Abstract
The Cdc48 AAA+ ATPase is an abundant and essential enzyme that unfolds substrates in multiple protein quality control pathways. The enzyme includes two conserved AAA+ ATPase motor domains, D1 and D2, that assemble as hexameric rings with D1 stacked above D2. Here, we report an ensemble of native structures of Cdc48 affinity purified from budding yeast lysate in complex with the adaptor Shp1 in the act of unfolding substrate. Our analysis reveals a continuum of structural snapshots that spans the entire translocation cycle. These data uncover elements of Shp1-Cdc48 interactions and support a ‘hand-over-hand’ mechanism in which the sequential movement of individual subunits is closely coordinated. D1 hydrolyzes ATP and disengages from substrate prior to D2, while D2 rebinds ATP and re-engages with substrate prior to D1, thereby explaining the dominant role played by the D2 motor in substrate translocation/unfolding.
Subject terms: Cryoelectron microscopy, Chaperones, Enzyme mechanisms, Ubiquitins
The Cdc48 enzyme is an abundant and essential enzyme that functions in many cellular pathways as a protein unfoldase. Here, the authors determine an ensemble of Cdc48 structures that capture snapshots of its unfolding action using a ‘hand-over-hand’ mechanism.
Introduction
The conserved AAA+ ATPase Cdc48 is an essential and abundant segregase that functions across many cellular pathways to separate proteins from various contexts, including organelle membranes, ribosomes, chromatin, and protein complexes1,2. Interactions with substrate adaptors allows Cdc48 to function in multiple pathways that have been characterized from yeast to human3. The human ortholog of Cdc48 (known as VCP or p97) is a major target of therapeutic development due to its importance in degenerative diseases and cancers4. Mutations in VCP cause multisystem proteinopathy-1 (MSP-1), a disease associated with the degeneration of brain, bone, and/or muscle, although the underlying mechanism of disease progression is poorly understood5. Additionally, VCP is upregulated in various types of cancer, and its inhibitors have been evaluated in clinical trials for cancer treatment6–8.
Cdc48 subunits comprise an N-terminal (N) domain followed by tandem AAA+ ATPase domains, D1 and D2, which are the motors that bind and hydrolyze ATP to drive substrate translocation and unfolding. The structures of D1 and D2 each comprise canonical AAA+ large α/β and small α subdomains that share sequence and structural similarity (hereafter simply referred to as ‘large’ and ‘small’ subdomains, respectively). Multiple structures show that Cdc48/VCP assembles as a homo-hexamer of stacked D1 and D2 rings, with each of the 12 large subdomains contributing conserved pore loop 1 (PL1) and pore loop 2 (PL2) residues that bind the unfolded substrate through an array of binding pockets that spans the central hexameric pore9–12. The N-domain sits atop the complex and is the primary binding site of substrate-recruiting adaptors.
Protein substrates are recruited to Cdc48 by a wide variety of adaptors that can also regulate subcellular localization and ATP hydrolysis rates. One of the most abundant Cdc48 adaptors in budding yeast is Shp1, which regulates protein phosphatase-1 (PP1) complex assembly and stability13,14. The human orthologs of Shp1, p47 and p37, function in post-mitotic Golgi reassembly15 and PP1 activation16,17, respectively. In human cells, the PP1 catalytic subunit is held in an inactive state through interactions with its binding partners SDS22 and Inhibitor-3 (I3), and is activated by unfolding of I3 in a VCP/p37-dependent manner16. The unfolding of I3 frees PP1 to bind downstream activating cofactors and allows it to perform its critical role in transcriptional regulation as a serine/threonine phosphatase. The relationship between Cdc48/VCP and PP1 is presumably conserved from yeast to human because the yeast orthologs of the PP1 complex are enriched in native pulldowns of Shp1 (human, PP1-SDS22-I3; yeast, Glc7-Sds22-Ypi1)9.
Insights into how AAA+ ATPases drive substrate unfolding have been derived from substrate-bound structures of Cdc48/VCP9–12 and related AAA+ unfoldases18–35. The leading model envisions unfolding driven by translocation of the substrate in an extended conformation through the hexamer pore by a ‘hand-over-hand’ (aka. ‘conveyor belt’) mechanism36,37. In this model, D1 and D2 form an ATP-stabilized helical spiral that presents a peptide-binding pocket with the optimal symmetry to bind substrate in an extended β-strand conformation, with an array of equivalent dipeptide binding sites formed at the interfaces of adjacent Cdc48 subunits. Typically, five hexamer subunits adopt the spiraling configuration and bind substrate, while the other subunit appears to be transitioning between the ends of the spiral. Progression along the unfolding/translocation cycle results from ATP hydrolysis-mediated dissociation of a Cdc48 subunit from the lagging end of the spiral while ATP binding mediates re-association at the leading end of the spiral to bind the next dipeptide of the substrate. Although we strongly favor this hand-over-hand mechanism, we note that alternatives have been suggested38–40.
Here, we determine an ensemble of cryo-EM structures of the Cdc48-Shp1 complex in the act of processing substrate. The structures belong to a series of nine sequential snapshots that represent a continuum of subunit movements during substrate translocation. The motion of each Cdc48 subunit is directly linked to its neighboring subunits through inter-subunit interactions. These structures provide a complete view of the entire protein translocation cycle and rationalize a model in which the binding and hydrolysis of ATP and the engagement and disengagement of substrate are highly coordinated.
Results and discussion
Overall structure
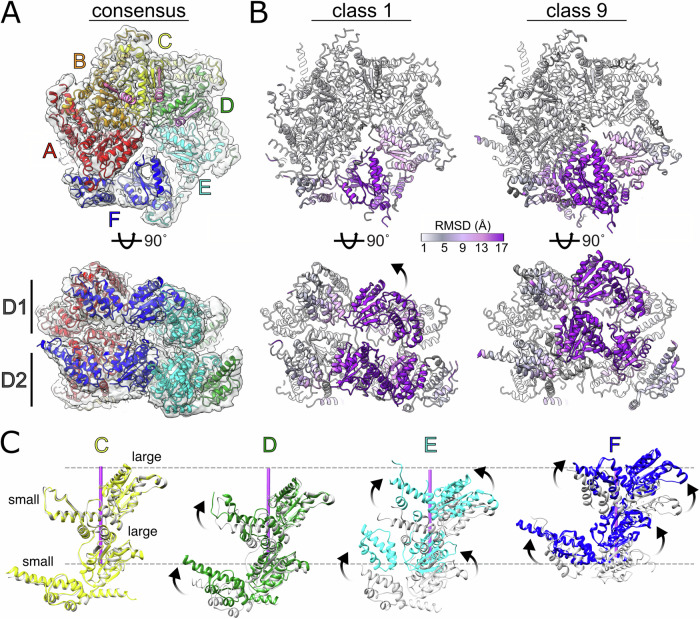
Native Cdc48-Shp1 complexes were purified by co-immunoprecipitation of FLAG-tagged Shp1 from budding yeast (S. cerevisiae) lysates and then stabilized through chemical crosslinking prior to cryo-EM specimen preparation, as described previously9. Active, substrate-bound complexes were enriched by stabilizing the particles with the ATP analog ADP•BeFx, which traps Cdc48 in the process of unfolding endogenous substrate. Cryo-EM images of the purified particles were recorded, and subsequent data processing produced an overall consensus charge density map at 2.9 Å resolution (Fig. 1A, Supplementary Fig. 1). The reconstruction closely resembled our previously reported 3.7 Å and 3.6 Å resolution structures of yeast9 and human11 complexes, respectively, in which the N-domain adopts the active-state ‘up’ conformation, and the D1 hexamer is stacked on top of D2 in a right-handed helical arrangement for subunits A-E, with subunit A being the topmost subunit that is engaged with substrate in both D1 and D2, while subunit F displays poor local resolution and is disengaged from the helical array and the substrate.
