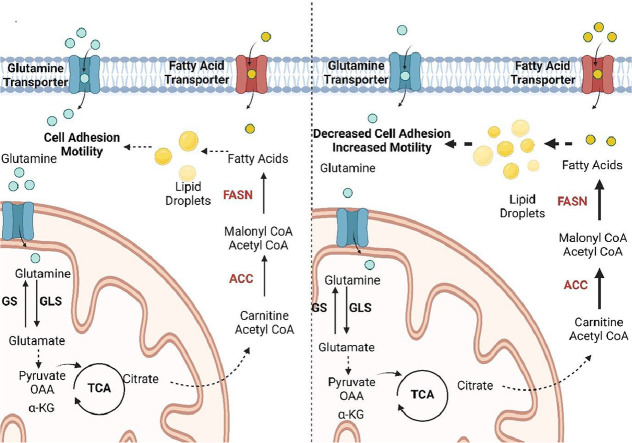
Highlights
-
•
Glutamine is differentially metabolized in primary versus metastatic tumors.
-
•
A three gene glutamine metabolism signature could predict prognosis.
-
•
Metastatic SW620 cells increased lipid uptake upon glutamine starvation.
-
•
Glutamine starvation led to increased motility in SW620 cells.
Keywords: L-Glutamine, Colorectal cancer, Lipid metabolism, Metabolic plasticity
Abstract
Introduction
Glutamine is a non-essential amino acid that is critical for cell growth. However, the differential metabolism of l-glutamine in metastatic versus primary colorectal cancer (CRC) has not been evaluated adequately.
Materials and methods
Differential expression of glutamine-related genes was determined in primary versus metastatic CRC. Univariate Cox regression and hierarchical clustering were used to generate a gene signature for prognostication. Untargeted metabolomics and 18O based fluxomics were used to identify differential metabolite levels and energy turnover in the paired primary (SW480) and metastatic (SW620) CRC cells. Western blot and qRT-PCR were used to validate differential gene expression. Subcellular localization of E-cadherin was determined by immunocytochemistry. Lipid droplets were visualized with Nile Red.
Results
The GO term “Glutamine metabolism” was significantly enriched in metastatic versus primary tumors. Supporting this, SW620 cells showed decreased membrane localization of E-cadherin and increased motility upon l-Glutamine withdrawal. A glutamine related signature associated with worse prognosis was identified and validated in multiple datasets. A fluxomics assay revealed a slower TCA cycle in SW480 and SW620 cells upon l-Glutamine withdrawal. SW620 cells, however, could maintain high ATP levels. Untargeted metabolomics indicated the preferential metabolism of fatty acids in SW620 but not SW480 cells. Lipids were mainly obtained from the environment rather than by de novo synthesis.
Conclusions
Metastatic CRC cells can display aberrant glutamine metabolism. We show for the first time that upon l-glutamine withdrawal, SW620 (but not SW480) cells were metabolically plastic and could metabolize lipids for survival and cellular motility.
Graphical abstract
Introduction
The metabolic state of transformed epithelial cells at the primary site, in the circulation and at the pre-metastatic niche are all known to be different [1]. Primary tumors in general show increased glycolytic flux and lactic acid release (Warburg effect), decreased oxidative phosphorylation (OXPHOS), and increased activation of the pentose phosphate pathway; the latter two events can also decrease reactive oxygen species production and increase cell survival. Epithelial to mesenchymal transition (EMT) is a series of genetic, epigenetic and metabolic changes accompanying the conversion of an epithelial phenotype of the primary tumor to a motile, less proliferating mesenchymal phenotype that can metastasize and develop chemoresistance [2]. Thus, the metabolic activity of the primary tumor and its metastatic counterpart is expected to be distinct. Additionally, for a metastatic cell to survive at the distant site, it needs to have enough metabolic plasticity to acclimatize to the metabolic idiosyncrasies of the tissue they home to [3].
A common feature of proliferative cancer cells is their ability to take up l-glutamine to support cell growth [4]. Glutamine catabolism maintains a steady supply of metabolites into the TCA cycle (anaplerosis), thus preserving TCA cycle integrity despite a continual efflux of metabolites to support growth [5]. l-glutamine is primarily utilized via its conversion to glutamate [via glutaminease1 (GLS1), rate limiting step] and then to α-ketoglutarate [via glutamate pyruvate transaminase 2 (GPT2)], which can then enter the TCA cycle and provide energy [6]. Such alterations can also affect signal transduction pathways by modulating the availability of metabolites necessary for post-translational modifications of key proteins [7].
Lipid metabolism entails lipid uptake from the environment, de novo synthesis, and catabolism. Deregulated lipid metabolism can affect many cellular characteristics such as plasma membrane composition, gene expression and signal transduction, in addition to energy generation via β‐oxidation [8]. The upregulation of lipogenic enzymes has been described in various cancer types and can contribute to enhanced proliferation by providing the biomass necessary for the formation of cell membrane and oncogenic signaling molecules [9].
We aimed to examine the differences in glutamine utilization at the transcriptome and metabolome level in primary and metastatic colorectal cancer (CRC). We have compared differences in the expression profile of glutamine metabolism related genes in publicly available paired and unpaired primary and metastatic CRC transcriptome data. We observed a stronger enrichment of glutamine metabolism related gene sets in metastatic tumors and suggest a 3 gene glutamine signature that can significantly predict worse overall survival (OS) and relapse free survival (RFS). Next, we used SW480 and SW620 cells that were derived from the same patient, but are representative of a primary tumor (SW480) and its metastatic counterpart (SW620) and also known to have distinct metabolic profiles [10] to identify differences in the metabolome when l-glutamine was withdrawn from the culture medium. We observed a remarkable increase in free fatty acids and cholesterol levels in SW620 cells compared to SW480 cells when l-glutamine was withdrawn, which were obtained from the environment rather than through de novo synthesis. We also observed by 18O-based fluxomics that the TCA cycle metabolites had a slower turnover rate in both cell lines under l-glutamine withdrawal. Additionally, SW620 cells showed decreased E-cadherin at the plasma membrane under l-glutamine withdrawal while 3D cultures generated in the presence of the GLS1 inhibitor CB-839 showed increased lobular budding compared to SW480 cells. Additionally, l-glutamine starved SW620 cells were more motile compared to SW480 cells. Overall, our data suggest that metastatic CRC cells show greater enrichment of glutamine metabolism compared to primary cells. However, in the absence of glutamine, metastatic cells can rewire their metabolism to rely on lipid metabolism, thereby continuing to survive and migrate.
