Abstract
Sequential lytic cycles driven by cascading transcriptional waves underlie pathogenesis in the apicomplexan parasite Toxoplasma gondii. This parasite’s unique division by internal budding, short cell cycle, and jumbled up classically defined cell cycle stages have restrained in-depth transcriptional program analysis. Here, unbiased transcriptome and chromatin accessibility maps throughout the lytic cell cycle are established at the single-cell level. Correlated pseudo-timeline assemblies of expression and chromatin profiles maps transcriptional versus chromatin level transition points promoting the cell division cycle. Sequential clustering analysis identifies functionally related gene groups promoting cell cycle progression. Promoter DNA motif mapping reveals patterns of combinatorial regulation. Pseudo-time trajectory analysis reveals transcriptional bursts at different cell cycle points. The dominant burst in G1 is driven largely by transcription factor AP2XII-8, which engages a conserved DNA motif, and promotes the expression of 44 ribosomal proteins encoding regulon. Overall, the study provides integrated, multi-level insights into apicomplexan transcriptional regulation.
Subject terms: Parasite genomics, Regulatory networks, Parasitic infection
Integration of gene expression and chromatin accessibility data for Toxoplasma gondii resolves transcriptional clusters to DNA motifs and identifies a G1 transcriptional burst associated with a ribosomal protein regulon driven by AP2XII-8.
Introduction
The beginning and end-point of the Toxoplasma gondii lytic cycle is an invasion competent tachyzoite harboring the apicomplexan phylum-defining complex of apical secretory organelles and cytoskeletal structures1. Two daughter cells per cell division round are produced by internal budding (endodyogeny)2. In this process, daughter buds encased by the cortical membrane cytoskeleton elements are assembled around the centrosome, which ultimately consumes the mother cell3. Apicomplexan cell division is strikingly different from mammalians and the canonical cell cycle phases poorly fit this process. Notably, daughter budding, which can be considered cytokinesis, occurs simultaneously with S and M phases (Fig. 1e)4. A canonical G2 phase is absent, whereas only G1 is clearly separated from all the other events. Recently, we encapsulated the apicomplexan cell division cycles in four distinct modules: (1) mother cytoskeleton disassembly, (2) DNA synthesis and chromosome segregation (D&S), (3) karyokinesis, and (4) daughter bud assembly (budding)5. Division modes within and across apicomplexan parasites differ by timing, recurrences of individual modules, and the number of repetitions, among which T. gondii endodyogeny is the simplest and most accessible model system.
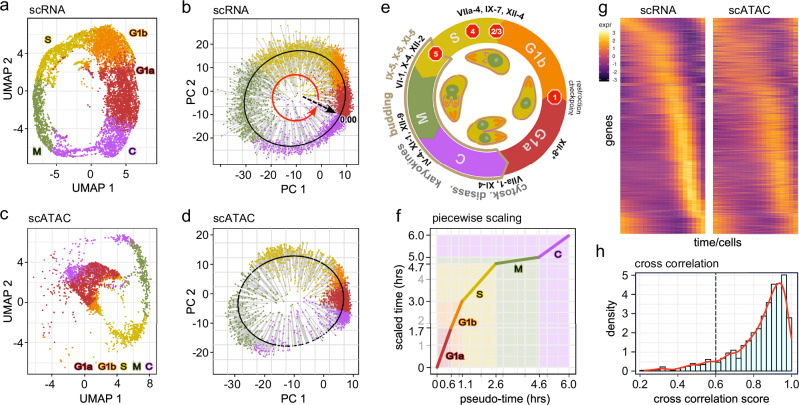
Fig. 1. scRNA-seq and scATAC-seq analysis of the tachyzoite lytic cell division cycle.
Asynchronously replicating intracellular RH strain tachyzoites were subjected to the 10x Genomics scRNA-seq (a, b) or scATAC-seq (c, d). Data were projected on UMAP (a, c) and the first two PCA (b, d) coordinates. Each dot represents a single tachyzoite. Derived cell division stage assignments were based on the definition put forward by Xue et al.6. In (b, d), the lytic cycle pseudo-time was reconstructed. The pseudo-time gene curve was constructed by mapping the cells ordered to 0–6 h, starting at the derived C-G1a transition. Each cell is color-shaded accordingly. e Schematic of the cell division cycle. Division by internal budding is illustrated in the inner circle and overlaps with S, M, and C phases. Stop signs illustrate the following checkpoints: (1) restriction; (2) DNA licensing; (3) centrosome duplication; (4) spindle assembly; (5) daughter assembly39. AP2 factors that have experimentally been validated are marked at the stages they act. * AP2XII-8 was established in this study. Panel modified from12. f Calibration of scRNA-seq derived pseudo-time against experimentally defined timeline of the cell division cycle stages (scaled time)4,6. g Pseudo-time heatmaps of single-cell transcriptome (left) and chromatin accessibility (right) for the 1238 genes displaying cyclic expression profiles. Rows: individual genes; columns: time (19 arbitrary time points) measured in different cells. Top to bottom organized by timing of peak gene expression, corresponding with the gene order in scRNA-seq. See Supplementary Fig. 1f for the reciprocal alignment by scATAC-seq peaks. Legend shows the range of scaled expressions. h Distribution of cross-correlation scores (CSS) representing the similarity of expression and accessibility curves across 1238 cyclic DEGs. The majority of genes show a CCS between their expression and accessibility profiles (CCS > 0.6). See also Supplementary Fig. 1 and Supplementary Data 1.
Cascading waves of just-in-time gene expression profiles promote progression through the division cycle2,6. The chromatin state defines the developmental stage and poises expression of the required genes throughout the cycle2,5,7. Temporal gene expression throughout the cell division cycle is mediated by transcription factors (TFs). Each TF controls a regulon, defined as a set of genes under the regulation of the same TF. The most abundant TFs in T. gondii belong to the Apetala2/ethylene response factor (AP2) family, which is the pivotal TF family driving the cascading transcriptional activities6,8,9. T. gondii harbors 67 AP2 TFs, the vast majority of which are expressed in the lytic cycle8,10–12.
Several studies have analyzed the transcriptome and chromatin state of apicomplexans. For example, transcriptome oscillations throughout the T. gondii tachyzoite lytic cycle have been determined by microarrays on synchronized parasites8, and later by single-cell RNA sequencing (scRNA-seq) on a small number of FACS sorted tachyzoites6. Moreover, bulk genome-wide studies on asynchronously cycling parasites have revealed that active promotor regions are marked by a complex pattern of histones H3 and H4 methylation and acetylation13–16. Together with MNase-seq data15 and recently applied Assay for Transp osase-Accessible Chromatin (ATAC) on bulk replicating tachyzoites17 this demonstrated that chromatin opening facilitates TF access and is a major regulatory mechanism underlying gene expression. However, there is no time-resolved, simultaneous expression and chromatin accessibility data available that permits the analysis of how different levels of control are integrated.
Here, we expand on the transcriptional insights by combining our recently developed experimental and computational tools18 with scRNA-seq and single-cell ATAC-seq (scATAC-seq) data on replicating tachyzoites (Supplementary Fig. 1a). Using a custom pseudo-time analysis pipeline, we constructed the oscillating transcriptome and chromatin landscape of T. gondii and showed that expression and chromatin opening are in general highly correlated. Through analysis of gene expression patterns and open chromatin regions, we identified major transition points in the T. gondii cell division cycle, revealing different organization levels of cell cycle checkpoint and progression regulations. Analysis of the cell cycle regulated AP2 TF expression profiles resolved four major clusters. An AP2 cluster peaking during the C/G1a transition was conjectured to fuel a major transcriptional burst during G1a. Functional analysis of AP2XII-8, an essential TF driving this burst, revealed its requirement for the expression of a ribosomal protein regulon. This was mediated by a DNA motif contained within a larger ribosomal protein motif, indicating that AP2XII-8 cooperates with other factors to regulate ribosome gene expression. To facilitate usage and access to our datasets and tools, an interactive web-app is provided where users can interact with the data and perform various analyses, including clustering and co-expression analysis (Supplementary Fig. 1b).
Results
Mapping T. gondii cell cycle progression by single-cell RNA and ATAC sequencing
We performed 10x Genomics scRNA-seq and scATAC-seq on RH strain T. gondii tachyzoites throughout the lytic cell division cycle. For scRNA-seq we acquired 12,735 cells with an average of 6220 reads/cell mapping to 488 genes/cell (median). scATAC-seq captured 3506 cells at a depth of 68,670 reads per cell with a median of 2911 fragments per cell (Supplementary Data 1). The Seurat R package19 was utilized to process and scale the data and perform dimensionality reduction. Figure 1a–d shows the Uniform Manifold Approximation and Projection (UMAP) and principal component analysis (PCA) plots of the scRNA-seq and scATAC-seq data. We used the previously DNA content inferred cell cycle stage assignments (G1a, G1b, S, M, C) and transferred them to our data using canonical correlation analysis19. This shows that the cell cycle progresses through a closed circular trajectory, sequentially traversing each cell cycle phase. This circular pattern is due to the periodic gene expression during the replication cycle.
To map the cell cycle progression, we performed a pseudo-time analysis by fitting a closed principal curve to the first two principal components (PCs) and orthogonally projected the cells onto this curve (Fig. 1b, d). The start of the cell cycle was set to the beginning of G1a, and cells were ordered along the trajectory within 19 evenly distributed time points. The arc-length was scaled from 0 to 6, reflecting the hours of the cell division cycle4 (Fig. 1f). We then applied a piecewise linear scaling to match the length of each pseudo-time phase to previously reported time-length of each phase4 (Supplementary Fig. 1c). Subsequently, we generated expression and accessibility time-series curves for each expressed gene (~6600 out of ~8200 total detected genes) using the scaled pseudo-time by fitting weighted periodic smoothing splines to the expression and accessibility data. A Fourier-based analysis was used to identify cyclic genes, defined as those displaying significant magnitudes of the dominant frequencies over one cycle. The analysis identified 4097 cyclically expressed genes and 3506 genes with cyclic chromatin accessibility, of which 2549 genes were dually cyclic in expression and accessibility curves (Supplementary Fig. 1d). Among the 2549 genes, we identified genes significantly overexpressed in at least one of the canonical phases (FC > 1.5, adj-p value < 0.05), resulting in 1238 unique cyclic differentially expressed genes (DEGs), out of 1620 total detected DEGs (Supplementary Fig. 1e and Supplementary Data 2).
