Abstract
Six distinct COI mitochondrial Haplotype Groups (HG) are morphologically, ecologically, and genetically characterized from the aquatic nematode family Tobrilidae. Collection locations included the extreme habitats of the Alkaline Lakes in the western Nebraska Sandhills and the contaminated stream, Johnson Creek, bordering the AltEn 2021 catastrophic pesticide release near the village of Mead in eastern Nebraska. Maximum likelihood and genetic distance metrics supported the genetic integrity of the haplotype groups. Discriminant function analysis of COI haplotype group datasets of combined morphological characters and soil chemistry attributes for both male and female Tobrilidae were classified correctly in all but one case. Scanning electron microscopy revealed new details about amphid apertures, male supplements, and spicules. Partial 18S gene phylogeny suggests that the genus Semitobrilus may not be a member of the subfamily Neotobrilinae, and three specimens in the 226 tobrilid dataset provide evidence of incongruence between COI and 18S derived phylogenies. Given the strong signal provided by the environmental chemistry data, tobrilid mitochondrial haplotypes may well have value as environmental indicators.
Keywords: Aquatic nematodes, DNA barcoding, extreme environments, Nebraska Sandhills, phylogeny, taxonomy, Tobrilidae
Nematodes of the order Triplonchida Cobb, 1919 are comprised of 10 families according to the review of Holovachov and Shoshin (2014). Included are familiar terrestrial families such as Tripylidae de Man, 1876, Prismatolaimidae Micoletsky, 1922, Diphtherophoridae Micoletsky, 1922, and Trichodoridae Thorne, 1935. Tobrilidae De Coninck, 1965 may be the geographically most widespread family in the order, with specimens found predominantly in fresh or brackish water and reported from all continents, including Antarctica (Tsalolikhin, 1981). Tobrilidae are broadly characterized within Triplonchida by a funnel- or cup-shaped stoma, with two teeth at the stoma base (except Quasitobrilus) (Zullini, 2006). There are 14 morphologically defined genera in Tobrilidae, divided into three subfamilies, with an estimated total of 100 species (Holovachov and Shoshin, 2014).
The greatest diversity of Tobrilidae has been reported from Lake Baikal, the world's oldest and deepest lake (Naumova and Gagarin, 2019a: Naumova and Gagarin, 2019b; Zullini, 2014). Six genera in Tobrilidae are considered endemic to Lake Baikal (Holovachov and Shoshin, 2014; Naumova and Gagarin, 2019b). Of the 100 morphologically described species in the family, only 12 have been reported from North America (Table 1). Evidence of extreme physiological adaptability within the family is inferred from their ability to withstand a wide range of salinity (Zullini, 2006). Some species have been considered indicators of high levels of contaminating metals (Kang et al., 2023) or anoxic conditions (Teiwes et al., 2007).
Table 1.
Tobrilidae (order Triplonchida) of North America (Holovachov and Shoshin, 2014)
| Sub-family | Tribe | Species |
|---|---|---|
| Neotobrilinae Tsalolikhin, 2001 | Neotobrilini Tsalolikhin, 1981 | Neotobrilus longus (Leidy, 1852) Tsalolikhin, 1981 |
| Neotobrilinae Tsalolikhin, 2001 | Neotobrilini Tsalolikhin, 1981 | Neotobrilus nicsmolae Abebe, Ferebee, Taylor, Mundo-Ocampo, Mekete & De Ley, 2013 |
| Neotobrilinae Tsalolikhin, 2001 | Neotobrilini Tsalolikhin, 1981 | Neotobrilus filipjevi (Ebsary, 1982) Tsalolikhin and Shoshin, 2009 |
| Neotobrilinae Tsalolikhin, 2001 | Neotobrilini Tsalolikhin, 1981 | Neotobrilus hopei (Loof and Riemann, 1976) Tsalolikhin, 1981 |
| Neotobrilinae Tsalolikhin, 2001 | Neotobrilini Tsalolikhin, 1981 | Semitobrilus pellucidus Bastian, 1865 |
| Neotobrilinae Tsalolikhin, 2001 | Neotobrilini Tsalolikhin, 1981 | Semitobrilus ebsaryi Tsalolikhin, 2000 |
| Neotobrilinae Tsalolikhin, 2001 | Epitobrilini Tsalolikhin, 2001 | Epitobrilus sablensis (Ebsary, 1982) Tsalolikhin, 2001 |
| Tobrilinae Tsalolikhin, 2001 | Tobrilini Filipjev, 1918 | Tobrilus gracilis Bastian, 1865 |
| Tobrilinae Tsalolikhin, 2001 | Tobrilini Filipjev, 1918 | Tobrilus aberrans (Schneider, 1925) Andrassy, 1959 |
| Tobrilinae Tsalolikhin, 2001 | Tobrilini Filipjev, 1918 | Tobrilus affinis Gagarin, 1996 |
| Tobrilinae Tsalolikhin, 2001 | Tobrilini Filipjev, 1918 | Eutobrilus graciliformis (Altherr and Delamare Deboutteville, 1972) Tsalolikhin, 1981 |
| Tobrilinae Tsalolikhin, 2001 | Tobrilini Filipjev, 1918 | Eutobrilus steineri (Micoletzky, 1925) Zullini, 2006 |
In this taxonomic study, we examined and compared specimens of Tobrilidae from the Alkaline Lakes of the western Nebraska Sandhills and an agrichemical-contaminated stream in eastern Nebraska (Johnsgard, 1995; Zahid et al., 2024). The Alkaline lakes overlay the Ogallala Aquifer, one of the world's largest underground reservoirs (Haacker, 2024). The five lakes in our study site are isolated from each other, are shallow in depth, and lack tributaries or an aboveground water source. They are maintained by groundwater and are considered evaporative lakes with pH values ranging from 7.5 to 10.5 (Gosselin, 1997; Gattoni et al., 2022). They are geochemically distinctive by their high levels of potassium-rich salts ranging over two orders of magnitude among lakes, depending on year and seasonal conditions. The potassium-to-sodium ratios in the highly alkaline lakes are approximately ten times higher than ocean water or fluids of the human body (Dunigan, 2024). Presumably, the nematodes that exist in the highest potassium levels, Bean, Border, and Kokjohn Lakes in this study, have unique physiological adaptations to regulate membrane potentials. Our goal in this study was to taxonomically characterize the tobrilid nematodes in the Alkaline Lakes by morphology, DNA barcodes, phylogeny, and habitat preferences. As in previous studies (Powers et al., 2016; Matczyszyn et al., 2022), we used discriminant function analysis (DFA) to test the accuracy of morphological characters and ecological attributes in correctly classifying unknown specimens within genetic groupings. We also compared tobrilids from the Alkaline Lakes to those collected from an eastern Nebraska stream that had been contaminated by a major agrichemical spill in 2021 (Zahid et al., 2024). Specimen information for all nematodes in this study, including the Nematode Identification (NID) number, GPS collection location, and GenBank accession number, is presented in Table 2.
Table 2.
Tobrilidae specimen information by Nematode Identification (NID) number, and molecular grouping by COI marker.
| NID # | COI Groupa | Genus or Sub-family | Gender/Stage |
Collection Location
|
GenBank accession #
|
|||
|---|---|---|---|---|---|---|---|---|
| County in NE | Site | Latitude; Longitude | Marker- 18S | Marker- COI | ||||
| 7157 | NA | Neotobrilus sp. | Female | Lancaster | Lincoln landfill | 40.871597; −96.654336 | PP854262 | |
| 11092 | NA | Neotobrilus sp. | Juvenile | Garden | Kokjohn Lake | 41.78259; −102.52226 | PP854263 | |
| 11093 | 2 | Neotobrilus sp. | Female | Garden | Kokjohn Lake | 41.78259; −102.52226 | PP855138 | |
| 11096 | NA | Neotobrilus sp. | Juvenile | Garden | Kokjohn Lake | 41.78259; −102.52226 | PP854264 | |
| 11097 | NA | Neotobrilus sp. | Juvenile | Garden | Kokjohn Lake | 41.78259; −102.52226 | PP854265 | |
| 11105 | NA | Neotobrilinae | Intersex | Garden | Gimlet Lake | 41.758702; −102.438185 | PP854246 | |
| 11890 | 2 | Neotobrilus sp. | Female | Garden | Bean Lake | 41.77776; −102.53009 | PP855143 | |
| 12093 | Singleton 1 | Neotobrilinae | Female | Garden | Border Lake | 41.79170; −102.53385 | PP854247 | PP855091 |
| 12103 | NA | Neotobrilinae | Female | Garden | Border Lake | 41.79170; −102.53385 | PP854248 | |
| 12127 | 1 | Tobrilus sp. | Female | Garden | Gimlet Lake | 41.75911; −102.43817 | PP854219 | PP855098 |
| 12129 | NA | Tobrilus sp. | Female | Garden | Gimlet Lake | 41.759109; −102.438171 | PP854227 | |
| 12138 | 1 | Tobrilus sp. | Female | Garden | Gimlet Lake | 41.75911; −102.43817 | PP854220 | PP855097 |
| 12141 | NA | Neotobrilus sp. | Female | Garden | Kokjohn Lake | 41.782081; −102.522884 | PP854266 | |
| 12144 | 2 | Neotobrilus sp. | Male | Garden | Kokjohn Lake | 41.78208; −102.52288 | PP854267 | PP855137 |
| 12168 | NA | Tobrilus sp. | Female | Garden | Island Lake | 41.733602; −102.410585 | PP854218 | |
| 12193 | 3 | Brevitobrilus sp. | Male | Garden | Island Lake | 41.73466; −102.41114 | PP854261 | PP855127 |
| 12194 | NA | Brevitobrilus sp. | Female | Garden | Island Lake | 41.734664; −102.411136 | PP854260 | |
| 12195 | NA | Tobrilus sp. | Female | Garden | Island Lake | 41.734664; −102.411136 | PP854208 | |
| 12204 | Singleton 2 | Neotobrilinae | Juvenile | Garden | Island Lake | 41.73466; −102.41114 | PP854243 | PP855101 |
| 12397 | NA | Brevitobrilus sp. | Male | Garden | Island Lake | 41.73481; −102.41120 | PP854259 | |
| 12400 | NA | Tobrilinae | Female | Garden | Island Lake | 41.73481; −102.41120 | PP854244 | |
| 12406 | 3 | Brevitobrilus sp. | Female | Garden | Island Lake | 41.73481; −102.41120 | PP854258 | PP855126 |
| 12408 | 3 | Brevitobrilus sp. | Female | Garden | Island Lake | 41.73481; −102.41120 | PP854257 | PP855124 |
| 12417 | 3 | Brevitobrilus sp. | Male | Garden | Island Lake | 41.73481; −102.41120 | PP854255 | PP855125 |
| 12420 | NA | Brevitobrilus sp. | Female | Garden | Island Lake | 41.73481; −102.41120 | PP854256 | |
| 12422 | NA | Neotobrilus sp. | Female | Garden | Border Lake | 41.79230; −102.53349 | PP854268 | |
| 12423 | NA | Neotobrilus sp. | Juvenile | Garden | Border Lake | 41.79230; −102.53349 | PP854269 | |
| 12424 | NA | Neotobrilus sp. | Female | Garden | Border Lake | 41.79230; −102.53349 | PP854270 | |
| 12425 | NA | Neotobrilus sp. | Juvenile | Garden | Border Lake | 41.79230; −102.53349 | PP854271 | |
| 12426 | 2 | Neotobrilus sp. | Juvenile | Garden | Border Lake | 41.79230; −102.53349 | PP854272 | PP855128 |
| 12427 | NA | Neotobrilus sp. | Male | Garden | Border Lake | 41.79230; −102.53349 | PP854273 | |
| 12428 | NA | Neotobrilus sp. | Juvenile | Garden | Border Lake | 41.79230; −102.53349 | PP854274 | |
| 12429 | NA | Neotobrilus sp. | Male | Garden | Border Lake | 41.79230; −102.53349 | PP854275 | |
