Abstract
Shigella flexneri is one of the agents most frequently linked to diarrheal illness in developing countries and often causes outbreaks in settings with poor hygiene or sanitary conditions. Travel is one of the means by which S. flexneri can be imported into developed countries, where this pathogen is not commonly seen. A robust and discriminatory subtyping method is needed for the surveillance of S. flexneri locally and regionally, and to aid in the detection and investigation of outbreaks. The PulseNet International network utilizes standardized pulsed-field gel electrophoresis (PFGE) protocols to carry out laboratory-based surveillance of foodborne pathogens in combination with epidemiologic data. A multicenter validation was carried out in nine PulseNet laboratories located in North and South America, Europe, and Asia, and it demonstrated that a new protocol is highly robust and reproducible for subtyping of S. flexneri. This protocol, already approved for PulseNet laboratories, applies NotI and XbaI as primary and secondary restriction enzymes, respectively, under electrophoresis conditions of initial switch time of 5 s to final switch time of 35 s, at 6 volts/cm.
Introduction
Shigellosis is a global human health problem. According to the World Health Organization (WHO), there are approximately 120 million cases of severe dysentery, the majority occurring in developing countries and affecting primarily young children (Nigoyi, 2005). Approximately 1.1 million people die from Shigella infections each year; 60% of these deaths occur in children under 5 years old. Over 580,000 cases are reported annually among travellers from industrialized countries (www.who.int/vaccine_research/diseases/diarrhoeal/en).
The infectious dose of Shigella spp. is very low; therefore, the disease may be easily transmitted from person to person or by ingestion of contaminated food and water, especially in settings with poor sanitation (Niyogi, 2005). The disease is believed to be transmitted strictly through contact to human fecal material (e.g., by infected food handlers, by consumption of sewage contaminated water or through its use for irrigation of fresh produce). Numerous outbreaks have been reported, in day-care centers and other closed-quarter institutions such as schools and cruise ships (Arvelo et al., 2009; CDC, 2005; Rooney et al., 2004; Shimosako et al., 2007). In addition, foodborne and waterborne outbreaks have been recognized (Arias et al., 2006; CDC, 1999; Frost et al., 1995; García-Fulgueiras et al., 2007; Reller et al., 2006).
S. flexneri, S. sonnei, and S. dysenteriae are the major species responsible for bacillary dysentery. S. sonnei is more common in industrialized countries,whereas S. flexneri ismore prevalent in less developed regions and is the most frequently isolated species worldwide (Kotloff et al., 1999; Niyogi, 2005; WHO, www.who.int/vaccine_research/diseases/diarrhoeal/en/).
However, this trend may vary from country to country and even within the same region (Al-Nimri et al., 2009; Vinh et al., 2009). Changes in dietary habits such as increased consumption of fresh produce and food imports have contributed to the emergence of S. flexneri–associated outbreaks in industrialized countries (Reller et al., 2006). Thus, there is a need for surveillance of S. flexneri at a global level if reliable reporting of shigellosis incidents is to occur according to the International Health Regulations supported by WHO (WHO, 2005).
PulseNet USA is the national molecular subtyping network for the surveillance of foodborne disease (Gerner-Smidt et al., 2006; Swaminathan et al., 2001). Since its establishment in 1996, the network has been instrumental in the rapid detection, investigation, and control of numerous outbreaks. The success of PulseNet USA led to the formation of an international network in 2003, now known as PulseNet International (Swaminathan et al., 2006). Today PulseNet International is comprised of six regional networks that utilize standardized methods and quality control procedures to subtype foodborne bacterial pathogens (www.pulsenetinternational.org). The use of highly standardized pulsed-field gel electrophoresis (PFGE) protocols allow inter- and intra-laboratory comparison of bacterial subtyping data (Cooper et al., 2006; Kam et al., 2008; Ribot et al., 2006). Unfortunately, the existing PulseNet protocol for S. sonnei was found to produce patterns with inadequate fragment resolution for S. flexneri. For this reason, the PulseNet S. flexneri Working Group was established to develop a standardized PFGE protocol optimized for this organism.
This study describes the development and validation of a standardized PFGE protocol for molecular subtyping of S. flexneri. The PulseNet S. flexneri Working Group is composed of nine laboratories: Instituto Nacional de Enfermedades Infecciosas-ANLIS “Carlos G. Malbrán” (INEI-ANLIS) of Argentina (co-coordinating laboratory), U.S. Centers for Disease Control and Prevention (CDC; co-coordinating laboratory), Instituto Adolfo Lutz of Brazil, National Microbiology Laboratory of Canada, Instituto de Salud Pública of Chile, Statens Serum Institute of Denmark, Global Disease Detection (GDD) Regional Center at the U.S. Naval Medical Research Unit 3 (NAMRU-3), Egypt, Central Public Health Laboratory of Oman, and Public Health Laboratory Center of Hong Kong, China.
Methods
Bacterial strains
A panel of 106 S. flexneri isolates (65 from CDC and 41 from INEI-ANLIS) were used in the initial evaluation of different protocol parameters in the coordinating laboratories. This panel included isolates classified as sporadic (not known to be epidemiologically related) from serotypes 1–6 and 12 isolates from three outbreaks (serotypes 2, 3, and 6) that occurred in Argentina. Isolates were stored at −70°C.