Transition points of accessibility and expression chart regulatory events
Considering the just-in-time gene expression principle, the peak expression of cyclic genes strongly correlates with when the gene product is needed, whereas the peak of chromatin accessibility should correlate with the transcriptional and/or epigenetic switches in cell cycle progression. To determine how these levels of control contribute to each cell cycle stage, we assembled the peak expression and accessibility times for each of the 1238 cyclic DEGs (Fig. 1g and Supplementary Fig. 1f), observing cascading expression/accessibility peaks that span the entire cell division cycle. Tight correlations between expression and accessibility curves, measured by the curve cross-correlation score (CCS), were detected for most (>90%) of the dually cyclic genes (Fig. 1h).
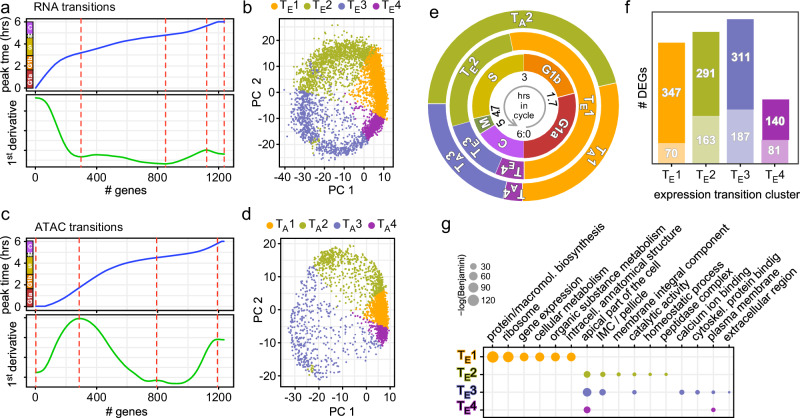
The non-linear progression of the peak expression times indicates that there are points of inflection that can define functional modules and switches in the cell division cycle (minor inflection points in close proximity to one another were merged together). To pinpoint these transitions, we calculated the sequential peak expression times of cyclic DEGs and identified the points of inflection (Fig. 2a). Utilizing these points as transition times, we found four distinct expression clusters: TE1–4 (Fig. 2b). Similar analysis was performed for DNA accessibility, which established four chromatin accessibility clusters: TA1–4 (Fig. 2c, d). When overlaid with the inferred cell cycle annotations6 (Fig. 2e inner circle), we observed that expression transitions corresponded well with C to G1a, G1b to S, and S to M transitions, but not with the M to C transition (Fig. 2e and Supplementary Fig. 1g, h). The TE1 cluster encompasses the entire G1a and G1b phases, whereas the ATAC-derived TA1 to TA2 transition marks the G1a to G1b switch, indicating regulation at chromatin level. The TE2 phase almost entirely matches the S phase, while the TE3 phase starts at the M phase and continues until mid C. On the other hand, the chromatin accessibility cluster TA2 encompasses G1b and S to the budding onset checkpoint (#5 Fig. 1e), whereas cluster TA3 starts slightly prior to TE3 and both end roughly at the same spot past the C midpoint. Finally, the TE4 and TA4 span the remainder up to G1a onset.
Fig. 2. Transition points mapping in scRNA-seq and scATAC-seq data.
Transcriptome (a) or chromatin accessibility (c) transitions identified from peak expression or chromatin accessibility of 1238 cyclic genes. Top panel shows the peak expression or chromatin accessibility time point for each gene ordered as in Fig. 1g or Supplementary Fig. 1f; bottom panel shows the first derivative. Red-dotted lines mark the transition points. The colored bars next to the y-axes in the top panels represent the cell division stages as defined in Fig. 1a–d. b, d RNA or ATAC transitions mapped to PCA of scRNA-seq or scATAC-seq data, respectively. TE1–4 and TA1–4 mark the cell populations falling within each transition cluster. The transition clusters are color-coded according to the labels to represent each phase. e Representative illustration of the biologically assigned stages (inner circle), the expression level transitions (TE1–4) (middle circle), and the chromatin accessibility transitions (TA1–4) (outer circle). f DEG analysis of each expression transition cluster TE1–4 defined in (a, b), relative to the other clusters. The total number of genes is composed of the upper number representing the number of genes with manually enhanced annotation on ToxoDB and the lower number in the lighter shade representing the genes annotated as hypothetical. g Functional enrichment for TE1–4 was performed by gene ontology (GO) analysis using ToxoDB annotations. Significant GO terms are shown (Benjamini <0.1). See also Supplementary Data 2.
Considering the cascading transcriptional events driving T. gondii’s replication, we theorized that the cell cycle switches were predominantly a reflection on expression transitions, with each transition propelled by a unique functional group of genes. To this end, we performed a differential gene expression analysis (DGEA) on the four expression-derived clusters, which identified 1347 unique DEGs (out of 1590 total DEGs: Fig. 2f and Supplementary Data 2), followed by gene ontology (GO) term analysis20,21. This revealed that TE1 contained genes typically associated with G1, whereas TE2–4 resolved in different functional clusters yet containing two daughter budding associated GO terms: apical part of the cell and IMC/pellicle (Fig. 2g). This reflects the extended budding throughout S-M-C.
Functional dissection by process and structure
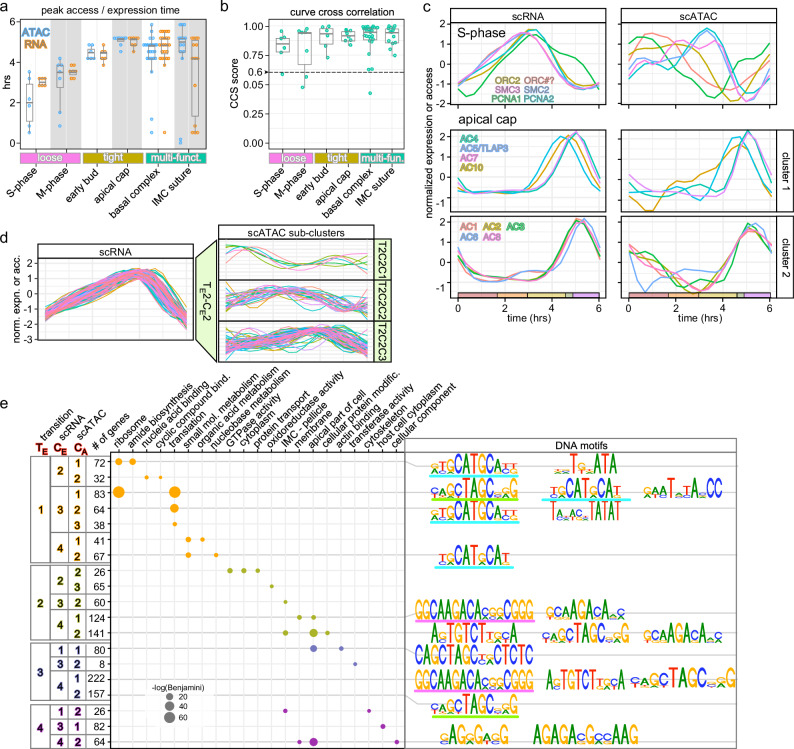
We assembled lists of genes acting in the same process and/or structure to probe our data for modules comprising functionally related gene sets. We selected processes and structures within the overlapping S-M-C and budding events (Fig. 3a–c and Supplementary Fig. 2). We differentiated three different correlations between expression and chromatin accessibility: loose (RNA peaks clustered; ATAC multiple peaks, or peaks hours offset from RNA peak), tight (peaks on both levels within 1.5 h window) and multi-functional (both RNA and ATAC peaks for the group spread out over hours) (Fig. 3a, b). We selected two representative gene sets for each of the variations. S- and M-phase genes represent loose correlation. Both gene sets display fairly tightly coordinated expression profiles (Fig. 3a, c and Supplementary Fig. 2a), yet, their chromatin accessibility profiles are scattered, with some peaking even in early G1 (Fig. 3a). When the whole curve shape is considered (CCS), several genes in each group appear below the 0.6 cut-off, confirming this “loose” pattern for the group (Fig. 3b). Overall, loose chromatin accessibility coordination despite a tight expression pattern suggests temporal control of TF activity.
Fig. 3. Dissection of functional gene sets expression and chromatin correlation.
Selected functional gene sets (details in Supplementary Fig. 2) representative of the different pattern (loose, tight, multi-fun(ct).) observed for peak times (a) and curve correlations between chromatin accessibility and expression profiles (b). Multi-fun(ct).: gene multi-functional sets in the same structure whose composition changes progressively through cell division. CCS > 0.6 indicates significant correlation. Box plots represent 75 percentile; horizontal bars indicate the median values; whisker represents the standard deviation. c Representative S-phase and apical cap gene sets, displaying loose and tight coordinated expression and chromatin patterns, respectively. S-phase: origin recognition complex (ORC2 and ORC#?)5, structural maintenance of chromosome factors SMC2 (condensin) and SMC3 (cohesin)60, and proliferating cell nuclear antigens 1 and 2 (PCNA1 and PCNA2)61. d Clustering analyses of DEGs in the TE2 transition cluster from Fig. 2f. For complete analysis results, see Supplementary Fig. 4. e GO term analysis using genes within each ATAC sub-cluster shown in Supplementary Fig. 4. Specific DNA motifs identified in the ATAC regions for genes in each sub-ATAC cluster are plotted. The gene clusters (dots) in the GO term plot are color-coded to indicate their transition stages, as shown in Fig. 2g. The GO terms were arranged (left to right) based on the Benjamini significance values. See also Supplementary Figs. 2, 4, 5 and Supplementary Data 3 and 4.
For the tight correlation we selected genes encoding early daughter bud (Fig. 3a, b and Supplementary Fig. 2b) and apical cap (Fig. 3a–c) proteins, which comprise distinct cytoskeletal elements assembled early in budding. The apical cap genes show tight peak times (4.5–5 h) that correspond at the expression and chromatin levels (Fig. 3c). Thus, this tight correlation suggests coordinated chromatin and expression controls for these genes.