| 12431 | NA | Neotobrilus sp. | Juvenile | Garden | Border Lake | 41.79230; −102.53349 | PP854276 | |
| 12432 | NA | Neotobrilus sp. | Juvenile | Garden | Border Lake | 41.79230; −102.53349 | PP854277 | |
| 12433 | NA | Neotobrilus sp. | Juvenile | Garden | Border Lake | 41.79230; −102.53349 | PP854278 | |
| 12434 | NA | Neotobrilus sp. | Juvenile | Garden | Border Lake | 41.79230; −102.53349 | PP854279 | |
| 12435 | NA | Neotobrilus sp. | Juvenile | Garden | Border Lake | 41.79230; −102.53349 | PP854280 | |
| 12442 | 2 | Neotobrilus sp. | Male | Garden | Border Lake | 41.79230; −102.53349 | PP854281 | PP855130 |
| 12444 | NA | Neotobrilus sp. | Male | Garden | Border Lake | 41.79230; −102.53349 | PP854282 | |
| 12522 | Singleton 3 | Neotobrilinae | Juvenile | Garden | Border Lake | 41.757717; −102.434 | PP854283 | PP855120 |
| 12523 | 2 | Neotobrilus sp. | Female | Garden | Border Lake | 41.757717; −102.434 | PP854284 | PP855136 |
| 12524 | 2 | Neotobrilus sp. | Juvenile | Garden | Border Lake | 41.757717; −102.434 | PP855133 | |
| 12525 | 2 | Neotobrilus sp. | Juvenile | Garden | Border Lake | 41.757717; −102.434 | PP854285 | PP855131 |
| 12528 | 2 | Neotobrilus sp. | Female | Garden | Border Lake | 41.79212; −102.53420 | PP854286 | PP855134 |
| 12533 | 2 | Neotobrilus sp. | Male | Garden | Border Lake | 41.79212; −102.53420 | PP854291 | PP855140 |
| 12537 | 2 | Neotobrilus sp. | Male | Garden | Border Lake | 41.79212; −102.53420 | PP854292 | PP855139 |
| 12538 | 2 | Neotobrilus sp. | Female | Garden | Border Lake | 41.79212; −102.53420 | PP854251 | PP855141 |
| 12539 | 2 | Neotobrilus sp. | Female | Garden | Border Lake | 41.79212; −102.53420 | PP854287 | PP855142 |
| 12540 | 2 | Neotobrilus sp. | Female | Garden | Border Lake | 41.79212; −102.53420 | PP854288 | PP855135 |
| 12554 | 4 | Neotobrilinae | Male | Garden | Gimlet Lake | 41.75543; −102.43447 | PP855086 | |
| 12556 | 1 | Tobrilinae | Female | Garden | Gimlet Lake | 41.75543; −102.43447 | PP855099 | |
| 12559 | 4 | Semitobrilus sp. | Juvenile | Garden | Gimlet Lake | 41.75543; −102.43447 | PP855059 | |
| 12561 | 4 | Semitobrilus sp. | Juvenile | Garden | Gimlet Lake | 41.75632; −102.43877 | PP855083 | |
| 12563 | 2 | Neotobrilus sp. | Juvenile | Garden | Bean Lake | 41.77068; −102.52883 | PP854250 | PP855144 |
| 12574 | 4 | Semitobrilus sp. | Female | Garden | Bean Lake | 41.77068; −102.52883 | PP854289 | PP855082 |
| 12575 | 2 | Neotobrilus sp. | Female | Garden | Bean Lake | 41.77068; −102.52883 | PP854254 | PP855132 |
| 12577 | NA | Neotobrilus sp. | Juvenile | Garden | Bean Lake | 41.77068; −102.52883 | PP854290 | |
| 12582 | 2 | Neotobrilus sp. | Immature Female | Garden | Bean Lake | 41.77068; −102.52883 | PP855129 | |
| 12586 | 1 | Tobrilus sp. | Female | Garden | Gimlet Lake | 41.75792; −102.43418 | PP854228 | PP855092 |
| 12587 | 1 | Tobrilus sp. | Male | Garden | Gimlet Lake | 41.75792; −102.43418 | PP854221 | PP855096 |
| 12588 | 1 | Tobrilus sp. | Male | Garden | Gimlet Lake | 41.75792; −102.43418 | PP854217 | PP855095 |
| 12591 | 1 | Tobrilus sp. | Female | Garden | Gimlet Lake | 41.75792; −102.43418 | PP854229 | PP855093 |
| 12592 | 4 | Semitobrilus sp. | Male | Garden | Gimlet Lake | 41.75792; −102.43418 | PP855081 | |
| 12593 | 4 | Semitobrilus sp. | Female | Garden | Gimlet Lake | 41.75792; −102.43418 | PP854234 | PP855084 |
| 12595 | 4 | Semitobrilus sp. | Male | Garden | Gimlet Lake | 41.75792; −102.43418 | PP854206 | PP855080 |
| 12596 | 4 | Semitobrilus sp. | Male | Garden | Gimlet Lake | 41.75792; −102.43418 | PP854207 | PP855079 |
| 12597 | NA | Semitobrilus sp. | Female | Garden | Gimlet Lake | 41.75792; −102.43418 | PP854235 | |
| 12598 | NA | Semitobrilus sp. | Male | Garden | Gimlet Lake | 41.75792; −102.43418 | PP854241 | |
| 12599 | 4 | Semitobrilus sp. | Male | Garden | Gimlet Lake | 41.75792; −102.43418 | PP855078 | |
| 12601 | 4 | Semitobrilus sp. | Male | Garden | Gimlet Lake | 41.75792; −102.43418 | PP855085 | |
| 12602 | 4 | Semitobrilus sp. | Female | Garden | Gimlet Lake | 41.75792; −102.43418 | PP855077 | |
| 12605 | 4 | Semitobrilus sp. | Female | Garden | Gimlet Lake | 41.75792; −102.43418 | PP854249 | PP855076 |
| 12606 | 1 | Tobrilus sp. | Juvenile | Garden | Gimlet Lake | 41.75792; −102.43418 | PP855094 | |
| 12607 | 1 | Tobrilus sp. | Female | Garden | Gimlet Lake | 41.75792; −102.43418 | PP854222 | PP855100 |
| 12613 | 4 | Semitobrilus sp. | Juvenile | Garden | Island Lake | 41.73257; −102.40362 | PP854240 | PP855075 |
| 12614 | 4 | Semitobrilus sp. | Juvenile | Garden | Island Lake | 41.73257; −102.40362 | PP854236 | PP855074 |
| 12615 | 4 | Semitobrilus sp. | Juvenile | Garden | Island Lake | 41.73257; −102.40362 | PP854237 | PP855073 |
| 12616 | 4 | Semitobrilus sp. | Juvenile | Garden | Island Lake | 41.73257; −102.40362 | PP854238 | PP855089 |
| 12617 | 4 | Semitobrilus sp. | Juvenile | Garden | Island Lake | 41.73257; −102.40362 | PP854242 | PP855072 |
| 12622 | NA | Neotobrilinae | Juvenile | Garden | Island Lake | 41.73257; −102.40362 | PP854245 | |
| 12623 | NA | Semitobrilus sp. | Juvenile | Garden | Island Lake | 41.73257; −102.40362 | PP854239 | |
| 12624 | 4 | Semitobrilus sp. | Juvenile | Garden | Island Lake | 41.73257; −102.40362 | PP855071 | |
| 12765 | 4 | Semitobrilus sp. | Juvenile | Garden | Island Lake | 41.73849; −102.40540 | PP855070 | |
| 12769 | 4 | Semitobrilus sp. | Female | Garden | Island Lake | 41.73849; −102.40540 | PP855069 | |
| 12770 | 4 | Semitobrilus sp. | Immature Female | Garden | Island Lake | 41.73849; −102.40540 | PP855068 | |
| 12773 | 4 | Semitobrilus sp. | Immature Female | Garden | Island Lake | 41.73849; −102.40540 | PP855067 | |
| 12774 | 4 | Semitobrilus sp. | Female | Garden | Island Lake | 41.73849; −102.40540 | PP855087 | |
| 12775 | 4 | Semitobrilus sp. | Male | Garden | Island Lake | 41.73849; −102.40540 | PP855066 | |
| 12776 | 4 | Semitobrilus sp. | Juvenile | Garden | Island Lake | 41.73849; −102.40540 | PP855065 | |
| 12778 | 4 | Semitobrilus sp. | Female | Garden | Island Lake | 41.73849; −102.40540 | PP855090 | |
| 12779 | 4 | Semitobrilus sp. | Female | Garden | Island Lake | 41.73849; −102.40540 | PP855064 | |
| 12780 | 4 | Semitobrilus sp. | Female | Garden | Island Lake | 41.73849; −102.40540 | PP855063 | |
| 12782 | 4 | Semitobrilus sp. | Female | Garden | Island Lake | 41.73849; −102.40540 | PP855062 | |
| 12786 | 4 | Semitobrilus sp. | Immature Female | Garden | Island Lake | 41.73849; −102.40540 | PP855061 | |
| 12827 | 4 | Semitobrilus sp. | Female | Garden | Gimlet Lake | 41.75767; −102.43452 | PP855060 | |
| 12836 | 4 | Semitobrilus sp. | Female | Garden | Gimlet Lake | 41.75767; −102.43452 | PP855088 | |
| 13994 | 6 | Neotobrilus sp. | Female | Saunders | Johnson Creek 1 | 41.23383; −96.46355 | PP855121 | |
| 14003 | 6 | Neotobrilus sp. | Male | Saunders | Johnson Creek 1 | 41.23383; −96.46355 | PP854252 | PP855122 |
| 14023 | 6 | Neotobrilus sp. | Female | Saunders | Johnson Creek 2 | 41.204850; −96.44180 | PP854253 | PP855123 |
| 14034 | 5 | Tobrilus sp. | Female | Saunders | Johnson Creek 3 | 41.19345; −96.42583 | PP854216 | PP855114 |
| 14036 | 5 | Tobrilus sp. | Male | Saunders | Johnson Creek 3 | 41.19345; −96.42583 | PP854230 | PP855109 |
| 14041 | 5 | Tobrilus sp. | Male | Saunders | Johnson Creek 3 | 41.19345; −96.42583 | PP854223 | PP855108 |
| 14042 | 5 | Tobrilus sp. | Female | Saunders | Johnson Creek 3 | 41.19345; −96.42583 | PP855112 | |
| 14044 | 5 | Tobrilus sp. | Male | Saunders | Johnson Creek 3 | 41.19345; −96.42583 | PP854215 | PP855110 |
| 14045 | 5 | Tobrilus sp. | Female | Saunders | Johnson Creek 3 | 41.19345; −96.42583 | PP854231 | PP855107 |
| 14047 | 5 | Tobrilus sp. | Female | Saunders | Johnson Creek 3 | 41.19345; −96.42583 | PP854224 | PP855115 |
| 14053 | 5 | Tobrilus sp. | Female | Saunders | Johnson Creek 3 | 41.19345; −96.42583 | PP854214 | PP855106 |
| 14054 | 5 | Tobrilus sp. | Male | Saunders | Johnson Creek 3 | 41.19345; −96.42583 | PP854232 | PP855105 |
| 14059 | 5 | Tobrilus sp. | Male | Saunders | Johnson Creek 4 | 41.19048; −96.42437 | PP854225 | PP855116 |
| 14061 | 5 | Tobrilus sp. | Male | Saunders | Johnson Creek 4 | 41.19048; −96.42437 | PP854213 | PP855117 |
| 14063 | 5 | Tobrilus sp. | Female | Saunders | Johnson Creek 4 | 41.19048; −96.42437 | PP854233 | PP855102 |
| 14065 | 5 | Tobrilus sp. | Female | Saunders | Johnson Creek 4 | 41.19048; −96.42437 | PP854226 | PP855103 |
| 14067 | 5 | Tobrilus sp. | Female | Saunders | Johnson Creek 4 | 41.19048; −96.42437 | PP854209 | PP855104 |
| 14071 | 5 | Tobrilus sp. | Male | Saunders | Johnson Creek 4 | 41.19048; −96.42437 | PP855113 | |
| 14072 | 5 | Tobrilus sp. | Male | Saunders | Johnson Creek 4 | 41.19048; −96.42437 | PP854212 | PP855118 |
| 14080 | 5 | Tobrilus sp. | Male | Saunders | Johnson Creek 5 | 41.17043; −96.40668 | PP854211 | PP855111 |
| 14081 | 5 | Tobrilus sp. | Female | Saunders | Johnson Creek 5 | 41.17043; −96.40668 | PP854210 | PP855119 |
NA indicates specimen appears only in 18S tree and has no COI clade designation.
Materials and Methods
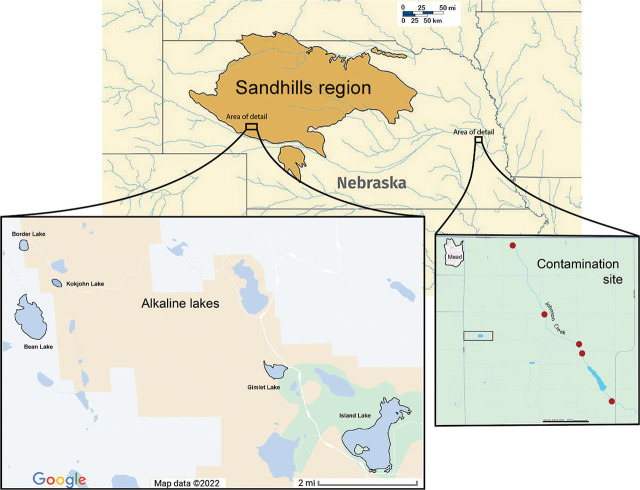
Soil collection and nematode isolation: Nematodes were collected from lake sediment and shoreline soil in 2019, 2020, and 2021 from five lake sites ranging in alkalinity within the Sandhills region of western Nebraska: Bean Lake, Border Lake, Gimlet Lake, Island Lake, and Kokjohn Lake. Additional samples were collected in 2022 from Johnson Creek near Mead, NE, a contaminated stream at an agricultural site approximately 400 miles east of the Alkaline Lakes (Fig. 1). From the Sandhills lakes in 2019, we collected three replicate samples per lake at about 10 meters from shore and in 2020 and 2021 collected four replicate samples per lake with a dredge from a kayak. For shoreline samples, 12–15 soil cores were taken along a 40-m transect at a depth of approximately 20 cm using an Oakfield Soil Corer with a 2.5-cm diameter (Gattoni et al., 2022, 2024). From Johnson Creek, we collected 500 ml of composite stream sediment by wading into the stream and taking 5–10 shovelfuls per site from five sites along 5 miles of the stream nearest the AltEn facility. All samples were stored at 8°C until nematodes were extracted from 200 cm3 of soil via the sieving and sugar centrifugation method (Jenkins, 1964).
