Abstract
Melanin is an amorphous, highly heterogeneous polymer found across all kingdoms of life. Although the properties of melanin can greatly vary, most forms are insoluble and strongly absorb light, appearing dark brown to black. Here, we describe a water-soluble form of melanin (peptidomelanin) secreted by the spores of Aspergillus niger (strain: melanoliber) during germination. Peptidomelanin is composed of an insoluble L-DOPA core polymer that is solubilized via short, copolymerized heterogeneous peptide chains forming a “corona” with a mean amino acid length of 2.6 ± 2.3. Based on in vitro experiments, we propose a biochemical copolymerization mechanism involving the hydroxylation of tyrosynylated peptides. Peptidomelanin is capable of chelating heavy metals such as lead, mercury, and uranium (as uranyl) in large quantities. Preliminary data indicates that peptidomelanin may have applications for the remediation of heavy metals in situ, including in agricultural settings.
Introduction
Melanin is an ancient pigment present across all kingdoms of life. It is a large, heterogeneous, amorphous polymer composed of phenolic and indole components1−8 (Figure S1), that appears dark brown or black under visible light. Its primary biological role is protection against ultraviolet9 and ionizing radiation,10 as well as thermoregulation.11 In pathogenic organisms, melanin also plays a role in virulence,12 as well as antibiotic and antifungal resistance.13 Despite its widespread importance, fundamental questions persist about melanin’s chemical structure. Its amorphicity, heterogeneity, and insolubility in aqueous solvents has made the structural and chemical characterization of the substance difficult. It is therefore simply categorized as a complex and diverse substance.
The vast majority of reports pertaining to melanin describe an insoluble polymer present as intracellular granules.14 Only a handful of reports describing soluble forms of melanin can be found in the literature. Aspergillus fumigatus, Madurella mycetomatis, Yarrowia lipolytica,15 and Bacillus thuringiensis,16 for example, secrete soluble melanin. Aspergillus nidulans,17Streptomyces cavourensis SV 2118 and Streptomyces lusitanus DMZ-319 produce both insoluble and soluble forms of melanin. Soluble melanin extracted from Inonotus hispidus,20 squid ink,21 and vinary waste22 possess excellent antioxidant activity while extracts from Inonotus obliquus(23) possess in vitro immunomodulatory activity. The chemical synthesis of soluble melanin possessing a larger number of carbonyl groups has also been reported.24
In this study we have discovered and biochemically characterized a novel form of soluble melanin that we term as peptidomelanin. Peptidomelanin was liberated into the surrounding medium during the germination of Aspergillus niger melanoliber spores. It was found be an amorphous, heterogeneous polymer containing an insoluble L-DOPA core. It was solubilized via a corona of short heterogeneous peptides with a mean residue length of 2.6 ± 2.3 amino acids anchored to the core polymer via peptide bonds. In vitro experiments using dansylated L-DOPA suggest a biochemical mechanism for the formation of peptidomelanin: via the hydroxylation of tyrosine residues within short peptides before their incorporation into the growing melanin polymer.
Based on existing reports describing the ability of amino acids25 and peptides26 to chelate heavy metals, we hypothesized that the short, heterogeneous peptide chains on peptidomelanin may also possess the ability to chelate heavy metals. Unlike other chemical contaminants, heavy metals cannot be easily removed from the soil, and will remain in the environment indefinitely unless remediated. Conventional soil remediation strategies greatly differ and are typically tailored to suit specific local conditions.27 Such strategies vary in cost with an upper bound of USD 100/m2.27 Of particular interest to this study are in situ immobilization techniques via chemical fixation. Immobilization of heavy metals involves their in situ conversion into insoluble or poorly soluble forms, thereby decreasing their mobility in water and their uptake by roots. EDTA,28 NTA,29 CaCO3, iron grit, conocarpus biochar, and phosphate rock30 have all been characterized as heavy metal immobilizers.
Interestingly, peptidomelanin was found to be a potent heavy metal chelating agent, possessing the ability to chelate large quantities of lead, mercury, and uranium (as uranyl). It was found to ameliorate the toxic effects of 100 ppm mercury on germinating wheat, improving seed mass and shoot length. The ease of preparation of peptidomelanin and the potential to produce it in situ lead us to believe that it may have applications in heavy metal remediation.
Here we describe the discovery and extraction of peptidomelanin, its biochemical composition, and its ability to chelate metals thereby ameliorating heavy metal toxicity for germinating seeds.
Results
We discovered secreted peptidomelanin in a strain of Aspergillus niger isolated from a soil sample in Mumbai, India. The initial discovery was serendipitous. We were attempting to extract insoluble fungal melanin from the spores of fungi isolated locally. While culturing one isolate in Sabouraud broth, we noticed the broth took on an unusually dark coloration. The broth appeared dark even after filtering through a 0.2 μm PVDF syringe filter, indicating that the dark coloration was not merely originating from the spores. At that point, we hypothesized that the coloration originated from a soluble pigment and pursued the line of inquiry described here.
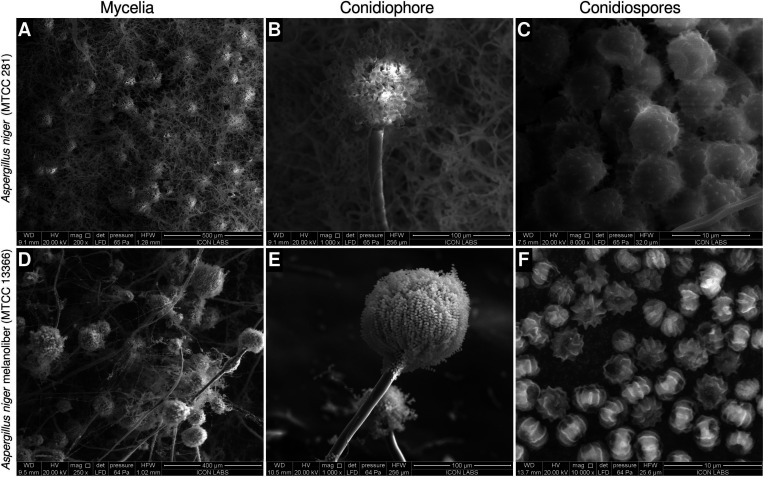
We named the strain Aspergillus niger melanoliber and deposited it in the Microbial Type Culture Collection (MTCC), Chandigarh (strain ID: MTCC 13366). Species identification was performed through ITS sequencing. The deposition certificate and strain identification report can be found in Figure S2. The ITS sequence was deposited into GenBank (accession ID: PP077302). A. niger melanoliber differs in morphology from the type strain of A. niger (MTCC 281/ATCC 9029). The type strain possesses a dense hyphal mass, abundant conidiophores, and large conidiospores (or simply, spores) with nondiscernible polar regions and evenly distributed echinulations (Figure 1A-C). A. niger melanoliber possesses a sparse hyphal mass, fewer large conidiophores, and small conodiospores with deep longitudinal striations originating from the poles (Figure 1D-F).
Figure 1.
Morphology of Aspergillus niger strains used in this study. (A–C) The A. niger type strain (MTCC 281). (A) Dense hyphal mass possessing abundant conidiophores. (B) Conodiophore possessing relatively fewer conidiospores (C) Large conidiospores with echinulations evenly distributed across their surface. Polar regions are not discernible. (D–F) A. niger melanoliber (MTCC 13366). (D) Sparse hyphal mass possessing fewer large conidiophores. (E) Conodiophore possessing a large number of small conidospores. (F) Small conidiospores possessing deep, longitudinal striations originating from discernible polar regions.
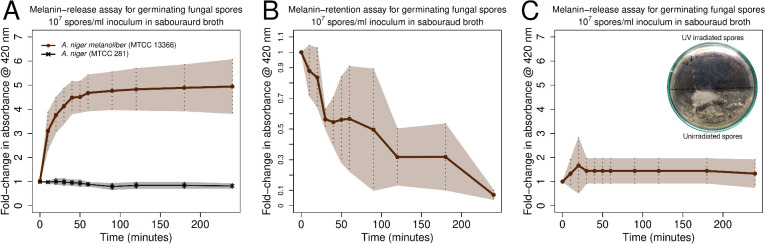
While germinating, A. niger melanoliber spores release peptidomelanin rapidly into Sabouraud broth, resulting in a ∼ 5-fold increase in absorbance (at 420 nm) in 1 h (Figure 2A). In contrast, spores of the type culture of A. niger (MTCC 281/ATCC 9029) do not release melanin into Sabouraud broth while germinating (Figure 2A). The melanin content within A. niger melanoliber spores decreases during germination (Figure 2B), indicating that peptidomelanin is not synthesized during germination; only existing peptidomelanin is released. Spores killed via UV-irradiation do not secrete peptidomelanin (Figure 2C). Therefore, peptidomelanin release is a biochemical process that requires a live organism to perform. Morphological changes associated with germination such as the disruption of the spore pellicle and hyphal growth occur at time points long after peptidomelanin release (Figure S3).
Figure 2.
Peptidomelanin release assay for Aspergillus spps. spores. (A) A. niger melanoliber (MTCC 13366) and A. niger (MTCC 281) spores were inoculated into Sabouraud broth. A. niger melanoliber rapidly released soluble peptidomelanin into the medium. In contrast, A. niger released no soluble peptidomelanin into the medium. (B) Peptidomelanin retention assay for A. niger melanoliber spores in Sabouraud broth. At predetermined time points, samples were isolated and fixed using 8% glutaraldehyde to halt peptidomelanin release. Samples were treated with 0.1 M NaOH to release retained peptidomelanin. Retained peptidomelanin decreased with time, indicating that new peptidomelanin is not synthesized during germination, and only existing peptidomelanin is released. 0.1 M NaOH did not degrade peptidomelain for the duration of incubation (Figure S4). (C) Melanin release assay for UV-irradiated A. niger melanoliber spores. Irradiated spores did not release peptidomelanin into the Sabouraud broth, demonstrating that only live spores release melanin. (Inset) Petri plate confirming UV irradiated spores do not germinate. All experiments were performed in quadruplicate. Lines represent means. The shaded area represents standard deviation.
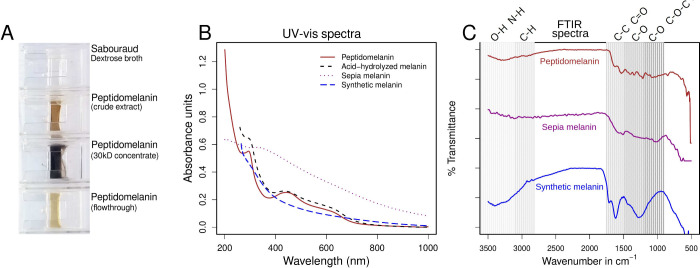
Peptidomelanin released into Sabouraud broth was precipitated at pH 2 (HCl-KCl buffer) and resuspended at pH 8.8 (Tris buffer) for further experiments. The buffer system was exchanged via dialysis through a 3.5 kDa membrane. Peptidomelanin concentrated using a 30 kDa centrifugal concentrator displayed a dark brown color, characteristic of melanin polymers (Figure 3A). Peptidomelanin displayed a UV–vis spectrum characteristic of L-DOPA melanin, absorbing across a UV–visible range of ≤200–650 nm but displaying a maxima in the UV region at ≤200 nm (Figure 3B). Peptidomelanin also possesses a broad local spectral maxima at 420–460 nm which was used for the colorimetric estimation of melanin content (Figure 2). Acid-hydrolyzed melanin (lacking an amino acid component) displays a similar spectrum, indicating that the melanin core polymer is responsible for the absorbance spectrum. Synthetic L-DOPA melanin also displays a similar spectrum, indicating that the melanin core polymer is composed of phenolic derivatives, most likely originating from L-DOPA. Sepia melanin displayed strong absorbance in the UV region while also displaying greater absorbance across the visual spectrum compared to the other samples tested.
Figure 3.
Spectral analysis of peptidomelanin. (A) Peptidomelanin appears dark brown in color. Peptidomelanin can be concentrated using a 30 kDa centrifugal concentrator. (B) UV–vis spectral overlay of four different melanin spectra. Peptidomelanin displays an absorbance spectrum characteristic of melanin. It displays the greatest absorbance in the UV region but strongly absorbs light in the visible spectrum as well, up to a wavelength of 650 nm, with absorption decreasing for longer wavelengths. Synthetic L-DOPA melanin displays a similar absorbance spectrum. Acid-hydrolyzed melanin (lacking a peptide component) also displays a similar spectrum. Sepia melanin displayed strong absorbance in the UV region while also displaying greater absorbance across the visual spectrum compared to the other samples. (C) FTIR spectra of peptidomelanin, sepia melanin, and synthetic L-DOPA melanin. All spectra have been vertically offset to ease comparison. All spectra appear somewhat similar. All display a broad dip between 3500 and 3100 cm–1 assignable to the O–H or N–H stretch vibration. Synthetic melanin displays and a sharp dip between 1800 to 1600 cm–1 assignable to C–C or C=O stretch vibration, while peptidomelanin and sepia melanin display shallower dips. Synthetic melanin displays strong signals in the 1600–1000 region, whose intensity is not matched in peptidomelanin and sepia melanin. This is possibly attributable to the simpler chemical composition of synthetic melanin.
