Abstract
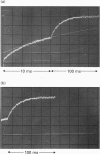
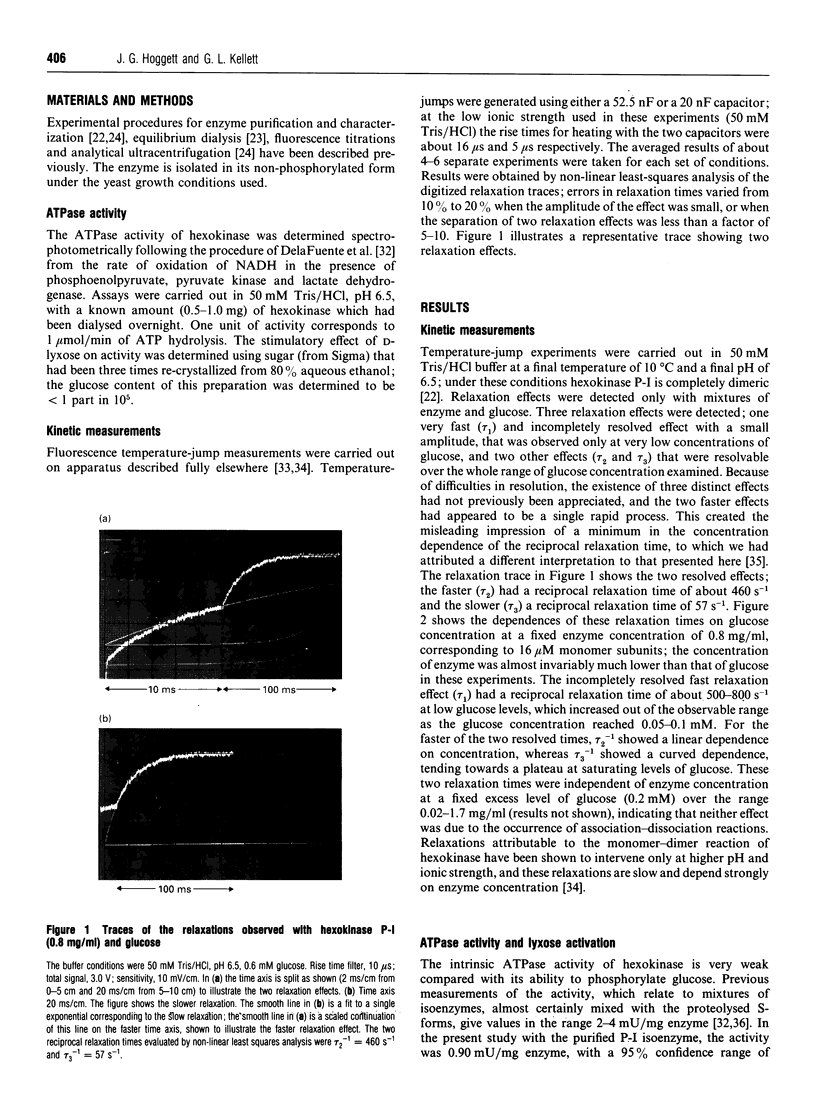
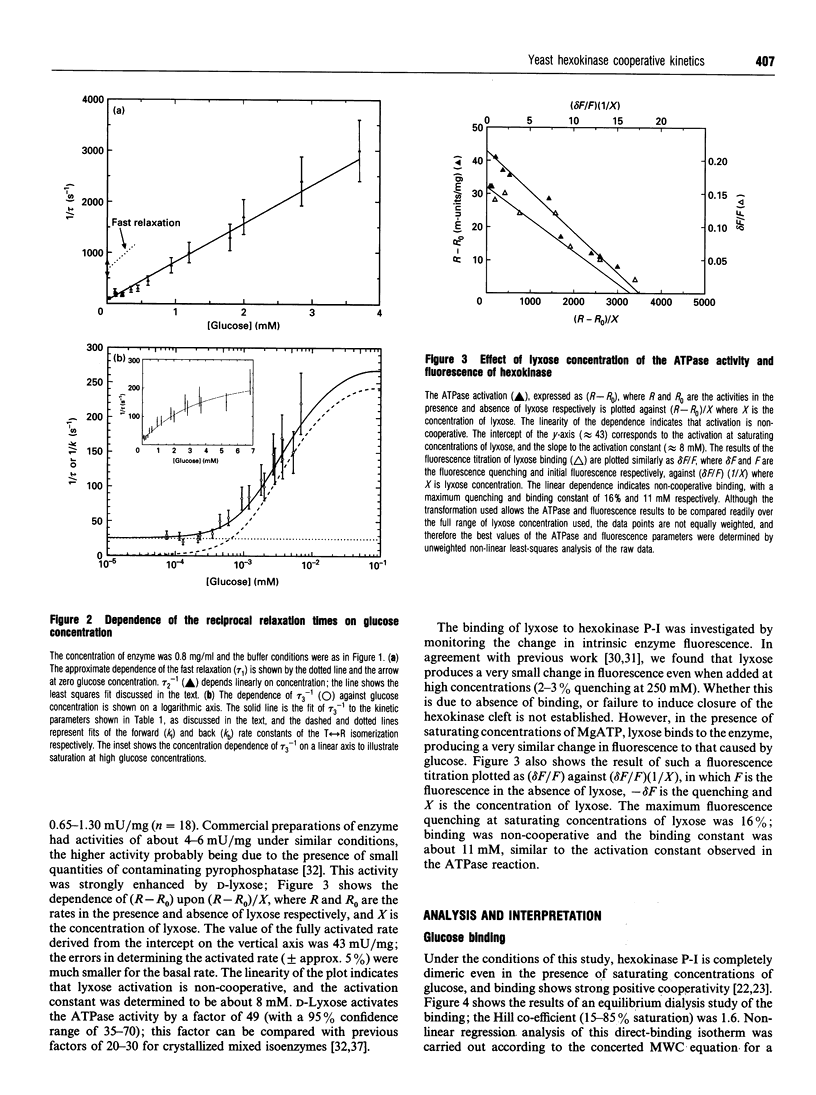
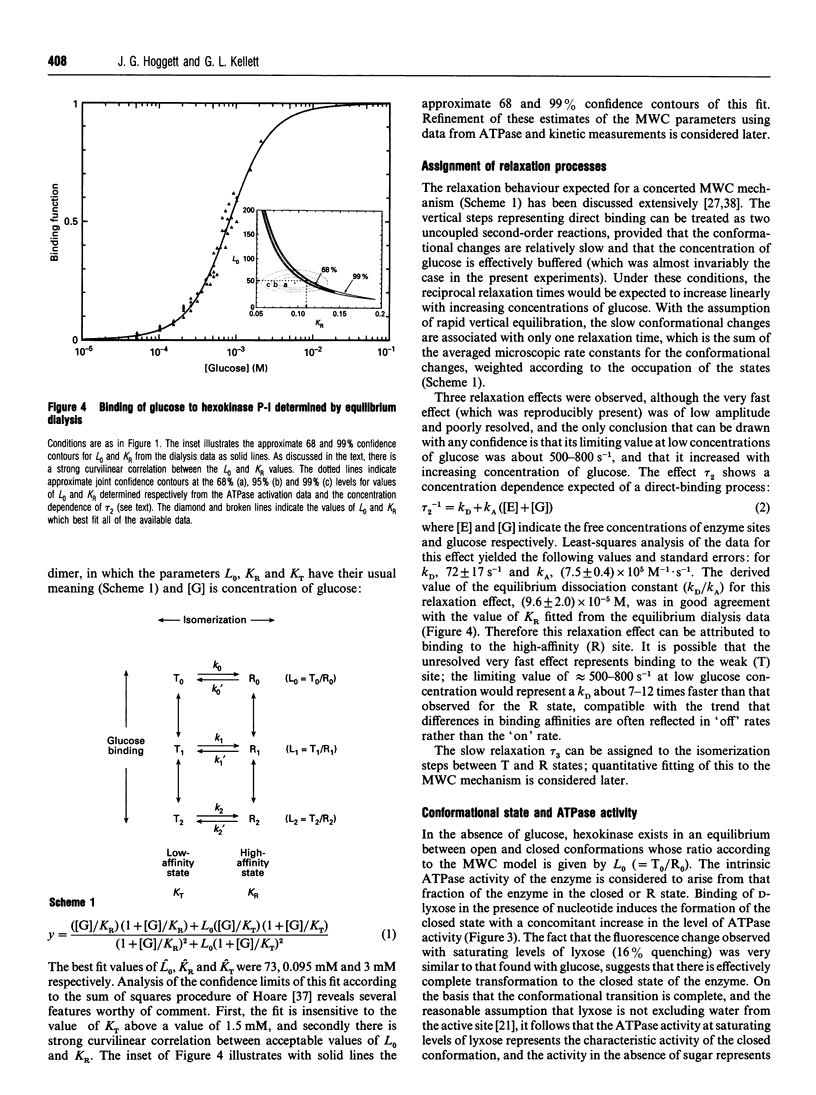
Kinetic studies of the cooperative binding of glucose to yeast hexokinase P-I at pH 6.5 have been carried out using the fluorescence temperature-jump technique. Three relaxation effects were observed: a fast low-amplitude effect which could only be resolved at low glucose concentrations (tau 1(-1) = 500-800 s-1), an intermediate effect (tau 2) which showed a linear dependence of reciprocal relaxation time on concentration, and a slow effect (tau 3) which showed a curved dependence on glucose concentration, increasing from approximately 28 s-1 at low concentrations to 250 s-1 at high levels. The findings are interpreted in terms of the concerted Monod-Wyman-Changeux mechanism, the two faster relaxations being assigned to binding to the R and T states, and the slow relaxation to isomerization between the states. Quantitative fitting of the kinetic data to the mechanism has been carried out using independent estimates of the equilibrium parameters of the model; these have been derived from equilibrium dialysis data and by determining the enhancement of the intrinsic ATPase activity of the enzyme by the non-phosphorylatable sugar lyxose, which switches the conformation of the enzyme to the active R state.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson C. M., Stenkamp R. E., McDonald R. C., Steitz T. A. A refined model of the sugar binding site of yeast hexokinase B. J Mol Biol. 1978 Aug 5;123(2):207–219. doi: 10.1016/0022-2836(78)90321-2. [DOI] [PubMed] [Google Scholar]
- Anderson C. M., Stenkamp R. E., Steitz T. A. Sequencing a protein by x-ray crystallography. II. Refinement of yeast hexokinase B co-ordinates and sequence at 2.1 A resolution. J Mol Biol. 1978 Jul 25;123(1):15–33. doi: 10.1016/0022-2836(78)90374-1. [DOI] [PubMed] [Google Scholar]
