Abstract
Biomarkers for classifying and grading gliomas have been extensively explored, whereas populations in public databases were mostly Western/European. Based on public databases cannot accurately represent Chinese population. To identify molecular characteristics associated with clinical outcomes of lower-grade glioma (LGG) and glioblastoma (GBM) in the Chinese population, we performed whole-exome sequencing (WES) in 16 LGG and 35 GBM tumor tissues. TP53 (36/51), TERT (31/51), ATRX (16/51), EFGLAM (14/51), and IDH1 (13/51) were the most common genes harboring mutations. IDH1 mutation (c.G395A; p.R132H) was significantly enriched in LGG, whereas PCDHGA10 mutation (c.A265G; p.I89V) in GBM. IDH1-wildtype and PCDHGA10 mutation were significantly related to poor prognosis. IDH1 is an important biomarker in gliomas, whereas PCDHGA10 mutation has not been reported to correlate with gliomas. Different copy number variations (CNVs) and oncogenic signaling pathways were identified between LGG and GBM. Differential genomic landscapes between LGG and GBM were revealed in the Chinese population, and PCDHGA10, for the first time, was identified as the prognostic factor of gliomas. Our results might provide a basis for molecular classification and identification of diagnostic biomarkers and even potential therapeutic targets for gliomas.
Introduction
Gliomas are the most common malignant brain tumors in the central nervous system (CNS) [1]. Gliomas are classified into grades I to IV by the World Health Organization (WHO), mainly based on histology and malignancy [2]. Recently, molecular characteristics have been added as an important criterion in the revised classification system [3, 4]. The traditional treatment for glioma includes surgical resection, radiotherapy, and chemotherapy based on temozolomide (TMZ) [5]. These treatment leads to a better prognosis, especially grades II and III (LGG, with median survival time of more than 7 years) [6]. However, glioblastoma (GBM, WHO IV) still has poor prognosis (with the 5-year survival rate of 5.8%) [7].
Isocitrate dehydrogenase (IDH) mutations are one of the most critical molecular markers affecting the diagnosis, prognosis and treatment of gliomas. In gliomas, IDH1 R132H is the most common mutation and the mutation is associated with slower progression and better prognosis. IDH1 mutations occur in over 70% of LGG and GBM that progress from LGG and IDH2 mutation occur in less than 5% in gliomas [8]. 2021 WHO classification systems of CNS includes three types of gliomas: IDH-mutant astrocytoma, IDH-mutant and 1p/19q-codeleted oligodendroglioma and IDH-wildtype GBM [4]. More and more molecular markers have been proven to play a crucial role in the classification, grading, prognosis, and treatment of gliomas. For example, IDH-wildtype astrocytic glioma carrying TERT promoter (TERTp) mutation, EGFR amplification, as well as gain-of-chromosome 7 and loss-of-chromosome 10 (+7/-10) are classified as GBM [4]. Although many studies have revealed the genomic landscape of gliomas based on Western/European populations predominantly, comparative genomic characteristics of LGG and GBM are yet to be displayed in Chinese patients [9–11]. Considering the impact of genetic backgrounds on the prognosis of gliomas and a lack of comparative genomics of LGG and GBM in the Chinese population, further comparison of the genomic characteristics between LGG and GBM in the Chinese population is essential. Here, we aim to analyze and compare genomic characteristics of 16 LGG and 35 GBM cases to explore both shared and grade-specific genomics of gliomas in Chinese patients, and thus offering new insight into potential molecular signatures and treatment targets.
Materials and methods
Selection of patients with gliomas and sample collection
Tumor tissues were obtained from 51 patients with primary gliomas from January 6, 2019 to July 1, 2020 in Liaocheng People’s Hospital and normal blood samples data was obtained from another study of our group [12]. These patients were histologically diagnosed with gliomas and underwent surgical resection. According to histopathological diagnosis, tumors were classified as grades II to IV. This study was approved by the medical ethics committee of Liaocheng People’s Hospital (No: 2019203, 6 January 2019) and strictly followed the guidelines of the Declaration of Helsinki. All involved patients had signed informed consent for participating in this study. Genomic DNA (gDNA) was extracted from fresh frozen tissues and peripheral blood samples using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The quality and quantity of gDNA was analyzed by NanoDrop 1000 (Thermo Scientific, Wilmington, USA) and Qubit 3.0 Fluorometer (Life Technologies, Carlsbad, USA).
Sanger sequencing and whole-exome sequencing
IDH1 (codon R132), IDH2 (codon R172) and TERT promoter (TERTp, C228T and C250T) mutations were detected by Sanger sequencing, using established primer (Table 1) [13, 14]. The primer sequences used for validation of PCDHGA10 (codon I89) were listed in Table 1. DNA fragments were sequenced by Sangon Biotech (Sangon, Shanghai, China).
Table 1. Primer sequence of 4 genes.
| Gene | Primer Sequence (5ʹ→3ʹ) |
|---|---|
| IDH1 | Forward: CTCCTGATGAGAAGAGGGTTG |
| Reverse: TGGAAATTTCTGGGCCATG | |
| IDH2 | Forward: TGGAACTATCCGGAACATCC |
| Reverse: AGTCTGTGGCCTTGTACTGC | |
| TERTp | Forward: AGTGGATTCGCGGGCACAGA |
| Reverse: GCAGCGCTGCCTGAAACTCG | |
| PCDHGA10 | Forward: AGGCTCTTTCGTGGGCAACATC |
| Reverse: GCATTCCCGCAGCGACATTTTC |
The whole exome was captured by SureSelect Human All Exon V6 Probes (Agilent, Santa Clara, USA) according to the manufacturer’s protocol. The qualified libraries were sequenced with Nextseq CN500 platform (Illumina, San Diego, USA) for 2×76 bp paired-end sequencing.
