Abstract
Diffuse-type gastric cancer (DGC) is a subtype of gastric cancer with aggressiveness and poor prognosis. It is of great significance to find sensitive drugs for DGC. In the current study, a total of 20 patient-derived organoids (PDOs) were analyzed for screening the therapeutic efficacy of small molecule kinases inhibitors on gastric cancers, especially the therapeutic difference between intestinal-type gastric cancer (IGCs) and DGCs. The IGCs are sensitive to multiple kinases inhibitors, while DGCs are resistant to most of these kinases inhibitors. It was found that DGCs showed drug-induced senescent phenotype after treatment by aurora kinases inhibitors (AURKi) Barasertib-HQPA and Danusertib. The cell diameter of cancer cells are increased with stronger staining of senescence-associated β-galactosidase (SA-β-GAL), and characteristic appearance of multinucleated giant cells. The senescent cancer cells secrete large amounts of chemokine MCP-1/CCL2, which recruit and induce macrophage to M2-type polarization in PDOs of DGC (DPDOs)-macrophage co-culture system. The up-regulation of local MCP-1/CCL2 can interact with MCP-1/CCL2 receptor (CCR2) expressed on macrophages and suppress their innate immunity to cancer cells. Overall, the special response of DGC to AURKi suggests that clinicians should select a sequential therapy with senescent cell clearance after AURKi treatment for DGC.
Keywords: Gastric cancer, Patient-derived organoids, Drug screening, AURK, Immunosuppressive
Highlights
-
•
High heterogeneity of cancers results in the diversity of therapeutic response.
-
•
Organoids are good models for evaluating drug sensitivity of cancers.
-
•
Co-cultivation system of organoids and immunocyte has been developed for interaction study.
1. Introduction
Gastric cancer (GC) is one of the most common malignancies worldwide. It is the fifth most common malignancies and the fourth-leading cause of cancer-related death [[1], [2], [3]]. The global burden of GC is expected to increase 62 % by 2040 [4]. GC shows great heterogeneity morphologically and genetically [5]. According to Lauren's classification, GC is divided into diffuse-type (DGC), intestinal-type (IDC) and mixed-type (MDC) [6]. The DGC often presents invasiveness, early metastasis, resistance to multiple therapeutic drugs, and poor prognosis [[7], [8], [9]]. Over the past few decades, the biological characteristic of GC has changed with declining incidence of IDC and increasing incidence of DGC [[10], [11], [12]]. In 2014, the Cancer Genome Atlas (TCGA) network proposed a breakthrough molecular classification. GC was divided into four molecular subtypes. Of which, the majority of DGC was classified as the genome-stable (GS) subtype, which often lacks therapeutic targets [13,14]. Therefore, there is an urgent demand to find sensitive drugs for DGC.
Patient-derived organoids (PDOs) are experimental models in vitro that can faithfully recapitulate disease characteristics of phenotype and genotype of their parental diseases. Since the establishment of small intestine organoids by Hans Clevers' team in 2009 [15], PDOs for GC [16], colorectal cancer [17], pancreatic cancer [18], liver cancer [19], and breast cancer [20] have been successfully established. PDOs have many advantages in revealing pathogenesis, drugs sensitivity, and personalized medicine [[21], [22], [23]].
Inducing cellular senescence is one of the developmental directions of anti-cancer drugs. Liu and coworkers found that AURKi can induce cellular senescence and slow down the growth of melanoma in a mouse model [24]. Cellular senescence refers to an irreversible state of cell growth arrest. Senescent cells can secrete a variety of growth factors, chemokines, and cytokines to form senescence-associated secretory phenotype (SASP). Some bioactive proteins may promote angiogenesis and epithelial-mesenchymal transition (EMT) of cancer cells, and attract inflammatory cells, including myeloid-derived suppressive cells (MDSCs), resulting in immunosuppressive microenvironment [24].
In the current study, the PDOs from GC are used for drug screening using a panel of kinase inhibitors. We find that PDOs of DGC (DPDOs) revealed drug-induced senescence response for AURKi Barasertib-HQPA and Danusertib. The senescent cancer cells present multinucleated giant cell transformation with SASP. By analyzing inflammatory factors, mononuclear macrophage chemokine-1 (MCP-1) is found significantly increased. MCP-1, also known as chemokine C–C motif ligand 2 (CCL2), is a member of the chemokine superfamily [25]. CCR2 is the receptor of MCP-1/CCL2, which is mainly expressed on mononuclear macrophages. Based on the binding of MCP-1/CCL2-CCR2, the activation of CCR2 signaling pathway can recruit mononuclear macrophages and change their polarization state. Some studies have uncovered that MCP-1/CCL2 can drive cellular senescence of mesenchymal stem cells [26]. Yousefzadeh and coworkers proposed that detection of circulating MCP-1/CCL2 can be used as potential biomarker to assess cellular senescence [27]. Moreover, activation of MCP-1/CCL2-CCR2 pathway is involved in carcinogenesis [28]. Jeong and colleagues reported that MCP-1/CCL2 secreted by stromal cells created immunosuppressive microenvironment and enhanced the invasiveness of DGC [29,30]. In addition, up-regulation of MCP-1/CCL2 can induce M2 polarization of mononuclear macrophages [31,32]. The co-culture model of PDOs and immune cells could provide an efficient mode to simulate cellular interactions and reactions in vitro [33]. Here, the co-culture model of PDOs-macrophage is used for exploring the interaction between PDOs and macrophages. It is clarified that AURKi induces cellular senescence of DPDOs, which secret high level of MCP-1/CCL2. Increased MCP-1/CCL2 induces mononuclear macrophages toward M2 polarization and attenuates their innate immune response to DGC.
2. Materials and methods
2.1. PDOs culture
A total of 20 PDOs were used in this study. The fresh tissues were obtained from surgical samples. The tissues were collected within 30 min after resection. Tissues were stored in Advanced DMEM/F12 medium (12634010, Thermo Fisher Scientific, USA), and washed at least 3 times by PBS containing penicillin-streptomycin (C0222, Beyotime, China) and puromycin (A1113802,Thermo Fisher Scientific, USA). After mincing, tissues were digested by tissue digestive medium containing 1–2 mg/mL collagenase I (40507ES60, Yeasen, China), 0.5–2.5 mg/mL collagenase IV (40510ES60, Yeasen, China), and 0.6–2.4 U/mL disperse enzyme II (40104ES60, Yeasen, China) at 37 °C for 1.5 h. The undigested tissue mass was discarded. The cell suspension was centrifuged at 1000 rpm for 5 min. The cell precipitation was mixed with the matrigel (356231, Corning, USA) at 1: 1 ratio, seeding in a 24-well plate with a 50 μL/well. After solidation at 37 °C for 30 min, 500 μL complete medium was added. The passaging, cryopreserving and recovering of PDOs were carried out according to our previous reports [34,35]. The components of complete medium include basic medium and cytokines. The basic medium consists of Advanced DMEM/F12, 1 % penicillin/streptomycin, 1 % primocin, 1 × HEPES, 1 × GlutaMAX, 1 × B27 (17504044, GIBCO, USA), 1 × N2 (17502048, GIBCO, USA), 10 nM gastrin I (G9145, Sigma-Aldrich, USA), 1 mM N-acetylcysteine (A7250, Sigma-Aldrich, USA), and 4 mM nicotinamide (N0636, Sigma-Aldrich, USA). The cytokines include 50 ng/mL EGF (AF-100-15, PeproTech, USA), 200 ng/mL FGF-basic (100-18B, Peprotech, USA), 2 μM TGFβ inhibitor A-83-01 (2939, Tocris Bioscience, UK), 100 ng/mL Wnt-3a (5036-WN, R&D Systems, USA), 500 ng/ml R-spondin 1 (120-38, PeproTech, USA), 100 ng/mL noggin (120-10C, PeproTech, USA), 5 μM p38 MAPK inhibitor SB202190 (S7067, Sigma-Aldrich, USA), 1 μM prostaglandin E2 (2296, Tocris bioscience, UK), and 10 μM RHOK inhibitor Y-27632 (Y0503, Sigma-Aldrich, USA). This study was approved by the Ethics Committee of Ruijin Hospital and the informed consents were obtained from the patients.
2.2. Drugs screening
A panel of kinase inhibitors was screened using the PDOs from four DPDOs and two from IDC (IPDOs). PDOs were digested into single-cell suspension, and centrifuged at 1000 rpm for 5 min. The cell precipitation was mixed with matrigel at 1:1 ratio, and seeded in a 96-well plate with a 5 μL/well (5 × 103 cells). After solidation at 37 °C for 30 min, 60 μL complete medium containing 1 μM drug was added. PDOs were cultured at 37 °C for 96 h. Then, 10 % of CCK8 solution (CK04,DOJINDO,Japan) was added, and incubated at 37 °C for 2 h. The OD450 value was measured by the enzyme-labeled instrument (EnVision,PerkinElmer,USA) for cell vitality analysis.
