Abstract
Background/Aim
The liver effectively restores both size and function following partial hepatectomy (PHx). Angiogenesis is crucial for the repair and regeneration of liver tissue post-PHx. Calcitonin gene-related peptide (CGRP) released from sensory nerves and its receptor—receptor activity-modifying protein 1 (RAMP1) are involved in angiogenesis. This study aimed to assess the role of RAMP1 signaling in angiogenesis during liver regeneration following PHx.
Materials and Methods
RAMP1 deficient (RAMP1–/–) and wild-type (WT) mice were subjected to PHx.
Results
RAMP1–/– mice demonstrated delayed liver regeneration, indicated by lower liver-to-body weight ratios compared to WT mice. This was associated with lower levels of Ki67+ hepatocytes and hepatic trophic growth factors. Additionally, RAMP1–/– mice exhibited lower levels of endothelial cell markers, including CD31, compared to WT mice. This reduction was associated with reduced levels of vascular endothelial growth factor (VEGF)-C, VEGF-D, and VEGF receptor 3 (VEGFR3). In WT mice with PHx, the administration of a VEGFR3 inhibitor reduced the liver-to-body weight ratio, Ki67+ hepatocytes, and VEGF-C/VEGFR3 expression levels in the liver compared to those in the vehicle-treated group.
Conclusion
The deletion of RAMP1 signaling suppresses liver regeneration and angiogenesis through VEGFR3. Specific activation of RAMP1 signaling may represent a potential therapeutic strategy for liver regeneration following PHx.
Keywords: Liver regeneration, RAMP1, angiogenesis
Surgical resection is the preferred treatment for liver tumors. Following partial hepatectomy (PHx), liver tissue regeneration occurs through extensive hepatocyte proliferation in the remaining liver lobe, which restores liver function and quality. Therefore, surgical resection of the liver relies on the regenerative potential of the remnant liver. Although this regenerative potential is crucial for patient survival (1), inadequate regeneration and restoration of liver remnants can result in post-hepatectomy liver failure, resulting in high morbidity and mortality in patients who undergo PHx (2,3).
To address the insufficiency of liver regeneration following PHx, it is crucial to understand the fundamental regulatory mechanisms involved (1). Post-PHx liver regeneration proceeds through three distinct phases: initiation, proliferation (progression), and termination (4). Assessments of post-PHx regenerative processes have primarily focused on the regulatory mechanisms of liver regeneration during the initiation and proliferation phases. The initial phase (inductive or initiative), occurring 0-3 days post-PHx, is characterized by extensive hepatocyte proliferation. The progression phase, spanning 4-8 days, involves both hepatocyte proliferation and the restoration of hepatic sinusoids through liver sinusoidal endothelial cell (LSEC) proliferation, which is known as angiogenesis (5,6). Hepatic angiogenesis or vascular reconstitution, which is driven by LSEC proliferation, plays a crucial role in liver regeneration post-PHx (7) and liver repair and restoration following acute liver damage from chemicals (8) and ischemia/reperfusion (9). Therefore, it is indispensable for liver regeneration post-PHx. However, the mechanisms underlying PHx-induced angiogenesis remain unclear.
The liver receives innervation from autonomic and sensory nerve fibers, which enter through the hilum and form plexuses along the portal vein and hepatic artery (10). The hepatic nervous system regulates liver function and regeneration and produces several neuropeptides, including calcitonin gene-related peptide (CGRP), a 37-amino acid neuropeptide (11). The action of CGRP is mediated by its receptor, which is composed of two heterodimer subunits: calcitonin receptor-like receptor (CLR) and receptor activity-modifying protein (RAMP) 1. RAMP1 regulates the specific binding of CGRP to CLR (11).
Accumulating evidence indicates that CGRP/RAMP1 signaling is closely associated with the immune system (12,13). CGRP-deficient mice exhibit aggravated concanavalin A (ConA)-induced acute liver injury by affecting T cell function (14). Additionally, RAMP1 signaling in resident liver macrophages (Kupffer cells) contributes to the mitigation of ConA-induced hepatitis (15). Moreover, CGRP/RAMP1 signaling may interact with regenerating hepatocytes in the PHx-livers. The deletion of RAMP1 signaling delays liver mass recovery and hepatocyte growth post-PHx (16). The activation of CGRP in isolated hepatocytes accelerates their proliferation.
Additionally, the CGRP/RAMP1 signaling pathway stimulates the production of pro-angiogenic factors, thereby facilitating angiogenesis. CGRP/RAMP1 signaling is crucial for angiogenesis in mice with tumor growth (17), skin wounds (18), and endometriosis (19). These findings hypothesized that RAMP1 signaling plays a crucial role in liver regeneration by facilitating angiogenesis. Therefore, this study aimed to assess the potential involvement of RAMP1 signaling in liver regeneration and angiogenesis post-PHx in mice.
Materials and Methods
Animals. Male RAMP1-deficient (RAMP1–/–) mice were generated as described previously (12). Male wild-type (WT) C57BL/6 mice (8-weeks-old, 20-23 g) were obtained from CLEA Japan (Tokyo, Japan). The mice were housed under sterile and temperature- and humidity-controlled environments with a 12-h light/dark cycle. All the animals had free access to food and water. All experimental protocols were reviewed and approved by the Institutional Animal Care and Use Committee (Approval no. 2023-061) and conducted following the guidelines established by the Science Council of Japan for animal experiments.