The multi-functional gene sets represent the basal complex (BC) and different aspects of the IMC skeleton. The BC has several sequential functions through the lytic cycle, which is accompanied by a sequential change in composition22,23. This is reflected in the relatively large window of peak times (Fig. 3a and Supplementary Fig. 2c). Despite an outlier, we do see good CCS for these gene sets. An additional gene set that our analysis surprisingly put in this group is the IMC suture proteins, which are assembled during daughter budding24,25. Here we see variation in expression peaks, whereas the curve CCSs are tight (Fig. 3a, b and Supplementary Fig. 2g). Taken together, our analysis predicts distinct functions that we will probe in future work.
Since transcriptional waves are AP2 driven6,8, we examined the expression and chromatin opening of the 32 cyclically expressed AP2s (Supplementary Fig. 3). They resolve in four expression clusters and largely align with the analysis of the microarray time-course gene-expression data8,12. In general, expression profiles are well mirrored in the chromatin accessibility profiles, with some exceptions, e.g., AP2XI-3 and AP2VIIb-3. Thus, although over 90% of cyclic genes show a cross correlation of expression and chromatin profiles (Fig. 1h), some functional gene clusters deviate from this rule, which is important to acknowledge when interpreting automatically clustered data, and hints at more complex expression control mechanisms.
Lytic cycle dissection by transcription and chromatin patterns
We agnostically dissected the single-cell data sets to gain further biological insights in coordinated gene clusters. We resolved the gene expression transitions groups of the 1347 DEGs by an automatic time-series clustering (Supplementary Data 3). Within each transition phase (TE1–4), the expression profiles of DEGs were further sorted into four distinct groups using a shape-based kernel method (e.g., Fig. 3d and Supplementary Fig. 4, left panels). The corresponding chromatin accessibilities for genes in each sub-cluster were assembled (Supplementary Fig. 4, middle panels). Concurring with previous observations that chromatin opening profiles were more divergent than the expression levels, we found an indispensable need to perform a second clustering analysis, resulting in scATAC sub-clusters (right panels in Fig. 3d and Supplementary Fig. 4). Thus, our data resolves at sequential levels: (1) scRNA transition; (2) expression sub-clusters; (3) chromatin sub-clusters.
To assess the functions of clusters within each ATAC sub-cluster, we performed GO term analysis (Fig. 3e and Supplementary Data 3). Results indicate cascading and partially overlapping functional modules within the T. gondii cell division process. This analysis is consistent with the overlapping cell division processes defined by concurrently expressed functional modules that we can resolve in our data sets. To permit data queries with any gene of interest, we created a web app facilitating searches and clustering based on curve shape, either on the transcription or chromatin level, as well as in combination (Supplementary Fig. 1b). To add yet another dimension to the above assessments and inferences, we mapped conserved DNA motifs under each ATAC sub-cluster peak (Fig. 3e and Supplementary Data 4). In parallel, we also performed motif searches on the entire gene sets in each TE1-4 transition cluster to capture higher levels of co-accessibility (Supplementary Fig. 5a), which mapped within 2 kb around the transcription start sites (TSSs; most even within 1 kb), strongly implying that these are cis-motifs acting on the nearest genes (Supplementary Fig. 5b). Several of these motifs represented previously identified TgAP2 DNA binding sites. For example, motif GCTAGC (motif 4 in TE2 and TE3) as well as motif CAAGACA (motif 21 in TE2 and TE3) have been reported as binding sites of AP2XI-526 (Supplementary Fig. 5a). Constitutively expressed AP2XI-5 cooperates with cyclic AP2X-5 and is known to regulate multiple S/M-phase genes26,27. Furthermore, TE4-motif 10 (AGAGACG) is a sequence element found in dense granule (GRA) and SAG1 promoters28.
Strikingly, very similar motifs were found in distinct clusters. For example, the core motif GCATGC is found in clusters TE1CE2CA1, TE1CE3CA1, TE1CE3CA2, and TE1CE4CA2 (Fig. 3e). This motif has been recognized to be present in 44% of T. gondii tachyzoite promoters, regardless of expression pattern29. Although the four clusters mapped here share this motif, each is unique in that it is either the only motif (TE1CE4CA2), or there is a combination with one or two unique motifs (T(G/T)NATA) in TE1CE2CA1, CAGCTAGCNG/A)G and GAATATA(C/A)CC in TE1CE3CA1, and TAN(A/T)(C/G)NTATAT in TE1CE3CA2). The GO-terms associated with each of these four clusters do in part overlap among TE1CE2CA1, TE1CE3CA1, and TE1CE3CA2, which share the ribosome and translation GO terms, but the GO-terms associated with TE1CE4CA2 revolve around nucleotide metabolism. Taken together, combinatorial DNA motifs permit fine tuning the expression of different functionally related gene sets.
AP2XII-8 is required for proper progression through the G1a phase
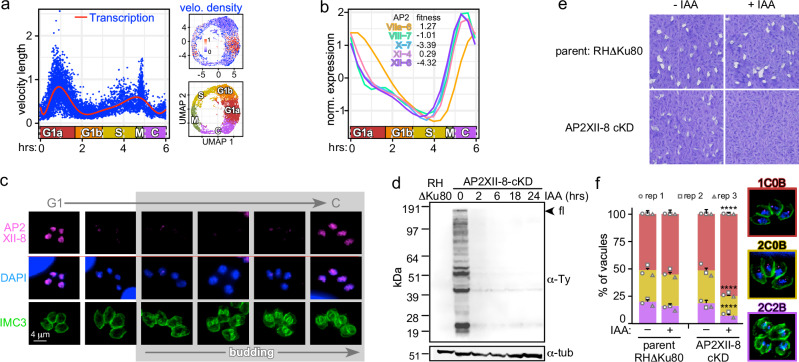
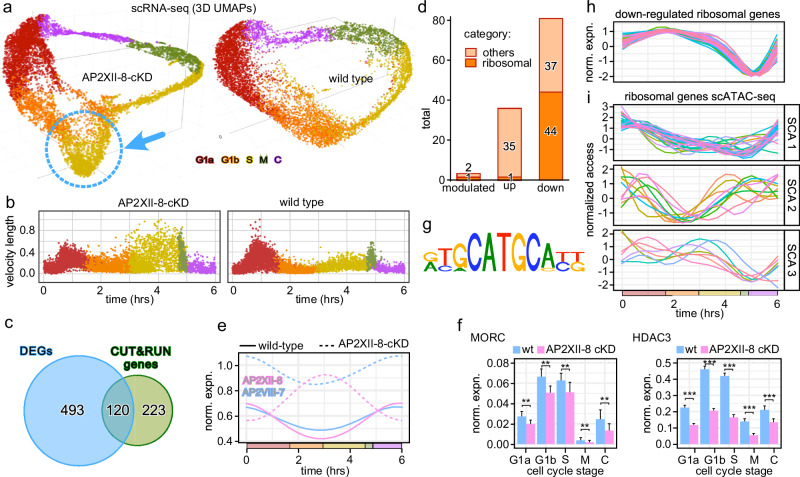
RNA velocity, derived from the kinetics of the ratio between spliced and incompletely spliced transcript, can be used as a measure of transcription rate30,31. Bursts in transcription rates point at major switches or checkpoints in transcriptional profiles31. We subjected the scRNA-seq data set to scVelo analysis31, revealing a major peak in G1a, next to a wider peak at the S-M-C transition (Fig. 4a). In particular, the G1a RNA velocity burst intrigued us as this stage has not been the focus of studies in Apicomplexa. We reasoned that this burst must be controlled by cyclic TgAP2s whose expression precedes G1a. TgAP2 cluster 4 comprised five TgAP2 TFs with such profiles (Supplementary Fig. 3). Two of these TgAP2s have a tachyzoite fitness score below −3, suggesting they are essential for lytic cycle progression32: AP2X-7 and AP2XII-8 (Fig. 4b). We had previously found that AP2XII-8 expression is enriched in a G1 entry temperature sensitive mutant33. Therefore, we selected AP2XII-8 to probe into the G1 transcription.
Fig. 4. Functional characterization of AP2XII-8 demonstrates it is essential for G1 progression.
a scVelo plot depicting RNA velocity length along the pseudo time. The calculated RNA velocity length can be viewed as a measure of active transcription. Each dot represents the velocity length of a cell. The red curve shows a fitted smoothing spline to the data. Inserts on the right show UMAPs of the density of RNA velocity (top) and the cell cycle stages as reference (bottom). b C-G1 peak expression profiles of the cluster 4 cyclically expressed AP2s (Supplementary Fig. 3). Fitness score for each of the five AP2s is listed. c Protein expression and localization of endogenously 5xTy tagged AP2XII-8 (strategy and validation: Supplementary Fig. 6) throughout the lytic cycle by IFA. AP2XII-8 was visualized with Ty MAb, DAPI highlights DNA and IMC3 antiserum tracks the cytoskeleton of mother and daughter buds. Representative of 2 biological replicates is shown. d AP2XII-8-mAID-5xTy knock-down kinetics assessed by western blot. Blot was probed with Ty MAb BB2 or α-tubulin as loading control. Full length (fl) AP2XII-8-mAID-5xTy has a predicted MW of 180.3 kDa. Representative of 2 biological replicates is shown. See Source Data for uncropped blots. Source data are provided as a Source Data file. e Plaque assay of AP2XII-8 depletion (8 days). Representative of 3 biological replicates is shown. f Phenotype quantifications in 24 h treated RHΔKu80 and AP2XII-8-cKD parasites. 1C0B 1 centrosome, 0 buds (G1), 2C0B 2 centrosomes, 0 buds (S), 2C2B 2 centrosomes, 2 buds (M/C). Centrosomes were detected using α-Centrin. At least 100 vacuoles were counted per biological replicate. Mean ± s.e. for n = 3 biological replicates. Statistics were performed using a two-sided standard two-sample z-test. ****p < 10−4. See also Supplementary Fig. 6 and Supplementary Data 8 and 10.
AP2XII-8 RNA expression peaks at the C-G1 transition (TE4-TE1) (Fig. 4b). We tagged the endogenous AP2XII-8 by CRISPR/Cas9 mediated insertion of a mini auxin-inducible degron (mAID) tag in frame with 5xTy epitopes (Supplementary Fig. 6). We tracked AP2XII-8 expression through the cell division cycle by immunofluorescence assay (IFA) co-stained with IMC3 as a daughter budding marker and DAPI as reporter for S-phase progression (nucleus size). AP2XII-8 localizes to the nucleus, and its temporal profile mimics its RNA expression: only detectable during the C-G1 transition (Fig. 4c). Next, we demonstrated that AP2XII-8 could be depleted from the parasites by adding IAA for as little as 2 h (Fig. 4d). Although by plaque assays we observed a minor fitness defect in the tagged strain under permissive condition (due to Tir1 introduction as reported using a similar strategy34), we did not observe any proliferation upon depletion, which confirms that AP2XII-8 is essential for the lytic cycle (Fig. 4e). Subsequently, AP2XII-8 depletion results in an accumulation of G1 stage parasites, supporting a role for AP2XII-8 in G1 progression (Fig. 4f). Thus, these findings pinpoint AP2XII-8 as an essential factor involved in the G1 transcriptional burst.