A panel of 17 S. flexneri isolates recovered in different geographic regions of Argentina was selected for the international validation phase (Table 1). The isolates were shipped in nutrient agar stabs to all the participating laboratories. Each laboratory was asked to propagate a single colony from each strain and freeze it for storage at −70°C. Salmonella enterica serovar Braenderup H9812 was used as the standard for the PFGE gels in accordance with PulseNet protocols (Hunter et al., 2005).
Table 1.
List of the 17 Isolates of Shigella flexneri Used for the Multicenter International Evaluation and Validation of Pulsed-Field Gel Electrophoresis Protocols
| Isolate | Year of isolation | Sporadic (S) or outbreak (O) | Serotype |
|---|---|---|---|
|
| |||
| AR-1 | 2006 | S | 1 |
| AR-2 | 2006 | S | 1 |
| AR-3 | 2004 | S | 2 |
| AR-4 | 2005 | S | 2 |
| AR-5 | 2005 | S | 2 |
| AR-6 | 2006 | O | 2 |
| AR-7 | 2006 | O | 2 |
| AR-8 | 2006 | S | 3 |
| AR-9 | 2006 | S | 3 |
| AR-10 | 2005 | O | 3 |
| AR-11 | 2005 | O | 3 |
| AR-12 | 2005 | S | 4 |
| AR-13 | 2007 | S | 5 |
| AR-14 | 2004 | S | 6 |
| AR-15 | 2004 | S | 6 |
| AR-16 | 2007 | O | 6 |
| AR-17 | 2007 | O | 6 |
The seven isolates used for Phase II/final standardization are shown in bold.
Phase I. Initial standardization and multicenter evaluation of PFGE protocol
The PulseNet PFGE protocol for S. sonnei was used as the basis for the development of the protocol for S. flexneri. Optimization focused primarily on the evaluation of alternative restriction endonucleases and testing of several electrophoresis conditions. A panel of rare-cutting restriction enzymes, including ApaI, AscI, SmaI, SpeI, SfiI, XbaI, and NotI, were evaluated using three reference strains to assess their ability to generate patterns with good levels of resolution and fragment distribution. The coordinating laboratories tested additional isolates from the initial panel of 106 S. flexneri isolates with the two most optimal enzymes, XbaI and NotI, using the electrophoresis conditions listed in Table 2A. The patterns were compared visually and using BioNumerics software.
Table 2.
(A) Electrophoresis Conditions Evaluated in Phase I with Shigella flexneri Strains; (B) Electrophoresis Conditions Tested with a Selection of Seven Strains of Shigella flexneri During Phase II/Final Standardization Trials
| (A) | ||
|---|---|---|
| PulseNet Protocol | Electrophoresis conditions a | Reference |
|
| ||
| Escherichia coli O157:H7, Shigella sonnei | 2.2–54.2 s for 18 h | Ribot et al., 2006. |
| Yersinia pestis modified | 1.8—25 s for 18 h | www.cdc.gov/pulsenet/protocols.htm |
| (B) | |
|---|---|
| Program | Electrophoresis conditions a |
|
| |
| Program 1 (P1) | Block 1: 2–10 s for 12h Block 2: 20—25 s for 5 h |
| Program 2 (P2) | Block 1: 2—5 s for 12h Block 2: 20—25s for 5h |
| Program 3 (P3) | 10—25 s for 17.5 h |
| Program 4 (P4) | 2—30 s for 17.5 h |
| Program 5 (P5) | 10—35 s for 17.5 h |
| Program 6 (P6) | 5—25 s for 17.5 h |
| Program 7 (P7) | 5—35 s for 17.5 h |
Run time indicated is approximate and was adjusted in each lab to achieve adequate migration of the standard strain DNA fragments.
The restriction enzymes XbaI (Fermentas, Lithuania) and NotI (Invitrogen, Carlsbad, CA) and the electrophoresis conditions of initial switch time (IST) of 1.8 s, final switch time (FST) of 25 s, and running time (RT) of 17–18 h were selected for the validation in the nine laboratories using the panel of 17 strains (Table 1). For both enzymes, 50 units were applied for each sample for 2-h digestion, although an amount of 30 units for 4-h digestion was also evaluated at INEI-ANLIS and found satisfactory. Each laboratory submitted gel images in TIFF format to CDC and INEI-ANLIS for analysis using BioNumerics software. The results from this validation indicated that the electrophoresis conditions used in Phase I were not optimal and required further evaluation.
Phase II. Final PFGE protocol development and multicenter validation
Further tests were performed at INEI-ANLIS and CDC in an effort to identify alternative electrophoresis conditions that would improve resolution of high and medium molecular weight (HMW and MMW, respectively) bands typically seen in NotI patterns. Seven additional electrophoresis conditions (Programs 1–7) were tested (Table 2B) using a subset of seven isolates (Table 1).
Once analysis was completed, Program 7 (P7; IST of 5 s, FST of 35 s, and RT of 17–18 h) was selected for the second phase validation. Considering the results and main issues observed in phase I, recommendations for improving gel quality (run length, DNA concentration, adequate plug preparation, and DNA restriction) were provided to all the laboratories.