Materials and methods
Cell culture SW480, SW620, HCT-116 and Caco-2 cells were cultured as described in Supplementary Materials and Methods. For l-Glutamine starvation, the cells were incubated for 24 h in l-Glutamine free [(-) LG] medium supplemented with 2 mM l-Glutamine where indicated [(+) LG].
Treatments (+) or (-) LG cells were treated with pre-optimized concentrations of 6-Aminonicotinamide (6-AN), AG-120, Etomoxir (Eto). More details are provided in Supplementary Materials and Methods.
RNA isolation RNA was isolated from 70 to 80 % confluent cells incubated in (+) or (-) LG medium and/or with the drugs using the RNA isolation kit (Macherey Nagel). Details of the primer sequences and PCR protocols are provided in Supplementary Materials and Methods.
Protein isolation and western blotting Proteins were isolated from 70 to 80 % confluent cells incubated in (+) or (-) LG medium and/or with the drugs using the MPER protein isolation kit (Promega), supplemented with protease and phosphatase inhibitors. Western blotting was carried out with 15–20 µg of protein using standard techniques. Further details and a list of antibodies are provided in Supplementary Materials and Methods.
Immunofluorescence staining 500,000 cells were counted and seeded onto glass bottom confocal dishes (SPL). After 24 h, the cells were incubated in the (+) or (-) LG medium for 24 h, fixed in 4 % paraformaldehyde (PFA) and processed for incubation in primary antibodies, followed by an Alexa Fluor conjugated secondary antibody. Where indicated, the fixed cells were also incubated with Alexa Fluor® 405 Phalloidin to detect F-Actin. For lipid droplet staining, 4 % PFA fixed cells were stained with the Nile Red. Counterstaining was carried out with DAPI. Further details are provided in Supplementary Materials and Methods.
Colony formation assay
500 cells were seeded in 12-well plates in complete medium and left overnight for attachment. The following day, the cells were washed with PBS and then incubated for 24 h in either (+) or (-) LG medium with the indicated treatments. The assay was carried out as described in Supplementary Materials and Methods.
Spheroid formation assay
SW480 and SW620 cells were seeded in 50,000 cells/well density to 96 well ultra-low attachment (ULA) plates (Corning) in a stem cell medium containing CB-839 (1 μM, Cayman Chemical) or vehicle for 10 days. Each individual spheroid formed in the 96 well ULA plate was imaged and evaluated for their shape and formation of lobular detachments. Further details are provided in Supplementary Materials and Methods.
Scratch wound healing assay
SW480 and SW620 cells were seeded as 1,500,000 cells/well to 6 well plates and left overnight for attachment. The following day, the cells were washed and a wound was generated using a sterile pipette tip. The cells were incubated in (+) and (-) LG media containing 0.5 μM mitomycin and the wounds were imaged at 0 h, 24 h and 48 h under an inverted light microscope.
Bioinformatics data analyses
Differential gene expression analysis of unpaired primary treatment naïve (n = 683) versus metastatic (n = 257) and 7 untreated matched primary versus metastatic lesion pairs (GSE131418) was conducted [11]. Log fold changes (LFC) were calculated for each pair for a total of 56 genes (112 probe sets) related to glutamine metabolism (Supplementary Table S1). Gene set enrichment analysis (GSEA) was conducted to assess molecular differences via GSEA 4.0.3 software using the Molecular Signature Database (MSigDB) [12]. 67 gene sets from the C5 gene ontology collection of MSigDB that included the keywords "glutamine," "glutamate," and "glutathione" were used for further analysis. Hierarchical clustering and heatmap visualization were carried out using Cluster 3.0 and Treeview software [13,14]. From GSE39582, 22 probesets (17 genes) with significant Cox p-values (<0.05) were selected to assess prognosis. The detailed protocols are provided in Supplementary Materials and Methods.
Metabolomics and fluxomic analyses
Metabolites from (+) and (-) LG medium treated SW480 and SW620 cells were extracted as described in Supplementary Methods. For fluxomics, the cells were labeled with 30 % 18O stable isotope (H2[18O]) for 5 min and the samples were processed for GC–MS (Q2010 Shimadzu) analysis [15]. Further details are provided in Supplementary Materials and Methods.
Statistical analyses
All experiments were carried out with at least 2–3 biological replicates with 3–4 technical replicates unless otherwise stated. Data analyses and graphing were carried out with Graphpad (v.9). The data were analyzed with t-test; p < 0.05 was considered to be significant. Individual statistical analyses for the in silico and metabolomics data are provided in the figure legends or in Supplementary Materials and Methods.
Results
Enrichment of glutamine metabolism in paired metastatic versus primary CRC
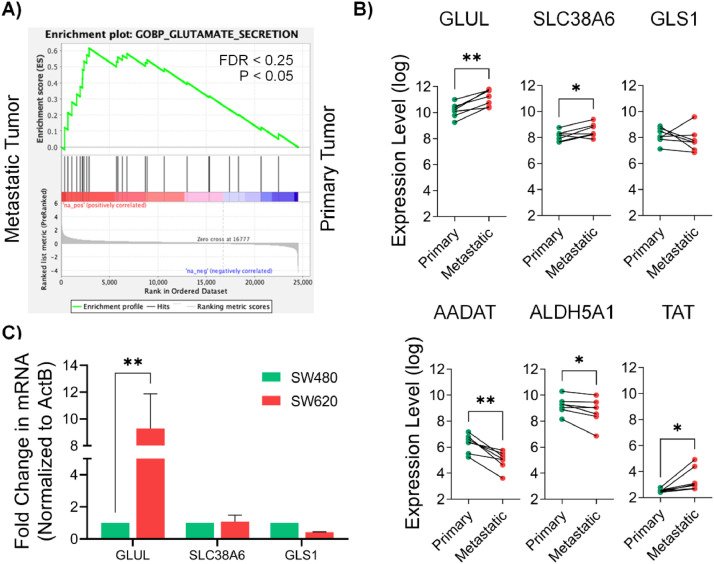
To analyze potential differences in glutamine metabolism between primary (P) and metastatic (M) CRC, we used transcriptome data from 7 treatment naïve P tumors and paired M lesions from the Moffitt Cancer Center (MCC) Cohort [11]. A GSEA indicated significant enrichment (p < 0.05, FDR<0.25) in the gene ontology (GO) term “Glutamate Secretion” in the M tumors compared to the matched P tumors (Fig. 1A). Next, we generated a glutamine metabolism related gene list (n = 56, Supplementary Table S1) in order to interrogate their differences in expression. We observed significant differences in the expression of 5 genes in the matched M versus P tumors (p < 0.05, Fig. 1B). The expression of glutamate-ammonia ligase (GLUL), SLC38A6 and TAT (tyrosine aminotransferase) were significantly increased while the expression of aminoadipate aminotransferase (AADAT) and succinic semialdehyde (ALDH5A1) were significantly decreased in the M tumors compared to the P tumors. The expression of GLS1 showed a trend for decrease, but did not reach statistical significance. To validate these differences in expression, we used the paired P and M CRC cell lines SW480 and SW620. We observed a significant increase in the expression of the key enzyme GLUL in SW620 cells (Fig. 1C); however, the difference in the expression of the other genes evaluated did not reach statistical significance. Taken together, we observed differential regulation of glutamine metabolism in the metastatic versus primary tumor samples.