Variant calling and annotation
Sequencing reads were mapped to human reference genome GRCh37/hg19 using Burrows-Wheeler Aligner (BWA) [15]. The duplicated reads were marked with Picard (http://broadinstitute.github.io/picard/) and realigned with Genome Analysis Toolkit (GATK) [16, 17]. All variants were annotated using ANNOVAR [18]. Variants were screened with 1000 Genomes Project (1KGP) [19], Exome Sequencing Project (ESP6500) (http://evs.gs.washington.edu/EVS/), The Exome Aggregation Consortium (ExAC) [20] and Catalogues of Somatic Mutations In Cancer (COSMIC) databases [21]. Variants were filtered out as follows: (1) variants presented in the 1KGP, ESP6500 or ExAC with a minor allele frequency of >1%; (2) base quality value of <20; (3) mutation reads depth of <10; (4) variant allele frequency (VAF) of < 5%; (5) synonymous variants.
CNVs analysis
CNVkit was used to analysis CNVs with default parameter settings [22]. Peripheral blood-derived germline DNA samples of 30 healthy subjects were used as normal references for calculating tumor CNVs, which were generated using the same sequencing method and analysis strategy. CNVs were reported if the log2 (CN) was above 1.5 or below 0.5. Then, GISTIC2.0 was used to identify regions of significantly recurring gains or losses [23]. q-values were calculated from p-values of respective genomic regions determined by permutation test. Regions with a q < 0.25 were considered as significantly recurring gains or losses.
Driver genes analysis
Cancer Gene Census (https://cancer.sanger.ac.uk/census/, CGC), Integrative Onco Genomics (https://www.intogen.org/search), BertVogelstein125 [24], SMG127 [25], and Comprehensive435 [26] were to identify known driver genes. Genes with high alteration frequencies identified at least one of the five data sources were considered as candidate driver genes [27].
Enrichment analysis and associated cancer pathway analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.kegg.jp/kegg/) and Gene Ontology (GO) database were used for signaling pathway analysis and functional enrichment analysis, respectively [28, 29]. Ten oncogenic signaling pathways were analyzed as previously described, including TP53, WNT, MYC, HIPPO, PI3K, NOTCH, Cell Cycle, NRF2, TGFβ, and RTK/RAS pathways [30]. Patients having one or more mutations or CNV involved genes in these pathways were considered to have altered pathways [31]. Highlighted pathways were visualized by PathwayMapper tool (http://pathwaymapper.org) [32].
Statistical analysis
Genetic mutations between LGG and GBM were compared with Fisher’s exact tests or Chi-square tests. 2-tailed p < 0.05 was considered significant. Kaplan-Meier estimate was illustrated for survival analysis, and log-rank test was used to determine the difference between two groups. The statistical analysis was performed using SPSS (Version23.0, Chicago, USA) or R v3.6.1 software.
Results
Clinical characteristics of patients with gliomas
Totally, 51 patients with gliomas, including 16 LGG and 35 GBM, were enrolled in this investigation (Table 2 and S1 Table). The median age was 51.41±13.39 years with a wide range (24–73 years). The majority of patients were male (n = 31, 60.78%). Most tumors were located in the frontal lobe (n = 17, 33.33%), temporal lobe (n = 17, 33.33%), followed by multiple regions (n = 15, 29.41%), parietal lobes (n = 1, 1.96%), and cerebellum (n = 1, 1.96%). IDH1 mutation (c.G395A; p.R132H) was more common in LGG (n = 7, 43.75%) than GBM (n = 6, 17.14%). TERTp mutations (C228T and C250T) were more common in GBM (n = 22, 62.86%) than LGG (n = 9, 56.25%). Forty-seven out of 51 patients had available follow-up data. The median overall survival (OS) time of LGG and GBM cohort were 30.4 and 13.0 months, respectively.
Table 2. Clinical and genetic characteristics of 51 patients with gliomas.
| Features | All (n = 51) | LGG (n = 16) | GBM (n = 35) |
|---|---|---|---|
| Sex | |||
| Male | 60.78% (31/51) | 50.00% (8/16) | 65.71% (23/35) |
| Female | 39.22% (20/51) | 50.00% (8/16) | 34.29% (12/35) |
| Age | |||
| Median (Range) | 51.41 (24–73) | 47.38 (24–64) | 53.26 (24–73) |
| Tumor location | |||
| Frontal lobe | 33.33% (17/51) | 37.50% (6/16) | 31.43% (11/35) |
| Temporal lobe | 33.33% (17/51) | 25.00% (4/16) | 37.14% (13/35) |
| Two or more lobes | 29.41% (15/51) | 37.50% (6/16) | 25.71% (9/35) |
| Other | 3.92% (2/51) | - | 5.71% (2/35) |
| IDH1 | |||
| Mutation | 25.49% (13/51) | 43.75% (7/16) | 17.14% (6/35) |
| Wild-type | 74.51% (38/51) (38/51) | 56.25% (9/16) | 82.86% (29/35) |
| TERTp | |||
| Mutation | 60.78% (31/51) | 56.25% (9/16) | 62.86% (22/35) |
| Wild-type | 39.22% (20/51) | 43.75% (7/16) | 37.14% (13/35) |
| Survival | |||
| Yes | 23.40% (11/47) | 60.00% (9/15) | 6.25% (2/32) |
| No | 76.60% (36/47) | 40.00% (6/15) | 93.75% (30/32) |
Mutational landscape of LGG and GBM patients
The average sequencing depth was 76.57× for 51 gliomas samples. 85.75% of target bases had a coverage of at least 30×. A total number of 5699 exonic mutations were detected in all patients, including 5330 missense mutations, 96 nonsense mutations, 57 frameshift Insertion–deletions (indels), 179 in-frame indels, and 37 unknown-function mutations (S1A Fig). Mutations with an unknown-function were removed, resulting in a median of 118 (range 96–134) variants, including 95.80% single nucleotide variants (SNV) and 4.20% indel mutations (S1B Fig). The major point mutations were C>T and T>C (S1C Fig).