Fig. 1. Structures of substrate-bound Cdc48 reveal relative motion among subunits.
A Cdc48 hexamer modeled into the 2.9 Å resolution consensus reconstruction. Subunit labels and colors are used throughout the manuscript. B 3DVA and 3D classification reveals structures with substantial variability in subunits E and F. Root mean square deviation (RMSD) measurements between Cα atoms of class 1 and 9 models shown as colorized ribbon. C Variable subunit motion observed in subunits D–F between classes 1 (gray) and 9 (color). Subunits A, B, and C positions are generally superimposable between classes 1 and 9 (subunits A and B not shown). Substrate axis shown as purple rod and its upper/lower limits denoted by dashed gray lines as a positional reference. Arrows in panels B and C indicate trajectory of motion.
3D variability analysis reveals the Cdc48 translocation pathway continuum
The poor local resolution in subunit F indicated its high degree of mobility relative to the rest of the hexamer. In order to resolve conformational heterogeneity in Cdc48, we applied 3D variability analysis (3DVA)41 to the 2.9 Å resolution dataset. This revealed a component that tracked the continuous motion of subunit F between the bottom and the top of the helical stack (Movie 1). Particles were split into ten classes along this component, nine of which were resolved to resolutions between 3.2 Å to 4.3 Å (Supplementary Figs. 2–4, Movies 2–3). The classes were assigned based on the relative position of subunit F along the translocation pathway: Class 1 displays subunit F in its lowest position relative to the helical array, and class 9 displays subunit F at the top of the helical array, re-engaged with substrate and subunit A in D2 but still disengaged in D1 (Fig. 1B, Movie 4). The intermediate classes 2-8 display subunit F along a sequential trajectory between classes 1 and 9. Across all classes, the N-domains were resolved in the ‘up’ state elevated above the D1 ring and did not display conformational variability relative to the D1 domain, although their weak density is indicative of high mobility.
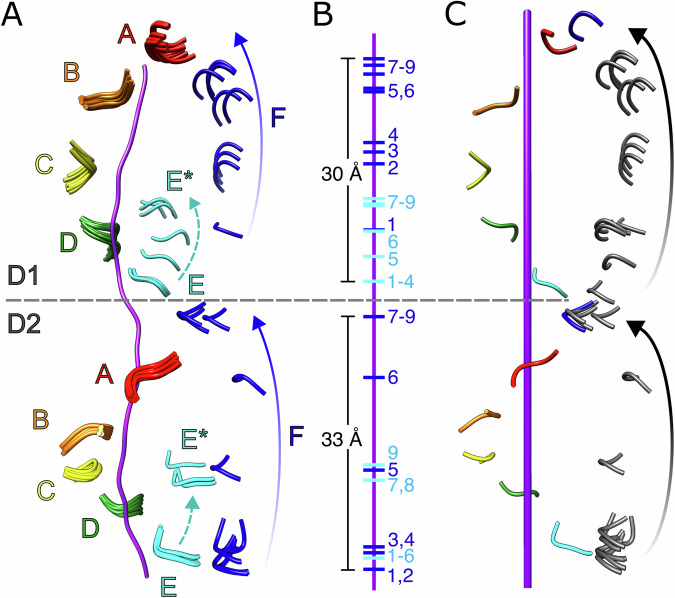
D1 and D2 for subunits A, B, and C were well resolved and closely superimposable across all classes (r.m.s.d < 1.0 Å, Fig. 1B, C). Subunit D displayed some small relative motion within the small ATPase domain, but the large subdomain was always part of the AAA+ helix and engaged with substrate (Fig. 1C). Although PL1 residues lacked precise density in classes that were disengaged from substrate, fitting of the large subdomains enabled positioning of PL1 (Fig. 2). We define the D1 and D2 domains as being substrate-engaged if the alpha carbon atom (Cα) of the nearest PL1 residue (D1, M288; D2, W561) is within 6 Å of a substrate Cα; in contrast, all substrate-disengaged domains had distances of at least 9 Å. Subunit E exhibited positional variability and disengagement from the AAA+ helix and substrate across some, but not all of the classes while, as expected, subunit F is always disengaged in D1, only engaged in classes 8 and 9 for D2, and is in a different position in all classes (Figs. 1C, 2A, B). This indicates that more than one subunit becomes disengaged from substrate at distinct stages along the translocation trajectory. The consensus class overlays best with classes 1–3, which have the largest number of contributing particles and show subunit E engaged with substrate and subunit F in an intermediate position along the translocation trajectory. In contrast, the disengagement of subunit E from substrate in latter classes suggests movement that follows the initial trajectory of subunit F (classes 5–9 in D1, 7–9 in D2, denoted as E* in Fig. 2A). In order to map the complete path taken by one subunit as it transitions from the bottom to the top of its respective helical stack, the positions of substrate-disengaged subunit E loops were mapped onto the path taken by subunit F by overlapping subunits A and B onto the positions of subunits B and C. This revealed a continuous path taken by PL1 between substrate disengagement at the bottom of the spiral through substrate re-engagement at the top of the spiral (Fig. 2C).
Fig. 2. Trajectory of pore loop-1 (PL1) motion during substrate translocation.
A Overlay of all nine models of PL1 (residues 287-289, D1; 560-562, D2) wrapped around substrate (purple). Subunit E residues disengaged from substrate are indicated as E*. B Position of PL1 models from subunits E and F relative to substrate. C Positioning of subunit E PL1 models along the path of subunit F reveals the continuous trajectory of a substrate-disengaged subunit from the bottom to the top of each helical stack. Substrate axis represented as purple rod. The end positions of the transitioning subunit inferred from extrapolation of the helical assembly are colored blue, with intermediate states inferred from class 1–9 conformations of subunits E* and F shown in gray.
D2 engages with substrate over a larger fraction of the translocation cycle than D1
Our analysis revealed that the trajectories for D1 and D2 are slightly out of phase at the start and end of the translation pathway, i.e., that the positions of subunits and their corresponding pore loops are different between D1 and D2 (Fig. 2, Movies 3–4). As subunit F progresses toward the top of the helical stack, subunit E disengages from the bottom and follows the translocation path of subunit F. At the start of the translocation trajectory, D1 of subunit E is engaged with the substrate in classes 1–4 and becomes disengaged in class 5, while D2 of subunit E does not disengage from substrate until class 7, indicating that D1 disengages from the substrate before D2. At the opposite end of the trajectory (class 9), D1 of subunit F is still disengaged from substrate while D2 has re-engaged to form a canonical substrate binding pocket with the corresponding pore loop residues from subunit A, indicating that D2 re-engages the substrate before D1 and that subunit F D2 has already assumed the position of subunit A in this class while D1 is still moving toward that position.
