Abstract
Humoral response to SARS-CoV-2 has been studied, predominantly the classical IgG and its subclasses. Although IgE antibodies are typically specific to allergens or parasites, a few reports describe their production in response to SARS-CoV-2 and other viruses. Here, we investigated IgE specific to receptor binding domain (RBD) of SARS-CoV-2 in a Brazilian cohort following natural infection and vaccination. Samples from 59 volunteers were assessed after infection (COVID-19), primary immunization with vectored (ChAdOx1) or inactivated (CoronaVac) vaccines, and booster immunization with mRNA (BNT162b2) vaccine. Natural COVID-19 induced IgE, but vaccination increased its levels. Subjects vaccinated with two doses of ChAdOx1 exhibited a more robust response than those immunized with two doses of CoronaVac; however, after boosting with BNT162b2, all groups presented similar IgE levels. IgE showed intermediate-to-high avidity, especially after the booster vaccine. We also found IgG4 antibodies, mainly after the booster, and they moderately correlated with IgE. ELISA results were confirmed by control assays, using IgG depletion by protein G and lack of reactivity with heterologous antigen. In our cohort, no clinical data could be associated with the IgE response. We advocate for further research on IgE and its role in viral immunity, extending beyond allergies and parasitic infections.
Keywords: SARS-CoV-2, IgE, IgG4, Avidity, COVID-19 vaccine, Sepharose 4B-Protein G
Subject terms: Infectious diseases, Vaccines
Introduction
Vaccines are known to trigger long-lasting IgG antibodies, with several biological properties, and SARS-CoV-2 vaccination is not exception, regardless of vaccine type1,2. Nonetheless, IgE antibodies are mainly elicited in parasitic infections or allergies. Generally, prolonged exposure to antigens, such as allergens or parasitic infections, leads to a class switch of IgE, located in the downstream region of the immunoglobulin (Ig) locus. This class of antibody is capable of activating mast cells and basophils, which degranulate, signaling inflammation1,3.
In humans, IgG4 is a particular antibody isotype, that, like IgE, is induced in an interleukin (IL)-4-rich microenvironment upon continuous antigenic stimuli, usually allergens. Unlike IgE, plasma cells need IL-10 to class-switch to IgG4; this IgG isotype does not activate Fc-mediated function, being mostly anti-inflammatory4,5. In allergy contexts, IgG4 competes with IgE to prevent it from triggering FcεR-mediated functions, thus it is not uncommon to find both IgE and IgG4 in response to the same antigen4,6.
Avidity is a parameter that reflects the multivalent binding strength between the antibody and the antigen7. IgG avidity to SARS-CoV-2 has been studied as a functional parameter that correlates with neutralizing antibodies and serves as a marker of vaccine-induced immune response8,9. IgE antibodies typically bind with high affinity to allergens; however, the avidity of this Ig class has not been extensively studied in the context of infectious diseases10.
Although uncommon, viral protein antigens can trigger IgE responses, as observed following respiratory syncytial virus and varicella zoster infections11,12, as well as Hepatitis B and Influenza vaccinations13,14. Moreover, the IgE class switch was described after in vitro immunization of human B cells with the Mumps-Measles-Rubella (MMR) vaccine15. Concerning SARS-CoV-2, two studies have described IgE following natural infection. Plüme et al. and Giménez-Orenga found that seric IgE correlated with the severity of COVID-19 infection, probably contributing to inflammation16,17.
As described in the literature, immunoglobulins IgG, IgM, and IgA mediate critical functions in infection and vaccination responses to SARS-CoV-2; however, the role of IgE is still unclear in these settings. In this study, we investigated IgE response following SARS-CoV-2 natural infection, inactivated or viral vector vaccination, and mRNA boosting. Employing the classic enzyme-linked immunosorbent assay (ELISA) and specific antibodies, we were able to detect the presence of IgE and IgG4 antibodies in the serum of vaccinated individuals. We then tested the functionality of IgE by avidity. To confirm the presence of IgE, we adapted the IgG removal process using the Sepharose-4B-protein G in an in-house assay.
Results
Population demographics
One Table 1 shows the demographic data of the studied population. As expected, in the first sampling, positive IgE indexes were found in individuals with documented COVID-19 infection but not in subjects without COVID-19 history. The majority of individuals with positive IgE index experienced symptoms (72%), but no symptom was specifically associated with IgE (p > 0.05).
Table 1.
Baseline characteristics.
| Groups | ||||||
|---|---|---|---|---|---|---|
| Demographics | Total 59 (100) | ChAd 9 (15) | ChAd-cov 9 (15) | Corona 22 (38) | Corona-cov 19 (32) | |
| Age; median (IQR), years | 50 (43–55) | 53 (37–55) | 47 (43–48) | 49 (42–54) | 54 (48–58) | |
| Age > 60 years; no. (%) | 7 (12) | 1 (11) | 0 (0) | 2 (9) | 4 (21) | |
| Symptoms, no. (%) | ||||||
| Cough | 18 (31) | 3 (33) | 0 (0) | 5 (23) | 10 (53) | |
| Fever/febrile | 9 (15) | 1 (11) | 0 (0) | 3 (14) | 5 (26) | |
| Shortness of breath | 5 (8) | 2 (22) | 0 (0) | 0 (0) | 3 (16) | |
| Myalgia | 15 (25) | 3 (33) | 0 (0) | 5 (23) | 7 (37) | |
| Headache | 22 (37) | 5 (56) | 0 (0) | 9 (41) | 8 (42) | |
| Anosmia/dysgeusia | 10 (17) | 1 (11) | 0 (0) | 0 (0) | 9 (47) | |
| Sore throat | 14 (24) | 3 (33) | 0 (0) | 3 (14) | 8 (42) | |
| Fatigue | 15 (25) | 4 (44) | 0 (0) | 4 (18) | 7 (37) | |
| Diarrhea | 8 (14) | 2 (22) | 0 (0) | 1 (5) | 5 (26) | |
| Asymptomatic | 22 (37) | 2 (22) | 9 (100) | 8 (37) | 3 (16) | |
| Pre-existing conditions, no. (%) | ||||||
| Comorbidity | 35 (59) | 5 (56) | 1 (11) | 17 (77) | 12 (63) | |
| Hypertension | 14 (24) | 3 (33) | 0 (0) | 5 (23) | 6 (32) | |
| Diabetes | 3 (5) | 1 (11) | 0 (0) | 0 (0) | 2 (11) | |
| Heart diseasesNE | 3 (5) | 0 (0) | 0 (0) | 0 (0) | 3 (16) | |
| Respiratory diseasesNE | 3 (8) | 1 (11) | 0 (0) | 10 (45) | 6 (32) | |
| Obesity | 4 (7) | 1 (11) | 0 (0) | 1 (5) | 2 (11) | |
| Immune-mediated diseasesNE | 7 (12) | 2 (22) | 0 (0) | 4 (18) | 1 (5) | |
| Smoke | 2 (3) | 1 (11) | 0 (0) | 1 (5) | 0 (0) | |
Absolute number of cases, percentage, and median interquartile (IQR) 25–75th.
NE Non-specific.