Figure 1.
Map of Nebraska with the Sandhills region outlined. Area of research on the alkaline lakes in this region is indicated in an enlarged map inset. A second enlarged map inset shows the area of research in eastern Nebraska on an agrichemical contamination site. Red circles indicate the location of sample collections from the stream sediment.
Lake sediments and shoreline soils were analyzed for biogeochemistry at Ward Laboratories, Inc. (Kearney, Nebraska). The summary is presented in Supplementary Tables S1 and S2.
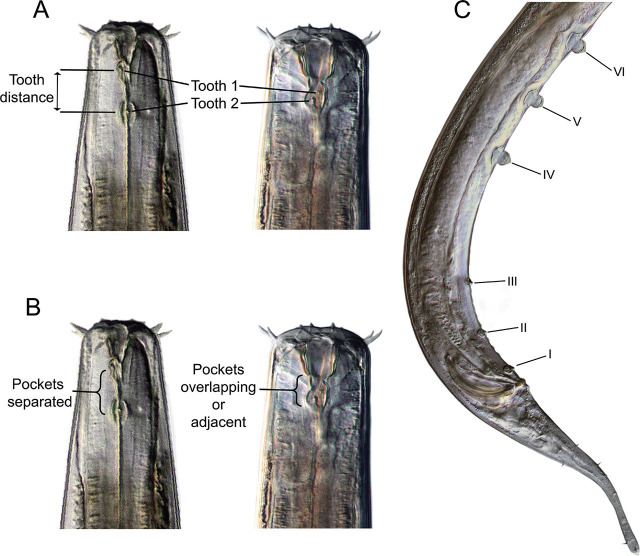
Nematode community analysis and morphological characterization: Abundance of tobrilid nematodes was estimated as a portion of the entire community, and if available, 25 individual specimens per sample were hand-picked, mounted on glass slides, heat-relaxed, and photographed. Measurements were recorded at ×400 and ×1,000 magnifications (Table 3a). Supplement numbering system follows the convention of Tsalolikhin and Shoshin (2009) (Fig. 2). Following microscopic analysis, slides were dismantled and immediately processed for DNA barcoding to preserve the linkage between DNA and morphology. Nematode images were taken of the full body, head, and tail with a Leica DC300 video camera mounted on a Leica DMLB light microscope with differential interference contrast. DNA was extracted from the photographed specimens by rupturing the nematode in an 18 mL droplet of sterilized water, which was stored at −20°C until PCR (Powers et al., 2014).
Table 3a.
Standard morphological measurements, abbreviations, and definitions.
| Gender | Character | Description (all measurements in μm) | |
|---|---|---|---|
| Females | Vulva | Length from anterior to vulva | |
| Females | Males | L | Overall body length |
| Females | Males | Tail | Portion of body from anus or cloaca to posterior terminus |
| Females | Males | a | Body length/greatest body diameter |
| Females | Males | b | Body length/distance from anterior to pharyngo-intestinal valve |
| Females | Males | c | Body length/tail Length |
| Females | Males | c′ | Tail length/body diameter at anus or cloaca |
| Females | V | % Vulva position/body length | |
| Females | VA/T | Distance from vulva to anus/tail length | |
| Females | Males | Tail % | % Tail length/body length |
| Females | Males | C set/lrw | Cephalic seta length/lip region width |
| Females | Males | L set/lrw | Labial seta length/lip region width |
| Females | Males | StomaL | Length of sclerotized portion of stoma (buccal cavity) |
| Females | Males | Dist | Distance between teeth |
| Males | Spic | Spicule length | |
| Males | Spic/L | % Spicule length/body length | |
| Males | Gub | Gubernaculum length | |
| Males | Cl-III | Distance between supplement III and cloaca | |
| Males | SR | Length of supplement row (I–VI) | |
| Males | SR/L | % Length of supplement row/total body length | |
Figure 2.
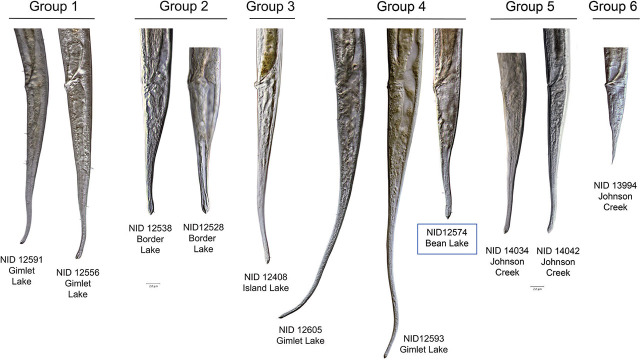
llustration of selected characters in Tobrilidae. A) Female heads (NID 12586, 13994) with teeth and tooth distance annotated. B) Female heads with differing tooth pocket arrangements. C) Male (NID 12533) posterior illustrating the numbering of supplements from the cloacal opening forward, with I designating the most posterior supplement and VII the most anterior.
The taxonomic keys, compendia, and references used to infer genus and species identities by morphology included Bongers (1989); Decraemer et al. (2019); Ebsary (1982); Gagarin and Naumova (2016); Holovachov and Shoshin (2014); Naumova and Gagarin (2017); Tsalolikhin and Shoshin (2009); and Zullini (2006).
DNA barcoding, phylogenetic tree construction, and reverse taxonomy: We applied “Reverse taxonomy” (Markmann and Tautz, 2005; Kanzaki et al., 2012), an approach that initially groups specimens based on position on a phylogenetic tree, followed by morphological assessment of nematodes that formed groups in the phylogenetic analysis. Importantly, each specimen on the COI phylogenetic tree was measured and photographed, and all light micrograph specimen images in the manuscript figures are represented on the COI tree. Nematode specimens were DNA barcoded, targeting the COI mitochondrial protein-coding gene and the 18S ribosomal DNA. The COI primers used were COI-JB3-Tob1 (JB3Tob1) –5′-TTTGGGCATCCTGAGGTTTATATTTTRA-3′(TSH, 2021 design based on JB3 (Bowles et al., 1992) and COI-R9-Tob1 (R9Tob1) – 5′-TGAAAATGAGCWACWACATAATAWGTRTC-3′(TSH, 2021 design based on COI-R9 (Powers et al., 2014)), which produce a 365-bp product once primers are trimmed. PCR was conducted in 0.5-mL thin-wall microcentrifuge tubes containing 30 μL of total volume consisting of 9 μL of the ruptured nematode template, 1.2 μL of double distilled water, 2.4 μL of both forward and reverse 20 μM primer solutions for a 1.6 μM final primer concentration, and 15 mL of JumpStart RED Taq ReadyMix (Sigma-Aldrich) at a 0.05U/mL final enzyme concentration. The initial hot start at 94°C for 5 min was followed by 35 cycles of 30 sec of denaturation at 94°C, annealing at 48°C for 30 sec, and extension at 72°C for 90 sec. The final extension occurred once at 72°C for 5 min. Successful PCR products were extracted with X-Tracta Tools (USA Scientific) prior to DNA sequencing from a 0.7% 1X TAE agarose gel, cleaned using Gel/PCR DNA Fragments Extraction Kit (IBI Scientific), and sent to Eton BioSciences, San Diego, CA for Sanger sequencing in both directions.
The 18S primers were 18s1.2a (5′-CGATCAGATACCGCCCTAG-3′) and 18sr2b (5′-TACAAAGGGCAGGGACGTAAT-3′), which produce a 593-bp product once primers are trimmed. 18s1.2a is the slightly re-designed 18s1.2 primer that was originally designed using consensus arthropod sequences (Mullin et al., 2003), while 18sr2b is a slightly redesigned reverse complement of primer rDNA2 from Vrain et al. (1992). This primer set amplifies approximately 630 bp of the 3’ portion of the 18S ribosomal DNA. PCR amplification of 5 uL of ruptured nematode template was conducted using the same conditions as the COI genetic marker except for annealing at 52°C, and 18S amplicon verification, cleaning, and sequencing were as described above. Forty-six of the 87 specimens used in the 18S phylogeny for this study are also represented on the COI phylogenetic tree.
To perform phylogenetic analyses and assess haplotype relationships among barcoded sequences of Tobrilidae, traces of barcode sequences of nematode specimens were edited using CodonCode Aligner Version 9.0 (http://www.codoncode.com). DNA sequences from this study were aligned with MEGAX v.10.2.6 to produce two separate (COI and 18S) alignments. Both alignments were subjected to analysis using a character-based maximum likelihood (ML) approach. ML trees were built using GTR+G (COI) and K2 + G + I (18S) models, both with 2,000 boot strap repetitions, and both with gap treatments using “Use all sites.” Each initial MEGAX alignment used MUSCLE with gap opening (−1,000) and gap extend (−500) penalties and UPGMB clustering method parameters. COI haplotype groups were generally defined by bootstrap values, a within-group distance that did not exceed 5%, and a distance to the nearest neighbor being greater than any within-group distance. Within- and between-COI clade genetic distances were calculated in MEGAX using p-distance with the assumption of Gamma Distributed Rates among Sites for 124 sequences, each with 393 nucleotide positions.
Discriminant Function Analyses: Discriminant Function Analysis (DFA) is a statistical method used to classify unknown individuals and the probability of their classification into pre-defined groups (Fisher, R. A., 1936; Lachenbruch and Goldstein, 1979; Jombart et al., 2010). Using DFA, we tested how well morphological characteristics or ecological (soil chemistry) attributes, or both morphological and soil chemistry attributes combined, classified female and male Tobrilidae into their respective COI haplotype groups (HGs).
Three datasets were created for both female and male Tobrilidae that were identified molecularly as belonging to specific COI HGs (Fig. 3) for use in our discriminant function analyses (DFA). The first dataset for both genders was soil chemistry attributes and the second dataset for both genders was morphological characters (Tables S1 and S2). The third dataset for both genders was a combination of both morphological characters and soil chemistry attributes.
Figure 3.
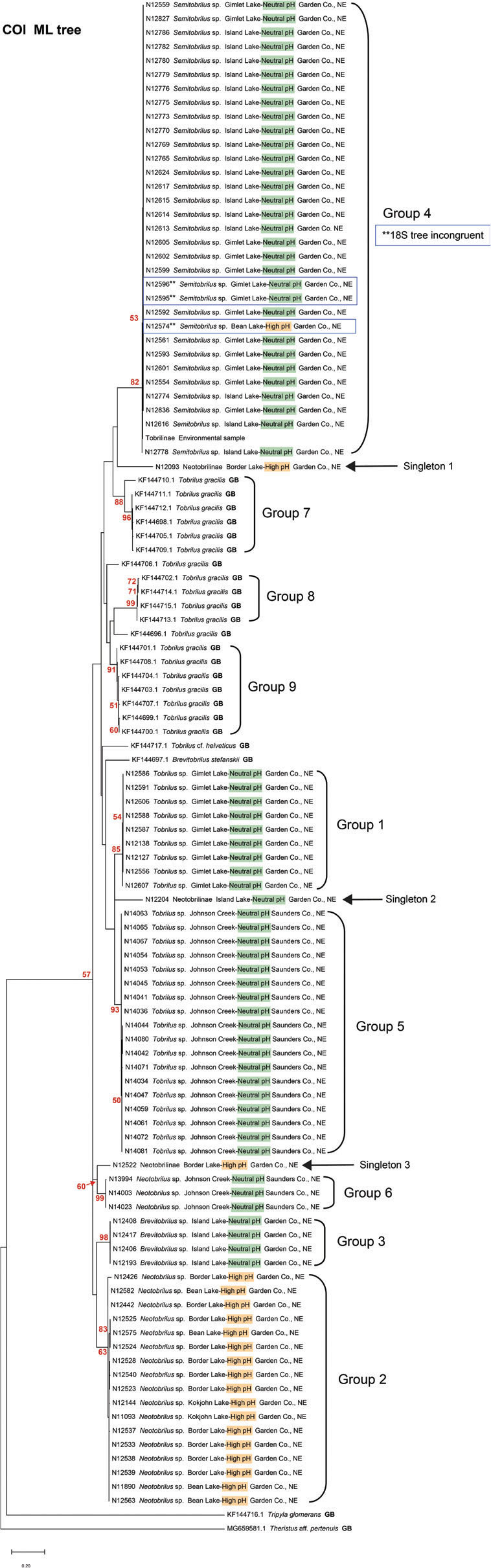
A maximum likelihood COI phylogenetic tree of 90 Nebraska specimens and 21 GenBank accessions. Highlighting in labels indicates lake pH. Green equals neutral, orange equals pH >8.5. Blue boxes around labels identify three specimens that display incongruence between COI and 18S trees. Nematode Identification numbers (NID) start with the prefix N followed by the number of the specimen.
