar spots on the surface or underside of leaf tissues, typically appearing as oval, fusiform, or irregular shapes, surrounded by yellow necrotic lesions [1]. Morphologically, the sexual morph is characterized by a globose perithecium; numerous, branched paraphyses that are slightly longer than asci; asci that are eight-spored, persistent, cylindrical to fusiform, short-pedicellate, and often have an apical ring; and ascospores that are 1–3 seriate, fusiform to narrowly oval, hyaline, and sometimes have a gelatinous sheath [3,8,9]. The asexual manifestations of Phyllachoraceae have been reported as a coelomycetous morph, demonstrating either spermatic or disseminative properties [10,11,12]. Phyllachora species predominantly infect the leaves, stems, and bracts of host plants, causing tissue necrosis or large-scale dieback through subepidermal to intracellular infections [13,14,15,16].
Poaceae is an important grassland plant resource. In a special investigation of diseases on grassland plants, we collected specimens with obvious tar spots from Pocaeae. In the present study, three newly discovered species of Phyllachora found in China are introduced, supported by morphological and phylogenetic analyses. In addition, in reviewing the original references of P. chloridis, P. graminis, and P. miscanthi, these were found to be relatively old and in Chinese or abbreviated. Plus, the illustrations were simple. In molecular identification, the ITS sequence is short, while the ITS, LSU, and SSU are incomplete. Therefore, three other known species of Phyllachora are redescribed with detailed descriptions and illustrations. Phyllachora represent potential threats to forage health, with untreated infestations posing risks to grassland management and safety. Therefore, further investigation of these Phyllachora species holds promise for improving the future safety and management of grasslands.
2. Materials and Methods
2.1. Specimen Collection and Herbarium Deposit
Diseased leaf tissues from different hosts were collected from Poaceae plants on grasslands in Chengdu and the Liangshan Yi Autonomous Prefecture, Sichuan Province, China, between August 2022 and June 2023. Each fresh sample was meticulously placed in a self-sealing bag and then transported to the laboratory for further analysis. All specimens were stored in a −20 °C ultra-low temperature refrigerator at the Herbarium of Sichuan Agricultural University, Chengdu, China (SICAU).
2.2. Morphological Studies
The ascomata were examined with a dissecting microscope NVT-GG (Shanghai Advanced Photoelectric Technology Co., Ltd., Shanghai, China) fitted with a VS-800C micro-digital camera (Shenzhen Weishen Times Technology Co., Ltd., Shenzhen, China). Further microscopic analysis of the ascomata, peridium, paraphyses, asci, and ascospores, among others, was performed using an BX53 compound microscope equipped (Olympus Corporation, Japan) with an SD1600AC digital camera in conjunction with CapStudio (version 3.8.10.0) software from Image Technology Company, Suzhou, China. Subsequently, the iodine reaction of the ascus wall was tested in Melzer’s reagent. A minimum of 20 measurements was taken for each feature using Tarosoft® Image Framework (version 0.9.7) software, developed by Tarosoft (R) in Nonthaburi, Thailand. The images were processed using Adobe Photoshop CC version 2022 software (Adobe Systems, San Jose, CA, USA). In addition, single ascospore isolations were performed according to the method described by Chomnunti et al. [17]. However, no spores had germinated.
2.3. DNA Extraction, Amplification, and Sequencing
The New Plant Genomic DNA Kit (Beijing Aidlab Biotechnologies Co., Ltd., Beijing, China) was used for total genomic DNA extraction from a single ascomata according to the manufacturer’s instructions. Amplifications of the ITS, LSU, and SSU gene fragments utilized three different primer pairs: ITS4/ITS5 for ITS [18], LROR/LR5 for LSU [19], and NS1/NS4 for SSU [18]. PCR reactions were performed according to the protocols provided by Golden Mix (Beijing TsingKe Biotech Co., Ltd., Beijing, China), including an initial denaturation at 98 °C for 2 min, followed by 30 cycles of denaturation at 98 °C for 10 s, annealing at 56 °C for 10 s, and extension at 72 °C for 10 s (for ITS and SSU) or 20 s (for LSU), with a final extension at 72 °C for 1 min. All PCR products were checked by electrophoresis in 2% agarose gels and sequenced at Hangzhou Youkang Biotech Co., Ltd., Chengdu, China, using forward and reverse primers.
2.4. Sequence Alignment and Phylogenetic Analyses
DNA sequences were aligned using Editseq 7.0.5.3 [20] to obtain consensus sequences. Sequences of Phyllachoraceae plant species for the establishment of multigene datasets were downloaded from GenBank (Table 1). The initial alignment of the sequences from individual loci was conducted using the MAFFT version 7 online service (https://mafft.cbrc.jp/alignment/server/, accession date: 1 April 2024), and then it was manually adjusted in BioEdit 7.0.5.3.
Table 1.
Samples used for multigene phylogenetic analysis a. GenBank Numbers b.
| Species | Location | Strain | Host (Family) | GenBank Accession Numbers | ||
|---|---|---|---|---|---|---|
| LSU | ITS | SSU | ||||
| Ascovaginospora stellipala T | Northern Wisconsin | P5-13A | Carex limosa (Cyperaceae) | U85088 | - | U85087 |
| Camarotella costaricensis | Panama | MM-21 | Acrocomia aculeata (Arecaceae) | KX430490 | KX451900 | KX451851 |
| Camarotella costaricensis | Panama | MM-149 | Acrocomia aculeata (Arecaceae) | KX430484 | KX451913 | KX451863 |
| Camarotella amarotella sp. | Panama | MM-27 | Arecaceae | KX430492 | KX451901 | KX451852 |
| Coccodiella calatheae T | Panama | MP5133 | Calathea crotalifera (Marantaceae) | MF460370 | MF460366 | MF460376 |