Animal procedures. An established mouse model of PHx (70%) was used as previously described (20). The mice were anesthetized through an intraperitoneal (i.p.) injection of mixed anesthetic agents containing 4.0 mg/kg of midazolam (Sandoz, a Novartis division, Basel, Switzerland), 0.75 mg/kg of medetomidine hydrochloride (Nippon Zenyaku Kogyo, Fukushima, Japan), and 5.0 mg/kg of butorphanol (Meiji Seika Pharma, Tokyo, Japan). A midline laparotomy was performed, and 4-0 vicryl sutures were applied around the base of the median and left lateral hepatic lobes, which were subsequently excised. Following surgery, the effects of medetomidine were reversed by administering an i.p. injection of atipamezole (0.75 mg/kg; Nippon Zenyaku Kogyo).
Experimental protocols. Under anesthesia with a midazolam-medetomidine-butorphanol mixture, the livers were excised at specified times post-PHx. A portion of each liver sample was placed in 10% formaldehyde and the rest was prepared for quantitative PCR analysis. Following this procedure, animals were euthanized through cervical dislocation.
In another set of experiments, animals were administered either the specific VEGFR3 kinase inhibitor SAR131675 (Selleck Chemicals, Houston, TX, USA) or vehicle, as described previously (21). SAR131675 (30 mg/kg) was dissolved in 0.2 ml phosphate-buffered saline (PBS) containing 1% dimethyl sulfoxide (DMSO) and administered subcutaneously daily for the first five days post-surgery.
Immunohistochemical staining. The excised liver tissues were immediately fixed in buffered 10% formaldehyde for histological analysis. Sections (4 μm thick) were prepared from paraffin-embedded tissues and subjected to immunostaining. For immunohistochemical staining, 4-μm thick paraffin sections were treated with Protein Block Serum-Free (Dako, Glostrup, Denmark), and subsequently incubated overnight at 4˚C with anti-mouse Ki67 (rabbit monoclonal antibody, 1:500; GeneTex, Irvine, CA, USA) antibodies. After washing in PBS, the sections were incubated with Mayer’s hematoxylin solution and universal Dako EnVision+ Dual Link System-HRP (DAB+) (Dako). Images of stained liver sections were captured using a microscope (Biozero BZ-700 Series; Keyence, Osaka, Japan). The number of Ki67+ hepatocytes was counted in five fields (200×) per animal using the ImageJ software, and the results were expressed as the percentage of Ki67+ hepatocytes.
Immunofluorescence analysis. Liver tissues were fixed in 4% periodate-lysine-paraformaldehyde overnight at 4˚C and subsequently incubated in 30% sucrose in 0.1 M phosphate buffer at 4˚C for three days. The tissues were embedded in Tissue-Tek optimal cutting temperature (O.C.T.) compound (Sakura Finetek, Torrance, CA, USA) and stored at –20˚C. Liver tissue sections were cut and blocked with 1% bovine serum albumin (BSA) in 0.5% Triton X-100 in PBS. The sections were incubated overnight at 4˚C with the following primary antibodies: anti-mouse CGRP (rabbit polyclonal antibody, 1:100; Sigma-Aldrich, St. Louis, MO, USA), anti-mouse lymphatic vessel endothelial hyaluronan receptor (LYVE)-1 (goat polyclonal antibody, 1:100; R&D Systems, Minneapolis, MN, USA), and anti-mouse VEGFR3 (rat polyclonal antibody, 1:100; R&D Systems). After washing with PBS, the sections were incubated at 4˚C overnight with the following secondary antibodies: Alexa Fluor 488-conjugated donkey anti-rabbit IgG, Alexa Fluor 488-conjugated donkey anti-rat IgG, Alexa Fluor 594-conjugated donkey anti-rabbit IgG, and Alexa Fluor 594-conjugated donkey anti-goat IgG (all from Molecular Probes, Eugene, OR, USA). Nuclei were detected using 4’,6-diamidino-2-phenylindole (DAPI). All images were obtained using a fluorescence microscope (Biozero BZ-700 Series; KEYENCE, Osaka, Japan).
Isolation of intrahepatic leukocytes. To effectively isolate intrahepatic leukocytes, the liver was perfused with Hank’s balanced salt solution through the portal vein under anesthesia. Subsequently, the excised livers were incubated in Roswell Park Memorial Institute (RPMI) medium containing 0.05% collagenase (Type IV; Sigma Chemical Co., St. Louis, MO, USA) for 20 min at 37˚C. The liver homogenates were filtered through a 70 μm cell strainer, and non-parenchymal cells were purified using density-gradient centrifugation on 33% Percoll (GE Healthcare Life Sciences, Piscataway, NJ, USA), as previously reported (22).
Flow cytometric analyses. Isolated non-parenchymal cells were incubated with an anti-mouse CD16/32 antibody (BioLegend, San Diego, CA, USA) to prevent non-specific binding. The cells were stained with a combination of the following reagents: Brilliant Violet 421-conjugated anti-mouse CD45 (30-F11) (BioLegend), PE/Cy7-conjugated anti-mouse CD31 (390) (BioLegend), and Alexa 488 anti-mouse LYVE-1 (ALY7) (Invitrogen, Carlsbad, CA, USA). For flow cytometric analysis, cells were initially gated on forward and side scatters, followed by gating on CD45– cells. Cells positive for 7-aminoactinomycin D (BioLegend) were excluded from the analysis. Samples were analyzed using a fluorescence-activated cell sorting (FACS) Verse cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Data were analyzed using Kaluza software v2.1 (Beckman Coulter, Brea, CA, USA).