2.3. Flow cytometry
The culture supernatants of PDOs were analyzed for human inflammatory factors (RayPlex® Human Inflammation Array Kit, China) using flow cytometry.
2.4. Co-culture of PDOs-macrophages
The human monocyte THP-1(1 × 106 cells) was cultured in 10 % FBS RPMI-1640 medium, and differentiated into macrophages after 24 h PMA stimulation (100 ng/mL). The macrophages were differentiated into M1 polarization by LPS (100 ng/mL) stimulation, or M2 polarization by IL-4 (100 ng/mL) stimulation. The macrophages of 300 μL (1 × 105 cells) were added to the upper chamber with 300 μL serum-free RPMI-1640 medium in the upper chamber. A 600 μL of 10 % FBS RPMI-1640 medium was added to the lower chamber and co-cultured at 37 °C for 48 h. The reagent of 1 % crystal violet was used for staining for 30 min. Wiping the cells in the upper chamber with a cotton swab, and taking photos for counting cells migrated to the lower chamber. The PDOs-macrophage co-culture was carried out by superimposing method. The THP-1(1 × 105 cells) was added to 96-well plates and differentiated into macrophages by 24 h stimulation of PMA (100 ng/mL). The PDOs of GC labeled by green fluorescent protein (GFP) were blown away. Then, about 300 PDOs were added to each well for co-culture in complete medium for 48 h. The LDH releasing in the supernatant was detected (C0016, Beyotime, China). The green fluorescence area of PDOs was calculated under the inverted microscope (AMF-4302-AU, USA).
2.5. ScRNA-seq analysis of senescence-related genes and immune microenvironment
The scRNA-seq dataset (GSE163558) was used for data mining. The tissue samples were come from six patients covering peri-carcinomatous tissues, primary gastric cancer, and metastatic lesions. A total of 52,700 single cells from six patients were analyzed.
2.6. Hematoxylin and eosin (HE) staining
The PDOs (diameter >100 μm) were collected into 1.5 mL eppendorf tube and centrifuged at 1500 rpm for 5 min, and then the PDOs were fixed with 4 % paraformaldehyde. The organoid cells were aggregated with 5 % agarose and embedded in paraffin for slice sectioning. Hematoxylin and eosin staining was carried out for morphological study.
2.7. Cytoskeleton staining by phalloidin
The PDOs (diameter >100 μm) were collected into 1.5 mL eppendorf tube and centrifuged at 1500 rpm for 5 min, and fixed with 4 % paraformaldehyde. Reagent of 0.5 % Triton X-100 was used for permeabilization. Then PDOs were incubated with 200 μL phalloidin (CoraLite® Plus 488-conjugated phalloidin antibody, PF00001, Proteintech, 1 : 40) for 1 h at room temperature, and washed with PBS for 3 times. Then the PDOs were stained with Hoechst (C1026,Beyotime, China) for 20 min. Images were taken using laser confocal microscopy (LEICA TCS SP8, LEICA, Germany).
2.8. Giemsa staining
The PDOs were digested using the TrypLE™ Express Enzyme for 30 min. The single-cell suspension was centrifuged at 1000 rpm for 5 min. The cell precipitation 20 μL (1 × 105/mL) was dropped onto a slide and dried at room temperature. The slide was fixed by methanol at room temperature for 10 min and then stained with Giemsa reagent (G1079, Servicebio, China) at 37 °C for 15 min. After cleaning and drying, the slide was soaked in xylene for 5 min, and sealed with neutral resin.
2.9. Senescence-associated β-galactosidase assay
Senescence-associated β-galactosidase (SA-β-Gal) (C0602, Beyotime, China) staining was carried out for evaluating cellular senescence of PDOs. PDOs were digested into single-cell suspension with TrypLE Express for 30 min. Then, the cells were collected in 1.5 mL eppendorf tube, and incubated with SA-β-Gal reagent at 37 °C overnight. At the next morning, PDOs were washed one time with PBS and dropped on slides with coverslips.
2.10. Enzyme-linked immunosorbent assay (ELISA)
The MCP-1 ELISA Kit (RK00052, Abclonal, China) was used to detect the MCP-1/CCL2 level of culture supernatant at OD 450 (EnVision, PerkinElmer, USA).
2.11. Lactate dehydrogenase release assay
The release of lactate dehydrogenase (LDH) is associated with apoptosis or necrosis of cells. LDH detection kit (C0016, Beyotime, China) was used to detect LDH level of the supernatant of co-culture system according to our previous report [36].
2.12. qRT-PCR
RNAs were extracted by TRIzol (R401-01-AA, Vazyme, China). Reverse transcription of 2 mg RNA into cDNA was performed using HiScript III RT SuperMix (R323-01, Vazyme, China). RT-PCR was performed using ChamQ Universal SYBR qPCR Master Mix (Q711-02, Vazyme, China) in a 10 μL reaction system. The gene primers related to macrophage polarization was designed and synthesized by Sangon Biotech (Shanghai, China). The primers used were as follows:
CD163-F: 5′-GAAGACAGAGACAGCGGCTT-3′
CD163-R: 5′-GGTATCTTAAAGGCTCACTGGGT-3′
CD80-F: 5′-GGGAAATGTCGCCTCTCTGAA-3′
CD80-R: 5′-CAAAACAGGCAGGGCTGATG-3′
GAPDH –F: 5′-ACCACCCTGTTGCTGTAGCCAA-3′
GAPDH –R: 5′-GTCTCCTCTGACTTCAACAGCG-3′
2.13. Knockdown of CCR2 by siRNA
The siRNAs for CCR2 was used for gene knockdown (Sangon Biotech, China). The interference effect of CCR2 was detected by Western Blot. Antibodies include anti-CCR2/CKR2 mAb (1 : 1000, A2385, Abclonal, China), HRP-conjugated goat anti-rabbit IgG (H + L) (1 : 3000, SA00001, Proteintech, USA), and HRP-conjugated anti-GAPDH (1 : 3000, HRP-60004, Proteintech, USA).
2.14. Statistical analysis
GraphPad Prism 6.0 (GraphPad Prism, RRID: SCR_002798) was used for statistical analysis. Shapro-Wilk test was used for normal test. The student's t-test (homogeneity of variance) or unpaired t-test with Welch's correction (heterogeneity of variance) was used for two group analysis. ANOVA or Brown-Forsythe ANOVA was used for multiple group analysis. Each experiment was repeated at least three times independently. P < 0.05 was considered statistically significant. Data were statistically analyzed by two-side.
3. Results
3.1. Drugs sensitivity difference between DPDOs and IPDOs
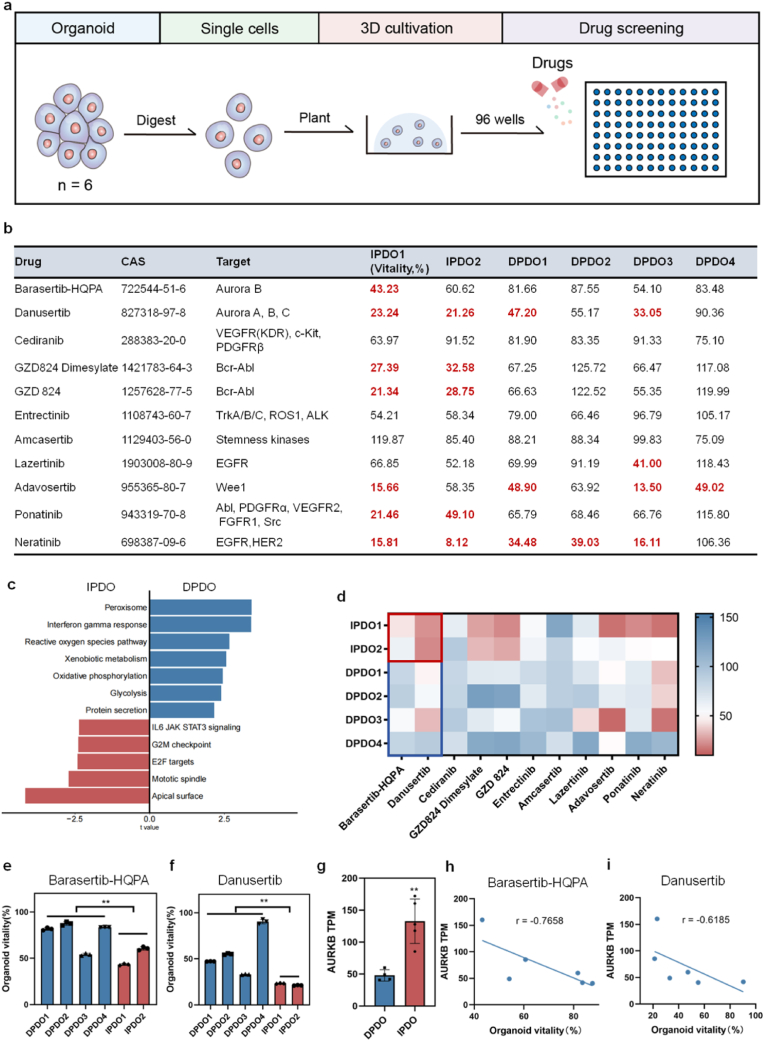
Drugs sensitivity of eleven kinases inhibitors were screened on PDOs from six GC cases (Fig. 1a). Taking the cell vitality less than 50 % as cut-off, two cases of IPDOs were highly sensitive to the kinases inhibitors, while four DPDOs were relatively not sensitive to these kinases inhibitors (Fig. 1b). To investigate the true causes of resistance to most kinase inhibitors of DGCs, we compared the transcriptomic sequencing data of IPDOs and DPDOs, and noticed that the top up-regulated pathways of DPDOs are peroxisome, interferon gamma response, reactive oxygen species pathway, xenobiotic metabolism, oxidative phosphorylation, glycolysis, and protein secretion, while the top up-regulated pathways of IPDOs are IL6 JAK STAT3 signaling, G2M checkpoint, E2F targets, mitotic spindle, and apical surface (Fig. 1c). We noticed that the sensitivity of IPDO1 and IPDO2 to AURKi Barasertib-HQPA (**P = 0.0013) and Danusertib (**P = 0.0018) were superior to that of four cases of DPDOs (Fig. 1d). There were significant differences in cell vitality between IPDOs and DPDOs (Fig. 1e and f). We examined the cell death ratio of organoids by AURKi treatment, and found that the cell death ratio of IPDO1 is significantly higher than that of DPDOs (Fig. S1). Upon transcriptomic analysis, the obvious down-regulation of AURKB was observed in DPDOs, compared to that in IPDOs (Fig. 1g). The expression levels of AURKB revealed higher correlation with cell vitality of organoids by Barasertib-HQPA(r = −0.7658) or Danusetib (r = −0.6185) treatment (Fig. 1h and i). There was no significant difference of AURKA expression (nsP = 0.87; Fig. S2a) and AURKC expression (nsP = 0.57; Fig. S2b) between DPDOs and IPDOs.
