Abstract
Ovarian cancer is a malignant tumor of ovary. It has the characteristics of difficult early diagnosis, poor late curative effect and high recurrence rate. It is the biggest disease that seriously threatens women’s health. Single cell sequencing technology refers to sequencing the genetic information carried by it at the single cell level to obtain the gene sequence, transcript, protein and epigenetic expression profile information of a certain cell type and conduct integrated analysis. It has unique advantages in the study of tumor occurrence and evolution, and can provide new methods for the study of ovarian cancer. This paper reviews the single cell sequencing technology and its application in ovarian cancer.
Keywords: Single cell sequencing, Ovarian cancer, Heterogeneity
Introduction
The incidence rate of ovarian cancer ranks the third in gynecological malignancies, accounting for 23% of all female genital tract tumors. It can occur at any age and has a rising trend year by year. At the time of treatment, 60%-70% of patients are in advanced stage (Shao and Chen 2013), and the 5-year survival rate is lower than 45% (Christine et al. 2019). The mortality rate is the highest among gynecological cancers. Ovarian cancer is a disease characterized by heterogeneity of histocytes. The sequence and expression information at the level of single cell is of great significance for studying the development and prognosis of diseases (Gao et al. 2020). Single cell sequencing technology provides a high resolution molecular phenotype identification method for a large number of single cancer cells, which can indicate the intratumoral heterogeneity of ovarian cancer, and is helpful for the diagnosis and treatment of diseases (Lovett, 2013).
Single cell sequencing technology
Single cell sequencing technology is a technology to sequence its genetic information at the single cell level, including the steps of single cell separation, dissolution, amplification and sequencing.
Single cell separation
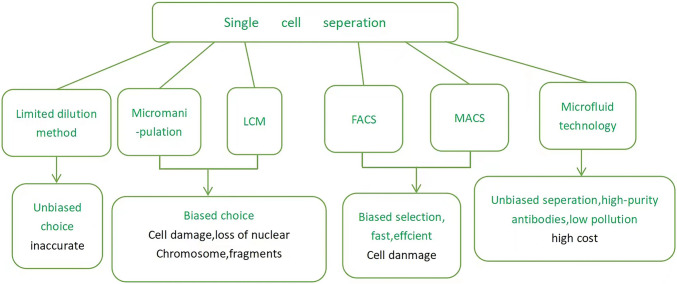
There are many methods for single cell separation, including limited dilution method, micromanipulation, laser capture microdissection, fluorescence activated cell sorting, magnetic activated cell sorting, microfluidic technology, etc. (Wang et al. 2020). Different methods have different application scope, results, advantages and disadvantages. Limited dilution method, that is, dilute the cell group with multiple to achieve separation effect (Hornburg et al., 2021). The micromanipulation method is to find the target cell under the microscope and suck it out with a micropipette (Zhang, 2022). Laser capture microdissection(LCM), is to use laser to melt the thermoplastic film covering the section under microscope imaging, and fix specific cell groups to obtain target cells (Zhang et al., 2018). Fluorescent activated cell sorting technology is to divide a cell group into multiple subpopulations according to specific fluorescent markers to purify and obtain target cells (Wang and Meng, 2020). Magnetic activated cell sorting method uses monoclonal antibodies that bind to magnetic particles to label cells and separate them from non-magnetic cells under the action of external magnetic field to achieve the purpose of separation and purification (Cui et al. 2021). Microfluid technology is a technology to separate cells through micro channels (Yu et al. 2022). For example, fluorescent activated cell sorting and magnetic activated cell sorting, as high-throughput technologies, can make biased selection according to the size and shape of cells or the expression of cell surface markers, which is fast and efficient, but also easy to damage cells (Sheng and Zong 2019). Microfluid technology can achieve unbiased separation and obtain high-purity antibodies with low pollution but high cost (Sheng and Zong 2019). As low flux technologies, micromanipulation and laser capture microdissection are biased choices based on cell morphology or fluorescence report gene expression, but they are easy to cause cell damage and loss of nuclear chromosome fragments (Sheng and Zong 2019). Limited dilution is an unbiased choice, simple but inaccurate (Liang 2019) (Fig. 1).
Fig. 1.
The method of single cell seperation. The single cell separation method and its advantages and disadvantages are shown in the figure, with green indicating advantages and black indicating disadvantages
Single cell dissolution
Cytolysis methods include physical method, chemical method and biological enzyme method to obtain genetic material. ①Physical methods are divided into heat shock and ultrasonic methods: Heat shock, namely repeated freeze–thaw method, is composed of freezing and thawing (Liu et al. 2020), which is more gentle than chemical method. Ultrasonic treatment is to heat cells with ultrasound (Bruno et al. 2021), but it is easy to cause DNA breakage. ②Chemical method:It is mainly the treatment method of lysis solution. By using lysis solution, cells can be broken and proteins can be dissolved. ③Biological enzyme method:Lysozyme or protease is used to dissolve cells. When extracting nucleic acid, chemical method and biological enzyme method should be used together.
Amplification of genetic material
Single cell whole genome amplification
Whole genome amplification is a non selective amplification of all genome sequences, aiming to significantly increase the total amount of DNA without sequence orientation (Wang and Yuan 2015). The most commonly used technique is multiple displacement amplification (MDA), that is, chain displacement amplification reaction is conducted with phi29DNA polymerase and hexamer with strong template binding under constant temperature (Huang et al. 2020). This technique is simple to operate, with long DNA fragments, low error rate,high efficiency and good DNA amplification coverage (Cai et al. 2021). However, if the amount of starting templates is low, it will lead to large amplification deviation. In addition,there are also several methods——①Degenerate oligonucleotide primer PCR technology (DOP-PCR) (Xu et al. 2019): First, use a lower temperature (≤ 25℃) for annealing, and then slowly raise the temperature to the primer extension temperature for extension. After several cycles, use a higher annealing temperature (≤ 55℃) for multi cycle conventional PCR reaction. Although simple, fast and cheap, the coverage rate is low and the loss rate is high (Sun et al., 2022). In recent years, the Russian Academy of Sciences() has updated this technology and formed iDOP PCR, namely the improved DOP PCR, which can achieve efficient amplification of low copy genomic DNA. Angie (Angie et al. 2016) et al. applied this technology to improve the STR map of human bone remains and blood stains. ②Ligation-mediated PCR (LM-PCR): The PCR reaction with the participation of the connector connection process is sensitive and high in yield, but the operation is cumbersome and the template DNA is easily lost. Ferrarini (Ferrarini et al. 2018) and others analyzed the single circulating tumor cells and white blood cells isolated from the blood of patients with prostate cancer or lung adenocarcinoma by simplifying the technology, and successfully conducted low flux sequencing. ③Primer extension pre-ampification (PEP) (Telenius et al. 1992): First, the primer was annealed at 37 ℃, then slowly heated to 55 ℃ for a long time, repeated multiple cycles, simple operation, but low yield and high error rate. ④Primer-based whole genome amplification (pWGA): The primer was synthesized on the template using phage T7gp4 primer enzyme at 37 ℃, and was amplified at constant temperature. The yield was high, but the fidelity was poor. ⑤Multiple annealing and looping-based amplification cycles (MALBAC) (Zhang et al. 2019): Academician Xie Xiaoliang studied in 2012 and found that the key is to add short special DNA molecules as primers, which are evenly amplified and have high coverage (Blagodatskikh et al., 2017), but are prone to amplification deviation when the copy number is very low,leading to false positive (Zhang et al., 2023). Yaxin Yao (Yao et al. 2018) and others applied this technology to assisted reproduction, hoping to achieve non-invasive free DNA detection instead of invasive embryo biopsy, and comprehensively analyze the quality of embryos from a more microscopic perspective, so as to better achieve eugenics and eugenics.The researchers used MALBAC technology to amplify sperm, and found that the reduction of recombination rate near the gene region was determined by molecular mechanism and was the result of unnatural selection. In addition, (Li et al. 2018) et al. found that hyperinsulinemia and hyperandrogenism may change some DNA methylation status in oocytes through single cell whole genome bisulfite sequencing (SC-WGBS), thus leading to the occurrence of polycystic ovary syndrome (PCOS).