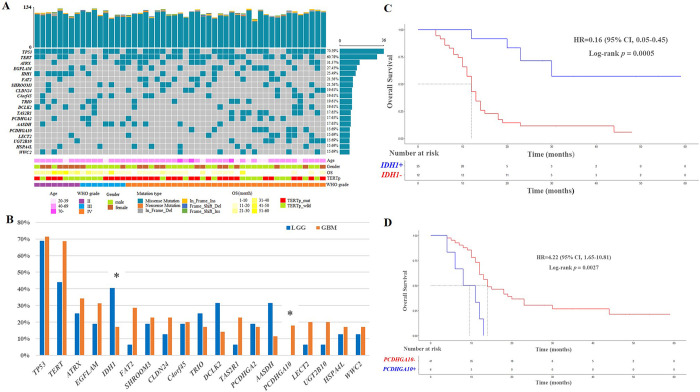
LGG and GBM shared 53 mutated genes, while 1800 and 3846 uniquely mutations were identified in LGG and GBM, respectively (S1D Fig). To demonstrate potential grade differences, those genes with a mutational frequency of > 15% were focused on. The top 5 mutated genes were TP53 (n = 36, 70.59%), TERT (n = 31, 60.78%), ATRX (n = 16, 31.37%), EFGLAM (n = 14, 27.45%), and IDH1 (n = 13, 25.49%) (Fig 1A). TP53, ATRX and IDH1 were the candidate driver gene identified in all of the five data sources (S2 Table). The mutational rates were different between LGG and GBM. IDH1, DCLK2, and AASDH were enriched in LGG, while FAT2, TAS2R1, PCDHGA10, LECT2, and UGT2B10 were enriched in GBM (Fig 1B). Mutations of IDH1 and PCDHGA10 were significantly different between LGG and GBM (p < 0.05, Fig 1B). IDH1 mutation (c.G395A; p.R132H) was enriched in LGG (43.75% vs. 17.14%), whereas PCDHGA10 mutation (c.A265G; p.I89V) in GBM (22.86% vs. 0%), which were consistent with Sanger sequencing (S2 Fig). Next, predictive values of IDH1 and PCDHGA10 mutations were analyzed. Patients with wild-type IDH1 had shorter OS compared to those with mutated IDH1 (14.29 months vs. 31.00 months, HR = 0.16; p = 0.0005; Fig 1C). Patients with mutated PCDHGA10 had shorter OS compared to those with wild-type PCDHGA10 (9.00 months vs. 19.95 months, HR = 4.22; p = 0.0027; Fig 1D).
Fig 1. Recurrent genetic alterations in 51 gliomas.
(A) Mutational landscape of gliomas. (B) Prevalence comparison of mutations in LGG and GBM (Chi-square test; *, p < 0.05). (C) OS of patients with IDH1 mutation. (D) OS of patients with PCDHGA10 mutation.
Comparing with the public database TCGA and cBioPortal, significant differences were observed in mutational frequencies of various genes, such as TERT, EGFLAM, IDH1, SHROOM3, and DCLK2 (S3 Fig).
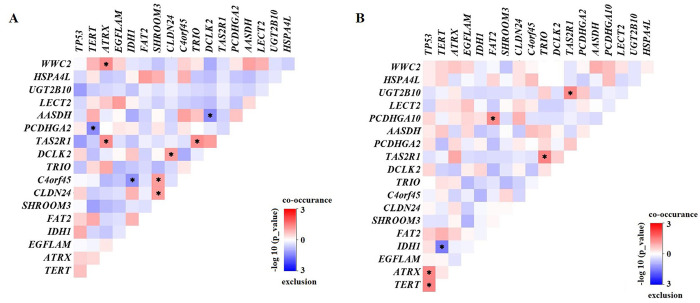
Then, mutual coexistence and exclusion of 19 genes were analyzed by correlation analysis. Interacted gene pairs were significantly different between LGG and GBM. For example, interactions of ATRX with WWC2 and TAS2R1 were coexisted in LGG (Fig 2A). Interactions of TP53 with ATRX and TERT, as well as interaction of PCDHGA10 and FAT2 were coexisted in GBM (Fig 2B).
Fig 2.
Concurrent and mutually exclusive somatic mutation patterns of mutated genes in LGG (A) and GBM (B). Significance was calculated using Fisher’s exact test, *p < 0.05.
CNVs in LGG and GBM
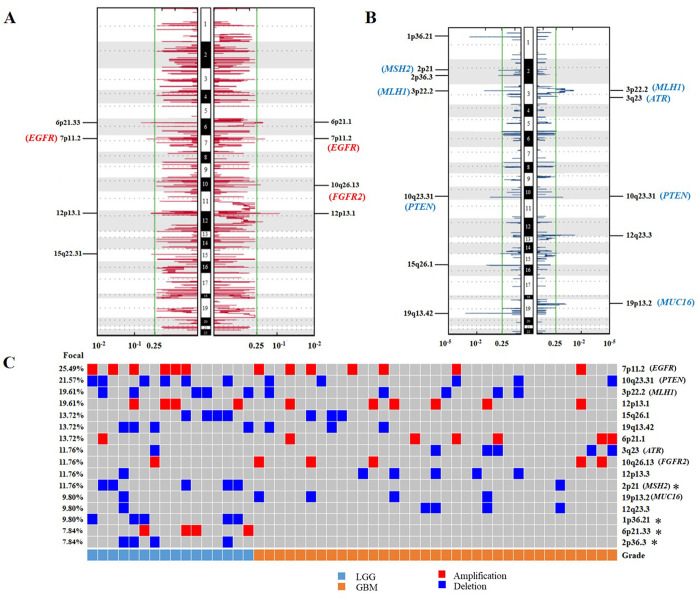
CNVs were analyzed by GISTIC2.0 in the LGG and GBM groups. We identified 6 significantly recurrent amplification regions (q < 0.25, Fig 3A) and 10 significantly recurrent deletion regions (q < 0.25, Fig 3B). Gain of chromosome 7 and loss of chromosome 10 were more common in gliomas (Fig 3C). In addition, according to potential driver genes in five data sources, we identified seven oncogenes or tumor-suppressor genes, including EGFR (7p11.2, 25.49%), PTEN (10q23.31, 21.57%), MLH1 (3p22.2, 19.61%), ATR (3q23, 11.76%), FGFR2 (10q26.13, 11.76%), MSH2 (2p21, 11.76%), and MUC16 (19p13.2, 9.50%) (S2 Table and Fig 3C). The LGG group had deletions in 2p21 (31.25% vs. 2.86%), 1p36.21 (31.25% vs. 0%), and 2p36.3 (25% vs. 0%), whereas amplification in 6p21.33 (25% vs. 0%) (p < 0.05, Fisher’s exact test, Fig 3C).
