Abstract
Benign prostatic hyperplasia (BPH), characterized by the non-malignant enlargement of the prostate, exhibits a pronounced association with inflammation resulting from androgen receptor (AR) deficiency. Ferroptosis, a cell death mechanism triggered by iron-dependent lipid peroxidation and closely linked to inflammation, has yet to be fully understood in the context of BPH. Using RNA sequencing, we observed a significant elevation of taurine-upregulated gene 1 (TUG1) long noncoding RNA (lncRNA) in BPH tissues compared to normal prostate tissue. High levels of TUG1 exhibited a discernible correlation with both prostate volume and the extent of inflammatory infiltration in BPH patients. The suppression of TUG1 not only led to a reduction in prostate size but also ameliorated AR-deficiency-induced prostatic hyperplasia. Mechanistically, a decrease in AR in prostate luminal cells prompted macrophage aggregation and the release of IL-1β, subsequently fostering the transcription of TUG1 via MYC. Induced TUG1, through competitive binding with miR-188-3p, facilitated the expression of GPX4, thereby diminishing intracellular ROS levels and impeding ferroptosis in prostate luminal cells. Notably, the ferroptosis inducer JKE-1674 alleviated inflammation-induced prostatic hyperplasia in vivo. Together, these findings suggest that AR deficiency crucially inhibits ferroptosis, promoting BPH via the TUG1/miR-188-3p/GPX4 signaling axis, and making ferroptosis induction a promising therapeutic strategy for BPH patients with AR deficiency.
Keywords: Benign prostatic hyperplasia, Androgen receptor, Inflammation, TUG1, Ferroptosis
1. Introduction
Benign prostatic hyperplasia (BPH) is a progressive condition primarily affecting elderly men, with its prevalence exceeding 80 % among those aged 70 and older [[1], [2], [3]]. Pathologically, BPH is characterized by benign proliferation of prostatic epithelial and stromal tissues, resulting in enlargement of the prostate's periurethral region (transition zone) [4]. This enlargement manifests clinically as bladder outlet obstruction, lower urinary tract symptoms (LUTS), and urethral infections, and can lead to severe complications like renal failure and cardiovascular issues in advanced stages [5]. Despite its widespread occurrence, the precise etiologic factors contributing to BPH are still not fully understood. Contemporary treatment strategies, which often involve α-blockers and 5-α-reductase inhibitors (5-ARIs), offer limited effectiveness, and necessitate long-term administration [6,7]. Therefore, advancing our understanding of the molecular dynamics involved in the onset and progression of BPH is critical for the development of more effective therapeutic approaches and improving patient outcomes.
Androgen and its receptor (AR) play important roles in prostate development and BPH pathogenesis [8,9]. Therapies targeting androgen, such as 5-alpha reductase inhibitors (5ARIs), are commonly used in BPH treatment [10]. However, the decline in serum androgen levels with aging is associated with an increased incidence of BPH, and 5ARI treatments are ineffective in some BPH patients [11,12]. These studies suggest that androgen and AR have complex functional roles in the development of BPH, and the underlying molecular mechanisms remain unclear. Pathological examinations often show leukocyte infiltration in BPH tissues, indicating chronic inflammation as a pivotal element in the pathogenesis of disease [13]. Androgens are known to modulate immune responses, attenuating inflammation-induced reactions [14]. Recent research has demonstrated a link between diminished AR expression in the luminal cells of BPH tissues and increased inflammation within the prostate [15]. Experimental models in mice have shown that a decrease in AR signaling in prostatic luminal cells intensifies BPH by encouraging immune cell infiltration, elevating inflammatory markers, and weakening epithelial cell integrity [15]. Although the association between reduced AR signaling and BPH is recognized, the underlying molecular mechanisms are yet to be fully elucidated.
Ferroptosis, an iron-dependent form of cell death characterized by lipid peroxide accumulation, is mediated by Glutathione peroxidase 4 (GPX4) [16,17]. This enzyme plays a crucial role in protecting lipids from oxidative stress by converting lipid peroxides into non-toxic lipid alcohols using glutathione, thus inhibiting ferroptosis [17]. GPX4 has been implicated in the proliferation and migration of prostate cancer cells, as well as in their resistance to therapy, and has shown an association with the onset of BPH [[18], [19], [20]]. The interplay between inflammatory processes and ferroptosis is intricate and multifaceted [21]. While ferroptosis is linked to various inflammatory conditions, including nonalcoholic steatohepatitis, neuroinflammation, and rheumatoid arthritis, inflammatory mediators can have diverse effects on ferroptosis, either promoting or inhibiting it [[22], [23], [24]]. The specific influence of localized inflammatory responses on ferroptosis within the context of BPH development, however, remains a topic of ongoing investigation.
In our study, we identified the long non-coding RNA (lncRNA) taurine-upregulated 1 (TUG1) as a crucial intermediary in connecting inflammation to suppressed ferroptosis, thereby contributing to the development of BPH. A deficiency of AR in prostate luminal cells leads to local macrophage infiltration and the subsequent release of IL-1β, which elevates TUG1 expression through the transcription factor MYC. TUG1, by competing with miR-188-3p, indirectly enhances GPX4 expression, thereby increasing cell tolerance to ferroptosis and promoting BPH progression. Our findings suggest that targeting TUG1 could mitigate AR-deficiency-induced BPH by inducing ferroptosis in prostate luminal cells, presenting a novel therapeutic approach for BPH that leverages this process.
2. Results
2.1. TUG1 upregulation correlates with the development and progression of BPH
In our quest to identify molecular differences associated with BPH pathogenesis, we performed RNA sequencing on prostate tissue from the transitional zone. This analysis included 6 patients diagnosed with BPH and 6 individuals with normal prostatic tissue (Fig. 1a and Supplementary Fig. 1a). All 6 control samples were exclusively sourced from male patients who had undergone radical cystectomy and did not have BPH or its associated complications, such as LUTS. The BPH cohort presented with international prostate symptom score (IPSS) scores above 15 and prostate volumes exceeding 90 cc, in contrast to the control group, which showed IPSS scores below 7 and prostate volumes less than 30 cc (Supplementary Fig. 1a). None of these subjects had been treated with 5-ARIs or α-receptor blockers, thereby eliminating potential treatment-related confounding effects. Comparative analysis of RNA sequencing data highlighted a significant upregulation of 60 genes and downregulation of 15 genes in BPH tissues (FC ≥ 2, P < 0.01; Fig. 1b–c), with TUG1 exhibiting the most pronounced differential expression. TUG1, a long non-coding RNA known for its non-protein-coding function, plays a pivotal role in maintaining male fertility, regulating pulmonary artery smooth muscle cell proliferation, and influencing tumor development [[25], [26], [27], [28], [29]]. To substantiate the relationship between TUG1 and BPH, we evaluated TUG1 expression levels in a larger cohort comprising 67 BPH samples and 20 normal prostate samples (Supplementary Table 1). Chromogenic in situ hybridization (CISH) and real-time quantitative PCR (RT-qPCR) revealed that TUG1 is predominantly expressed in the prostate luminal cells, with markedly higher levels in BPH tissues compared to the normal prostate (Fig. 1d–e and Supplementary Fig. 1b).
Fig. 1.
TUG1 upregulation in BPH tissues correlates with disease progression. a Diagram of sampling locations for BPH (n = 6) and control samples (n = 6) used for RNA-seq. The diagram on the right represents the BPH specimens, and the left represents control specimens sourced from the prostatic transitional zone post-radical cystoprostatectomy in males (none LUTS, prostate volumes <30 cc, and IPSS score <7). b Volcano plot and heatmap depict the transcriptional profiles of genes with significant differential expression between BPH and control tissues (FC ≥ 2, P < 0.01). c Quantitative comparison of TUG1 gene expression in BPH versus control tissues using RNA-seq datasets. d CISH staining analysis of the TUG1 expression in BPH and control tissues. Scale bars: 50 μm. e RT-qPCR analysis of the expression levels of TUG1 between BPH (n = 67) and control (n = 20) samples in our validation cohort. f H&E, CISH (TUG1), and IHC (CD45 and Ki-67) staining analyses in the different areas of the BPH tissues. Scale bars: 50 μm. g-h Comparative evaluation of the proportion of CD45+ (g) and Ki-67+ luminal cells (h) in regions with high or low TUG1 expression in BPH tissues. i Representative FACS plots of small and large volume human prostate fractions into leukocytes (CD45+), epithelial cells (CD45-/EpCAM+), and stromal cells (CD45-/EpCAM−). j-k Correlation analysis between TUG1 expression levels and the frequency of leukocytes (j) or prostate gland volume (k) in BPH tissues. l Univariate and multivariate analyses exploring the association of TUG1 expression with clinical and histopathological characteristics in BPH patients. ***P < 0.001; n.s no significant.
The progression of BPH, objectively determined by prostate volume and IPSS score, correlates with inflammatory states and cellular proliferation within prostate tissues [30]. We assessed these parameters by immunohistochemistry (IHC) staining CD45 and Ki-67 in prostate tissue. Interestingly, regions with elevated TUG1 expression in luminal cells corresponded to increased leukocyte infiltration (CD45+ cells), although the luminal cell proliferation indices remained unchanged (Fig. 1f–h). Flow cytometry analysis of CD45+ cells in 67 BPH patient samples demonstrated that leukocyte infiltration in BPH tissues correlates positively not only with prostate volume and IPSS score but also significantly with TUG1 expression levels (Fig. 1i–j and Supplementary Figs. 1c–1d). Univariate analysis pointed to a correlation between TUG1 expression and various clinical parameters: prostate volume, IPSS score, intra-prostatic inflammation, and serum testosterone levels (Fig. 1k–l and Supplementary Figs. 1e–1f). However, multivariate logistic regression analysis identified TUG1 expression as being independently associated only with prostate volume in BPH patients (Fig. 1l). Notably, no association was observed between TUG1 expression in BPH tissues and cellular proliferation indices (Fig. 1l and Supplementary Fig. 1g). These findings underscore the upregulation of TUG1 in BPH development and progression, aligning positively with local prostate inflammation and increased prostate volume.
2.2. TUG1 deletion in luminal cells inhibit age-related prostate enlargement
TUG1, a lncRNA highly conserved between humans and mice, plays an ambiguous role in various tissues [[25], [26], [27], [28], [29]]. Through RT-qPCR analysis of 25 primary mouse tissues and RNA-sequencing (RNA-seq) data of normal human prostate tissue, we confirmed that TUG1 is predominantly expressed in the prostate, ranking within the top 5 % of the most abundant genes, similar to the expression levels of the prostate-specific gene AR (Fig. 2a). Data from the Human Protein Atlas (HPA), encompassing 40 human tissues, also indicated elevated TUG1 expression in the human prostate, ranking third in terms of abundance (Supplementary Fig. 2a). HPA single-cell sequencing data of human prostate tissue pinpointed the primary expression of TUG1 to a specific type of luminal cells (Fig. 2b). Anatomically, the murine prostate comprises anterior prostate (AP), ventral prostate (VP), and dorsolateral prostate (DLP) lobes (Fig. 2c). RT-qPCR and FISH analyses specifically identified the predominant expression of Tug1 in DLP luminal cells (Fig. 2d–e).
Fig. 2.
