Abstract
Tick-borne rickettsioses are considered among the oldest known vector-borne zoonotic diseases. Among the rickettsiae, Rickettsia africae is the most reported and important in Africa, as it is the aetiological agent of African tick bite fever (ATBF). Studies describing the prevalence of R. africae in southern Africa are fragmented, as they are limited to small geographical areas and focused on Amblyomma hebraeum and Amblyomma variegatum as vectors. Amblyomma spp. ticks were collected in Angola, Mozambique, South Africa, Zambia and Zimbabwe during the sampling period from March 2020 to September 2022. Rickettsia africae was detected using the ompA gene, while characterisation was conducted using omp, ompA, ompB and gltA genes. In total, 7734 Amblyomma spp. ticks were collected and were morphologically and molecularly identified as Amblyomma eburneum, A. hebraeum, Amblyomma pomposum and A. variegatum. Low levels of variability were observed in the phylogenetic analysis of the R. africae concatenated genes. The prevalence of R. africae ranged from 11.7% in South Africa to 35.7% in Zambia. This is one of the largest studies on R. africae prevalence in southern Africa and highlights the need for the inclusion of ATBF as a differential diagnosis when inhabitants and travellers present with flu-like symptoms in the documented countries.
Keywords: African tick bite fever, tick-borne diseases, prevalence, zoonotic disease, tick-borne pathogen, wildlife
1. Introduction
Rickettsia species are highly fastidious, obligate intracellular Gram-negative bacteria classified in the family Rickettsiaceae [1]. Tick-borne rickettsioses are considered among the oldest known vector-borne zoonotic diseases [2]. Most pathogenic species of Rickettsia are categorised into three groups, namely the typhus group (TG), transitional group (TRG), and the spotted fever group (SFG), while the ancestral group (AG) is still under debate [3]. The SFG contain 23 species, of which 16 are pathogenic, including Rickettsia conorii, the causative agent of Mediterranean spotted fever [4], R. conorii subsp. raoultii and Rickettsia slovaca, which causes rickettsial lymphadenopathy [5] and Rickettsia rickettsii, which causes rocky mountain spotted fever [6]; all of which are associated with ticks as the main vectors. By contrast, the TG contains only two species, Rickettsia prowazekii and Rickettsia typhi, which are associated with lice or fleas as their vectors, respectively [7,8,9]. Finally, the TRG contains both arthropod-restricted symbionts and human pathogens transmitted by fleas, ticks and mites [10,11]. Among the rickettsiae, Rickettsia africae is the most frequently reported and significant in Africa, as it is the aetiological agent of African tick bite fever (ATBF) [12].
The symptoms of ATBF resemble those of malaria. It is an acute, flu-like illness with symptoms comprising fever, myalgia, severe headache, nausea and fatigue. In most cases, an inoculation eschar is present, though it may go unnoticed on darker skin tones, with regional lymphadenitis, cutaneous rash and, in rare cases, aphthous stomatitis [13]. Whereas flu-like symptoms are common, some cases may present with some but not all ailments, while other cases may be asymptomatic. Severe disease is rare, with no records of life-threatening complications or deaths associated with the disease.
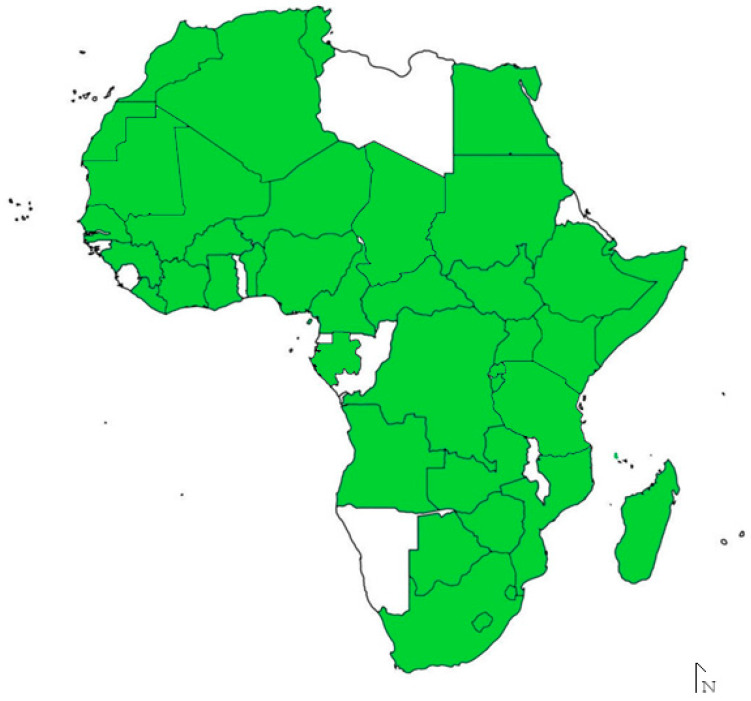
Amblyomma spp. ticks are known to be the primary vectors of R. africae and hence the distribution of R. africae is reliant on the distribution of the tick vectors [13] (Figure 1). Therefore, the seasonality of R. africae is also dependent on the seasonality of the vectors [14,15]. Amblyomma variegatum (Fabricius, 1794) [16] is the most widespread vector, found in central, western and eastern Africa as well as the West Indies; its New World distribution comprises the islands of Guadeloupe, Martinique, St. Kitts and Nevis, and Antigua following introductions from Africa [7]. Accordingly, R. africae is largely restricted to the African continent [17,18,19], although the disease it causes, ATBF, has been detected in many countries globally due to increases in tourism to sub-Saharan Africa and R. africae dispersal via the tick vector on livestock or wild hosts [20,21]. In endemic areas, the infection rate of ticks with R. africae can reach up to 100%, and among travellers, ATBF is the second most commonly identified cause of systemic febrile illness, following malaria [22,23].
Figure 1.
Distribution of Rickettsia africae in Africa based on detection in either vector ticks or human hosts. White shaded areas indicate countries where R. africae has not been detected or investigated. Image was constructed with information obtained from Genbank, Mazhetese, et al. [21], Pillay, et al. [24] and Zhang, et al. [25].
Several gaps are evident in the literature focusing on R. africae, including a lack of information on the role of mammalian hosts in the maintenance of R. africae and the prevalence rates of infection between male and female ticks and its effects on pathogen transmission. Regarding the role of the mammalian hosts, conflicting hypotheses on the natural reservoirs for R. africae can be found in the literature, as data on the role of mammalian hosts in the maintenance of R. africae is scant and contradictory. Kelly, et al. [26] reported that 26–90% of cattle in Zimbabwe had antibodies against R. africae, although none displayed any clinical symptoms. During their study, they isolated Rickettsia-like organisms from Amblyomma hebraeum ticks and cultured these isolates. Eight naïve cattle and three male guinea pig subjects were inoculated with these isolates. Kelly, et al. [26] observed seroconversion in all eight of the naïve cattle breeds and all three of the guinea pig subjects, suggestive of the possible maintenance of R. africae in cattle. It is key to note that the conclusion of Kelly, et al. [26] on the maintenance of R. africae in cattle was based solely on the seroconversion of naïve cattle. It has also been reported that R. africae serological assays have low specificity [20]. To date, no studies have been conducted using DNA methods to determine whether cattle play a role in the maintenance of R. africae. The other possibility is that Amblyomma spp. does not require a mammalian reservoir. Although Amblyomma spp. are well documented to transmit R. africae transovarially, no studies are currently available on the efficiency of the transovarial transmission across several tick generations [21]. Thus, insufficient evidence is available on the maintenance of R. africae in the field.
Knowledge on the prevalence rate of R. africae in male and female ticks could contribute to the disease epidemiology of R. africae, which has been mostly absent in the literature to date. Male ticks are more likely to stay attached to hosts when compared to female ticks; however, males have been documented to detach from host during nights, changing predilection site or even hosts [27]. Both male and female Amblyomma spp. are known to be aggressive hunters with high biting ratios. The biting ratio alongside the attachment duration could impact pathogen transmission. Therefore, documenting and understanding the difference in infection rate between male and female ticks are crucial for a comprehensive understanding of R. africae transmission and epidemiology.
