ABSTRACT
Small GTPases switch between GDP- and GTP-bound states during cell signaling. The ADP-ribosylation factor (ARF) family of small GTPases is involved in vesicle trafficking. Although evolutionarily well conserved, little is known about ARF and ARF-like GTPases in plants. We characterized biochemical properties and cellular localization of the essential small ARF-like GTPase TITAN 5 (TTN5; also known as HALLIMASCH, ARL2 and ARLC1) from Arabidopsis thaliana, and two TTN5 proteins with point mutants in conserved residues, TTN5T30N and TTN5Q70L, that were expected to be unable to perform nucleotide exchange and GTP hydrolysis, respectively. TTN5 exhibited very rapid intrinsic nucleotide exchange and remarkably low GTP hydrolysis activity, functioning as a non-classical small GTPase being likely present in a GTP-loaded active form. We analyzed signals from YFP–TTN5 and HA3–TTN5 by in situ immunolocalization in Arabidopsis seedlings and through use of a transient expression system. Colocalization with endomembrane markers and pharmacological treatments suggests that TTN5 can be present at the plasma membrane and that it dynamically associates with membranes of vesicles, Golgi stacks and multivesicular bodies. Although TTN5Q70L mirrored wild-type TTN5 behavior, the TTN5T30N mutant differed in some aspects. Hence, the unusual rapid nucleotide exchange activity of TTN5 is linked with its membrane dynamics, and TTN5 likely has a role in vesicle transport within the endomembrane system.
Keywords: TTN5, ARL2, Endomembrane, GTPase, Plasma membrane, Vesicle
Summary: The small ARF-like GTPase TTN5 has unusual rapid nucleotide exchange activity. It is likely present in a GTP-loaded form in cells where it is linked with membrane dynamics and vesicle transport within the endomembrane system.
INTRODUCTION
Regulatory processes in signal transduction rely heavily on guanine nucleotide-binding proteins of the GTPase family. After identifying oncogenes (HRAS, KRAS and NRAS), the RAS superfamily of small GTPases emerged, encompassing conserved members across eukaryotes. This family is divided into five mammalian subfamilies: the Rat sarcoma (RAS), RAS homologs (RHO), RAS-like proteins in the brain (RAB), Ras-related nuclear proteins (RAN) and ADP-ribosylation factor (ARF) subfamilies (Bos, 1988; Kahn et al., 1992; Ahmadi et al., 2017). In Arabidopsis thaliana, only four families exist, the Rho of plants (ROP), RAB, RAN and ARF subfamilies (Vernoud et al., 2003). These subfamilies are classified by sequence identity, with conserved sequence motifs playing essential regulatory roles in cells (Kahn et al., 1992). Many mammalian small GTPases act as molecular switches in signal transduction, switching from inactive GDP-loaded to active GTP-loaded forms, enabling differential protein complex formations or acting in tethering complexes to target membranes. With typically low intrinsic GDP-to-GTP exchange and GTP hydrolysis activity, small GTPases require guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) for regulation. GEFs are potentially recruited to inactive GTPases to their site of action and accelerate GDP-GTP exchange leading to GTPase activation. The GTP-loaded GTPases exert their function via direct effector interaction (Sztul et al., 2019; Nielsen, 2020; Adarska et al., 2021) until inactivation occurs through GAP-stimulated GTP hydrolysis. ARF GTPases are often involved in vesicle-mediated endomembrane trafficking in mammalian cells and yeast (Just and Peränen, 2016). In plants, small GTPase activities and their cellular functions are not well understood. Although Arabidopsis has 12 ARF and seven ARF-like (ARL) and associated SAR1 proteins, plant ARF proteins are poorly described (Singh et al., 2018). The best-studied plant ARF GTPases, SAR1 and ARF1, act in anterograde and retrograde vesicle transport between the endoplasmic reticulum (ER) and the Golgi. SAR1 is involved in COPII-mediated trafficking from the ER to Golgi, whereas ARF1 participates in the COPI pathway (Singh et al., 2018; Nielsen, 2020). Another ARF-like protein, ARL1, might function in endosome-to-Golgi trafficking (Latijnhouwers et al., 2005; Stefano et al., 2006). These roles of ARF1 and SAR1 in vesicle formation are well conserved in eukaryotes, suggesting that other plant ARF members might also function in the endomembrane system. A recent study has shown Golgi-related localization for some ARF and ARF-like proteins (Niu et al., 2022), promoting a general involvement of ARF proteins in the endomembrane system.
TITAN 5 [TTN5; also known as HALLIMASCH (HAL), ARL2 and ARLC1] is essential for plant development, and was initially identified in two independent screens for abnormal embryo mutants. ttn5 loss-of-function mutants arrest soon after cell division of the fertilized egg cell, indicating a fundamental, potentially housekeeping, role in cellular activities (Mayer et al., 1999; McElver et al., 2000; Lloyd and Meinke, 2012). The TTN5 sequence is closely related to that of human ARL2 (hsARL2), which has high nucleotide dissociation rates, up to 4000-fold faster than RAS (Hanzal-Bayer et al., 2005; Veltel et al., 2008). HsARL2 is associated with various cellular functions, including microtubule development (Bhamidipati et al., 2000; Fleming et al., 2000; Radcliffe et al., 2000; Antoshechkin and Han, 2002; Tzafrir et al., 2002; Mori and Toda, 2013), adenine nucleotide transport in mitochondria (Sharer et al., 2002) and control of phosphodiesterase activity in cilia (Ismail et al., 2011; Fansa and Wittinghofer, 2016), and yeast and Caenorhabditis homologs have been identified (Radcliffe et al., 2000; Antoshechkin and Han, 2002). With regard to TTN5, the cellular roles remain unknown. Its molecular function and the GTPase characteristics of TTN5 have not yet been demonstrated.
Here, we show, by means of stopped-flow fluorimetry kinetic assays, that TTN5 is a functional small GTPase with conserved GTP hydrolysis and rapid nucleotide exchange characteristics. Fluorescence microscopy combined with pharmacological treatments suggests that TTN5 might be located at the plasma membrane (PM) and within the endomembrane system. Our study enables future investigation of cellular and physiological functions of this small GTPase.
RESULTS
TTN5 exhibits the atypical characteristics of rapid nucleotide exchange and slow GTP hydrolysis
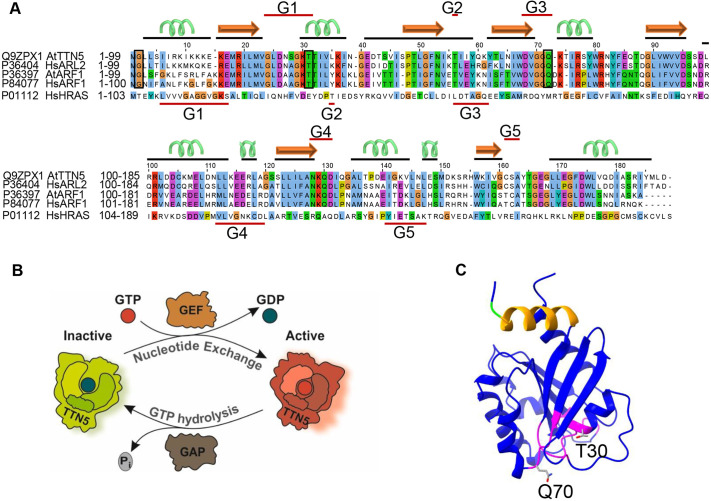
TTN5 has higher sequence similarity with HsARL2 than it does with Arabidopsis ARF or ARL proteins (Fig. 1A) (McElver et al., 2000; Vernoud et al., 2003). Its ubiquitous gene expression and regulation during plant development, particularly in the root epidermis, as revealed in public RNA-seq datasets of organ and single cell analysis of roots (Fig. S1A,B, and see Materials and Methods), reflect its crucial role. TTN5 is strongly expressed during early embryo development where cell division, elongation and differentiation take place (Fig. S1C), suggesting that it has function in fundamental processes, especially when cells grow and divide.
Fig. 1.
TTN5 a predicted functional small ARF-like GTPase with nucleotide exchange capacity. (A) Sequence alignment of TTN5 with HsARL2, HsARF1, HsHRAS and AtARF1 created with Jalview (Waterhouse et al., 2009). Conserved G-motifs (G1–G5; red lines) are defined for TTN5 and HRAS. The TTN5 secondary structure is depicted by black lines and corresponding cartoon (α-helix green; β-sheet orange). Conserved residues in ARF and ARL proteins are highlighted by boxes (G2, and mutated T30 and Q70). TTN5T30N is expected to have a low nucleotide exchange capacity, whereas TTN5Q70L is expected to have a low GTPase hydrolysis activity. (B) Model of the predicted GTPase nucleotide exchange and hydrolysis cycle of TTN5. TTN5 switches from an inactive GDP-loaded to an active GTP-loaded form. GDP to GTP nucleotide exchange and GTP hydrolysis might be aided by GEFs and GAPs. (C) Predicted protein structural model of TTN5; GTP-binding pocket (magenta); N-terminal amphipathic helix (orange); conserved G2 (green); mutagenized T30 and Q70 (sticks). The model was generated with AlphaFold (Jumper et al., 2021), and adapted with UCSF ChimeraX 1.2.5 (Goddard et al., 2018).
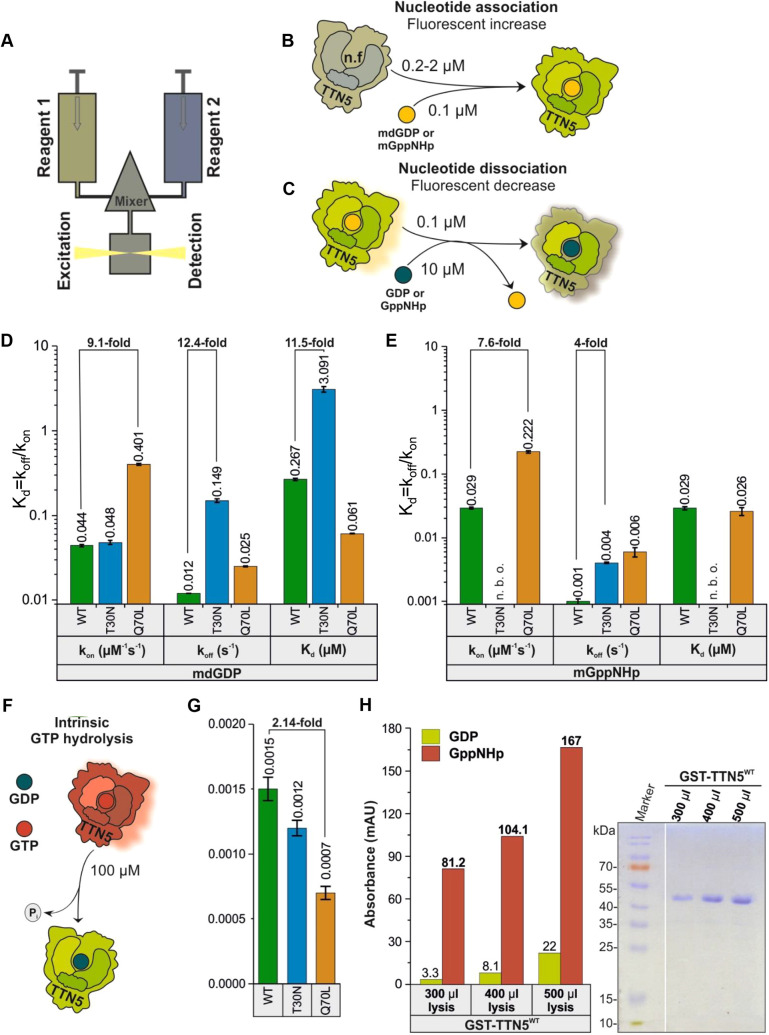
Based on sequence similarity and structural predictions, TTN5 is presumed to function as a molecular switch. (Fig. 1B,C). Although HsARL2 shows fast GDP-GTP exchange characteristics (Hanzal-Bayer et al., 2005; Veltel et al., 2008), it was unclear whether plant TTN5 shared these characteristics. We here characterized the nucleotide binding and GTP hydrolysis properties of TTN5WT and two mutants, TTN5T30N and TTN5Q70L, using heterologously expressed proteins and in vitro biochemical assays (experimental workflow illustrated in Fig. S2A–E), as previously established for human GTPases (Eberth and Ahmadian, 2009). TTN5T30N was chosen because it is assumed to preferentially bind GEFs, sequestering them from their proper context, whereas TTN5Q70L was chosen because it is thought to be defective in hydrolyzing GTP; equivalent mutants have been frequently used and characterized dominant-negative and constitutively active (Scheffzek et al., 1997; Zhou et al., 2006; Newman et al., 2014). We monitored the real-time kinetics of interactions of fluorescent guanine nucleotides using stopped-flow fluorimetry suited for very rapid enzymatic reactions (Fig. 2A–C). 2-deoxy-3-O-N-methylanthraniloyl-deoxy-GDP (mdGDP) and GppNHp (mGppNHp), a non-hydrolyzable GTP analog, were used to mimic GDP and GTP binding to TTN5. This approach allowed us to monitor real-time kinetics and determine and quantify nucleotide association (kon) and dissociation (koff) characteristics of small GTPases, such as has been done for HsARL2 and HsARL3 (Hillig et al., 2000; Hanzal-Bayer et al., 2005; Veltel et al., 2008; Zhang et al., 2018). The kon value is defined as the rate of nucleotide binding to GTPases, to form the GTPase–nucleotide complex (Fig. 2B), whereas the koff value describes the rate of nucleotide dissociation from GTPases (Fig. 2C). TTN5 proteins were able to bind both nucleotides, except for mGppNHp binding by TTN5T30N (Figs S3A–F, S4A–E). TTN5Q70L revealed the highest kon value for mGDP binding (0.401 µM−1s−1), being 9-fold higher compared to that of TTN5WT (0.044 µM−1s−1) and TTN5T30N (0.048 µM−1s−1), respectively (Fig. 2D, Fig. S3D–F). kon values for mGppNHp binding were about half for TTN5WT (0.029 µM−1s−1) and TTN5Q70L (0.222 µM−1s−1) compared to those for mGDP binding (Fig. 2E; Fig. S4C,D). The differences in kon for the respective nucleotide binding were small. However, TTN5Q70L showed a 7.5-fold faster mGppNHp binding than TTN5WT. Remarkably, we were not able to monitor mGppNHp association with TTN5T30N but observed its dissociation (koff=0.026 s−1; Fig. 2E). To confirm the binding capability of TTN5T30N with mGppNHp, we measured the mGppNHp fluorescence in real-time before and after titration of nucleotide-free TTN5T30N and binding occurred too fast to resolve the association rate (Fig. S4B).
Fig. 2.