- Bennett W. S., Jr, Steitz T. A. Glucose-induced conformational change in yeast hexokinase. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4848–4852. doi: 10.1073/pnas.75.10.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett W. S., Jr, Steitz T. A. Structure of a complex between yeast hexokinase A and glucose. II. Detailed comparisons of conformation and active site configuration with the native hexokinase B monomer and dimer. J Mol Biol. 1980 Jun 25;140(2):211–230. doi: 10.1016/0022-2836(80)90103-5. [DOI] [PubMed] [Google Scholar]
- Blázquez M. A., Lagunas R., Gancedo C., Gancedo J. M. Trehalose-6-phosphate, a new regulator of yeast glycolysis that inhibits hexokinases. FEBS Lett. 1993 Aug 23;329(1-2):51–54. doi: 10.1016/0014-5793(93)80191-v. [DOI] [PubMed] [Google Scholar]
- DelaFuente G., Lagunas R., Sols A. Induced fit in yeast hexokinase. Eur J Biochem. 1970 Oct;16(2):226–233. doi: 10.1111/j.1432-1033.1970.tb01075.x. [DOI] [PubMed] [Google Scholar]
- Derechin M., Rustum Y. M., Barnard E. A. Dissociation of yeast hexokinase under the influence of substrates. Biochemistry. 1972 May 9;11(10):1793–1797. doi: 10.1021/bi00760a009. [DOI] [PubMed] [Google Scholar]
- Easterby J. S., Rosemeyer M. A. Purification and subunit interactions of yeast hexokinase. Eur J Biochem. 1972 Jul 13;28(2):241–252. doi: 10.1111/j.1432-1033.1972.tb01907.x. [DOI] [PubMed] [Google Scholar]
- Feldman I., Kramp D. C. Fluorescence-quenching study of glucose binding by yeast hexokinase isoenzymes. Biochemistry. 1978 Apr 18;17(8):1541–1547. doi: 10.1021/bi00601a029. [DOI] [PubMed] [Google Scholar]
- Fröhlich K. U., Entian K. D., Mecke D. The primary structure of the yeast hexokinase PII gene (HXK2) which is responsible for glucose repression. Gene. 1985;36(1-2):105–111. doi: 10.1016/0378-1119(85)90074-5. [DOI] [PubMed] [Google Scholar]
- Hoare D. G. A curve-fitting procedure utilizing a small computer for equations containing three or four unknown constants. Anal Biochem. 1972 Apr;46(2):604–615. doi: 10.1016/0003-2697(72)90332-6. [DOI] [PubMed] [Google Scholar]