Fig. 1.
Drug sensitive screening using a panel of kinases inhibitors on IPDOs and DPDOs. (a) Flow chart of drug screening for kinases inhibitors in PDOs of GC. (b) A panel of kinases inhibitors, CAS number, and cell vitality of PDOs are presented. (c) The GSVA pathway analysis for IPDOs and DPDOs. (d) The heatmap shows sensitivity difference between IPDOs and DPDOs to kinases inhibitors. The color column on the right side represents drug sensitivity. The red indicates sensitivity and the blue indicates resistance. (e) The bar chart shows cell vitality difference to Barasertib-HQPA between IPDOs and DPDOs (**P = 0.0013). (f) The bar chart shows cell vitality difference to Danusertib between IPDOs and DPDOs (**P = 0.0018). (g) The transcriptomic levels of AURKB gene in IPDOs and DPDOs (**P = 0.002). (h) The correlation analysis of AURKB level and cell vitality upon Barasertib-HQPA treatment (r = −0.7658). (i) The correlation analysis of AURKB level and cell vitality upon Danusertib treatment (r = −0.6185).
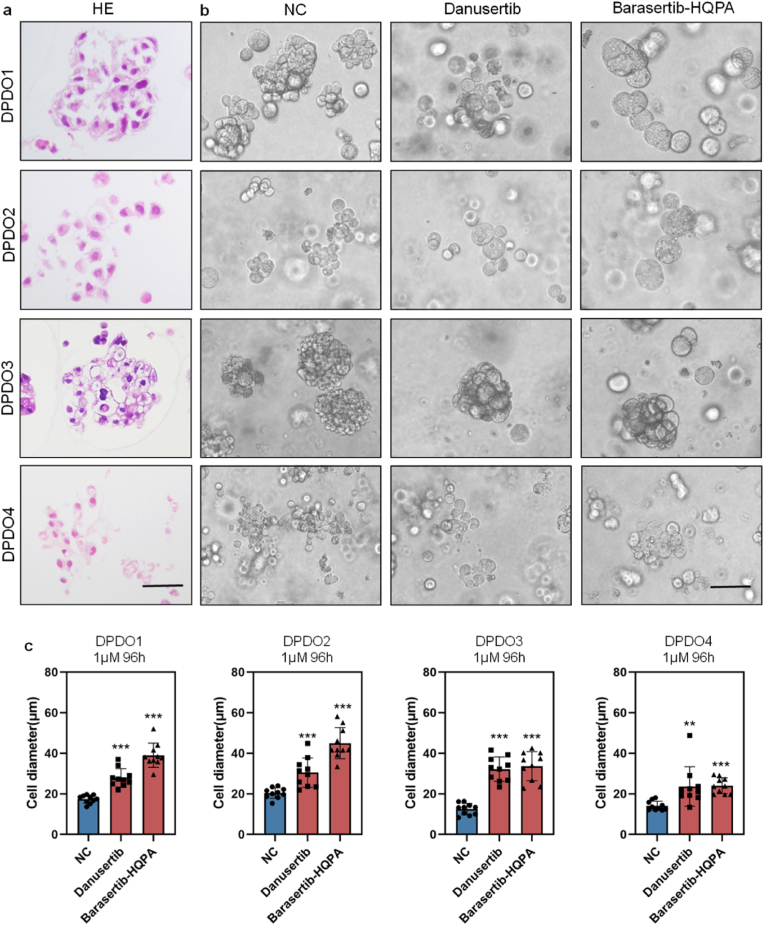
Histologically, four DPDOs showed the characteristics of poorly adhesive arrangement without obvious glandular structure (Fig. 2a,Fig. S3). After treating by low concentration (1 μM) of Barasertib-HQPA and Danusertib for 96 h, the cell diameter of DPDO was significantly increased, with no obvious cell lysis and necrosis (Fig. 2b). To clearly present the morphological feature, we measured single-cell diameter of DPDOs after drug treatment. Compared to controls, the cell diameter of Danusertib group in DPDO1 was significantly increased (17.43 ± 1.98 μm vs 27.90 ± 4.53 μm, ***P < 0.0001). The cell diameter of Barasertib-HQPA group was significantly increased too (17.43 ± 1.98 μm vs 38.99 ± 5.95 μm, ***P < 0.0001). Similarly, compared to controls, the cell diameter of DPDO2 in Danusertib group was significantly increased (20.36 ± 2.49 μm vs 30.49 ± 6.86 μm, ***P < 0.0001). The cell diameter of Barasertib-HQPA group also significantly increased (20.36 ± 2.49 μm vs 44.95 ± 7.29 μm, ***P < 0.0001). The cell diameter of DPDO3 in Danusertib group was significantly increased (12.47 ± 2.81 μm vs 32.18 ± 5.98 μm, ***P < 0.0001). The cell diameter of Barasertib-HQPA group was also increased (12.47 ± 2.81 μm vs 33.64 ± 7.15 μm, ***P < 0.0001). The cell diameter of DPDO4 in Danusertib group was significantly increased (14.06 ± 2.29 μm vs 23.63 ± 9.70 μm, **P = 0.007). The cell diameter of Barasertib-HQPA group also significantly increased (14.06 ± 2.29 μm vs 24.01 ± 3.91 μm, ***P < 0.0001) (Fig. 2c).
Fig. 2.
The specific response of DPDOs to AURKi Barasertib-HQPA and Danusertib. (a) In HE staining of PDOs slice, four DPDOs shows the morphology of DGC without obvious glandular structure and poorly adhesive arrangement. (b) Compared to NC group, the cell diameter of Danusertib group or Barasertib-HQPA group is significantly increased, with no cell lysis and necrosis. (c) By Danusertib and Barasertib-HQPA treatment (1 μΜ, 96 h), the cell diameter of four DPDOs is significantly increased compared to NC group, n = 10,scale bar = 30 μm; **P < 0.01; ***P < 0.001.