| Coccodiella melastomatum | Venezuela | CMU78543 | Miconia sp. (Melastomataceae) | - | - | U78543 |
| Coccodiella miconiae | Northern Wisconsin | ppMP1342 | Melastomataceae | KX430506 | MF460365 | KX451871 |
| Coccodiella miconiicola | Panama | TH-571 | Ossaea micrantha (Melastomataceae) | KX430512 | - | KX451880 |
| Coccodiella miconiicola | Panama | CBMAP-H290A | Miconia sp. (Melastomataceae) | MF460373 | MF460368 | MF460379 |
| Coccodiella miconiicola | Ecuador | SO-15 | Graffenrieda sp. (Melastomataceae) | MF460374 | MF460369 | MF460380 |
| Coccodiella toledoi | Ecuador | MM-165 | Melastomataceae | KX430488 | KX451917 | KX451865 |
| Neophyllachora cerradensis | Brazil | UB21823 | Myrcia torta (Myrtaceae) | - | KC683470 | - |
| Neophyllachora cerradensis T | Brazil) | UB21908 | Myrcia pinifolia (Myrtaceae) | - | KC683471 | - |
| Neophyllachora myrciae | Brazil | UB21292 | Myrcia pallens (Myrtaceae) | - | KC683463 | - |
| Neophyllachora myrciae | Brazil | UB22192 | Myrcia variabilis (Myrtaceae) | - | KC683476 | - |
| Neophyllachora myrciariae T | Brazil | UB21781 | Myrciaria delicatula (Myrtaceae) | - | KC683469 | - |
| Neophyllachora subcircinans | Brazil | UB09748 | Psidium australe (Myrtaceae) | - | KC683441 | - |
| Neophyllachora subcircinans | Brazil Paraguay | UB21347 | Psidium guineense (Myrtaceae) | - | KC683466 | - |
| Neophyllachora subcircinans | Brazil Paraguay | UB21747 | Psidium australe (Myrtaceae) | - | KC683467 | KC902622 |
| Neophyllachora truncatispora | Brazil | UB14083 | Myrcia camapuanensis (Myrtaceae) | - | KC683448 | KC902614 |
| Phyllachora arthraxonis | China: Yunnan | MHYAU: 072 | Arthraxon hispidus (Poaceae) | MG269803 | MG269749 | - |
| Phyllachora arundinellae | China: Yunnan | MHYAU: 108 | Arundinella setosa (Poaceae) | MG269815 | MG269761 | - |
| Phyllachora capillipediicola | China: Yunnan | MHYAU 20089 | Poaceae | MG356698 | KY498084 | - |
| Phyllachora capillipediicola | China: Yunnan | MHYAU: 20090 | Poaceae | MG356699 | KY498115 | - |
| Phyllachora chloridis T | Thailand | MFLU 15-0173 | Chloris sp. (Poaceae) | MF197499 | KY594026 | MF197505 |
| Phyllachora chloridis | Thailand | MFLU 16-2980 | Chloris sp. (Poaceae) | MF197500 | KY594027 | MF197506 |
| Phyllachora chloridis (P. chloridis-virgatae) | China: Yunnan | MHYAU 20136 | Chloris virgata (Poaceae) | MG356685 | KY498122 | - |
| Phyllachora chloridis (P. chloridis-virgatae) | China: Yunnan | MHYAU: 20058 | Chloris virgata (Poaceae) | MG356683 | KY498102 | - |
| Phyllachora chloridis (P. chloridis-virgatae) | China: Yunnan | MHYAU 20137 | Chloris virgata (Poaceae) | MG356686 | KY498092 | - |
| Phyllachora chloridis | China: Sichuan | SICAU 24-0053 | Chloris virgata (Poaceae) | PP785310 | PP785321 | PP785299 |
| Phyllachora chloridis | China: Sichuan | SICAU 24-0054 | Chloris virgata (Poaceae) | PP785311 | PP785322 | PP785300 |
| Phyllachora chrysopogonicola T | Thailand | MFLU 16-2096 | Chrysopogon zizanioides (Poaceae) | MF372146 | MF372145 | - |
| Phyllachora cynodonticola T | Thailand | MFLU 16-2977 | Cynodon sp. (Poaceae) | MF197501 | KY594024 | MF197507 |
| Phyllachora cynodonticola | Thailand | MFLU 16-2978 | Imperata sp. (Poaceae) | MF197502 | KY594025 | MF197508 |
| Phyllachora cynodontis | China: Yunnan | MHYAU 20042 | Cynodon dactylon (Poaceae) | KY498080 | KY471328 | - |
| Phyllachora cynodontis | China: Yunnan | MHYAU: 20043 | Cynodon dactylon (Poaceae) | KY498081 | KY471329 | - |
| Phyllachora cynodontis | China: Yunnan | MHYAU 20131 | Cynodon dactylon (Poaceae) | KY498079 | KY471327 | - |
| Phyllachora cynodontis | China: Yunnan | MHYAU 20130 | Cynodon dactylon (Poaceae) | KY498083 | KY471331 | - |
| Phyllachora chongzhouensis | China: Sichuan | SICAU 24-0044 | Phragmites australis (Poaceae) | PP785312 | PP785323 | PP785301 |
| Phyllachora chongzhouensis | China: Sichuan | SICAU 24-0045 | Phragmites australis (Poaceae) | PP785313 | PP785324 | PP785302 |
| Phyllachora dendrocalami-hamiltoniicola | China: Yunnan | MHYAU 221 | Dendrocalamus hamiltonii (Poaceae) | MK614118 | - | - |
| Phyllachora dendrocalami-membranacei | China: Yunnan | MHYAU 220 | Dendrocalamus membranaceus (Poaceae) | MK614117 | MK614102 | - |
| Phyllachora dendrocalami-membranacei | China: Yunnan | MHYAU 222 | Dendrocalamus membranaceus (Poaceae) | MK614119 | MK614103 | - |
| Phyllachora flaccidudis | China | IFRD9445 | Cenchrus flaccidus (Poaceae) | ON072101 | ON075524 | ON072097 |
| Phyllachora graminis | Germany | RoKi3084 | Arrhenatherum elatius (Poaceae) | KX430507 | - | KX451872 |
| Phyllachora graminis | Germany | NY-K-Phg2 | Poaceae | MW774239 | - | - |
| Phyllachora graminis | Germany | NY-K-Phg1 | Poaceae | MW774238 | - | - |
| Phyllachora graminis | China: Sichuan | SICAU 24-0051 | Lolium perenne (Poaceae) | PP785306 | PP785317 | PP785295 |
| Phyllachora graminis | China: Sichuan | SICAU 24-0052 | Lolium perenne (Poaceae) | PP785307 | PP785318 | PP785296 |
| Phyllachora heterocladae T | China: Sichuan | MFLU 18-1221 | Phyllostachys heteroclada (Poaceae) | MK296472 | MK305902 | MK296468 |
| Phyllachora huiliensis | China: Sichuan | SICAU 24-0048 | Bothriochloa ischaemum (Poaceae) | PP785308 | PP785319 | PP785297 |
| Phyllachora huiliensis | China: Sichuan | SICAU 24-0049 | Bothriochloa ischaemum (Poaceae) | PP785309 | PP785320 | PP785298 |
| Phyllachora imperatae | China: Yunnan | MHYAU: 014 | Imperata cylindrica (Poaceae) | MG269800 | MG269746 | - |
| Phyllachora indosasae | China: Yunnan | MHYAU 125 | Indosasa hispida (Poaceae) | MG195662 | MG195637 | - |
| Phyllachora isachnicola T | China: Yunnan | MHYAU: 179 | Isachne albens (Poaceae) | MH018563 | MH018561 | - |
| Phyllachora isachnicola | China: Yunnan | MHYAU: 180 | Isachne albens (Poaceae) | MH018564 | MH018562 | - |
| Phyllachora keralensis | China: Yunnan | MHYAU: 20082 | Poaceae | MG269792 | KY498106 | MH992447 |