An FTIR spectral comparison of peptidomelanin with sepia melanin and synthetic L-DOPA melanin revealed that all substances display somewhat similar spectra (Figure 3C). All spectra contain a broad dip between 3500 and 3100 cm–1 assignable to the O–H or N–H stretch vibration (shallower for peptidomelanin and sepia melanin, deeper for synthetic melanin). Synthetic melanin displays a sharp dip between 1800 to 1600 cm–1 assignable to C–C or C=O stretch vibration, while peptidomelanin and sepia melanin display shallower dips in this region. Unlike synthetic melanin, peptidomelanin and sepia melanin do not display strong signals in the 1600–1000 cm–1 region, possibly due to their more complex chemical composition. Synthetic melanin is entirely composed of polymerized L-DOPA residues, while peptidomelanin possesses a large peptide component. Sepia melanin possesses sulfur, possibly alluding to a minor pheomelanin component.31 Notably, the 1000 cm–1 signal observed in synthetic melanin, corresponding to C–O or C–O–C stretch vibrations is absent in sepia melanin and peptidomelanin. The overall spectra for all 3 samples are characteristic of polymers possessing phenolic educts, such as those belonging to the L-DOPA melanin family. Allomelanin extracted from the black knot fungus,32 melanin extracted from Pseudomonas sp. WH00155,33 and melanin extracted from Bacillus cereus (Bc58)34 display similar spectra.
Tyrosinases are enzymes capable of oxidizing a broad range of phenols and catechols into quinones.35−37 In the context of melanin biosynthesis, tyrosinase synthesizes melanin from l-tyrosine through the DOPA pathway.38 Melanin synthesis inhibition experiments using tropolone lead to the formation of albino colonies lacking any pigmentation (Figure S5). As tropolone is a tyrosinase inhibitor,39 these observations indicate that the melanin core polymer is composed of phenolic derivatives, most likely originating from l-tyrosine.
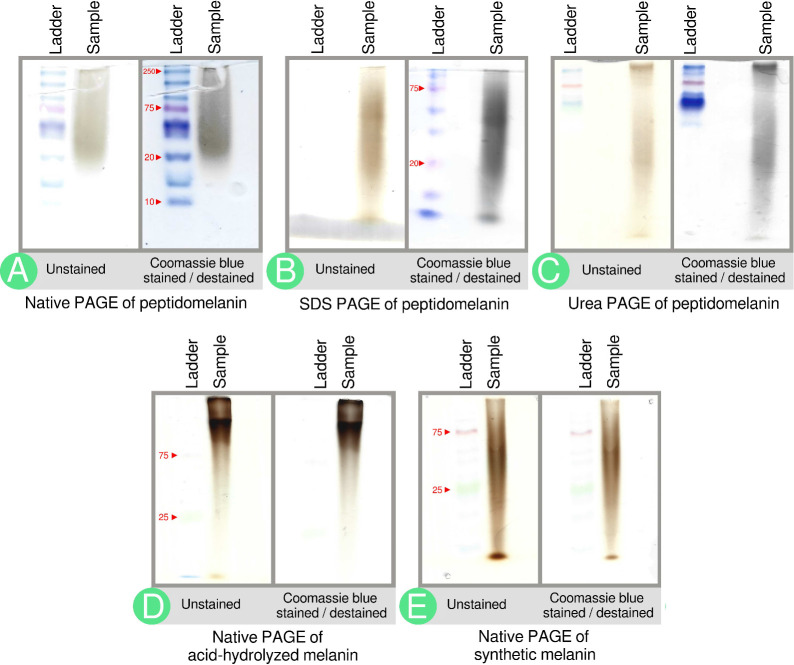
Preliminary experiments using polyacrylamide gel electrophoresis (PAGE) and agarose gels provided structural and biochemical insights pertaining to peptidomelanin. Peptidomelanin runs as a smear, rather than a discrete band, on native PAGE, SDS PAGE, and urea PAGE gels (Figure 4A-C). This indicates that peptidomelanin is an amorphous polymer with molecular weights in the kilodalton range. Synthetic L-DOPA melanin, a known amorphous polymer, also runs as a smear on a native PAGE gel (Figure 4E). The native PAGE peptidomelanin smear is stained by Coomassie blue (Figure 4A), indicating that it possesses an amino acid component that we later determined to be composed of short peptides rather than long proteins. Peptidomelanin smears on SDS and Urea PAGE gels (Figure 4B,C) are also stained by Coomassie blue, indicating that they retain their amino acid component even under strong denaturing conditions. This indicates that the amino acid component is covalently bound to peptidomelanin. Peptidomelanin was acid-hydrolyzed using 6 N HCl under a N2 atmosphere. The melanin hydrolysate was insoluble and had to be resolubilized in water by dissolving in DMSO and dialysing against deionized water. The resulting substance runs slower on a native PAGE gel but still forms a smear. The smear does not readily uptake Coomassie blue, indicating that acid-hydrolysis removed the amino acid component (Figure 4D). This substance is therefore referred to simply as acid-hydrolyzed melanin. The synthetic L-DOPA melanin smear run on a native PAGE, known to lack a peptide component, also does not readily uptake Coomassie blue (Figure 4E).
Figure 4.
PAGE gels indicate the biochemical composition of peptidomelanin. (A) Native PAGE of peptidomelanin. A brown smear from ∼20 to ≥250 kDa is observed. Coomassie blue staining darkens the smear, indicating the presence of an amino acid component. (B) SDS PAGE and (C) 8 M Urea PAGE of peptidomelanin. Coomassie blue staining reveals that the amino acid component does not separate from the melanin component, indicating a covalent linkage between the two components (ladder not resolved). The observation of smears rather than discrete bands on gels A–C confirms that peptidomelanin is an amorphous polymer. (D) Native PAGE of resolubilized acid-hydrolyzed melanin displays a smear that runs slower on a gel. The smear was only marginally stained by Coomassie blue. Both these observations indicate the loss of the amino acid component upon acid-hydrolysis. (E) Native PAGE of synthetic L-DOPA melanin. The smear does not uptake Coomassie blue as it lacks an amino acid component.
The solubility of peptidomelanin, and the corresponding insolubility of acid-hydrolyzed melanin lacking an amino acid component, confirm that amino acids form a “corona” that solubilizes the insoluble core polymer.
Transmission electron microscopy (TEM) performed on peptidomelanin revealed a median particle diameter of 618940 nm in solution, corresponding to particles with a median molecular weight of 9730327 MDa (Figure S6). While our TEM instrument had a resolution of 5 nm, PAGE experiments were able to detect molecules in the kilodalton range. This indicates that peptidomelanin adopts a wide range of molecular sizes and assemblages from the kilodalton to megadalton range. It should be noted that the megadalton-sized particles observed under TEM could be composed of kilodalton-sized subunits observed during PAGE.
Peptidomelanin does not possess surface-accessible chitin, as evidenced by the inability of the native PAGE peptidomelanin smear to uptake Calcofluor White (Figure S7A). Peptidomelanin does not possess nucleic acids, as evidence by the inability of an agarose gel peptidomelanin smear to uptake ethidium bromide (Figure S7B). The elemental composition of peptidomelanin was obtained using SEM-EDS (Table S3, Data set S1). The absence of phosphorus confirms the absence of nucleic acids. Peptidomelanin was found to contain K, Na, and Ca, indicating it possesses some ability to chelate these metals.
The amino acid component of peptidomelanin was confirmed via LC-MS of the acid-hydrolysate. Peptidomelanin was acid-hydrolyzed using 6 N HCl under a N2 atmosphere. The resulting insoluble acid-hydrolyzed melanin was centrifuged to obtain an insoluble pellet. The supernatant was evaporated to remove HCl and the resulting residue was resuspended in deionized water. LC-MS was performed on this solution to obtain its amino acid composition (Table 1). The percentage amino acid composition of peptidomelanin was calculated by simply dividing the total mass of amino acids observed during LC-MS by the total mass of peptidomelanin prior to acid-hydrolysis (eq 1), and was found to be 22.98 ± 1.84%. LC-MS reports and analyses are provided in the Supporting Information (Data set S2, Text S1, and Table S1).
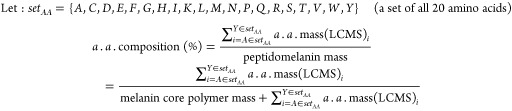 |
1 |
Table 1. Amino Acid Composition of the Peptidomelanin Acid Hydrolysate Acquired via LC-MS (DNA Labs India, Hyderabad)a.
| Absolute mass % | Relative mass % | |
|---|---|---|
| Amino acid | (mean ± SD) | (mean ± SD) |
| Glycine | 7.73 ± 0.82 | 33.62 ± 0.92 |
| Glutamate** | 2.49 ± 0.05 | 10.85 ± 0.61 |
| Proline | 2.45 ± 0.09 | 10.69 ± 0.46 |
| Alanine | 2.39 ± 0.12 | 10.42 ± 0.27 |
| Aspartate* | 2.26 ± 0.19 | 9.83 ± 0.06 |
| Arginine | 1.42 ± 0.10 | 6.19 ± 0.05 |
| Leucine | 0.90 ± 0.02 | 3.93 ± 0.21 |
| Lysine | 0.78 ± 0.07 | 3.39 ± 0.03 |
| Serine | 0.61 ± 0.01 | 2.67 ± 0.16 |
| Phenylalanine | 0.51 ± 0.03 | 2.23 ± 0.04 |
| Threonine | 0.47 ± 0.18 | 2.03 ± 0.62 |
| Histidine | 0.25 ± 0.11 | 1.08 ± 0.40 |
| Tyrosine | 0.21 ± 0.01 | 0.93 ± 0.03 |
| Valine | 0.18 ± 0.01 | 0.80 ± 0.01 |
| Methionine | 0.16 ± 0.01 | 0.69 ± 0.03 |
| Isoleucine | 0.13 ± 0.02 | 0.55 ± 0.15 |
| Cysteine | 0.01 ± 0.00 | 0.06 ± 0.01 |
| Tryptophan*** | 0.01 ± 0.00 | 0.04 ± 0.00 |
| Asparagine* | 0.00 ± 0.00 | 0.01 ± 0.01 |
| Glutamine** | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Total | 22.98 ± 1.84 | 100 |
19 of 20 canonical amino acids were detected in the peptidomelanin acid-hydrolysate. Absolute mass % refers to the mass of each amino acid as a percentage of the total mass of peptidomelanin. The sum total of the absolute mass % of all amino acids is 22.98 ± 1.84%, which is the total % amino acid composition of peptidomelanin. Relative mass % refers to the mass of each amino acid as a percentage of the total mass of all amino acids present. It should be noted that some amino acids cannot be quantified via acid hydrolysis. Amide groups like those found on asparagine* and glutamine** would be converted into carboxyl groups in hot hydrochloric acid, leading to an over-representation of aspartate* and glutamate**. Tryptophan*** is readily degraded under acid hydrolytic conditions and would therefore be underrepresented.