Single cell transcriptome amplification
Transcriptome amplification, that is, reverse transcription of mRNA into cDNA through PCR. At present, Smart-seq and Smart-seq2 technologies are commonly used (Zong et al. 2012). Smart-seq technology uses poly T bases as downstream primers to start reverse transcription, uses reverse transcriptase SMART scribe RTase to extend several C bases beyond the template at the end of transcription, and transcribes three G base upstream primers of the cDNA band for amplification. Smart-seq2 technology is to use the chain replacement and continuous synthesis characteristics of Phi29DNA polymerase to efficiently amplify cyclized cDNA. In addition to the above two technologies, there also have some technologies:——①CEL-Seq2: The process is ploy T capture, in vitro transcription, RNA interruption, reverse transcription, and sequencing. It is an amplification technology using linear RNA (Li et al. 2019). ②Drop-Seq: Based on microfluidic technology, a single cell is encapsulated in a droplet for analysis, with low cost and high throughput (Yanai and Hashimshony 2019). ③MATQ-seq:It is a quantitative single-cell total RNA sequence sequencing method using multiple annealing and dC tailing, with high sensitivity (Sukhbaatar et al. , 2017; Sheng et al. 2017).
Single cell sequencing
Single cell transcriptome sequencing
Single cell transcriptome sequencing is a new technology to sequence the transcriptome at the single cell level. It can study the gene expression in a single cell, and solve the problem of cell heterogeneity that cannot be solved by tissue sample sequencing (Orr and Edwards 2018; Cao et al. 2021; Gawad et al. 2016). It is especially suitable for stem cells, tumor cells and cell populations in early embryonic development with high heterogeneity, and it can be used in diseases, immunity, tumors, tissues, organs Plays an important role in the study of development. Zhang Baoyue (Bageritz and Raddi , 2019) et al. used microfluidic devices to reduce the loss of cells and improve the reverse transcription efficiency, thus improving the sequencing efficiency accordingly, especially when the number of cells in the sample is limited. Li Ting (Zhang et al. 2019) et al. using transcriptome sequencing technology, it was found that intrauterine hypoxia caused changes in gene expression of rat offspring oocytes, which affected the metabolic process of lipid and insulin, and may be related to metabolic diseases.
Single cell whole genome sequencing
The principle of single cell whole genome sequencing technology is to amplify a small amount of whole genome DNA of isolated single cells, obtain a complete genome with high coverage, and then conduct high-throughput sequencing to reveal the difference of cell population and the relationship between cell evolution (Stuart and Satija 2019; Quainoo et al. 2017; Buermans and Dunnen 2014; Liu et al. 2019). Academician Xie Xiaoliang used this technology to sequence sperm in 2012, and found that 5% of the sperm genome is aneuploid, which is of great significance in the study of male infertility (Picelli et al., 2014). (Li et al. 2019) et al. used single cell whole genome sequencing to find that the copy number variation of sub giant alkali basal cells occurred in the development of cerebral cortical nerves. YANG (Rohrback et al. 2018) et al. Through the whole genome sequencing, it was found that the tumor cells integrating HPV in POU5F1B still survived after radiotherapy, and had different characteristics before and after radiotherapy. The minor cell subsets before radiotherapy became the major cell subsets after radiotherapy. (Yang et al. 2022) et al. developed direct nuclear labeling and RNA sequencing (DNTR seq), which can identify secondary subclones of leukemia patients, and can also link single cells with large-scale genomes to analyze heterogeneous tumors and other complex tissues. For example, Li (Zachariadis et al., 2020) et al. sequenced the whole genome of gallbladder small cell neuroendocrine carcinoma and determined new biomarkers, which are of great value for the prognosis and treatment of patients with lymphatic metastasis. CRISPR/Cas9 system designed an sgRNA library and transferred it to target cells to screen for anti-cancer drugs,which were used to identify whole genomes for gene function loss. It was found that targeted silencing of related genes reduced the vitality of cancer cells,providing a method for discovering potential therapeutic targets (Silva et al. 2011; Prince et al. 2019).
Ovarian cancer
Ovarian cancer is a malignant tumor derived from the germinal epithelium of the ovary. In the early stage, it is often asymptomatic. In the late stage, the main symptoms are abdominal distension, abdominal mass, ascites, and other gastrointestinal symptoms (Chen and Meng 2021). Functional tumors may have irregular vaginal bleeding or postmenopausal bleeding.According to the pathological and molecular genetic characteristics, epithelial ovarian cancer can be divided into type I and type II. Type I is mostly stage I with good prognosis, including low-grade serous carcinoma, low-grade endometrioid carcinoma, mucinous carcinoma and clear cell carcinoma (Zhang and Gu , 2020). Type II mostly showed progressive stage with poor prognosis, mainly including high-grade serous carcinoma and high-grade endometrioid carcinoma (Zhang and Gu , 2020). At present, it is believed that the histological origin of epithelial ovarian cancer is diverse:High grade serous carcinoma of the ovary may occur after the formation of intraepithelial carcinoma of the fallopian tube falls off and is planted on the surface of the ovary (Scott et al., 2022). Low grade serous carcinoma may also occur after the normal fallopian tube epithelium falls off to the surface of the ovary and invaginates to form an inclusion cyst. Endometriosis may be the histological source of clear cell carcinoma of ovary, endometrioid carcinoma and seromucinous carcinoma (McGranahan and Swanton , 2017).
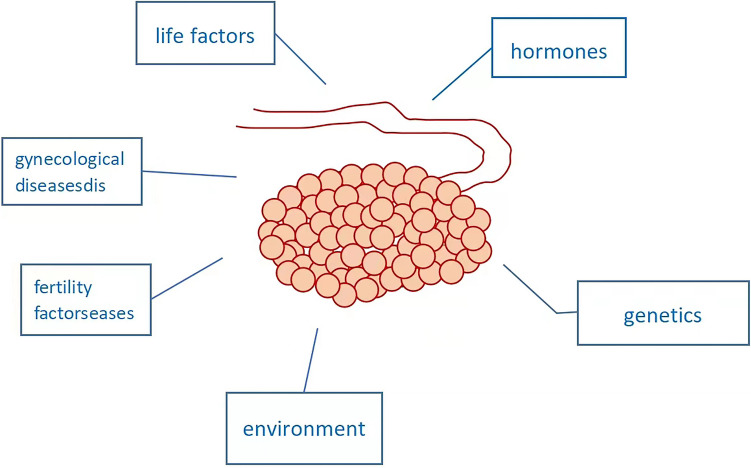
The etiology of ovarian cancer is still unclear, which may be related to genetics, hormones, gynecological diseases, fertility factors, environment and life factors (Pan 2012) (Fig. 2). Women with BRCA1 or BRCA2 mutations are 10–40 times more likely to develop ovarian cancer, and the onset age is often earlier (Zha , 2022). Studies have shown that about 50% of high-grade serous ovarian cancer has BRCA1 or BRCA2 gene mutations (Li et al., 2016). The serum CA125 level in 80% of patients increased, but nearly half of the early cases did not (Vasan et al. 2019). Therefore, it is not only used for early diagnosis, but more used for disease monitoring and efficacy evaluation. Yang Jinfang (Kroeger and Drapkin 2017) et al. used salivary mRNA biomarkers and serum carcinoembryonic antigen (CEA) in the detection of ovarian cancer for the first time. Through the control experiment on 60 healthy people and 60 patients with ovarian cancer, it was verified that the combination of the two can improve the detection accuracy, with the sensitivity of 85% and specificity of 88.3%. Huang Min (Yang et al., 2021) et al. found that the combination of ITIH4 and hyaluronic acid can stabilize the ECM and inhibit the invasion and metastasis of ovarian cancer cells. The decreased expression of ITIH4 makes the ECM easy to be destroyed by hydrolysis, thus promoting the growth, invasion and metastasis of cancer cells. At the same time, the m/z 3272 fragment of ITIH4 is up-regulated in ovarian cancer (Zhang et al., 2021), which may be a new marker for the diagnosis of ovarian cancer. Li Mojuan (Huang et al. 2019) et al. found that Ezrin may play a promoter role in ovarian cancer, and its up-regulation can promote the growth, metastasis and invasion of SKOV3 and CaOV3 in cancer cell lines, which may be a new biological marker for the diagnosis of ovarian cancer in the future and can be used for the evaluation of prognosis.