Biochemical properties of TTN5 suggest its presence in a GTP-loaded active form in cells. (A) Schematic illustration of the stopped-flow fluorescence device for monitoring nucleotide-binding kinetics of heterologously expressed and purified TTN5 protein (Fig. S2A–D). It consists of two motorized thermostated syringes, a mixing chamber and a fluorescence detector. Two different reagents, 1 and 2, with one containing a fluorescent reporter group (mdGDP or mGppNHp to mimic GDP and GTP) are rapidly mixed and transferred to a fluorescence detection cell. (B) Schematic illustration of nucleotide association. Nucleotide-free TTN5 (for preparation see Fig. S2E) was rapidly mixed with mdGDP. A fluorescence increase is expected upon association of mdGDP with TTN5. Similar measurements were performed with mGppNHp. (C) Schematic illustration of intrinsic nucleotide dissociation. mdGDP-bound TTN5 is mixed with a molar excess of GDP. A fluorescence decrease is expected upon mdGDP dissociation from TTN5 and binding of free unlabeled GDP. Similar measurements were performed with mGppNHp. (D,E) Kinetics of association and dissociation of fluorescent nucleotides mdGDP (D) or mGppNHp (E) with TTN5 proteins (WT, TTN5T30N, TTN5Q70L). Association rate constants (kon in µM−1s−1) were determined from the plot of increasing observed rate constants (kobs in s−1) against corresponding TTN5 protein concentrations (as denoted in A,B; for full data, see Figs S3A–F, S4A–E). Intrinsic dissociation rates (koff in s−1) were determined from the plot of fluorescent decrease upon exchange from mdGDP-bound or mGppNHp-bound TTN5 to GDP-bound TTN5 (as denoted in A,C; for full data, see Figs S3G–I, S4F–H). The nucleotide affinity (dissociation constant Kd in µM) of the corresponding TTN5 proteins was calculated from the koff/kon ratio. When mixing mGppNHp with nucleotide-free TTN5T30N, no binding was observed (n.b.o.) under these experimental conditions. kon and koff values presented as bar graphs are calculated from the average of four to six measurements and presented as mean±s.d. (F,G) GTP hydrolysis of TTN5 proteins determined by HPLC. (F) Schematic illustration of GTP hydrolysis measurement. (G) GTP-bound TTN5 proteins incubated at different time points before injecting them on a reversed-phase HPLC system. Evaluated data (Fig. S5) resulted in determination of GTP hydrolysis rates (kcat in s−1). Each bar represents the kcat value obtained from a single experiment per condition, comprising six data points. Error bars indicate the standard errors of the fitted values as determined by Origin software. (H) TTN5 accumulation in a GTP-loaded form by HPLC values and Coomassie Blue-stained SDS-PAGE. GST–TTN5WT (46.5 kDa) was purified from bacterial cell lysates at three different volumes in the presence of free GppNHp. Presence of much higher amounts of GppNHp-bound versus GDP-bound GST-TTN5 protein indicates that TTN5 rapidly exchanged bound nucleotide and accumulated in this state. This was a confirmatory experiment performed once.
We next measured the dissociation (koff) of mdGDP and mGppNHp from TTN5 proteins with excess amounts of GDP and GppNHp, respectively (Fig. 2C) with interesting differences (Fig. 2D,E; Figs S3G–I,S4F–H). First, TTN5WT showed a 100-fold faster koff value (mGDP, 0.012 s−1) (Fig. 2D; Fig. S3G), compared to classical small GTPases, including RAC1 (Haeusler et al., 2006) and HRAS (Gremer et al., 2011), but very similar to the koff value of HsARF3 (Fasano et al., 2022). Second, koff values for mGDP and mGppNHp were in a similar range for TTN5WT (mGDP, 0.012 s−1; mGppNHp, 0.001 s−1) and TTN5Q70L (mGDP, 0.025 s−1; mGppNHp, 0.006 s−1), but koff values differed 10-fold between the two nucleotides in TTN5WT (Fig. 2D,E; Figs S3G,I, S4F,H). Thus, mGDP dissociated from proteins 10-fold faster than mGppNHp. Third, mGDP dissociation from TTN5T30N (0.149 s−1) was 12.5-fold faster than that of TTN5WT and 37-fold faster than mGppNHp dissociation of TTN5T30N (0.004 s−1; Fig. 2D,E; Figs S3H, S4G). Mutants of CDC42, RAC1, RHOA, ARF6, RAD, GEM and RAS GTPases that are equivalent to TTN5T30N display decreased nucleotide binding affinity and therefore tend to remain in a nucleotide-free state, complexed with their cognate GEFs (Erickson et al., 1997; Ghosh et al., 1999; Radhakrishna et al., 1999; Jung and Rösner, 2002; Kuemmerle and Zhou, 2002; Wittmann et al., 2003; Nassar et al., 2010; Huang et al., 2013; Chang and Colecraft, 2015; Fisher et al., 2020; Shirazi et al., 2020). Given that TTN5T30N exhibited fast nucleotide dissociation, these results suggest that TTN5T30N might act in either a dominant-negative or fast-cycling manner as reported for other GTPase mutants (Fiegen et al., 2004; Wang et al., 2005; Fidyk et al., 2006; Klein et al., 2006; Soh and Low, 2008; Sugawara et al., 2019; Aspenström, 2020).
The dissociation constant (Kd) is calculated from the ratio koff/kon, which inversely indicates the affinity of interactions between proteins and nucleotides (higher Kd=lower affinity). Interestingly, TTN5WT binds mGppNHp (0.029 µM) 10-fold tighter than mGDP (0.267 µM), a difference, which was not observed for TTN5Q70L (mGppNHp, 0.026 µM; mGDP, 0.061 µM; Fig. 2D,E). The lower affinity of TTN5WT for mdGDP compared to mGppNHp brings us closer to the hypothesis that classifies TTN5 as a non-classical GTPase prone to remain in the active, GTP-bound state (Jaiswal et al., 2013). The Kd value for the TTN5T30N–mGDP interaction was 11.5-fold higher (3.091 µM) than for TTN5WT, suggesting that this mutant exhibited faster nucleotide exchange and lower affinity for nucleotides and might behave in a dominant-negative manner in signal transduction, similar to what occurs with other GTPases with a T30N exchange (Vanoni et al., 1999).
To get hints on TTN5 functionalities during the GTPase cycle, it is crucial to determine its ability to hydrolyze GTP. Accordingly, the catalytic rate of intrinsic GTP hydrolysis (kcat), was determined by incubating GTP-bound TTN5 proteins and analyzing the samples at various time points (Fig. 2F; Fig. S5). Determined kcat values were quite remarkable in two respects (Fig. 2G). First, all TTN5 proteins, TTN5WT, TTN5T30N and TTN5Q70L, showed quite similar kcat values (0.0015 s−1, 0.0012 s−1, 0.0007 s−1; Fig. 2G; Fig. S5). TTN5Q70L GTP hydrolysis activity was unexpectedly high given that such glutamine mutations typically impair hydrolysis and result in constitutively active GTPases (Hodge et al., 2020; Matsumoto et al., 2021). Second, the kcat value of TTN5WT (0.0015 s−1) although comparatively low to other GTPases (Jian et al., 2012; Esposito et al., 2019), was 8-fold lower than the determined koff value for mGDP dissociation (0.012 s−1; Fig. 2E). This means that a fast intrinsic GDP/GTP exchange versus a slow GTP hydrolysis can have drastic effects on TTN5 activity in resting cells, given that TTN5 can accumulate in its GTP-bound form, unlike classical GTPases (Jaiswal et al., 2013). To investigate this scenario, we pulled down GST–TTN5 protein in the presence of an excess amount of GppNHp and measured the nucleotide-bound form of GST–TTN5. Isolated GST–TTN5 bound to increasing amounts of GppNHp, indicating that the bound nucleotide is rapidly exchanged for free nucleotide (here GppNHp; Fig. 2H), which is in contrast to what is seen for classical GTPases, which remain in their inactive GDP-bound forms under the same experimental conditions (Walsh et al., 2019; Hodge et al., 2020).
In summary, TTN5 contains conserved regions required for nucleotide binding and binds nucleotides. Interestingly, the slow intrinsic GTP hydrolysis rates in combination with high GDP dissociation rates indicates that TTN5 tends to exist in a GTP-loaded form, as opposed to the classical GTPases. This might have drastic effects on TTN5 activity in cells under resting conditions (Jaiswal et al., 2013). By contrast, the TTN5Q70L mutant, which we originally suspected would be constitutively active, still has intrinsic GTPase activity, whereas the T30N variant exhibits a low affinity for mGDP. Therefore, we propose that TTN5 exhibits typical functions of a small GTPase based on in vitro biochemical activity studies, including guanine nucleotide association and dissociation, and emphasize its divergence among the ARF GTPases because of its kinetics.
TTN5 may be a highly dynamic protein and localize to different intracellular compartments
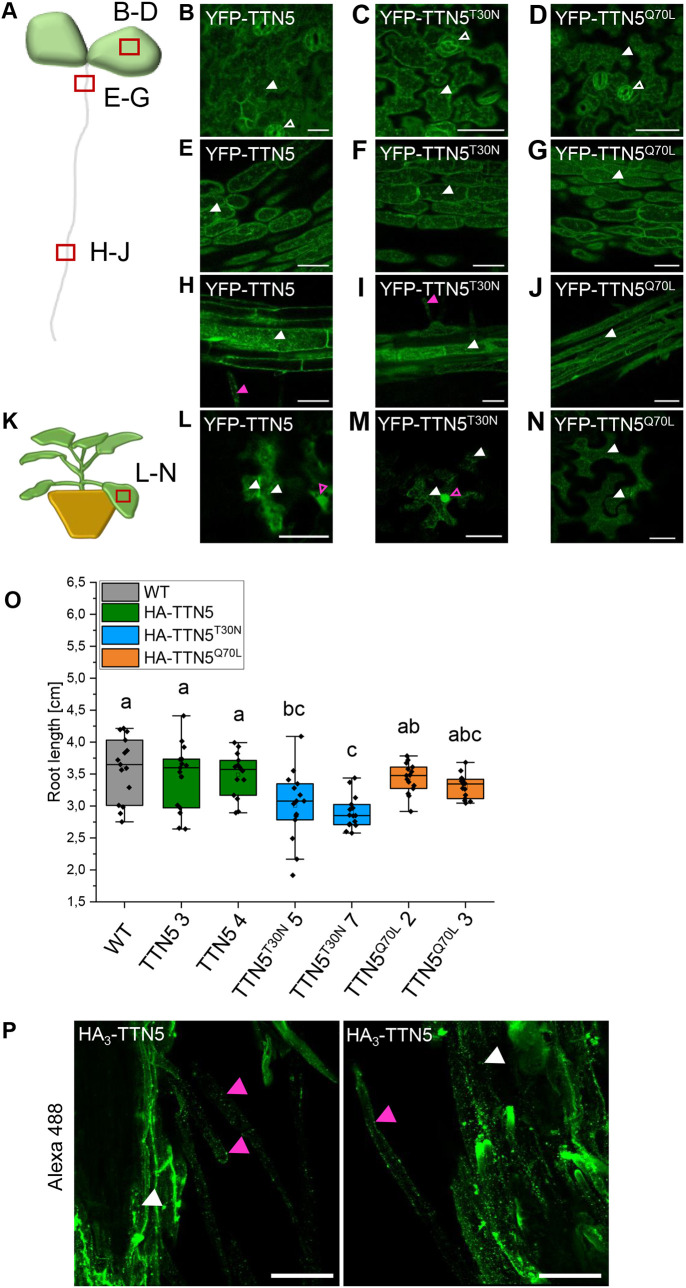
Several eukaryotic ARF GTPases function in vesicle transport and are located at various membranous sites linked with the endomembrane compartments (Vernoud et al., 2003). Localization had not been comprehensively studied for TTN5. To obtain hints as to where in a cell TTN5 localizes to, we first created transgenic Arabidopsis lines constitutively expressing YFP-tagged TTN5 (pro35S::YFP–TTN5) and its two mutant forms (pro35S::YFP–TTN5T30N, pro35S::YFP–TTN5Q70L) and investigated the localization in 6-day-old seedlings in the epidermis of cotyledons, hypocotyls, root hair zone and in root tips (Fig. 3A; Fig. S6A). Microscopy observations were made in different planes of the tissues, for example, inside the cells across the vacuoles (Fig. S6) and underneath the PM at the cell peripheries (Fig. 3). We chose to investigate YFP–TTN5 in the epidermis, as TTN5 transcripts were detected there in plants (Fig. S1B). YFP signals in epidermal cotyledon cells of YFP–TTN5 seedlings were detected in nuclei and cytoplasm and/or in close proximity to the PM (Fig. S6B). Similar localization patterns were found for mutant YFP–TTN5 signals (Fig. S6C,D). YFP signals in YFP–TTN5, YFP–TTN5T30N and YFP–TTN5Q70L seedlings were also present in a similar pattern in stomata (Fig. 3B–D). In hypocotyls, an intracellular YFP signal was observed in nuclei and in close proximity to or at the PM with all three YFP–TTN5 forms (Fig. S6E–G). Investigation of the root hair zone showed YFP signals in the cytoplasm and at the PM of root hairs (Fig. S6H–J). In the root tip, YFP signal was detectable inside the cytoplasm and in nuclei (Fig. S6K). The pattern was similar for YFP–TTN5T30N and YFP–TTN5Q70L (Fig. S6L,M). Fluorescence signal in YFP–TTN5, YFP–TTN5T30N and YFP–TTN5Q70L seedlings inside the cytoplasm was confined to punctate structures, indicating that fluorescence was present in vesicle-like structures together with free signal. This localization pattern was also present in leaf epidermal cells of the cotyledons (Fig. 3B–D), in the hypocotyls (Fig. 3E–G) and in cells of the root hair zones and in root hairs (Fig. 3H–J). These observed structures point to an association of TTN5 with vesicle and endomembrane trafficking. A closer inspection of the dynamics of these structures in the leaf epidermis of cotyledons showed high mobility of fluorescent signals (Movies 1–3) as well as in hypocotyl cells (Movie 4). Interestingly, the mobility of these punctate structures differed within the YFP–TTN5T30N hypocotyl cells, but not in the leaf epidermis cells (Movie 5, compare with Fig. S2), which was not the case for the YFP–TTN5Q70L mutant (Movie 6, compare with Fig. S5). We detected that movement of YFP–TTN5T30N was slow or completely arrested for approximately half of the cells within the hypocotyl epidermis compared to movements for YFP–TTN5 and YFP–TTN5Q70L (Movies 4–6). This loss of fluorescence signal mobility in YFP–TTN5T30N seedlings might be a consequence of a missing effector interaction. We did not observe the blocked mobility for fluorescence signals in cells expressing YFP–TTN5, YFP–TTN5T30N or YFP–TTN5Q70L in the root elongation zone (Movies 7–9). No mobility of YFP fluorescence signal was visible in root tip cells for any YFP–TTN5 form (Movies 10–12).
Fig. 3.
TTN5 might be present in punctate structures in seedlings. Microscopy observations of YFP fluorescence were made in a plane underneath the PM at the cell peripheries. (A) Schematic representation of an Arabidopsis seedling with indicated imaged positions (red rectangle). (B–J) Analysis of YFP–TTN5, YFP–TTN5T30N and YFP-TTN5Q70L Arabidopsis seedlings via fluorescent confocal microscopy. (B–D) Fluorescence signals observed in stomata (empty white arrowhead) and in the epidermis of cotyledons in punctate structures (filled white arrowhead). (E–G) Localization in hypocotyls showed a similar pattern of punctate structures. (H–J) Signals were present in punctate structures in the root hair zone and root hairs (filled magenta arrowhead). (K) Schematic representation of a N. benthamiana plant, used for transient expression with indicated imaged position (red rectangle). (L–N) YFP signals in N. benthamiana leaf epidermal cells expressing YFP–TTN5, YFP–TTN5T30N and YFP–TTN5Q70L. Signals were present in punctate structures (white arrowheads) and in nuclei (empty magenta arrowheads). Experiments were repeated twice with two seedlings (n=2) or one plant (n=1). (O) Root length measurement of 10-day-old HA3–TTN5, HA3–TTN5T30N and HA3–TTN5Q70L Arabidopsis seedlings in comparison with wild-type (WT). Only HA3–TTN5T30N showed slightly reduced root length compared to WT. Analysis was conducted in replicates (n=14). The box represents the 25–75th percentiles, and the median is indicated. The whiskers show the 5–95th percentiles. One-way ANOVA with Tukey post-hoc test was performed. Different letters indicate groups that have a statistical significance difference at P<0.05. (P) Maximum intensity projection of whole-mount immunostaining of HA3–TTN5 roots in the differentiation zone (anti-HA primary antibody, Alexa-488-labeled secondary antibody). Alexa Fluor 488 signals were present in punctate structures in root cells (filled white arrowhead) and root hairs (filled magenta arrowhead), which is comparable to what was seen for YFP signals (H–J). Experiment was repeated three times with two seedlings (n=2). Scale bars: 50 µm.