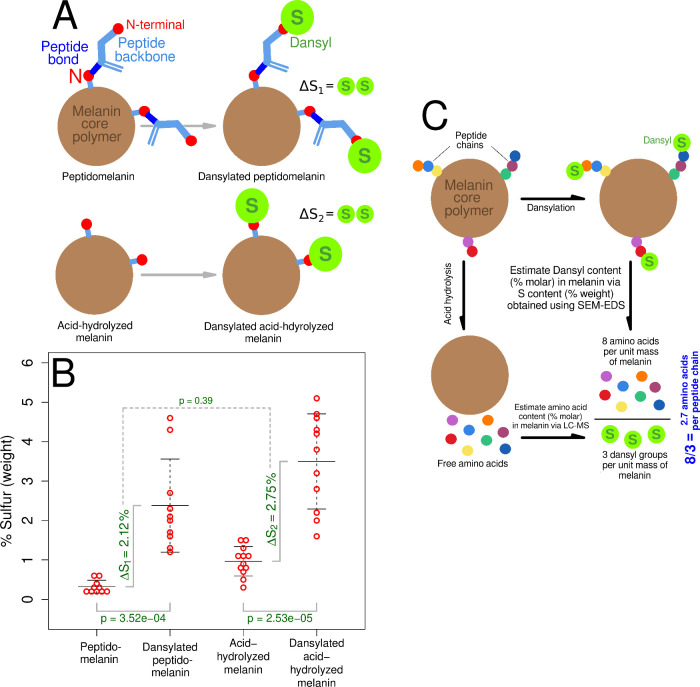
Despite possessing amino acids, papain-hydrolyzed peptidomelanin did not display peptide fragments in the molecular weight range of 400 to 2000 Da (Figure S8, Figure S9). This indicates that the amino acids in peptidomelanin are polymerized as short peptides too small to be cleaved by papain, rather than as long proteins. We used dansyl chloride in order to determine the mean amino acid length of the peptides comprising peptidomelanin. Dansyl chloride is a sulfur-containing fluorophore that can be covalently linked to the N-terminal of amino acids and peptides. We successfully dansylated peptidomelanin and acid-hydrolyzed melanin (Figure 5A,B). Dansylated peptidomelanin remained soluble whereas acid-hydrolyzed melanin did not. We subjected four substances: peptidomelanin, dansylated peptidomelanin, acid-hydrolyzed melanin, and dansylated acid-hydrolyzed melanin, to SEM-EDS in order to quantify their sulfur content (% weight). The chemical nature of these substances is depicted in Figure 6A. The sulfur originating from the dansyl group alone (ΔS1) can be quantified using SEM-EDS by normalizing and subtracting the sulfur content (% weight) of dansylated peptidomelanin from the sulfur content (% weight) of peptidomelanin (Figure 6B). We found ΔS1 to be 2.12 ± 2.03% (% weight). The dansyl content (% molar) could then be calculated from ΔS1 (where % molar = % weight of substance/MW). As dansyl groups covalently link to N-terminals in a 1:1 molar stoichiometric ratio with a 100% reaction efficiency (Text S2, Table S2, Figure S15), the dansyl content (% molar) corresponds to the N-terminal content (% molar). The amino acid content (% molar) could be calculated from LC-MS data (Table 1). The mean number of amino acids per peptide chain was then simply the amino acid content (% molar) divided by the N-terminal content (% molar), or the number of amino acids for every N-terminal. This value was found to be 2.6 ± 2.3 amino acids. Figure 6C illustrates the experiments and calculations performed. All calculations are provided in Text S2 (Equations S1–S10). All raw SEM-EDS data is provided in Data set S3.
Figure 5.
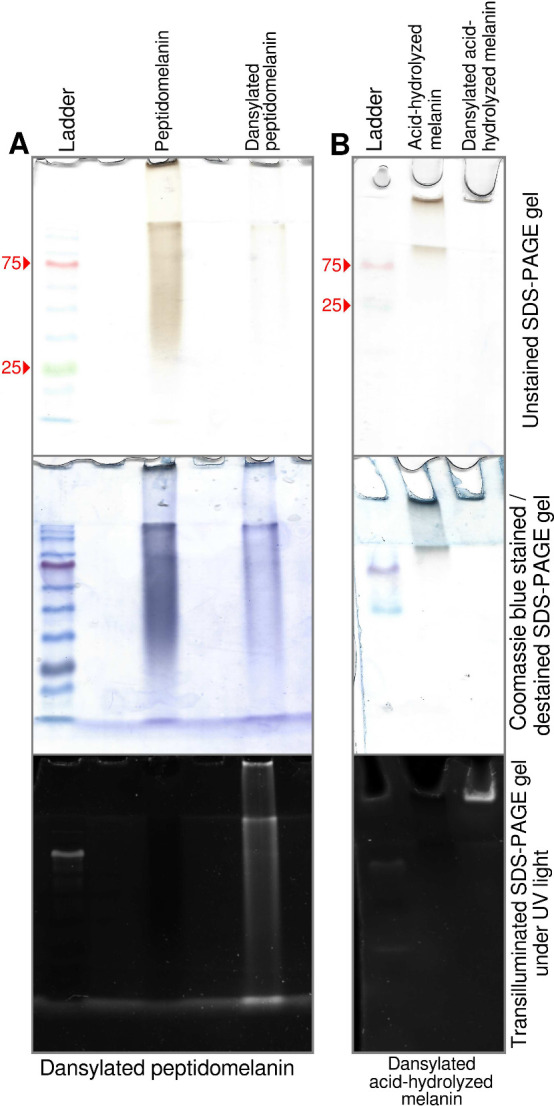
Dansylation of peptidomelanin and its derivatives. Dansyl chloride is a sulfur-containing fluorophore that can be covalently linked to the N-terminal of amino acids and peptides. (A) SDS PAGE of peptidomelanin and dansylated peptidomelanin. Both substances are stained by Coomassie blue. However, only dansylated peptidomelanin displays fluorescence under UV transillumination. (B) SDS PAGE of resolubilized acid-hydrolyzed melanin and dansylated acid-hydrolyzed melanin. Both substances are poorly stained by Coomassie blue as they lack their amino acid component. Only dansylated acid-hydrolyzed melanin displays fluorescence under UV transillumination. Dansylated acid-hydrolyzed peptidomelanin could not be resolubilized and therefore does not migrate on an SDS PAGE gel.
Figure 6.
Dansylation experiments using peptidomelanin and its derivatives. (A) Schematic showing all the substances under study. Dansyl chloride is a sulfur-containing fluorophore that can be covalently linked to the N-terminal of amino acids and peptides. Peptidomelanin will possess an amino acid component, whereas acid-hydrolyzed melanin will not. (B) SEM-EDS was used to estimate the sulfur (weight %) content of all four substances depicted in panel A. Sulfur acts as a proxy for the dansyl group. The increase in sulfur (ΔS1 = 2.12 ± 2.03%) between peptidomelanin and dansylated peptidomelanin represents sulfur originating only from dansyl linked to N-terminals. Likewise, the increase in sulfur (ΔS2 = 2.75%) between acid-hydrolyzed melanin and dansylated acid-hydrolyzed melanin represents sulfur originating only from dansyl linked to N-terminals. ΔS1 is not significantly different from ΔS2 (p = 0.39, 2-sided p-value), indicating that peptides are linked to the melanin core polymer via a peptide bond (see text). Error bars represent the standard deviation from the mean. (C) The mean peptide chain length can be calculated using data from dansylation experiments (panel B) as well as LC-MS amino acid quantification experiments (Table 1). Placeholder data is used here to explain the rationale. The actual mean peptide chain length was calculated to be 2.6 ± 2.3 amino acids. All calculations are provided in Text S2.
Each short peptide chain possesses an individual sequence. This can be inferred simply by observing that the number of canonical amino acids detected in peptidomelanin (19 of 20, Table 1) is larger than the mean number of amino acids per peptide chain (2.6 ± 2.3). Therefore all the amino acids observed cannot possibly be contained within a single chain. The dansylated amino acid mixture from acid-hydrolyzed dansylated peptidomelanin was subjected to electrophoresis in order to resolve the dansylated N-terminal amino acid (Figure S10). We observed a broad smear, indicating the presence of multiple dansylated N-terminal amino acids which further confirms that every peptide chain possesses an individual sequence.
The difference in sulfur content originating only from the dansyl group for dansylated peptidomelanin (ΔS1 = 2.12 ± 2.03%) and dansylated acid-hydrolyzed melanin (ΔS2 = 2.75 ± 1.54%) is not statistically significant (p = 0.98, Figure 6B, Text S2). Therefore, the total number of dansylatable N-terminals (% molar) remains very similar in both peptidomelanin and acid-hydrolyzed melanin despite the latter lacking a peptide component. From this, we infer that the loss of the peptide N-terminus during acid-hydrolysis is compensated for by the appearance of an N-terminus on the surface of acid-hydrolyzed melanin in a ∼ 1:1 ratio. This indicates that peptides are linked to the melanin core polymer via a peptide bond, which will appear as an N-terminus after hydrolysis.
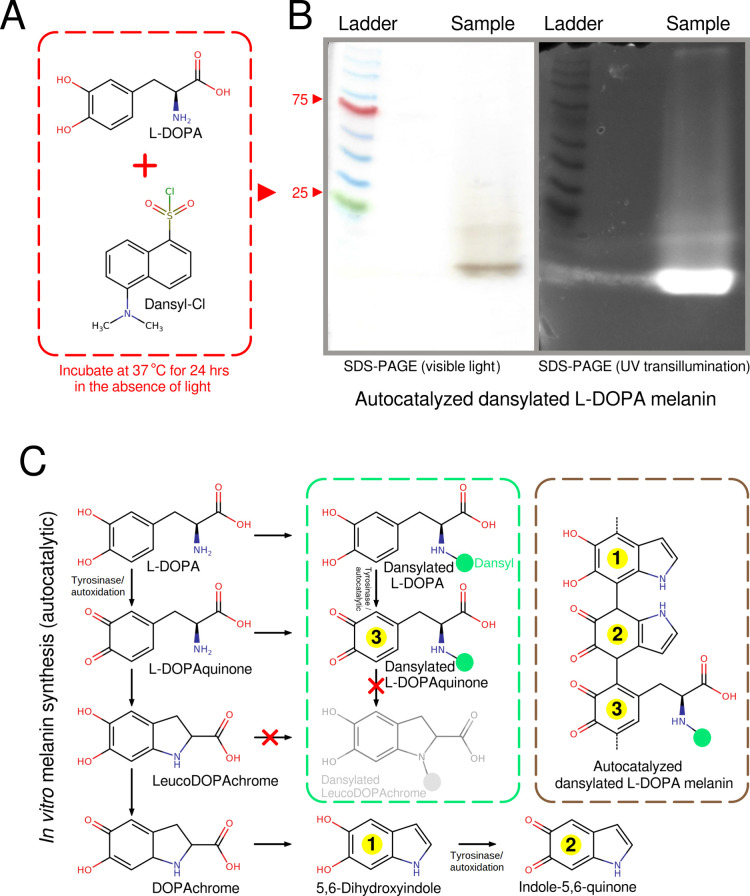
As a peptide bond is formed between the backbones of two amino acids, we hypothesized that tyrosine could serve as a “bridge” copolymerizing the peptides to the melanin core polymer. Tyrosine is hydroxylated to L-DOPA (L-3,4-dihydroxyphenylalanine) via tyrosinase during the first step of eumelanin synthesis via the DOPA pathway40 (Figure 7C). Short peptides could be incorporated into the growing melanin polymer via the hydroxylation of endogenous tyrosine residues. The resulting L-DOPA would still remain bound to the rest of the peptide via a peptide bond, and therefore contain an “occupied” N-terminus. Under these circumstances L-DOPA would not be able to undergo intramolecular addition (cyclization) into leucoDOPAchrome and its metabolites along the DOPA pathway (Figure 7C). We performed a simple experiment to determine whether L-DOPA containing an occupied N-terminus would still polymerize into melanin. L-DOPA was dansylated under conditions that allowed dansyl chloride to react with primary amines (Figure 7A), thereby occupying the N-terminus and chemically simulating a short peptide chain. We found that dansylated L-DOPA was incorporated into melanin that was autoxidized in vitro (Figure 7B). This experiment demonstrates that it is biochemically possible for short peptides to be incorporated into melanin via endogenous L-DOPA. Further confirmatory experiments are required too determine whether a similar process occurs in vivo in A. niger melanoliber.
Figure 7.
In vitro formation of melanin from dansylated L-DOPA shows that it is possible for L-DOPA containing an occupied N-terminus to be incorporated into melanin. (A) L-DOPA and dansyl chloride were incubated under conditions that allowed for the dansylation of primary amines. These conditions also allowed for the in vitro autoxidation of L-DOPA into melanin. (B) An SDS PAGE gel confirms that the autoxidized L-DOPA melanin is linked to dansyl. (C) Reactions along the melanin biosynthetic pathway that can form eumelanin from L-DOPA via nonenzymatic or autodixation reactions.40 Tyrosinase can increase reaction rates41 but is not essential for the in vitro formation of eumelanin. Only L-DOPA and L-DOPAquinone possess dansylatable N-terminals (primary amines). Metabolites from leucoDOPAchrome onward only possess secondary amines and cannot be dansylated under our reaction conditions. Likewise, dansylated L-DOPAchrome, lacking a primary amine group, cannot undergo intramolecular addition (cyclization) into dansylated leucoDOPAchrome. Therefore, L-DOPA and L-DOPAquinone containing dansylated/occupied N-terminals can be incorporated into melanin.