Real-time quantitative (RT-PCR). Total RNA was extracted from the liver tissues and homogenized using RNAiso Plus (Takara Bio, Shiga, Japan). Single-stranded cDNA was synthesized from 1 μg of total RNA through reverse transcription using the ReverTra Ace qPCR RT kit (Toyobo, Osaka, Japan). Quantitative PCR was performed using TB Green Premix Ex Taq II (Tli RNaseH Plus; Takara Bio). The gene-specific primers used in these experiments are listed in Table I. The data were normalized to the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels within the same sample.
Table I. Primers used for reverse transcription and quantitative PCR.
Statistical analyses. All results are presented as the mean±standard deviation (SD). All statistical analyses were performed using GraphPad Prism version 8 (GraphPad Software, La Jolla, CA, USA). Data were compared between two groups using an unpaired two-tailed Student’s t-test and between multiple groups using one-way analysis of variance (ANOVA), followed by Tukey’s post-hoc test. Statistical significance was set at p<0.05.
Results
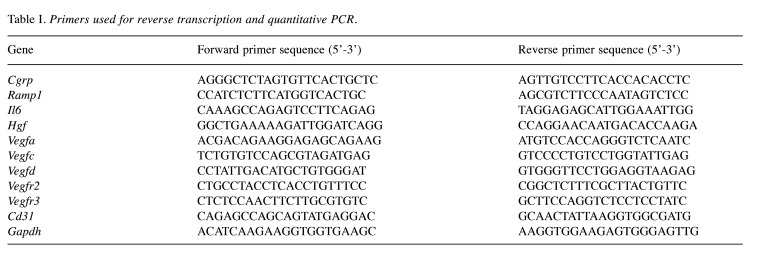
Deletion of RAMP1 signaling suppressed liver regeneration post-PHx. To determine the role of RAMP1 signaling in liver regeneration, WT and RAMP1–/– mice were subjected to PHx. Compared with the WT mice, RAMP1–/– mice exhibited a lower liver/body weight ratio on days 5 and 7 post-PHx (Figure 1A). Because hepatocyte proliferation is primarily responsible for the recovery of liver mass post-PHx, we determined the number of Ki67+ hepatocytes, a marker of cellular proliferation (Figure 1B). As illustrated in Figure 1B and C, the number of Ki67+ hepatocytes in the liver on days 2, 3, 5, and 7 was lower in RAMP1–/– mice than that in WT mice. Additionally, we measured the levels of hepatic trophic growth factors, including interleukin (IL)-6 and hepatocyte growth factor (HGF). The mRNA levels of IL-6 in RAMP1–/– mice on days 1 and 2 were lower than those in WT mice (Figure 1C). Additionally, HGF mRNA levels in RAMP1–/– mice on day 3 were lower than those in WT mice (Figure 1D). These results indicate that deficiency in RAMP1 signaling impairs liver regeneration post-PHx.
Figure 1. Impaired liver regeneration post-partial hepatectomy (PHx) in receptor activity-modifying protein 1 (RAMP1)–/– mice. (A) Liver-to-body weight ratio in wild type (WT) and RAMP1–/– mice post-PHx. Data are expressed as the mean±standard deviation (SD) (n=5-7 mice per group). *p<0.05. (B) Percentage of Ki67+ hepatocytes in the livers of WT and RAMP1–/– mice post-PHx. Data are expressed as the mean±SD (n=4-5 mice per group). *p<0.05. (C) Representative images of livers immunostained for Ki67 in WT and RAMP1–/– mice post-PHx. Scale bars indicate 100 μm. (D) Interleukin (IL)-6 and hepatocyte growth factor (HGF) mRNA levels in the livers of WT and RAMP1–/– mice post-PHx. Data are expressed as the mean±SD (n=4-7 mice per group). *p<0.05.
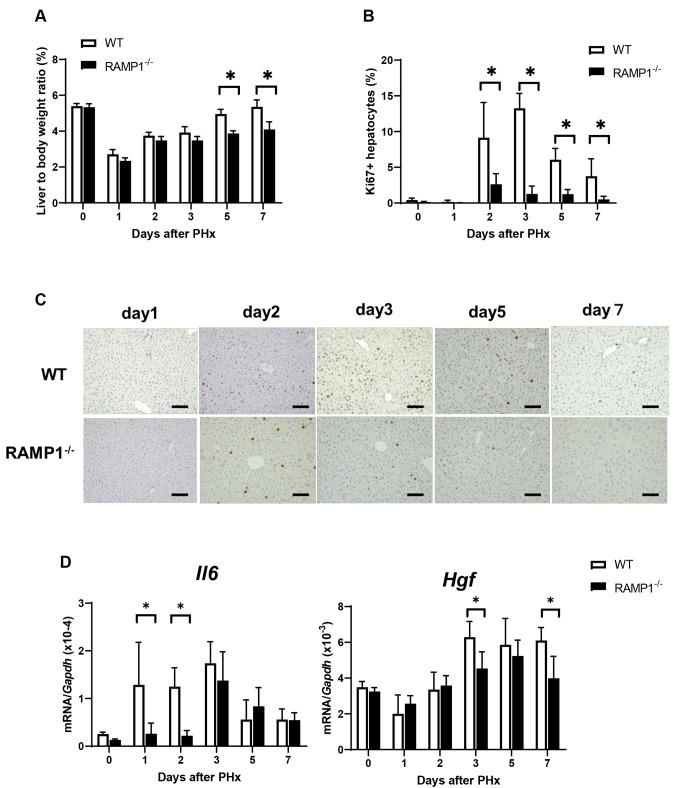
Expression of CGRP and RAMP1. The aforementioned results indicate that RAMP1 signaling is involved in hepatocyte proliferation during liver regeneration. Therefore, we assessed the expression levels of CGRP and RAMP1 in the liver. Immunofluorescence analysis revealed that CGRP nerve fibers were distributed around the portal and lymphatic vessels in the portal area of the WT mice (Figure 2A). The mRNA levels of Cgrp on days 3 and 7 in WT mice were higher than those in RAMP1–/– mice (Figure 2B). As expected, no expression of Ramp1 was observed in RAMP1–/– mice. The mRNA levels of Ramp1 in WT mice were transiently reduced on day 1, followed by an increase on days 5 and 7 (Figure 2B).