Despite the number of reports that have detected R. africae in several vectors, studies describing the prevalence of R. africae in southern Africa are fragmented, limited to small geographical areas and focused on the primary vectors only (Amblyomma hebraeum (Koch, 1844) and A. variegatum) [28,29,30,31,32,33,34,35]. Recent studies list 21 Amblyomma spp. occurring in south-eastern Africa, most of which have limited to no information on infection rates with R. africae. The paucity of information on prevalence rates is compounded by a lack of data on R. africae strains from these neglected vectors.
The characterisation and systematics of R. africae has relied on the use of several molecular markers. Common markers include the outer-membrane protein OmpA (ompA) [1], outer-membrane protein OmpB (ompB) [36] and citrate synthase (gltA) [37]. Less common markers encompass the 17 kDa surface antigen (omp) [38], surface cell antigen 4 (sca4) [39], 16S rRNA [40] and the internal transcribed spacer (ITS) [41]. Rarely used markers include dksA-xerC, mppA-purC and rpmE-tRNAfMet, which are all intergenic spacers [4]. However, it is standard practice to use a combination of several of the above-mentioned markers [24]. In recent years, the popularity of whole genome sequencing has increased due to its lower cost, yet the first and only whole genome sequence of R. africae became available in 2009 [42]. Rickettsiae are known for their high level of collinearity, with the main markers exhibiting homogeneity ranging from 82.2% to 99.8%, which hinders intra-species differentiation [43].
Although intraspecific variability of SFG rickettsiae is generally low [43], a greater level of heterogeneity has been reported when using the ompA and ompB molecular markers as compared to the intergenic spacers [30,44]. Intra-species variation was more prominent in studies that have applied three or more markers [45,46,47]. In this study, we investigated the prevalence of R. africae in Amblyomma spp. ticks collected in southern Africa and characterised phylogenetic relationships of R. africae using the ompA, ompB, gltA and omp genetic markers.
2. Materials and Methods
2.1. Sample Collection and Identification
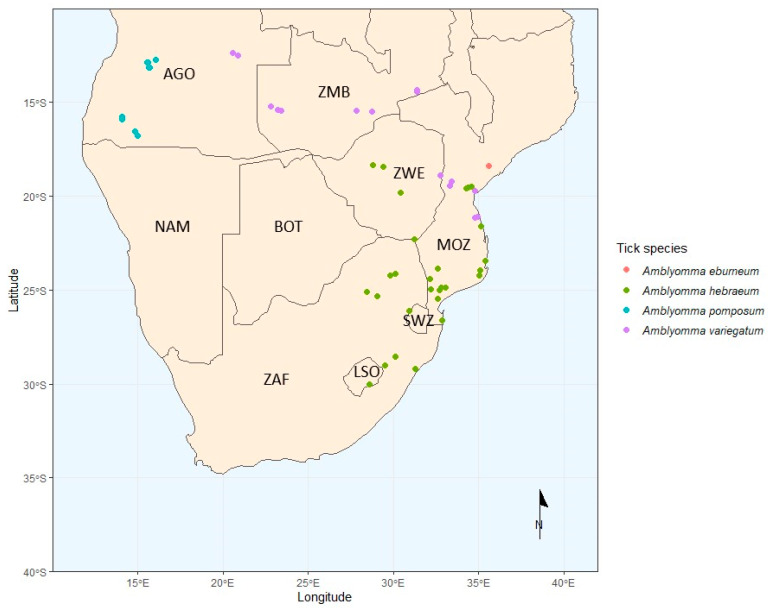
Amblyomma spp. ticks from Smit, et al. [48] were used for this study. These were collected in Angola, Zambia and Zimbabwe, from randomly sampled cattle and goats, while in Mozambique, Amblyomma spp. were collected from both cattle and legally hunted wildlife (Figure 2). Samples were also used from Dlamkile, et al. [49], who collected Amblyomma spp. ticks from randomly sampled cattle in South Africa from 2020 to 2022.
Figure 2.
Map of southern Africa, illustrating the Amblyomma spp. collection points during the 2020–2022 sampling period performed by Smit et al. [41]. The colour of the dots represents the species of Amblyomma spp. ticks collected at the respective sampling points. Country codes: AGO—Angola, BOT—Botswana, LSO—Lesotho, MOZ—Mozambique, NAM—Namibia, SWZ—Eswatini, ZAF—South Africa, ZMB—Zambia, and ZWE—Zimbabwe.
Morphological identification of the species level was conducted using identification keys from Walker et al. [50] and Voltzit and Keirans [51], while morphological characteristics were documented in Smit, et al. [48].
2.2. DNA Extraction and Amplification
DNA was extracted from halved individual ticks using the Chelex 100 resin method as described in Smit, et al. [52]. Tick identification was confirmed in Smit, et al. [48] by molecular characterisation targeting the 12S rRNA [53], 16S rRNA [54], cytochrome oxidase I (coi) [55] and cytochrome B (cytB) [56] molecular markers.
With the use of Cannon and Roe [57], the sample size for the tick collection in Mozambique was calculated as the number of ticks required to detect at least one infected tick, at a confidence level of 95%, assuming an infection rate of 10% and an infinite population. This resulted in 132 ticks required to provide statistically significant results. A subsample of 160 ticks per species was screened for R. africae using the ompA gene as described by Mazhetese, et al. [58].
Rickettsia africae characterisation was conducted by amplifying four gene regions: omp, ompA, ompB and gltA (Table 1). In total, 50 Amblyomma ticks positive for R. africae were selected to ensure five positive ticks per species per country. Each gene amplification consisted of a 25 µL reaction comprising 10 µL Phusion Flash PCR Master Mix (1X Final concentration), 0.5 µL each of the corresponding primers (Table 1) (final concentration of 0.5 µM per primer), 7 µL double-distilled water and 2 µL sample DNA. The PCR cycling conditions for omp, ompB and gltA were as follows: initial denaturation at 98 °C for 10 s, followed by 35 cycles of 92 °C for 1 s, 50 °C for 5 s, and 68 °C for 15 s. Final annealing was conducted at 72 °C for 2 min with a hold stage at 4 °C. The ompA cycling conditions were an initial denaturation step at 98 °C for 10 s, followed by 30 cycles of 92 °C for 1 s, 51 °C for 5 s, and 68 °C for 15 s. Final annealing was conducted at 70 °C for 1 min with a hold stage at 4 °C. A sample from a previous study which was proven to be R. africae by sequencing was used as a positive control for each amplification. A no-template (double-distilled water) negative control was included in each amplification.
Table 1.
Primers used for amplification of specific targeted regions.
| Target Region | Primer Name | Annealing Temperature (°C) | Sequence (5′–3′) | Expected Fragment Length (bp) | Reference |
|---|---|---|---|---|---|
| ompA | Rr190.70F | 51 | ATG GCG AAT ATT TCT CCA AAA | 632 | Mazhetese, et al. [58] |
| Rr190.701R | GTT CCG TTA ATG GCA GCA TCT | ||||
| ompB | ompBF | 50 | ACA TK*G TTA TAC ARA GTG Y*TA ATG C | 444 | Pillay and Mukaratirwa [47] |
| ompBR | CCG TCA TTT CCA ATA ACT AAC TC | ||||
| omp | 17kDaF | 50 | AAT GAG TTT TAT ACT TTA CAA AAT TCT AAA AAC CA | 450 | Pillay and Mukaratirwa [47] |
| 17kDaR | CAT TGT TCG TCA GGT TGG CG | ||||
| gltA | gltAF | 50 | ACC TAT ACT TAA AGC AAG TAT Y*GG T | 1234 | Pillay and Mukaratirwa [47] |
| gltAR | TCT AGG TCT GCT GAT TTT TTG TTC A |
* Degenerate bases ensure binding to all R. africae strains. Keto (K) can accommodate guanine (G) or thymine (T), while pyrimidine (Y) can accommodate cytosine (C) or thymine (T).
PCR products were separated on a 1.5% agarose gel to confirm whether a sample tested positive for R. africae and that the band was the expected size to be sent for sequencing. The gel was visualised using the Bio-Rad gel documentation system with assisted visualisation programming. Samples that produced visible single bands were sent to the Central Analytical Facility (CAF), Stellenbosch, South Africa, for Sanger sequencing in both directions.
