Abstract
Background
Cabbage Fusarium wilt (CFW) is a devastating disease caused by the soil-borne fungus Fusarium oxysporum f. sp. conglutinans (Foc). One of the optimal measures for managing CFW is the employment of tolerant/resistant cabbage varieties. However, the interplay between plant genotypes and the pathogen Foc in shaping the rhizosphere microbial community, and the consequent influence of these microbial assemblages on biological resistance, remains inadequately understood.
Results
Based on amplicon metabarcoding data, we observed distinct differences in the fungal alpha diversity index (Shannon index) and beta diversity index (unweighted Bray–Curtis dissimilarity) within the rhizosphere of the YR (resistant to Foc) and ZG (susceptible to Foc) cabbage varieties, irrespective of Foc inoculation. Notably, the Shannon diversity shifts in the resistant YR variety were more pronounced following Foc inoculation. Disease-resistant plant variety demonstrate a higher propensity for harboring beneficial microorganisms, such as Pseudomonas, and exhibit superior capabilities in evading harmful microorganisms, in contrast to their disease-susceptible counterparts. Furthermore, the network analysis was performed on rhizosphere-associated microorganisms, including both bacteria and fungi. The networks of association recovered from YR exhibited greater complexity, robustness, and density, regardless of Foc inoculation. Following Foc infection in the YR rhizosphere, there was a notable increase in the dominant bacterium NA13, which is also a hub taxon in the microbial network. Reintroducing NA13 into the soil significantly improved disease resistance in the susceptible ZG variety, by directly inhibiting Foc and triggering defense mechanisms in the roots.
Conclusions
The rhizosphere microbial communities of these two cabbage varieties are markedly distinct, with the introduction of the pathogen eliciting significant alterations in their microbial networks which is correlated with susceptibility or resistance to soil-borne pathogens. Furthermore, we identified a rhizobacteria species that significantly boosts disease resistance in susceptible cabbages. Our results indicated that the induction of resistance genes leading to varied responses in microbial communities to pathogens may partly explain the differing susceptibilities of the cabbage varieties tested to CFW.
Video Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-024-01883-0.
Keywords: Rhizosphere microbes, Cabbage, Fusarium oxysporum f. sp. Conglutinans, Disease resistance, Fusarium wilt, Co-occurrence networks, Plant defence
Background
Cabbage (Brassica oleracea L.), belonging to the Cruciferae family, stands as the most extensively cultivated vegetable worldwide. In 2022, the production value of cabbage reached a substantial US$9.9 billion in China, as reported by the Food and Agriculture Organization [1]. However, the future of cabbage production faces potential threats from both traditional and emerging pathogens, particularly Fusarium oxysporum. Cabbage Fusarium wilt (CFW) is a soil-borne disease caused by pathogen Fusarium oxysporum f. sp. Conglutinans (Foc), which can remain in soil for years or even decades [2]. It infiltrates cabbage roots, obstructing xylem vessels, which leads to leaf wilting and, in some cases, the demise of the entire plant. This is accompanied by stunted growth and ultimately results in plant death. Over recent decades, CFW has been a major cause of significant agricultural losses to cabbage crops globally, impacting food production and the agricultural economy [3–5]. In managing soil-borne diseases, employing resistant cabbage varieties is recognized as one of the key strategies in the control of cabbage Fusarium wilt (CFW). Through extensive breeding programs, Chinese breeders have successfully identified several cabbage varieties with inherent resistance to CFW. Among these, ZhongGan21 (ZG) stands out as a popular variety, acclaimed for its high yield and favorable organoleptic qualities, yet it exhibits a marked susceptibility to CFW. In contrast, the YR Zhonggan21 (YR) variety, a derivative of ZG, demonstrates notable resistance to this disease. This resistance is attributed to the introduction of specific disease resistance genes (FOC1) into YR, resulting in a variety whose genetic makeup closely mirrors that of its progenitor, ZG, while offering enhanced resilience against CFW [6, 7].
The rhizosphere microbiota endows host plants with many advantages, encompassing enhanced nutrient assimilation, augmented stress resilience, and fortified defenses against soil-borne pathogens [8, 9]. Central to this interaction is the soil within the rhizosphere, serving as a primary reservoir of microorganisms. Here, plant genotypes exert influence through the secretion of specific exudates, thereby sculpting the microbial composition at the soil-root interface [10, 11]. The plant genotypes emerge as pivotal determinants in the specification and functional attributes of the microorganisms thriving within the rhizosphere [12–15]. Plants adeptly modulate their rhizosphere microbiota, selectively fostering or inhibiting specific members of the local microbial consortium. This modulation serves as an initial line of defense against soil-borne pathogens through various mechanisms [16]. Upon pathogen challenge, plants simultaneously deploy a “cry for help” strategy, enlisting beneficial microorganisms to colonize their root systems and counteract pathogen attacks [8]. Beneficial microorganisms can inhibit pathogen invasion either by directly suppressing pathogens or by activating the plant’s immune system. Plants employ complex immune responses to defend against pathogen attacks, involving key signaling pathways mediated by salicylic acid (SA), ethylene (ET), and jasmonic acid (JA). Salicylic acid (SA) plays a crucial role in systemic acquired resistance (SAR), with SA accumulation leading to the activation of defense genes, thereby enhancing resistance against a broad spectrum of pathogens. Ethylene (ET) is another important hormone in plant defense, involved in both local and systemic induced resistance (ISR). ET signaling, often in conjunction with jasmonic acid (JA), regulates the expression of defense-related genes. Beneficial microorganisms, such as plant growth-promoting rhizobacteria (PGPR) and arbuscular mycorrhizal fungi (AMF), enhance plant immunity by activating these signaling pathways. They typically induce ISR through the SA, JA, and ET pathways, providing robust protection against various pathogens [17, 18].
The functional dynamics of a microbial community transcend the mere aggregation of individual member capabilities. This complexity emanates from intricate interactions and shared evolutionary paths, collectively enhancing the microbiome’s activity beyond that of any solitary species [19]. Probing these interactions is pivotal for a comprehensive understanding of the microbiome’s functionality. Network analysis has emerged as an instrumental approach in microbiome research. It elucidates symbiotic relationships within microbial communities, uncovers critical microbial links essential for community stability and composition, and evaluates the impact of these interactions and the network's overall performance on host health. In microbial co-occurrence networks, species interactions manifest as positive (cooperative or mutually beneficial), negative (competitive), or neutral associations [20, 21]. Network analysis further aids in identifying pivotal species, termed hub taxa, which significantly influence both the function and composition of the microbiome [22]. Moreover, species not classified as hub taxa can still significantly contribute within these complex networks [23].
Examining how the structure, composition, and functional network of rhizosphere microbial communities change before and after inoculating different cabbage genotypes with Foc can give crucial hints to understanding the impacts of both genotypes and pathogens on these microbial communities. This information also sheds light on the role of microbial communities in varying disease susceptibility phenotypes in cabbage. More precise disease control measures, such as resistance breeding based on beneficial microbial communities of disease-resistant varieties, can be developed based on this information. Our hypothesis posits that (1) distinct variations exist in the rhizosphere microbial communities associated with two cabbage genotypes exhibiting differential susceptibility to CFW, and (2) there is a potential relationship between differences in microbial community composition, structure, functional network, and susceptibility to CFW. To validate these hypotheses, we characterized the microbial composition and tracked the changes before and after CFW inoculation in a CFW-resistant variety YR and a susceptible variety ZG. Concurrent network analyses were conducted to elucidate the influence of genotype and pathogen challenge on microbial community dynamics, and to determine the contributions of these microbial communities to the resistance against CFW. Furthermore, we investigated the protective roles of beneficial rhizobacteria through strain isolation, plate inhibition assays, and pot experiments. Finally, we assessed the impact of key microbial taxa on plant defense mechanisms by quantifying the transcript levels of defense-related genes within the salicylic acid (SA), jasmonic acid (JA), and Ethylene (ET) signaling pathways. In our study, we focused on the genes WRKY70, CTR1, PR1, and PR4 due to their significant roles in plant defense mechanisms. WRKY70 is a transcription factor that negatively regulates the SA biosynthesis and positive regulators of SA-mediated defense; CONSTITUTIVE TRIPLE RESPONSE 1(CTR1), a rapidly accelerated fibrosarcoma (raf)-like Serine/Threonine protein kinase that negatively regulates the ET signaling pathway; PATHOGENESIS-RELATED PROTEIN 1 (PR1) is a well-known marker for the SA signaling pathway, with its expression strongly induced upon pathogen attack to enhance resistance; PATHOGENESIS-RELATED PROTEIN 4 (PR4) is a marker for the JA/ET signaling pathway, encoding an antifungal protein crucial for defending against fungal pathogens and enhancing overall immunity [24–26]. Additionally, these genes play important roles in regulating plant immunity against Fusarium pathogens, further strengthening tomato defense mechanisms [27–29]
Material and methods
Cultivation of cabbage and collection of rhizosphere samples
The seeds of the resistant variety YR and the susceptible variety ZG were surface-sterilized using 0.5% sodium hypochlorite solution for 5 min, subsequently followed by three successive rinses with sterile ddH2O. These sterilized seeds were then sown in a carefully prepared soil mixture, consisting of natural soil, vermiculite, and commercial substrate in a proportion of 2:3:4, ensuring optimal conditions for growth. The plants were inoculated with the Foc race 1 representative strain FGL-03–6 after developing two true leaves. This involved immersing the plants with a conidial suspension (1 × 106 spores/ml) for 20 min by submerging the roots. As a control, uninoculated plants received water treatment. Additionally, unplanted soil pots, referred to as bulk soil controls, were included and maintained under conditions similar to those of the YR and ZG plants. In total, we set up five treatments: YR without Foc inoculation, ZG without Foc inoculation, YR with Foc inoculation, ZG with Foc inoculation, and bulk soil. Each treatment included 30 pots (10 × 10 × 12 cm), each containing one plant. Rhizosphere soil samples were collected 15 days post-inoculation. Five biological replicates were taken from each treatment, with each replicate consisting of a composite sample obtained by mixing three randomly selected individual samples. To obtain the rhizosphere soil, we carefully uprooted the plants and gently tapped them to remove soil adhering to the roots, following the method described by Wei et al. [30]. This soil was designated as rhizosphere soil. The same method was employed to collect bulk soil from the unplanted pots, ensuring the sampling depth matched the planted pots. Briefly, we removed the top 0–3 cm layer of soil, using the remaining soil as bulk soil. Each replicate consisted of a composite sample obtained by mixing three individual samples. The samples used for DNA extraction were stored at – 80 °C, while those for microbial cultivation form YR inoculated with Foc were preserved at 4 °C.
