Abstract
In pathogenic Bacillota, spores can form an infectious particle and can take up a central role in the environmental persistence and dissemination of disease. A poorly understood aspect of spore-mediated infection is the fibrous structures or ‘endospore appendages’ (ENAs) that have been seen to decorate the spores of pathogenic Bacilli and Clostridia. Current methodological approaches are opening a window on these long enigmatic structures. Using cryoID, Alphafold modelling and genetic approaches we identify a sub-class of robust ENAs in a Bacillus paranthracis foodborne outbreak strain. We demonstrate that L-ENA are encoded by a rare three-gene cluster (ena3) that contains all components for the self-assembly of ladder-like protein nanofibers of stacked heptameric rings, their anchoring to the exosporium, and their termination in a trimeric ‘ruffle’ made of a complement C1Q-like BclA paralogue. The role of ENA fibers in spore-spore interaction and the distribution of L-ENA operon as mobile genetic elements in B. cereus s.l. strains suggest that L-ENA fibers may increase the survival, spread and virulence of these strains.
Subject terms: Cryoelectron microscopy, Biopolymers in vivo, Bacterial structural biology, Supramolecular assembly
The pathogenic Bacillus paranthracis NVH 0075-95 produces pili-like (L-ENA) structures on its spore surface to mediate spore-spore contacts. Using cryoEM, a molecular model for the fiber architecture and the adhesive tip fibrillum is proposed.
Introduction
Endospores formed by bacteria in the Bacillota (Firmicutes) phylum are among the most durable lifeforms found in nature. This cryptobiotic way of living can bridge long periods that are not conducive to the survival of the vegetative state, waiting for environmental stimuli to trigger germination and terminate the state of hibernation. Their remarkable resilience can in part be traced back to the extraordinary robust layers that encase the genomic DNA while maintaining the permeability for germinants that signal when conditions are again conducive for metabolically active life1,2. For the endospore-forming bacteria of the genera Bacillus and Clostridium, the genome is housed within a partially dehydrated central core that is encapsulated by a thick peptidoglycan layer, referred to as the cortex. The cortex, in turn, is covered by at least four consecutive protein layers that make up the spore coat: i.e., the basement layer, the inner and outer coat, and the crust, comprising about 70 different proteins1,3. Many species have an additional layer called the exosporium, which forms a flexible, glycosylated, paracrystalline sac-like structure that surrounds the spore, defining an interstitial volume between the crust and the exosporium4. As the outermost spore layer, the exosporium mediates endospore´s connection to the environment and the infected host5,6. An important role in this interaction is played by the bacterial collagen-like protein of Anthracis (BclA), which covers the spore surface as a dense decoration called the hairy nap6. It plays an immune regulatory function by binding complement factor H (CHF) thereby inhibiting downstream complement activation7.
A poorly understood component of the spore surface is micrometer-long filamentous, pilus-like structures seen in virulent Bacillus strains, dubbed endospore appendages (ENA). First reported and classified for Clostridium spores in the late 1960s8–10, followed by B. cereus in the 1970s11, ENAs can vary greatly in length, diameter, and overall shape. They are present on spores of notable human and animal pathogens such as B. anthracis11, B. cereus11, C. botulinum10, and C. sordellii12, and seem mostly absent in saprophytic species, including B. subtilis. Pilus-like structures are well known from vegetative cells13. In Gram-positive bacteria, pili are predominantly formed by the sortase-mediated covalent coupling and cell-wall anchoring of secreted LPxTG motif containing subunits via isopeptide bonds14–16. These pili are primarily found in pathogenic species (i.e., Corynebacterium diphtheriae, Staphylococcus spp., Enterococcus spp., Streptococcus spp., and Bacillus spp.), where they serve an adhesive function, by binding to host tissues and/or mediating self-contact in multicellular communities or biofilms. Endospores represent an important dissemination form in pathogenic Bacillaceae, so similar to pili on vegetative cells, ENAs may mediate cell-cell and cell-tissue interaction.
Despite observations for more than six decades, their physico-chemical robustness made ENAs resist traditional proteomic identification methods (N-terminal sequencing; peptide fingerprinting)17 preventing the genetic study of their function. This stalemate was recently broken by the virtue of cryoID, the direct molecular identification of proteinaceous structures through 3D reconstruction by cryo-electron transmission microscopy (cryoEM)18,19. Spores of the food-poisoning outbreak strain Bacillus paranthracis NVH 0075-95 were found to display two structurally distinct ENAs, based on the typical patterning seen in nsTEM images dubbed ‘staggered’ and ‘ladder’-like or ‘S’- and ‘L-ENA’, respectively18,19. CryoID studies of S-ENA revealed it represents a pilus superfamily composed of two major subunits, Ena1A and Ena1B. Both proteins consist of a single DUF3992 domain comprising a β-jellyroll fold with an N-terminal extension. Ena1A and 1B subunits self-organize into micrometers-long helical ultrastructures by lateral β-sheet augmentation, fortified by lateral and longitudinal inter-molecular disulfide bridges. Strikingly, S-ENAs terminate into 3–5 flexible, 2 nm diameter, tip fibrillae (hereafter referred to as ruffles) for which a genetic identification or structural elucidation remains absent. Phylogenomic analysis showed that ena1/2 clusters are ubiquitous in pathogenic Bacilli (>90% occurrence in B. cereus s.l.) suggesting that S-ENAs could be hitherto overlooked virulence factors. Biomechanical studies suggest S-ENAs are implicated in self-adherence and the clustering of spores, albeit by an unknown mechanism20.
As mentioned above, the spores of NVH 0075-95 are decorated with a second type of appendages, dubbed L-ENA. They are shorter (typically <1 µm), 7 nm diameter ENAs that are tethered to the exosporium layer and are decorated with a single ruffle at their distal terminus which has been identified as a major driver in spore-spore aggregation21. In this contribution, we use a combined cryoEM-genome mining approach to identify the major structural subunit of L-ENA (hereafter referred to as Ena3A) and solve the helical fiber ultrastructure to a resolution of 3.3 Å. We show that ena3A is embedded in a three-gene cluster that holds all components for L-ENA assembly and anchoring to the exosporium.
Results
L-ENA fibers constitute a sub-class of disulfide cross-linked endospore appendages
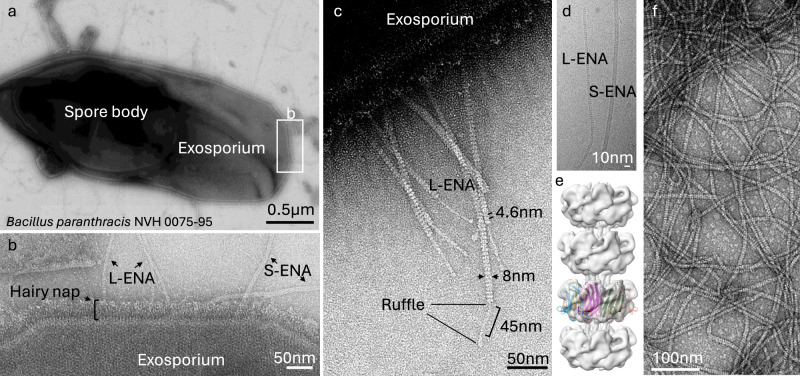
The endospores of the food-poisoning outbreak strain B. paranthracis NVH 0075-95 are decorated with two types of ENA fibers, i.e., S-ENA and L-ENA (Fig. 1a, b). Close inspection of the exosporium layer reveals that L-ENA fibers protrude from the brush-like hairy nap layer (Fig. 1b) seemingly tethered to the exosporium. L-ENA fibers have an apparent diameter of 8 nm and exhibit a ladder-like uranyl staining pattern in nsTEM with a typical recurring distance of 4.6 nm between consecutive ladder segments. L-ENAs consistently terminate in a single 2 nm diameter tip fibrillum or ruffle, that consists of a stalk region (45 nm in length) and a terminal knob domain (Fig. 1c). In preparation of cryoEM data collection, we produced a sample enriched in ENA via shear-induced dislodging from the spore surface followed by a series of purification steps as described in ref. 18. Next, a 1568 movie cryoEM dataset (60 frames, 0.766 Å/pixel) was collected on a JEOL CRYO ARM 300 microscope, yielding a 5.8 Å resolution (FSC = 0.143 criterion) reconstructed cryoEM-volume of L-ENA fibers with C7 symmetry (see Fig. 1e and Supplementary Fig. 1). The refined helical parameters are a twist of 17.04° and a rise of 43.82 Å. We attribute the limited resolution of the final reconstruction to (i) the sparsity of images that contained L-ENA fibers (414 micrographs, i.e., 26% of the total dataset), and (ii) the innate flexibility of the L-ENA fibers (discussed below). The limited resolution precluded us to de novo build an atomic model of L-ENA but allowed us to identify the fold of the constituent monomers. Indeed, an exploratory rigid body docking exercise using Ena1B as template model (extracted from PDB: 7A02) taught us that the L-ENA subunits appeared structurally homologous to Ena1A/B and therefore likely represented members of the DUF3992 family. A careful search of the NVH 0075-95 genome (GCA_027945115.1) with HMMsearch 3.022 using a hidden Markov model generated from a dataset of Ena1/2 protein sequences as query led to the identification of a fourth gene outside the ena1a-c cluster encoding for a DUF3992 domain-containing protein. As such, we reasoned that WP_017562367 represented the candidate L-ENA subunit and ordered its coding sequence as a synthetic gene, and cloned it into pET28a for cytoplasmic expression in E. coli C43 (DE3), similar to the approach taken for recombinant S-ENA production18. To probe for the presence of putative L-ENA fibers, the insoluble fraction (after lysozyme treatment and 1% SDS extraction) of the E. coli lysate was analyzed using nsTEM. The obtained micrographs confirmed the presence of micron-sized, 7 nm diameter fibers with a characteristic ladder-like pattern at 4.6 nm intervals (Fig. 1f). Notably, the recEna3A fibers did not have any ruffles on their termini. This result served as an initial confirmation that WP_017562367 is the major subunit of L-ENAs found on B. paranthracis NVH 0075-95. To reflect the fact that WP_017562367 is a member of a ena gene cluster, we propose to name it ena3a, following the convention used for the ena1a-c (e.g., B. cereus type S-ENA) and ena2a-c (e.g., B. thuringiensis type S-ENA)18.