Quinone intermediates formed during the in vitro autoxidation of L-DOPA were found to be largely nonreactive against methionine (Figure S11, Data set S4). Methionine served as a model amino acid, possessing both a nucleophile (−NH2) and an electrophile (−COOH) in its backbone. Methionine can also easily be detected via SEM-EDS through its sulfur content. The paucity of incorporated methionine eliminates the possibility of the large-scale incorporation of peptides into the growing melanin polymer, or modifications of their amino acids, via reactions with quinone groups.
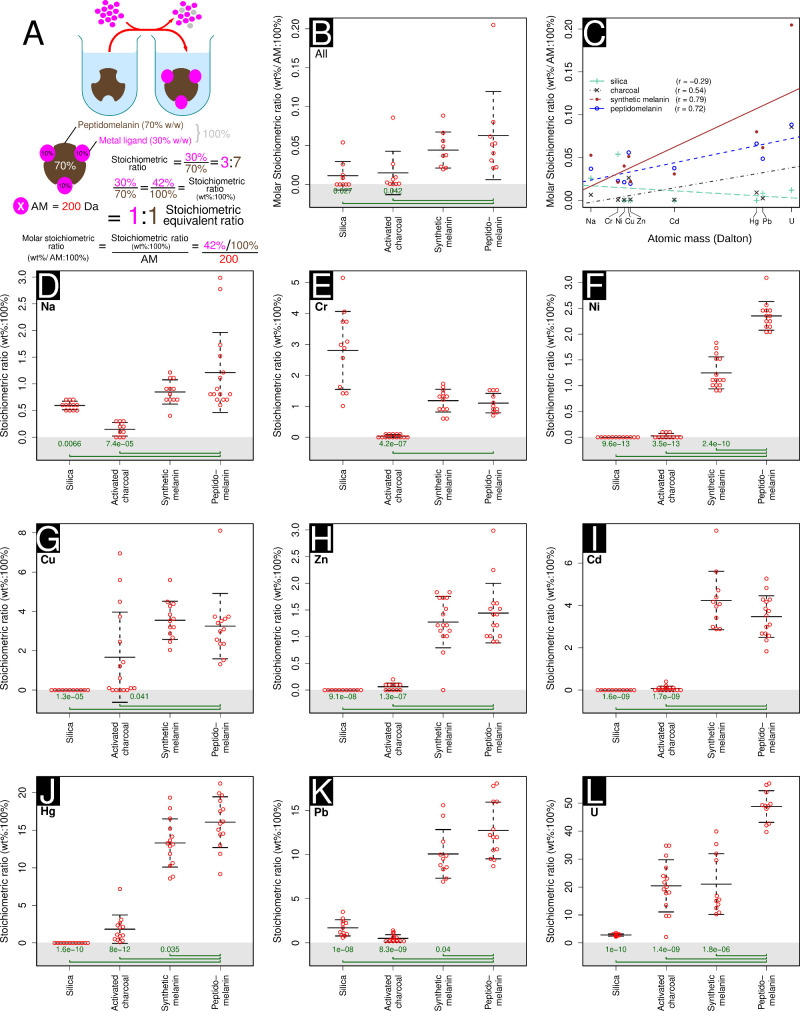
We assessed the ability of peptidomelanin to chelate metals. The ability of other forms of melanin to chelate heavy metals42−44 and the presence of amino acids on peptidomelanin capable of forming complexes with metals lead us to believe peptidomelanin would possess similar abilities. We performed experiments to determine the metal:substance stoichiometric ratios (wt%:100%) and molar stoichiometric ratios (wt%/AM:100%), as depicted in Figure 8A, for 4 substances against 9 metals. The substances chosen were (1) peptidomelanin; (2) synthetic L-DOPA melanin, serving as a positive control; (3) activated charcoal, serving as standard metal chelating agent45−48 to benchmark against peptidomelanin and (4) silica, expected to act as an inert substrate. The metals chosen ranged from light to heavy, and were: sodium, chromium, nickel, copper, zinc, cadmium, mercury, lead, and uranium (as uranyl) (Figure 8). A solution containing excess metal salt was introduced to a solution containing any one of these four substances and allowed to react. Excess metal salt was then removed via dialysis or pelleting, depending on the solubility of the metal complex (Figure 8A). The elemental compositions of all metal complexes were assayed using SEM-EDS.
Figure 8.
Metal-binding stoichiometric assays used to determine which substance tested has the greatest capacity to chelate metal ions. Four substances were tested: silica, activated charcoal, synthetic L-DOPA melanin, and peptidomelanin. (A) A visual depiction of our protocol. An excess of metal salt is added to all substances tested. The unbound metal salt was then removed via dialysis or centrifuging and washing, depending on the solubility of the metal complex. SEM-EDS was used to estimate the percentage metal in all complexes. Placeholder data is used to describe how the stoichiometric ratio (wt%:100%), stoichiometric equivalent ratio, and molar stoichiometric ratio are calculated (AM: atomic mass). (B) The mean metal-binding stoichiometric ratio of all substances tested across 9 metals, ranging from sodium to uranium (as uranyl). Peptidomelanin can chelate significantly larger ratios of metal compared to silica and activated charcoal. Synthetic melanin does not display a significant difference in metal chelation compared to peptidomelanin. Here, the molar stoichiometric ratio is used to normalize our data across metals with different molecular weights. (C) The ratios of metal chelated by peptidomelanin, synthetic melanin, and activated charcoal is positively correlated with the atomic mass of the metal. Silica displays no strong correlation. Regression lines for all substances tested are shown. (D–L) Metal-binding stoichiometric assays used to determine which substance tested has the greatest capacity to chelate a given metal ion. The means obtained from these experiments were used as data for the summary statistics in panels B and C. The individual metal ions tested were: (D) sodium, from NaCl, (E) chromium, from CrCl3.6H2O, (F) nickel, from Ni(NO3)2, (G) copper, from CuSO4.5H2O, (H) zinc, from ZnSO4, (I) cadmium, from Cd(NO3)2, (J) mercury, from HgCl2, (K) lead, from Pb(NO3)2, and (L) Uranium, from UO2(CH3COO–).2H2O. For panels D to L, ≥10 SEM-EDS spectra were collected per sample (technical replicates). For panels B and D–L, statistical significance testing was performed using the Welch 2-sample t test. All substances were compared to peptidomelanin. P-values ≤0.05 are shown. Error bars represent the standard deviation from the mean. Raw data is provided in Data set S5.
Peptidomelanin was observed to chelate significantly larger ratios of metals compared to silica and activated charcoal (Figure 8B). Synthetic melanin did not display a significant difference in metal chelation ratios compared to peptidomelanin. Silica did not chelate heavy metals. However, it is interesting to note that both peptidomelanin and synthetic melanin were able to chelate larger ratios of metals compared to activated charcoal, which is extensively used in agricultural settings and was expected to chelate heavy metals.45−48 The ratios of metal chelated by peptidomelanin, synthetic melanin, and activated charcoal is positively correlated with the atomic mass (AM) of the metal (Figure 8C). For example, peptidomelanin chelates a smaller ratio of AM-adjusted sodium (0.05 wt %/AM:100%, AM = 23 Da) compared to uranium (as uranyl) (0.20 wt %/AM:100%, AM = 238 Da). Silica displays no strong correlation. This phenomenon may simply be explained by the fact that heavier metals possess higher coordination numbers49 and would therefore be more easily chelated than lighter metals.
Stoichiometric data for individual metals ranging from sodium to uranium (as uranyl) is provided in Figure 8D–L. Both peptidomelanin and synthetic melanin were able to chelate all metals tested, including agriculturally relevant heavy metals (chromium to uranium), in significantly larger stoichiometric ratios compared to activated charcoal. For example, peptidomelanin chelates mercury, lead and uranium (as uranyl) at stoichiometric ratios (wt%:100%) of greater than 10% (Figure 8J,K,L). Due to the ease of peptidomelanin preparation and solvation, we tested its ability to remediate substrates contaminated with mercury expecting to observe an improvement in seed germination and growth. All raw SEM-EDS data for metal binding experiments is provided in Data set S5.
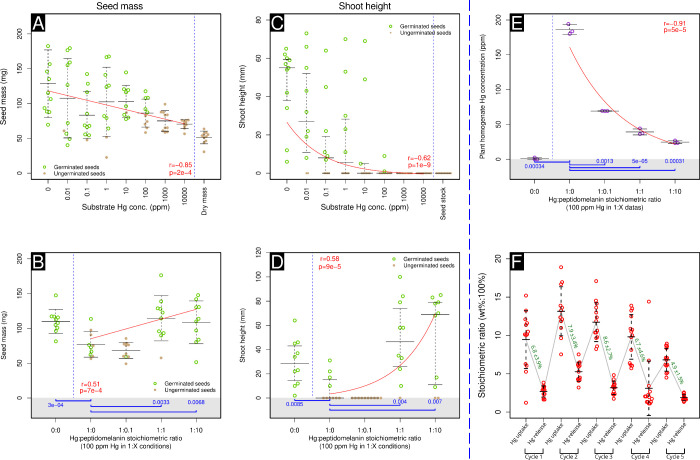
We performed germination experiments on wheat seeds planted in silica substrate intentionally contaminated with mercury. Mercury from HgCl2 was prepared at concentrations ranging from 0.01 ppm to 10,000 ppm, at 10-fold concentration intervals. An uncontaminated control was also prepared. Ten wheat seeds per concentration were introduced into this substrate and incubated outdoors at ambient temperature and lighting conditions for 7 days (Table S4). At the end of this time-interval, we observed a significant decrease in both seed mass and shoot length with increasing concentrations of mercury (Figure 9A,C). Wheat seed mass was found to be inversely correlated with increasing mercury concentration (r = −0.40, log–linear correlation, Figure 9A). Wheat shoot length was found to be inversely correlated with increasing mercury concentration (r = −0.62, log–log correlation, Figure 9C). These observations were expected as mercury is known to be toxic to plants.50,51
Figure 9.
Wheat germination dose–response experiments show that peptidomelanin can ameliorate soil mercury toxicity in agricultural contexts. (A) Wheat seed mass shows a moderate inverse correlation (log–linear) with mercury concentration (r = −0.40, p = 2 × 10–4) across a range of 0 ppm to 10,000 ppm mercury. (B) When germinated in substrate containing 100 ppm mercury, wheat seed mass shows a moderate positive correlation (log–linear) with increasing concentrations of peptidomelanin (r = 0.51, p = 7 × 10–4). Wheat seeds germinated in substrate containing 100 ppm mercury completely neutralized by peptidomelanin (at 1:1 and 1:10 stoichiometric equivalent ratios) display significantly larger seed masses compared to wheat seeds germinated in 100 ppm mercury alone (1:0). (C) When germinated in substrate containing 100 ppm mercury, wheat shoot length shows a moderate inverse correlation (log–log) with increasing concentrations of peptidomelanin (r = −0.62, p = 1 × 10–9). (D) Shoot lengths of wheat seeds grown in substrate containing 100 ppm mercury completely neutralized by peptidomelanin (at 1:1 and 1:10 stoichiometric equivalent ratios) display a moderate positive correlation (log–log) with increasing concentrations of peptidomelanin (r = 0.58, p = 9 × 10–5). (E) ICP-OES quantification of the mercury concentration of wheat seed homogenates. When germinated in substrate containing 100 ppm mercury, plant tissue mercury concentrations show a strong inverse correlation (log–log) with increasing concentrations of peptidomelanin (r = −0.91, p = 5 × 10–5). Wheat seeds germinated in substrate containing 100 ppm mercury either partially or completely neutralized by peptidomelanin (at 1:0.1, 1:1, and 1:10 stoichiometric equivalent ratios) display significantly lower tissue mercury concentrations compared to wheat seeds germinated in 100 ppm mercury alone (1:0). (F) Peptidomelanin reuse experiment. We assayed peptidomelanin’s ability to bind mercury after 5 cycles of binding and removal via EDTA chelation. Peptidomelanin shows good reuse characteristics for 4 cycles. Mercury was quantified via SEM-EDS. Green values indicate the change in mercury stoichiometric ratio (wt%:100%) between mercury binding and removal (mean and standard deviation). For panels B, D, and E, statistical significance testing was performed using the Welch 2-sample t test (p-values in blue). All substances were compared to wheat germinated in 100 ppm mercury in the absence of peptidomelanin. P-values ≤0.05 are shown. For panels A–E, “r” (in red) represents the Pearson’s correlation coefficient. For panels A–E, “p” (in red) represents the p-value of our regression model against the null model (a horizontal line) using an ANOVA. All data points, and not just the means/medians, were used to create regression models. For panels A,B, E and F, error bars represent the standard deviation from the mean. For panels C and D, error bars represent the interquartile range. Here, the median and quartiles were considered as shoot lengths do not follow a Gaussian distribution. Raw data (images) are provided in Figure S12.
