Abstract
Nucleocytoplasmic large DNA viruses (NCLDVs; also called giant viruses), constituting the phylum Nucleocytoviricota, can infect a wide range of eukaryotes and exchange genetic material with not only their hosts but also prokaryotes and phages. A few NCLDVs were reported to encode genes conferring resistance to beta‑lactam, trimethoprim, or pyrimethamine, suggesting that they are potential vehicles for the transmission of antibiotic resistance genes (ARGs) in the biome. However, the incidence of ARGs across the phylum Nucleocytoviricota, their evolutionary characteristics, their dissemination potential, and their association with virulence factors remain unexplored. Here, we systematically investigated ARGs of 1416 NCLDV genomes including those of almost all currently available cultured isolates and high-quality metagenome-assembled genomes from diverse habitats across the globe. We reveal that 39.5% of them carry ARGs, which is approximately 37 times higher than that for phage genomes. A total of 12 ARG types are encoded by NCLDVs. Phylogenies of the three most abundant NCLDV-encoded ARGs hint that NCLDVs acquire ARGs from not only eukaryotes but also prokaryotes and phages. Two NCLDV-encoded trimethoprim resistance genes are demonstrated to confer trimethoprim resistance in Escherichia coli. The presence of ARGs in NCLDV genomes is significantly correlated with mobile genetic elements and virulence factors.
Subject terms: Antimicrobial resistance, Viral evolution, Metagenomics
Nucleocytoplasmic large DNA viruses, or ‘giant viruses’, infect a wide range of eukaryotes and can exchange genetic material not only with their hosts but also with bacteria and phages. Here, the authors show that many giant viruses carry diverse antibiotic resistance genes, which are associated with mobile genetic elements and genes encoding potential virulence factors.
Introduction
Due to its impact on human health, antibiotic resistance was identified as one of the top 10 threats to global health by the World Health Organization in 20191. Antibiotic resistance can arise through point mutation or horizontal gene transfer (HGT), with the latter often being cited as a key driver of its rapid spread2. HGT mainly takes three forms in prokaryotes: conjugation, transduction, or transformation2. Of these, transduction, the phage-mediated transfer of genetic material, is considered a route for ARG exchange among prokaryotes. Genes conferring resistance to beta-lactam antibiotics, glycopeptides, macrolides, peptide antibiotics, and tetracyclines, have been detected in phages from a variety of environments3–9. Moreover, the functionality of phage-encoded beta-lactam resistance genes from aquatic environments was verified using heterologous expression systems based on E. coli7,9.
Prokaryotes do not only exchange genetic material with other prokaryotes and phages, but also acquire genetic material from eukaryotes10–12. One of the notable examples of eukaryote-to-prokaryote HGT is that a gene conveying mupirocin resistance in bacteria was suggested to be transferred from an early evolved eukaryote11. As with prokaryotes, eukaryotes exchange genetic material with their viruses13,14. A comprehensive analysis of HGT between 201 eukaryotic and 108,842 viral taxa revealed that NCLDVs probably infecting all major eukaryotic microbial lineages were the main contributors to genetic exchange across eukaryotic diversity15. In addition, there was evidence that a number of genes found in giant viruses were shared with prokaryotes or phages16. Given the complex cross-kingdom HGT mentioned above, it is conceivable that some giant viruses are potential effective vehicles of DNA transfer between eukaryotes and prokaryotes.
To date, there are only three studies that have reported ARGs of NCLDVs. In a pioneering study, Muller et al. found that six NCLDVs of the family Marseilleviridae harbored a gene encoding dihydrofolate reductase individually. Moreover, the authors demonstrated that one of the six dihydrofolate reductase genes was capable of conferring resistance to trimethoprim and pyrimethamine while expressed in Saccharomyces cerevisiae17. In a follow-up study, Colson et al. documented the presence of a beta-lactamase gene in 21 NCLDV genomes and revealed that the product of beta-lactamase gene of the giant Tupanvirus exhibited beta-lactam hydrolyzing activity after expression in E. coli18. Recently, genes encoding dihydrofolate reductase or beta-lactamase were observed in eight NCLDV genomes reconstructed from permafrost metagenomes19. These previous findings indicate that NCLDVs are potential vehicles for the transmission of ARGs in the biome. However, the incidence of ARGs across the phylum Nucleocytoviricota, their evolutionary characteristics, their dissemination potential, and their association with virulence factors have not yet been explored.