To evaluate the Arabidopsis data and to better visualize YFP–TTN5, we expressed YFP–TTN5 constructs transiently in Nicotiana benthamiana leaf epidermis cells. Fluorescence signals in YFP–TTN5-, YFP–TTN5T30N- and YFP-TTN5Q70L-expressing cells were all localized at or in close proximity to the PM and in several cytosolic punctate structures, apart from nuclei, similar to what was seen in Arabidopsis cotyledons, hypocotyls and root hair zones (Fig. 3K–N; Fig. S6N–Q). Additionally, YFP signals were detected in a net-like pattern typical for ER localization (Fig. 3M,N). This indicates that the fluorescent signal localization is similar in both Arabidopsis epidermis cells and N. benthamiana leaf epidermis.
It should be noted that the 35S promoter-driven YFP–TTN5 constructs did not complement the ttn5-1 embryo-lethal phenotype (Fig. S7A,B). Western blot analysis with anti-GFP antibody using YFP–TTN5 Arabidopsis seedlings revealed three weak YFP bands ranging between 26 and 35 kDa, besides the expected and strong 48 kDa YFP–TTN5 band (Fig. S7C). We cannot explain the presence of these small protein bands. They might correspond to free YFP, proteolytic products or potentially to proteins produced from aberrant transcripts with perhaps alternative translation start or stop sites. On the other hand, a 35S promoter-driven triple hemagglutinin-tagged HA3–TTN5 did complement the ttn5-1 embryo-lethal phenotype (Fig. S7D,E). Western blot analysis with anti-HA antibody performed with HA3–TTN5 seedlings showed a single, correctly sized band, but no band that was 13 to 18 kDa smaller (Fig. S7D). Hence, the inability of YFP–TTN5 to complement the embryo-lethal phenotype is presumably due to the relatively large YFP tag in comparison with the small GTPase and smaller HA3 tag. Interestingly, HA3–TTN5T30N seedlings presented a shorter root length phenotype, which might be due to the atypical biochemical TTN5T30N characteristics, whereas HA3–TTN5 and HA3–TTN5Q70L seedlings had no obvious phenotypic difference from wild-type (Fig. 3O).
To verify that the localization patterns observed with YFP–TTN5 constructs are representative of a functional TTN5, we performed immunofluorescence staining against the HA3 tag in HA3–TTN5 roots and compared the localization patterns (Fig. 3P). Alexa Fluor 488-labeled anti-HA antibody staining reflected HA3–TTN5 localization, and signals were visible in root cells and root hairs as expected. Signals were mostly present in punctate structures close to the PM and in the cytosol (Fig. 3P), fitting the fluorescence signals obtained with YFP–TTN5.
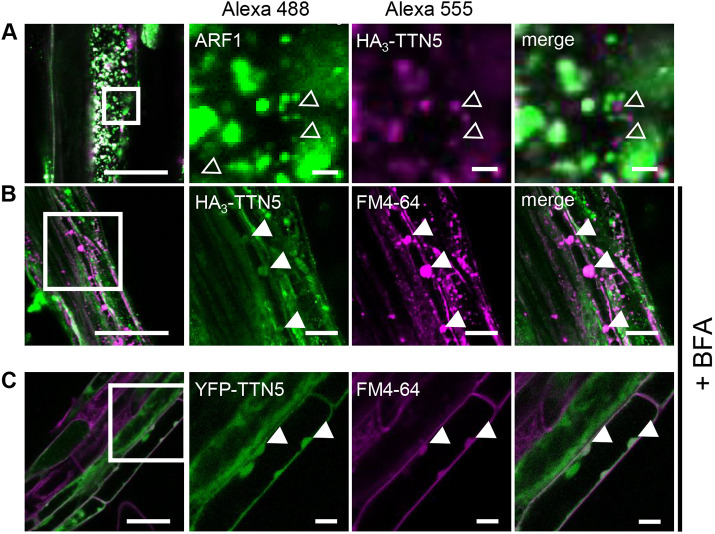
For a more detailed investigation of the HA3–TTN5 subcellular localization, we performed co-immunofluorescence staining with an Alexa Fluor 488-labeled anti-ARF1 antibody, which recognizes the Golgi and trans-Golgi network (TGN), together with Alexa Fluor 555-labeled HA3–TTN5 (Robinson et al., 2011; Singh et al., 2018) (Fig. 4A). ARF1–Alexa Fluor 488 staining was clearly visible in punctate structures representing presumably Golgi stacks (Fig. 4A) (Singh et al., 2018). Similar structures were seen with HA3–TTN5–Alexa Fluor 555 staining, but these did not colocalize with the ARF1-labeled structures, although they were in close proximity to each other (Fig. 4A). We hypothesized that the HA3–TTN5 structures might be connected to intracellular trafficking steps and performed brefeldin A (BFA) treatment, a commonly used tool in cell biology for preventing dynamic membrane trafficking events and vesicle transport involving the Golgi. BFA is a fungal macrocyclic lactone that leads to a loss of cis-cisternae and accumulation of Golgi stacks, known as BFA-induced compartments, and Golgi–ER fusion (Ritzenthaler et al., 2002; Wang et al., 2016). For better BFA body identification, we simultaneously used the membrane dye FM4-64, which can emit fluorescence in a lipophilic membrane environment. FM4-64 marks the PM only a few minutes after application to the cell, and can then be endocytosed; in the presence of BFA, accumulates in BFA bodies (Bolte et al., 2004). We observed BFA bodies positive for both HA3–TTN5–Alexa Fluor 488 and FM4-64 signals (Fig. 4B). Similar patterns were observed for YFP–TTN5-derived signals in YFP–TTN5-expressing roots (Fig. 4C). Hence, HA3–TTN5 and YFP–TTN5 are present in similar subcellular membrane compartments.
Fig. 4.
Whole-mount immunolocalization hints at TTN5 presence in BFA bodies. (A,B) Colocalization of HA3–TTN5 seedlings by whole-mount immunostaining. (A) Detection of HA3–TTN5 (anti-HA primary antibody, Alexa Fluor 555-labeled secondary antibody) with Golgi and TGN marker ARF1 (anti-ARF1 primary antibody, Alexa Fluor 488-labeled secondary antibody). Both fluorescence signals were detected in vesicle-like structures in root cells in close proximity to each other but mostly not colocalizing. The experiment was repeated twice with three seedlings (n=3). (B) Detection of HA3–TTN5 (anti-HA primary antibody, Alexa Fluor 488-labeled secondary antibody) and staining with membrane dye FM4-64 after BFA treatment (10 mM FM4-64 FX and 72 µM BFA for 1 h). Alexa Fluor 488 signals colocalized with FM4-64 in BFA bodies in root cells. The experiment was repeated three times with three seedlings (n=3). (C) YFP fluorescence in YFP–TTN5 seedlings, co-analyzed with FM4-64 after BFA treatment. YFP fluorescence signals colocalized with FM4-64 in BFA bodies similar to what was seen in B. The experiment was performed once with three independent YFP–TTN5 lines (n=3). Colocalization indicated by filled white arrowheads, non-colocalized HA3–TTN5 Alexa Fluor 488-labeled signals is indicated with empty white arrowheads. Scale bars: 50 µm (overview images on left), 10 µm (magnifications).
In HA3–TTN5 immunostaining, we did not observe any staining in nuclei or ER (Figs 3P, 4A,B), in contrast to the fluorescence signals in YFP–TTN5-expressing cells. This might indicate that either the nuclear and ER signals seen with YFP–TTN5 correspond to the smaller proteins detected, or that immunostaining was not suited to detect this localization. Hence, we focused on analysis of the area where there were overlapping localization patterns between fluorescence with YFP-labeled TTN5 and HA3-TTN5 immunostaining, such as the specific signal patterns seen for punctate membrane structures.
Taken together, our results show that signals of YFP–TTN5 and HA3–TTN5 were located in multiple membrane compartments in the epidermis of different Arabidopsis organs and of N. benthamiana leaves, including particular ring-like punctate structures and vesicles. Fluorescence signals in YFP–TTN5- and YFP-TTN5Q70L-expressing seedlings displayed high mobility in cells, as expected from an active GTPase functioning in dynamic processes, such as vesicle trafficking. In contrast, fluorescence signals for YFP–TTN5T30N were less mobile, consistent with the root length phenotype conferred by HA3–TTN5T30N, speaking in favor of the observed TTN5T30N kinetics, with a very fast nucleotide exchange rate and nucleotide affinity loss. Altogether, the TTN5 intracellular localization indicates that TTN5 might have multiple cellular functions as an active GTPase as it can associate with different intracellular structures of the endomembrane system.
TTN5 might associate with components of the cellular endomembrane system
The overlapping localization of HA3–TTN5 and YFP–TTN5 signals prompted us to better resolve the membrane structures and compartments of the highly dynamic endomembrane system. Well-established fluorescent markers and pharmacological treatments help to determine the nature of individual components in cells in parallel to colocalization studies with proteins of interest, such as TTN5. We conducted colocalization experiments in N. benthamiana leaf epidermis as the fluorescence signals were comparable to those for Arabidopsis cotyledons and root epidermis. Moreover, it represents an established system for functional association of fluorescent proteins with multiple endomembrane components and optimal identification of membrane structures (Brandizzi et al., 2002; Hanton et al., 2009).
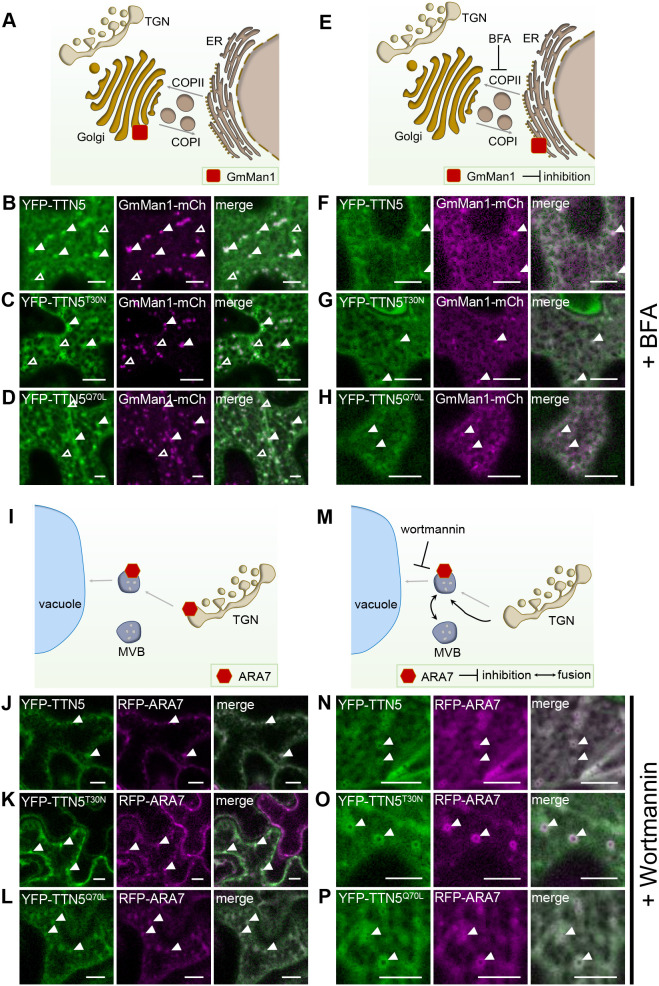
At first, we further investigated the ER–Golgi connection as a characteristic site of association with small GTPases, like the tested ARF1, involved in COPI vesicle transport (Just and Peränen, 2016). We used the soybean (Glycine max) protein α-1,2 mannosidase 1 (GmMan1) as a marker; this protein is a glycosidase that acts on glycoproteins at the cis-Golgi, facing the ER (Fig. 5A) and is visible as nearly round punctuate structures throughout the whole cell (Nelson et al., 2007; Wang et al., 2016). Fluorescence signals in leaf discs transiently expressing YFP–TTN5 and its mutant variants partially colocalized with GmMan1–mCherry signals at Golgi stacks (Fig. 5B–D). We also observed YFP fluorescence in form of circularly shaped ring structures with a fluorescence-depleted center; such structures can be of vacuolar origin as described for similar fluorescent rings for ANNI–GFP (Tichá et al., 2020). Furthermore, quantitative analysis reflected the visible colocalization of GmMan1 marker and YFP fluorescence with Pearson coefficients 0.63 (YFP–TTN5), 0.65 (YFP–TTN5T30N) and 0.68 (YFP–TTN5Q70L) (Fig. S8A; similar obtained overlap coefficients), indicating a strong correlation between the signals. We performed an additional object-based analysis to compare overlapping YFP fluorescence in YFP–TTN5-expressing leaves with GmMan1–mCherry signals (YFP/mCherry ratio) and vice versa (mCherry/YFP ratio). We detected 24% overlapping YFP fluorescence signals for TTN5 with Golgi stacks, whereas in YFP-TTN5T30N and YFP-TTN5Q70L-expressing leaves, signals only shared 16 and 15% overlap with GmMan1–mCherry-positive Golgi stacks (Fig. S8B). Some YFP signals did not colocalize with the GmMan1 marker; these signals were more prominent in leaves expressing YFP–TTN5T30N and less prominent for leaves expressing YFP-TTN5Q70L compared to YFP–TTN5 expression (Fig. 5B–D). Indeed, we identified 48% GmMan1–mCherry signal overlapping with YFP-positive structures in YFP–TTN5Q70L leaves, whereas this was 43% and 31% for YFP fluorescence signals in YFP–TTN5 and YFP–TTN5T30N-expressing leaves, respectively (Fig. S8B), indicating a smaller amount of GmMan1-positive Golgi stacks colocalizing with YFP signals for YFP–TTN5T30N. Hence, the GTPase-active TTN5 forms are likely present at cis-Golgi stacks at higher levels compared to TTN5T30N.
Fig. 5.