2.3. Phylogenetic Analysis
Contigs of forward and reverse sequences were constructed using CLC main workbench version 23.0.2 (developed by CLC Bio, http://www.clcbio.com (accessed on 1 August 2024)). These contigs of each individual sample for each individual gene were manually corrected on CLC main workbench version 23.0.2. Manual corrections consisted of base calling, where base inconsistency between the forward and reverse sequences were corrected. For R. africae characterisation, individual genes and a concatenated matrix were used for processing. Reference sequences were obtained from the GenBank database and used for comparison (https://www.ncbi.nlm.nih.gov/genbank/) (accessed on 17 February 2024) (Supplementary Table S1). For each of the individual genes (omp, ompA, ompB and gltA), R. conorii was used as the outgroup. For the concatenated matrix, R. conorii subsp. raoultii was used as an outgroup since it was one of the few Rickettsia strains for which all genes were available from the same sample on GenBank. Orientation of the assembled contigs was evaluated, and these contigs were aligned alongside the reference sequences with the use of the online version of MAFFT version 7 (developed by http://mafft.cbrc.jp/alignment/server/index.html) (accessed on 17 February 2024) with default parameters. The aligned matrix was manually viewed, edited and truncated using MEGA 11. The best-fit model was determined using the jModelTest2 [59] on the CIPRES Science Gateway (https://www.phylo.org/ (accessed on 1 August 2024)) platform. The file format was changed depending on program requirements, using FaBox version 1.61 (https://users-birc.au.dk/palle/php/fabox/index.php) (accessed on 17 February 2024). Bayesian analysis was performed in MrBayes version 3 [60]. The Hasegawa–Kishino-Yano (HKY) model with gamma rates and invariable sites was used for the concatenated R. africae tree. Individual gene tree models are listed in the figure descriptions. Five Monte Carlo Markov Chains (MCMC) were run for 5,000,000 iterations, saving every 1000th tree, while the first 25% of drafted trees were discarded. Tracer version 1.6 [61] was used for the inspection of estimated sample size (ESS) (>200) and parameter sampling using graphical plots indicating parameter stabilisation. The resulting tree was visualised and edited in iTOL version 6.8 [62].
2.4. Statistical Analysis
The statistical significance of R. africae prevalence between the males and females of each species was calculated using the numbers collected for each respective sex, and the number of positives for each respective sex. The statistical significance of R. africae prevalence between the Amblyomma spp. in each country was calculated using the total number collected in a specific country and the total number of positives for each species. A χ2 analysis was conducted in Microsoft Excel version 2402 to evaluate the independence between infection rate and sex of tick species, as well as to evaluate the independence between infection rate and tick species. If the expected number was lower than five, then the Fisher’s exact test was used instead. A Bonferroni correction was applied, resulting in a p-value threshold for acceptance of 0.004 (0.05/12 tests).
2.5. Ethical Considerations
This study obtained approval from the Research and Animal Ethics Committee of the University of Pretoria (REC 121-20). Furthermore, consent was granted by the Department of Agriculture, Land Reform, and Rural Development (DALRRD) in South Africa, under Section 20 of the Animal Diseases Act 1984 (Act no. 35 of 84) (12/11/1/1 (1937SS)).
3. Results
In total, 7734 adult Amblyomma ticks were collected in southern Africa and were morphologically identified as Amblyomma eburneum (Gerstäcker, 1873) [63] (n = 208), A. hebraeum (n = 4758), Amblyomma pomposum (Dönitz, 1909) [64] (n = 191) and A. variegatum (n = 2577) [48].
Amplification success varied amongst the genes; ompA had a success rate of 78% (39/50), while ompB had a success rate of 86% (43/50), omp had a success rate of 74% (37/50), and gltA had a success rate of 82% (41/50). Although 37 samples did amplify using the omp gene, only 29 produced high-quality sequences, while the remaining nine were of low quality and could not produce contigs. The gltA locus also experienced low sequence quality, where 41 samples amplified but only 36 were of good quality and formed contigs. All samples that produced visible single bands for ompA and ompB generated sequences that were of high quality, all of which formed contigs. All sequences were deposited in GenBank (Supplementary Table S1).
Individual gene trees are located in Supplementary Data S2. The Bayesian analysis of the ompA gene illustrated a clear clustering of all the sequences obtained in this study with those obtained from GenBank, with the exception of three R. africae sequences obtained from Hyalomma spp., which formed their own clade (probability of 0.98) (Supplementary Data S2: Figure S1). The ompB analysis depicted the majority of the sequences from this study clustering together with reference sequences from GenBank (Supplementary Data S2: Figure S2). Nine sequences were observed branching from the main clade with a high probability (0.98). Of these nine sequences, seven were collected from wildlife (Syncerus caffer) in Mozambique; the other two sequences were derived from A. variegatum collected from livestock (Bos taurus) in Angola and Mozambique. The Bayesian analysis of the omp gene clustered all the sequences of this study alongside references obtained from GenBank (Supplementary Data S2: Figure S3). Four branches were observed extending from the main clade; however, three of these nodes were not supported (probability lower than 0.95). Two of these separations are located on long branches; when the sequences were compared to those on GenBank, identities of 96% or higher were observed with R. africae 17 kDa sequences. The gltA topology displayed greater variation (Supplementary Data S2: Figure S4). Three separate clades were observed for R. africae; the first contained two R. africae sequences from Hyalomma and one sequence obtained from Gorilla gorilla (gorilla), the second clade contained one R. africae sequence from Amblyomma compressum, and the last clade contains all the sequences from this study and several reference sequences from GenBank. Two long branches were observed separating from the main clade; however, these nodes were not supported (probability lower than 0.95).
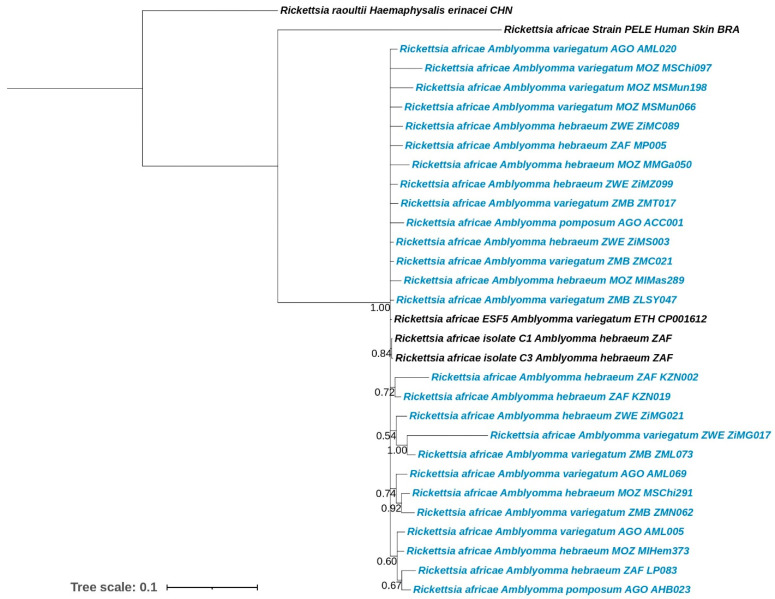
The Bayesian analysis of the phylogeny based on the concatenated genes indicated very little variation between the sequences obtained from this study (Figure 3). The majority of the samples from this study clustered with reference sequences from Pillay and Mukaratirwa [47] (C1 and C3) from South Africa, and with the R. africae whole genome sequence from an A. variegatum tick from Ethiopia. Variation in the R. africae sequences was observed for isolates obtained from humans. Several R. africae samples branched separately from the main clade; however, these nodes were not supported (probability lower than 0.95).
Figure 3.
Bayesian inference analysis of the concatenated Rickettsia africae genes (omp, ompA, ompB and gltA). Analysis was performed using the HKY model with gamma rates and invariable sites. Posterior probability values are indicated at each branch node. Sequences from this study are in blue. Country of origin is indicated in the sample ID as: AGO—Angola, BRA—Brazil, CHN—China, ETH—Ethiopia, MOZ—Mozambique, ZAF—South Africa, ZMB—Zambia, and ZWE—Zimbabwe.