AP2XII-8 target identification by CUT&RUN and scRNA-seq
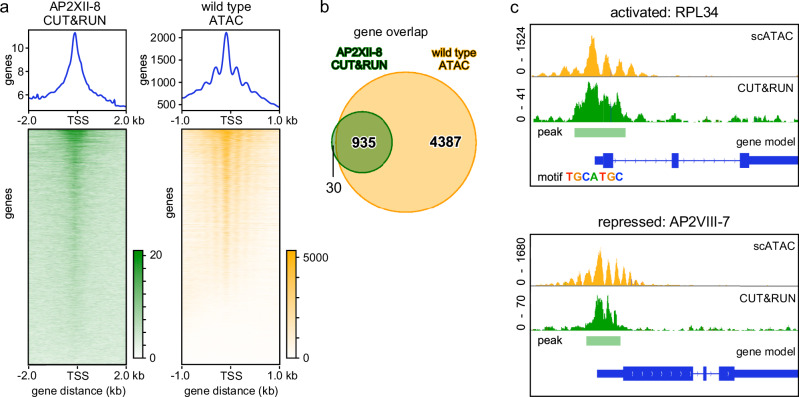
To resolve how AP2XII-8 promotes G1 progression we sought to determine the genes under its control. We applied Cleavage Under Targets and Release Using Nuclease (CUT&RUN) to determine the sites in the genome occupied by AP2XII-8. Although Ty epitope mediated CUT&RUN has been applied before to T. gondii35, we first optimized the procedure and determined that 200 million parasites with 2 μg BB2 antibody had the optimal detection capacity and comparable detection power in our H3K4me3 control as reported by ChIP-seq14,36 (Supplementary Fig. 7a–f). We processed the CUT&RUN data using three negative controls and identified 2029 binding events (peaks) with high stringency (adj_p value < 0.05). 88% of CUT&RUN and 94% of ATAC peaks fall within 2 kb distance of the TSS, consistent with a role in transcriptional regulation (Fig. 5a). We merged the nearby CUT&RUN peaks and assigned the long peaks to the nearest downstream gene, creating a list of 965 genes targeted by AP2XII-8 (Supplementary Data 5). The binding sites of 935 (97%) of these genes fall within the peaks determined from the scATAC-seq data (5322 genes; Fig. 5b and Supplementary Data 6). To discover the AP2XII-8 tightly controlled targets, we filtered out genes detected in any of the negative controls, and narrowed down to 343 genes that exhibit ~2-fold stronger read count intensity compared to the rest of the genes (Supplementary Fig. 7g). Robust overlaps between the CUT&RUN and ATAC profiles can be seen in the representative chromosome maps of two different genes, where AP2XII-8 is detected in the promoter (Fig. 5c).
Fig. 5. Identification of AP2XII-8 direct target genes by CUT&RUN.
a Display of AP2XII-8 CUT&RUN (left) and wild-type ATAC-peaks ranked by peak height (top to bottom) around the transcription start site (TSS). 2029 AP2XII-8 CUT&RUN peaks were mapped upon concatenating the highly significant (adj_p value < 0.05) binding events using 3 data sets (AP2XII-8 vs. 3 negative controls. p values were calculated using a one-sided test based on the Poisson distribution and adjusted for multiple testing using the Benjamini–Hochberg method. Gradient bars represent signal intensity. Supplementary Fig. 7). b Venn diagram shows the overlap of 965 genes to which CUT&RUN peaks were assigned with the 5322 genes with an ATAC profile from the scATAC-seq data considered as bulk (constitutive and cyclic at all time points combined). c Two representative examples of genes with a CUT&RUN and ATAC profile around their TSSs. RPL34 (TGGT1_227600) is activated by AP2XII-8 and contains a TGCATGC in its promoter (see Fig. 6) AP2VIII-7 (TGGT1_269010) is suppressed by AP2XII-8 and is devoid of motif TGCATGC. Green: AP2XII-8 CUT&RUN data; yellow: RH scATAC-seq results. See also Supplementary Fig. 7.
To determine the direct functional targets of AP2XII-8 (i.e., genes that AP2XII-8 has binding affinity to as well as modulations in expression upon binding), we generated scRNA-seq data upon 2 h AP2XII-8 depletion. Comparing the unperturbed and AP2XII-8 depleted parasites in 3D UMAPs shows a distinct bulge at the G1b-S interface in the KD condition (Fig. 6a). Furthermore, in the scVelo profile upon 2 h AP2XII-8 depletion shows a dampened G1a burst, strongly supporting AP2XII-8 is a major contributor to the G1a transcriptional burst (Fig. 6b). In addition, the velocity of S- and M-phase cells becomes more scattered (noise), corresponding with diverging bulge in the 3D UMAP. Under the theoretical assumptions that genes directly controlled by AP2XII-8 reside in the up and down-regulated genes, we performed a DEG analysis, comparing the AP2XII-8 KD vs. WT data in each cell division phase. This identified a total of 613 DEGs (Supplementary Data 5), of which 120 overlapped with the 343 CUT&RUN identified target genes (Fig. 6c and Supplementary Fig. 7g). Of the 120 genes, referred to as direct functional targets of AP2XII-8 hereinafter, 81 are downregulated (activated by AP2XII-8), 36 are upregulated (repressed by AP2XII-8), while 3 are both down- and upregulated in different phases (modulated) (Fig. 6d). Among all the direct functional targets, there are two AP2 TF, AP2VIII-7 and AP2XII-8 itself, that are AP2XII-8 regulated genes. Coincidentally, both AP2VIII-7 and AP2XII-8 display a similar C-G1a expression peak profile (Fig. 4b). Upon 2 h AP2XII-8 protein depletion, AP2XII-8 transcripts peak at the G1b-S transition, while the AP2VIII-7 transcriptional profile maintains its overall curve shape but increases its expression level for approximately 2-fold (Fig. 6e). More interestingly, the previously identified T. gondii sexual stage repressor complex, MORC/HDAC315, is also influenced by AP2XII-8 transcriptional activity. Both MORC and HDAC3 show mild G1b-S transcriptional peaks and are the repressing targets of AP2XII-8, however, only MORC is directly engaged by AP2XII-8 (Fig. 6f and Supplementary Data 5). Since MORC recruits HDAC3 to limit chromatin accessibility to the merozoite-specific genes following interaction with various AP2s, this raises an interesting question of how different AP2s interactively regulate each other and how these AP2s cooperate with other developmental stage-specific factors to promote T. gondii lytic cycle, and this finding here will serve as a starting point for future studies.
Fig. 6. AP2XII-8 perturbed scRNA-seq identifies an AP2XII-8 mediated ribosome regulon.
a 3D UMAPs of scRNA-seq datasets collected upon 2 h KD of AP2XII-8-cKD (left) and wild-type (right, same data as in Fig. 1a) parasites. The dotted blue circle marked the bulge cell population. Cells at different divisions are color-coded per the labels provided. b scVelo plots depicting RNA velocity length along the pseudo time for the AP2XII-8-cKD (2 h) and wild-type parent lines. The calculated RNA velocity length can be viewed as a measure of active transcription. Each dot represents the velocity length of a cell and its color corresponds to the derived cell cycle stage. c Venn diagram of AP2XII-8-dependent DEGs intersected with the AP2XII-8 CUT&RUN genes. d Differentiation of the 120 genes at the Venn intersection (c) by their mode of regulation. modulated means either up or downregulated dependent on the cell cycle stage. RP encoding genes are highlighted. e Two AP2 factors under direct control of AP2XII-8 change expression patterns upon AP2XII-8 depletion. f MORC and HDAC3 change of gene expression upon AP2XII-8 depletion. Bars indicate mean expression across cells in the indicated phase (x-axes). Two-tailed t-test performed on normalized expression data (not raw counts). **p < 10−2; ***p < 10−3. Whiskers indicate standard deviation. See Supplementary Fig. 1g for the number of cells. g Motif search performed on the 81 AP2XII-8 downregulated genes under the CUT&RUN peaks revealed the presence of one DNA motif. h Wild-type, unperturbed pseudo-time expression profiles of the 44 ribosomal genes activated by AP2XII-8. i Wild-type, unperturbed pseudo-time chromatin accessibility profiles of the 44 ribosomal genes activated by AP2XII-8 automatically clustered in three sub-clusters (SCA1-3). See also Supplementary Figs. 7 and 8 and Supplementary Data 5.
Motif searches under the CUT&RUN peaks of activated genes identified a single motif: (G/A)TGCATGCGA(T/C((T/G) (Fig. 6g). Of the 120 genes with CUT&RUN peaks which expression changes upon AP2XII-8 depletion 42, 5, and 2 of the down-regulated, upregulated and modulated genes, respectively, contain this motif. This same or very similar motif was identified in the TE1 phase cluster (Fig. 3e: TE1CE2CA1, TE1CE3CA1, TE1CE3CA2, and TE1CE4CA2; Supplementary Fig. 5a: motifs 6 and 16 in TE1). Thus, this agnostically detected motif in the scRNA and scATAC analysis can now be associated with AP2XII-8.
AP2XII-8 controls a ribosome protein regulon
Of the 81 genes activated by AP2XII-8, 44 encode ribosomal proteins (RPs) (Fig. 6d). The motif engaged by AP2XII-8 was previously mapped in the promoter regions of ribosomal proteins (RPs)37: it is contained within [T/C]GCATGC[G/A] (Supplementary Fig. 8a and Supplementary Data 4). This more extended motif was named Toxoplasma RP promoter element 2 (TRP-2)37.
The tightly clustered pseudo-time course expression profiles of the AP2XII-8 activated RP genes show depletion in M-phase and a broad peak starting at C and around the G1a-G1b transition (Fig. 6h). Their corresponding chromatin accessibility profiles resolved in three clusters (sub-cluster accessibility: SCA1−3; Fig. 6i), suggesting an additional epigenetic level of regulation.