Our finding that D2 spends more time than D1 engaged with the substrate is consistent with better resolved substrate density in D2 than in D1 (Supplementary Fig. 5). Presumably, D2 binds substrate more tightly because it contains conserved large hydrophobic residues at critical positions in PL1 (W561 and Y562), while D1 displays more flexible and smaller residues at the equivalent positions (M288 and A289). These observations indicate that D2 grips substrate more tightly consistent with the finding that catalytically inactivating mutants in D1 retain some substrate processing activity while the corresponding mutations in D2 completely abrogate translocation42. Notably, the apparent weaker binding in D1 appears to be functionally important because mutations of M288 to canonical large hydrophobic residues causes a lethal phenotype in yeast43.
Small and large ATPase subdomain motions are asynchronous and linked to movement of adjacent subunits
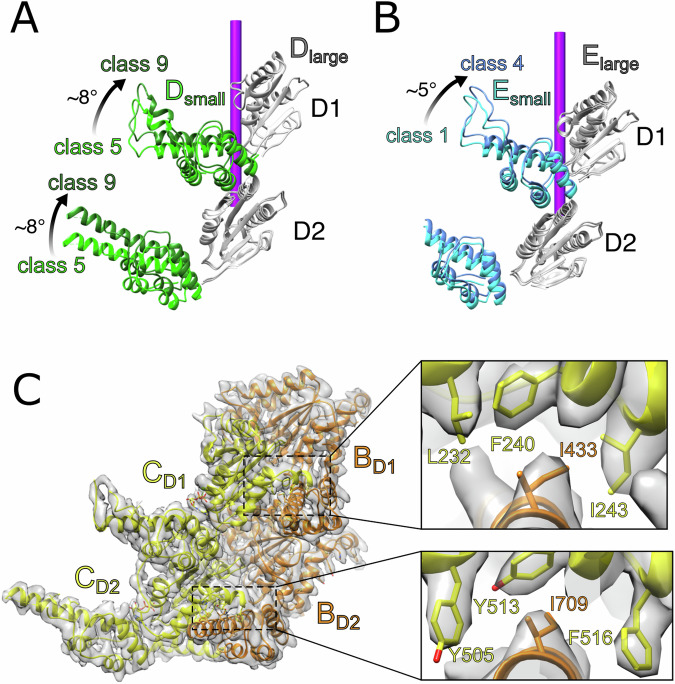
The small ATPase subdomains of subunit D display variable local resolutions between classes 1–9 as they begin to move behind subunit E and were modeled separately from their large subdomains. The subunit D large subdomain remains essentially stationary through all classes, including when subunit E is disengaged from the substrate in classes 5–9, whereas the subunit D small subdomain undergoes an ~8° rotation in both D1 and D2 (Fig. 3A). The large and small subdomains of subunit E were also fitted separately, with both large subdomains and the D2 small subdomain remaining stationary through classes 1–4 while the D1 small subdomain undergoes a ~5° rotation (Fig. 3B). This difference in motion between D1 and D2 of subunit E explains how D1 moves before D2 and illustrates how the two rings move asynchronously.
Fig. 3. Flexible motions within individual subunits during substrate translocation.
A The small subdomains of subunit D in both D1 and D2 move independently from their large subdomains between classes 5–9. B The small subdomain of subunit E from D1 is relatively mobile between classes 1–4 while its large subdomain and the D2 subdomains remain stationary. C Inter-subunit contacts between neighboring large and small ATPase subdomains (BC interface shown). The large subdomains form hydrophobic binding pockets comprising F240, I243, and L232 of D1 and Y505, Y513, and F516 of D2. A conserved isoleucine (D1, I433; D2, I709) is inserted into each binding pocket from the neighboring subunit. This inter-subunit interaction appears to be preserved during subunit movement.
The relative motions of small and large subdomains within a subunit appear to be driven by inter-subunit contacts between the small subdomain of one subunit and the large subdomain of the following subunit, which are largely preserved as subunits move along the translocation pathway. As clearly defined for subunits B and C in the center of the helical stack, the primary interaction for D1 involves I422 of the B subunit small subdomain inserting into a hydrophobic pocket formed by L232, F240, and I243 of the C subunit large subdomain (Fig. 3C). For D2, contacts include an analogous interaction between subunit B small subdomain I709 and subunit C large subdomain Y505, Y513, and F516. These interactions appear to be maintained throughout the entire translocation pathway, although the poor local resolution in mobile subunits does not allow definitive visualization in all cases. Thus, the leading subunit’s large subdomain appears to drag the small subdomain of the following subunit, linking the movement of adjacent subunits (Supplementary Fig. 6). A similar interaction was described in maintaining the interface of neighboring large and small subdomains of Vps4, suggesting that these inter-subunit contacts may be a conserved feature of AAA+ enzymes18.
The conformations modeled in classes 1–9 span the entire substrate translocation cycle and support the hand-over-hand mechanism in which Cdc48 walks along the substrate to pull unfolded substrate through the central pore. Previous structures of substrate-bound Cdc48 report that a single Cdc48 subunit of the homohexamer disengages from substrate during the ‘hand-over-hand’ cycle9,10. Here, we find that at some stages of the reaction cycle, two subunits are fully disengaged (Fig. 2), and the movement of three subunits (D, E, and F) are coordinated through maintenance of the inter-domain interactions between adjacent subunits (Movie 3). Most of the motion occurs within subunits E and F, and the inter-subunit interaction between the large subdomain of E and the small subdomain of D causes concomitant motion within subunit D.
Nucleotide states are coupled to subunit movement
Our reconstructions allow assignment of nucleotide density as corresponding to ATP, ADP, or apo (Supplementary Figs. 7–11, Supplementary Table 1). In all classes, the D1 and D2 active sites of subunits A, B, and C display nucleotide density corresponding to ATP (ADP•BeFx) coordinated with Mg2+ (Supplementary Fig. 7). Walker B glutamates (D1, E315; D2, E588), which are thought to activate a water molecule for nucleophilic attack on the ATP gamma phosphate44, lack clear density, perhaps because of the sensitivity of negatively charged residues to electron irradiation45. Nevertheless, in all active sites displaying ADP•BeFx density the nucleotide and Walker B glutamate main chain superimposed closely with other well-defined structures of AAA+ ATPase-ATP complexes (Supplementary Fig. 8). This is consistent with the possibility that Walker B glutamates are competent to catalyze ATP hydrolysis in all the ADP•BeFx (ATP)-bound states in classes 1–9.
Arginine finger residues in D1 (R369, R372) and D2 (R645, R648) are provided by the following subunit to coordinate nucleotide phosphates and promote ATP hydrolysis by stabilizing the pentavalent double negatively charged transition state46. Their side chains are well defined for active sites that bind ADP•BeFx in the consensus structure, with the guanidium group within hydrogen-bonding distance to a gamma phosphate oxygen atom (i.e., a fluorine in BeFx) (Supplementary Fig. 7). For active sites that bind ADP•BeFx in classes that lack resolution to visualize the arginine side chain, the arginine main chain and bound nucleotide are superimposable with the high-resolution structures, consistent with guanidinium-phosphate coordination. As with the Walker B geometry, this is consistent with all of the ATP (ADP•BeFx)-bound active sites adopting a catalytically competent conformation.
