Abstract
A fluorescence method is described for direct measurement of the interaction between methanol dehydrogenase (MDH) and its electron acceptor cytochrome cL. This has permitted a distinction to be made between factors affecting electron transfer and those affecting the initial binding or docking process. It was confirmed that the initial interaction is electrostatic, but previous conclusions with respect to the mechanism of EDTA inhibition have been modified. It is proposed that the initial 'docking' of MDH and cytochrome cL is by way of ionic interactions between lysyl residues on its surface and carboxylate groups on the surface of cytochrome cL. This interaction is not inhibited by EDTA, which we suggest acts by binding to nearby lysyl residues, thus preventing movement of the 'docked' cytochrome to its optimal position for electron transfer, which probably involves interaction with the hydrophobic funnel in the surface of MDH.
Full text
PDF
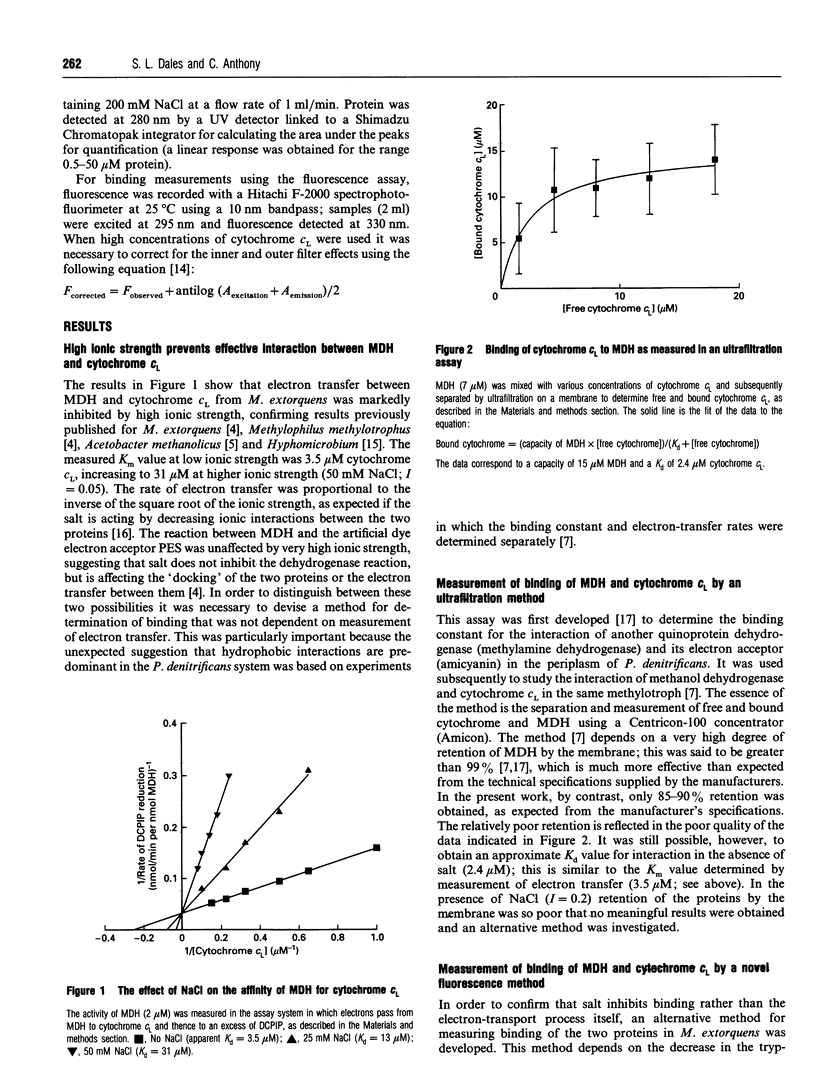
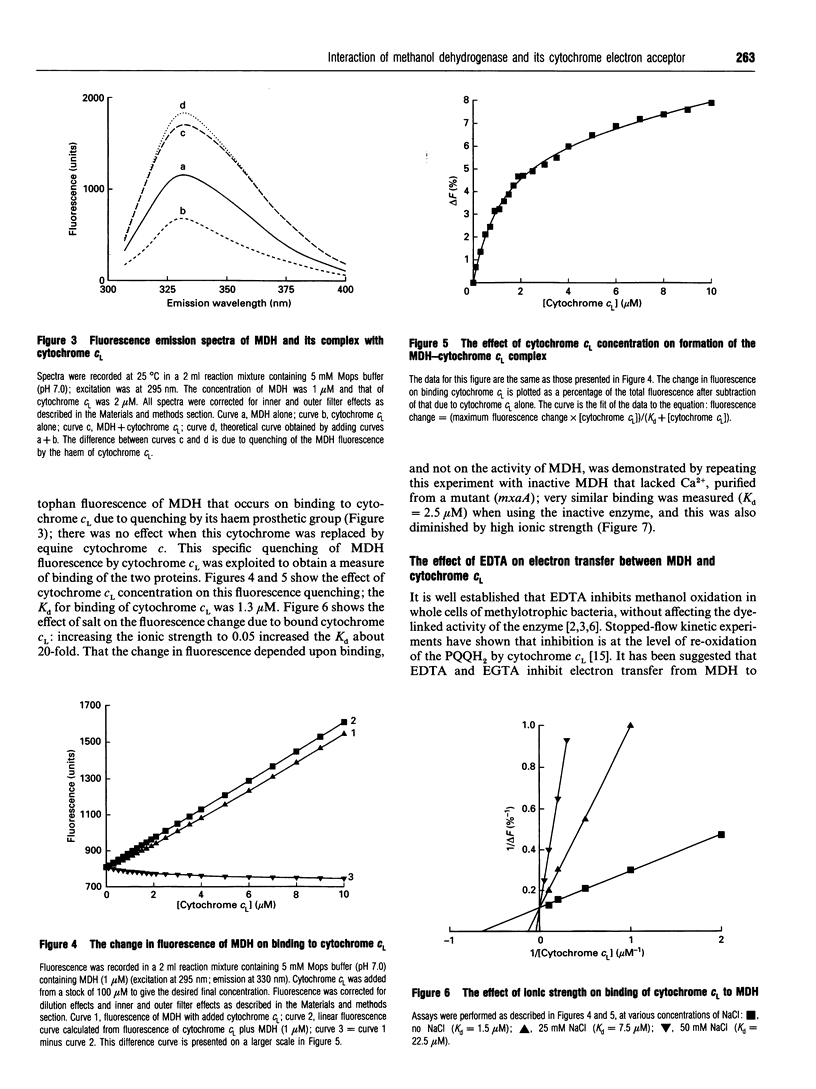
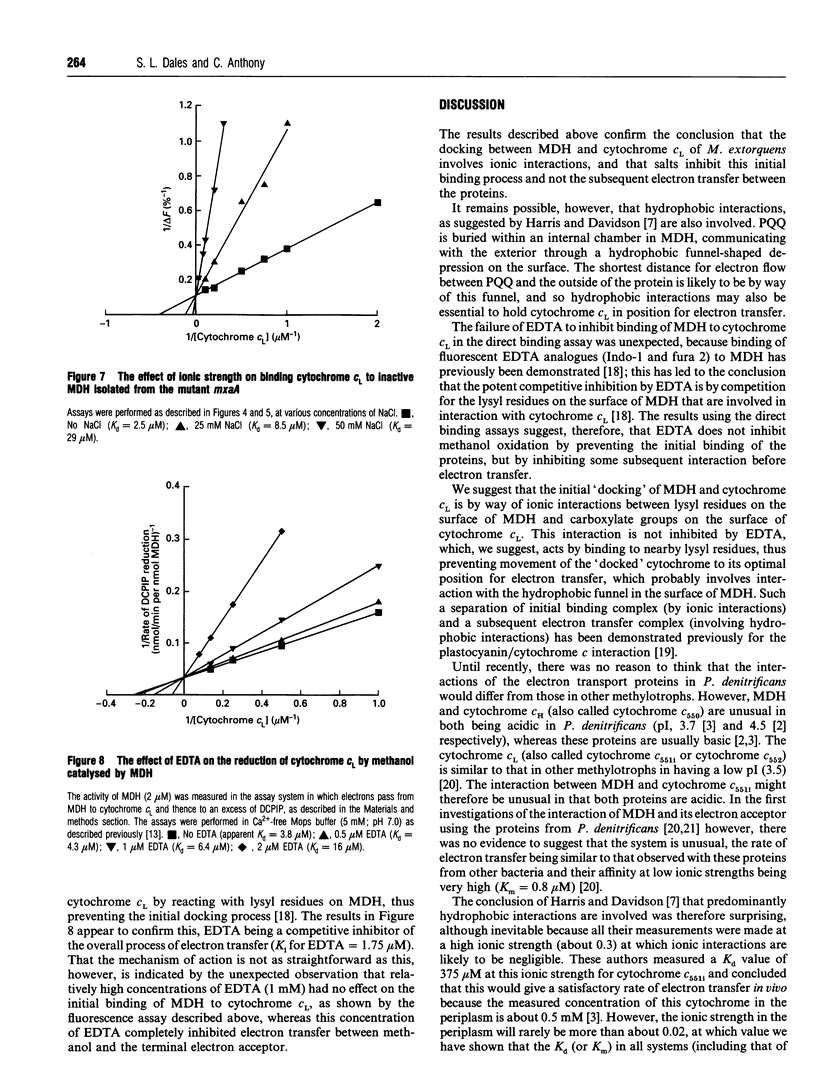

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anthony C. Bacterial oxidation of methane and methanol. Adv Microb Physiol. 1986;27:113–210. doi: 10.1016/s0065-2911(08)60305-7. [DOI] [PubMed] [Google Scholar]
- Anthony C., Ghosh M., Blake C. C. The structure and function of methanol dehydrogenase and related quinoproteins containing pyrrolo-quinoline quinone. Biochem J. 1994 Dec 15;304(Pt 3):665–674. doi: 10.1042/bj3040665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C. The c-type cytochromes of methylotrophic bacteria. Biochim Biophys Acta. 1992 Jan 30;1099(1):1–15. [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. 