Disrupting Tug1 in mouse prostate luminal cells inhibits age-related glandular enlargement. a Analysis of Tug1 expression across various mouse tissues and its abundance in BPH tissues. The primary figure is derived from RT-qPCR analysis of Tug1 expression across various mouse tissues, with the top right figure showing Tug1 abundance using RNA-seq data from BPH tissues. b Examination of TUG1 expression in different cellular populations within human prostate tissues by using single-cell data from the HPA database. c A schematic representation of the anatomy of the mouse prostate. AP, anterior prostate; DLP, dorsolateral prostate; VP, ventral prostate. d RT-qPCR analysis of Tug1 expression in the AP, DLP, and VP of the mouse prostate. e FISH combined with IF analysis for the localization and quantification of Tug1 in different regions of the mouse prostate. Scale bars: 50 μm. f Construction and characterization of the mouse model with prostate luminal cell-specific knockout of Tug1. The primary figure outlines the construction and analysis process for the mouse model, with the top right figure presenting RT-qPCR analysis of Tug1 expression in the AP, DLP, and VP of the prostate in Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice at 12 months g Representative images of prostate tissues and H&E staining of the AP, DLP, and VP from Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice at 4, 8, and 12 months. The dashed line area represents the prostate tissue of mice. Scale bars: 1 cm for gross; 50 μm for H&E staining. h-i Comparison of the relative expansion (h) or weight (i) of the AP, DLP, and VP of the mouse prostate between Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice at 4, 8, and 12 months. *P < 0.05; **P < 0.01; ***P < 0.001; n.s no significant.
To explore the biological role of Tug1 in murine prostate luminal cells, we created Pbsn-Cre4; Tug1fl/fl mice by crossing a Tug1 conditional line with a Pbsn-Cre4 transgenic mouse line (Fig. 2f, Supplementary Figs. 2b–2c). RT-qPCR data demonstrated that Cre recombinase, driven by the Pbsn promoter, selectively targets Tug1 for depletion in prostate luminal cells (Fig. 2f). To ascertain whether Tug1 deletion in murine prostate luminal cells affects other organs, we compared body weights and urogenital system organ characteristics between Tug1 knockout and wild-type mice. Our findings revealed no significant differences in growth patterns, reproductive organ morphology, bladder and testis weights, or histological structures (Supplementary Figs. 2d–2j). Notably, Tug1 depletion in luminal cells markedly inhibited age-related glandular epithelial expansion in the DLP region (Fig. 2g–h), leading to a significant reduction in the weight of the DLP region in knockout mice compared to wild types (Fig. 2i). Conversely, Tug1 depletion in the AP and VP regions did not significantly affect epithelial expansion or prostate weight (Fig. 2h–i). Furthermore, Ki-67 staining analysis revealed no significant differences in the proliferative capacities of luminal cells between the Tug1 knockout and wild-type groups (Supplementary Figs. 2k–2l). These findings align with clinical observations showing no correlation between Tug1 and prostate epithelial cell proliferation. Collectively, these results suggest that Tug1 depletion in luminal cells specifically inhibits the age-related expansion of the DLP region in the murine prostate, independently of changes in luminal cell proliferation.
2.3. AR absence in prostate luminal cells promotes TUG1 expression through IL-1β/MYC signaling
The absence of AR in prostate luminal cells has been recently identified as a key mechanism related to the development of BPH [15]. In our BPH validation cohort, a significant reduction in AR expression was confirmed compared to control groups, negatively correlating with prostate volume in BPH patients (Supplementary Figs. 3a–3c). Interestingly, a significant negative correlation between AR expression levels and TUG1 in BPH was also observed (Fig. 3a). Given that AR deficiency-induced BPH development is related to the release of inflammatory/cytokine factors, and the expression of TUG1 in BPH tissue is associated with the degree of inflammatory infiltration, we used RT-qPCR to analyze the correlation between TUG1 expression levels and six AR-regulated inflammatory/cytokine factors (IL-1α, IL-1β, IL-6, TNF-α, CCL2, and CXCL10) in BPH tissues. The results showed a significant positive correlation between TUG1 expression and IL-1β mRNA in BPH tissues, further validated at the protein level through enzyme-linked immunosorbent assay (ELISA) (Fig. 3b–c, Supplementary Figs. 3d–3h). Furthermore, compared to normal prostate tissue, IL-1β expression levels were significantly elevated in BPH tissues, correlating positively with patient prostate volume (Supplementary Figs. 3i–3j). Single-cell sequencing data from the Human Protein Atlas (HPA) database of prostate tissues revealed that IL-1β is primarily derived from macrophages (Fig. 3d). The role of macrophage-mediated inflammatory response in promoting BPH development has been confirmed by multiple studies [31,32]. Using immunohistochemical staining for the macrophage marker CD68, we also confirmed a significant increase in macrophage infiltration in BPH tissues compared to normal prostate tissue (Supplementary Figs. 3k–3l). Additionally, a negative correlation was found between CD68 positive cell rate and AR expression levels in BPH tissues, while a positive correlation was observed with TUG1 expression (Fig. 3e and Supplementary Figs. 3m–3n).
Fig. 3.
Downregulation of AR promotes TUG1 expression via IL-1β/MYC signaling in prostate luminal cells. a-c Correlation analyses between TUG1 expression and AR mRNA levels (a), IL1B mRNA levels (b), and IL-1β protein concentrations (c) in BPH tissues from our validation cohort. d Analysis of IL1B expression across different cellular populations within human prostate tissues using single-cell data from the HPA database. e CISH combined with IHC staining for the quantification of TUG1, AR, CD68, and IL-1β in BPH tissues. f Schematic representation of the experimental approach for AR inhibitor treatment in Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice. g-h Analysis of DLP weight (g) and H&E staining (h) in the prostate of Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice treated with PBS or MDV3100. i IF staining for AR and CD68 in the prostate of Pbsn-Cre4 Tug1wt/wt mice treated with PBS or MDV3100. j RT-qPCR analysis of Il1b and Tug1 expression in the prostate of Pbsn-Cre4 Tug1wt/wt mice treated with PBS or MDV3100. k FISH combined with IF for the quantification of Tug1 and AR expression in the prostate of Pbsn-Cre4 Tug1wt/wt mice treated with PBS or MDV3100. l Integrative analysis of the JASPAR, PROMO, and AnimalTFDB databases along with TUG1-related genes to identify transcription factors involved in TUG1 regulation. m RT-qPCR analysis of Tug1 expression in RWPE-1 and BPH-1 cells transfected with siRNA targeting MYC or SP1, followed by treatment with PBS or IL-1β. n RT-qPCR analysis of TUG1 expression in RWPE-1 and BPH-1 cells treated with IL-1β and the MYC inhibitor 10058-F4. o IHC staining for MYC in the prostate of Pbsn-Cre4 Tug1wt/wt mice treated with PBS or MDV3100. p Identification of MYC binding motifs within the human TUG1 promoter region. q-r ChIP-qPCR analysis of the MYC binding site at the TUG1 promoter region in RWPE-1 (q) and BPH-1 (r) cells. s-t Luciferase reporter assays of wild-type (WT) and mutant (MUT) TUG1 promoters in RWPE-1 (s) and BPH-1 (t) cells transfected with MYC siRNA and treated with IL-1β. All scale bars: 50 μm *P < 0.05; **P < 0.01; ***P < 0.001; n.s no significant.
To further clarify the regulatory relationship between AR, macrophages, IL-1β, and TUG1, we treated prostate luminal cells (RWPE-1) and BPH cells (BPH-1) with IL-1β or co-cultured them with macrophages (RAW 264.7). Both IL-1β and macrophages were found to promote TUG1 expression in RWPE-1 and BPH-1 cells, while an IL-1β inhibitor (AS101) reversed the promotional effect of macrophages on TUG1 expression in these cells (Supplementary Figs. 3o–3r). Additionally, Tug1-specific knockout/wild-type mice treated with an AR inhibitor MDV3100 (Enzalutamide), or placebo were analyzed for changes in prostate weight and the degree of macrophage infiltration (Fig. 3f). The results indicated that treatment with an AR inhibitor in wild-type adult mice promoted enlargement in the DLP region of the mouse prostate and increased infiltration of macrophages, as well as the expression of Il1b and Tug1 in prostate luminal cells (Fig. 3g–k and Supplementary Fig. 3s). Tug1 knockout in prostate luminal cells inhibited the enlargement effects induced by AR deficiency in adult mice (Fig. 3g–k).
Considering that macrophages can promote TUG1 expression in prostate luminal cells by secreting IL-1β, we investigated the potential intermediate transcription factors by which IL-1β regulates TUG1. Using JASPAR, PROMO, and AnimalTFDB databases, we analyzed transcription factors potentially binding to the TUG1 promoter region (2000 bp in front of the transcription start site), and in combination with genes significantly correlated with TUG1 expression in BPH tissues, identified two potential transcription factors: MYC and SP1 (Fig. 3l). To ascertain whether these two transcription factors mediate the regulatory effect of IL-1β on TUG1, we interfered with MYC or SP1 expression in RWPE-1 and BPH cells treated with IL-1β. The results showed that decreasing MYC could hinder promotional effect of IL-1β on TUG1 expression in prostate luminal cells, while SP1 interference had no such effect (Fig. 3m). The application of a MYC inhibitor, 10058-F4, also reversed the promotional effect of IL-1β on TUG1 expression (Fig. 3n). Overexpression of MYC in RWPE-1 and BPH cells significantly promoted TUG1 expression (Supplementary Figs. 3t–3u). IHC staining and RT-qPCR analysis showed a significant upregulation of Myc in AR-inhibited mouse prostate luminal cells (Fig. 3o and Supplementary Fig. 3v). MYC, an important transcription factor regulating cell functions, promotes the expression of target genes by recognizing specific sequences in the promoter region. We identified two potential MYC binding motifs in the TUG1 promoter region (Fig. 3p). Chromatin immunoprecipitation quantitative real-time PCR (CHIP-qPCR) experiments further confirmed that MYC in RWPE-1 and BPH cells could bind to site 1 but not to site 2 (Fig. 3q–r and Supplementary Fig. 3w). Additionally, we constructed luciferase reporter gene plasmids for the wild-type and mutant of site 1 (Supplementary Fig. 3x). Overexpression of MYC or IL-1β treatment in RWPE-1 and BPH cells significantly enhanced the luciferase activity of the wild-type site 1, while downregulation of MYC not only inhibited the luciferase activity of the wild-type site 1 but also hindered the promotional effect of IL-1β on this activity (Fig. 3s–t and Supplementary Figs. 3y–3z). However, IL-1β treatment, MYC interference, or MYC overexpression had no effect on the luciferase activity of the mutant site 1 (Fig. 3s–t and Supplementary Figs. 3y–3z). These results suggest that the absence of AR in prostate epithelial cells can promote transcription and expression of TUG1 by inducing macrophage infiltration and release of the inflammatory factor IL-1β, thereby elevating MYC levels in prostate luminal cells.