| Phyllachora keralensis | China: Yunnan | MHYAU: 20083 | Poaceae | MG269793 | KY498088 | - |
| Phyllachora maydis | USA | BPI 893231 | Zea mays (Poaceae) | - | KU184459 | - |
| Phyllachora maydis | USA | BPI 910560 | Zea mays (Poaceae) | - | MG881846 | - |
| Phyllachora maydis | USA | BPI638583 | Zea mays (Poaceae) | - | OL342922 | - |
| Phyllachora maydis | USA | BPI638576 | Zea mays (Poaceae) | - | OL342921 | - |
| Phyllachora maydis | USA | BPI638565 | Zea mays (Poaceae) | - | OL342920 | - |
| Phyllachora miscanthi | China: Yunnan | MHYAU: 167 | Miscanthus sinensis (Poaceae) | MG195669 | MG195644 | - |
| Phyllachora miscanthi | China: Yunnan | MHYAU: 157 | Miscanthus sinensis (Poaceae) | MG195668 | MG195643 | - |
| Phyllachora miscanthi | China: Sichuan | SICAU 24-0050 | Miscanthus floridulus (Poaceae) | PP785305 | PP785316 | PP785294 |
| Phyllachora neidongensis | China: Sichuan | SICAU 24-0046 | Themeda triandra (Poaceae) | PP785314 | PP785325 | PP785303 |
| Phyllachora neidongensis | China: Sichuan | SICAU 24-0047 | Themeda triandra (Poaceae) | PP785315 | PP785326 | PP785304 |
| Phyllachora oplismeni-compositi | China: Yunnan | MHYAU: 170 |
Oplismenus compositus (Poaceae) | MG195673 | MG195648 | - |
| Phyllachora panicicola | China: Yunnan | MHYAU: 024 |
Panicum khasianum (Poaceae) | MG195674 | MG195649 | - |
| Phyllachora panicicola T | China | MFLU 16-2979 | Panicum sp. (Poaceae) | MF197503 | KY594028 | MF197504 |
| Phyllachora pogonatheri | China: Yunnan | MHYAU:071 | Pogonatherum paniceum (Poaceae) | MG269802 | MG269748 | - |
| Phyllachora pogonatheri | China: Yunnan | MHYAU: 070 | Pogonatherum crinitum (Poaceae) | MG269801 | MG269747 | - |
| Phyllachora qualeae | Unknown | UB 21159 | Qualea multiflora (Vochysiaceae) | - | KU682781 | - |
| Phyllachora qualeae | Unknown | UB 21771 | Qualea multiflora (Vochysiaceae) | - | KU682780 | - |
| Phyllachora sandiensis T | China: Shaanxi | IFRD9446 | Cenchrus flaccidus (Poaceae) | ON075528 | ON075525 | ON072098 |
| Phyllachora sacchari-spontanei | China: Yunnan | MHYAU: 087 | Saccharum spontaneum (Poaceae) | MG195670 | MG195645 | - |
| Phyllachora sinobambusae | China: Yunnan | MHYAU 085 | Sinobambusa tootsik (Poaceae) | MG195655 | MG195630 | - |
| Phyllachora sphaerocaryi T | China: Yunnan | MHYAU 178 | Sphaerocaryum malaccense (Poaceae) | MK614114 | MK614100 | - |
| Phyllachora sphaerocaryi | China: Yunnan | MHYAU: 178 | Sphaerocaryum malaccense (Poaceae) | - | MH018560 | - |
| Phyllachora thysanolaenae T | Thailand | MFLU 16-2071 | Thysanolaena maxima (Poaceae) | - | - | MF372147 |
| Phyllachora vulgata voucher | USA | BPI_640260 | Muhlenbergia mexicana (Poaceae) | - | OP831215 | - |
| Phyllachora xinpingensis | China: Yunnan | IFRD9465 | Chrysopogon aciculatus (Poaceae) | OP359416 | OP359398 | - |
| Phyllachora yuanjiangensis | China: Yunnan | IFRD9466 | Arundinella setosa (Poaceae) | OP359417 | OP359399 | OP359400 |
| Phyllachora yushaniae-falcatiauritae | China: Yunnan | MHYAU 123 | Yushania falcatiaurita (Poaceae) | MG195656 | MG195631 | - |
| Phyllachora yushaniae-polytrichae | China: Yunnan | MHYAU 122 | Yushania polytricha (Poaceae) | MG195657 | MG195632 | MH992455 |
| Phyllachora yushaniae-polytrichae | China: Yunnan | MHYAU 158 | Yushania polytricha (Poaceae) | MG195658 | MG195633 | - |
| Polystigma pusillum | Costa Rica | MM-113 | Andira inermis (Fabaceae) | KX430474 | KX451907 | KX451858 |
| Polystigma pusillum | Costa Rica | MM-147 | Andira inermis (Fabaceae) | KX430483 | - | KX451862 |
| Polystigma pusillum | Panama | MM-19 | Andira inermis (Fabaceae) | KX430489 | KX451899 | KX451850 |
| Telimena bicincta | Costa Rica | MM-108 | Picramnia antidesma (Picramniaceae) | KX430473 | KX451906 | KX451857 |
| Telimena bicincta | Costa Rica | MM-133 | Picramnia antidesma (Picramniaceae) | KX430478 | KX451910 | KX451861 |
a The newly generated sequences are indicated in bold. “T” marked ex-type or ex-epitype strains. b “-” means sequence unavailable.
The phylogenetic relationships among the taxa were inferred using both maximum likelihood (ML) and Bayesian inference (BI) methods within Phylosuite software, version 1.2.3. [21]. Telimena bicincta (MM-108 and MM-133) was chosen as the outgroups. Maximum likelihood phylogenies were inferred using IQ-TREE [22] with an edge-linked partition model for 10,000 ultrafast bootstraps [22]. ModelFinder [23] was employed to select the best-fit partition model (edge-linked) based on the BIC criterion. The best-fit model according to BIC was TIM2e+I+G4 for ITS, LSU, and SSU. The Bayesian inference phylogenies were inferred using MrBayes under the partition model (2 parallel runs, 2,000,000 generations), within the initial 25% of the sampled data discarded as burn-in, and the best nucleotide substitution model for each locus was identified using ModelFinder of Phylosuite [21]. The best-fit model according to AIC was SYM+FQ+G4 for ITS, GTR+F+G4 for LSU, and TN+F+G4 for SSU. The resulting trees were visualized using FigTree v.1.4.3 [21], which is available at http://tree.bio.ed.ac.uk/software/figtree (accessed on 10 April 2024), and they were further refined in Adobe Illustrator CS6 2023 (v.27.6.0).
3. Results
3.1. Phylogenetic Analysis
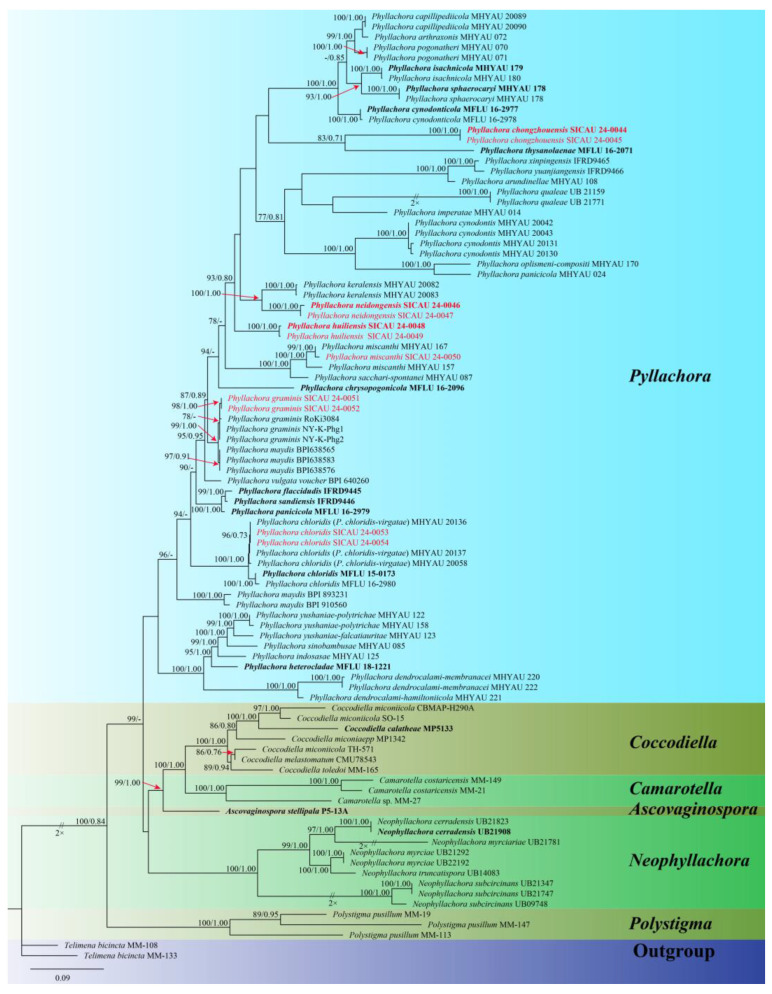
Based on the ITS, LSU, and SSU sequence data, the molecular phylogenetic relationships were analyzed using six genera (Ascovaginospora, Camarotella, Coccodiella, Neophyllachora, Phyllachora, and Polystigma) within the Phyllachoraceae. Concatenated sequences from the three genes were obtained from 80 strains of Phyllachoraceae, resulting in a dataset with 4343 characters (LSU = 1176, ITS = 1237, SSU = 1929), including gaps. The best-scoring maximum likelihood consensus tree (lnL = −35,249.614) is depicted in Figure 1.