In this work, we conducted a comprehensive analysis of ARGs across 1416 giant virus genomes. This genome collection encompassed nearly all currently available genomes of cultured isolates and high-quality metagenome-assembled genomes (MAGs) sourced from diverse habitat types worldwide. To ensure the robustness of our results, we analyzed isolate genomes and MAGs separately, where applicable, to account for the potential effects of contaminating DNA sequences. The ARG profiles of over 40,277 phage genomes were analyzed for comparison. Gene trees of representative NCLDV-encoded ARGs were constructed to elucidate their evolutionary relationships with their counterparts in prokaryotes, eukaryotes, and phages. The functionality of two selected NCLDV-encoded ARGs was verified in E. coli. Additionally, we annotated mobile genetic elements (MGEs) of NCLDVs to explore their possible correlation with ARG carriage. Finally, virulence factors (VFs) of NCLDVs were annotated and their co-occurring patterns with ARGs were examined.
Results
Overview of viral genomes analyzed in this study
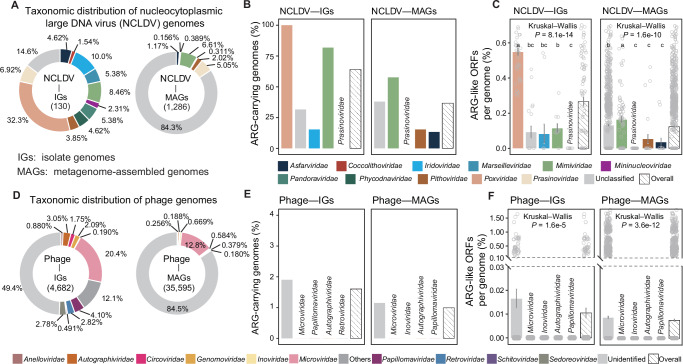
The NCLDV genomes analyzed in this study comprised 130 isolate genomes and 1,286 MAGs (Fig. 1A and Supplementary Data 1). These viral genomes could be classified into at least 11 known NCLDV families. As to the isolate genomes, the top-3 dominant families included Poxviridae (accounting for 32.3% of all isolate genomes), Iridoviridae (10.0%), and Mimiviridae (8.46%). As to the MAGs, Mimiviridae (6.61%), Prasinoviridae (5.05%), and Pithoviridae (2.02%) were the top-3 dominant families. Seven families (i.e., Asfarviridae, Coccolithoviridae, Marseilleviridae, Mimiviridae, Phycodnaviridae, Pithoviridae, and Prasinoviridae; Supplementary Data 1) were represented commonly by both the isolate genomes and MAGs. When habitats of the investigated NCLDV genomes were taken into account, 55.4% of the isolate genomes were host-associated and 90.9% of the MAGs were from either freshwater or marine environments (Supplementary Fig. 1A and Supplementary Data 1).
Fig. 1. Taxonomic overview of viral genomes and their antibiotic resistance gene (ARG) carriage.