TTN5 might be associated with the endomembrane system in N. benthamiana pavement cells. YFP signals were detected in N. benthamiana pavement cells transiently expressing YFP–TTN5, YFP–TTN5T30N and YFP–TTN5Q70L with specific endomembrane markers via fluorescence confocal microscopy. (A) Schematic representation of GmMan1 localization at the cis-Golgi site. (B–D) Partial colocalization of the YFP signal with the Golgi marker GmMan1–mCherry at cis-Golgi stacks (filled white arrowheads). Additionally, YFP fluorescence signals were detected in non-colocalizing punctate structures with depleted fluorescence in the center (empty white arrowheads). (E) Schematic representation of GmMan1 localization at the ER upon BFA treatment. BFA blocks ARF GEF proteins, leading to a loss of Golgi cis-cisternae and formation of BFA-induced compartments due to an accumulation of Golgi stacks up to redistribution of the Golgi to the ER by fusion (Renna and Brandizzi, 2020). (F–H) GmMan1–mCherry and YFP fluorescence were present in the ER and in colocalizing punctate structures upon BFA treatment (36 µM for ∼30–60 min). (I) Schematic representation of ARA7 localization at the TGN and MVBs. (J–L) Colocalization of YFP signals with the MVB marker RFP–ARA7. (M) Schematic representation of ARA7 localization in swollen MVBs upon wortmannin treatment. Wortmannin inhibits PI3K function leading to TGN/EE fusion to swollen MVBs (Renna and Brandizzi, 2020). (N–P) ARA7-RFP colocalized with YFP signal in swollen MVBs upon wortmannin treatment (10 µM for ∼30–10 min). Colocalization indicated with filled arrowheads; non-colocalized YFP signal is indicated with with empty arrowheads. Corresponding colocalization analysis data is presented in Fig. S8. Experiments were repeated three times with two plants (n=2). Scale bars: 10 μm.
Next, we evaluated the Golgi localization by BFA treatment, resulting in a corresponding redistribution of GmMan1–mCherry (Ritzenthaler et al., 2002; Wang et al., 2016) (Fig. 5E). We found that upon BFA treatment, the GmMan1–mCherry signal was present in the ER and in BFA-induced compartments with partially matching localization of YFP signal of YFP–TTN5 constructs, suggesting a connection of TTN5 to Golgi localization (Fig. 5F–H). Hence, colocalization with GmMan1–mCherry and BFA treatment is indicative of YFP signals localizing to Golgi stacks upon YFP–TTN5 expression, whereas there was lower association of the YFP–TTN5T30N mutant form with this membrane compartment.
Second, we investigated localization to the endocytic compartments, endosomes of the TGN and multivesicular bodies (MVBs) using RFP–ARA7 (RABF2B), a small RAB-GTPase present there (Kotzer et al., 2004; Lee et al., 2004; Stierhof and El Kasmi, 2010; Ito et al., 2016) (Fig. 5I). These compartments play a role in sorting proteins between the endocytic and secretory pathways, with MVBs developing from the TGN and representing the final stage in transport to the vacuole (Valencia et al., 2016; Heucken and Ivanov, 2018). Colocalization studies revealed that the YFP signal in YFP–TTN5-expressing leaves was present at RFP–ARA7-positive MVBs (Fig. 5J). Noticeably, overlaps between RFP–ARA7 and YFP fluorescence signals upon TTN5T30N expression were lower than for the other TTN5 forms (Fig. 5J–L; Fig. S8C,D). We obtained a Pearson coefficient for YFP fluorescence from YFP–TTN5 or YFP–TTN5Q70L expression with RFP–ARA7 of 0.78, whereas a coefficient of only 0.59 was obtained with YFP–TTN5T30N, confirming the visual observation (Fig. S8C; similar obtained overlap coefficients). Object-based analysis showed that, RFP–ARA7-positive structures had an overlap with YFP fluorescence in YFP–TTN5-expressing leaves of 29%, and even more with YFP–TTN5Q70L (75%) unlike with YFP–TTN5T30N (21%) (Fig. S8D). Based on this, signals of YFP–TTN5Q70L and YFP–TTN5 tended to colocalize better with ARA7-positive compartments than YFP–TTN5T30N.
To test MVB localization, we treated plant cells with wortmannin, a fungal metabolite that inhibits phosphoinositide 3-kinase (PI3K) function and thereby causes swelling of the MVBs (Cui et al., 2016), a common approach to study endocytosis events (Fig. 5M). RFP–ARA7-expressing cells showed the typical wortmannin-induced formation of doughnut-like shaped MVBs (Jaillais et al., 2008). YFP fluorescence in YFP–TTN5-expressing leaves partially colocalized with these structures (Fig. 5N–P) indicating that fluorescence signals from expression of YFP–TTN5 and its two mutants are present in MVBs. YFP signals in YFP–TTN5Q70L-expressing leaf discs were located even to a greater extent to MVBs than for YFP–TTN5 and much more than for YFP–TTN5T30N-expressing cells, suggesting an active role for YFP–TTN5Q70L in MVBs, for example in the lytic degradation pathway or the recycling of proteins, similar to the role of ARA7 (Kotzer et al., 2004).
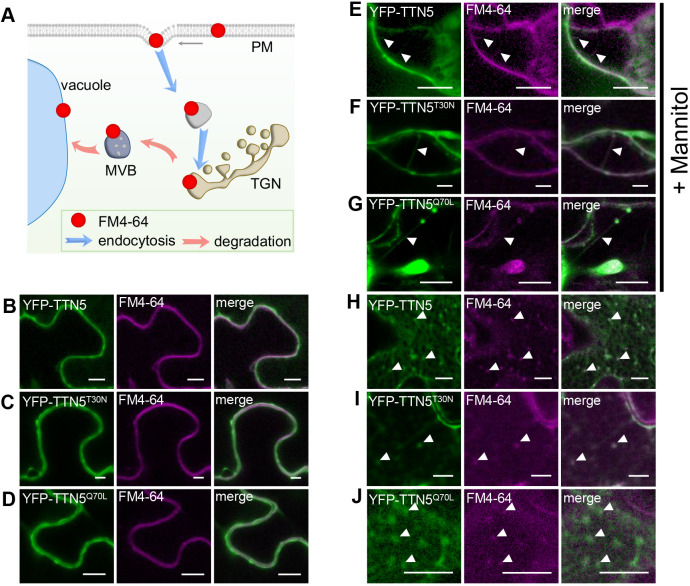
Finally, to investigate a possible connection of TTN5 with the PM, we determined the colocalization of YFP signals from YFP–TTN5 constructs with FM4-64 (Fig. 6A). Fluorescence signals for all three YFP–TTN5 forms colocalized with FM4-64 at the PM in a similar manner (Fig. 6B–D). To further investigate PM localization, we performed mannitol-induced plasmolysis. YFP signals for all YFP–TTN5 constructs were located similarly to FM4-64-stained Hechtian strands, thread-like structures attached to the apoplast visible upon plasmolysis and surrounded by PM (Fig. 6E–G).
Fig. 6.
TTN5 might colocalize with endocytosed PM material. (A) Schematic representation of the progressive stages of lipophilic membrane dye FM4-64 localization and internalization in cells. After infiltration, it first localizes in the PM, and later in intracellular vesicles and membrane compartments, reflecting the endocytosis process (Bolte et al., 2004). (B–J) YFP fluorescence colocalized with FM4-64 in N. benthamiana leaf epidermal cells as observed by confocal microscopy, following transient expression of YFP–TTN5, YFP–TTN5T30N and YFP–TTN5Q70L. (B–D) YFP signals colocalized with FM4-64 at the PM. (E–G) PM localization of YFP fluorescence was evaluated after mannitol-induced plasmolysis (1 M for ∼15–30 min). Formation of Hechtian strands is a sign of PM material and fluorescence staining there (filled arrowheads). (H–J) Internalized FM4-64 was present in vesicle-like structures that showed YFP signals. Colocalization indicated with filled arrowheads. Experiments were repeated three times with two plants (n=2). Scale bars: 10 μm.
In summary, these colocalization experiments show that YFP signals upon YFP–TTN5 expression are found in different membrane sites of the endomembrane system, including the Golgi, MVBs and PM. We hypothesize that, similar to other ARF proteins, this pattern indicates that TTN5 participates in a highly dynamic vesicle trafficking process. Indeed, recorded dynamic YFP signal movement of YFP–TTN5 and YFP–TTN5Q70L in N. benthamiana pavement cells colocalized with GmMan1–mCherry signals, and revealed high motion over time, whereas this was less the case for the YFP–TTN5T30N construct (Movies 13–15).
One potential cellular trafficking route is the degradation pathway to the vacuole. We, therefore, investigated fluorescence localization upon transient expression of YFP–TTN5 together with FM4-64 in late endosomal compartments, which might be involved in vacuolar targeting. FM4-64 is used as a marker for membranes of late endosomal compartment and vacuole targeting, because following PM visualization FM4-64-stained endocytic vesicles become apparent at later stages, as well as tonoplast staining (Ueda et al., 2001; Emans et al., 2002; Dhonukshe et al., 2007; Ivanov and Vert, 2021). Next to YFP colocalization in YFP–TTN5-expressing leaves with FM4-64 at the PM, we detected colocalization with intracellular fluorescent compartments; a similar expression was seen for both mutant forms (Fig. 6H–J). This indicates that YFP–TTN5 might be involved in targeting of endocytosed PM material, irrespective of the mutations.
In summary, YFP signals upon YFP–TTN5 and YFP–TTN5Q70L expression were dynamic and colocalized with endomembrane structures, whereas fluorescence signal in YFP–TTN5T30N-expressing leaf discs tended to be less mobile and dynamic and colocalized less with such structures.
DISCUSSION
This work provides evidence that the small ARF-like GTPase TTN5 has very rapid intrinsic nucleotide exchange capacity with a conserved nucleotide-switching mechanism. TTN5 might primarily be present in a GTP-loaded active form in cells as a dynamic protein with respect to its localization to membrane structures, potentially associating it with vesicle transport and different endomembrane processes (Fig. 7). The active TTN5Q70L mutant was capable of nucleotide switching and appeared to be mostly similarly localized to wild-type TTN5. The TTN5T30N mutant, on the other hand, has a lower nucleotide exchange capacity, and differed significantly in localization properties and its dynamics, albeit depending on cell types, and conferred a root length phenotype. Therefore, the GTP-bound state that we presume for TTN5 is most likely crucial for correct protein localization and dynamics.
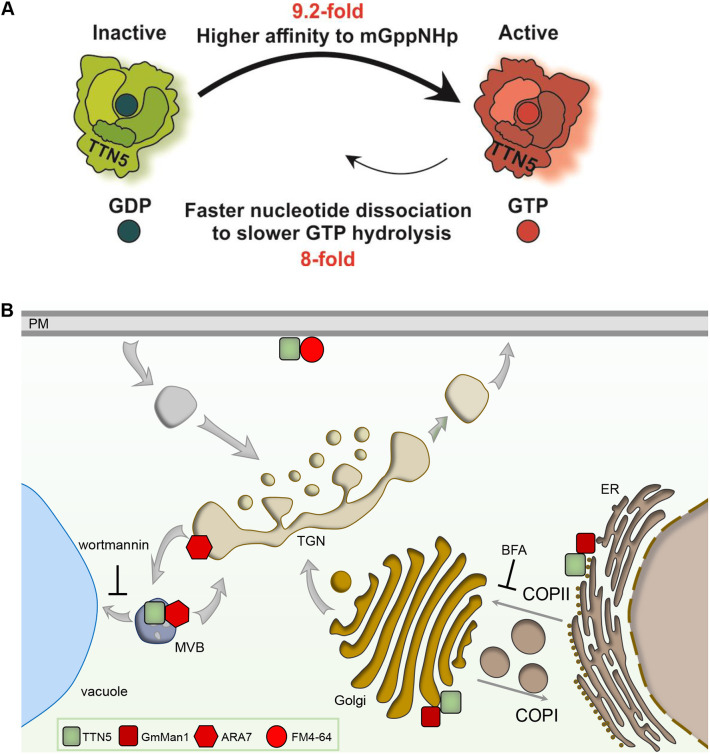
Fig. 7.
Schematic models summarizing TTN5 kinetic GTPase activities and potential localization within cells. (A) Model of the predicted GTPase nucleotide exchange and hydrolysis cycle of TTN5 based on the biochemical investigation. The TTN5 affinity for mGppNHp is 9.2-fold higher than it is for mGDP resulting in fast switching from inactive GDP-loaded to active GTP-loaded form. mGppNHp dissociation is 8-fold faster than GTP hydrolysis, but both processes were much slower than nucleotide association. TTN5 kinetics identified TTN5 as a non-classical GTPase that tends to stay in a GTP-loaded form even under resting conditions. (B) Presumed TTN5 locations within the cell. TTN5 (green square) can be present at the PM similar as FM4-64 (red circle) or in the endomembrane compartments of the TGN or MVB as found by ARA7-colocalization (red hexagon). Additionally, TTN5 might colocalize with GmMan1-positive (red square) Golgi stacks.
TTN5 exhibits characteristic GTPase functions
TTN5 is classified as an ARL2 homolog based on its high sequence identity with human HsARL2 (McElver et al., 2000), which is reinforced by structural prediction of a nucleotide-binding pocket. TTN5, TTN5T30N and TTN5Q70L can all bind guanine nucleotides. kon values for TTN5T30N and TTN5 were nearly identical, indicating that the mutation has no effect on GDP-binding characteristics, as would usually expected in the absence of a GEF. The TTN5Q70L kon value was clearly higher compared to that of the wild type, indicating that this mutant can bind GDP faster. Compared to other Ras superfamily members, these values are in the range of HRAS (Hanzal-Bayer et al., 2005) and around ten times slower than the fast association of RAC1 (Jaiswal et al., 2013). Well-studied RAS proteins, like RAC1, RAC2 and RAC3, have an intrinsic nucleotide exchange reaction rates of ∼40,000 s−1 (Haeusler et al., 2006), whereas TTN5 has a remarkably fast rate of nucleotide exchange that is very similar to that of HsARL2 (Hanzal-Bayer et al., 2005; Veltel et al., 2008). This suggests that TTN5 quickly replaces GDP for GTP and transforms from an inactive to an active state presumably without the need of GEF interaction. This could also be an explanation for what is seen with TTN5Q70L. Small GTPases with mutations in the glutamine residue of switch II region (e.g. Q71 for HsARF1 and ARL1, and Q61 for HRAS) are constitutively active (Zhang et al., 1994; Van Valkenburgh et al., 2001; Karnoub and Weinberg, 2008). Accordingly, TTN5Q70L is likely to exchange GDP rapidly to GTP and switch itself to stay in an active form, as suggested by the fast-intrinsic nucleotide exchange rate. Interestingly, TTN5T30N resulted in an even higher dissociation rate constant (koff.). The calculated Kd confirmed the higher nucleotide-binding affinity for GDP of TTN5 and TTN5Q70L compared with TTN5T30N. Reports on HsARL2, HsARF6 and HsARL4D show that their corresponding T30N mutants led to a similar decreased affinity for GDP (Macia et al., 2004; Hanzal-Bayer et al., 2005; Li et al., 2012).
Interestingly, a comparison of mdGDP with mGppNHp revealed that all three versions had higher GTP affinity than with GDP, with the highest for TTN5Q70L. These high GTP affinities in combination with fast GDP exchange rates and extremely slow hydrolysis pinpoint TTN5 as being GTP-loaded, even in resting conditions, which is very unusual. This atypical behavior has already been reported for a few non-classical RHO GTPases like RHOD and RIF (Jaiswal et al., 2013). This unusual GTP-bound active state along with it lacking N-myristoylation and the phylogenetic distance (Boisson et al., 2003; Vernoud et al., 2003) strengthens that there are major differences between TTN5 and other ARF proteins. The similarity in binding affinity between wild-type and TTN5Q70L is consistent with a previous report on HsARL2 (Hanzal-Bayer et al., 2005). Additionally, an equivalent ratio of nucleotide affinity was found between HRAS and HRASQ61L, but with a much higher affinity, more typical for small GTPases (Der et al., 1986). Given that Q70 is important for GAP-stimulated GTP hydrolysis (Cherfils and Zeghouf, 2013), we assume nucleotide exchange activity is unaffected.