Our results from the R. africae conventional PCR showed that the prevalence in Angola was 24.4% (108/443) (Supplementary Table S2). Amblyomma pomposum had a prevalence of 41.9% (80/191), of which 25% of females (1/4) and 42.2% of males (79/187) tested positive. The A. variegatum from Angola had a prevalence of 11.1% (28/252), of which 25% of females (4/16) and 10.2% of males (24/236) tested positive. There was no association between R. africae infection rates and A. pomposum sex (p = 0.087), or between R. africae infection rates and A. variegatum sex (p = 0.641). However, there was an association between R. africae infection and tick species in Angola (X2(1, n = 443) = 55.813, p < 0.001), with higher infection rates observed in A. pomposum (Supplementary Table S2).
In Mozambique, the overall prevalence of R. africae was 25.7% (301/1170), of which the prevalence in ticks collected from livestock was 28.6% (244/854) and in ticks collected from wildlife, 18.0% (57/316) (Supplementary Table S2). In livestock, two Amblyomma spp. were identified in Mozambique, A. hebraeum and A. variegatum. The prevalence of R. africae in A. hebraeum from livestock was 33.5% (181/541), of which 19.3% (33/171) of females and 40% (148/370) of males tested positive; this difference was statistically significant (X2(1, n = 541) = 22.513, p < 0.001). The prevalence of R. africae in A. variegatum from livestock was 20.1% (63/313), of which 17.9% (14/78) of females and 20.9% (49/235) of males tested positive; this difference was not significant (X2(1, n = 313) = 0.307, p = 0.58). However, there was a statistical association between R. africae infection rate and the two tick species found on livestock from Mozambique (X2(1, n = 854) = 17.261, p = 0.002).
In the wildlife from Mozambique, two Amblyomma species were identified as A. eburneum and A. variegatum. The prevalence of R. africae in A. eburneum from wildlife was 14.1% (49/235), of which 19.5% (8/41) of females and 12.2% (14/115) of males tested positive; this difference was not significant (p = 0.472). The prevalence of R. africae in A. variegatum from wildlife was 21.9% (35/160), of which 15.1% (8/53) of females and 25.2% (27/107) of males tested positive; this difference was also not significant (X2(1, n = 160) = 2.132, p = 0.144). There was no statistical association between R. africae infection rates and the two tick species in wildlife from Mozambique (X2(1, n = 316) = 3.226, p = 0.072); nor was there a significant association in the R. africae prevalence between livestock Amblyomma spp. and wildlife Amblyomma spp. (X2(1, n = 1170) = 13,393, p = 0.004).
The infection rates by sex of the A. hebraeum from South Africa were not documented in the previous study [49] and thus no comparison can be made between the sexes for this country. The overall prevalence of R. africae was 11.4% (48/421) (Supplementary Table S2).
The overall R. africae prevalence in A. variegatum from Zambia was 35.7% (202/566) (Supplementary Table S2). Females had a prevalence of 37.5% (12/32) and males had a prevalence of 35.6% (190/534); this difference was not statistically significant (X2(1, n = 566) = 0.048, p = 0.826).
The overall R. africae prevalence from the ticks collected in Zimbabwe was 28.7% (99/345) (Supplementary Table S2). Both A. hebraeum and A. variegatum were collected in Zimbabwe. The prevalence in A. hebraeum was 29.3% (98/334), where 29% (38/131) of females tested positive and 29.6% (60/203) of males tested positive (X2(1, n = 334) = 0.012, p = 0.914). Too few A. variegatum (11 in total) were collected in Zimbabwe for a valid statistical analysis by sex. No statistically significant differences in the R. africae prevalence between the species were observed in Zimbabwe (p = 0.189).
4. Discussion
In this study, we investigated the prevalence of R. africae in Amblyomma ticks from southern Africa. Overall, four Amblyomma spp. were studied: A. eburneum, A. hebraeum, A. pomposum and A. variegatum. Based on molecular findings, insufficient evidence is available to differentiate A. pomposum from A. variegatum [48]. However, based on morphological descriptions and behavioural features, Smit, et al. [48] still regarded A. pomposum and A. variegatum as separate species, which requires further investigation. Since the taxonomy of the genus Amblyomma has remained unchanged, A. pomposum and A. variegatum are regarded as distinct species in this manuscript.
The phylogenetic analysis of the R. africae ompA gene indicated homogeneity, except for three sequences obtained from Hyalomma in previous studies [65,66], branching from the main clade with high probability. This separation observed between R. africae obtained from Amblyomma spp. and Hyalomma spp. has been documented in several articles such as Pillay and Mukaratirwa [47], Pillay, et al. [67] and Thekisoe, et al. [68]. This highlights that the variability of R. africae depends on tick origin. It should be noted that the R. africae vector competency of Hyalomma spp. has not been validated; thus, the sequences of R. africae isolates from Hyalomma spp. may represent residual DNA from co-feeding with other tick species or from an infected blood meal. The ompB analysis depicted a clear separation between the majority of the sequences and nine sequences generated during this study (probability of 0.98), of which eight were from Mozambique and one from Angola. Of these nine sequences that branched from the main clade, seven were collected from wildlife. Few reference sequences could be used for the ompB analysis due to several large gaps observed in the alignment resulting in inaccurate topologies. Pillay and Mukaratirwa [47], from which the primer sequences were obtained, noted that ompB alongside gltA provided the best intraspecific resolution. This contradicts what we observed for the ompB analysis where all the sequences, aside from the nine that formed a distinct branch, cluster together regardless of vector species or geographic location. We did, however, observe high levels of resolution using the gltA and 17 kDA genes, with clear separation between R. africae from different vectors such as Hyalomma, A. compressum and the Amblyomma spp. from this study. No separation based on geographical location was observed in any of the individual gene topologies.
Phylogenetical analysis of the R. africae concatenated sequences from this study (omp, ompA, ompB and gltA) illustrated low levels of variability, in which most of the R. africae sequences from A. pomposum, A. hebraeum and A. variegatum clustered together regardless of the country or type of host. This is to be expected due to the high level of collinearity within members of the SFG [9]. The homogeneity in R. africae observed in our study was also reported by Kimita, et al. [45], Iweriebor, et al. [44] and Pillay, et al. [67]. Kimita, et al. [45] highlighted the greater resolution obtained from the concatenation of these genes as compared to using each gene separately during phylogenetic analysis. Several sequences from this study were excluded from the concatenated analysis, since the samples did not have all four genes available for each isolate or clone (Supplementary Table S1). There were limited sequences to which we could compare since few are available for the omp locus on GenBank. Several other studies have also reported problematic amplification of the abovementioned loci, which could be due to variation in primer binding sites or sensitivity of the assays to inhibitors [44]. This high level of collinearity in R. africae provides a stable framework for vaccine development. However, most of the current studies focus solely on structural genes, such as the ones used in this study. To advance vaccine development, research should explore functional and phenotypic markers, as well as their expression levels, associated with virulence, cell invasion and pathogen maintenance.
The role of mammalian hosts in the maintenance of R. africae is still disputed with no molecular evidence in support of mammalian infection, as opposed to exposure measured by serology [21]. Although, multiple pathogens are known to be acquired by Amblyomma spp. ticks during ingestion of a blood meal from an infected host, such as Ehrlichia ruminantium [69] and Theileria mutans [70], several studies have determined a low prevalence in the cattle but a high prevalence in attached ticks. Diobo, et al. [71] conducted a study in Côte d’Ivoire, where they collected ticks and blood samples from cattle in five areas. Screening was performed of both the ticks and blood samples using the poT15-dam2 gene of R. africae. Diobo, et al. [71] noted a 43.67% positivity rate for R. africae in ticks collected from cattle, while no positives were detected in blood samples. Barradas, et al. [31] reported similar results during their study in Huambo, Angola. From the 98 blood samples they collected from cattle, none tested positive for R. africae using the ompB gene region. From the 116 ticks collected, 5 (4.3%) tested positive for R. africae. These findings support the hypothesis that mammalian hosts are not reservoirs for R. africae. If cattle were the reservoirs, one would expect to see high infection rates in their blood when detecting high infection rates in the ticks. However, current documentation appears to support that cattle become infected only after infected ticks feed on them. Ultimately, the role of mammals as reservoirs for R. africae should be investigated. Nevertheless, with no experimental evidence, we cannot fully exclude the possibility that the R. africae detected in the ticks in this study originated from the bloodmeal.
