Abstract
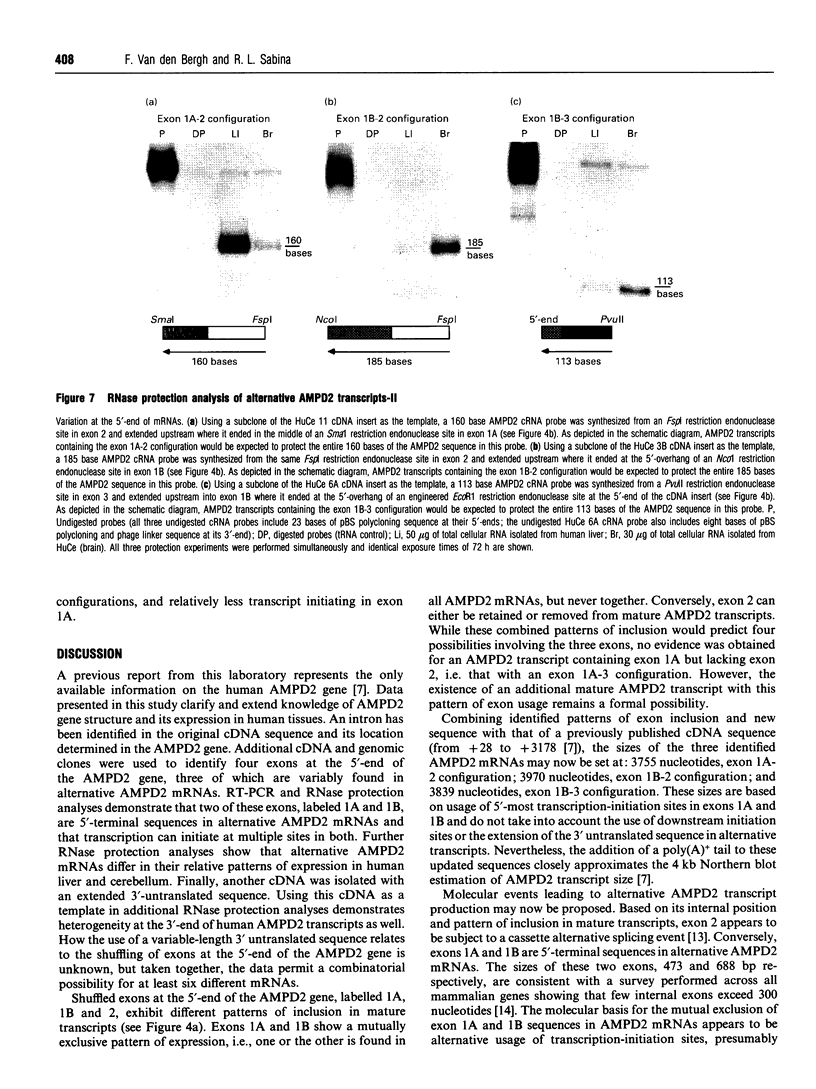
AMP deaminase (AMPD) is a highly regulated enzymic activity and multiple isoforms of this enzyme are coded for by a multigene family in mammalian species, including man. Isoform L (liver) is the main activity present in adult human liver and is the protein product of the AMPD2 gene, which is widely expressed in non-muscle tissues and cells. A previous report described almost the full-length cDNA sequence and part of the human AMPD2 gene and also presented Northern blot evidence for multiple transcripts in brain. This study was performed to further characterize the AMPD2 gene and its expression in human tissues. AMPD2 genomic and human cerebellum cDNA clones were isolated, sequenced and used as probes in RNase protection analyses which together demonstrated the following: (1) an intervening sequence near the 5'-end of the published AMPD2 cDNA, which affects the predicted N-terminal amino acid sequence of isoform L; (2) alternative transcripts resulting from exon shuffling at, or near, the 5'-end of the AMPD2 gene that exhibit tissue-specific patterns of relative abundance; (3) predicted usage of three different initiation codons to confer variable N-terminal extensions on isoform L polypeptides; and (4) an extension of a 3' untranslated sequence in some AMPD2 transcripts. In addition, reverse transcriptase PCR and additional RNase protection analyses were used to map the 5'-ends of two mutually-exclusive exon 1 sequences, both of which contain multiple transcription-initiation sites. These results are discussed in relation to predicted isoform L diversity across human tissues and cells.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bausch-Jurken M. T., Mahnke-Zizelman D. K., Morisaki T., Sabina R. L. Molecular cloning of AMP deaminase isoform L. Sequence and bacterial expression of human AMPD2 cDNA. J Biol Chem. 1992 Nov 5;267(31):22407–22413. [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Andreadis A., Nadal-Ginard B. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Hawkins J. D. A survey on intron and exon lengths. Nucleic Acids Res. 1988 Nov 11;16(21):9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol. 1987;155:397–415. doi: 10.1016/0076-6879(87)55027-3. [DOI] [PubMed] [Google Scholar]
- Mahnke-Zizelman D. K., Sabina R. L. Cloning of human AMP deaminase isoform E cDNAs. Evidence for a third AMPD gene exhibiting alternatively spliced 5'-exons. J Biol Chem. 1992 Oct 15;267(29):20866–20877. [PubMed] [Google Scholar]
- Morisaki H., Morisaki T., Newby L. K., Holmes E. W. Alternative splicing: a mechanism for phenotypic rescue of a common inherited defect. J Clin Invest. 1993 May;91(5):2275–2280. doi: 10.1172/JCI116455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara N., Goto H., Watanabe T. Isozymes of rat AMP deaminase. Biochim Biophys Acta. 1975 Oct 22;403(2):530–537. doi: 10.1016/0005-2744(75)90081-9. [DOI] [PubMed] [Google Scholar]
- Ogasawara N., Goto H., Watanabe T. Isozymes of rat brain AMP deaminase: developmental changes and characterizations of five forms. FEBS Lett. 1975 Oct 15;58(1):245–248. doi: 10.1016/0014-5793(75)80270-5. [DOI] [PubMed] [Google Scholar]
- Ogasawara N., Goto H., Yamada Y., Watanabe T., Asano T. AMP deaminase isozymes in human tissues. Biochim Biophys Acta. 1982 Feb 2;714(2):298–306. doi: 10.1016/0304-4165(82)90337-3. [DOI] [PubMed] [Google Scholar]
- Ogasawara N., Goto H., Yamada Y., Watanabe T. Distribution of AMP deaminase isozymes in various human blood cells. Int J Biochem. 1984;16(3):269–273. doi: 10.1016/0020-711x(84)90099-5. [DOI] [PubMed] [Google Scholar]
- Ogasawara N., Goto H., Yamada Y., Watanabe T. Distribution of AMP-deaminase isozymes in rat tissues. Eur J Biochem. 1978 Jun 15;87(2):297–304. doi: 10.1111/j.1432-1033.1978.tb12378.x. [DOI] [PubMed] [Google Scholar]
- Sabina R. L., Morisaki T., Clarke P., Eddy R., Shows T. B., Morton C. C., Holmes E. W. Characterization of the human and rat myoadenylate deaminase genes. J Biol Chem. 1990 Jun 5;265(16):9423–9433. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Chen J. K., Feng S., Dalgarno D. C., Brauer A. W., Schreiber S. L. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994 Mar 11;76(5):933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]