2.4. TUG1 augments ferroptosis resistance in prostate luminal cells
To further explore the role of TUG1 in regulating downstream signaling pathways contributing to BPH, we performed Gene ontology (GO) and Gene set enrichment analysis (GSEA) analyses using RNA-seq data from BPH and normal prostate tissues. These analyses revealed a significant link between BPH onset and the inhibition of ferroptosis signaling (Fig. 4a–b). MTS assays demonstrated that primary prostate luminal cells from BPH tissues were more resistant to Erastin-induced ferroptosis than those from normal tissues (Fig. 4c–d), aligning with in vitro findings from BPH and normal prostate luminal cell lines (Fig. 4e–f). A positive association was also noted between TUG1 expression and Erastin resistance in both primary cells and cell lines (Supplementary Fig. 4a). This leads us to hypothesize that TUG1 contributes to BPH by mitigating ferroptosis in prostate luminal cells. In BPH-1 and RWPE-1 cell lines with either TUG1 knockdown or overexpression, TUG1 inhibition heightened Erastin sensitivity, while its overexpression diminished it (Fig. 4g–h and Supplementary Figs. 4d–4e). MTS assays on mouse-derived primary prostate luminal cells showed enhanced Erastin sensitivity following Tug1 knockout (Fig. 4i–j). Moreover, TUG1 overexpression curbed Erastin-induced cell death, which, when spurred by TUG1 interference or Tug1 knockout, was effectively counteracted by the ferroptosis inhibitors Ferrostatin-1 and Liproxstatin-1 (Fig. 4k–m and Supplementary Figs. 4f–4g). These findings indicate that TUG1 enhances the resilience of prostate luminal cells to ferroptosis, thereby fortifying their resistance to cell death.
Fig. 4.
TUG1 regulates the sensitivity of prostate luminal cells to ferroptosis. a GO enrichment analysis of genes differentially expressed between BPH and normal prostate tissue, focusing on KEGG Pathway and WikiPathway databases. b GSEA of the "WP_FERROPTOSIS" gene set comparing BPH to normal prostate tissue. c-d Evaluation of cell viability (c) and IC50 determination (d) in primary prostate luminal cells derived from BPH and normal prostate tissue following treatment with gradient concentrations (2−1 μM–26 μM) of Erastin for 24 h. e-f Assessment of cell viability (e) and IC50 (f) in BPH-1, RWPE-1, and RWPE-2 cells treated with gradient concentrations (2−1 μM–26 μM) of Erastin for 24 h. g-h Analysis of cell viability (g) and IC50 (h) in BPH-1 and RWPE-1 cells transfected with control shRNA or TUG1 shRNA and treated with gradient concentrations (2−1 μM–26 μM) of Erastin for 24 h. i-j Cell viability assessment (i) and IC50 analysis (j) in primary prostate luminal cells from Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice treated with gradient concentrations (2−1 μM–26 μM) of Erastin for 24 h. k-m Measurement of cell death in BPH-1 (k) and RWPE-1 (l) cells expressing control shRNA or TUG1 shRNA, and in primary prostate luminal cells (m) from Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice, following treatment with Erastin (20 μM) in the absence or presence of Ferrostatin-1 (2 μM) or Liproxstatin-1 (1 μM) for 24 h. n-p Analysis of MDA production in BPH-1 (n) and RWPE-1 (o) cells expressing control shRNA or TUG1 shRNA, and in primary prostate luminal cells (p) from Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice, treated with DMSO or Erastin (20 μM). q-s Detection of lipid ROS in BPH-1 (q) and RWPE-1 (r) cells expressing control shRNA or TUG1 shRNA, and in primary prostate luminal cells (s) from Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice, treated with DMSO or Erastin (20 μM). t-v Iron detection assay for intracellular Fe2+ in BPH-1 (t) and RWPE-1 (u) cells expressing control shRNA or TUG1 shRNA, and in primary prostate luminal cells (v) from Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice, treated with DMSO or Erastin (20 μM). *P < 0.05; **P < 0.01; ***P < 0.001; n.s no significant.
Ferroptosis, a novel iron-dependent form of programmed cell death, is driven by lethal accumulation of lipid peroxides [33]. Recognizing the pivotal roles of Fe2+ and reactive oxygen species (ROS) in ferroptosis and given that malondialdehyde (MDA) is a primary, well-studied byproduct of polyunsaturated fatty acid peroxidation, we assessed their concentrations in Erastin-treated prostate luminal cells. The data revealed a marked increase in MDA, ROS, and Fe2+ levels in cells with TUG1 downregulation or Tug1 knockout, in contrast to cells with TUG1 overexpression (Fig. 4n–v and Supplementary Figs. 4h–4m). In essence, TUG1 appears to suppress cellular lipid peroxidation, enhancing the tolerance of prostate luminal cells to ferroptosis.
2.5. TUG1 modulates ferroptosis in prostate luminal cells through GPX4
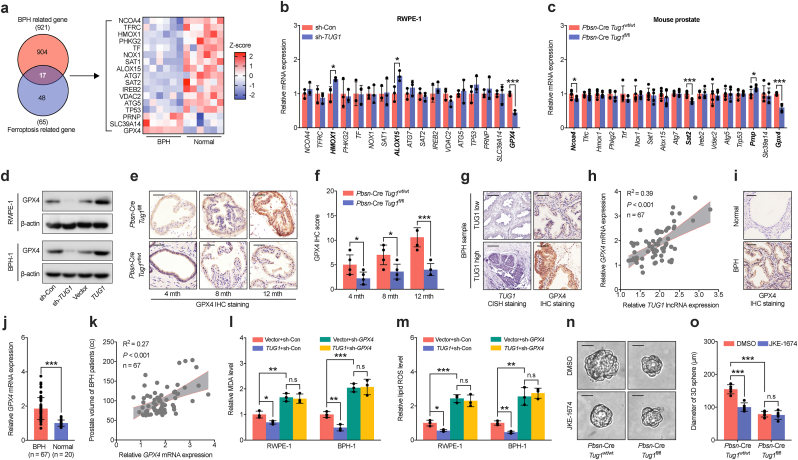
To explore the role of TUG1 in modulating ferroptosis in prostate luminal cells, we analyzed 921 genes related to BPH and 65 genes regulating ferroptosis, identifying 17 potential candidates (Fig. 5a). GPX4 was the only gene significantly downregulated in RWPE-1, BPH-1, and mouse prostate luminal cells with disruption TUG1 or knockout of Tug1 (Fig. 5b–d and Supplementary Fig. 5a). Conversely, overexpression of TUG1 in RWPE-1 and BPH-1 cells significantly upregulated GPX4 (Fig. 5d and Supplementary Fig. 5b). IHC staining and RT-qPCR analysis indicated an age-related increase in Gpx4 expression in adult mouse prostate luminal cells, which knockout of the Tug1 gene notably reduced (Fig. 5e–f and Supplementary Fig. 5c). Furthermore, expression of TUG1 in BPH tissues correlated positively with GPX4 levels (Fig. 5g–h and Supplementary Fig. 5d).
Fig. 5.
Regulation of ferroptosis inhibition in prostatic luminal cells by TUG1 through GPX4. a Analysis of the intersection between genes associated with BPH (n = 925) and genes related to ferroptosis (n = 65), identifying 17 overlapping genes (left). Their expression patterns in BPH and normal prostatic tissues are examined based on RNA sequencing data (right). b-c RT-qPCR analysis of the expression of 17 candidate genes in RWPE-1 cells (b) post-transfection with control shRNA or TUG1 shRNA, and in prostate tissues (c) from Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice. d Western blot analysis of GPX4 protein levels in RWPE-1 and BPH-1 cells transfected with control shRNA, TUG1 shRNA, empty vector, or TUG1 overexpression vector. e-f IHC staining for GPX4 (e) and subsequent scoring analysis (f) in prostates from Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice at 4, 8, and 12 months g CISH combined with IHC staining for the quantification of TUG1 and GPX4 in BPH tissues. h Correlation analyses between TUG1 expression and GPX4 mRNA levels in BPH tissues from our validation cohort. i-j IHC staining (i) and RT-qPCR analysis (j) of GPX4 expression in BPH and control prostate tissues from our validation cohort. k Correlation analysis between GPX4 expression levels and prostate gland volume in BPH tissues from our validation cohort. l-m Analysis of MDA production (l) and lipid ROS levels(m) in BPH-1 and RWPE-1 cells expressing empty vector or TUG1 overexpression vector, combined with GPX4 shRNA. n-o 3D culture of prostate luminal cells (n) and analysis of their diameters (o) from Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice treated with DMSO or JKE-1674. All scale bars: 50 μm *P < 0.05; **P < 0.01; ***P < 0.001; n.s no significant.
GPX4, crucial in controlling ferroptosis, prevents its occurrence by reducing lipid peroxides [34]. Notably, GPX4 levels were not only higher in BPH tissues compared to normal prostate tissues but also correlated with prostate size, IPSS score, and inflammatory status in BPH patients, though not with the proliferative capacity of prostate luminal epithelial cells (Fig. 5i–k and Supplementary Figs. 5e–5j). To elucidate the function of GPX4 in the regulation of ferroptosis by TUG1, we interfered with GPX4 expression in RWPE-1 and BPH- 1 cells under stable overexpression of TUG1. The findings indicate that GPX4 deficiency not only alleviates MDA and ROS reduction caused by TUG1 but also obstructs the enhancement of cell survival by TUG1 (Fig. 5l–m and Supplementary Figs. 5k–5l). JKE-1674, a selective and potent inhibitor of GPX4, is established to trigger ferroptosis in cells. Our study showed that reducing expression of TUG1 did not impact the responsiveness of RWPE-1 and BPH-1 cells to ferroptosis induced by JKE-1674 (Supplementary Figs. 5m–5p). Additionally, by using an in vitro 3D culture of primary mouse prostate luminal cells, we found that JKE-1674 induced ferroptosis in wild-type cells of Tug1, but not in cells with knockout of Tug1 (Fig. 5n–o). These results collectively demonstrate that TUG1 distinctly regulates ferroptosis in prostate luminal cells through GPX4.
2.6. miR-188-3p as a mediator of TUG1 regulatory influence on GPX4
Growing evidence indicates that LncRNAs regulate target genes via competing endogenous RNA (ceRNA) mechanisms by interacting with miRNAs [35]. To investigate the ceRNA function of TUG1 in diminishing ferroptosis in prostate luminal cells, we employed LncBook and LncRNASNP to identify miRNAs that might bind to TUG1. Considering the role of GPX4 as a gene regulated by TUG1 in ferroptosis, we used miRDB, miRWalk, and TargetScan to predict potential miRNA binding sites in GPX4. Our comparative analysis singled out miR-188-3p as the only miRNA common to both TUG1 and GPX4 (Fig. 6a). In BPH tissue samples, the expression of miR-188-3p was significantly reduced compared to normal prostate tissue, inversely correlating with GPX4 expression and prostate volume (Supplementary Figs. 6a–6c). Suppressing miR-188-3p in RWPE-1 and BPH-1 cells resulted in increased expression of GPX4 (Supplementary Figs. 6d–6e). Further validating the role of miR-188-3p in the regulation of GPX4 by TUG1, we treated RWPE-1 and BPH-1 cells overexpressing TUG1 with miR-188-3p mimics, observing that heightened levels of miR-188-3p suppressed GPX4 expression and nullified the enhancing effect of TUG1 on GPX4 (Fig. 6b–d).
Fig. 6.