Table 3a describes the morphological characters examined for females only, males only, and both males and females. The following soil chemistry attributes are described in Table 3b: soil pH, % OM, NO3, K, SO4, Zn, Fe, Mn, Cu, Ca, Mg, Na, B, CEC, % H, % K, % Ca, % Mg, % Na, Cl-, and P.
Table 3b.
Soil sample chemistry analyses, abbreviations, and definitions.
| Parameter | Methodology |
|---|---|
| Soil pH | Soil pH-soil pH 1:1 (1 volume of soil in 1 volume of water) |
| % OM | % Organic Matter-Measure of organic matter in soil by % LOI |
| NO3 | Nitrate ppm-Nitrate, KCl extractable |
| K | Potassium ppm-extracted by ammonium acetate |
| SO4 | Sulfate ppm-extracted by Mehlich S-III |
| Zn | Zinc ppm-DTPA (diethylenetriaminepentaacetic acid) micronutrient extraction method |
| Fe | Iron ppm-DTPA (diethylenetriaminepentaacetic acid) micronutrient extraction method |
| Mn | Manganese ppm-DTPA (diethylenetriaminepentaacetic acid) micronutrient extraction method |
| Cu | Copper ppm-DTPA (diethylenetriaminepentaacetic acid) micronutrient extraction method |
| Ca | Calcium ppm-extracted by ammonium acetate |
| Mg | Magnesium ppm-extracted by ammonium acetate |
| Na | Sodium ppm-extracted by ammonium acetate |
| B | Boron ppm-hot water extracted |
| CEC | CEC (meq/100g)-Cation Exchange Capacity: Sum (in meq) of the 4 cations(K+Ca+Mg+Na+)/100g soil |
| % H | %H Sat-%Base Saturation |
| % K | %K Sat-%Base Saturation |
| % Ca | %Ca Sat-%Base Saturation |
| % Mg | %Mg Sat-%Base Saturation |
| % Na | %Na Sat-%Base Saturation |
| Cl- | Chloride ppm-Chloride |
| P | Phosphorus ppm-Phosphorus, extracted by Mehlich P-III |
For the three female and three male datasets, the variables (the morphological characters or soil chemistry attributes) were initially inspected for missing values. If a variable was missing in more than 40% of the observations, such as when a morphological character was obscured, or the specimen was not in the appropriate developmental stage, those variables were dropped from further analysis. If the missing percentage was less than 40%, the missing values were replaced with the average of the remaining values. Correlations were then checked for the remaining variables using Pearson's Correlations Coefficient. Correlations were checked for elimination of highly correlated variables from the discriminant analysis. When two variables were highly correlated (greater than 0.8 or lower than −0.8), only one of the variables was retained for further analysis. The variables selected included a consideration of morphological characteristics specific to sex and important soil attributes such as K for the alkaline lake sites.
For each DFA analysis, the list of variables was determined by the correlation matrix. The dataset was normalized and then used in a stepwise variable selection using Akaike's Information Criterion (AIC) (Akaike, 1973). The stepAIC function in R v.4.1.2 (R Core Team, 2021) determined the final set of variables that can distinguish between the HGs from the model with the lowest AIC. Finally, using the selected features from the stepAIC method, linear discriminant analysis (LDA) was used to segment the HGs based on the continuous variables with equal prior probabilities (otherwise known as ‘priors’) for each HG. Three specimens that exhibited COI-18s incongruities were excluded from the morphological analyses used as the basis for the discriminant function analysis.
Scanning Electron Microscopy (SEM) preparation: Living nematodes were initially fixed in cold 4% glutaraldehyde for 24 hours, then rinsed in 0.1 M sodium cacodylate buffer before fixation in 2% osmium tetroxide for 8–12 hours. Nematodes were again rinsed in cacodylate buffer, followed by dehydration in a progressively increasing concentration of cold ETOH solutions until the nematodes were in 100% ETOH. Nematodes were critical point-dried, coated with silver, and mounted on an SEM stub before viewing on a Hitachi S4700 field-emission scanning electron microscope (SEM) at the Morrison Microscopy Core Research Facility of the Nebraska Center for Biotechnology http://biotech.unl.edu/microscopy.
Results
A total of 226 tobrilid specimens were microscopically examined during this study, 184 from the Alkaline Lake region and 42 from Johnson Creek in eastern Nebraska. Both eastern and western regions of Nebraska contained members of the subfamilies Tobrilinae and Neotobrilinae. Depending on specimen quality, sex, and stage, detailed morphometrics were recorded for a subset of these specimens.
Phylogenetic trees and genetic distances: A maximum likelihood COI phylogenetic tree of 90 Nebraska specimens and 21 GenBank accessions places the Alkaline Lake and Johnson Creek Tobrilidae into 6 haplotype groups (Fig. 3). Haplotype groups (HGs) 1 through 4 exclusively included specimens collected in the Alkaline Lakes, while HGs 5 and 6 are comprised of specimens collected in Johnson Creek. The three high-pH lakes, Bean, Border, and Kokjohn, contained only members of HG 2. The two lakes with more neutral pH, Gimlet and Island Lakes, each have a haplotype group found exclusively in their waters. Gimlet Lake was the only lake with HG 1, and HG 3 was only collected in Island Lake. HG 4 was found in both Island and Gimlet Lakes. The Sandhills Alkaline Lake locality and Johnson Creek in eastern Nebraska had representative specimens of the subfamilies Tobrilinae (HGs 1 and 5) and Neotobrilinae (HGs 2, 3, 4, and 6). Each of the six haplotype groups was supported by a combination of: a) medium to high bootstrap values (e.g., 65–100), b) low within-group mean genetic distance estimates (0.0%–0.54%), and c) relatively high between-group pairwise distance measurements that ranged from 6.04% to 16.44% (Table 4). Included in the COI tree were three clades of GenBank sequences identified as Tobrilus gracilis from Europe (Ristau et al., 2013). These European specimens comprised three haplotype groups on the tree that were well-supported by bootstrap values (88–99), had low levels of within-group mean genetic distances (0.46%–2.74%), and exhibited large between-group distances when compared among the three European populations (11.76%–18.36%). The between-group mean distance estimates for European versus Nebraskan haplotype groups ranged from (10.71%–17.34%). None of the three European haplotype groups identified as Tobrilus gracilis exhibited a genetically close relationship with the Nebraskan COI haplotype groups.
Table 4.
Within- and between-group mean distances of 9 tobrilid COI groups. Within-group distance is shown in bold type.
| Group 4 | Group 1 | Group 5 | Group 6 | Group 3 | Group 2 | Group 7 | Group 9 | Group 8 | |
|---|---|---|---|---|---|---|---|---|---|
| Group 4 | 0.000639 | ||||||||
| Group 1 | 0.159155 | 0.000651 | |||||||
| Group 5 | 0.165664 | 0.060499 | 0.002543 | ||||||
| Group 6 | 0.157512 | 0.126478 | 0.111416 | 0.001826 | |||||
| Group 3 | 0.158968 | 0.123860 | 0.124103 | 0.096937 | 0 | ||||
| Group 2 | 0.164354 | 0.131401 | 0.133042 | 0.094518 | 0.093262 | 0.005378 | |||
| Group 7 | 0.141845 | 0.141270 | 0.126408 | 0.131963 | 0.140716 | 0.151729 | 0.027397 | ||
| Group 9 | 0.141204 | 0.110544 | 0.107067 | 0.117939 | 0.117579 | 0.124228 | 0.130528 | 0.007567 | |
| Group 8 | 0.173377 | 0.128000 | 0.145053 | 0.158447 | 0.133743 | 0.144301 | 0.183562 | 0.117613 | 0.004566 |
Morphological characteristics of haplotype groups: Measurements were obtained from most of the adult specimens in COI HGs 1–6 from Nebraska (Table 5). Notable diagnostic characters and observations include the following:
Tooth distance and location of “pockets”. HGs 1 and 5 have the typical characteristics of the subfamily Tobrilinae, with two overlapping or adjacent pockets located just posterior to the relatively large, sclerotized buccal cavity. Each pocket contains a single small tooth (Figs. 2, 4). Distance between the teeth in HG 1 averages 5.7 μm in females and 3.0 μm in males, while in HG 5, it averages 2.0 μm in females and 2.1 μm in males. Teeth in HGs 2, 3, 4, and 6 are located in distinctly separate non-overlapping pockets one behind the other, posterior to the stoma base, characteristic of the subfamily Neotobrilinae. Tooth distances average 12.4 μm (females) and 11.8 μm (males) in HG 2.
-
Male supplements. (Figs. 2, 5)
Supplements among the six haplotype groups vary in their internal and external complexity, number, and spacing. Members of HG 1 and 5 have submerged supplements that do not protrude more than 1–2 μm above the cuticle surface. All HG 1 males examined have 6 supplements; most HG 5 specimens also have 6 supplements, but a few possess 7. In HG 4, the supplements are also 6 in number, submerged or flush with the body contour. HGs 2, 3, and 6 all have 6 protruding supplements, with the 3 more anterior ones larger and echinate and the posterior 3 smaller and less prominent. The largest gap in supplement spacing for HG 1 is between the cloacal opening and supplement; supplement I was measured from the cloacal opening forward, with I designating the most posterior supplement and VI [or VII] the most anterior (Fig. 2). Supplements IV and V are the closest together. In HG 2 males, supplement I is close to the cloacal opening and the largest gap is between supplements III and IV, separating the 3 larger anterior supplements from the 3 smaller, more posterior ones. HG 3 shows a similar pattern, but the distance between I and II is generally less than in HG 2. For HG 4, I and II are also closer together, and the largest gap is seen between supplements V and VI. HG 6 shows a very similar pattern to that of HG 2, but HG 6 is represented by a single male specimen. HG 5 is very similar to HG 1 in supplement spacing but with a larger gap separating V and VI.
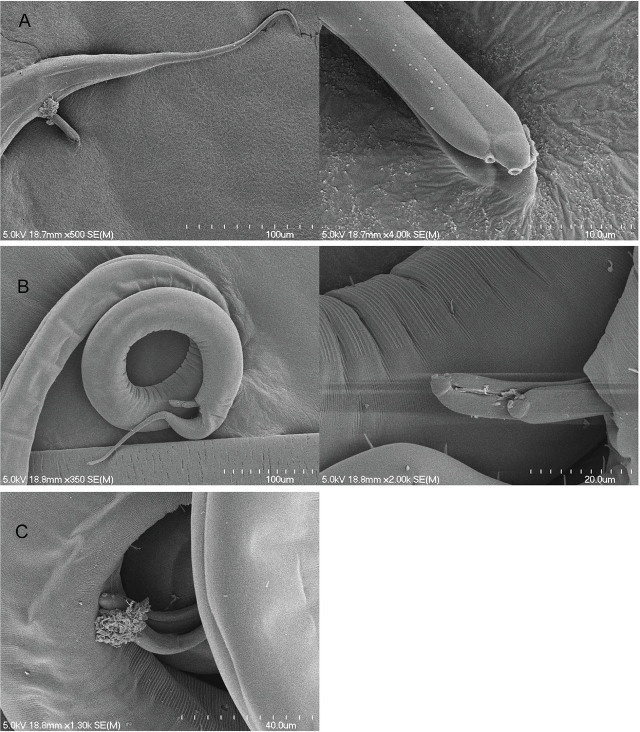
Micropapillae. The presence or absence, number, and arrangement of intersupplementary structures, called micropapillae (Figs. 6, 7), are other taxonomic characters used to differentiate members of Tobrilidae. The term “micropapillae” may be a misnomer as pointed out by Tsalolikhin and Shoshin (2009). These structures are not shortened somatic setae but appear as thickened transverse annuli between the supplements, best seen in SEM (Figs. 6, 7). There is significant variation among the six haplotype groups in expression of this character. HGs 1, 3, and 5 lack micropapillae, but the ventral cuticle between the supplements in males of HG 5 is finely annulated in appearance along the entirety of the supplement range. In HG 4, numerous ventral pores are observed - with light microscopy - along the entire length of the body from the cloacal opening to the head region. Micropapillae are present in HGs 2 and 6 but differ in number. In HG 2, there are 5 to 10 between supplements I and II, 4 or 5 between II and III, and 4 between III and IV. In HG 6, there are 4 micropapillae between I and II and between II and III and only one anterior to supplement III.
Spicule length. Average spicule length (measured along the arc) in HG 4 was more than twice as long as in any of the other five groups (Fig. 4), averaging 131.5 μm or 5.4% of total body length. HG 6 has conspicuously shorter spicules with an average length of 35.4 μm (1.9% of body length). HG 1 has slightly longer spicules: 43.3 μm long on average, or 2.0% of body length. HGs 2 (60.6 μm; 3.4%), 3 (57.2 μm; 2.7%), and 5 (60.3 μm; 4.7%) fall between these extremes. A newly observed feature of the spicules can be seen in Figure 7. On the distal tip of each spicule is a conspicuous pore of unknown function. The diameter of the pore is approximately 1.0 μm. This feature was seen in four specimens of HG 4 but not observed in other groups. Spicules were more commonly everted in HG 4, possibly because the spicule length in this group is generally twice that of the other groups.