The Arabidopsis genome encodes only the two large of five mammalian ARF-GEF subgroups, BFA-inhibited GEF (BIG) and the Golgi BFA-resistance factor 1 (GBF/GNOM) family (Memon, 2004; Wright et al., 2014; Brandizzi, 2018), but no TTN5 GEF protein has been reported. Potential interactions with GEFs are of high interest given the potential role of TTN5 as a co-GEF, similar to what is seen for HsARL3 and HsARL2, where their effector BART stabilizes the active GTPase (ElMaghloob et al., 2021). Especially, interactions at the nucleotide-binding site, which are prevented in the TTN5T30N mutant, will be of great interest to study further for TTN5.
Taken together, the categorization as a non-classical GTPase has three implications. First, very slow hydrolysis rates predict the existence of a TTN5 GAP. Second, TTN5T30N might function as a dominant-negative mutant and, in the presence of a GEF, it cannot bind GDP. Third, the TTN5Q70L hydrolysis rate is not decreased.
TTN5 might act in the endomembrane system
The localization data on YFP– and HA3–TTN5 suggest that it might be localized at different cellular membrane compartments, typical for the ARF-like family (Memon, 2004; Sztul et al., 2019), and supports potential involvement of TTN5 in endomembrane trafficking. Even though YFP–TTN5 did not complement the ttn5-1 embryonic lethality, we made several observations that suggest that the YFP–TTN5 signals seen at various membrane sites are meaningful. YFP–TTN5 might not complement due to differences in TTN5 levels and interactions in some cell types, which were hindered specifically for YFP–TTN5 but not HA3–TTN5. In a previous study, overexpression of ARF1 did not affect the intracellular localization compared to endogenous tagged-ARF1 but did cause the formation of tubulated structures (Bottanelli et al., 2017). Alhough constitutively driven, YFP–TTN5 expression might be delayed or insufficient at early embryonic stages resulting in the lack of embryonic lethal complementation. On the other hand, the fast nucleotide exchange activity might be hindered by the large YFP compared to the small HA3 tag, given that HA3–TTN5 rescued the embryo lethality. The lack of complementation represents a challenge for determining the localization of small GTPases with rapid nucleotide exchange in plants. Despite these limitations, we made relevant observations that made us believe that YFP signals in YFP–TTN5-expressing cells at membrane sites are meaningful. First, using pharmacological treatments and colocalization with organellar markers, we noted that various particular membrane compartments showed YFP signals, such as the punctate small ring-like structures, resembling previously reported ANNI–GFP staining (Tichá et al., 2020), the large wortmannin-induced ring-like structures and the BFA bodies, all of which are meaningful for vesicle transport and PM protein regulation processes (Wang et al., 2009; Suo et al., 2021). Furthermore, fluorescence signals obtained with YFP–TTN5 constructs also depended on T30 and Q70 residues; particularly YFP–TTN5T30N had partly quite distinct fluorescence localization patterns, reduced mobility in certain cells and differing degrees of colocalization with the utilized markers. Next to this, HA3–TTN5T30N seedlings showed reduced root growth which might be due to similar reasons. Given that TTN5T30N has a very fast nucleotide exchange rate and less affinity to nucleotides compared to TTN5, these differing YFP fluorescence patterns of YFP–TTN5T30N at membrane sites and the effect on root growth are not unexpected. Hence, we considered these specific YFP localizations at membrane sites as valid, especially when supported by HA3–TTN5 immunodetection.
Following up, colocalization analysis showed that both cis-Golgi and MVB-positive structures colocalized with a higher proportion with YFP signals from YFP–TTN5Q70L than from YFP–TTN5T30N. This could indicate the site of TTN5 action, considering our knowledge of ARF family activation in other organisms with high TTN5 sequence similarity. Small GTPases are usually recruited or move to their place of action upon interaction with their specific GEF and nucleotide exchange-dependent activation (Sztul et al., 2019; Nielsen, 2020; Adarska et al., 2021). Although our biochemical data implies no need for a typical GTPase–GEF interaction for activation, it can be still important for localization. Most of effector–GTPase interactions take place with GTPases in their GTP-bound form (Sharer and Kahn, 1999; Hanzal-Bayer et al., 2005). One exception is the role of ARL2–Alp41-GDP in microtubule dynamics by interaction with Cofactor D–Alp1D (Bhamidipati et al., 2000; Mori and Toda, 2013). Another possibility is a hindrance of dimerization by the T30N mutation. ARF1 protein dimer formation is important for the formation of free vesicles (Beck et al., 2009; Beck et al., 2011) and is associated with cell mobility, which was disturbed in YFP–TTN5T30N-expressing cells. Colocalization of YFP fluorescence upon YFP–TTN5 expression with ARA7-positive structures, even in the wortmannin-induced swollen state, might indicate that TTN5 has similar functions to ARA7. ARA7 is involved in endocytic cargo transport to the vacuole, for example, endocytosis of PM material (Ueda et al., 2001; Sohn et al., 2003; Kotzer et al., 2004; Ebine et al., 2011). Colocalization of FM4-64-labeled intracellular structures with fluorescence in YFP–TTN5-expressing cells might indicate the TTN5 has a role in endocytosis and the possible degradation pathway into the vacuole. Our data on colocalization with the different markers support the hypothesis that TTN5 might have functions in vesicle trafficking.
A potential explanation of YFP localization to similar compartments upon YFP–TTN5 and YFP–TTN5Q70L expression compared to fluorescence signal of YFP–TTN5T30N expression can be made based on a special feature of TTN5 in the ARF family. ARF GTPases are mostly myristoylated on G2, which is essential for their membrane binding. TTN5, as well as HsARL2 and HsARL3, lack this myristoylation, although G2 is present (Boisson et al., 2003; Kahn et al., 2006). HsARL2 and HsARL3 are still able to bind membranes, probably only through their N-terminal amphipathic helix, as was established for SAR1, with HsARL2 membrane binding efficiency being nucleotide independent (Lee et al., 2005; Kapoor et al., 2015). We suggest similar behavior for TTN5, as detected YFP signals localized to membranous compartments. Based on the varying colocalization degrees, with signals of YFP–TTN5T30N construct being less prominent at the Golgi and MVBs compared to YFP–TTN5 and YFP–TTN5Q70L, we hypothesize that different membrane localization could be associated with a nucleotide- or nucleotide exchange-dependent process. In a nucleotide-free or GDP-bound state, TTN5 might be predominantly present close to the PM, whereas in an active GTP-bound state, which according to enzyme kinetics is expected to be predominant, it would be dynamically linked with the endomembrane system. Interestingly, with respect to the intracellular dynamics, we observed that the TTN5T30N mutant had a different behavior in different organ types. This could be due to differing GEFs. Likewise, it is conceivable that constitutively expressed TTN5 has different effector binding partners.
The broad diversity of biological functions for proteins that have related sequences to TTN5, such that they are associated with a variety of signaling cascades, is also reflected by these related proteins having very different protein partners. Few orthologs of HsARL2 interaction partners are present in Arabidopsis. It is therefore exceedingly interesting to identify interacting proteins to determine whether TTN5 performs similar functions to HsARL2 or what other role it might play, especially with regard to the potential GTP dependence of TTN5 essential function, which fits with already known functions of other ARF GTPases (Sztul et al., 2019; Nielsen, 2020; Adarska et al., 2021). In addition, ARF proteins are affected by a similar set of GEFs and GAPs, indicating an interconnected network of ARF signaling. ARF double knockdowns have revealed specific phenotypes, suggesting redundancy in the ARF family (Volpicelli-Daley et al., 2005; Kondo et al., 2012; Nakai et al., 2013; Adarska et al., 2021). Investigation of the role of TTN5 within the ARF family might reveal a missing link in ARF signaling and cell traffic.
Conclusion
In this study, we identified TTN5 as a functional ARF-like GTPase that not only shares sequence similarity with HsARL2, but also the very fast nucleotide exchange capacity, in contrast to other characterized ARF and ARL proteins. TTN5 has a fast nucleotide dissociation and a slow GTP hydrolysis rate, and a higher affinity for GTP than GDP. Thus, TTN5 is a non-classical GTPase that most likely accumulates in a GTP-bound state in cells in line with certain cellular phenotypes and protein localization data. The nucleotide exchange capacity affected the localization and dynamics of YFP-tagged TTN5 forms, and the association of TTN5 with the endomembrane system. In the future, identification of a potential TTN5 GEF, GAP and effector proteins as well as other interaction partners, and particularly potential PM target proteins as cargo for vesicle transport, will be of great interest to clarify potential roles of TTN5 in endomembrane trafficking and whole-plant physiological contexts.
MATERIALS AND METHODS
Arabidopsis plant material and plant growth conditions
The Arabidopsis ttn5-1 (Stock Number: CS16077) mutant was previously described (McElver et al., 2000). Heterozygous seedlings were selected by genotyping using the primers TTN5 intron1 fwd and pDAP101 LB1 (Table S1). For pro35S::YFP-TTN5 and pro35S::HA3-TTN5 constructs, TTN5 coding sequences was amplified with B1 and B2 attachment sites for Gateway cloning (Life Technologies) using the primer TITAN5 n-ter B1 and TITAN5 stop B2 (Table S1). The obtained PCR fragments were cloned via BP reaction (Life Technologies) into pDONR207 (Invitrogen). pro35S::YFP-TTN5 and pro35S::HA3-TTN5 constructs were created via LR reaction (Life Technologies) with the destination vector pH7WGY2 (Karimi et al., 2005) (VIB-UGent Center for Plant Systems Biology, Vector ID:1_48) and pALLIGATOR2 (Bensmihen et al., 2004), respectively. Agrobacteria were transformed with obtained constructs and used for stable Arabidopsis transformation (method adapted from Clough and Bent, 1998). Arabidopsis seeds were sterilized with sodium hypochlorite solution (6% sodium hypochlorite and 0.1% Triton X-100) and stored for 24 h at 4°C for stratification. Seedlings were grown upright on half-strength Hoagland agar medium [1.5 mM Ca(NO3)2, 0.5 mM KH2PO4, 1.25 mM KNO3, 0.75 mM MgSO4, 1.5 µM CuSO4, 50 µM H3BO3, 50 µM KCl, 10 µM MnSO4, 0.075 µM (NH4)6Mo7O24, 2 µM ZnSO4, 50 μM FeNaEDTA and 1% sucrose, pH 5.8, supplemented with 1.4% Plant agar (Duchefa)] in growth chambers (CLF Plant Climatics) under long-day conditions (16 h light at 21°C, 8 h darkness at 19°C). Seedlings were grown for 6 days (6-day system) or 10 days (10-day system) or 17 days with the last 3 days on fresh plates (2-week system).
Seed clearing was undertaken by incubating seeds in a chloral hydrate-glycerol clearing solution [chloral hydrate:glycerol:water, 8:1:2 (g:ml:ml)] for 4 h up to overnight. Imaging was done using an Axio Imager.M2 (Zeiss).
Root length measurement were performed using JMicroVision with the image analysis toolbox for measuring and quantifying components of high-definition images [version 1.3.4; https://jmicrovision.github.io; Roduit, N.]
Nicotiana benthamiana plants were grown on soil for 2–4 weeks in a greenhouse facility under long-day conditions (16 h of light, 8 h of darkness).
Point mutant generation of TTN5
pDONR207:TTN5 was used as a template for site-directed TTN5 mutagenesis. Primers T5T30Nf and T5T30Nr (Table S1) were used to amplify the entire vector generating the TTN5T30N coding sequence and primers TQ70Lf and T5Q70Lr (Table S1) were used to amplify the entire vector generating the TTN5Q70L coding sequence. The PCR amplifications were run using the following conditions: 95°C, 30 s; 18 cycles of 95°C for 30 s, 55°C for 1 min, and 72°C for 8 min; then 72°C for 7 min. The completed reaction was treated with 10 units of DpnI endonuclease for 1 h at 37°C and then used for Escherichia coli transformation. Successful mutagenesis was confirmed by Sanger sequencing.
In vitro GTPase activity assays
An overview of protein expression and purification is shown in Fig. S2A. Recombinant pGEX-4T-1 bacterial protein expression vectors (Amersham, Germany) containing coding sequences for TTN5, TTN5T30N and TTN5Q70L were transferred into E. coli BL21 (DE3) Rosetta strain (Invitrogen, Germany). Following induction of GST–TTN5 fusion protein expression according to standard procedures (Hemsath et al., 2005), cell lysates were obtained after cell disruption with a probe sonicator (Bandelin sonoplus ultrasonic homogenizer, Germany) using a standard buffer [300 mM NaCl, 3 mM dithiothreitol (DTT), 10 mM MgCl2, 0.1 mM GDP, 1% glycerol and 50 mM Tris-HCl, pH 7.4]. GST fusion proteins were purified by loading total bacterial lysate on a preequilibrated glutathione Sepharose column (Sigma, Germany) using fast performance liquid chromatography system (Cytiva, Germany) (Step 1, affinity-purified GST-TTN5 protein fraction). GST-tagged protein fractions were incubated with thrombin (Sigma, Germany) at 4°C overnight for cleavage of the GST tag (Step 2, GST cleavage) and applied again to the affinity column (Step 3, yielding TTN5 protein fraction). Purified proteins were concentrated using 10 kDa ultra-centrifugal filter Amicon (Merck Millipore, Germany). The quality and quantity of proteins were analyzed by SDS-PAGE (Bio-Rad), a UV/Vis spectrometer (Eppendorf, Germany) and high-performance liquid chromatography (HPLC) using a reversed-phase C18 column (Sigma, Germany) and a pre-column (Nucleosil 100 C18, Bischoff Chromatography) as described previously (Eberth and Ahmadian, 2009) (Fig. S2B–D).
Nucleotide-free TTN5 protein was prepared from the TTN5 protein fraction (Eberth and Ahmadian, 2009) as illustrated in Fig. S2E. 0.5 mg TTN5 protein was combined with 1 U of agarose bead-coupled alkaline phosphatase (Sigma-Aldrich, Germany) for degradation of bound GDP to GMP and Pi in the presence of a 1.5-fold molar excess of non-hydrolyzable GTP analog GppCp (Jena Bioscience, Germany). After confirmation of GDP degradation by HPLC, 0.002 U snake venom phosphodiesterase (Sigma-Aldrich, Germany) per mg TTN5 was added to cleave GppCp to GMP, G and Pi. The reaction progress of degradation of nucleotides was analyzed by HPLC using 30 µM TTN5 in a 30 µl injection volume (Beckman Gold HPLC, Beckman Coulter). After completion of the reaction, in order to remove the agarose bead-coupled alkaline phosphatase, the solution was centrifuged for 10 min at 10,000 g, 4°C, which was followed by snap freezing and thawing cycles to inactivate the phosphodiesterase. mdGDP (2-deoxy-3-O-N-methylanthraniloyl GDP)- and mGppNHp 2′/3′-O-(N-methyl-anthraniloyl)-guanosine-5′-[(β,γ)-imido]triphosphate)-bound TTN5, TTN5T30N and TTN5Q70L were prepared by incubation of nucleotide-free forms with fluorescent nucleotides (Jena Bioscience, Germany) in a molar ratio of 1 to 1.2. The solution was purified from the excess amount of mdGDP and mGppNHp by using prepacked gel-filtration NAP-5 Columns (Cytiva, Germany) to remove unbound nucleotides. Protein and nucleotide concentration were determined using the Bradford reagent (Sigma-Aldrich, Germany) and HPLC, respectively.