Statistical analysis
TIFF files were analyzed using BioNumerics v. 4.6 (Applied Maths, Sint-Martens-Latem, Belgium), following normalization with PulseNet universal size standard S. Braenderup H9812 (run in three lanes in a 10-well gel and four lanes in a 15-well gel). Dendograms comparing the patterns from each laboratory were constructed for every isolate using Dice coefficient and unweighted pair group method with arithmetic mean (UPGMA) and a band position tolerance window of 1.5% as established by PulseNet.
The analysis of the different running conditions was also made by visual comparisons to assess the fragment distribution with each condition.
Results
Phase I. Initial standardization and multicenter evaluation of PFGE protocol
The plug preparation steps from the PulseNet Protocol for S. sonnei (Ribot et al., 2006) were successfully applied to process S. flexneri isolates. DNA restriction of S. flexneri was initially evaluated with XbaI and BlnI as recommended for S. sonnei. The BlnI profiles contained a large number of DNA fragments, which greatly complicated pattern analysis. Out of six restriction enzymes evaluated for use in combination with XbaI (ApaI, AscI, SmaI, and SpeI [Fig. 1]; SfiI and NotI [data not shown]), NotI was selected for further testing because it generated a manageable number (10–25) of evenly distributed DNA fragments (Fig. 2). A modified version of the Yersinia pestis electrophoresis conditions, 1.8–25 s, were selected for the multicenter evaluation because it yielded better resolution and distribution of DNA fragments with both enzymes (Fig. 2).
FIG. 1.
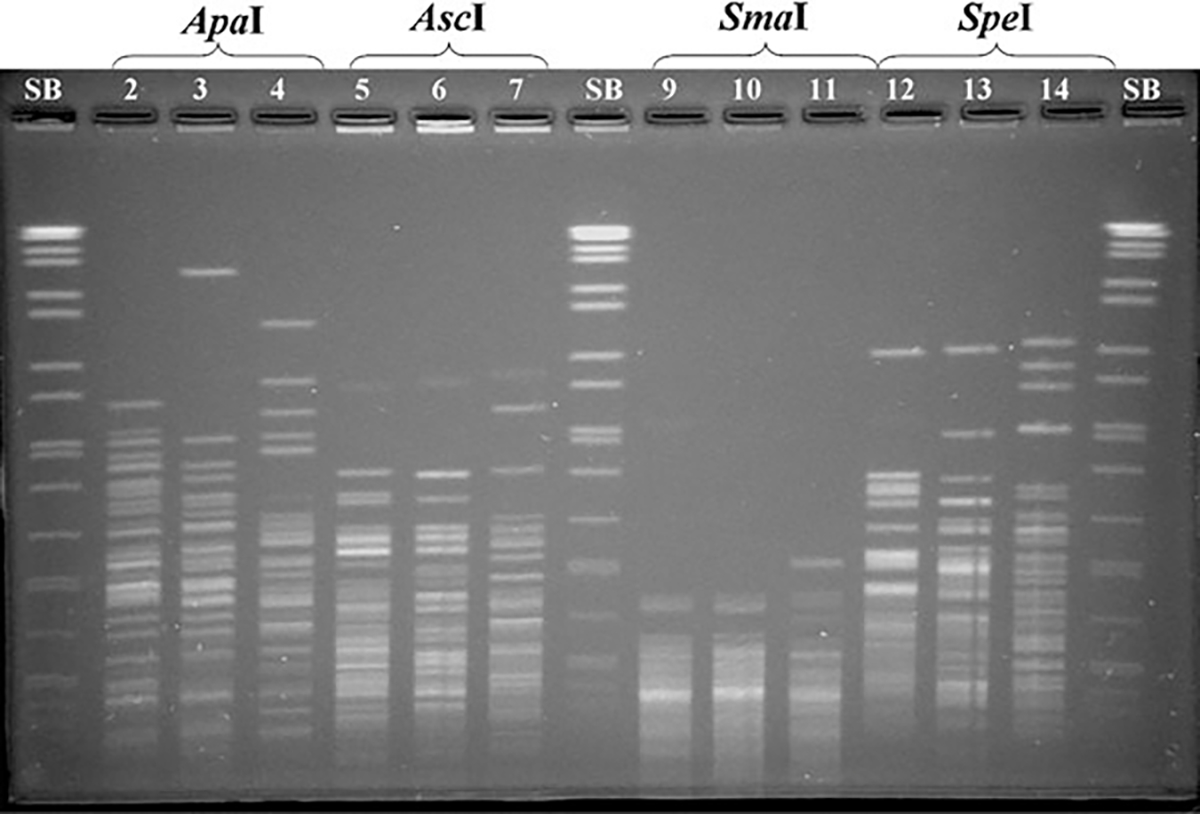
Pulsed-field gel electrophoresis of three Shigella flexneri isolates using different restriction enzymes: ApaI in lines 2–4; AscI in lines 5–7; SmaI in lines 9–11; SpeI in lines 12–14. SB: PN universal size standard Salmonella enterica serotype Braenderup (H9812) in lines 1, 8, and 15.
FIG. 2.