Figure 1.
Phylogenetic tree generated from maximum likelihood analysis using the concatenated sequences of the ITS, LSU, and SSU loci of the genera in Phyllachoracea. Notes are marked with maximum likelihood bootstrap proportions ≥ 70% (left) and Bayesian inference posterior probability values ≥ 0.7 (right) (MLBP/BIPP). Some inter-section support is marked with red arrows, ex-type or ex-epitype strains are highlighted in bold, and study species are indicated in red.
The SICAU 24-0044, SICAU 24-0045, SICAU 24-0048, SICAU 24-0049, SICAU 24-0046, SICAU 24-0047 are clustered within the genus Phyllachora, and since the ML and BI phylogenetic trees exhibited similar topologies, only the ML tree (Figure 1) is presented. Three new species were identified as P. chongzhouensis, P. neidongensis, and P. huiliensis. The three-gene phylogenetic analysis (Figure 1) suggests that the collections of P. chongzhouensis (SICAU 24-0044 and SICAU 24-0045) cluster together in a distinct clade, a sister to P. thysanolaenae (MFLU 16-2071), with MLBS/BYPP values of 83%/0.71. Additionally, the species P. neidongensis (SICAU 24-0046 and SICAU 24-0047) is a sister to P. keralensis (MHYAU:20083 and MHYAU:20082), with MLBS/BYPP values of 100%/0.99. Meanwhile, P. huiliensis (SICAU 24-0048 and SICAU 24-0049) formed a separate branch, which is a sister to P. neidongensis (SICAU 24-0046 and SICAU 24-0047), with MLBS/BYPP values of 93%/0.80. In addition, SICAU 24-0050 clustered with the strains of P. miscanthi, showing 100% MLBS and 1.00 BYPP. SICAU 24-0051 and SICAU 24-0052, clustered with the strains of P. graminis, with 78% MLBS. Collections SICAU 24-0053 and SICAU 24-0054, while different, also clustered with the strains of P. chloridis, showing 96% MLBS and 0.73 BYPP.
3.2. Taxonomy
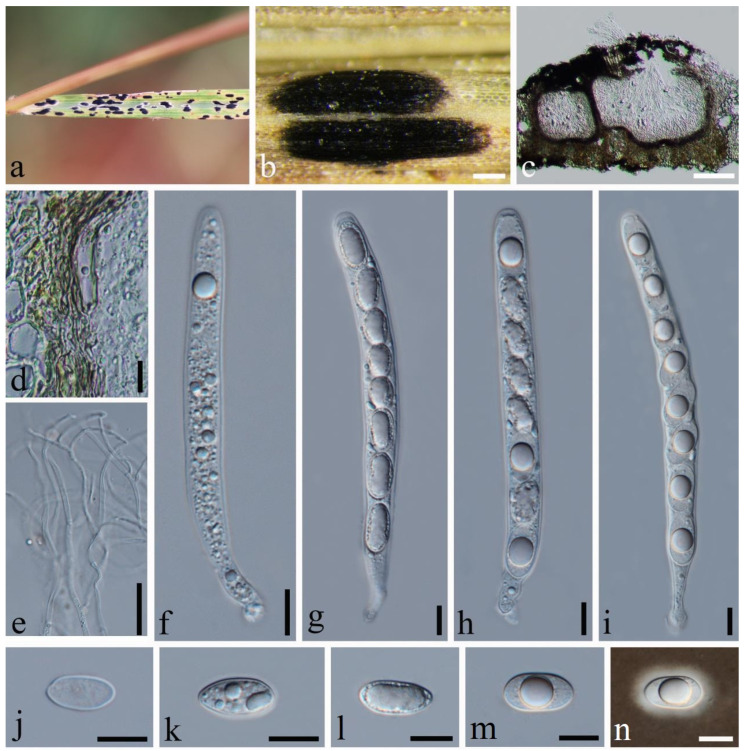
Phyllachora chloridis Dayar., R.G. Shivas & K.D. Hyde. Mycosphere, 8(10): 1606 (2017). Figure 2.
Figure 2.
Phyllachora chloridis (SICAU 24-0053). (a,b) Black spots on leaves. (c) Vertical cross-section of ascoma. (d) Peridium. (e) Paraphyses. (f–h) Asci. (i–l) Ascospores. Scale bars: 300 μm (b), 100 μm (c), 10 μm (d–h), 5 μm (i–l).
=Phyllachora chloridis-virgatae J.C. Li, H.X. Wu, Y.Y. Li, X.H. Hao, J.Y. Song, Suwannarach & Wijayawardene, Journal of Fungi 8(5): 520 (2022).
Index Fungorum number: IF552804.
Description: Parasite associated with leaves of Chloris virgata (Poaceae), causing tar spot on leaves. Tar spots: 1.4–2.7 × 0.4–0.8 mm ( = 2.0 × 0.6 mm, n = 30) on both sides of the leaf, black, carbonaceous, fusiform, or cymbiform spots, scattered and shiny. Sometimes, there is a pale-yellow-to-yellow stripe at the edge of the tar spot. Sexual morph: Ascomata: 109–264 × 202–354 μm ( = 178 × 288 μm, n = 30), embedded within the leaf tissue, occupying the entire thickness of the leaf, often developing adjacent to neighboring ascomata and constrained by host vascular tissue, suboblate to subglobose, sometimes irregular, without obvious ostioles. Peridium: 18–40 μm wide, approximately 6–10 layers, dark brown to black; the darker cells are the outer layer, and the large, slightly paler cells are the inner layer. Paraphyses: 1.6–2.7 μm wide, numerous, persistent, filiform, unbranched, septate, slightly longer than the asci. Asci: 81–120 × 8–15 μm ( = 99 × 11 μm, n = 50), eight-spored, long, cylindrical, apex obtuse to rounded, negative staining with Melzer’s reagent. Ascospores: 11–18 × 5–9 μm ( = 14 × 7 μm, n = 50), uniseriate, sometimes overlapping, and angled; the cells are hyaline, ellipsoidal, and occasionally ovoid, one-celled, with some containing one or two large fat globules at the center. Asexual morph: Not observed.
Material examined: China, Sichuan Province, Liangshan Yi Autonomous Prefecture, Huili City, Xinfa Township (26.17′39.95″ N, 102.17′4.03″ E, alt. 1879 m), on leaves of Chloris virgata Sw., 28 September 2022, Qirong Sun & Chunlin Yang, SQR202209009 (SICAU 24-0053). GenBank accession numbers: ITS = PP785321, LSU = PP785310, SSU = PP785299; ibid. SQR2022090099 (SICAU 24-0054), GenBank accession numbers: ITS = PP785311, LSU = PP785322, SSU = PP785300.
Notes: According to the phylogenetic analysis, SICAU 24-0053 and SICAU 24-0054 are closely related to Phyllachora chloridis, with strong statistical support (100% MLBS, 1.00 BYPP). P. chloridis and P. chloridis-virgatae have the same host, and there was no difference in their molecular comparison [2,24]; so, they are considered to be the same species. Specimens SICAU 24-0053 and SICAU 24-0054, compared to the P. chloridis holotype (MFLU 15-0173), showed no differences in the ITS, LSU, and SSU sequences (all 100% identical) and matched the morphological description provided by Dayarathne et al. [2]. In conclusion, the collections SICAU 24-0053 and SICAU 24-0054 were classified within P. chloridis.
Phyllachora chongzhouensis Q.R. Sun, X.L. Xu & C.L. Yang, sp. nov. Figure 3.
Figure 3.
