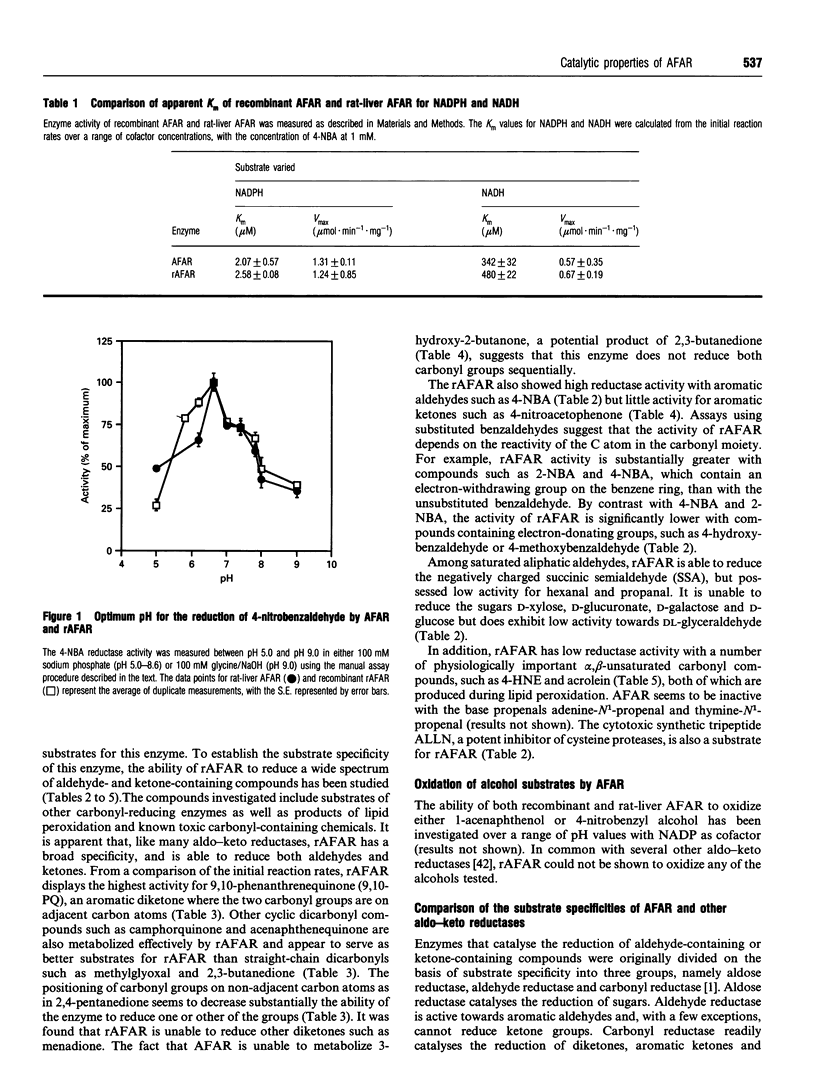
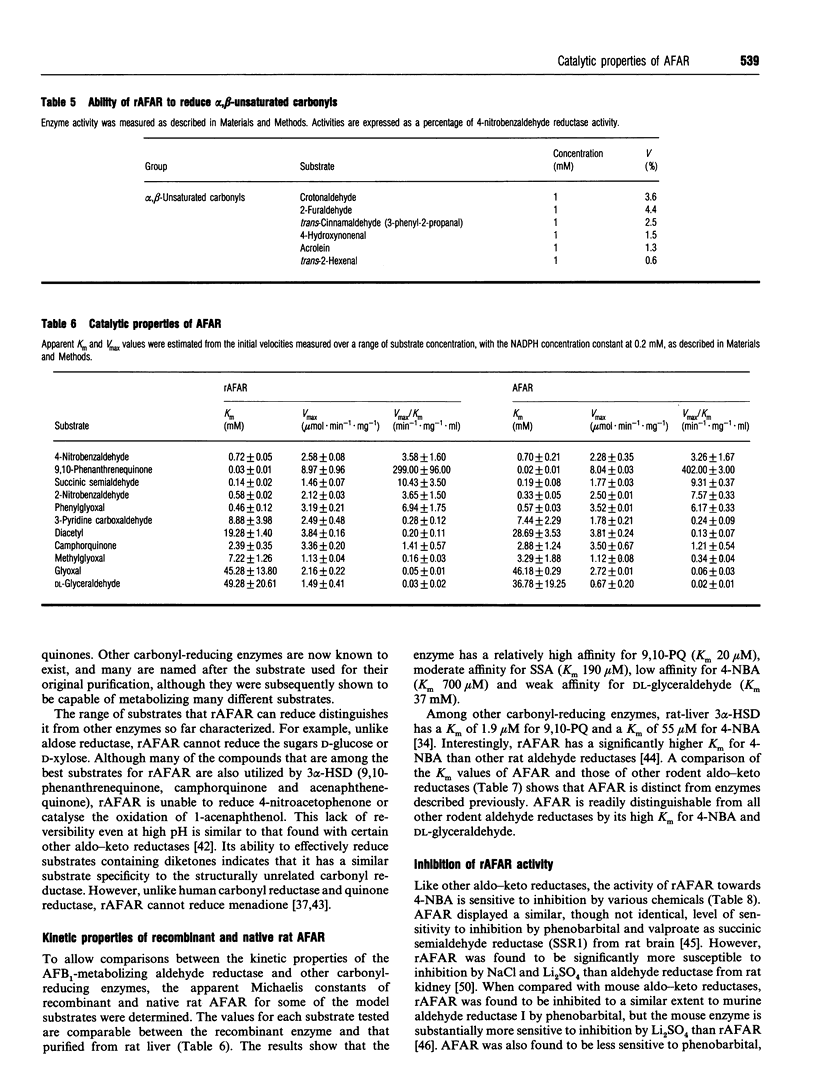
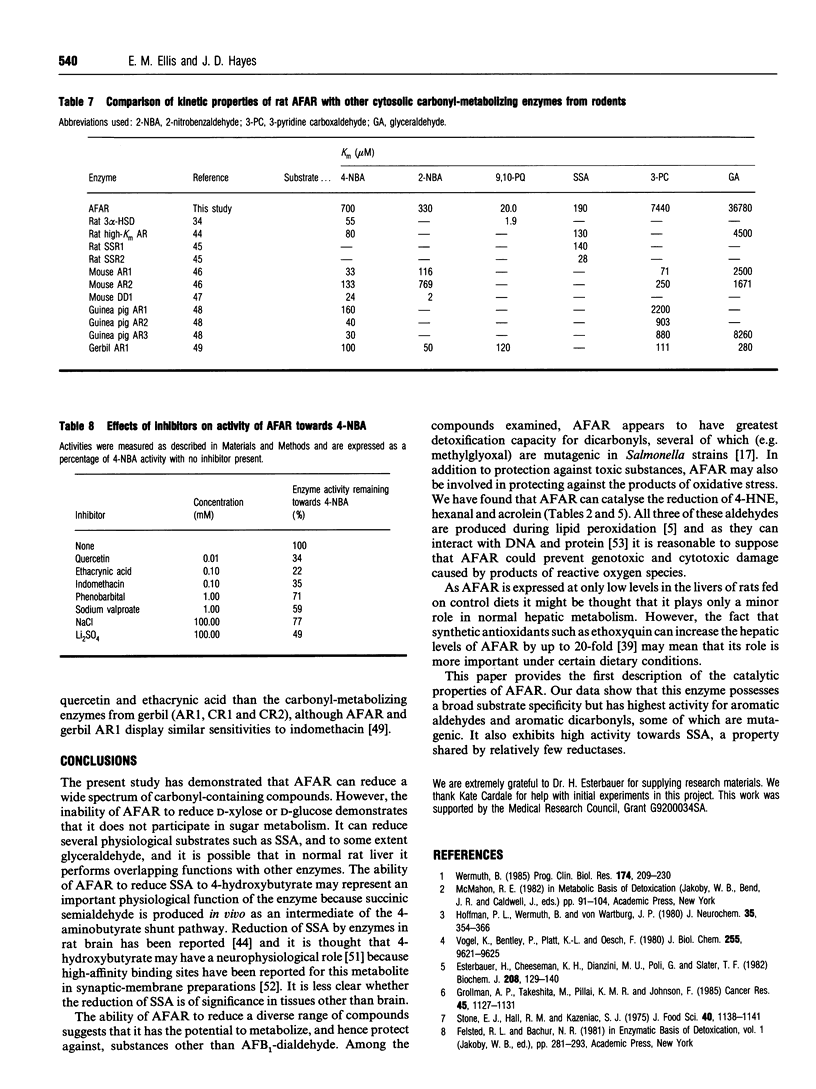
Abstract
The enzyme from rat liver that reduces aflatoxin B1-dialdehyde exhibits a unique catalytic specificity distinct from that of other aldo-keto reductases. This enzyme, designated AFAR, displays high activity towards dicarbonyl-containing compounds with ketone groups on adjacent carbon atoms; 9,10-phenanthrenequinone, acenaphthenequinone and camphorquinone were found to be good substrates. Although AFAR can also reduce aromatic and aliphatic aldehydes such as succinic semialdehyde, it is inactive with glucose, galactose and xylose. The enzyme also exhibits low activity towards alpha,beta-unsaturated carbonyl-containing compounds. Determination of the apparent Km reveals that AFAR has highest affinity for 9,10-phenanthrenequinone and succinic semialdehyde, and low affinity for glyoxal and DL-glyceraldehyde.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Belinsky M., Jaiswal A. K. NAD(P)H:quinone oxidoreductase1 (DT-diaphorase) expression in normal and tumor tissues. Cancer Metastasis Rev. 1993 Jun;12(2):103–117. doi: 10.1007/BF00689804. [DOI] [PubMed] [Google Scholar]
- Benavides J., Rumigny J. F., Bourguignon J. J., Cash C., Wermuth C. G., Mandel P., Vincendon G., Maitre M. High affinity binding sites for gamma-hydroxybutyric acid in rat brain. Life Sci. 1982 Mar 15;30(11):953–961. doi: 10.1016/0024-3205(82)90624-5. [DOI] [PubMed] [Google Scholar]
- Bohren K. M., Bullock B., Wermuth B., Gabbay K. H. The aldo-keto reductase superfamily. cDNAs and deduced amino acid sequences of human aldehyde and aldose reductases. J Biol Chem. 1989 Jun 5;264(16):9547–9551. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Carper D., Nishimura C., Shinohara T., Dietzchold B., Wistow G., Craft C., Kador P., Kinoshita J. H. Aldose reductase and p-crystallin belong to the same protein superfamily as aldehyde reductase. FEBS Lett. 1987 Aug 10;220(1):209–213. doi: 10.1016/0014-5793(87)80905-5. [DOI] [PubMed] [Google Scholar]
