Abstract
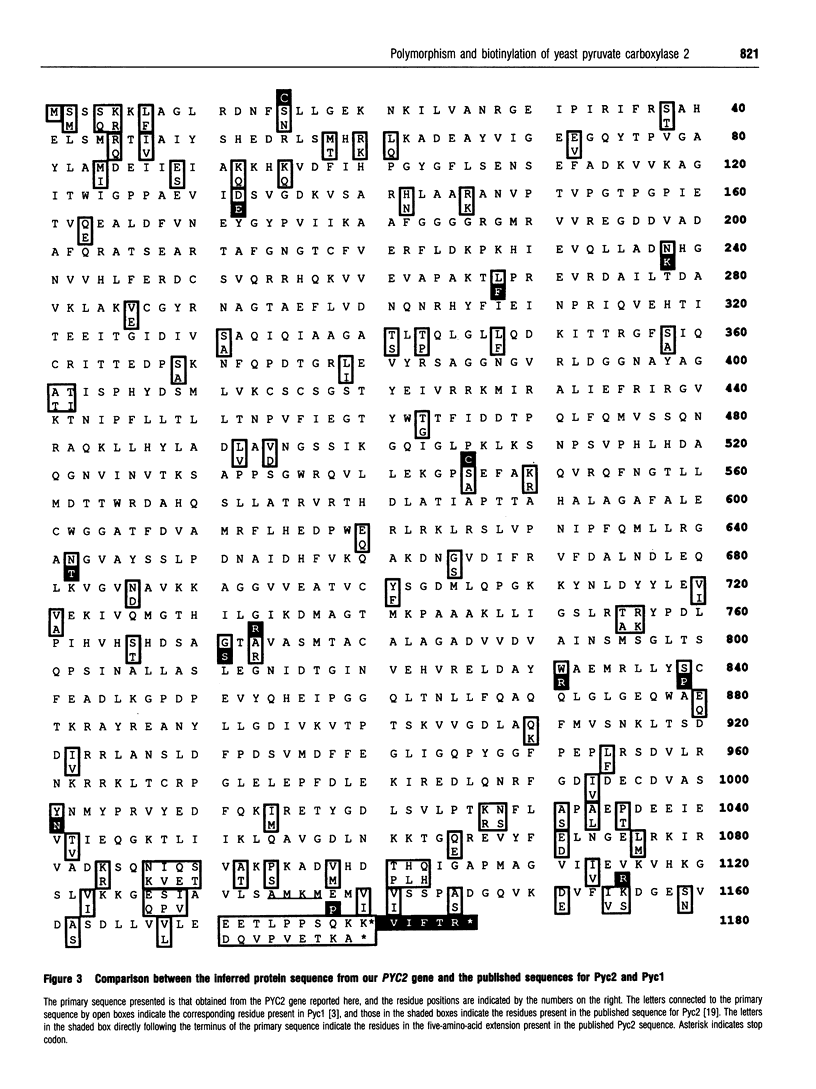
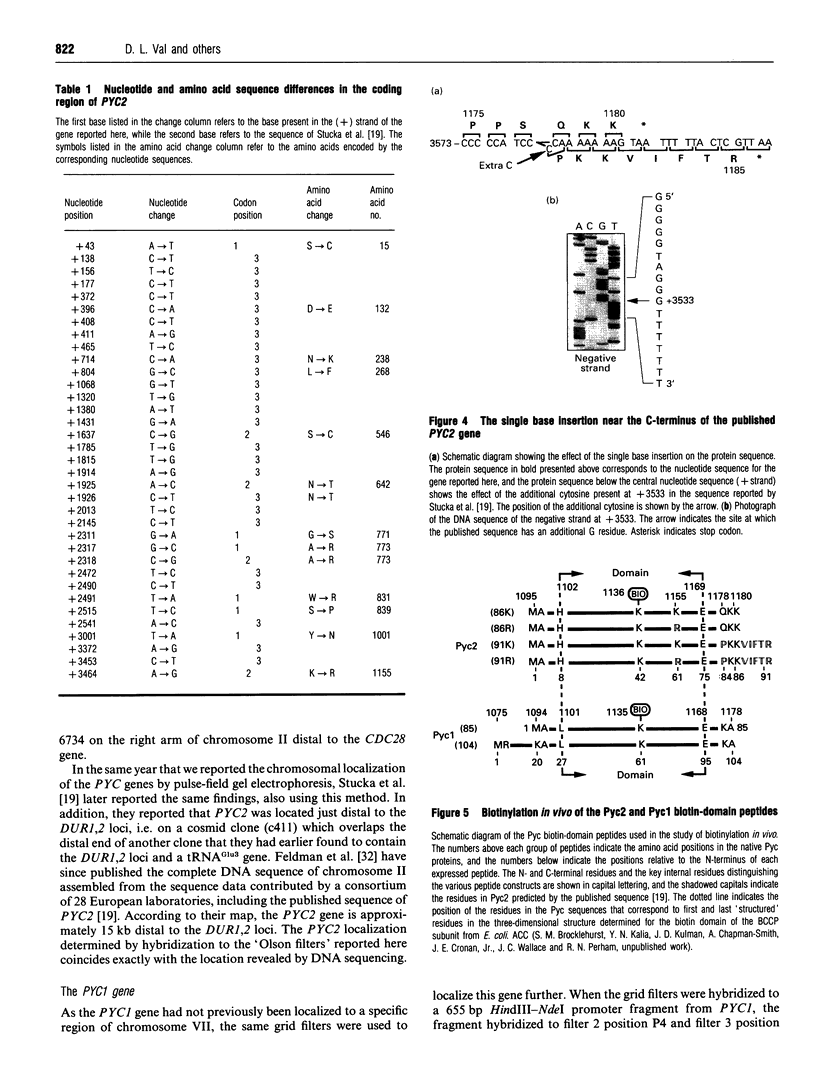
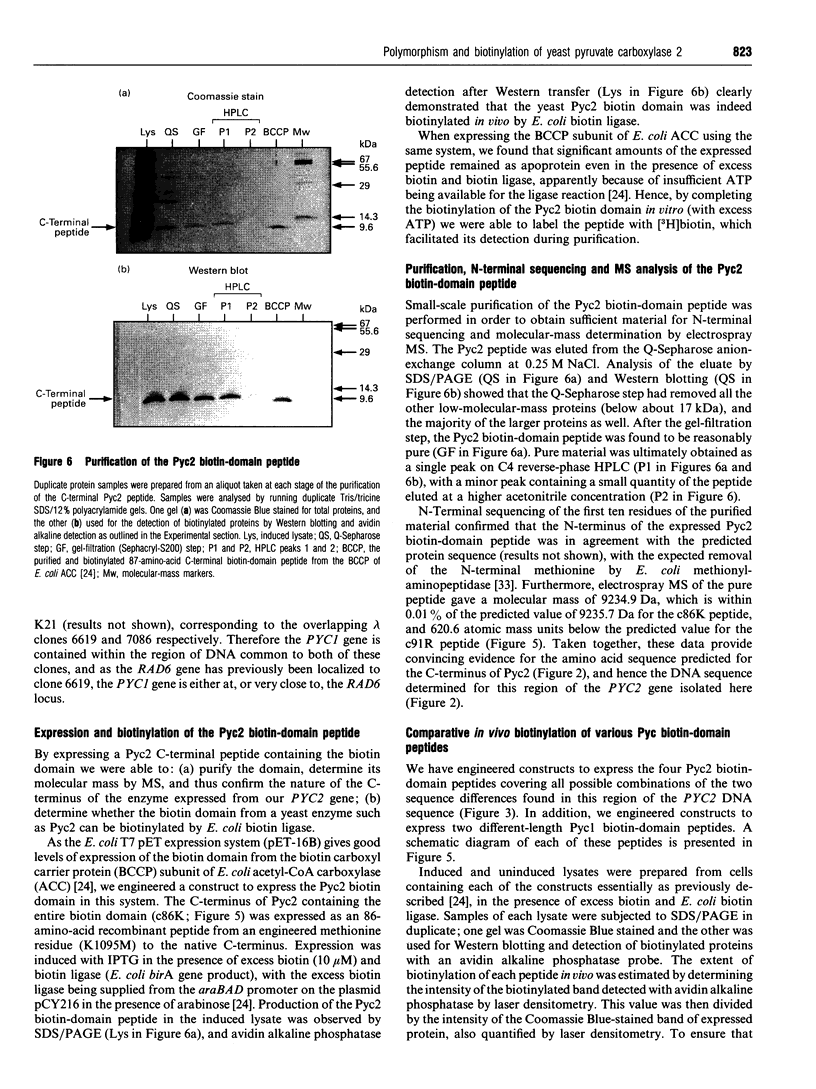
In Saccharomyces cerevisiae there are two isoenzymes of pyruvate carboxylase (Pyc) encoded by separate genes designated PYC1 and PYC2. We report the isolation and sequencing of a PYC2 gene, and the localization of both genes on the physical map of S. cerevisiae. Comparison with the previously reported sequence [Stucka, Dequin, Salmon and Gancedo (1991) Mol. Gen. Genet. 229, 307-315] revealed significant differences within the open reading frame. The most notable difference was near the 3' end, where we found a single base deletion reducing the open reading frame by 15 bases. We have confirmed the C-terminus of Pyc2 encoded by the gene isolated here by expressing and purifying an 86-amino-acid biotin-domain peptide. In addition, we investigated the effects of the two changes in the Pyc2 biotin domain (K1155R substitution and Q1178P/five-amino-acid extension) on the extent of biotinylation in vivo by Escherichia coli biotin ligase, and compared the biotinylation of peptides containing these changes with that of two different-length Pyc1 biotin-domain peptides. The K1155R substitution had very little effect on biotinylation, but the five-amino-acid C-terminal extension to Pyc2 and the N-terminal extension to Pycl both improved biotinylation in vivo.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Attwood P. V., Keech D. B. Pyruvate carboxylase. Curr Top Cell Regul. 1984;23:1–55. doi: 10.1016/b978-0-12-152823-2.50005-2. [DOI] [PubMed] [Google Scholar]
- Bordo D., Argos P. Suggestions for "safe" residue substitutions in site-directed mutagenesis. J Mol Biol. 1991 Feb 20;217(4):721–729. doi: 10.1016/0022-2836(91)90528-e. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brocklehurst S. M., Perham R. N. Prediction of the three-dimensional structures of the biotinylated domain from yeast pyruvate carboxylase and of the lipoylated H-protein from the pea leaf glycine cleavage system: a new automated method for the prediction of protein tertiary structure. Protein Sci. 1993 Apr;2(4):626–639. doi: 10.1002/pro.5560020413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M. A., Olah T. A., Weitzmann C. J., Cooperman B. S. Protein estimation by the product of integrated peak area and flow rate. Anal Biochem. 1989 Nov 1;182(2):295–299. doi: 10.1016/0003-2697(89)90597-6. [DOI] [PubMed] [Google Scholar]
- Cronan J. E., Jr Biotination of proteins in vivo. A post-translational modification to label, purify, and study proteins. J Biol Chem. 1990 Jun 25;265(18):10327–10333. [PubMed] [Google Scholar]
- Dardel F., Davis A. L., Laue E. D., Perham R. N. Three-dimensional structure of the lipoyl domain from Bacillus stearothermophilus pyruvate dehydrogenase multienzyme complex. J Mol Biol. 1993 Feb 20;229(4):1037–1048. doi: 10.1006/jmbi.1993.1103. [DOI] [PubMed] [Google Scholar]
- Fall R. R., Vagelos P. R. Acetyl coenzyme A carboxylase. Proteolytic modification of biotin carboxyl carrier protein. J Biol Chem. 1973 Mar 25;248(6):2078–2088. [PubMed] [Google Scholar]
- Feldmann H., Aigle M., Aljinovic G., André B., Baclet M. C., Barthe C., Baur A., Bécam A. M., Biteau N., Boles E. Complete DNA sequence of yeast chromosome II. EMBO J. 1994 Dec 15;13(24):5795–5809. doi: 10.1002/j.1460-2075.1994.tb06923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K., Okamura-Ikeda K., Motokawa Y. Lipoylation of H-protein of the glycine cleavage system. The effect of site-directed mutagenesis of amino acid residues around the lipoyllysine residue on the lipoate attachment. FEBS Lett. 1991 Nov 18;293(1-2):115–118. doi: 10.1016/0014-5793(91)81164-4. [DOI] [PubMed] [Google Scholar]
- Green J. D., Laue E. D., Perham R. N., Ali S. T., Guest J. R. Three-dimensional structure of a lipoyl domain from the dihydrolipoyl acetyltransferase component of the pyruvate dehydrogenase multienzyme complex of Escherichia coli. J Mol Biol. 1995 Apr 28;248(2):328–343. doi: 10.1016/s0022-2836(95)80054-9. [DOI] [PubMed] [Google Scholar]