If we consider the IQR50 as high IgE levels, immune-mediated disease was not a predictor for high IgE (p = 0.344 for natural infection IgE levels, p = 0.915 for ChAdOx1 or CoronaVac vaccine-IgE response and p = 0.124 for BNT162b2). However, it should be emphasized that this was a limited data collection. The immune-mediated conditions described by the volunteers were hypothyroidism, rheumatoid arthritis, sinusitis, and rhinitis. No volunteer in this particular analysis indicated any hypersensitivity during the interviews following the administration of the SARS-CoV-2 vaccination schedule.
Specific IgE anti-RBD levels are present in vaccine response
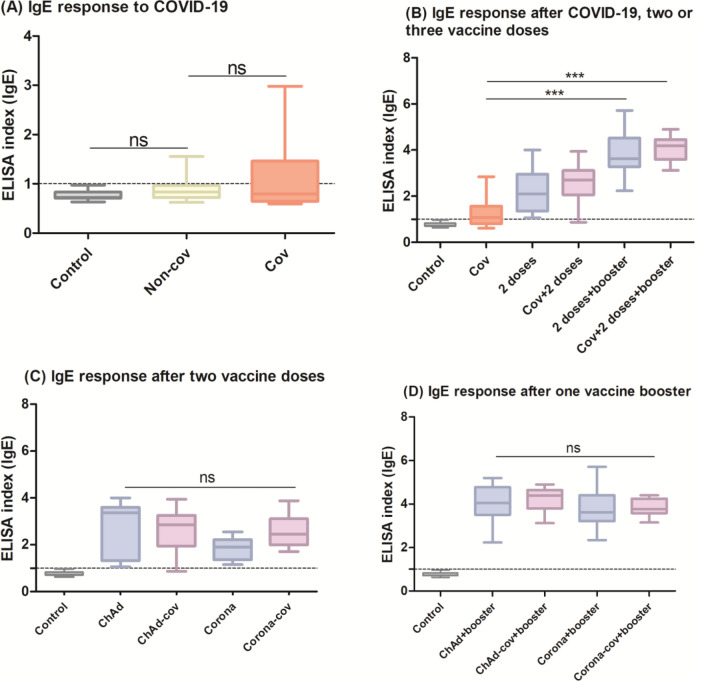
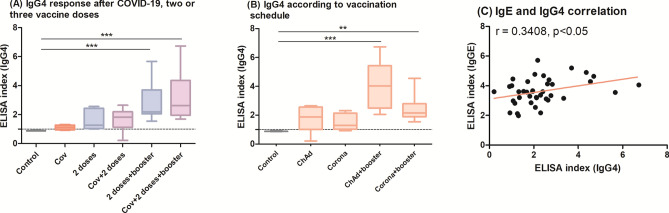
Previous studies have demonstrated IgE antibodies following SARS-CoV-2 infection16,17; therefore, we analyzed samples comparing subjects with (Cov) and without (Non-cov) documented COVID-19. Although we could not find a statistical difference, some individuals exhibited IgE above the cutoff (23% of the Cov subjects) (Fig. 1A). Comparing the natural infection and vaccine response, IgE levels are higher after two vaccine doses, with a further increase after the boosting. For volunteers who were infected by SARS-CoV-2 before the vaccination, IgE indexes were slightly augmented. We found that two vaccine doses plus one booster induced a higher IgE index than natural infection, regardless of having a COVID-19 history (p < 0.001 for both cases) (Fig. 1B).
Fig. 1.
ELISA index of IgE-RBD: (A) sera collected from subjects with (Cov, n = 28) and without (Non-cov, n = 31) COVID-19 before the vaccines were available; (B) comparison of IgE levels following natural infection (Cov), two vaccine doses with (Cov + 2 doses, n = 28) or without (2 doses, n = 31) previous infection, and booster vaccine with (Cov + 2 doses + booster, n = 28) or without (2 doses + booster, n = 31) previous infection, regardless of whether the first two doses were from CoronaVac or ChAdOx1 vaccines; (C) IgE response to each vaccine regimen after two viral-vector (ChAd, n = 9) or inactivated (Corona, n = 22) vaccine doses, with or without previous SARS-CoV-2 infection (ChAd-cov, n = 9/Corona-cov, n = 19); (D) IgE levels following mRNA boost in volunteers who had received viral-vector (ChAd + booster, n = 9) or inactivated (Corona + booster, n = 22) vaccines, with or without a COVID-19 history (ChAd-cov + booster, n = 9/Corona-cov + booster, n = 19). The index refers to the ratio between the optical density (OD) of the sample and the cutoff value. The dotted line shows the cutoff of anti-RBD IgE. The groups were compared using the Kruskal–Wallis test followed by Dunn’s post hoc test. Control group = 30 pre-pandemic sera.
When examining the groups according to vaccination schedule (ChAd, ChAd-cov, Corona, Corona-cov), specific anti-RBD IgE was elevated after the first two vaccine doses, especially in individuals vaccinated with ChAdOx1. Evaluating the gain of anti-RBD IgE with vaccine administration in individuals with or without a prior diagnosis, values were more pronounced in individuals who had COVID-19, although without statistical significance (Fig. 1C). Nonetheless, booster vaccination increased IgE indexes across all groups regardless of vaccine type or previous COVID-19, and no statistical difference was found comparing the four groups (Fig. 1D).
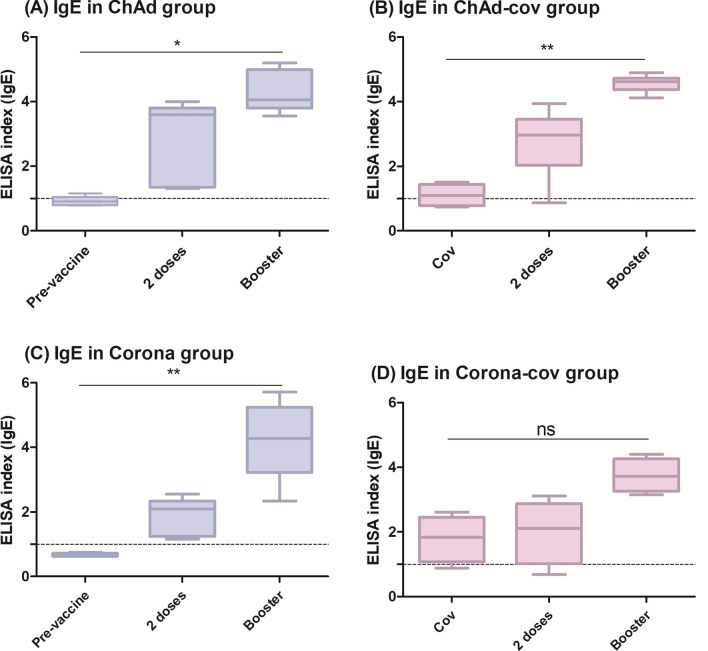
Analysis of the kinetics within each group revealed that the IgE index becomes statistically significant only after the booster dose (p < 0.05 for ChAd, p < 0.01 for ChAd-cov, and p < 0.01 for Corona). However, IgE levels increase with each subsequent dose (Fig. 2).
Fig. 2.
Kinetics analysis of IgE response, showing that the index is higher in subjects previously diagnosed with COVID-19 and rises after each vaccine dose, especially after the booster, regardless of group: (A) ChAd (n = 9), (B) ChAd-cov (n = 9), (C) Corona (n = 22), or (D) Corona-cov (n = 19). The ELISA index refers to the ratio between the optical density (OD) of the sample and thecutoff value. The dotted line shows the cutoff of anti-RBD IgE. The responses at each dose were compared using the Friedman test followed by Dunn’s post hoc test.