Figure 2. Hepatic expression of calcitonin gene-related peptide (CGRP) and receptor activity-modifying protein 1 (RAMP1) during liver regeneration post-partial hepatectomy (PHx). (A) Immunofluorescence staining for CGRP (green) and LYVE-1 (red) in the livers of WT mice on day 0. Cell nuclei are stained with 4’,6-diamidino-2-phenylindole (DAPI) (blue). PV, portal vein. Scale bar: 100 μm. (B) Cgrp and Ramp1 mRNA levels in the livers of WT and RAMP1–/– mice post-PHx. Data are expressed as the mean±standard deviation (SD) (n=5-6 mice per group). *p<0.05, #p<0.05 vs. day 0.
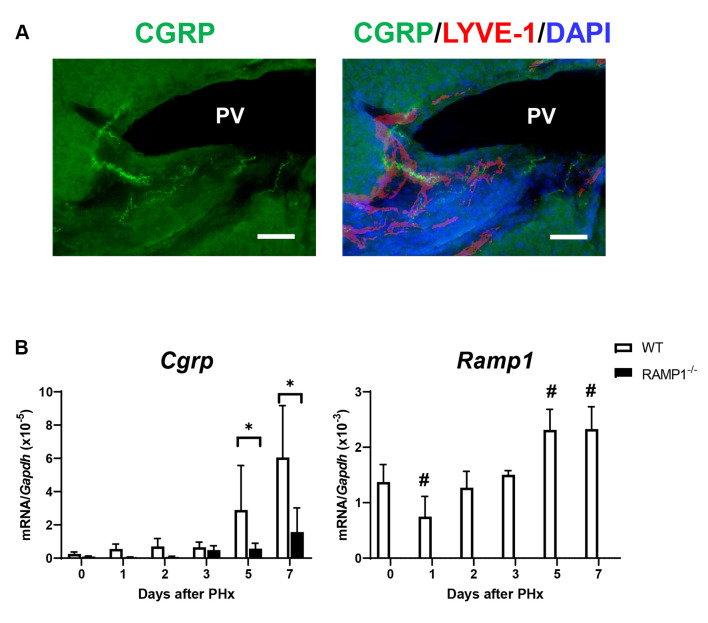
Deletion of RAMP1 signaling reduced angiogenesis in the liver post-PHx. Angiogenesis contributes to liver regeneration post-PHx. Therefore, we determined the mRNA levels of Cd31 and Vegfr2, which are markers of LSECs in the liver. The mRNA levels of Cd31 and Vegfr2 in the liver of RAMP1–/– mice on day 5 were lower than those in WT mice (Figure 3A). Additionally, flow cytometry analysis revealed that the percentage of LSECs (defined as CD31+/LYVE-1+ cells) in RAMP1–/– mice was lower than that in WT mice (Figure 3B). These results indicate that RAMP1 deficiency impairs angiogenesis during liver regeneration post-PHx.
Figure 3. Hepatic endothelial cell expression in wild type (WT) and receptor activity-modifying protein 1 (RAMP1)–/– mice post-partial hepatectomy (PHx). (A) mRNA levels of CD31 and vascular endothelial growth factor receptor-2 (VEGFR2) in livers of WT and RAMP1–/– mice after PHx. Data are expressed as the mean±standard deviation (SD) (n=5-6 mice per group). *p<0.05. (B) Representative dot plots of liver sinusoidal endothelial cells (LSECs) (CD31high/LYVE-1high cells) gated on CD45- cells and percentage of LSECs in WT and RAMP1–/– mice on day 5. Data are expressed as the mean±SD (n=3 mice per group). *p<0.05.