All kinetic fluorescence measurements including nucleotide association and dissociation reactions were monitored on a stopped-flow instrument system SF-61, HiTech Scientific (TgK Scientific Limited, UK) and SX20 MV (Applied Photophysics, UK) at 25°C using nucleotide exchange buffer (10 mM K2HPO4/KH2PO4, pH 7.4, 5 mM MgCl2, 3 mM DTT, 30 mM Tris-HCl, pH 7.5) (Eberth and Ahmadian, 2009). Fluorescence was detected at 366 nm excitation and 450 nm emission using 408 nm cut-off filter for mant-nucleotides (Hemsath and Ahmadian, 2005).
To determine the intrinsic nucleotide exchange rate, koff, 0.2 µM mdGDP- and mGppNHp-bound proteins were combined with a 200-fold molar excess of 40 µM non-fluorescent GDP in two different set of experiments, respectively. The decay of the fluorescence intensity representing mdGDP and mGppNHp dissociation and replacement by non-fluorescent nucleotide were recorded over time (Fig. S2G). Moreover, to determine the nucleotide association rate, kon, of mdGDP and mGppNHp to the nucleotide-free GTPase, 0.2 µM fluorescent nucleotides were mixed with different concentrations of nucleotide-free TTN5 variants. The increase in the fluorescent intensity was obtained by analyzing the conformational change of fluorescent nucleotides after binding to the proteins (Fig. S2H).
The data provided by the stopped-flow assay, were applied to obtain the observed rate constants. Dissociation rate constants or nucleotide exchange rates (koff in s−1) and pseudo-first-order rate constants or observed rate constants (kobs in s−1) at the different concentrations of the protein were obtained by non-linear curve fitting using Origin software (version 2021b). The slopes obtained from plotting kobs against respective concentrations of proteins were used as the second-order association rate constants (kon in µM−1s−1). The equilibrium constant of dissociation (Kd in µM) was calculated from the ratio of koff/kon. In order to investigate the intrinsic GTP-hydrolysis rate of TTN5 variants, the HPLC method is used as described previously (Eberth and Ahmadian, 2009). As an accurate strategy, HPLC provides the nucleotide contents over time. The GTPase reaction rates were determined by mixing 100 μM nucleotide-free GTPase and 100 μM GTP at 25°C in a standard buffer without GDP. The GTP contents were measured at different times and the data were fitted with Origin software to get the observed rate constant.
Nicotiana benthamiana leaf infiltration
N. benthamiana leaf infiltration was performed with the Agrobacterium (Agrobacterium radiobacter) strain C58 (GV3101) carrying the respective constructs for confocal microscopy (LSM 780, Zeiss). Agrobacteria cultures were grown overnight at 28°C, centrifuged for 5 min at 4°C at 5000 g, resuspended in infiltration solution (5% sucrose, a pinch of glucose, 0.01% Silwet Gold, 150 µM Acetosyringone) and incubated for 1 h at room temperature. The bacterial suspension was adjusted to an OD600=0.4 and infiltrated into the abaxial side of N. benthamiana leaves.
Subcellular localization of fluorescent protein fusions
Cloning of YFP-tagged TTN5 constructs is described in the section ‘Arabidopsis plant material and growth conditions’ above. Localization studies were carried out by laser-scanning confocal microscopy (LSM 780 or LSM880, Zeiss) with a 40× C-Apochromat water immersion objective. YFP constructs and Alexa Fluor 488 stainings were excited at 488 nm and detected at 491–560 nm. mCherry, Alexa 555 and FM4-64 fluorescence was excited at 561 nm and detected at 570–633 nm.
Wortmannin (10 µM, Sigma-Aldrich), BFA (36 µM, Sigma-Aldrich) and plasma membrane dyes FM4-64 (165 µM, Thermo Fisher Scientific) were infiltrated into N. benthamiana leaves or used for incubation bath of Arabidopsis seedlings. FM4-64 was detected after 5 min incubation. Wortmannin and BFA were incubated for 25 min before checking the treatment effect. Plasmolysis was induced by incubating leaf discs in 1 M mannitol solution for 15 min. Signal intensities were increased for better visibility.
RFP–ARA7 clones were a gift from Dr Thierry Gaude (Ecole Normale Supérieure, Lyon, France).
Whole-mount immunostaining
Whole-mount immunostaining by immunofluorescence was performed according to Pasternak et al. (2015). Briefly, Arabidopsis seedlings were grown in the standard condition in Hoagland medium for 4–6 days. Methanol or formaldehyde (4%) was used to fix the seedlings. The seedlings were transferred to a glass slide and resuspended in 1× microtubule-stabilizing buffer (MTSB). Seedlings were digested with 2% Driselase dissolved in 1× MTSB at 37°C for 40 mins. Following digestion, a permeabilization step was performed by treating the seedlings with permeabilization buffer (3% IGEPAL C630, 10% DMSO in 1× MTSB buffer) at 37°C for 20 mins. Then blocking was performed with a buffer consisting of 5% BSA for 30 min at room temperature. They were incubated overnight with different primary antibodies (detailed information is listed below). After two washes with 1× MTSB, seedlings were incubated with a respective Alexa Fluor secondary antibody for 2 h at 37°C. After five steps of washing with 1x PBS, coverslips were mounted on slides with the antifade reagent (Prolong glass Antifade Mountant with NucBlue Stain, Invitrogen, P36985). Fluorescence microscopy was conducted as described in the previous section.
Immunodetection was conducted with following antibody combinations: HA detection was performed using anti-HA antibody (1:100 dilution, rabbit Abcam ab9110 or chicken AGRISERA, AS20 4463, Lot: 2303) followed by Alexa Fluor 488 or Alexa Fluor 555-labeled secondary antibodies (1:200 anti-rabbit-IgG, Thermo Fisher Scientific, A32731, Lot: 2541675 and 1:500 goat anti-chicken-IgY, Thermo Fisher Scientific A32932). ARF1 (Golgi and TGN marker) was detected using primary anti-ARF1 antibody (1:200 dilution, rabbit, Agrisera, AS08 325, Lot: 2208), in combination with Alexa Fluor™ Plus 488-labeled secondary antibody.
For initiation of BFA bodies, seedlings were first treated with BFA (72 µM, Sigma-Aldrich) and fixable plasma membrane dye FM4-64 FX (10 mM, Thermo Fisher Scientific, F34653) for 1 h, before formaldehyde fixation.
Immunoblot detection
After total protein extraction from Arabidopsis plants grown for 6 days or in the 2-week system, sample separation by SDS-PAGE and immunodetection were performed as previously described (Le et al., 2015). In brief, plant material was ground under liquid nitrogen and proteins were extracted with SDG buffer (62 mM Tris-HCl, pH 8.6, 2.5% SDS, 2% DTT, 10% glycerol). Samples were separated on 12% SDS-PAGE gels. Following electrophoresis, the proteins were transferred to a Protran nitrocellulose membrane (Amersham).
Membranes were blocked for 1 h in 5% milk-TBST solution (20 mM Tris-HCl, pH 7.4, 180 mM NaCl and 0.1% Tween 20), followed by 1 h antibody incubation (anti-GFP, monoclonal mouse antibody, Roche, catalog no. 11814460001, 1:1000). After three washes with TBST for 10 min each, membranes were incubated in secondary antibody (anti-mouse-IgG conjugated to HRP, polyclonal goat antibody, Sigma-Aldrich, cat. no. SAB3701159, 1:5000) for 1 h. HA detection was performed with a directly coupled anti-HA antibody (anti-HA–HRP, high-affinity monoclonal rat antibody, 3F10, Roche, catalog no. 12013819001, 1:1000). Immunodetection was performed after three washes with TBST for 10 min each, using the enhanced chemiluminescence system (GE Healthcare) and the FluorChem Q System for quantitative western blot imaging (ProteinSimple) with the AlphaView software. Full images of uncropped western blot from this study can be found in Fig. S9.
JACoP based colocalization analysis
Colocalization analysis was carried out with the ImageJ (Schneider et al., 2012) Plugin Just Another Colocalization Plugin (JACoP) (Bolte and Cordelières, 2006) and a comparison of Pearson's and Overlap coefficients was performed. Object-based analysis was performed for punctate structures (method adapted from Ivanov et al., 2014). Colocalization for both channels was calculated based on the distance between geometrical centers of signals and presented as a percentage. Analysis was done in three replicates each (n=3).
Structure prediction
TTN5 structure prediction was performed by AlphaFold (Jumper et al., 2021). The molecular graphic was edited with UCSF ChimeraX (1.2.5, Goddard et al., 2018), developed by the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco, with support from the National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases.
In silico tool for gene expression analysis
RNA-seq data was analyzed from previously published studies and was visualized with the AtGenExpress eFP at https://bar.utoronto.ca/eplant/ (Nakabayashi et al., 2005; Schmid et al., 2005; Ryu et al., 2019; Waese et al., 2017).
Accession numbers for sequence data
Sequence data used in this article can be found in the TAIR and GenBank data libraries under accession numbers: ARA7 (TAIR: AT4G19640), ARF1 (TAIR: AT1G23490), GmMan1 (Uniprot: Q0PKY2) and TTN5 (TAIR: AT2G18390).
Statistical analysis
One-way ANOVA was used for statistical analysis and performed in OriginPro 2019. Fisher's least significant difference or a Tukey's test was chosen as a post-hoc test with P<0.05.
Supplementary Material
Acknowledgements
We thank Gintaute Matthäi and Elke Wieneke for excellent technical and Natalie Köhler for experimental assistance. Thanks to Anna Sergeeva, Madhumita Narasimhan and Alexander Hertle for advice and help with whole-mount immunolocalization. We thank Stefanie Weidtkamp-Peters and Sebastian Hänsch for assistance and Ksenia Krooß (NFDI4BIOIMAGE, DFG project ID 501864659) for help with microscopy, metadata annotation and upload to the BioImage Archive repository, all members of the Center for Advanced Imaging (CAi) at Heinrich Heine University. Funding for instrumentation was through Zeiss LSM780+4channel FLIM extension (Picoquant): DFG-INST 208/551-1 FUGG and Zeiss LSM 880 Airyscan Fast DFG-INST 208/746-1 FUGG.
Footnotes
Author contributions
Conceptualization: I.M., M.R.A., R.I., P.B.; Formal analysis: I.M., A.M., M.R.A., R.I.; Investigation: I.M., A.M., S.K.S., P.C., R.I.; Writing - original draft: I.M.; Writing -review & editing: I.M., A.M., M.R.A., R.I., P.B.; Visualization: I.M., A.M.; Supervision: M.R.A., R.I., P.B.; Funding acquisition: M.R.A., P.B.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) Project no. 267205415-SFB 1208, project B05 to P.B.; Ba1610/7-2 to P.B.; Germany′s Excellence Strategy – EXC-2048/1 – project ID 390686111. Support was provided by DFG AH 92/8-3 to A.M. and M.R.A. Open access funding provided by Heinrich Heine University Duesseldorf. Deposited in PMC for immediate release.
Data availability
Microscopic images are available at BioImage Archive with the accession number S-BIAD1241 (doi:10.6019/S-BIAD1241).
Contributor Information
Rumen Ivanov, Email: Rumen.Ivanov@hhu.de.
Petra Bauer, Email: Petra.Bauer@hhu.de.