IgE antibodies showed intermediary-to-high avidity towards RBD
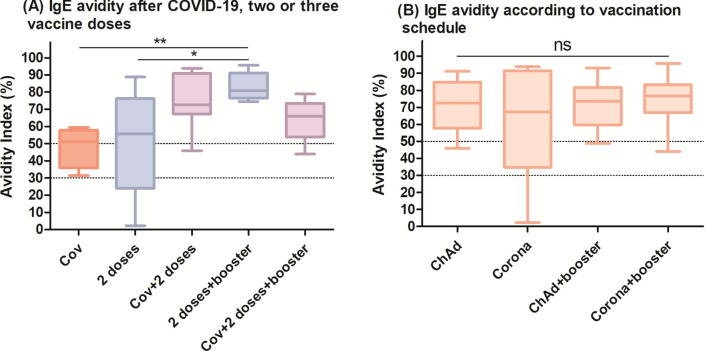
Considering functionality, we performed an avidity assay to describe the binding strength of IgE for the RBD antigen. Our data suggest that having COVID-19 induced IgE of intermediary avidity, whereas vaccines mostly led to high-avidity IgE (Fig. 3). To note, only samples which provided an O.D. ≥ 0.2 were assayed for avidity, implicating in lower n, described in the legend.
Fig. 3.
(A) Vaccination after previous infection and two vaccine doses plus one booster induced IgE of higher avidity than natural infection. Each group had the following n: Cov = 4, 2 doses = 5, Cov + 2 doses = 9, 2 doses + booster = 12, Cov + 2 doses + booster = 11. (B) However, when the avidity results of IgE triggered by CoronaVac and ChAdOx1 were compared, no differences were observed. Each group had the following n: ChAd = 8, Corona = 6, ChAd + booster = 9, Corona + booster = 14. The avidity index refers to the OD of the well treated with KSCN/OD of the well without it, converted in percentage. The dotted lines show the classification of avidity as low (< 30%), intermediate (30–49%) or high (≥ 50). The groups were compared using the Kruskal–Wallis test followed by Dunn’s post hoc test.
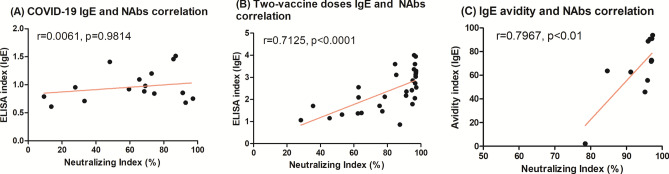
To speculate about neutralization—another functional parameter, we performed a correlation analysis to check if IgE data would correlate with neutralizing antibodies (NAbs) after natural infection and two vaccine doses from previously published data, using a surrogate neutralization assay based on an inhibition ELISA (cPass SARS-CoV-2 Neutralization Antibody Detection Kit, GenScript) (Fig. 4). Briefly, this kit measures the capacity of a sera sample to inhibit the binding between the Receptor binding domain (RBD) and the angiotensin converting enzyme 2 (ACE-2), the virus receptor18. Natural infection NAbs did not show correlation with IgE (r = 0.0061, p = 0.9814), whereas two vaccine doses-NAbs correlated strongly (r = 0.7125, p < 0.0001), as well as IgE-avidity (r = 0.7967, p < 0.01).If the IgE levels after ChAdOx1 and CoronaVac vaccines are analyzed separately, significant but more discrete correlations are observed (r = 0.5192, p > 0.05 for ChAdOx1 and r = 0.3284, p > 0.05 for CoronaVac, graphs available in Supplementary material). Notably, we did not have that many paired samples tested for both IgE levels, avidity and neutralization, implicating a lower statistical power during the analysis. Additionally, since we did not have data on neutralization after the booster, we could not perform this analysis either. It is also important to highlight that such data are only suggestive, and further investigation would be required to prove that IgE could be neutralizing.
Fig. 4.
Spearman’s correlation suggested that (A) neutralizing antibodies did not correlate with IgE levels after COVID-19 (n = 17 pairs) but correlated strongly with (B) IgE levels after two doses of vaccine (n = 30 pairs) and with (C) IgE avidity (n = 13 pairs). NAbs indexes were first reported in Silva et al. (reference 18) and were replotted in this manuscript for correlation analysis. Given that not all samples studied here had matched NAbs results, number of pairs is lower than the n assayed in other experiments, and ChAd, ChAd-cov, Corona and Corona-cov groups were pooled together for analysis, to yield more statistical power. These data was obtained using a surrogate neutralization assay based on an inhibition ELISA (cPass SARS-CoV-2 Neutralization Antibody Detection Kit, GenScript), which measures the capacity of a sera sample to inhibit RBD and ACE-2 binding.
High IgE levels are moderately followed by IgG4 isotype
IgE is typically induced by Th2 microenvironment, which may be propitiated by Alum19. However, we observed IgE in individuals immunized with ChAdOx1 and an enhancement after BNT162b2, both vaccines that do not use Alum as an adjuvant. Since IgE and IgG4 are both induced in strong Th2 environments and it has been previously reported that viral antigens may trigger both immunoglobulins11,20, we tested the samples for IgG4 to better investigate Th2 antibodies (Fig. 5). While COVID-19 did not induce detectable levels of this IgG subclass, two vaccine doses increased its levels; but only after the booster IgG4 index was statistically higher than the pre-pandemic control, irrespective of previous SARS-CoV-2 infection (p < 0.001 for 2 doses + booster and for Cov + 2 doses + booster). When the vaccine schedules were studied separately, we confirmed that an mRNA booster was needed to achieve a higher IgG4 titer than control (p < 0.001 for ChAdOx1 + booster and p < 0.01 for CoronaVac + booster). Despite that, we found only a moderate correlation between IgE and IgG4 levels (r = 0.3408, p < 0.05).
Fig. 5.
(A) Increased IgG4 isotype was mainly detected after the BNT162b2 booster. Each group had the following n: Control = 30, Cov = 28, 2 doses = 31, Cov + 2 doses = 27, 2 doses + booster = 31, Cov + 2doses + booster = 27. (B) Comparing vaccination schedule, IgG4 was particularly higher in people who received ChAdOx1 + BNT162b2. Each group had the following n: Control = 30, ChAd = 18, Corona = 41, ChAd + booster = 18, Corona + booster = 41. (C) IgE and IgG4 levels showed a modest correlation (n = 40 pairs). The ELISA index refers to the ratio between the optical density (OD) of the sample and the cutoff value. The dotted line shows the cutoff of IgG4 anti-RBD. (A,B) The groups were compared using the Kruskal–Wallis test followed by Dunn’s post hoc test; (C) Spearman’s test was used to study the correlation between IgE and IgG4 levels.