We then attempted to remediate the effects of mercury toxicity on germinating wheat seeds planted in silica substrate intentionally contaminated with 100 ppm mercury using different stoichiometric equivalent ratios of peptidomelanin. Here, a stoichiometric equivalent (Figure 8A) is defined as the mass of peptidomelanin needed to completely chelate and neutralize one unit mass of mercury (data obtained from Figure 8J). Silica substrate containing mercury:peptidomelanin stoichiometric equivalent ratios of 0:0 (growth control), 1:0 (toxicity control), 1:0.1, 1:1, and 1:10 were prepared and planted with wheat seeds. Incubation was performed outdoors at ambient temperature and lighting conditions for 7 days (Table S4). At the end of this time-interval, we observed a statistically significant increase in seed mass of seeds planted in 1:1 and 1:10 mercury:peptidomelanin stoichiometric equivalent ratios (Figure 9B), compared to the seeds planted in only 100 ppm mercury (0:1). The increase in seed mass was positively correlated with the increasing stoichiometric equivalent ratio of melanin (r = 0.51, log–linear correlation, Figure 9B).
Likewise, we observed a statistically significant increase in the shoot lengths of seeds planted in 1:1 and 1:10 mercury:peptidomelanin stoichiometric equivalent ratios (Figure 9D), compared to the seeds planted in only 100 ppm mercury (0:1). The increase in shoot length was positively correlated with the increasing stoichiometric equivalent ratio of melanin (r = 0.58, log–log correlation, Figure 9D).
ICP-OES was used to quantify the concentration of mercury in germinating wheat seeds planted in all mercury:peptidomelanin stoichiometric equivalent ratios tested. We observed a statistically significant decrease in mercury concentrations of seeds across 1:0.1, 1:1 and 1:10 mercury:peptidomelanin stoichiometric equivalent ratios (Figure 9E), compared to seeds planted in only 100 ppm mercury (0:1). The internal mercury concentration was strongly and inversely correlated with the increasing stoichiometric equivalent ratio of melanin (r = −0.91, log–log correlation, Figure 9E). This indicates that peptidomelanin present in the silica substrate is able to chelate a sufficient quantity of mercury, preventing it from being absorbed by germinating wheat seeds. Peptidomelanin was found to localize within the root epidermis but did not penetrate into the cortex (Figure S13).
Overall, wheat germination experiments indicate that peptidomelanin may have applications in the remediation of agricultural soils contaminated with heavy metals. Pictures of all wheat seeds and seedlings experimented upon can be found in Figure S12. The ICP-OES report for seedling mercury concentration can be found in Data set S6. Raw data for the peptidomelanin reuse experiment can be found in Data set S7.
Peptidomelanin can be reused in agricultural roles or in any other role, as determined by peptidomelanin reuse experiments. Briefly, peptidomelanin was incubated in excess mercury (HgCl2) and washed, ensuring that only mercury bound to peptidomelanin was retained. Bound mercury was removed via EDTA chelation, freeing peptidomelanin for reuse. Five such cycles were performed, and the mercury concentration bound to peptidomelanin was estimated via SEM-EDS twice per cycle: before and after EDTA chelation (Figure 9F). We observed that peptidomelanin displayed good reusability over 4 cycles, with the stoichiometric ratio (wt%:100%) of mercury extracted from the polymer using EDTA ranging from 6.7 to 7.9%. A lower stoichiometric ratio (wt%:100%) of mercury was extracted during the fifth cycle (4.9%), indicating that the mercury chelation efficiency of peptidomelanin will degrade upon continuous use.
Discussion
In this study we report the discovery of peptidomelanin, a novel biopolymer secreted during the germination of Aspergillus niger melanoliber spores possessing the ability to chelate heavy metals. Although previous reports describing soluble melanins isolated from varying sources exist in the literature,15−24 none describe the substances’ biochemical composition in sufficient detail. To the best of our knowledge, no previous reports describing the biochemical composition of peptidomelanin exist in the literature.
Based on our experimental data, we can draw the following inferences about the biochemical composition and properties of peptidomelanin: 1: Peptidomelanin is an amorphous polymer with molecular weights of particles in the kilodalton to megadalton range, as determined by PAGE (Figure 4A-C) and TEM (Figure S6). 2: Peptidomelanin is composed of a phenolic core polymer as it possesses a similar FTIR spectrum to sepia melanin and synthetic L-DOPA melanin (Figure 3C). The FTIR spectrum of peptidomelanin is also similar to natural samples of L-DOPA melanin.32−34 Melanin synthesis is also inhibited by tropolone (Figure S5), indicating the core polymer is composed of phenolic derivatives most likely originating from tyrosine.
3: The peptide “corona” is responsible for the solubility of peptidomelanin (Figure 4). 4: The peptide component constitutes ∼23% of peptidomelanin, as determined by LC-MS (Table 1). It should be noted that some amino acids, such as tryptophan, will be readily degraded during acid hydrolysis. Other amino acids possessing amides (−CONH2) will be converted to acid forms (−COOH) during acid hydrolysis. As acid hydrolysis is the only technique available to us to study the amino acid composition of peptidomelanin, such losses are not quantifiable. 5: The mean length of these peptide chains is 2.6 ± 2.3 amino acids. This was determined by a combination of the aforementioned LC-MS data (Table 1) and the elemental composition (SEM-EDS) of dansylated peptidomelanin (Figure 6C). Although the calculated error margin was fairly large, our confidence interval ranges from 0.3 to 4.9. This still indicates that petidomelanin is composed of peptides with a length of less than 5 residues. 6: Peptide chains do not possess a single sequence. This was determined by cellulose thread electrophoresis of the supernatant of acid-hydrolyzed dansylated peptidomelanin (Figure S10), as well as simply observing the number of amino acids present in peptidomelanin (19 of 20, Table 1). 7: Amino acid chains are covalently linked to peptidomelanin via a peptide bond, as inferred from the elemental composition (SEM-EDS) of dansylated peptidomelanin (Figure 6B).
8: Based on the phenolic composition of the peptidomelanin core polymer, and based on the linkage of peptide chains to peptidomelanin via a peptide bond, we hypothesize that tyrosine residues (the precursor of L-DOPA) may serve as “bridges”: copolymerizing the peptide component to the L-DOPA core polymer. During sporogenesis, the simplest biosynthetic approach to produce peptidomelanin would be to proteolytically cleave excess cytoplasmic proteins and incorporate the resulting peptides into the growing peptidomelanin polymer via tyrosyl side chains. As tyrosinases are known to possess promiscuous proteolytic activity,52 both of the aforementioned processes could be catalyzed by a single enzyme. In vitro melanin autoxidation experiments using dansylated L-DOPA, with dansyl chemically simulating a short polypeptide chain, confirmed that the substance can be incorporated into a growing melanin polymer. However, further in vivo experiments are required to confirm if a similar biochemical process occurs in A. niger melanoliber. To the best of our knowledge, no previous study has reported the heteropeptidyl nature of soluble melanin, nor proposed any mechanism for its copolymerization.
9: Metal ions and oxides can be chelated to peptidomelanin via functional groups on both the melanin core L-DOPA polymer and peptide chains. This was determined via the metal-chelating ability of both peptidomelanin and synthetic L-DOPA melanin (Figure 8). Our model of the biochemical composition of peptidomelanin is illustrated in Figure 10.
Figure 10.
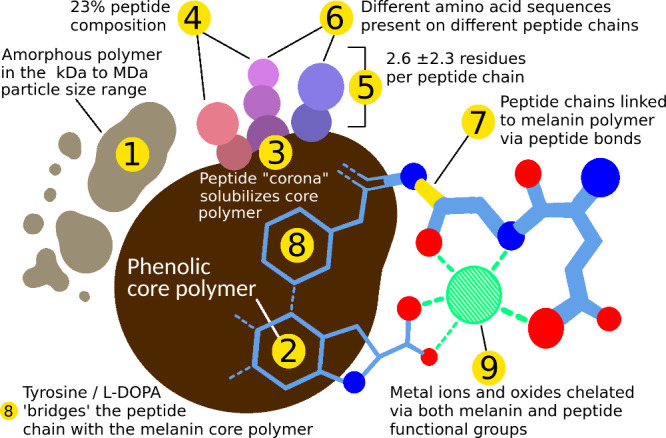
Our model of the biochemical composition of peptidomelanin. Claims 1–9 are supported by experimental data as described in the text.
Peptidomelanin was observed to ameliorate the toxicity of 100 ppm mercury during the germination of wheat, improving its seed mass and shoot length (Figure 9), indicating potential agricultural applications for the substance. Future in situ experiments will be performed to assay the efficacy of peptidomelanin in this role.
Methods
Unless otherwise specified, all experiments were performed at room temperature (24 °C).
Scanning Electron Microscopy–Energy-Dispersive X-ray Spectroscopy (SEM-EDS)
SEM/SEM-EDS experiments were performed using a FEI (Field Electron and Ion Company) Quanta 200 scanning electron microscope at Icon Laboratories Pvt. Ltd., Mumbai. Samples were observed at 20 kV and under a low vacuum mode with a chamber pressure of 65 Pascal.
Transmission Electron Microscopy
Transmission electron microscopy (TEM) was performed at Icon Laboratories Pvt. Ltd., Mumbai. 100 μL of a 0.1 mg/mL peptidomelanin sample was incubated with 900 μL of 100 mM HgCl2 for 10 min at room temperature. A low concentration of peptidomelanin is required to prevent precipitation in the presence of mercury. This mixture was then dialyzed twice against 1 L of deionized water (Milli-Q) through SnakeSkin 3.5 kDa molecular weight cutoff dialysis tubing. Dialysis removed excess (unbound) mercury, leaving behind peptidomelanin “stained” with mercury. Thirty μL of mercury-stained peptidomelanin was diluted with 4 mL of deionized water (Milli-Q). 0.2 μL was pipetted onto a carbon-coated copper TEM grid (200 mesh). The sample was dried under an infrared lamp prior to TEM visualization.
TEM experiments were performed using a FEI (Field Electron and Ion Company) Tecnai G2 Spirit BioTwin transmission electron microscope, with a resolution limit of 5 nm. Images were acquired at 100 kV using a tungsten “W” filament. Images were acquired on an Olympus soft imaging solutions Veleta CCD (charge-coupled device) camera. Images were interpreted on the Tecnai imaging and analysis software. Images were acquired at magnifications of 30–300 kX.
Images containing peptidomelanin
particles were analyzed using
Fiji/ImageJ.53 Pixel-to-nanometer conversions
were performed using the scale bars present as insets in all TEM micrograms.
Images were converted to 8-bit grayscale to find the edges of individual
polymer molecules. Edges were detected using a threshold of 128. Particle
areas were then calculated from the number of pixels constituting
every particle, with a 100 nm2 minimum threshold to avoid
background noise. Particle diameters were calculated from area using 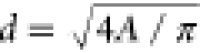 . As the particles are highly irregular,
the diameter is simply a useful abstraction of the data. As the particle
size distribution is non-Gaussian (Figure S6C), we reported a medianQ3Q1 value (618940 nm).
. As the particles are highly irregular,
the diameter is simply a useful abstraction of the data. As the particle
size distribution is non-Gaussian (Figure S6C), we reported a medianQ3Q1 value (618940 nm).
The median molecular weight of peptidomelanin particles observed under TEM can be estimated from their median diameter (618940 nm), and the reported density of melanin (1.37 g/mL54) using eq 2.
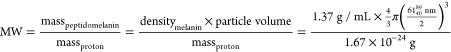 |
2 |
Peptidomelanin Exudation Assay
Both A. niger strains were inoculated on Sabouraud agar plates and incubated at 37 °C for 5 days in the absence of light to yield a dense sporulating fungal mass. This fungal mass was harvested using a nichrome loop and added to 5 mL of Sabouraud broth until the absorbance at 420 nm reached 0.6. A further 10 mL of Sabouraud broth was added to make a total of 15 mL. Absorbance readings at 420 nm were recorded at t = 0, 10, 20, 30, 40, 50, 60 min, 1.5, 2, 3, and 4 h intervals. Absorbance was recorded using a Systronics Digital colorimeter type 112 (Sr. No. 12275).