If mammalian hosts are not reservoirs for R. africae, we can then speculate that R. africae is a vertically transmitted symbiont, highly dependent on the selected vector due to its intracellular life cycle. This relationship with the vector may be the key factor driving the molecular variation in the R. africae genome as observed in the individual gene trees, where R. africae from different tick species clustered separately in some cases. Thus, a potential explanation for the formation of discrete clades in the individual gene trees could be attributed to vector–pathogen symbiosis. Indeed, Thu, et al. [72] noted a strong association between rickettsial genotypes and their host tick species in Japan. For R. africae, evolutionary divergence may be occurring between vector species of ticks.
Several studies have highlighted that tick-borne pathogens in the microbiome of the ticks interact with each other [73,74,75]. Macaluso, et al. [73] found that when Dermacentor variabilis ticks had a rickettsial infection, the transovarial transmission of a second Rickettsia species was inhibited. Ponnusamy, et al. [76] noted that the Amblyomma americanum microbiome is altered based on the geographical distribution of the ticks, which in turn affects the composition and diversity of rickettsial species present. Importantly, although not detected here, African Amblyomma spp. may harbour rickettsiae other than R. africae [77,78]. However, no studies have been specifically performed to determine how combinations of organisms such as different Rickettsia spp. in the tick microbiome contribute to each other’s evolution.
The prevalence of R. africae in Amblyomma spp. ticks ranged from 11.7% in South Africa to 35.7% in Zambia. The literature on the prevalence of R. africae in southern Africa is fragmented and limited to small geographical areas. In Angola, the overall prevalence was 24.4% and was divided between A. pomposum (72.1%) and A. variegatum (12.5%). Previous studies reported a prevalence of 45.5% [31] and 100% [79], respectively; although the sample sizes were very small (eleven and six, respectively), and Palomar, et al. [79] grouped their Amblyomma ticks into two pools, of which both tested positive. Our prevalence is much lower than other studies in the area; however, this could be attributed to their much smaller sample size. In this study, we found an overall prevalence of 25.7% in Mozambique (including Amblyomma spp. ticks collected from both wildlife and livestock), 33.5% for A. hebraeum, and 20.1% for A. variegatum from livestock. Magaia, et al. [30] reported a prevalence of 80% in A. hebraeum and 67% in A. variegatum ticks in Mozambique, whereas Matsimbe, et al. [28] documented a prevalence of only 4.5% in A. variegatum. Our results are within the expected range, and it is important to consider methodological differences between studies in terms of gene targets for PCR and the use of conventional or quantitative assays.
There are no previous records of R. africae sequences from Amblyomma spp. ticks collected from wildlife in southern Africa. South Africa is the most well-represented country in the literature regarding R. africae prevalence in livestock ticks from southern Africa, with the prevalence in vectors ranging from 11.8% to 62.2% [44,47,58,80,81]. Our prevalence of 11.4% is on the lower end of what is expected for the country. In Zambia, only A. variegatum ticks were collected and the overall prevalence of R. africae was 35.7%. Previous studies in Zambia recorded widely different infection rates of 36.8% [31], 80.0% [35] and 15.6% [34]. In these studies, the sample sizes for Amblyomma spp. ticks were relatively small (19, 30 and 46, respectively). The prevalence for Zambia in the current study is within the range of other studies from the same areas. One study in Zimbabwe recorded a prevalence of R. africae of 40.0% (64/166) in A. hebraeum and 0% (n = 9) in A. variegatum [29]. In our study, an overall prevalence of 28.7% was found, with a prevalence of 29.3% in A. hebraeum and 9.1% in A. variegatum. Thus, our prevalence estimate is much lower in A. hebraeum but higher in A. variegatum than what was reported previously, although we can only compare to this one study. Several factors such as seasonality, sampling timeframe, and sample size may have influenced this discrepancy between the observed R. africae prevalences in Zimbabwe.
Statistically significant differences were observed between sexes in A. hebraeum ticks collected from livestock in Mozambique, where males had an R. africae prevalence of 40% compared to females with a prevalence of 19.3%. The observed higher prevalence in these males could be attributed to their migratory habits. Observations made by Stachurski [27] noted that A. variegatum males migrate during the night from predilection sites and from hosts in the same kraal. This migratory behaviour alongside cofeeding could increase the chances of males having higher infection rates for R. africae compared to the females, assuming that vertical and transstadial transmission in the wild is not efficient in this species.
In Angola, A. pomposum was statistically more likely to be infected with R. africae than A. variegatum. Amblyomma pomposum was restricted to the central western regions in Angola, placing travellers and indigenous inhabitants in these areas more at risk of contracting ATBF. In the livestock of Mozambique, A. hebraeum was more likely to be infected with R. africae than was A. variegatum. No statistical differences in the infection rates of R. africae between Amblyomma spp. were observed in other countries. This suggests that in all other countries, A. hebraeum and A. variegatum have a similar likelihood of transmitting R. africae, given equal biting rates. This is not unexpected, since A. hebraeum and A. variegatum are both considered to be the main vectors of R. africae in Africa [13].
With the high R. africae prevalence observed in the collected Amblyomma ticks in each country and the aggressive behaviour of these ticks, it is interesting to observe that there are few to no reports of ATBF or TBF in the local populations in southern Africa. Silva-Ramos and Faccini-Martínez [82] conducted a systematic review of the R. africae published case reports in humans, and found 108 documented cases, of which eight cases were in indigenous individuals, and the remaining infections were in travellers to southern Africa. Of these 100 cases of travellers, 80% were European, with South Africa being the most visited country.
Cattle are regarded as the primary host for all the collected Amblyomma species [15,51], while A. eburneum has also been documented to prefer African buffalo (Syncerus caffer) [51]. The majority of the rural communities are reliant on subsistence livestock farming, where the younger portion of the population are used as cattle herders. Although the primary hosts of the Amblyomma spp. are regarded as livestock, Amblyomma spp. are documented to parasitise humans, placing those in direct contact with their livestock at greater risk to be preyed upon [83], and therefore to contract R. africae. We can speculate that in impoverished communities in Africa, where R. africae is endemic, locals are exposed to the pathogen at a young age and develop immunity. Later, in their adult years, if they are infected with R. africae, symptoms are generally mild for which no medical attention is sought. For cases where medical attention is sought, they are neither documented as case reports nor published as clinical cases. Ultimately, very little or no documentation of the R. africae prevalence in individuals from R. africae-endemic areas is available.
5. Conclusions
This is one of the largest studies on R. africae prevalence in southern Africa. The prevalence in this study ranged from 11.7% to 35.7%, highlighting the need for these areas to include ATBF as a differential diagnosis when inhabitants and travellers present with flu-like symptoms. Limited research has been conducted on R. africae for many African countries, leading to probable underreported cases of ATBF. This study is the first in southern Africa to obtain sequence of R. africae from Amblyomma spp. ticks collected from wildlife and make these data publicly available. More research should be conducted on R. africae from ticks feeding on wildlife to clarify the phylogenetic placing of these sequences.
Acknowledgments
A special thank you to Delta Safaris, Mungari, who allowed us to collect ticks and hosted us in their camp, and to all field technicians and state vets who assisted in the collection of ticks in all the represented countries.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms12081663/s1, Table S1: GenBank accession numbers; Table S2: Rickettsia africae infection rates; Figures S1–S4: Individual gene phylogenetic trees (ompA, ompB, omp, gltA).
Author Contributions
A.S.: Methodology, Investigation—Fieldwork, Investigation—Laboratory analysis, Data Curation, Writing—Original Draft, Visualisation; F.C.M.: Investigation—Fieldwork, Writing—Review & Editing; S.H.W.: Investigation—Laboratory analysis, Writing—Review & Editing; C.M.: Investigation—Fieldwork, Writing—Review & Editing; G.S.: Investigation—Fieldwork, Writing—Review & Editing; S.M.: Investigation—Fieldwork, Writing—Review & Editing; H.R.V.: Visualisation, Writing—Review & Editing; Z.D.: Investigation—Fieldwork, Writing—Review & Editing; W.H.S.: Writing—Review & Editing; D.M.-L.: Methodology, Resources, Writing—Review & Editing, Supervision, Funding acquisition; B.L.M.: Writing—Review & Editing, Supervision; L.N.: Conceptualisation, Methodology, Resources, Writing—Review & Editing, Supervision, Funding acquisition. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, and further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflicts of interests.