Heterologous controls and IgG-depleted samples confirm anti-RBD-IgE
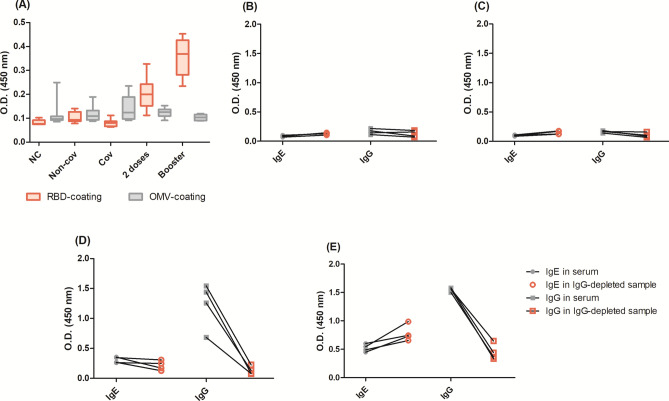
Provided that sera samples were diluted at 1:5 to detect IgE and IgG4 antibodies, two different assays were conducted to confirm that the results were specific and not related to sera background or IgG cross-reaction. Comparing the IgE levels of samples tested in microplates coated with SARS-CoV-2 RBD versus N. meningitidis OMV, we observed similar results for samples that had lower IgE levels: negative controls and pre-vaccine with (Cov) and without (Non-cov) natural infection. When the second dose and booster samples were assayed, the Optical density (OD) of the RBD plate was higher, suggesting that the results were not related to the sera background (Fig. 6A). Another option to confirm that the ELISA signal is specific to IgE antibodies was to deplete sera samples from IgG, proving that this antibody class was not cross-detected (Fig. 6B–E). Negative control, pre-vaccine, and two dose vaccines maintained the same OD, whereas booster doses even showed slightly higher OD, probably due to the lack of competition for antigen binding with IgG. In parallel, the OD of IgG from samples collected after two and booster vaccine doses decreased, proving that the purification reduced IgG from the samples.
Fig. 6.
Control tests: (A) sera background comparing the optical density (OD) of IgE-detection incubating sera at 1:5 dilution in microplates coated with SARS-CoV-2 RBD or N. meningitidis OMVs. Each group (negative control, Non-cov, Cov, 2 doses and booster) had an n = 8. For another control assay, we depleted IgG from the samples, to test if, once depleted, the IgE optical signal (using anti-IgE-ε chain) would be maintained after depletion whereas the IgG optical signal (using anti-IgG-Fc) would decrease after depletion. Four samples were used: (B) collected before the pandemic as negative controls, (C) collected before COVID-19 vaccines were available. In these cases, there were no specific antibodies in the samples, and the O.D.s, before and after depletion, were almost the same. Four samples (D) collected after two vaccine doses, and (E) collected after the booster dose. In these cases, there was both IgG and IgE in the samples, so the IgE O.D. was either the same or even enhanced, probably due lack of IgG competition for RBD, while IgG O.D. decrease, proving that the depletion worked.
In addition, blank wells were tested for each assay in quadruplicates. The blank mean values were 0.0569 ± 0.009 for the plate only (without secondary antibody); 0.0852 ± 0.009 for the RBD-coated plate plus anti-IgE, and 0.0728 ± 0.012 for the OMV-coated plate plus anti-IgE.
Discussion
Surprisingly, we observed an IgE-mediated immune response to both SARS-CoV-2 infection and vaccination using ChAdOx1, CoronaVac and BNT162b2 vaccines. Plüme et al. and Giménez-Orenga have reported the presence of IgE antibodies following natural infection16,17, which was particularly evident in severe cases, and proposed that IgE-mediated mechanisms might contribute to inflammation in COVID-19. However, in our cohort, participants were asymptomatic or experienced mild symptoms and exhibited detectable IgE antibodies as well.
It is noteworthy that to detect IgE, which is less abundant than IgM and IgG, the assay’s sensibility needs to be enhanced. Following Plume and collaborators16, we used sera samples diluted at a 1:5 ratio, a secondary antibody specific to the ε chain from IgE, and TMB—a sensitive colorimetric substrate. To gain specificity, we calculated the cutoff using a 99% confidence interval21; moreover, appropriate controls were used throughout all the experimentation, and the results were confirmed by the lack of reactivity for heterologous antigen and by the agreement with IgG-depleted samples’ results (Fig. 6).
Atopic individuals are known to exhibit increased IgE secretion22; however, our study suggests that a history of immune-mediated disease did not contribute to elevated IgE index values. This may be attributed to the small number and heterogeneity of immune-mediated conditions among the individuals in our cohort (n = 7), and studies with larger populations are warranted to address this point with greater confidence. In addition, none of the subjects in this study reported hypersensitivity after having COVID-19 or being vaccinated, although other studies have shown otherwise23,24.
A relationship between respiratory viral infections and atopy has been suggested before. An interesting investigation using an experimental model demonstrated FcεR expression by dendritic cells in mice’s lungs after viral infection with Sendai virus, which was followed by mucous cell metaplasia, consistent with asthma presentation. These findings support the hypothesis that respiratory viral infections in individuals genetically predisposed to Th2 responses may facilitate asthma25.
In the case of SARS-CoV-2, we could not find mechanistic studies, but it was verified that patients with chronic rhinosinusitis took twice as long to clear SARS-CoV-2 compared with matched controls26, and elevation in total IgE was observed in pediatric patients with long-COVID; most of them were sensitized against aeroallergens27. Other allergy-related symptoms following COVID-19, such as skin lesions, were reported during the pandemic, but it seems that they were either unspecific to the infection or the causality relationship remains elusive28. Even though results from our cohort do not support an association between atopy and enhancement of anti-RBD IgE levels, evidence suggesting the contrary led us to believe that further investigation is needed to elucidate this issue.
IgG4 and IgE are located in the upstream region of the Ig locus, and B cells require prolonged antigenic stimulation to undergo class-switching to these antibodies, supported by Th2 cytokines1,20. Aluminum hydroxide is known to induce a strong Th2 environment that supports this response19; however, in our cohort, ChAdOx1, which lacks Alum, induced even higher IgE and IgG4 indexes than CoronaVac, an Alum-adjuvanted vaccine. This suggests a role of the active immunogen in eliciting IgE and IgG4 responses. Similarly, previous studies have demonstrated IgE-responses independent of aluminum salts, as observed with Flu and MMR vaccines14,15. The overall IgE levels seem to reflect the IgG response observed in the cohort from where the samples of this study were obtained18: subjects with a COVID-19 history before vaccination showed slightly elevated antibody levels after the two ChAdOx1 or CoronaVac doses; ChAdOx1 induced more antibodies than CoronaVac, suggesting it to be more immunogenic; and a BNT162b2 booster increased antibody levels in all groups (with or without COVID-19, ChAdOx1, or CoronaVac).
The role played by IgG avidity in COVID-19 has been investigated. A study proposed that high affinity antibodies would be required to interfere in the RBD-ACE-2 binding, thus neutralizing the virus7, which was supported by laboratory results8. A comparison between infection and vaccination responses showed that the latter induces IgG of higher avidity, suggesting better functionality9, a result also observed for IgE in this study. Unfortunately, we could not find more studies addressing IgE avidity to SARS-CoV-2 or other pathogens to discuss this aspect much further. However, it is important to highlight that, for allergenic responses, higher affinity to the epitope may predict FcεR activation and IgE-mediated function10. This would be especially relevant in inflammatory contexts of health and disease.