3.2. Drug-induced SASP in DPDOs
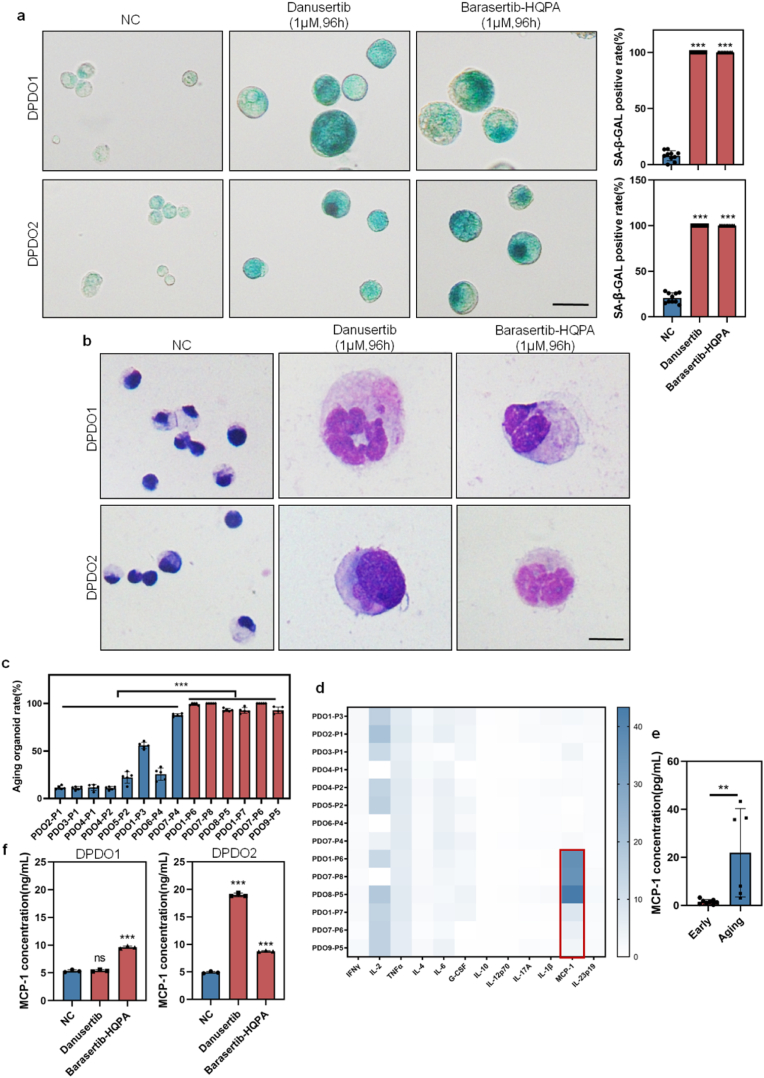
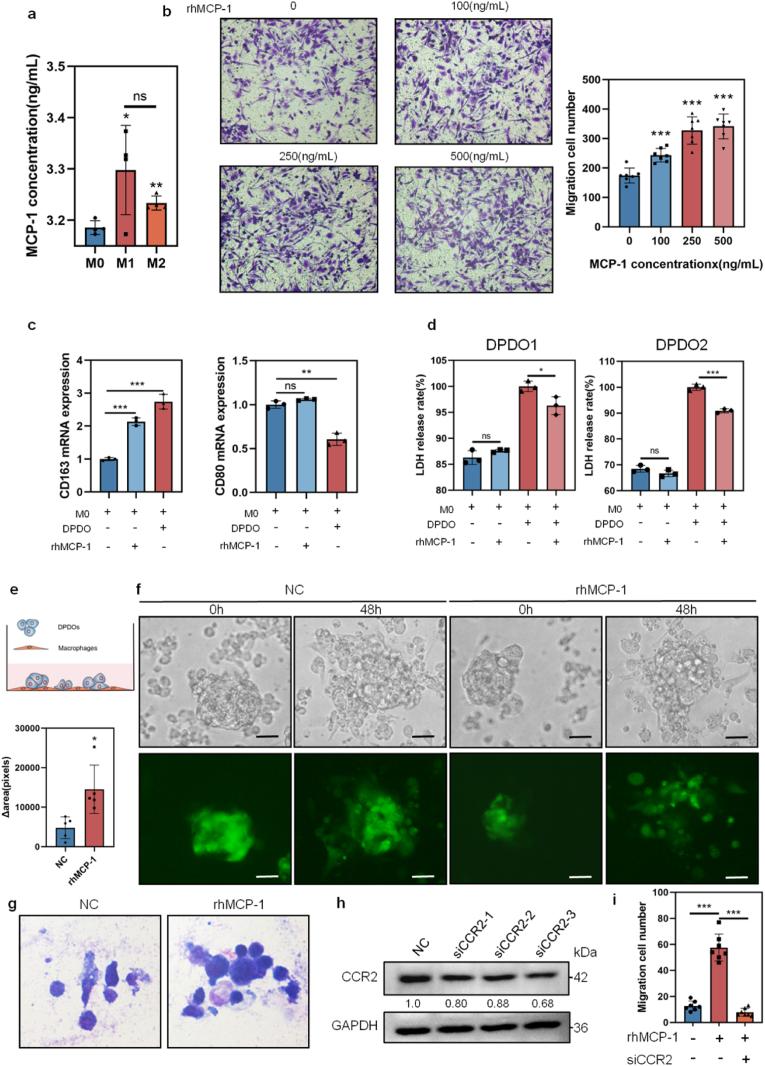
We examined the SA-β-GAL staining for DPDOs and found that the positive rate of SA-β-GAL staining in Barasertib-HQPA group or Danusertib group (1 μΜ, 96 h) is significantly higher than that in NC group (DPDO1 case: Danusertib vs NC is 100 ± 0 % vs 7.89 ± 4.26 %, ***P < 0.0001; Barasertib-HQPA vs NC is 100 ± 0 % vs 7.89 ± 4.26 %, ***P < 0.0001). (DPDO2 case: Danusertib vs NC is 100 ± 0 % vs 20.67 ± 5.36 %, ***P < 0.0001; Barasertib-HQPA vs NC is 100 ± 0 % vs 20.67 ± 5.36 %, ***P < 0.0001) (Fig. 3a). We examined the morphological feature of DPDOs on smears by Giemsa staining and found that compared to NC group, cancer cells in Danusertib and Barasertib-HQPA-treated groups (1 μM, 96 h) revealed giant cell changes with increased nuclear number and cell diameter, which is consistent with the phenotype of cellular senescence (Fig. 3b).
Fig. 3.
Drug-induced cellular senescence in DPDOs with up-regulation of MCP-1/CCL2. (a) The positive rate of SA-β-GAL staining is increased in both DPDO1 and DPDO2 after Danusertib or Barasertib-HQPA treatment (1 μM,96 h). n = 10,scale bar = 30 μm. (b) Compared to NC group, enlarged cancer cells with multinucleated giant cell changes are observed in Danusertib or Barasertib-HQPA treated group by Giemsa staining. scale bar = 20 μm (c) The bar chart shows increased positive rate of SA-β-GAL in six aging PDOs relative to eight early passage PDOs. (d) The heatmap shows inflammatory factors concentration of culture supernatants from active PDOs and aging PDOs. The red box marks the up-regulation of MCP-1/CCL2. (e) The bar chart presents the concentration difference of MCP-1/CCL2 between aging PDOs and active PDOs. (f) The bar chart shows concentration difference of MCP-1/CCL2 in the culture supernatants of NC group and Danusertib or Barasertib-HQPA-treated groups (1 μM, 96 h) in DPDO1 and DPDO2 cases. n = 3; nsP> 0.05; **P < 0.01; ***P < 0.001.
To verify the correlation between cellular senescence of PDOs and inflammatory factors of culture supernatants, we detected the SA-β-GAL positive rate and inflammatory factors in different passaging PDOs in our living biobank [34]. The positive rate of SA-β-GAL was increased along with passaging (early PDOs vs aging PDOs: 29.45 ± 26.26 % vs 96.26 ± 3.48 %, **P < 0.0001)(Fig. 3c). We analyzed the levels of a panel of inflammatory factors (IFN-γ, IL-2, TNF-α, IL-4, IL-6, G-CSF, IL-10, IL-12p70, IL-17A, IL-1β, MCP-1, and IL-23p19) in the culture supernatants and found that the change of MCP-1/CCL2 was significant (early PDOs vs aging PDOs: 1.49 ± 0.89 vs 21.93 ± 15.79 pg/mL, **P = 0.0079)(Fig. 3d–e). Upon ELISA analysis, the level of MCP-1/CCL2 in culture supernatant in Danusertib group or Barasertib-HQPA group (1 μM,96 h) showed heterogeneity. In the DPDO1 case, the concentration of MCP-1/CCL2 in Danusertib group was not significantly increased (NC vs Danusertib: 5.34 ± 0.19 pg/mL vs 5.40 ± 0.16 pg/mL, P = 0.74), while the concentration of MCP-1/CCL2 in Barasertib-HQPA group was significantly increased (NC vs Barasertib-HQPA:5.34 ± 0.19 pg/mL vs 9.63 ± 0.19 pg/mL, ***P < 0.0001). In the DPDO2 case, the concentration of MCP-1/CCL2 in both Danusertib and Barasertib-HQPA groups was significantly increased (NC vs Danusertib: 5.34 ± 0.19 pg/mL vs 18.99 ± 0.28 pg/mL, ***P < 0.0001; NC vs Barasertib-HQPA: 5.34 ± 0.19 pg/mL vs 8.76 ± 0.09 pg/mL, ***P < 0.0001) (Fig. 3f).