Fig. 1. Negative stain transmission electron microscopy images of the endospores of the food-poisoning outbreak strain B. paranthracis NVH 0075-95.
a Composite montage of a single endospore with a central spore body and the surrounding exosporial sack, b High-magnification image of the exosporium decorated with the hairy nap and two types of endospore appendages (L- and S-ENA), c L-ENA fibers anchored to the exosporium, exhibit a ‘ladder-like’ pattern (4.6 nm intervals), are terminally decorated with single tip fibrillae, i.e., ruffles. Ruffles consist of a 45 nm long stalk that terminates into a globular head domain, d cryoEM image of isolated ENA fibers, e reconstructed cryoEM-volume of L-ENA fibers (5.8 Å at FSC = 0.143) with rigid body docking of 7 copies of Ena1B19-117, f nsTEM image of recEna3A produced in and purified from the cytoplasm of E. coli. The data shown are representative of experiments made independently in triplicate.
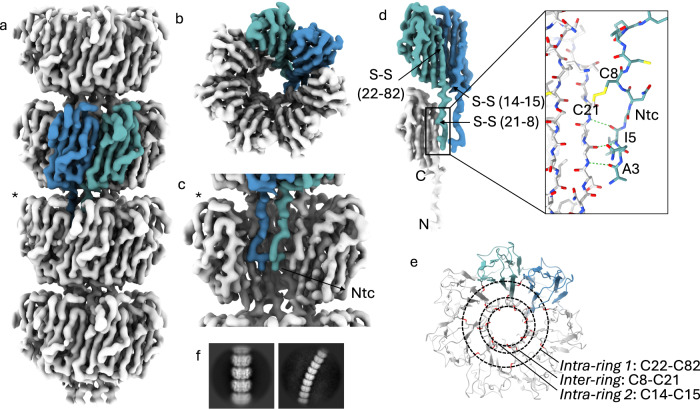
Given the relative ease of production and purification of recEna3A fibers in comparison to the L-ENA purification from the natural source, we proceeded to collect a 10886 movies cryoEM dataset of vitrified recEna3A fibers (Supplementary Fig. 2) leading to a 3.3 Å global resolution (FSC = 0.143 criterion) cryoEM-volume after helical refinement using C7 symmetry in CryoSPARC v4.0.3 (Fig. 2). The refined helical parameters are a twist of 18.5° and a rise of 44.97 Å. The recEna3A cryoEM-volume reveals an axial stacking of heptameric Ena3A rings that rotate 18.5° clockwise relative to each other (Fig. 2a). Each ring is composed of seven Ena3A molecules that interface laterally via β-sheet augmentation. The rings encircle a central, hollow volume which we refer to as the ring lumen (Fig. 2b). Consecutive rings interlock with each other using the N-terminal extensions of the upper ring docking into the lumen of the ring below (Fig. 2c). We refer to the first 14 N-terminal residues as the N-terminal connector (i.e., Ntc) in analogy to the N-termini of Ena1A/B subunits that interlock from above with subunits i-9 and i-10 of the neighboring helical staircase18. Note that the orientation of the L-ENA fiber in Fig. 2 is such that the top rings represent the distal, ruffled end of the fiber, and that the bottom rings form the pointy end of the fibers, proximal to the spore surface. Close inspection of the cryoEM map revealed covalent cross-links between neighboring Ena3A subunits at three distinct locations (Fig. 2d). After manual building of the final L-ENA model, we identified these contacts to be 3 types of inter-molecular disulfide bridges. Two disulfide bridges (i.e., Cys22-Cys82 and Cys14-Cys15) serve to reinforce the lateral contacts within a single ring, whereas one disulfide bridge (i.e., Cys8-Cys21) confers longitudinal coupling between the lumen and the Ntcs of consecutive rings. Given the heptameric nature of the structure, each ring segment will contain 21 inter-molecular disulfide bridges that are organized along three concentric rings centered on the fiber axis (Fig. 2e).
Fig. 2. Cryo-EM volume of recEna3A L-ENA fibers.
a Helical ultrastructure of L-ENA determined at 3.32 Å global resolution (cut-off criterion at FSC = 0.143 Å−1) revealing an axial stacking of heptameric Ena3A rings that rotate 18.5° clockwise relative to each other. Two neighboring Ena3A monomers are colored in blue and cyan, b top-view of a single ring with a highlight of the β-sheet augmentation between the blue and cyan subunit, c zoom-in of a single ring - for clarity, two subunits were removed from the ring highlighted with an asterisk in (a) - showing the docking of the N-terminal connectors that covalently tether the ring docked above, d Highlight of the lateral intra-ring contacts: two types of disulfide bridges (Cys14-Cys15; Cys22-Cys82) exist within two neighboring subunits. In turn, each subunit connects to a subunit in the ring below via their Ntc (Cys8-Cys21; inset in stick representation), e On-axis top-view of the L-ENA fiber model in cartoon with disulfide bridges shown in red; f 2D class averages of straight and curved L-ENA segments covering 4 and 9 rings, respectively.
Next, we tested the stability of recombinant L-Ena fibers by subjecting the fibers to a series of physical (autoclaving) and/or chemical treatments (8 M urea, SDS, proteinase K, formic acid (FA)). Following the respective treatments, we performed nsTEM imaging of the recovered fibers and determined the 2D class average to gauge the intactness of the fibers (Supplementary Fig. 4). Remarkably, the 2D class average images of all treatments (apart from the 100% FA) are very similar to the 2D class averages obtained from ex vivo purified fibers. We were not able to produce 2D class averages for the 100% FA-treated sample, likely indicating that the Ena3A monomers had (partially) unfolded. Interestingly, the FA-treated fibers did not depolymerize, suggesting that disulfide bonds had not been reduced. We conclude that the extensive hydrogen bonding between the Ena3A subunits combined with the covalent cross-links underlies the physico-chemical robustness of the L-ENA fibers.
L-ENA marries this stability with remarkable flexibility as judged from the regions of high local curvature in the nsTEM micrographs and 2D class average images obtained during cryoEM processing (Fig. 1f). To illustrate the point further, we manually selected and extracted curved L-ENA fiber segments from the motion-corrected micrographs and performed 2D classification (Supplementary Fig. 3a). To resolve the underlying structural heterogeneity, we performed a 3D variability analysis using CryoSPARC (Supplementary Fig. 3b). The resulting volume series (processed in ChimeraX 1.423,24 and exported as Supplementary Movie 1) provides further molecular insights into the L-ENA flexibility. Ena3A rings exhibit a rocking motion normal to the fiber axis with the respective hinge points centered on the Ntcs. Hence, L-ENA flexibility can be traced back to (i) the spatial separation between consecutive rings thereby providing the possibility for local ring displacement without introducing steric clashes, (ii) combined with the intrinsic flexibility of the N-terminal connectors.
