Abstract
Understanding the role of chemokine receptors in health and disease has been of increasing interest in recent years. Chemokine receptor CXCR4 has been extensively studied because of its defined role in immune cell trafficking, HIV infection, inflammatory diseases, and cancer progression. We have developed high affinity rigidified CXCR4 antagonists that incorporate metal ions to optimize the binding interactions with the aspartate side chains at the extracellular surface of the CXCR4 chemokine receptor and increase the residence time. Cross- and side-bridged tetraazamacrocylic complexes offer significant advantages over the non-bridged molecular structures in terms of receptor affinity, potential for radiolabelling, and use in therapeutic applications. Our investigation has been extended to the influence of the ring size on bridged tetraazamacrocyclic compounds with the addition of two novel chelators (bis-cross-bridged homocyclen and bis-cross-bridged cyclen) to compare to the bis-bridged cyclam, along with novel metal complexes formed with copper(II) or zinc(II). The in vitro biological assays showed that all of the zinc(II) complexes are high affinity antagonists with a marked increase in CXCR4 selectivity for the bis-cross-bridged cyclen complex, whereas the properties of the copper(II) complexes are highly dependent on metal ion geometry. X-ray crystal structural data and DFT computational studies allow for the rationalisation of the relative affinities and the aspartate residue interactions on the protein surface. Changing the ring size from 14-membered can increase the selectivity for the CXCR4 receptor whilst retaining potent inhibitory activity, improving the key pharmacological characteristics.
Keywords: CXCR4, AMD3100, Plerixafor, cyclam, macrocycle, cancer therapeutics, molecular imaging
1. Introduction
Chemokine receptors, belonging to the family of G protein-coupled receptors (GPCRs), are involved in a wide variety of diseases including cancer and HIV/AIDS as well as inflammatory disorders such as multiple sclerosis, rheumatoid arthritis, and inflammatory bowel disease [1,2,3,4,5]. Their endogenous ligands, chemokines, are small, water-soluble proteins (8–12 kDa) that bind to their cognate receptor and thereby elicit a cellular response. In general, chemokines and chemokine receptors are promiscuous in their interactions. However, chemokine receptor CXCR4 has a sole natural ligand, CXCL12, and, in cancer, this interaction can facilitate metastasis to secondary sites, promote tumour growth, and therapy resistance [6,7,8,9]. CXCR4 has been reported to be overexpressed in more than 23 different human cancers including non-small cell lung, breast, ovarian, prostate, pancreatic, and colorectal cancer [7,10,11,12,13,14,15].
Multiple SAR studies have been carried out to produce selective small molecule chemokine receptor antagonists [16]. Research has also indicated that cancer progression can be mediated by such small molecule receptor antagonists [9,17]. Furthermore, the expression level of CXCR4 has been linked to cancer progression and resistance to radio-/chemotherapy, indicating that it may also be a valuable prognostic biomarker for use in the molecular imaging of cancer growth, metastasis, and relapse [9,14,18,19,20]. Radiolabelled CXCR4 binding ligands have recently been developed for targeted radionuclide imaging and radioisotope therapy [21,22,23].
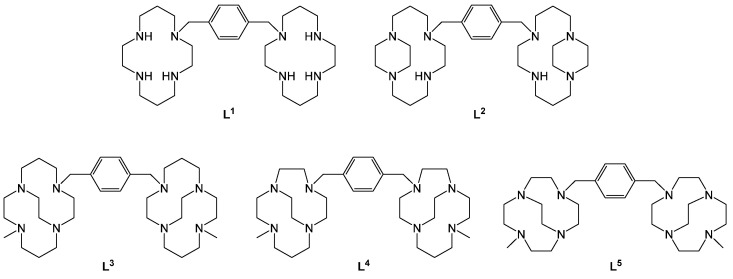
It has previously been shown that saturated tetraazamacrocyclic structures bind to CXCR4 with a remarkably high affinity. AMD3100 (Plerixafor™, L1 in Figure 1) was initially discovered as a highly specific CXCR4 inhibitor for blocking the HIV cell entry process and was eventually licensed for clinical use in 2008 as a hematopoietic stem cell mobilising agent in patients with multiple myeloma or lymphoma [24]. The high affinity binding interaction of azamacrocyclic antagonists, which are protonated at physiological pH, is achieved via multiple H-bonds and electrostatic interactions with amino acid residues on the accessible extracellular surface of the CXCR4 receptor [25]. Site-directed mutagenesis studies have shown that bis-azamacrocyclic compounds have particularly strong interactions with Asp171 in transmembrane loop IV and Asp262 in transmembrane loop VI. H-bonding interactions with Glu288 and Tyr45 as well as π-interactions with Trp94 and Tyr116 have also been identified [26,27,28].
Figure 1.
Chemical structures of ligands described in this work.
Macrocyclic chelators such as the 14-membered cyclam ring in AMD3100 can form stable complexes with transition metal ions, which are a good size match with the chelator cavity size. It is proposed that these metal complexes interact with the carboxylate amino acid side chains on the surface of CXCR4 via coordination interactions [29]. The different electronic arrangements of the metal ion result in a range of preferred coordination numbers and optimised geometries. For first row transition metal complexes of cyclam, a flat configuration with all four nitrogens in the same plane and the six-membered chelate rings occupying the chair conformation is most commonly observed, however, it has been proposed that a folded configuration is optimal for binding to the CXCR4 chemokine receptor [30,31,32].
Configurational fixing has been identified as a mechanism to strengthen coordinate bonds with carboxylate functional groups present on the extracellular surface of CXCR4. We used this approach to optimize the interactions of the metal complexes of macrocyclic chelators with CXCR4. This was achieved by adding ethylene bridges between adjacent (side-bridge) or non-adjacent (cross-bridge) nitrogen atoms in the cyclam rings such as L2 and L3, respectively [33,34,35,36,37]. Furthermore, we demonstrated that there is a significant increase in both potency and residence time at the binding site on the receptor’s extracellular surface [34,38].
In the present study, we expanded the library of configurationally restricted bis-tetraazamacrocylic CXCR4 antagonists and studied their zinc(II) and copper(II) complexes to probe the effect of ring size on the receptor binding characteristics including affinity and selectivity [39,40]. This study will facilitate robust compound selection of a lead drug candidate for translation to in vivo studies, based on the understanding of the coordination interactions to rationalize the results of the in vitro biological assays.
2. Materials and Methods
1,4,7,10-Tetraazacyclododecane (cyclen) and 1,4,7,10-tetraazatridecane (homocyclen) were purchased from CheMatech (France). All other reagents were purchased from Sigma Aldrich (USA). Elemental analyses were performed by Quantitative Technologies Inc. Electrospray mass spectra were collected on a Shimadzu LCMS 2020 electrospray mass spectrometer. NMR spectra were obtained on a Varian Bruker AVANCE II 300 MHz NMR spectrometer. Single crystal X-ray diffraction data were collected using an imaging plate diffractometer operating with Mo radiation. Routine data collection and processing procedures were adopted. Several compounds used in this study were synthesised using the following procedures in the literature: L1 [41], CuL1 and ZnL2 [42], L2 and CuL2 [33], ZnL2 [43], L3 and CuL3 [34].
2.1. Synthesis of Ligands L1–L5
Ligands L1, L2, and L3 were synthesised using the following procedures in the literature: L1 [41], L2 [33], and L3 [34]. Synthesis of homocyclen-glyoxal (1) and cyclen-glyoxal (2) was carried out as previously reported by Weisman et al. [44]. Synthesis of p-linked cyclen-glyoxal (4) was carried out as previously reported by Le Baccon et al. [45].
2.1.1. Synthesis of p-Linked Homocyclen-Glyoxal (3)
Homocyclen-glyoxal (1) (5.120 g, 0.025 mol) was dissolved in anhydrous acetonitrile (50 mL), then α,α’-dibromo-p-xylene (2.998 g, 0.012 mol) was added and stirred for 24 h. The pale-yellow solid was filtered off and rinsed with acetonitrile, then diethyl ether before drying under vacuum. Yield: 7.406 g (88%). 13C [1H] NMR (75 MHz, D2O): δ 17.65 (N-β-CH2), 42.00 (N-α-CH2), 45.93 (N-α-CH2), 46.40 (N-α-CH2), 48.03 (N-α-CH2), 50.09 (N-α-CH2), 52.19 (N-α-CH2), 53.83 (N-α-CH2), 58.85 (N-α-CH2), 59.60 (N-α-CH2), 69.68 (Caminal), 81.58 (Caminal), 128.63 (Caromatic), 132.58 (Caromatic). Elemental analysis: calculated (%) for C30H48N8Br2•2H2O: C, 50.28; H, 7.31; N, 15.64. Found: C, 49.93; H, 7.34; N, 15.34. HRMS (ESI): calculated for [C30H48N8Br]+, 599.3180; found 599.3162.
2.1.2. Synthesis of 5
p-Xylene linked glyoxal-bridged homocyclen (3) was added to anhydrous acetonitrile (500 mL) with stirring. Iodomethane (162 g, 1.1 mol) was added to the suspension in one portion and the mixture was stirred for 5 days at room temperature under nitrogen. A second portion of iodomethane (0.83 g, 5.8 mmol) was added to the mixture, and then the mixture was stirred at room temperature for a further 5 days. At reaction completion, excess iodomethane was removed by bubbling nitrogen through the suspension for 30 min. The yellow solid was filtered off, washed with diethyl ether (5 × 20 mL), and dried in vacuo to obtain the methylated intermediate (5) as an orange solid (8.7 g, 74%). 13C[1H] NMR (75 MHz, D2O): δ 18.90 (N-β-CH2), 42.16 (N-α-CH2), 45.35 (N-α-CH2), 46.46 (N-α-CH2), 48.25 (N-α-CH3), 50.49 (N-α-CH2), 51.57 (N-α-CH2), 52.33 (N-α-CH2), 59.57 (N-α-CH2), 60.41 (N-α-CH2), 65.16 (N-α-CH2), 76.47 (Caminal), 78.40 (Caminal), 129.00 (Caromatic), 133.58 (Caromatic). Elemental analysis: calculated (%) for C32H54N8I4•0.5C4H10O•2H2O: C, 36.09; H, 5.61; N, 9.90. Found: C, 35.93; H, 5.31; N, 9.74.