Fig. 2.
Possible causes of ovarian cancer
The principle of primary treatment for ovarian cancer is surgery, supplemented by radiotherapy, chemotherapy and other comprehensive treatment (Li et al. 2021). The thoroughness of primary surgery is closely related to the prognosis. If the treatment is inappropriate in the early stage, the recurrence rate can reach about 20%, and the recurrence rate of ovarian cancer after the initial treatment is 70–85% (Duan et al. 2018). The recurrence is usually seen in 2–3 years after surgery. Once the recurrence occurs, the prognosis is poor. 18F-FDG PET/CT combined with CA125 plays an important role in the diagnosis of postoperative recurrence and metastasis of ovarian cancer. The recurrent and metastatic lesions showed elevated 18F-FDG uptake, and PET/CT showed hypermetabolic lesions (Li et al. 2021; Vistad et al. 2017). Except for stage IA and IB, mucinous carcinoma or low-grade serous carcinoma and endometrioid carcinoma that have undergone comprehensive staging surgery, other patients need chemotherapy. Common chemotherapy drugs include cisplatin, carboplatin, paclitaxel, cyclophosphamide, etc. Platinum combined with paclitaxel is the “gold standard” first-line chemotherapy scheme (Paola et al. 2016) 0.80% of the patients responded well to the platinum based initial treatment (Xu and Xu 2022), but there are still patients who have multiple recurrences (Liu et al. 2020), and the interval between recurrences is getting shorter and shorter, eventually leading to death, and most of these patients died of platinum resistant tumors. Studies have shown that AT rich interaction domain 1A (ARID1A) leads to multiple drug resistance of ovarian cancer by up regulating the expression of multidrug resistance related protein 2 (MRP2) (Li et al. 2017). This discovery provides a new method to solve the problem of chemotherapy resistance of ovarian cancer.
4 Application of single cell sequencing technology in ovarian cancer
Single cell sequencing technology is widely used in the study of tumor cell heterogeneity, the discovery of mutation sites, the discovery of tumor cell clonal evolution and the discovery of related new markers, which contributes to the precise and personalized treatment of tumors. Single cell sequencing has played a great role in colorectal cancer (Li et al., 2022), gastric cancer (Willems et al., 2023), liver cancer (Huang et al., 2022), lung cancer (Tian and Li , 2022), breast cancer (Yu et al. 2021) and other cancer research. The occurrence of ovarian cancer is caused by continuous DNA changes in a single cell or a single cell clone, which leads to abnormal cell development and loss of normal function (Shih and Kurman 2004). According to the characteristics of molecular genetics, ovarian cancer is mainly divided into two types. Type I tumors are characterized by KRAS, BRAF, PIK3CA, CTNNB1 and PTEN gene mutations and high-frequency microsatellite instability. Type II is characterized by p53 gene mutation. The information generated from RNA level analysis is helpful to understand the pathogenesis of ovarian cancer, discover new biomarkers and therapeutic targets, and further contribute to the development of new drugs (Luo et al. 2018).
Revealing the mechanism of tumor drug resistance
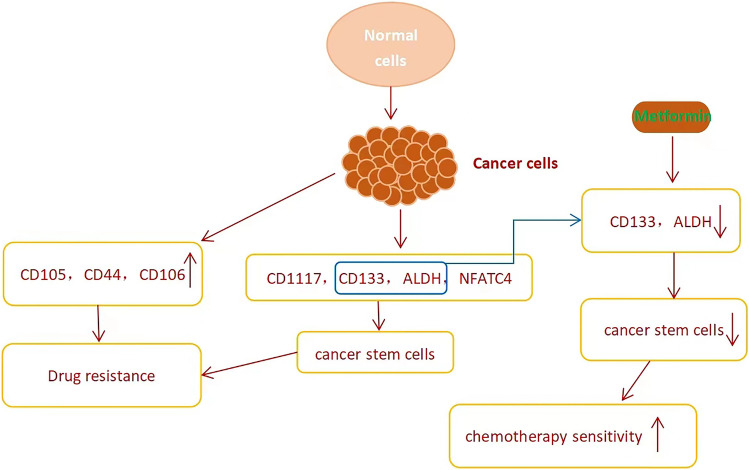
Tumor drug resistance refers to the characteristics that some tumor cell genes are mutated after the tumor is alleviated by anticancer drugs, making the tumor cells no longer sensitive to the anticancer drugs. Chemotherapy did not significantly improve the overall survival rate of patients with ovarian cancer, mainly because the cancer cells had tolerance to chemotherapy drugs. Drug resistance of tumor is the main factor limiting the cure of cancer (Etiology 2021). Tian Min (Gov et al. 2017) et al. analyzed AXL, a member of TYRO3-AXL-MER (TAM) receptor tyrosine kinase (RTK) family, through KEGG, and found that its expression in drug-resistant ovarian cancer cell A2780/DDP was significantly higher than that in the parent, which suggested that AXL mediated glycolysis was related to the resistance of ovarian cancer to cisplatin. Zhang (Tian et al. 2020) et al. found that the high expression of CD105, CD44 or CD106 is related to tumor drug resistance. These three may be used as surface markers and therapeutic targets of potential stem cell related cells in ovarian cancer, and also have some value in judging the prognosis.Bhardwaj (Zhang et al. 2019) et al. found that the differential expression of lncRNAs in ovarian cancer was related to the invasion, metastasis and drug resistance of cancer cells. It is reported that lncRNAs have the potential to treat cancer (Cowan et al. 2016; Li et al. 2015; Akrami et al. 2013). Its expression disorder can promote the invasion and metastasis of cancer cells and increase drug resistance. More and more studies have shown that tumor stem cells are associated with drug resistance and tumor recurrence. Therefore, eliminating tumor stem cells may help increase chemotherapy sensitivity (Shen et al. 2021; Chae and Kim 2018; Clevers 2011; Merlos-Suarez et al. 2011). The surface markers that can be used to identify ovarian cancer stem cells include CD117 (Yang et al. 2022), CD133 (Keyvani et al., 2019), ALDH (Baba et al. 2009), and nuclear factor of activated T cells, cytoplasmic 4 (NFATC4) (Nwani et al., 2019; Cole et al. 2020). Metformin can reduce tumor stem cells by reducing ALDH + CD133 + CSCs like cells in ovarian cancer (Hermann et al. 2007; Brown et al. 2020), thereby increasing chemotherapy sensitivity (Fig. 3). Kurat et al. used a whole genome CRISPR library to identify genes for acquired resistance to cytarabine(Ara-C) and discovered a new source of Ara-C resistance in acute myeloid leukemia(AML) (Yang and Zhang 2023).