Fig. 1.
Enrichment of glutamine metabolism in paired metastatic versus primary tumor samples. (A) GSEA of 7 drug naïve primary and paired metastatic lesions showing significant enrichment of the GO term Glutamate Secretion. (B) Expression of 6 genes related to glutamine metabolism in matched primary vs metastatic tumors (n = 7). (C) Expression of 3 of the genes measured by qRT-PCR using SW480 and SW620. ACTB served as the internal control. Data are represented as mean ± SEM (*p < 0.05, **p < 0.01, ***p < 0.001, paired t-test). For abbreviations, please see the text.
Enrichment of glutamine metabolism in non-paired primary versus metastatic CRC
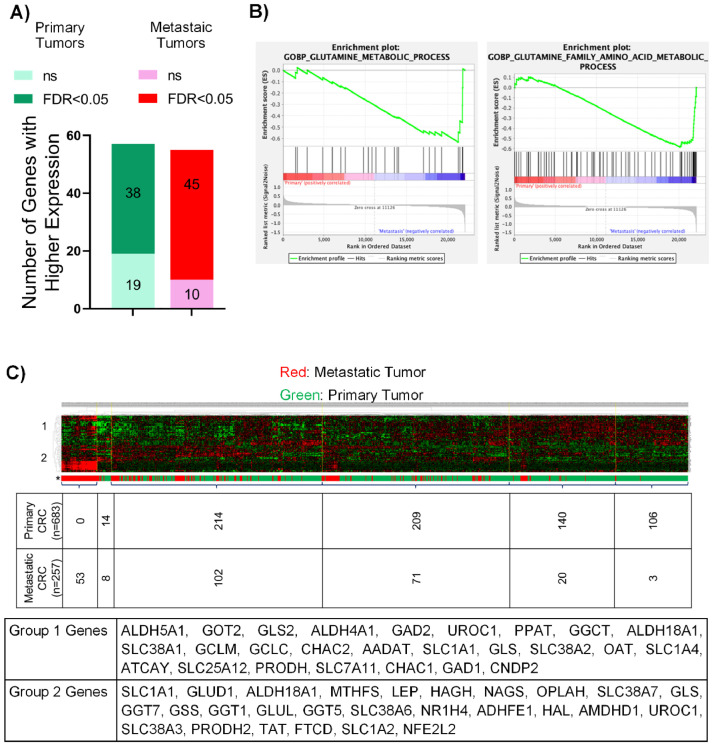
To further analyze the molecular differences in glutamine metabolism, the expression of genes related to glutamine metabolism (Supplementary Table S1) was compared between a larger set of non-paired P (n = 683) vs M CRC tumors (n = 257) [11]. We observed that 83 of the 112 probe sets examined showed significant difference; of these, 45 and 38 probe sets were highly expressed in M and P tumors, respectively (Fig. 2A). Genes with a log fold change (LFC)>1 that showed significantly higher expression in the M tumors are listed in Supplementary Table S2. Use of the same GSEA analysis described above with the non-paired tumors revealed no enrichment in P lesions while the terms “Glutamine Metabolic Process” and “Glutamine Family Amino Acid Metabolic Process” were significantly enriched (p < 0.05, FDR<0.25) in M CRC (Fig. 2B). A hierarchical clustering based on the expression of glutamine-related genes (Supplementary Table S1) indicated that non-paired P and M tumors showed distinct expression patterns. Most of the M tumors were clustered on the left side of the plot, while P tumors were clustered more homogeneously on the right (Fig. 2C, Chi-square p value<0.001). We could separate the genes that showed primarily higher expression in P tumors (group 1) and those with high expression in M tumors (group 2). Of note, the non-paired P and M tumors from the larger non-paired cohort also showed differential expression of the same genes (high expression of TAT, GLUL, SLC38A6 and low expression of AADAT and ALDH5A1) as the paired P and M tumors.
Fig. 2.
Enrichment of glutamine metabolism in non-paired primary versus metastatic CRC (A) The number of glutamine metabolism related genes that show high expression in P and M tumors. Differences between primary and metastasis samples was analyzed by t-test, Benjamini-Hochberg method was applied for correction. (B) GSEA of non-paired primary vs metastatic tumors using the key words “Glutamine”, “Glutamate” and “Glutathione”. In this analysis, all available M tumors regardless of the treatment status were used (C) Hierarchical clustering showing distinct separation of non-paired primary vs metastatic CRC, Chi-square p value<0.001. Due to the existence of multiple probe sets for a specific gene, some gene names exist in both Group 1 and Group 2 genes. Numbers given below heat maps indicated the number of tumor samples.