In addition to subunits A, B, and C, ATP density is also present in early classes of subunit D (classes 1–5 in D1 and classes 1–6 in D2, Supplementary Fig. 9) as well as in later classes of subunit F (classes 7–9 in D2, Supplementary Fig. 11). The transition in density from ATP to ADP in the later classes of subunit D (class 6 in D1 and class 7 in D2) indicates that ATP hydrolysis occurs in this subunit at the DE interface (Supplementary Fig. 9). Given the superimposable, apparently active conformation at all the ATP-binding active sites, it is not clear why hydrolysis should be favored at this specific location. One possibility is suggested by the motion of the subunit E large subdomains along the translocation pathway, which may result in a structurally modest but catalytically critical repositioning of the finger arginine to allow catalysis, as has been suggested for Vps422. Another possibility for preferential ATP hydrolysis at subunit D is that the highly constrained environment at the subunit A, B, and C active sites creates an unfavorable environment for phosphate release after ATP hydrolysis. In contrast to the AB, BC and CD interfaces, the nucleotide-binding pocket at the subunit DE interface active site becomes widened as subunit E progresses along the translocation path (from class 6 in D1 and from class 7 in D2), which may provide a favorable environment for phosphate release. This is reflected in the increased distance as the arginine finger residues from subunit E move away from the nucleotide at the subunit D active site from 3.0–3.2 Å in ATP-like bound states to >6 Å in ADP/apo states (Supplementary Figs. 9, 12). This widened gap persists at the EF interface with distances always exceeding 10 Å (Supplementary Fig. 10).
Nucleotide density at the subunit E active site appears as ADP in classes 1–4, which likely represents hydrolyzed nucleotide that is poised for release from the widened EF interface. Indeed, nucleotide is no longer observed (i.e., becomes empty/apo, or too low resolution to discern) in classes 5–9 in D1 and classes 7–9 in D2 (Supplementary Table 1, Supplementary Fig. 10). Likewise, subunit F nucleotide density is variable, being absent in all classes for D1 and in classes 1–6 for D2, but corresponding to ADP•BeFx•Mg2+ in D2 of classes 7–9 as it approaches subunit A and begins to reconnect to the Cdc48 helical array (Supplementary Fig. 11).
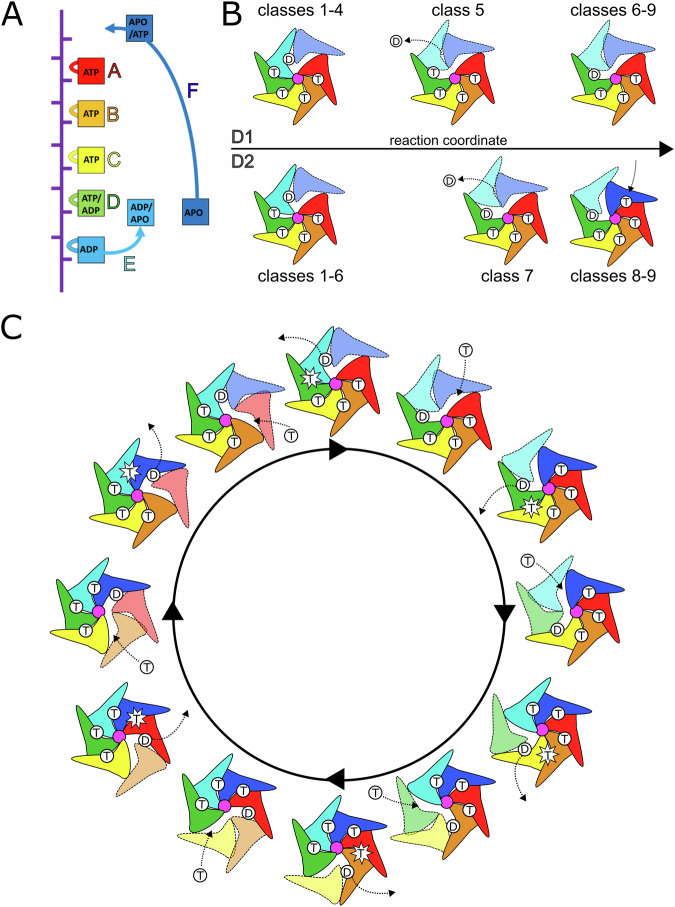
Taken together, the ensemble of classes establishes the relationship between the nucleotide hydrolysis cycle and substrate translocation (Fig. 4A). Subunits A-D are tightly engaged with substrate and their inter-subunit interfaces (AB, BC, and CD) are also compact in ATP-bound states (Fig. 4, Supplementary Table 1). A progression from substrate-engaged to disengaged state is observed for subunit E, which also correlates with a transition of nucleotide state from ATP to ADP at the DE interface (subunit D active site). This transition may be a result of a widening of the DE interface as subunit E and its nucleotide-coordinating arginine fingers peel away from subunit D, thereby promoting ATP hydrolysis and phosphate release at this interface. Opening of the active site as subunit E moves along the translocation pathway allows release of ADP, while at the end of the translocation cycle subunit F re-engages substrate and binds ATP and subunit A at the top of the helical stack, thereby restoring the compacted interface. This subunit therefore assumes the position of the ‘new’ subunit A and the translocation cycle advances by one subunit.
Fig. 4. Cdc48 translocation model.
A Nucleotide state assignments of individual Cdc48 subunits, combined for D1 and D2 and showing the range of nucleotides in the 9 classes visualized along the translocation pathway. B D1 and D2 do not move in lockstep with each other. D1 hydrolyzes ATP before D2. D2 binds ATP before D1. Substrate-disengaged subunits shown in lighter shades and separated from substrate (purple). T, ATP; D, ADP. C Complete translocation ATPase cycle of the Cdc48 hexamer. Applies to both D1 and D2.
Native Cdc48-Shp1 complexes are enriched with PP1 and ubiquitylated substrate
Our structure analysis also revealed insights into Shp1 and substrate interactions with Cdc48. Our previous work indicated that native yeast Cdc48-Shp1 purifications were enriched with components of the protein phosphatase-1 (PP1) complex, which includes Glc7, Sds22, and Ypi19. These results were consistent with studies of human proteins indicating that the orthologous p97-p37 complex unfolds human Ypi1 (Inhibitor-3, I3) to allow the formation of PP1 holoenzymes16,17. This prompted us to investigate if PP1 components are a substrate of the Cdc48-Shp1 complex in yeast. To test this, we immobilized native Shp1-associated complexes from budding yeast lysates using ADP•BeFx to trap bound substrates. Samples were then eluted with excess ATP to outcompete ADP•BeFx and permit substrate unfolding and release from the immobilized resin. Mass spectrometry proteomics of the ATP eluate revealed an enrichment of PP1 components compared to controls (Supplementary Fig. 13). Diglycine ubiquitin signatures were not detected on PP1 peptides, suggesting that Ypi1 unfolding is ubiquitin-independent as has been reported for I316. Therefore, ubiquitin-independent unfolding of Ypi1/I3 by Cdc48-Shp1 / VCP-p37 appears to be conserved from yeast to human. Additionally, ubiquitin (Ubi4) was enriched in the ATP elution sample, indicating that the Cdc48-Shp1 complex also processes ubiquitylated substrates that are distinct from PP1. An enrichment of ubiquitin was also observed in our recent characterization of the homologous human p97-p47 complex11 and other studies indicate that p47 associates with substrates containing various ubiquitin modifications, including mono-ubiquitin47, K63-linked48, and K11/K48 branched chains49. Interestingly, p47 does not appear to be associated with PP150, which suggests that yeast Shp1 acts on both ubiquitin-dependent and independent substrates while human p37 and p47 have specialized roles that have diverged from the ancestral Shp1.