3.3. MCP-1/CCL2 expression analysis in scRNA-seq
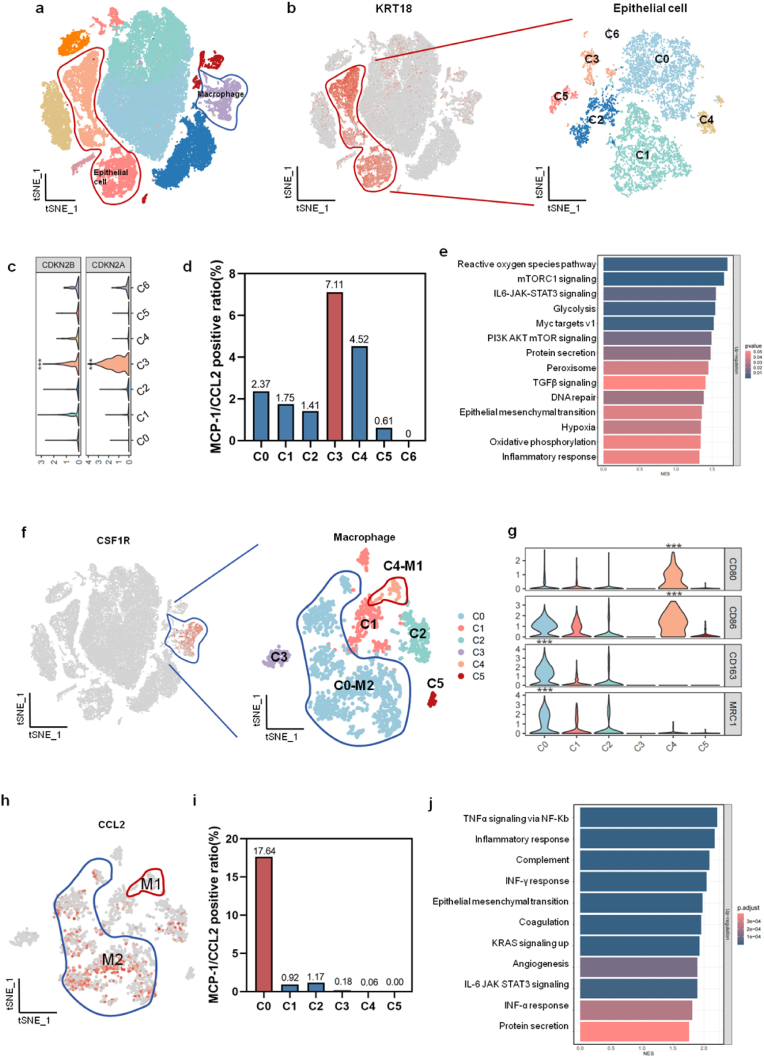
To clarify the relationship between cell types and the expression of MCP-1/CCL2, we analyzed the transcriptomic features of scRNA-seq using an external dataset (GSE163558). We divided 52,700 single cells into 11 clusters (C0 to C10)(Fig. 4a). Using Keratin 18 (KRT18) as the epithelial marker, the epithelial cells were further divided into seven clusters (C0 to C6)(Fig. 4b). The cell cycle-inhibiting genes CDKN2A (average log2FC = 2.0662, ***P < 0.0001) and CDKN2B (average log2FC = 0.3344, ***P < 0.0001) were obviously increased in C3 cluster than other clusters, indicating its senescent feature (Fig. 4c). We compared the expressing levels of MCP-1/CCL2 of cellular clusters and found that the positive proportion of MCP-1/CCL2 expression was highest in the C3 cluster (7.11 %)(Fig. 4d). By gene set enrichment analysis (GSEA), the up-regulated genes of C3 cluster were enriched in pathways of reactive oxygen species,protein secretion,EMT, hypoxia, oxidative phosphorylation, and inflammatory response (Fig. 4e).
Fig. 4.
The relationship between cell types and the expression of MCP-1/CCL2 in scRNA-seq analysis. (a) In tSNE plot of scRNA-seq analysis, the cells were divided into eleven clusters, including epithelial cells (red line marked) and macrophage (blue line marked). (b) Using KRT18 as the epithelial marker gene, the epithelial cells are further divided into seven clusters (C0 to C6). (c) Senescence associated genes of CDKN2A and CDKN2B are mainly expressed in the C3 cluster. (d) Among C0 to C6 clusters, the proportion of MCP-1/CCL2 expressing cells is the highest (7.11 %) in the C3 cluster. (e) In GSEA analysis, up-regulated genes of C3 are enriched in pathways of reactive oxygen species,mTORC1 signaling,IL6-JAK-STAT3 signaling,and so on. (f) Using CSF1R as the marker gene, the macrophages are further divided into six clusters (C0 to C5). (g) In violin plot, the M1 polarization cells (C4) express high levels of CD80 and CD86, while the M2 polarization cells (C0) express high levels of CD206 and CD163. (h) The gene expression feature of MCP-1/CCL2 in macrophages. (i) Among six clusters of macrophages, the proportion of MCP-1/CCL2 expressing cell is the highest (17.64 %) in the C0 cluster. (j) By GSEA analysis, the up-regulated genes of C0/M2 cluster are enriched in pathways of TNFα signaling via NF-κB,inflammatory response,complement,and so on.***P < 0.001.
Taking colony stimulating factor 1 receptor (CSF1R) as the marker of macrophage, we divided macrophages into six subclusters (C0 to C5) (Fig. 4f). The up-regulation of CD80 (average log2FC = 1.0358, ***P < 0.0001) and CD86 (average log2FC = 0.4528, ***P < 0.0001) was defined as cluster of M1 polarization (C4). The up-regulation of CD206 (average log2FC = 1.1585, ***P < 0.0001) and CD163 (average log2FC = 1.4598, ***P < 0.0001) was defined as the cluster of M2 polarization (C0) (Fig. 4h). We compared the proportion of MCP-1/CCL2 expressing cells in C0 to C5, and found that the proportion of MCP-1/CCL2 positive expression was the highest (17.64 %) in C0-M2 (Fig. 4i). By GSEA analysis, the up-regulated genes of C0-M2 were enriched in pathways of TNFα signaling via NF-κB,inflammatory response,INF-γ response,EMT, angiogenesis,IL-6 JAK STAT3 signaling,INF-α response,and protein secretion (Fig. 4j). It suggested that the high level of MCP-1/CCL2 might be related to the M2 polarization of macrophages.
3.4. Up-regulation of MCP-1/CCL2 weakens the killing effect of macrophages
To clarify the effect of senescent cancer cells on macrophages, we established a co-culture system of macrophage and PDOs. The THP-1 cells were induced to M0 macrophages by PMA (100 ng/mL, 24 h). Then M0 macrophages were further induced to M1 by LPS stimulation or M2 by IL-4 stimulation. We examined the concentrations of MCP-1/CCL2 in culture supernatant of macrophages in different differentiation states by ELISA and found that the concentration of MCP-1/CCL2 in M1 macrophage (3.2981 ± 0.0752 ng/mL, *P = 0.0429) and M2 macrophage (3.2336 ± 0.0118 ng/mL, **P = 0.0024) were higher than that in M0 macrophages (3.1856 ± 0.0117 ng/mL) (Fig. 5a), indicating that the basic concentration of MCP-1/CCL2 in macrophages at different states was about 3 ng/mL. Using human recombinant MCP-1 (rhMCP-1: 0, 100, 250, and 500 ng/mL) induction, the migration of macrophages showed a concentration-dependent increasing (NC vs above concentrations: 174 ± 23 vs 243 ± 21; 327 ± 43; 341 ± 39, respectively, ***P = 0.0002, ***P < 0.0001 and ***P < 0.0001) (Fig. 5b). After rhMCP-1 (100 ng/mL) incubation on DPDO1 or co-culture M0 with DPDO1 for 48 h, the CD163 (M2 marker) expression was significantly increased by RT-PCR (NC vs rhMCP-1 DPDO1: 1.00 ± 0.04 vs 2.13 ± 0.10, ***P = 0.0001; NC vs co-culture group: 1.00 ± 0.04 vs 2.74 ± 0.18, ***P = 0.0002), while the expression of CD80 (M1 marker) did not significantly changed in rhMCP-1 DPDO1 group (NC vs rhMCP-1 DPDO1: 1.00 ± 0.03 vs 1.06 ± 0.01, nsP = 0.08), but was decreased in the co-culture group (NC vs co-culture group: 1.00 ± 0.03 vs 0.61 ± 0.06, **P = 0.001)(Fig. 5c). It means that increased MCP-1/CCL1 promoted M2 polarization, but not M1 of macrophages.
Fig. 5.