Three gene signature reveals poor clinical outcome with elevated expression
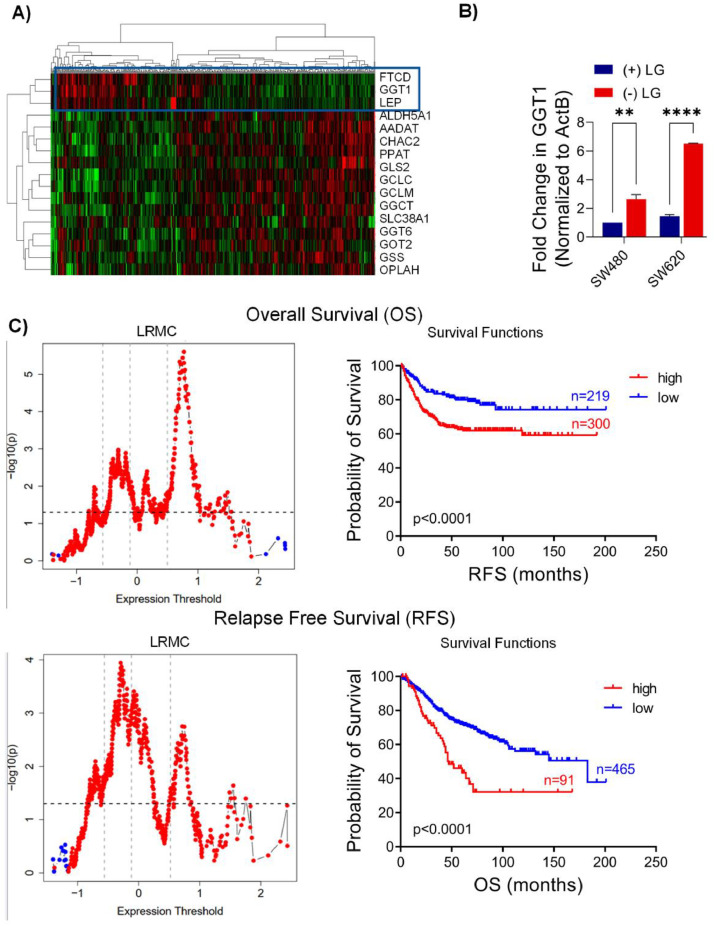
To determine whether any of the glutamine related genes are of prognostic significance, we carried out a univariate Cox regression analysis for 124 probe sets annotating the 56 glutamine metabolism related genes in GSE39582, which includes 556 primary tumors with non-zero overall survival data [16]. Among these, 22 probe sets (17 genes) had Cox p < 0.05 and were used for further analyses. Hierarchical clustering analysis revealed three genes (FTCD, GGT and LEP) that clustered separately and generated a clear sub-group among the tumors (Fig. 3A). To validate the expression of these genes in P and M, we used the SW480 and SW620 CRC cell lines as models for P and M tumors, respectively. We grew these cells both in the presence and absence of l-glutamine in the culture medium. The expression of GGT was higher in SW620 cells compared to SW480 cells and increased further in SW620 cells when glutamine was withdrawn (Fig. 3B). The expression of FTCD and LEP were below the level of detection by RT-qPCR in both cell lines.
Fig. 3.
Three Gene Glutamine Signature Reveals Poor Clinical Outcomes. (A) Hierarchical clustering of CRC tumors based on expression of 17 genes linearly associated with OS. (B) Confirmation of the differential expression of GGT1 with qRT-PCR in SW480 and SW620 cells grown +/- l-glutamine medium. ACTB served as the internal control. Data are represented as mean ± SEM (**p < 0.01, ***p < 0.001). (C) Log rank tests at all possible cut-off values and Kaplan–Meier graphs showed that high mean Z score of FTCD, GGT1 and LEP was associated significantly with worse OS and RFS in GSE39582.
The mean Z score of these three genes was strongly associated with poor overall survival (OS) and relapse free survival (RFS) as shown by log rank tests carried out at all possible cut-offs using GSE39582 (Fig. 3C). Multivariate analyses with parameters that were shown to be prognostic in univariate analyses in GSE39582 previously (mismatch repair status, KRAS or BRAF mutation, TNM stage [17]) showed that the mean Z score was independent of clinical confounding factors when OS was used as the clinical outcome, but not when RFS was used (Table 1). An independent analysis of TCGA COAD cohort (n = 425) could clearly validate a worse OS in patients with a high mean Z score (Supplementary Figure S1).
Table 1.
Multivariate analysis of the mean Z score of the three genes (FTCD, GGT and LEP).
| Overall Survival (OS) | ||||
|---|---|---|---|---|
| 95 % CI |
||||
| p | HR | Lower | Upper | |
| Mismatch repair status (proficient, deficient) | 0.377 | 1.262 | 0.753 | 2.114 |
| TNM stagea | <0.001 | 1.8 | 1.451 | 2.232 |
| KRAS or BRAF mutationb | 0.093 | 1.315 | 0.955 | 1.811 |
| Three gene mean Z score | <0.001 | 1.47 | 1.226 | 1.762 |
| Relapse Free Survival (RFS) | ||||
|---|---|---|---|---|
| 95 % CI |
||||
| p | HR | Lower | Upper | |
| Mismatch repair status (proficient, deficient) | 0.01 | 2.461 | 1.239 | 4.886 |
| TNM stagea | <0.001 | 2.314 | 1.84 | 2.911 |
| KRAS or BRAF mutationb | 0.122 | 1.3 | 0.933 | 1.812 |
| Three gene mean Z score | 0.148 | 1.287 | 0.914 | 1.812 |
Treated as a continuous variable (0, 1, 2, 3, 4).
KRAS or BRAF mutation: Samples are considered as wild type if only both genes are wild type, other situations are considered mutated.
Alterations in the metabolome of SW620 and SW480 cells in response to l-Glutamine starvation
We next carried out an untargeted metabolomics analysis to evaluate the metabolome of SW480 and SW620 cells when l-Glutamine was withdrawn. A substantial difference in the metabolite profiles between the two cell lines in +/- l-glutamine medium was observed as confirmed by PLS-DA (Supplementary Figure S2). l-Glutamine starved SW620 cells had significantly higher concentrations of free fatty acids and cholesterol, compared to l-Glutamine starved SW480 cells (Fig. 4A, the full list of metabolites (n = 100) is shown in Supplementary Table S5). Of note, the levels of these free fatty acids were similar in the two cell lines grown in complete medium (Fig. 4A). A GSEA was carried out with the full list of GO terms (Biological Process) in GSE131418. The top 25 enriched gene sets for P and M tumors showed a remarkable number of GO terms related to lipid metabolism in the M tumors, while the P tumors showed enrichment in GO terms related to proliferation (Supplementary Tables S3 and S4, lipid metabolism related GO terms are annotated in red). These data suggest that M tumors may preferentially carry out lipid metabolism to accommodate either biomass generation or increased energy demand for dissemination [9].
Fig. 4.