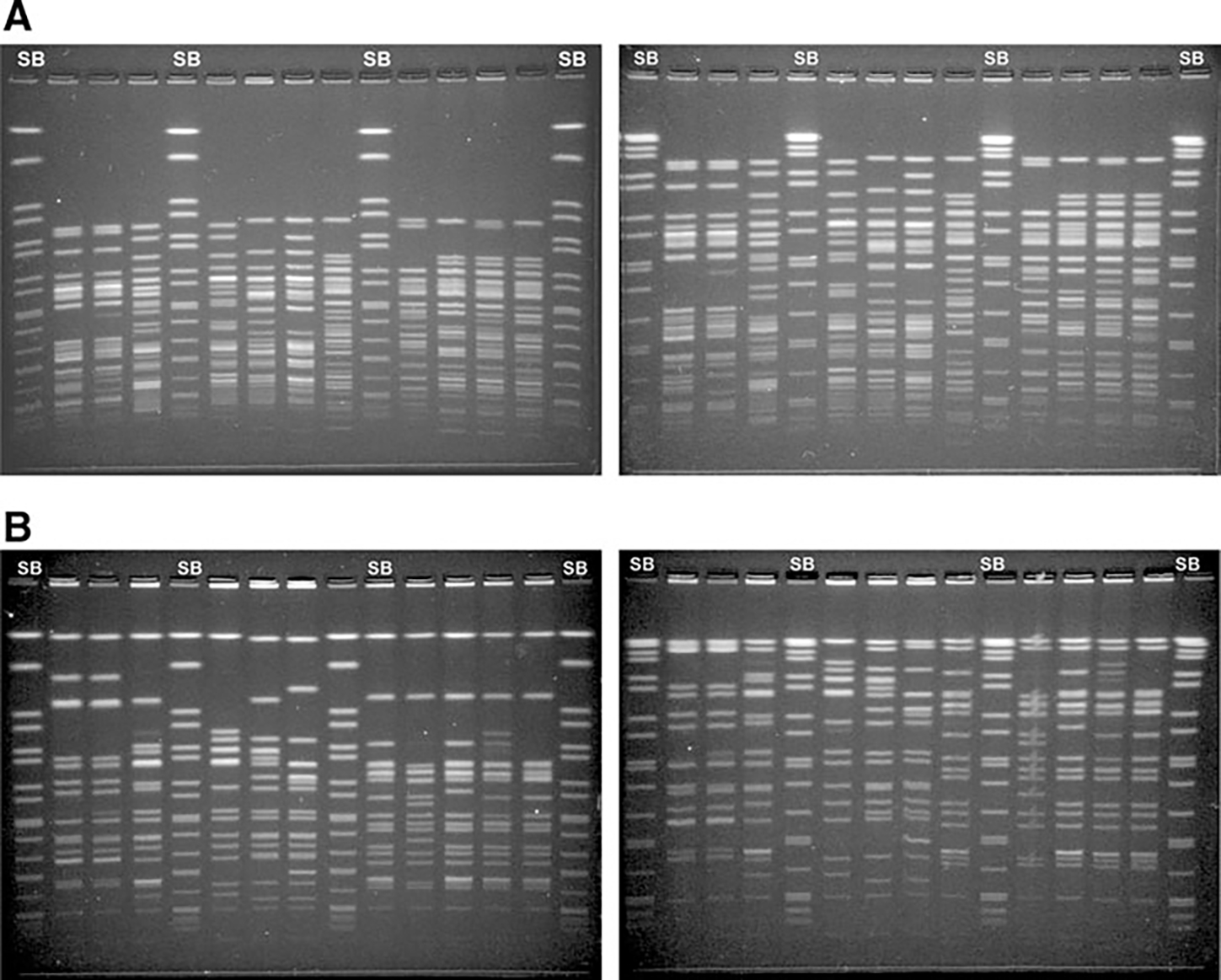
Pulsed-field gel electrophoresis (PFGE) of Shigella flexneri strains using two different electrophoresis conditions. SB, Standard strain Salmonella enterica serotype Braenderup H9812. (A) XbaI-PFGE. (B) NotI-PFGE. (Left panel A,B) Conditions of the standardized PulseNet Shigella sonnei protocol, 2.2–54.2 s. (Right panel A,B) Conditions of the standardized PulseNet Yersinia pestis modified protocol, 1.8–25 s.
Results from two out of the nine laboratories were excluded from analysis. In one case, the results were received after the analysis stage was concluded. In the second case, the results showed a high degree of variance from the expected PFGE profiles. We were unable to determine the actual cause of these differences (storage practices and/or culture conditions) but concluded that they were not likely to be related to the PFGE protocol itself. Exclusion of these results from the analysis did not impede our ability to assess the reproducibility or robustness of the protocol. In the analysis of patterns from seven laboratories, the XbaI enzyme produced an average of 22.8 bands per strain. The majority of pattern differences observed for a given isolate were the result of variation in the fragments’ resolution, mostly in the lower portion of the gel. Occasionally differences were also seen that appeared to be the result of genetic changes due to culturing of the strains. Other deviations from the predicted patterns were observed but were unable to ascertain if these were the result of incomplete restriction or genetic variation. The number of band differences did not appear to be linked to a specific serotype.