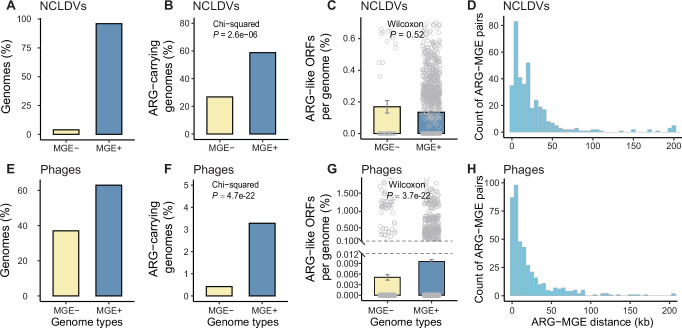
Taxonomic distribution of the genomes of (A) nucleocytoplasmic large DNA viruses (NCLDVs) and (D) phages analyzed in this study. For NCLDVs, the 11 currently known families are shown. For phages, the 10 most abundant families (according to the number of genomes in individual families) are displayed, and the rest families are referred to as “Others”. Possibility of ARG-carriage in (B) NCLDVs and (E) phages. Genomic potential of ARG-carriage of (C) NCLDVs and (F) phages. For clarity, only the most abundant four families, the unclassified genomes (as a whole), and the overall patterns are displayed in (B–F), with data on all viral families being presented in Supplementary Data 7 and 8. Viral family names are displayed above the bars where colors cannot be clearly recognized. Lower-case letters above the bars in (C) and (F) represent significantly different groups assessed with two-sided Wilcoxon rank-sum test, and P-values indicate the overall difference among all families assessed with Kruskal–Wallis test. Data presented in (C) and (F) were mean values ± standard error of the mean (SEM). Each data point represents an individual genome in the corresponding group. The number (n) of genomes in each group can be found in Supplementary Data 7 for (C) and Supplementary Data 8 for (F). The y-axis was truncated to zoom in on values below 0.03% in (F). ORF is the abbreviation for open reading frame, IG is for isolate genome, and MAG is for metagenome-assembled genome. Source data are provided as a Source Data file on Github107.
The phage genomes analyzed in this study consisted of 4682 isolate genomes and 35,595 MAGs (Fig. 1D and Supplementary Data 2). These phage genomes could be assorted to at least 78 known phage families. As to the isolate genomes, the three most dominant families were Microviridae (20.4%), Papillomaviridae (4.10%), and Autographiviridae (3.05%). Regarding the MAGs, the top-3 dominant families were Microviridae (12.8%), Inoviridae (0.669%), and Autographiviridae (0.584%). Twenty-five families (including Microviridae, Papillomaviridae, Autographiviridae, Retroviridae, and Genomoviridae, etc.; Supplementary Data 2) were common to both the isolate genomes and MAGs. As to habitats, 58.4% of the isolate genomes and 53.9% of the MAGs originated from host-associated environments, while 28.2% of the MAGs were found in aquatic environments, including both freshwater and marine settings (Supplementary Fig. 1D and Supplementary Data 2).
Taxonomic and habitat distribution of ARG-carrying viral genomes
A total of 749 ARG-like open reading frames (ORFs) were found in NCLDVs (Supplementary Data 3) and 453 in phages (Supplementary Data 4). Additionally, 181 and 67 potential efflux pump genes were found in NCLDVs (Supplementary Data 5) and phages (Supplementary Data 6), respectively. Given that efflux pumps often serve multiple purposes, with antibiotic elimination typically being one collateral activity, we did not include them in the subsequent analyses as ARGs.
A considerable proportion (39.5%, Supplementary Data 7) of the 1416 examined NCLDV genomes harbored ARG-like ORFs (referred to as ARGs hereafter). Among the investigated isolate genomes, 63.1% carried ARGs (referred to as possibility of ARG carriage, see Methods for more details), being 1.7 times as high as that of the MAGs (36.5%, Fig. 1B). Although nine out of the 11 known NCLDV families were ARG carriers, their possibility of ARG carriage varied widely (Supplementary Data 7). The top-3 dominant families represented by the isolates had varying possibility of ARG carriage (Fig. 1B): Poxviridae (100%), Mimiviridae (81.8%), and Iridoviridae (15.4%). A similar trend was seen for the MAGs, with Mimiviridae having the highest (57.6%) and Prasinoviridae the lowest (0.00%) possibility of ARG carriage (Fig. 1B). As to habitats, freshwater isolates had the highest possibility of ARG carriage (80.0%, Supplementary Fig. 1B), followed by host-associated isolates (63.9%), soil MAGs (50.0%), marine MAGs (41.3%), and tailings MAGs (35.3%).
In contrast, phages exhibited a much lower possibility (1.06%) of ARG carriage. Among the 78 known phage families, only four carried ARGs, and none of the top-4 dominant families, represented by either the isolates or MAGs, harbored any ARG (Fig. 1E). Regarding habitats, built environment MAGs had a notably higher possibility (9.29%, Supplementary Fig. 1E) of ARG carriage compared to those of other habitats (<4.40%).