2.1.3. Synthesis of 6
p-Xylene linked glyoxal-bridged cyclen (4) was added to anhydrous acetonitrile (300 mL) with stirring. Iodomethane (5 g, 7.7 mmol) was added to the suspension in one portion, and then the mixture was stirred for 14 days at room temperature under nitrogen. At the end of the reaction, excess iodomethane was removed by bubbling nitrogen through the suspension for 30 min. The yellow solid was filtered off, washed with diethyl ether (5 × 20 mL), and dried in vacuo to obtain the methylated intermediate (6) as a white solid (7.06 g, 89%). 13C[1H] NMR (75 MHz, D2O): δ 41.77 (N-α-CH2), 42.06 (N-α-CH2), 45.17 (N-α-CH2), 45.46 (N-α-CH2), 45.74 (N-α-CH2), 53.97 (N-α-CH3), 58.17 (N-α-CH2), 59.27 (N-α-CH2), 60.30 (N-α-CH2), 64.04 (N-α-CH2), 77.03 (Caminal), 77.28 (Caminal), 128.37 (Caromatic), 132.80(Caromatic). Elemental analysis: calculated (%) for C30H50N8I3Br: C, 36.64; H, 5.12; N, 11.39. Found: C, 36.30; H, 4.93; N, 11.16. MS (ESI): m/z = 775 [M-2I]+; m/z = 647 [M-3I]+.
2.1.4. Synthesis of L4
The methylated intermediate (5) (8.7 g, 8.2 mmol) was dissolved in ethanol (300 mL), and sodium borohydride (18.7 g, 0.49 mol) was slowly added to the suspension as a solid. The clear solution was stirred at room temperature for 14 days and concentrated in vacuo. Water (200 mL) was added, the pH was adjusted to 14 (KOH pellets), and the basic solution was extracted with benzene (5 × 200 mL). The combined organic fractions were dried over MgSO4, filtered, and evaporated in vacuo to yield L4 as a yellow oil (3.82 g, 84%). 13C[1H] NMR (75 MHz, D2O): δ 26.54 (N-β-CH2), 43.53 (N-α-CH3), 52.24 (N-α-CH2), 52.69 (N-α-CH2), 53.74 (N-α-CH2), 54.61 (N-α-CH2), 56.02 (N-α-CH2), 56.14 (N-α-CH2), 56.40 (N-α-CH2), 57.23 (N-α-CH2, broad; 2 unresolved peaks), 58.27 (N-α-CH2), 60.44 (N-α-CH2), 128.61 (Caromatic), 138.82 (Caromatic). Elemental analysis: calculated (%) for C32H58N8•0.67C6H6•1.67H2O: C, 67.88; H, 10.34; N, 17.59. Found: C, 67.70; H, 9.95; N, 17.30. MS (ESI): m/z = 555 [M]+.
2.1.5. Synthesis of L5
The methylated intermediate (6) (7 g, 6.8 mmol) was dissolved in ethanol (300 mL), and sodium borohydride (7.7 g, 0.2 mol) was slowly added to the suspension as a solid. The clear solution was stirred at room temperature for 5 days and concentrated in vacuo. Water (200 mL) was added, the pH was adjusted to 14 (KOH pellets), and the basic solution was extracted with benzene (5 × 200 mL). The combined organic fractions were dried over MgSO4, filtered, and evaporated in vacuo to yield L5 as a yellow oil (2.86 g, 80%). 13C[1H] NMR (75 MHz, D2O): δ 42.84 (N-α-CH3), 55.60 (N-α-CH2), 56.00 (N-α-CH2), 56.58 (N-α-CH2), 57.38 (N-α-CH2), 59.12 (N-α-CH2), 60.10 (N-α-CH2), 127.81 (Caromatic), 138.09 (Caromatic). MS (ESI): m/z = 527 [M+H]+.
2.1.6. Preparation of HCl Salts of L4 and L5
The ligand HCl salt was made by bubbling HCl gas through an ethanol solution, filtering the yellow solid that formed, and washing with ethanol and ether before drying under vacuum. Elemental analysis (L4): calculated (%) for C32H58N8•7HCl•6H2O: C, 41.86; H, 8.45; N, 12.20. Found: C, 42.16; H, 8.56; N, 12.07. Elemental analysis (L5): calculated (%) for C30H54N8•7HCl•5H2O: C, 41.32; H, 8.21; N, 12.85. Found: C, 41.42; H, 8.36; N, 12.58.
2.2. Synthesis of Cu(II) and Zn(II) Complexes of L1–L5
Metal complexes of L1, L2, and L3 were synthesised using the following procedures in the literature: CuL1 and ZnL1 [42], CuL2 [33], ZnL2 [43], and CuL3 [34].
2.2.1. Synthesis of Zn2L3
In an inert atmosphere glove box, L3 (0.583 g, 1 mmol) and anhydrous zinc(II) acetate (0.367 g, 2 mmol) were dissolved in methanol (20 mL) and stirred overnight. The metal complex solution was removed from the glovebox. NH4PF6 (5 equiv.) was dissolved in methanol (5 mL), filtered, and added to the zinc complex. The mixture was stored in a freezer overnight to facilitate precipitation. The white precipitate was then collected on a glass frit by filtration, washed with methanol then diethyl ether, and dried under vacuum, yielding Zn2L3 (0.961 g, 93%). Elemental analysis: calculated (%) for [C34H58N8Zn2](CH3COO)2(PF6)2•0.7NH4PF6: C, 36.93; H, 5.77; N, 9.86. Found: C, 36.77; H, 5.98; N, 9.93. MS (ESI): m/z = 416 [Zn2L3(CH3COO)2]2+.
2.2.2. Synthesis of Cu2L4
In an inert atmosphere glove box, L4 (0.555 g, 1 mmol) and anhydrous copper(II) acetate (0.363 g, 2 mmol) were dissolved in methanol (30 mL) and stirred for 1 day. The resulting blue solution was removed from the glovebox and filtered to remove trace solids. NH4PF6 (5 equiv.) was dissolved in methanol (10 mL) and added to the copper complex. Precipitation occurred immediately and was completed by standing in a freezer overnight. The blue powder was filtered and washed with methanol and diethyl ether, yielding Cu2L4 (0.360 g, 30%). Elemental analysis: calculated (%) for [C32H58N8Cu2](CH3COO)1.5(PF6)2.5•1.5CH3OH: C, 37.12; H, 5.85; N, 9.49. Found: C, 37.38; H, 5.85; N, 9.18. MS (ESI): m/z = 770 [HCuL4(CH3COO)2(CH3OH)]+, m/z = 400 [Cu2L4(CH3COO)2]2+.
2.2.3. Synthesis of Zn2L4
In an inert atmosphere glove box, L4 (0.555 g, 1 mmol) and anhydrous zinc(II) acetate (0.367 g, 2 mmol) were dissolved in methanol (30 mL) and stirred for 1 day. The resulting yellow solution was removed from the glovebox and filtered to remove trace solids. NH4PF6 (5 equiv.) was dissolved in methanol (10 mL) and added to the zinc complex. Precipitation occurred immediately and was completed by standing in a freezer overnight. The powder was filtered, yielding Zn2L4 (0.443 g, 41%). Elemental analysis: calculated (%) for [C30H58N8Zn2](CH3COO)2.3(PF6)1.7•1H2O: C, 40.48; H, 6.21; N, 10.32. Found: C, 40.15; H, 6.54; N, 10.62. MS (ESI): m/z = 402 [Zn2L4(CH3COO)2]2+.
2.2.4. Synthesis of Cu2L5
In an inert atmosphere glove box, L5 (0.527 g, 1 mmol) and anhydrous copper(II) acetate (0.363 g, 2 mmol) were dissolved in acetonitrile (30 mL) and stirred for 1 day. The solvent was evaporated, yielding a blue oil. The blue residue was dissolved in methanol (10 mL). NH4PF6 (5 equiv.) was dissolved in methanol (10 mL) and added to the copper complex. Precipitation occurred immediately and was completed by standing in a freezer overnight. The blue powder was filtered and washed with methanol and diethyl ether, yielding Cu2L5 (1.06 g, 89%). Elemental analysis: calculated (%) for [C30H54N8Cu2](CH3COO)2(PF6)2•0.8NH4PF6: C, 34.25; H, 5.34; N, 10.34. Found: C, 34.25; H, 5.18; N 9.95. MS (ESI): m/z = 386 [Cu2L5(CH3COO)2]2+.
2.2.5. Synthesis of Zn2L5
In an inert atmosphere glove box, L5 (0.527 g, 1 mmol) and anhydrous zinc(II) acetate (0.367 g, 2 mmol) were dissolved in acetonitrile (30 mL) and stirred for 1 day. The solvent was evaporated, yielding a yellow oil. The residue was dissolved in methanol (10 mL). NH4PF6 (5 equiv.) was dissolved in methanol (10 mL) and added to the zinc complex. Precipitation occurred immediately and was completed by standing in a freezer overnight. The powder was filtered and washed with methanol and diethyl ether, yielding Zn2L5 (1.06 g, 98%). Elemental analysis: calculated (%) for [C30H54N8Zn2](CH3COO)2(PF6)2•1H2O: C, 37.68; H, 5.77; N, 10.34. Found: C, 37.79; H, 5.77; N, 10.20. MS (ESI): m/z = 388 [Zn2L5(CH3COO)2]2+.
2.3. Biological Evaluation
2.3.1. General
Buffy coat preparations from healthy donors were obtained from the Blood Bank in Leuven, Belgium. For the chemokine binding studies, peripheral blood mononuclear cells (PBMCs) activated with phytohemagglutinin (PHA; Sigma, Belgium) were used. Human T-lymphocytic SUP-T1 cells and TZM-bl cells were obtained from the American Type Culture Collection (Rockville, MD) and maintained in RPMI-1640 supplemented with 10% foetal bovine serum (FBS). Human glioblastoma U87.MG and U87.CD4 cells were kindly provided by Dr. D. Littman [46,47] and used to generate cells expressing CXCR4 or CCR5 [48]. The fluorescent chemokines CXCL12AF647 and CCL5AF647 were purchased from Almac. The CXCR4 inhibitor AMD3100 and the CCR5 inhibitor maraviroc were a kind gift of Dr. Gary Bridger, affiliated at that time with AnorMED (Langley, Canada). HIV-1 NL4-3 (CXCR4-using, X4) and HIV-1 BaL (CCR5-using, R5) were obtained through the AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH).