Indirect promotion of GPX4 expression in prostate luminal cells by TUG1 through miR-188-3p binding. a A Venn diagram illustrates miR-188-3p as a shared microRNA molecule capable of interacting with both TUG1 and GPX4 according to LncBook and LncRNASNP, miRDB, miRWalk, and TargetScan database. b-c RT-qPCR analyses of GPX4 expression in RWPE-1 (b) and BPH-1 (c) cells following transfection with control mimic or miR-188-3p mimic, in combination with empty vector or TUG1 overexpression vector. d Western blot analysis evaluates the GPX4 protein levels in RWPE-1 and BPH-1 cells transfected with control mimic or miR-188-3p mimic, together with empty vector or TUG1 overexpression vector. e FISH staining analysis of the co-localization of TUG1 and miR-188-3p in RWPE-1 and BPH-1 cells. Scale bars: 20 μm. f Bioinformatics analysis identified potential binding sites of miR-188-3p on TUG1 and GPX4, leading to the construction of corresponding wild-type and mutant luciferase reporter plasmids. g Evaluation of the transcriptional activity of wild-type (WT) and mutant (MUT) TUG1 promoters in RWPE-1 cells treated with control mimic or miR-188-3p mimic, utilizing a luciferase reporter assay. h Luciferase reporter assays of the wild-type (WT) and mutant (MUT) GPX4 3′-UTR in RWPE-1 cells transfected with control mimic or miR-188-3p mimic, combined with TUG1 WT or MUT overexpression vectors. i Biotin-coupled probe RNA pull-down assays assess the enrichment of TUG1 and the GPX4 3′-UTR in RWPE-1 and BPH-1 cells. j-k RIP assays using an anti-Ago2 antibody to investigate the interaction between miR-188-3p and GPX4 in RWPE-1 (j) and BPH-1 (k) cells transfected with control shRNA, TUG1 shRNA, empty vector, or TUG1 overexpression vector, and in prostate tissues (l) from Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice. m FISH staining analysis for the co-localization of miR-188-3p and GPX4 in prostate luminal cells derived from Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice. Scale bars: 50 μm *P < 0.05; **P < 0.01; ***P < 0.001; n.s no significant.
To confirm the ceRNA interaction of TUG1 with miR-188-3p in upregulating GPX4, we demonstrated co-localization of TUG1 and miR-188-3p in the cytoplasm of RWPE-1 and BPH-1 cells using FISH (Fig. 6e). Knockdown TUG1 or knockout of Tug1 increased the levels of miR-188-3p in RWPE-1, BPH-1, and mouse prostate cells, while overexpression of TUG1 decreased them (Supplementary Figs. 6f–6h). In BPH tissues, an inverse correlation was observed between TUG1 and miR-188-3p (Supplementary Fig. 6i). Bioinformatics analysis identified binding sites of miR-188-3p on both TUG1 and the 3′-untranslated region (3′-UTR) of GPX4, prompting us to construct corresponding wild-type and mutant luciferase reporter vectors (Fig. 6f). Elevated levels of miR-188-3p in RWPE-1 and BPH-1 cells significantly reduced luciferase activity in vectors containing the wild-type TUG1 and the 3′UTRs of GPX4, a reduction absent in vectors with mutated binding sites (Fig. 6g–h and Supplementary Fig. 6j). Additionally, overexpression of TUG1 (rather than mutant form) partially reversed the inhibition of luciferase activity from the 3′UTR of GPX4 by miR-188-3p, suggesting TUG1 sequesters miR-188-3p from binding to the 3′UTR of GPX4 (Fig. 6h and Supplementary Fig. 6k).
RNA pulldown experiments with biotinylated miR-188-3p confirmed its direct interaction with TUG1 and the 3′UTRs of GPX4 in RWPE-1 and BPH-1 cells (Fig. 6i). Given that miRNAs modulate mRNA translation and degradation in an Ago2-dependent manner, RNA immunoprecipitation (RIP) experiments in RWPE-1 and BPH-1 cells showed significant enrichment of the 3′UTR of GPX4 in Ago2 immunoprecipitation in cells with TUG1 knockdown, an enrichment reduced in cells overexpressing TUG1 (Fig. 6j–k). Similarly, in mouse prostate luminal cells with Tug1 knockout, the 3′UTR of Gpx4 showed higher enrichment in Ago2 immunoprecipitation compared to wild-type cells (Fig. 6l). Furthermore, FISH in mouse prostate tissues indicated increased co-localization of miR-188-3p with the 3′UTR of Gpx4 in prostate luminal cells with Tug1 knockout compared to wild-type (Fig. 6m). In summary, these results suggest that TUG1 positively regulates GPX4 expression by sponging miR-188-3p.
2.7. miR-188-3p attenuates ferroptosis in prostatic luminal cells through GPX4-dependent regulation of TUG1
To investigate the role of miR-188-3p in modulating ferroptosis within prostatic luminal cells, we assessed alterations in cellular sensitivity to Erastin after administering either miR-188-3p mimics or inhibitors to RWPE-1 and BPH-1 cell lines. Our findings reveal that upregulation of miR-188-3p markedly increases the susceptibility of cells to Erastin, enhancing Erastin-induced cell mortality (Fig. 7a–b). In contrast, reducing miR-188-3p levels significantly diminishes this susceptibility, curtailing Erastin lethality (Fig. 7a–b). Nonetheless, MTS and colony formation assay indicate that modulation of miR-188-3p expression does not affect the response of cell to JKE-1674, a GPX4-specific inhibitor (Supplementary Figs. 7a–7h), suggesting miR-188-3p involvement in ferroptosis regulation through GPX4.
Fig. 7.
miR-188-3p as an intermediate mediator for TUG1-dependent inhibition of ferroptosis in prostate luminal cells via GPX4. RWPE-1 and BPH-1 cells were allocated into eight groups and subjected to transfection with control mimic, miR-188-3p mimic, control inhibitor, or miR-188-3p inhibitor. Additionally, cells were co-transfected with TUG1 overexpression vector in combination with control mimic, miR-188-3p mimic, miR-188-3p mimic plus empty vector, or miR-188-3p mimic plus GPX4 overexpression vector. a-b Cell viability (a) and the IC50 (b) were assessed in the transfected cells treated with gradient concentrations (2−1 μM–26 μM) of Erastin for 24 h. c-d Colony formation assays were conducted for the transfected RWPE-1 and BPH-1 cells treated with Erastin (20 μM). e-f Cell death was quantitatively assessed in the transfected RWPE-1 (e) and BPH-1 (f) cells following exposure to Erastin (20 μM). g-h 3D cultures of prostate luminal cells transfected with control inhibitor or miR-188-3p inhibitor were analyzed for their diameters (g), using cells derived from either Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice following treatment with Erastin (20 μM). Scale bars: 50 μm. i-j Levels of MDA production (i) and lipid ROS (j) were measured in the transfected RWPE-1 and BPH-1 cells (mentioned above) following treatment with Erastin (20 μM). *P < 0.05; **P < 0.01; ***P < 0.001; n.s no significant.
To elucidate miR-188-3p mediation of TUG1 influence on ferroptosis via GPX4, TUG1-overexpressing RWPE-1 and BPH-1 cells received treatment with miR-188-3p mimics. The treatment counteracted the enhanced resistance to Erastin conferred by TUG1 overexpression, with subsequent GPX4 overexpression reversing this effect, reinstating the Erastin resistance of cell (Fig. 7a–b). Further verification through colony formation and cell apoptosis assays confirmed miR-188-3p as a critical intermediary in the GPX4 suppression of ferroptosis mediated by TUG1 in these cells (Fig. 7c–f). Additionally, 3D culture experiments with primary mouse prostatic luminal cells showed that Tug1 knockout augmented Erastin sensitivity, an effect that miR-188-3p inhibition could abolish, highlighting miR-188-3p pivotal role in modulating this sensitivity (Fig. 7g–h). Furthermore, RWPE-1 and BPH-1 cells transfected with miR-188-3p mimics exhibited a significant increase in Erastin-triggered ferroptosis signals, such as ROS and MDA production (Fig. 7i–j). In contrast, suppression of miR-188-3p led to decreased ROS and MDA levels (Fig. 7i–j). TUG1 overexpression could attenuate Erastin impact on ROS and MDA accumulation, a process that miR-188-3p could reverse, and which GPX4 overexpression could ultimately neutralize (Fig. 7i–j). These observations indicate that TUG1 indirectly governs GPX4 expression via miR-188-3p, thereby deterring ferroptosis in prostatic luminal cells.
2.8. IL-1β release induced by AR deficiency modulates the ferroptosis of prostate epithelial cells through the MYC/TUG1/miR-188-3p/GPX4 signaling pathway
Deficiency of AR in prostate luminal cells leads to the infiltration of macrophages and the secretion of IL-1β, which plays a crucial role in regulating the expression of TUG1. Our investigation explores its role in the attenuation of the ferroptosis signal mediated by TUG1 in these cells. Using RT-qPCR, IHC, and FISH staining, our findings show that MDV3100 reduces AR activity in the luminal cells of the wild-type mouse prostate, leading to decreased levels of miR-188-3p and increased expression of Gpx4. However, these phenomena are not detected in Tug1 knockout cells (Fig. 8a–b and Supplementary Figs. 8a–8b). Furthermore, MDV3100 decreases the inducers of ferroptosis, such as MDA, ROS, and Fe2+, in wild-type cells, while deletion of Tug1 counters the inhibition of ferroptosis by MDV3100 (Fig. 8c–e and Supplementary Figs. 8a–8b). Treatment with IL-1β in RWPE-1 and BPH-1 cells leads to decreased levels of miR-188-3p, increased expression of GPX4, and reduced inducers of ferroptosis (Fig. 8f and Supplementary Figs. 8c–8g). Disruption of the MYC gene hinders the effect of IL-1β, which can be reversed by overexpression of TUG1 (Fig. 8f and Supplementary Figs. 8c–8g). This suggests that AR deficiency regulates TUG1 through the IL-1β/MYC signaling pathway, subsequently affecting the miR-188-3p/GPX4 axis and ferroptosis in prostate luminal cells.
Fig. 8.
Release of IL-1β triggered by AR deficiency inhibits ferroptosis in prostate epithelial cells through the MYC/TUG1/miR-188-3p/GPX4 pathway a-b RT-qPCR analysis was conducted to assess the expression levels of miR-188-3p and GPX4 (a), while IHC staining was utilized to detect the GPX4 protein level (b) in the prostates of Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice treated with PBS or MDV3100. Scale bars: 50 μm. c-e The levels of MDA production (c), lipid ROS (d), and intracellular Fe2+ (e) were quantified in the prostates of Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice treated with PBS or MDV3100. f Western blot analysis was performed to evaluate the GPX4 protein level in RWPE-1 and BPH-1 cells treated with PBS or IL-1β, and co-transfected with control shRNA, MYC shRNA, TUG1 shRNA, MYC shRNA plus empty vector, or MYC shRNA plus TUG1 overexpression vector. g-i Transillumination images of regenerated prostate tissues (g), their corresponding weights (h), and H&E staining (i) were obtained from Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice mixed with UGSM cells transfected with empty vector or Il1b overexpression vector. Scale bars: 1 cm for (g); 50 μm for (i). j-l RT-qPCR analysis of miR-188-3p (j) and Gpx4 expression (k) and IHC staining to detect GPX4 protein levels (l) were performed in the regenerated prostate tissues from prostate luminal cells of Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice mixed with UGSM cells transfected with empty vector or Il1b overexpression vector. Scale bars: 50 μm. m-o Measurements of MDA production (m), lipid ROS (n), and intracellular Fe2+ (o) were conducted in the regenerated prostate tissues from prostate luminal cells of Pbsn-Cre4 Tug1wt/wt or Pbsn-Cre4 Tug1fl/fl mice mixed with UGSM cells transfected with empty vector or Il1b overexpression vector. p-r Transillumination images of regenerated prostate tissues (p), their corresponding weights (q), H&E staining, and IHC staining of GPX4 (r) were conducted on prostate luminal cells of Pbsn-Cre4 Tug1wt/wt mice mixed with UGSM cells transfected with Il1b overexpression vector, followed by treatment with PBS or JKE-1674. Scale bars: 5 mm for (p); 50 μm for (r). s-t Representative images of prostate tissues, H&E staining, and IHC staining of GPX4 (s), and their corresponding weights (t) were obtained from the DLP of 12-month-old mice treated with PBS or JKE-1674. The dashed line area represents the prostate tissue of mice. Scale bars: 1 cm for gross; 50 μm for H&E staining. u A schematic diagram illustrates the mechanism by which AR deficiency-induced TUG1 suppresses ferroptosis and promotes BPH through the miR-188-3p/GPX4 signaling pathway. *P < 0.05; **P < 0.01; ***P < 0.001; n.s no significant.