Fig. 3.
The high expression of CD105,CD44,CD106,CD117,CD133,ALDH,and NFATC4 is related to tumor drug resistance. So do cancer stem cells. Metformin can reduce cancer stem cells by reducing CD133 and ALDH,thereby increasing chemotherapy sensitivity
Characterize tumor heterogeneity
Tumor heterogeneity refers to the changes in molecular biology or genes in the progeny cells of a tumor after multiple divisions and proliferation during its growth process, resulting in differences in tumor growth rate, invasion ability, drug sensitivity, prognosis and other aspects (Wright et al., 2017; Baslan and Hicks 2017). Due to the existence of tumor heterogeneity, different subgroups have different sensitivity to chemotherapy drugs. Sensitive subgroups gradually disappear under the action of drugs, while insensitive subgroups continue to multiply, leading to tumor recurrence or metastasis (Prensner et al. 2011). At the same time, the lack of quantity and quality of tumor blood vessels, the change of hydrostatic pressure and microenvironment make it impossible for drugs to completely enter some cancer cells, making the tumor gradually evolve into drug resistant type (Bai and Cui 2020). In addition, tumor microenvironment can also fully protect cancer cells by secreting factors, so heterogeneous microenvironment is the key to tumor therapy. Li Guiming (Bai et al., 2021) et al. sequenced single-cell RNA from colorectal cancer samples and found that the proliferative activity of effector T cells in young colorectal cancer (yCRC) was significantly lower than that in old colorectal cancer (oCRC). There is more MIF expression in yCRC, which causes B cell aggregation through the MIF-CXCR4 pathway, leading to stronger inhibition of yCRC on the formation of immune microenvironment than oCRC, which can explain why yCRC is more aggressive than oCRC. Ovarian cancer is highly heterogeneous. The higher the heterogeneity, the worse the prognosis. At the same time, it is also because of its high heterogeneity, leading to the occurrence of drug resistance and treatment failure (Baslan and Hicks 2017). Single cell sequencing technology provides a method to characterize the heterogeneity of ovarian cancer, and provides an opportunity to determine key molecular characteristics, prognosis, and therapeutic response (Bhardwaj et al. 2021; DuFort et al. 2016; Armingol et al. 2020). According to the characteristics of molecular genetics, ovarian cancer is mainly divided into two types. Type I tumors are characterized by KRAS, BRAF, PIK3CA, CTNNB1 and PTEN gene mutations and high-frequency microsatellite instability.Type II is characterized by p53 gene mutation. Benjamin et al. (Li and Sheng 2016) carried out single-cell RNA sequencing on 22 ascites samples, and found that tumor fibroblasts expressed more diverse in non-malignant cells. JAK/STAT signal pathway is the main pathway to regulate cytokines (Izar et al. 2020). The activation of this pathway promoted the occurrence of tumor drug resistance. In recent years, JAK inhibitors have played an important role in cancer treatment. Guo Jiapei et al. (Xin et al. 2020) found that artemisinin can induce hepatocyte apoptosis by inhibiting JAK/STAT3 pathway. Khalafallah (Boris et al. 2018) et al. had a detailed understanding of the heterogeneity in glioblastoma through single cell RNA sequencing. It is reported that the heterogeneity of glioblastoma mainly involves three signal pathways: RTK/RAS/PI3K, P53/MDM2/MDM4 and RB/CDK4/INK4A (Guo et al. 2017), which provided a new idea for the treatment and improvement of the prognosis of the disease.
Assist in disease treatment
In addition to the above applications, single cell sequencing technology can also be used to identify new targets and therapeutic approaches for ovarian cancer, thereby changing its therapeutic mode (Khalafallah et al., 2020). Hu (Talukdar et al., 2021) et al. first characterized human oviduct epithelial tissue through the deep single cell RNA sequencing technology Smart Seq2, found and verified six new cell subtypes, found that the proportion of EMT (epithelial mesenchymal transformation) cell status in the “decomposition” analysis was significantly negatively correlated with the prognosis, and defined an EMT high subtype with poor prognosis. This will enlighten the prognosis analysis of ovarian cancer and the development of treatment methods for EMT high subtype tumors. Liu (Hu et al., 2020) et al. found that the expression of MYH9 is related to tumor stage, metastasis, residue, prognosis and recurrence. Overexpression of this gene is an independent predictor of poor prognosis of ovarian cancer and may become a target for future ovarian cancer treatment. Some studies have confirmed that knockout of MYH9 gene can pass Wnt/ β- Catenin pathway inhibits the proliferation, migration and invasion of ovarian cancer cells (Cameron et al. 2014). Ying Chen (Liu et al. , 2019) et al. found that the expression of non coding RNA PVT1 was up-regulated in ovarian cancer, and was related to the advanced stage of patients, indicating a poor prognosis. At the same time, its interaction with a histone lysine N-methyltransferase EZH2 could inhibit the expression of miR-214 in cancer cells, thus further promoting cancer progress. Hornburg (Chen et al., 2018) et al. analyzed the tumor cells of 42 patients with ovarian cancer by single cell RNA sequencing and CD8 IHC, and found that the dysfunction of CD8 + GZMB cells was later than that of CD8 + GZMK T cells,which provided ideas for improving immunotherapy of ovarian cancer.
Conclusion and prospect
Single cell sequencing technology provides a new method for the research of ovarian cancer, and provides great help for people to overcome various problems in the diagnosis and treatment of ovarian cancer. Although it is only a few years since its birth, it has made rapid development and remarkable achievements. It not only plays an important role in the development of human medicine, but also plays a great role in promoting animal medicine and plant medicine. However,there are certain limitations to the current use of scRNA-seq in clinical research:①The difficulty in obtaining human histological specimen. ②Extensive data analysis and interpretation require interdisciplinary collaboration among experts from multiple fields.③Expensive and time-conmusing. Although there are still many deficiencies in single cell sequencing technology, these deficiencies will be gradually improved in the future with the development of science and technology and the continuous improvement of people's understanding. And single cell sequencing technology will play a greater role in tumor, microbiology, neuroscience, immunology and other research fields.
Acknowledgements
Not applicable.
Authors’ contributions
QY drafted and revised the manuscript. JT designed and supervised the study. QC,SS,YW and NL designed the figure. All authors have read and approved the final manuscript.