- Hirel P. H., Schmitter M. J., Dessen P., Fayat G., Blanquet S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles J. R. The mechanism of biotin-dependent enzymes. Annu Rev Biochem. 1989;58:195–221. doi: 10.1146/annurev.bi.58.070189.001211. [DOI] [PubMed] [Google Scholar]
- Kondo H., Shiratsuchi K., Yoshimoto T., Masuda T., Kitazono A., Tsuru D., Anai M., Sekiguchi M., Tanabe T. Acetyl-CoA carboxylase from Escherichia coli: gene organization and nucleotide sequence of the biotin carboxylase subunit. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9730–9733. doi: 10.1073/pnas.88.21.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H., Uno S., Komizo Y., Sunamoto J. Importance of methionine residues in the enzymatic carboxylation of biotin-containing peptides representing the local biotinyl site of E. coli acetyl-CoA carboxylase. Int J Pept Protein Res. 1984 Jun;23(6):559–564. doi: 10.1111/j.1399-3011.1984.tb03127.x. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Li S. J., Cronan J. E., Jr The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. J Biol Chem. 1992 Jan 15;267(2):855–863. [PubMed] [Google Scholar]
- Lim F., Morris C. P., Occhiodoro F., Wallace J. C. Sequence and domain structure of yeast pyruvate carboxylase. J Biol Chem. 1988 Aug 15;263(23):11493–11497. [PubMed] [Google Scholar]
- Lim F., Rohde M., Morris C. P., Wallace J. C. Pyruvate carboxylase in the yeast pyc mutant. Arch Biochem Biophys. 1987 Oct;258(1):259–264. doi: 10.1016/0003-9861(87)90343-2. [DOI] [PubMed] [Google Scholar]
- MUNKRES K. D., RICHARDS F. M. THE PURIFICATION AND PROPERTIES OF NEUROSPORA MALATE DEHYDROGENASE. Arch Biochem Biophys. 1965 Mar;109:466–479. doi: 10.1016/0003-9861(65)90391-7. [DOI] [PubMed] [Google Scholar]
- McAllister H. C., Coon M. J. Further studies on the properties of liver propionyl coenzyme A holocarboxylase synthetase and the specificity of holocarboxylase formation. J Biol Chem. 1966 Jun 25;241(12):2855–2861. [PubMed] [Google Scholar]