Up-regulation of MCP-1/CCL2 mediates immunosuppressive effect on DPDOs-macrophage co-culture system. (a) The bar chart shows concentration of MCP-1/CCL2 in supernatants of M0, M1, and M2 macrophages. n = 3. (b) The migrated cells of macrophages reveal concentration-dependent increasing upon rhMCP-1 incubation. n = 3. (c) Upon rhMCP-1 (100 ng/mL) treatment and co-culture of macrophages-DPDO1 for 48 h, the mRNA expression of CD163 is increased, while the mRNA expression of CD80 is not significantly changed. (d) By rhMCP-1 incubation (100 ng/mL, 48 h) on M0-DPDO1 and M0-DPDO2, the LDH contents of supernatants are decreased. n = 3. (e) The diagram of co-culture of macrophages and PDOs. (f) Upon rhMCP-1 (100 ng/mL) incubation for 48 h, the numbers of cancer cells are increased. The upper lane is light images and the lower lane is images labeled by GFP. Scale bar = 30 μm (g) The images from Giemsa staining present decreased cellular disruption upon of rhMCP-1 incubation (100 ng/mL, 48 h) on DPDO1, compared to NC group. (h) The knockdown efficacy of CCR2 gene by Western blot. (i) The migrated cells stimulated by MCP-1/CCL2 was reversed by knockdown of CCR2. nsP > 0.05,*P < 0.05,***P < 0.001.
LDH release is an important parameter of cell disruption. We found that rhMCP-1 alone did not result in LDH release from macrophages (Fig. 5d). We examined the LDH releasing of supernatant of macrophages-DPDOs co-culture upon rhMCP-1 induction, and found that LDH releasing was decreased in DPDO1 case (NC vs rhMCP-1: 100 ± 0.82 % vs 96.29 ± 1.40 %,*P = 0.03). The LDH releasing was also decreased in DPDO2 case (NC vs rhMCP-1: 100 ± 0.96 % vs 90.89 ± 0.64 %,***P = 0.0004) (Fig. 5d). It means that increased MCP-1/CCL2 in microenvironment weakens the killing effect of macrophages on cancer cells. In addition, we labeled DPDO1 with green fluorescence protein (GFP) and then co-cultured with M0 macrophages (Fig. 5e). Upon rhMCP-1 (100 ng/mL) incubation for 48 h, the GFP area of DPDO1 was significantly increased, compared to NC group (Δ area,NC vs rhMCP-1 incubation: 5077 ± 1606 vs 16980 ± 5866,*P = 0.0119)(Fig. 5f). By Giemsa staining, cancer cells in NC group showed fragmentation, indicating attacked by macrophages, while the cancer cells in rhMCP-1-incubation group was relatively intact (Fig. 5g).
CCR2 is a receptor of MCP-1/CCL2, which mainly expressed on macrophages. We knocked down the CCR2 expression of M0 by siRNA and detected the effect upon MCP-1/CCL2 incubation (Fig. 5h). The migrated cells stimulated by MCP-1/CCL2 was reversed by knockdown of CCR2 (NC vs rhMCP-1 incubation vs siCCR2-rhMCP-1 incubation: 13 ± 3 vs 58 ± 10 (***P < 0.0001); vs 8 ± 3(***P < 0.0001) (Fig. 5i).
4. Discussion
In the field of cancer therapy, it was once believed that cellular senescence-induction for anti-cancer strategy could be an alternative direction [37,38]. However, cancer cells might develop senescence but do not die [39]. With better understanding of the biology of senescence, it has been found that senescent cells can produce a large amount of cytokines and inflammatory factors, forming the senescence-associated secretory phenotype [40,41]. These cytokines may affect tumor microenvironment. The role of senescence of GC cells is not clear.
DGC is a clinically challenging cancer subtype. It often presents invasive growth, early metastasis, and non-sensitive to therapeutic drugs [8,9]. In 2014, The TCGA network proposed a molecular classification of GC, which divided GC into four subtypes. The most DGC was classified as the genome-stable (GS) subtype with limited therapeutic targets [13].
PDO is a new model emerged recent years. PDO cells maintained the structure and cellular features, as well as the drug sensitivity of their parental tissues [[42], [43], [44]]. We have constructed a living biobank for gastrointestinal cancers, which cover various subtypes of GC [34]. In the current study, we screened a panel of kinases inhibitors using PDOs from intestinal-type GC (IPDOs) and diffuse-type GC (DPDOs). Compared to IPDOs, DPDOs are more resistant to multiple kinases inhibitors. Upon transcriptomic analysis, the top up-regulated pathways of DPDOs were enriched in peroxisome, interferon gamma response, reactive oxygen species pathway, xenobiotic metabolism, oxidative phosphorylation, glycolysis, and protein secretion. Huang and coworkers reported that peroxisomes disruption relates to the resistance to MAPK inhibitor [45]. Bansal et al. proposed that high level of xenobiotic also relates the therapeutic resistance [46]. We noticed that DPDOs presented drug-induced cellular senescence with increased cell diameter, strong positive SA-β-GAL staining, and multinucleated giant cell changes upon treatment by AURKi Danusertib [47] and Barasertib-HQPA [48]. Upon transcriptomic analysis, the significant down-regulation of AURKB was observed in DPDOs, compared to that in IPDOs, which may attribute to decreased sensitivity to AURKi. By analyzing the inflammatory factors in PDO culture supernatants, we confirmed that the supernatants of senescent PDOs showed SASP. The content of chemokine MCP-1/CCL2 was significantly increased. We further analyzed the correlation of MCP-1/CCL2 expression and cellular types in scRNA-seq dataset and found that a portion of senescent cancer cells produced MCP-1/CCL2, which attributes to the M2 polarization of macrophages in tumor microenvironment.
Macrophages are important sentinel guards of innate immune system. Under normal physiological condition, macrophages can be induced and aggregated to clear foreign pathogens or transformed cells upon chemotactic effect of inflammatory. If the senescent cancer cells secrete inflammatory factors to promote clearance of senescent cells, it may be a beneficial effect. According to the current study, although the up-regulation of MCP-1/CCL2 can induce macrophage aggregation, but the MCP-1/CCL2 promoted M2 polarization of macrophage, forming an immunosuppressive microenvironment. As a result, the killing function of macrophages is weakened.
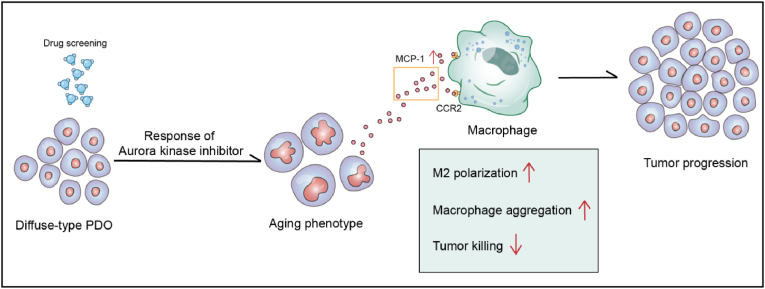
There are some limitations in the current study. Firstly, we are not sure whether AURKi influenced telomere reverse transcriptase (TERT). TERT was reported as an important factor for endless proliferation in cancers [49]. Secondly, although we found that AURKi can lead to cellular senescence but we did not know the subsequent therapeutic strategy. So, in the next step, we should explore the clearance strategies of senescent cells. Recently, several studies on other types of malignancy have revealed that the senescent cancer cells cannot be ignored, because it is likely to become the source of recurrence and metastasis. Several teams are actively explored the intervention strategies for targeting senescent cells [50]. However, there were no reports about drug-induced senescence phenotype in GC study. Our results suggested that the up-regulation of MCP-1/CCL2 by senescent cancer cells could induce macrophages aggregation and M2 polarization, resulting in a reduced killing effect on cancer cells (Fig. 6). The AURKi-induced cellular senescence of DPDOS is our new finding. Increased MCP-1/CCL2 in SASP cancer cells can promote M2 polarization of macrophages, which is the adverse effect of senescent cancer cells. Therefore, the sequential therapy with AURKi plus clearance of senescent cells should be considered for DGC treatment. The strategy for eliminating senescent cancer cells is our research direction in the future.
Fig. 6.
The diagram of drug screening on PDOs of GC. The PDOs from DGC show drug-induced cellular senescence upon AURKi treatment. The senescent cancer cells produce MCP-1/CCL2 and attenuate innate immune response of macrophages in DGC.
Disclosure statement
No potential conflict of interest was reported by author(s).
Data availability statement
The results/data/figures in this manuscript have not been published elsewhere, nor are they under consideration by another publisher.
CRediT authorship contribution statement
Ruixin Yang: Writing – original draft, Methodology, Conceptualization. Wingyan Kwan: Investigation. Yutong Du: Investigation. Ranlin Yan: Investigation. Lu Zang: Investigation. Chen Li: Investigation. Zhenggang Zhu: Resources, Conceptualization. Io Hong Cheong: Writing – review & editing, Conceptualization. Zisis Kozlakidis: Writing – review & editing, Conceptualization. Yingyan Yu: Writing – review & editing, Writing – original draft, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by grants from the Shanghai Science and Technology Committee (Grant No. 20DZ2201900), the National Natural Science Foundation of China (Grant No. 82072602, 82372933, and 82173222), the Innovation Foundation of Translational Medicine of Shanghai Jiao Tong University School of Medicine (Grant No.TM202001), and the Collaborative Innovation Center for Clinical and Translational Science by Chinese Ministry of Education & Shanghai Municipal Government (Grant No. CCTS-2022202 and CCTS-202302). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.canlet.2024.217106.