In general, NotI-PFGE profiles consisted of fewer bands (average 14.3) that were more evenly distributed throughout the patterns than XbaI profiles. The majority of the resolution issues observed with NotI-PFGE were in the top portion of the patterns. As with XbaI, some of the NotI patterns showed some differences compared to the profiles obtained by the coordinating laboratories but no correlation was found between these differences and specific serotype. Other issues associated with weak bands of low molecular weight DNA fragments and incomplete restriction, were observed with both enzymes, although the latter was more frequently associated with XbaI. Finally, the ability of multiple laboratories to accurately cluster outbreak related strains was evaluated. Overall, outbreak strains restricted with NotI clustered more closely together (with average similarities of 99.1, 95.9, and 97.1 among outbreak isolates of serotypes 2, 3, and 6, respectively, considering the similarity observed in each laboratory between outbreak isolates) than those generated with XbaI (with average similarities of 94.7, 95.9, and 92.1 among outbreak isolates of serotypes 2, 3, and 6, respectively).
Phase II. Final PFGE protocol development and multicenter validation
Results from the phase I trials suggested that NotI could be a better choice as primary enzyme. However, the electrophoresis conditions of Y. pestis modified protocol were not completely satisfactory for use with this enzyme because high molecular weight bands were slightly compressed. To resolve this problem, seven new electrophoresis conditions (P1–P7, Table 2B) were tested using a subset of seven strains (Table 1). Results of these tests showed that P1, P2, and P4 did not improve the resolution of HMW or MMW bands (data not shown). On the other hand, P3 and P6 showed improvements for three of the seven strains for MMW bands, and P5 and P7 improved the resolution of HMW bands significantly in five of the seven strains (Fig. 3A).
FIG. 3.
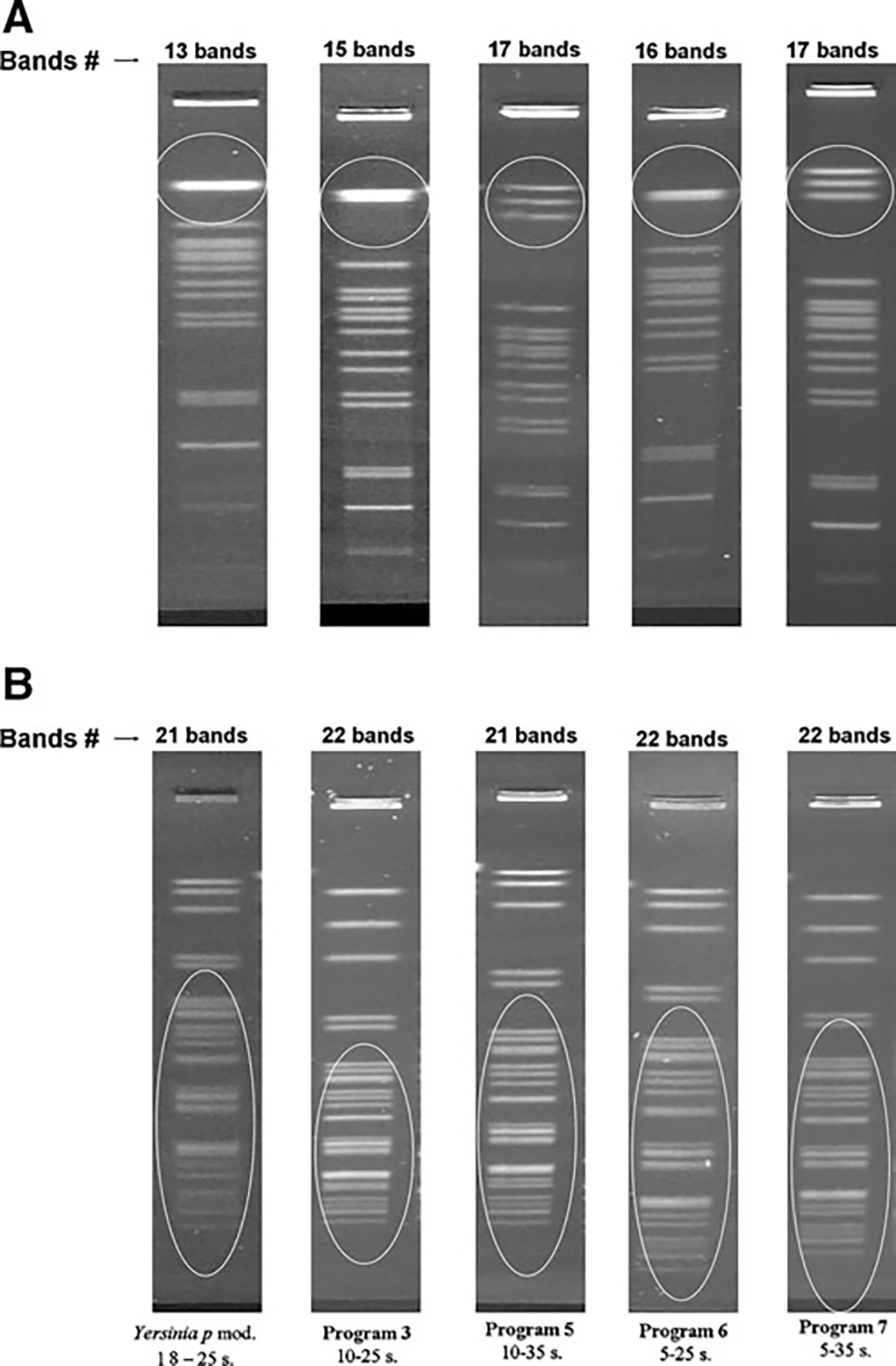
Visual comparison of the effect of five different electrophoresis conditions (Yersinia pestis mod and Programs 3, 5, 6, and 7) in a representative isolate of Shigella flexneri. (A) NotI-PFGE patterns. (B) XbaI-PFGE patterns.