- Mossé M. O., Linder P., Lazowska J., Slonimski P. P. A comprehensive compilation of 1001 nucleotide sequences coding for proteins from the yeast Saccharomyces cerevisiae (= ListA2) Curr Genet. 1993 Jan;23(1):66–91. doi: 10.1007/BF00336752. [DOI] [PubMed] [Google Scholar]
- Murtif V. L., Samols D. Mutagenesis affecting the carboxyl terminus of the biotinyl subunit of transcarboxylase. Effects on biotination. J Biol Chem. 1987 Aug 25;262(24):11813–11816. [PubMed] [Google Scholar]
- Pares S., Cohen-Addad C., Sieker L., Neuburger M., Douce R. X-ray structure determination at 2.6-A resolution of a lipoate-containing protein: the H-protein of the glycine decarboxylase complex from pea leaves. Proc Natl Acad Sci U S A. 1994 May 24;91(11):4850–4853. doi: 10.1073/pnas.91.11.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perham R. N. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes: a paradigm in the design of a multifunctional protein. Biochemistry. 1991 Sep 3;30(35):8501–8512. doi: 10.1021/bi00099a001. [DOI] [PubMed] [Google Scholar]
- Reed K. E., Cronan J. E., Jr Escherichia coli exports previously folded and biotinated protein domains. J Biol Chem. 1991 Jun 25;266(18):11425–11428. [PubMed] [Google Scholar]
- Reed L. J., Hackert M. L. Structure-function relationships in dihydrolipoamide acyltransferases. J Biol Chem. 1990 Jun 5;265(16):8971–8974. [PubMed] [Google Scholar]
- Riles L., Dutchik J. E., Baktha A., McCauley B. K., Thayer E. C., Leckie M. P., Braden V. V., Depke J. E., Olson M. V. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics. 1993 May;134(1):81–150. doi: 10.1093/genetics/134.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M., Grisafi P., Botstein D. Structure and function of the yeast URA3 gene: expression in Escherichia coli. Gene. 1984 Jul-Aug;29(1-2):113–124. doi: 10.1016/0378-1119(84)90172-0. [DOI] [PubMed] [Google Scholar]