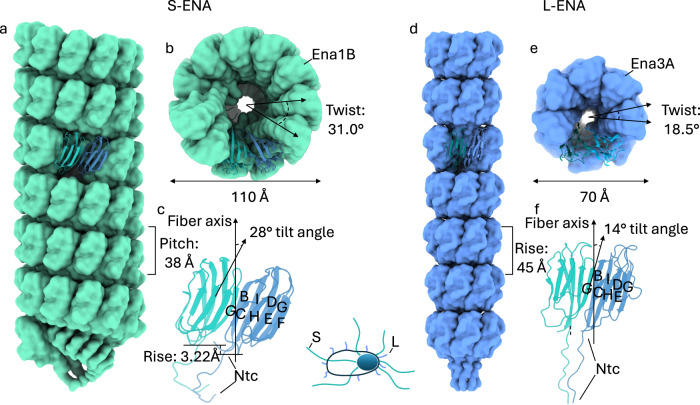
Ena3A subunits consist of a typical jellyroll fold25 comprised of two juxtaposed β-sheets containing strands BIDG and CHEF (Fig. 3). As mentioned earlier, the jellyroll domain is preceded by a flexible 14-residue Ntc that mediates inter-ring coupling. The backbone (420 atoms) root-mean-squared displacement (RMSD) between Ena1B and Ena3A is 3.7 Å even though the sequence identity is only 28.4% (Supplementary Fig. 5a–c). Despite the structural similarities at the fold level, S- and L-ENA have a markedly different quaternary architecture (Fig. 3a–c). Ena1B subunits interact laterally via β-sheet augmentation (Supplementary Fig. 5d, e) with a non-zero (i.e., 3.2 Å) vertical offset, making a 28° angle relative to the fiber axis. This in turn leads to a helical stacking of Ena1B monomers yielding an S-ENA fiber with a diameter of 110 Å. Ena3A subunits form a similar dimer interface (see the residues marked with an asterisk in Supplementary Fig. 5b) that follows the same register between the interfacing strands G and C (Supplementary Fig. 5f, g). Contrary to Ena1B though, neighboring Ena3A subunits lie within the same plane (i.e., no vertical offset) thereby producing closed ring-like structures. We attribute this difference in axial displacement to the relative tilts that the subunits make with respect to the fiber axis, i.e., 28° and 14° for Ena1B and Ena3A, respectively (Fig. 3c, f). L-ENA fiber biogenesis therefore likely proceeds via docking and covalent locking (via the Cys8-Cys21 disulfide) of fully formed rings, whereas S-ENA fibers elongate via integration of successive monomers.
Fig. 3. Structural comparison between the S- and L-ENA fiber architectures.
a Side and b on-axis view of an S-ENA fiber (pdb-id: 7A02) composed of Ena1B subunits. Helical parameters: Rise: 3.22 Å, Twist: 31.0°, Pitch: 38 Å, c Dimeric contacts of Ena1B subunits via β-sheet augmentation at the interface between the G and C strands. Subunits are tilted 28° with respect to the fiber axis. This out-of-plane interaction leads to a helical stacking of Ena1B monomers, d Side and e on-axis view of an L-ENA fiber (pdb-id: 8PDZ) composed of Ena3A subunits. Helical parameters: Rise = pitch: 45 Å, Twist: 18.5°, f Dimeric contacts of Ena3A subunits via β-sheet augmentation at the interface between the G and C strands. Although subunits are tilted 14° with respect to the fiber axis, their contacts remain in-plane yielding a heptameric ring.
Distribution and expression of the ena3A gene cluster
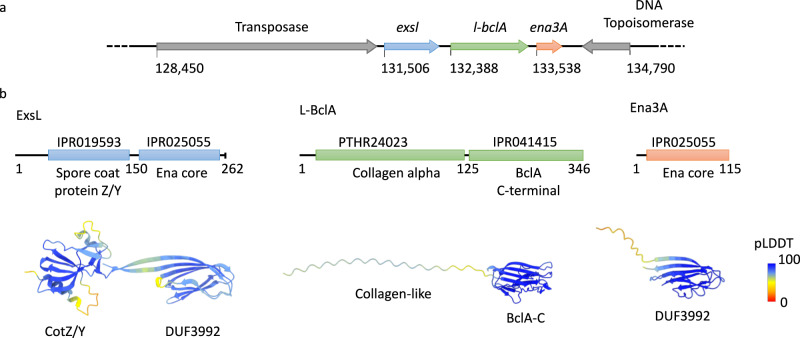
Inspection of the GCA_027945115 genome reveals that ena3A (PGS39_28750) is embedded in a three-gene cluster on the NZ_CP116205 plasmid, hereafter referred to as the ena3 gene cluster (Fig. 4). In this cluster, ena3A is preceded by the genes PGS39_28740 and PGS39_28745. For reasons discussed below, we suggest naming these two genes, i.e. exsL and l-bclA, respectively. The ena3 gene cluster is rare: it was found in only 62 organisms through a remote search of the entire NCBI RefSeq non-redundant protein database using cblaster (CAGECAT v. 1.0). Assemblies of sufficient quality were downloaded and appended to a representative database of genomes of the Bacillus genus. The proportion of genomes carrying an ena3 gene cluster was indeed low; only 9.5% (n = 62/656) of the Bacillus cereus group sensu lato genomes, of which 51 were B. cereus (n = 51/126), four B. thuringiensis (n = 4/52), one B. anthracis (n = 1/63), one B. paranthracis (n = 1/4), one B. toyonensis (n = 1/204), two B. mobilis (n = 2/5), and two Bacillus spp. (2/4) (Supplementary Fig. 6; see also https://microreact.org/project/uzm4JFrrsCPZeRnMpRqvvf-supplementary-figure-9-ena3-paper). Most genomes had one copy of the gene cluster (n = 53), while nine strains had paralogs, carrying two (n = 5, B. cereus1, Bacillus spp.2.), three (n = 2, B. cereus), four (n = 1, B. thuringiensis) and five (n = 1, B. thuringiensis) copies. The ena3 gene cluster was not found in B. subtilis or other saprophytic Bacilli. Six of the 14 human clinical isolates carrying ena3 were isolated from bloodborne infections according to available metadata on NCBI.
Fig. 4. Expression of L-ENA genes is concomitant with sporulation.
a Chromosomal organization of exsL, l-bclA, and ena3A, and primers used in standard PCR using cDNA template; b The L-ENA genes l-bclA and ena3A form an operon. Agarose gel electrophoresis (1%) of PCR products conducted on cDNA from NVH 0075-95 culture (4, 8, 12, and 16 h). Primer combinations and expected product sizes are shown in (a). Genomic DNA (gDNA) from NVH 0075-95 and cDNA prepared from its isogenic Δena1ABC/Δena3A mutant were used as positive and negative controls for the PCR, respectively; c Expression of L-ENA genes relative to rpoB during vegetative growth and sporulation. Data are presented as mean values with error bars representing the standard deviation of three independent experiments, each with three technical replicates. Source data are provided as a Source Data file.
Thirteen genomes with an L-ENA gene cluster were complete and closed, and the genomic location of the gene cluster could be inspected. The ena3 gene cluster was located either on the chromosome (n = 7 genomes), on plasmids (n = 1 genome) or were found as paralogs on one or more plasmids and on the chromosome (n = 5 genomes).
The ena3 gene cluster of NVH 0075-95 is flanked by an incomplete topoisomerase downstream and a complete Tn3 family transposase upstream (Fig. 6). This gene synteny was found in only two other strains in the cblaster/clinker analysis: B. cereus AFS093282 (NZ_NVMQ01000017.1) and B. pacificus strain BC444B (accession NZ_JAOPRQ010000006.1). In other strains, the L-ENA gene cluster was flanked by frameshifted versions of Tn3 family transposases, tyrosine- type recombinase/integrase, and site-specific integrases. A few strains had a shorter Tn3 family transposase upstream of the L-ENA gene clusters. Taken together, the L-ENA gene cluster is/or has been located on a transposon and is part of the mobilome of the B. cereus s.l. group, accounting for the polyphyletic distribution of this gene cluster in the population.
Fig. 6. Genetic organization of the L-ENA gene cluster.
a the ena3A gene is embedded in a three-gene cluster, preceded on the CP116205.1 plasmid by two genes here annotated as exsL and l-bclA, b Domain organization of the L-ENA proteins as identified by Interpro (10.1093/nar/gkac993). ExsL: consists of an N-terminal spore coat protein Z/Y domain and a C-terminal Ena-core domain (DUF3992), L-BclA: consists of an N-terminal collagen-like domain and a C-terminal BclA-C domain, Ena3A: consists of a single Ena-core domain (DUF3992). AlphaFold2 predictions are shown in cartoon representation, color-coded according to the pLDDT score.