Fig 3. Differential CNVs between LGG and GBM.
(A) Amplification regions in LGG (left) and GBM (right). (B) Deletion regions in LGG (left) and GBM (right). (C) Percentage of CNVs located in different chromosomal regions in LGG and GBM. The green line indicates the cut-off of significance (q = 0.25). Significance was calculated using Fisher’s exact test, *p < 0.05.
Enrichment analysis and oncogenic signaling pathways
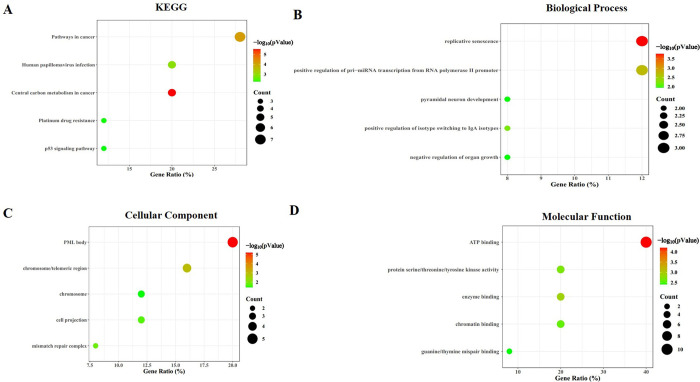
To understand the biological distributions of the frequently altered genes, we performed KEGG and GO analyses. KEGG analysis revealed that the frequently altered genes were highly enriched in “pathway in cancer” (Fig 4A). GO analysis indicated that mutated genes were mainly involved in replicative senescence and positive regulation of pri−miRNA transcription from RNA polymerase II promoter in biological processes (Fig 4B); PML body and chromosome/telomeric region in cellular components (Fig 4C) and ATP binding in molecular functions (Fig 4D).
Fig 4. The significantly enriched KEGG pathways and GO annotations of frequently altered genes in gliomas cases.
(A) KEGG pathway. (B) Biological processes. (C) Cellular components. (D) Molecular functions.
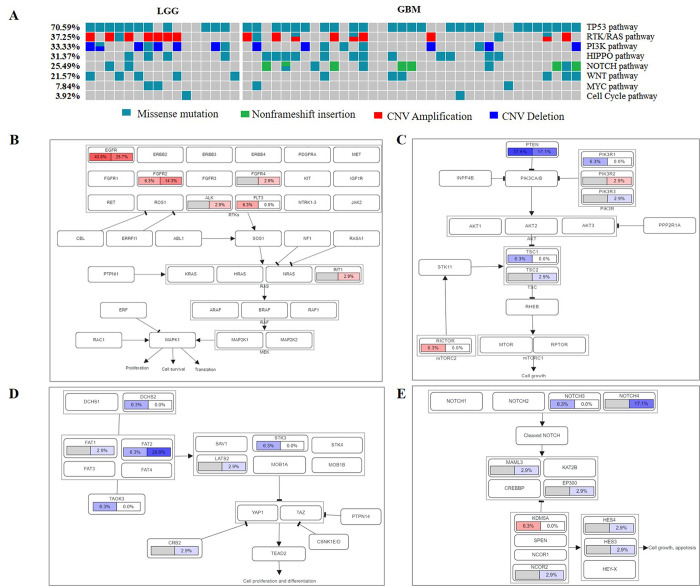
To compare oncogenic signaling pathways by genetic variations, we analyzed mutations and CNVs in LGG and GBM. Notably, TP53 (70.59%), RTK/RAS (37.25%), PI3K (33.33%), HIPPO (31.37%), and NOTCH (25.49%) pathways were frequently altered in gliomas. However, NRF2 and TGFB related genes were not found (Fig 5A). The most frequently mutated gene in the TP53 signaling pathway was TP53. There were differences in oncogenic signaling pathways between LGG and GBM. Interestingly, alterations in RTK/RAS (50% LGG vs. 31.43% GBM) and PI3K (50% LGG vs. 25.71% GBM) pathways preferentially occurred in LGG. Notably, alterations in HIPPO (34.29% GBM vs. 25% LGG) and NOTCH (28.57% GBM vs. 12.5% LGG) pathways were more common in GBM. RTK/RAS pathway was mainly caused by oncogene EGFR (43.8% LGG vs. 25.7% GBM, Fig 5B), while PI3K pathway activation was attributed to tumor suppressor gene PTEN (37.5% LGG vs. 17.1% GBM, Fig 5C). HIPPO pathway was modified by tumor suppressor gene FAT2 (28.6% GBM vs. 6.3% LGG, Fig 5D), while NOTCH pathway was regulated by tumor suppressor gene NOTCH4 (17.1% GBM vs. 0% LGG, Fig 5E).
Fig 5. The frequencies of oncogenic signaling pathways altered in patients with gliomas.
(A) The frequencies of oncogenic signaling pathways altered in gliomas. (B) RTK/RAS pathway. (C) PI3K pathway. (D) HIPPO pathway. (E) NOTCH pathway. Pathways were labeled with LGG on the left whereas GBM on the right. Red represents an oncogene while blue represents a tumor suppressor gene.
Discussion
In this study, we performed WES to compare molecular characteristics between 16 LGG with 35 GBM patients in the Chinese population. We have highlighted significant differences in genetic landscapes between LGG and GBM.