On average, NCLDV genomes had 0.136% of their total ORFs annotated as ARGs (referred to as genomic potential of ARG carriage, see Methods for more details; Supplementary Data 7). Variations existed among NCLDV families in genomic potential of ARG carriage (Supplementary Data 7). For the top-3 dominant families represented by the isolates, Poxviridae had a significantly higher genomic potential of ARG carriage (0.547%) compared to Mimiviridae (0.113%) and Iridoviridae (0.081%) (Kruskal–Wallis test: n = 94, P = 8.1e-14; Fig. 1C). Among the MAGs, Mimiviridae had the highest genomic potential of ARG carriage (0.162%), followed by Pithoviridae (0.053%) and Asfarviridae (0.034%) (Kruskal–Wallis test: n = 1,275, P = 1.6e-10; Fig. 1C). Regarding habitats, host-associated (0.342%) and freshwater isolates (0.278%) exhibited the highest genomic potential of ARG carriage (Supplementary Fig. 1C).
In contrast, phages showed a much lower genomic potential of ARG carriage. Overall, phage genomes had only 0.008% of their total ORFs annotated as ARGs (Supplementary Data 8). None of the top-4 dominant phage families carried any ARG (Fig. 1F). Built environments and tailings, where only MAGs were available, stood out as the habitats whose phages had the highest genomic potential of ARG carriage (0.071% and 0.030%, respectively; with the others below 0.020%; Supplementary Fig. 1F).
Diversity and composition of ARGs in viral genomes
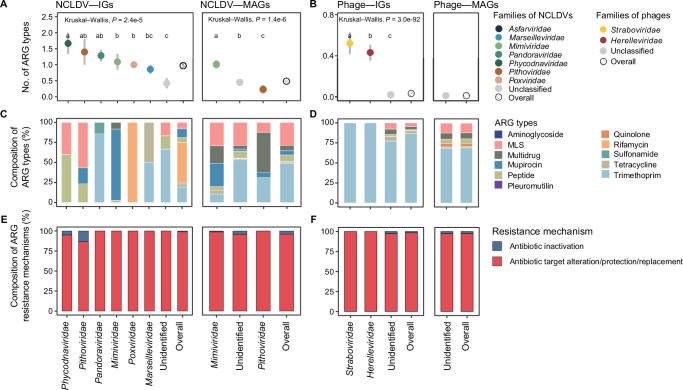
A total of 12 ARG types were identified in NCLDVs, consisting of 19 antimicrobial resistance gene families (as defined in the CARD database20; Supplementary Data 3). On average, NCLDVs harbored 0.48 ARG types per genome. There were significant differences between NCLDV families in the number of ARG types encoded by them, with Phycodnaviridae (average = 1.67 in isolates), Pithoviridae (1.40 in isolates), Pandoraviridae (1.29 in isolates), and Mimiviridae (1.09 in isolates and 1.01 in MAGs) encoding the highest number of ARG types among all known NCLDV families (Kruskal–Wallis test: n = 97, P = 2.4e-5 for isolates; n = 1195, P = 1.4e-6 for MAGs; Fig. 2A).
Fig. 2. Diversity and composition of ARGs in viral genomes.
Number of ARG types detected in the genomes of different taxonomic groups in (A) NCLDVs and (B) phages. Data are presented as mean values ± SEM. The unit of study is one genome. The number (n) of genomes in each group can be found in Supplementary Data 7 for (A) and Supplementary Data 8 for (B). Lower-case letters above the bars represent significantly different groups assessed with two-sided Wilcoxon rank-sum test, and P-values indicate the overall difference among all families assessed with Kruskal–Wallis test. Composition of ARG types in different families in (C) NCLDVs and (D) phages. Composition of ARG resistance mechanisms in different families in (E) NCLDVs and (F) phages. For clarity, only viral families with more than five ARGs, the unclassified genomes (as a whole), and the overall patterns of NCLDVs and phages are displayed. MLS is the abbreviation of macrolides, lincosamides, and streptogramines. Source data are provided as a Source Data file on Github107.