Funding Statement
This research was funded by AgriSETA; the Meat Industry Trust; and the University of Pretoria doctorate research bursary. This work was also supported by the Belgian Directorate-General for Development Cooperation (DGD) within the DGD-ITM Framework Agreement 4.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Fournier P.-E., Roux V., Raoult D. Phylogenetic analysis of spotted fever group rickettsiae by study of the outer surface protein rOmpA. Int. J. Syst. Evol. Microbiol. 1998;48:839–849. doi: 10.1099/00207713-48-3-839. [DOI] [PubMed] [Google Scholar]
- 2.Yssouf A., Socolovschi C., Kernif T., Temmam S., Lagadec E., Tortosa P., Parola P. First molecular detection of Rickettsia africae in ticks from the Union of the Comoros. Parasit. Vectors. 2014;7:444. doi: 10.1186/1756-3305-7-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shpynov S., Fournier P.-E., Pozdnichenko N., Gumenuk A., Skiba A. New approaches in the systematics of rickettsiae. New Microbes New Infect. 2018;23:93–102. doi: 10.1016/j.nmni.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fournier P.-E., Zhu Y., Ogata H., Raoult D. Use of highly variable intergenic spacer sequences for multispacer typing of Rickettsia conorii strains. J. Clin. Microbiol. 2004;42:5757–5766. doi: 10.1128/JCM.42.12.5757-5766.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santibáñez S., Portillo A., Ibarra V., Santibáñez P., Metola L., García-García C., Palomar A.M., Cervera-Acedo C., Alba J., Blanco J.R., et al. Epidemiological, clinical, and microbiological characteristics in a large series of patients affected by Dermacentor-Borne-Necrosis-Erythema-Lymphadenopathy from a unique centre from Spain. Pathogens. 2022;11:528. doi: 10.3390/pathogens11050528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDade J.E., Newhouse V.F. Natural history of Rickettsia rickettsii. Annu. Rev. Microbiol. 1986;40:287–309. doi: 10.1146/annurev.mi.40.100186.001443. [DOI] [PubMed] [Google Scholar]
- 7.Parola P., Paddock C.D., Raoult D. Tick-borne rickettsioses around the world: Emerging diseases challenging old concepts. Clin. Microbiol. Rev. 2005;18:719–756. doi: 10.1128/CMR.18.4.719-756.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merhej V., Raoult D. Rickettsial evolution in the light of comparative genomics. Biol. Rev. 2011;86:379–405. doi: 10.1111/j.1469-185X.2010.00151.x. [DOI] [PubMed] [Google Scholar]
- 9.Diop A., Raoult D., Fournier P.-E. Rickettsial genomics and the paradigm of genome reduction associated with increased virulence. Microbes Infect. 2018;20:401–409. doi: 10.1016/j.micinf.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Gillespie J.J., Beier M.S., Rahman M.S., Ammerman N.C., Shallom J.M., Purkayastha A., Sobral B.S., Azad A.F. Plasmids and rickettsial evolution: Insight from Rickettsia felis. PLoS ONE. 2007;2:e266. doi: 10.1371/journal.pone.0000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beliavskaia A., Tan K.K., Sinha A., Husin N.A., Lim F.S., Loong S.K., Bell-Sakyi L., Carlow C.K.S., AbuBakar S., Darby A.C., et al. Metagenomics of culture isolates and insect tissue illuminate the evolution of Wolbachia, Rickettsia and Bartonella symbionts in Ctenocephalides spp. fleas. Microb. Genom. 2023;9:001045. doi: 10.1099/mgen.0.001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly P.J., Beati L., Mason P.R., Matthewman L.A., Roux V., Raoult D. Rickettsia africae sp. nov., the etiological agent of African tick bite fever. Int. J. Syst. Evol. Microbiol. 1996;46:611–614. doi: 10.1099/00207713-46-2-611. [DOI] [PubMed] [Google Scholar]
- 13.Jensenius M., Fournier P.-E., Kelly P., Myrvang B., Raoult D. African tick bite fever. Lancet Infect. Dis. 2003;3:557–564. doi: 10.1016/S1473-3099(03)00739-4. [DOI] [PubMed] [Google Scholar]
- 14.Walker J.B., Olwage A. The tick vectors of Cowdria ruminantium (Ixodoidea, Ixodidae, genus Amblyomma) and their distribution. Onderstepoort J. Vet. Res. 1987;54:353–379. [PubMed] [Google Scholar]
- 15.Petney T.N., Horak I.G., Rechav Y. The ecology of the African vectors of heartwater, with particular reference to Amblyomma hebraeum and Amblyomma variegatum. Onderstepoort J. Vet. Res. 1987;54 [PubMed] [Google Scholar]
- 16.Fabricius J.C. Entomologia Systematica. Impensis CG Proft; Copenhagen, Denmark: 1794. [Google Scholar]
- 17.Eldin C., Mediannikov O., Davoust B., Cabre O., Barré N., Raoult D., Parola P. Emergence of Rickettsia africae, Oceania. Emerg. Infect. Dis. 2011;17:100. doi: 10.3201/eid1701.101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waner T., King R., Keysary A., Eremeeva M.E., Din A.B., Mumcuoglu K.Y., Atiya-Nasagi Y. Rickettsia africae and Candidatus Rickettsia barbariae in ticks in Israel. Am. J. Trop. Med. Hyg. 2014;90:920. doi: 10.4269/ajtmh.13-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogel H., Foley J., Fiorello C.V. Rickettsia africae and novel rickettsial strain in Amblyomma spp. ticks, Nicaragua, 2013. Emerg. Infect. Dis. 2018;24:385. doi: 10.3201/eid2402.161901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fournier P., Roux V., Caumes E., Donzel M., Raoult D. Outbreak of Rickettsia africae infections in participants of an adventure race in South Africa. Clin. Infect. Dis. 1998;27:316–323. doi: 10.1086/514664. [DOI] [PubMed] [Google Scholar]
- 21.Mazhetese E., Magaia V., Taviani E., Neves L., Morar-Leather D. Rickettsia africae: Identifying gaps in the current knowledge on vector-pathogen-host interactions. J. Infect. Dev. Ctries. 2021;15:1039–1047. doi: 10.3855/jidc.13291. [DOI] [PubMed] [Google Scholar]
- 22.Socolovschi C., Huynh T.P., Davoust B., Gomez J., Raoult D., Parola P. Transovarial and trans-stadial transmission of Rickettsiae africae in Amblyomma variegatum ticks. Clin. Microbiol. Infect. 2009;15:317–318. doi: 10.1111/j.1469-0691.2008.02278.x. [DOI] [PubMed] [Google Scholar]
- 23.Mediannikov O., Trape J.-F., Diatta G., Parola P., Fournier P.-E., Raoult D. Rickettsia africae, western Africa. Emerg. Infect. Dis. 2010;16:571. doi: 10.3201/eid1603.090346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pillay A., Manyangadze T., Mukaratirwa S. Prevalence of Rickettsia africae in tick vectors collected from mammalian hosts in sub-Saharan Africa: A systematic review and meta-analysis. Ticks Tick. Borne Dis. 2022;13:101960. doi: 10.1016/j.ttbdis.2022.101960. [DOI] [PubMed] [Google Scholar]
- 25.Zhang E.Y., Kalmath P., Abernathy H.A., Giandomenico D.A., Nolan M.S., Reiskind M.H., Boyce R.M. Rickettsia africae infections in sub-Saharan Africa: A systematic literature review of epidemiological studies and summary of case reports. Trop. Med. Int. Health. 2024;29:541–583. doi: 10.1111/tmi.14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly P.J., Mason P.R., Manning T., Slater S. Role of cattle in the epidemiology of tick-bite fever in Zimbabwe. J. Clin. Microbiol. 1991;29:256–259. doi: 10.1128/jcm.29.2.256-259.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stachurski F. Attachment kinetics of the adult tick Amblyomma variegatum to cattle. Med. Vet. Entomol. 2006;20:317–324. doi: 10.1111/j.1365-2915.2006.00633.x. [DOI] [PubMed] [Google Scholar]