The most frequently mutated genes were TP53, TERT, ATRX, EFGLAM, and IDH1 in 51 Chinese patients with gliomas. Except for EFGLAM, the other genes were biomarkers for gliomas [9, 33–35]. EGFLAM was related to poor prognosis in GBM as previously described [36]. Notably, mutations of IDH1 and PCDHGA10 were significantly different between LGG and GBM. IDH1 mutation (c.G395A; p.R132H) was enriched in LGG and patients with IDH1 mutation had better long-term survival. IDH mutations were identified in LGG and secondary GBM, as an important biomarker for longer OS in LGG [37]. By contrast, PCDHGA10 mutation (c.A265G; p.I89V) was significantly enriched in GBM but not in LGG (22.86% vs. 0%). In addition, PCDHGA10 mutation correlated with shorter OS time. PCDHGA10 might be a potential biomarker for prognosis in gliomas. PCDHGA10 mutation (c.A265G; p.I89V) has not been reported to be associated with gliomas, which has been reported in other tumors (bladder cancer, gastric adenomas, and gastrointestinal stromal tumors) [38–40]. PCDHGA10 other mutations (such as c.G1765A, p.G589S; c.A395G/T, D132G/V) have been reported in astrocytoma grade IV [41, 42]. PCDHGA10 mutational frequency was low in the TCGA GBM (0.8%, 3/374) and cBioPortal GBM (1.01%, 4/397). PCDHGA10 is a member of Pcdh-γ gene clusters. There has been accumulating evidence that members of PCDH family act as tumor suppressor genes in several types of cancer [43–47]. For example, knockdown of PCDHGA9 promoted migration and invasion of gastric cancer cells, while PCDHGA9 overexpression inhibited proliferation and metastasis of gastric cancer cells [43]. PCDHGA10 is significantly upregulated in lung squamous cell carcinoma (LUSC) and a high level of PCDHGA10 expression is associated with a poorer prognosis. PCDHGA10 might be a potential molecular marker in LUSC [48]. However, the potential role of PCDHGA10 in tumorigenesis of gliomas has not been reported. Therefore, further studies are required to investigate the pathogenic functions of PCDHGA10 in GBM, especially in Chinese population.
Using CNVs analysis, 6 amplification regions and 10 deletion regions were defined as significantly recurrent CNVs. Oncogenes EGFR, FGFR2 and MUC16, as well as tumor-suppressor genes PTEN, MLH1, ATR, and MSH2 were identified according to driver genes in five data sources. EGFR amplification and PTEN deletion have values in diagnosis, response to therapy and prognosis in molecular subgroups of gliomas [49]. EGFR amplification occurred in 40–60% of GBM, which may serve as an attractive therapeutic target in GBM [50]. Approximately 70% of GBM had PTEN loss [51], however, PTEN loss as a prognostic factor has not been verified and remains controversial [52]. EGFR and PTEN could alter receptor tyrosine kinase (RTK)/PI3K/AKT/mTOR pathway and promote tumor progression [53]. In our study, LGG had deletions in 1p36.21, 2p21 and 2p36.3, whereas amplification in 6p21.33. Notably, 1p36.21 deletion was related to poor prognosis in astrocytoma and neuroblastoma [54, 55].
Enrichment analysis revealed that the frequently altered genes were involved in complex pathways in cancer (e.g., TP53, RTK/RAS and PI3K pathway), biological processes (e.g., replicative senescence and positive regulation of pri−miRNA transcription), cellular components (e.g., ML body and chromosome/telomeric region), and molecular functions (e.g., ATP binding and enzyme binding). In patients with gliomas, the most common mutations were harbored in P53 (70.59%), RTK/RAS (37.25%), PI3K (33.33%), HIPPO (31.37%) and NOTCH (25.49%) pathways. 78% of GBM had alterations in the p53 pathway [51], which might promote LGG progression to GBM [56]. Dysfunctional RTK/RAS pathway was attributed to EGFR, whereas PI3K pathway attributed to PTEN. Mutated EGFR and PTEN activate PI3K pathway, leading to activation of downstream AKT/mTOR cascade [57]. AKT/mTOR pathway promotes proliferation, survival and migration [58]. Dysregulated HIPPO pathway was mainly caused by mutated FAT2. FAT2 may be involved in cancer aggressiveness, which remains inconclusive [59]. Pathogenic NOTCH pathway may regulate tumor initiation, progression, and recurrence of gliomas. However, the role of NOTCH pathway in gliomas development is debated [60].
In our study, genomic profiles of gliomas were similar to public databases like TCGA and cBioPortal, such as TP53, IDH1, ATRX, EGFR and PTEN. However, most of gene mutation frequencies were significant differences between our cohort and public databases. Due to different human lineages have genetic heterogeneity, which was shaped by many factors, including evolutionary history, environmental exposures, and lifestyle practices. In addition, the sample size of our cohort was relatively small, which may cause uncertainty frequency counting of gene alterations.
Several limitations should be mentioned in this study. First, the sample size of this retrospective cohort was relatively small. Furthermore, this study was based on single omics, which might lack adequate validation information. In addition, PCDHGA10 might be related to the prognosis of GBM, however, additional experiments are required to explore pathologic functions of PCDHGA10 in gliomas. Hence, we should conduct multi-center studies with larger sample sizes to validate our findings with multi-omics platforms.
Conclusion
This study reveals comprehensive genomic characteristics of LGG and GBM in Chinese patients, which may provide a better understanding of differential molecular signatures between LGG and GBM. In addition, PCDHGA10 mutation might be a novel biomarker for poor prognosis in GBM. The discovery of unique molecular biomarkers could contribute to glioma classification, help predict prognosis and provide therapeutic options. These results need to be confirmed by comprehensive studies with larger sample sizes, especially for a prognostic role of PCDGHA10 in GBM.
Supporting information
(A) Number of each type of mutation. (B) Number of SNV and Indels in all mutations. (C) Distribution of point mutation types in SNV. (D) The Venn diagram showed the number of co-mutated and uniquely-mutated genes in LGG and GBM patients.