To determine whether IL-1β, through TUG1, inhibits ferroptosis in prostate luminal cells and thereby contributes to prostate enlargement in vivo, we established a prostate regeneration tissue model. This model involved isolating prostate luminal cells and urogenital sinus mesenchyme cells (UGSM) from adult mice. UGSM cells, overexpressing Il1b or a control vector, were combined with either Tug1-deficient or wild-type prostate luminal cells and transplanted beneath the renal capsule of immunodeficient SCID/Beige male mice (Supplementary Fig. 8h). The results reveal that Il1b-overexpressing UGSM cells significantly enhance the capabilities for prostate tissue regeneration compared to controls, an effect that is attenuated by Tug1 knockout (Fig. 8g–i). RT-qPCR and IHC staining show that overexpression of Il1b in UGSM cells reduces the expression of miR-188-3p, increases Gpx4 levels, and raises the inducers of ferroptosis in the regenerated tissue, effects do not present in Tug1-deficient cells (Fig. 8j–o). Evaluating the therapeutic efficacy of the ferroptosis inhibitor JKE-1674 in both the model and aged mice showed that JKE-1674 significantly reduces prostate tissue regeneration and Gpx4 expression stimulated by Il1b-overexpressing UGSM cells, in addition to reducing the size of the prostate and Gpx4 levels in aged mice (Fig. 8p–t and Supplementary Figs. 8i–8k). In summary, these insights reveal that the reduction of AR in prostate luminal cells regulates the MYC/TUG1/miR-188-3p/GPX4 pathway via IL-1β, suppressing ferroptosis and leading to prostate enlargement. Therefore, inducing ferroptosis in prostate luminal cells emerges as a novel therapeutic target for the management of BPH.
3. Discussion
Androgen and AR-mediated signaling are crucial in prostate development [8], yet their role in prostate enlargement and the emergence of BPH in middle-aged and elderly men remains elusive. Recent evidence implicates the disruption of AR signaling in luminal cells, leading to compromised epithelial barrier function and prostatic inflammation, as a significant contributor to BPH pathogenesis [15]. Our study identified the LncRNA TUG1 as markedly upregulated in BPH tissues versus normal counterparts, with its expression levels closely linked to increased prostate volume and worsened clinical symptoms (IPSS). Notably, AR deficiency in prostate luminal cells may trigger macrophage infiltration and IL-1β release, thereby elevating TUG1 expression via MYC. TUG1, acting through the ceRNA mechanism, indirectly upregulates GPX4 by binding miR-188-3p, inhibiting ferroptosis in prostate luminal cells and fostering BPH development (Fig. 8u).
Existing studies highlights TUG1 regulatory roles across various biological contexts, including male fertility, retinal development, pulmonary arterial hypertension, and cancer progression [[25], [26], [27], [28], [29]]. Our findings in BPH provide further insights into the biological significance of TUG1. Recent research have elucidated LncRNA involvement in BPH progression through mechanisms such as luminal cell proliferation, epithelial-mesenchymal transition, and autophagy [36,37]. Yan et al. research linked autophagy-related ferroptosis to BPH occurrence [20]. TUG1 plays a critical role in modulating cellular ferroptosis signaling, exhibiting divergent effects across various diseases. Specifically, TUG1 is positively associated with ferroptosis in leukocytes during the onset of pre-eclampsia [38], whereas it contributes to liver fibrosis by suppressing ferroptosis in hepatic stellate cells [39]. However, the potential of LncRNAs, including TUG1, to modulate BPH development via ferroptosis regulation remains unexplored. Our results reveal that TUG1 is markedly overexpressed in prostate tissues, particularly in luminal cells, leading to benign prostate enlargement through the inhibition of ferroptosis rather than by promoting cell proliferation. This discovery offers novel insights into the pathogenesis of BPH. Additionally, the ferroptosis inducer JKE-1764 has shown efficacy in promoting luminal cell ferroptosis and attenuating prostate enlargement, suggesting a novel therapeutic avenue for BPH.
The subsequent challenge involves elucidating the molecular mechanisms underlying the upregulation of TUG1 during the development of BPH. AR reduction in luminal cells disrupts tight junctions, triggering a harmful cycle of internal prostate inflammation closely linked to BPH [15]. Our study further clarified the contributions of AR deficiency and internal prostate inflammation, notably macrophage infiltration, to BPH development. However, the specific mechanisms through which these inflammatory factors influence benign prostate enlargement have yet to be fully understood. Cellular ferroptosis is intricately linked to the immune and inflammatory responses of the body [21]. Inflammatory mediators such as TNF-α facilitate cystine uptake and glutathione synthesis, safeguarding fibroblasts against ferroptosis, a process implicated in the pathogenesis of rheumatoid arthritis [40]. Similarly, IL-6 has been shown to drive tumor progression in head and neck squamous cell carcinoma by fostering resistance to ferroptosis [41]. Moreover, recent research has verified that IL-1β, released by macrophages, can boosts NADPH production of gastric cancer, which subsequently sustains sufficient iron-sulfur cluster maintenance and protects tumor cells from ferroptosis [42]. Despite the broad impact of reduced AR expression on the release of numerous inflammatory mediators, our research uniquely demonstrates that AR deficiency upregulates the transcription factor MYC in prostate luminal cells via IL-1β from macrophages. The interaction of MYC with specific sequences in the promoter region of TUG1 not only stimulates its transcription but also suppresses ferroptosis signaling, thereby contributing to the onset of BPH. Given that MYC is an oncogene targeted by numerous experimental drugs [43], although these targeted therapies have not yet been approved for clinical use, our insights highlight a potential new therapeutic approach for BPH management.
LncRNAs are crucial in the regulation of gene expression, utilizing the ceRNA mechanism to indirectly influence miRNA target genes, as well as directly binding to proteins or DNA, thereby impacting a wide range of physiological and pathological processes [44]. Recent studies have illuminated the multifaceted regulatory functions of TUG1, which extends to modulating downstream genes and their associated signaling pathways. For example, TUG1 maintains the integrity of blood-testis barrier by directly interacting with the EZH2 protein, resulting in enhanced transcription of CCL2 [45]. Additionally, TUG1 influences energy metabolism in podocytes under diabetic conditions by binding to DNA sequences in the promoter region of the PGC-1α gene, promoting its expression [46]. Predominantly operating through the ceRNA mechanism, TUG1 'sponges' miRNAs, thereby indirectly augmenting the expression of specific miRNA target genes, including but not limited to miR-222-3p/CELF1, miR-194/FXR, miR-376b-3p/DPD, and miR-455-3p/AMPKβ2 [29,[47], [48], [49]]. In this research, the identification of miR-188-3p as a target miRNA interacting with TUG1 has been established. By sponging miR-188-3p, TUG1 promotes the expression of GPX4, thereby enhancing the resistance of prostate luminal cells to ferroptosis. While previous reports have documented the competitive binding of TUG1 with miR-188-3p, it was previously associated with the regulation of FGF5, not GPX4 [50]. The role of the miR-188-3p/GPX4 pathway in cell ferroptosis, regulated by non-coding RNAs distinct from TUG1 [51,52], highlights the complexity of LncRNA-mediated regulatory networks. The specific activation of the TUG1-miR-188-3p-GPX4 signaling axis during the development of BPH illustrates the sophisticated diversity of LncRNA regulatory mechanisms, offering new insights into the molecular underpinnings of BPH.
In conclusion, analysis of differential expression profiles between BPH and normal prostate tissues reveals TUG1 as a novel LncRNA linked to BPH development and symptoms. The AR deficiency-induced macrophage accumulation and IL-1β release elevate TUG1 expression via MYC. TUG1 interaction with miR-188-3p, indrectly promoting GPX4 expression, underscores a novel pathway inhibiting ferroptosis in luminal cells, thereby contributing to benign prostate enlargement. Findings illuminate the intricate interplay among AR, inflammatory factors, LncRNAs, and ferroptosis in BPH pathogenesis, proposing ferroptosis inducers as a viable therapeutic strategy for BPH patients.
4. Materials and methods
4.1. Prostate sample collection
In the present study, prostate tissue samples were collected from two distinct cohorts comprising samples of BPH and normal prostate tissues. The initial cohort consisted of 6 BPH samples and an equal number of normal prostate samples. The BPH specimens were procured from patients undergoing open simple prostatectomy to address large volume prostates, whereas the normal prostate tissues were exclusively obtained from male patients following radical cystectomy. The second cohort contained 67 BPH samples alongside 20 normal prostate samples. Patients undergoing holmium laser enucleation of the prostate provided the BPH samples, and the normal prostate samples were also exclusively derived from post-radical cystectomy. Exclusion criteria for BPH samples included the presence of cancer within the transition zone, hypogonadism, and any treatment involving testosterone or 5ARIs. All collected prostate tissues, designated for research purposes, were harvested from the transition zone after surgical procedures. A portion of these samples was preserved in liquid nitrogen for RNA extraction, while others were allocated for the preparation of frozen and paraffin-embedded sections to facilitate pathological morphology studies. The demographic and clinical features of participants in the first and second cohorts are delineated in Supplementary Fig. 1a and Supplementary Table 1, respectively. This research received ethical approval from the Ethics Committee of Shanghai Ninth People's Hospital. In accordance with the approved protocols, all participants provided written informed consent, and their data were processed and analyzed with strict confidentiality.
4.2. Mouse models and prostate regeneration assays
In compliance with the ethical standards detailed in the National Institutes of Health Guide for the Care and Use of Laboratory Animals, this study provided humane care for all animals involved. The experiment protocol was sanctioned by the Animal Care and Utilization Committee at Shanghai Jiao Tong University School of Medicine, incorporating principles of randomization and a single-blind approach. Utilizing the CRISPR-Cas9 system provided by Cyagen Company (China) Tug1−/− and LoxP-flanked Tug1 mice were engineered. By crossing Tug1floxp/floxp with Pbsn-Cre mice (Cyagen), prostate-specific conditional Tug1 knockout mice were generated. The genotyping of these mice was conducted through PCR, analyzing genomic DNA extracted from tail biopsy samples. The sequences for the genotyping primers are listed in Supplementary Table 2. The study further investigated the suppression of AR signaling by administering MDV3100, also known as Enzalutamide (Selleck, USA), diluted in 10 % DMSO, 45 % polyethylene glycol 400 (Sigma-Aldrich, USA), and 45 % normal saline, biweekly at a dose of 10 mg/kg to mice aged 8–12 months using oral gavage with an 18-gauge rounded gavage needle (Sigma-Aldrich). To explore the effects of ferroptosis inducers on prostate enlargement, JKE-1674 (25 mg/kg, Selleck) dissolved in 10 % ethanol and 90 % PEG-400, was administered to mice on alternate days for one month using oral gavage with an 18-gauge rounded gavage needle. Following the procedures outlined by Xin et al. [53], prostate regeneration assays were executed. In this process, 1 × 105 dissociated prostate cells from either wild-type or Tug1 knockout mice, aged 8 months, were amalgamated with 1 × 105 murine UGSM cells in type I collagen sourced from rat tails. These composite cells were implanted under the renal capsules of immunodeficient male SCID/Bg mice. The grafts were then monitored over an 8-week period to evaluate the capacity for prostate regeneration.