To assess the full extent of AP2XII-8 regulated RPs, we performed a motif search for all RPs that have CUT&RUN signal assigned to their promoters regardless of differential expression status (76 out of 137 total RPs). To this end, we extended the CUT&RUN peaks by 250 nucleotides on each side. This identified the two previously reported motifs, matching TRP-1 (TCGGCTTATATTCGG) and TRP-2 ([T/C]GCATGC[G/A]) (Supplementary Fig. 8a)37. Of the 76 RPs with CUT&RUN signal, 67 had at least one of the motifs present with majority of RPs having TRP-2 (Supplementary Fig. 8b). Analysis of the distribution of distance between successive motifs shows that all tend to co-occur in relatively close distance to each other, whereas TRP-1 and TRP-2 tend to co-occur close to each other on opposite strands, hinting at possible homo- and/or hetero-dimerization (Supplementary Fig. 8c–e). Consistent with a role in transcriptional regulation, both motifs were strongly enriched within 1 kb of the TSS (Supplementary Fig. 8f). In conclusion, our analysis maps AP2XII-8 to the control of a subset of RPs through engaging a more narrowly defined DNA motif than the previously mapped TRPs. This is consistent with the presence of additional, combinatorial controllers of RP gene expression.
Discussion
Combined single-cell transcriptome and chromatin landscape analysis of the T. gondii lytic cycle facilitated the establishment of highly resolved pseudo-timelines at the two major levels of gene regulation. The scRNA and scATAC atlases are accessible through a newly developed web-app that permits data interrogation and visualization in several ways (https://umbibio.math.umb.edu/toxosc/; Supplementary Fig. 1b). Chromatin opening within 2 kb of the TSS corresponds very well to the cyclic expression pattern of 90% of the genes (Fig. 1). The data depth followed by subsequent RNA and chromatin level clustering analysis as well as DNA motif mapping permitted resolution of transcriptional regulation of the just-in-time gene expression beyond previous studies6,8,17.
Against the background of known cell cycle checkpoints and the functional modules of apicomplexan cell division (Fig. 1e)2,5,38,39, we mapped transition points between gene clusters at the chromatin and transcriptional level (Fig. 2). The C-G1a transition was consistently present across all levels, even though this is not a formal cell cycle checkpoint. The other checkpoints and cell biological module boundaries were more fluid. Some transitions were only detectable at the chromatin level (e.g., the restriction checkpoint), while others only at the transcriptional level (e.g., spindle assembly checkpoint) (Fig. 2e). Furthermore, there was one transition that did not align with a cell cycle checkpoint: the TE3–TE4 transition in the middle of C-phase, which does closely align with the TA3–TA4 transition. This transition is not clear in the PCA plot, but the scRNA UMAP plot shows two visually distinct cell groups in C-phase: the first is the tail coming out of M-phase, whereas the second half seamlessly associates with the G1a phase (Fig. 1a). Noting that the assignment of S-phase is based on 2N DNA content6, these cells are in the middle of the unique intertwined apicomplexan cell division event. At this point, T. gondii tachyzoites again execute multiple modules simultaneously: karyokinesis, mother cytoskeleton disassembly, and completion of daughter bud assembly (Fig. 1e). Indeed, most cortical cytoskeleton assembly genes peak at the 5–5.5 h mark (BC, alveolins, GAPMs, glideosome, apical cap, sutures) (Supplementary Fig. 2 and Fig. 3c). Currently, the genes responsible for karyokinesis and mother cytoskeleton disassembly are largely unknown but must be contained within the roughly 40% hypothetical genes within TE3 and/or TE4, thereby guiding our future work in revealing this biology (Fig. 2f). In conclusion, our analysis makes predictions about the level of checkpoint regulation, the modular regulation of transitions, and points to gene clusters executing the currently poorly understood biological processes. These findings provide numerous starting points for future dissections of apicomplexan specific biology.
The mapping of DNA motifs under the ATAC peaks of coordinated gene sets identified several known as well as novel patterns, leading to novel insights into the combinatorial regulation of gene expression. The first progressive insight is that we map the AP2XII-8 engaged TGCATGCA motif to several gene clusters in G1 (Fig. 3e and Supplementary Fig. 5a). This widespread motif has been reported before in T. gondii8,17,29, and here we mapped it as the AP2XII-8 RP regulon motif (Fig. 6 and Supplementary Fig. 8a). Furthermore, this regulon is essential for G1 progression and is part of the G1 transcriptional burst.
The AP2XII-8 motif is contained within the previously mapped, much longer TRP-2 motif (Supplementary Fig. 8a)37. This suggests additional factors likely associated with this motif, which are potentially in a complex with AP2XII-8. ApiAP2s are capable of homo- or hetero-dimerization and can act cooperatively or competitively as repressors and/or activators on the same gene12,16,27,40,41. Furthermore, TRP-2 is found in genes well-beyond RPs, whereas TRP-1 is more narrowly present in RP promoters37. Hence, other protein complex combinations could engage (part of) the same TRP-2 motif. Besides the RP regulon demonstrating cis-regulation by AP2XII-8, 223 AP2XII-8 promoter region binding sites are transcriptionally inactive (Fig. 6c and Supplementary Data 5). This could indicate that genes are co-regulated by additional TgAP2 factors, either in (an alternative) heterodimeric complex or as an additional, dominantly acting TgAP2 in the promoter of these genes. In addition, TgAP2s are encountered in complex with epigenetic factors7,42–44. For example, TgAP2s acting as repressors of non-tachyzoite genes are in a complex with MORC and HDAC3 to maintain a silent but poised state7,16,45. Interestingly, both MORC and HDAC3 are upregulated upon AP2XII-8 depletion, whereas MORC is directly engaged by AP2XII-8, but HDAC3 is just below the cut-off in the CUT&RUN data (Fig. 6f and Supplementary Data 5). In future work we will explore how these mechanisms mesh with AP2XII-8.
The G1-S phase bulge in the 3D UMAP upon AP2XII-8 depletion comprises AP2IX-10, a stress factor normally not expressed in the lytic cycle46, and several ApiAP2 factors that in the normal cycle peak in S-phase (Supplementary Fig. 3: AP2s IV-5, VIIa-3, VIIa-7, X-9 and XII-9). None of these are engaged by AP2XII-8 in the CUT&RUN data but are consistent with a transcriptional signature of S-phase entry. However, AP2XII-8 depletion prevents G1-S progression on the cell biological level (Fig. 4f). Interestingly, AP2XII-8 contains the DNA-motif it engages in its own promoter and appears under the control of a feedback loop (Fig. 6e) Collectively, these data hint at other factors in parallel with AP2XII-8 driving G1-progression. Indeed, we mapped such a putative factor with a severe fitness score, AP2X-7, which displays the same expression cluster as AP2XII-8 (Fig. 4b).
Aligning with our T. gondii data, time course chromatin accessibility and gene expression measurements of synchronized erythrocytic cycle Plasmodium falciparum demonstrated that the chromatin opening upstream of transcribed genes corresponds very well with the temporal/developmental changes in transcriptional levels9. Indeed, chromatin accessibility displays a slightly wider temporal peak than transcriptional activity in our data (Fig. 1g). Although at that point, no time-resolved insights are available, active promotor regions are marked by a complex pattern of histones H3 and H4 methylation and acetylation in both P. falciparum47 and T. gondii13. Overlaying, integrating, and further time resolving these pattens as shown here will advance the understanding of how transcription is organized and meshes at different levels.
In summary, the combination of transcription and chromatin opening data synergizes into a deep understanding of the transcriptional network driving the unique T. gondii cell division cycle. Furthermore, the new methods and analytical approaches can be applied to other biological lytic cycle processes, or to any other comparative cell cycle studies.
Methods
Parasites and host cell culture
All parasite strains used for this study are listed in Supplementary Data 7. T. gondii tachyzoite cultures were maintained in human telomerase reverse transcriptase (hTERT)-immortalized human foreskin fibroblasts (HFFs)48. Immunofluorescence assays (IFAs), were performed using primary HFFs. Selection for stable parasite transfectants was performed under 1 µM pyrimethamine and stable lines were cloned by limiting dilution. Protein knock-downs using the minimal Auxin-Inducible Degron (mAID) system were performed under 500 µM auxin (IAA: 3-indoleacetic acid in 100% ethanol; Sigma-Aldrich).
Plasmid cloning and parasite strain generation
All primers, synthetic DNA (Twist BioScience), and plasmids used are provided in Supplementary Data 8.
An AP2XII-8 targeted protospacer sequence (P1 primer) was incorporated into BsaI-HF digested pU6-universal plasmid. The resultant plasmid was denoted as pU6-AP2XII-8.
Plasmid mAID-5xTy/DHFR/OsTir1_N was constructed by Gibson assembly (NEBuilder HiFi DNA Assembly Master Mix; NEB) of the following fragments: 1. mAID CDS PCR amplified (TruFi DNA Polymerase; Azura Genomics) from plasmid plinker-AID-Ty-HXGPRT-LoxP using primers P8 and P9; 2. the codon diversified sequence of 5xTy epitope tag including the 3’UTR from DHFR was PCR amplified from pTWIST_5xTY_3’-dhfr using primers P10 and P11; 3. the DHFR promoter was amplified from plasmid DHFR-TetO7‐sag4‐Ty using primers P12 and P13; 4. the DHFR-TS(m2m3) CDS and T2A skip peptide were amplified from plasmid DHFR_T2A_5xV5 using primers P14 and P15; 5. the OsTir1 CDS was amplified from RHΔKu80/Tir1 strain genomic DNA using primers P16 and P17; 6. the HXGPRT 3’-UTR was amplified from a plasmid plinker-AID-Ty-HXGPRT-LoxP using primers P18 and P19.
Subsequently, an AvrII restriction enzyme site was introduced in the above plasmid between DHFR-TS and T2A to generate plasmid mAID-5xTy/DHFR/OsTir1, by Gibson assembling the following fragments: (1) plasmid backbone containing mAID-5xTy and the DHFR promoter generated by BglII, AatII and NotI digestion of plasmid mAID-5xTy/DHFR/OsTir1_N; (2) PCR amplified DHFR-TS CDS from plasmid mAID-5xTy/DHFR/OsTir1_N using primers P25 and P26, which introduces the AvrII site; (3) PCR amplified fragment containing T2A, OsTir1 and HXGPRT 3’-UTR containing the additional AvrII site from plasmid mAID-5xTy/DHFR/OsTir1_N using primers P27 and P28. The resulting plasmid was validated by various restriction enzyme combinations, PCR (primer pairs P22 + P23 and P22 + P24), Sanger sequencing (primers P6, P21, P21, P22, P23 and P24), and whole plasmid sequencing through Plasmidsaurus.