Funding
The role and mechanism of LncRNA CACNA1G-AS1 in ovarian cancer.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Akrami R, Jacobsen A, Hoell J, Schultz N, Sander C, Larsson E (2013) Comprehensive analysis of long non-coding RNAs in ovarian cancer reveals global patterns and targeted DNA amplification. PLoS ONE 8(11):e80306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambers A, Turnbough M, Benjamin R, Gill-King H, King J, Sajantila A, Bruce B (2016) Modified DOP-PCR for improved STR typing of degraded DNA from human skeletal remains and bloodstains. Leg Med 18:7–12 [DOI] [PubMed] [Google Scholar]
- Armingol E, Officer A, Harismendy O, Lewis NE. Deciphering cell–cell interactions and communication from gene expression. Nat Rev Genet. 2020;22(2). [DOI] [PMC free article] [PubMed]
- Baba T, Convery PA, Matsumura N, Whitaker RS, Kondoh E, Perry T, Huang Z, Bentley RC, Mori S, Fujii S, Marks JR, Berchuck A, Murphy SK (2009) Epigenetic regulation of CD133 and tumorigenicity of CD133+ ovarian cancer cells. Oncogene. 28(2):209–18 [DOI] [PubMed] [Google Scholar]
- Bageritz J,Raddi G. Single-Cell RNA Sequencing with Drop-Seq. Methods in molecular biology (Clifton, N.J.),2019,1979. [DOI] [PubMed]
- Bai RL, Cui JW (2020) Tumor heterogeneity-the challenge of accurate clinical diagnosis and treatment. Chin J Clin Oncol 47(21):1082–1087 [Google Scholar]
- Bai X, Li YX, Zeng XM, Zhao Q, Zhang Z (2021) Single-cell sequencing technology in tumor research. Clin Chim Acta 518:101–9 [DOI] [PubMed] [Google Scholar]
- Baslan T, Hicks J (2017) Unravelling biology and shifting paradigms in cancer with single-cell sequencing. Nat Rev Cancer 17(9):557–569 [DOI] [PubMed] [Google Scholar]
- Bhardwaj V,Tan Y Q,Wu M M,Ma L,Zhu T,Lobie P E.,Pandey V. Long non-coding RNAs in recurrent ovarian cancer: Theranostic perspectives. Cancer Letters, 2021;502(prepublish). [DOI] [PubMed]
- Biotechnology - DNA Libraries; Research Results from Russian Academy of Sciences Update Knowledge of DNA Libraries [Improved DOP-PCR (iDOP-PCR): A robust and simple WGA method for efficient amplification of low copy number genomic DNA]. Biotech Week, 2017. [DOI] [PMC free article] [PubMed]
- Blagodatskikh KA, Kramarov VM, Barsova EV, Garkovenko AV, Shcherbo DS, Shelenkov AA, Ustinova VV, Tokarenko MR, Baker SC, Kramarova TV, Ignatov KB (2017) Improved DOP-PCR (iDOP-PCR): A robust and simple WGA method for efficient amplification of low copy number genomic DNA. PLoS One. 12(9):e0184507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boris W,Shobhana T,Zenas C,Wang J H,Timothy K.S. Single-cell sequencing in ovarian cancer:a new frontier in precision medicine. Current Opinion in Obstetrics and Gynecology, 2018. [DOI] [PubMed]
- Brown JR, Chan DK, Shank JJ, Griffith KA, Fan H, Szulawski R, Yang K, Reynolds RK, Johnston C, McLean K,Uppal S,Liu J R,Cabrera L,Taylor S E,Orr B C,Modugno F,Mehta P,Bregenzer M,Mehta G,Shen H,Coffman L G,Buckanovich R J.Phase II clinical trial of metformin as a cancer stem cell-targeting agent in ovarian cancer. JCI insight,2020;5(11). [DOI] [PMC free article] [PubMed]
- Bruno A, de Mora D, Freire-Paspuel B, Rodriguez AS, ParedesEspinosa MB, Olmedo M, Sanchez M, Romero J, Paez M, Gonzalez M, Orlando A, GarciaBereguiain MA (2021) Analytical and clinical evaluation of a heat shock SARS-CoV-2 detection method without RNA extraction for N and E genes RT-qPCR. Int J Infect Dis 109:315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buermans HP, den Dunnen JT (2014) Next generation sequencing technology: Advances and applications. Biochim Biophys Acta 1842(10):1932–1941 [DOI] [PubMed] [Google Scholar]
- Cai YH, Wang JJ, Zou K (2020) The Progresses of Spermatogonial Stem Cells Sorting Using Fluorescence-Activated Cell Sorting. Stem Cell Rev Rep 16(1):94–102 [DOI] [PubMed] [Google Scholar]
- Cameron W. B, Roel G. W. V, Aaron M, Benito C, Houtan N, Sofie R. S, Zheng S Y, Debyani C, J. Zachary S, Samuel H. B, Rameen B, Brady B, Chang-Jiun Wu, Giannicola G, Ilya S, Jill B-S, Zou L H, Rahulsimham V, Sachet A. S, Giovanni C, W. K. Yung, Zhang W, Carrie S, Tom M, Kenneth A, Darell D. B, Erwin G. V M, Michael P, Andrew S, Keith L. B, Jennif. The Somatic Genomic Landscape of Glioblastoma. Cell, 2014;157(3).
- Cao C, Ma Q, Mo S, Shu G, Liu Q, Ye J, Gui Y (2021) Single-Cell RNA Sequencing Defines the Regulation of Spermatogenesis by Sertoli-Cell Androgen Signaling. Front Cell Dev Biol. 9(9):763267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae YC, Kim JH (2018) Cancer stem cell metabolism: target for cancer therapy. BMB Rep 51(7):319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZB, Meng YG (2021) Research progress in the relationship between BRCA gene and diagnosis and treatment of ovarian cancer. Int J Obstet Gynecol 48(02):144–148 [Google Scholar]
- Chen Y, Du H, Bao LW, Liu WX (2018) LncRNA PVT1 promotes ovarian cancer progression by silencing miR-214. Cancer Biol Med 15(3):238–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christine S, Christine R, Suzy L (2019) Ovarian Cancer: An Integrated Review. Semin Oncol Nurs 35(2):151–156 [DOI] [PubMed] [Google Scholar]
- Clevers H (2011) The cancer stem cell: premises, promises and challenges. Nat Mad 17:313–319 [DOI] [PubMed] [Google Scholar]
- Cole AJ, Iyengar M, Panesso-Gómez S, O'Hayer P, Chan D, Delgoffe GM, Aird KM, Yoon E, Bai S, Buckanovich RJ. NFATC4 promotes quiescence and chemotherapy resistance in ovarian cancer. JCI insight, 2020;5(7). [DOI] [PMC free article] [PubMed]
- Cowan R, Chi D, Kehoe S, Kehoe S, Nankivell M (2016) Primary surgery or neoajuvant chemotherapy in advanced ovarian cancer:the debate continues. Am Soc Clin Oncol Educ Book 36:153–162 [DOI] [PubMed] [Google Scholar]
- Cui J X, Liu L, Li D H,Pu X F. Research progress in the application of external field separation technology and microfluidic technology in the separation of micro/nanoscales. Se Pu.2021;39(11):1157–1170. Chinese. [DOI] [PMC free article] [PubMed]
- Duan M, Hao J, Cui S, Worthley DL, Zhang S, Wang Z, Shi J, Liu L, Wang XY, Ke A, Cao Y, Xi RB, Zhang XM, Zhou J, Fan J, Li C, Gao Q (2018) Diverse modes of clonal evolution in HBV-related hepatocellular carcinoma revealed by single-cell genome sequencing. Cell Res 28(3):359–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuFort C C, DelGiorno K E, Carlson M A,Ryan J.O,Zhao C M,Huang Z D,Curtis B. T,Robert J.C,Christopher D.T,J. Scott B,Paolo P. P,Gregory I.F,H. Michael S,Sunil R. Hingorani.Interstitial Pressure in Pancreatic Ductal Adenocarcinoma Is Dominated by a Gel-Fluid Phase. Biophysical Journal, 2016;110(9). [DOI] [PMC free article] [PubMed]
- Chakri R. Etiology, Risk and Metrics in Ovarian Cancer. Cancer Science & Therapy, 2021;13(12).