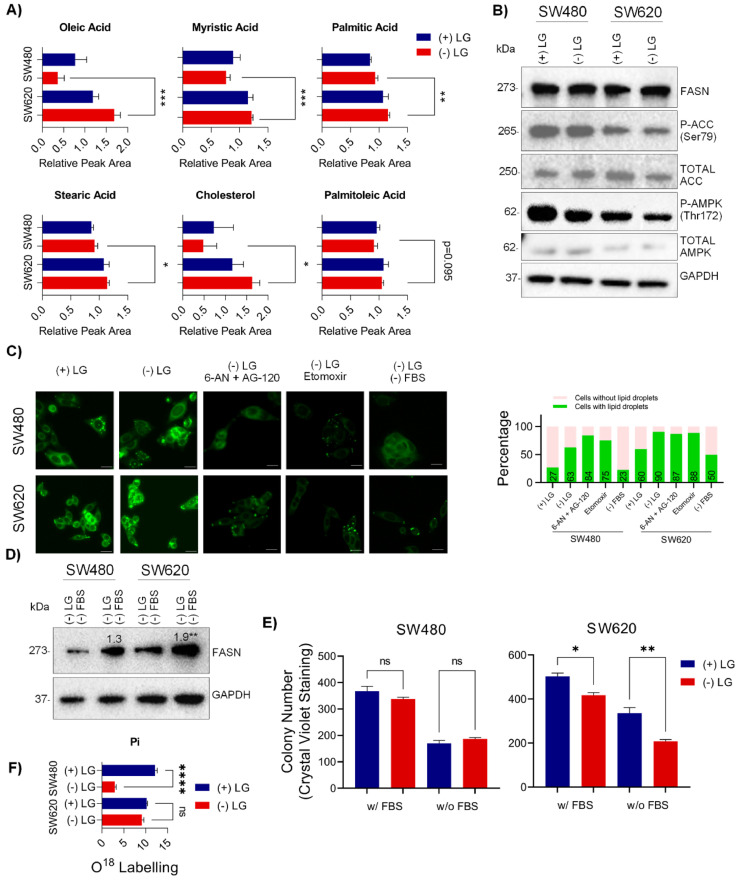
Elevation of Fatty Acid Synthesis and/or Uptake in Response to l-Glutamine Starvation in SW480 and SW620 cells. (A) Untargeted metabolomics analysis of SW480 and SW620 cells showing fatty acid levels (B) Western blot assay showing the activation of lipid metabolism related proteins. GAPDH served as the loading control. (C) Lipid droplet staining in SW480 and SW620 cells with various treatments (left panel). Scale bar 100 µm. Percentages of cells containing lipid droplets (n = 200) is shown in the right panel. (D) Western blot of SW480 and SW620 cells +/- serum. The densitometric analysis of 2 independent replicates is shown in the figure. (E) Colony formation assay with serum withdrawal +/- l-glutamine. (F) Fluxomic assay with 18O showing the turnover of inorganic phosphate (Pi). Data are represented as mean ± SEM (ns: non-significant, **p < 0.01). (+) and (-) LG indicates the culture medium with and without l-glutamine, respectively.
L-Glutamine starved SW620 cells showed hypophosphorylation of the key lipid metabolism kinase Acetyl CoA Carboxylase (ACC) at S79 and its upstream kinase AMPK (T172) along with a high but steady expression of Fatty acid synthase (FASN) (Fig. 4B). These results suggest enhanced fatty acid synthesis or uptake in response to l-Glutamine starvation in SW620 cells compared to SW480 cells. We therefore evaluated whether the overall lipid content of SW480 and SW620 cells was altered using Nile Red. We observed a higher proportion of lipid droplets in SW620 cells compared to SW480 cells grown in complete medium (Fig. 4C), suggesting that SW620 cells relied more on lipid metabolism. Upon l-glutamine starvation almost all of SW620 cells (90 %) remained lipid droplets positive compared to only 60 % in SW480 cells (Fig. 4C). We also observed comparable levels of lipids in HCT-116 and Caco-2 cells grown in complete medium. In the absence of l-glutamine, the lipid levels remained unchanged in Caco-2 cells, while HCT-116 cells showed a modest increase (Supplementary Figure S3). HCT-116 cells are known to be aggressive and develop hepatic metastases efficiently in immunodeficient mice. In contrast, Caco-2 cells are derived from a relatively differentiated tumor and do not metastasize easily [18].
Inhibition of fatty acid oxidation with Etomoxir or blockage of NAPDH generation (essential for fatty acid synthesis) using a combination of the pentose phosphate pathway inhibitor 6-AN and the isocitrate dehydrogenase inhibitor AG-120 could not ameliorate the enhanced lipid droplet formation in SW480 or SW620 cells (Fig. 4C), suggesting that the cells were capable of obtaining lipids from the environment. The primary source of lipids for these cells would therefore be the serum. Indeed, the withdrawal of serum could decrease lipid droplet formation of SW480 cells; however, the amount of lipid droplets remained relatively higher in SW620. Interestingly, serum withdrawal in SW620 cells was accompanied by an increase in FASN levels (Fig. 4D). These results indicate that both SW620 and SW480 can increase the fatty acid uptake from the environment to survive upon l-Glutamine starvation. When the external source of lipids was withdrawn (through serum starvation), SW620 cells (unlike SW480 cells) were capable of activating a compensatory lipid synthesis, which is reflected by increased lipid droplet accumulation. A colony formation assay exhibited that serum withdrawal led to fewer colonies being formed in both SW480 cells and SW620 cells. However, serum withdrawal plus l-glutamine starvation led to the formation of fewer colonies in SW620 cells compared to SW480 cells (Fig. 4E).
SW480 and SW620 cells are grown in Leibovitz l-15 medium which uses galactose (rather than glucose) as the primary source of carbons. The use of galactose entails expenditure of 1 extra ATP molecule requiring a greater reliance on TCA cycle and OXPHOS [19]. Withdrawal of l-glutamine would abrogate the availability of α-ketoglutarate, a major anaplerotic molecule that feeds into the TCA cycle and would lead to a loss of energy generation via OXPHOS. To substantiate this, we carried out a fluxomic analysis utilizing 18O to evaluate the turnover of the TCA cycle metabolites. As expected, we observed a decrease in the turnover of several of the TCA cycle intermediates in both SW480 and SW620 cells upon l-glutamine withdrawal (Supplementary Figure S4, Supplementary Table S6). The turnover of inorganic phosphate (Pi) was also significantly lower in SW480 cells grown in the absence of l-glutamine, very likely due to an inefficient OXPHOS. However, the turnover of Pi did not show any significant change in the SW620 cells grown in the absence or presence of l-glutamine (Fig. 4F). Cytosolic Pi levels reflect the demand for ATP synthesis by mitochondrial ATP synthase [20]. Thus, SW620 cells are very likely metabolically more plastic and may be able to generate ATP from galactose (Leloir pathway) available in the culture medium.