- Hoggett J. G., Kellett G. L. Kinetics of the monomer-dimer reaction of yeast hexokinase PI. Biochem J. 1992 Oct 15;287(Pt 2):567–572. doi: 10.1042/bj2870567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggett J. G., Kellett G. L., Tickner E. L. A comparison of the binding of glucose of dimeric yeast hexokinase P-I and P-II isoenzymes [proceedings]. Biochem Soc Trans. 1977;5(3):776–778. doi: 10.1042/bst0050776. [DOI] [PubMed] [Google Scholar]
- Hoggett J. G., Kellett G. L. Yeast hexokinase. A fluorescence temperature-jump study of the kinetics of the binding of glucose to the monomer forms of hexokinases P-I and P-II. Eur J Biochem. 1976 Sep 15;68(2):347–353. doi: 10.1111/j.1432-1033.1976.tb10821.x. [DOI] [PubMed] [Google Scholar]
- Hoggett J. G., Kellett G. L. Yeast hexokinase: substrate-induced association--dissociation reactions in the binding of glucose to hexokinase P-II. Eur J Biochem. 1976 Jun 15;66(1):65–77. doi: 10.1111/j.1432-1033.1976.tb10426.x. [DOI] [PubMed] [Google Scholar]
- Hoggett J. G., Kellett G. L. Yeast hexokinase: substrate-induced association--dissociation reactions in the binding of glucose to hexokinase P-II. Eur J Biochem. 1976 Jun 15;66(1):65–77. doi: 10.1111/j.1432-1033.1976.tb10426.x. [DOI] [PubMed] [Google Scholar]
- Jentoft J. E., Neet K. E., Stuehr J. E. Relaxation spectra of yeast hexokinases. Isomerization of the enzyme. Biochemistry. 1977 Jan 11;16(1):117–121. doi: 10.1021/bi00620a019. [DOI] [PubMed] [Google Scholar]
- Kirschner K., Gallego E., Schuster I., Goodall D. Co-operative binding of nicotinamide-adenine dinucleotide to yeast glyceraldehyde-3-phosphate dehydrogenase. I. Equilibrium and temperature-jump studies at pH 8-5 and 40 degrees C. J Mol Biol. 1971 May 28;58(1):29–50. doi: 10.1016/0022-2836(71)90230-0. [DOI] [PubMed] [Google Scholar]
- Koshland D. E. Application of a Theory of Enzyme Specificity to Protein Synthesis. Proc Natl Acad Sci U S A. 1958 Feb;44(2):98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D. E., Jr, Némethy G., Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966 Jan;5(1):365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- Kriegel T. M., Rush J., Vojtek A. B., Clifton D., Fraenkel D. G. In vivo phosphorylation site of hexokinase 2 in Saccharomyces cerevisiae. Biochemistry. 1994 Jan 11;33(1):148–152. doi: 10.1021/bi00167a019. [DOI] [PubMed] [Google Scholar]