DNA extraction and PCR amplification
Genomic DNA was extracted from 0.25 g of soil samples, harnessing the E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, U.S.) in strict accordance with the manufacturer’s protocol. Each sample was extracted three times. The integrity of the isolated DNA was confirmed via 1% agarose gel electrophoresis. Furthermore, to determine the concentration and purity of the extracted DNA, we employed the NanoDrop 2000 UV–vis spectrophotometer (Thermo Scientific, Wilmington, USA), ensuring high-quality genomic material for subsequent analyses (A260/280: 1.8–2.0). Each biological replicate’s DNA was obtained from a mixture of DNA extracted from three technical replicates of the same biological sample. The extracted DNA was subsequently diluted to 1 ng/µl using sterile distilled water to ensure optimal conditions for PCR amplification. Targeting the key regions within microbial genomes, we amplified the V3-V4 hypervariable region of the 16S rRNA gene for bacteria and the ITS1 region for fungi. This was achieved using specific primer pairs: 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′) for bacterial samples, and ITS1-1F-F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS1-1F-R (′5-GCTGCGTTCTTCATCGATGC-3′) for fungal samples. The PCR reaction mixture consisted of 15 µl Phusion® High-Fidelity PCR Master Mix, 2 µM of each forward and reverse primer, and 10 ng of template DNA. The PCR protocol involved an initial denaturation at 98 ℃ for 1 min, followed by 30 cycles of 98 ℃ for 10 s (denaturation), 50 ℃ for 30 s (annealing), and 72 ℃ for 30 s (extension). This was concluded with a final extension at 72 ℃ for 5 min. The PCR amplicons were confirmed by electrophoresis on a 2% agarose gel, mixing equal volumes of loading buffer (containing SYB green) with the PCR products. Following amplification, the PCR products were amalgamated in equidensity ratios and subsequently purified employing the QIAGEN Gel Extraction method to ensure the integrity and purity of the samples. For sequencing, libraries were meticulously prepared using the TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, USA), adhering closely to the manufacturer’s guidelines and incorporating index codes for precise sample identification. The quality of the resultant library was assessed using the Qubit® 2.0 Fluorometer (Thermo Scientific) and the Agilent Bioanalyzer 2100 system. Sequencing was performed on the Illumina NovaSeq platform, generating 250 bp paired-end reads.
16S rRNA gene and ITS read processing
In this study, we processed the bacterial 16S rRNA gene and fungal ITS sequences employing a suite of bioinformatics tools: fastp [31], USEARCH v10.0 [32], vsearch [33], and QIIME v1.9.1 [34]. Initially, we trimmed primer sequences and discarded low-quality reads with Q-scores below 20. Subsequently, we merged the paired 16S rRNA gene amplicon sequencing reads using vsearch parameters (–fastq_mergepairs, –fastq_minovlen 12, –fastq_minmergelen 360, –fastq_maxns 5). This was followed by quality filtering and removal of forward and reverse primer sequences (–fastx_filter, -fastq_maxee 0.5, vsearch). We identified biological reads at 100% sequence similarity utilizing unoise3 [32] under default settings. Each zero-radius operational taxonomic unit (zOTU) received a taxonomic classification using usearch (-sintax with -strand both and -sintax_cutoff 0.6) against the RDP database (RDP 16S v16) [35]. Bacterial zOTUs assigned to Mitochondria, Cyanobacteria, and Chloroplast were excluded. From the remaining sequences, we constructed a zOTU table (-zotus). The ITS amplicon data underwent a similar process. Reads were merged (–fastq_mergepairs, –fastq_minovlen 12, –fastq_minmergelen 200, –fastq_maxmergelen 350, –fastq_maxns 5, vsearch), with subsequent removal of primer sequences and quality filtering (–fastx_filter, -fastq_maxee 0.5, vsearch). We identified biological reads at 100% sequence similarity using unoise3 with default parameters. Fungal taxonomic assignment was conducted using usearch (-sintax with -strand both and -sintax_cutoff 0.6) against the UNITE database (v8.3) [36]. The reserved sequences were then used to construct the zOTU table. The bacterial and fungal zOTU tables were rarefied to 6796 and 7178 reads, respectively, using QIIME 1.91 (single_rarefaction.py).
Co-occurrence network analysis
To investigate the interaction between the microbial community species of CFW-resistant variety YR and susceptible variety ZG and change patterns in both after Foc inoculation, we used a random matrix theory (RMT) approach to construct intra-kingdom (bacterial-bacterial and fungal-fungal) and inter-kingdom (bacterial-fungal) networks for YR (control and Foc-inoculated) and ZG (control and Foc-inoculated), respectively. Network construction was conducted with the Molecular Ecological Network Analyses pipeline (MENAP, http://ieg4.rccc.ou.edu/mena/) [37–40] with the following settings: only fill 0.01 in blanks with paired valid values; data was taken centered log-ratio transform; use Pearson correlation similarity matrix; decrease the cutoff from the top for calculation order; and for scan speed, Regress Poisson distribution only. The global properties of the networks were calculated in MENAP. The networks were graphed using Cytoscape [41].
Bacteria isolation and antifungal assays
To isolate the widest possible diversity of bacteria, we used different culture medium: Luria–Bertani Agar (LB), Reasoner’s 2A Agar (R2A), Nutrient Agar (NA), and Bacillus Polymyxa Agar (BP). We took 1 g rhizosphere soil from YR inoculated with Foc in 20 ml PBS buffer and mixed it thoroughly, kept it for 1 min, took 0.5 ml of the supernatant, and then diluted it 100 times with PBS buffer. Then, 20 μl was taken and applied to culture medium. The plates were placed in an incubator at 25 °C for 2–5 days. Bacterial colonies were then selected based on morphology, color, and margins and further purified by spreading on new plates. The isolated strains were suspended in 25% glycerol and preserved at – 80 °C for long-term storage. Subsequently, these strains were cultured on agar plates for further analysis. FGL03-6 was cultured in potato dextrose agar (PDA) to full plate size, and then round pieces of about 1 cm diameter were taken on the PDA culture medium for inhibition assay.
Bacterial identification and whole genome sequencing of NA13
The 16S rRNA gene from the bacterial strains was amplified using primers 27F (5′-AGAGTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) (Additional file 1: Table S1). The amplification products were subjected to 1% agarose gel electrophoresis, followed by sequencing at Beijing Tsingke Biotech Co., Ltd. Species identification was performed using the NCBI nucleotide collection (nr/nt) database, utilizing the BLASTn tool for sequence comparison and analysis [42]. Whole-genome sequencing of the strain NA13 was carried out using the PromethION platform at GrandOmics Biosciences Co., Ltd., leveraging Oxford Nanopore Technologies (ONT). Sequences with an average quality score of 10 or lower were discarded, resulting in a final dataset of 1.71 Gb comprising 150,579 high-quality reads. Subsequently, a de novo hybrid assembly of these long reads obtained from the PromethION platform was conducted employing Canu (version 1.6) [43]. The assembled sequences have been deposited in the NCBI database, under GenBank accession number SAMN38604125. Annotation of NA13’s whole genome sequence was carried out using the prokaryotic gene prediction software Prokka 1.14.6 [44]. In the analysis and prediction of secondary metabolites within the NA13 genome, the AntiSMASH bacterial version was utilized (accessible at https://antismash.secondarymetabolites.org/#!/start) [45]. Functional annotation of the genome was performed using the Clusters of Orthologous Group (COG) databases, with blastP employed under an E-value threshold of < 1e-5 [46]. Subsequently, a circular map of the genome was constructed utilizing shinyCircos-V2.0 [47].
Glasshouse experiments to assess effects of NA13 on ZG defense
The study exploring the biological mechanisms responsible for the NA13-mediated enhancement of ZG’s resistance to Foc was conducted under controlled greenhouse conditions. The experimental setup employed sterilized commercial nursery soil specifically prepared for cabbage cultivation. The experimental design included planting trials in 10 × 10 pots, with treatments including Foc, Foc + NA13, NA13, and control. ZG seeds were subjected to sterilization using 75% ethanol for 1 min, followed by sodium hypochlorite treatment for three minutes, and then rinsed with sterile distilled water. The sterilized seeds were germinated at 28 °C in an incubator for 2 days, prior to being sown in sterilized commercial growing substrates. Following a 30-day period of seedling growth, the roots were immersed in an NA13 bacterial suspension (OD600 = 0.5, 4 × 108 CFUs/ml) for 20 min. Subsequently, the seedlings were transplanted into pots. To sustain NA13 exposure, the seedlings were inoculated at 5-day intervals using a drenching method with the NA13 suspension (OD600 = 0.5, 4 × 108 CFUs/ml, 5 ml). In total, we performed three root drench treatments with NA13, specifically on days 5, 10, and 15 post-transplant. Foc spore suspensions (1 × 106 spores/ml) were inoculated into the plants seven days subsequent to the initial NA13 strain inoculation, employing the root-drenching method. Samples were harvested 13 days after Foc inoculation. As a control, an equivalent volume of water was applied to the treatments where bacterial inoculation was omitted. Throughout the experimental period, the ZG seedlings were cultivated in an incubator set at 28 °C with a photoperiod of 12 h light and 12 h darkness. At harvest, the roots were carefully separated from the soil, enveloped in aluminum foil, and subsequently stored at – 80 °C for total RNA extraction.
Quantification of defense-related genes in the root
RNA was extracted using the RNAprep Pure Plant Kit, adhering strictly to the manufacturer’s instructions. RNA concentrations were subsequently quantified using a Qubit™ fluorometer, ensuring the RNA’s purity and quality met the standards required for downstream applications. For cDNA synthesis, 1 μg of RNA was utilized per 20-μl reaction, employing random hexamers and oligo (dT) primers as provided in the Tetro cDNA Synthesis Kit. The study then focused on the qRT-PCR setup, selecting 4 defense genes relevant to the ET, SA and JA signaling pathways (Additional file 1: Table S1), previously characterized in earlier research [48–51]. Quantification was conducted using the SYBR Green qRT-PCR kit on a ViiA™ 7 sequence detection system, following the kit’s for reaction mix and cDNA sample preparation protocol. Actin was chosen as the house-keeping gene (Salicylic acid suppression of clubroot in broccoli [52]. Finally, qRT-PCR data were analyzed using the software available with the ViiA™ 7 system, calculating the relative expression levels of the defense genes and normalizing them using the house-keeping gene Actin (Additional file 1: Table S1).
Statistical analysis
Statistical analyses were primarily performed within the R computing environment, except as noted otherwise. Variations in sample proportions were highlighted using extended error bars, which denote a 95% confidence interval. For the calculation of alpha diversity indices, including the Shannon index, we utilized QIIME (alpha_diversity.py). To determine the statistical significance of variations among different rhizosphere soil samples, the Kruskal–Wallis test was applied (krus.test function in R, P < 0.05). Principal coordinates analysis (PCoA), implemented via the cmdscale function in R and based on the Bray–Curtis dissimilarity matrix derived from a rarefied zOTU table, facilitated the examination of beta diversity and community structure differences in rhizosphere microbiomes. Additionally, PERMANOVA analysis, incorporating 999 permutations (Adonis function, vegan package in R), was conducted to ascertain the influence of genotype and Foc inoculation on the explained variance. A linear modeling approach was utilized to evaluate the variations in microbial taxa abundance among resistant-sensitive cabbage varieties, both pre- and post-Foc inoculation. The results were visualized using the R package ggplot2 [53]. Treatment effects on defence gene expression were analyzed using one-way ANOVA and Tukey post hoc tests.
Results
Characterization of amplicon sequencing datasets
High-throughput sequencing of all samples yielded 10,243,240 bacterial and 10,844,440 fungal raw reads. Post-clustering, a refined set of 640,691 bacterial and 413,035 fungal high-quality reads were retained. We retained zero-radius operational taxonomic units (zOTUs) that appeared at least 3 times across all samples and had a read count of no less than 20. Following the exclusion of low abundance zOTUs and subsequent resampling, these reads were categorized into 336 bacterial and 137 fungal zOTUs. Rarefaction analysis indicated that our sequencing was sufficient to reveal the true diversity of the samples (Additional file 2: Fig. S1). The resampled zOTU table was used for subsequent community analysis (Additional file 3: Table S2).