- Ferrarini A,Forcato C,Buson G,Tononi P,Del M V,Terracciano M,Bolognesi C,Fontana F,Medoro G,Neves R,Möhlendick B,Rihawi K,Ardizzoni A,Sumanasuriya S,Flohr Pe,Lambros M,de Bono J,Stoecklein N H,Manaresi N.A streamlined workflow for single-cells genome-wide copy-number profiling by low-pass sequencing of LM-PCR whole-genome amplification products. PloS one, 2018;13(3). [DOI] [PMC free article] [PubMed]
- Gao SF, Xiao X, Zhang LY, Ni L, Liu LY, Han L, Yang J (2020) Application of Single Cell Sequencing Technology in Reproductive Research. Chin J Cell Biol 42(12):2234–2243 [Google Scholar]
- Gawad C,Koh W,Quake S.Single-cell genome sequencing: current state of the science. Nature reviews. Genetics, 2016;17(3). [DOI] [PubMed]
- Gov E,Kori M,Arga K Y. RNA-based ovarian cancer research from 'a gene to systems biomedicine' perspective. Systems biology in reproductive medicine, 2017;63(4). [DOI] [PubMed]
- Guo JP, Guo B, Li LL, Liu DX, Shi J, Wu JH (2017) Artemisinin induces apoptosis of hepatocellular carcinoma cells by inhibiting JAK2/STAT3 signal pathway. J Shanxi Med Univ 48(02):97–100 [Google Scholar]
- Hermann P C, Huber S L, Herrler T, Alexandra A, Joachim W.E, Markus G, Christiane J. B, Christopher H. Distinct Populations of Cancer Stem Cells Determine Tumor Growth and Metastatic Activity in Human Pancreatic Cancer. Cell Stem Cell, 2007;1(3). [DOI] [PubMed]
- Hornburg M,Desbois M,Lu S,Guan Y H,Lo A A.,Kaufman S,Elrod A,Lotstein A,DesRochers T M.,Munoz R J L.,Wang X W,Giltnane J,Mayba O,Turley S J.,Bourgon R,Daemen A,Wang Y L.Single-cell dissection of cellular components and interactions shaping the tumor immune phenotypes in ovarian cancer. Cancer Cell,2021,39(7). [DOI] [PubMed]
- Hu Z Y,Mara A,Abdulkhaliq A,Nina W,Matteo M,Shi T Y,Zhong Z,Laura S G,Salma ES,Mohammad K,Garry M,Feng Y,Kenta M,Zheng Y Y,Kay C,Stephen D,Sunanda D,Leticia C,Riccardo G C,Hooman S,Vikram R,David M P,Stephanie J,Vincenzo C,Tatjana SS,Christopher Y,Ahmed A A.The Repertoire of Serous Ovarian Cancer Non-genetic Heterogeneity Revealed by Single-Cell Sequencing of Normal Fallopian Tube Epithelial Cells. Cancer Cell,2020,37(2). [DOI] [PubMed]
- Huang Z H,Wu C,Liu X K,Lu S,You L M,Guo F Y,Stalin A,Zhang J Y,Zhang F Q,Wu Zh S,Tan Y Y,Fan X T,Huang J Q,Zhai Y Y,Shi R,Chen M L,Wu C F,Li H Y,Wu J R. Single-Cell and Bulk RNA Sequencing Reveal Malignant Epithelial Cell Heterogeneity and Prognosis Signatures in Gastric Carcinoma. Cells,2022,11(16). [DOI] [PMC free article] [PubMed]
- Huang M, Zhang W, Zhao BB, Li L (2019) Relationship between inter-a-trypsin inhibitor heavy chain 4 and ovarian cancer. Chin J Cancer Res 31(6):955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Yi DR, Jin FJ, Ye YQ (2020) Research progress of single cell separation methods and instruments. J Instrum 41(05):140–153 [Google Scholar]
- Izar B, Tirosh I, Stover EH, Wakiro I et al (2020) A single-cell landscape of high-grade serous ovarian cancer. Nat Med 26(8):1271–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyvani V, Farshchian M, Esmaeili SA, Yari H, Moghbeli M, Nezhad SK, Abbaszadegan MR. Ovarian cancer stem cells and targeted therapy. Journal of ovarian research,2019,12(1). [DOI] [PMC free article] [PubMed]
- Khalafallah A M,Huq S,Jimenez A E,Serra R,Bettegowda C,Mukherjee D. "Zooming in" on Glioblastoma: Understanding Tumor Heterogeneity and its Clinical Implications in the Era of Single-Cell Ribonucleic Acid Sequencing. Neurosurgery,2020. [DOI] [PubMed]
- Kroeger P T,Drapkin R. Pathogenesis and heterogeneity of ovarian cancer. Current opinion in obstetrics & gynecology,2017;29(1). [DOI] [PMC free article] [PubMed]
- Kurata M, Rathe SK, Bailey NJ, Aumann NK, Jones JM, Veldhuijzen GW, Moriarity BS, Largaespada DA (2016) Using genome-wide CRISPR library screening with library resistant DCK to find new sources of Ara-C drug resistance in AML. Sci Rep 6:36199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L,Sheng K.Research progress and clinical significance of tumor heterogeneity in ovarian cancer. Progress in Obstetrics and Gynecology,2016;25(04):308–310.
- Li M J,Xiong D,Huang H,Wen Z Y. Ezrin Promotes the Proliferation, Migration, and Invasion of Ovarian Cancer Cells. Biomedical and Environmental Sciences, 2021;34(2). [DOI] [PubMed]
- Li T,Liu Y,Yue S J,Liao Z C,Luo Z Q,Wang M J,Gao C D,Ding Y,Lin Z L. Analyzing the Effects of Intrauterine Hypoxia on Gene Expression in Oocytes of Rat Offspring by Single Cell Transcriptome Sequencing. Frontiers in genetics, 2019;10. [DOI] [PMC free article] [PubMed]
- Li Q N,Guo L,Hou Y,Ou X H,Liu Z H,Sun Q Y. The DNA methylation profile of oocytes in mice with hyperinsulinaemia and hyperandrogenism as detected by single-cell level whole genome bisulphite sequencing (SC-WGBS) technology. Reproduction, fertility, and development, 2018;30(12). [DOI] [PubMed]
- Li M, Li XQ, Yan SJ, et al.~(18) Application of F-Deoxyglucose PET/CT combined with CA125 in the diagnosis of recurrence and metastasis of ovarian cancer. J Chongqing Med Univ. 2017;42(12):1635–1638.