Next, we set out to track the expression of L-ENA genes as a function of time. First, we construct an overall sporulation timeline that will serve as a reference frame for a qPCR analysis of the ena3a gene cluster. In brief, under the culturing conditions we employed, there was no asymmetric septation within the first five hours of cultivation. Asymmetric septation first began to appear at the 6th hour, and by the 8th hour, nearly all the cells had formed asymmetric septa (Supplementary Fig. 7). By the 9th hour, the processes of asymmetric division and engulfment had reached completion. From the 10th hour onward, the appearance of phase-bright bodies became increasingly prominent, and by the 12th hour, the cell population was predominantly composed of cells containing phase-bright bodies (Supplementary Fig. 7). This trend continued until the 14th hour, during which the majority exhibited very bright bodies, and spore maturation continued, with matured spores beginning to be released by the16th hour. To determine the expression of L-ENA genes (exsL, l-bclA, and ena3A), NVH 0075-95 was cultured in sporulation medium for 16 h, and cDNA prepared from culture samples collected at four-hour intervals after inoculation (4, 8, 12 and 16 h) and analyzed by PCR. Using primers that specifically amplify open reading frames across exsL→l-bclA and l-bclA→ena3A (Fig. 4a), we detected a ~587 bp PCR product across l-bclA→ena3A, but not for exsL→l-bclA (Fig. 4b). This suggests that l-bclA and ena3A are expressed bicistronically. Notably, no l-bclA-ena3A transcript was detected during the first 8 h of cultivation, which represents the vegetative growth phase (Fig. 4b). Consistent with the PCR result, the data from a qPCR analysis indicated that the L-ENA genes are overexpressed exclusively during the sporulation phase (12 and 16 h) (Fig. 4c; Supplementary Fig. 7). Notably, a ~13000, ~1200 and ~40-fold increase in the expression of ena3A, l-bclA and exsL, respectively, was evident in the samples collected at 12 h after inoculation. Intrigued by the approximately tenfold difference in mRNA levels between the bicistronic genes l-bclA and ena3A, we conducted a new set of qPCR reactions with different primers but obtained similar results. Notably, literature suggests the occurrence of segmental decay patterns within polycistronic operons, as well as mRNA decay from the 5′ end (in addition to the 3′ end) in B. cereus26, which could explain our observations.
The ena3 gene cluster consists of the L-ENA subunit, the exosporium anchoring protein ExsL, and the complement C1Q-like ruffle protein L-BclA
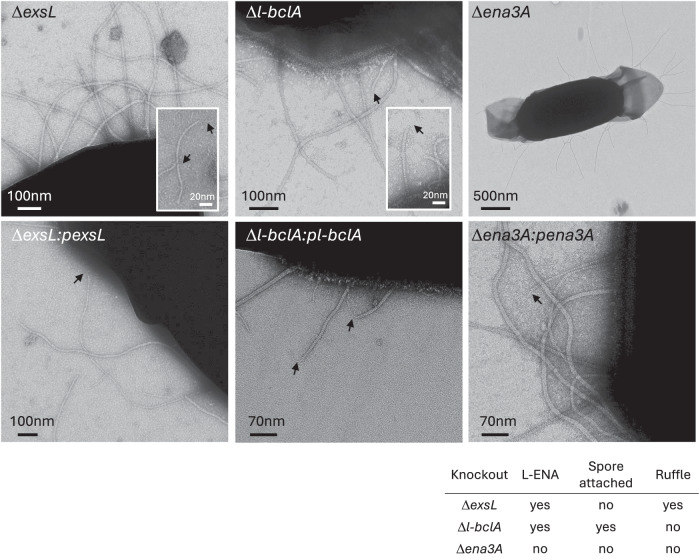
The gene ena3A is found in a cluster with exsL and l-bclA. In pursuit of the biological roles of exsL and l-bclA, we made individual knockout strains (ΔexsL and Δl-bclA) and investigated their respective endospores by nsTEM (Fig. 5). ExsL-depleted spores are devoid of L-ENA fibers coupled to the exosporium but are otherwise morphologically identical to wild-type NVH 0075-95 spores. Careful inspection of the grid areas leads to the identification of detached L-ENAs in the spore supernatant, with ruffles present, suggesting that ExsL mediates the connection of L-ENA to the exosporium. Conversely, Δl-bclA spores have L-ENAs present on the exosporium, but the fibers lack the distal ruffle. We do note that the ruffles remained present on the S-ENA fibers, suggesting that l-bclA specifically encodes for the L-ENA ruffle protein. As expected, ena3a− spore samples were completely devoid of L-ENA fibers, be it spore attached or detached. Complementation of the ΔexsL, Δl-bclA, and Δena3A mutants with a low copy plasmid (pHT304) containing the respective genes, restored the corresponding spore phenotypes to that of the wild-type strain (Fig. 5).
Fig. 5. Phenotypic read-out of L-ENA mutants.
nsTEM micrographs of the respective L-ENA operon deletion mutants. The data shown are representative of experiments made independently in triplicate.
A primary sequence analysis of ExsL (WP_048548726.1) using Interpro27 shows that it is composed of an N-terminal spore coat protein Z (PF10612) domain and a C-terminal Ena-core (DUF3992) domain (see the domain organization and corresponding AF2 prediction in Fig. 6b). To gain further insights into the molecular mechanisms of L-ENA anchoring and ruffle formation, we performed Alphafold2 (AF2) modeling28–30. First, we tested for the plausibility of a Ena3A-ExsL complex formation. In Supplementary Fig. 8 we show the AF2 multimer v1.2 model of an Ena3A-ExsL dimer, which had an overall pLDDT score of 82.6 and a ptmscore of 0.73, which is in support of the hetero-dimer hypothesis. As expected, Ena3A interfaces with ExsL via its C-terminal Ena-core domain in a manner that mimics the Ena3A dimer interface found in the L-ENA structure (Supplementary Fig. 8e, f). This is somewhat surprising given the low sequence identity (17.4%) between the Ena3A and the ExsL Ena-core domain (i.e. residues 157-262). Inspection of the pairwise sequence alignment (Supplementary Fig. 8d) between Ena3A and ExsL learns that key Ena3A residues (C82, S84, and T86) involved in lateral subunit contacts are conserved in ExsL (C224, S226, and T228). In fact, AF2 predicts a disulfide bridge between ExsL cys224 and Ena3A cys22, which mimics the intra-ring L-ENA S-S bridge between cys22 and cys82 (Supplementary Fig. 8e, f; Fig. 2d, e). Despite the low sequence identity in Ena3A and the ExsL Ena-core, the Ena3A-Ena3A and Ena3A-ExsL contacts are highly equivalent (Supplementary Fig. 8e, f), in line with the general mechanism of β-sheet augmentation, which is primarily driven by shape complementarity and backbone H-bonding and is relatively insensitive to the amino acid sequence in the paring β-strands31.
In addition, we looked at the AF2 prediction of a putative ExsL-ExsY dimer. ExsY (WCA20099.1) is one of the major constituents of the B. cereus exosporium and shares structural similarities to the C-terminal CotZ domain of the exsL gene in the ena3 gene cluster, albeit at low sequence identity, i.e., 30.8% (Supplementary Fig. 9a, b, f). Our assumption based on the ΔexsL phenotype and the ExsL-Ena3A AF2 model was that ExsL could also be a potential binding partner of ExsY and, in doing so, act as an exosporium-embedded anchor point for L-ENA fibers. Indeed, the AF2 multimer model of a ExsL-ExsY dimer (with a relatively high pLDDT score of 80.2 and a ptmscore of 0.71) shows an ExsL-ExsY coupling via the N-terminal ExsL1-156 domain with ExsY, lending further credence to the supposition that ExsL serves as a connective bridge between the paracrystalline exosporium and L-ENA. In analogy to the ExsL-Ena3A dimer, the predicted ExsL-ExsY contact mimics the homomeric ExsY contacts (Supplementary Fig. 9c, d, e, g) that we observe in a high confidence AF2 multimer prediction of an ExsY hexamer (pLDDT = 81.3; ptmscore = 0.86), i.e., the base building unit of the exosporium lattice.
In turn, L-BclA (WP_271292911.1) contains an N-terminal Collagen triple helix repeat domain (PF01391) followed by a BclA-C-terminal domain (PF18573; part of the C1q_TNF clan (Pfam CL0100)). BclA is the major glycoprotein of the hairy nap found to cover the exosporium of most B. cereus s.l. species32,33. Interestingly, this same architecture is found in vertebrate complement component C1q34 (uniprot: P02745). In Supplementary Fig. 10a, b, we present the AF2 multimer model of an L-BclA94-336 homotrimer that consists of the C-terminal trimerization domain and a segment of the collagen-like triple helix. The corresponding pLDDT value of 94 and ptmscore of 0.9 support the accuracy of the predicted fold, as well as the supposed trimeric stoichiometry in analogy to the crystal structure (PDB: 1WCK) of BclA-C of B. anthracis. Based on this model, we predict the lateral dimension of the C-terminal domain to be approximately 4 nm, and for the triple collagen helix, we project an average length of 2.8 Å per residue (see discussion below; Supplementary Fig. 10a). The all-atom RMSD between L-BclA-CTD and BclA-C is 3.55 Å despite the low sequence identity of 22.1%, demonstrative of significant structural homology (Supplementary Fig. 10c). Tan and Turnbough showed that the attachment of BclA to the exosporium is dependent on (i) proteolytic removal of the first 19 residues from the N-terminus and (ii) the presence of an N-terminal submotif –hereafter referred to as the exosporium leader sequence (interpro: IPR0212010; Supplementary Fig. 10d) - in front of the collagen-like region35. Inspection of the N-terminal region of L-BclA demonstrates the absence of an exosporium leader sequence, which indicates that this collagen-like protein is likely not targeted to the exosporium.