Phyllachora chongzhouensis (SICAU 24-0044, holotype). (a,b) Black spots on leaves. (c) Vertical cross-section of ascoma. (d) Peridium. (e) Paraphyses. (f–h) Asci. (i–m) Ascospores. Scale bars: 400 μm (b), 50 μm (c), 10 μm (d–m).
Index Fungorum number: IF902115.
Etymology: Refer to the collection site, Chongzhou City, Sichuan Province, China.
Holotype: SICAU 24-0044
Description: Parasite on the leaves of Phragmites australis (Poaceae), causing tar spots on leaves. Tar spots: 1.2–2.8 × 0.3–0.7 mm ( = 1.7 × 0.5 mm, n = 30) on leaf surfaces, black, carbonaceous, elliptical to irregular shapes, often protruding above the leaf surface in a domed manner, and leaves are infested on both sides, solitary to gregarious. Sexual morph: Ascomata: 82–199 × 129–306 μm ( = 116 × 188 μm, n = 30), spindle-shaped, aligned, immersed in the leaf tissue, occupying the entire leaf thickness, and often developed next to the adjacent ascomata, confined by the host’s vascular tissue. Peridium: 16–27 μm wide, around 8–15 layers thick, from brown to black, containing host epidermal cells and sporadically encompassing the cuticle and inner layers. Paraphyses: 2.7–5.1 μm wide, numerous, persistent, filiform, unbranched, septate, slightly longer than the asci. Asci: 86–142 × 14–29 μm ( = 111 × 19 μm, n = 50), eight-spored, cylindrical, apex obtuse to rounded, pedicellate at posterior end, walls uniform in thickness, negative staining with Melzer’s reagent. Ascospores: 13–28 × 9–15 μm ( = 21 × 12 μm, n = 50), uniseriate, sometimes overlapping and oblique, hyaline; some are ellipsoidal, occasionally ovoid, one-celled, and ripe with a central depression. Asexual morph: Not observed.
Material examined: China, Sichuan Province, Chongzhou City, Jiayu Yangma Wetland Park (31°6′26.87″ N, 103°44′32.96″ E, alt. 730 m), on leaves of Phragmites australis (Cav.) Trin. ex Steud., 30 April 2024, Qirong Sun, SQR202404002 (SICAU 24-0044, holotype). GenBank accession numbers: ITS = PP785323, LSU = PP785312, SSU = PP785301; ibid SQR2024040022 (SICAU 24-0045), GenBank accession numbers: ITS = PP785324, LSU = PP785313, SSU = PP785302.
Notes: Multi-locus phylogenetic analyses utilizing a concatenated ITS, LSU, and SSU sequence dataset revealed that the new species P. chongzhouensis is related to P. thysanolaenae in a subclade with 83% MLBS and 0.71 BYPP statistical support (Figure 1). However, P. chongzhouensis (SICAU 24-0044) exhibits distinctly different morphological characteristics compared to P. thysanolaenae (MFLU 16-2071). Specifically, P. chongzhouensis features larger asci (86–142 × 14–29 μm vs. 100–126 × 13–18 μm) and wider asci walls (9–15 μm vs. 4–5 μm). Moreover, the ascospores of P. chongzhouensis are ellipsoidal with a central depression, whereas the ascospores of P. thysanolaenae (MFLU 16-2071) are cylindrical–fusiform and surrounded by a mucilaginous sheath [25]. In the comparison of the SSU sequences, there is a 7.37% nucleotide difference between P. chongzhouensis (SICAU 24-0044) and its phylogenetically affiliated P. thysanolaenae (MFLU 16-2071). Additionally, there are no available ITS and LSU sequences for comparison. These findings on the morphological features and molecular phylogenetic characteristics strongly support the proposal for establishing P. chongzhouensis as a new species, as recommended by Hyde et al. [26].
Phyllachora graminis (Pers.) Fuckel, Jahrb. Nassauischen Vereins Naturk. 23-24: 216 (1870), Figure 4.
Figure 4.
Phyllachora graminis (SICAU 24-0051). (a,b) Black spots on leaves. (c) Vertical cross-section of ascoma. (d) Peridium. (e) Paraphyses. (f–i) Asci. (j–n) Ascospores. Scale bars: 300 μm (b), 50 μm (c), 10 μm (d–n).
Index Fungorum number: IF200927.
Description: Parasite on leaves of Lolium perenne (Poaceae), causing tar spots on leaves. Tar spots: 0.9–1.9 × 0.3–0.6 mm ( = 1.3 × 0.4 mm, n = 20) on both sides of the leaf, black, carbonaceous, elliptical to irregular, solitary to gregarious, with a halo around the periphery. Sexual morph: Ascomata: 102–299 × 109–273 μm ( = 196 × 172 μm, n = 20), immersed within the leaf tissue, occupying the entire leaf thickness, often developing adjacent to neighboring ascomata and confined by host vascular tissue, ellipsoid to spherical, sometimes irregular. Peridium: 7–17 μm wide, approximately 8–10 layers, dark brown to black; the darker cells are the outer layer, and the large, slightly paler cells are the thin-walled inner layer. Paraphyses: 1.2–1.9 μm wide, numerous, persistent, filiform, unbranched, septate, slightly longer than asci. Asci: 64–101 × 6–10 μm ( = 85 × 8 μm, n = 50), eight-spored, long, cylindrical, slightly curved, apex obtuse to rounded, negative staining with Melzer’s reagent. Ascospores: 8–15 × 4–7 μm ( = 11 × 6 μm, n = 50), uniseriate, sometimes overlapping and oblique, hyaline, ellipsoidal, one-celled, some with fat globules in the center, some middle depressions. Asexual morph: Not observed.
Material examined: China, Sichuan Province, Chengdu City, Qionglai Jiguan Townships (30°17′2.13″ N 103°15′48.56″ E, alt. 551 m), on leaves of Lolium perenne Linn., 23 May 2023, Qirong Sun & Liping Gao, SQR202305038 (SICAU 24-0051). GenBank accession numbers: ITS = PP785317, LSU = PP785306, SSU = PP785295; China, Sichuan Province, Chengdu City, Jinjiang District, (30°33′53.46″ N, 104°9′10.08″ E, alt. 755 m), on leaves of Lolium perenne Linn., 5 June 2023, Qirong Sun & Liping Gao, SQR202305052 (SICAU 24-0052). GenBank accession numbers: ITS = PP785318, LSU = PP785307, SSU = PP785296.
Notes: The collections SICAU 24-0051 and SICAU 24-0052 clustered together with the known species Phyllachora graminis with a 78% ML bootstrap support value (Figure 1). Nucleotide comparisons of SICAU 24-0051 showed high homology with the sequences of P. graminis (RoKi3084). In the LSU and SSU regions, the similarities are 99.82% (549/550, 1 gap) and 100% (420/420, 0 gaps), respectively. And the same similarities were observed in SICAU 24-0052. Furthermore, morphological analysis of the new collection aligns with the description of P. graminis provided by Orton [27]. Based on comprehensive evidence, SICAU 24-0051 and SICAU 24-0052 can be classified within P. graminis.
Phyllachora huiliensis Q.R. Sun & C.L. Yang, sp. nov., Figure 5.
Figure 5.
Phyllachora huiliensis (SICAU 24-0048, holotype). (a,b) Black spots on leaves. (c) Vertical cross-section with orifices. (d) Peridium. (e) Paraphyses. (f–i) Asci. (j–m) Ascospores. Scale bars: 400 μm (b), 50 μm (c), 10 μm (d–i), 5 μm (j–m).
Index Fungorum number: IF902117.
Etymology: Refer to the collection site: Huili City, Sichuan Province, China.
Holotype: SICAU 24-0048.