- MONOD J., WYMAN J., CHANGEUX J. P. ON THE NATURE OF ALLOSTERIC TRANSITIONS: A PLAUSIBLE MODEL. J Mol Biol. 1965 May;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Mayes E. L., Hoggett J. G., Kellett G. L. The binding of glucose to native and proteolytically modified yeast hexokinase PI. Eur J Biochem. 1983 Jun 1;133(1):127–134. doi: 10.1111/j.1432-1033.1983.tb07437.x. [DOI] [PubMed] [Google Scholar]
- Ohning G. V., Neet K. E. 6-(p-toluidinyl)naphthalene-2-sulfonic acid as a fluorescent probe of yeast hexokinase: conformational states induced by sugar and nucleotide ligands. Biochemistry. 1983 Jun 7;22(12):2986–2995. doi: 10.1021/bi00281a031. [DOI] [PubMed] [Google Scholar]
- Olsen L. R., Reed G. H. The structure of the MnIIADP-nitrate-lyxose complex at the active site of hexokinase. Arch Biochem Biophys. 1993 Jul;304(1):242–247. doi: 10.1006/abbi.1993.1345. [DOI] [PubMed] [Google Scholar]
- Peters B. A., Neet K. E. Yeast hexokinase PII. Conformational changes induced by substrates and substrate analogues. J Biol Chem. 1978 Oct 10;253(19):6826–6831. [PubMed] [Google Scholar]
- Rose M., Albig W., Entian K. D. Glucose repression in Saccharomyces cerevisiae is directly associated with hexose phosphorylation by hexokinases PI and PII. Eur J Biochem. 1991 Aug 1;199(3):511–518. doi: 10.1111/j.1432-1033.1991.tb16149.x. [DOI] [PubMed] [Google Scholar]
- Rossi A. Is hysteresis of yeast hexokinase A due to isomerization or dissociation of the enzyme molecule? Braz J Med Biol Res. 1993 Jul;26(7):673–675. [PubMed] [Google Scholar]
- Rudolph F. B., Fromm H. J. A study on the kinetics and mechanism of D-lyxose and D-xylose activation of the adenosine triphosphatase activity associated with yeast hexokinase. J Biol Chem. 1971 Apr 10;246(7):2104–2110. [PubMed] [Google Scholar]
- Schmidt J. J., Colowick S. P. Chemistry and subunit structure of yeast hexokinase isoenzymes. Arch Biochem Biophys. 1973 Oct;158(2):458–470. doi: 10.1016/0003-9861(73)90537-7. [DOI] [PubMed] [Google Scholar]