- Li G M, Xiao G Z, Qin P F,Wan X Y,Fu Y J,Zheng Y H,Luo M Y,Ren D L,Liu S P,Chen H X,Lin H C.Single-Cell RNA Sequencing Reveals Heterogeneity in the Tumor Microenvironment between Young-Onset and Old-Onset Colorectal Cancer[J].Biomolecules,2022,12(12). [DOI] [PMC free article] [PubMed]
- Li M L,Liu F T,Zhang Y J,Wu X S,Wu W G,Wang X A,Zhao S,Liu S B,Liang H B, Zhang F,Ma Q,Xiang S S,Li H F,Jiang L,Hu Y P,Gong W,Zhang Y,Ma T L,Zhang K,Liu Y,Liu Y B. Whole-genome sequencing reveals the mutational landscape of metastatic small-cell gallbladder neuroendocrine carcinoma (GB-SCNEC). Cancer Letters,2016,391. [DOI] [PubMed]
- Li J, Han L, Roebuck P et al (2015) TANRIC: An Interactive Open Platform to Explore the Function of lncRNAs in Cancer. Cancer Res 75(18):3728–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang GT (2019) Overview of Single Cell Sequencing Technology. Biology Teaching 44(8):5–7 [Google Scholar]
- Liu L Y,Yi J J,Deng X J,Yuan J H,Zhou B X,Lin Z Q,Zeng Z Y. MYH9 overexpression correlates with clinicopathological parameters and poor prognosis of epithelial ovarian cancer.[J]. Oncology letters,2019,18(2). [DOI] [PMC free article] [PubMed]
- Liu Y, Jeraldo P, Herbert W,McDonough S,Eckloff B,Schulze-M D,de Vera J-P,Cockell C,Leya T,Baqué M,Jen J,Walther-A M.Whole genome sequencing of cyanobacterium Nostoc sp. CCCryo 231–06 using microfluidic single cell technology. iScience. 2022;25(5):104291. [DOI] [PMC free article] [PubMed]
- Liu L Y, Ning Y X, Yi J J, Yuan J H,Fang W Y,Lin Z Q,Zeng Z Y.miR-6089/MYH9/β-catenin/c-Jun negative feedback loop inhibits ovarian cancer carcinogenesis and progression. Biomed Pharmacother. 2020;125:109865. [DOI] [PubMed]
- Liu J, Jiao X, Gao Q (2020) Neoadjuvant chemotherapy-related platinum resistance in ovarian cancer. Drug Discov Today 25(7):1232–1238 [DOI] [PubMed] [Google Scholar]
- Lovett M.The applications of single-cell genomics.[J]. Human molecular genetics,2013,22(R1). [DOI] [PMC free article] [PubMed]
- Luo Q Y,Wu X W,Zhang Y P,et al. ARID1A ablation leads to multiple drug resistance in ovarian cancer via transcriptional activation of MRP2. Cancer Letters, 2018;427. [DOI] [PubMed]
- McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future.Cell,2017,168(4). [DOI] [PubMed]
- Merlos-Suarez A, Barriga FM, Jung P, Mar I, María VC, David R, Marta S, Xavier H-M, da Victoria S-D, Purificación M, Hans C, Elena S, Ramón M, Eduard B (2011) The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8:511–524 [DOI] [PubMed] [Google Scholar]
- Nwani NG, Condello S, Wang Y, Swetzig WM, Barber E, Hurley T, Matei D. A Novel ALDH1A1 inhibitor targets cells with stem cell characteristics in ovarian cancer.Cancers,2019,11(4). [DOI] [PMC free article] [PubMed]
- Orr B, Edwards RP. Diagnosis and Treatment of Ovarian Cancer[J].Hematol Oncol Clin North Am. 2018;32(6):943–964. [DOI] [PubMed]
- Pan F. Xie Xiaoliang, academician of the American Academy of Sciences: The dawn of single-cell whole-genome sequencing. China Science Daily, 2012–12–27(001).
- Paola M,Elena I,Federico F,Luigi G,Maria P.Imaging biomarkers in ovarian cancer: the role of 18F-FDG PET/CT. The Quarterly Journal of Nuclear Medicine and Molecular Imaging,2016;60(2). [PubMed]
- Picelli S, Faridani OR, Björklund AK,Winberg G,Sagasser S,Sandberg R.Full-length RNA-seq from single cells using Smart-seq2. Nature protocols,2014,9(1). [DOI] [PubMed]
- Prensner J R, Iyer M K, Balbin O A, Dhanasekaran S M,Cao Q,Brenner J C,Laxman B,Asangani I A,Grasso C S,Kominsky H D,Cao X H,Jing X J,Wang X J,Siddiqui J,Wei J T,Robinson D,Iyer H K,Palanisamy N,Maher C A,Chinnaiyan A M.Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression.Nature biotechnology,2011;29(8). [DOI] [PMC free article] [PubMed]
- Prince GMSH, Yang TY, Lin H, Chen MC (2019) Mechanistic insight of cyclin-dependent kinase 5 in modulating lung cancer growth. Chin J Physiol 62(6):231–240 [DOI] [PubMed] [Google Scholar]
- Reproduction. Heredity, 2018;40(08):620-631.
- Quainoo S, Coolen JP, van Hijum SA, Huynen MA, Melchers WJ, van Schaik W, Wertheim HF (2017) Whole-Genome Sequencing of Bacterial Pathogens: the Future of Nosocomial Outbreak Analysis. Clin Microbiol Rev 30(4):1015–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrback S,April C,Kaper F,Rivera R R,Liu C S,Siddoway B,Chun J. Submegabase copy number variations arise during cerebral cortical neurogenesis as revealed by single-cell whole-genome sequencing. Proceedings of the National Academy of Sciences of the United States of America,2018;115(42). [DOI] [PMC free article] [PubMed]
- Scott M,Lawrenson K,Kim K,Strickland K,Medeiros F,Cerda V,Walsh C.Pathways endometriosis associated ovarian cancer, a histological origin?. Gynecologic Oncology,2022,164(1).
- Shao T, Chen XW (2013) Research progress in etiology hypothesis, risk factors and epidemiology of ovarian cancer. Chinese Journal of Clinicians (electronic Version) 7(19):8894–8897 [Google Scholar]
- Shen MJ, Olsthoorn RCL, Zeng Y, Bakkum T, Kros A, Boyle AL (2021) Magnetic-Activated Cell Sorting Using Coiled-Coil Peptides: An Alternative Strategy for Isolating Cells with High Efficiency and Specificity. ACS Appl Mater Interfaces 13(10):11621–11630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng K, Zong C (2019) Single-Cell RNA-Seq by Multiple Annealing and Tailing-Based Quantitative Single-Cell RNA-Seq (MATQ-Seq). Methods Mol Biol 1979:57–71 [DOI] [PubMed] [Google Scholar]
- Sheng K, Cao W, Niu Y, Deng Q, Zong C (2017) Effective detection of variation in single-cell transcriptomes using MATQ-seq. Nat Methods 14(3):267–270 [DOI] [PubMed] [Google Scholar]
- Shih IM, Kurman RJ (2004) Ovarian tumorigenesis: a proposed model based on morphological and molecular genetic analysis. Am J Pathol 164(5):1511–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva IA, Bai S, McLean K, Yang K, Griffith K, Thomas D, Ginestier C, Johnston C, Kueck A, Reynolds RK, Wicha MS, Buckanovich RJ (2011) Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res 71(11):3991–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Satija R (2019) Integrative single-cell analysis. Nat Rev Genet 20(5):257–272 [DOI] [PubMed] [Google Scholar]
- Sukhbaatar N,Bachmayr-H A,Auer K,Aust S,Deycmar S,Horvat R,Pils D.Two different, mutually exclusively distributed, TP53 mutations in ovarian and peritoneal tumor tissues of a serous ovarian cancer patient: indicative for tumor origin?. Cold Spring Harbor molecular case studies,2017,3(4). [DOI] [PMC free article] [PubMed]
- Sun C R,Jia N,Huang X L,Xiao F,Zhou J,Zhang Y,Fu J,Xu Z,Qu D,Cui X D,Wang Y. Real-time multiple cross displacement amplification assay for rapid and sensitive detection of Haemophilus influenzae [J]. Frontiers in Cellular and Infection Microbiology,2022,12. [DOI] [PMC free article] [PubMed]
- Talukdar S,Chang Z,Winterhoff B,Starr T K. Single-Cell RNA Sequencing of Ovarian Cancer: Promises and Challenges.[J]. Advances in experimental medicine and biology,2021,1330. [DOI] [PubMed]
- Telenius H, Carter NP, Bebb CE,Nordenskjöld M,Ponder B A,Tunnacliffe A. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics,1992;13(3). [DOI] [PubMed]
- Tian B L,Li Q. Single-Cell Sequencing and Its Applications in Liver Cancer[J]. Frontiers in Oncology,2022,12. [DOI] [PMC free article] [PubMed]
- Tian M,Chen X S,Li L Y,Wu H Z,Zeng D,Wang X L,Zhang Y,Xiao S S,Cheng Y. Inhibition of AXL enhances chemosensitivity of human ovarian cancer cells to cisplatin via decreasing glycolysis. Acta pharmacologica Sinica,2020;42(7). [DOI] [PMC free article] [PubMed]
- Vasan N, Baselga J, Hyman D M. A view on drug resistance in cancer. Nature,2019;575(7782). [DOI] [PMC free article] [PubMed]
- Vistad I, Bjørge L, Solheim O, Fiane B,Sachse K,Tjugum J,Skrøppa S,Bentzen A G,Stokstad T,Iversen G A,Salvesen H B,Kristensen G B,Dørum A.A national, prospective observational study of first recurrence after primary treatment for gynecological cancer in Norway. Acta obstetricia et gynecologica Scandinavica, 2017;96(10). [DOI] [PubMed]
- Wang R Y,Meng Q.Progress and Application of Laser Capture Microcutting Technology[J].Journal of Chongqing Medical University,2020,45(01):29–31.