- Sakai A., Chibazakura T., Shimizu Y., Hishinuma F. Molecular analysis of POP2 gene, a gene required for glucose-derepression of gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 1992 Dec 11;20(23):6227–6233. doi: 10.1093/nar/20.23.6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samols D., Thornton C. G., Murtif V. L., Kumar G. K., Haase F. C., Wood H. G. Evolutionary conservation among biotin enzymes. J Biol Chem. 1988 May 15;263(14):6461–6464. [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Shenoy B. C., Xie Y., Park V. L., Kumar G. K., Beegen H., Wood H. G., Samols D. The importance of methionine residues for the catalysis of the biotin enzyme, transcarboxylase. Analysis by site-directed mutagenesis. J Biol Chem. 1992 Sep 15;267(26):18407–18412. [PubMed] [Google Scholar]
- Stucka R., Dequin S., Salmon J. M., Gancedo C. DNA sequences in chromosomes II and VII code for pyruvate carboxylase isoenzymes in Saccharomyces cerevisiae: analysis of pyruvate carboxylase-deficient strains. Mol Gen Genet. 1991 Oct;229(2):307–315. doi: 10.1007/BF00272171. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Takeshita S., Sato M., Toba M., Masahashi W., Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61(1):63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- Wada K., Fujibayashi Y., Yokoyama A. Copper(II)[2,3-butanedionebis(N4-methylthiosemicarbazone)], a stable superoxide dismutase-like copper complex with high membrane penetrability. Arch Biochem Biophys. 1994 Apr;310(1):1–5. doi: 10.1006/abbi.1994.1132. [DOI] [PubMed] [Google Scholar]
- Walker M. E., Val D. L., Rohde M., Devenish R. J., Wallace J. C. Yeast pyruvate carboxylase: identification of two genes encoding isoenzymes. Biochem Biophys Res Commun. 1991 May 15;176(3):1210–1217. doi: 10.1016/0006-291x(91)90414-3. [DOI] [PubMed] [Google Scholar]
- Wallis N. G., Perham R. N. Structural dependence of post-translational modification and reductive acetylation of the lipoyl domain of the pyruvate dehydrogenase multienzyme complex. J Mol Biol. 1994 Feb 11;236(1):209–216. doi: 10.1006/jmbi.1994.1130. [DOI] [PubMed] [Google Scholar]