Phages encoded a total of nine ARG types, encompassing 18 antimicrobial resistance gene families (Supplementary Data 4). On average, phages carried mere 0.011 ARG types per genome (Fig. 2B), which was much lower than that of NCLDVs. As to isolates, the average ARG types encoded by Straboviridae (0.52) and Herelleviridae (0.43) were significantly higher than those of other families (Kruskal–Wallis test: n = 2378, P = 3.0e-92; Fig. 2B). In contrast, no known families of the MAGs carried enough ARG counts (>5 ARGs) for analysis of the possible differences among families. Similar patterns were found when the numbers of ARGs carried by individual viral families were compared (Supplementary Fig. 2).
As to the composition of ARG types, NCLDV isolates predominantly carried rifampin (51.2%) and trimethoprim (19.5%) resistance genes (Fig. 2C). Specifically, rifampin resistance was exclusively carried by Poxviridae. Trimethoprim was a major ARG type for Pandoraviridae (85.7%) and Marseilleviridae (50.0%). Additionally, Mimiviridae exhibited an apparent tendency to carry mupirocin resistance genes (88.9%). Other families, including Pithoviridae and Phycodnaviridae, mainly harbored macrolide-lincosamide-streptogramin and peptide resistance genes. The identifiable ARG-carrying families represented by NCLDV MAGs included Mimiviridae and Pithoviridae, and they both were characterized by a high dominance of multidrug (21.7% and 50.0%, respectively), macrolide-lincosamide-streptogramin (29.2% and 12.5%, respectively), and trimethoprim (10.5% and 31.3%, respectively) resistance genes (Fig. 2C).
Both isolates and MAGs of phages were found to have trimethoprim resistance genes as the most dominant ARG type (86.7% in isolates and 68.6% in MAGs), followed in decreasing order by macrolide-lincosamide-streptogramin (4.56% in isolates and 12.0% in MAGs) and multidrug (4.00% in isolates and 7.46% in MAGs) resistance genes (Fig. 2D). Among the families represented by isolates, Straboviridae and Herelleviridae carried exclusively trimethoprim resistance genes (Fig. 2D).
Regarding resistance mechanisms, antibiotic target alteration/protection/replacement were the most predominant mechanism for both NCLDVs (overall accounting for 96.5% of the ARGs, Fig. 2E) and phages (97.5%, Fig. 2F). The only exception was the NCLDV family Pithoviridae represented by isolates, which harbored a relatively high proportion (16.7%) of antibiotic inactivation genes (Fig. 2E).
The most frequently detected antimicrobial resistance gene family in viral genomes was the dfr genes, with 303 occurrences in NCLDVs and 325 occurrences in phages respectively (Table 1). The dfr genes encode dihydrofolate reductases that can be targeted by the antibiotic trimethoprim. Bacterial trimethoprim resistance mediated by the dfr genes can arise through mutations of the native dfr genes21, or through acquiring a resistant homolog, or simply an additional dfr gene to increase the production of dihydrofolate reductase22. Other major gene families included the F-subtype ATP-binding cassette genes, ileS genes, and glycopeptide resistance cluster-associated genes, etc. (Table 1). F-subtype ATP-binding cassette proteins belong to the ATP-binding cassette superfamily but lack transmembrane domains. They are associated with ribosomes, and some can confer resistance by binding to ribosomes and inducing conformational changes, thereby leading to drug release in bacteria23. The ileS genes encode isoleucyl-tRNA synthetases, a target of the antibiotic mupirocin. Bacterial mupirocin resistance can arise from mutation of the native ileS genes or by acquiring an alternate ileS homolog that is inherently resistant24. The detected glocopeptides resistance associated genes in this study, such as vanT and vanH, are general units found on multiple vancomycin resistance operons, which function by allowing the restructuring of peptidoglycan precursors to end in D-Ala-D-Lac, resulting in decreased vancomycin binding affinity25. In addition, the antibiotic inactivation mechanisms are mainly employed by streptogramin vat acetyltransferase genes, which catalyze the transfer of an acetyl group from acetyl-CoA to the secondary alcohol of streptogramin A compounds, thus inactivating virginiamycin-like antibiotics26.