Given that IgE specific to SARS-CoV-2 antigens has not been extensively investigated, it would be interesting to add related antibodies aiming at a more comprehensive analysis. From what is known from allergy models—the main source of IgE investigations, IgG4 could be a related antibody. This is because both are located in distant downstream regions of the Ig locus, the class switch to IgE and IgG4 share similar Th2 stimuli1,20, and concomitant IgE and IgG4 to viral antigens have been studied before11. In the context of COVID-19, IgG4 has been suggested as a marker of severe disease27,29,30 but, concerning immunization, IgG4 levels have been detected after booster doses of mRNA vaccines31–33, agreeing with our results. IgG4 is mostly non-inflammatory, thus the main concern regarding it would be the lack of Fc mediated-functions, such as antibody dependent cellular phagocytosis (ADCP), complement deposition (ADCD), and cellular cytotoxicity (ADCC), conferred by IgG1 and IgG3 subclasses34. IgG4 could contribute to protection by neutralizing SARS-CoV-2, thus further investigations are warranted to elucidate this issue35,36, which is supported by the generally excellent binding affinity of IgG44. Our results show only a modest correlation (r = 0.3408) between IgE and IgG4 levels, thus we are unable to confirm that the induction of both isotypes occurred because of a similar Th2 microenvironment. Therefore, cellular and genetic profiling would be required to support this assumption more comprehensively4.
The literature reports several virus-induced IgE responses to viral infections or vaccines11–14,16,17. Whether these antibodies participate in the viral immune response or have any pathogenic role remains unclear. As previously discussed, viral infections and atopy might influence each other, exacerbating symptoms25,27. On the other hand, anti-HIV IgE antibodies, despite not demonstrating neutralizing activity, were able to inhibit HIV proliferation in vitro, a finding hypothesized to be mediated by cytotoxicity37. More recently, IgE-mediated cytotoxicity induced the death of pancreatic cancer cells38. Taken together, these studies suggest a potentially protective role for IgE unrelated to parasites.
Unfortunately, our study did not include similar functional assay, but the ELISA index of IgE-RBD antibodies exhibited a significant correlation with the neutralizing index, and the vaccine-induced IgE had mostly high avidity, suggesting a good binding strength to SARS-CoV-2. The neutralizing antibody data we had available employed an assay that does not distinguish antibody classes, thus we assumed it would be worth investigating whether IgE antibodies contribute to neutralization or participate in other immunological mechanisms that could protect against SARS-CoV-2. Given the potency of IgG for neutralization, this assumption could only be confirmed by a controlled experiment, purifying IgE from the sera and testing its ability to neutralize SARS-CoV-2 alone, as it has been done for IgG and IgM39. Also, it is worth emphasizing that the neutralization data was based on a inhibition ELISA kit and, even though it presented excellent correlations with pseudovirus and live-virus neutralization assays40,41, it may also be influenced by sera titration42.
The study population consisted only of women, which may limit the generalization of its findings. The literature measuring total IgE levels according to sex is inconclusive43, with some evidence pointing to higher IgE in men compared to women at different ages44,45. We tested samples of three male subjects from the larger cohort that originated the population of this study, but we did not use such data for analysis to avoid biases due to the low number (n). These individuals were 38, 51, and 54 years old; two of them had comorbidities (both had hypertension and obesity), none of them presented immune-mediated disease, only one had COVID-19 before vaccination, and all of them received two doses of CoronaVac and one BNT162b2 booster. None of them presented positive IgE before the vaccines, even the one that had COVID-19 (mean value of 0.678 ± 0.021). After two doses of CoronaVac, the mean IgE index was 1.323 ± 0.071 and, upon BNT162b2 booster, it increased to 3.817 ± 1.054. Although the IgE index after two CoronaVac doses was somewhat lower than the female median, after the booster, it was similar to what we observed for women. Further studies with a proportional female/male ratio could support these observations.
Our study has some limitations: the sample size was relatively small, and only the female population was assessed; therefore, the data should be interpreted with caution. Moreover, we were unable to measure total IgE levels in the samples or conduct an in-depth functional characterization for IgE, describing virus neutralization and potential to induce cytotoxicity, for example, besides avidity. However, it is important to note that the samples were thoroughly characterized using molecular and various serology assays, control tests were performed, and this is not the first report to demonstrate IgE antibodies specific to viral antigens.
Conclusion
With the methodologies used, our study corroborated previous evidence for an IgE-specific response to SARS-CoV-2 and documented the role of vaccination in increasing such IgE levels. To date, characteristics that predispose individuals to this response have not been identified, but its overall kinetic profile appears to follow the same IgG pattern: vectored vaccines induce a stronger response than inactivated ones; mRNA boosters bring anti-RBD IgE to similar levels, regardless of the primary vaccine; IgE presents high avidity to a key SARS-CoV-2 antigen; and IgG4 levels are present, but only moderately correlated with IgE. Further research is warranted to fully elucidate the IgE response in SARS-CoV-2 infection and immunization.
Material and methods
Ethical statement, study population and clinical characteristics
A total of 148 sera samples were provided by 59 health professional volunteers between 2020 and 2022. The volunteers were part of a cohort of healthcare workers from the Adolfo Lutz Institute Central and Regional Laboratories (respectively located in São Paulo-SP and Santo André-SP). The following inclusion criteria were applied: having a molecular test for COVID-19; receiving two doses of either ChAdOx1 (AstraZeneca/Oxford) or CoronaVac (Sinopharm/Instituto Butantan) plus a BNT162b2 (Pfizer) booster; attending the sample collection before the vaccine, after the 2nd dose, and after the booster; having previous serology for SARS-CoV-2-IgG; presenting a similar timeframe between collections to diminish temporality bias. As the majority of individuals who met these criteria were women, we focused our analysis on a female population. For natural infection assessment, we analyzed sera samples from confirmed COVID-19 (Cov) cases collected 24 (10–32) days after diagnosis by RT-qPCR in 2020, as described previously18. These samples were compared with specimens obtained from volunteers without documented COVID-19 (Non-cov). Cov and Non-cov volunteers were discriminated through molecular and serological tests, which were routinely performed between March and December 2020, regardless of the presence of symptoms during this period. This period precedes the emergence of variants and the advent of vaccination in Brazil. The COVID-19 cases studied here were asymptomatic or experienced mild clinical symptoms and did not require hospitalization or intensive medical care.
To assess the effect of vaccination, the groups were categorized based on the vaccine schedule:
ChAd (n = 9)—two doses of vectored vaccine (ChAdOx1, AstraZeneca/Oxford) administered in an 85 (85–86) day interval, plus one booster of RNA vaccine (BNT162b2, Pfizer) 185 days (183–187) after the 2nd dose.
ChAd-cov (n = 9)—confirmed COVID-19 256 (245–257) days before receiving two doses of vectored vaccine (ChAdOx1, AstraZeneca/Oxford) administered in a 90 day interval, plus one booster of RNA vaccine (BNT162b2, Pfizer) 189 days (188–190) after the 2nd dose.
Corona (n = 22)—two doses of inactivated vaccine (CoronaVac, Sinopharm/Instituto Butantan) administered in a 22 (21–23) day interval, plus one booster of RNA vaccine (BNT162b2, Pfizer) 214 (213–217) days after the 2nd dose.
Corona-cov (n = 19)—confirmed COVID-19 257 (213–277) days before receiving an inactivated vaccine (CoronaVac, Sinopharm/Instituto Butantan) administered in a 22 (22–23) day interval, plus one booster of RNA vaccine (BNT162b2, Pfizer) 215 (213–218) days after the 2nd dose.