Deprivation of l-Glutamine induces distinct functional outcomes in SW480 and SW620
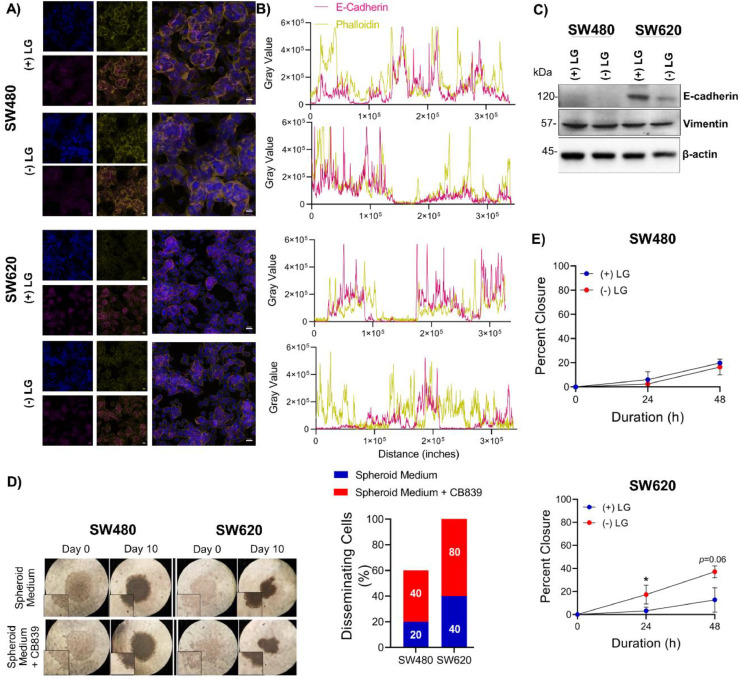
We next queried whether SW480 vs SW620 showed any changes in cell-adhesion markers when grown in +/- l-glutamine medium. Indeed, we observed that l-Glutamine starvation led to a loss of association of E-Cadherin with the membrane (colocalization with phalloidin) only in SW620 cells (Fig. 5A & B) along with an overall decrease in E-Cadherin expression in SW620 cells grown in the absence of l-glutamine (Fig. 5C). We did not observe any difference in the expression of the mesenchymal marker Vimentin in either SW480 or SW620 cells (Fig. 5C).
Fig. 5.
Deprivation of l-Glutamine induces distinct functional outcomes in SW480 and SW620. (A) Representative images of IF staining for E-cadherin (magenta) and Phalloidin (yellow). Scale bar corresponds to 20 µm. (B) Colocalization analysis of Phalloidin and E-Cadherin. (C) Western blot analysis of SW480 and SW620 cells treated with (+) and (-) LG medium. β-actin served as the loading control. (D) Representative light microscopy images of two independent replicates of spheroid formation assays. (E) Wound healing assay of SW480 and SW620 cells treated with (+) and (-) LG medium for 24–48 h (t-test).
We next carried out a spheroid formation assay to determine the effects of l-Glutamine withdrawal on cell-cell adhesion. Since the specific composition of the spheroid medium precluded the withdrawal of l-glutamine, we incubated the spheroids with the glutaminase inhibitor CB-839 (Telaglenastat), which inhibits the conversion of glutamine to glutamate [21]. We observed a higher proportion of lobular budding from CB-839 treated SW620 cell spheroids compared to the SW480 counterparts (Fig. 5D) suggesting that l-glutamine withdrawal led to a loss of cell-cell adhesion in SW620 cells.
To further substantiate our findings, we carried out a scratch wound healing assay of SW620 and SW480 cells grown in the absence or presence of l-glutamine medium. We observed no significant change in the motility of SW480 cells in the presence or absence of l-glutamine whereas SW620 cells showed increased motility when grown in the absence of l-glutamine at all time points (Fig. 5E, Supplementary Figure S5).
Discussion
During the process of dissemination, cancer cells frequently undergo metabolic rewiring in order to survive the nutrient and energy deficit. Using an in silico approach, we first aimed to understand whether glutamine metabolism is important for the metastasis of CRC. For this, we curated a list of 56 genes related to glutamine metabolism and observed their differential expression in both paired and non-paired M and P CRC tumors. Among the significantly upregulated genes were critical enzymes such as GLUL, which catalyzes the conversion of glutamate and ammonia to glutamine; SLC38A6, which transports l-glutamine; and TAT, which uses glutamate as a nitrogen donor for tyrosine synthesis [22]. We observed a decrease in the expression of AADAT, which catalyzes a reversible transamination reaction between l-2-aminoadipate and 2-oxoglutarate to produce 2-oxoadipate and l-glutamate [23], as well as ALDH5A1, which converts the neurotransmitter γ-Aminobutyric acid (GABA, generated from the decarboxylation of glutamate) into succinate, which can then enter the TCA cycle [24].
Univariate Cox regression analysis showed that glutamine metabolism was related to patient prognosis in CRC and a higher score generated on the basis of three genes was associated with worse prognosis across multiple datasets. These three genes included FTCD (Formimidoyltransferase cyclodeaminase) which carries out the enzymatic transamination of several amino acids into glutamate, GGT (Gamma-glutamyltransferase-1) which catalyzes the cleavage of glutathione into glutamate and LEP (Leptin) which regulates glutamine transport [[25], [26], [27]]. Of note, all three genes were classified under Group 2 (Fig. 2C); genes that were highly enriched in the M tumors.
Several studies have reported that serum levels of glutamine are lower in CRC compared to healthy controls [28,29]. Moreover, low serum glutamine levels could act as an independent prognostic biomarker for poor prognosis (disease free survival, DFS) in CRC [30]. We therefore aimed to understand whether glutamine withdrawal could affect metabolism and energy utilization in SW480 and SW620 cells. We observed that upon withdrawal of l-glutamine from the culture medium for 24 h, the metastatic SW620 cells retained the turnover of inorganic phosphate (Pi) while the primary SW480 cells showed a decrease, suggesting that SW620 cells could maintain ATP synthesis, unlike SW480 cells.