Sperm. HGs 2 and 6 have flagellate spermatozoa, observed in both males and in the filled spermathecae of females. Sperm cells in HGs 1, 3, 4, and 5 are spherical to ovoid, without flagella.
Vaginal musculature. Musculature associated with the vaginal canal is generally categorized in Tobrilidae as weakly, moderately, or strongly developed, and the presence or absence of concentric bands or rings of sphincter muscles has been considered a useful diagnostic character, providing the specimens being examined are fully mature. Musculature in HGs 1, 3, and 5 is weakly to moderately developed and lacks any concentric muscle bands. HGs 2, 4, and 6 are all characterized by strongly developed muscles arranged in a multi-layered, bulb-like configuration (Fig. 8).
Paravulval structures. HG 5 females are distinguished by the presence of slightly raised and thickened cuticular “pads” anterior and posterior to the vulval opening. These are absent in HGs 1, 2, 3, 4 (although one mature female specimen had corrugations or folds in the ventral cuticle anterior and posterior to the vulva), and 6.
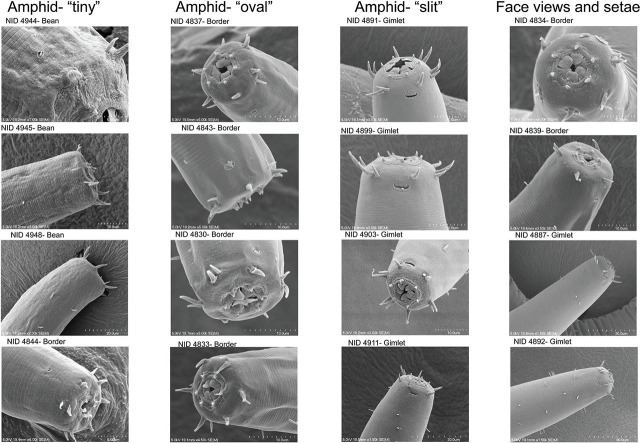
Labial and cephalic setae. In general, labial and cephalic setae in all six haplotype groups are fairly short, not exceeding 35% of the lip region width (lrw) in any of the groups, with the range of seta lengths forming a continuum across the clades (Fig. 9). HGs 1 and 5 have comparatively longer setae at 5.6–8.5 μm (18.3–32.5% lrw) for HG 1 and 5.2–7.4 μm (20.6–34.1% lrw) for HG 5. Setae in HGs 2 (4.5–6.2 μm; 18.1–29.9%) and 4 (5.0–6.9 μm; 18.5–24.9%) are somewhat shorter, and HGs 3 and 6 display the shortest head setae: 2.3–4.6 μm in HG 3 (18.1–29.9%) and 3.7–4.2 μm in HG 6 (25.3–32.1%). In specimens of HG 2 from Bean and Border Lakes, SEM face and lateral profiles (Fig. 9) show single cephalic setae positioned directly anterior to the amphid apertures, approximately 5–6 μm distant. In members of HG 4 from Gimlet Lake, the single cephalic setae on the lateral sides of the body are positioned closer to the amphid aperture, approximately 2–3 μm, and diagonal with respect to the amphid location.
Amphid shape. Two variations in amphid shape were observed in the SEM examination. HG 2 specimens from Border Lake have distinctly rounded amphid apertures, 1.0–2.0 μm in diameter. HG 2 amphids have continuous circular margins, as seen in Figure 9. The HG 4 amphids shown from Gimlet Lake are more laterally flattened in shape and do not have a continuous margin around the aperture. Instead, the anterior-most portion of the aperture appears pectinate, resembling a comb-like fringe. Some of the circular amphids of HG 2 appear to have an internal pectinate structure that is nearly flush with the external cuticle and continuous around the amphid aperture margin.
Body length. In general, female specimens are longer than males except in HGs 3 and 6, where males are slightly longer: 1281 μm on average for HG 6 males vs. an average length of 1267 μm for females and 2157 μm vs. 2033 μm for males and females of HG 3, respectively. Average body length for the sampled populations (males and females together) in HG 2 ranges from 1794 to 1999 μm, from 1918 to 2079 μm in HG 5, from 2169 to 2393 μm in HG 1, and from 2179 to 2493 μm in HG 4 (Fig. 10).
Tail length. Average tail length is greater in females than in males across all haplotype groups. Tails of HG 4 females are distinctly longer (average 361 μm) than those of other groups, with HGs 1 and 3 (267 μm and 258 μm, respectively) also fairly long. Shorter tails are seen in HG 6 (204 μm), HG 2 (189 μm), and HG 5 (123 μm). A similar pattern is seen in males: HG 4 (281 μm) again has conspicuously longer tails, followed by HG 3 (182 μm), HG 2 (159 μm), HG 1 (155 μm), HG 6 (133 μm), and HG 5 (122 μm) (Fig. 11).
Subterminal setae. HGs 1, 4, and 6 apparently lack subterminal setae, while HGs 2, 3, and 5 have this feature. In HG 5, the STS is consistently closer to the tail terminus (97.2% of tail length) than in HG 3 (96.3%) or HG 2 (94.1%).
Table 5.
Comparisons of selected morphological characteristics of the main groups (mean ± standard deviation with range of values)
|
Group 1
Tobrilus Female (n=7) |
Group 2
Neotobrilus Female (n=9) |
Group 3
Brevitobrilus Female (n=4) |
Group 4
Semitobrilus Female (n=4) |
Group 5
Tobrilus Female (n=17) |
Group 6
Neotobrilus Female (n=3) |
|
|---|---|---|---|---|---|---|
| L | 2366.7 ± 236.6 (2073–2863) | 1999.3 ± 301.7 (1449–2341) | 2032.8 ± 197.9 (1727–2229) | 2729.6 ± 192.1 (2415–2937) | 2079.3 ± 209.8 (1769–2739) | 1225.4 ± 157.7 (1087–1446) |
| a | 31.2 ± 2.1 (29.7–36.1) | 31.5 ± 5.9 (21.8–41.1) | 35.8 ± 2.3 (32.8–39.0) | 33.7 ± 1.1 (32.4–35.2) | 29.7 ± 2.4 (24.1–33.5) | 26.4 ± 1.9 (24.2–28.8) |
| b | 5.6 ± 0.2 (5.4–6.0) | 5.4 ± 0.5 (4.1–6.0) | 6.4 ± 0.3 (6.1–6.7) | 6.7 ± 0.2 (6.4–6.9) | 5.5 ± 0.4 (4.5–6.0) | 4.9 ± 0.2 (4.6–5.2) |
| c | 8.9 ± 0.7 (7.4–9.6) | 10.6 ±1.0 (8.4–11.6) | 7.9 ± 0.4 (7.1–8.2) | 7.1 ± 0.2 (6.9–7.3) | 10.0 ± 2.5 (8.5–19.8) | 9.5 ± 1.8 (7.7–12.0) |
| c′ | 6.8 ± 0.7 (6.0–8.0) | 4.6 ±0.4 (4.2–5.3) | 7.4 ± 0.5 (6.9–8.1) | 7.8 ± 0.4 (7.2–8.2) | 5.8 ± 0.8 (2.8–6.8) | 5.2 ± 1.2 (3.4–6.1) |
| V | 44.7% ± 2.4% (42.0%–48.5%) | 46.1% ± 3.8% (43.6%–56.4%) | 41.0% ± 1.2% (39.6%–43.0%) | 48.7% ± 1.6% (46.3%–50.6%) | 42.5% ± 3.9% (32.4%–47.1%) | 42.1% ± 1.2% (41.2%–43.8%) |
| Tail | 265.1 ± 19.3 (239–297) | 188.5 ± 17.6 (154 –210) | 257.8 ± 13.5 (242 –275) | 385.7 ± 23.9 (350–414) | 216.2 ± 34.4 (97–264) | 131.4 ± 11.7 (121–148) |
| V-A/T | 3.9 ± 0.4 (3.2–4.4) | 4.7 ± 0.8 (2.7–5.5) | 3.6 ± 0.2 (3.3–3.8) | 2.6 ± 0.1 (2.5–2.8) | 4.7 ± 1.3 (3.7–9.7) | 4.5 ± 1.1 (3.5–6.0) |
| LRW | 30.7 ± 2.7 (28–36) | 28.9 ± 2.9 (24–34) | 24.3 ± 2.6 (21–28) | 28.9 ± 1.7 (27–31) | 26.8 ± 2.9 (22–31) | 15.4 ± 1.3 (14–17) |
| Set L | 5.9 ± 2.4 (2–8) | 6.1 ± 2.3 (4–10) | 4.6 ± 1.1 (3–5) | 8.2 ± 0.7 (8–9) | 6.7 ± 1.2 (5–8) | 4.5 ± 1.2 (3–5) |
| Set L/LRW | 19.5% ± 8.4% (4.9%–28.9%) | 21.6% ± 9.3% (13.8%–38.9%) | 19.0% ± 5.1% (12.5%–25.0%) | 28.4% ± 2.1% (25.2%–30.9%) | 25.0% ± 3.6% (17.6%–31.8%) | 28.7% ± 6.8% (19.6%–36.1%) |
| Stoma L | 12.3 ± 1.2 (10–14) | 9.7 ± 1.2 (8–12) | 8.5 ± 0.5 (8–9) | 7.5 ± 1.5 (6–10) | 13.1 ± 1.2 (10–15) | 8.3 ± 0.7 (7–9) |
| Ant tooth pos | 17.8 ± 4.8 (9–23) | 20.3 ± 4.7 (15–29) | 19.9 ± 4.9 (15–26) | 17.2 ± 2.3 (15–20) | 20.9 ± 1.7 (18–24) | 11.6 ± 1.4 (10–13) |
| Post tooth pos | 24.2 ± 2.4 (19–27) | 32.7 ± 3.8 (25–39) | 29.1 ± 4.6 (24–35) | 24.0 ± 4.9 (19–32) | 23.0 ± 2.2 (20–27) | 22.7 ± 2.3 (19–24) |
| Tooth dist | 6.3 ± 3.8 (2–13) | 12.4 ± 2.7 (8–16) | 11.3 ± 1.2 (10–12) | 6.7 ± 3.2 (5–12) | 2.1 ± 0.8 (1–4) | 11.0 ± 1.0 (10–12) |
|
Group 1
Tobrilus Male (n=2) |
Group 2
Neotobrilus Male (n=7) |
Group 3
Brevitobrilus Male (n=3) |
Group 4
Semitobrilus Male (n=4) |
Group 5
Tobrilus Male (n=19) |
Group 6
Neotobrilus Male (n=1) |
|
|---|---|---|---|---|---|---|