A notable feature of collagen-like proteins is that the number of residues that comprise the collagen-stalk region in the primary sequence translates proportionally to the axial dimension of the folded, trimeric entity simply due to the extended nature of the collagen fold33. To that end, we searched the GCA_027945115.1 genome for the ‘local’ orthologue of BclA (uniprot: Q81JD7) and compared its primary sequence to L-BclA (Supplementary Fig. 10e). BclANVH 0075-95 (WCA20088.1) has a collagen-like region of 125 residues, whereas the corresponding domain of L-BclA spans 173 residues. Based on the calibration discussed in Supplementary Fig. 9a, this translates into a predicted fully extended length of a folded trimeric entity of 39 nm and 52 nm for BclA and L-BclA, respectively. These predictions are in agreement with experimental measurements (average height of the hairy nap: ~36 nm and average length of L-ENA ruffle: ~50 nm) from nsTEM micrographs (Supplementary Fig. 10f). To further understand the topology of the L-ENA/L-BclA complex, we performed a single round of 2D classification based on particles that were manually picked from the cryoEM dataset that was collected on ex vivo isolated ENA fibers, focusing on L-ENA termini that were decorated with ruffles (Fig. 7; Supplementary Fig. 10g). This class average shows that the ruffle docks into the lumen of the Ena3A ring at the apex of the L-ENA fiber, i.e., mimicking the inter-ring docking mechanism that exists along the body of the fiber. We, therefore, compared the Ntc of Ena3A to the N-terminus of L-BclA (Supplementary Fig. 10d). Although no particular sequence homology could be detected, both sequences contain a double cysteine motif at positions 8 and 13, respectively. As cysteine 8 mediates the coupling of the Ena3A Ntc to the ring lumen, we speculate that the L-BclA-C13C14 motif could be involved in a similar coupling mechanism.
Fig. 7. Mesoscale model of an L-ENA fiber projecting from the exosporium.
L-ENA fibers are helical, protein ultrastructures composed of heptameric Ena3A rings that stack axially. At the tip that is distal from the spore, the L-ENA fiber terminates into a bacterial collagen-like ruffle protein, i.e., L-BclA. The L-ENA fiber protrudes through the hairy nap layer while being tethered to the exosporium via a dedicated anchoring protein ExsL. ExsL is expected to couple to or be integrated in the 2D crystal lattice of the exosporium. Note that for clarity, the L-ENA fibers are depicted shorter here.
Discussion
Although prominently present on the surfaces of many endospores36 and described since the 1970s for Bacillus11, the genetic identity and biological role of ENAs have remained poorly understood19. Using cryoID, Alphafold modeling, and genetic approaches we were able to identify (this work and18) two members of a sub-class (DUF3992-based) of robust ENAs found on the surface of Bacillus paranthracis spores. We showed that the endospores of the food-poisoning outbreak strain B. paranthracis NVH 0075-95 are decorated with two types of filamentous protein structures, i.e., the endospore appendages S- and L-ENA. Moreover, we have shown that both S- and L-ENA fibers are decorated with respectively, 3–5 or a single tip fibrillae (or ruffles) at their terminus. While the genetic identification of the S-ENA ruffles is still undergoing, the work presented here identifies L-ENA ruffles as L-BclA, a member of the family of bacterial collagen-like proteins (CLPs). It is well established that Firmicutes endospores display bacterial CLPs on their surface37–39, but the biological function remains unclear for most of these proteins39. The most well-known CLP within the Bacillus clade is BclA32, the major glycoprotein40 of the hairy nap41 that is targeted to the exosporium via an N-terminal exosporium leader sequence42,43. BclA has been shown to recruit complement factor H (CFH) to the spore surface7, and its recruitment onto the exosporium facilitates complement C3 degradation, which in turn inhibits downstream complement activation, and ultimately promotes spore persistence. Here we have shown that contrary to BclA, L-BclA is not coupled to the exosporium - this is reflected in the absence of an exosporium leader sequence at the N-terminus of L-BclA. Based on the morphological similarities between S- and L-ENA tip fibrillae (i.e. a ~2 nm wide flexible fibrillum terminating in a knob-like domain) in combination with the l-bclA deletion phenotype (L-ENA ruffle: absent, S-ENA: present), we hypothesize that the tip fibrillum of S-ENA is likely to be encoded by a separate CLP also lacking an exosporium leader sequence, and are searching for putative ‘S-BclA’ candidates among a list of CLP genes we identified in the B. paranthracis NVH 0075-95 genome. This suggests that each ENA subtype is decorated by its own cognate ruffle protein hinting at a functional diversification across the ENA subclasses. Similar specialization of bacterial pili towards a specific target or class of surfaces has been reported, for e.g., sortase-mediated and chaperone-user pili44. Virulent strains can carry multiple pilus operons, most of which display a single, functionally diverse adhesin subunit at the tip of the pilus45. The strategic spatial positioning of pilus-presented adhesins promotes efficient attachment and there is a high probability that the ruffles of Ena’s exert related functions.
Following on the first structural and molecular characterization of ENAs18, Jonsmoen et al. showed that ENAs are involved in spore aggregation21. While the molecular underpinnings of the spore-spore coupling mechanism remained unclear at the time, we recently identified L-BclA as the ruffle species that is involved in establishing inter-spore interactions21. Biofilm formation in the B. cereus s.l. is well recognized46 and instantiates as floating pellicles, submerged biofilms attached to abiotic surfaces, and on living tissues47. Spores tend to accumulate towards the later stages of biofilm development and can –depending on the strain– constitute up to 90% of the total biofilm cell count48. Our structural data allows us to frame a molecular model for the development of L-ENA-mediated spore interaction networks. Although the exact role of spores in biofilms is still an area of active research, it can reasonably be expected that spore-spore interactions will contribute to virulence by affecting (i) biofilm resilience through fortification (e.g., reinforced core protected from stressors), (ii) development of a spore reservoir with sustained spore shedding, and (iii) establishment of spore clumps representing the minimal infectious dose (i.e. strength in numbers). Given that the ena3A gene cluster is found in pathogenic Bacilli, we anticipate that L-ENA fibers could be considered as a sub-class of secondary effect virulence factors through their impact on spore aggregation, and by extension biofilm formation. The aggregation of spores by ENA fibers, a mechanism that may facilitate the survival of spores in the environment, provide additional protection from immune responses, and increase the local infection dose, suggests that L-ENA fibers might increase the survival, spread, and virulence of these strains. Additional research is necessary to fully understand the role of ENAs in these processes. What remains unclear is whether the tip fibrillum (L-BclA) recognizes a specific target on the spore surface, be it a sugar, protein or proteoglycan or whether the interaction is based on aspecific interaction, similar to the hydrophobic CsuE adhesin found on the archaic Csu pili of Acinetobacter baumanni that is also implicated in the early stages of microcolony development leading up to biofilm formation49.
From a structural perspective, L-ENA fibers are remarkable products of macromolecular self-assembly, whose assemblage and surface display require a mere three genes: exsL, ena3A, and l-bclA. Each Ena3A subunit is cross-linked to its neighbor via 3 disulfide bridges, which totals to 21 covalent connections for each heptameric unit. Consequently, L-ENA fibers are able to withstand extreme physico-chemical stressors (heat, strong acids, chaotropes, detergent, and desiccation) and are among the most stable protein structures found in nature. In its biological context, however, a protein fiber will only be as strong as its weakest link. It is therefore expected that these high tensile strength ENAs require an anchoring mechanism of comparable durability. The phenotype of ΔexsL spores (i.e. spores with detached L-ENA) combined with the AF2 modeling of the putative ExsL-Ena3A and ExsL-ExsY heteromers suggests that the anchoring complex is directly integrated into the exosporium 2D lattice. The exosporium of the B. anthracis/cereus/thuringiensis group is a flexible, disulfide cross-linked, protein 2D lattice that is predominantly composed of ExsY hexameric units50. The AF2 model for the ExsL-ExsY dimer suggests that ExsL could be embedded within the ExsY lattice by mimicking the homotypic ExsY contacts, including the formation of an inter-molecular disulfide bridge. Similarly, the AF2 model for an ExsL-Ena3A dimer is reminiscent of the Ena3A contacts found in the L-ENA cryoEM model – this in turn also includes one of the intra-ring Ena3A disulfide bridges. Based on this, we hypothesize that ExsL acts as a bridging moiety that anchors the L-ENA pilus onto the spore surface by maintaining an unbroken chain of disulfide cross-links across the various contact points (i.e., ExsY → ExsL; ExsL → Ena3A). The experimental determination of the respective stoichiometries and the elucidation of the complex epitopes will be the subject of further study.