YR and ZG have distinctly different rhizosphere microbiota
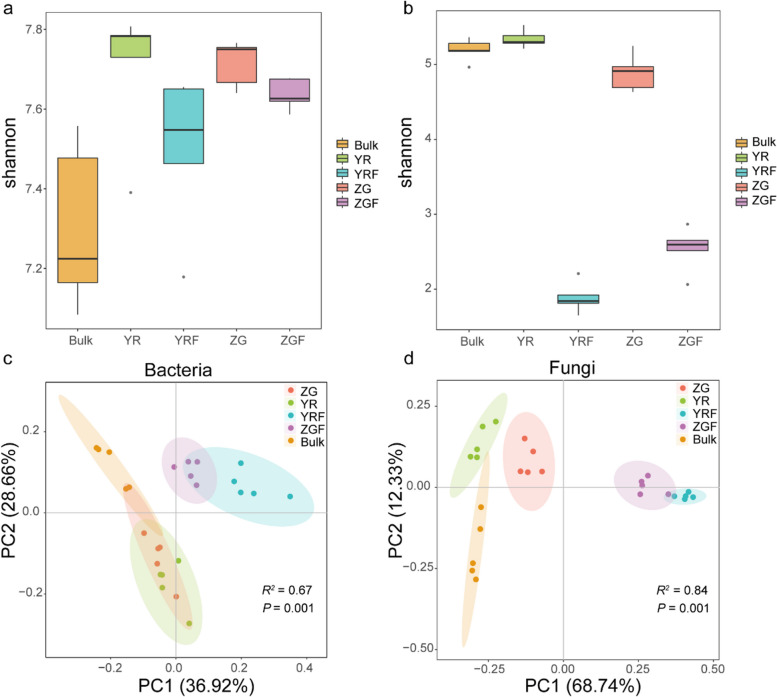
To characterize the effect of host selection on the microbiome of the plant and the pattern of microbial changes between rhizosphere in CFW-resistant and susceptible varieties after inoculation with the pathogen Foc, we analyzed the alpha diversity of rhizosphere bacterial and fungal communities of cabbage using the Shannon index. Our analysis revealed that the presence of FOC1 resistance genes had a significant impact on the fungal community diversity (Kruskal–Wallis and Wilcoxon rank sum tests, P < 0.05), but did not significantly affect the bacterial community diversity in the YR (resistant) and ZG (susceptible) varieties. Meanwhile, the bacterial Shannon diversity in ZG was significantly higher than in bulk soil (Kruskal–Wallis and Wilcoxon rank sum tests, P < 0.05), while the diversity in YR was not significantly different. However, while YR exhibited a higher bacterial Shannon index than ZG, this difference did not reach statistical significance (Wilcoxon rank sum test, P > 0.05; Fig. 1a). Post-inoculation with Foc, a decrease in Shannon diversity was observed in both YR and ZG, with a more pronounced reduction in YR (from 7.783 to 7.547) compared to ZG (from 7.750 to 7.626), although this change was not significant (Wilcoxon rank sum test, P > 0.05; Fig. 1a). In terms of fungal community diversity, YR displayed significantly higher Shannon index values than both ZG and bulk soil (Wilcoxon rank sum test, P < 0.05), whereas the difference between YR and bulk soil, ZG and bulk soil was not significant (Fig. 1b; Additional file 4: Table S3; Wilcoxon rank sum test, P > 0.05). Following Foc inoculation, a similar pattern of change was noted in the fungal Shannon index as observed with bacteria, with YR showing a more pronounced alteration than ZG.
Fig. 1.
a, b Shannon diversity of bacterial and fungal communities in rhizosphere soil. c, d Bacterial and fungal community principal coordinates analysis based on bray–curtis dissimilarity matrix. Bulk, YR, YRF, ZG, and ZGF indicates bulk soil, YR Zhonggan21, YR Zhonggan21 inoculated with Fusarium oxysporum f. sp. Conglutinans (Foc), Zhonggan21 and Zhonggan21 inoculated with Foc, respectively
Furthermore, we evaluated the impact of the resistance gene FOC1 and the fungal pathogen Foc on the structure of the microbial community in the cabbage rhizosphere. With regard to beta diversity, a Bray–Curtis dissimilarity matrix was calculated. Overall, dissimilarities in the microbial community between YR, ZG, YR inoculated with Foc and ZG inoculated with Foc among samples were displayed using principal coordinates analysis (PCoA). We found that the bacterial and fungi communities cluster along the first principal-coordinate analysis axis according to the genotype and Foc challenge, indicating that disease resistance gene FOC1 and fungal pathogen Foc considerably influenced bacterial and fungal community composition (Fig. 1c, d; PERMANOVA, P < 0.05; Additional file 5: Table S4). In addition, the distance between YR inoculated with Foc and Foc-uninoculated YR was further than the distance between Foc-inoculated ZG and Foc-uninoculated ZG. The combination of this result and the Shannon index result (Fig. 1 a and b) suggests that the microbiomes in the resistant cabbage YR rhizospheres may be able to respond more actively to challenges when confronted with pathogenic Foc compared to those in ZG.
Specific differences in microbiome between rhizosphere soil of YR and ZG
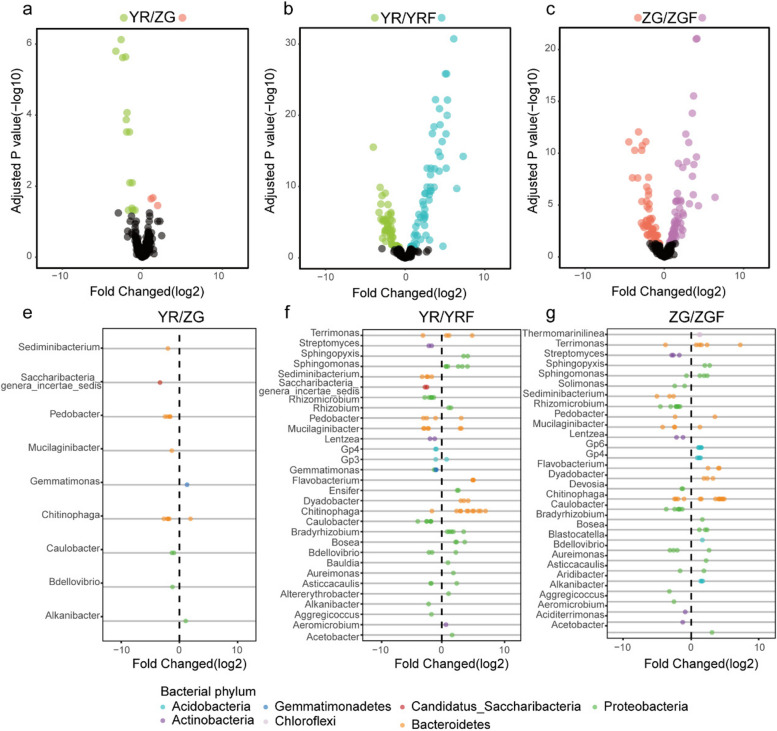
To more comprehensively delineate the alterations within the microbiota, we examined the disparities at the zOTU level among four distinct rhizosphere microbiota groups: YR, ZG, YR inoculated with Foc, and ZG inoculated with Foc. We analyzed the enrichment of zOTUs according to their taxonomy using volcano and scatter plots. We found that YR was enriched with more bacterial zOTUs compared to ZG, while ZG was enriched with more fungal zOTUs compared to YR (Figs. 2a and 3a; Additional file 6: Table S5). In the YR rhizosphere, an enrichment of bacterial zOTUs spanning diverse genera was observed. Notably, the relative abundance of several genera, potentially harboring beneficial bacteria—including Bdellovibrio (+ 2.3-fold), Caulobacter (+ 4.3-fold), Chitinophaga (+ 14.4-fold), Mucilaginibacter (+ 2.5-fold), Pedobacter (+ 15.6-fold), Saccharibacteria (+ 9.8-fold), and Sediminibacterium (+ 3.9-fold)—was significantly elevated compared to that in the ZG rhizosphere (FDR adjusted P < 0.05). ZG was also enriched with some potentially beneficial bacteria from the genera Gemmatimonas (+ 2.5-fold) and Chitinophaga (+ 3.7-fold) (FDR adjusted P < 0.05), but the abundance and richness of potentially beneficial bacteria were lesser than YR. (Fig. 2e). Compared to YR inoculated without Foc, 63 zOTUs were enriched in the rhizosphere of YR inoculated with Foc, while 53 zOTUs were notably absent (Additional file 6: Table S5; FDR adjusted P < 0.05). Meanwhile, the zOTUs enriched in the rhizosphere of YR inoculated with Foc predominantly belonged to the phyla Bacteroidetes, Proteobacteria, Actinobacteria, and Acidobacteria (Fig. 2b, Additional file 7: Fig. S2). In the more detailed genus level of taxonomic resolution, the majority of these enriched zOTUs belong to beneficial genera Pseudomonas (+ 25.1-fold), Chitinophaga (+ 247.5-fold), Flavobacterium (+ 215.6-fold), Rhizobium (+ 6.3-fold), Acetobacter (+ 3.2-fold), Altererythrobacter (+ 2.2-fold), Asticcacaulis (+ 5.6-fold), Aureimonas (+ 3.7-fold), Bdellovibrio (+ 4.9-fold), Blastococcus (+ 1.7-fold), Bosea (+ 24.4-fold), Bradyrhizobium (+ 29.3-fold), Dyadobacter (+ 211.3-fold), Ensifer (+ 12.7-fold), Mucilaginibacter (+ 17.7-fold), Pedobacter (+ 9.2-fold), Pseudolabrys (+ 2.5-fold), Rhodoplanes (+ 2.6-fold), Rhodopseudomonas (+ 8.6-fold), Sphingomonas (+ 212.8-fold), and Terrimonas (+ 37.1-fold) (Fig. 2f). Concurrently, compared to ZG inoculated without Foc, the rhizosphere of ZG inoculated with Foc exhibited a differential pattern, with 59 zOTUs being enriched and 59 zOTUs being absent (Fig. 2c; FDR-adjusted P < 0.05). At the phylum level, these enriched zOTUs predominantly fell within the Bacteroidetes, Proteobacteria, Acidobacteria, and Chloroflexi phyla. Most of these zOTUs belonged to the beneficial genus Pseudomonas (+ 16.5-fold), Chitinophaga (+ 212.8-fold), Flavobacterium (+ 28.7-fold), Acetobacter (+ 6.5-fold), Asticcacaulis (+ 3.1-fold), Aureimonas (+ 3.8-fold), Bdellovibrio (+ 5.0-fold), Bosea (+ 9.8-fold), Bradyrhizobium (+ 2.2-fold), Dyadobacter (+ 14.5-fold), Ensifer (+ 2.4-fold), Gp4 (+ 17.0-fold), Gp6 (+ 25.7-fold), Mucilaginibacter (+ 2.2-fold), Pedobacter (+ 8.5-fold), Rhodopseudomonas (+ 5.0-fold), Sphingomonas (+ 28.8-fold), and Terrimonas (+ 211.5-fold) (Fig. 2g).