| L | 2168.7 ± 54.4 (2114–2223) | 1794.1 ± 152.5 (1579–2026) | 2156.7 ± 183.1 (1899–2307) | 2517.3 ± 161.9 (2244–2663) | 1918.3 ± 142.2 (1623–2146) | 1281.3 ± 0.0 (1281–1281) |
| a | 37.2 ± 3.5 (33.8–40.7) | 35.5 ± 3.3 (30.1–39.1) | 39.4 ± 0.1 (39.3–39.5) | 37.3 ± 3.2 (32.8–41.3) | 34.5 ± 3.3 (26.8–39.4) | 29.6 ± 0.0 (29.6–29.6) |
| b | 5.4 ± 0.3 (5.1–5.6) | 5.3 ± 0.3 (4.8–5.6) | 6.5 ± 0.4 (5.9–6.9) | 6.7 ± 0.3 (6.3–7.0) | 5.5 ± 0.3 (4.3–6.0) | 5.6 ± 0.0 (5.6–5.6) |
| c | 14.0 ± 0.1 (14.0–14.1) | 13.7 ± 3.1 (11.3–20.8) | 11.9 ± 0.6 (11.2–12.6) | 8.9 ± 0.5 (8.5–9.6) | 14.5 ± 1.1 (12.9–16.6) | 10.5 ± 0.0 (10.5–10.5) |
| c′ | 4.2 ± 0.1 (4.1–4.3) | 3.5 ± 0.6 (2.1–4.1) | 4.7 ± 0.1 (4.6–4.8) | 5.3 ± 0.2 (5.1–5.7) | 3.8 ± 0.4 (2.8–4.8) | 3.8 ± 0.0 (3.8–3.8) |
| T | 66.4% ± 2.6% (63.8%–69.0%) | 63.6% ± 2.4% (61.3%–67.8%) | 66.6% ± 0.5% (66.3%–67.3%) | 64.2% ± 1.7% (61.8%–66.3%) | 66.6% ± 3.6% (59.0%–77.0%) | 68.7% ± 0.0% (68.7%–68.7%) |
| Tail | 154.6 ± 4.5 (150–159) | 134.8 ± 20.3 (90–158) | 181.8 ± 11.8 (169 –197) | 281.9 ± 14.9 (264–305) | 132.7 ± 10.9 (100–147) | 122.3 ± 0.0 (122–122) |
| LRW | 26.1 ± 0.7 (25–27) | 25.1 ± 2.9 (22–29) | 20.2 ± 1.5 (18–22) | 26.0 ± 1.2 (25–27) | 22.3 ± 1.8 (18–25) | 13.2 ± 0.0 (13–13) |
| Set L | 8.5 ± 1.0 (7–9) | 6.2 ± 1.6 (3–8) | 3.3 ± 0.3 (3–4) | 6.7 ± 0.8 (5–7) | 7.2 ± 0.9 (5–9) | 4.2 ± 0.0 (4–4) |
| Set L/LRW | 32.5% ± 4.7% (27.8%–37.3%) | 24.2% ± 7.8% (9.4%–31.5%) | 16.4% ± 0.5% (15.9%–17.0%) | 25.8% ± 3.6% (20.2%–30.3%) | 32.6% ± 3.9% (23.9% –39.2%) | 32.1% ± 0.0% (32.1%–32.1%) |
| Stoma L | 12.2 ± 0.2 (12–12) | 8.3 ± 0.5 (8–9) | 9.4 ± 5.3 (5–17) | 7.3 ± 1.4 (5–9) | 12.1 ± 1.4 (9–17) | 8.0 ± 0.0 (8–8) |
| Ant tooth pos | 21.0 ± 0.9 (20–22) | 16.5 ± 1.8 (14–19) | 13.8 ± 1.4 (12–16) | 16.3 ± 2.3 (13–19) | 19.0 ± 1.1 (17–21) | 10.0 ± 0.0 (10–10) |
| Post tooth pos | 24.0 ± 0.9 (23–25) | 28.3 ± 2.1 (25–32) | 23.8 ± 0.6 (23–24) | 22.5 ± 2.9 (18–25) | 21.0 ± 1.4 (18–24) | 24.4 ± 0.0 (24–24) |
| Tooth dist | 3.0 ± 0.0 (3–3) | 11.8 ± 2.2 (7–14) | 9.8 ± 1.1 (9–11) | 6.2 ± 1.9 (5–9) | 2.0 ± 0.8 (1–3) | 14.4 ± 0.0 (14–14) |
| Spicule | 43.3 ± 0.7 (43–44) | 60.6 ± 8.4 (50–74) | 57.2 ± 4.6 (52–63) | 135.8 ± 4.5 (130–143) | 35.4 ± 1.9 (32–40) | 60.3 ± 0.0 (60–60) |
| Gubernaculum | 21.7 ± 1.0 (21–23) | 19.1 ± 4.7 (11–25) | 19.8 ± 1.6 (18–22) | 37.7 ± 5.8 (30–45) | 13.3 ± 2.8 (7–17) | 25.6 ± 0.0 (26–26) |
| Supplement # | 6 | 6 | 6 | 6 | 5–7 | 6 |
| Supp prominence | submerged | protruding | protruding | submerged | submerged | protruding |
| Supp rel size | equal | unequal | unequal | equal | equal | unequal |
| Cl-I | 28.2% ± 0.8% (27.4%–28.9%) | 4.4% ± 0.6% (3.5%–5.5%) | 5.0% ± 0.3% (4.7%–5.3%) | 22.7% ± 1.5% (20.3%–24.4%) | 30.1% ± 4.7% (23.9%–42.0%) | 5.0% ± 0.0 (5.0%–5.0%) |
| I-II | 24.6% ± 4.0% (20.6%–28.6%) | 11.2% ± 1.0% (9.5%–12.6%) | 6.8% ± 1.0% (5.5%–7.9%) | 11.6% ± 1.1% (10.3%–12.9%) | 23.0% ± 2.0% (20.1%–27.4%) | 12.7% ± 0.0% (12.7%–12.7%) |
| II-III | 22.8% ±1.7% (21.1%–24.5%) | 12.2% ± 1.7% (9.8%–15.2%) | 14.5% ± 1.2% (13.5%–16.3%) | 23.8% ± 2.0% (20.5%–25.3%) | 19.9% ± 1.8% (17.3%–24.6%) | 12.0% ± 0.0% (12.0%–12.0%) |
| III-IV | 21.7% ± 3.7% (18.0%–25.4%) | 34.7% ± 5.2% (25.8%–41.2%) | 39.7% ± 4.6% (34.5%–45.6%) | 9.0% ± 0.7% (8.1%–9.8%) | 19.2% ± 2.1% (14.4%–22.3%) | 31.3% ± 0.0% (31.3%–31.3%) |
| IV-V | 14.4% ± 0.9% (13.5%–15.3%) | 17.3% ± 6.5% (0.0%–19.8%) | 17.9% ± 1.0% (16.5%–18.9%) | 18.3% ± 2.1% (15.9%–21.5%) | 14.9% ± 5.2% (8.6%–31.8%) | 19.0% ± 0.0% (19.0%–19.0%) |
| V-VI | 16.5% ± 6.9% (9.5%–23.4%) | 27.0% ± 11.1% (18.6%–46.0%) | 21.0% ± 1.8% (18.5%–22.9%) | 37.2% ± 5.2% (31.0%–45.2%) | 23.4% ± 5.5% (10.0%–30.8%) | 24.9% ± 0.0% (24.9%–24.9%) |
| Cl-III | 156.5 ± 3.9 (153–160) | 77.1 ± 5.1 (70–85) | 67.8 ± 5.7 (62–75) | 181.6 ± 27.0 (150–224) | 143.6 ± 7.1 (131–155) | 65.1 ± 0.0% (65–65 |
| Spic/III | 0.3 ± 0.0 (0.3–0.3) | 0.8 ± 0.1 (0.6–0.9) | 0.9 ± 0.1 (0.7–0.9) | 0.8 ± 0.1 (0.6–0.9) | 0.2 ± 0.0 (0.2–0.3) | 0.9 ± 0.0 (1–1) |
| SR/L | 9.6% ± 0.7% (8.9%–10.3%) | 15.7% ± 2.0% (13.7%–20.0%) | 12.0% ± 0.5% (11.3%–12.5%) | 12.4% ± 1.0% (11.3%–13.8%) | 10.3% ± 0.8% (9.1%–12.1%) | 17.1% ± 0.0 (17.1%–17.1%) |
Figure 4.
Comparison of the heads of specimens from each of the six Haplotype Groups, with their identification number and lake from which they were collected. Stoma shape, pockets, and tooth distance are featured. Blue box indicates incongruent specimen between COI and 18S trees.
Figure 5.
Comparison of male spicules and supplements from haplotype groups one through six. The nematode identification (NID) number and collection location are shown for each. Groups 1, 4, and 5 have supplements that do not conspicuously protrude from the cuticle surface. Groups 2, 3, and 6 have protruding supplements. Group 4 has notably long spicules compared to other groups. The blue box around the label for NID 12595 denotes a specimen incongruent for COI and 18S trees.
Figure 6.
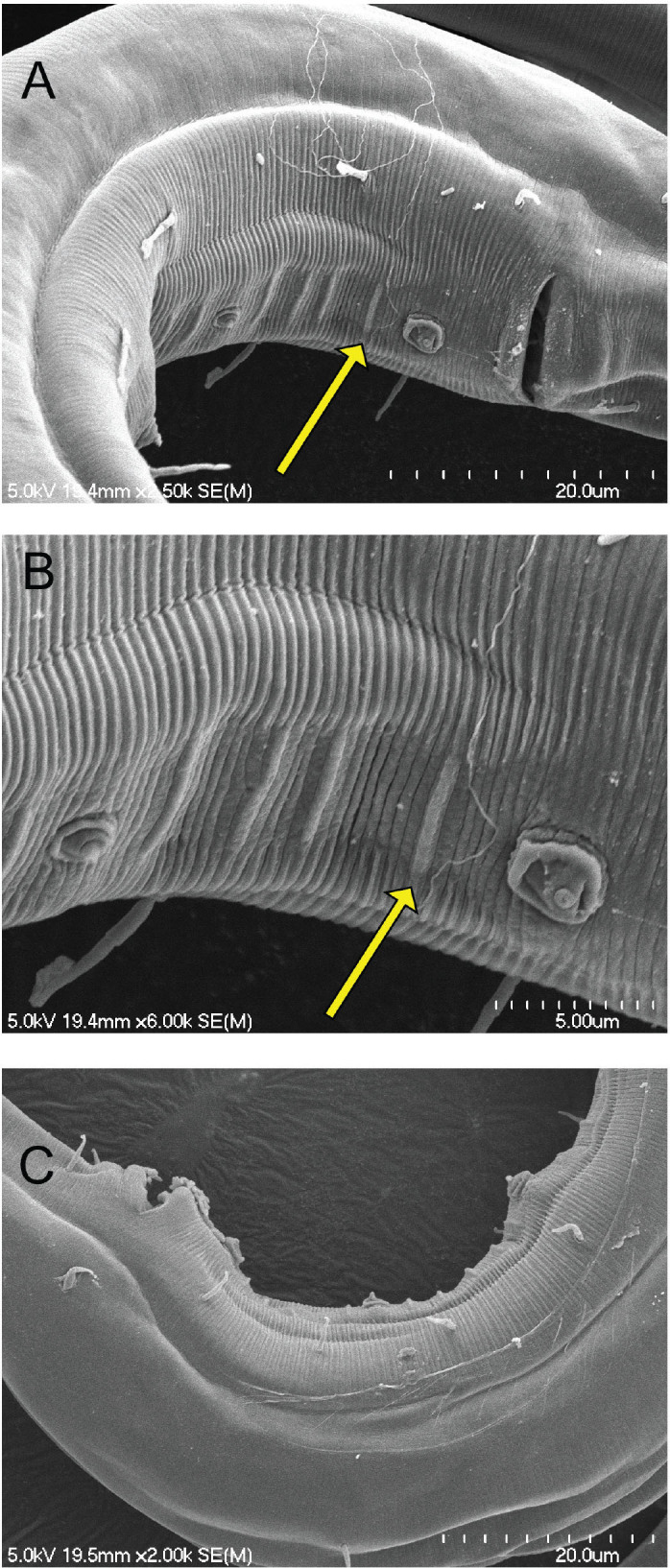
Scanning electron micrographs of the male posterior region showing the intersupplementary structures known as micropapillae. A, B) NID 4829 from Border Lake, micropapillae in ventral view appear as four ridges between supplements; C) NID 4841 from Border Lake, micropapillae in lateral view appear as small bumps or papillae.
Figure 7.
Scanning electron micrographs of Group 4 male spicules, all from Gimlet Lake, displaying possible pores at the tip. A) NID 4913 tail and close-up of spicules; B) NID 4926 posterior and close-up of spicules; C) NID 4899 spicules.
Figure 8.
Comparison of the musculature associated with the vaginal canal in haplotype groups one through six. The Tobrilinae, Groups 1 and 5, display weak or poorly developed vaginal musculature. Group 3 appears to have moderately developed musculature, and Groups 2, 4, and 6 are all characterized by strongly developed muscles arranged in a multi-layered, bulb-like configuration. Blue box around NID 12574 indicates incongruent specimen with respect to the COI and 18S trees and has a strong, bulb-like musculature characteristic of Group 2.
Figure 9.
Scanning electron micrographs of the head, amphid and cephalic setae of selected specimens from alkaline lakes in the Nebraska Sandhills. Amphid shapes were categorized as tiny, oval or slit-like. Slit-like amphids were positioned closest to cephalic setae. Inner labial setae are notably shorter than the outer labial and cephalic setae, which are of similar lengths.
Figure 10.
Entire body images of female and male specimens from each of the six haplotype groups. The nematode identification (NID) number and collection location are shown for each. Blue box indicates incongruent specimen.
Figure 11.
Comparison of female tails from each of the six haplotype groups. The nematode identification (NID) number and collection location is shown for each. Blue box in Group four indicates specimen, NID 12574, considered incongruous between the COI and 18S phylogenies, and which displays differing morphology from the other two individuals.