Substrate and Shp1 contacts with the Cdc48 N domains
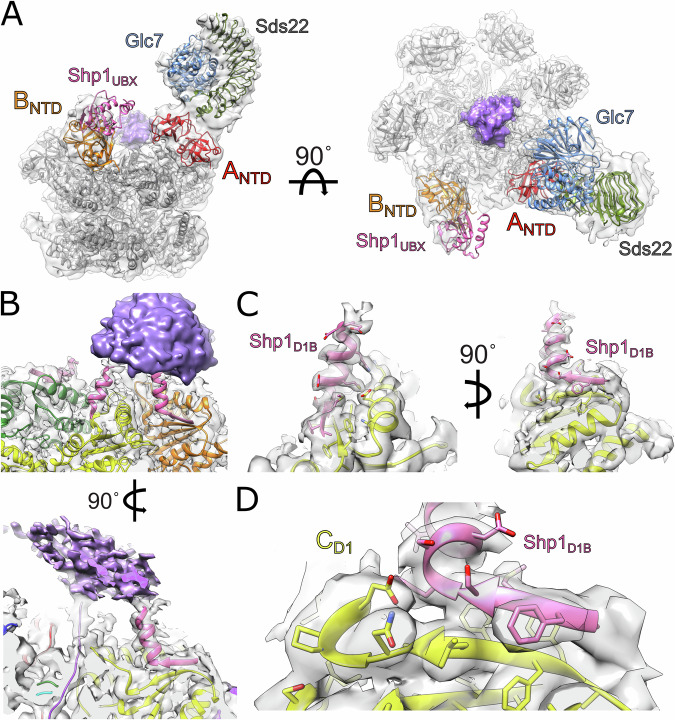
The Cdc48 N-terminal domains (NTDs) are associated with low-resolution density in the ‘up’ position seen in all our reconstructions, consistent with their known conformational variability in this active-state conformation51. We therefore performed focused classification to probe for additional features associated with these domains. This revealed a sub-class that contained a large density attached to the NTD of subunit A into which the crystal structure of the human PP1-SDS22 (Glc7-Sds22) subcomplex fit as a rigid body (Fig. 5A)52. The subunit B N-domain was associated with density corresponding to the Shp1 UBX domain, as previously reported9,11,53. This structure is consistent with a recent report of reconstituted human PP1 complex bound to p97 and p37 in which density for SDS22 and PP1 were observed on an N-domain of p97 of one subunit and p37 UBX density on an N-domain of a following subunit54. Ypi1 is not apparent within the modeled density, though it may be part of unassignable low-resolution density observed above the central pore that connects to the substrate modeled in the translocation pore (Supplementary Fig. 14). Focused classification over other subunits (B-F) did not resolve PP1 density, probably because these classes contained too few particles and/or those subunit NTDs displayed greater mobility.
Fig. 5. Substrate and adaptor densities associated with Cdc48.
A Rigid-body fitting of PP1 (Glc7-Sds22, PDB 6OBN) and UBX (PDB 6OPC) crystal structures into Cdc48 reconstruction containing PP1 density. Unassigned density in purple above the central pore of the hexamer likely represents a mixture of adaptor and substrate density. B Model and density of Shp1 D1B (Cdc48 ATPase Domain 1 binding beta-strand) motif associated with D1 large subdomains of subunits B, C, and D. Putative unstructured substrate/adaptor density colored in purple, as in A. C, D Close-up model and density of Shp1 D1B motif associated with subunit C.
Shp1 contacts the D1 large ATPase subdomain
The D1 large subdomain includes a solvent-exposed β-sheet that is well resolved in subunits B, C, and D. In the consensus structure, these subunits display additional density that is associated with the outermost β-strand of the D1 large subdomain and connected with poorly resolved substrate density over the central pore opening (residues 276-279, Fig. 5B). The density over subunit C was of sufficient quality to build 14 additional residues that include a four-residue β-strand that extends the β-sheet, followed by ten additional α-helical residues directed toward the extra density above the central pore of the hexamer (Fig. 5C).
To determine the source of this density, we employed ColabFold55 to predict potential interactions between the D1 large subdomain and Shp1 as well as other possible adaptors and substrates5. The best scoring model showed a specific region of Shp1 (residues 125-138) that fit precisely as a rigid body within the extra density (Supplementary Fig. 15). Densities at subunits B and D are consistent with the model built at subunit C, and it seems probable that equivalent structures are present at subunits A, E, and F but are not apparent due to the poorer local resolution for those subunits or being otherwise averaged out from low occupancy density. This additional interaction between this D1-binding motif (D1B) of Shp1 and D1 likely strengthens adaptor binding through avidity by complementing binding of the Shp1 UBX domain with the Cdc48 NTD. It may also promote optimal positioning of the Shp1-bound substrate to feed it into the central pore of the hexamer. Interestingly, the D1B motif is weakly conserved in p47 and missing entirely in p37, suggesting that other motifs provide additional binding interfaces in higher eukaryotes (Supplementary Fig. 16). In support of this, a recent structure of reconstituted VCP-p47 complex showed density attributed to the p47 SEP domain attached to the solvent-exposed VCP D1 domain56, and another structure showed that the SHP box of p37 interacts with the NTD of VCP54. Thus, it appears that the multi-domain arrangement among Shp1 and its related adaptors enables Cdc48/VCP binding primarily through its UBX domain and positioning of the substrate over the central pore through its other D1B, SEP, and SHP motifs/domains. Future studies will be needed to test these hypotheses and validate the role of the D1B motif.
Structures of multiple substrate-bound AAA+ complexes determined in recent years support a hand-over-hand mechanism of substrate translocation. The structures show four or five AAA+ subunits wrapping around an unfolding substrate in a helical arrangement, with pore loop residues forming an array of substrate-binding pockets that binds consecutive dipeptides in an extended beta-strand conformation. The remaining one or two subunits that are not part of the helical stack are disengaged from substrate and on a path between the ends of the AAA+ helix. Our study builds on these insights by resolving multiple conformations of Cdc48 along the substrate translocation pathway (Movie 1). Nine discrete classes reconstructed along the trajectory revealed how translocation is driven by a cycle of ATP hydrolysis, phosphate release, ADP dissociation, and ATP binding, and shows that D1 and D2 do not move in lockstep. Specifically, D1 hydrolyzes ATP, releases phosphate and dissociates from substrate before D2, while D2 rebinds ATP and re-engages with substrate before D1. The longer substrate engagement by D2 suggests that its pulling force is stronger than D1, which is supported by biochemical observations that D2 is the dominant motor in Cdc4842,57. The movement of successive subunits is coupled by inter-subunit contacts, with up to three consecutive subunits moving along the translocation pathway simultaneously (Movie 3).
Our work also demonstrates the utility of extensive cryo-EM image classification to characterize the remarkable compositional heterogeneity of complexes isolated from native sources. In addition to multiple states along the translocation trajectory, this approach uncovered interactions of the PP1 substrate and Shp1 adaptor with NTDs of adjacent Cdc48 subunits, as recently reported for the human VCP-p37-PP1 complex54. This approach also revealed an uncharacterized structural motif of Shp1 that was bound to D1 in interactions that appear different from human p37 and p47, thus underscoring the structural diversity of Cdc48/VCP-adaptor interactions. Our findings further illuminate the translocation mechanism of Cdc48 and other AAA+ ATPases, and provides insights to the role of Shp1 and other adaptors in Cdc48 processes.