Peer review history
The peer review history is available online at https://journals.biologists.com/jcs/lookup/doi/10.1242/jcs.262315.reviewer-comments.pdf
References
- Adarska, P., Wong-Dilworth, L. and Bottanelli, F. (2021). ARF GTPases and their ubiquitous role in intracellular trafficking beyond the Golgi. Front. Cell Dev. Biol. 9, 679046. 10.3389/fcell.2021.679046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadi, Y., Ghorbanihaghjo, A. and Argani, H. (2017). The balance between induction and inhibition of mevalonate pathway regulates cancer suppression by statins: a review of molecular mechanisms. Chem. Biol. Interact. 273, 273-285. 10.1016/j.cbi.2017.06.026 [DOI] [PubMed] [Google Scholar]
- Antoshechkin, I. and Han, M. (2002). The C. elegans evl-20 gene is a homolog of the small GTPase ARL2 and regulates cytoskeleton dynamics during cytokinesis and morphogenesis. Dev. Cell 2, 579-591. 10.1016/S1534-5807(02)00146-6 [DOI] [PubMed] [Google Scholar]
- Aspenström, P. (2020). Fast-cycling Rho GTPases. Small GTPases 11, 248-255. 10.1080/21541248.2017.1391365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, R., Adolf, F., Weimer, C., Bruegger, B. and Wieland, F. T. (2009). ArfGAP1 activity and COPI vesicle biogenesis. Traffic 10, 307-315. 10.1111/j.1600-0854.2008.00865.x [DOI] [PubMed] [Google Scholar]
- Beck, R., Prinz, S., Diestelkötter-Bachert, P., Röhling, S., Adolf, F., Hoehner, K., Welsch, S., Ronchi, P., Brügger, B., Briggs, J. A. G.et al. (2011). Coatomer and dimeric ADP ribosylation factor 1 promote distinct steps in membrane scission. J. Cell Biol. 194, 765-777. 10.1083/jcb.201011027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensmihen, S., To, A., Lambert, G., Kroj, T., Giraudat, J. and Parcy, F. (2004). Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett. 561, 127-131. 10.1016/S0014-5793(04)00148-6 [DOI] [PubMed] [Google Scholar]
- Bhamidipati, A., Lewis, S. A. and Cowan, N. J. (2000). ADP ribosylation factor-like protein 2. (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J. Cell Biol. 149, 1087-1096. 10.1083/jcb.149.5.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson, B., Giglione, C. and Meinnel, T. (2003). Unexpected protein families including cell defense components feature in the N-myristoylome of a higher eukaryote. J. Biol. Chem. 278, 43418-43429. 10.1074/jbc.M307321200 [DOI] [PubMed] [Google Scholar]
- Bolte, S. and Cordelières, F. P. (2006). A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213-232. 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Bolte, S., Talbot, C., Boutte, Y., Catrice, O., Read, N. D. and Satiat-Jeunemaitre, B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214, 159-173. 10.1111/j.0022-2720.2004.01348.x [DOI] [PubMed] [Google Scholar]
- Bos, J. L. (1988). The ras gene family and human carcinogenesis. Mutat. Res. 195, 255-271. 10.1016/0165-1110(88)90004-8 [DOI] [PubMed] [Google Scholar]
- Bottanelli, F., Kilian, N., Ernst, A. M., Rivera-Molina, F., Schroeder, L. K., Kromann, E. B., Lessard, M. D., Erdmann, R. S., Schepartz, A., Baddeley, D.et al. (2017). A novel physiological role for ARF1 in the formation of bidirectional tubules from the Golgi. Mol. Biol. Cell 28, 1676-1687. 10.1091/mbc.e16-12-0863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi, F. (2018). Transport from the endoplasmic reticulum to the Golgi in plants: where are we now? Semin. Cell Dev. Biol. 80, 94-105. 10.1016/j.semcdb.2017.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi, F., Snapp, E. L., Roberts, A. G., Lippincott-Schwartz, J. and Hawes, C. (2002). Membrane protein transport between the endoplasmic reticulum and the golgi in tobacco leaves is energy dependent but cytoskeleton independent: evidence from selective photobleaching. Plant Cell 14, 1293-1309. 10.1105/tpc.001586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, D. D. and Colecraft, H. M. (2015). Rad and Rem are non-canonical G-proteins with respect to the regulatory role of guanine nucleotide binding in Ca(V)1.2 channel regulation. J. Physiol. 593, 5075-5090. 10.1113/JP270889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils, J. and Zeghouf, M. (2013). Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93, 269-309. 10.1152/physrev.00003.2012 [DOI] [PubMed] [Google Scholar]
- Clough, S. J. and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735-743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cui, Y., Shen, J., Gao, C., Zhuang, X., Wang, J. and Jiang, L. (2016). Biogenesis of plant prevacuolar multivesicular bodies. Mol. Plant 9, 774-786. 10.1016/j.molp.2016.01.011 [DOI] [PubMed] [Google Scholar]
- Der, C. J., Finkel, T. and Cooper, G. M. (1986). Biological and biochemical properties of human rasH genes mutated at codon 61. Cell 44, 167-176. 10.1016/0092-8674(86)90495-2 [DOI] [PubMed] [Google Scholar]
- Dhonukshe, P., Aniento, F., Hwang, I., Robinson, D. G., Mravec, J., Stierhof, Y.-D. and Friml, J. (2007). Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17, 520-527. 10.1016/j.cub.2007.01.052 [DOI] [PubMed] [Google Scholar]
- Eberth, A. and Ahmadian, M. R. (2009). In vitro GEF and GAP assays. Curr. Protoc. Cell Biol. 43, 14.19.11-14.19.25. 10.1002/0471143030.cb1409s43 [DOI] [PubMed] [Google Scholar]
- Ebine, K., Fujimoto, M., Okatani, Y., Nishiyama, T., Goh, T., Ito, E., Dainobu, T., Nishitani, A., Uemura, T., Sato, M. H.et al. (2011). A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 13, 853-859. 10.1038/ncb2270 [DOI] [PubMed] [Google Scholar]
- ElMaghloob, Y., Sot, B., McIlwraith, M. J., Garcia, E., Yelland, T. and Ismail, S. (2021). ARL3 activation requires the co-GEF BART and effector-mediated turnover. Elife 10, e64624. 10.7554/eLife.64624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emans, N., Zimmermann, S. and Fischer, R. (2002). Uptake of a fluorescent marker in plant cells is sensitive to brefeldin A and wortmannin. Plant Cell 14, 71-86. 10.1105/tpc.010339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson, J. W., Cerione, R. A. and Hart, M. J. (1997). Identification of an actin cytoskeletal complex that includes IQGAP and the Cdc42 GTPase. J. Biol. Chem. 272, 24443-24447. 10.1074/jbc.272.39.24443 [DOI] [PubMed] [Google Scholar]
- Esposito, A., Ventura, V., Petoukhov, M. V., Rai, A., Svergun, D. I. and Vanoni, M. A. (2019). Human MICAL1: activation by the small GTPase Rab8 and small-angle X-ray scattering studies on the oligomerization state of MICAL1 and its complex with Rab8. Protein Sci. 28, 150-166. 10.1002/pro.3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fansa, E. K. and Wittinghofer, A. (2016). Sorting of lipidated cargo by the Arl2/Arl3 system. Small GTPases 7, 222-230. 10.1080/21541248.2016.1224454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano, G., Muto, V., Radio, F. C., Venditti, M., Mosaddeghzadeh, N., Coppola, S., Paradisi, G., Zara, E., Bazgir, F., Ziegler, A.et al. (2022). Dominant ARF3 variants disrupt Golgi integrity and cause a neurodevelopmental disorder recapitulated in zebrafish. Nat. Commun. 13, 6841. 10.1038/s41467-022-34354-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidyk, N., Wang, J. B. and Cerione, R. A. (2006). Influencing cellular transformation by modulating the rates of GTP hydrolysis by Cdc42. Biochemistry 45, 7750-7762. 10.1021/bi060365h [DOI] [PubMed] [Google Scholar]
- Fiegen, D., Haeusler, L. C., Blumenstein, L., Herbrand, U., Dvorsky, R., Vetter, I. R. and Ahmadian, M. R. (2004). Alternative splicing of Rac1 generates Rac1b, a self-activating GTPase. J. Biol. Chem. 279, 4743-4749. 10.1074/jbc.M310281200 [DOI] [PubMed] [Google Scholar]
- Fisher, S., Kuna, D., Caspary, T., Kahn, R. A. and Sztul, E. (2020). ARF family GTPases with links to cilia. Am. J. Physiol. Cell Physiol. 319, C404-C418. 10.1152/ajpcell.00188.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming, J. A., Vega, L. R. and Solomon, F. (2000). Function of tubulin binding proteins in vivo. Genetics 156, 69-80. 10.1093/genetics/156.1.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, P. M., Ghosh-Choudhury, N., Moyer, M. L., Mott, G. E., Thomas, C. A., Foster, B. A., Greenberg, N. M. and Kreisberg, J. I. (1999). Role of RhoA activation in the growth and morphology of a murine prostate tumor cell line. Oncogene 18, 4120-4130. 10.1038/sj.onc.1202792 [DOI] [PubMed] [Google Scholar]
- Goddard, T. D., Huang, C. C., Meng, E. C., Pettersen, E. F., Couch, G. S., Morris, J. H. and Ferrin, T. E. (2018). UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14-25. 10.1002/pro.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremer, L., Merbitz-Zahradnik, T., Dvorsky, R., Cirstea, I. C., Kratz, C. P., Zenker, M., Wittinghofer, A. and Ahmadian, M. R. (2011). Germline KRAS mutations cause aberrant biochemical and physical properties leading to developmental disorders. Hum. Mutat. 32, 33-43. 10.1002/humu.21377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler, L. C., Hemsath, L., Fiegen, D., Blumenstein, L., Herbrand, U., Stege, P., Dvorsky, R. and Ahmadian, M. R. (2006). Purification and biochemical properties of Rac1, 2, 3 and the splice variant Rac1b. Methods Enzymol. 406, 1-11. 10.1016/S0076-6879(06)06001-0 [DOI] [PubMed] [Google Scholar]
- Hanton, S. L., Matheson, L. A., Chatre, L. and Brandizzi, F. (2009). Dynamic organization of COPII coat proteins at endoplasmic reticulum export sites in plant cells. Plant J. 57, 963-974. 10.1111/j.1365-313X.2008.03740.x [DOI] [PubMed] [Google Scholar]
- Hanzal-Bayer, M., Linari, M. and Wittinghofer, A. (2005). Properties of the interaction of Arf-like protein 2 with PDEdelta. J. Mol. Biol. 350, 1074-1082. 10.1016/j.jmb.2005.05.036 [DOI] [PubMed] [Google Scholar]
- Hemsath, L. and Ahmadian, M. R. (2005). Fluorescence approaches for monitoring interactions of Rho GTPases with nucleotides, regulators, and effectors. Methods 37, 173-182. 10.1016/j.ymeth.2005.05.014 [DOI] [PubMed] [Google Scholar]
- Hemsath, L., Dvorsky, R., Fiegen, D., Carlier, M. F. and Ahmadian, M. R. (2005). An electrostatic steering mechanism of Cdc42 recognition by Wiskott-Aldrich syndrome proteins. Mol. Cell 20, 313-324. 10.1016/j.molcel.2005.08.036 [DOI] [PubMed] [Google Scholar]
- Heucken, N. and Ivanov, R. (2018). The retromer, sorting nexins and the plant endomembrane protein trafficking. J. Cell Sci. 131, 2. 10.1242/jcs.203695 [DOI] [PubMed] [Google Scholar]
- Hillig, R. C., Hanzal-Bayer, M., Linari, M., Becker, J., Wittinghofer, A. and Renault, L. (2000). Structural and biochemical properties show ARL3-GDP as a distinct GTP binding protein. Structure 8, 1239-1245. 10.1016/S0969-2126(00)00531-1 [DOI] [PubMed] [Google Scholar]
- Hodge, R. G., Schaefer, A., Howard, S. V. and Der, C. J. (2020). RAS and RHO family GTPase mutations in cancer: twin sons of different mothers? Crit. Rev. Biochem. Mol. Biol. 55, 386-407. 10.1080/10409238.2020.1810622 [DOI] [PubMed] [Google Scholar]
- Huang, X., Shen, Y., Zhang, Y., Wei, L., Lai, Y., Wu, J., Liu, X. and Liu, X. (2013). Rac1 mediates laminar shear stress-induced vascular endothelial cell migration. Cell Adh. Migr. 7, 472-478. 10.4161/cam.27171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail, S. A., Chen, Y. X., Rusinova, A., Chandra, A., Bierbaum, M., Gremer, L., Triola, G., Waldmann, H., Bastiaens, P. I. and Wittinghofer, A. (2011). Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat. Chem. Biol. 7, 942-949. 10.1038/nchembio.686 [DOI] [PubMed] [Google Scholar]
- Ito, E., Uemura, T., Ueda, T. and Nakano, A. (2016). Distribution of RAB5-positive multivesicular endosomes and the trans-Golgi network in root meristematic cells of Arabidopsis thaliana. Plant Biotechnol. 33, 281-286. 10.5511/plantbiotechnology.16.0218a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, R. and Vert, G. (2021). Endocytosis in plants: peculiarities and roles in the regulated trafficking of plant metal transporters. Biol. Cell 113, 1-13. 10.1111/boc.202000118 [DOI] [PubMed] [Google Scholar]
- Ivanov, R., Brumbarova, T., Blum, A., Jantke, A.-M., Fink-Straube, C. and Bauer, P. (2014). SORTING NEXIN1 is required for modulating the trafficking and stability of the Arabidopsis IRON-REGULATED TRANSPORTER1. Plant Cell 26, 1294-1307. 10.1105/tpc.113.116244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais, Y., Fobis-Loisy, I., Miège, C. and Gaude, T. (2008). Evidence for a sorting endosome in Arabidopsis root cells. Plant J. 53, 237-247. 10.1111/j.1365-313X.2007.03338.x [DOI] [PubMed] [Google Scholar]
- Jaiswal, M., Fansa, E. K., Dvorsky, R. and Ahmadian, M. R. (2013). New insight into the molecular switch mechanism of human Rho family proteins: shifting a paradigm. Biol. Chem. 394, 89-95. 10.1515/hsz-2012-0207 [DOI] [PubMed] [Google Scholar]
- Jian, X., Gruschus, J. M., Sztul, E. and Randazzo, P. A. (2012). The pleckstrin homology. (PH) domain of the Arf exchange factor Brag2 is an allosteric binding site. J. Biol. Chem. 287, 24273-24283. 10.1074/jbc.M112.368084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., Tunyasuvunakool, K., Bates, R., Žídek, A., Potapenko, A.et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583-589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, A. and Rösner, H. (2002). RAC1-dependent regulation of cholinergically induced lamellar protrusive activity is independent of MAPKinase and attenuated by active p-JNK. Neuroreport 13, 2443-2446. 10.1097/00001756-200212200-00014 [DOI] [PubMed] [Google Scholar]
- Just, W. W. and Peränen, J. (2016). Small GTPases in peroxisome dynamics. Biochim. Biophys. Acta. – Mol. Cell Res. 1863, 1006-1013. 10.1016/j.bbamcr.2016.01.004 [DOI] [PubMed] [Google Scholar]
- Kahn, R. A., Der and G, C. J. and Bokoch, M. (1992). The ras superfamily of GTP-binding proteins: guidelines on nomenclature. FASEB J. 6, 2512-2513. 10.1096/fasebj.6.8.1592203 [DOI] [PubMed] [Google Scholar]
- Kahn, R. A., Cherfils, J., Elias, M., Lovering, R. C., Munro, S. and Schurmann, A. (2006). Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J. Cell Biol. 172, 645-650. 10.1083/jcb.200512057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, S., Fansa, E. K., Möbitz, S., Ismail, S. A., Winter, R., Wittinghofer, A. and Weise, K. (2015). Effect of the N-Terminal Helix and Nucleotide Loading on the Membrane and Effector Binding of Arl2/3. Biophys. J. 109, 1619-1629. 10.1016/j.bpj.2015.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M., De Meyer, B. and Hilson, P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10, 103-105. 10.1016/j.tplants.2005.01.008 [DOI] [PubMed] [Google Scholar]
- Karnoub, A. E. and Weinberg, R. A. (2008). Ras oncogenes: split personalities. Nat. Rev. Mol. Cell Biol. 9, 517-531. 10.1038/nrm2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, S., Franco, M., Chardin, P. and Luton, F. (2006). Role of the Arf6 GDP/GTP cycle and Arf6 GTPase-activating proteins in actin remodeling and intracellular transport. J. Biol. Chem. 281, 12352-12361. 10.1074/jbc.M601021200 [DOI] [PubMed] [Google Scholar]
- Kondo, Y., Hanai, A., Nakai, W., Katoh, Y., Nakayama, K. and Shin, H. W. (2012). ARF1 and ARF3 are required for the integrity of recycling endosomes and the recycling pathway. Cell Struct. Funct. 37, 141-154. 10.1247/csf.12015 [DOI] [PubMed] [Google Scholar]
- Kotzer, A. M., Brandizzi, F., Neumann, U., Paris, N., Moore, I. and Hawes, C. (2004). AtRabF2b. (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J. Cell Sci. 117, 6377-6389. 10.1242/jcs.01564 [DOI] [PubMed] [Google Scholar]
- Kuemmerle, J. F. and Zhou, H. (2002). Insulin-like growth factor-binding protein-5. (IGFBP-5) stimulates growth and IGF-I secretion in human intestinal smooth muscle by Ras-dependent activation of p38 MAP kinase and Erk1/2 pathways. J. Biol. Chem. 277, 20563-20571. 10.1074/jbc.M200885200 [DOI] [PubMed] [Google Scholar]