- Schulze I. T., Colowick S. P. The modification of yeast hexokinases by proteases and its relationship to the dissociation of hexokinase into subunits. J Biol Chem. 1969 May 10;244(9):2306–2316. [PubMed] [Google Scholar]
- Shoham M., Steitz T. A. The 6-hydroxymethyl group of a hexose is essential for the substrate-induced closure of the cleft in hexokinase. Biochim Biophys Acta. 1982 Aug 10;705(3):380–384. doi: 10.1016/0167-4838(82)90260-6. [DOI] [PubMed] [Google Scholar]
- Stachelek C., Stachelek J., Swan J., Botstein D., Konigsberg W. Identification, cloning and sequence determination of the genes specifying hexokinase A and B from yeast. Nucleic Acids Res. 1986 Jan 24;14(2):945–963. doi: 10.1093/nar/14.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A., Anderson W. F., Fletterick R. J., Anderson C. M. High resolution crystal structures of yeast hexokinase complexes with substrates, activators, and inhibitors. Evidence for an allosteric control site. J Biol Chem. 1977 Jul 10;252(13):4494–4500. [PubMed] [Google Scholar]
- Tickner E. L., Hoggett J. G., Kellett G. L. The cooperative binding of glucose to yeast hexokinase PI dimer. Biochem Biophys Res Commun. 1976 Oct 4;72(3):808–815. doi: 10.1016/s0006-291x(76)80205-7. [DOI] [PubMed] [Google Scholar]
- Vojtek A. B., Fraenkel D. G. Phosphorylation of yeast hexokinases. Eur J Biochem. 1990 Jun 20;190(2):371–375. doi: 10.1111/j.1432-1033.1990.tb15585.x. [DOI] [PubMed] [Google Scholar]
- Walsh R. B., Clifton D., Horak J., Fraenkel D. G. Saccharomyces cerevisiae null mutants in glucose phosphorylation: metabolism and invertase expression. Genetics. 1991 Jul;128(3):521–527. doi: 10.1093/genetics/128.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womack F. C., Colowick S. P. Proton-dependent inhibition of yeast and brain hexokinases by aluminum in ATP preparations. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5080–5084. doi: 10.1073/pnas.76.10.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]