2. The methanol-oxidizing enzyme of Pseudomonas sp. M 27. Biochem J. 1964 Sep;92(3):614–621. doi: 10.1042/bj0920614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H. T., Anthony C. The interaction of methanol dehydrogenase and cytochrome cL in the acidophilic methylotroph Acetobacter methanolicus. Biochem J. 1991 Nov 15;280(Pt 1):139–146. doi: 10.1042/bj2800139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H. T., Anthony C. The mechanism of inhibition by EDTA and EGTA of methanol oxidation by methylotrophic bacteria. FEMS Microbiol Lett. 1992 Sep 15;75(2-3):231–234. doi: 10.1016/0378-1097(92)90409-h. [DOI] [PubMed] [Google Scholar]
- Cox J. M., Day D. J., Anthony C. The interaction of methanol dehydrogenase and its electron acceptor, cytochrome cL in methylotrophic bacteria. Biochim Biophys Acta. 1992 Feb 13;1119(1):97–106. doi: 10.1016/0167-4838(92)90240-e. [DOI] [PubMed] [Google Scholar]
- Davidson V. L., Graichen M. E., Jones L. H. Binding constants for a physiologic electron-transfer protein complex between methylamine dehydrogenase and amicyanin. Effects of ionic strength and bound copper on binding. Biochim Biophys Acta. 1993 Aug 16;1144(1):39–45. doi: 10.1016/0005-2728(93)90028-e. [DOI] [PubMed] [Google Scholar]
- Day D. J., Anthony C. Soluble cytochromes c of methanol-utilizing bacteria. Methods Enzymol. 1990;188:298–303. doi: 10.1016/0076-6879(90)88046-d. [DOI] [PubMed] [Google Scholar]
- Dijkstra M., Frank J., Jr, Duine J. A. Studies on electron transfer from methanol dehydrogenase to cytochrome cL, both purified from Hyphomicrobium X. Biochem J. 1989 Jan 1;257(1):87–94. doi: 10.1042/bj2570087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M., Anthony C., Harlos K., Goodwin M. G., Blake C. The refined structure of the quinoprotein methanol dehydrogenase from Methylobacterium extorquens at 1.94 A. Structure. 1995 Feb 15;3(2):177–187. doi: 10.1016/s0969-2126(01)00148-4. [DOI] [PubMed] [Google Scholar]