(TIF)
(A) Hot spots of mutations in the IDH1 gene. (B) Sanger sequencing results of the novel mutation of the IDH1 gene. (C) Hot spots of mutations in the PCDHGA10 gene. (D) Sanger sequencing results of the novel mutation of the PCDHGA10 gene.
(TIF)
Comparison of mutation frequencies between our study and cBioPortal database (A) and TCGA database (B).
(TIF)
(XLSX)
(XLSX)
Acknowledgments
The authors thank all patients and their families for participating in this study.
Data Availability
The data reported in this study are available in the CNGB Nucleotide Sequence Archive (CNSA: https://db.cngb.org/cnsa; accession number CNP0002254 and CNP0002252).
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018;392(10145):432–46. Epub 2018/08/01. doi: 10.1016/S0140-6736(18)30990-5 . [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. Epub 2007/07/10. doi: 10.1007/s00401-007-0243-4 ; PubMed Central PMCID: PMC1929165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louis DN, Perry A, Reifenberger G, Deimling Av, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–20. doi: 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 4.Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8):1231–51. Epub 2021/06/30. doi: 10.1093/neuonc/noab106 ; PubMed Central PMCID: PMC8328013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stupp R, Brada M, van den Bent MJ, Tonn JC, Pentheroudakis G. High-grade glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25 Suppl 3:iii93–101. Epub 2014/05/02. doi: 10.1093/annonc/mdu050 . [DOI] [PubMed] [Google Scholar]
- 6.Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47(5):458–68. Epub 2015/04/08. doi: 10.1038/ng.3273 . [DOI] [PubMed] [Google Scholar]
- 7.Tan AC, Ashley DM, López GY, Malinzak M, Friedman HS, Khasraw M. Management of glioblastoma: State of the art and future directions. CA Cancer J Clin. 2020;70(4):299–312. Epub 2020/06/02. doi: 10.3322/caac.21613 . [DOI] [PubMed] [Google Scholar]
- 8.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–73. Epub 2009/02/21. doi: 10.1056/NEJMoa0808710 ; PubMed Central PMCID: PMC2820383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng C, Wang J, Li M, Wang H, Lou F, Cao S, et al. Comprehensive molecular characterization of Chinese patients with glioma by extensive next-generation sequencing panel analysis. Cancer Manag Res. 2021;13:3573–88. Epub 2021/05/07. doi: 10.2147/CMAR.S291681 ; PubMed Central PMCID: PMC8092857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Jin Y, Zou Q, Shi X, Wu Q, Lin Z, et al. Integrated genomic and transcriptomic analysis suggests KRT18 mutation and MTAP are key genetic alterations related to the prognosis between astrocytoma and glioblastoma. Ann Transl Med. 2021;9(8):713. Epub 2021/05/15. doi: 10.21037/atm-21-1317 ; PubMed Central PMCID: PMC8106028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin H, Yu Z, Tian T, Shen G, Chen W, Fan M, et al. Integrative genomic and transcriptomic analysis of primary malignant gliomas revealed different patterns between grades and somatic mutations related to glioblastoma prognosis. Front Mol Biosci. 2022;9:873042. Epub 2022/07/23. doi: 10.3389/fmolb.2022.873042 ; PubMed Central PMCID: PMC9294235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang D, Gu L, Zheng J, Zhang Q, Xu Q, Li R, et al. Germline VWF/MPRIP and somatoplasm FGA variants synergically confer susceptibility to non-traumatic osteonecrosis of the femoral head. Sci Rep. 2023;13(1):3112. Epub 2023/02/23. doi: 10.1038/s41598-023-30260-4 ; PubMed Central PMCID: PMC9946931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juratli TA, Kirsch M, Robel K, Soucek S, Geiger K, von Kummer R, et al. IDH mutations as an early and consistent marker in low-grade astrocytomas WHO grade II and their consecutive secondary high-grade gliomas. J Neurooncol. 2012;108(3):403–10. Epub 2012/03/14. doi: 10.1007/s11060-012-0844-1 . [DOI] [PubMed] [Google Scholar]
- 14.Pesenti C, Paganini L, Fontana L, Veniani E, Runza L, Ferrero S, et al. Mass spectrometry-based assay for the molecular diagnosis of glioma: concomitant detection of chromosome 1p/19q codeletion, and IDH1, IDH2, and TERT mutation status. Oncotarget. 2017;8(34):57134–48. Epub 2017/09/17. doi: 10.18632/oncotarget.19103 ; PubMed Central PMCID: PMC5593631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England). 2009;25(14):1754–60. Epub 2009/05/20. doi: 10.1093/bioinformatics/btp324 ; PubMed Central PMCID: PMC2705234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43(1110):11.0.1-.0.33. Epub 2014/11/29. doi: 10.1002/0471250953.bi1110s43 ; PubMed Central PMCID: PMC4243306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. Epub 2010/07/21. doi: 10.1101/gr.107524.110 ; PubMed Central PMCID: PMC2928508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. Epub 2010/07/06. doi: 10.1093/nar/gkq603 ; PubMed Central PMCID: PMC2938201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. Epub 2015/10/04. doi: 10.1038/nature15393 ; PubMed Central PMCID: PMC4750478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karczewski KJ, Weisburd B, Thomas B, Solomonson M, Ruderfer DM, Kavanagh D, et al. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res. 2017;45(D1):D840–d5. Epub 2016/12/03. doi: 10.1093/nar/gkw971 ; PubMed Central PMCID: PMC5210650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47(D1):D941–d7. Epub 2018/10/30. doi: 10.1093/nar/gky1015 ; PubMed Central PMCID: PMC6323903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12(4):e1004873. Epub 2016/04/23. doi: 10.1371/journal.pcbi.1004873 ; PubMed Central PMCID: PMC4839673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. Epub 2011/04/30. doi: 10.1186/gb-2011-12-4-r41 ; PubMed Central PMCID: PMC3218867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–58. Epub 2013/03/30. doi: 10.1126/science.1235122 ; PubMed Central PMCID: PMC3749880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–9. Epub 2013/10/18. doi: 10.1038/nature12634 ; PubMed Central PMCID: PMC3927368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamborero D, Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Kandoth C, Reimand J, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci Rep. 2013;3:2650. Epub 2013/10/03. doi: 10.1038/srep02650 ; PubMed Central PMCID: PMC3788361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Guo Y, Cheng Z, Tian C, Chen Y, Chen R, et al. Whole-exome sequencing of rectal neuroendocrine tumors. Endocrine-related cancer. 2023;30(9). Epub 2023/01/17. doi: 10.1530/erc-22-0257 ; PubMed Central PMCID: PMC10450454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51(D1):D587–d92. Epub 2022/10/28. doi: 10.1093/nar/gkac963 ; PubMed Central PMCID: PMC9825424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43(Database issue):D1049–56. Epub 2014/11/28. doi: 10.1093/nar/gku1179 ; PubMed Central PMCID: PMC4383973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell. 2018;173(2):321–37.e10. Epub 2018/04/07. doi: 10.1016/j.cell.2018.03.035 ; PubMed Central PMCID: PMC6070353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Shan C, Wu S, Cheng B, Fan C, Cai L, et al. Genomic profiling identified novel prognostic biomarkers in Chinese midline glioma patients. Front Oncol. 2020;10:607429. Epub 2021/03/23. doi: 10.3389/fonc.2020.607429 ; PubMed Central PMCID: PMC7968371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bahceci I, Dogrusoz U, La KC, Babur Ö, Gao J, Schultz N. PathwayMapper: a collaborative visual web editor for cancer pathways and genomic data. Bioinformatics (Oxford, England). 2017;33(14):2238–40. Epub 2017/03/24. doi: 10.1093/bioinformatics/btx149 ; PubMed Central PMCID: PMC5859976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–63. Epub 2016/01/30. doi: 10.1016/j.cell.2015.12.028 ; PubMed Central PMCID: PMC4754110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–98. Epub 2015/06/11. doi: 10.1056/NEJMoa1402121 ; PubMed Central PMCID: PMC4530011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin H, Sa JK, Bae JS, Koo H, Jin S, Cho HJ, et al. Clinical targeted next-Generation sequencing panels for detection of somatic variants in gliomas. Cancer Res Treat. 2020;52(1):41–50. Epub 2019/05/18. doi: 10.4143/crt.2019.036 ; PubMed Central PMCID: PMC6962483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, Zhang J, Hong L, Zhou Y. EGFLAM correlates with cell proliferation, migration, invasion and poor prognosis in glioblastoma. Cancer Biomark. 2019;24(3):343–50. Epub 2019/03/05. doi: 10.3233/CBM-181740 ; PubMed Central PMCID: PMC6484271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabha N, Knobbe CB, Maganti M, Al Omar S, Bernstein M, Cairns R, et al. Analysis of IDH mutation, 1p/19q deletion, and PTEN loss delineates prognosis in clinical low-grade diffuse gliomas. Neuro Oncol. 2014;16(7):914–23. Epub 2014/01/29. doi: 10.1093/neuonc/not299 ; PubMed Central PMCID: PMC4057130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Every MJ, Dancik G, Paramesh V, Gurda GT, Meier DR, Cash SE, et al. Genomic case report of a low grade bladder tumor metastasis to lung. BMC urology. 2018;18(1):74. Epub 2018/09/05. doi: 10.1186/s12894-018-0386-8 ; PubMed Central PMCID: PMC6122771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim CH, Cho YK, Kim SW, Choi MG, Rhee JK, Chung YJ, et al. The chronological sequence of somatic mutations in early gastric carcinogenesis inferred from multiregion sequencing of gastric adenomas. Oncotarget. 2016;7(26):39758–67. Epub 2016/10/26. doi: 10.18632/oncotarget.9250 ; PubMed Central PMCID: PMC5129968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pang Y, Xie F, Cao H, Wang C, Zhu M, Liu X, et al. Mutational inactivation of mTORC1 repressor gene DEPDC5 in human gastrointestinal stromal tumors. Proc Natl Acad Sci U S A. 2019;116(45):22746–53. Epub 2019/10/23. doi: 10.1073/pnas.1914542116 ; PubMed Central PMCID: PMC6842588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45(10):1141–9. Epub 2013/08/07. doi: 10.1038/ng.2734 ; PubMed Central PMCID: PMC3799953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shlien A, Campbell BB, de Borja R, Alexandrov LB, Merico D, Wedge D, et al. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet. 2015;47(3):257–62. Epub 2015/02/03. doi: 10.1038/ng.3202 . [DOI] [PubMed] [Google Scholar]
- 43.Weng J, Xiao J, Mi Y, Fang X, Sun Y, Li S, et al. PCDHGA9 acts as a tumor suppressor to induce tumor cell apoptosis and autophagy and inhibit the EMT process in human gastric cancer. Cell Death Dis. 2018;9(2):27. Epub 2018/01/20. doi: 10.1038/s41419-017-0189-y ; PubMed Central PMCID: PMC5833845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Peng K, Xie R, Zheng J, Guo J, Wei R, et al. Protocadherin γ-A7 is down-regulated in colorectal cancer and associated with the prognosis in patients with wild-type KRAS. Hum Pathol. 2019;83:14–21. Epub 2018/08/20. doi: 10.1016/j.humpath.2018.08.007 . [DOI] [PubMed] [Google Scholar]