2.3.2. HIV Infection Assay
TZM-bl cells [49] were seeded in transparent 96-well plates at 1 × 104 cells per well in DMEM (Dulbecco’s modified Eagle medium; Life Technologies, Waltham, MA, USA) with 10% FBS and 10 mM HEPES. Subsequently, compounds were added, and the cell/sample mixture was incubated at 37 °C. After 30 min, the virus was added at 100 pg p-24 per well. After 48 h of incubation, the assay plates were analysed. For the analysis, steadylite plus substrate solution (PerkinElmer, Waltham, MA, USA) was added to the assay plates. The luminescent signal of the lysed cell suspension was analysed in white 96-well plates on a Spectra-Max L luminescence microplate reader (Molecular Devices, Sunnyvale, CA, USA) after a 10 min incubation period in the dark. Luciferase activity induced by HIV-1 Tat protein expression was measured as an assessment of the amount of HIV replication.
2.3.3. Anti-CXCR4 Antibody (Clone 12G5) Binding Inhibition Assay
SUP-T1 cells were washed in phosphate buffered saline (PBS) with 2% FBS and resuspended at 3 × 106 cells/mL in the same buffer solution. For each sample, 0.3 × 106 cells were pre-incubated with the sample in PBS/FBS 2% for 30 min at 4 °C. The cells were washed once in PBS/FBS 2% before the addition of the monoclonal antibody (mAb) against CXCR4 (clone 12G5; BD Biosciences, San Jose, CA, USA). After incubation (30 min at 4 °C), the cells were washed, resuspended in PBS containing 1% paraformaldehyde, and analysed on a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA, USA).
2.3.4. Chemokine (CXCL12AF647/CCL5AF647) Binding Inhibition Assay
Human PBMCs were washed once with assay buffer (Hanks’ balanced salt solution with 20 mM HEPES buffer and 0.2% bovine serum albumin, pH 7.4) and then incubated for 15 min at room temperature with the sample diluted in assay buffer at the indicated concentrations. Subsequently, CXCL12AF647 (25 ng/mL) or CCL5AF647 (50 ng/mL) was added to the compound-incubated cells. The cells were incubated for 30 min at room temperature. Thereafter, the cells were washed twice in assay buffer, fixed in 1% paraformaldehyde in PBS, and analysed on the FL4 channel of a FACSCalibur flow cytometer equipped with a 635 nm red diode laser (Becton Dickinson, San Jose, CA, USA). The percentage of inhibition of CXCL12AF647 or CCL5AF647 binding was calculated according to the formula: [1 − ((MFI − MFINC)/(MFIPC − MFINC))] × 100, where MFI is the mean fluorescence intensity of the cells incubated with CXCL12AF647 or CCL5AF647 in the presence of the inhibitor, MFINC is the mean fluorescence intensity measured in the negative control (i.e., autofluorescence of unlabelled cells), and MFIPC is the mean fluorescence intensity of the positive control (i.e., cells exposed to CXCL12AF647 or CCL5AF647 alone).
2.3.5. Measurement of Intracellular Chemokine-Induced Calcium Mobilisation
CXCR4- or CCR5-positive U87 glioblastoma/astrocytoma cells were loaded with the fluorescent calcium indicator Fluo-3 acetoxymethyl (Molecular Probes) at 4 µM in the assay buffer (Hanks’ balanced salt solution with 20 mM HEPES buffer and 0.2% bovine serum albumin, pH 7.4) for 45 min at room temperature. After thorough washing with the assay buffer, cells were pre-incubated for 10 min at 37 °C in the same buffer with the compounds of interest. The intracellular calcium mobilisation in response to CXCL12 (CXCR4 chemokine ligand) or CCL3L1 (CCR5 chemokine ligand) was then measured at 37 °C by monitoring the fluorescence as a function of time simultaneously in all the wells by using a fluorometric imaging plate reader (FLIPR; Molecular Devices, Sunnyvale, CA, USA).
2.3.6. Evaluation of Cellular Cytotoxicity
Cytotoxicity of the compounds was evaluated after 48 h of incubation at 37 °C in PHA-stimulated PBMCs (5 × 105 cells per well in culture medium). Each condition was tested in duplicate. Cytotoxicity was evaluated microscopically, and in addition, the cell viability was assessed using the colorimetric CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, Fitchburg, WI, USA), which is based on the reduction of the tetrazolium salt MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] to formazan by metabolically active cells. Absorbance at 490 nm was measured with the Versa-Max ELISA microplate reader (Molecular Devices, Sunnyvale, CA, USA). The cytotoxic concentration 50 (CC50) of each compound was calculated based on the absorbance of negative (i.e., cells without compound) and positive (i.e., only culture medium) control samples.
2.4. X-ray Crystallography
Single-crystal X-ray diffraction data were collected in a series of ω-scans using a Stoe IPSD2 image plate diffractometer utilising monochromated Mo radiation (λ = 0.71073 Å). Standard procedures were employed for the integration and processing of the data using X-RED [50]. Samples were coated in a thin film of perfluoro-polyether oil and mounted at the tip of a glass fibre located on a goniometer. Data were collected from crystals held at 150 K in an Oxford Cryosystems nitrogen gas cryostream. Crystal structures were solved using routine automatic direct methods implemented within SHELXS-97 [51]. Completion of structures was achieved by performing least-squares refinement against all unique F2 values using SHELXL2018 [51]. All non-H atoms were refined with anisotropic displacement parameters. Hydrogen atoms were placed using a riding model. Disorder, where present, was treated conservatively with standard methods. In each structure, the core was well-determined, but the unbound anions were much less well-determined. Further information is provided in the Supplementary Materials.
2.5. Computational Methods
Single macrocycle components of the Zn(II) and Cu(II) complexes of sL1–sL5, with water and/or acetate bound, were computationally modelled to obtain an estimate of the thermodynamics of binding. All quantum mechanical calculations were performed using Gaussian 09 software (RevA.02) [52]. Density functional theory (DFT) calculations were performed utilising the M06 functional with an ultrafine integration grid, and the all electron cc-pVDZ basis set was used on all atoms [53]. Full geometry optimisations and vibrational frequency calculations were performed in the gas phase. All calculations of the Zn(II) complexes were run in the singlet electronic state, while the Cu(II) complexes were run as doublet states.
3. Results and Discussion
In previous studies, we characterised copper(II) and zinc(II) complexes of L2 (Figure 1) and showed that the zinc(II) complex is a more potent CXCR4 antagonist than the copper(II) complex. A single configuration of the cyclam ring (trans-II) was observed in both the solid state and solution (using NMR) for [Zn2L2]4+, and so the potency could be correlated with the coordination sphere around the metal centre [33]. X-ray crystallography, DFT computational studies, and in vitro analyses of the biological properties have shown that the weaker bonding for [Cu2L2]4+ at the “free” coordination site for protein binding reduces potency [43]. We then reported the bis-cross-bridged cyclam derivative L3 (Figure 1) and its copper(II) complex, which showed an increase in rigidity and higher affinity for binding to the aspartate residues at the extracellular surface of the CXCR4 chemokine receptor than [Cu2L2]4+ (Table 1 and Table 2) [34]. The complexes [Zn2L2]4+ and [Cu2L3]4+ were identified as the lead compounds from our previous studies, showing higher affinity and increased residence time in vitro relative to non-bridged compounds. To complete the study and provide a more complete understanding of the key molecular properties, the [Zn2L3]4+ complex was synthesised along with cross-bridged derivatives where the ring size was varied (L4 and L5) to determine the impact of cavity size on the binding affinity, biological activity, and receptor selectivity. Further X-ray structural characterisation of [Cu2L2]4+ is also presented. The aim is to optimise the coordination interactions and the affinity for the CXCR4 receptor to improve the pharmacological profile of this class of compounds.
Table 1.
Anti-HIV and cellular toxicity evaluation of CXCR4 antagonists.
| Compound | Anti-HIV Activity a IC50 (nM) |
Toxicity b CC50 (µM) |
|
|---|---|---|---|
| NL4-3 (X4) | BaL (R5) | PMBCs | |
| Cu2L1 | 10.1 ± 1.6 | >20,000 | >100 |
| Zn2L1 | 0.73 ± 0.34 | >15,000 | >90 |
| Cu2L2 | 12.9 ± 2.8 | >20,000 | >100 |
| Zn2L2 | 2.9 ± 1.2 | >15,000 | >100 |
| Cu2L3 | 0.74 ± 0.60 | >5000 | >40 |
| Zn2L3 | 6.2 ± 2.5 | >1000 | >100 |
| Cu2L4 | 0.87 ± 0.32 | >2000 | >10 |
| Zn2L4 | 4.8 ± 1.5 | >2000 | >90 |
| Cu2L5 | 31.6 ± 2.4 | >20,000 | >30 |
| Zn2L5 | 16.9 ± 0.9 | >20,000 | >100 |
| AMD3100 | 0.42 ± 0.14 | >20,000 | >100 |
| Maraviroc | >20,000 | 2.9 ± 1.2 | >100 |
a Concentration required to inhibit the replication of the HIV-1 strains NL4-3 (X4) and BaL (R5) in TZM-bl cells by 50%. b Concentration required to reduce cell (PMBCs) viability by 50%. Data are presented as the mean ± SEM of two to five experiments.
Table 2.