The AP2XII-8-cKD strain was generated by co-transfecting the RHΔKu80 strain with 50 µg of plasmid pU6-AP2XII-8 and 4 µg of PCR repair cassette amplified from plasmid mAID-5xTy/DHFR/OsTir1 using primers P2 and P3. Parasite clones were validated by diagnostic PCR (Supplementary Fig. 6).
(Immuno-) fluorescence microscopy
Intracellular parasites grown overnight in a 6-well plate containing coverslips confluent with HFF cells were fixed with 100% methanol and blocked for 30 min with 0.5% BSA in 1x PBS. The primary antisera (IMC3, 1:2000; Ty (BB2), 1:500; Centrin (Millipore;clone 20H5), 1:1000) and secondary antisera (Thermo Fisher Scientific; mouse A594, 1:400; rabbit A488, 1:400) were diluted in blocking solution and applied for 1 h at RT, followed by three 5 min washes in 1x PBS. DNA staining was performed using 4’,6-diamidino-2-phenylindole (DAPI) in the first wash after the last antibody. Images were acquired with a DeltaVision Ultra microscope and subsequent processing was carried out with FIJI software. For the quantitative analyses for the cell cycle arrest study, at least 100 vacuoles were examined per experiment.
Western blot
Intracellularly replicating parasites were harvested through 26G needle passage and filtered (3 µm pore polystyrene filter), washed 3 times in 1X PBS before lysis in resuspension buffer (50 mM Tris-HCl pH 7.3, 150 mM NaCl, 1:50 diluted Protease Inhibitor Cocktail [Sigma-Aldrich, 1% SDS). An equivalent of 2 × 107 parasites whole protein lysate was loaded on a NuPAGE 4–12% Bis-Tris protein gel (Invitrogen) followed by transfer onto a PVDF membrane (Thermo Scientific). The blot was blocked in 6% milk in 1X PBS (for α-Ty1 Tag BB2, 1:5000; Thermo Fisher Scientific) or 5% milk 1% BSA (for α-tubulin MAb, 1:2000; DSHB) and probed with primary and secondary antibodies (Mouse Immunoglobulins/HRP, 1:10,000; Agilent) listed in Supplementary Data 9. Blots were washed 3 × 10 min in PBST (1X PBS with 0.1% Tween 20) following each antibody incubation. Ty signal was developed by SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) whereas tubulin signal was developed by Immobilon Western Chemiluminescent HRP Substrate (Millipore). Chemiluminescent detection was performed on a LI-COR Odyssey M imager. Blots were stripped for re-probing using stripping buffer (62.5 mM Tris-HCl pH 6.8, 2% SDS, and 0.8% (v/v) β-mercaptoethanol) at 50 °C for 20 min followed by 3 × 10 min PBST washes.
scRNA-seq
RHΔKu80::ptub-YFP2/sagCAT (WT) or AP2XII-8-cKD parasites were grown overnight in hTERT cells and harvested by 26G needle passage and 3 µm pore polystyrene filtration. Parasite concentrations were adjusted to 2.5 × 106/ml and 4 ml (107 tachyzoites) were inoculated on T25 flasks confluent with HFFs. Two T25 flasks were used per time point (27, 30, and 33 hpi for WT; 27, 30 hpi for AP2XII-8-cKD; IAA was applied for 2 h to AP2XII-8-cKD parasites prior harvest) and intracellular tachyzoites harvested by washing monolayers 3X with ice-cold 1X PBS, scraping the parasites-contained HFFs in ice-cold Ed1 media, and releasing parasites by 26G needle passage. For each time point 5 × 106 tachyzoites were each pelleted at 1000 × g, 4 °C for 15 min and resuspended in 1 ml of ice-cold resuspension buffer (1X Hank’s Balanced Salt Solution (Thomas Scientific), 0.04% BSA (MilliporeSigma-Omnipour)). Parasites were pelleted at 1000 × g, 4 °C for 15 min and resuspended in 60 µl of ice-cold resuspension buffer. Recounted parasites were pooled across the time points at 106/ml for at an end volume of 150 µl.
scRNA-seq library preparation followed the Chromium Next GEM Single-Cell 3’ Reagent Kits v3.1 protocol (10x Genomics; WT) or Chromium Next GEM Single-Cell 3ʹ Reagent Kits v3.1 (Dual Index) (10x Genomics; AP2XII-8-cKD), aiming for 10,000 cell recovery. The quality and concentration of the generated scRNA-seq library was assessed using an Agilent 2200 TapeStation. The WT library was sequenced on an Illumina NextSeq 550 High-Output Kit (150 cycles) flow cell at the Tufts University Core Facility (TUCF) with the following sequencing parameters: Read 1 (28 cycles), i7 Index (8 cycles), i5 Index (0 cycles), and Read 2 (91 cycles). The AP2XII-8-cKD library was sequenced on an Illumina NextSeq 550 Mid-Output Kit (150 cycles) flow cell at TUCF with the following sequencing parameters: Read 1 (28 cycles), i7 Index (10 cycles), i5 Index (10 cycles), and Read 2 (90 cycles).
scATAC-seq
RHΔKu80::ptub-YFP2/sagCAT inocula were prepared as for scRNA-seq. A total of 5 × 106 mechanically released tachyzoites per time point were each pelleted at 1000 × g, 4 °C for 15 min. Without disturbance, the pellet was washed with 100 µl ice-cold 1X PBS followed by 1000 × g, 4 °C for 15 min. Cells were lysed using 100 µl of ice-cold ATAC lysis buffer (Active Motif), followed immediately by addition of 1 ml pre-chilled wash buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1% BSA, 0.1% Tween-20). Nuclei were pelleted at 500 × g, 4 °C for 5 min and resuspended in 190 µl ice-cold resuspension buffer (as for scRNA-seq). Each sample was resuspended to 4 × 106/ml and 100 µl of each nuclei sample was pooled.
The scATAC-seq library was produced following the Chromium Next GEM Single-Cell ATAC Reagent Kits v1.1 protocol (10x Genomics) aiming for 10,000 nuclei recovery. The quality and concentration of the generated scATAC-seq library was assessed on an Agilent 2100 Bioanalyzer and sequenced on a NovaSeq 6000 platform at the Bauer Core Facility (Harvard University) using the following run parameters: Read 1N (50 cycles), i7 Index (8 cycles), i5 Index (16 cycles), and Read 2N (50 cycles).
CUT&RUN
Intracellularly replicating AP2XII-cKD or RHΔKu80 parasites were harvested 30 h post-inoculation by 26G needle passage and pelleted by centrifugation at 1000 × g, 4 °C for 15 min (same spin condition for subsequent washes). Parasites were washed following the ChIC/CUT&RUN Kit Version 3 (EpiCypher) user manual before binding to Ty1 BB2 MAb, Ty1 Diagenode MAb, H3K4me3 rabbit antibodies, or Mouse IgG1 Isotype Control (antibody details in Supplementary Data 9 and Supplementary Fig. 7a–d). Subsequent chromatin digestions and CUT&RUN DNA generations were performed following the user manual (EpiCypher), with 5 ng CUT&RUN-enriched DNA carried over to the library preparation step.
The CUT&RUN libraries were produced utilizing the CUT&RUN Library Prep Kit (EpiCypher, Inc, Catalog No. 14-1001) User Manual Version 1.0, with modifications pointed out in the manual tailored specifically to the transcription factor applications. The qualities and concentrations of the generated CUT&RUN libraries were assessed using an Agilent 4150 TapeStation. The libraries were multiplexed following the Primer Selection Guide (EpiCypher, Inc) and sequenced on the Illumina MiSeq Standard V3 150 cycles (75 bp paired-end) flow cells at the Tufts University Core Facility (TUCF).
RNA- and ATAC-seq read alignment
Reference genome and gene annotations for T. gondii (ME49 strain, release 59) were downloaded from https://toxodb.org/49. Custom references were assembled using the 10X genomics pipelines (cellranger-7.0.1 and cellranger-atac-2.1.0)50 using the cellranger mkref command. Subsequently, the raw RNA-seq and ATAC-seq reads were mapped against the reference genome using cellranger count and cellranger-atac count commands respectively with the default parameters.
scRNA-seq data processing
Downstream data analysis was performed in R (version 4.2.2). Gene expression count data was processed using the Seurat R package (version 4.3.0)19. Lowly expressed genes and cells with a few number of detected reads were filtered from downstream analysis using Seurat function CreateSeuratObject with parameters min.cells = 5, and min.features = 100. Expression data was normalized and scaled using NormalizeData with parameter normalization.method = “LogNormalize”, FindVariableFeatures with nfeatures = 6000, and ScaleData. Dimensionality reduction was performed using Principal Component Analysis (PCA) and Uniform Manifold Approximation (UMAP) as implemented in Seurat using runPCA() and runUMAP() with parameter dims = 1:10. Clustering analysis was carried out using KNN graph based technique using FindNeighbors and FindClusters functions with parameters dims = 1:10, reduction = “pca”. The data set was down sampled to include 8000 cells. Cell labels were inferred using the published scRNA-seq data6 with Seurat functions FindTransferAnchors() and TransferData().
scATAC-seq data processing
The chromatin accessibility peak-by-cell matrix data was processed in R using the Seurat R package (version 4.3.0)19. Cells and peaks with low counts were filtered out using Seurat function CreateChromatinAssay with parameters min.cells = 5 and min.features = 100. Significantly enriched peaks were selected for further downstream analysis (peak_region_fragments >200, peak_region_fragments <6000, pct_reads_in_peaks >40), nucleosome_signal <4 and TSS.enrichment >2). Data was normalized with Seurat function RunTFIDF. Dimensionality reduction was performed using singular value decomposition (SVD) and Uniform Manifold Approximation (UMAP) as implemented in Seurat functions RunSVD with default parameters and RunUMAP with parameters reduction = “lsi” and dims = 1:30. Clustering analysis was performed with functions FindNeighbors and FindClusters with parameters reduction = “lsi”, dims = 1:30 and algorithm = “SLM algorithm”. Gene activity matrix was then generated from the chromatin assay using GeneActivity function extending the regions to 600 and 200 bp up- and down-stream of the TSS. The gene activity was utilized to create a Seurat object followed by normalization and clustering analysis similar to the scRNA-seq data. The scRNA-seq and scATAC-seq data were integrated using Seurat functions SelectIntegrationFeatures, FindIntegrationAnchors and IntegrateData with scRNA-seq data used as reference for integration.