- 28.Matsimbe A.M., Magaia V., Sanches G.S., Neves L., Noormahomed E., Antunes S., Domingos A. Molecular detection of pathogens in ticks infesting cattle in Nampula province, Mozambique. Exp. Appl. Acarol. 2017;73:91–102. doi: 10.1007/s10493-017-0155-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandara S. The Distribution of Amblyomma variegatum and A. hebraeum in Zimbabwe and Their Infection with Ehrlichia ruminantium and Rickettsia africae. University of Pretoria; Pretoria, South Africa: 2018. [Google Scholar]
- 30.Magaia V., Taviani E., Cangi N., Neves L. Molecular detection of Rickettsia africae in Amblyomma ticks collected in cattle from Southern and Central Mozambique. J. Infect. Dev. Ctries. 2020;14:614–622. doi: 10.3855/jidc.11625. [DOI] [PubMed] [Google Scholar]
- 31.Barradas P.F., Mesquita J.R., Ferreira P., Gärtner F., Carvalho M., Inácio E., Chivinda E., Katimba A., Amorim I. Molecular identification and characterization of Rickettsia spp. and other tick-borne pathogens in cattle and their ticks from Huambo, Angola. Ticks Tick. Borne Dis. 2021;12:101583. doi: 10.1016/j.ttbdis.2020.101583. [DOI] [PubMed] [Google Scholar]
- 32.Chitanga S., Chibesa K., Sichibalo K., Mubemba B., Nalubamba K.S., Muleya W., Changula K., Simulundu E. Molecular detection and characterization of Rickettsia species in Ixodid ticks collected from cattle in Southern Zambia. Front. Vet. Sci. 2021;8:684487. doi: 10.3389/fvets.2021.684487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ledger K.J., Innocent H., Lukhele S.M., Dorleans R., Wisely S.M. Entomological risk of African tick-bite fever (Rickettsia africae infection) in Eswatini. PLOS Negl. Trop. Dis. 2022;16:e0010437. doi: 10.1371/journal.pntd.0010437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu Y., Simuunza M., Kajihara M., Ndebe J., Saasa N., Kapila P., Furumoto H., Lau A.C.C., Nakao R., Takada A., et al. Detection of tick-borne bacterial and protozoan pathogens in ticks from the Zambia–Angola border. Pathogens. 2022;11:566. doi: 10.3390/pathogens11050566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phiri B.S.J., Kattner S., Chitimia-Dobler L., Woelfel S., Albanus C., Dobler G., Küpper T. Rickettsia spp. in Ticks of South Luangwa Valley, Eastern Province, Zambia. Microorganisms. 2023;11:167. doi: 10.3390/microorganisms11010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roux V., Raoult D. Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmpB (ompB) Int. J. Syst. Evol. Microbiol. 2000;50:1449–1455. doi: 10.1099/00207713-50-4-1449. [DOI] [PubMed] [Google Scholar]
- 37.Roux V., Rydkγna E., Eremeeva M., Raoult D. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int. J. Syst. Bacteriol. 1997;47:252–261. doi: 10.1099/00207713-47-2-252. [DOI] [PubMed] [Google Scholar]
- 38.Bouyer D.H., Stenos J., Crocquet-Valdes P., Moron C.G., Popov V.L., Zavala-Velazquez J.E., Foil L.D., Stothard D.R., Azad A.F., Walker D.H. Rickettsia felis: Molecular characterization of a new member of the spotted fever group. Int. J. Syst. Evol. Microbiol. 2001;51:339–347. doi: 10.1099/00207713-51-2-339. [DOI] [PubMed] [Google Scholar]
- 39.Blanc G., Ngwamidiba M., Ogata H., Fournier P.-E., Claverie J.-M., Raoult D. Molecular evolution of rickettsia surface antigens: Evidence of positive selection. Mol. Biol. Evol. 2005;22:2073–2083. doi: 10.1093/molbev/msi199. [DOI] [PubMed] [Google Scholar]
- 40.Parola P., Inokuma H., Camicas J.-L., Brouqui P., Raoult D. Detection and identification of spotted fever group Rickettsiae and Ehrlichiae in African ticks. Emerg. Infect. Dis. 2001;7:1014–1018. doi: 10.3201/eid0706.010616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jado I., Escudero R., Gil H., Jiménez-Alonso M.I., Sousa R., García-Pérez A.L., Rodríguez-Vargas M., Lobo B., Anda P. Molecular method for identification of Rickettsia species in clinical and environmental samples. J. Clin. Microbiol. 2006;44:4572–4576. doi: 10.1128/JCM.01227-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fournier P.-E., El Karkouri K., Leroy Q., Robert C., Giumelli B., Renesto P., Socolovschi C., Parola P., Audic S., Raoult D. Analysis of the Rickettsia africae genome reveals that virulence acquisition in Rickettsia species may be explained by genome reduction. BMC Genom. 2009;10:166. doi: 10.1186/1471-2164-10-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fournier P.-E., Dumler J.S., Greub G., Zhang J., Wu Y., Raoult D. Gene sequence-based criteria for identification of new Rickettsia isolates and description of Rickettsia heilongjiangensis sp. nov. J. Clin. Microbiol. 2003;41:5456–5465. doi: 10.1128/JCM.41.12.5456-5465.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iweriebor B.C., Nqoro A., Obi C.L. Rickettsia africae an agent of African tick bite fever in ticks collected from domestic animals in Eastern Cape, South Africa. Pathogens. 2020;9:631. doi: 10.3390/pathogens9080631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimita G., Mutai B., Nyanjom S.G., Wamunyokoli F., Waitumbi J. Phylogenetic variants of Rickettsia africae, and incidental identification of” Candidatus Rickettsia moyalensis” in Kenya. PLoS Negl. Trop. Dis. 2016;10:e0004788. doi: 10.1371/journal.pntd.0004788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halajian A., Palomar A.M., Portillo A., Heyne H., Romero L., Oteo J.A. Detection of zoonotic agents and a new Rickettsia strain in ticks from donkeys from South Africa: Implications for travel medicine. Travel. Med. Infect. Dis. 2018;26:43–50. doi: 10.1016/j.tmaid.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 47.Pillay A.D., Mukaratirwa S. Genetic diversity of Rickettsia africae isolates from Amblyomma hebraeum and blood from cattle in the Eastern Cape province of South Africa. Exp. Appl. Acarol. 2020;82:529–541. doi: 10.1007/s10493-020-00555-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smit A., Mulandane F., Labuschagne M., Wójick S.H., Malabwa C., Sili G., Mandara S., Dlamkile Z., Stoltsz W.H., Vineer H.R., et al. Intra-and interspecific variation of Amblyomma ticks from southern Africa. Parasites Vectors. :2024. in press. [Google Scholar]
- 49.Dlamkile Z., Neves L., Morar-Leather D., Brandt C., Pretorius A., Steyn H., Liebenberg J. Characterisation of South African field Ehrlichia ruminantium using multilocus sequence typing. Onderstepoort J. Vet. Res. 2023;90:1–8. doi: 10.4102/ojvr.v90i1.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker A., Bouattour A., Camicas J., Estrada-Pena A., Horak I., Latif A., Pegram R., Preston P. Ticks of domestic animals in Africa: A Guide to Identification of Species. Bioscience Reports; Edinburgh, UK: 2003. [Google Scholar]
- 51.Voltzit O., Keirans J. A review of African Amblyomma species (Acari, Ixodida, Ixodidae) Acarina. 2003;11:135–214. [Google Scholar]
- 52.Smit A., Mulandane F.C., Wojcik S.H., Horak I.G., Makepeace B.L., Morar-Leather D., Neves L. Sympatry of Amblyomma eburneum and Amblyomma variegatum on African buffaloes and prevalence of pathogens in ticks. Ticks Tick. Borne Dis. 2023;14:102247. doi: 10.1016/j.ttbdis.2023.102247. [DOI] [PubMed] [Google Scholar]