Description: Parasite on leaves of Bothriochloa ischaemum (Poaceae), causing tar spots on leaves. Tar spots: 0.1–2.2 × 0.2–0.6 mm ( = 1.3 × 0.4 mm, n = 20) on the upper leaf surface, fusiform or cymbiform, amphigenous, solitary to gregarious, sometimes with orifices. Sexual morph: Ascomata: 86–175 × 132–224 μm ( = 123 × 171 μm, n = 30), immersed in the leaf tissue, occupying the entire leaf thickness, often developing next to the adjacent ascomata and confined by the host vascular tissue, suboblate to subglobose, sometimes irregular. Peridium: 17–27 μm wide, approximately 6–10 layers, dark brown; the darker cells are the outer layer, and the large, slightly paler cells are the thin-walled inner layer. Paraphyses: 1.8–3.1 μm wide, numerous, persistent, filiform, unbranched, septate, slightly longer than asci. Asci: 74–112 × 7–12 μm ( = 92 × 9 μm, n = 50), eight-spored, long, cylindrical, apex obtuse to rounded, negative staining with Melzer’s reagent. Ascospores: 11–17 × 5–8 μm ( = 13 × 6 μm, n = 50), uniseriate, occasionally overlapping, hyaline, and ellipsoidal or ovoid cells, some of which contain one or two large lipid droplets centrally. Asexual morph: Not observed.
Material examined: China, Sichuan Province, Liangshan Yi Autonomous Prefecture, Huili City, Neidong Township (26°35′5.99″ N 102°20′50.30″ E, alt. 2143 m), on leaves of Bothriochloa ischaemum (L.) Keng, 24 July 2023, Qirong Sun & Chunlin Yang, SQR202307002 (SICAU 24-0048, holotype). GenBank accession numbers: ITS = PP785319, LSU = PP785308, SSU = PP785297; ibid SQR2023070022 (SICAU 24-0049), GenBank accession numbers: ITS = PP785320, LSU = PP785309, SSU = PP785298.
Notes: In the phylogenetic tree, the collections of Phyllachora huiliensis cluster in an independent subclade within Phyllachora (Figure 1). Morphologically, P. huiliensis (SICAU 24-0048, holotype) displays distinct characteristics compared to P. neidongensis (SICAU 24-0046, holotype). P. huiliensis has smaller ascomata (86–175 × 132–224 μm vs. 145–481 × 196–480 μm) than the latter. Furthermore, the paraphyses of P. neidongensis are branched, and they have gelatinous sheaths around the ascospores, which is not observed in P. huiliensis. Nucleotide comparisons reveal notable differences between P. huiliensis (SICAU 24-0048, holotype) and P. neidongensis (SICAU 24-0046, holotype). The nucleotide differences are 19.46% (86/443, 0 gaps), 9.13% (68/745, 0 gaps), and 1.81% (17/937, 0 gaps) in the ITS, LSU, and SSU regions, respectively. Hence, we describe P. huiliensis as a new species in Phyllachora, as recommended by Maharachchimbura et al. [3].
Phyllachora miscanthi Syd. & P. Syd., Annales Mycologici 15 (3-4): 227 (1917), Figure 6.
Figure 6.
Phyllachora miscanthi (SICAU 24-0050). (a,b) Black spots on leaves. (c) Vertical cross-section of ascoma. (d) Peridium. (e) Paraphyses. (f–i) Asci. (j–m) Ascospores. Scale bars: 300 μm (b), 100 μm (c), 10 μm (d–m).
Index Fungorum number: IF165328.
Description: Parasite on leaves of Miscanthus floridulus (Poaceae), causing tar spot on leaves. Tar spots: 1.4–2.7 × 0.4–0.8 mm ( = 2.0 × 0.6 mm, n = 30) on both sides of the leaf surface, black, carbonaceous, ellipse to irregularity, amphigenous, solitary to gregarious. Sexual morph: Ascomata: 195–614 × 178–399 μm ( = 178 × 288 μm, n = 20), immersed in the leaf tissue, occupying the entire leaf thickness, often developing next to adjacent ascomata and confined by host vascular tissue, irregular in shape, and no obvious ostiolate. Peridium: 7–18 μm wide, approximately 10–13 layers, dark brown to black; the darker cells are the outer layer, and the large, slightly paler cells are the thin-walled inner layer. Paraphyses: 1.7–2.5 μm wide, numerous, persistent, filiform, unbranched, septate, slightly longer than the asci. Asci: 83–135 × 14–27 μm ( = 106 × 20 μm, n = 50), eight-spored, long, cylindrical, sometimes wider in the middle, blunt to rounded at the apex, with a stalk at the base, negative staining with Melzer’s reagent. Ascospores: 19–29 × 7–11 μm ( = 23 × 9 μm, n = 50), large, uniseriate, elliptical, with the other end more pointed, single or double arrangement. Asexual morph: Not observed.
Material examined: China, Sichuan Province, Chengdu City, Qionglai City, Haihong Community (30°17′2.13″ N, 103°15′48.56″ E, alt. 551 m), on leaves of Miscanthus floridulus (Lab.) Warb. ex Schum. et Laut., 23 May 2023, Qirong Sun & Liping Gao, SQR202305037 (SICAU 24-0050). GenBank accession numbers: ITS = PP785316, LSU = PP785305, SSU = PP785294.
Notes: Phylogenetic analyses revealed that SICAU 24-0050 forms a sub-branch within Phyllachora miscanthi (99% MLBS and 1.00 BYPP) (Figure 1), specifically close to P. miscanthi (MHYAU:167). Sequence comparisons demonstrated a high similarity in their LSU (99.6%, 455/457, 2 gaps) and SSU (99.6%, 790/793, 3 gaps). Furthermore, the morphological analysis of the new collection aligns with the description of P. miscanthi provided by Zhang Z. Y. et al. [28]. Based on comprehensive evidence, SICAU 24-0050 can be classified within P. miscanthi.
Phyllachora neidongensis Q.R. Sun & C.L. Yang, sp. nov., Figure 7.
Figure 7.
Phyllachora neidongensis (SICAU 24-0046, holotype). (a,b) Black spots on leaves. (c) Vertical cross-section of ascoma. (d) Peridium. (e) Paraphyses. (f–i) Asci. (j–m) Ascospores. (n) Ascospore with mucilaginous sheath. Scale bars: 200 μm (b), 100 μm (c), 10 μm (d–n).
Index Fungorum number: IF902116.
Etymology: Refer to the collection site, Neidong Township, Huili City, Sichuan Province, China.
Holotype: SICAU 24-0046.
Description: Parasite on leaves of Themeda triandra (Poaceae), causing tar spots on leaves. Tar spots: 1.1–2.5 × 0.4–1.0 mm ( = 1.6 × 0.6 mm, n = 30) on the upper leaf surface, fusiform or cymbiform, solitary to gregarious, black, carbonaceous, with a yellow halo of discolored host tissue. Sexual morph: Ascomata: 145–481 × 196–480 μm ( = 327 × 346 μm, n = 30), immersed within the leaf tissue, occupying the entire thickness, often developing adjacent to the neighboring ascomata and confined by the host vascular tissue; the structures are suboblate to subglobose, occasionally irregular in shape, and lack obvious ostioles. Peridium: 14–35 μm wide, approximately 6–8 layers, dark brown; the darker cells are the outer layer, and the large, slightly paler cells are the thin-walled inner layer. Paraphyses: 1.1–2.8 μm wide, numerous, persistent, filiform, branched, septate, slightly longer than the asci. Asci: 77–135 × 8–14 μm ( = 110 × 11 μm, n = 50), eight-spored, long, cylindrical, pedicellate at the posterior end, walls uniform in thickness, negative staining with Melzer’s reagent. Ascospores: 12–19 × 7–10 μm ( = 16 × 8 μm, n = 50), uniseriate, with cells occasionally overlapping and oblique; the hyaline, ellipsoidal-to-ovoid cells consist of a single cell type, some of which contain one or two large lipid droplets centrally located within the cell, all surrounded by a gelatinous sheath. Asexual morph: Not observed.