In vitro evaluation of the CXCR4 antagonists in the competition binding and signalling experiments.
| Compound | Competition Binding Assay a IC50 (nM) |
Ca2+ Signalling Assay b IC50 (nM) |
|||
|---|---|---|---|---|---|
| 12G5 | CXCL12 | CCL5 | CXCL12 | CCL3L1 | |
| Cu2L1 | 50.1 ± 7.2 | 5.1 ± 0.9 | >1000 | 113 ± 12.7 | >20,000 |
| Zn2L1 | 5.9 ± 2.4 | 0.41 ± 0.03 | >1000 | 4.9 ± 2.9 | >2000 |
| Cu2L2 | 465 ± 155 | 8.9 ± 1.1 | >1000 | 185 ± 75 | >20,000 |
| Zn2L2 | 1.4 ± 0.4 | 0.43 ± 0.00 | 255 ± 58 | 5.6 ± 1.9 | 120 ± 28 |
| Cu2L3 | 1.12 ± 0.15 | 0.54 ± 0.03 | 741 ± 145 | 3.1 ± 0.3 [23] | >1000 |
| Zn2L3 | 1.9 ± 0.3 | 0.90 ± 0.23 | >1000 | 17.5 ± 11.7 | >5000 |
| Cu2L4 | 1.3 ± 0.4 | 0.53 ± 0.03 | 702 ± 219 | 11.9 ± 8.3 | >10,000 |
| Zn2L4 | 3.4 ± 0.2 | 2.1 ± 0.1 | >1000 | 66.1 ± 0.9 | >20,000 |
| Cu2L5 | 1.9 ± 0.6 | 0.83 ± 0.02 | >1000 | 11.3 ± 4.7 | >15,000 |
| Zn2L5 | 8.6 ± 4.7 | 0.49 ± 0.01 | >1000 | 10.3 ± 6.6 | >20,000 |
| AMD3100 | 83.7 ± 8.3 | 18.0 ± 4.2 [37] | >1000 | 203.5 ± 19.4 [23] | >20,000 |
| Maraviroc | - | >1000 | 10.3 ± 3.5 | >20,000 | 3.1 ± 0.6 |
a Concentration required to inhibit the binding of PE-conjugated anti-CXCR4 mAb (12G5) in T-lymphocytes or the binding of AF647-labelled chemokines CXCL12 or CCL5 in PMBCs by 50%. b Concentration required to inhibit chemokine-induced Ca2+ signalling in CXCR4- and CCR5-transfected cells by 50%. Data are presented as the mean ± SEM of two to three experiments.
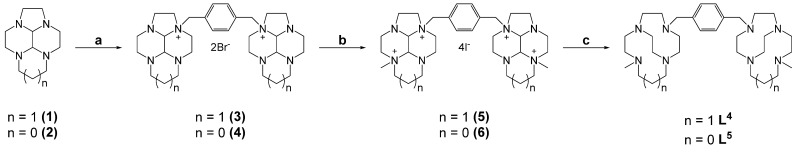
3.1. Synthesis of Ligands and Metal Complexes
The synthesis of the novel chelators L4 and L5 and their copper(II) and zinc(II) complexes followed our previously reported route for the synthesis of the chelator L3 and were achieved on a multi-gram scale with all steps with a >70% yield. We have previously reported the synthesis of para-substituted ethyl-bridged cyclams [34]. The synthetic methods were applied to the 13-membered tetraazamacrocycle homocyclen to form L4 and the 12-membered cyclen to form L5. The ligands were formed by reacting the macrocycle glyoxal condensates (1 and 2) with α,α’-dibromo-p-xylene to form bis-tetraazamacrocycles 3 and 4. The intermediates were subsequently alkylated with methyl iodide (5 and 6), followed by reduction with sodium borohydride to form the target chelators (Scheme 1). Copper(II) and zinc(II) complexes were formed in an analogous method to that described previously, in methanol and using acetate salts of the metal ions [34].
Scheme 1.
Synthesis of para-linked bis-tetraazamacrocycles homocyclen (L4) and cyclen (L5). (a) p-Dibromoxylene; (b) CH3I; (c) NaBH4.
3.2. Crystallographic and Spectroscopic Characterisation of Copper(II) Complexes
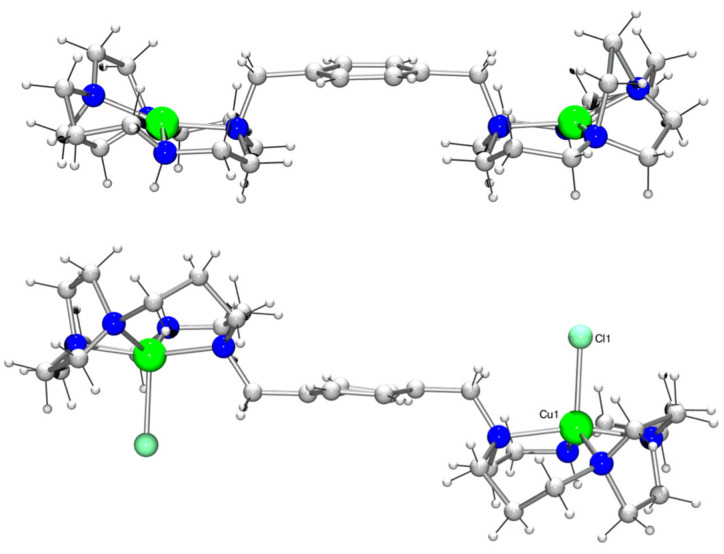
Single crystals of both [Cu2L2](ClO4)4 and [Cu2L2Cl2]Cl2 (Figure 2) were grown via ether diffusion into a methanolic solution. The crystal structure of [Cu2L2](ClO4)4 contained two macrocyclic units and was in a trans-II configuration. The copper(II) ions in each ring were not crystallographically equivalent to each other, but their environments were very similar. They both adopted a tetrahedrally distorted square planar geometry, with Cu−N bond lengths ranging from 1.879(14) to 2.073(17) Å in one ring and 1.871(16) to 2.237(15) Å in the second. The metal–nitrogen bond lengths in [Cu2L2]4+ were, as expected, somewhat shorter than in [Zn2L2(OAc)2]2+, and in addition, there was no significant interaction between the copper(II) centre and the counter anion. The Cu(1)…O(16) and Cu(2)…O(5) distances were 3.16(2) and 2.58(2) Å, respectively (Figure S1 and Tables S1–S3). An example of a mono-side-bridged copper(II) complex reported by Kaden and co-workers had two independent copper(II) environments in the asymmetric unit [54]. The Cu−N bond lengths were similar to [Cu2L2]4+, being in the range of 1.979(4) to 2.016(4) Å. Analogous to [Cu2L2]4+, the copper(II) was also four-coordinate in the solid state. The macrocyclic framework was in a trans-II configuration, where the methyl and hydrogen arms sat trans to each other. In each of the two crystal structures, the copper(II) ion sat in the plane of the four nitrogen atoms, the deviation from the mean plane being 0.070 and 0.007 Å, respectively.
Figure 2.
X-ray crystal structures of [Cu2L2](ClO4)4 (top) and [Cu2L2Cl2]Cl2 (bottom), non-coordinating anions omitted for clarity (see Supplementary Materials for further details and the ORTEP plots).
The analysis of the crystal structure of [Cu2L2Cl2]2+ was complicated because the asymmetric unit lay on a crystallographic mirror plane that did not correspond with a mirror plane in the molecule (Figure 2). This generated two orientations for the molecule in equal amounts. The copper(II) centre was five-coordinate and can best be described as being in a square-based pyramidal coordination environment, the base consisting of the four macrocyclic nitrogen atoms with a bound chloride counter-anion in the apical position (Cu(1)…Cl(1) distance is 2.4133(14) Å). The copper(II)–nitrogen bond lengths were in the range of 1.938(7) to 2.141(6) Å (Figure S2 and Tables S4–S6), which was similar to [Cu2L2]4+. Again, the macrocycle adopted the equivalent of a trans-II configuration in the solid state (Figure 2). The metal–metal distance of 11.5637(14) Å was, as expected, longer than in [Cu2L2]4+ (10.913(2) Å in [Cu2L2]4+), as the two macrocycles were located in the equivalent of a trans position relative to each other. In the structure of [Cu2L2](ClO4)4, the metal centres were cis to each other with a three point angle of 151.31(9)°, whereas in [Cu2L2Cl2]Cl2, the metal centres were trans to each other with a three point angle of exactly 180° due to the crystallographic inversion centre. This distance was similar to that found in [Zn2L2(OAc)2]2+ (11.6584(10) Å) [33]. The angle between the two macrocycles could then be determined. The same three planes were defined and resulted in an angle of ca. 180°, similar to [Zn2L2(OAc)2]2+, with any deviation being attributed to the disorder in the structure. The angle between the plane of the carbon–nitrogen bond and the plane of the phenyl moiety was 88.200(99)°.
UV/Vis was used to investigate the solution behaviour of [Cu2L2]4+, showing a broad band at 557 nm [43]. This is a higher value than that usually observed for square planar copper(II) cyclam complexes, which can be attributed to the tetragonal distortion of the copper(II) centre. A more appropriate comparison is with the mono-side-bridged cyclam copper complex reported by Kowallick et al., which had an absorption maximum at 556 nm in aqueous solution [54]. Five-coordinate copper(II) complexes generally exhibit a band at around 600 nm [55]. There was no significant shift in the band at 557 nm upon the addition of sodium acetate (2 M sodium acetate; 560 nm), suggesting a preference for a four-coordinate copper(II) species in solution.
3.3. DFT Calculations
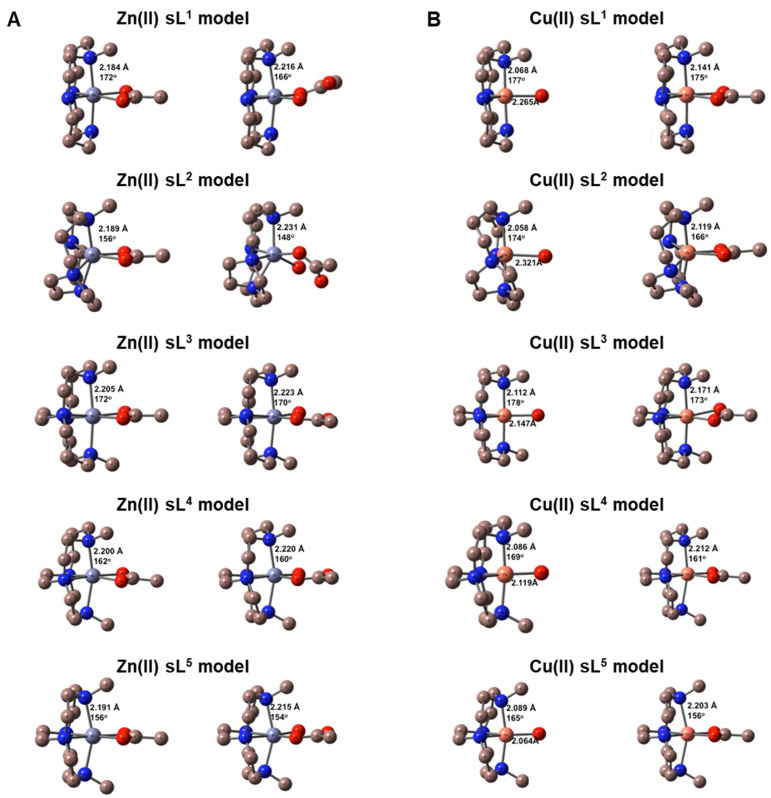
To obtain an estimate of the strength of water and/or acetate binding to the zinc(II) and copper(II) complexes of L1 through L5, DFT calculations were performed on single ring models of these metal complexes, which had the chelators designated as sL1 through sL5 (Figure 3). It was anticipated that increased affinity for acetate coordination over a water molecule would correspond to increased receptor affinity. This mimics the coordination interaction with aspartate or glutamate on the chemokine receptor protein surface.