Peak gene assignment
All peaks detected by cellranger 10x pipeline (5444 total peaks) and gene annotation file were used to assign peaks to genes using bedtools51. Peaks located entirely within the gene/coding region (exon-2 to exon-n) were filtered out from further analysis (427 total). The remaining peaks were assigned to the nearest downstream gene with distance cut-off <3000 bp. Multiple peaks assigned to the same gene were merged. In total 5322 genes were uniquely assigned to a peak.
Pseudo-time analysis
Pseudo-time analysis was performed as previously described18. Briefly, an elliptic curve was fitted to the first two principal components in each scRNA- and scATAC-seq data using Ellipsefit function from MyEllipsefit R package (https://github.com/MarkusLoew/MyEllipsefit). The elliptic curve was used as a prior to fit a closed principal curve to each data set using principal_curve function from the R package princurve with the parameter smoother = “periodic_lowess“52. Cells were orthogonally projected on to the principal curve and were ordered according to arc-length distance from the beginning of the curve. The transferred cell cycle phases were used to adjust the start time of the trajectory to match the start of G1a phase. A piecewise linear scaling was performed to scale each phase to match the actual length as previously reported4.
Marker analysis
Differential expression analysis (DEA) was performed using FindAllMarkers function from the Seurat R package with parameters only.pos = TRUE. This analysis was carried out to identify DEGs of each inferred cell cycle phase. Fold change cutoff >1.5 and adjusted p value < 0.05 were used to determine significant DEGs. The same function and cutoffs were used to identify DEGs of each detected transition group.
Fitting gene curves
The expression and accessibility of genes along the reconstructed pseudo-time were estimated by fitting a smoothing spline to expression and accessibility profile of each gene with smooth.spline function and smoothing parameter λ = 0.1 and weight = 1/3 for 0 expression values. The fitted splines were sampled at regular intervals. Timing of peak expression and accessibility per gene were calculated by examining the local maxima of the fitted curves.
Curve cross correlation analysis
Reconstructed time course expression and accessibility were used to measure the similarity (correlation) of the two curves in corresponding genes using ccf function in R53.
Transition points along the cell cycle
A smoothing spline was fitted to the calculated peak expression and peak accessibility of cell cycle regulated genes with the smooth.spline function and smoothing parameter λ = 0.005. The smoothing parameter was set by trial and error. The transition points in the peak expression and peak accessibility curves were identified by examining the inflection points of the fitted curves. The detected points of inflection were used to calibrate transition time during cell cycle.
Time course clustering
Time course clustering of gene expression curves was performed as previously described18. Briefly, gene expression and accessibility curves were individually utilized to build n × N time course matrices, with rows representing genes and columns representing expression/accessibility time points. Each data was z-score scaled and a Dynamic Time-Warping (DTW) clustering algorithm was used to measure the similarities between the time series. In this analysis we used the function tsclust from dtwclust R package (https://github.com/asardaes/dtwclust) with the following parameters: type = hierarchical, control = hierarchical_control(method = “complete”), args = tsclust_args(dist = list(window.size = 4L). The number of clusters was set by empirically trial and error.
RNA velocity calculations
For each data set, loom files were generated using velocyto command line tool30. Loom files were processed in python using the scanpy library54 and spliced/unspliced counts were estimated. The scvelo python package31 was used to calculate velocity lengths using the dynamic model.
CUT&RUN data analysis
Paired-end CUT&RUN reads were mapped with BWA-mem (version 0.7.17)55 against the T. gondii ME49 reference genome. Peaks were called relative to the negative control with MACS256 using the following command and parameters: macs2 callpeak -p 0.05 -m 2 50 -t AP2XII-8.sorted.bam -c AP2XII-8-RH_S1.sorted.bam --nomodel -f BAM --extsize 120. Peaks were assigned to closest downstream genes using bedtools51. Peaks located entirely within the gene/coding region (exon-2 to exon-n) were filtered out from further analysis. The remining peaks were assigned to the nearest downstream gene with distance cut-off <2000 bp. Multiple peaks assigned to the same gene were merged. No distance criteria between the peaks were applied.
Gene set enrichment
Gene Ontology Enrichment Analysis (GOEA) was performed using available GO on ToxoDB.org (version 64) and significant GO terms (Benjamini <0.1) were determined.
Motif search
Motif search was performed under the ATAC peaks of genes within each ATAC sub-cluster (Fig. 3e). Moreover, this analysis was performed under the ATAC regions of DEGs in each expression-derived transition cluster (Fig. 2f and Supplementary Fig. 5a). CUT&RUN peaks were searched to identify potential binding sites of AP2XII-8 TF. This analysis was restricted to gene groups with at least 50 sequences. Motif discovery analysis was performed using the STREME command line tool57 from the MEME-Suite package58. A second motif discovery software, BAMM (https://bammmotif.mpibpc.mpg.de/)59 was also employed for additional rigor. For both software, default parameters were used. Significant motifs (p value < 0.05) from both methods were rederived and merged using a custom script as follows. All discovered motifs were pairwise aligned using the pairwiseAlignment function from the R Biostrings package. A similarity matrix was constructed by min-max normalization of pairwise alignment score followed by hierarchical clustering using the hclust command with Euclidian distance. The resulting three was used to cluster similar motifs (tree cutoff 0.6), which identified 21 clusters of similar motifs across the two software. Next, we ran a multiple sequence alignment on each cluster using the msa function form the R msa package and generated a PWM from which consensus sequences were calculated and visualized using the seqLogo function from the R seqLogo package. All discovered motifs along with their significance are presented in Supplementary Data 4. Motif co-occurrence analysis was performed on streme result59.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We would like to thank VEuPathDB for the readily accessible T. gondii genome assemblies and their functional annotations. We would also like to thank David Roos for helpful suggestion regarding the analysis, particularly relating to grouping genes based on expression/accessibility profile similarities. This study was supported by the National Institute of Health grants AI150090 (K.Z. and M.J.G.), and AI167570 (K.Z., M.T.D. and M.J.G.). We would like to acknowledge the 1S10OD032203-01 (Tufts University Core Facility Genomics Core) for the support of the included NGS analysis. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
K.Z., M.T.D. and M.J.G. conceived the approach, K.Z. and M.J.G. co-wrote the manuscript. J.L. performed all wet-lab T. gondii experiments, except select CUT&RUN and CUT&Tag experiments performed by Y.W., and established scRNA-seq and scATAC-seq in collaboration with C.D.K. Y.R., A.A., N.S. and K.Z. performed data analysis. D.D. and K.Z. developed periodic spline fits. All authors edited and proofread the final manuscript.
Data availability
The scRNA-seq, scATAC-seq and CUT&RUN data (fastq) generated in this study have been deposited to the Sequence Read Archive (SRA) under the accession number PRJNA1002574. These sequencing data are available to the public. An interactive web-application for visualization and exploration of our data set can be found here: https://umbibio.math.umb.edu/toxosc/. Source data are provided with this paper.
Code availability
The analysis R code is available on GitHub: https://github.com/umbibio/scToxoplasmaCDC (10.5281/zenodo.8219739).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jingjing Lou, Yasaman Rezvani.
These authors jointly supervised this work: Kourosh Zarringhalam, Marc-Jan Gubbels.
Contributor Information
Kourosh Zarringhalam, Email: kourosh.zarringhalam@umb.edu.
Marc-Jan Gubbels, Email: gubbelsj@bc.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51011-7.
References
- 1.Leander, B. S. & Keeling, P. J. Morphostasis in alveolate evolution. Trends Ecol. Evol.18, 395–402 (2003). 10.1016/S0169-5347(03)00152-6 [DOI] [Google Scholar]
- 2.Gubbels, M. J. et al. Fussing about fission: defining variety among mainstream and exotic apicomplexan cell division modes. Front. Cell Infect. Microbiol.10, 269 (2020). 10.3389/fcimb.2020.00269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson-White, B. R. et al. Cytoskeleton assembly in Toxoplasma gondii cell division. Int. Rev. Cell Mol. Biol.298, 1–31 (2012). 10.1016/B978-0-12-394309-5.00001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radke, J. R. et al. Defining the cell cycle for the tachyzoite stage of Toxoplasma gondii. Mol. Biochem. Parasitol.115, 165–175 (2001). 10.1016/S0166-6851(01)00284-5 [DOI] [PubMed] [Google Scholar]
- 5.Gubbels, M. J., Coppens, I., Zarringhalam, K., Duraisingh, M. T. & Engelberg, K. The modular circuitry of apicomplexan cell division plasticity. Front. Cell Infect. Microbiol.11, 670049 (2021). 10.3389/fcimb.2021.670049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue, Y. et al. A single-parasite transcriptional atlas of Toxoplasma Gondii reveals novel control of antigen expression. eLife9, e54129 (2020). 10.7554/eLife.54129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farhat, D. C. & Hakimi, M. A. The developmental trajectories of Toxoplasma stem from an elaborate epigenetic rewiring. Trends Parasitol.38, 37–53 (2022). 10.1016/j.pt.2021.07.016 [DOI] [PubMed] [Google Scholar]
- 8.Behnke, M. S. et al. Coordinated progression through two subtranscriptomes underlies the tachyzoite cycle of Toxoplasma gondii. PLoS ONE5, e12354 (2010). 10.1371/journal.pone.0012354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toenhake, C. G. et al. Chromatin accessibility-based characterization of the gene regulatory network underlying plasmodium falciparum blood-stage development. Cell Host Microbe23, 557–569.e559 (2018). 10.1016/j.chom.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oberstaller, J., Pumpalova, Y., Schieler, A., Llinas, M. & Kissinger, J. C. The Cryptosporidium parvum ApiAP2 gene family: insights into the evolution of apicomplexan AP2 regulatory systems. Nucleic Acids Res.42, 8271–8284 (2014). 10.1093/nar/gku500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lorenzi, H. et al. Local admixture of amplified and diversified secreted pathogenesis determinants shapes mosaic Toxoplasma gondii genomes. Nat. Commun.7, 10147 (2016). 10.1038/ncomms10147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zarringhalam, K., Ye, S., Lou, J., Rezvani, Y. & Gubbels, M. J. Cell cycle-regulated ApiAP2s and parasite development: the Toxoplasma paradigm. Curr. Opin. Microbiol.76, 102383 (2023). 10.1016/j.mib.2023.102383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gissot, M., Kelly, K. A., Ajioka, J. W., Greally, J. M. & Kim, K. Epigenomic modifications predict active promoters and gene structure in Toxoplasma gondii. PLoS Pathog.3, e77 (2007). 10.1371/journal.ppat.0030077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sindikubwabo, F. et al. Modifications at K31 on the lateral surface of histone H4 contribute to genome structure and expression in apicomplexan parasites. eLife6, e29391 (2017). 10.7554/eLife.29391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farhat, D. C. et al. A MORC-driven transcriptional switch controls Toxoplasma developmental trajectories and sexual commitment. Nat. Microbiol.5, 570–583 (2020). 10.1038/s41564-020-0674-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antunes, A. V. et al. In vitro production of cat-restricted Toxoplasma pre-sexual stages. Nature625, 366–376 (2024). 10.1038/s41586-023-06821-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulahannan, N. et al. Genomic insights into host and parasite interactions during intracellular infection by Toxoplasma gondii. PLoS ONE17, e0275226 (2022). 10.1371/journal.pone.0275226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rezvani, Y. et al. Comparative single-cell transcriptional atlases of Babesia species reveal conserved and species-specific expression profiles. PLoS Biol.20, e3001816 (2022). 10.1371/journal.pbio.3001816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stuart, T. et al. Comprehensive integration of single-cell data. Cell177, 1888–1902.e1821 (2019). 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet25, 25–29 (2000). 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The Gene Ontology Consortium et al. The Gene Ontology knowledgebase in 2023. Genetics22410.1093/genetics/iyad031 (2023).