- 53.Beati L., Keirans J.E. Analysis of the systematic relationships among ticks of the genera Rhipicephalus and Boophilus (Acari: Ixodidae) based on mitochondrial 12S ribosomal DNA gene sequences and morphological characters. J. Parasitol. 2001;87:32–48. doi: 10.1645/0022-3395(2001)087[0032:AOTSRA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 54.Black W.C., Piesman J. Phylogeny of hard-and soft-tick taxa (Acari: Ixodida) based on mitochondrial 16S rDNA sequences. Proc. Natl. Acad. Sci. USA. 1994;91:10034–10038. doi: 10.1073/pnas.91.21.10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 56.Simon C., Frati F., Beckenbach A., Crespi B., Liu H., Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. SocAm. 1994;87:651–701. doi: 10.1093/aesa/87.6.651. [DOI] [Google Scholar]
- 57.Cannon R., Roe R. Livestock Disease Surveys: A Field Manual for Veterinarians. Australia Australian Gvt Publishing Service; Canberra, Australia: 1982. [Google Scholar]
- 58.Mazhetese E., Lukanji Z., Byaruhanga C., Neves L., Morar-Leather D. Rickettsia africae infection rates and transovarial transmission in Amblyomma hebraeum ticks in Mnisi, Bushbuckridge, South Africa. Exp. Appl. Acarol. 2022;86:407–418. doi: 10.1007/s10493-022-00696-w. [DOI] [PubMed] [Google Scholar]
- 59.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 61.Rambaut A., Suchard M., Drummond A. Tracer. University of Edinburgh; Edinburgh, UK: 2014. version 1.6.0. [Google Scholar]
- 62.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gerstäcker A. Gliderthiere (Insekten, Arachniden, Myriapoden und Isopoden) C.J. Winter’sche Verlaghandlung; Leipzig, Germany: Heidelberg, Germany: 1873. [Google Scholar]
- 64.Dönitz W. Über das Zeckengenus Amblyomma (On the Tick Genus Amblyomma) Sitzungsber Ges Naturforsch Freunde Berl. 1909;8:440–482. [Google Scholar]
- 65.Abdel-Shafy S., Allam N.A., Mediannikov O., Parola P., Raoult D. Molecular detection of spotted fever group rickettsiae associated with ixodid ticks in Egypt. Vector Borne Zoonotic Dis. 2012;12:346–359. doi: 10.1089/vbz.2010.0241. [DOI] [PubMed] [Google Scholar]
- 66.Orkun Ö., Karaer Z., Çakmak A., Nalbantoğlu S. Spotted fever group rickettsiae in ticks in Turkey. Ticks Tick. Borne Dis. 2014;5:213–218. doi: 10.1016/j.ttbdis.2012.11.018. [DOI] [PubMed] [Google Scholar]
- 67.Pillay A., Nyangiwe N., Mukaratirwa S. Low genetic diversity and population structuring of Amblyomma hebraeum and Rickettsia africae from coastal and inland regions in the Eastern Cape Province of South Africa. Med. Vet. Entomol. 2023;37:275–285. doi: 10.1111/mve.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thekisoe O., Ramatla T., Ringo A., Mnisi S., Mphuthi N., Mofokeng L., Lekota K., Xuan X. Molecular detection of Rickettsia africae from Amblyomma hebraeum ticks in Mafikeng city of North West Province, South Africa. Res. Vet. Sci. 2023;164:105027. doi: 10.1016/j.rvsc.2023.105027. [DOI] [PubMed] [Google Scholar]
- 69.Gueye A., Mbengue M., Diouf A. Epidémiologie de la cowdriose au Sénégal. I. Etude de la transmission et du taux d’infection d’Amblyomma variegatum (Fabricius, 1794) dans la région des Niayes. Rev. Elev. Méd. Vét. Pays Trop. 1993;46:441–447. doi: 10.19182/remvt.9440. [DOI] [PubMed] [Google Scholar]
- 70.Warnecke M., Schein E., Voigt W., Uilenberg G., Young A. Development of Theileria mutans (Theiler, 1906) in the gut and the haemolymph of the tick Amblyomma variegatum (Fabricius, 1794) Z Parasitenkd. 1980;62:119–125. doi: 10.1007/BF00927858. [DOI] [PubMed] [Google Scholar]
- 71.Diobo F.N., Yao P.K., Diaha A.C., Adjogoua V.E., Faye H.K. Detection of Rickettsia africae in ticks and cattle in Côte d’Ivoire by real-time PCR. J. Appl. Biosci. 2021;166:17242–17251. [Google Scholar]
- 72.Thu M.J., Qiu Y., Matsuno K., Kajihara M., Mori-Kajihara A., Omori R., Monma N., Chiba K., Seto J., Gokuden M., et al. Diversity of spotted fever group rickettsiae and their association with host ticks in Japan. Sci. Rep. 2019;9:1500. doi: 10.1038/s41598-018-37836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Macaluso K.R., Sonenshine D.E., Ceraul S.M., Azad A.F. Rickettsial infection in Dermacentor variabilis (Acari: Ixodidae) inhibits transovarial transmission of a second Rickettsia. J. Med. Entomol. 2002;39:809–813. doi: 10.1603/0022-2585-39.6.809. [DOI] [PubMed] [Google Scholar]
- 74.Budachetri K., Kumar D., Crispell G., Beck C., Dasch G., Karim S. The tick endosymbiont Candidatus Midichloria mitochondrii and selenoproteins are essential for the growth of Rickettsia parkeri in the Gulf Coast tick vector. Microbiome. 2018;6:141. doi: 10.1186/s40168-018-0524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bonnet S.I., Pollet T. Update on the intricate tango between tick microbiomes and tick-borne pathogens. Parasite Immunol. 2021;43:e12813. doi: 10.1111/pim.12813. [DOI] [PubMed] [Google Scholar]
- 76.Ponnusamy L., Gonzalez A., Van Treuren W., Weiss S., Parobek C.M., Juliano J.J., Knight R., Roe R.M., Apperson C.S., Meshnick S.R. Diversity of Rickettsiales in the microbiome of the lone star tick, Amblyomma americanum. Appl. Environ. Microbiol. 2014;80:354–359. doi: 10.1128/AEM.02987-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mutai B.K., Wainaina J.M., Magiri C.G., Nganga J.K., Ithondeka P.M., Njagi O.N., Jiang J., Richards A.L., Waitumbi J.N. Zoonotic surveillance for rickettsiae in domestic animals in Kenya. Vector Borne Zoonotic Dis. 2013;13:360–366. doi: 10.1089/vbz.2012.0977. [DOI] [PubMed] [Google Scholar]
- 78.Kisten D., Brinkerhoff J., Tshilwane S.I., Mukaratirwa S. A pilot study on the microbiome of amblyomma hebraeum tick stages infected and non-infected with rickettsia africae. Pathogens. 2021;10:941. doi: 10.3390/pathogens10080941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Palomar A.M., Molina I., Bocanegra C., Portillo A., Salvador F., Moreno M., Oteo J.A. Old zoonotic agents and novel variants of tick-borne microorganisms from Benguela (Angola), July 2017. Parasites Vectors. 2022;15:140. doi: 10.1186/s13071-022-05238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Halajian A., Palomar A.M., Portillo A., Heyne H., Luus-Powell W.J., Oteo J.A. Investigation of Rickettsia, Coxiella burnetii and Bartonella in ticks from animals in South Africa. Ticks Tick. Borne Dis. 2016;7:361–366. doi: 10.1016/j.ttbdis.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 81.Jongejan F., Berger L., Busser S., Deetman I., Jochems M., Leenders T., de Sitter B., van der Steen F., Wentzel J., Stoltsz H. Amblyomma hebraeum is the predominant tick species on goats in the Mnisi Community Area of Mpumalanga Province South Africa and is co-infected with Ehrlichia ruminantium and Rickettsia africae. Parasit. Vectors. 2020;13:172. doi: 10.1186/s13071-020-04059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Silva-Ramos C.R., Faccini-Martínez Á.A. Clinical, epidemiological, and laboratory features of Rickettsia africae infection, African tick-bite fever: A systematic review. Infez. Med. 2021;29:366. doi: 10.53854/liim-2903-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Onyiche T.E., Labruna M.B., Saito T.B. Unraveling the epidemiological relationship between ticks and rickettsial infection in Africa. Front. Trop. Dis. 2022;3:952024. doi: 10.3389/fitd.2022.952024. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, and further inquiries can be directed to the corresponding authors.