Contributor Information
Zisis Kozlakidis, Email: kozlakidisz@iarc.fr.
Yingyan Yu, Email: yingan3y@sjtu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Usui Y., Taniyama Y., Endo M., Koyanagi Y.N., Kasugai Y., Oze I., Ito H., Imoto I., Tanaka T., Tajika M., Niwa Y., Iwasaki Y., Aoi T., Hakozaki N., Takata S., Suzuki K., Terao C., Hatakeyama M., Hirata M., Sugano K., Yoshida T., Kamatani Y., Nakagawa H., Matsuda K., Murakami Y., Spurdle A.B., Matsuo K., Momozawa Y. Helicobacter pylori, homologous-recombination genes, and gastric cancer. N. Engl. J. Med. 2023;388:1181–1190. doi: 10.1056/NEJMoa2211807. [DOI] [PubMed] [Google Scholar]
- 3.Feng W., Ma C., Rao H., Zhang W., Liu C., Xu Y., Aji R., Wang Z., Xu J., Gao W.Q., Li L. Setd2 deficiency promotes gastric tumorigenesis through inhibiting the SIRT1/FOXO pathway. Cancer letters. 2023;579 doi: 10.1016/j.canlet.2023.216470. [DOI] [PubMed] [Google Scholar]
- 4.Thrift A.P., Wenker T.N., El-Serag H.B. Global burden of gastric cancer: epidemiological trends, risk factors, screening and prevention. Nat. Rev. Clin. Oncol. 2023;20:338–349. doi: 10.1038/s41571-023-00747-0. [DOI] [PubMed] [Google Scholar]
- 5.Smyth E.C., Nilsson M., Grabsch H.I., van Grieken N.C., Lordick F. Gastric cancer. Lancet. 2020;396:635–648. doi: 10.1016/S0140-6736(20)31288-5. [DOI] [PubMed] [Google Scholar]
- 6.Grabsch H.I., Tan P. Gastric cancer pathology and underlying molecular mechanisms. Dig. Surg. 2013;30:150–158. doi: 10.1159/000350876. [DOI] [PubMed] [Google Scholar]
- 7.Piessen G., Messager M., Leteurtre E., Jean-Pierre T., Mariette C. Signet ring cell histology is an independent predictor of poor prognosis in gastric adenocarcinoma regardless of tumoral clinical presentation. Ann. Surg. 2009;250:878–887. doi: 10.1097/SLA.0b013e3181b21c7b. [DOI] [PubMed] [Google Scholar]
- 8.Lordick F., Janjigian Y.Y. Clinical impact of tumour biology in the management of gastroesophageal cancer. Nat. Rev. Clin. Oncol. 2016;13:348–360. doi: 10.1038/nrclinonc.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ge S., Xia X., Ding C., Zhen B., Zhou Q., Feng J., Yuan J., Chen R., Li Y., Ge Z., Ji J., Zhang L., Wang J., Li Z., Lai Y., Hu Y., Li Y., Li Y., Gao J., Chen L., Xu J., Zhang C., Jung S.Y., Choi J.M., Jain A., Liu M., Song L., Liu W., Guo G., Gong T., Huang Y., Qiu Y., Huang W., Shi T., Zhu W., Wang Y., He F., Shen L., Qin J. A proteomic landscape of diffuse-type gastric cancer. Nat. Commun. 2018;9:1012. doi: 10.1038/s41467-018-03121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marrelli D., Pedrazzani C., Morgagni P., de Manzoni G., Pacelli F., Coniglio A., Marchet A., Saragoni L., Giacopuzzi S., Roviello F. Changing clinical and pathological features of gastric cancer over time. Br. J. Surg. 2011;98:1273–1283. doi: 10.1002/bjs.7528. [DOI] [PubMed] [Google Scholar]
- 11.Wu H., Rusiecki J.A., Zhu K., Potter J., Devesa S.S. Stomach carcinoma incidence patterns in the United States by histologic type and anatomic site. Cancer Epidemiol. Biomarkers Prev. 2009;18:1945–1952. doi: 10.1158/1055-9965.EPI-09-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henson D.E., Dittus C., Younes M., Nguyen H., Albores-Saavedra J. Differential trends in the intestinal and diffuse types of gastric carcinoma in the United States, 1973-2000: increase in the signet ring cell type. Arch. Pathol. Lab Med. 2004;128:765–770. doi: 10.5858/2004-128-765-DTITIA. [DOI] [PubMed] [Google Scholar]
- 13.Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeoh K.G., Tan P. Mapping the genomic diaspora of gastric cancer. Nat. Rev. Cancer. 2022;22:71–84. doi: 10.1038/s41568-021-00412-7. [DOI] [PubMed] [Google Scholar]
- 15.Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 16.Bartfeld S., Bayram T., van de Wetering M., Huch M., Begthel H., Kujala P., Vries R., Peters P.J., Clevers H. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148:126–136.e126. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato T., Stange D.E., Ferrante M., Vries R.G., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 18.Boj S.F., Hwang C.I., Baker L.A., Chio I.I., Engle D.D., Corbo V., Jager M., Ponz-Sarvise M., Tiriac H., Spector M.S., Gracanin A., Oni T., Yu K.H., van Boxtel R., Huch M., Rivera K.D., Wilson J.P., Feigin M.E., Öhlund D., Handly-Santana A., Ardito-Abraham C.M., Ludwig M., Elyada E., Alagesan B., Biffi G., Yordanov G.N., Delcuze B., Creighton B., Wright K., Park Y., Morsink F.H., Molenaar I.Q., Borel Rinkes I.H., Cuppen E., Hao Y., Jin Y., Nijman I.J., Iacobuzio-Donahue C., Leach S.D., Pappin D.J., Hammell M., Klimstra D.S., Basturk O., Hruban R.H., Offerhaus G.J., Vries R.G., Clevers H., Tuveson D.A. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broutier L., Mastrogiovanni G., Verstegen M.M., Francies H.E., Gavarró L.M., Bradshaw C.R., Allen G.E., Arnes-Benito R., Sidorova O., Gaspersz M.P., Georgakopoulos N., Koo B.K., Dietmann S., Davies S.E., Praseedom R.K., Lieshout R., Jnm I.J., Wigmore S.J., Saeb-Parsy K., Garnett M.J., van der Laan L.J., Huch M. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23:1424–1435. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sachs N., de Ligt J., Kopper O., Gogola E., Bounova G., Weeber F., Balgobind A.V., Wind K., Gracanin A., Begthel H., Korving J., van Boxtel R., Duarte A.A., Lelieveld D., van Hoeck A., Ernst R.F., Blokzijl F., Nijman I.J., Hoogstraat M., van de Ven M., Egan D.A., Zinzalla V., Moll J., Boj S.F., Voest E.E., Wessels L., van Diest P.J., Rottenberg S., Vries R.G.J., Cuppen E., Clevers H. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172:373–386.e310. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Koh V., Chakrabarti J., Torvund M., Steele N., Hawkins J.A., Ito Y., Wang J., Helmrath M.A., Merchant J.L., Ahmed S.A., Shabbir A., Yan So J.B., Yong W.P., Zavros Y. Hedgehog transcriptional effector GLI mediates mTOR-Induced PD-L1 expression in gastric cancer organoids. Cancer Lett. 2021;518:59–71. doi: 10.1016/j.canlet.2021.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan H.H.N., Siu H.C., Law S., Ho S.L., Yue S.S.K., Tsui W.Y., Chan D., Chan A.S., Ma S., Lam K.O., Bartfeld S., Man A.H.Y., Lee B.C.H., Chan A.S.Y., Wong J.W.H., Cheng P.S.W., Chan A.K.W., Zhang J., Shi J., Fan X., Kwong D.L.W., Mak T.W., Yuen S.T., Clevers H., Leung S.Y. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell. 2018;23:882–897.e811. doi: 10.1016/j.stem.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 23.Harada K., Sakamoto N., Ukai S., Yamamoto Y., Pham Q.T., Taniyama D., Honma R., Maruyama R., Takashima T., Ota H., Takemoto Y., Tanabe K., Ohdan H., Yasui W. Establishment of oxaliplatin-resistant gastric cancer organoids: importance of myoferlin in the acquisition of oxaliplatin resistance. Gastric Cancer. 2021;24:1264–1277. doi: 10.1007/s10120-021-01206-4. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y., Hawkins O.E., Vilgelm A.E., Pawlikowski J.S., Ecsedy J.A., Sosman J.A., Kelley M.C., Richmond A. Combining an aurora kinase inhibitor and a death receptor ligand/agonist antibody triggers apoptosis in melanoma cells and prevents tumor growth in preclinical mouse models. Clin. Cancer Res. 2015;21:5338–5348. doi: 10.1158/1078-0432.CCR-15-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun C., Li X., Guo E., Li N., Zhou B., Lu H., Huang J., Xia M., Shan W., Wang B., Li K., Weng D., Xu X., Gao Q., Wang S., Hu J., Lu Y., Mills G.B., Chen G. MCP-1/CCR-2 axis in adipocytes and cancer cell respectively facilitates ovarian cancer peritoneal metastasis. Oncogene. 