- Ciaccio P. J., Tew K. D. cDNA and deduced amino acid sequences of a human colon dihydrodiol dehydrogenase. Biochim Biophys Acta. 1994 Jun 28;1186(1-2):129–132. doi: 10.1016/0005-2728(94)90144-9. [DOI] [PubMed] [Google Scholar]
- Deyashiki Y., Ogasawara A., Nakayama T., Nakanishi M., Miyabe Y., Sato K., Hara A. Molecular cloning of two human liver 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase isoenzymes that are identical with chlordecone reductase and bile-acid binder. Biochem J. 1994 Apr 15;299(Pt 2):545–552. doi: 10.1042/bj2990545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis E. M., Judah D. J., Neal G. E., Hayes J. D. An ethoxyquin-inducible aldehyde reductase from rat liver that metabolizes aflatoxin B1 defines a subfamily of aldo-keto reductases. Proc Natl Acad Sci U S A. 1993 Nov 1;90(21):10350–10354. doi: 10.1073/pnas.90.21.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterbauer H., Cheeseman K. H., Dianzani M. U., Poli G., Slater T. F. Separation and characterization of the aldehydic products of lipid peroxidation stimulated by ADP-Fe2+ in rat liver microsomes. Biochem J. 1982 Oct 15;208(1):129–140. doi: 10.1042/bj2080129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grollman A. P., Takeshita M., Pillai K. M., Johnson F. Origin and cytotoxic properties of base propenals derived from DNA. Cancer Res. 1985 Mar;45(3):1127–1131. [PubMed] [Google Scholar]
- Guzelian P. S. Comparative toxicology of chlordecone (Kepone) in humans and experimental animals. Annu Rev Pharmacol Toxicol. 1982;22:89–113. doi: 10.1146/annurev.pa.22.040182.000513. [DOI] [PubMed] [Google Scholar]
- Hayes J. D., Judah D. J., Neal G. E. Resistance to aflatoxin B1 is associated with the expression of a novel aldo-keto reductase which has catalytic activity towards a cytotoxic aldehyde-containing metabolite of the toxin. Cancer Res. 1993 Sep 1;53(17):3887–3894. [PubMed] [Google Scholar]
- Hoffman P. L., Wermuth B., von Wartburg J. P. Human brain aldehyde reductases: relationship to succinic semialdehyde reductase and aldose reductase. J Neurochem. 1980 Aug;35(2):354–366. doi: 10.1111/j.1471-4159.1980.tb06272.x. [DOI] [PubMed] [Google Scholar]