An untargeted metabolomics study indicated high levels of fatty acids in SW620 cells (but not SW480 cells) grown under glutamine withdrawal, suggesting higher metabolic plasticity in the M cells when glutamine was limiting. Supporting this, a very recent publication has reported that under l-glutamine starvation, the ATF4 target TRIM2 could increase the activity of CPT1A, the rate limiting enzyme of FAO, to enable cell survival [31]. A comparison of GO gene sets between non-paired primary and metastatic tumors also supported the preferential enrichment of lipid metabolism in M tumors. As a source of high energy, lipid metabolism has been shown to promote cancer survival and metastasis by enhancing antioxidative and antiapoptotic mechanisms, therapy resistance, and inhibition of immune surveillance [32]. Both increased uptake and synthesis of lipids has been associated with the metastatic program [9]. Moreover, high expression of FASN could enhance peritoneal metastasis of ovarian cancer partly through the induction of epithelial mesenchymal transition (EMT) [33]. EMT, which is commonly characterized by the loss of E-Cadherin expression, is known to be crucial in tumorigenesis through enhancing metastasis, chemoresistance and tumor stemness [34]. We observed a remarkable decrease in the overall protein level of E-cadherin along with a decrease in its association with the plasma membrane in SW620 (but not SW480) cells grown under l-glutamine starvation. When grown in 3D as spheroids, SW620 cells treated with the GLS1 inhibitor CB-839 showed enhanced lobular budding, most likely stemming from loose of cell-to-cell adhesion due to the loss of E-cadherin [35]. A similar decrease in E-cadherin has been also been reported in SW480 and DLD1 cells grown in low glutamine suggesting that even a partial decrease in glutamine availability can affect epithelial markers in CRC cells [30]. The low cell-cell adhesion was also supported by an increase in the motility of SW620 (but not SW480) cells grown in the absence of l-glutamine.
The lack of any direct in vivo evidence of increased lipid metabolism in metastatic cancer cells grown under l-glutamine restriction can be considered as a limitation of the current study. Since l-glutamine is a non-essential amino acid, it can be synthesized in the body and therefore its removal from the diet may not mimic the glutamine-free environment that we were able to generate in vitro. Nonetheless, a low glutamate environment could have been created with the use of drugs such as CB-839 (Telaglenastat), DON or BPTES that inhibit the conversion of l-glutamine to glutamate [36]. However, it is worth mentioning that CB-839 may also have pleiotropic effects in cells such as the inhibition of autophagy. DON is known to be highly toxic while BPTES has poor bioavailability [36]. Another limitation is that we were unable to validate the expression of the signature genes in multiple CRC cell lines as their expression was very low in most CRC cell lines [Human Protein Atlas (https://www.proteinatlas.org/)]. We were also unable to identify the specific transporter that may aid in the uptake of fatty acids, which is why we were unable to treat the cells with a specific drug to inhibit the uptake.
Mortality in CRC stems primarily from metastatic disease [37]. Most metastatic CRC (mCRC) is associated with the overexpression of EGFR, which can be targeted by anti-EGFR monoclonal antibodies that are used in combination with fluoropyrimidine-based chemotherapy regimens [37]. Additionally, multiple receptor tyrosine kinase (RTK) inhibitors such as regorafenib can be used to target additional RTKs that may be activated in advanced CRC, although use of this drug has been associated with adverse events and thereby discontinuation [38]. A small subset of advanced CRC patients with microsatellite instability (MSI) or mismatch repair deficiency may benefit from immune checkpoint inhibitors [39,40]. Thus, there is still a clear need for new therapeutic options, especially for patients with advanced CRC. Better understanding of the differences in metabolic profiles of primary versus metastatic tumors may help design new therapies that can specifically target metastatic cells.
Conclusions
Overall, our study showed that glutamine metabolism can differ in primary versus metastatic CRC tumors. We identified a three gene signature that could significantly predict prognosis across multiple datasets. Unlike SW480 cells, SW620 cells showed stronger metabolic plasticity upon glutamine withdrawal by accumulating lipids, primarily from the environment and a loss of epithelial markers and increased motility upon l-glutamine withdrawal. These data suggest that loss of l-glutamine may lead to poor prognosis through a compensatory accumulation of lipids for budding and enhanced cellular motility.
Ethical approval statement
No ethical approval was necessary for this research.
Availability of data and materials
Codes used in this study can be made available upon request.
Funding
The study was funded by the TÜBİTAK project (120Z229) to SB and AEGT. AEGT was supported by BIDEB 2211. ÇE was supported by YÖK 100/2000.
CRediT authorship contribution statement
Aliye Ezgi Güleç Taşkıran: Writing – original draft, Investigation, Formal analysis, Data curation. Diren Arda Karaoğlu: Investigation, Formal analysis, Data curation. Cemil Can Eylem: Writing – original draft, Investigation, Formal analysis, Data curation. Çağdaş Ermiş: Investigation. İsmail Güderer: Data curation. Emirhan Nemutlu: Writing – original draft, Supervision, Formal analysis. Seçil Demirkol Canlı: Writing – original draft, Supervision, Investigation, Formal analysis, Data curation. Sreeparna Banerjee: Writing – original draft, Supervision, Resources, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to acknowledge Prof Dr Tulin Guray, METU, for useful discussions.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2024.102078.
Appendix. Supplementary materials
References
- 1.Dorsch M., et al. Statins affect cancer cell plasticity with distinct consequences for tumor progression and metastasis. Cell Rep. 2021;37(8) doi: 10.1016/J.CELREP.2021.110056. Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schild T., Low V., Blenis J., Gomes A.P. Unique metabolic adaptations dictate distal organ-specific metastatic colonization. Cancer Cell. 2018;33(3):347–354. doi: 10.1016/j.ccell.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Earle W.R., Evans V.J., Hawkins N.M., Peppers E.V., Westfall B.B. Effect of glutamine on the growth and metabolism of liver cells in vitro. J. Natl. Cancer Inst. 1956;17(2):131–138. Aug. [PubMed] [Google Scholar]
- 5.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y., et al. Colorectal cancers utilize glutamine as an anaplerotic substrate of the TCA cycle in vivo. Sci. Rep. 2019;9(1):19180. doi: 10.1038/s41598-019-55718-2. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomes A.P., Blenis J. A nexus for cellular homeostasis: the interplay between metabolic and signal transduction pathways. Curr. Opin. Biotechnol. 2015;34:110–117. doi: 10.1016/j.copbio.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu Y., et al. Lipid metabolism in cancer progression and therapeutic strategies. MedComm. 2020;2(1):27–59. doi: 10.1002/mco2.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.X. Luo et al., Emerging roles of lipid metabolism in cancer metastasis, vol 16, no 76. 2017. doi: 10.1186/s12943-017-0646-3. [DOI] [PMC free article] [PubMed]