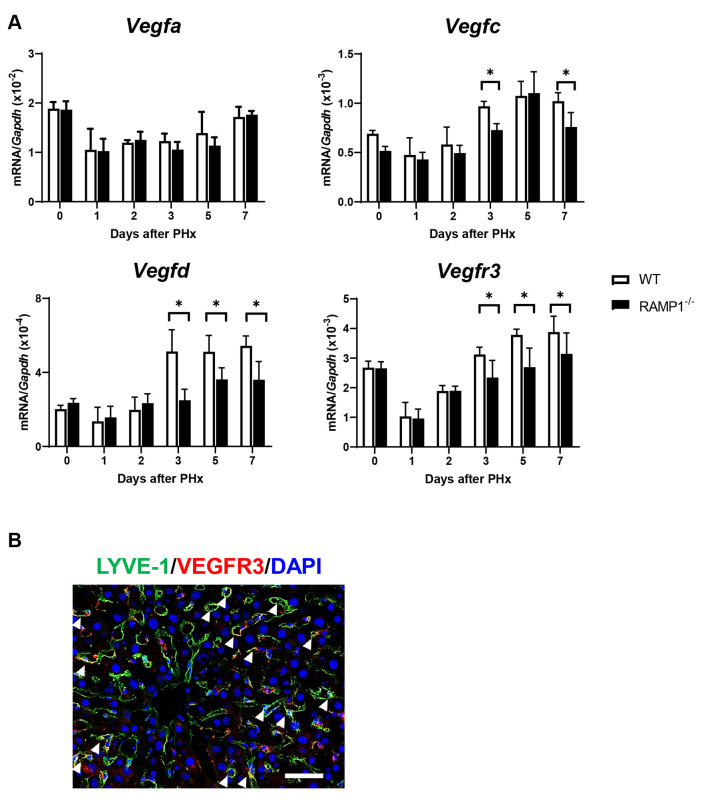
Deletion of RAMP1 signaling down-regulated the expression of VEGF-C, VEGF-D, and VEGFR3. To understand how RAMP1 signaling regulates angiogenesis (hepatic vascular reconstitution), we assessed the mRNA levels of pro-angiogenic factors, including Vegfa, which plays a crucial role in angiogenesis. However, there was no significant difference in Vegfa mRNA levels between the two genotypes (Figure 4A). However, the mRNA levels of Vegfc and Vegfd from days 3 to 7 in RAMP1–/– mice were lower than those in the WT mice. Correspondingly, the mRNA levels of Vegfr3, which is a receptor for both VEGF-C and VEGF-D, were lower in RAMP1–/– mice compared to WT mice. Additionally, immunofluorescence analysis revealed that VEGFR3 was co-localized with LYVE-1+ cells (Figure 4B), indicating that VEGFR3 was expressed by LSECs (5).
Figure 4. Hepatic expression of pro-angiogenic factors in wild type (WT) and receptor activity-modifying protein 1 (RAMP1)–/– mice post-partial hepatectomy (PHx). (A) mRNA levels of vascular endothelial growth factor-A (VEGFA), VEGFC, VEGFD, and vascular endothelial growth factor receptor-3 (VEGFR3) in the livers of WT and RAMP1–/– mice post-PHx. Data are expressed as the mean±standard deviation (SD) (n=4-6 mice per group). *p<0.05. (B) Immunofluorescence staining of LYVE-1 (green) and VEGFR3 (red) in the livers of WT mice on day 7. Cell nuclei are stained with 4’,6-diamidino-2-phenylindole (DAPI) (blue). Arrowheads indicate merged cells. Scale bar: 100 μm.
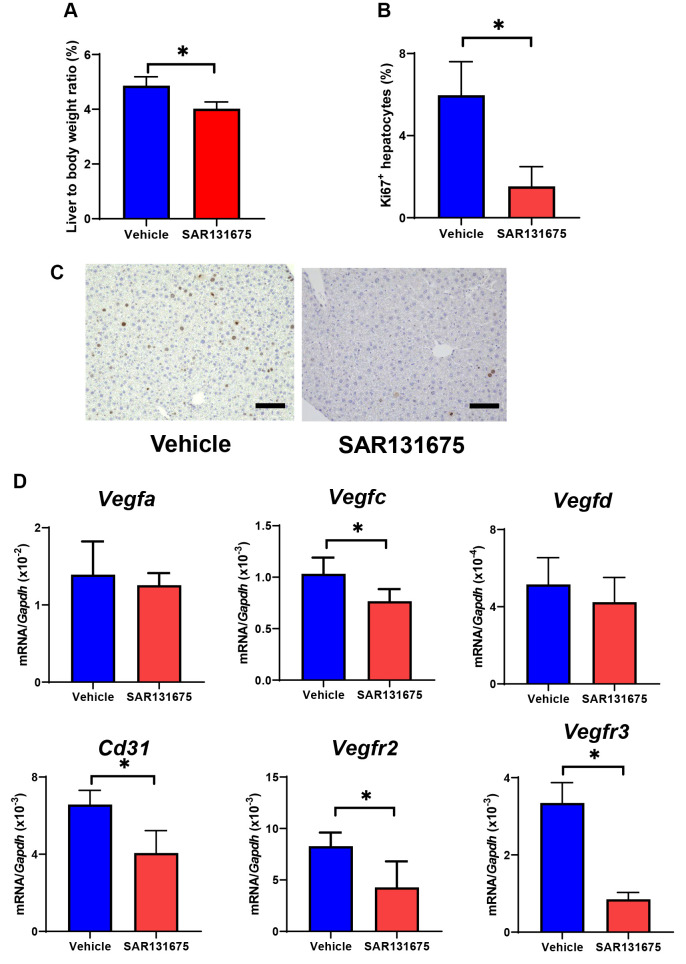
VEGFR3 inhibition impaired liver regeneration and angiogenesis post-PHx. Additionally, we assessed whether VEGFR3 was involved in liver regeneration and angiogenesis post-PHx. Therefore, we treated WT mice with the VEGFR3 inhibitor SAR131675, which reduced the liver/body weight ratio and percentage of Ki67+hepatocytes on day 5 (Figure 5). Moreover, SAR131675 reduced the mRNA levels of Vegfc, Cd31, Vegfr2, and Vegfr3 on day 7.