- Inoue S., Sharma R. C., Schimke R. T., Simoni R. D. Cellular detoxification of tripeptidyl aldehydes by an aldo-keto reductase. J Biol Chem. 1993 Mar 15;268(8):5894–5898. [PubMed] [Google Scholar]
- Judah D. J., Hayes J. D., Yang J. C., Lian L. Y., Roberts G. C., Farmer P. B., Lamb J. H., Neal G. E. A novel aldehyde reductase with activity towards a metabolite of aflatoxin B1 is expressed in rat liver during carcinogenesis and following the administration of an anti-oxidant. Biochem J. 1993 May 15;292(Pt 1):13–18. doi: 10.1042/bj2920013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazu T., Shinoda M., Nakayama T., Deyashiki Y., Hara A., Sawada H. Aldehyde reductase is a major protein associated with 3-deoxyglucosone reductase activity in rat, pig and human livers. Biochem J. 1991 Nov 1;279(Pt 3):903–906. doi: 10.1042/bj2790903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinweber F. J., Greenough R. C., Schwender C. F., Kaplan H. R., Di Carlo F. J. Bunolol metabolism by cell-free preparations of human liver: biosynthesis of dihydrobunolol. Xenobiotica. 1972 Mar;2(2):191–202. doi: 10.3109/00498257209111050. [DOI] [PubMed] [Google Scholar]
- Lindahl R. Aldehyde dehydrogenases and their role in carcinogenesis. Crit Rev Biochem Mol Biol. 1992;27(4-5):283–335. doi: 10.3109/10409239209082565. [DOI] [PubMed] [Google Scholar]
- Marnett L. J., Hurd H. K., Hollstein M. C., Levin D. E., Esterbauer H., Ames B. N. Naturally occurring carbonyl compounds are mutagens in Salmonella tester strain TA104. Mutat Res. 1985 Jan-Feb;148(1-2):25–34. doi: 10.1016/0027-5107(85)90204-0. [DOI] [PubMed] [Google Scholar]
- Molowa D. T., Wrighton S. A., Guzelian P. S. Purification and characterization of aldo-keto reductases from gerbil liver: immunochemical evidence for related proteins in other mammalian species. Arch Biochem Biophys. 1986 Dec;251(2):487–494. doi: 10.1016/0003-9861(86)90356-5. [DOI] [PubMed] [Google Scholar]
- Ohta M., Tanimoto T., Tanaka A. Characterization of aldose reductase and aldehyde reductase from the medulla of rat kidney. Chem Pharm Bull (Tokyo) 1990 Jun;38(6):1639–1643. doi: 10.1248/cpb.38.1639. [DOI] [PubMed] [Google Scholar]