2020;39:1681–1695. doi: 10.1038/s41388-019-1090-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin H.J., Lee H.J., Heo J., Lim J., Kim M., Kim M.K., Nam H.Y., Hong G.H., Cho Y.S., Choi S.J., Kim I.G., Shin D.M., Kim S.W. Senescence-associated MCP-1 secretion is dependent on a decline in BMI1 in human mesenchymal stromal cells. Antioxidants Redox Signal. 2016;24:471–485. doi: 10.1089/ars.2015.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yousefzadeh M.J., Schafer M.J., Noren Hooten N., Atkinson E.J., Evans M.K., Baker D.J., Quarles E.K., Robbins P.D., Ladiges W.C., LeBrasseur N.K., Niedernhofer L.J. Circulating levels of monocyte chemoattractant protein-1 as a potential measure of biological age in mice and frailty in humans. Aging Cell. 2018;17 doi: 10.1111/acel.12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu M., Wang Y., Xia R., Wei Y., Wei X. Role of the CCL2-CCR2 signalling axis in cancer: mechanisms and therapeutic targeting. Cell Prolif. 2021;54 doi: 10.1111/cpr.13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang H., Brekken R.A. Location matters: profiling diffuse type gastric cancer at the single-cell level. Clin. Cancer Res. : an official journal of the American Association for Cancer Research. 2021;27:6284–6286. doi: 10.1158/1078-0432.CCR-21-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeong H.Y., Ham I.H., Lee S.H., Ryu D., Son S.Y., Han S.U., Kim T.M., Hur H. Spatially distinct reprogramming of the tumor microenvironment based on tumor invasion in diffuse-type gastric cancers. Clin. Cancer Res. : an official journal of the American Association for Cancer Research. 2021;27:6529–6542. doi: 10.1158/1078-0432.CCR-21-0792. [DOI] [PubMed] [Google Scholar]
- 31.Wang X., Wang J., Zhao J., Wang H., Chen J., Wu J. HMGA2 facilitates colorectal cancer progression via STAT3-mediated tumor-associated macrophage recruitment. Theranostics. 2022;12:963–975. doi: 10.7150/thno.65411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X., Yao W., Yuan Y., Chen P., Li B., Li J., Chu R., Song H., Xie D., Jiang X., Wang H. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. 2017;66:157–167. doi: 10.1136/gutjnl-2015-310514. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y., Liu X., Ainiwan Y., Li M., Pan J., Chen Y., Xiao Z., Wang Z., Xiao X., Tang J., Zeng G., Liang J., Su X., Kungulli R., Fan Y., Lin Q., Liya A., Zheng Y., Chen Z., Xu C., Zhang H., Chen G. Axl as a potential therapeutic target for adamantinomatous craniopharyngiomas: based on single nucleus RNA-seq and spatial transcriptome profiling. Cancer letters. 2024;592 doi: 10.1016/j.canlet.2024.216905. [DOI] [PubMed] [Google Scholar]
- 34.Yang R., Du Y., Kwan W., Yan R., Shi Q., Zang L., Zhu Z., Zhang J., Li C., Yu Y. A quick and reliable image-based AI algorithm for evaluating cellular senescence of gastric organoids. Cancer Biol Med. 2023;20:519–536. doi: 10.20892/j.issn.2095-3941.2023.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang R., Xiang Z., Yan R., Kwan W., Zang L., Zhu Z., Qi Y., Xu Y., Zhang X., Gao H., Yu Y. Benchmark for establishment of organoids from gastrointestinal epithelium and cancer based on available consumables and reagents. Chin. J. Cancer Res. 2023;35:636–644. doi: 10.21147/j.issn.1000-9604.2023.06.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang Z., Zhou Z., Song S., Li J., Ji J., Yan R., Wang J., Cai W., Hu W., Zang L., Zhu Z., Zhang Z., Li M., Yu Y. Dexamethasone suppresses immune evasion by inducing GR/STAT3 mediated downregulation of PD-L1 and Ido1 pathways. Oncogene. 2021;40:5002–5012. doi: 10.1038/s41388-021-01897-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L., Lankhorst L., Bernards R. Exploiting senescence for the treatment of cancer. Nat. Rev. Cancer. 2022;22:340–355. doi: 10.1038/s41568-022-00450-9. [DOI] [PubMed] [Google Scholar]
- 38.Qiao N., Lyu Y., Liu F., Zhang Y., Ma X., Lin X., Wang J., Xie Y., Zhang R., Qiao J., Zhu H., Chen L., Fang H., Yin T., Chen Z., Tian Q., Chen S. Cross-sectional network analysis of plasma proteins/metabolites correlated with pathogenesis and therapeutic response in acute promyelocytic leukemia. Front. Med. 2024;18:327–343. doi: 10.1007/s11684-023-1022-x. [DOI] [PubMed] [Google Scholar]
- 39.Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 40.Birch J., Gil J. Senescence and the SASP: many therapeutic avenues. Genes Dev. 2020;34:1565–1576. doi: 10.1101/gad.343129.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao S.G., Jackson J.G. SASP: tumor suppressor or promoter? Yes. Trends Cancer. 2016;2:676–687. doi: 10.1016/j.trecan.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 42.Yang R., Yu Y. Patient-derived organoids in translational oncology and drug screening. Cancer Lett. 2023;562 doi: 10.1016/j.canlet.2023.216180. [DOI] [PubMed] [Google Scholar]
- 43.Weng G., Tao J., Liu Y., Qiu J., Su D., Wang R., Luo W., Zhang T. Organoid: bridging the gap between basic research and clinical practice. Cancer letters. 2023;572 doi: 10.1016/j.canlet.2023.216353. [DOI] [PubMed] [Google Scholar]
- 44.Li H., Liu H., Chen K. Living biobank-based cancer organoids: prospects and challenges in cancer research. Cancer Biol Med. 2022;19:965–982. doi: 10.20892/j.issn.2095-3941.2021.0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang F., Cai F., Dahabieh M.S., Gunawardena K., Talebi A., Dehairs J., El-Turk F., Park J.Y., Li M., Goncalves C., Gagnon N., Su J., LaPierre J.H., Gaub P., Joyal J.S., Mitchell J.J., Swinnen J.V., Miller W.H., Jr., Del Rincon S.V. Peroxisome disruption alters lipid metabolism and potentiates antitumor response with MAPK-targeted therapy in melanoma. J. Clin. Invest. 2023;133 doi: 10.1172/JCI166644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bansal A., Simon M.C. Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol. 2018;217:2291–2298. doi: 10.1083/jcb.201804161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carpinelli P., Ceruti R., Giorgini M.L., Cappella P., Gianellini L., Croci V., Degrassi A., Texido G., Rocchetti M., Vianello P., Rusconi L., Storici P., Zugnoni P., Arrigoni C., Soncini C., Alli C., Patton V., Marsiglio A., Ballinari D., Pesenti E., Fancelli D., Moll J. PHA-739358, a potent inhibitor of Aurora kinases with a selective target inhibition profile relevant to cancer. Mol Cancer Ther. 2007;6:3158–3168. doi: 10.1158/1535-7163.MCT-07-0444. [DOI] [PubMed] [Google Scholar]
- 48.Tao Y., Zhang P., Girdler F., Frascogna V., Castedo M., Bourhis J., Kroemer G., Deutsch E. Enhancement of radiation response in p53-deficient cancer cells by the Aurora-B kinase inhibitor AZD1152. Oncogene. 2008;27:3244–3255. doi: 10.1038/sj.onc.1210990. [DOI] [PubMed] [Google Scholar]
- 49.Kumar N., Sethi G. Telomerase and hallmarks of cancer: an intricate interplay governing cancer cell evolution. Cancer letters. 2023;578 doi: 10.1016/j.canlet.2023.216459. [DOI] [PubMed] [Google Scholar]
- 50.Bharti V., Watkins R., Kumar A., Shattuck-Brandt R.L., Mossing A., Mittra A., Shen C., Tsung A., Davies A.E., Hanel W., Reneau J.C., Chung C., Sizemore G.M., Richmond A., Weiss V.L., Vilgelm A.E. BCL-xL inhibition potentiates cancer therapies by redirecting the outcome of p53 activation from senescence to apoptosis. Cell Rep. 2022;41 doi: 10.1016/j.celrep.2022.111826. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The results/data/figures in this manuscript have not been published elsewhere, nor are they under consideration by another publisher.