4.3. Cell culture, organoid establishment, and reagents
The BPH-1 cell line was acquired from the Shanghai Institutes for Biological Sciences. Human embryonic kidney 293T (HEK-293T) and RWPE-1 cell lines were obtained from the American Type Culture Collection. Human primary prostate luminal cells were isolated directly from normal and BPH prostate tissues. RWPE-1 and primary prostate luminal cells were cultured in Keratinocyte Serum Free Medium (K-SFM) supplemented with 0.05 mg/ml bovine pituitary extract and 5 ng/ml human recombinant epidermal growth factor. BPH-1 cells were maintained in RPMI-1640 medium, whereas HEK-293T cells were cultured in DMEM; both media were supplemented with 10 % FBS. The establishment and culture of 3D prostate luminal cell organoids followed the protocol previously described by the group of Hans Clevers [54]. Cultures were incubated at 37 °C in a humidified atmosphere with 5 % CO2 and routinely tested for mycoplasma contamination using the LookOut Mycoplasma qPCR Detection Kit (Sigma-Aldrich). Chemical compounds including MDV3100, 10058-F4, Erastin, Ferrostatin-1, Liproxstatin-1, and JKE-1674 were sourced from Selleck Chemicals and solubilized in DMSO. Recombinant human IL-1 beta protein (Proteintech, China) was reconstituted in sterile 1 × PBS (pH 7.4) with 0.1 % endotoxin-free recombinant human serum albumin.
4.4. Plasmid construction and cell transfection
The promoter region of TUG1 (spanning −2000 bp to 0 bp), the full-length TUG1, the 3′-UTR of GPX4, and their respective mutant variants were cloned into the pGL4 plasmid (Promega). Lentivector-mediated constructs targeting TUG1 and GPX4, as well as a non-targeting control, were engineered and synthesized by the Department of Biochemistry and Molecular Cell Biology at Shanghai Jiao Tong University School of Medicine. The full-length TUG1 was specifically engineered into a pLVX-puro vector. For viral production and subsequent infection, HEK293T cells were transfected using a mixture of 3.5 μg of the requisite plasmids (either shRNAs or overexpression vectors), 2.25 μg of psPAX, and 4.25 μg of pMD2.G, alongside 30 μl of polyethylenimine. This transfection occurred when the cells reached 80%–90 % confluence. Forty-eight hours post-transfection, the medium containing the virus was harvested, filtered through a 0.45 μm filter, and used to infect target cells in the presence of 4 μg/ml polybrene (Sigma-Aldrich). Following a 48-h period of viral infection, cells were subjected to puromycin selection for 72 h. Various RNA molecules including the nonspecific control mimic, miR-188-3p mimic, control inhibitor, miR-188-3p inhibitor, MYC siRNA, SP1 siRNA, and a negative control siRNA were sourced from GenePharma (China) and introduced into cells employing Lipofectamine 3000 reagent (Thermo Fisher Scientific, USA), adhering to the manufacturer's protocol. The sequences for these components are detailed in Supplementary Table 2.
4.5. Cell viability and colony formation assays
Prostate cells, subjected to gradient concentrations of ferroptosis inducers, were seeded into 96-well plates at a density of 1000 cells per well and incubated at 37 °C for 24 h. Subsequently, MTS solution (Promega) was applied to each well, followed by further incubation at 37 °C for 1 h. The absorbance at 490 nm was determined using a BioTek Microplate Reader (BioTek, USA). In the colony formation assay, prostate cells were plated in 6-well plates at a density of 500 cells per well and maintained in culture for two weeks until the formation of visible colonies. These colonies were fixed with 4 % paraformaldehyde and stained with 0.1 % crystal violet, enabling their visualization and enumeration.
4.6. Immunoblot analysis
Total protein was extracted from cells employing RIPA buffer (Beyotime, China), supplemented with a protease inhibitor cocktail (Sigma-Aldrich). The protein concentrations were determined using BCA Protein Assay Kits (Thermo Fisher Scientific). Western blotting procedures were carried out and visualized following previously established methods [55]. Detailed information regarding the antibodies used can be found in Supplementary Table 2.
4.7. CISH, FISH, IHC, IF, and H&E staining procedures
CISH, FISH, IHC, IF, and H&E staining of prostate tissues were conducted following previously established protocols [55,56]. Details regarding the primary and secondary antibodies utilized are provided in Supplementary Table 2. The semiquantitative evaluation of CISH and IHC staining involved an assessment of staining intensity (I: 0 = negative, 1 = weak, 2 = moderate, 3 = intense) and the percentage of positively-stained cells (P: 0–5% = 0, 6–35 % = 1, 36–70 % = 2, >70 % = 3). A final quantitative score (Q) was calculated by multiplying the intensity score (I) by the percentage score (P). This scoring was independently conducted by two experienced pathologists in a double-blind manner to ensure unbiased results.
4.8. RT-qPCR analysis
Total RNA from prostate tissues or cells was isolated using Trizol reagent (Invitrogen, USA). cDNA was synthesized from 1 μg of RNA employing reverse transcriptase and oligo(dT) primers (Takara, Japan), following the manufacturer's guidelines. qPCR was performed in triplicate using Fast-SYBR® Green Master Mix on an Applied Biosystems ViiA™ 7 Real-Time PCR System (Applied Biosystems, USA) to evaluate the expression levels of various genes. The GAPDH housekeeping gene served as an endogenous control to normalize the amount of cDNA between samples. Relative gene expression was calculated using the 2−ΔΔCt method [57]. Primer sequences utilized for RT-qPCR are listed in Supplementary Table 2.
4.9. RNA-seq and pathway analysis
Total RNA was extracted utilizing RNA Isolation Kits (Agilent, USA), and quality control assessments were conducted using Qubit 4.0 (Thermo Fisher Scientific). Following the removal of rRNA via oligo(dT), library preparation was performed using the Ultra™ II Directional RNA Library Prep Kit (NEB, USA) according to the manufacturer's instructions, and sequencing was executed on the Illumina HiSeq X Ten platform (Illumina, USA). The sequencing reads were aligned to the human genome GRCh38 using HISAT2. Expression levels of each transcript were quantified as reads per kilobase of exon model per million mapped reads (RPKM). The R Bioconductor DESeq2 package was employed to identify differentially expressed genes.
For the GO analysis of BPH and normal prostate tissues, we utilized the Metascape tool (http://metascape.org) [58] applying default settings. We selected all marker genes exhibiting an average log-fold change greater than 0.5 and adjusted P values less than 0.01. Functional enrichment analysis was conducted using the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway and WikiPathway databases. GSEA was performed with the fgsea R package, employing the Molecular Signatures Database WikiPathway gene sets. We set the permutation type to phenotype and executed 1000 permutations. The P values for both over-representation analysis and GSEA were adjusted for false discovery rates (FDRs), and a gene set was deemed significant if the FDR was below 0.05.
4.10. MDA, lipid ROS, and Fe2+ measurement and flow cytometry
The relative concentration of MDA in cell lysates was quantified using the Lipid Peroxidation Assay Kit (Sigma-Aldrich), following the manufacturer's guidelines. Absorbance was measured at 532 nm using a BioTek Microplate Reader. The level of lipid ROS was assessed by flow cytometry utilizing BODIPY-C11 dye (Thermo Fisher Scientific). Cells were incubated with BODIPY-C11 (2.5 μM/mL) at 37 °C for 30 min in the dark, then washed twice with PBS, and analyzed by flow cytometry to quantify intracellular lipid ROS. A minimum of 10,000 cells per sample was evaluated, with data analysis conducted using FlowJo Software. The measurement of intracellular Fe2+ levels was performed employing an iron assay kit (Abcam), adhering to the manufacturer's protocol. Cells were harvested, washed with ice-cold PBS, homogenized, and centrifuged at 4 °C to eliminate insoluble material. The supernatant was collected, and an iron reducer was added to each sample, followed by incubation at room temperature for 30 min. Subsequently, 100 μL of iron probe was introduced to each sample, mixed, and incubated for 1 h at room temperature. Measurements were taken immediately at 593 nm using a BioTek Microplate Reader. Following digestion of BPH tissue into a single-cell suspension, the cells were incubated with fluorescence-conjugated CD45 and EpCAM antibodies at 4 °C for 30 min. Details regarding the antibodies utilized for flow cytometry analysis are provided in Supplementary Table 2. Flow cytometry analyses and sorting were executed using the LSRFortessa™ X-20 system (BD Biosciences, USA), with data analysis performed using FlowJo Software.
4.11. ELISA
We quantified human IL-1β concentrations in BPH tissue lysates utilizing a sandwich ELISA protocol on Nunc MaxiSorp 96-well plates (Sigma-Aldrich). Capture and detection anti-IL-1β was acquired from Sigma-Aldrich, and the streptavidin-HRP conjugate was procured from Sigma-Aldrich. Preparation of the o-phenylenediamine dihydrochloride substrate solution (Sigma-Aldrich) followed the manufacturer's guidelines. IL-1β concentrations were ascertained by measuring absorbance at 450 nm using a BioTek Microplate Reader.
4.12. Dual-luciferase reporter assay
RWPE-1 and BPH-1 cells, seeded in 24-well plates, underwent transfection with firefly and renilla luciferase plasmids via Lipofectamine 3000 (Thermo Fisher Scientific). At 48 h post-transfection, after PBS washes, we measured luciferase activity using the Dual-Luciferase Reporter Assay System (Promega), adhering to the manufacturer's protocol. Firefly luciferase activity was normalized to renilla luciferase activity, serving as an internal control.
4.13. ChIP-qPCR assay
In RWPE-1 and BPH-1 cells, we performed ChIP assays to investigate MYC binding to the TUG1 promoter region, using the Pierce™ Magnetic ChIP Kit (Thermo Fisher Scientific) in line with the manufacturer's recommendations. ChIP assays utilized anti-MYC and mouse anti-IgG purchased from Abcam. We analyzed the enrichment of specific DNA fragments via qPCR, with primer sequences provided in Supplementary Table S2.
4.14. RIP and RNA pull-down assays
The RIP assay was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Sigma-Aldrich), adhering to the manufacturer's protocol. In brief, prostate cells were collected and lysed using RIP lysis buffer. Subsequently, the lysates were incubated with magnetic beads conjugated to either an anti-Ago2 antibody or a control IgG antibody (Abcam) at 4 °C for 6 h. Following incubation, the beads were washed and treated with proteinase K to digest proteins. The RNA bound to the immunocomplexes was then purified and subjected to RT-qPCR analysis. For the RNA pull-down assay, the procedure was carried out using biotinylated miR-188-3p probes and the Pierce Magnetic RNA-Protein Pull-Down Kit, following the manufacturer's guidelines. Prostate cell lysates, obtained through sonication and lysis, were incubated with probe-coated beads overnight at 4 °C. After washing, the RNA complexes attached to the beads were eluted, purified, and their presence confirmed by RT-qPCR. Primers related to both RIP and RNA pull-down assays are detailed in Supplementary Table S2.