Figure 3.
DFT calculated structures of single ring models of the macrocyclic complexes with zinc(II) and copper(II). (A) [ZnsL(OAc)]+ on the left and [ZnsL(OAc)(H2O)]+ on the right. Values shown are the average Zn–N bond lengths and the axial N–Zn–N angle. (B) [CusL(H2O)]2+ on the left and [CusL(OAc)]+ on the right. Values shown are the average Cu–N bond lengths, the axial N–Cu–N angle, and the Cu–O bond length.
M06/cc-pVDZ calculations were performed on all complexes. Figure 3A shows the DFT geometry optimised structures for [ZnsL(OAc)]+ on the left and [ZnsL(OAc)(H2O)]+ on the right ([ZnsL(H2O)2]2+ data are shown in Figure S3), while Figure 3B shows the structures for [CusL(H2O)]2+ on the left and [CusL(OAc)]+ on the right. In each of these figures, the bonding parameters listed are the average M(II)–N bond lengths in angstroms (Å) and the axial N–M(II)–N angle in degrees (°). Based on our previous studies, zinc(II) complexes tend to have uniformly high activity in biological assays, and assuming that the two metal ions independently form coordinate bonds with different amino acid residues on the extracellular surface of CXCR4, single ring complexes were modelled. For the zinc(II) complex, there was particular interest in the influence of the macrocycle ring size on the inclusion of a water molecule along with the monodentate acetate in the coordination sphere. In correlation with the X-ray structural data that have previously been reported, a strong preference for the inclusion of a water molecule in the coordination sphere was observed for the cross-bridged complexes [56]. For zinc(II), it was more favourable to have the water molecule present in all of the structures, except for the side-bridged macrocyclic chelator present in sL2, which was more stable with the bidentate acetate present. The Gibbs free energies of binding, as listed in Table 3, specify the order of interaction strength as: sL3 > sL4 > sL5 > sL1 > sL2. This is consistent with the X-ray dataset previously reported [56].
Table 3.
Change in Gibbs energy (298 K) for single ring chelator model reactions. Results were obtained from the M06/cc-pVDZ calculations.
| Single Ring Chelator Model | ΔG298 (kJ/mol) [CusL(H2O)]2+ + OAc– → [CusL(OAc)]+ + H2O |
ΔG298 (kJ/mol) [ZnsL(OAc)]+ + H2O → [ZnsL(OAc)(H2O)]+ |
ΔG298 (kJ/mol) [ZnsL(H2O)2]2+ + OAc– → [ZnsL(OAc)(H2O)]+ + H2O |
|---|---|---|---|
| s L1 | −822.2 | −14.9 | −825.8 |
| s L2 | −800.4 | 4.0 | − |
| s L3 | −833.5 | −25.1 | −830.6 |
| s L4 | −845.9 | −23.5 | −840.4 |
| s L5 | −866.7 | −22.5 | −849.1 |
Similarly for the copper(II) complexes, the optimised structures were analogous to the reported crystal structures of related complexes. The [CusL(H2O)]2+ complexes were five-coordinate while the [CusL(OAc)]+ complexes were six-coordinate, with the two oxygen atoms of the coordinated acetate occupying cis equatorial positions in an anisotropic bidentate fashion for sL1, sL2, and sL3, while the two Cu(II)–O bond lengths were similar in the sL4 and sL5 complexes. It was favourable for the acetate anion to replace the coordinated water molecule in all complexes with the Gibbs energy changes listed in Table 3. However, similar to the zinc(II) complexes, the sL2 ligand had the least favourable reaction energy, with the order of interaction being: sL5 > sL4 > sL3 > sL1 > sL2. Interestingly, the acetate binding energies tracked the Cu(II)–OH2 bond length: as Cu(II)–O increased in [CusL(H2O)]2+, the acetate binding strength decreased. The Cu(II)–O values are also shown in Figure 3B. The calculations for copper(II) were designed to determine whether the acetate interaction correlated to the affinity recorded in the biological assays. Binding of the acetate over water was strongly preferred in all cases. The copper(II) complex of the side-bridged macrocycle (sL2) in [Cu2L2]4+ had the lowest binding energy for acetate, which matched with both the reduced potency of this CXCR4 antagonist and the UV/Vis solution studies. The calculations validated the observed increase in affinity for the cross-bridged compounds (Table 1 and Table 2). There was a slight increase in the binding energy for acetate as the ring size decreased, with the most favourable reaction for acetate in replacing a bound water observed for cross-bridged cyclen sL5 (Table 3). In general, these calculations indicate that [Cu2L5]4+ should have the most favourable interaction with the CXCR4 receptor, although there were no major differences between the three cross-bridged derivatives for this parameter.
3.4. Biological Evaluation
A series of binding assays were performed to assess the in vitro biological activity and function of the small molecules targeting CXCR4. Data demonstrating HIV-1-CXCR4 entry blocking, interference with the CXCL12-CXCR4 axis, anti-CXCR4 antibody competition, and receptor specificity are shown in Table 1 and Table 2.
3.4.1. Assessment of Cellular Cytotoxicity for the Investigated Compounds
The cellular cytotoxicity of all compounds was evaluated in PHA-stimulated PBMCs. The cytotoxic concentration 50 (i.e., CC50; the concentration that reduces cell viability by 50%) was determined, based on the microscopic evaluation of the PBMCs and quantification of their cell viability using the MTS assay. CC50 values were close to or higher than 100 µM for most of the investigated metal complexes (Cu2L1, Zn2L1, Cu2L2, Zn2L2, Zn2L3, Zn2L4, and Zn2L5) as well as for the reference compounds AMD3100 and Maraviroc. Cu2L4 had the lowest CC50 value in PBMCs (>10 µM). Furthermore, CC50 values of the copper(II) complexes of L3 and L5 were moderate to high, situated around 50 to 40 µM, respectively (Table 1). The CC50 values indicate low toxicity in relation to the anticipated dosing levels.
3.4.2. Antiviral Activity against HIV-1 NL4-3 (X4) and BaL (R5) Replication
CXCR4 is one of the two main co-receptors involved in the infection process of HIV-1 [57,58]. In an initial set of experiments, the compounds were evaluated for their anti-HIV-1 activity. The effect of the compounds on the replication of the CXCR4-using HIV-1 strain NL4-3 (X4) and the CCR5-using HIV-1 strain BaL (R5) was determined in a HIV Tat-regulated luciferase reporter gene assay in the TZM-bl cells. IC50 values for the inhibition of the HIV-1 strain NL4-3 (X4) were in the low nanomolar range (0.73–31.6 nM). Inhibition of the HIV-1 strain BaL (R5) was less efficient, with IC50 values varying greater than 1 µM (Table 1), highlighting the specificity of the different derivatives to the CXCR4 receptor. IC50 values of AMD3100, a CXCR4-specific inhibitor, and Maraviroc, a CCR5-specific inhibitor, were 0.42 ± 0.14 nM and 2.9 ± 1.2 nM for X4 and R5 HIV-1 replication inhibition, respectively (Table 1). Efficient inhibition of CXCR4-using (X4) HIV-1 was observed for all compounds, with a lower potency observed for the copper(II) complexes of L1, L2, and L5 compared to their zinc(II) counterparts. In contrast, a higher potency for the copper(II) cross-bridged complexes (L3 and L4) was observed compared to the corresponding zinc(II) complexes. The least active compound (Cu2L5) had an IC50 value of 31.6 ± 2.4 nM. None of the complexes exhibited any increase in anti-HIV activity compared to AMD3100. All metal complexes showed weak, if any, activity against the R5 HIV-1 strain (IC50 > 1 µM), suggesting selectivity for CXCR4 over CCR5 (Table 1).
3.4.3. Inhibition of Anti-CXCR4 Monoclonal Antibody (Clone 12G5) Binding
The inhibitory potency of the compounds was further evaluated through competition binding experiments using the anti-CXCR4 monoclonal antibody (clone 12G5). The 12G5 antibody has been reported to block the entry process of HIV-1 via CXCR4 [59]. Successful blocking of the binding of the 12G5 antibody to CXCR4 using the small molecule inhibitor AMD3100 has previously been reported [60], confirming that this antibody can be a useful tool to study the interactions of the novel metal complexes reported herein with CXCR4. The results are shown in Table 2 and Figure S4. All of the compounds have promising efficacy as CXCR4 inhibitors, with IC50 values in the nanomolar range, ranging from 465 nM to 1.12 nM. Amongst the different metal complexes, Zn2L2, Cu2L3, Zn2L3, Cu2L4, and Cu2L5 showed the highest potency with IC50 values < 2 nM. (Table 2).
3.4.4. Potent Antagonism of Natural Chemokine Ligand Interaction to CXCR4 and Receptor Selectivity Against CCR5
Binding affinity and selectivity towards CXCR4 were further evaluated through competition binding assays using natural chemokine ligands, tagged with Alexa Fluor 647, as previously reported [61,62]. The CXCR4 binding affinity of the different compounds was determined through their ability to inhibit interactions between CXCR4 and CXCL12AF647. Selectivity towards CXCR4 was evaluated in a similar assay using CCR5, another co-receptor involved in HIV entry mechanism, and its fluorescent-labelled natural ligand CCL5AF647. All of the metal complexes showed a dose-dependent inhibition of CXCL12AF647 binding to CXCR4, with low nanomolar IC50 values ranging from 0.41 to 8.9 nM (Table 2 and Figure S5). Interestingly, apart from Zn2L4 (2.1 ± 0.1 nM), all of the other zinc(II) complexes had very similar high potency (<1 nM). The reference CXCR4 inhibitor, AMD3100, was less effective, with an IC50 value of 18.0 ± 4.2 nM (Table 2). In competition binding experiments using CCR5/CCL5AF647, IC50 values > 1 µM were observed for all but three compounds (Zn2L2, Cu2L3, Cu2L4), indicating selectivity towards CXCR4. Although lower, the IC50 of Zn2L2, Cu2L3 and Cu2L4 still remained in the medium-high nanomolar range with values of 255 ± 58 nM, 741 ± 145 nM, and 702 ± 219 nM, respectively (Table 2 and Figure S5). As a reference, the CCR5-specific inhibitor, Maraviroc, displayed an IC50 value of 10.3 ± 3.5 nM in this assay (Table 2). In this competition binding assay using CXCL12AF647, variation in the ring size had more of an effect on the copper(II) complexes in comparison with the zinc(II) complexes, which, apart from Zn2L4, displayed similar activity (Zn2L1, Zn2L2, Zn2L3, Zn2L5). Interestingly, Zn2L3, Zn2L4, and Zn2L5 displayed improved selectivity for CXCR4 (>1 µM for CCR5) compared to the previous lead compound Zn2L2 (255 ± 58 nM for CCR5). All metal complexes studied displayed higher activity compared to AMD3100.