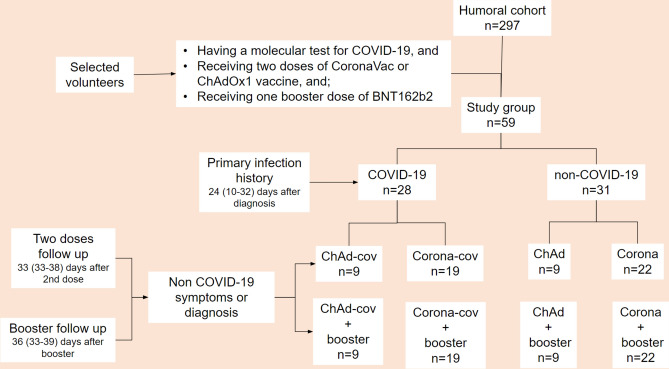
For each volunteer, blood samples were collected before vaccination (mean 22, 15–28 days), after the 2nd dose (mean 33, 26–48 days), and after the booster dose (mean 36, 33–39 days). To reduce variables that could distort results regarding the humoral response, we selected only volunteers who were diagnosed with COVID-19 during the first wave of the disease in Brazil and without information or suspicion of diagnosis during the interval between doses or between serological assessments after vaccination. Figure 7 illustrates the workflow for the population selection and sampling times.
Fig. 7.
Workflow of the selection and sampling of the study population. The population studied here originated from a 297-volunteer cohort of healthcare workers, who were followed since the beginning of the COVID-19 pandemic in Brazil. We selected volunteers who: had undergone molecular testing for COVID-19 before the vaccines; had received two doses of ChAdOx1 or CoronaVac and one BNT162b2 booster; did not have a history of COVID-19 symptoms or diagnosis between vaccine doses; attended the blood collection after each vaccination. Using these criteria, our study population (n = 59) was divided into four different groups according to the primary vaccine received (ChAd or Corona) and COVID-19 history (cov): ChAd-cov, ChAd, Corona-cov and Corona.
Serum samples of people living with HIV (viral load suppression or < 20 copies/mL) and a TCD4 + count > 200 cells/mL) or other infectious diseases, contracted before the COVID-19 pandemic served as negative controls (control group, n = 30).
This study was registered and approved by the institutional ethics committee of Adolfo Lutz Institute (CAAE 31924420.8.0000.0059), and written informed consent was provided by all participants. Additionally, all experiments were performed following relevant guidelines and regulations.
ELISA-RBD
IgE and IgG4 anti-RBD were detected using an in-house ELISA, following established protocols from the literature with some modifications29,46. Individual sera samples were heat-inactivated in a water bath at 56ºC for 30 min on the day before46. High-binding plates (Costar) were coated with 1 μg/mL of recombinant RBD, diluted in Carbonate-Bicarbonate buffer (pH 9.6), and left at 4 °C overnight. The plates were blocked with 5% PBS-skimmed milk (La Sereníssima®) for 2 h at 37 °C. After that, individual sera were diluted at 1:5 and incubated at 37 °C for 2 h. HRP labeled-anti-human IgE-ε chain (Sigma-Aldrich, product no. A-9667) or biotinylated-anti-human-IgG4 (Sigma-Aldrich, product no. B-3648), diluted at 1:1,000, were used for a 2 h incubation at 37 °C. For IgG4 plates, streptavidin-HRP (Zymed, product no. 43-4323) was incubated at 37 °C for 1 h. The reactions were developed with tetramethylbenzidine (TMB) (Sigma-Aldrich) at 37ºC for 20 min and stopped with 1N sulfuric acid (H2SO4).
The cutoff was established following Frey et al.21, using 30 pre-pandemic sera as negative controls and a 99% confidence interval, aiming to increase the specificity of the assay. The results were expressed as an ELISA index (EI), which was calculated as follow: OD of the sample/cutoff. Indexes ≥ 1 were considered positive and the results were arbitrarily classified as high if IgE was ≥ IQR50 or low if IgE was < IQR50.
Avidity-ELISA-RBD
The protocol described above was performed with one modification: after sera and before secondary antibody, plates were incubated at room temperature (RT) (20–25 °C), for 20 min with the chaotropic agent potassium thiocyanate (KSCN) 1.5 M47. The avidity index (AI, %) was calculated as the ratio between the OD of the sample with KSCN/OD without KSCN, and considered low (< 30%), intermediary (30–49%), or high (≥ 50%)48. Because samples with low O.D., thus, non-detectable antibodies, are not suitable for avidity-ELISA49, the assay was not carried out for Control and Non-cov groups, which did not had specific antibodies, and for samples presenting an O.D. ≤ 0.200 in other groups.
Heterologous-ELISA
As the antibodies analyzed in this study are present at low concentrations in blood (150 ng/ml of IgE versus 10 mg/ml of IgG), sera samples were tested at 1:5 dilution. To check if the signal obtained was specific, control assays were employed: heterologous-antigen ELISA and IgG depletion.
For heterologous ELISA, the protocol described above was performed testing the IgE of 40 randomly selected samples (8 negative controls; 8 pre-vaccine/Non-cov; 8 pre-vaccine/Cov; 8 after two vaccine doses; 8 after booster dose) in a plate coated with outer membrane vesicles (OMV) of Neisseria meningitidis B:4:P1.9 strain, as heterologous antigen50.
IgG depletion
IgG was depleted from 16 randomly selected sera samples (4 negative controls, 4 pre-vaccine, 4 after two vaccine doses, and 4 after booster dose) following an in-house protocol, using protein G immobilized in agarose (Sigma-Aldrich, product no. P-3296). The test was conducted in a microplate, as described in Haslund-Gourley and collaborators39. Sera samples were diluted at 1:5 in phosphate-buffered saline (PBS, pH 7.4) for an 80 μl final volume. Next, 20 μl of protein G was added and the microplate was placed on an orbital shaker at 75 rpm in ice for 1 h (L.E.D. Orbit, Lab-Line Instruments Inc.). Afterward, the plate was centrifuged at 1000 g and 4 °C for 1 min. 80 μl of supernatant was collected from each well and these steps were repeated two more times to ensure that IgG titers would decrease. After each step, antibody concentration was assessed by ELISA. After the third round of purification, the depleted samples were assayed for IgE and IgG anti-RBD, in parallel with sera samples. The IgE-ELISA protocol followed the one described above and for IgG-ELISA, the same protocol was employed in sera diluted at 1:50, using the secondary antibody anti-human IgG-Fc (product no. A-0170, Sigma-Aldrich) at 1:10,000 dilution. Given that the depleted samples were already diluted at 1:5 in PBS, to compare proportional dilutions, we used pure depleted samples for IgE (1/1*1/5 = 1/5) and depleted samples diluted at 1:10 for IgG (1/10*1/5 = 1/50).
Statistical analysis
Categorical variables were compared by Pearson’s χ2 test or Fisher’s exact test, as appropriate. Continuous variables were presented as range, median, and interquartile range (IQR 25th–75th). The Kruskal–Wallis test followed by Dunn’s post hoc test was used to compare the groups. Repeated measures were analyzed using the Friedman test. A two-sided p ≤ 0.05 was considered significant. Statistical analyses were performed using STATA v. 14 (StataCorp LLCP) and GraphPad Prism v. 5 (GraphPad Software Inc) softwares.
Supplementary Information
Acknowledgements
The authors would like to thank Dr Florian Krammer (Mount Sinai Hospital, New York, NY, USA) for donating the plasmid to produce the RBD to Dr Carlos Roberto Prudencio, and Dr Leyva Cecilia Vieira de Melo (Parasitology Center, Adolfo Lutz Institute, São Paulo, SP, Brazil) for donating the anti-human-IgG-Fc antibody.