Methods
Isolation of Cdc48-Shp1 complexes from yeast cell lysate
Cdc48-Shp1 complexes were purified as described previously9. In brief, a C-terminal 3xFLAG tag was introduced to the endogenous locus of the Shp1 gene in budding yeast by homologous recombination (strain BY4741). Liquid yeast cell cultures were grown and harvested at mid-log phase (OD600 ~ 1.5). Cells were pelleted, washed, and then flash frozen in liquid nitrogen. Frozen cells were lysed into powder by a freezer mill (SPEX). Powder lysates were resuspended in co-IP buffer (100 mM KOAc, 10 mM Mg2+, 10% glycerol, 0.1% Igepal CA-630, supplemented with protease inhibitor cocktail) and then clarified by centrifugation. Clarified supernatant was used as the basis of native complex purification through rapid affinity binding, washing, and elution termed ‘lysate-to-grid’ in which cryo-EM specimens are prepared within three hours of thawing cell lysate using cold co-IP buffer (full protocol in Cooney et al.58). Competitive elutions were performed using excess 3xFLAG peptide (a gift from Michael Kay, University of Utah) resuspended in co-IP buffer without glycerol or detergent. Eluates were stabilized by chemical crosslinking by treating samples with 0.1% glutaraldehyde for 10 min, followed by quenching with excess Tris to a final concentration of ~30 mM. The crosslinked eluates were used immediately for downstream cryo-EM specimen preparation.
Cryo-EM specimen preparation
Specimens were prepared on glow discharged UltraAuFoil R1.2/1.3 Au300 mesh grids (SPT Labtech). Glow discharging was performed for 60 seconds at 25 mA using a Pelco easiGlow unit (Ted Pella, Inc.). 3.5 μl of sample were applied to the grid, followed by blotting and vitrification using a Mk. II Vitrobot (Thermo Fisher Scientific) with a blot time of 5 s into liquid ethane.
Data collection and image reconstruction
Cryo-EM movies were recorded on a 300 kV Titan Krios G3 with a K3 direct detector using SerialEM59 with a dose rate of 19.3 e/pix/sec at a nominal magnification of 81,000x (corresponding to a super-resolution pixel size of 0.54 Å/pixel). Each movie contained 40 frames recorded over a span of 2.0 seconds each, corresponding to a total electron exposure of ~45 e/Å2. A total of 5913 movies were recorded. Patch motion correction and CTF estimation were performed in cryoSPARC and micrographs were 2x binned to their physical pixel size of 1.08 Å/pixel60,61.
Cryo-EM data processing
All data processing was performed in CryoSPARC60. Templates were generated using multiple rounds of 2D classification for particles identified by blob picking from a small subset of the movies collected (249 movies). These templates were used in template picking of the entire data set. This led to selection of 4,690,491 particles, which were reduced to 1,456,150 particles after multiple rounds of 2D classification. Particles were subsequently re-extracted with a box size of 288 pixels from 5847 movies, and further rounds of 2D classification resulted in 539,957 particles. Ab initio classification followed by heterogeneous refinement in CryoSPARC led to the sorting of 325,377 particles into a well-resolved class. Non-uniform refinement of these particles resulted in a 2.9 Å reconstruction of the Cdc48 complex (Supplementary Fig. 1).
3DVA analysis
3DVA analysis41 of the particles contributing to the 2.9 Å consensus structure revealed a component that displayed variability of mobile subunits and was used in a 3DVA Display job to cluster the particles into 10 classes, of which 9 could be reconstructed using non-uniform refinement, with the 10th class failing to reconstruct due to limited particle number. Inspection of the 9 reconstructed classes revealed asymmetric substrate-bound structures with variable positioning of mobile subunits. The mobile subunits had limited local resolution, so another round of 3DVA and 3DVA display was performed on all classes. After non-uniform refinement of the resulting clusters, resolution improvements were observed in 6 classes by excluding additional particles (Supplementary Fig. 2). This refinement led to final resolutions ranging between 3.2–4.3 Å among all classes (Supplementary Figs. 2, 3, 13).
Particles were re-extracted with a larger box size of 360 pixels to include additional volume for substrate and adaptor interactions with the N-domain. 3DVA analysis of particles was then performed using a large mask over the N-domain and part of D1 (Supplementary Fig. 11), and particles containing density on the N-terminus of subunit A were selected from clustering in 3DVA display. This was followed by multiple rounds of 2D classification, ab initio refinement, heterogeneous refinement, and further 3DVA analysis to isolate particles that contained this density. This led to 7727 particles containing extra density, with non-uniform refinement yielding a reconstruction at 4.2 Å resolution.
Model building, refinement, and validation
Model building and refinement were performed for each of the class reconstructions. Models of the D1 and D2 ATPase domains were built and subjected to atomic refinement using Phenix 1.2.162 and Coot v0.9.8.163. Substrate was built as described9. For subunits with ADP•BeFx modeled, magnesium was restrained to the OG1 of the final Thr in the Walker A motif at a distance of 2.038 Å. Low resolution information limited the refinements of transitioning subunits, which were restrained to rigidly build initial coordinates. Pore loops for subunit E and F structures that lacked density were modeled based on the well-defined structures of subunit B and optimized to minimize clash and improve geometry. Other residues without clear density have been reduced to stub atoms or were omitted from the model altogether.
Rigid body fitting of Sds22/PP1 used PDB 6OBN52 in UCSF Chimera for Sds22/PP1 density. All figures and movies were prepared in UCSF Chimera64. A summary of model refinement statistics is provided in Supplementary Table 2.
ColabFold structure prediction
An initial search for interactors with the outer beta strand of the D1 large subdomain was performed by performing predictions over ~100 residue sections of Glc7, Sds22, Ypi1, Doa1, and Shp1 using ColabFold55. The best scoring interaction was between Shp1 and Cdc48, and a refined prediction was made in ColabFold using sequences for residues 125-139 of Shp1 and the Cdc48 D1 large subdomain (residues 219-379). Template mode was selected as pdb70 and 24 recycles were performed. This prediction fit well as a rigid body into the density for subunit C (Supplementary Fig. 15) of the consensus structure and was used as a starting model for further refinement in Phenix62.