- 45.Jao TM, Fang WH, Ciou SC, Yu SL, Hung YL, Weng WT, et al. PCDH10 exerts tumor-suppressor functions through modulation of EGFR/AKT axis in colorectal cancer. Cancer Lett. 2021;499:290–300. Epub 2020/12/04. doi: 10.1016/j.canlet.2020.11.017 . [DOI] [PubMed] [Google Scholar]
- 46.Lin Y, Ge X, Zhang X, Wu Z, Liu K, Lin F, et al. Protocadherin-8 promotes invasion and metastasis via laminin subunit γ2 in gastric cancer. Cancer Sci. 2018;109(3):732–40. Epub 2018/01/13. doi: 10.1111/cas.13502 ; PubMed Central PMCID: PMC5834795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waha A, Güntner S, Huang TH, Yan PS, Arslan B, Pietsch T, et al. Epigenetic silencing of the protocadherin family member PCDH-gamma-A11 in astrocytomas. Neoplasia (New York, NY). 2005;7(3):193–9. Epub 2005/04/01. doi: 10.1593/neo.04490 ; PubMed Central PMCID: PMC1501138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song S, Lu R, Chen Y, Feng Y. PCDHGA10 as a potential biomarker of lung squamous cell carcinoma based on bioinformatics and experimental verification. Molecular biotechnology. 2024. Epub 2024/05/10. doi: 10.1007/s12033-024-01178-7 . [DOI] [PubMed] [Google Scholar]
- 49.Brito C, Azevedo A, Esteves S, Marques AR, Martins C, Costa I, et al. Clinical insights gained by refining the 2016 WHO classification of diffuse gliomas with: EGFR amplification, TERT mutations, PTEN deletion and MGMT methylation. BMC Cancer. 2019;19(1):968. Epub 2019/10/19. doi: 10.1186/s12885-019-6177-0 ; PubMed Central PMCID: PMC6798410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muñoz-Hidalgo L, San-Miguel T, Megías J, Monleón D, Navarro L, Roldán P, et al. Somatic copy number alterations are associated with EGFR amplification and shortened survival in patients with primary glioblastoma. Neoplasia (New York, NY). 2020;22(1):10–21. Epub 2019/11/22. doi: 10.1016/j.neo.2019.09.001 ; PubMed Central PMCID: PMC6864306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mao H, Lebrun DG, Yang J, Zhu VF, Li M. Deregulated signaling pathways in glioblastoma multiforme: molecular mechanisms and therapeutic targets. Cancer Invest. 2012;30(1):48–56. Epub 2012/01/13. doi: 10.3109/07357907.2011.630050 ; PubMed Central PMCID: PMC3799884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nikiforova MN, Hamilton RL. Molecular diagnostics of gliomas. Arch Pathol Lab Med. 2011;135(5):558–68. Epub 2011/04/30. doi: 10.5858/2010-0649-RAIR.1 . [DOI] [PubMed] [Google Scholar]
- 53.Ohgaki H, Kleihues P. Genetic alterations and signaling pathways in the evolution of gliomas. Cancer Sci. 2009;100(12):2235–41. Epub 2009/09/10. doi: 10.1111/j.1349-7006.2009.01308.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belirgen M, Berrak SG, Ozdag H, Bozkurt SU, Eksioglu-Demiralp E, Ozek MM. Biologic tumor behavior in pilocytic astrocytomas. Childs Nerv Syst. 2012;28(3):375–89. Epub 2012/01/17. doi: 10.1007/s00381-011-1676-6 . [DOI] [PubMed] [Google Scholar]
- 55.Koh KN, Lee JY, Lim J, Shin J, Kang SH, Suh JK, et al. Genetic alterations detected by targeted next-generation sequencing and their clinical implications in neuroblastoma. Anticancer Res. 2020;40(12):7057–65. Epub 2020/12/09. doi: 10.21873/anticanres.14733 . [DOI] [PubMed] [Google Scholar]
- 56.Ludwig K, Kornblum HI. Molecular markers in glioma. J Neurooncol. 2017;134(3):505–12. Epub 2017/02/25. doi: 10.1007/s11060-017-2379-y ; PubMed Central PMCID: PMC5568999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jhanwar-Uniyal M, Dominguez JF, Mohan AL, Tobias ME, Gandhi CD. Disentangling the signaling pathways of mTOR complexes, mTORC1 and mTORC2, as a therapeutic target in glioblastoma. Adv Biol Regul. 2022;83:100854. Epub 2022/01/09. doi: 10.1016/j.jbior.2021.100854 . [DOI] [PubMed] [Google Scholar]
- 58.Jhanwar-Uniyal M, Wainwright JV, Mohan AL, Tobias ME, Murali R, Gandhi CD, et al. Diverse signaling mechanisms of mTOR complexes: mTORC1 and mTORC2 in forming a formidable relationship. Adv Biol Regul. 2019;72:51–62. Epub 2019/04/24. doi: 10.1016/j.jbior.2019.03.003 . [DOI] [PubMed] [Google Scholar]
- 59.Li L, Fu LQ, Wang HJ, Yan ZL, Yu XC, Wang YY. FAT2 is a novel independent prognostic factor for the poor prognosis of gastric carcinoma. Int J Clin Exp Pathol. 2017;10(12):11603–9. Epub 2017/12/01. ; PubMed Central PMCID: PMC6966061. [PMC free article] [PubMed] [Google Scholar]
- 60.Parmigiani E, Taylor V, Giachino C. Oncogenic and tumor-suppressive functions of NOTCH signaling in glioma. Cells. 2020;9(10):2304. Epub 2020/10/21. doi: 10.3390/cells9102304 ; PubMed Central PMCID: PMC7602630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Number of each type of mutation. (B) Number of SNV and Indels in all mutations. (C) Distribution of point mutation types in SNV. (D) The Venn diagram showed the number of co-mutated and uniquely-mutated genes in LGG and GBM patients.
(TIF)
(A) Hot spots of mutations in the IDH1 gene. (B) Sanger sequencing results of the novel mutation of the IDH1 gene. (C) Hot spots of mutations in the PCDHGA10 gene. (D) Sanger sequencing results of the novel mutation of the PCDHGA10 gene.
(TIF)
Comparison of mutation frequencies between our study and cBioPortal database (A) and TCGA database (B).
(TIF)
(XLSX)
(XLSX)
Data Availability Statement
The data reported in this study are available in the CNGB Nucleotide Sequence Archive (CNSA: https://db.cngb.org/cnsa; accession number CNP0002254 and CNP0002252).