Fig. 2.
a–c Enrichment (positive) and depletion (negative) of bacterial zOTUs between different plant treatments. Each point represents an individual zOTU. e–g Top 30 genera of enriched or depleted zOTUs between different plant treatments. Colors of points indicate phylum classification
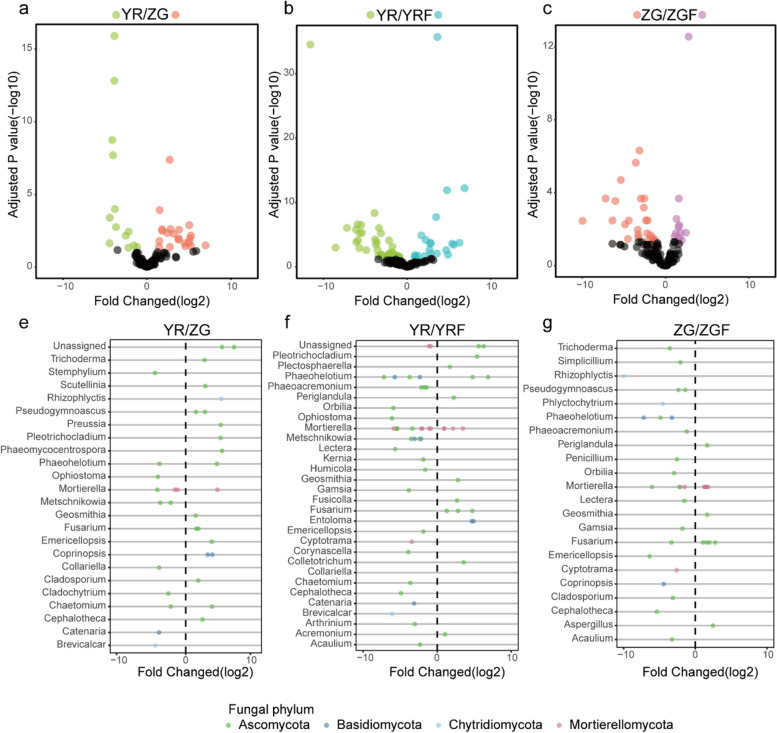
Fig. 3.
a–c Enrichment (positive) and depletion (negative) of fungal zOTUs between different plant treatments. Each point represents an individual zOTU. e–g Top 30 genera of enriched or depleted zOTUs between different plant treatments. Colors of points indicate phylum classification
Comparing the differences in fungal zOTU abundance between YR and ZG, opposite trends were observed. Compared to YR, more fungal zOTUs were enriched in ZG, and these zOTUs belonged to a wider range of genera than those enriched in YR compared to ZG (Fig. 3a; Additional file 6: Table S5). Several potentially pathogenic fungi from the genera Fusarium (+ 6.4-fold), Cladosporium (+ 3.4-fold), Rhizophlyctis (+ 25.1-fold), Pseudogymnoascus (+ 9.3-fold), Pleotrichocladium (+ 25.0-fold), Phaeomycocentrospora (+ 25.2-fold), and Geosmithia (+ 2.7-fold) were significantly enriched in ZG, while several potentially pathogenic fungi were also enriched in YR, including Ophiostoma (zOTU_138, + 16.3-fold), Metschnikowia (zOTU_133, zOTU160, + 17.6-fold), Cladochytrium (zOTU_57, + 5.7-fold), and Stemphylium (zOTU_60, + 22.6-fold) (FDR adjusted P < 0.05). In addition, some potentially beneficial fungi were also enriched in YR and ZG. Catenaria (zOTU_78, + 22.6-fold), Chaetomium (zOTU_40, + 4.5-fold), and Mortierella (zOTU_65, zOTU_50, zOTU_51, + 22.8-fold) were significantly enriched in the YR compared to ZG. Trichoderma (zOTU_112, + 6.5-fold), Preussia (zOTU_188, + 25.1-fold), Scutellinia (zOTU_69, + 7.0-fold), Coprinopsis (zOTU_110, zOTU_124, + 23.2-fold), Chaetomium (zOTU_148, + 13.7-fold), Scutellinia (zOTU_69, + 7.1-fold), and Mortierella (zOTU_171, + 22.6-fold) were significantly enriched in the ZG compared to YR (FDR adjusted P < 0.05; Fig. 3e). After inoculation with Foc, YR and ZG exhibited similar enrichment patterns. Compared to YR inoculated without Foc, YR inoculated with Foc enriched 21 zOTUs while 39 zOTUs were absent. Similarly, compared to ZG inoculated without Foc, ZG inoculated with Foc enriched 12 zOTUs while 25 zOTUs were absent. (Fig. 3 b and c; Additional file 6: Table S5). Interestingly, these absent zOTUs belonged to the potentially pathogenic genera Ophiostoma, Stemphylium, Metschnikowia, Humicola, Arthrinium, Kernia, Lectera, Phaeoacremonium, and Acaulium (Fig. 3 f and g). The results of the differential analysis of bacterial and fungal abundance indicated that the resistant variety YR tended to recruit more beneficial microorganisms compared to the susceptible variety ZG while reducing the accumulation of pathogenic organisms to respond to the Foc challenge.
Characterization of YR and ZG rhizosphere microbiome co-occurrence networks
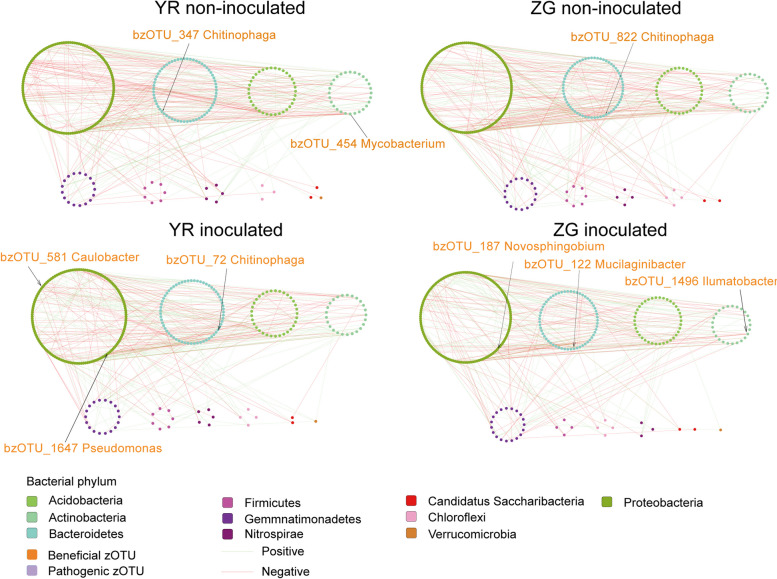
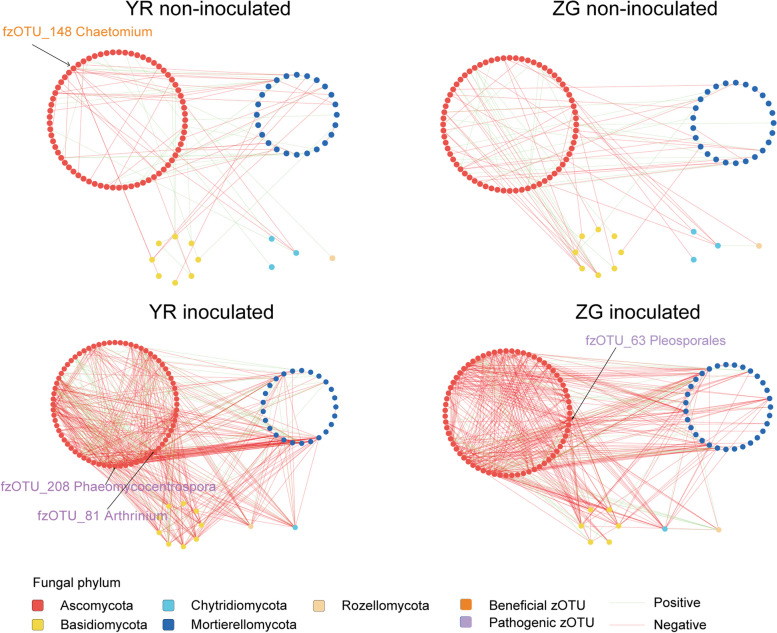
To explore how disease resistance gene FOC1 and pathogen Foc affect the structure of microbial co-occurrence in cabbage, we analyzed bacterial-bacterial and fungal-fungal intra-kingdom networks as well as bacterial-fungal inter-kingdom networks. Through intra-kingdom network analysis of the YR bacterial and fungal networks, without Foc inoculation, it was found that the bacterial network contained 293 nodes and 420 edges, while the fungal network comprised 105 nodes and 125 edges. In the intra-kingdom co-occurrence networks of the ZG bacterial and fungal communities without Foc inoculation, the bacterial network comprised 297 nodes and 394 edges, while the fungal network included 105 nodes and 115 edges. After inoculation with Foc, the intra-kingdom bacterial and fungal networks for YR contained 298 nodes and 390 edges, and 108 nodes and 536 edges, respectively. Meanwhile, the intra-kingdom bacterial and fungal networks for ZG consisted of 298 nodes and 368 edges, and 106 nodes and 460 edges, respectively (Figs. 4 and 5; Additional file 8: Table S6). Regardless of whether Foc was inoculated or not, the difference in the number of nodes and edges within the intra-kingdom co-occurrence networks demonstrated that the connectivity between zOTUs in the YR bacterial networks were consistently stronger than in the ZG bacterial networks, and similarly, the connectivity in the YR fungal networks were stronger than in the ZG fungal networks. Additionally, the YR bacterial networks were more complex than the ZG bacterial networks, and the YR fungal network were more complex than the ZG fungal networks. On the other hand, we observed that both before and after Foc inoculation, the YR bacterial networks demonstrated higher average degrees (2.867 pre-inoculation, 2.617 post-inoculation) and densities (0.01 pre-inoculation, 0.009 post-inoculation) compared to the ZG bacterial networks. Similarly, the YR fungal networks exhibited higher average degrees (2.381 pre-inoculation, 9.926 post-inoculation) and densities (0.023 pre-inoculation, 0.093 post-inoculation) than those in the ZG fungal networks, both before and after Foc inoculation (Additional file 8: Table S6). The zOTUs classified as “Nodes with max degree” were identified as hub zOTUs [54]. Before and after inoculation with Foc, there was a change in the hub nodes with the max degree within the YR bacterial network. Initially characterized by Mycobacterium (bzOTU_454) and Chitinophaga (bzOTU_347), the network shifted to include Caulobacter (bzOTU_581), Pseudomonas (bzOTU_1647), and Chitinophaga (bzOTU_72). In the ZG bacterial network, the hub node Chitinophaga (bzOTU_822) was replaced by Novosphingobium (bzOTU_187), Ilumatobacter (bzOTU_1496), and Mucilaginibacter (bzOTU_122) following inoculation with Foc. Notably, the potentially beneficial taxa Chitinophaga (bzOTU_822) and Pseudomonas (bzOTU_1647) enriched in the YR inoculated with Foc were also identified as hub taxa in the network. Thus, we considered Chitinophaga (bzOTU_822) and Pseudomonas (bzOTU_1647) as key taxa for YR to respond to the challenge from pathogenic Foc. In the YR fungal network, the hub node with the max degree changed from Chaetomium (fzOTU_148) before Foc inoculation to Arthrinium (fzOTU_81), Phaeohelotium (fzOTU_83), Acaulium (fzOTU_210), and Phaeomycocentrospora (fzOTU_208) after inoculation with Foc. In the ZG fungal network, the hub nodes with the max degree changed from Entoloma (fzOTU_145, fzOTU_90) before Foc inoculation to Pleosporales (fzOTU_63) and Phaeohelotium (fzOTU_89) after inoculation with Foc. (Figs. 4 and 5).
Fig. 4.
Bacterial co-occurrence networks from rhizosphere structural communities of YR and ZG in control and Foc-inoculated plants
Fig. 5.