Mass spectrometry
Mass spectrometry methods were as described previously with minor adaptations9. Co-IP eluates from Shp1-FLAG in the presence of 1 mM ADP•BeFx or 5 mM ATP, to elute putative substrate, were processed separately for proteomics analysis (n = 3 for each sample). For each sample, snap-frozen eluates were thawed with the addition of 100 μL of 6 M guanidinium/HCl in 100 mM Tris/HCl pH 8.5 containing protease inhibitor cocktail (Sigma). Samples were washed with 6 M guanidine/HCl 100 mM Tris/HCl, pH 8.5 using centrifugal filters (30 kD MWCO). Cysteines were reduced using 10 mM TCEP and alkylated using 30 mM chloroacetamide. The guanidine solution was removed using two 25 mM ammonium bicarbonate (pH 8) washes. Proteins were resuspended in 25 mM ammonium bicarbonate (pH 8) and digested overnight at 37 °C with 0.2 μg of MS-Grade trypsin (Pierce). Trypsin reactions were quenched using 1 mM PMSF, and trypsin was removed by centrifugal filtration. Samples were placed in mass spectrometry vials, dried using a vacuum concentrator, and then resuspended at 1 μg/μL in 3% acetonitrile, 0.1% formic acid. Mass spectrometry data were collected using an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) coupled to Ultimate 3000 liquid chromatography (LC) pump (Thermo Fisher Scientific). Peptides were separated using an EASY-Spray PepMap RSLC C18 column (Thermo Fisher Scientific). The mobile phase consisted of buffer A (0.1% formic acid in optima water) and buffer B (optima water and 0.1% formic acid in 80% acetonitrile). The peptides were eluted at 300 nL/min with the following gradients over 2 h: 3–25% B for 80 min; 25–35% B for 20 min; 35–45% B for 8 min; 45–85% B for 2 min and 85% for 8 min. Data were acquired using the top speed method (3 s cycle). A full scan mass spectrum at resolution of 120,000 at 200 m/z mass was acquired in the Orbitrap with a target value of 4e5 and a maximum injection time of 50 ms. Peptides with charge states of 2-6 were selected from the top abundant peaks by the quadrupole for collisional dissociation (CID with normalized energy 30) MS/MS, and the fragment ions were detected in the linear ion trap with target AGC value of 1e4, a maximum injection time of 35 ms, and a dynamic exclusion time of 60 s. Precursor ions with ambiguous charge states were not fragmented. PEAKS Studio software (version 10) was used for de novo sequencing and database searching to identify proteins in the raw MS data, and to quantify, filter (quality-control), and normalize the quantitation data for each protein65. Peptides were identified from MS/MS spectra by searching against the Swiss-Prot Saccharomyces cerevisiae (strain ATCC 204508 / S288c) database (downloaded August 2020) with a reverse sequence decoy concatenated database66. Variables for the search were as follows: enzyme was set as trypsin with one missed cleavage site. Carbamidomethylation of cysteine was set as a fixed modification while N-terminal acetylation and methionine oxidation were set as variable modifications. A false positive rate of 0.01 was set as the maximum for peptides and proteins. The minimum length of peptide was set to 7 amino acids. At least 2 peptides were required for protein identification. The precursor mass error of 20 ppm was set for the precursor mass, and the mass error was set as 0.3 Da for the MSMS. Label-free quantitation was enabled with MS1 tolerance ±20 ppm and a MS2 tolerance ±50 ppm. The probability of each protein being a Shp1 interactor was calculated by comparing 3 replicates of each co-IP using the SAINT software package67.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by grants to PSS (NIH R35 GM133772), CPH (NIH U54 AI170856 and R01 GM112080), JCP (NIH R01 AG066874), and IC (NIH F31 CA254427). We thank the University of Utah Arnold and Mabel Beckman Center for Cryo-EM and Center for High Performance Computing for support in cryo-EM and computation, respectively. We also thank the Fritz B. Burns Biological Mass Spectrometry Facility at Brigham Young University for support in mass spectrometry.
Author contributions
P.S.S. and C.P.H. conceived the study; P.S.S. and I.C. designed the research; I.C., H.L.S., K.C., O.N.F., R.C., and J.C.P. performed experiments; all authors analyzed the results. P.S.S. and C.P.H. wrote the manuscript. All authors contributed to manuscript revision and approved the final version.
Peer review
Peer review information
Nature Communications thanks Helmut Bergler, Andrew Truman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Cryo-EM reconstructions and atomic models have been deposited in the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB), respectively, under the following accession numbers: EMD-42076, PDB 8UB4 (Consensus); EMD-41992, PDB 8U7T (Class 1); EMD-42038, PDB 8U9P (Class 2); EMD-42057, PDB 8UAA (Class 3); EMD-42025, PDB 8U8I (Class 4); EMD-42032, PDB 8U9C (Class 5); EMD-42039, PDB 8U9Q (Class 6); EMD-42042, PDB 8U9Z (Class 7); EMD-42043, PDB 8UA0 (Class 8); EMD-42047, PDB 8UA1 (Class 9). The mass spectrometry data generated in this study have been deposited in the ProteomeXchange (PRIDE) database under accession code PXD048280.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ian Cooney, Heidi L. Schubert.
Change history
12/13/2024
A Correction to this paper has been published: 10.1038/s41467-024-55349-w
Contributor Information
Christopher P. Hill, Email: chris@biochem.utah.edu
Peter S. Shen, Email: peter.shen@biochem.utah.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51835-3.
References
- 1.van den Boom, J. & Meyer, H. VCP/p97-mediated unfolding as a principle in protein homeostasis and signaling. Mol. Cell69, 182–194 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Ye, Y., Tang, W. K., Zhang, T. & Xia, D. A Mighty “Protein Extractor” of the Cell: Structure and Function of the p97/CDC48 ATPase. Front Mol. Biosci.4, 39 (2017). [DOI] [PMC free article] [PubMed]
- 3.Braxton, J. R. & Southworth, D. R. Structural insights of the p97/VCP AAA+ ATPase: How adapter interactions coordinate diverse cellular functionality. J. Biol. Chem.299, 105182 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kilgas, S. & Ramadan, K. Inhibitors of the ATPase p97/VCP: from basic research to clinical applications. Cell Chem. Biol.30, 3–21 (2023). [DOI] [PubMed] [Google Scholar]
- 5.Kimonis, V. E., Fulchiero, E., Vesa, J. & Watts, G. VCP disease associated with myopathy, Paget disease of bone and frontotemporal dementia: review of a unique disorder. Biochim Biophys. Acta1782, 744–748 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Anderson, D. J. et al. Targeting the AAA ATPase p97 as an approach to treat cancer through disruption of protein homeostasis. Cancer Cell28, 653–665 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roux, B. et al. Targeting acute myeloid leukemia dependency on VCP-mediated DNA repair through a selective second-generation small molecule inhibitor. Sci. Transl. Med.13, eabg1168 (2021). [DOI] [PMC free article] [PubMed]
- 8.Le Moigne, R. et al. The p97 Inhibitor CB-5083 Is a Unique Disrupter of Protein Homeostasis in Models of Multiple Myeloma. Mol. Cancer Ther.16, 2375–2386 (2017). [DOI] [PubMed]
- 9.Cooney, I. et al. Structure of the Cdc48 segregase in the act of unfolding an authentic substrate. Science (1979)365, 502–505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Twomey, E. C. et al. Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Science (1979)365, eaax1033 (2019). [DOI] [PMC free article] [PubMed]
- 11.Xu, Y. et al. Active conformation of the p97-p47 unfoldase complex. Nat. Commun.13, 2640 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan, M. et al. Mechanistic insight into substrate processing and allosteric inhibition of human p97. Nat. Struct. Mol. Biol.28, 614–625 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Cheng, Y.-L. & Chen, R.-H. Assembly and quality control of the protein phosphatase 1 holoenzyme involves the Cdc48–Shp1 chaperone. J. Cell Sci.128, 1180–1192 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Böhm, S. & Buchberger, A. The budding yeast Cdc48Shp1 complex promotes cell cycle progression by positive regulation of protein phosphatase 1 (Glc7). PLoS One8, e56486 (2013). [DOI] [PMC free article] [PubMed]