Peptidomelanin Retention Assay
The aforementioned A. niger melanoliber sporulating fungal mass was suspended in 5 mL of Sabouraud broth until the absorbance at 420 nm reached 0.6. A further 10 mL of Sabouraud broth was added to make a total volume of 15 mL. Absorbance readings at 420 nm were recorded at t = 0, 10 min, 20 min, 30 min, 40 min, 50 min, 60 min, 1.5, 2, 3, and 4 h intervals. At every time interval, 1 mL of this mixture was pipetted out and 0.2 mL of 8% glutaraldehyde was immediately added to fix the spores and stop melanin release. Each sample was then centrifuged at 10,000 rpm (11,200 RCF) for 10 min at 4 °C. The pellet was then resuspended in 100 μL of 0.1 M NaOH to extract melanin from the spores and incubated for 1 h. 0.1 M NaOH did not degrade peptidomelain for the duration of incubation (Figure S4). Absorbance at 420 nm for each sample was measured in 96 well plates.
UV Irradiation
The aforementioned A. niger melanoliber sporulating fungal mass was suspended in 2 mL of saline in a quartz cuvette until the absorbance at 420 nm reached 0.1. This cuvette was irradiated using a HiMedia Hi-UV Intense transilluminator. Spores were incubated overnight (12 h) under UV light (312 nm) at an intensity of 3300 μW/cm2. A 100 μL aliquot of irradiated spores was streaked on a Sabouraud agar plate in a semicircle. The other semicircle was streaked using 100 μL of A. niger melanoliber spores (absorbance of 0.1 at 420 nm) that were not irradiated. This plate was incubated at 37 °C for 24 h in the absence of light. The remaining irradiated spores were used for peptidomelanin exudation experiments.
UV–Vis Spectroscopy
UV–vis spectroscopy was performed using a Shimadzu UV–vis Spectrophotometer UV - 1900i spectrophotometer. Absorbance values were collected from 200 to 1000 nm, with a resolution of 0.5 nm. Spectra were collected 5 times per sample and averaged. The spectra of peptidomelanin and sepia melanin (Nortindal Sea Products, S.L.) were recorded in deionized water (Milli-Q). The spectra of synthetic L-DOPA melanin (Merck: M8631–100MG, CAS: 8049–97–6) and acid-hydrolyzed melanin were recorded in DMSO. As per the manufacturer’s description (https://www.sigmaaldrich.com/IN/en/product/sigma/m8631, accessed on 17th July, 2024), synthetic L-DOPA was prepared by the oxidation of tyrosine using hydrogen peroxide.
FTIR Spectroscopy
FTIR spectra for peptidomelanin, sepia melanin, and synthetic L-DOPA melanin were recorded on a Bruker Alpha II Compact FT-IR Spectrometer. Attenuated total reflectance (ATR) spectroscopy was performed in transmission mode across a wavenumber range of 3500 cm–1 to 500 cm–1, with a resolution of 2 cm–1. Sixteen scans were collected and averaged per sample. Peptidomelanin and sepia melanin were dialyzed against deionized water (Milli-Q) and dried overnight at 80 °C to convert into a dry powder prior to FTIR spectra collection. Synthetic L-DOPA melanin was also analyzed as a dry powder.
Peptidomelanin Extraction
A. niger melanoliber was cultured on Sabouraud dextrose agar. Growth at 37 °C for 5 days in the absence of light yielded a dense mass of spores. Agar from one Petri plate (10 cm) containing spores was homogenized and mixed with 40 mL Sabouraud dextrose broth to induce peptidomelanin release. This mixture was incubated at 37 °C for 2 h in the absence of light. The mixture was then centrifuged at 7500 rpm (6300 RCF) for 10 min at 24 °C, and the supernatant containing peptidomelanin was subjected to syringe filtration (PVDF, 0.2 μm pore size). Peptidomelanin in the filtrate was acid precipitated using a 1:5 ratio of peptidomelanin:buffer, where the buffer used was 100 mM HCl-KCl (pH 2). This mixture was immediately centrifuged at 7500 rpm (6300 RCF) for 10 min at 24 °C. The peptidomelanin filtrate was washed thrice using the same volume of HCl-KCl buffer (pH 2.0) while centrifuging at 7500 rpm (6300 RCF) for 10 min at 24 °C. After washing, the peptidomelanin concentrate (pellet) was dissolved in 2 mL of 100 mM Tris buffer (pH 8.8) and stored at 4 °C. Peptidomelanin remains stable for months in Tris buffer under these storage conditions. Before use, the melanin concentrate was dialyzed twice against 2 L of deionized water (Milli-Q) through SnakeSkin 3.5 kDa molecular weight cutoff dialysis tubing. Based on the dry weight, the mass of peptidomelanin extracted per plate ranged between 6 and 10 mg (3–5 mg/mL).
Peptidomelanin could be concentrated using a 30 kDa centrifugal concentrator (Amicon Ultra Centrifugal Filters, UFC503008, with regenerated cellulose membranes). Centrifugation at 10,000 rpm (11,200 RCF)/15 min at 24 °C to concentrate peptidomelanin. The concentrate was washed thrice using deionized water.
Polyacrylamide Gel Electrophoresis (PAGE)
Native PAGE, SDS-PAGE and urea-PAGE experiments were performed using the running (resolving) gel, stacking gel, and running buffer compositions provided in Table 2.
Table 2. Composition of Gels Used for Native PAGE, SDS-PAGE and Urea-PAGEa.
| Native PAGE | SDS-PAGE | Urea-PAGE | |
|---|---|---|---|
| Running (resolving) gel | |||
| Deionized water (Milli-Q) | 3.7 mL | 3.7 mL | Make up to 10 mL |
| 30% acrylamide | 3.3 mL | 3.3 mL | 3.3 mL |
| 10% bis-acrylamide | 330 μL | 330 μL | 330 μL |
| Tris-HCl (1.5M, pH 8.8) | 2.5 mL | 2.5 mL | 2.5 mL |
| 10% SDS | 100 μL | ||
| Urea (6 M final conc.) | 3.6 g | ||
| 10% ammonium persulfate | 100 μL | 100 μL | 100 μL |
| TEMED | 6 μL | 6 μL | 6 μL |
| Stacking gel | |||
| Deionized water (Milli-Q) | 2.7 mL | 2.7 mL | Make up to 4 mL |
| 30% acrylamide | 670 μL | 670 μL | 670 μL |
| 10% bis-acrylamide | 67 μL | 67 μL | 67 μL |
| Tris-HCl (1.5 M, pH 6.8) | 500 μL | 500 μL | 500 μL |
| 10% SDS | 40 μL | ||
| Urea (6 M final conc.) | 1.4 g | ||
| 10% ammonium persulfate | 40 μL | 40 μL | 40 μL |
| TEMED | 4 μL | 4 μL | 4 μL |
| Running buffer (10×) | |||
| Tris base | 30.3 g | 30.3 g | 30.3 g |
| Glycine | 144 g | 144 g | 144 g |
| SDS | 10 g | ||
| Urea (6 M final conc.) | 360 g | ||
| Deionized water (Milli-Q) | Make up to 1 L | Make up to 1 L | Make up to 1 L |
SDS refers to sodium dodecyl sulfate. TEMED refers to tetramethylethylenediamine.
All PAGE gels were run at 100 V for 1 h. The HiMedia MBT092 prestained protein ladder was used as a molecular weight marker. PAGE gels were stained using Coomassie brilliant blue. The staining solution possessed the following composition: Coomassie brilliant blue: 50 mg, methanol: 50 mL, glacial acetic acid: 10 mL, deionized water (Milli-Q): 40 mL. PAGE gels were placed in staining solution for 12 h. PAGE gels were destained using destaining solution possessing the following composition: methanol: 50 mL, glacial acetic acid: 10 mL, deionized water (Milli-Q): 40 mL.
Peptidomelanin Acid-Hydrolysis
An aqueous solution of peptidomelanin (500 μL) was mixed with 500 μL of 6 N HCl containing 0.5% phenol. This reaction mixture was subjected to acid-hydrolysis at 80 °C for 24 h under an inert N2 atmosphere. After the incubation period, the reaction mixture was centrifuged at 7500 rpm (6300 RCF) for 15 min at 24 °C to separate the components. The pellet and supernatant (containing hydrolyzed peptides) were separated and stored. Excess HCl was removed from the supernatant via heating at 80 °C for 24 h or until complete removal of the HCl. The solid residue was resuspended in deionized water (Milli-Q) and used for amino acid estimation via LC-MS (refer Text S1, Table S1, and Data set S2 for all data).
Papain Hydrolysis
Papain (Merck, P4762–25 mg) at 0.1 mg/mL was prepared in 100 mM Tris (pH 7.5) (stock-P). BSA at 10 mg/mL was prepared in deionized water (Milli-Q) (stock-B). Peptidomelanin at 4.5 mg/mL was prepared in 100 mM tris (pH 7.5) (stock-M). Three reaction mixtures were also prepared: (1) Stock-P (450 μL) + stock-B (5 μL) + deionized water (45 μL); (2) stock-M (100 μL) + deionized water (900 μL); (3) stock-P (900 μL) + stock-M (100 μL). Reaction mixture 1 served as a positive control. Reaction mixture 2 served as a negative control. Reaction mixtures were incubated at 55 °C in a water bath for 4 h. Papain was deactivated by heating all mixtures at 95 °C for 15 min. All reaction mixtures were centrifuged at 10,000 rpm (11,200 RCF) for 10 min at 4 °C and the supernatants were analyzed using matrix-assisted laser desorption/ionization (MALDI).
Matrix-Assisted Laser Desorption/Ionization (MALDI)
MALDI was performed using the Rapiflex MALDI-TOF Tissuetyper instrument (Bruker Daltonics) at the Liquid Chromatography Mass Spectrometry (LCMS) Facility, Indian Institute of Science (IISc), Bangalore. A matrix was prepared with 50% acetonitrile (ACN) and 0.1% Trifluoroacetic acid (TFA). A mixture containing 1 μL of each sample and matrix (1:1 proportion) was spotted on the target plate. A MTP 384 Polished Plate (ID: 211 1002808) was used as a target plate. Samples were crystallized using the dry droplet method. The instrument was configured to operate in reflector mode with positive polarity, acquiring 34,000 shots for data collection. The calibration was performed using the Peptide Calibration Standard (Bruker Daltonics, 8206195) mono spectrum and reference list, for accurate mass calibration and analysis of peptide signals in each sample. The FlexControl software was used to acquire the data.
Dansylation
Dansylation was peformed using a standard protocol.55 Dansyl chloride was procured from Merck (D2625–1G). A dansyl chloride stock solution containing 2.5 mg/mL dansyl chloride in acetone was prepared. The reaction mixture for dansylating peptidomelanin and acid-hydrolyzed melanin was prepared as follows: 10 μL of a melanin stock (4.5 mg/mL) + 90 μL sodium bicarbonate (0.4 M) + 100 μL Dansyl chloride stock. This reaction mixture was incubated at 37 °C for 3 h in the absence of light. The dansylated reaction mixture was then dialyzed twice against 2 L of deionized water (Milli-Q) through SnakeSkin 3.5 kDa molecular weight cutoff dialysis tubing to remove small molecules.
The reaction mixture for dansylating L-DOPA (Figure 7B) was prepared as follows: 10 mg dansyl chloride in 5 mL acetone +10 mg L-DOPA in 5 mL sodium bicarbonate (0.4 M). This reaction mixture was incubated at 37 °C for 24 h in the absence of light to allow for simultaneous dansylation and autoxidation. A longer incubation period was necessary to allow L-DOPA to fully polymerize into melanin. The contents were concentrated using a 30 kDa centrifugal concentrator (Amicon Ultra Centrifugal Filters, UFC503008, with regenerated cellulose membranes) before loading on an SDS-PAGE gel.
The dansylation reaction efficiency (%) was estimated using eudiometry, as described in the Supporting Information (Text S2, Table S2, Figure S15). We observed that the reaction proceeded to completion when incubated at 37 °C for 3 h in the absence of light (dansylation reaction efficiency (%) = 102 ± 19%). This is expected as dansylation reactions have been reported to proceed to completion for all canonical amino acids55 under similar reaction conditions.
Quinone Off-Target Reaction Assays
One mg L-DOPA was dissolved in 1 mL deionized water (Milli-Q) and incubated at 37 °C for 24 h in the absence of light. The L-DOPA polymer was dialyzed twice against 2 L of deionized water (Milli-Q) through SnakeSkin 3.5 kDa molecular weight cutoff dialysis tubing to remove unreacted monomers. The polymer was then heated at 80 °C to evaporate remaining water. SEM-EDS was performed on the residue to estimate sulfur content.