When these programs were tested using XbaI, P3 showed better distribution of the bands in all the strains tested. However, this program did not show significant improvements in the resolution of low molecular weight (LMW) bands, the most numerous and difficult to analyze in XbaI profiles (Fig. 3B). Furthermore, this program did not improve the resolution of HMW bands obtained with NotI, and the improvements observed for XbaI were minor (duplets with weak or very close bands) and likely to be linked to slight variation in the electrophoresis run length and not easily reproducible. In summary, P7 electrophoresis conditions improved the overall resolution of NotI patterns without negatively affecting the XbaI patterns (Figs. 3 and 4). Considering these results, P7 was selected for the second round of international multicenter validation.
FIG. 4.
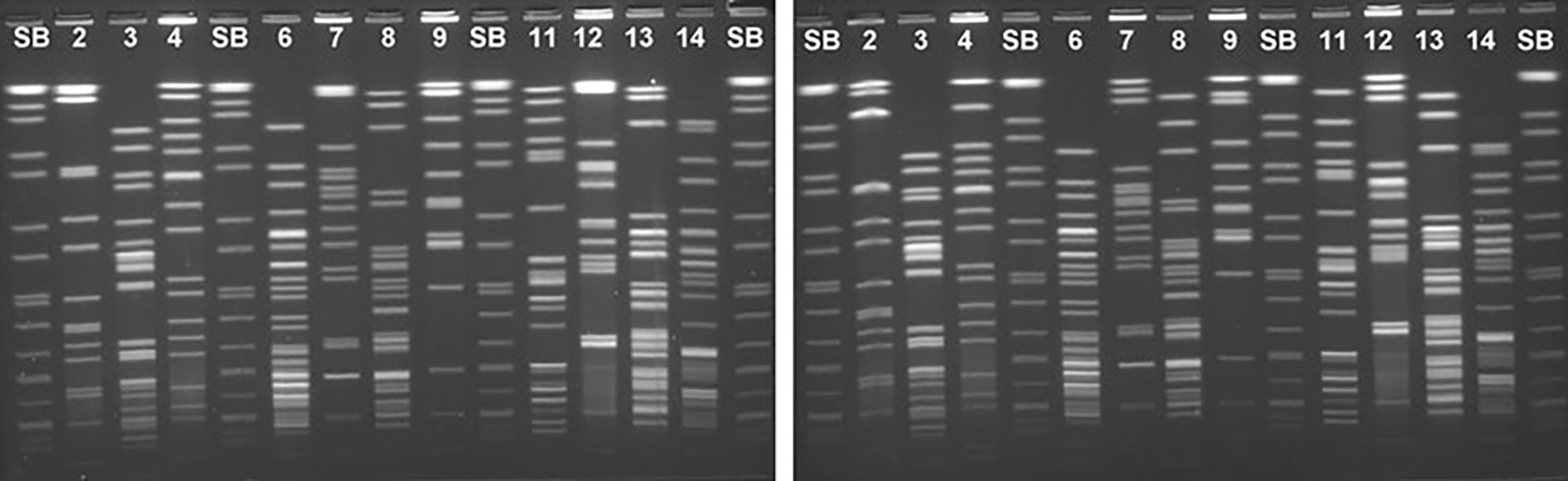
Two electrophoresis conditions tested with five representative Shigella flexneri strains run in a single gel with the restriction enzymes XbaI (lines 3, 6, 8, 11, 13, and 14) and NotI (lines 2, 4, 7, 9, and 12). SB, PulseNet universal size standard Salmonella enterica serotype Braenderup (H9812). (Left panel) Program 3, 10–25 s. (Right panel) Program 7, 5–35 s.
As in the previous interlaboratory comparison with Y. pestis–modified conditions, P7 also yielded several pattern differences that were attributed to mutations or not explainable, mainly in the same laboratory that had had the highest rate of mutational events previously. Therefore, the results from this laboratory were excluded from analysis, and eight of the nine laboratories were considered for this second validation round.
PFGE analysis of XbaI patterns showed an average of 21.8 bands per strain. Fewer pattern differences were observed with these conditions, suggesting that P7 electrophoresis conditions produced data that was easier to analyze. Similar to what was seen within phase I, NotI patterns contained a lower number of bands than XbaI (average 15.1 bands), which resulted in more efficient resolution that made pattern analysis easier and less prone to errors (Fig. 4).
In summary, these results, together with the improved gel quality from the laboratories, supported a higher robustness and reproducibility of the P7 in comparison with the Y. pestis– modified electrophoresis conditions for a S. flexneri PFGE protocol.
Discussion
S. flexneri is a common cause of diarrheal illness in developing countries and has caused outbreaks of disease in closed communities in industrialized areas (Nigoyi, 2005). Implementation of a standardized PFGE protocol for S. flexneri subtyping would be useful for the detection of clusters of illness and the identification of transmission pathways and infection sources, particularly important in regions where this organism is endemic. This is of great importance particularly in the context of the International Health Regulations’ recommendations for the investigation of foodborne diseases.