- Parekh H. K., Sladek N. E. NADPH-dependent enzyme-catalyzed reduction of aldophosphamide, the pivotal metabolite of cyclophosphamide. Biochem Pharmacol. 1993 Sep 14;46(6):1043–1052. doi: 10.1016/0006-2952(93)90669-n. [DOI] [PubMed] [Google Scholar]
- Pawlowski J. E., Huizinga M., Penning T. M. Cloning and sequencing of the cDNA for rat liver 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase. J Biol Chem. 1991 May 15;266(14):8820–8825. [PubMed] [Google Scholar]
- Pawlowski J. E., Penning T. M. Overexpression and mutagenesis of the cDNA for rat liver 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase. Role of cysteines and tyrosines in catalysis. J Biol Chem. 1994 May 6;269(18):13502–13510. [PubMed] [Google Scholar]
- Penning T. M., Smithgall T. E., Askonas L. J., Sharp R. B. Rat liver 3 alpha-hydroxysteroid dehydrogenase. Steroids. 1986 Apr-May;47(4-5):221–247. doi: 10.1016/0039-128x(86)90094-2. [DOI] [PubMed] [Google Scholar]
- Riley R. J., Workman P. DT-diaphorase and cancer chemotherapy. Biochem Pharmacol. 1992 Apr 15;43(8):1657–1669. doi: 10.1016/0006-2952(92)90694-e. [DOI] [PubMed] [Google Scholar]
- Rivett A. J., Smith I. L., Tipton K. F. Purification of the high-Km aldehyde reductase from rat brain and liver and from ox brain. Biochem J. 1981 Aug 1;197(2):473–481. doi: 10.1042/bj1970473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumigny J. F., Maitre M., Cash C., Mandel P. Specific and non-specific succinic semialdehyde reductases from rat brain: isolation and properties. FEBS Lett. 1980 Aug 11;117(1):111–116. doi: 10.1016/0014-5793(80)80924-0. [DOI] [PubMed] [Google Scholar]
- Sawada H., Hara A., Kato F., Nakayama T. Purification and properties of reductases for aromatic aldehydes and ketones from guinea pig liver. J Biochem. 1979 Oct;86(4):871–881. doi: 10.1093/oxfordjournals.jbchem.a132619. [DOI] [PubMed] [Google Scholar]
- Sawada H., Hara A., Nakayama T., Nakagawa M., Inoue Y., Hasebe K., Zhang Y. P. Mouse liver dihydrodiol dehydrogenases. Identity of the predominant and a minor form with 17 beta-hydroxysteroid dehydrogenase and aldehyde reductase. Biochem Pharmacol. 1988 Feb 1;37(3):453–458. doi: 10.1016/0006-2952(88)90214-6. [DOI] [PubMed] [Google Scholar]