Figure 5. Effect of the vascular endothelial growth factor receptor-3 (VEGFR3) inhibitor SAR131675 on liver regeneration and angiogenic factors post-partial hepatectomy (PHx). (A) Liver-to-body weight ratio in wild type (WT) mice treated with vehicle or SAR131675 on day 5. Data are expressed as the mean±standard deviation (SD) (n=5 mice per group). *p<0.05. (B) Percentage of Ki67+ hepatocytes in the livers of WT treated with vehicle or SAR131675. Data are expressed as the mean±SD (n=5 mice per group). *p<0.05. (C) Representative photos of Ki67-immunostaining in liver tissue. Scale bars indicate 100 μm. (D) mRNA levels of vascular endothelial growth factor-A (VEGFA), VEGFC, VEGFD, and endothelial cell markers including CD31, vascular endothelial growth factor receptor-2 (VEGFR2), and VEGFR3. Data are expressed as the mean±SD (n=5 mice per group). *p<0.05.
Discussion
The successful regeneration of the liver post-PHx requires hepatocyte proliferation and hepatic vascular reconstitution. Because CGRP/RAMP1 signaling is associated with angiogenesis, RAMP1 signaling is involved in liver regeneration by affecting angiogenesis. In this study, we demonstrated that, compared to WT mice post-PHx, RAMP1-deficient mice exhibited lower liver-to-body weight ratios, suppressed hepatocyte proliferation, and delayed liver regeneration. Additionally, RAMP1 deficiency suppressed microvascular reconstitution in the remnant liver, as indicated by reduced hepatic expression of VEGF-C, VEGF-D, and VEGFR3. Inhibition of VEGFR3 resulted in impaired liver regeneration and microvascular reconstitution post-PHx. These results indicate that the activation of RAMP1 signaling stimulates liver regeneration and angiogenesis post-PHx.
CGRP is released from the sensory neurons that enter the liver through the hilum and form plexuses along the portal vein and hepatic artery (10). In the liver, CGRP-positive nerves are present in the connective tissues in the peri-portal and intra-lobular area (23). Consistent with this, immunofluorescence analysis demonstrated that CGRP+ nerve fibers are distributed along the portal veins and lymphatic vessels and terminate in the periportal regions. The mRNA levels of Cgrp and Ramp1 in WT mice increased on days 5 and 7, indicating that CGRP/RAMP1 is involved in the pro-angiogenic phase following PHx.
The crucial role of angiogenesis in liver regeneration post-PHx has been reported previously (6). Reconstitution of the hepatic vasculature, followed by massive hepatocyte proliferation, is essential for liver regeneration post-PHx (6). LSEC proliferation is crucial during the angiogenic phase of liver regeneration post-PHx (5,7). This study demonstrated that RAMP1 signaling contributes to angiogenesis during liver regeneration post-PHx. Previously, we demonstrated the role of RAMP1 signaling in angiogenesis under numerous pathological conditions (17-19). Additionally, we demonstrated that macrophage-derived VEGF-A was involved in angiogenesis. Evidence indicates that numerous pro-angiogenic factors mediate LSEC proliferation, and the significance of pro-angiogenic signaling has been highlighted in liver regeneration post-PHx (24). For example, LSECs secrete pro-angiogenic factors, including HGF and wingless-type MMTV integration site family member 2 (Wnt2), through the VEGF-A/VEGFR2 pathway, resulting in orchestrated liver regeneration in mice (5). Additionally, LSEC-derived angiopoietin-2 facilitates LSEC proliferation during liver regeneration post-PHx (25) and chemical-induced acute liver injury (8). This study demonstrated that the activation of RAMP1 signaling induced an increase in the expression of VEGF-C, VEGF-D, and their receptor VEGFR3 during liver regeneration post-PHx. Because LSECs express VEGFR3 in conjunction with VEGFR2 (5), it is reasonable to indicate that VEGF-C and VEGF-D bind to VEGFR3 in LSCEs to stimulate LSEC proliferation, thereby contributing to the restoration of liver mass size and function. To support this notion, this study demonstrated that a VEGFR3 inhibitor impaired liver regeneration and angiogenesis, as indicated by reduced liver mass recovery and hepatocyte proliferation, and diminished endothelial cell growth factor and LSEC markers. Additionally, we demonstrated that the VEGF-C/VEGFR3 pathway facilitates liver repair following hepatic ischemia-reperfusion injury in mice (26).
This study indicated that RAMP1 signaling facilitates liver regeneration and angiogenesis by inducing VEGF-C and VEGF-D expression. However, the precise molecular mechanisms by which RAMP1 signaling induces the hepatic expression of VEGF-C and VEGF-D during liver regeneration post-PHx remain unclear. Additionally, the sources of VEGF-C and VEGF-D in this study require further clarification, although our previous study indicated that liver macrophages generate VEGF-C and VEGF-D (26). Consistent with this, VEGF-C is produced by intestinal villous macrophages (27,28). Moreover, RAMP1 is expressed in liver macrophages during immune-mediated hepatitis in mice (15). An in vitro study demonstrated that CGRP enhances VEGF-C in cultured bone marrow-derived macrophages (29) and peritoneal macrophages (18) in a RAMP signaling-dependent manner. These findings indicate that liver macrophages produce VEGF-C and VEGF-D through RAMP1 signaling to stimulate angiogenesis in the remnant liver. However, further studies are required to clarify whether RAMP1 signaling in liver macrophages produces VEGF-C and VEGF-D to facilitate LSEC proliferation.