Methods
Negative stain transmission electron microscopy
Negative stain TEM (nsTEM) imaging of bacterial spores and recEna3A filaments was done using formvar/carbon-coated copper grids (Electron Microscopy Sciences) with a 400-hole mesh. The grids were glow discharged (ELMO; Agar Scientific) with 4 mA plasma current for 45 s. 3 μl of a bacterial spore suspension or recEna3A protein solution was applied onto the glow-discharged grids and left to adsorb for 1 min. The solution was dry blotted, followed by three washes with 15 μl Milli-Q. Next, 15 μl drops of 2% uranyl acetate were applied three times for 10 s, 2 s, and 1 min respectively, with a blotting step in between each application. The excess uranyl acetate was then dry blotted with Whatman type 1 paper. All grids were screened with a 120 kV JEOL 1400 microscope equipped with LaB6 filament and TVIPS F416 CCD camera.
Cryo-electron transmission microscopy
QUANTIFOIL® holey Cu 400 mesh grids with 2‐µm holes and 1‐µm spacing were glow discharged in vacuum using plasma current of 5 mA for 1 min (ELMO; Agar Scientific). 3 µl of 0.6 mg/ml graphene oxide (GO) solution was applied onto the grid and incubated 1 min for absorption at room temperature. Excess GO was blotted using a Whatman grade 1 filter paper and left to dry under ambient conditions. For cryo‐plunging, 3 µl of fiber suspension was applied on the GO‐coated grids at 100% humidity and room temperature in a Gatan CP3 cryo‐plunger. After 1 min of absorption, the grid was machine‐blotted with Whatman grade 2 filter paper for 3.5 s from both sides and plunge frozen into liquid ethane at −176 °C. Grids were stored in liquid nitrogen until data collection. Two datasets were collected for ex vivo and recEna3A appendages with slight changes in the collection parameters. High‐resolution cryoEM movies were recorded on a JEOL CRYO ARM 300 microscope equipped with omega energy filter and a K2 or K3 direct electron detector run in counting mode. For the ex vivo Ena, the microscope was equipped with a K2 summit detector and had the following settings: 300 keV, 100 mm aperture, 30 frames/image, 62.5 e−/Å2, total exposure 2.315‐s exposure, and 0.82 Å/pixel. The recEna3A dataset was recorded using a K3 detector, at a pixel size of 0.782 Å/pix, and a total exposure of 64.66 e−/Å2 accrued over 60 frames/movie.
Generation of deletion mutants and complementation constructs
Deletion mutants in B. paranthracis NVH 0075-95 were generated following the markerless gene replacement method described previously18,51. The gene replacement constructs contained the start and the stop codons of the respective genes to be deleted flanked by upstream and downstream homologous sequences. The upstream and downstream homologous sequences were 806 bp and 677 bp for ena3A, 727 bp and 750 bp for l-bclA, and 701 bp and 728 bp for exsL, respectively. Synthetic constructs (Synbio Technologies LLC) were cut out from the pUC57-Amp vector by digesting with EcoRI, and the gene replacement DNA fragment were cloned into EcoRI digested pMAD-I-SceI shuttle plasmid52. The gene replacement constructs, i.e., pMAD-I-SceI-Δena3A, pMAD-I-SceI-Δl-bclA, and pMAD-I-SceI-ΔexsL were then used to transform NVH 0075-95 or the Δena1ABC triple mutant18. Details of competent cell preparation, transformation, and screening are described18,19.
For complementation experiments, DNA fragment containing a 300 bp region upstream of the start codon of the l-bclA-ena3A operon and open reading frames of the respective genes was ordered (Synbio Technologies LLC) or amplified by PCR and cloned in the low copy number plasmid pHT304 at EcoRI restriction site.
Culturing and RNA extraction
B. paranthracis NVH 0075-95 and B. paranthracis NVH 0075-95 ΔenaABC/Δena3A quadruple mutant (control) were cultured in sporulation medium as previously described18 with a slight modification. Here, we used nutrient broth (oxoid) in place of bacto medium (Difco). For RNA extraction, a sample of 20 ml culture was withdrawn every four hours (4 h, 8 h, 12 h, and 16 h) from three independent cultures, centrifuged, and cell pellets frozen immediately at −80 °C until RNA extraction. Similarly, for fluorescence microscopy analysis, we collected 1 ml of the sample every hour and stained it with 10 µM of the membrane dye FM4-64 (MedChemExpress, # HY-103466), followed by a 2-min incubation at room temperature. 3 µl of the stained sample was applied to a thin layer of 1.5% agarose (w/v) (Sigma Aldrich, # A9539) solidified on a 10-well multitest slide (MP Biomedicals, # 096041805E). The slide was incubated for 1 min at room temperature in a biological safety cabinet to immobilize the sample and subsequently covered with a standard coverslip. Images were captured using the 100× phase-contrast objective (Plan Apo λ, Nikon) of the Nikon Eclipse Ti2 fluorescence microscope equipped with a DS-Qi2 camera. Exposure times were set to 1 s and 200 ms for the 100× phase-contrast and TRITC, respectively.
RNA was extracted using Purelink RNA minikit (Ambion, Life Technologies) according to the manufacturer’s protocol with a slight modification. Briefly, cell pellets were thawed on ice followed by addition of 200 µl of lysozyme solution (10 mM Tris-HCl (pH 8), 0.1 mM EDTA, 10 mg/ml lysozyme) and incubation for 5 min at RT. A volume of 1 µl of 10% SDS was added to the cell suspension and vortexed, after which 150 µl was transferred to new RNAse-free tubes. Following addition of 350 µl of lysis buffer, the cells were lysed by bead beating (FastPrep®-24, MP Biomedicals) for a total of two minutes at Speed 6, with 1 min cooling on ice every 30 s. The lysates were centrifuged for 5 min (2600 × g), and the supernatants were transferred to new RNAse-free tubes. The remaining steps were carried out as described in the manufacturer’s protocol. RNA concentration was measured using Nanodrop 1000 (Thermo Scientific), and 1.5 µg RNA was treated with Turbo DNAase (Ambion) in 20 µl reaction volume following the manufacturer’s instruction.
Quantitative real-time PCR (qRT-PCR) and PCR
Complementary DNA (cDNA) was synthesized from 500 ng DNAse-treated RNA using the QuantiTect reverse transcription kit (Qiagen) according to the manufacturer’s instructions. qRT-PCR was carried out using the PowerUp™ SYBR™ Green Master Mix kit (Applied Biosystems) and the AriaMx Real-Time PCR system (Agilent Technologies) following the manufacturers’ instructions. The expression of exsL, l-bclA, ena3A, and rpoB (housekeeping gene) was analyzed from cDNA of each of the three independent cultures sampled after 4, 8, 12, and 16 h after inoculation and the relative gene expression ratio was calculated according to the Pfaffl formula53. Primers were used at 300 nM concentration, and their efficiencies were calculated from the slope of the standard curve using the formula (10−1/slope-1) × 100, and were found to be between 95% and 101%. No template was added to the negative control samples. The Ct values obtained for the 4-h samples were used as the calibrator sample in the data analysis, and rpoB served as an internal control gene. Each cDNA sample was analyzed in triplicate, and the experiment was repeated independently. The cycling conditions of the qRT-PCR were: 50 °C/2 min, 95 °C/2 min; and 40 cycles of 95 °C/15 s, 60 °C/1 min.
To determine if exsL, bclA, and ena3A are co-expressed, standard PCR was conducted using the cDNAs as templates. Primers that span across the indicated genes (Table S4), and DreamTaq PCR master mix (Thermo Scientific™) were used. Purified genomic DNA (gDNA) from NVH 0075-95 and cDNA from NVH 0075-95 ΔenaABC/Δena3A (4 h and 16 h) were used as positive and negative controls, respectively.
Sequencing strain B. paranthracis NVH 0075-95
To achieve a closed, updated, and high-quality genome of B. paranthracis NVH 0075-95, the strain was subjected to hybrid assembly of long- and short-read sequences. Genomic DNA from NVH 0075-95 was prepared from bacteria grown overnight on blood agar plates. All sequencing was performed by Novogene Co. (London, UK) using a combination of Pacific Biosciences (PacBio) RS II single-molecule real-time (SMRT) sequencing platform with a with a SMRTbell template library and an Illumina NovaSeq 6000 platform (2 × 150 bp) with an insert size of 300 bp.
A closed genome was achieved using the assembly pipeline Unicycler v. 0.4.8.0+galaxy354 conducting a short-read-first hybrid assembly. Genome contiguity, completeness, and correctness were assessed using Quast v. 5.0.2+galaxy355, Bandage 0.8.1+galaxy356, BUSCO v. 5.3.2+galaxy0 (mode genome, gene predictor prodigal, lineage dataset bacillales_odb10)57 and post-assembly corrections using Pilon (58) wrapped in the assembler Unicycler v. 0.4.8.0+galaxy3. Species validation was done on the Type Strain Genome Server (TYGS)59,60. Plasmids and contigs were predicted with PlasFlow v. 1.1.0 (threshold >0.6, remaining default settings)61, and the genome was annotated using the online NCBI Prokaryotic Genome Annotation Pipeline (PGAP)62–64. Data is available at NCBI under accession number GCA_027945115.1.