- Latijnhouwers, M., Hawes, C., Carvalho, C., Oparka, K., Gillingham, A. K. and Boevink, P. (2005). An Arabidopsis GRIP domain protein locates to the trans-Golgi and binds the small GTPase ARL1. Plant J. 44, 459-470. 10.1111/j.1365-313X.2005.02542.x [DOI] [PubMed] [Google Scholar]
- Le, C. T. T., Brumbarova, T., Ivanov, R., Stoof, C., Weber, E., Mohrbacher, J., Fink-Straube, C. and Bauer, P. (2015). ZINC FINGER OF ARABIDOPSIS THALIANA12. (ZAT12) interacts with FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR. (FIT) linking iron deficiency and oxidative stress responses. Plant Physiol. 170, 540-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, G. J., Sohn, E. J., Lee, M. H. and Hwang, I. (2004). The Arabidopsis rab5 homologs rha1 and ara7 localize to the prevacuolar compartment. Plant Cell Physiol. 45, 1211-1220. 10.1093/pcp/pch142 [DOI] [PubMed] [Google Scholar]
- Lee, M. C., Orci, L., Hamamoto, S., Futai, E., Ravazzola, M. and Schekman, R. (2005). Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell 122, 605-617. 10.1016/j.cell.2005.07.025 [DOI] [PubMed] [Google Scholar]
- Li, C.-C., Wu, T.-S., Huang, C.-F., Jang, L.-T., Liu, Y.-T., You, S.-T., Liou, G.-G. and Lee, F.-J. S. (2012). GTP-binding-defective ARL4D alters mitochondrial morphology and membrane potential. PLoS One 7, e43552. 10.1371/journal.pone.0043552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, J. and Meinke, D. (2012). A comprehensive dataset of genes with a loss-of-function mutant phenotype in Arabidopsis. Plant Physiol. 158, 1115-1129. 10.1104/pp.111.192393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia, E., Luton, F., Partisani, M., Cherfils, J., Chardin, P. and Franco, M. (2004). The GDP-bound form of Arf6 is located at the plasma membrane. J. Cell Sci. 117, 2389-2398. 10.1242/jcs.01090 [DOI] [PubMed] [Google Scholar]
- Matsumoto, S., Taniguchi-Tamura, H., Araki, M., Kawamura, T., Miyamoto, R., Tsuda, C., Shima, F., Kumasaka, T., Okuno, Y. and Kataoka, T. (2021). Oncogenic mutations Q61L and Q61H confer active form-like structural features to the inactive state. (state 1) conformation of H-Ras protein. Biochem. Biophys. Res. Commun. 565, 85-90. 10.1016/j.bbrc.2021.05.084 [DOI] [PubMed] [Google Scholar]
- Mayer, U., Herzog, U., Berger, F., Inzé, D. and Jürgens, G. (1999). Mutations in the PILZ group genes disrupt the microtubule cytoskeleton and uncouple cell cycle progression from cell division in Arabidopsis embryo and endosperm. Eur. J. Cell Biol. 78, 100-108. 10.1016/S0171-9335(99)80011-9 [DOI] [PubMed] [Google Scholar]
- McElver, J., Patton, D., Rumbaugh, M., Liu, C.-M., Yang, L. J. and Meinke, D. (2000). The TITAN5 gene of Arabidopsis encodes a protein related to the ADP ribosylation factor family of GTP binding proteins. Plant Cell 12, 1379-1392. 10.1105/tpc.12.8.1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memon, A. R. (2004). The role of ADP-ribosylation factor and SAR1 in vesicular trafficking in plants. Biochim. Biophys. Acta 1664, 9-30. 10.1016/j.bbamem.2004.04.005 [DOI] [PubMed] [Google Scholar]
- Mori, R. and Toda, T. (2013). The dual role of fission yeast Tbc1/cofactor C orchestrates microtubule homeostasis in tubulin folding and acts as a GAP for GTPase Alp41/Arl2. Mol. Biol. Cell 24, 1713-1718. 10.1091/mbc.e12-11-0792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi, K., Okamoto, M., Koshiba, T., Kamiya, Y. and Nambara, E. (2005). Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J. 41, 697-709. 10.1111/j.1365-313X.2005.02337.x [DOI] [PubMed] [Google Scholar]
- Nakai, W., Kondo, Y., Saitoh, A., Naito, T., Nakayama, K. and Shin, H. W. (2013). ARF1 and ARF4 regulate recycling endosomal morphology and retrograde transport from endosomes to the Golgi apparatus. Mol. Biol. Cell 24, 2570-2581. 10.1091/mbc.e13-04-0197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar, N., Singh, K. and Garcia-Diaz, M. (2010). Structure of the dominant negative S17N mutant of Ras. Biochemistry 49, 1970-1974. 10.1021/bi9020742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, B. K., Cai, X. and Nebenführ, A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126-1136. 10.1111/j.1365-313X.2007.03212.x [DOI] [PubMed] [Google Scholar]
- Newman, L. E., Zhou, C. J., Mudigonda, S., Mattheyses, A. L., Paradies, E., Marobbio, C. M. and Kahn, R. A. (2014). The ARL2 GTPase is required for mitochondrial morphology, motility, and maintenance of ATP levels. PLoS One 9, e99270. 10.1371/journal.pone.0099270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, E. (2020). The small GTPase superfamily in plants: a conserved regulatory module with novel functions. Annu. Rev. Plant Biol. 71, 247-272. 10.1146/annurev-arplant-112619-025827 [DOI] [PubMed] [Google Scholar]
- Niu, F., Ji, C., Liang, Z., Guo, R., Chen, Y., Zeng, Y. and Jiang, L. (2022). ADP-ribosylation factor D1 modulates Golgi morphology, cell plate formation, and plant growth in Arabidopsis. Plant Physiol. 190, 1199-1213. 10.1093/plphys/kiac329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak, T., Tietz, O., Rapp, K., Begheldo, M., Nitschke, R., Ruperti, B. and Palme, K. (2015). Protocol: an improved and universal procedure for whole-mount immunolocalization in plants. Plant Methods 11, 50. 10.1186/s13007-015-0094-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe, P. A., Vardy, L. and Toda, T. (2000). A conserved small GTP-binding protein Alp41 is essential for the cofactor-dependent biogenesis of microtubules in fission yeast. FEBS Lett. 468, 84-88. 10.1016/S0014-5793(00)01202-3 [DOI] [PubMed] [Google Scholar]
- Radhakrishna, H., Al-Awar, O., Khachikian, Z. and Donaldson, J. G. (1999). ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J. Cell Sci. 112, 855-866. 10.1242/jcs.112.6.855 [DOI] [PubMed] [Google Scholar]
- Renna, L. and Brandizzi, F. (2020). The mysterious life of the plant trans-Golgi network: advances and tools to understand it better. J. Microsc. 278, 154-163. 10.1111/jmi.12881 [DOI] [PubMed] [Google Scholar]
- Ritzenthaler, C., Nebenführ, A., Movafeghi, A., Stussi-Garaud, C., Behnia, L., Pimpl, P., Staehelin, L. A. and Robinson, D. G. (2002). Reevaluation of the effects of brefeldin A on plant cells using tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. Plant Cell 14, 237-261. 10.1105/tpc.010237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, D. G., Scheuring, D., Naramoto, S. and Friml, J. (2011). ARF1 localizes to the golgi and the trans-golgi network. Plant Cell 23, 846-849. 10.1105/tpc.110.082099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, K. H., Huang, L., Kang, H. M. and Schiefelbein, J. (2019). Single-Cell RNA Sequencing Resolves Molecular Relationships Among Individual Plant Cells. Plant Physiol. 179, 1444-1456. 10.1104/pp.18.01482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek, K., Ahmadian, M. R., Kabsch, W., Wiesmüller, L., Lautwein, A., Schmitz, F. and Wittinghofer, A. (1997). The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277, 333-339. 10.1126/science.277.5324.333 [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davison, T. S., Henz, S. R., Pape, U. J., Demar, M., Vingron M., Schölkopf, B., Weigel, D. and Lohmann J. U. (2005). A gene expression map of Arabidopsis thaliana development. Nat Genet. 37, 501-506. 10.1038/ng1543 [DOI] [PubMed] [Google Scholar]
- Schneider, C. A., Rasband, W. S. and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671-675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharer, J. D. and Kahn, R. A. (1999). The ARF-like 2. (ARL2)-binding protein, BART. Purification, cloning, and initial characterization. J. Biol. Chem. 274, 27553-27561. 10.1074/jbc.274.39.27553 [DOI] [PubMed] [Google Scholar]
- Sharer, J. D., Shern, J. F., Van Valkenburgh, H., Wallace, D. C. and Kahn, R. A. (2002). ARL2 and BART enter mitochondria and bind the adenine nucleotide transporter. Mol. Biol. Cell 13, 71-83. 10.1091/mbc.01-05-0245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirazi, F., Jones, R. J., Singh, R. K., Zou, J., Kuiatse, I., Berkova, Z., Wang, H., Lee, H. C., Hong, S., Dick, L.et al. (2020). Activating KRAS, NRAS, and BRAF mutants enhance proteasome capacity and reduce endoplasmic reticulum stress in multiple myeloma. Proc. Natl. Acad. Sci. USA 117, 20004-20014. 10.1073/pnas.2005052117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, M. K., Richter, S., Beckmann, H., Kientz, M., Stierhof, Y.-D., Anders, N., Fäßler, F., Nielsen, M., Knöll, C., Thomann, A.et al. (2018). A single class of ARF GTPase activated by several pathway-specific ARF-GEFs regulates essential membrane traffic in Arabidopsis. PLoS Genet. 14, e1007795. 10.1371/journal.pgen.1007795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh, U. J. and Low, B. C. (2008). BNIP2 extra long inhibits RhoA and cellular transformation by Lbc RhoGEF via its BCH domain. J. Cell Sci. 121, 1739-1749. 10.1242/jcs.021774 [DOI] [PubMed] [Google Scholar]
- Sohn, E. J., Kim, E. S., Zhao, M., Kim, S. J., Kim, H., Kim, Y. W., Lee, Y. J., Hillmer, S., Sohn, U., Jiang, L.et al. (2003). Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell 15, 1057-1070. 10.1105/tpc.009779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano, G., Renna, L., Hanton, S. L., Chatre, L., Haas, T. A. and Brandizzi, F. (2006). ARL1 plays a role in the binding of the GRIP domain of a peripheral matrix protein to the Golgi apparatus in plant cells. Plant Mol. Biol. 61, 431-449. 10.1007/s11103-006-0022-y [DOI] [PubMed] [Google Scholar]
- Stierhof, Y. D. and El Kasmi, F. (2010). Strategies to improve the antigenicity, ultrastructure preservation and visibility of trafficking compartments in Arabidopsis tissue. Eur. J. Cell Biol. 89, 285-297. 10.1016/j.ejcb.2009.12.003 [DOI] [PubMed] [Google Scholar]
- Sugawara, R., Ueda, H. and Honda, R. (2019). Structural and functional characterization of fast-cycling RhoF GTPase. Biochem. Biophys. Res. Commun. 513, 522-527. 10.1016/j.bbrc.2019.04.018 [DOI] [PubMed] [Google Scholar]
- Suo, Y., Hu, F., Zhu, H., Li, D., Qi, R., Huang, J. and Wu, W. (2021). BIG3 and BIG5 redundantly mediate vesicle trafficking in Arabidopsis. Biomolecules 11, 732. 10.3390/biom11050732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztul, E., Chen, P.-W., Casanova, J. E., Cherfils, J., Dacks, J. B., Lambright, D. G., Lee, F.-J. S., Randazzo, P. A., Santy, L. C., Schürmann, A.et al. (2019). ARF GTPases and their GEFs and GAPs: concepts and challenges. Mol. Biol. Cell 30, 1249-1271. 10.1091/mbc.E18-12-0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichá, M., Richter, H., Ovečka, M., Maghelli, N., Hrbáčková, M., Dvořák, P., Šamaj, J. and Šamajová, O. (2020). Advanced microscopy reveals complex developmental and subcellular localization patterns of ANNEXIN 1 in Arabidopsis. Front. Plant Sci. 11, 1153. 10.3389/fpls.2020.01153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir, I., McElver, J. A., Liu, C.-M., Yang, L. J., Wu, J. Q., Martinez, A., Patton, D. A. and Meinke, D. W. (2002). Diversity of TITAN functions in Arabidopsis seed development. Plant Physiol. 128, 38-51. 10.1104/pp.010911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, T., Yamaguchi, M., Uchimiya, H. and Nakano, A. (2001). Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20, 4730-4741. 10.1093/emboj/20.17.4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia, J. P., Goodman, K. and Otegui, M. S. (2016). Endocytosis and endosomal trafficking in plants. Annu. Rev. Plant Biol. 67, 309-335. 10.1146/annurev-arplant-043015-112242 [DOI] [PubMed] [Google Scholar]
- Van Valkenburgh, H., Shern, J. F., Sharer, J. D., Zhu, X. and Kahn, R. A. (2001). ADP-ribosylation factors. (ARFs) and ARF-like 1. (ARL1) have both specific and shared effectors: characterizing ARL1-binding proteins. J. Biol. Chem. 276, 22826-22837. 10.1074/jbc.M102359200 [DOI] [PubMed] [Google Scholar]
- Vanoni, M., Bertini, R., Sacco, E., Fontanella, L., Rieppi, M., Colombo, S., Martegani, E., Carrera, V., Moroni, A., Bizzarri, C.et al. (1999). Characterization and properties of dominant-negative mutants of the ras-specific guanine nucleotide exchange factor CDC25(Mm). J. Biol. Chem. 274, 36656-36662. 10.1074/jbc.274.51.36656 [DOI] [PubMed] [Google Scholar]
- Veltel, S., Kravchenko, A., Ismail, S. and Wittinghofer, A. (2008). Specificity of Arl2/Arl3 signaling is mediated by a ternary Arl3-effector-GAP complex. FEBS Lett. 582, 2501-2507. 10.1016/j.febslet.2008.05.053 [DOI] [PubMed] [Google Scholar]
- Vernoud, V., Horton, A. C., Yang, Z. and Nielsen, E. (2003). Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 131, 1191-1208. 10.1104/pp.013052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli-Daley, L. A., Li, Y., Zhang, C. J. and Kahn, R. A. (2005). Isoform-selective effects of the depletion of ADP-ribosylation factors 1-5 on membrane traffic. Mol. Biol. Cell 16, 4495-4508. 10.1091/mbc.e04-12-1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waese, J., Fan, J., Pasha, A., Yu, H., Fucile, G., Shi, R., Cumming, M., Kelley, L. A., Sternberg, M. J., Krishnakumar, V.et al. (2017). ePlant: visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell 29, 1806-1821. 10.1105/tpc.17.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, T. G., Li, Y., Wersäll, A. and Poole, A. W. (2019). Small GTPases in platelet membrane trafficking. Platelets 30, 31-40. 10.1080/09537104.2018.1535703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. B., Wu, W. J. and Cerione, R. A. (2005). Cdc42 and Ras cooperate to mediate cellular transformation by intersectin-L. J. Biol. Chem. 280, 22883-22891. 10.1074/jbc.M414375200 [DOI] [PubMed] [Google Scholar]
- Wang, J., Cai, Y., Miao, Y., Lam, S. K. and Jiang, L. (2009). Wortmannin induces homotypic fusion of plant prevacuolar compartments*. J. Exp. Bot. 60, 3075-3083. 10.1093/jxb/erp136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Liu, F., Ren, Y., Wang, Y., Liu, X., Long, W., Wang, D., Zhu, J., Zhu, X., Jing, R.et al. (2016). GOLGI TRANSPORT 1B regulates protein export from the endoplasmic reticulum in rice endosperm cells. Plant Cell 28, 2850-2865. 10.1105/tpc.16.00717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M. and Barton, G. J. (2009). Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189-1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann, T., Bokoch, G. M. and Waterman-Storer, C. M. (2003). Regulation of leading edge microtubule and actin dynamics downstream of Rac1. J. Cell Biol. 161, 845-851. 10.1083/jcb.200303082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, J., Kahn, R. A. and Sztul, E. (2014). Regulating the large Sec7 ARF guanine nucleotide exchange factors: the when, where and how of activation. Cell. Mol. Life Sci. 71, 3419-3438. 10.1007/s00018-014-1602-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. J., Rosenwald, A. G., Willingham, M. C., Skuntz, S., Clark, J. and Kahn, R. A. (1994). Expression of a dominant allele of human ARF1 inhibits membrane traffic in vivo. J. Cell Biol. 124, 289-300. 10.1083/jcb.124.3.289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Yang, Z., Lin, T., Wu, Y., Zou, T., yang, J. and Zhang, Y. (2018). ARL2 regulates trafficking and expression of isoprenylated proteins and is crucial for development of photoreceptor outer segments. Invest. Ophthalmol. Vis. Sci. 59, 963-963. [Google Scholar]
- Zhou, C., Cunningham, L., Marcus, A. I., Li, Y. and Kahn, R. A. (2006). Arl2 and Arl3 regulate different microtubule-dependent processes. Mol. Biol. Cell 17, 2476-2487. 10.1091/mbc.e05-10-0929 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.