Study limitations. First, RAMP1 is expressed in macrophages and hepatocytes (16). The deletion of RAMP1 signaling impaired liver regeneration post-PHx (16), which is consistent with our results. Additionally, direct stimulation of isolated hepatocytes with CGRP facilitated hepatocyte mediated by CGRP receptor signaling. These results indicate that RAMP1 signaling in hepatocytes regulates vascular reconstitution during liver regeneration. Second, the role of RAMP1 signaling in macrophages requires further assessment. Third, this study results did not include any data from patients who underwent PHx. Further human studies are required to understand the effects of RAMP1 signaling on liver regeneration post-PHx.
Conclusion
This study demonstrated that RAMP1 deficiency impaired liver regeneration and angiogenesis following PHx. This study indicates that reduced expression of VEGF-C, VEGF-D, and VEGFR3 is involved in delayed angiogenesis during liver regeneration following PHx. RAMP1 activation facilitates liver regeneration following PHx.
Conflicts of Interest
The Authors declare that there are no conflicts of interest in relation to this study.
Authors’ Contributions
SN: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft; YI: Formal analysis, Investigation, Writing – review & editing. NM, YK, KH, and MK: Data curation, Investigation, Methodology; KT: Methodology, Resources; YK and HA: Supervision, Validation, Writing – review & editing.
Acknowledgements
The Authors thank Michiko Ogino and Kyoko Yoshikawa for the technical assistance.
Funding
This research was supported by a research grant (22K16544) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology.
References
- 1.Michalopoulos GK, Bhushan B. Liver regeneration: biological and pathological mechanisms and implications. Nat Rev Gastroenterol Hepatol. 2021;18(1):40–55. doi: 10.1038/s41575-020-0342-4. [DOI] [PubMed] [Google Scholar]
- 2.Rahbari NN, Garden OJ, Padbury R, Brooke-smith M, Crawford M, Adam R, Koch M, Makuuchi M, Dematteo RP, Christophi C, Banting S, Usatoff V, Nagino M, Maddern G, Hugh TJ, Vauthey J, Greig P, Rees M, Yokoyama Y, Fan ST, Nimura Y, Figueras J, Capussotti L, Büchler MW, Weitz J. Posthepatectomy liver failure: A definition and grading by the International Study Group of Liver Surgery (ISGLS) Surgery. 2011;149(5):713–724. doi: 10.1016/j.surg.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Golriz M, Majlesara A, El Sakka S, Ashrafi M, Arwin J, Fard N, Raisi H, Edalatpour A, Mehrabi A. Small for Size and Flow (SFSF) syndrome: An alternative description for posthepatectomy liver failure. Clin Res Hepatol Gastroenterol. 2016;40(3):267–275. doi: 10.1016/j.clinre.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 4.Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 2004;5(10):836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 5.Ding BS, Nolan DJ, Butler JM, James D, Babazadeh AO, Rosenwaks Z, Mittal V, Kobayashi H, Shido K, Lyden D, Sato TN, Rabbany SY, Rafii S. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. 2010;468(7321):310–315. doi: 10.1038/nature09493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rafii S, Butler JM, Ding BS. Angiocrine functions of organ-specific endothelial cells. Nature. 2016;529(7586):316–325. doi: 10.1038/nature17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Rudder M, Dili A, Stärkel P, Leclercq IA. Critical role of LSEC in post-hepatectomy liver regeneration and failure. Int J Mol Sci. 2021;22(15):8053. doi: 10.3390/ijms22158053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otaka F, Ito Y, Goto T, Kojo K, Tanabe M, Hosono K, Majima M, Koizumi W, Amano H. Recovery of liver sinusoidal endothelial cells following monocrotaline-induced liver injury. In Vivo. 2021;35(5):2577–2587. doi: 10.21873/invivo.12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito Y, Hosono K, Amano H. Responses of hepatic sinusoidal cells to liver ischemia-reperfusion injury. Front Cell Dev Biol. 2023;11:1171317. doi: 10.3389/fcell.2023.1171317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller BM, Oderberg IM, Goessling W. Hepatic nervous system in development, regeneration, and disease. Hepatology. 2021;74(6):3513–3522. doi: 10.1002/hep.32055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hay DL, Christopoulos G, Christopoulos A, Poyner DR, Sexton PM. Pharmacological discrimination of calcitonin receptor: receptor activity-modifying protein complexes. Mol Pharmacol. 2005;67(5):1655–1665. doi: 10.1124/mol.104.008615. [DOI] [PubMed] [Google Scholar]