- Sladek N. E., Manthey C. L., Maki P. A., Zhang Z., Landkamer G. J. Xenobiotic oxidation catalyzed by aldehyde dehydrogenases. Drug Metab Rev. 1989;20(2-4):697–720. doi: 10.3109/03602538909103572. [DOI] [PubMed] [Google Scholar]
- Smithgall T. E., Harvey R. G., Penning T. M. Spectroscopic identification of ortho-quinones as the products of polycyclic aromatic trans-dihydrodiol oxidation catalyzed by dihydrodiol dehydrogenase. A potential route of proximate carcinogen metabolism. J Biol Chem. 1988 Feb 5;263(4):1814–1820. [PubMed] [Google Scholar]
- Stolz A., Rahimi-Kiani M., Ameis D., Chan E., Ronk M., Shively J. E. Molecular structure of rat hepatic 3 alpha-hydroxysteroid dehydrogenase. A member of the oxidoreductase gene family. J Biol Chem. 1991 Aug 15;266(23):15253–15257. [PubMed] [Google Scholar]
- Takahashi M., Fujii J., Teshima T., Suzuki K., Shiba T., Taniguchi N. Identity of a major 3-deoxyglucosone-reducing enzyme with aldehyde reductase in rat liver established by amino acid sequencing and cDNA expression. Gene. 1993 May 30;127(2):249–253. doi: 10.1016/0378-1119(93)90728-l. [DOI] [PubMed] [Google Scholar]
- Tipton K. F., Singer T. P. Advances in our understanding of the mechanisms of the neurotoxicity of MPTP and related compounds. J Neurochem. 1993 Oct;61(4):1191–1206. doi: 10.1111/j.1471-4159.1993.tb13610.x. [DOI] [PubMed] [Google Scholar]
- Tomlinson D. R., Stevens E. J., Diemel L. T. Aldose reductase inhibitors and their potential for the treatment of diabetic complications. Trends Pharmacol Sci. 1994 Aug;15(8):293–297. doi: 10.1016/0165-6147(94)90010-8. [DOI] [PubMed] [Google Scholar]
- Tulsiani D. R., Touster Resolution and partial characterization of two aldehyde reductases of mammalian liver. J Biol Chem. 1977 Apr 25;252(8):2545–2550. [PubMed] [Google Scholar]
- Turner A. J., Whittle S. R. Biochemical dissection of the gamma-aminobutyrate synapse. Biochem J. 1983 Jan 1;209(1):29–41. doi: 10.1042/bj2090029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel K., Bentley P., Platt K. L., Oesch F. Rat liver cytoplasmic dihydrodiol dehydrogenase. Purification to apparent homogeneity and properties. J Biol Chem. 1980 Oct 25;255(20):9621–9625. [PubMed] [Google Scholar]