One mg L-DOPA was mixed with 0.75 mg methionine (1:1 molar ratio) in 1 mL deionized water (Milli-Q) and incubated at 37 °C for 24 h in the absence of light. The L-DOPA polymer was dialyzed twice against 2 L of deionized water (Milli-Q) through SnakeSkin 3.5 kDa molecular weight cutoff dialysis tubing to remove unreacted monomers. The polymer was then heated at 80 °C to evaporate remaining water. SEM-EDS was performed on the residue to estimate sulfur content.
One mg L-DOPA was dissolved in 500 μL of 0.4 M sodium bicarbonate. 1.4 mg of dansyl-Cl was dissolved in 500 μL of acetone. Both solutions were mixed (1:1 molar ratio) and incubated at 37 °C for 24 h in the absence of light. The L-DOPA polymer was dialyzed twice against 2 L of deionized water (Milli-Q) through SnakeSkin 3.5 kDa molecular weight cutoff dialysis tubing to remove unreacted monomers. The polymer was then heated at 80 °C to evaporate remaining water. SEM-EDS was performed on the residue to estimate sulfur content.
The sulfur (% weight) obtained from SEM-EDS was normalized to report molar ratios of dansyl or methionyl groups covalently linked to L-DOPA using a generalized formula. Let substance A (molecular weight = MWA Da) copolymerize with substance B (molecular weight = MWB Da). Let substance A possess SA% sulfur (% weight), as determined by its empirical formula. Let substance B possess no sulfur. Let substances A and B copolymerize into AB. Let AB possess SAB% sulfur, as observed under SEM-EDS. The A:B molar ratio (%) is given in eq 3:
 |
3 |
It should be noted that the molar ratio of dansyl covalently linked to L-DOPA generated using this protocol cannot be used to estimate the efficiency of dansylation. This is because most amide (−NH2) groups on L-DOPA will be cyclized into secondary amines during the autoxidation of L-DOPA into melanin, and will be unavailable for dansylation.
Metal Binding Stoichiometric Assays
100 mM stock solutions of the following metal salts were prepared: NaCl, CrCl3.6H2O, Ni(NO3)2, CuSO4·5H2O, ZnSO4, Cd(NO3)2, HgCl2, Pb(NO3)2, and UO2(CH3COO–)·2H2O.
For all metals: a reaction mixture of 200 μL of fungal peptidomelanin/synthetic melanin (4.5 mg/mL) + 800 μL of metal salt stock was prepared. Incubation was performed at room temperature (24 °C) for 10 min before centrifuging at 10,000 rpm (11,200 RCF) for 10 min at 24 °C. Metal complexes that were soluble (only sodium) were dialyzed twice against 2 L of deionized water (Milli-Q) through SnakeSkin 3.5 kDa molecular weight cutoff dialysis tubing to remove excess salt. Metal complexes that were insoluble (all other metals) were centrifuged at 7500 rpm (6300 RCF) for 10 min at 24 °C and washed with 40 mL of deionized water (Milli-Q) twice. All complexes were then heated at 80 °C for 12 h to evaporate remaining water. SEM-EDS was performed on all residues to estimate metal content.
Activated charcoal (HiMedia Hi-AR) and silica SiO2, extra pure, Loba Chemie Pvt. Ltd. Product code: 0571901000) were used as controls. Activated charcoal was boiled in deionized water (Milli-Q) to suspend it in solution. 200 μL of 4.5 mg/mL suspensions of both substances were incubated with 800 μL of the aforementioned metal salt stock solutions. Incubation was performed at room temperature (24 °C) for 10 min before centrifuging at 10,000 rpm (11,200 RCF) for 10 min at 24 °C. These suspensions were washed twice through a 0.2 μm PVDF syringe filter, using 40 mL of deionized water (Milli-Q). PVDF membranes were then heated at 80 °C for 12 h to evaporate remaining water. SEM-EDS was performed on activated charcoal and silica particles mounted on PVDF membranes to estimate metal content.
Mercury Dose–Response Titration Experiments on Germinating Wheat
Silica (SiO2, extra pure, Loba Chemie Pvt. Ltd. Product code: 0571901000) was used as an inert substrate for mercury dose–response titration experiments on germinating wheat. 1.25 g SiO2 and 5 g H2O were added together to form 6.25 g of substrate per condition. Tricyclazole (0.5 μg/mL) was added to all conditions to prevent fungal growth. Mercury (from HgCl2) concentrations of 0, 0.01, 0.1, 1, 10, 100, 1000, 10,000 ppm were prepared in this substrate as described in Table S5. Ten wheat seeds were planted into every condition and incubated outdoors for 7 days under ambient conditions (Table S4) and were watered daily. Seed mass and shoot lengths were recorded at the end of this time interval.
Mercury:Peptidomelanin Dose–Response Titration Experiments on Germinating Wheat
Silica substrate contaminated with 100 ppm mercury was prepared as described in the previous subsection. Mercury:peptidomelanin stoichiometric equivalent ratios of 0:0, 1:0, 1:0.1, 1:1, and 1:10 were prepared as described in Table S6. Ten wheat seeds were planted into every condition and incubated outdoors for 7 days under ambient conditions (Table S4) and watered daily. Seed mass and shoot lengths were recorded at the end of this time interval. Samples were prepared for ICP-OES at Ramaiah Advanced Testing Lab (Bangalore) as described in Table S7.
Peptidomelanin Reuse Assay
The protocol used for this assay is complicated and is best described diagrammatically in Figure S14.
Acknowledgments
The authors thank the Research and Development Cell (R&D cell), St. Xavier’s College (Mumbai) for providing a seed grant to the Department of Microbiology, St. Xavier’s College (Mumbai) to fund this project. RK acknowledges the financial support received from the Department of Biotechnology, Ministry of Science and Technology, Government of India, under the DBT-Junior Research Fellowship (DBT-JRF) Programme. The authors thank Prof. Siddhartha P. Sarma (Molecular Biophysics Unit, Indian Institute of Science) for the insightful discussions and laboratory space. The authors thank Mr. Buddy White for his invaluable support, which was crucial to starting this project. We would like to thank Mr. Kiran Rambhau Bhotkar (Assistant Manager - Application Support, SEM-EDS), Mrs. Sunita Samgir (Senior Executive - Application support, SEM-EDS), and Mr. Venkatesh Thogiti (Senior Executive- Application support, TEM) from Icon Laboratories Pvt. Ltd., Mumbai, for their excellent work as our electron microscopy technicians. We thank the Liquid Chromatography Mass Spectrometry (LCMS) Facility for performing MALDI experiments. We thank Ranjini S., Adidev Ajithkumar, Dorothy Martin, Gargi Pandey, Srinidhi G. Santhanakrishnan, Divyani Singh, Neha Debbarma, Namrata Pandey, Nyaipriya Devi, and Rishika Pandey for their technical assistance.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.4c03704.
Figure S1: A sampling from the literature of the structural diversity of melanin polymers; Figure S2: Deposition of Aspergillus niger melanoliber into the Microbial Type Culture Collection (Chandigarh, India); Figure S3: Timeline of events during the germination of A. niger melanoliber (MTCC 13366); Figure S4: The effect of sodium hydroxide (NaOH) on the short-term stability of peptidomelanin; Figure S5: Melanin biosynthesis inhibition experiments using tropolone; Figure S6: Particle size estimation of peptidomelanin using transmission electron microscopy (TEM); Figure S7: Peptidomelanin does not contain surface-accessible chitin or nucleic acids; Figure S8: The principle behind the papain hydrolysis experiments; Figure S9: Papain hydrolysis of peptidomelanin reveals the absence of proteins, despite the presence of amino acids in peptidomelanin acid-hydrolysate; Figure S10: Cellulose thread electrophoresis; Figure S11: Quinone intermediates do not react with amino acids during the in vitro autoxidation of L-DOPA to form melanin; Figure S12: Raw data for wheat germination dose–response experiments; Figure S13: Transverse sections of wheat root germinated in silica substrate; Figure S14: Protocol for determining the reuse potential of peptidomelanin after 5 cycles of mercury binding and removal via EDTA chelation; Figure S15: Eudiometric estimation of the dansylation reaction efficiency (%); Text S1: Raw data pertaining to the amino acid composition of peptidomelanin; Text S2: SEM-EDS data normalization and processing for peptidomelanin, dansylated peptidomelanin, acid-hydrolyzed melanin, and dansylated acid-hdyrolyzed melanin; Table S1: LC-MS quantification of the amino acid composition of peptidomelanin; Table S2: Raw data for eudiometric dansylation experiments, performed in order to determine the dansylation reaction efficiency (%); Table S3: Elemental composition of peptidomelanin acquired via SEM-EDS (Icon Laboratories, Mumbai); Table S4: Meteorological data collected at the Kempegowda station (Bangalore, India) during the months of November–December, 2023; Table S5: Mercury dose–response titration experiments on germinating wheat: experimental setup; Table S6: Mercury dose–response titration experiments on germinating wheat: experimental setup; Table S7: ICP-OES Hg quantification in germinating wheat: experimental setup; Data set S1: SEM-EDS raw data for the elemental composition of peptidomelanin; Data set S2: LC-MS reports for the amino acid composition of peptidomelanin; Data set S3: SEM-EDS raw data for the sulfur estimation (% weight) of peptidomelanin, dansylated peptidomelanin, acid-hydrolyzed melanin, and dansylated acid-hydrolyzed melanin. Note that the SEM-EDS raw data for peptidomelanin is identical in Data sets S1 and S3; Data set S4: SEM-EDS raw data for experiments to confirm that quinone intermediates do not react with amino acids during the in vitro polymerization of L-DOPA melanin. Data set S5: SEM-EDS raw data for metal-chelating stoichiometric assays; Assays were performed using 4 substances: silica, activated charcoal, synthetic L-DOPA melanin, and peptidomelanin. Assays were performed using 9 metals: sodium, chromium, nickel, copper, zinc, cadmium, mercury, lead, and uranium (as uranyl); Data set S6: ICP-OES report for mercury concentration in wheat seeds and seedlings, provided by Ramaiah Advanced Testing Lab (Bangalore); Data set S7: SEM-EDS raw data for the peptidomelanin reuse experiment, for 5 peptidomelanin reuse cycles (ZIP)
Accession Codes
GenBank accession ID PP077302: ITS sequence for A. niger melanoliber (MTCC 13366).