- 10.Lin C.S., Liu L.T., Ou L.H., Pan S.C., Lin C.I., Wei Y.H. Role of mitochondrial function in the invasiveness of human colon cancer cells. Oncol. Rep. 2018;39(1):316–330. doi: 10.3892/or.2017.6087. [DOI] [PubMed] [Google Scholar]
- 11.Kamal Y., Schmit S.L., Hoehn H.J., Amos C.I., Frost H.R. Transcriptomic differences between primary colorectal adenocarcinomas and distant metastases reveal metastatic colorectal cancer subtypes. Cancer Res. 2019;79(16):4227–4241. doi: 10.1158/0008-5472.CAN-18-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Hoon M.J.L., Imoto S., Nolan J., Miyano S. Open source clustering software. Bioinformatics. 2004;20(9):1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 14.Saldanha A.J. Java Treeview - Extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 15.Eylem C.C., et al. Gas chromatography-mass spectrometry based 18O stable isotope labeling of Krebs cycle intermediates. Anal. Chim. Acta. 2021;1154 doi: 10.1016/j.aca.2021.338325. [DOI] [PubMed] [Google Scholar]
- 16.Marisa L., et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10(5) doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demirkol S., et al. A combined ULBP2 and SEMA5A expression signature as a prognostic and predictive biomarker for colon cancer. J. Cancer. 2017;8(7):1113–1122. doi: 10.7150/jca.17872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang C.Te, et al. High-throughput microscopy reveals the impact of multifactorial environmental perturbations on colorectal cancer cell growth. Gigascience. 2021;10(4) doi: 10.1093/gigascience/giab026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marroquin L.D., Hynes J., Dykens J.A., Jamieson J.D., Will Y. Circumventing the crabtree effect: replacing media glucose with galactose increases susceptibility of hepG2 cells to mitochondrial toxicants. Toxicol. Sci. 2007;97(2) doi: 10.1093/toxsci/kfm052. [DOI] [PubMed] [Google Scholar]
- 20.Sun X., Li Z., Wang X., He J., Wu Y. Inorganic phosphate as ‘bioenergetic messenger’ triggers M2-type macrophage polarization. Adv. Sci. 2024 doi: 10.1002/advs.202306062. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y., et al. Colorectal cancers utilize glutamine as an anaplerotic substrate of the TCA cycle in vivo. Sci. Rep. 2019;9(1):19180. doi: 10.1038/s41598-019-55718-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehere P., et al. Tyrosine aminotransferase: biochemical and structural properties and molecular dynamics simulations. Protein Cell. 2010;1(11):1023–1032. doi: 10.1007/s13238-010-0128-5. Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buchli R., Alberati-Giani D., Malherbe P., Köhler C., Broger C., Cesura A.M. Cloning and functional expression of a soluble form of kynurenine/α -aminoadipate aminotransferase from rat kidney. J. Biol. Chem. 1995;270(49):29330–29335. doi: 10.1074/jbc.270.49.29330. [DOI] [PubMed] [Google Scholar]
- 24.Xia J., Li S., Liu S., Zhang L. Aldehyde dehydrogenase in solid tumors and other diseases: potential biomarkers and therapeutic targets. MedComm. 2023;4(1):e195. doi: 10.1002/mco2.195. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watkins D., Venditti C.P., Rosenblatt D.S. Rosenberg's Molecular and Genetic Basis of Neurological and Psychiatric Disease. Elsevier; 2020. Vitamins: cobalamin and folate; pp. 687–697. ch. 51. [DOI] [Google Scholar]
- 26.Guvenc Tuna B., Cleary M.P., Demirel P.B., Dogan S. Leptin signaling in liver tissue of a transgenic breast cancer mouse model. Cureus. 2020;12(1):1–10. doi: 10.7759/cureus.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanigan M.H. Gamma-glutamyl transpeptidase: redox regulation and drug resistance. Adv. Cancer. Res. 2014;122:103–141. doi: 10.1016/B978-0-12-420117-0.00003-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu J., et al. Colorectal cancer detection using targeted serum metabolic profiling. J. Proteome Res. 2014;13(9) doi: 10.1021/pr500494u. [DOI] [PubMed] [Google Scholar]
- 29.Tan B., et al. Metabonomics identifies serum metabolite markers of colorectal cancer. J. Proteome Res. 2013;12(6) doi: 10.1021/pr400337b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun H., Zhang C., Zheng Y., Liu C., Wang X., Cong X. Glutamine deficiency promotes recurrence and metastasis in colorectal cancer through enhancing epithelial–mesenchymal transition. J. Transl. Med. 2022;20(1):330. doi: 10.1186/s12967-022-03523-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao K., Liu K., Wang Z., Zhao K., Mei Y. TRIM2 promotes metabolic adaptation to glutamine deprivation via enhancement of CPT1A activity. FEBS J. 2024 doi: 10.1111/febs.17218. Jul. [DOI] [PubMed] [Google Scholar]
- 32.Martin-Perez M., Urdiroz-Urricelqui U., Bigas C., Benitah S.A. Lipid metabolism in metastasis and therapy. Curr. Opin. Syst. Biol. 2021;28 doi: 10.1016/j.coisb.2021.100401. [DOI] [Google Scholar]
- 33.Jiang L., et al. Up-regulated FASN expression promotes transcoelomic metastasis of ovarian cancer cell through epithelial-mesenchymal transition. Int. J. Mol. Sci. 2014;15(7) doi: 10.3390/ijms150711539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loh C.Y., et al. The e-cadherin and n-cadherin switch in epithelial-to-mesenchymal transition: signaling, therapeutic implications, and challenges. Cells. 2019;8(10) doi: 10.3390/CELLS8101118. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S., Matsumoto K., Lish S.R., Cartagena-Rivera A.X., Yamada K.M. Budding epithelial morphogenesis driven by cell-matrix versus cell-cell adhesion. Cell. 2021;184(14):3702–3716. doi: 10.1016/J.CELL.2021.05.015. e30Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cyriac R., Lee K. Glutaminase inhibition as potential cancer therapeutics: current status and future applications. J. Enzyme Inhib. Med. Chem. 2024;39(1) doi: 10.1080/14756366.2023.2290911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gmeiner W.H. Recent advances in therapeutic strategies to improve colorectal cancer treatment. Cancers. 2024;16(5):1029. doi: 10.3390/cancers16051029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rizzo A., Nannini M., Novelli M., Dalia Ricci A., Di Scioscio V., Pantaleo M.A. Dose reduction and discontinuation of standard-dose regorafenib associated with adverse drug events in cancer patients: a systematic review and meta-analysis. Ther. Adv. Med. Oncol. 2020;12 doi: 10.1177/1758835920936932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Golshani G., Zhang Y. Advances in immunotherapy for colorectal cancer: a review. Therap. Adv. Gastroenterol. 2020;13 doi: 10.1177/1756284820917527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guven D.C., et al. The association between albumin levels and survival in patients treated with immune checkpoint inhibitors: a systematic review and meta-analysis. Front. Mol. Biosci. 2022;9 doi: 10.3389/fmolb.2022.1039121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Codes used in this study can be made available upon request.