Distribution of L-ena
Cblaster65, a Python toolkit for detecting collocated genes, was run remotely through the web application CAGECAT v.1.0 (settings: -max gap 200 bp, n of unique query seq=3, min coverage 50%, min identity 30%, maximum e-value 0.01) on the NCBI RefSeq non-redundant protein database, using amino acid sequence of WP_017562367.1 (Ena3A), WP_048548723.1 (BclA), WP_048548726.1 (ExsL) and WP_058548727.1 (Tn3 family transposase) as query. The genomic region spanning WP_017562367.1, WP_048548723.1, WP_048548726.1 is hereafter referred to as “the gene cluster”. To investigate the proportion of organisms with the gene cluster within the B. cereus s.l. group, publicly available genomes yielding hits in the cblaster search (see Source Data file sheet Supplementary Fig. 6) were downloaded from NCBI RefSeq database (n = 656), NCB (https://www.ncbi.nlm.nih.gov/refseq/; Table EV1) and appended to a representative database of assemblies of B. cereus s.l. group. In addition, 136 B. subtilis were included for comparison. Assemblies were quality-checked using QUAST, and only genomes of the correct size (~4.9–6.2 Mb) and a GC content of ~35% were included in the downstream analysis. Pairwise tBLASTn searches were performed (BLAST+ v. 2.11.066, e-value 1e-10, max_hspr 1, default settings) to search for homo- and orthologs of the following query protein sequences from strain NVH 0075-95: WP_017562367.1, WP_048548723.1, and WP_048548726.1. Proteins were considered as orthologs or homologs when they matched the query protein with high coverage (>70%) and moderate sequence identity (>30%), and when the whole gene cluster was present in the genome with corresponding synteny as the NVH 0075-95 strain. Genomic location (chromosome or plasmid) was inspected manually for complete and closed genomes. Mashtree v. 0.5767 was used to infer whole genome clustering for the B. cereus s.l. group, using the accurate option (--min-depth 0). Clustering and metadata (hits for query proteins) were visualized in Microreact web browser68.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank Dirk Reiter at the VIB-VUB Facility for Bio Electron Cryogenic Microscopy (BECM) and Yohannes Beyene Mekonnen at NMBU for technical assistance. This work was funded by VIB, NMBU, EOS Excellence in Research Program by FWO through grant G0G0818N to H.R. and G043021N to M.S. M.A. recognizes the Grant from the Norwegian Research Council (NFR): 335029 - FORSKER22.
Author contributions
M.S., M.A., and H.R. designed the project. M.S. performed cryogenic freezing, nsTEM, and cryo-EM imaging and data processing with assistance from M.F. and B.P. E.D.Z. conducted the gene expression studies, fluorescence microscopy studies, designed and generated the knockout strains and complementation constructs, and prepared the spores for analysis with support from K.O.S. A.K.L. conducted all sequencing and genomic analysis, M.S. wrote the manuscript with contributions from N.V.G. and all authors.
Peer review
Peer review information
Nature Communications thanks Grace Taylor-Joyce and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The ex vivo L-ENA and the recombinant Ena3A cryo-EM maps were deposited to EMDB under entry IDs EMD-17579 and EMD-17627, respectively. The atomic model for recEna3A was deposited to the PDB under ID 8PDZ. The genome assembly of Bacillus paranthracis NVH 0075-95 is available at NCBI under GCA_027945115.1. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mike Sleutel, Email: Mike.Sleutel@vub.be.
Han Remaut, Email: Han.Remaut@vub.be.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51804-w.
References
- 1.McKenney, P. T., Driks, A. & Eichenberger, P. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat. Rev. Microbiol.11, 33–44 (2013). 10.1038/nrmicro2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao, Y. et al. Bacterial spore germination receptors are nutrient-gated ion channels. Science380, 387–391 (2023). 10.1126/science.adg9829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henriques, A. O., Charles, J. & Moran, P. Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol.61, 555–588 (2007). 10.1146/annurev.micro.61.080706.093224 [DOI] [PubMed] [Google Scholar]
- 4.Rodenburg, C. M., McPherson, S. A., Turnbough, C. L. Jr. & Dokland, T. Cryo-EM analysis of the organization of BclA and BxpB in the Bacillus anthracis exosporium. J. Struct. Biol.186, 181–187 (2014). 10.1016/j.jsb.2014.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panessa-Warren, B. J., Tortora, G. T. & Warren, J. B. High resolution FESEM and TEM reveal bacterial spore attachment. Microsc. Microanal.13, 251–266 (2007). 10.1017/S1431927607070651 [DOI] [PubMed] [Google Scholar]
- 6.Kailas, L. et al. Surface architecture of endospores of the Bacillus cereus/anthracis/thuringiensis family at the subnanometer scale. Proc. Natl Acad. Sci. USA108, 16014–16019 (2011). 10.1073/pnas.1109419108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang, Y. et al. Bacillus anthracis spore surface protein BclA mediates complement factor H binding to spores and promotes spore persistence. PLoS Pathog.12, e1005678 (2016). 10.1371/journal.ppat.1005678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker, P. D. The spore antigens of Clostridium sporogenes, Cl. bifermentans and Cl. sordellii. J. Pathol. Bacteriol.85, 41–49 (1963). 10.1002/path.1700850105 [DOI] [PubMed] [Google Scholar]
- 9.Hodgkiss, W. & Ordal, Z. J. Morphology of the spore of some strains of Clostridium botulinum type E. J. Bacteriol.91, 2031–2036 (1966). 10.1128/jb.91.5.2031-2036.1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodgkiss, W., Ordal, Z. J. & Cann, D. C. The morphology and ultrastructure of the spore and exosporium of some Clostridium species. J. Gen. Microbiol.47, 213–225 (1967). 10.1099/00221287-47-2-213 [DOI] [PubMed] [Google Scholar]
- 11.Hachisuka, Y. & Kuno, T. Filamentous appendages of Bacillus cereus spores. Jpn J. Microbiol.20, 555–558 (1976). 10.1111/j.1348-0421.1976.tb01025.x [DOI] [PubMed] [Google Scholar]
- 12.Rode, L. J., Pope, L., Filip, C. & Smith, L. D. Spore appendages and taxonomy of Clostridium sordellii. J. Bacteriol.108, 1384–1389 (1971). 10.1128/jb.108.3.1384-1389.1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukaszczyk, M., Pradhan, B. & Remaut, H. In Bacterial Cell Walls and Membranes (ed. Kuhn, A.) 369–413 (Springer International Publishing, Cham, 2019).
- 14.Hendrickx, A. P. A., Budzik, J. M., Oh, S.-Y. & Schneewind, O. Architects at the bacterial surface — sortases and the assembly of pili with isopeptide bonds. Nat. Rev. Microbiol.9, 166–176 (2011). 10.1038/nrmicro2520 [DOI] [PubMed] [Google Scholar]
- 15.Mazmanian, S. K., Liu, G., Ton-That, H. & Schneewind, O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science285, 760–763 (1999). 10.1126/science.285.5428.760 [DOI] [PubMed] [Google Scholar]
- 16.Ton-That, H. & Schneewind, O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol.50, 1429–1438 (2003). 10.1046/j.1365-2958.2003.03782.x [DOI] [PubMed] [Google Scholar]
- 17.Kozuka, S. & Tochikubo, K. Properties and origin of filamentous appendages on spores of Bacillus cereus. Microbiol. Immunol.29, 21–37 (1985). 10.1111/j.1348-0421.1985.tb00799.x [DOI] [PubMed] [Google Scholar]
- 18.Pradhan, B. et al. Endospore Appendages: a novel pilus superfamily from the endospores of pathogenic Bacilli. EMBO J.40, e106887 (2021). 10.15252/embj.2020106887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zegeye, E. D. & Aspholm, M. Efficient Electrotransformation of Bacillus thuringiensis for Gene Manipulation and Expression. Curr. Protoc. 2, 1-15 (2022). 10.1002/cpz1.588 [DOI] [PubMed] [Google Scholar]
- 20.Jonsmoen, U. L. et al. Endospore pili: flexible, stiff, and sticky nanofibers. Biophys. J.122, 2696–2706 (2023). 10.1016/j.bpj.2023.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonsmoen, U. L. et al. The role of endospore appendages in spore-spore contacts in pathogenic bacilli. Preprint at bioRxiv10.1101/2024.04.22.590507 (2024).