Author contributions
Conceptualization and design were performed by Amanda Izeli Portilho, Valéria de Oliveira Silva, Luis Fernando de Macedo Brígido and Elizabeth De Gaspari. Volunteers recruitment and data collection were performed by Valéria Oliveira Silva, Elaine Monteiro Matsuda, Ivana Barros de Campos e Isabela Penteriche de Oliveira. Antigen production and equipment facility (centrifuge) were provided by Hernan Hermes Monteiro Da Costa and Carlos Roberto Prudencio. Execution of laboratory tests were performed by Amanda Izeli Portilho, Valéria Oliveira Silva, Rosemeire Yamashiro and Elizabeth De Gaspari. Samples management were performed by Valéria Oliveira Silva. Statistical analysis was performed by Amanda Izeli Portilho and Valéria Oliveira Silva. The manuscript was written by Amanda Izeli Portilho and Valéria Oliveira Silva. Elaine Monteiro Matsuda, Luis Fernando Macedo Brigido and Elizabeth De Gaspari revised the original draft. All authors contributed to the study, commented on previous versions of the manuscript, and approved the final manuscript.
Funding
This study was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo/FAPESP (grants numbers 2018/04202-0, 2018/14384-9, 2021/11936-3, 2022/05566-1); Conselho Nacional de Desenvolvimento Científico e Tecnológico/CNPq (grants numbers 305301/2022-5, MS-DIAHV 24/2019 process 442776/2019-5); Financiadora de Estudos e Projetos (Grant Number 01160075); Fundo de Investimento para Educação Sanitária e Imunização em Massa contra Doenças Transmissíveis/FESIMA (Grants Numbers 011/2021, 59/2021) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior/CAPES (finance code 001).
Data availability
The data from this manuscript may be made available upon reasonable request to the corresponding author (Professor Elizabeth De Gaspari, PhD).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-71047-5.
References
- 1.Collins, A. M. & Jackson, K. J. L. A temporal model of human IgE and IgG antibody function. Front. Immunol.4, 235 (2013). 10.3389/fimmu.2013.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krammer, F. The role of vaccines in the COVID-19 pandemic: What have we learned? Semin. Immunopathol.45, 451–468 (2023). 10.1007/s00281-023-00996-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bax, H. J., Keeble, A. H. & Gould, H. J. Cytokinergic IgE action in mast cell activation. Front. Immunol.3, 229 (2012). 10.3389/fimmu.2012.00229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aalberse, R. C., Platts-Mills, T. A. & Rispens, T. The developmental history of IgE and IgG4 antibodies in relation to atopy, eosinophilic esophagitis, and the modified Th2 response. Curr. Allergy Asthma Rep.16, 45 (2016). 10.1007/s11882-016-0621-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aalberse, R. C., Stapel, S. O., Schuurman, J. & Rispens, T. Immunoglobulin G4: An odd antibody. Clin. Exp. Allergy39, 469–477 (2009). 10.1111/j.1365-2222.2009.03207.x [DOI] [PubMed] [Google Scholar]
- 6.Uversky, V. N., Redwan, E. M., Makis, W. & Rubio-Casillas, A. IgG4 Antibodies induced by repeated vaccination may generate immune tolerance to the SARS-CoV-2 spike protein. Vaccines11, 991 (2023). 10.3390/vaccines11050991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauer, G. High avidity of vaccine-induced immunoglobulin G against SARS-CoV-2: Potential relevance for protective humoral immunity. Explor. Immunol.2, 133–156 (2021). [Google Scholar]
- 8.Nakagama, Y. et al. Antibody avidity maturation following recovery from infection or the booster vaccination grants breadth of SARS-CoV-2 neutralizing capacity. J. Infect. Dis.227, 780–787 (2023). 10.1093/infdis/jiac492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Struck, F. et al. Vaccination versus infection with SARS-CoV-2: Establishment of a high avidity IgG response versus incomplete avidity maturation. J. Med. Virol.93, 6765–6777 (2021). 10.1002/jmv.27270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazebrouck, S., Canon, N. & Dreskin, S. C. The effector function of allergens. Front. Allergy3, 818732 (2022). 10.3389/falgy.2022.818732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bui, R. H. D. et al. Virus-specific IgE and IgG4 antibodies in serum of children infected with respiratory syncytial virus. J. Pediatr.110, 87–90 (1986). [DOI] [PubMed] [Google Scholar]
- 12.Smith-Norowitz, T. A. et al. Long term persistence of IgE anti-varicella zoster virus in pediatric and adult serum post chicken pox infection and after vaccination with varicella virus vaccine. Int. J. Biomed. Sci.5, 353–358 (2009). 10.59566/IJBS.2009.5353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith-Norowitz, T. A. et al. IgE anti hepatitis B virus surface antigen antibodies detected in serum from inner city asthmatic and non asthmatic children. Hum. Immunol.75, 378–382 (2014). 10.1016/j.humimm.2013.12.011 [DOI] [PubMed] [Google Scholar]
- 14.Smith-Norowitz, T. A. et al. Long term persistence of IgE anti-influenza virus antibodies in pediatric and adult serum post vaccination with influenza virus vaccine. Int. J. Med. Sci.8, 239–244 (2011). 10.7150/ijms.8.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imani, F. & Kehoe, K. E. Infection of human B lymphocytes with MMR vaccine induces IgE class switching. Clin. Immunol.100, 355–361 (2001). 10.1006/clim.2001.5073 [DOI] [PubMed] [Google Scholar]
- 16.Plūme, J. et al. Early and strong antibody responses to SARS-CoV-2 predict disease severity in COVID-19 patients. J. Transl. Med.20, 176 (2022). 10.1186/s12967-022-03382-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giménez-Orenga, K. et al. HERV-W ENV antigenemia and correlation of increased anti-SARS-CoV-2 immunoglobulin levels with post-COVID-19 symptoms. Front. Immunol.13, 1020064 (2022). 10.3389/fimmu.2022.1020064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silva, V. O. et al. Inhibition of receptor-binding domain—ACE2 interaction after two doses of sinovac’s CoronaVac or AstraZeneca/Oxford’s AZD1222 SARS-CoV-2 vaccines. J. Med. Virol.94, 1217–1223 (2022). 10.1002/jmv.27396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, P., Zou, Y. & Hu, Z. Advances in aluminum hydroxide-based adjuvant research and its mechanism. Hum. Vaccines Immunother.11, 477–488 (2015). 10.1080/21645515.2014.1004026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeannin, P., Delneste, Y., Lecoanet-Henchoz, S., Gretener, D. & Bonnefoy, J. Y. Interleukin-7 (IL-7) enhances class switching to IgE and IgG4 in the presence of T cells via IL-9 and sCD23. Blood91, 1355–1361 (1998). 10.1182/blood.V91.4.1355 [DOI] [PubMed] [Google Scholar]
- 21.Frey, A., Di, J. & Zurakowski, D. A statistically defined endpoint titer determination method for immunoassays. J. Immunol. Methods221, 35–41 (1998). 10.1016/S0022-1759(98)00170-7 [DOI] [PubMed] [Google Scholar]
- 22.Sanjuan, M. A., Sagar, D. & Kolbeck, R. Role of IgE in autoimmunity. J. Allergy Clin. Immunol.137, 1651–1661 (2016). 10.1016/j.jaci.2016.04.007 [DOI] [PubMed] [Google Scholar]