Fungal co-occurrence networks from rhizosphere structural communities of YR and ZG in control and Foc-inoculated plants
The inter-kingdom co-occurrence network suggested that FOC1 and pathogen Foc affect the microbial network structure of cabbage. Consistent with the intra-kingdom networks, the YR network has more nodes, more edges, and a higher average degree than the ZG network, which also indicates that the YR network is more complex than the ZG network and is a highly connected network (Additional file 8: Table S6). After inoculating Foc, the network edges, average degree, and density increased in both the resistant (YR) and susceptible (ZG) varieties. Meanwhile, the YR network after Foc inoculation had more edges, higher average degree, and density compared to ZG inoculated with Foc, which indicated that the disease-resistant variety YR had a more complex, robust, and density network after the Foc challenge. Moreover, in the YR network inoculated with Foc compared to the YR network inoculated without Foc, the hub node changed from Flavobacterium (bzOTU_620) to Mortierella (fzOTU_171), Paramyrothecium (fzOTU_170), and Metschnikowia (fzOTU_109). Similarly, in the ZG network inoculated with Foc compared to the ZG network inoculated without Foc, the hub node changed from Nocardioides (bzOTU_737) to Alternaria (fzOTU_164), Stemphylium (fzOTU_60), and Fusarium (fzOTU_128) (Additional file 9: Fig. S3).
Bacterial isolation, identification, and whole genome sequencing
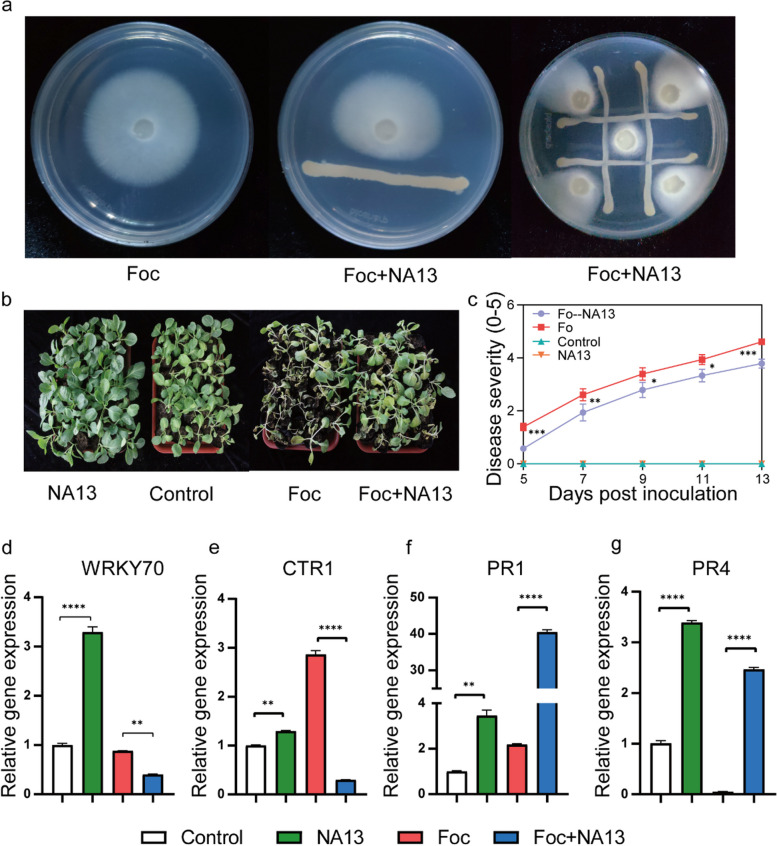
We attempted to isolate Chitinophaga spp. and Pseudomonas spp. and investigated their contribution to Foc resistance in cabbage. In total, we isolated 136 bacterial strains with a direct inhibitory effect on Foc from the rhizosphere of cabbage variety YR inoculated with Foc, using different medium. We successfully isolated Pseudomonas brassicacearum NA13, which has considerable Foc-inhibiting properties (Fig. 7a). Meanwhile, 16S rRNA gene sequences of Pseudomonas brassicacearum NA13 had a 98.64% nucleotide similarity match with the sequences of bzOTU_1647 (Additional file 10: Fig. S4), indicating that NA13 was the bacterium that enriched after YR inoculation with Foc.
Fig. 7.
a Strong inhibition of the NA13 on Fusarium oxysporum f. sp. Conglutinans (Foc) growth on a PDA medium. b, c Effects of NA13 inoculation on cabbage growth and survival. d–g Effects of NA13 and Foc inoculations on the transcription of genes associated with ethylene (ET), jasmonic acid (JA), and salicylic acid (SA) signaling pathways. NA13 inoculation with NA13 only, Control no inoculation, Foc inoculation with Foc only, NA13 + Foc dual inoculation with NA13 and Foc
We performed whole-genome sequencing of NA13 and predicted a genome size of 6,609,035 bp containing 9223 coding sequences (CDS), 185 tRNA genes, and 21 rRNA operons (Fig. 6). The NA13 genome was then analyzed with antiSMASH to predict secondary metabolites that may be associated with antimicrobial activity. The analysis revealed 13 secondary metabolite gene clusters in Table S7. Cluster 7, Cluster 10 and Cluster 11 exhibited 100% similarity to the secondary metabolites 2,4-diacetylphloroglucinol, syringomycin and hydrogen cyanide, respectively (Additional file 11: Fig. S5, Additional file 12: Table S7). We predicted gene function in the NA13 genome using the COG approach. The genome of NA13 is significantly enriched in genes encoding proteins involved with amino acid transport and metabolism and inorganic ion transport and metabolism (Additional file 13: Fig. S6). Genes coding for bacterial flagellin, chitin deacetylase, chitin, and Type I to Type V secretion systems were detected, as were genes fliR and flhB, which code for effector proteins that interact with the plant immune system and induce the plant’s primary responses (Additional file 14: Table S8) [55].
Fig. 6.
Circular map of the Pseudomonas brassicacearum NA13 genome. The distribution of the circle from the outermost to the center is, (i) scale marks of the genome; (ii) protein-coding genes on the forward strand; (iii) protein-coding genes on the reverse strand; (iv) tRNA (blue) and rRNA (red) on the forward strand; (v) tRNA (blue) and rRNA (red) genes on the reverse strand; (vi) GC content; (vii) GC skew. Protein-coding genes are color coded according to their COG categories
NA13 promoted plant growth and defense
To investigate whether NA13 affects the defence response of CFW susceptible varieties against Foc, we pre-treated the susceptible variety ZG with NA13 in a greenhouse experiment and then inoculated it with the Foc strain FGL03-6. Overall, treatments inoculated with NA13 had higher plant survival compared to no inoculation during Foc infestation (Fig. 7 b and c). Further, we were interested in exploring whether NA13 indirectly regulates the expression of plant defence genes besides directly suppressing Foc to promote cabbage survival. Our research indicated that in the presence of Foc in soil, the NA13 strain exhibited a notable ability to enhance the activation of both Ethylene (ET), Jasmonic Acid (JA) and Salicylic Acid (SA) signaling pathways in cabbage (Fig. 7d, e). This response was evidenced by the differential expression of key genes associated with these pathways. For instance, the transcription factor WRKY70, pivotal in plant defense, exhibited downregulation of 2.2-fold (Fig. 7d). In contrast, CTR1, a key regulator of the ET signaling pathway, showed a significant decrease in expression by 9.3-fold (Fig. 7e). More strikingly, the gene PR1, a marker of the SA signaling pathway and systemic acquired resistance, was upregulated dramatically, showing an 18.4-fold increase (Fig. 7f). In contrast, PR4, a marker of the ET/JA signaling pathway associated with enhanced defense responses, exhibited an even more pronounced increase of 49.2-fold (Fig. 7g). These findings collectively underscore the potent influence of NA13 treatments on cabbage’s defense mechanisms, specifically through the modulation of ET, JA, and SA signaling pathways (Fig. 7d–g).
Discussion
The rhizosphere microbiota is important for enhancing the adaptation and productivity of plant hosts and plays a key role in plant resistance to biotic and abiotic stresses [8, 56, 57]. It has attracted significant attention in recent decades. It is known that different plant genotypes can produce different rhizosphere microbiomes [58, 59]. However, there is a lack of detailed study on the variability of rhizosphere microbial communities within the same plant species attributable to distinct disease resistance genes. Additionally, the differential responses of rhizosphere microbiota in cabbage varieties, resistant versus susceptible, to Foc infection, and the correlation between the levels of CFW resistance in cabbage and its rhizosphere microbial composition, remain underexplored. This study established that genotypic differences, as determined by a single disease resistance gene (FOC1), along with inoculation with Foc, significantly influenced the rhizosphere microbiota of both the resistant variety YR and susceptible variety ZG. Notably, the microbial community diversity in YR underwent a more pronounced alteration following Foc inoculation compared to ZG, as evidenced by both alpha and beta diversity analyses.
As plants grow, they deliver large amounts of nutrients to their roots, which are released into the soil through the root system. These root exudates selectively recruit beneficial microbes from the soil to colonize their root system; this phenomenon is more pronounced when plants are under stress [8, 60]. Our findings indicated a significant association between the FOC1 gene and the enrichment of specific taxa within the rhizosphere microbiota. Notably, a greater number of bacterial zOTUs were enriched in YR compared to ZG, and these enriched bacterial zOTUs are classified as potentially beneficial bacteria. For instance, when exposed to the pathogen Rhizoctonia solani, sugar beet roots attract Chitinophaga into the endosphere, suppressing the fungal pathogen [61]. Sphingomonas can promote plant growth [62]. Conversely, a higher number of fungal zOTUs were enriched in ZG, encompassing both potentially pathogenic as well as beneficial. Intriguingly, beneficial bacteria predominated among the bacterial zOTUs enriched in both YR and ZG, with a notably higher enrichment observed in YR. Similarly, pathogenic fungi were the dominant component in the enriched fungal zOTUs of both YR and ZG, with ZG exhibiting a greater enrichment of pathogenic fungal species compared to YR. Although potentially pathogenic fungi were found in the YR, fungal zOTU belongs to the genus Fusarium, is well known for including phytopathogenic species, and was only enriched in ZG [63].
After inoculation with Foc, the YR variety exhibited an enrichment of 63 bacterial zOTUs, whereas 53 bacterial zOTUs were notably absent, compared to the condition before inoculation. In contrast, the ZG variety displayed an equal number of both enriched and absent bacterial zOTUs post-inoculation, compared to the condition before inoculation. Interestingly, most of these zOTUs enriched by YR and ZG belonged to genera beneficial to plants, which also indicated that YR has a stronger ability to recruit beneficial bacteria compared to ZG. Additionally, following inoculation with Foc, the YR variety demonstrated an enrichment of 21 fungal zOTUs and a concurrent absence of 39 fungal zOTUs, compared to the condition before inoculation. In comparison, the ZG variety exhibited an enrichment of 12 fungal zOTUs, alongside the absence of 25 fungal zOTUs, compared to the condition before inoculation. Furthermore, the zOTUs absent in each variety, which predominantly belong to pathogenic fungal genera, revealed a notable tendency in the YR variety to reduce the recruitment of pathogenic fungi following infestation with Foc. This observation implies that both the resistant variety YR and the susceptible variety ZG exhibit a tendency to recruit beneficial microbes while concurrently reducing the recruitment of pathogenic fungi in response to infestation by the pathogenic fungus Foc. This phenomenon has also been confirmed in similar studies [64, 65]. Additionally, Boer et al. investigated the antagonistic effect of individual and mixed strains of four bacteria on plant pathogens and found that more bacteria in the soil could lead to stronger competition with pathogens for resources [66]. In contrast, the presence of pathogenic fungi other than the genus Fusarium may facilitate the development of CFW. For example, Rhizophlyctis, which is enriched in ZG and can degrade cellulose, may promote the colonization of cabbage roots by pathogenic fungi. [67]. Correspondingly, Khoury and Alcorn observed that infection by Rhizoctonia solani in two cotton varieties resulted in root lesions, a condition that may potentially compromise the effectiveness of physical barriers in these plants, thereby facilitating colonization by Verticillium albo-atrum [68].