In the absence of a specific protocol for S. flexneri, the PFGE protocol for S. sonnei has been used by PulseNet for subtyping this organism since 1999. However, due to the complexity and poor resolution of these patterns, it has been challenging to accurately analyze data from multiple laboratories. From previous experience, it was known that plug preparation steps outlined in the S. sonnei protocol worked well for processing S. flexneri strains. However, the S. sonnei electrophoresis conditions yielded patterns that were difficult to analyze. All PulseNet PFGE protocols list restriction enzymes as “primary” or “secondary.” The primary enzyme (default) is the enzyme used in routine analysis of isolates including cluster detection (Swaminathan et al., 2001). The secondary enzyme is primarily used to further examine clusters of isolates with indistinguishable or closely related primary enzyme profiles. The main purpose is to increase the discriminatory power of PFGE and disprove genetic relatedness of epidemiologically unrelated isolates. In this study, NotI and XbaI were found to be the most suitable for macrorestriction analysis of this microorganism because they produced a manageable number of fragments within the migration field. In general, NotI-PFGE patterns contained fewer fragments that were more evenly distributed than the typical bands in XbaI-PFGE profiles. As a result, NotI patterns were also much easier to analyze and less prone to the band marking errors (Fig. 2). In addition, NotI data showed better correlation than XbaI in clustering known outbreak strains and because of this it was decided to use NotI as the primary enzyme in this protocol.
After feedback from the participating laboratories, a series of recommendations were made to improve gel quality and reproducibility of results in the next validation phase. Program 7 (IST, 5 s; FST, 35 s) was selected because it provided increased reproducibility and higher resolution of fragments. The results from the second multicenter validation showed that compression of the NotI HMW fragments was significantly reduced with the P7. The average number of differences due to resolution issues was reduced compared with Y. pestis modified electrophoresis conditions. Furthermore, the XbaI patterns generated with these electrophoresis conditions were much easier to analyze compared to those obtained in the previous validation. Thus, no further changes were introduced to the protocol and the ramping conditions IST: 5 s – FST: 35 s were selected by the validation group as the official electrophoresis parameters. Using the same electrophoresis conditions for both enzymes provides two main benefits; 1) it allows laboratories to load plug slices restricted with these enzymes on the same gel, which saves time and money, and 2) it simplifies the use of the protocol making it more user friendly.
Lastly, the data obtained with the proposed protocol showed a high degree of concordance with the epidemiological information available for the isolates tested. The sporadic strains of the six serotypes of S. flexneri were discriminated using the validated PFGE protocol, and outbreak strains were closely related. International validation of protocols like the one described herein are critical to the implementation of molecular tools in the public health setting by ensuring that methods are capable of producing reproducible and accurate data that can be compared with laboratories around the globe.
Many laboratories have S. flexneri fingerprints generated using the electrophoresis conditions for S. sonnei. Unfortunately, these patterns will not be comparable with those obtained under the new conditions; therefore, each laboratory will have to evaluate whether a selection of relevant isolates should be re-run under the new conditions to use as background information (i.e., a number of isolates associated with outbreaks or representing the predominant patterns and serotypes).
Conclusion
Here we describe the standardization and international validation of a PFGE protocol for subtyping of S. flexneri isolates. This protocol was derived from the PulseNet standardized protocol for S. sonnei with modifications made to increase its robustness, reproducibility, and ease of use. The selection of restriction enzymes NotI and XbaI and the use of different electrophoresis conditions highlight the changes instituted in this protocol. A multicenter validation conducted in nine PulseNet laboratories located in different regions of the world (North and South America, Europe, and Asia) demonstrated that the protocol is robust and reproducible. Also, the collaborative study identified quality measures that are recommended to ensure adequate band resolution and patterns comparison (i.e., verifying correct electrophoresis length and restriction conditions). This validated PFGE protocol applies NotI and XbaI as primary and secondary restriction enzymes, respectively, under electrophoresis conditions of IST of 5 s to FST of 35 sec, at 6 volts/cm. It is now preferred for subtyping of S. flexneri in the PulseNet networks and is publicly available at www.pulsenetinternational.org/protocols.
Footnotes
Disclosure Statement
No competing financial interests exist.