Combined morphological and ecological analysis of haplotype groupings as determined by discriminant function analysis:
Females
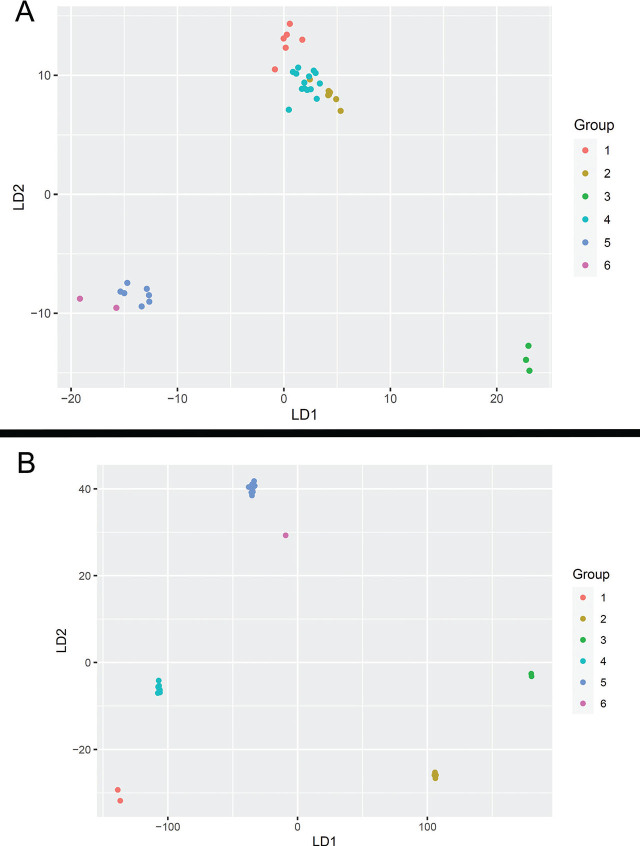
Female Combined – 14 variables were selected using the AIC method and then used for LDA analysis. Six of these variables included morphological characters, and the remaining eight variables were soil chemistry attributes. The most significant morphological characters and soil attributes are presented in Table S1. All 14 variables were used for LDA with almost equal priors. There were 6 groups, the first 4 with a prior of 0.17 and the remaining two groups with a prior of 0.16. There were five linear discriminants (LD) needed to properly discriminate among the haplotype groups, although the first two discriminants accounted for almost 95% of the variation. All the groups were classified correctly into their haplotype groups using the five LDs except 1 specimen in HG 4, which was classified as HG 2 instead (Fig. 12A).
Female Morphological – The seven variables selected using the AIC method for LDA analysis were L, b, c, V, VA/T, L set/lrw, and Dist. All seven variables were used for LDA with almost equal priors. Since there were 6 groups, the first 4 groups had a prior of 0.17, and the remaining two groups had a prior of 0.16. Five linear discriminants (LD) were needed to properly discriminate among the haplotype groups. At least three LDs were needed to account for 90% of the variation in the data. The groups were classified fairly correctly except for a few specimens in HG 2 and HG 4. For HG 1, out of 6, one specimen was classified as HG 5 and the remaining as HG 1. For HG 4, out of 15, one specimen was classified as HG 2 and the remaining as HG 4.
Female Soil Chemistry – The four variables selected using the AIC method for LDA analysis were K, Cu, Mg, and P (Table S1). All four variables were used for LDA with almost equal priors. Since there were 6 groups, the first 4 groups had a prior of 0.17, and the remaining two groups had a prior of 0.16. Five linear discriminants (LD) were needed to properly discriminate among the groups. The first two discriminants accounted for almost 95% of the variation. The haplotype groups were classified correctly only for HG 1 and HG 3; all remaining HGs had some misclassifications. Specimens in HG 2 were classified as HG 3 and HG 4, specimens in HG 4 were classified as HG 3 and HG 6, specimens in HG 5 were classified as HG 1 and HG 6, and finally, specimens in HG 6 were also classified as HG 1 and HG 5.
Figure 12.
Linear discriminant analysis (LDA) of haplotype groupings using combined morphological and ecological attributes for A) females; B) males.
Males
Male Combined – 16 variables were selected using the AIC method for LDA analysis. Nine of these variables were morphological characters, and the remaining seven were soil chemistry attributes. The significant morphological characters were L, a, c, C set/lrw, Stoma L, Dist, Cl-III, SR, and SR/L, and important soil attributes were Soil pH, K, Mg, CEC, % Mg, % OM, and P (Table S2). All 16 variables were used for LDA with almost equal priors. Since there are 6 groups, the first 4 groups have a prior of 0.17, and the remaining two groups have a prior of 0.16. Five linear discriminants (LD) were needed to properly discriminate between the haplotype groups. The first two LDs account for almost 98% of the variation. All the groups were classified correctly into their haplotype groups using the five LDs (Fig. 12B).
Male Morphological - Three variables selected using the AIC method for LDA analysis were c, C set/lrw, and Cl-III (Table S2). All three variables were used for LDA with almost equal priors. Since there are 6 groups, the first 4 groups have a prior of 0.17, and the remaining two groups have a prior of 0.16. Three linear discriminants (LD) were needed to properly discriminate between the haplotype groups. The first two LDs accounted for 85% variation in the data. The groups are classified fairly correctly except for one specimen in HG 2, which is classified as HG 3.
Male Soil Chemistry - Four variables selected using the AIC method for LDA analysis were % OM, K, Mg, and % Mg. All four variables were used for LDA with almost equal priors. Since there are 6 groups, the first 4 groups have a prior of 0.17, and the remaining two groups have a prior of 0.16. Four linear discriminants (LD) were needed to properly discriminate between the haplotype groups. The first two discriminants accounted for almost 98% of the variation. The groups were classified correctly only for HG 1, HG 3, and HG 6. The remaining HGs 2, 4, and 5 have some misclassifications. A specimen in HG 2 was classified as HG 3, in HG 4 as HG 1, and in HG 5 classified as HG 6.
Relationships inferred from 18S Ribosomal DNA: In the 18S tree (Fig. 13), HGs 2, 3, and 6 were well supported as a clade (86 bootstrap support), with HG 3 recognized as a subgroup embedded within HG 2. HGs 1, 4, and 5 were more strongly supported as part of a larger clade with 99 bootstrap support. The 18S sequences from HGs 1 and 5, representative of specimens from western and eastern Nebraska, respectively, were identical. HG 4 was a discrete subgroup within this clade.
Figure 13.
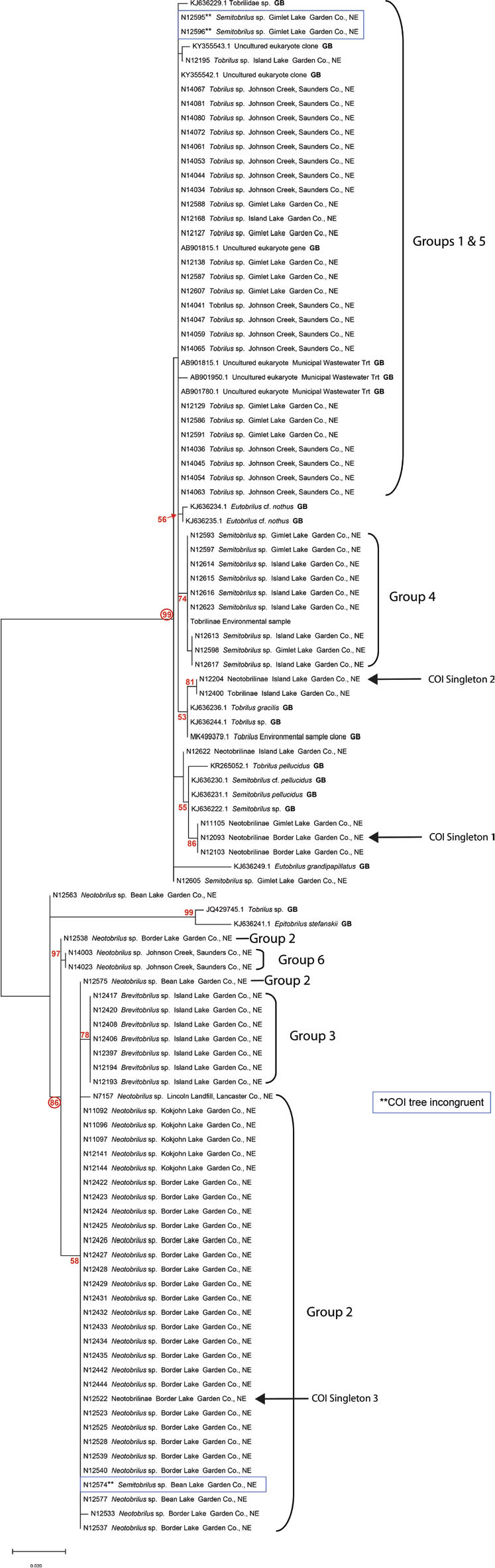
A maximum likelihood 18S phylogenetic tree of 88 Nebraska specimens and 19 GenBank accessions. Tobrilus Groups 1 and 5 are identical for the 18S barcode, and they are united in a strongly supported clade (99 bootstrap value) with Semitobrilus Group 4 and GenBank accessions identified as Tobrilus and Semitobrilus. Blue boxes identify specimens incongruous between COI and 18S trees.
A nonconformity between COI and 18S trees: Three of the specimens in the tobrilid dataset examined with both COI and 18S markers revealed a nonconformity between the two genetic markers. Each of the three nonconforming specimens were members of the COI HG 4 (Fig. 3). Two of the specimens, NID 12595 and 12596, both males, were collected from Gimlet Lake, and the third specimen, NID 12574, a female, was collected from Bean Lake. NID 12574 was the only member of HG 4 collected from a high pH lake. It also stood out among other members of HG 4 in possessing a short tail (198 μm), which conforms to members of HG 2 (154–210 μm), in contrast to the long tails characteristic of HG 4 (350–414 μm) (Fig. 11). Similarly, other morphological measurements of NID 12574 such as body and stoma length, V, b, c, tooth distance and a bulb-like vaginal musculature (Fig. 8) all aligned with HG 2 and not HG 4. Its placement on the 18S tree was firmly within a clade populated by HG 2 specimens (Fig. 13). The two male specimens, NID 12595 and 12596, were grouped together with HG 1 and 5 in the 18S tree. Their morphological measurements, however, were entirely consistent with other HG 4 specimens.
Relationship to described species of tobrilids from North America: The study of Abebe et al., 2013 provides a comparison of the four Neotobrilus species known from North America. The HG 2 Neotobrilus most closely compares to Neotobrilus nicsmolae Abebe et al., 2013. The body lengths of both males and females are larger in HG 2. In N. nicsmolae, the amphid diameter appears larger, the amphid is positioned more posteriorly on the body, and somatic setae appear more prominent and numerous than in HG 2. Two Semitobrilus species, S. pellucidus (Bastian, 1865) Tsalolikhin, 1981 and S. ebsaryi Tsalolikhin, 2000, have been reported from North America. Semitobrilus pellucidus has spicules that range between 80–90 μm versus 130–143 for HG 4. Semitobrilus ebsaryi, formerly Tobrilus longicaudatus (Schneider, 1923) Andrassy, 1959, is morphologically the closest to HG 4, with a shorter spicule of 108–122 μm and a slightly sigmoid vaginal lumen, which is a variable character in HG 4. The genus Tobrilus, according to Zullini (2006), is restricted to Europe and Asia. The Nebraskan Tobrilus species, HGs 1 and 5, clearly belong to the genus Tobrilus and likely represent new species.
Discussion
The Alkaline Lakes of the western Sandhills region of Nebraska constitute an extreme environment. The three Alkaline Lakes at the upper end of the pH and potassium alkalinity spectrum (Bean Lake, Border Lake, and Kokjohn Lake) contain no vertebrate organisms, few macroinvertebrates, and diatoms which, based on their presence in tobrilid guts, are the primary food source of the nematodes. Members of Tobrilidae were prominent in each of the five Alkaline Lakes, with a single COI haplotype group found in the three high pH lakes, and three additional COI haplotype groups distributed between the two neutral pH lakes.
Sediment samples collected in eastern Nebraska from Johnson Creek provide evidence that tobrilid nematodes can survive in different types of extreme environments. In 2021, Johnson Creek was the site of a major contamination event that washed 4 million gallons of highly contaminated sludge containing concentrated seed coat pesticides directly into the creek. The resulting aquatic nematode community was reduced to species mixtures of Tobrilidae and Monhysteridae. Attempts to identify the tobrilid genera and species from the Alkaline Lakes and Johnson Creek have proved challenging.
The most comprehensive morphological key to the genera in Triplonchida is by Zullini (2006). All the specimens examined in this study readily key to Tobrilidae based on the characteristics of the wide or funnel-shaped stoma with two teeth at its base (Figs. 2, 4). The first couplet within Tobrilidae splits the family into Tobrilinae and Neotobrilinae based on fused or adjacent pockets at the stoma base with two teeth 0–8 μm apart (Tobrilinae), versus well-separated, discrete pockets, 6–25 μm apart, posterior to the buccal cavity, each with a single tooth (Neotobrilinae) (Fig. 4). The COI HGs 1 and 5 morphologically key to the genus Tobrilus Andrássy, 1959 based on overlapping stomatal pockets with teeth no more than 0–6 μm apart, relatively short cephalic setae and small supplements in the male that do not protrude above the cuticle surface (Fig. 5). All of the cephalic setae we observed would be considered relatively short, with a length that rarely exceeds 20–30% of the body width at their location. SEM images of the cephalic region of specimens from the Alkaline Lakes show a general pattern of short labial papillae and more prominent cephalic setae and outer labial setae (Fig. 9).