- Wallin R., Gebhardt O., Prydz H. NAD(P)H dehydrogenase and its role in the vitamin K (2-methyl-3-phytyl-1,4-naphthaquinone)-dependent carboxylation reaction. Biochem J. 1978 Jan 1;169(1):95–101. doi: 10.1042/bj1690095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Fujii Y., Nakayama K., Ohkubo H., Kuramitsu S., Kagamiyama H., Nakanishi S., Hayaishi O. Structural similarity of bovine lung prostaglandin F synthase to lens epsilon-crystallin of the European common frog. Proc Natl Acad Sci U S A. 1988 Jan;85(1):11–15. doi: 10.1073/pnas.85.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wermuth B. Aldo-keto reductases. Prog Clin Biol Res. 1985;174:209–230. [PubMed] [Google Scholar]
- Wermuth B., Bohren K. M., Heinemann G., von Wartburg J. P., Gabbay K. H. Human carbonyl reductase. Nucleotide sequence analysis of a cDNA and amino acid sequence of the encoded protein. J Biol Chem. 1988 Nov 5;263(31):16185–16188. [PubMed] [Google Scholar]
- Wermuth B. Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to prostaglandin 9-ketoreductase and xenobiotic ketone reductase. J Biol Chem. 1981 Feb 10;256(3):1206–1213. [PubMed] [Google Scholar]
- Williams J. B., Lu A. Y., Cameron R. G., Pickett C. B. Rat liver NAD(P)H:quinone reductase. Construction of a quinone reductase cDNA clone and regulation of quinone reductase mRNA by 3-methylcholanthrene and in persistent hepatocyte nodules induced by chemical carcinogens. J Biol Chem. 1986 Apr 25;261(12):5524–5528. [PubMed] [Google Scholar]
- Wilson D. K., Bohren K. M., Gabbay K. H., Quiocho F. A. An unlikely sugar substrate site in the 1.65 A structure of the human aldose reductase holoenzyme implicated in diabetic complications. Science. 1992 Jul 3;257(5066):81–84. doi: 10.1126/science.1621098. [DOI] [PubMed] [Google Scholar]
- Winters C. J., Molowa D. T., Guzelian P. S. Isolation and characterization of cloned cDNAs encoding human liver chlordecone reductase. Biochemistry. 1990 Jan 30;29(4):1080–1087. doi: 10.1021/bi00456a034. [DOI] [PubMed] [Google Scholar]