References
- Al-Nimri S, Miller WA, Byrne BA, Guibert G, Chen L. A unified approach to molecular epidemiology investigations: Tools and patterns in California as a case study for endemic shigellosis. BMC Infect Dis 2009;9:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias C, Sala MR, Domínguez A, Bartolomé R, Benavente A, Veciana P, Pedrol A, G Hoyo; Outbreak Working Group. Waterborne epidemic outbreak of Shigella sonnei gastroenteritis in Santa María de Palautordera, Catalonia, Spain. Epidemiol Infect 2006;134:598–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvelo W, Hinkle CJ, Nguyen TA, Weiser T, Steinmuller N, Khan F, Gladbach S, Parsons M, Jennings D, Zhu BP, Mintz E, Bowen A. Transmission risk factors and treatment of pediatric shigellosis during a large daycare center-associated outbreak of multidrug resistant Shigella sonnei: Implications for the management of shigellosis outbreaks among children. Pediatr Infect Dis J 2009;28:976–980. [DOI] [PubMed] [Google Scholar]
- [CDC] Centers for Disease Control and Prevention. Outbreaks of Shigella sonnei infection associated with eating fresh parsley. United States and Canada, July–August 1998. MMWR Morb Mortal Wkly Rep 1999;48:285–289. [PubMed] [Google Scholar]
- [CDC] Centers for Disease Control and Prevention. Outbreaks of multidrug-resistant Shigella sonnei gastroenteritis associated with day care centers—Kansas, Kentucky, and Missouri. MMWR Morb Mortal Wkly Rep 2005;55:1068–1071. [PubMed] [Google Scholar]
- Cooper KLF, Luey CKY, Bird M, Terajima J, Nair GB, Kam KM, Arakawa E, Safa A, Cheung DT, Law CP, Watanabe H, Kubota K, Swaminathan B, Ribot EM. Development and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping of Vibrio cholerae. Foodborne Pathog Dis 2006;3:51–58. [DOI] [PubMed] [Google Scholar]
- Frost JA, McEvoy MB, Bentley CA, Andersson Y. An outbreak of Shigella sonnei infection associated with consumption of iceberg lettuce. Emerg Infect Dis 1995;1:26–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fulgueiras A, Sánchez S, Guillén JJ, Marsilla B, Aladueña A, Navarro C. A large outbreak of Shigella sonnei gastroenteritis associated with consumption of fresh pasteurized milk cheese. Eur J Epidemiol 2007;17:533–538. [DOI] [PubMed] [Google Scholar]
- Gerner-Smidt P, Hise K, Kincaid J, Hunter S, Rolando S, Hyytiä-Trees E, Ribot EM, Swaminathan B; Pulsenet Taskforce. PulseNet USA: A five-year update. Foodborne Pathog Dis 2006;3:9–19. [DOI] [PubMed] [Google Scholar]
- Hunter SB, Vauterin P, Lambert-Fair MA, Van Duyne MS, Kubota K, Graves L, Wrigley D, Barrett T, Ribot E. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: Converting the national databases to the new size standard. J Clin Microbiol 2005;43:1045–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam KM, Luey CY, Parsons MB, Cooper KLF, Nair GB, Alam M, Islam MA, Cheung DTL, Chu YW, Ramamurthy T, Pazhani GP, Bhattacharya SK, Watanabe H, Terajima J, Arakawa E, Ratchtrachenchai OA, Huttayananont S, Ribot EM, Gerner-Smidt P, Swaminathan B; Vibrio parahaemolyticus PulseNet PFGE Protocol Working Group. Evaluation and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping Vibrio parahaemolyticus: An international multicenter collaborative study. J Clin Microbiol 2008;46:2766–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, Adak GK, Levine MM. Global burden of Shigella infections: Implications for vaccine development and implementation of control strategies. Bull World Health Organ 1999;77:651–666. [PMC free article] [PubMed] [Google Scholar]
- Niyogi SK. Shigellosis. J Microbiol 2005;43:133–143. [PubMed] [Google Scholar]
- Reller ME, Nelson JM, Mølbak K, Ackman DM, Schoonmaker-Bopp DJ, Root TP, Mintz ED. A large, multiple restaurant outbreak of Infection with Shigella flexneri serotype 2a traced to tomatoes. Clin Infect Dis 2006;42:163–169. [DOI] [PubMed] [Google Scholar]
- Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 2006;3:59–67. [DOI] [PubMed] [Google Scholar]
- Rooney RM, Bartram JK, Cramer EH, Mantha S, Nichols G, Suraj R, Todd EC. A review of outbreaks of foodborne disease associated with passenger ships: Evidence for risk management. Public Health Rep 2004;119:427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimosako J, Onaka T, Yamaguchi M, Yokota M, Nakamura T, Fujii F, Matsumoto E, Shibata H, Fukuda M, Tanaka T. An outbreak of extended-spectrum beta-lactamase (ESBL)–producing Shigella sonnei at a day care nursery in Sakai City, 2006. Jpn J Infect Dis 2007;60:408–409. [PubMed] [Google Scholar]
- Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV; CDC PulseNet Task Force. PulseNet: The molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg Infect Dis 2001;7:382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan B, Gerner-Smidt P, Ng LK, Lukinmaa S, Kam KM, Rolando S, Gutiérrez EP, Binsztein N. Building PulseNet International: An interconnected system of laboratory networks to facilitate timely public health recognition and response to foodborne disease outbreaks and emerging foodborne diseases. Foodborne Pathog Dis 2006;3:36–50. [DOI] [PubMed] [Google Scholar]
- Vinh H, Nhu NT, Nga TV, Duy PT, Campbell JI, Hoang NV, Boni MF, My PV, Parry C, Nga TT, Van Minh P, Thuy CT, Diep TS, Phuong le T, Chinh MT, Loan HT, Tham NT, Lanh MN, Mong BL, Anh VT, Bay PV, Chau NV, Farrar J, Baker S. A changing picture of shigellosis in southern Vietnam: Shifting species dominance, antimicrobial susceptibility and clinical presentation. BMC Infect Dis 2009;9:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [WHO] World Health Organization. International Health Regulations, 2nd ed. Geneva, WHO, 2005. [Google Scholar]