Material examined: China, Sichuan Province, Liangshan Yi Autonomous Prefecture, Huili City, Neidong Township (26°35′46.14″ N 102°20′41.96″ E, alt. 2093 m), on leaves of Themeda triandra Forsk., 26 September 2022, Qirong Sun & Chunlin Yang. SQR202209010 (SICAU 24-0046, holotype). GenBank accession numbers: ITS = PP785325, LSU = PP785314, SSU = PP785303; ibid SQR2022090100 (SICAU 24-0047), GenBank accession numbers: ITS = PP785326, LSU = PP785315, SSU = PP785304.
Notes: The three-gene phylogenetic analyses show that Phyllachora neidongensis is related to P. keralensis (99%MLBS, 1.00 BYPP). Microscopically, P. neidongensis exhibits larger ascomata (145–481 × 196–480 μm vs. 71–96 × 74–165 μm), longer asci (77–135 × 8–14 μm vs. 49–79 × 11–14 μm), and larger ascospores (12–19 × 7–10 μm vs. 9–13 × 6–7 μm) than P. keralensis, as described by Teng et al. [29]. Additionally, the paraphyses of P. neidongensis are branched, while those of the former are not. Nucleotide comparisons reveal significant differences between P. neidongensis (SICAU 24-0046) and P. keralensis (MHYAU:20083), viz. 23.04% (91/395, 0 gap), and 6.40% (49/499, 0 gap) in the ITS and LSU, respectively. Therefore, P. neidongensis strain SICAU 24-0046 was proposed as a new species.
4. Discussion
Phyllachora species are thermophilic, more likely to occur in hot summer conditions, and are mainly found in tropical and subtropical regions [12,30,31,32]. To date, 84 species of Phyllachora have been recognized in China, and they are mainly distributed in southern China [33,34,35,36], viz., Yunnan, Guangxi, Guangdong, and Sichuan Provinces. Yunnan Province is the most studied [37]. Only 22 species have been identified at the molecular level, and most of their host plants are Poaceae, which are most frequently found in high-temperature and high-humidity environments [1,2,9,24]. Thus, we inferred that the hot and humid summer climate and rich plant diversity in Sichuan Province facilitate the occurrence and infiltration of Phyllachora species. However, there are very few studies on Phyllachora fungi, with most focusing only on morphological identification, such as P. cynodontis, P. graminis, P. lespedezae, P. quadraspora, and P. sacchari [33,34,36]. These fungi infect more than 10 types of host plants, including Lespedeza bicolor (Fabaceae), Cynodon dactylon, Lolium perenne, Phyllostachys sulphurea, Eleusine indica, and Miscanthus sinensis (Poaceae) [33,34,36]. Until now, only one Phyllachora heterocladae species on Phyllostachys heteroclada has been identified with both morphological and molecular analyses [9]. Most Phyllachora species studies lack the support of comprehensive morphological and phylogenetic analyses.
In this study, the host plants of Phyllachora were Bothriochloa ischaemum, Phragmites australis, Themeda triandra, Chloris virgata, Lolium perenne, and Miscanthus floridulus, in which B. ischaemum, P. australis, and T. triandra were newly discovered. It is well known that Phyllachora species are often recognized as obligate fungi, but P. virgatae and P. jiaensis have been reported from the same host (Chloris virgata) on the grass resources of China [1]. Similarly, P. miscanthi and P. graminis have been found on Lolium perenne in Chengdu and Ya’an City, Sichuan Province [33]. Therefore, Phyllachora species cannot be easily distinguished based on different host plants, and detailed morphological characteristics and molecular analyses are essential to reveal new taxa and enhance understanding of their diversity.
In Sichuan Province, China, the vast plateau grasslands, which harbor diverse ecosystems and rich forage species, have esthetic, ecological, and economic value. However, the persistence of irrational management practices and the effects of climate change have led to periodic, widespread outbreaks of grassland diseases. Species of the genus Phyllachora, in particular, are emerging as major pathogens affecting grassland plant diseases [14,15]. During our investigation, a large area of Phragmites australis in Yangma Wetland Park, Chongzhou City, was infested with tar spot, characterized by dense black spots on yellow, withered, and dead leaves. This has also been observed in other wetland parks in Chengdu. In addition, the incidence of tar spot on Chloris virgata, located in Huili City, was also serious in this survey. In previous studies, Liu et al. [36] proposed that the species in genus Phyllachora are important plant pathogenic fungi, which are extremely harmful to herbages, as exemplified by Phyllachora maydis in the United States, causing serious effects on the quality and yield [13,14,15,16]. Despite these challenges, research on fungal diseases remains scarce. Therefore, comprehensive research is urgently needed to trace the disease type on grasslands and to protect their ecological security.
Acknowledgments
Xiu-Lan Xu and Zhen Zeng express gratitude for project funding from the Chengdu Park City Construction Administration Bureau.
Author Contributions
Methodology, Q.-R.S. and Y.D.; formal analysis, Q.-R.S. and F.-H.W.; resources, X.-L.X., C.-L.Y. and L.-P.G.; data curation, Q.-R.S., F.L., Y.-Q.Y. and Y.D., writing—original draft preparation, Q.-R.S.; writing—review and editing, Q.-R.S., F.L., X.-L.X., Y.-G.L. and C.-L.Y.; supervision, F.-H.W., Y.-Q.Y. and Y.D.; project administration, X.-L.X., C.-L.Y. and Z.Z.; funding acquisition, X.-L.X. and Z.Z. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequence data are available in NCBI GenBank following the accession numbers in the manuscript. All species data are available in Index Fungorum.