- 15.Kondo, H. et al. p47 is a cofactor for p97-mediated membrane fusion. Nature388, 75–78 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Weith, M. et al. Ubiquitin-independent disassembly by a p97 AAA-ATPase complex drives PP1 holoenzyme formation. Mol. Cell72, 766–777.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 17.van den Boom, J. et al. Targeted substrate loop insertion by VCP/p97 during PP1 complex disassembly. Nat. Struct. Mol. Biol. 2021 28:1228, 964–971 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Han, H., Monroe, N., Sundquist, W. I., Shen, P. S. & Hill, C. P. The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets. Elife6, e31324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su, M. et al. Mechanism of Vps4 hexamer function revealed by cryo-EM. Sci. Adv.3, e1700325 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, L., Myasnikov, A., Pan, X. & Walter, P. Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction. Elife9, e54031 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zehr, E. A., Szyk, A., Szczesna, E. & Roll-Mecak, A. Katanin grips the β-tubulin tail through an electropositive double spiral to sever microtubules. Dev. Cell52, 118–131.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monroe, N., Han, H., Shen, P. S., Sundquist, W. I. & Hill, C. P. Structural basis of protein translocation by the Vps4-Vta1 AAA ATPase. Elife6, e24487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han, H. et al. Structure of Vps4 with circular peptides and implications for translocation of two polypeptide chains by AAA+ ATPases. Elife8, e44071 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puchades, C. et al. Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science (1979)358, eaao0464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de la Peña, A. H., Goodall, E. A., Gates, S. N., Lander, G. C. & Martin, A. Substrate-engaged 26 S proteasome structures reveal mechanisms for ATP-hydrolysis–driven translocation. Science (1979)362, eaav0725 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong, Y. et al. Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature565, 49–55 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ripstein, Z. A., Huang, R., Augustyniak, R., Kay, L. E. & Rubinstein, J. L. Structure of a AAA+ unfoldase in the process of unfolding substrate. Elife6, e25754 (2017). [DOI] [PMC free article] [PubMed]
- 28.Gates, S. N. et al. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science (1979)357, 273–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deville, C. et al. Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase. Sci. Adv. 3, e1701726 (2017). [DOI] [PMC free article] [PubMed]
- 30.Yu, H. et al. ATP hydrolysis-coupled peptide translocation mechanism of Mycobacterium tuberculosis ClpB. Proc. Natl. Acad. Sci. USA115, E9560–E9569 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White, K. I., Zhao, M., Choi, U. B., Pfuetzner, R. A. & Brunger, A. T. Structural principles of SNARE complex recognition by the AAA+ protein NSF. Elife7, e38888 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho, C. et al. Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone. Nat. Commun.10, 5764 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alfieri, C., Chang, L. & Barford, D. Mechanism for remodelling of the cell cycle checkpoint protein MAD2 by the ATPase TRIP13. Nature559, 274–278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo, Y. H. et al. Cryo-EM structure of the essential ribosome assembly AAA-ATPase Rix7. Nat. Commun.10, 513 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puchades, C. et al. Unique structural features of the mitochondrial AAA+ protease AFG3L2 reveal the molecular basis for activity in health and disease. Mol. Cell75, 1073–1085.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puchades, C., Sandate, C. R. & Lander, G. C. The molecular principles governing the activity and functional diversity of AAA+ proteins. Nat. Rev. Mol. Cell Biol.21, 43–58 (2020). [DOI] [PMC free article] [PubMed]
- 37.Gates, S. N. & Martin, A. Stairway to Translocation: AAA+ motor structures reveal the mechanisms of ATP‐dependent substrate translocation. Protein Sci.10.1002/pro.3743 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fei, X. et al. Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate. Elife9, e52774 (2020). [DOI] [PMC free article] [PubMed]
- 39.Sauer, R. T., Fei, X., Bell, T. A. & Baker, T. A. Structure and function of ClpXP, a AAA+ proteolytic machine powered by probabilistic ATP hydrolysis. Crit. Rev. Biochem Mol. Biol.57, 188–204 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cordova, J. C. et al. Stochastic but highly coordinated protein unfolding and translocation by the ClpXP proteolytic machine. Cell158, 647–658 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Punjani, A. & Fleet, D. J. 3D variability analysis: resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol.213, 107702 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Bodnar, N. O. & Rapoport, T. A. Molecular mechanism of substrate processing by the Cdc48 ATPase complex. Cell169, 722–735.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esaki, M., Md Islam, T., Tani, N. & Ogura, T. Deviation of the typical AAA substrate-threading pore prevents fatal protein degradation in yeast Cdc48. Sci. Rep.7, 5475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, X. & Wigley, D. B. The ‘glutamate switch’ provides a link between ATPase activity and ligand binding in AAA+ proteins. Nat. Struct. Mol. Biol. 2008 15:1115, 1223–1227 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartesaghi, A., Matthies, D., Banerjee, S., Merk, A. & Subramaniam, S. Structure of β-galactosidase at 3.2-Å resolution obtained by cryo-electron microscopy. Proc. Natl. Acad. Sci. USA111, 11709–11714 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nadanaciva, S., Weber, J., Wilke-Mounts, S. & Senior, A. E. Importance of F 1 -ATPase Residue α-Arg-376 for Catalytic Transition State Stabilization. Biochemistry38, 15493–15499 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Meyer, H. H., Wang, Y. & Warren, G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J.21, 5645–5652 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shibata, Y. et al. p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO. Nat. Commun. 2012 3:13, 1–13 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Yau, R. G. et al. Assembly and function of heterotypic ubiquitin chains in cell-cycle and protein quality control. Cell171, 918–933.e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kracht, M. et al. Protein phosphatase-1 complex disassembly by p97 is initiated through multivalent recognition of catalytic and regulatory subunits by the p97 SEP-domain adapters. J. Mol. Biol.432, 6061–6074 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Banerjee, S. et al. 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science (1979)351, 871–875 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choy, M. S. et al. SDS22 selectively recognizes and traps metal-deficient inactive PP1. Proc. Natl. Acad. Sci. USA116, 20472–20481 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dreveny, I. et al. Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47. EMBO J.23, 1030–1039 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van den Boom, J., Marini, G., Meyer, H. & Saibil, H. R. Structural basis of ubiquitin-independent PP1 complex disassembly by p97. EMBO J.42, e113110 (2023). [DOI] [PMC free article] [PubMed]
- 55.Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 2022 19:619, 679–682 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conicella, A. E. et al. An intrinsically disordered motif regulates the interaction between the p47 adaptor and the p97 AAA+ ATPase. Proc. Natl. Acad. Sci. USA117, 26226–26236 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song, C., Wang, Q. & Li, C. C. H. ATPase activity of p97-valosin-containing protein (VCP). D2 mediates the major enzyme activity, and D1 contributes to the heat-induced activity. J. Biol. Chem.278, 3648–3655 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Cooney, I. et al. Lysate-to-grid: Rapid Isolation of Native Complexes from Budding Yeast for Cryo-EM Imaging. Bio Protoc13, e4596 (2023). [DOI] [PMC free article] [PubMed]
- 59.Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol.152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol.192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol.74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr.66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pettersen, E. F. et al. UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput Chem.25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Zhang, J. et al. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteom.11, M111.010587 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bairoch, A. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res.28, 45–48 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Teo, G. et al. SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J. Proteom.100, 37–43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Cryo-EM reconstructions and atomic models have been deposited in the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB), respectively, under the following accession numbers: EMD-42076, PDB 8UB4 (Consensus); EMD-41992, PDB 8U7T (Class 1); EMD-42038, PDB 8U9P (Class 2); EMD-42057, PDB 8UAA (Class 3); EMD-42025, PDB 8U8I (Class 4); EMD-42032, PDB 8U9C (Class 5); EMD-42039, PDB 8U9Q (Class 6); EMD-42042, PDB 8U9Z (Class 7); EMD-42043, PDB 8UA0 (Class 8); EMD-42047, PDB 8UA1 (Class 9). The mass spectrometry data generated in this study have been deposited in the ProteomeXchange (PRIDE) database under accession code PXD048280.