Table 1.
Most prevalently detected antimicrobial resistance gene families in nucleocytoplasmic large DNA virus (NCLDV) and phage genomes
| AMR gene familya | ARG type | Mechanism | No. detected in NCLDVs | No. detected in phages |
|---|---|---|---|---|
| trimethoprim resistant dihydrofolate reductase dfr | trimethoprim | antibiotic target replacement | 303 | 325 |
| F-subtype ATP-binding cassette protein | multiple antibiotics | antibiotic target protection | 219 | 46 |
| antibiotic-resistant isoleucyl-tRNA synthetase (ileS) | mupirocin | antibiotic target alteration | 52 | 0 |
| streptogramin vat acetyltransferase | MLS | antibiotic inactivation | 27 | 5 |
| glycopeptide resistance gene cluster;vanH | glycopeptide | antibiotic target alteration | 24 | 6 |
| glycopeptide resistance gene cluster;vanT | glycopeptide | antibiotic target alteration | 20 | 0 |
| tetracycline-resistant ribosomal protection protein | tetracycline | antibiotic target protection | 16 | 5 |
| pmr phosphoethanolamine transferase | polymyxin | antibiotic target alteration | 12 | 9 |
| glycopeptide resistance gene cluster;vanS | glycopeptide | antibiotic target alteration | 11 | 2 |
aAMR gene families were adopted from CARD database. AMR gene families with more than 10 ORFs detected in NCLDVs were shown.
There existed apparent differences among viral families in the composition of antimicrobial resistance gene families they harbored (Supplementary Fig. 3). For instance, Mimiviridae contained the most diverse array of gene families. Phycodnaviridae and Pithoviridae both encoded multiple types of F-subtype ATP-binding cassette protein genes, glycopeptide resistance genes, and streptogramin vat acetyltransferase genes. In contrast, only two antimicrobial resistance gene families were carried by Pandoraviridae (Supplementary Fig. 3).
Evolutionary characteristics of selected NCLDV ARGs
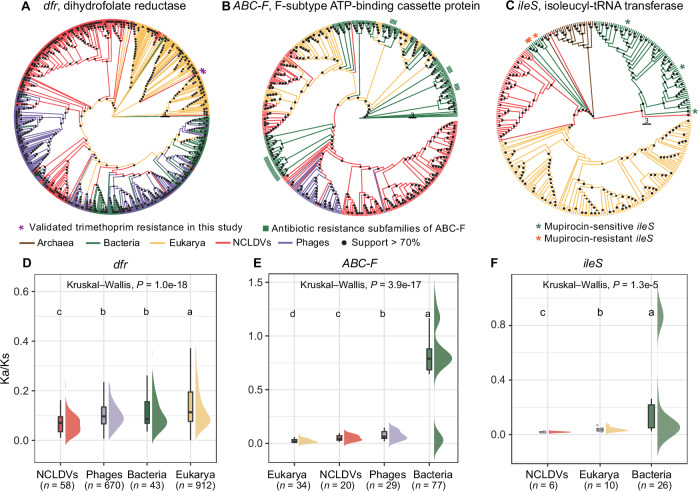
The evolutionary relationships between the top-3 dominant antimicrobial resistance gene families (Table 1) of NCLDVs and their homologs of eukaryotes, prokaryotes, and phages were analyzed. As to the most dominant gene family (i.e., the dfr genes encoding dihydrofolate reductases), NCLDV sequences did not form a monophyletic clade. Instead, they exhibited diverse potential origins (Fig. 3A). Firstly, it was frequently observed that certain sequences of NCLDVs showed closer phylogenetic relationships with phage sequences, forming distinct clades at multiple instances. Secondly, some NCLDV sequences were inserted within the eukaryotic clade. Lastly, complex evolutionary relationships existed between phages and prokaryotes in dihydrofolate reductase, and some NCLDV sequences were inserted within the phage/prokaryotic clades. The multiple alignments comparing dihydrofolate reductase sequences of NCLDVs with those of bacteria showed conservatism at the trimethoprim binding sites (Supplementary Fig. 4).