3.4.5. Inhibition of Chemokine Induced Signalling via CXCR4 or CCR5
The potency of the different metal complexes was investigated in a functional downstream receptor signalling assay. The inhibitory effect of the tested compounds was examined by Ca2+ mobilisation studies with the natural ligands CXCL12 and CCL3L1 for the chemokine receptors CXCR4 and CCR5, respectively. The Ca2+ ions released from the endoplasmic reticulum were measured over time in CXCR4- or CCR5-transfected U87 glioblastoma cells via their specific interaction with the fluorescent Ca2+ indicator Fluo-3 [63]. The IC50 values measured for the inhibition of CXCL12-CXCR4 signalling were in the low nanomolar range (3.1–185 nM), and all compounds were more potent than AMD3100 (203.5 ± 19.4 nM) in this assay (Table 2). In the case of CCR5, IC50 values measured were all in the micromolar range, except for Zn2L2 (120 ± 28 nM), which appeared to be acting as a dual antagonist of CXCR4 and CCR5 in this experimental setup (Table 2). A strong correlation was seen between the IC50 values of CXCL12-induced calcium signalling via CXCR4 and CXCL12-CXCR4 binding (Figure 4A).
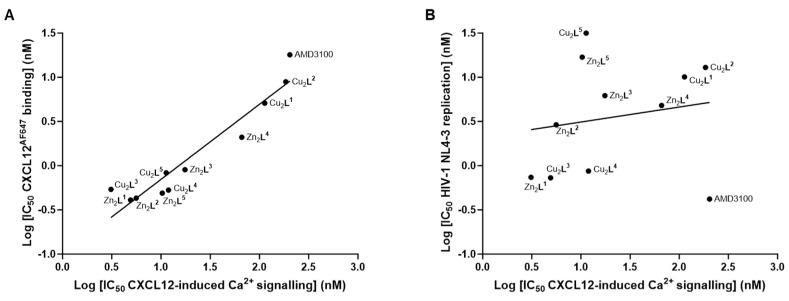
Figure 4.
Correlation of the inhibitory effects of the experimental compounds regarding CXCL12-induced calcium signalling with CXCL12AF647 binding (correlation coefficient r = 0.95) (A) or HIV-1 NL4-3 (X4) replication (correlation coefficient r = 0.17) (B).
In this receptor functional assay (CXCL12-induced calcium signalling via CXCR4), varying the ring size of the copper(II) complexes (L4/L5) did not impact significantly on the high affinity for CXCR4 compared to the previous lead compound Cu2L3; however, it resulted in a 10-fold increase in selectivity for CXCR4 over CCR5. Similar results were observed with Zn2L5, which retained a high affinity for CXCR4 (10.3 ± 6.6 nM) compared to the previous Zn2L2 (5.6 ± 1.9 nM) lead candidate, but with a >100-fold increase in selectivity for CXCR4 (Table 2). Zn2L2 was the only compound with a sub-micromolar activity in the CCL3L1-induced calcium signalling assay, with an IC50 value of 120 ± 28 nM (Table 2). The latter observation could imply that significant off-target effects will be observed if using the Zn2L2 compound as a prognostic tracer or therapy for CXCR4 overexpressing cells in vivo. A correlation plot showed consistency between the CXCR4 chemokine binding assay and calcium signalling assay (r = 0.95, Figure 4A). However, no correlation was found between the CXCL12-induced calcium signalling assay and anti-HIV properties (r = 0.17, Figure 4B), indicating that an alternate optimisation route is required to develop antiviral compounds over chemokine signal blocking compounds. Further correlation plots are available in Figure S6.
4. Conclusions
Structural investigation of the influence of macrocycle ring size on a range of metal ion complexes demonstrated that in similar compounds, subtle effects on the coordination geometry and positioning of the metal ion in the cavity dictate the biological efficacy and target selectivity.
AMD3100 effectively inhibited HIV infection in vitro, but was inferior for blocking the CXCL12 signalling pathway, which is relevant to the growth and dissemination of multiple cancers. All of the metal complexes investigated in this work compared favourably with AMD3100, with Cu2L2 having an overall similar profile. The advantage of configurational restriction is clear from a coordination geometry perspective, especially with the copper(II) complexes, as the cross-bridged compounds had much higher activity than the unrestricted or side-bridged compounds. The flexible coordination geometry of the d10 zinc(II) ion showed little difference in the compound potency across the zinc(II) compounds tested, with all showing significant improvement over AMD3100. However, the zinc(II) complexes of L3, L4, and L5 also showed a significant improvement in the selectivity for CXCR4 (over CCR5) whilst still retaining a similar high affinity and activity against CXCL12 binding compared to previous lead compounds. This is important for drug development progression where selectivity is as important as affinity/activity. Based on the observations reported in the study, with high importance placed on the binding selectivity, we identified Zn2L5 as a new drug candidate to take forward to target the CXCL12–CXCR4 axis in cancer therapy studies. We also plan to continue with our investigation and development of Cu2L3 in cancer imaging and therapy. It is noteworthy that zinc(II) metallodrugs have applications as cancer therapeutics alongside other medicinal applications [64].
Whilst the trend observed in the calculations can be rationalised (as the metal ion is pushed out of the ring, it will interact more strongly with the acetate/aspartate residues), it is not consistent with the biological assay results. This is likely due to factors such as the relative positioning of the two metal centres for optimum interactions, and possibly, the reduced stability of the copper(II) complexes of cross-bridged homocyclen and cyclen. Zinc(II) and copper(II) are ideal comparators as the d9 copper(II) ion has distinct geometric preferences (and stabilises distortion for octahedral and square-based pyramidal geometry), whereas the d10 zinc(II) ion is more flexible in the coordination sphere that can be adopted. This was reflected in the assay results. However, the optimised copper(II) complexes could be radiolabelled with copper-64 (PET imaging radioisotope) or copper-67 (therapeutic/SPECT imaging radioisotope), offering a route to the development of CXCR4-targeted theranostics for cancer imaging and therapy. In addition to the biological assay data, it has previously been shown that complex stability for copper(II) is higher for the cross-bridged cyclam compared to the cross-bridged homocyclen and cross-bridged cyclen [36], suggesting that the metal ion would be more likely to be retained in vivo. This is in line with our previously reported data showing that the configuration of Cu2L3 leads to increased affinity for CXCR4, stronger interactions, and longer residence time at the receptor [34]. We have also recently shown that L3 can be successfully and stably radiolabelled with copper-64, resulting in high and specific CXCR4-mediated tumour uptake [23]. Due to the improved selectivity observed, further consideration should be given for the radiolabelling and stability evaluation of Cu2L5 to determine whether this complex could become a candidate for theranostic development. For applications targeting the CXCL12–CXCR4 signalling axis in cancer therapy, Zn2L5 provides a further option for comparison to our current lead compound, Cu2L3.
Acknowledgments
The authors wish to thank the EPSRC National Mass Spectrometry Service at Swansea as well as Sandra Claes, Evelyne Van Kerckhove, and Eric Fonteyn for their excellent technical assistance in the framework of the biological validation of the experimental compounds in this article.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pharmaceutics16081000/s1, Figure S1: ORTEP representation of [Cu2L2](ClO4)4; Figure S2: ORTEP representation of [Cu2L2Cl2]Cl2; Figure S3: Structures of [ZnsL(H2O)2]2+ from the DFT calculations; Figure S4: Inhibition of anti-CXCR4 mAb (clone 12G5) binding by the different experimental compounds; Figure S5: Binding inhibition of the fluorescent labelled chemokines CXCL12AF647 and CCL5AF647; Figure S6: Correlation plots between the biological assays; Table S1: Crystal data and structure refinement for [Cu2L2](ClO4)4; Table S2: Bond lengths for [Cu2L2](ClO4)4; Table S3: Bond angles for [Cu2L2](ClO4)4; Table S4: Crystal data and structure refinement for [Cu2L2Cl2]Cl2; Table S5: Bond lengths for [Cu2L2Cl2]Cl2; Table S6: Bond angles for [Cu2L2Cl2]Cl2.