- 12.Tsujikawa K, Yayama K, Hayashi T, Matsushita H, Yamaguchi T, Shigeno T, Ogitani Y, Hirayama M, Kato T, Fukada S, Takatori S, Kawasaki H, Okamoto H, Ikawa M, Okabe M, Yamamoto H. Hypertension and dysregulated proinflammatory cytokine production in receptor activity-modifying protein 1-deficient mice. Proc Natl Acad Sci U S A. 2007;104(42):16702–16707. doi: 10.1073/pnas.0705974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russo AF, Hay DL. CGRP physiology, pharmacology, and therapeutic targets: migraine and beyond. Physiol Rev. 2023;103(2):1565–1644. doi: 10.1152/physrev.00059.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tiegs G, Hentschel J, Wendel A. A T cell-dependent experimental liver injury in mice inducible by concanavalin A. J Clin Invest. 1992;90(1):196–203. doi: 10.1172/JCI115836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue T, Ito Y, Nishizawa N, Eshima K, Kojo K, Otaka F, Betto T, Yamane S, Tsujikawa K, Koizumi W, Majima M. RAMP1 in Kupffer cells is a critical regulator in immune-mediated hepatitis. PLoS One. 2018;13(11):e0200432. doi: 10.1371/journal.pone.0200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laschinger M, Wang Y, Holzmann G, Wang B, Stöß C, Lu M, Brugger M, Schneider A, Knolle P, Wohlleber D, Schulze S, Steiger K, Tsujikawa K, Altmayr F, Friess H, Hartmann D, Hüser N, Holzmann B. The CGRP receptor component RAMP1 links sensory innervation with YAP activity in the regenerating liver. FASEB J. 2020;34(6):8125–8138. doi: 10.1096/fj.201903200R. [DOI] [PubMed] [Google Scholar]
- 17.Toda M, Suzuki T, Hosono K, Hayashi I, Hashiba S, Onuma Y, Amano H, Kurihara Y, Kurihara H, Okamoto H, Hoka S, Majima M. Neuronal system-dependent facilitation of tumor angiogenesis and tumor growth by calcitonin gene-related peptide. Proc Natl Acad Sci USA. 2008;105(36):13550–13555. doi: 10.1073/pnas.0800767105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurashige C, Hosono K, Matsuda H, Tsujikawa K, Okamoto H, Majima M. Roles of receptor activity-modifying protein 1 in angiogenesis and lymphangiogenesis during skin wound healing in mice. FASEB J. 2014;28(3):1237–1247. doi: 10.1096/fj.13-238998. [DOI] [PubMed] [Google Scholar]
- 19.Honda M, Ito Y, Hattori K, Hosono K, Sekiguchi K, Tsujikawa K, Unno N, Majima M. Inhibition of receptor activity-modifying protein 1 suppresses the development of endometriosis and the formation of blood and lymphatic vessels. J Cell Mol Med. 2020;24(20):11984–11997. doi: 10.1111/jcmm.15823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3(7):1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 21.Hosono K, Kojo K, Narumiya S, Majima M, Ito Y. Prostaglandin E receptor EP4 stimulates lymphangiogenesis to promote mucosal healing during DSS-induced colitis. Biomed Pharmacother. 2020;128:110264. doi: 10.1016/j.biopha.2020.110264. [DOI] [PubMed] [Google Scholar]
- 22.Goto T, Ito Y, Satoh M, Nakamoto S, Nishizawa N, Hosono K, Naitoh T, Eshima K, Iwabuchi K, Hiki N, Amano H. Activation of iNKT cells facilitates liver repair after hepatic ischemia reperfusion injury through acceleration of macrophage polarization. Front Immunol. 2021;12:754106. doi: 10.3389/fimmu.2021.754106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoyanova II, Gulubova MV. Immunocytochemical study on the liver innervation in patients with cirrhosis. Acta Histochem. 2000;102(4):391–402. doi: 10.1078/0065-1281-00568. [DOI] [PubMed] [Google Scholar]
- 24.Kostallari E, Shah VH. Angiocrine signaling in the hepatic sinusoids in health and disease. Am J Physiol Gastrointest Liver Physiol. 2016;311(2):G246–G251. doi: 10.1152/ajpgi.00118.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J, Srivastava K, Wieland M, Runge A, Mogler C, Besemfelder E, Terhardt D, Vogel MJ, Cao L, Korn C, Bartels S, Thomas M, Augustin HG. Endothelial cell-derived angiopoietin-2 controls liver regeneration as a spatiotemporal rheostat. Science. 2014;343(6169):416–419. doi: 10.1126/science.1244880. [DOI] [PubMed] [Google Scholar]
- 26.Nakamoto S, Ito Y, Nishizawa N, Goto T, Kojo K, Kumamoto Y, Watanabe M, Majima M. Lymphangiogenesis and accumulation of reparative macrophages contribute to liver repair after hepatic ischemia–reperfusion injury. Angiogenesis. 2020;23(3):395–410. doi: 10.1007/s10456-020-09718-w. [DOI] [PubMed] [Google Scholar]
- 27.Suh SH, Choe K, Hong SP, Jeong SH, Mäkinen T, Kim KS, Alitalo K, Surh CD, Koh GY, Song JH. Gut microbiota regulates lacteal integrity by inducing VEGF-C in intestinal villus macrophages. EMBO Rep. 2019;20(4):e46927. doi: 10.15252/embr.201846927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hosono K, Yamashita A, Tanabe M, Ito Y, Majima M, Tsujikawa K, Amano H. Deletion of RAMP1 signaling enhances diet-induced obesity and fat absorption via intestinal lacteals in mice. In Vivo. 2024;38(1):160–173. doi: 10.21873/invivo.13422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsuru S, Ito Y, Matsuda H, Hosono K, Inoue T, Nakamoto S, Kurashige C, Mishima T, Tsujikawa K, Okamoto H, Majima M. RAMP1 signaling in immune cells regulates inflammation-associated lymphangiogenesis. Lab Invest. 2020;100(5):738–750. doi: 10.1038/s41374-019-0364-0. [DOI] [PubMed] [Google Scholar]