- 23.Greenhawt, M. et al. The risk of allergic reaction to SARS-CoV-2 vaccines and recommended evaluation and management: A systematic review, meta-analysis, GRADE assessment, and international consensus approach. J. Allergy Clin. Immunol. Pract.9, 3546–3567 (2021). 10.1016/j.jaip.2021.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouhpayeh, H. & Ansari, H. Adverse events following COVID-19 vaccination: A systematic review and meta-analysis. Int. Immunopharmacol.109, 108906 (2022). 10.1016/j.intimp.2022.108906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grayson, M. H. et al. Induction of high-affinity IgE receptor on lung dendritic cells during viral infection leads to mucous cell metaplasia. J. Exp. Med.204, 2759–2769 (2007). 10.1084/jem.20070360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Recalde-Zamacona, B. et al. Chronic rhinosinusitis is associated with prolonged SARS-CoV-2 RNA shedding in upper respiratory tract samples: A case-control study. J. Intern. Med.289, 921–925 (2021). 10.1111/joim.13237 [DOI] [PubMed] [Google Scholar]
- 27.Körner, R. W., Bansemir, O. Y., Franke, R., Sturm, J. & Dafsari, H. S. Atopy and elevation of IgE, IgG3, and IgG4 may be risk factors for Post COVID-19 condition in children and adolescents. Children10, 1598 (2023). 10.3390/children10101598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomsitz, D., Biedermann, T. & Brockow, K. Skin manifestations reported in association with COVID-19 infection. JDDG J. Ger. Soc. Dermatol.19, 530–534 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Moura, A. D. et al. Assessment of avidity related to IgG subclasses in SARS-CoV-2 Brazilian infected patients. Sci. Rep.11, 17642 (2021). 10.1038/s41598-021-95045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Della-Torre, E. et al. Serum IgG4 level predicts COVID-19 related mortality. Eur. J. Intern. Med.93, 107–109 (2021). 10.1016/j.ejim.2021.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irrgang, P. et al. Class switch toward noninflammatory, spike-specific IgG4 antibodies after repeated SARS-CoV-2 mRNA vaccination. Sci. Immunol.8, eade2798 (2023). 10.1126/sciimmunol.ade2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiszel, P. et al. Class switch towards spike protein-specific IgG4 antibodies after SARS-CoV-2 mRNA vaccination depends on prior infection history. Sci. Rep.13, 13166 (2023). 10.1038/s41598-023-40103-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buhre, J. S. et al. mRNA vaccines against SARS-CoV-2 induce comparably low long-term IgG Fc galactosylation and sialylation levels but increasing long-term IgG4 responses compared to an adenovirus-based vaccine. Front. Immunol.13, 1020844 (2023). 10.3389/fimmu.2022.1020844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalkeri, R. et al. Altered IgG4 antibody response to repeated mRNA versus recombinant protein SARS-CoV-2 vaccines. J. Infect.88, 6–8 (2024). 10.1016/j.jinf.2024.106119 [DOI] [PubMed] [Google Scholar]
- 35.Kadkhoda, K. COVID-19: An immunopathological view. mSphere5, e00344-e420 (2020). 10.1128/mSphere.00344-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pillai, S. Is it bad, is it good, or is IgG4 just misunderstood?. Sci. Immunol.8, 3–5 (2023). 10.1126/sciimmunol.adg7327 [DOI] [PubMed] [Google Scholar]
- 37.Pellegrino, M. G. et al. HIV type 1-specific IgE in serum of long-term surviving children inhibits HIV type 1 production in vitro. AIDS Res. Hum. Retrovir.18, 363–372 (2002). 10.1089/088922202753519142 [DOI] [PubMed] [Google Scholar]
- 38.Fu, S. L. et al. Immunoglobulin E antibodies from pancreatic cancer patients mediate antibody-dependent cell-mediated cytotoxicity against pancreatic cancer cells. Clin. Exp. Immunol.153, 401–409 (2008). 10.1111/j.1365-2249.2008.03726.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haslund-Gourley, B. S. et al. IgM N-glycosylation correlates with COVID-19 severity and rate of complement deposition. Nat. Commun.15, 404 (2024). 10.1038/s41467-023-44211-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray, M. J. et al. Validation of a commercially available indirect assay for SARS-CoV-2 neutralising antibodies using a pseudotyped virus assay. J. Infect.82, 170–177 (2021). 10.1016/j.jinf.2021.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graninger, M. et al. Comprehensive comparison of seven SARS-CoV-2-specific surrogate virus neutralization and anti-spike IgG antibody assays using a live-virus neutralization assay as a reference. Microbiol. Spectr.11, e0231422 (2023). 10.1128/spectrum.02314-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papenburg, J. et al. Evaluation of a commercial culture-free neutralization antibody detection kit for severe acute respiratory syndrome-related coronavirus-2 and comparison with an antireceptor-binding domain enzyme-linked immunosorbent assay. Open Forum. Infect. Dis8, ofab220 (2021). 10.1093/ofid/ofab220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan, S. R. et al. Determinants of serum immunoglobulin levels: A systematic review and meta-analysis. Front. Immunol.12, 664526 (2021). 10.3389/fimmu.2021.664526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Omenaas, E., Bakke, P., Elsayed, S., Hanoa, R. & Gulsvik, A. Total and specific serum IgE levels in adults: Relationship to sex, age and environmental factors. Clin. Exp. Allergy24, 530–539 (1994). 10.1111/j.1365-2222.1994.tb00950.x [DOI] [PubMed] [Google Scholar]
- 45.Barbee, R. A., Halonen, M., Lebowitz, M. & Burrows, B. Distribution of IgE in a community population sample: correlations with age, sex, and allergen skin test reactivity. J. Allergy Clin. Immunol.68, 106–111 (1981). 10.1016/0091-6749(81)90167-6 [DOI] [PubMed] [Google Scholar]
- 46.Stadlbauer, D. et al. SARS-CoV-2 seroconversion in humans: A detailed protocol for a serological assay, antigen production, and test setup. Curr. Protoc. Microbiol.57, 1–15 (2020). 10.1002/cpmc.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vermont, C. L. et al. Antibody avidity and immunoglobulin G isotype distribution following immunization with a monovalent meningococcal B outer membrane vesicle vaccine. Infect. Immun.70, 584–590 (2002). 10.1128/IAI.70.2.584-590.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chackerian, B., Lowy, D. R. & Schiller, J. T. Conjugation of a self-antigen to papillomavirus-like particles allows for efficient induction of protective autoantibodies. J. Clin. Invest.108, 415–423 (2001). 10.1172/JCI11849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dimitrov, J. D., Lacroix-Desmazes, S. & Kaveri, S. V. Important parameters for evaluation of antibody avidity by immunosorbent assay. Anal. Biochem.418, 149–151 (2011). 10.1016/j.ab.2011.07.007 [DOI] [PubMed] [Google Scholar]
- 50.De Gaspari, E. & Zollinger, W. Expression of class 5 antigens by meningococcal strains obtained from patients in Brazil and evaluation of two new monoclonal antibodies. Braz. J. Infect. Dis.5, 143–153 (2001). 10.1590/S1413-86702001000300007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data from this manuscript may be made available upon reasonable request to the corresponding author (Professor Elizabeth De Gaspari, PhD).