Author Contributions
Conceptualisation S.J.A., T.J.H., and D.S.; Methodology, S.J.A., T.J.H., D.S., D.P.L., T.V.L. and T.J.P.; Validation, I.R., B.P.B. and T.V.L.; Formal analysis, T.J.P., D.P.L., T.D., T.V.L. and S.J.A.; Investigation, I.R., T.D., B.P.B., T.A., A.N.C., N.L.F., A.K., D.L.M. (Danny L. Maples), R.D.M., D.L.M. (Dallas L. Matz), G.M. and R.U.; Resources, T.J.P., D.P.L., S.J.A., T.J.H. and D.S.; Data curation, T.J.P. and D.P.L.; Writing—original draft preparation, I.R., B.P.B., A.K., T.D., S.J.A., T.J.H. and D.S.; Writing—review and editing, I.R., B.P.B., A.K., T.D., S.J.A., T.J.H. and D.S.; Visualisation, I.R., B.P.B., T.D., T.J.P., D.P.L., S.J.A. and T.V.L.; Supervision, S.J.A., T.J.H. and D.S.; Project administration, S.J.A., T.J.H. and D.S.; Funding acquisition, S.J.A., T.J.H. and D.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are contained within the article and in the Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This work was supported by Yorkshire Cancer Research (award reference number HEND376), the Medical Research Council (grant number MC_PC_18049), and the Engineering and Physical Sciences Research Council (EP/V055836/2). This work, in part, was supported by funding from the KU Leuven (GOA/10/014, PF/10/018, and C22/17/008) and the Foundation of Scientific Research (FWO nos. G.0485.08 and G.0528.12). We acknowledge the Health Research Award for project no. HR13–157 from the Oklahoma Centre for the Advancement of Science and Technology. This project was supported by the National Centre for Research Resources and the National Institute of General Medical Sciences of the National Institutes of Health through grant no. 8P20M103447.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Murdoch C., Finn A. Chemokine receptors and their role in inflammation and infectious diseases. Blood. 2000;95:3032–3043. doi: 10.1182/blood.V95.10.3032. [DOI] [PubMed] [Google Scholar]
- 2.Nagarsheth N., Wicha M.S., Zou W.P. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017;17:559–572. doi: 10.1038/nri.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adlere I., Caspar B., Arimont M., Dekkers S., Visser K., Stuijt J., de Graaf C., Stocks M., Kellam B., Briddon S., et al. Modulators of CXCR4 and CXCR7/ACKR3 Function. Mol. Pharmacol. 2019;96:737–752. doi: 10.1124/mol.119.117663. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Cuesta E.M., Santiago C.A., Vallejo-Diaz J., Juarranz Y., Rodriguez-Frade J.M., Mellado M. The Role of the CXCL12/CXCR4/ACKR3 Axis in Autoimmune Diseases. Front. Endocrinol. 2019;10 doi: 10.3389/fendo.2019.00585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miao M., De Clercq E., Li G.D. Clinical significance of chemokine receptor antagonists. Expert Opin. Drug Metab. Toxicol. 2020;16:11–30. doi: 10.1080/17425255.2020.1711884. [DOI] [PubMed] [Google Scholar]
- 6.Zeelenberg I.S., Ruuls-Van Stalle L., Roos E. The chemokine receptor CXCR4 is required for outgrowth of colon carcinoma micrometastases. Cancer Res. 2003;63:3833–3839. [PubMed] [Google Scholar]
- 7.Domanska U.M., Kruizinga R.C., Nagengast W.B., Timmer-Bosscha H., Huls G., de Vries E.G., Walenkamp A.M. A review on CXCR4/CXCL12 axis in oncology: No place to hide. Eur. J. Cancer. 2013;49:219–230. doi: 10.1016/j.ejca.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Hattermann K., Mentlein R. An infernal trio: The chemokine CXCL12 and its receptors CXCR4 and CXCR7 in tumor biology. Ann. Anat. Anat. Anz. Off. Organ Anat. Ges. 2013;195:103–110. doi: 10.1016/j.aanat.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 9.Yang Y., Li J., Lei W., Wang H., Ni Y., Liu Y., Yan H., Tian Y., Wang Z., Yang Z., et al. CXCL12-CXCR4/CXCR7 Axis in Cancer: From Mechanisms to Clinical Applications. Int J Biol Sci. 2023;19:3341–3359. doi: 10.7150/ijbs.82317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cojoc M., Peitzsch C., Trautmann F., Polishchuk L., Telegeev G.D., Dubrovska A. Emerging targets in cancer management: Role of the CXCL12/CXCR4 axis. OncoTargets Ther. 2013;6:1347–1361. doi: 10.2147/OTT.S36109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domanska U.M., Timmer-Bosscha H., Nagengast W.B., Oude Munnink T.H., Kruizinga R.C., Ananias H.J., Kliphuis N.M., Huls G., De Vries E.G., de Jong I.J., et al. CXCR4 inhibition with AMD3100 sensitizes prostate cancer to docetaxel chemotherapy. Neoplasia. 2012;14:709–718. doi: 10.1593/neo.12324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gagliardi F., Narayanan A., Reni M., Franzin A., Mazza E., Boari N., Bailo M., Zordan P., Mortini P. The role of CXCR4 in highly malignant human gliomas biology: Current knowledge and future directions. Glia. 2014;62:1015–1023. doi: 10.1002/glia.22669. [DOI] [PubMed] [Google Scholar]
- 13.Marechal R., Demetter P., Nagy N., Berton A., Decaestecker C., Polus M., Closset J., Deviere J., Salmon I., Van Laethem J.L. High expression of CXCR4 may predict poor survival in resected pancreatic adenocarcinoma. Br J Cancer. 2009;100:1444–1451. doi: 10.1038/sj.bjc.6605020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trautmann F., Cojoc M., Kurth I., Melin N., Bouchez L.C., Dubrovska A., Peitzsch C. CXCR4 as biomarker for radioresistant cancer stem cells. Int. J. Radiat. Biol. 2014;90:687–699. doi: 10.3109/09553002.2014.906766. [DOI] [PubMed] [Google Scholar]
- 15.Werner L., Guzner-Gur H., Dotan I. Involvement of CXCR4/CXCR7/CXCL12 Interactions in Inflammatory bowel disease. Theranostics. 2013;3:40–46. doi: 10.7150/thno.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahirovic Y.A., Pelly S., Jecs E., Miller E.J., Sharma S.K., Liotta D.C., Wilson L.J. Small molecule and peptide-based CXCR4 modulators as therapeutic agents. A patent review for the period from 2010 to 2018. Expert Opin. Ther. Pat. 2020;30:87–101. doi: 10.1080/13543776.2020.1707186. [DOI] [PubMed] [Google Scholar]
- 17.Xu C., Zhao H., Chen H.T., Yao Q.H. CXCR4 in breast cancer: Oncogenic role and therapeutic targeting. Drug Des. Dev. Ther. 2015;9:4953–4964. doi: 10.2147/dddt.s84932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hermann P.C., Huber S.L., Herrler T., Aicher A., Ellwart J.W., Guba M., Bruns C.J., Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Panneerselvam J., Jin J.K., Shanker M., Lauderdale J., Bates J., Wang Q., Zhao Y.D., Archibald S.J., Hubin T.J., Ramesh R. IL-24 Inhibits Lung Cancer Cell Migration and Invasion by Disrupting The SDF-1/CXCR4 Signaling Axis. Plos One. 2015;10 doi: 10.1371/journal.pone.0122439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bao S., Darvishi M., H Amin A., Al-Haideri M.T., Patra I., Kashikova K., Ahmad I., Alsaikhan F., Al-qaim Z.H., Al-Gazally M.E., et al. CXC chemokine receptor 4 (CXCR4) blockade in cancer treatment. J. Cancer Res. Clin. Oncol. 2023;149:7945–7968. doi: 10.1007/s00432-022-04444-w. [DOI] [PubMed] [Google Scholar]
- 21.Renard I., Archibald S.J. CXCR4-targeted metal complexes for molecular imaging. In: Sadler P.J., VanEldik R., editors. Medicinal Chemistry. Volume 75. 2020. pp. 447–476. [Google Scholar]
- 22.Mayerhoefer M.E., Archibald S.J., Messiou C., Staudenherz A., Berzaczy D., Schoder H. MRI and PET/MRI in hematologic malignancies. J. Magn. Reson. Imaging. 2020;51:1325–1335. doi: 10.1002/jmri.26848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burke B., Miranda C., Lee R., Renard I., Nigam S., Clemente G., D’Huys T., Ruest T., Domarkas J., Thompson J., et al. Copper-64 PET imaging of the CXCR4 chemokine receptor using a cross-bridged cyclam bis-tetraazamacrocyclic antagonist. J. Nucl. Med. 2019 doi: 10.2967/jnumed.118.218008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Clercq E. The bicyclam AMD3 100 story. Nat. Rev. Drug Discov. 2003;2:581–587. doi: 10.1038/nrd1134. [DOI] [PubMed] [Google Scholar]
- 25.Amendola V., Bergamaschi G., Fabbrizzi L., Licchelli M., Mangano C. The interaction of Mozobil (TM) with carboxylates. Org. Biomol. Chem. 2016;14:905–912. doi: 10.1039/C5OB01704A. [DOI] [PubMed] [Google Scholar]
- 26.Gerlach L.O., Skerlj R.T., Bridger G.J., Schwartz T.W. Molecular interactions of cyclam and bicyclam non-peptide antagonists with the CXCR4 chemokine receptor. J Biol Chem. 2001;276:14153–14160. doi: 10.1074/jbc.M010429200. [DOI] [PubMed] [Google Scholar]
- 27.Rosenkilde M.M., Gerlach L.-O., Jakobsen J.S., Skerlj R.T., Bridger G.J., Schwartz T.W. Molecular Mechanism of AMD3100 Antagonism in the CXCR4 Receptor: TRANSFER OF BINDING SITE TO THE CXCR3 RECEPTOR*. J. Biol. Chem. 2004;279:3033–3041. doi: 10.1074/jbc.M309546200. [DOI] [PubMed] [Google Scholar]
- 28.Wong R.S., Bodart V., Metz M., Labrecque J., Bridger G., Fricker S.P. Comparison of the potential multiple binding modes of bicyclam, monocylam, and noncyclam small-molecule CXC chemokine receptor 4 inhibitors. Mol. Pharmacol. 2008;74:1485–1495. doi: 10.1124/mol.108.049775. [DOI] [PubMed] [Google Scholar]
- 29.Gerlach L.O., Jakobsen J.S., Jensen K.P., Rosenkilde M.R., Skerlj R.T., Ryde U., Bridger G.J., Schwartz T.W. Metal ion enhanced binding of AMD3100 to Asp(262) in the CXCR4 receptor. Biochemistry. 2003;42:710–717. doi: 10.1021/bi0264770. [DOI] [PubMed] [Google Scholar]