Competitive and cooperative interactions between microbial species can influence plant health [9, 69, 70]. In this study, we found that FOC1 also has a significant influence on the rhizosphere microbial co-occurrence network of cabbage. Both before and after Foc inoculation, the microbial interaction network of the YR variety exhibited increased complexity, as evidenced by a greater number of nodes and edges. It has been demonstrated that soil microbial communities with more intricate networks (with more nodes and edges) confer greater benefits to plants compared to simpler networks [71]. Complex networks help to better respond to environmental changes or suppress soil-borne pathogens. For example, the microbial network of healthy soil is more complex than that of soil susceptible to bacterial wilt [72]. Additionally, inoculation of grassland soil into sterilized greenhouse soil increased the complexity of the chrysanthemum rhizosphere microbial network and suppressed the development of pathogenic fungi Olpidium [73].
The average degree and network density are important indicators of the network’s effectiveness. After inoculation with Foc, the average degree and network density were increased in both varieties, but the YR network had a higher average degree and network density than the ZG network both before and after inoculation. YR networks with higher average degrees and network density indicated high connectivity and community efficiency. The combination of higher average degrees and network density indicated that microbial communities were more responsive to environmental perturbations, and highly connected networks provided greater functional redundancy [74–76]. This was in alignment with the results of our alpha and beta diversity analyses. In this sense, the success of pathogen invasion can be reduced if the rhizosphere microbial community is highly connective and efficient [77]. Agler et al. introduced the concept of “microbial hubs” to describe the presence of highly interconnected species within plant microbial networks [78]. Their findings proposed that these extensively interconnected species played a crucial role in plant health, serving as intermediaries between plants and their microbiome. Microbial hubs may play a pivotal role in sustaining disease-suppressive soil conditions, enhancing nutrient absorption, augmenting the efficacy of biocontrol agents, and facilitating the mediation of defense signals among plants [78–80]. There were two hub taxa identified as Mycobacterium and Chitinophaga in the YR bacterial network (Fig. 4), with Mycobacterium and Chitinophaga being considered potentially beneficial bacteria [81]. In contrast, there was only one hub taxa identified as Chitinophaga in the ZG bacterial network (Fig. 4). After inoculation with Foc, the hub taxa of YR and ZG were replaced by Caulobacter (bzOTU_581), Pseudomonas (bzOTU_1647), Chitinophaga (bzOTU_72) and Novosphingobium (bzOTU_187), Ilumatobacter (bzOTU_1496), Mucilaginibacter (bzOTU_122), respectively (Fig. 4). Meanwhile, Pseudomonas (bzOTU_1647) and Chitinophaga (bzOTU_72) were enriched in YR after inoculation with Foc, while hub taxa in the ZG network were not enriched in ZG after inoculation with Foc. Consequently, we hypothesized that Pseudomonas (bzOTU_1647) and Chitinophaga (bzOTU_72) played a critical role in the functional network after YR inoculation with Foc.
We have successfully isolated Pseudomonas brassicacearum NA13 from Foc-inoculated YR and sequenced the whole genome. Our results indicated that the NA13 strain plays a beneficial role both in inhibiting the growth of the pathogen Foc and in promoting plant defense responses. The genomic characterization of Pseudomonas brassicacearum NA13 was associated with plant immune responses and the transport of bacterial secondary metabolites, which may contribute to the protection of cabbage against Foc infestation. The rhizosphere microbiota are abundant in microbe-associated molecular patterns (MAMPs) that initiate the primary layer of plant immune defense, limiting pathogen proliferation. MAMPs such as bacterial flagellin, chitin deacetylase, and chitin activate strong, tissue-specific immune responses in the roots of Arabidopsis thaliana. These responses operate through pathways independent of salicylic acid (SA) and jasmonic acid (JA) signaling, demonstrating a highly specialized defense mechanism [82]. NA13 undoubtedly possesses a range of components acting as MAMPs (e.g., flagellin). Meanwhile, there were clusters of related genes that code for the secondary metabolites syringomycin, 2,4-diacetylphloroglucinol and hydrogen cyanide, which are well-known antifungal compounds [83, 84]. Concurrently, the presence of NA13 was observed to induce a robust defense response in plants. In the JA/ET and SA signaling pathways, the marker genes PR4 (JA/ET) and PR1 (SA) were significantly upregulated, indicating a robust activation of the plant's defense mechanisms. Intriguingly, WRKY70, a negative regulator within the SA biosynthesis, along with negative regulators CTR1 of the ET signaling pathway, demonstrated a marked downregulation. In addition to these findings, it has been well established that root-associated microbes trigger Induced Systemic Resistance (ISR), a defense mechanism that is distinct from the SA pathway and often associated with JA and ET signaling pathways [85]. ISR activation by beneficial microbes like NA13 enhances the plant's ability to fend off a wide range of pathogens through a priming effect, where the plant’s immune system is put on alert and responds more robustly to subsequent pathogen attacks. This modulation of gene expression represents a noteworthy shift in the plant's immune defense strategy, underlining the pivotal role of NA13 in orchestrating these changes. The significant alterations in the expression of these defense signaling pathway marker genes signify the activation of plant immune defenses, highlighting the intricate interplay between microbial presence and plant physiological responses. This indicated that (1) NA13 is capable of activating the plant’s immune response to pathogens, and (2) this bacterium could use secondary metabolites to have beneficial effects on the plant, such as directly inhibiting the growth of pathogens.
The resistance gene FOC1 was classified as a TIR-NBS-LRR type R gene [6]. In plant immune responses, the activation of R genes triggers downstream immune signaling, which regulates plant resistance to pathogens. For instance, the TIR domain of the tobacco N gene is essential and sufficient for its association with the pathogen-derived elicitor p50, conferring resistance to tobacco mosaic virus [86]. The plant defense system relies on the mitogen-activated protein kinase (MAPK) signaling cascade, various transcription factors including NAC, MYB, and WRKY, pathogenesis-related (PR) genes, and the signaling networks for hormones such as salicylic acid (SA), jasmonic acid (JA), ethylene (ET), gibberellin (GA), and abscisic acid (ABA), all of which function both independently and in concert [87–90]. Zhou et al. demonstrated that the TIR-NBS-LRR type R gene GmTNL16 participates in soybean defense against Phytophthora via the JA and SA pathways [91]. Nevertheless, the intricate interactions and regulatory mechanisms among beneficial microbes such as NA13, the resistance gene FOC1, the pathogen Foc, and the plant immune system remain to be fully elucidated. Further research is essential to uncover the complexities of these interactions and their implications for plant immunity.
These findings not only provided deeper insights into the molecular mechanisms governing plant immunity but also underscored the potential of manipulating such pathways for enhancing disease resistance in crops. In summary, our results suggested that NA13 may play a key role in the three-way interaction with host plant immunity and fungal pathogen via a mechanism that enhances plant defence and inhibit Foc growth.
Conclusions
This study revealed that differences in specific resistance genes between CFW resistant and susceptible cabbage genotypes, along with inoculation with the Foc pathogen, significantly influenced the structure and composition of the rhizosphere microbial community. We successfully established a correlation between microbial communities and differential resistance levels to CFW. The FOC1 gene induced specific taxa in resistant variety YR and susceptible variety ZG, particularly an increase in beneficial bacteria in YR. Furthermore, the rhizosphere microbes in YR exhibited a more proactive response to Foc inoculation, effectively enhancing the recruitment of beneficial bacteria while simultaneously inhibiting the proliferation of harmful fungi. This work contributes to a better understanding of the phylogenetic mechanisms underlying the recruitment of beneficial rhizosphere microbiota and highlights the importance of considering microbial communities in the selection of CFW-resistant varieties. This opens up new avenues for breeding crops that more effectively utilize microbiota for protection and improved disease management.
Supplementary Information
Additional file 1: Table S1. Primer sequences used in this study.
Additional file 2: Figure S1. Alpha rarefaction plots for bacteri and fungi were generated at the species level using the “observed number of zOTUs”
Additional file 3: Table S2. Metadata, zOTU representative sequences, taxonomy annotation and zOTU table.
Additional file 4: Table S3. Alpha diversity estimators of samples.
Additional file 5: Table S4. PERMANOVA by adonis of all bacterial 16S and fungal ITS samples.
Additional file 6: Table S5. Differential abundance of zOTUs between YR and ZG.
Additional file 7: Figure S2. Relative abundance of major bacterial (a) and fungal phyla (b) in the soil samples.
Additional file 8: Table S6. Topology properties of the intra- and inter-kingdom networks.
Additional file 9: Figure S3. Inter-kingdom co-occurrence networks from rhizosphere structural communities of YR and ZG in control and Foc-inoculated plants.
Additional file 10: Figure S4. Alignment of the 16S rRNA sequences of the isolate NA13 and zOTU_1647.
Additional file 11: Figure S5. Alignment of the genome sequences of the isolate NA13 with three identified secondary metabolite regions (a) BGC0000437 (Syringomycin), (b) BGC0002345 (hydrogen cyanide) and (c) BGC0000281 (2,4-Diacetylphloroglucinol).
Additional file 12: Table S7. Secondary metabolism gene cluster types in the genome of NA13.
Additional file 13: Figure S6. Clusters of Orthologous Group annotations for the Pseudomonas brassicacearum NA13 genome.
Additional file 14: Table S8. List of genes encoding proteins involved in bacterial flagellin, chitin deacetylase, chitin, and Type I to Type V secretion systems.
Acknowledgements
Not applicable.
Authors’ contribution
ZM, BX, and JL conceived and directed the research. XP and SC performed all the lab experiments. YJ, LJ, XZ, and LS coordinated bacterial isolation. YY, YL and JZ contributed to pathogen inoculation. XP and JL contributed to sequencing analysis. XP and RAAK wrote the initial manuscript. RAAK, ZM, BX, and JL review and edit the final version. ZM, BX, and JL coordinated the research. All authors have read and approved the manuscript.
Funding
This study was supported by the National Key R&D Program of China (2022YFD1401200), National Natural Science Foundation of China (NSFC 31571962), and National Key R&D Program of China (2023YFC2604900).
Availability of data and materials
The sequences for 16S rRNA gene, ITS, and bacterium NA13 genome are deposited in the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/) under the accession number PRJNA908139, PRJNA908211, and SAMN38604125, respectively. All other data are presented directly in the manuscript or provided in the supplementary files attached to this submission.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhenchuan Mao, Email: maozhenchuan@caas.cn.
Bingyan Xie, Email: xiebingyan@caas.cn.
Jian Ling, Email: lingjian@caas.cn.