Conflicts of Interest
The authors declare no conflicts of interests.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Li J.C., Wu H.X., Li Y., Li X.H., Song Y.J., Suwannarach N., Wijayawardene N.N. Taxonomy, Phylogenetic and Ancestral Area Reconstruction in Phyllachora, with Four Novel Species from Northwestern China. J. Fungi. 2022;8:520. doi: 10.3390/jof8050520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dayarathne M., Maharachchikumbura S., Jones E.B.G., Goonasekara I.D., Bulgakov T.S., Al-Sadi A.M., Lumyong S., McKenzie E.H.C. Neophyllachora gen. nov. (Phyllachorales), three new species of Phyllachora from Poaceae and resurrection of Polystigmataceae (Xylariales) Mycosphere. 2017;8:1598–1625. doi: 10.5943/mycosphere/8/10/2. [DOI] [Google Scholar]
- 3.Maharachchikumbura S., Hyde K.D., Jones E., Mckenzie E., Wijayawardene N.N. Families of Sordariomycetes. Fungal Divers. 2016;79:1–317. doi: 10.1007/s13225-016-0369-6. [DOI] [Google Scholar]
- 4.Hongsanan S., Maharachchikumbura S.S., Hyde K.D., Samarakoon M.C., Jeewon R., Zhao Q., Al-Sadi A., Bahkali A.H. An updated phylogeny of Sordariomycetes based on phylogenetic and molecular clock evidence. Fungal Divers. 2017;84:25–41. doi: 10.1007/s13225-017-0384-2. [DOI] [Google Scholar]
- 5.Wijayawardene N.N., Hyde K.D., Dai D.Q., Sánchez-García M., Goto B.T., Saxena R.K., Erdoğdu M., Selçuk F., Rajeshkumar K.C., Aptroot A., et al. Outline of Fungi and fungus-like taxa -2021. Mycosphere. 2022;13:53–453. doi: 10.5943/mycosphere/13/1/2. [DOI] [Google Scholar]
- 6.Guterres D.C., Dos Santos M.D.D.M., Furlanetto C., Pinho D.B., Barreto R.W., Dianese J.C. Filling a gap in the taxonomy of phyllachoroid fungi: Proposition of Neopolystigma, gen. nov., and the new family Neopolystigmataceae. Mycologia. 2022;114:900–913. doi: 10.1080/00275514.2022.2092365. [DOI] [PubMed] [Google Scholar]
- 7.Theissen F., Sydow H. Die Dothideales. Annales Mycologici. 1915;13:147–746. [Google Scholar]
- 8.Dos Santos M.D.M., Noronha Fonseca M.E., Silva Boiteux L., Câmara P.E.A.S., Dianese J.C. ITS phylogeny and taxonomy of Phyllachora species on native Myrtaceae from the Brazilian Cerrado. Mycologia. 2016;108:1141–1164. doi: 10.3852/16-025. [DOI] [PubMed] [Google Scholar]
- 9.Yang C.L., Xu X.L., Liu Y.G., Hyde K.D., Mckenzie E. A new species of Phyllachora (Phyllachoraceae, Phyllachorales) on Phyllostachys heteroclada from Sichuan, China. Phytotaxa. 2019;392:186–196. doi: 10.11646/phytotaxa.392.3.2. [DOI] [Google Scholar]
- 10.Hyde K.D. Refined families of Sordariomycetes. Mycosphere. 2020;11:305–1059. doi: 10.5943/mycosphere/11/1/7. [DOI] [Google Scholar]
- 11.Hirst J.M. Ainsworth and Bisby’s: Dictionary of the Fungi. Eighth Edition By D.L. Hawksworth, P.M. Kirk, B.C. Sutton and D.N. Pegler. Wallingford, UK: CAB INTERNATIONAL (1995), pp. 616. Exp. Agric. 1997;33:247–252. [Google Scholar]
- 12.Cannon P.F. A Revision of Phyllachora and Some Similar Genera on the Host Family Leguminosae. CABI; Wallingford, UK: 1991. pp. 1–302. [Google Scholar]
- 13.Mottaleb K.A., Loladze A., Sonder K., Kruseman G., Vicente F.S. Threats of Tar Spot Complex disease of maize in the United States of America and its global consequences. Mitig. Adapt. Strat. Glob. Change. 2019;24:281–300. doi: 10.1007/s11027-018-9812-1. [DOI] [Google Scholar]
- 14.Mccoy A.G., Romberg M.K., Zaworski E.R., Robertson A.E., Phibbs A., Hudelson B.D., Smith D.L., Beiriger R.L., Raid R.N., Byrne J.M., et al. First Report of Tar Spot on Corn (Zea mays) Caused by Phyllachora maydis in Florida, Iowa, Michigan, and Wisconsin. Plant Dis. 2018;102:1851. doi: 10.1094/PDIS-02-18-0271-PDN. [DOI] [Google Scholar]
- 15.Dalla Lana F., Plewa D.E., Phillippi E.S., Garzonio D., Hesterman R., Kleczewski N.M., Paul P.A. First Report of Tar Spot of Maize (Zea mays), Caused by Phyllachora maydis, in Ohio. Plant Dis. 2019;103:1780. doi: 10.1094/PDIS-01-19-0070-PDN. [DOI] [Google Scholar]
- 16.Telenko D.E.P., Ross T.J., Shim S., Wang Q., Singh R. Draft Genome Sequence Resource for Phyllachora maydis—An Obligate Pathogen That Causes Tar Spot of Corn with Recent Economic Impacts in the United States. Mol. Plant-Microbe Interact. 2020;33:884–887. doi: 10.1094/MPMI-03-20-0075-A. [DOI] [PubMed] [Google Scholar]
- 17.Chomnunti P., Hongsanan S., Aguirre-Hudson B., Tian Q., Peršoh D., Dhami M.K., Alias A.S., Xu J., Liu X., Stadler M., et al. The sooty moulds. Fungal Divers. 2014;66:1–36. doi: 10.1007/s13225-014-0278-5. [DOI] [Google Scholar]
- 18.White T.J., Bruns S., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Michael A., David H., editors. PCR Protocols. Academic Press; London, UK: 1990. pp. 315–322. [Google Scholar]
- 19.Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 21.Zhang D., Gao F., Jakovlić I., Zou H., Zhang J., Li W.X., Wang G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020;20:348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen L., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X., Wu S., Wang C., Feng Y., Zhao C., Chen Z.-Q., Yu J.-F., Luo R., Promputtha I., Sun D.-F. Two new species of Phyllachora (Phyllachoraceae; Phyllachorales) on bamboo from China. Phytotaxa. 2019;425:78–86. doi: 10.11646/phytotaxa.425.2.2. [DOI] [Google Scholar]
- 25.Tamakaew N. Tar spot fungi from Thailand. Mycosphere. 2017;8:1054–1058. doi: 10.5943/mycosphere/8/8/6. [DOI] [Google Scholar]
- 26.Hyde K.D., Norphanphoun C., Ma J., Yang H.D., Zhang J.Y., Du T.Y., Gao Y., Gomes De Farias A.R., He S.C., He Y.K., et al. Mycosphere notes 387–412—Novel species of fungal taxa from around the world. Mycosphere. 2023;14:663–744. doi: 10.5943/mycosphere/14/1/8. [DOI] [Google Scholar]
- 27.Orton C.R. Graminicolous Species of Phyllachora in North America. Mycologia. 2018;36:18–53. doi: 10.1080/00275514.1944.12017527. [DOI] [Google Scholar]
- 28.Zhang Z.Y., Fan J.H., Li X.L., Zeng X.X., Zhang H. 2005 Taxonomic study on the genus Phyllachora in China. J. Northwest Agric. For. Univ. 2005;201:129–131. [Google Scholar]
- 29.Teng X.Q., Li C.Y., He M.H., Zhang T., Yang G.H., Gong L.W., Zhang Z.Y. Four new records of the genus Phyllachora in China. Mycology. 2010;6:918–919. [Google Scholar]
- 30.Kirk P.M., Cannon P.F., Minter D.W., Stalpers J.A. Dictionary of Fungi. 10th ed. CABI; London, UK: 2008. p. 527. [Google Scholar]
- 31.Piepenbring M., Hofmann T.A., Kirschner R., Mangelsdorff R., Trampe T. Diversity patterns of Neotropical plant parasitic microfungi. Ecotropica. 2011;17:27–40. [Google Scholar]
- 32.Mardones M., Trampe-Jaschik T., Oster S., Elliott M., Urbina H., Schmitt I., Piepenbring M. Phylogeny of the order Phyllachorales (Ascomycota, Sordariomycetes): Among and within order relationships based on five molecular loci. Persoonia-Mol. Phylogeny Evol. Fungi. 2017;39:74–90. doi: 10.3767/persoonia.2017.39.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin Y. Preliminary report on the study of nevus of grasses in Tibet and Sichuan; Proceedings of the Joint Annual Meeting of the Chinese Society of Phytophthora and Mycology; Hangzhou, China. 2–5 November 2017; pp. 42–43. [Google Scholar]
- 34.Teng X.Q. Study on the species diversity of Mollicutes in China. Agric. Sci. Inf. 2020;20:27–29. [Google Scholar]
- 35.Liu N., Li M. Phyllachora jianfengensis sp. nov. from China. Mycotaxon. 2016;130:1039–1043. doi: 10.5248/130.1039. [DOI] [Google Scholar]
- 36.Liu N. Taxonomic Study of the Genus Phyllachora in China. Institute of Microbiology, Chinese Academy of Sciences; Beijing, China: 2007. [Google Scholar]
- 37.Li J., Wu H., Song J. Two new Phyllachora species in Southwest China. Phytotaxa. 2023;578:275–285. doi: 10.11646/phytotaxa.578.3.5. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequence data are available in NCBI GenBank following the accession numbers in the manuscript. All species data are available in Index Fungorum.