4.15. Bioinformatic analysis
To predict transcription factors binding to the TUG1 promoter region, we employed the JASPAR, PROMO, and AnimalTFDB databases. The JASPAR was employed to further investigate the specific binding sequence of MYC on the TUG1 promoter. The LncRNASNP and LncBook databases were employed to predict microRNAs (miRNAs) that bind to TUG1. Concurrently, miRwalk, miRDB, and TargetScan were utilized to forecast miRNAs targeting the 3′-UTR of GPX4. The expression levels of TUG1 and IL1B in various tissues and cell types were analyzed using the online HPA database (https://www.proteinatlas.org, TISSUE or SINGLE CELL sub-sections) [59,60].
4.16. Statistical analysis
Data are presented as mean ± standard deviation (SD), reflecting results from at least three independent experiments. Group differences were evaluated using the Student's t-test (two-tailed) and one-way analysis of variance (ANOVA), followed by Tukey's post hoc test for multiple comparisons. Linear regression analysis was employed to examine the correlation between two variables, with the goodness of fit indicated by R2. The Chi-square test and multivariate logistic regression analyses were utilized to assess risk variables associated with TUG1 expression in the prostate. All statistical analyses were conducted using SPSS software version 23.0, with a P-value <0.05 deemed indicative of statistical significance.
Funding
This research was funded by the Top-level Clinical Discipline Project of Shanghai Pudong (No. PWYgf 2021–06), the National Natural Science Foundation of China (No. 82170788), the National Natural Science Foundation for Young Scientists (No. 82200855), and the Fundamental Research Program Funding of the Ninth People's Hospital affiliated to Shanghai Jiao Tong University School of Medicine (JYZZ194).
5. Ethics approval and consent to Participate
Ethical approval for this study was granted by the Ethics Committee of the Shanghai Ninth People's Hospital, affiliated with Shanghai Jiao Tong University School of Medicine (SH9H-2021-TK369-1). This study was conducted in strict adherence to the ethical principles outlined in the Declaration of Helsinki.
CRediT authorship contribution statement
Ming Zhan: Writing – original draft, Supervision, Conceptualization. Huan Xu: Methodology, Data curation. Guopeng Yu: Investigation, Data curation. Qi Chen: Methodology. Ruifeng Yang: Methodology, Data curation. Yanbo Chen: Resources, Funding acquisition. Jianchao Ge: Formal analysis. Zhong Wang: Supervision, Resources, Funding acquisition. Ruimeng Yang: Writing – original draft, Supervision. Bin Xu: Writing – original draft, Supervision, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this paper.
Acknowledgments
We express our sincere gratitude to the staff, patients, and all participants involved in this study for their unwavering dedication and invaluable contributions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2024.103298.
Contributor Information
Zhong Wang, Email: zhongwang2000@sina.com.
Ruimeng Yang, Email: ruyang@coh.org.
Bin Xu, Email: chxb2004@shsmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Data availability
The RNA-seq data is available in Supplementary Table S3. The datasets utilized and analyzed during this study are accessible from the corresponding authors upon reasonable request.
References
- 1.Bushman W. Etiology, epidemiology, and natural history of benign prostatic hyperplasia. Urol Clin North Am. 2009;36(4):403–415. doi: 10.1016/j.ucl.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Vuichoud C., Loughlin K.R. Benign prostatic hyperplasia: epidemiology, economics and evaluation. Can. J. Urol. 2015;22(Suppl 1):1–6. [PubMed] [Google Scholar]
- 3.Lim K.B. Epidemiology of clinical benign prostatic hyperplasia. Asian J Urol. 2017;4(3):148–151. doi: 10.1016/j.ajur.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chughtai B., Forde J.C., Thomas D.D., Laor L., Hossack T., Woo H.H., Te A.E., Kaplan S.A. Benign prostatic hyperplasia. Nat Rev Dis Primers. 2016;2 doi: 10.1038/nrdp.2016.31. [DOI] [PubMed] [Google Scholar]
- 5.Cornu J.N., Oelke M., Parsons K.F. Benign prostatic hyperplasia and lower urinary tract symptoms. N. Engl. J. Med. 2012;367(17):1668. doi: 10.1056/NEJMc1209907. author reply 1668-9. [DOI] [PubMed] [Google Scholar]
- 6.Kim E.H., Larson J.A., Andriole G.L. Management of benign prostatic hyperplasia. Annu. Rev. Med. 2016;67:137–151. doi: 10.1146/annurev-med-063014-123902. [DOI] [PubMed] [Google Scholar]
- 7.Dahm P., Brasure M., MacDonald R., Olson C.M., Nelson V.A., Fink H.A., Rwabasonga B., Risk M.C., Wilt T.J. Comparative effectiveness of newer Medications for lower urinary tract symptoms Attributed to benign prostatic hyperplasia: a Systematic review and Meta-analysis. Eur. Urol. 2017;71(4):570–581. doi: 10.1016/j.eururo.2016.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francis J.C., Swain A. Prostate Organogenesis. Cold Spring Harb Perspect Med. 2018;8(7) doi: 10.1101/cshperspect.a030353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho C.K., Habib F.K. Estrogen and androgen signaling in the pathogenesis of BPH. Nat. Rev. Urol. 2011;8(1):29–41. doi: 10.1038/nrurol.2010.207. [DOI] [PubMed] [Google Scholar]
- 10.Azzouni F., Mohler J. Role of 5alpha-reductase inhibitors in benign prostatic diseases. Prostate Cancer Prostatic Dis. 2012;15(3):222–230. doi: 10.1038/pcan.2012.1. [DOI] [PubMed] [Google Scholar]
- 11.Harman S.M., Metter E.J., Tobin J.D., Pearson J., Blackman M.R., Baltimore A. Longitudinal Study of, Longitudinal effects of aging on serum total and free testosterone levels in healthy men. Baltimore Longitudinal Study of Aging. J. Clin. Endocrinol. Metab. 2001;86(2):724–731. doi: 10.1210/jcem.86.2.7219. [DOI] [PubMed] [Google Scholar]
- 12.Bartsch G., Rittmaster R.S., Klocker H. Dihydrotestosterone and the concept of 5alpha-reductase inhibition in human benign prostatic hyperplasia. Eur. Urol. 2000;37(4):367–380. doi: 10.1159/000020181. [DOI] [PubMed] [Google Scholar]
- 13.De Nunzio C., Presicce F., Tubaro A. Inflammatory mediators in the development and progression of benign prostatic hyperplasia. Nat. Rev. Urol. 2016;13(10):613–626. doi: 10.1038/nrurol.2016.168. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt M., Naumann H., Weidler C., Schellenberg M., Anders S., Straub R.H. Inflammation and sex hormone metabolism. Ann. N. Y. Acad. Sci. 2006;1069:236–246. doi: 10.1196/annals.1351.021. [DOI] [PubMed] [Google Scholar]
- 15.Zhang B., Kwon O.J., Henry G., Malewska A., Wei X., Zhang L., Brinkley W., Zhang Y., Castro P.D., Titus M., Chen R., Sayeeduddin M., Raj G.V., Mauck R., Roehrborn C., Creighton C.J., Strand D.W., Ittmann M.M., Xin L. Non-cell-autonomous regulation of prostate epithelial Homeostasis by androgen receptor. Mol Cell. 2016;63(6):976–989. doi: 10.1016/j.molcel.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie Y., Hou W., Song X., Yu Y., Huang J., Sun X., Kang R., Tang D. Ferroptosis: process and function. Cell Death Differ. 2016;23(3):369–379. doi: 10.1038/cdd.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y., Wan Y., Jiang Y., Zhang L., Cheng W. GPX4: the hub of lipid oxidation, ferroptosis, disease and treatment. Biochim. Biophys. Acta Rev. Canc. 2023;1878(3) doi: 10.1016/j.bbcan.2023.188890. [DOI] [PubMed] [Google Scholar]
- 18.Viswanathan V.S., Ryan M.J., Dhruv H.D., Gill S., Eichhoff O.M., Seashore-Ludlow B., Kaffenberger S.D., Eaton J.K., Shimada K., Aguirre A.J., Viswanathan S.R., Chattopadhyay S., Tamayo P., Yang W.S., Rees M.G., Chen S., Boskovic Z.V., Javaid S., Huang C., Wu X., Tseng Y.Y., Roider E.M., Gao D., Cleary J.M., Wolpin B.M., Mesirov J.P., Haber D.A., Engelman J.A., Boehm J.S., Kotz J.D., Hon C.S., Chen Y., Hahn W.C., Levesque M.P., Doench J.G., Berens M.E., Shamji A.F., Clemons P.A., Stockwell B.R., Schreiber S.L. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547(7664):453–457. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M.E., Chen J., Lu Y., Bawcom A.R., Wu J., Ou J., Asara J.M., Armstrong A.J., Wang Q., Li L., Wang Y., Huang J., Chen M. RB1-deficient prostate tumor growth and metastasis are vulnerable to ferroptosis induction via the E2F/ACSL4 axis. J. Clin. Invest. 2023;133(10) doi: 10.1172/JCI166647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y., Zhou Y., Liu D., Wang Z., Qiu J., Zhang J., Chen P., Zeng G., Guo Y., Wang X., DiSanto M.E., Zhang X. Glutathione Peroxidase 3 induced mitochondria-mediated apoptosis via AMPK/ERK1/2 pathway and resisted autophagy-related ferroptosis via AMPK/mTOR pathway in hyperplastic prostate. J. Transl. Med. 2023;21(1):575. doi: 10.1186/s12967-023-04432-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X., Kang R., Kroemer G., Tang D. Ferroptosis in infection, inflammation, and immunity. J. Exp. Med. 2021;218(6) doi: 10.1084/jem.20210518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He F., Zhang P., Liu J., Wang R., Kaufman R.J., Yaden B.C., Karin M. ATF4 suppresses hepatocarcinogenesis by inducing SLC7A11 (xCT) to block stress-related ferroptosis. J. Hepatol. 2023;79(2):362–377. doi: 10.1016/j.jhep.2023.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui Y., Zhang Y., Zhao X., Shao L., Liu G., Sun C., Xu R., Zhang Z. ACSL4 exacerbates ischemic stroke by promoting ferroptosis-induced brain injury and neuroinflammation. Brain Behav. Immun. 2021;93:312–321. doi: 10.1016/j.bbi.2021.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Zhou R., Chen Y., Li S., Wei X., Hu W., Tang S., Ding J., Fu W., Zhang H., Chen F., Hao W., Lin Y., Zhu R., Wang K., Dong L., Zhao Y., Feng X., Chen F., Ding C., Hu W. TRPM7 channel inhibition attenuates rheumatoid arthritis articular chondrocyte ferroptosis by suppression of the PKCalpha-NOX4 axis. Redox Biol. 2022;55 doi: 10.1016/j.redox.2022.102411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewandowski J.P., Dumbovic G., Watson A.R., Hwang T., Jacobs-Palmer E., Chang N., Much C., Turner K.M., Kirby C., Rubinstein N.D., Groff A.F., Liapis S.C., Gerhardinger C., Bester A., Pandolfi P.P., Clohessy J.G., Hoekstra H.E., Sauvageau M., Rinn J.L. The Tug1 lncRNA locus is essential for male fertility. Genome Biol. 2020;21(1):237. doi: 10.1186/s13059-020-02081-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S., Cao W., Gao S., Nie X., Zheng X., Xing Y., Chen Y., Bao H., Zhu D. TUG1 regulates pulmonary arterial smooth muscle cell proliferation in pulmonary arterial hypertension. Can. J. Cardiol. 2019;35(11):1534–1545. doi: 10.1016/j.cjca.2019.07.630. [DOI] [PubMed] [Google Scholar]