References
- 1.FAOSTAT (2022) Value of agricultural production database. Available via URL: https://www.fao.org/faostat/en/#data/QV. Accessed Jun 3 2024
- 2.Snyder WC, Hansen HN. The species concept in Fusarium. Am J Bot. 1940;27:64. 10.2307/2436688. 10.2307/2436688 [DOI] [Google Scholar]
- 3.Li M, Zhang T, Li X, Yan H. Fusarium wilt disease on curcifer vegetable and its pathogenic identification. Plant Prot. 2003;29:44–5. [Google Scholar]
- 4.Zhang Y, Zheng J, Xie B, Li J, Wu X, Shi Y, et al. Identification on pathogen of cabbage wilt disease. Acta Phytopathologica Sinaca. 2008;38:337345. https://www.cabdirect.org/cabdirect/abstract/20083235441
- 5.Kashiwa T, Inami K, Teraoka T, Komatsu K, Arie T. Detection of cabbage yellows fungus Fusarium oxysporum f. sp. conglutinans in soil by PCR and real-time PCR. Journal of General Plant Pathology. 2016;82:240–7. 10.1007/s10327-016-0668-5
- 6.Lv H, Fang Z, Yang L, Zhang Y, Wang Q, Liu Y, et al. Mapping and analysis of a novel candidate Fusarium wilt resistance gene FOC1 in Brassica oleracea. BMC Genomics. 2014;15:1094. 10.1186/1471-2164-15-1094. 10.1186/1471-2164-15-1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LÜ Honghao, YANG Limei, FANG Zhiyuan, ZHANG Yangyong, ZHUANG Mu, LIU Yumei, et al. A new Fusarium wilt-resistant, early maturing and high-quality spring cabbage cultivar‘YR Zhonggan 21.’ Acta Horticulturae Sinica. 2021; 10.1016/j.tplants.2020.03.014
- 8.Liu H, Brettell LE, Qiu Z, Singh BK. Microbiome-mediated stress resistance in plants. Trends Plant Sci. 2020;25:733–43. 10.1016/j.tplants.2020.03.014. 10.1016/j.tplants.2020.03.014 [DOI] [PubMed] [Google Scholar]
- 9.Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 1979;2011(332):1097–100. 10.1126/science.1203980. 10.1126/science.1203980 [DOI] [PubMed] [Google Scholar]
- 10.Jones DL, Nguyen C, Finlay RD. Carbon flow in the rhizosphere: carbon trading at the soil–root interface. Plant Soil. 2009;321:5–33. 10.1007/s11104-009-9925-0. 10.1007/s11104-009-9925-0 [DOI] [Google Scholar]
- 11.Mendes R, Garbeva P, Raaijmakers JM. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev. 2013;37:634–63. 10.1111/1574-6976.12028. 10.1111/1574-6976.12028 [DOI] [PubMed] [Google Scholar]
- 12.Berg G, Smalla K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol. 2009;68:1–13. 10.1111/j.1574-6941.2009.00654.x. 10.1111/j.1574-6941.2009.00654.x [DOI] [PubMed] [Google Scholar]
- 13.Bulgarelli D, Garrido-Oter R, Münch PC, Weiman A, Dröge J, Pan Y, et al. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe. 2015;17:392–403. 10.1016/j.chom.2015.01.011. 10.1016/j.chom.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91–5. 10.1038/nature11336 [DOI] [PubMed]
- 15.Marschner P, Yang C-H, Lieberei R, Crowley DE. Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol Biochem. 2001;33:1437–45. 10.1016/S0038-0717(01)00052-9. 10.1016/S0038-0717(01)00052-9 [DOI] [Google Scholar]
- 16.Berg G, Köberl M, Rybakova D, Müller H, Grosch R, Smalla K. Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol Ecol. 2017;93. 10.1093/femsec/fix050 [DOI] [PubMed]
- 17.van Loon LC. Plant responses to plant growth-promoting rhizobacteria. Eur J Plant Pathol. 2007;119:243–54. 10.1007/s10658-007-9165-1. 10.1007/s10658-007-9165-1 [DOI] [Google Scholar]
- 18.Dey M, Ghosh S. Arbuscular mycorrhizae in plant immunity and crop pathogen control. Rhizosphere. 2022;22: 100524. 10.1016/j.rhisph.2022.100524. 10.1016/j.rhisph.2022.100524 [DOI] [Google Scholar]
- 19.Rogers GB, Hoffman LR, Carroll MP, Bruce KD. Interpreting infective microbiota: the importance of an ecological perspective. Trends Microbiol. 2013;21:271–6. 10.1016/j.tim.2013.03.004. 10.1016/j.tim.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: networks, competition, and stability. Science. 1979;2015(350):663–6. 10.1126/science.aad2602. 10.1126/science.aad2602 [DOI] [PubMed] [Google Scholar]
- 21.Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10:538–50. 10.1038/nrmicro2832. 10.1038/nrmicro2832 [DOI] [PubMed] [Google Scholar]
- 22.Banerjee S, Schlaeppi K, van der Heijden MGA. Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol. 2018;16:567–76. 10.1038/s41579-018-0024-1. 10.1038/s41579-018-0024-1 [DOI] [PubMed] [Google Scholar]
- 23.Šikić M, Lančić A, Antulov-Fantulin N, Štefančić H. Epidemic centrality—is there an underestimated epidemic impact of network peripheral nodes? Eur Phys J B. 2013;86:440. 10.1140/epjb/e2013-31025-5. 10.1140/epjb/e2013-31025-5 [DOI] [Google Scholar]
- 24.Li J, Zhong R, Palva ET. WRKY70 and its homolog WRKY54 negatively modulate the cell wall-associated defenses to necrotrophic pathogens in Arabidopsis. PLoS ONE. 2017;12: e0183731. 10.1371/journal.pone.0183731. 10.1371/journal.pone.0183731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–41. 10.1016/0092-8674(93)90119-B. 10.1016/0092-8674(93)90119-B [DOI] [PubMed] [Google Scholar]
- 26.Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, et al. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci. 1998;95:15107–11. 10.1073/pnas.95.25.15107. 10.1073/pnas.95.25.15107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du N, Guo H, Fu R, Dong X, Xue D, Piao F. Comparative transcriptome analysis and genetic methods revealed the biocontrol mechanism of Paenibacilluspolymyxa NSY50 against tomato Fusarium wilt. Int J Mol Sci. 2022;23:10907. 10.3390/ijms231810907. 10.3390/ijms231810907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abbasi S, Safaie N, Sadeghi A, Shamsbakhsh M. Streptomyces strains induce resistance to Fusarium oxysporum f. sp. lycopersici Race 3 in tomato through different molecular mechanisms. Front Microbiol. 2019;10. 10.3389/fmicb.2019.01505 [DOI] [PMC free article] [PubMed]
- 29.Werner BT, Koch A, Šečić E, Engelhardt J, Jelonek L, Steinbrenner J, et al. Fusarium graminearum DICER-like-dependent sRNAs are required for the suppression of host immune genes and full virulence. PLoS ONE. 2021;16: e0252365. 10.1371/journal.pone.0252365. 10.1371/journal.pone.0252365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei Z, Yang X, Yin S, Shen Q, Ran W, Xu Y. Efficacy of Bacillus-fortified organic fertiliser in controlling bacterial wilt of tomato in the field. Appl Soil Ecol. 2011;48:152–9. 10.1016/j.apsoil.2011.03.013. 10.1016/j.apsoil.2011.03.013 [DOI] [Google Scholar]
- 31.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90. 10.1093/bioinformatics/bty560. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. 10.1093/bioinformatics/btq461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 33.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4: e2584. 10.7717/peerj.2584. 10.7717/peerj.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. 10.1038/nmeth.f.303. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–42. 10.1093/nar/gkt1244. 10.1093/nar/gkt1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kõljalg U, Larsson K, Abarenkov K, Nilsson RH, Alexander IJ, Eberhardt U, et al. UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 2005;166:1063–8. 10.1111/j.1469-8137.2005.01376.x. 10.1111/j.1469-8137.2005.01376.x [DOI] [PubMed] [Google Scholar]
- 37.Deng Y, Jiang Y-H, Yang Y, He Z, Luo F, Zhou J. Molecular ecological network analyses. BMC Bioinformatics. 2012;13:113. 10.1186/1471-2105-13-113. 10.1186/1471-2105-13-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao N, Zhou A, Kempher ML, Zhou BY, Shi ZJ, Yuan M, et al. Disentangling direct from indirect relationships in association networks. Proceedings of the National Academy of Sciences. 2022;119. 10.1073/pnas.2109995119 [DOI] [PMC free article] [PubMed]
- 39.Zhou J, Deng Y, Luo F, He Z, Yang Y. Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO 2. mBio. 2011;2. 10.1128/mbio.00122-11 [DOI] [PMC free article] [PubMed]
- 40.Zhou J, Deng Y, Luo F, He Z, Tu Q, Zhi X. Functional molecular ecological networks. mBio. 2010;1. 10.1128/mbio.00169-10 [DOI] [PMC free article] [PubMed]
- 41.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. http://www.genome.org/cgi/doi/10.1101/gr.1239303 [DOI] [PMC free article] [PubMed]
- 42.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5-9. 10.1093/nar/gkn201. 10.1093/nar/gkn201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k -mer weighting and repeat separation. Genome Res. 2017;27:722–36. http://www.genome.org/cgi/doi/10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed]
- 44.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. 10.1093/bioinformatics/btu153. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 45.Blin K, Shaw S, Augustijn HE, Reitz ZL, Biermann F, Alanjary M, et al. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023;51:W46–50. 10.1093/nar/gkad344 [DOI] [PMC free article] [PubMed]
- 46.Galperin MY, Wolf YI, Makarova KS, Vera Alvarez R, Landsman D, Koonin EV. COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 2021;49:D274–81. 10.1093/nar/gkaa1018. 10.1093/nar/gkaa1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Jia L, Tian G, Dong Y, Zhang X, Zhou Z, et al. shinyCircos‐V2.0: Leveraging the creation of Circos plot with enhanced usability and advanced features. iMeta. 2023;2. 10.1002/imt2.109 [DOI] [PMC free article] [PubMed]
- 48.Liu M, Wu F, Wang S, Lu Y, Chen X, Wang Y, et al. Comparative transcriptome analysis reveals defense responses against soft rot in Chinese cabbage. Hortic Res. 2019;6:68. 10.1038/s41438-019-0149-z. 10.1038/s41438-019-0149-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan Y, Qin L, Su H, Yang S, Wei X, Wang Z, et al. Transcriptome and coexpression network analyses reveal hub genes in Chinese Cabbage (Brassica rapa L. ssp. pekinensis) during different stages of Plasmodiophora brassicae infection. Front Plant Sci. 2021;12. 10.3389/fpls.2021.650252 [DOI] [PMC free article] [PubMed]
- 50.Lu L, Monakhos SG, Lim YP, Yi SY. Early defense mechanisms of Brassica oleracea in response to attack by Xanthomonas campestris pv. campestris. Plants. 2021;10:2705. 10.3390/plants10122705 [DOI] [PMC free article] [PubMed]
- 51.Wu J, Zhao Q, Yang Q, Liu H, Li Q, Yi X, et al. Comparative transcriptomic analysis uncovers the complex genetic network for resistance to Sclerotinia sclerotiorum in Brassica napus. Sci Rep. 2016;6:19007. 10.1038/srep19007. 10.1038/srep19007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lovelock DA, Donald CE, Conlan XA, Cahill DM. Salicylic acid suppression of clubroot in broccoli (Brassicae oleracea var. italica) caused by the obligate biotroph Plasmodiophora brassicae. Australasian Plant Pathology. 2013;42:141–53. 10.1007/s13313-012-0167-x
- 53.Villanueva RAM, Chen ZJ. ggplot2: elegant graphics for data analysis (2nd ed.). Measurement ( Mahwah N J). 2019;17:160–7. 10.1111/j.1467-985X.2010.00676_9.x