- Wang M, Yuan W (2015) Microalgal cell disruption in a high-power ultrasonic flow system. Bioresour Technol 193:171–177 [DOI] [PubMed] [Google Scholar]
- Wang Q, Wang Z, Zhang Z et al (2020) Overview of Single Cell Sequencing Technology. Chinese Journal of Medicinal Guide 22(7):433–439 [Google Scholar]
- Willems A,Panchy N,Hong T. Using Single-Cell RNA Sequencing and MicroRNA Targeting Data to Improve Colorectal Cancer Survival Prediction[J]. Cells,2023,12(2). [DOI] [PMC free article] [PubMed]
- Wright J D, Chen L, Hou J Y, Burke W M,Tergas A I,Ananth C V,Neugut A I,Hershman D L.Association of Hospital Volume and Quality of Care With Survival for Ovarian Cancer.Obstetrics and gynecology,2017,130(3). [DOI] [PMC free article] [PubMed]
- Xin P, Xu X, Deng C,Liu S,Wang Y Z,Zhou X G,Ma H X,Wei D H,Sun S Q. The role of JAK/STAT signaling pathway and its inhibitors in diseases. International Immunopharmacology, 2020;80(C). [DOI] [PubMed]
- Xu B L,Xu H L.The clinical effect and safety of paclitaxel+lobplatin chemotherapy in the treatment of advanced ovarian cancer[J].Clinical Medicine Research and Practice,2022;7(33):103–105.
- Xu XL, Wu LJ, Yan RX (2019) Single cell whole genome amplification technology and application. Progress in Biochemistry and Biophysics 46(04):342–352 [Google Scholar]
- Yanai I,Hashimshony T. CEL-Seq2-Single-Cell RNA Sequencing by Multiplexed Linear Amplification. Methods in molecular biology (Clifton, N.J.),2019;1979. [DOI] [PubMed]
- Yang X, Zhang B (2023) A review on CRISPR/Cas: a versatile tool for cancer screening, diagnosis, and clinic treatment. Funct Integr Genomics 23(2):182 [DOI] [PubMed] [Google Scholar]
- Yang J F,Xiang C P,Liu J M. Clinical significance of combining salivary mRNAs and carcinoembryonic antigen for ovarian cancer detection. Scandinavian Journal of Clinical and Laboratory Investigation,2021,81(1). [DOI] [PubMed]
- Yang D,Zhang W Y,Liu Y,Liang J Q,Zhang T Q,Bai Y B,Hao W J,Ma K X,Lu D N,Chen J.Single-cell whole-genome sequencing identifies human papillomavirus integration in cervical tumour cells prior to and following radiotherapy. Oncology letters,2018;15(6). [DOI] [PMC free article] [PubMed]
- Yang L, Xie HJ, Li YY, Wang X, Liu XX, Mai J (2022) Molecular mechanisms of platinum-based chemotherapy resistance in ovarian cancer (Review). Oncol Rep 47(4):82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y X,La Y F,Di R,Liu Q Y,Hu W P,Wang X Y,Chu M X.Comparison of Different Single Cell Genome Amplification Methods and Application of MALBAC in Assisted [DOI] [PubMed]
- Yu M J,Tan J J,Wang J H. Research Progress of Single Cell Sequencing in Lung Cancer.[J]. Zhongguo fei ai za zhi = Chinese journal of lung cancer,2021;24(4). [DOI] [PMC free article] [PubMed]
- Yu T, Wang YY, Fei J, Huang J, Hu ZB (2022) Study on the detection performance of three single-cell whole-genome amplification methods for 1–4 Mb copy number variation. Journal of Molecular Diagnosis and Therapy 14(09):1549–1553 [Google Scholar]
- Zachariadis V,Cheng H T,Andrews N,Enge M. A Highly Scalable Method for Joint Whole-Genome Sequencing and Gene-Expression Profiling of Single Cells. Molecular Cell,2020,80. [DOI] [PubMed]
- Zha T Y.BRCA gene detection: “compass” for prevention and treatment of ovarian cancer. Jiangsu Health Care,2022,(07):21.
- Zhang C P.Summary of single cell sequencing technology and its application[J].Henan Science,2022,40(09):1390–1397.
- Zhang Q,Gu M L. Single-cell sequencing and its application in breast cancer. Yi chuan = Hereditas,2020,42(3). [DOI] [PubMed]
- Zhang Y Z,Fu B Q,Niu C Y,Gao Y H Wang J.Development of single cell sequencing technology[J].Acta Metrologica Sinica,2023,44(01):149–156.
- Zhang X L,Li C L,Fan Z,Sun S T,Zhao T,Liang J N,Wang Q,Liu Y W.Single cell sorting method based on micromanipulation[J].journal of Chinese electron microscopy society,2018,37(01):97–100.
- Zhang J,Yuan B Z,Zhang H D,Li H X. Human epithelial ovarian cancer cells expressing CD105, CD44 and CD106 surface markers exhibit increased invasive capacity and drug resistance. Oncology letters,2019;17(6). [DOI] [PMC free article] [PubMed]
- Zhang B Y,Xu H,Huang Y Q,Shu W L,Feng H T,Gai J,Zhong J F,Chen Y. Improving single-cell transcriptome sequencing efficiency with a microfluidic phase-switch device. The Analyst,2019;144(24). [DOI] [PMC free article] [PubMed]
- Zhang M, Cheng S, Jin Y,Wang Y. Roles of CA125 in diagnosis, prediction, and oncogenesis of ovarian cancer.[J].BBA - Reviews on Cancer,2021,1875(2). [DOI] [PubMed]
- Zhang L, Cui X, Schmitt K,Hubert R,Navidi W,Arnheim N.Whole genome amplification from a single cell: implications for genetic analysis. Proceedings of the National Academy of Sciences of the United States of America,1992;89(13). [DOI] [PMC free article] [PubMed]
- Zong C, Lu S, Chapman AR,Xie X.S. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science, 2012;338(6114). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.