- 27.Young T.L., Matsuda T., Cepko C.L. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr. Biol. 2005;15(6):501–512. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki M.M., Iijima K., Ogami K., Shinjo K., Murofushi Y., Xie J., Wang X., Kitano Y., Mamiya A., Kibe Y., Nishimura T., Ohka F., Saito R., Sato S., Kobayashi J., Yao R., Miyata K., Kataoka K., Suzuki H.I., Kondo Y. TUG1-mediated R-loop resolution at microsatellite loci as a prerequisite for cancer cell proliferation. Nat. Commun. 2023;14(1):4521. doi: 10.1038/s41467-023-40243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y.H., Wu M.H., Huang Y.H., Yeh C.T., Cheng M.L., Chi H.C., Tsai C.Y., Chung I.H., Chen C.Y., Lin K.H. Taurine up-regulated gene 1 functions as a master regulator to coordinate glycolysis and metastasis in hepatocellular carcinoma. Hepatology. 2018;67(1):188–203. doi: 10.1002/hep.29462. [DOI] [PubMed] [Google Scholar]
- 30.Vela Navarrete R., Garcia Cardoso J.V., Barat A., Manzarbeitia F., Lopez Farre A. BPH and inflammation: pharmacological effects of Permixon on histological and molecular inflammatory markers. Results of a double blind pilot clinical assay. Eur. Urol. 2003;44(5):549–555. doi: 10.1016/s0302-2838(03)00368-3. [DOI] [PubMed] [Google Scholar]
- 31.Sheng J., Yang Y., Cui Y., He S., Wang L., Liu L., He Q., Lv T., Han W., Yu W., Hu S., Jin J. M2 macrophage-mediated interleukin-4 signalling induces myofibroblast phenotype during the progression of benign prostatic hyperplasia. Cell Death Dis. 2018;9(7):755. doi: 10.1038/s41419-018-0744-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Lin W.J., Izumi K., Jiang Q., Lai K.P., Xu D., Fang L.Y., Lu T., Li L., Xia S., Chang C. Increased infiltrated macrophages in benign prostatic hyperplasia (BPH): role of stromal androgen receptor in macrophage-induced prostate stromal cell proliferation. J. Biol. Chem. 2012;287(22):18376–18385. doi: 10.1074/jbc.M112.355164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang X., Stockwell B.R., Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021;22(4):266–282. doi: 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang W.S., Stockwell B.R. Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 2016;26(3):165–176. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tong S., Mo M., Hu X., Wu L., Chen M., Zhao C. MIR663AHG as a competitive endogenous RNA regulating TGF-beta-induced epithelial proliferation and epithelial-mesenchymal transition in benign prostate hyperplasia. J. Biochem. Mol. Toxicol. 2023;37(9) doi: 10.1002/jbt.23391. [DOI] [PubMed] [Google Scholar]
- 37.Zhou L., Li Y., Li J., Yao H., Huang J., Li C., Wang L. Decoding ceRNA regulatory network and autophagy-related genes in benign prostatic hyperplasia. Int. J. Biol. Macromol. 2023;225:997–1009. doi: 10.1016/j.ijbiomac.2022.11.162. [DOI] [PubMed] [Google Scholar]
- 38.Lekva T., Michelsen A.E., Roland M.C.P., Norwitz E.R., Estensen M.E., Olstad O.K., Akkouh I.A., Henriksen T., Bollerslev J., Aukrust P., Ueland T. Increased ferroptosis in leukocytes from preeclamptic women involving the long non-coding taurine upregulated gene 1 (TUG1) J. Intern. Med. 2024;295(2):181–195. doi: 10.1111/joim.13732. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X., Zhao L., Ying K., Xu J., Huang Y., Zhu R., Ding Y., Cai W., Wu X., Miao D., Xu Q., Zeng Y., Yu F. TUG1 protects against ferroptosis of hepatic stellate cells by upregulating PDK4-mediated glycolysis. Chem. Biol. Interact. 2023;383 doi: 10.1016/j.cbi.2023.110673. [DOI] [PubMed] [Google Scholar]
- 40.Wu J., Feng Z., Chen L., Li Y., Bian H., Geng J., Zheng Z.H., Fu X., Pei Z., Qin Y., Yang L., Zhao Y., Wang K., Chen R., He Q., Nan G., Jiang X., Chen Z.N., Zhu P. TNF antagonist sensitizes synovial fibroblasts to ferroptotic cell death in collagen-induced arthritis mouse models. Nat. Commun. 2022;13(1):676. doi: 10.1038/s41467-021-27948-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li M., Jin S., Zhang Z., Ma H., Yang X. Interleukin-6 facilitates tumor progression by inducing ferroptosis resistance in head and neck squamous cell carcinoma. Cancer Lett. 2022;527:28–40. doi: 10.1016/j.canlet.2021.12.011. [DOI] [PubMed] [Google Scholar]
- 42.Han Y., Zhang Y.Y., Pan Y.Q., Zheng X.J., Liao K., Mo H.Y., Sheng H., Wu Q.N., Liu Z.X., Zeng Z.L., Yang W., Yuan S.Q., Huang P., Ju H.Q., Xu R.H. IL-1beta-associated NNT acetylation orchestrates iron-sulfur cluster maintenance and cancer immunotherapy resistance. Mol Cell. 2023;83(11):1887–1902 e8. doi: 10.1016/j.molcel.2023.05.011. [DOI] [PubMed] [Google Scholar]
- 43.Wang C., Zhang J., Yin J., Gan Y., Xu S., Gu Y., Huang W. Alternative approaches to target Myc for cancer treatment. Signal Transduct Target Ther. 2021;6(1):117. doi: 10.1038/s41392-021-00500-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Statello L., Guo C.J., Chen L.L., Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021;22(2):96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang S., Qian Z., Ge X., Li C., Xue M., Liang K., Ma R., Ouyang L., Zheng L., Jing J., Cao S., Zhang Y., Yang Y., Chen Y., Ma J., Yao B. LncRNA Tug1 maintains blood-testis barrier integrity by modulating Ccl2 expression in high-fat diet mice. Cell. Mol. Life Sci. 2022;79(2):114. doi: 10.1007/s00018-022-04142-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Long J., Badal S.S., Ye Z., Wang Y., Ayanga B.A., Galvan D.L., Green N.H., Chang B.H., Overbeek P.A., Danesh F.R. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J. Clin. Invest. 2016;126(11):4205–4218. doi: 10.1172/JCI87927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang M., Li F., Liu Y., Gu Z., Zhang L., Lee J., He L., Vatsalya V., Zhang H.G., Deng Z., Zhang X., Chen S.Y., Guo G.L., Barve S., McClain C.J., Feng W. Probiotic-derived nanoparticles inhibit ALD through intestinal miR194 suppression and subsequent FXR activation. Hepatology. 2023;77(4):1164–1180. doi: 10.1002/hep.32608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B., Huang N., Wei S., Xv J., Meng Q., Aschner M., Li X., Chen R. lncRNA TUG1 as a ceRNA promotes PM exposure-induced airway hyper-reactivity. J. Hazard Mater. 2021;416 doi: 10.1016/j.jhazmat.2021.125878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tasaki Y., Suzuki M., Katsushima K., Shinjo K., Iijima K., Murofushi Y., Naiki-Ito A., Hayashi K., Qiu C., Takahashi A., Tanaka Y., Kawaguchi T., Sugawara M., Kataoka T., Naito M., Miyata K., Kataoka K., Noda T., Gao W., Kataoka H., Takahashi S., Kimura K., Kondo Y. Cancer-specific targeting of taurine-upregulated gene 1 enhances the effects of Chemotherapy in pancreatic cancer. Cancer Res. 2021;81(7):1654–1666. doi: 10.1158/0008-5472.CAN-20-3021. [DOI] [PubMed] [Google Scholar]
- 50.Zhang P., Li Y.N., Tu S., Cheng X.B. SP1-induced lncRNA TUG1 regulates proliferation and apoptosis in islet cells of type 2 diabetes mellitus via the miR-188-3p/FGF5 axis. Eur. Rev. Med. Pharmacol. Sci. 2021;25(4):1959–1966. doi: 10.26355/eurrev_202102_25096. [DOI] [PubMed] [Google Scholar]
- 51.Yang W., Zhao J., Zhao Y., Li W., Zhao L., Ren Y., Ou R., Xu Y. Hsa_circ_0048179 attenuates free fatty acid-induced steatosis via hsa_circ_0048179/miR-188-3p/GPX4 signaling. Aging (Albany NY) 2020;12(23):23996–24008. doi: 10.18632/aging.104081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin J., Wang Y., Zheng D., Liang M., He Q. A novel identified Circular RNA, mmu_mmu_circRNA_0000309, involves in Germacrone-mediated Improvement of diabetic nephropathy through regulating ferroptosis by targeting miR-188-3p/GPX4 signaling Axis. Antioxid Redox Signal. 2022;36(10–12):740–759. doi: 10.1089/ars.2021.0063. [DOI] [PubMed] [Google Scholar]
- 53.Xin L., Ide H., Kim Y., Dubey P., Witte O.N. In vivo regeneration of murine prostate from dissociated cell populations of postnatal epithelia and urogenital sinus mesenchyme. Proc Natl Acad Sci U S A. 2003;100(Suppl 1):11896–11903. doi: 10.1073/pnas.1734139100. Suppl 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drost J., Karthaus W.R., Gao D., Driehuis E., Sawyers C.L., Chen Y., Clevers H. Organoid culture systems for prostate epithelial and cancer tissue. Nat. Protoc. 2016;11(2):347–358. doi: 10.1038/nprot.2016.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang R.M., Song S.Y., Wu F.Y., Yang R.F., Shen Y.T., Tu P.H., Wang Z., Zhang J.X., Cheng F., Gao G.Q., Liang J., Guo M.M., Yang L., Zhou Y., Zhao S.X., Zhan M., Song H.D. Myeloid cells interact with a subset of thyrocytes to promote their migration and follicle formation through NF-kappaB. Nat. Commun. 2023;14(1):8082. doi: 10.1038/s41467-023-43895-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang R.M., Zhan M., Xu S.W., Long M.M., Yang L.H., Chen W., Huang S., Liu Q., Zhou J., Zhu J., Wang J. miR-3656 expression enhances the chemosensitivity of pancreatic cancer to gemcitabine through modulation of the RHOF/EMT axis. Cell Death Dis. 2017;8(10):e3129. doi: 10.1038/cddis.2017.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 58.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karlsson M., Zhang C., Mear L., Zhong W., Digre A., Katona B., Sjostedt E., Butler L., Odeberg J., Dusart P., Edfors F., Oksvold P., von Feilitzen K., Zwahlen M., Arif M., Altay O., Li X., Ozcan M., Mardinoglu A., Fagerberg L., Mulder J., Luo Y., Ponten F., Uhlen M., Lindskog C. A single-cell type transcriptomics map of human tissues. Sci. Adv. 2021;7(31) doi: 10.1126/sciadv.abh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C.A., Odeberg J., Djureinovic D., Takanen J.O., Hober S., Alm T., Edqvist P.H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J.M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., Ponten F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220) doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data is available in Supplementary Table S3. The datasets utilized and analyzed during this study are accessible from the corresponding authors upon reasonable request.