Taxonomic uncertainty increases among the specimens morphologically identified as members of the subfamily Neotobrilinae, HGs 2, 3, 4, and 6. HGs 2 and 6 most closely conform to the genus Neotobrilus Tsalolikhin, 1981 based on the strong, bulb-like vaginal musculature, protruding echinate supplements, and tooth distances exceeding 11 μm. HG 3 has tooth distances of approximately 10 μm but lacks strong vaginal musculature, and the protruding supplements are not echinate. In most characteristics, it conforms to the genus Brevitobrilus Tsalolikhin, 1981. HG 4 appears to combine characteristics of both subfamilies. In respect to its heavily muscular vagina and well-separated pockets, it conforms to Neotobrilinae. The supplements, however, do not protrude from the cuticle, and the tooth distance of 6–12 μm is intermediate and overlapping with both subfamilies. Notably in most HG 4 female specimens, the vagina is slightly anteriorly directed, and males have exceptionally long spicules. Collectively these features best fit the genus Semitobrilus Tsalolikhin, 1981 (Tsalolikhin, 2009).
Characterization of the haplotype groups by partial COI and 18S sequences provided additional phylogenetic insight and some surprises. The COI gene sequences, due to a relatively rapid mutation rate, strongly supported the existence of haplotype groups, but relationships among haplotype groups remained obscure. The 18S sequences did permit grouping of COI-derived haplotype groups but also raised the possibility of past hybridization between nematodes with distinctly different mitochondrial lineages. Three specimens, one female and two males, had the COI sequence indicative of mitochondrial haplotype HG 4, but were members of different 18S clades and did not cluster with the other HG 4 members in the 18S phylogeny. The incongruence was further supported by morphological evidence, in which the female (NID 12574) with the mitochondrial HG 4 haplotype, had the morphological features of HG 2 (Fig. 11). Both male specimens had the morphological features of HG 4, which matched their COI haplotype group but fell into the 18S clade of Tobrilus Group 1.
Discriminant function analysis was most successful when male or female morphological characteristics were used in combination with the environmental chemistry data. Morphology alone or chemistry alone did not achieve the level of discrimination of the combined analyses.
Given the strong signal provided by the environmental chemistry data, it is reasonable to speculate that tobrilid haplotypes may have value as environmental indicators. Improved taxonomy will aid the effort, with a needed emphasis on improved keys for genus and species diagnosis. This study identified some unique characteristics relative to the spicules, inter-supplement cuticles, and amphids. These observations need to be extended across additional species and linked to diagnostic molecular markers. It should be noted that while the COI marker strongly delineates 6 distinct Nebraskan tobrilid groupings, relationships based on 18S suggest that the conventional subfamily groupings may require reconsideration. Specifically, Haplotype group 4, identified as belonging to the genus Semitobrilus, clusters with the Tobrilinae, and is supported by a trio of GenBank accessions of Semitobrilus specimens. Traditionally, Semitobrilus is classified in the subfamily Neotobrilinae (Zullini, 2006).
Supplementary Material
Supplementary Material Details
Supplementary Material Details
Acknowledgments
This work would not have been possible without the support of the Crescent Lake National Wildlife Refuge, the University of Nebraska Eastern Nebraska Research, Extension and Education Center (ENREEC), the UNL Water Center, NSF Proposals DEB 2327477 and DEB 2327478, David Dunigan of the Sandhills Lake Collaboration Initiative (SALCI), John DeLong and Jon Garbisch of the Cedar Point Biological Station, Liz VanWormer and Judy Wu-Smart of Nebraska OneHealth and Mary Harner of the Platte Basin Timelapse Project. We are very grateful to the UNL Statistical Cross-disciplinary Collaboration and Consulting Lab for providing statistical support, and for the participation of the following on our collecting trips: Abigail Borgmeier, Innocent Byiringiro, Roger Carlson, Ethan Freese, Kaitlin Gattoni, Eli Gendron, Jay Ghosh, Daniel Gschwentner, Parr McQueen, and Maddie Rawlings.
Literature Cited
- Abebe E., Ferebee B., Taylor T., Mundo-Ocampo M., Mekete T., De Ley P.. Neotobrilus nicsmolae n. sp. (Tobrilidae: Nematoda) and Chronogaster carolinensis n. sp. (Chronogasteridae: Nematoda) from Lake Phelps, North Carolina. Journal of Nematology. 2013;45(1):66–77. [PMC free article] [PubMed] [Google Scholar]
- Akaike H. Petrov B. N., Csäki F. 2nd International Symposium on Information Theory. Budapest: Akademiai Ki à do; 1973. Information theory and the maximum likelihood principle. , eds. [Google Scholar]
- Bongers T. van de Haar Jan. The nematodes of the Netherlands. London: British Museum of Natural History; 1989. Translation: [Google Scholar]
- Bowles J., Blair D., McManus D. P.. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Molecular Biochemical Parasitology. 1992;54(2):165–173. doi: 10.1016/0166-6851(92)90109-w. [DOI] [PubMed] [Google Scholar]
- Decraemer W., Eisendle-Flöckner U., Abebe E. Thorp J. H., Rogers D. C. Thorp and Covich's Freshwater Invertebrates. Fourth Ed. Amsterdam: Elsevier; 2019. Phylum Nematoda; pp. 269–299. [Google Scholar]
- Dunigan D. Norby M. M., Diamond J., Sutherlen A., Fritz S. C., Hachiya K., Norby D. A., Forsberg M. The Nebraska Sandhills. Lincoln, NE: University of Nebraska Press; Sandhills Alkaline Lakes; p. 117. [Google Scholar]
- Ebsary B. A.. Canadian species of Tobrilus (Nematoda: Tobrilidae) with description of three new species. Canadian Journal of Zoology. 1982;60(12):3048–3062. [Google Scholar]
- Fisher R. A.. The use of multiple measurements in taxonomic problems. Annals of Eugenics. 1936;7(2):179–188. [Google Scholar]
- Gagarin V. G., Naumova T. V.. Tobrilus methanus sp. n. and Tripyla posolskii sp. n. (Nematoda, Triplonchida) from Lake Baikal, Russia. Zootaxa. 2016;4196(1):95–106. doi: 10.11646/zootaxa.4196.1.5. https://doi.org/10.11646/zootaxa.4196.1.5. [DOI] [PubMed] [Google Scholar]
- Gattoni K., Gendron E. M. S., Borgmeier A., McQueen J. P., Mullin P. G., Powers K., Powers T. O., Porazinska D. L.. Context-dependent role of abiotic and biotic factors structuring nematode communities along two environmental gradients. Molecular Ecology. 2022;31:3903–3916. doi: 10.1111/mec.16541. https://doi.org/10.1111/mec.16541. [DOI] [PubMed] [Google Scholar]
- Gattoni K., Gendron E. M., Powers K., Powers T. O., Harner M. J., Porazinska D. L.. Effects of drought-induced stress on nematode communities in aquatic and terrestrial habitats of the Nebraska Sandhills. Frontiers in Ecology and Evolution. 2024;12:1305930. [Google Scholar]
- Gosselin D. C.. Major-ion chemistry of compositionally diverse lakes, Western Nebraska, USA: Implications for paleoclimatic interpretations. Journal of Paleolimnology. 1997;17:33–49. https://doi.org/10.1023/A:1007908909148. [Google Scholar]
- Haacker E. Norby M. M., Diamond J., Sutherlen A., Fritz S. C., Hachiya K., Norby D. A., Forsberg M. The Nebraska Sandhills. Lincoln, NE: University of Nebraska Press; 2024. Groundwater: How the High Plains aquifer shapes the Sandhills; pp. 102–104. [Google Scholar]
- Holovachov O., Shoshin A. Schmidt-Rhaesa A. Handbook of Zoology. Gastrotricha, Cycloneuralia, Gnathifera, Vol. 2: Nematoda. Berlin: de Gruyter; 2014. Order Triplonchida Cobb, 1919; pp. 251–276. [Google Scholar]
- Jenkins W. R.. A rapid centrifugal-flotation technique for separating nematodes from soil. Plant Disease Reporter. 1964;48(9):692. [Google Scholar]
- Johnsgard P. A. This fragile land: A natural history of the Nebraska Sandhills. Lincoln: University of Nebraska Press; 1995. [Google Scholar]
- Jombart T., Devillard S., Balloux F.. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genetics. 2010;11:94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Zheng S., Wan T., Wang L., Yang Q., Zhang J.. Nematode as a biomonitoring model for evaluating ecological risks of heavy metals in sediments from an urban river. Ecological Indicators. 2023;147:110013. [Google Scholar]
- Kanzaki N., Giblin-Davis R. M., Scheffrahn R. H., Taki H., Esquivel A., Davies K. A., Herre E. A.. Reverse taxonomy for elucidating diversity of insect-associated nematodes: A case study with termites. PLoS One. 2012:7. doi: 10.1371/journal.pone.0043865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachenbruch P. A., Goldstein M.. Discriminant analysis. Biometrics. 1979:69–85. https://doi.org/10.2307/2529937. [Google Scholar]
- Markmann M., Tautz D.. Reverse taxonomy: An approach towards determining the diversity of meiobenthic organisms based on ribosomal RNA signature sequences. Philosophical Transactions of the Royal Society B: Biological Sciences. 2005;360(1462):1917–1924. doi: 10.1098/rstb.2005.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matczyszyn J. N., Harris T., Powers K., Everhart S. E., Powers T. O.. Ecological and morphological differentiation among COI haplotype groups in the plant parasitic nematode species. Journal of Nematology. 2022;54(1):20240025. doi: 10.2478/jofnem-2022-0009. https://doi.org/10.2478/jofnem-2022-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin P. G., Harris T. S., Powers T. O.. Systematic status of Campydora Cobb, 1920 (Nematoda: Campydorina) Nematology. 2003;5(5):699–711. https://doi.org/10.1163/156854103322746878. [Google Scholar]
- Naumova T. V., Gagarin V. G.. Tobrilus saprophagus sp. n. and Epitobrilus interstitialis sp. n. (Nematoda, Triplonchida) from Lake Baikal, Russia. Zootaxa. 2017;4353(1):133–145. doi: 10.11646/zootaxa.4353.1.8. [DOI] [PubMed] [Google Scholar]
- Naumova T. V., Gagarin V. G.. Two new nematode species of the genus Tobrilus Andrássy, 1959 (Nematoda, Triplonchida) from Lake Baikal, Russia. European Journal of Taxonomy. 2019a;579:1–13. https://doi.org/10.5852/ejt.2019.579. [Google Scholar]
- Naumova T. V., Gagarin V. G.. Review of the free-living nematode (Nematoda) fauna of Lake Baikal. Zootaxa. 2019b;4608(1):20240025. doi: 10.11646/zootaxa.4608.1.5. [DOI] [PubMed] [Google Scholar]
- Powers T. O., Bernard E. C., Harris T. S., Higgins R., Olson M., Lodema M., Matczyszyn J., Mullin P., Sutton L., Powers K. S.. Species discovery and diversity in Lobocriconema (Criconematidae: Nematoda) and related plant-parasitic nematodes from North American ecoregions. Zootaxa. 2016;4085(3):20240025. doi: 10.11646/zootaxa.4085.3.1. [DOI] [PubMed] [Google Scholar]
- Powers T. O., Bernard E. C., Harris T. S., Higgins R., Olson M., Lodema M., Mullin P., Sutton L., Powers K. S.. COI haplotype groups in Mesocriconema (Nematoda: Criconematidae) and their morphospecies associations. Zootaxa. 2014;3827(2):101–146. doi: 10.11646/zootaxa.3827.2.1. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2021. https://www.R-project.org/ [Google Scholar]
- Ristau K., Steinfartz S., Traunspurger W.. First evidence of cryptic species diversity and significant population structure in a widespread freshwater nematode morphospecies (Tobrilus gracilis) Molecular Ecology. 2013;22(17):4562–4575. doi: 10.1111/mec.12414. https://doi.org/10.1111/mec.12414. [DOI] [PubMed] [Google Scholar]
- Teiwes M., Bergtold M., Traunspurger W.. Factors influencing the vertical distribution of nematodes in sediments. Journal of Freshwater Ecology. 2007;22(3):429–439. [Google Scholar]
- Tsalolikhin S.Y.. A revision of the genus Tobrilus (Nematoda, Tobrilidae) Zoologicheskii Zhurnal. 1981;60(9):1302–1313. [Google Scholar]
- Tsalolikhin S. Y., Shoshin Y.. Review of the genus Neotobrilus Tsalolikhin, 1981 (Nematoda, Enoplida: Tobrilidae) Russian Journal of Nematology. 2009;17(1):59–72. [Google Scholar]
- Vrain T. C., Wakarchuk D. A., Levesque A. C., Hamilton R. I.. Intraspecific rDNA restriction fragment length polymorphism in the Xiphinema americanum group. Fundamental and Applied Nematology. 1992;15(6):563–573. [Google Scholar]
- Zahid M., Taiba J., Cox K., Khan A. S., Uhing T., Rogan E.. Pesticide residues in adults living near a bioenergy plant with 85,000 tons of contaminated wetcake. Chemosphere. 2024;349:140941. doi: 10.1016/j.chemosphere.2023.140941. [DOI] [PubMed] [Google Scholar]
- Zullini A. Abebe E., Andrássy I., Traunspurger W. Freshwater nematodes: Ecology and taxonomy. Wallingford, UK: CABI Publishing; 2006. Order Triplonchida; pp. 293–325. [Google Scholar]
- Zullini A.. Is a biogeography of freshwater nematodes possible? Nematology. 2014;16(1):1–8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material Details
Supplementary Material Details