Author Contributions
¶ S.B., S.S., B.M.B. contributed equally to this work. R.K., A.S., and D.N. conceived the experiment(s), R.K., S.B., S.S., B.B., D.S., L.R., P.P., M.K.Y., S.N., A.S., H.B., E.K., and D.N. conducted the experiment(s), R.K., R.G., and D.N. analyzed the results. All authors reviewed the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Revskaya E.; et al. Compton scattering by internal shields based on melanin-containing mushrooms provides protection of gastrointestinal tract from ionizing radiation. Cancer Biotherapy and Radiopharmaceuticals 2012, 27, 570–576. 10.1089/cbr.2012.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Płonka P.; Grabacka M. Melanin synthesis in microorganisms: biotechnological and medical aspects. Acta Biochim. Pol. 2019, 53, 429. 10.18388/abp.2006_3314. [DOI] [PubMed] [Google Scholar]
- Jalmi P.; Bodke P.; Wahidullah S.; Raghukumar S. The fungus Gliocephalotrichum simplex as a source of abundant, extracellular melanin for biotechnological applications. World J. Microbiol. Biotechnol. 2012, 28, 505–512. 10.1007/s11274-011-0841-0. [DOI] [PubMed] [Google Scholar]
- Andersen N.; et al. Structure, chemistry, and biosynthesis of the melanins. Fortschr. Chem. Org. Naturst. 1974, 521–582. [DOI] [PubMed] [Google Scholar]
- Zajac G.; et al. The fundamental unit of synthetic melanin: a verification by tunneling microscopy of X-ray scattering results. Biochimica et Biophysica Acta (BBA)-General Subjects 1994, 1199, 271–278. 10.1016/0304-4165(94)90006-X. [DOI] [PubMed] [Google Scholar]
- Tran-Ly A. N.; Reyes C.; Schwarze F. W.; Ribera J. Microbial production of melanin and its various applications. World J. Microbiol. Biotechnol. 2020, 36, 1–9. 10.1007/s11274-020-02941-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aamir M.; Singh V. K.; Dubey M. K.; Meena M.; Kashyap S. P.; Katari S. K.; Upadhyay R. S.; Umamaheswari A.; Singh S.; et al. In silico prediction, characterization, molecular docking, and dynamic studies on fungal SDRs as novel targets for searching potential fungicides against Fusarium wilt in tomato. Front. Pharmacol. 2018, 9, 1038. 10.3389/fphar.2018.01038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero R. J.; Casadevall A. Melanin. Current biology 2020, 30, R142–R143. 10.1016/j.cub.2019.12.042. [DOI] [PubMed] [Google Scholar]
- Brenner M.; Hearing V. J. The protective role of melanin against UV damage in human skin. Photochemistry and photobiology 2008, 84, 539–549. 10.1111/j.1751-1097.2007.00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadachova E.; Casadevall A. Ionizing radiation: how fungi cope, adapt, and exploit with the help of melanin. Curr. Opin. Microbiol. 2008, 11, 525–531. 10.1016/j.mib.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalida A.; Negro J. J.; Galván I. Melanin-based color variation in the bearded vulture suggests a thermoregulatory function. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology 2008, 149, 87–91. 10.1016/j.cbpa.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Nosanchuk J. D.; Casadevall A. Impact of melanin on microbial virulence and clinical resistance to antimicrobial compounds. Antimicrob. Agents Chemother. 2006, 50, 3519–3528. 10.1128/AAC.00545-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosanchuk J. D.; Casadevall A. The contribution of melanin to microbial pathogenesis. Cellular microbiology 2003, 5, 203–223. 10.1046/j.1462-5814.2003.00268.x. [DOI] [PubMed] [Google Scholar]
- Riley P. A. Melanin. The international journal of biochemistry & cell biology 1997, 29, 1235–1239. 10.1016/S1357-2725(97)00013-7. [DOI] [PubMed] [Google Scholar]
- Almeida-Paes R.; Nosanchuk J. D.; Zancope-Oliveira R. M.. Fungal melanins: biosynthesis and biological functions. In Melanin: Biosynthesis, Functions, and Health Effects; Nova Science Publishers: New York, 2012; pp 77–107. [Google Scholar]
- Aghajanyan A. E.; Hambardzumyan A. A.; Hovsepyan A. S.; Asaturian R. A.; Vardanyan A. A.; Saghiyan A. A. Isolation, purification and physicochemical characterization of water-soluble Bacillus thuringiensis melanin. Pigment cell research 2005, 18, 130–135. 10.1111/j.1600-0749.2005.00211.x. [DOI] [PubMed] [Google Scholar]
- Rowley B.; Pirt S. Melanin production by Aspergillus nidulans in batch and chemostat cultures. Microbiology 1972, 72, 553–563. 10.1099/00221287-72-3-553. [DOI] [PubMed] [Google Scholar]
- Wibowo J. T.; et al. Characterization of an insoluble and soluble form of melanin produced by Streptomyces cavourensis SV 21, a sea cucumber associated bacterium. Marine Drugs 2022, 20, 54. 10.3390/md20010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhusudhan D.; Mazhari B. B. Z.; Dastager S. G.; Agsar D. Production and cytotoxicity of extracellular insoluble and droplets of soluble melanin by Streptomyces lusitanus dmz-3. BioMed. Res. Int. 2014, 2014, 1. 10.1155/2014/306895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; et al. Differences between water-soluble and water-insoluble melanin derived from Inonotus hispidus mushroom. Food Chemistry: X 2022, 16, 100498. 10.1016/j.fochx.2022.100498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X.; et al. Preparation of water-soluble melanin from squid ink using ultrasound-assisted degradation and its anti-oxidant activity. Journal of Food Science and Technology 2014, 51, 3680–3690. 10.1007/s13197-013-0937-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghajanyan A. E.; et al. Development of the technology for producing water-soluble melanin from waste of vinary production and the study of its physicochemical properties. European Food Research and Technology 2022, 248, 485–495. 10.1007/s00217-021-03894-9. [DOI] [Google Scholar]
- Wold C. W.; Gerwick W. H.; Wangensteen H.; Inngjerdingen K. T. Bioactive triterpenoids and water-soluble melanin from Inonotus obliquus (Chaga) with immunomodulatory activity. Journal of functional foods 2020, 71, 104025. 10.1016/j.jff.2020.104025. [DOI] [Google Scholar]
- Bronze-Uhle E. S.; Piacenti-Silva M.; Paulin J. V.; Battocchio C.; Graeff C. F. d. O.. Synthesis of water-soluble melanin. arXiv, August 29, 2015. 10.48550/arXiv.1508.07457. [DOI]
- Kocaman A. Combined interactions of amino acids and organic acids in heavy metal binding in plants. Plant Signal. Behav. 2023, 18, 2064072. 10.1080/15592324.2022.2064072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R.; Rai J. Phytochelatins: peptides involved in heavy metal detoxification. Applied biochemistry and biotechnology 2010, 160, 945–963. 10.1007/s12010-009-8565-4. [DOI] [PubMed] [Google Scholar]
- Liu L.; Li W.; Song W.; Guo M. Remediation techniques for heavy metal-contaminated soils: Principles and applicability. Science of the total environment 2018, 633, 206–219. 10.1016/j.scitotenv.2018.03.161. [DOI] [PubMed] [Google Scholar]
- Lo I. M.; Yang X. EDTA extraction of heavy metals from different soil fractions and synthetic soils. Water, Air, and Soil Pollution 1999, 109, 219–236. 10.1023/A:1005000520321. [DOI] [Google Scholar]
- Naghipour D.; Gharibi H.; Taghavi K.; Jaafari J. Influence of EDTA and NTA on heavy metal extraction from sandy-loam contaminated soils. Journal of Environmental Chemical Engineering 2016, 4, 3512–3518. 10.1016/j.jece.2016.07.034. [DOI] [Google Scholar]
- Derakhshan Nejad Z.; Jung M. C.; Kim K.-H. Remediation of soils contaminated with heavy metals with an emphasis on immobilization technology. Environmental geochemistry and health 2018, 40, 927–953. 10.1007/s10653-017-9964-z. [DOI] [PubMed] [Google Scholar]
- Mbonyiryivuze A.; Nuru Z. Y.; Diop Ngom B.; Mwakikunga B.; Mokhotjwa Dhlamini S.; Park E.; Maaza M. Morphological and chemical composition characterization of commercial sepia melanin. Am. J. Nanomater. 2015, 3 (1), 22–27. 10.12691/ajn-3-1-3. [DOI] [Google Scholar]
- Singla S.; et al. Isolation and characterization of allomelanin from pathogenic black knot fungus- a sustainable source of melanin. ACS omega 2021, 6, 35514–35522. 10.1021/acsomega.1c05030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiran G. S.; Jackson S. A.; Priyadharsini S.; Dobson A. D.; Selvin J. Synthesis of Nm-PHB (nanomelanin-polyhydroxy butyrate) nanocomposite film and its protective effect against biofilm-forming multi drug resistant Staphylococcus aureus. Sci. Rep. 2017, 7, 9167. 10.1038/s41598-017-08816-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Cai J.; Deng Y.; Chen Y.; Ren G. Characterization of melanin produced by a wild-type strain of Bacillus cereus. Frontiers of Biology in China 2007, 2, 26–29. 10.1007/s11515-007-0004-8. [DOI] [Google Scholar]
- Panzella L.; Napolitano A. Natural and bioinspired phenolic compounds as tyrosinase inhibitors for the treatment of skin hyperpigmentation: Recent advances. Cosmetics 2019, 6, 57. 10.3390/cosmetics6040057. [DOI] [Google Scholar]
- Passi S.; Nazzaro-Porro M. Molecular basis of substrate and inhibitory specificity of tyrosinase: phenolic compounds. British Journal of Dermatology 1981, 104, 659–665. 10.1111/j.1365-2133.1981.tb00752.x. [DOI] [PubMed] [Google Scholar]
- Jackson H.; Kendal L. The oxidation of catechol and homocatechol by tyrosinase in the presence of amino-acids. Biochem. J. 1949, 44, 477. 10.1042/bj0440477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenman H. C.; Greer E. M.; McGrail C. W. The role of melanins in melanotic fungi for pathogenesis and environmental survival. Applied microbiology and biotechnology 2020, 104, 4247–4257. 10.1007/s00253-020-10532-z. [DOI] [PubMed] [Google Scholar]
- Pal A. K.; Gajjar D. U.; Vasavada A. R. DOPA and DHN pathway orchestrate melanin synthesis in Aspergillus species. Med. Mycol. 2014, 52, 10–18. 10.3109/13693786.2013.826879. [DOI] [PubMed] [Google Scholar]
- Sansinenea E.; Ortiz A. Melanin: a photoprotection for Bacillus thuringiensis based biopesticides. Biotechnology letters 2015, 37, 483–490. 10.1007/s10529-014-1726-8. [DOI] [PubMed] [Google Scholar]
- Fenoll L.; et al. Tyrosinase kinetics: discrimination between two models to explain the oxidation mechanism of monophenol and diphenol substrates. International Journal of Biochemistry & Cell Biology 2004, 36, 235–246. 10.1016/S1357-2725(03)00234-6. [DOI] [PubMed] [Google Scholar]
- Oh J.-J.; Kim J. Y.; Kim Y. J.; Kim S.; Kim G.-H. Utilization of extracellular fungal melanin as an eco-friendly biosorbent for treatment of metal-contaminated effluents. Chemosphere 2021, 272, 129884. 10.1016/j.chemosphere.2021.129884. [DOI] [PubMed] [Google Scholar]
- Thaira H.; Raval K.; Manirethan V.; Balakrishnan R. M. Melanin nano-pigments for heavy metal remediation from water. Sep. Sci. Technol. 2019, 54, 265–274. 10.1080/01496395.2018.1443132. [DOI] [Google Scholar]
- Manirethan V.; Raval K.; Rajan R.; Thaira H.; Balakrishnan R. M. Kinetic and thermodynamic studies on the adsorption of heavy metals from aqueous solution by melanin nanopigment obtained from marine source: Pseudomonas stutzeri. Journal of environmental management 2018, 214, 315–324. 10.1016/j.jenvman.2018.02.084. [DOI] [PubMed] [Google Scholar]
- Gaballa A.; et al. Characterization of the roles of activated charcoal and Chelex in the induction of PrfA regulon expression in complex medium. PLoS One 2021, 16, e0250989. 10.1371/journal.pone.0250989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadeer R.; Khalid N. Removal of cadmium from aqueous solutions by activated charcoal. Separation science and Technology 2005, 40, 845–859. 10.1081/SS-200028819. [DOI] [Google Scholar]
- Motojima K.; Tachikawa E.; Kamiyama H.; Imahashi T. Removal of radiocobalt in waste water by activated charcoal using oxine as a chelating agent. Annals of Nuclear Energy 1978, 5, 5–12. 10.1016/0306-4549(78)90055-5. [DOI] [Google Scholar]
- Dawar K.; et al. Evaluating the Efficacy of Activated Carbon in Minimizing the Risk of Heavy Metals Contamination in Spinach for Safe Consumption. ACS omega 2023, 8, 24323–24331. 10.1021/acsomega.3c01573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown I. What factors determine cation coordination numbers?. Acta Crystallographica Section B: Structural Science 1988, 44, 545–553. 10.1107/S0108768188007712. [DOI] [Google Scholar]
- Qu R.; Han G.; Liu M.; Li X. The mercury behavior and contamination in soil profiles in Mun River Basin, Northeast Thailand. International journal of environmental research and public health 2019, 16, 4131. 10.3390/ijerph16214131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; et al. Status of mercury accumulation in agricultural soils across China (1976–2016). Ecotoxicology and Environmental Safety 2020, 197, 110564. 10.1016/j.ecoenv.2020.110564. [DOI] [PubMed] [Google Scholar]
- Biundo A.; Braunschmid V.; Pretzler M.; Kampatsikas I.; Darnhofer B.; Birner-Gruenberger R.; Rompel A.; Ribitsch D.; Guebitz G. M. Polyphenol oxidases exhibit promiscuous proteolytic activity. Commun. Chem. 2020, 3, 62. 10.1038/s42004-020-0305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J.; et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostert A. B. Melanin, the what, the why and the how: An introductory review for materials scientists interested in flexible and versatile polymers. Polymers 2021, 13, 1670. 10.3390/polym13101670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapuhi Y.; Schmidt D. E.; Lindner W.; Karger B. L. Dansylation of amino acids for high-performance liquid chromatography analysis. Analytical biochemistry 1981, 115, 123–129. 10.1016/0003-2697(81)90534-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.