- 30.Hunter T.M., McNae I.W., Liang X.Y., Bella J., Parsons S., Walkinshaw M.D., Sadler P.J. Protein recognition of macrocycles: Binding of anti-HIV metallocyclams to lysozyme. Proc. Natl. Acad. Sci. USA. 2005;102:2288–2292. doi: 10.1073/pnas.0407595102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang X.Y., Parkinson J.A., Weishaupl M., Gould R.O., Paisey S.J., Park H.S., Hunter T.M., Blindauer C.A., Parsons S., Sadler P.J. Structure and dynamics of metallomacrocycles: Recognition of zinc xylyl-bicyclam by an HIV coreceptor. J. Am. Chem. Soc. 2002;124:9105–9112. doi: 10.1021/ja0260723. [DOI] [PubMed] [Google Scholar]
- 32.Liang X.Y., Weishaupl M., Parkinson J.A., Parsons S., McGregor P.A., Sadler P.J. Selective recognition of configurational substates of zinc cyclam by carboxylates: Implications for the design and mechanism of action of anti-HIV agents. Chem.-A Eur. J. 2003;9:4709–4717. doi: 10.1002/chem.200304808. [DOI] [PubMed] [Google Scholar]
- 33.Valks G.C., McRobbie G., Lewis E.A., Hubin T.J., Hunter T.M., Sadler P.J., Pannecouque C., De Clercq E., Archibald S.J. Configurationally Restricted Bismacrocyclic CXCR4 Receptor Antagonists. J. Med. Chem. 2006;49:6162–6165. doi: 10.1021/jm0607810. [DOI] [PubMed] [Google Scholar]
- 34.Khan A., Nicholson G., Greenman J., Madden L., McRobbie G., Pannecouque C., De Clercq E., Ullom R., Maples D.L., Maples R.D., et al. Binding Optimization through Coordination Chemistry: CXCR4 Chemokine Receptor Antagonists from Ultrarigid Metal Complexes. J. Am. Chem. Soc. 2009;131:3416–3417. doi: 10.1021/ja807921k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cain A.N., Freeman T.N.C., Roewe K.D., Cockriel D.L., Hasley T.R., Maples R.D., Allbritton E.M.A., D’Huys T., van Loy T., Burke B.P., et al. Acetate as a model for aspartate-based CXCR4 chemokine receptor binding of cobalt and nickel complexes of cross-bridged tetraazamacrocycles. Dalton Trans. 2019;48:2785–2801. doi: 10.1039/C8DT04728F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith R., Huskens D., Daelemans D., Mewis R.E., Garcia C.D., Cain A.N., Freeman T.N.C., Pannecouque C., De Clercq E., Schols D., et al. CXCR4 chemokine receptor antagonists: Nickel(II) complexes of configurationally restricted macrocycles. Dalton Trans. 2012;41:11369–11377. doi: 10.1039/c2dt31137b. [DOI] [PubMed] [Google Scholar]
- 37.Alzahrani S.O., McRobbie G., Khan A., D’Huys T., Van Loy T., Walker A.N., Renard I., Hubin T.J., Schols D., Burke B.P., et al. Trans-IV restriction: A new configuration for metal bis-cyclam complexes as potent CXCR4 inhibitors. Dalton Trans. 2024;53:5616–5623. doi: 10.1039/D3DT01729J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baghdadi N.E., Burke B.P., Alresheedi T., Nigam S., Saeed A., Almutairi F., Domarkas J., Khan A., Archibald S.J. Multivalency in CXCR4 chemokine receptor targeted iron oxide nanoparticles. Dalton Trans. 2021;50:1599–1603. doi: 10.1039/D0DT02626C. [DOI] [PubMed] [Google Scholar]
- 39.McRobbie G. Ph.D. Thesis. University of Hull; UK: 2009. Configurationally Restricted Bis-Tetraazamacrocyclic Complexes: Chemokine Receptor Antagonists. [Google Scholar]
- 40.D’Huys T. Ph.D. Thesis. KU Leuven (Katholieke Universiteit Leuven); Leuven, Belgium: 2018. Biological Characterization of Novel CXCR4 and CXCR7/ACKR3 Receptor Inhibitors. [Google Scholar]
- 41.Brandes S., Gros C., Denat F., Pullumbi P., Guilard R. New facile and convenient synthesis of bispolyazamacrocycles using Boc protection. Determination of geometric parameters of dinuclear copper(II) complexes using ESR spectroscopy and molecular mechanics calculations. Bull. Soc. Chim. Fr. 1996;133:65–73. [Google Scholar]
- 42.Bridger G.J., Skerlj R.T., Thornton D., Padmanabhan S., Martellucci S.A., Henson G.W., Abrams M.J., Yamamoto N., Devreese K., Pauwels R., et al. Synthesis and Structure-Activity-Relationships of Phenylenebis(Methylene)-Linked Bis-Tetraazamacrocycles That Inhibit HIV Replication - Effects of Macrocyclic Ring Size and Substituents on the Aromatic Linker. J. Med. Chem. 1995;38:366–378. doi: 10.1021/jm00002a019. [DOI] [PubMed] [Google Scholar]
- 43.McRobbie G., Valks G.C., Empson C.J., Khan A., Silversides J.D., Pannecouque C., De Clercq E., Fiddy S.G., Bridgeman A.J., Young N.A., et al. Probing key coordination interactions: Configurationally restricted metal activated CXCR4 antagonists. Dalton Trans. 1039 doi: 10.1039/b705800d. [DOI] [PubMed] [Google Scholar]
- 44.Weisman G.R., Ho S.C.H., Johnson V. Tetracyclic tetraamines by glyoxal-macrocyclic tetraamine condensation. Tetrahedron Lett. 1980;21:335–338. doi: 10.1016/S0040-4039(01)85466-6. [DOI] [Google Scholar]
- 45.Le Baccon M., Chuburu F., Toupet L., Handel H., Soibinet M., Dechamps-Olivier I., Barbier J.-P., Aplincourt M. Bis-aminals: Efficient tools for bis-macrocycle synthesis. New J. Chem. 2001;25:1168–1174. doi: 10.1039/b103995b. [DOI] [Google Scholar]
- 46.Björndal A., Deng H., Jansson M., Fiore J.R., Colognesi C., Karlsson A., Albert J., Scarlatti G., Littman D.R., Fenyö E.M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schols D., Esté José A., Cabrera C., De Clercq E. T-Cell-Line-Tropic Human Immunodeficiency Virus Type 1 That Is Made Resistant to Stromal Cell-Derived Factor 1α Contains Mutations in the Envelope gp120 but Does Not Show a Switch in Coreceptor Use. J. Virol. 1998;72:4032–4037. doi: 10.1128/JVI.72.5.4032-4037.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Princen K., Hatse S., Vermeire K., De Clercq E., Schols D. Establishment of a novel CCR5 and CXCR4 expressing CD4+ cell line which is highly sensitive to HIV and suitable for high-throughput evaluation of CCR5 and CXCR4 antagonists. Retrovirology. 2004;1:2. doi: 10.1186/1742-4690-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montefiori D.C. Measuring HIV Neutralization in a Luciferase Reporter Gene Assay. In: Prasad V.R., Kalpana G.V., editors. Methods in Molecular Biology. Volume 485. 2009. pp. 395–405. [DOI] [PubMed] [Google Scholar]
- 50.X-AREA v 1.64. STOE & Cie GmbH; Darmstadt, Germany: 2012. [Google Scholar]
- 51.Sheldrick G.M. A short history of SHELX. Acta Crystallogr. Sect. A: Found. Crystallogr. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 52.Gaussian, Inc.; Wallingford CT, USA: 2009. [Google Scholar]
- 53.Zhao Y., Truhlar D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 2008;120:215–241. doi: 10.1007/s00214-007-0310-x. [DOI] [Google Scholar]
- 54.Kowallick R., Neuburger M., Zehnder M., Kaden T.A. Metal complexes with macrocyclic ligands.45. Axial coordination tendency in reinforced tetraazamacrocyclic complexes. Helv. Chim. Acta. 1997;80:948–959. doi: 10.1002/hlca.19970800326. [DOI] [Google Scholar]
- 55.Silversides J.D., Allan C.C., Archibald S.J. Copper (II) cyclam-based complexes for radiopharmaceutical applications: Synthesis and structural analysis. Dalton Trans. 2007:971–978. doi: 10.1039/b615329a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maples R.D., Cain A.N., Burke B.P., Silversides J.D., Mewis R.E., D’Huys T., Schols D., Linder D.P., Archibald S.J., Hubin T.J. Aspartate-Based CXCR4 Chemokine Receptor Binding of Cross-Bridged Tetraazamacrocyclic Copper(II) and Zinc(II) Complexes. Chem. -A Eur. J. 2016;22:12916–12930. doi: 10.1002/chem.201601468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schols D., Este J.A., Henson G., DeClercq E. Bicyclams, a class of potent anti-HIV agents, are targeted at the HIV coreceptor Fusin/CXCR-4. Antivir. Res. 1997;35:147–156. doi: 10.1016/S0166-3542(97)00025-9. [DOI] [PubMed] [Google Scholar]
- 58.Schols D., Struyf S., VanDamme J., Este J.A., Henson G., DeClercq E. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J. Exp. Med. 1997;186:1383–1388. doi: 10.1084/jem.186.8.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKnight A., Wilkinson D., Simmons G., Talbot S., Picard L., Ahuja M., Marsh M., Hoxie J.A., Clapham P.R. Inhibition of human immunodeficiency virus fusion by a monoclonal antibody to a coreceptor (CXCR4) is both cell type and virus strain dependent. J. Virol. 1997;71:1692–1696. doi: 10.1128/jvi.71.2.1692-1696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carnec X., Quan L., Olson W.C., Hazan U., Dragic T. Anti-CXCR4 monoclonal antibodies recognizing overlapping epitopes differ significantly in their ability to inhibit entry of human immunodeficiency virus type 1. J. Virol. 2005;79:1930–1933. doi: 10.1128/JVI.79.3.1930-1933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hatse S., Princen K., Liekens S., Vermeire K., De Clercq E., Schols D. Fluorescent CXCL12(AF647) as a novel probe for nonradioactive CXCL12/CXCR4 cellular interaction studies. Cytom. Part A. 2004;61A:178–188. doi: 10.1002/cyto.a.20070. [DOI] [PubMed] [Google Scholar]
- 62.Schoofs G., Van Hout A., D’Huys T., Schols D., Van Loy T. A Flow Cytometry-based Assay to Identify Compounds That Disrupt Binding of Fluorescently-labeled CXC Chemokine Ligand 12 to CXC Chemokine Receptor 4. J. Vis. Exp. 2018 doi: 10.3791/57271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Princen K., Hatse S., Vermeire K., De Clercq E., Schols D. Evaluation of SDF-1/CXCR4-induced Ca2+ signaling by fluorometric imaging plate reader (FLIPR) and flow cytometry. Cytom. Part A. 2003;51A:35–45. doi: 10.1002/cyto.a.10008. [DOI] [PubMed] [Google Scholar]
- 64.Wiesmann N., Tremel W., Brieger J. Zinc oxide nanoparticles for therapeutic purposes in cancer medicine. J. Mater. Chem. B. 2020;8:4973–4989. doi: 10.1039/D0TB00739K. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and in the Supplementary Materials.