- 22.Mistry, J., Finn, R. D., Eddy, S. R., Bateman, A. & Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res.41, e121–e121 (2013). 10.1093/nar/gkt263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci.30, 70–82 (2021). 10.1002/pro.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci.27, 14–25 (2018). 10.1002/pro.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richardson, J. S. The anatomy and taxonomy of protein structure. Adv. Protein Chem.34, 167–339 (1981). 10.1016/S0065-3233(08)60520-3 [DOI] [PubMed] [Google Scholar]
- 26.Kristoffersen, S. M. et al. Global mRNA decay analysis at single nucleotide resolution reveals segmental and positional degradation patterns in a Gram-positive bacterium. Genome Biol.13, R30 (2012). 10.1186/gb-2012-13-4-r30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paysan-Lafosse, T. et al. InterPro in 2022. Nucleic Acids Res.51, D418–D427 (2023). 10.1093/nar/gkac993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richard, E. et al. Protein complex prediction with AlphaFold-Multimer. Preprint at bioRxiv10.1101/2021.10.04.463034 (2022).
- 30.Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods19, 679–682 (2022). 10.1038/s41592-022-01488-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Remaut, H. & Waksman, G. Protein-protein interaction through beta-strand addition. Trends Biochem. Sci.31, 436–444 (2006). 10.1016/j.tibs.2006.06.007 [DOI] [PubMed] [Google Scholar]
- 32.Sylvestre, P., Couture-Tosi, E. & Mock, M. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol. Microbiol.45, 169–178 (2002). 10.1046/j.1365-2958.2000.03000.x [DOI] [PubMed] [Google Scholar]
- 33.Sylvestre, P., Couture-Tosi, E. & Mock, M. Polymorphism in the collagen-like region of the Bacillus anthracis BclA protein leads to variation in exosporium filament length. J. Bacteriol.185, 1555–1563 (2003). 10.1128/JB.185.5.1555-1563.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rety, S. et al. The crystal structure of the Bacillus anthracis spore surface protein BclA shows remarkable similarity to mammalian proteins. J. Biol. Chem.280, 43073–43078 (2005). 10.1074/jbc.M510087200 [DOI] [PubMed] [Google Scholar]
- 35.Thompson, B. M. & Stewart, G. C. Targeting of the BclA and BclB proteins to the Bacillus anthracis spore surface. Mol. Microbiol.70, 421–434 (2008). 10.1111/j.1365-2958.2008.06420.x [DOI] [PubMed] [Google Scholar]
- 36.Hachisuka, Y., Kozuka, S. & Tsujikawa, M. Exosporia and appendages of spores of Bacillus species. Microbiol. Immunol.28, 619–624 (1984). 10.1111/j.1348-0421.1984.tb00714.x [DOI] [PubMed] [Google Scholar]
- 37.Díaz-González, F. et al. Protein composition of the outermost exosporium-like layer of Clostridium difficile 630 spores. J. Proteom.123, 1–13 (2015). 10.1016/j.jprot.2015.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McElroy, K., Mouton, L., Du Pasquier, L., Qi, W. & Ebert, D. Characterisation of a large family of polymorphic collagen-like proteins in the endospore-forming bacterium Pasteuria ramosa. Res. Microbiol.162, 701–714 (2011). 10.1016/j.resmic.2011.06.009 [DOI] [PubMed] [Google Scholar]
- 39.Qiu, Y., Zhai, C., Chen, L., Liu, X. & Yeo, J. Current insights on the diverse structures and functions in bacterial collagen-like proteins. ACS Biomater. Sci. Eng.9, 3778–3795 (2021). [DOI] [PubMed]
- 40.Maes, E. et al. Glycosylation of BclA glycoprotein from Bacillus cereus and Bacillus anthracis exosporium is domain-specific. J. Biol. Chem.291, 9666–9677 (2016). 10.1074/jbc.M116.718171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bozue, J. A., Welkos, S. & Cote, C. K. The Bacillus anthracis Exosporium: what’s the big “hairy” deal? Microbiol. Spectr.3, 1–15 (2015). [DOI] [PubMed]
- 42.Tan, L. & Turnbough, C. L. Jr. Sequence motifs and proteolytic cleavage of the collagen-like glycoprotein BclA required for its attachment to the exosporium of Bacillus anthracis. J. Bacteriol.192, 1259–1268 (2010). 10.1128/JB.01003-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chattopadhyay, D., Walker, D. R., Rich-New, S. T., Kearney, J. F. & Turnbough, C. L. Jr. Crystal structure and induced stability of trimeric BxpB: implications for the assembly of BxpB-BclA complexes in the exosporium of Bacillus anthracis. mBio14, e0117223 (2023). 10.1128/mbio.01172-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conover, M. S. et al. Inflammation-induced adhesin-receptor interaction provides a fitness advantage to uropathogenic E. coli during chronic infection. Cell Host Microbe20, 482–492 (2016). 10.1016/j.chom.2016.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lukaszczyk, M., Pradhan, B. & Remaut, H. The biosynthesis and structures of bacterial pili. Subcell. Biochem.92, 369–413 (2019). 10.1007/978-3-030-18768-2_12 [DOI] [PubMed] [Google Scholar]
- 46.Lin, Y., Briandet, R. & Kovács, Á. T. Bacillus cereus sensu lato biofilm formation and its ecological importance. Biofilm4, 100070 (2022). 10.1016/j.bioflm.2022.100070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang, Y., Flint, S. H. & Palmer, J. S. Bacillus cereus spores and toxins - The potential role of biofilms. Food Microbiol.90, 103493 (2020). 10.1016/j.fm.2020.103493 [DOI] [PubMed] [Google Scholar]
- 48.Wijman, J. G., de Leeuw, P. P., Moezelaar, R., Zwietering, M. H. & Abee, T. Air-liquid interface biofilms of Bacillus cereus: formation, sporulation, and dispersion. Appl. Environ. Microbiol.73, 1481–1488 (2007). 10.1128/AEM.01781-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pakharukova, N. et al. Structural basis for Acinetobacter baumannii biofilm formation. Proc. Natl Acad. Sci. USA115, 5558–5563 (2018). 10.1073/pnas.1800961115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terry, C. et al. Molecular tiling on the surface of a bacterial spore - the exosporium of the Bacillus anthracis/cereus/thuringiensis group. Mol. Microbiol.104, 539–552 (2017). 10.1111/mmi.13650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janes, B. K. & Stibitz, S. Routine markerless gene replacement in Bacillus anthracis. Infect. Immun.74, 1949–1953 (2006). 10.1128/IAI.74.3.1949-1953.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindbäck, T. et al. CodY, a pleiotropic regulator, influences multicellular behaviour and efficient production of virulence factors in Bacillus cereus. Environ. Microbiol.14, 2233–2246 (2012). 10.1111/j.1462-2920.2012.02766.x [DOI] [PubMed] [Google Scholar]
- 53.Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res.29, e45 (2001). 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput. Biol.13, e1005595 (2017). 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mikheenko, A., Prjibelski, A., Saveliev, V., Antipov, D. & Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics34, i142–i150 (2018). 10.1093/bioinformatics/bty266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wick, R. R., Schultz, M. B., Zobel, J. & Holt, K. E. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics31, 3350–3352 (2015). 10.1093/bioinformatics/btv383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A. & Zdobnov, E. M. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol.38, 4647–4654 (2021). 10.1093/molbev/msab199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE9, e112963 (2014). 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meier-Kolthoff, J. P., Carbasse, J. S., Peinado-Olarte, R. L. & Göker, M. TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res.50, D801–D807 (2021). 10.1093/nar/gkab902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meier-Kolthoff, J. P. & Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun.10, 2182 (2019). 10.1038/s41467-019-10210-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krawczyk, P. S., Lipinski, L. & Dziembowski, A. PlasFlow: predicting plasmid sequences in metagenomic data using genome signatures. Nucleic Acids Res.46, e35–e35 (2018). 10.1093/nar/gkx1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tatusova, T. et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res.44, 6614–6624 (2016). 10.1093/nar/gkw569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haft, D. H. et al. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res.46, D851–D860 (2017). 10.1093/nar/gkx1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li, W. et al. RefSeq: expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res.49, D1020–D1028 (2020). 10.1093/nar/gkaa1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gilchrist, C. L. M. et al. cblaster: a remote search tool for rapid identification and visualization of homologous gene clusters. Bioinforma. Adv.1, vbab016 (2021). 10.1093/bioadv/vbab016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol.215, 403–410 (1990). 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 67.Katz, L. S. et al. Mashtree: a rapid comparison of whole genome sequence files. J. Open Source Softw. 4, 1–6 (2019). [DOI] [PMC free article] [PubMed]
- 68.Argimón, S. et al. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb. Genomics2, e000093 (2016). 10.1099/mgen.0.000093 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The ex vivo L-ENA and the recombinant Ena3A cryo-EM maps were deposited to EMDB under entry IDs EMD-17579 and EMD-17627, respectively. The atomic model for recEna3A was deposited to the PDB under ID 8PDZ. The genome assembly of Bacillus paranthracis NVH 0075-95 is available at NCBI under GCA_027945115.1. Source data are provided with this paper.