- Harris T. K., Davidson V. L. Binding and electron transfer reactions between methanol dehydrogenase and its physiologic electron acceptor cytochrome c-551i: a kinetic and thermodynamic analysis. Biochemistry. 1993 Dec 28;32(51):14145–14150. doi: 10.1021/bi00214a011. [DOI] [PubMed] [Google Scholar]
- Harris T. K., Davidson V. L., Chen L., Mathews F. S., Xia Z. X. Ionic strength dependence of the reaction between methanol dehydrogenase and cytochrome c-551i: evidence of conformationally coupled electron transfer. Biochemistry. 1994 Oct 25;33(42):12600–12608. doi: 10.1021/bi00208a010. [DOI] [PubMed] [Google Scholar]
- Peerey L. M., Brothers H. M., 2nd, Hazzard J. T., Tollin G., Kostić N. M. Unimolecular and bimolecular oxidoreduction reactions involving diprotein complexes of cytochrome c and plastocyanin. Dependence of electron-transfer reactivity on charge and orientation of the docked metalloproteins. Biochemistry. 1991 Sep 24;30(38):9297–9304. doi: 10.1021/bi00102a023. [DOI] [PubMed] [Google Scholar]
- Richardson I. W., Anthony C. Characterization of mutant forms of the quinoprotein methanol dehydrogenase lacking an essential calcium ion. Biochem J. 1992 Nov 1;287(Pt 3):709–715. doi: 10.1042/bj2870709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S., Boyd G., Mathews F. S., Xia Z. X., Dai W. W., Zhang Y. F., Davidson V. L. The active site structure of the calcium-containing quinoprotein methanol dehydrogenase. Biochemistry. 1993 Dec 7;32(48):12955–12958. doi: 10.1021/bi00211a002. [DOI] [PubMed] [Google Scholar]
- Xia Z. X., Dai W. W., Xiong J. P., Hao Z. P., Davidson V. L., White S., Mathews F. S. The three-dimensional structures of methanol dehydrogenase from two methylotrophic bacteria at 2.6-A resolution. J Biol Chem. 1992 Nov 5;267(31):22289–22297. [PubMed] [Google Scholar]