- 54.Kudjordjie EN, Sapkota R, Steffensen SK, Fomsgaard IS, Nicolaisen M. Maize synthesized benzoxazinoids affect the host associated microbiome. Microbiome. 2019;7:59. 10.1186/s40168-019-0677-7. 10.1186/s40168-019-0677-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCann HC, Guttman DS. Evolution of the type III secretion system and its effectors in plant–microbe interactions. New Phytol. 2008;177:33–47. 10.1111/j.1469-8137.2007.02293.x. 10.1111/j.1469-8137.2007.02293.x [DOI] [PubMed] [Google Scholar]
- 56.Vannier N, Agler M, Hacquard S. Microbiota-mediated disease resistance in plants. PLoS Pathog. 2019;15: e1007740. 10.1371/journal.ppat.1007740. 10.1371/journal.ppat.1007740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chialva M, Lanfranco L, Bonfante P. The plant microbiota: composition, functions, and engineering. Curr Opin Biotechnol. 2022;73:135–42. 10.1016/j.copbio.2021.07.003. 10.1016/j.copbio.2021.07.003 [DOI] [PubMed] [Google Scholar]
- 58.Simonin M, Dasilva C, Terzi V, Ngonkeu ELM, Diouf D, Kane A, et al. Influence of plant genotype and soil on the wheat rhizosphere microbiome: evidences for a core microbiome across eight African and European soils. FEMS Microbiol Ecol. 2020;96. 10.1093/femsec/fiaa067 [DOI] [PubMed]
- 59.Pérez-Jaramillo JE, Carrión VJ, Bosse M, Ferrão LFV, de Hollander M, Garcia AAF, et al. Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. ISME J. 2017;11:2244–57. 10.1038/ismej.2017.85. 10.1038/ismej.2017.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abedini D, Jaupitre S, Bouwmeester H, Dong L. Metabolic interactions in beneficial microbe recruitment by plants. Curr Opin Biotechnol. 2021;70:241–7. 10.1016/j.copbio.2021.06.015. 10.1016/j.copbio.2021.06.015 [DOI] [PubMed] [Google Scholar]
- 61.Carrión VJ, Perez-Jaramillo J, Cordovez V, Tracanna V, de Hollander M, Ruiz-Buck D, et al. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science. 1979;2019(366):606–12. 10.1126/science.aaw9285. 10.1126/science.aaw9285 [DOI] [PubMed] [Google Scholar]
- 62.Oberholster T, Vikram S, Cowan D, Valverde A. Key microbial taxa in the rhizosphere of sorghum and sunflower grown in crop rotation. Sci Total Environ. 2018;624:530–9. 10.1016/j.scitotenv.2017.12.170. 10.1016/j.scitotenv.2017.12.170 [DOI] [PubMed] [Google Scholar]
- 63.Furuya H, Tubaki K, Matsumoto T, Fuji S, Naito H. Deleterious effects of fungi isolated from paddy soils on seminal root of rice. J Gen Plant Pathol. 2005;71:333–9. 10.1007/s10327-005-0208-1. 10.1007/s10327-005-0208-1 [DOI] [Google Scholar]
- 64.Kwak M-J, Kong HG, Choi K, Kwon S-K, Song JY, Lee J, et al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat Biotechnol. 2018. 10.1038/nbt.4232. 10.1038/nbt.4232 [DOI] [PubMed] [Google Scholar]
- 65.Yin J, Zhang Z, Zhu C, Wang T, Wang R, Ruan L. Heritability of tomato rhizobacteria resistant to Ralstonia solanacearum. Microbiome. 2022;10:227. 10.1186/s40168-022-01413-w. 10.1186/s40168-022-01413-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Boer W, Wagenaar A-M, Klein Gunnewiek PJA, Van Veen JA. In vitro suppression of fungi caused by combinations of apparently non-antagonistic soil bacteria. FEMS Microbiol Ecol. 2007;59:177–85. 10.1111/j.1574-6941.2006.00197.x. 10.1111/j.1574-6941.2006.00197.x [DOI] [PubMed] [Google Scholar]
- 67.Eichorst SA, Kuske CR. Identification of cellulose-responsive bacterial and fungal communities in geographically and edaphically different soils by using stable isotope probing. Appl Environ Microbiol. 2012;78:2316–27. 10.1128/AEM.07313-11. 10.1128/AEM.07313-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khoury FY. Influence of Rhizoctonia solani on the susceptibility of cotton plants to Verticillium albo-atrum and on root carbohydrates. Phytopathology. 1973;63:352. 10.1094/Phyto-63-352 [DOI] [Google Scholar]
- 69.Hassani MA, Durán P, Hacquard S. Microbial interactions within the plant holobiont. Microbiome. 2018;6:58. 10.1186/s40168-018-0445-0. 10.1186/s40168-018-0445-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chepsergon J, Moleleki LN. Rhizosphere bacterial interactions and impact on plant health. Curr Opin Microbiol. 2023;73: 102297. 10.1016/j.mib.2023.102297. 10.1016/j.mib.2023.102297 [DOI] [PubMed] [Google Scholar]
- 71.Tao J, Meng D, Qin C, Liu X, Liang Y, Xiao Y, et al. Integrated network analysis reveals the importance of microbial interactions for maize growth. Appl Microbiol Biotechnol. 2018;102:3805–18. 10.1007/s00253-018-8837-4. 10.1007/s00253-018-8837-4 [DOI] [PubMed] [Google Scholar]
- 72.Qi G, Ma G, Chen S, Lin C, Zhao X. Microbial network and soil properties are changed in bacterial wilt-susceptible soil. Appl Environ Microbiol. 2019;85. 10.1128/AEM.00162-19 [DOI] [PMC free article] [PubMed]
- 73.Ma H, Pineda A, Hannula SE, Kielak AM, Setyarini SN, Bezemer TM. Steering root microbiomes of a commercial horticultural crop with plant-soil feedbacks. Appl Soil Ecol. 2020;150: 103468. 10.1016/j.apsoil.2019.103468. 10.1016/j.apsoil.2019.103468 [DOI] [Google Scholar]
- 74.Mougi A, Kondoh M. Diversity of interaction types and ecological community stability. Science. 1979;2012(337):349–51. 10.1126/science.1220529. 10.1126/science.1220529 [DOI] [PubMed] [Google Scholar]
- 75.Hernandez DJ, David AS, Menges ES, Searcy CA, Afkhami ME. Environmental stress destabilizes microbial networks. ISME J. 2021;15:1722–34. 10.1038/s41396-020-00882-x. 10.1038/s41396-020-00882-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shaw GT-W, Liu A-C, Weng C-Y, Chen Y-C, Chen C-Y, Weng FC-H, et al. A network-based approach to deciphering a dynamic microbiome’s response to a subtle perturbation. Sci Rep. 2020;10:19530. 10.1038/s41598-020-73920-5 [DOI] [PMC free article] [PubMed]
- 77.Wei Z, Yang T, Friman V-P, Xu Y, Shen Q, Jousset A. Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat Commun. 2015;6:8413. 10.1038/ncomms9413. 10.1038/ncomms9413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Agler MT, Ruhe J, Kroll S, Morhenn C, Kim S-T, Weigel D, et al. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016;14: e1002352. 10.1371/journal.pbio.1002352. 10.1371/journal.pbio.1002352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brachi B, Filiault D, Whitehurst H, Darme P, Le Gars P, Le Mentec M, et al. Plant genetic effects on microbial hubs impact host fitness in repeated field trials. Proceedings of the National Academy of Sciences. 2022;119. 10.1073/pnas.2201285119 [DOI] [PMC free article] [PubMed]
- 80.Shi Y, Delgado-Baquerizo M, Li Y, Yang Y, Zhu Y-G, Peñuelas J, et al. Abundance of kinless hubs within soil microbial networks are associated with high functional potential in agricultural ecosystems. Environ Int. 2020;142: 105869. 10.1016/j.envint.2020.105869. 10.1016/j.envint.2020.105869 [DOI] [PubMed] [Google Scholar]
- 81.Berendsen RL, Pieterse CMJ, Bakker PAHM. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–86. 10.1016/j.tplants.2012.04.001. 10.1016/j.tplants.2012.04.001 [DOI] [PubMed] [Google Scholar]
- 82.Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, Werck-Reichhart D, et al. Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell. 2010;22:973–90. 10.1105/tpc.109.069658. 10.1105/tpc.109.069658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chauhan V, Mazumdar S, Pandey A, Kanwar SS. Pseudomonas lipopeptide: an excellent biomedical agent. MedComm – Biomaterials and Applications. 2023;2. 10.1002/mba2.27
- 84.Stepanov AA, Poshvina DV, Vasilchenko AS. 2,4-Diacetylphloroglucinol modulates Candida albicans virulence. Journal of Fungi. 2022;8:1018. 10.3390/jof8101018. 10.3390/jof8101018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, Bakker PAHM. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol. 2014;52:347–75. 10.1146/annurev-phyto-082712-102340. 10.1146/annurev-phyto-082712-102340 [DOI] [PubMed] [Google Scholar]
- 86.Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh-Kumar SP. A novel role for the tir domain in association with pathogen-derived elicitors. PLoS Biol. 2007;5: e68. 10.1371/journal.pbio.0050068. 10.1371/journal.pbio.0050068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM. Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol. 2012;28:489–521. 10.1146/annurev-cellbio-092910-154055. 10.1146/annurev-cellbio-092910-154055 [DOI] [PubMed] [Google Scholar]
- 88.Ramamoorthy R, Jiang S-Y, Kumar N, Venkatesh PN, Ramachandran S. A Comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 2008;49:865–79. 10.1093/pcp/pcn061. 10.1093/pcp/pcn061 [DOI] [PubMed] [Google Scholar]
- 89.van Loon LC, Rep M, Pieterse CMJ. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol. 2006;44:135–62. 10.1146/annurev.phyto.44.070505.143425. 10.1146/annurev.phyto.44.070505.143425 [DOI] [PubMed] [Google Scholar]
- 90.Zhang S, Klessig DF. MAPK cascades in plant defense signaling. Trends Plant Sci. 2001;6:520–7. 10.1016/S1360-1385(01)02103-3. 10.1016/S1360-1385(01)02103-3 [DOI] [PubMed] [Google Scholar]
- 91.Zhou L, Deng S, Xuan H, Fan X, Sun R, Zhao J, et al. A novel TIR-NBS-LRR gene regulates immune response to Phytophthora root rot in soybean. Crop J. 2022;10:1644–53. 10.1016/j.cj.2022.03.003. 10.1016/j.cj.2022.03.003 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Primer sequences used in this study.
Additional file 2: Figure S1. Alpha rarefaction plots for bacteri and fungi were generated at the species level using the “observed number of zOTUs”
Additional file 3: Table S2. Metadata, zOTU representative sequences, taxonomy annotation and zOTU table.
Additional file 4: Table S3. Alpha diversity estimators of samples.
Additional file 5: Table S4. PERMANOVA by adonis of all bacterial 16S and fungal ITS samples.
Additional file 6: Table S5. Differential abundance of zOTUs between YR and ZG.
Additional file 7: Figure S2. Relative abundance of major bacterial (a) and fungal phyla (b) in the soil samples.
Additional file 8: Table S6. Topology properties of the intra- and inter-kingdom networks.
Additional file 9: Figure S3. Inter-kingdom co-occurrence networks from rhizosphere structural communities of YR and ZG in control and Foc-inoculated plants.
Additional file 10: Figure S4. Alignment of the 16S rRNA sequences of the isolate NA13 and zOTU_1647.
Additional file 11: Figure S5. Alignment of the genome sequences of the isolate NA13 with three identified secondary metabolite regions (a) BGC0000437 (Syringomycin), (b) BGC0002345 (hydrogen cyanide) and (c) BGC0000281 (2,4-Diacetylphloroglucinol).
Additional file 12: Table S7. Secondary metabolism gene cluster types in the genome of NA13.
Additional file 13: Figure S6. Clusters of Orthologous Group annotations for the Pseudomonas brassicacearum NA13 genome.
Additional file 14: Table S8. List of genes encoding proteins involved in bacterial flagellin, chitin deacetylase, chitin, and Type I to Type V secretion systems.
Data Availability Statement
The sequences for 16S rRNA gene, ITS, and bacterium NA13 genome are deposited in the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/) under the accession number PRJNA908139, PRJNA908211, and SAMN38604125, respectively. All other data are presented directly in the manuscript or provided in the supplementary files attached to this submission.