- 22.Engelberg, K., Bechtel, T., Michaud, C., Weerapana, E. & Gubbels, M. J. Proteomic characterization of the Toxoplasma gondii cytokinesis machinery portrays an expanded hierarchy of its assembly and function. Nat. Commun.13, 4644 (2022). 10.1038/s41467-022-32151-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gubbels, M. J. et al. Toxoplasma gondii’s basal complex: the other apicomplexan business end is multifunctional. Front. Cell Infect. Microbiol.12, 882166 (2022). 10.3389/fcimb.2022.882166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen, A. L. et al. Novel components of the Toxoplasma inner membrane complex revealed by BioID. MBio6, e02357-14 (2015). 10.1128/mBio.02357-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, A. L. et al. Novel insights into the composition and function of the Toxoplasma IMC sutures. Cell Microbiol.1910.1111/cmi.12678 (2017). [DOI] [PMC free article] [PubMed]
- 26.Walker, R. et al. Toxoplasma transcription factor TgAP2XI-5 regulates the expression of genes involved in parasite virulence and host invasion. J. Biol. Chem.288, 31127–31138 (2013). 10.1074/jbc.M113.486589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lesage, K. M. et al. Cooperative binding of ApiAP2 transcription factors is crucial for the expression of virulence genes in Toxoplasma gondii. Nucleic Acids Res.46, 6057–6068 (2018). 10.1093/nar/gky373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mercier, C. et al. Common cis-acting elements critical for the expression of several genes of Toxoplasma gondii. Mol. Microbiol.21, 421–428 (1996). 10.1046/j.1365-2958.1996.6501361.x [DOI] [PubMed] [Google Scholar]
- 29.Markus, B. M., Waldman, B. S., Lorenzi, H. A. & Lourido, S. High-resolution mapping of transcription Initiation in the asexual stages of Toxoplasma gondii. Front. Cell Infect. Microbiol.10, 617998 (2020). 10.3389/fcimb.2020.617998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.La Manno, G. et al. RNA velocity of single cells. Nature560, 494–498 (2018). 10.1038/s41586-018-0414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bergen, V., Lange, M., Peidli, S., Wolf, F. A. & Theis, F. J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol.38, 1408–1414 (2020). 10.1038/s41587-020-0591-3 [DOI] [PubMed] [Google Scholar]
- 32.Sidik, S. M. et al. A genome-wide CRISPR screen in toxoplasma identifies essential apicomplexan genes. Cell166, 1423–1435.e1412 (2016). 10.1016/j.cell.2016.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szatanek, T. et al. Cactin is essential for G1 progression in Toxoplasma gondii. Mol. Microbiol.84, 566–577 (2012). 10.1111/j.1365-2958.2012.08044.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilde, M. L. et al. Characterisation of the OTU domain deubiquitinase complement of Toxoplasma gondii. Life Sci. Alliance6, e202201710 (2023). 10.26508/lsa.202201710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waldman, B. S. et al. Identification of a master regulator of differentiation in Toxoplasma. Cell180, 359–372.e316 (2020). 10.1016/j.cell.2019.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nardelli, S. C. et al. Genome-wide localization of histone variants in Toxoplasma gondii implicates variant exchange in stage-specific gene expression. BMC Genomics23, 128 (2022). 10.1186/s12864-022-08338-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Poppel, N. F., Welagen, J., Vermeulen, A. N. & Schaap, D. The complete set of Toxoplasma gondii ribosomal protein genes contains two conserved promoter elements. Parasitology133, 19–31 (2006). 10.1017/S0031182006009954 [DOI] [PubMed] [Google Scholar]
- 38.Alvarez, C. A. & Suvorova, E. S. Checkpoints of apicomplexan cell division identified in Toxoplasma gondii. PLoS Pathog.13, e1006483 (2017). 10.1371/journal.ppat.1006483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White, M. W. & Suvorova, E. S. Apicomplexa cell cycles: something old, borrowed, lost, and new. Trends Parasitol.34, 759–771 (2018). 10.1016/j.pt.2018.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong, D. P., Radke, J. B. & White, M. W. Opposing transcriptional mechanisms regulate toxoplasma development. mSphere210.1128/mSphere.00347-16 (2017). [DOI] [PMC free article] [PubMed]
- 41.White, M. W., Radke, J. R. & Radke, J. B. Toxoplasma development—turn the switch on or off? Cell Microbiol.16, 466–472 (2014). 10.1111/cmi.12267 [DOI] [PubMed] [Google Scholar]
- 42.Fleck, K., McNutt, S., Chu, F. & Jeffers, V. An apicomplexan bromodomain protein, TgBDP1, associates with diverse epigenetic factors to regulate essential transcriptional processes in Toxoplasma gondii. MBio14, e0357322 (2023). 10.1128/mbio.03573-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris, M. T. et al. A novel GCN5b lysine acetyltransferase complex associates with distinct transcription factors in the protozoan parasite Toxoplasma gondii. Mol. Biochem. Parasitol.232, 111203 (2019). 10.1016/j.molbiopara.2019.111203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, J. et al. Lysine acetyltransferase GCN5b interacts with AP2 factors and is required for Toxoplasma gondii proliferation. PLoS Pathog.10, e1003830 (2014). 10.1371/journal.ppat.1003830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srivastava, S., Holmes, M. J., White, M. W. & Sullivan, W. J. Jr Toxoplasma gondii AP2XII-2 contributes to transcriptional repression for sexual commitment. mSphere8, e0060622 (2023). 10.1128/msphere.00606-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Primo, V. A. Jr et al. The extracellular milieu of toxoplasma’s lytic cycle drives lab adaptation, primarily by transcriptional reprogramming. mSystems6, e0119621 (2021). 10.1128/mSystems.01196-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saraf, A. et al. Dynamic and combinatorial landscape of histone modifications during the intraerythrocytic developmental cycle of the malaria parasite. J. Proteome Res.15, 2787–2801 (2016). 10.1021/acs.jproteome.6b00366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roos, D. S., Donald, R. G., Morrissette, N. S. & Moulton, A. L. Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Methods Cell Biol.45, 27–63 (1994). 10.1016/S0091-679X(08)61845-2 [DOI] [PubMed] [Google Scholar]
- 49.Gajria, B. et al. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res.36, D553–D556 (2008). 10.1093/nar/gkm981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun.8, 14049 (2017). 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quinlan, A. R. BEDTools: the swiss-army tool for genome feature analysis. Curr. Protoc. Bioinforma.47, 11.12.1–34 (2014). 10.1002/0471250953.bi1112s47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hastie, T. & Stuetzle, W. Principal curves. J. Am. Stat. Assoc.84, 502–516 (1989). 10.1080/01621459.1989.10478797 [DOI] [Google Scholar]
- 53.Brockwell, P. J. & Davis, R. A. Time Series: Theory and Methods 2nd edn (Springer-Verlag, 1991).
- 54.Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol.19, 15 (2018). 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics25, 1754–1760 (2009). 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol.9, R137 (2008). 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bailey, T. L. STREME: accurate and versatile sequence motif discovery. Bioinformatics37, 2834–2840 (2021). 10.1093/bioinformatics/btab203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bailey, T. L., Johnson, J., Grant, C. E. & Noble, W. S. The MEME Suite. Nucleic Acids Res.43, W39–W49 (2015). 10.1093/nar/gkv416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kiesel, A. et al. The BaMM web server for de-novo motif discovery and regulatory sequence analysis. Nucleic Acids Res.46, W215–W220 (2018). 10.1093/nar/gky431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jessberger, R. The many functions of SMC proteins in chromosome dynamics. Nat. Rev. Mol. Cell Biol.3, 767–778 (2002). 10.1038/nrm930 [DOI] [PubMed] [Google Scholar]
- 61.Guerini, M. N., Que, X., Reed, S. L. & White, M. W. Two genes encoding unique proliferating-cell-nuclear-antigens are expressed in Toxoplasma gondii. Mol. Biochem. Parasitol.109, 121–131 (2000). 10.1016/S0166-6851(00)00240-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The scRNA-seq, scATAC-seq and CUT&RUN data (fastq) generated in this study have been deposited to the Sequence Read Archive (SRA) under the accession number PRJNA1002574. These sequencing data are available to the public. An interactive web-application for visualization and exploration of our data set can be found here: https://umbibio.math.umb.edu/toxosc/. Source data are provided with this paper.
The analysis R code is available on GitHub: https://github.com/umbibio/scToxoplasmaCDC (10.5281/zenodo.8219739).