Fig. 3. Evolutionary characteristics of representative NCLDV-encoded ARGs.
Phylogenetic trees of (A) dihydrofolate reductase, (B) F-subtype ABC protein, and (C) isoleucyl-tRNA transferase from NCLDVs, eukaryotes, prokaryotes, and phages. Dfr, ABC-F ATP-binding cassette genes, and ileS are the top three most prevalent antimicrobial resistance gene families detected in NCLDVs. Nodes with support values > 70% are labeled with black dots. Information on antibiotic resistance phenotypes are adopted from ref. 96 for ABC-F and from ref. 52 for ileS genes. Roots of the trees are determined using the midpoint rooting method. Ka/Ks ratio of (D) dfr, (E) ABC-F, and (F) ileS sequences in NCLDVs, phages, eukaryotes, and prokaryotes, respectively. Lower-case letters above the bars in (D) to (F) represent significantly different groups assessed with two-sided Wilcoxon rank-sum test, and P-values indicate the overall difference among all families assessed with Kruskal–Wallis test. The unit of study is one sequence pair. The boxes represent 25th percentile, median and 75th percentile of the data, and the whiskers show the minimum or maximum value of the data. Data points are shown when n ≤ 10. Source data are provided as a Source Data file on Github107.
Within NCLDVs, certain F-subtype ATP-binding cassette (ABC-F) sequences have been identified as vga-type ABC-F, msr-type ABC-F, or sal-type ABC-F, while others annotated by profile HMMs could not be pinpointed to a more precise subfamily (Supplementary Data 3). Therefore, we constructed a gene tree encompassing the broader ABC-F family of proteins and marked antibiotic resistance-associated subfamilies on the tree (Fig. 3B). The analysis revealed that ABC-F proteins of NCLDVs primarily clustered into two distinct groups on the gene tree. Specifically, the majority of NCLDV sequences formed a monoclade along with phage sequences, sharing a close evolutionary relationship with a subset of the antibiotic resistance subfamilies of ABC-F proteins of bacteria. Additionally, a smaller subset of ABC-F proteins of NCLDVs were dispersed within the clade of eukaryotic sequences.
The third most frequently detected antimicrobial resistance gene family (i.e., the ileS genes) in NCLDVs were absent in phage genomes (Table 1). Mupirocin was reported to selectively inhibit certain bacterial and archaeal ileS-encoding enzymes while sparing eukaryotic counterparts, rendering them inherently mupirocin-resistant27. Moreover, prior studies showed that certain bacterial mupirocin-resistant ileS genes likely originated from eukaryotes27,28. We found that mupirocin-sensitive bacterial ileS genes primarily clustered within the clade dominated by bacterial sequences (Fig. 3C). In contrast, mupirocin-resistance bacterial ileS genes exhibited closer evolutionary relationships with eukaryotic ileS genes, with giant virus ileS genes being positioning intermediately to resistant bacterial ileS and eukaryotic ileS genes. This positioning indicated a potential bridging role of NCLDVs in the possible transmission between eukaryotic and resistant bacterial ileS genes.
We further inferred selection pressures on the three abovementioned ARG families of NCLDVs, phages, eukaryotes, bacteria, and archaea by calculating the ratios of non-synonymous (Ka) to synonymous (Ks) substitution (referred to as Ka/Ks ratios hereafter) of the relevant genes respectively. All analyzed genes of different taxonomic groups were found to undergo negative selection pressures, as evidenced by their Ka/Ks ratios being markedly lower than one (Fig. 3D–F). Despite this, the strength of selection pressure on a given ARG could vary considerably among different taxonomic groups. The average Ka/Ks ratio (0.075) for the dfr genes of NCLDVs was significantly lower than those of other three groups (0.104 in phages, 0.120 in bacteria, and 0.146 in eukaryotes; Kruskal–Wallis test: n = 1683, P = 1.0e-18; Fig. 3D). The results indicated a tendency toward functional conservatism and stability in the evolution of the dfr genes of NCLDVs, providing a possible explanation for its high incidence in NCLDVs. The Ka/Ks ratios of two other genes of NCLDVs were also notably low (0.050 for the ABC-F genes and 0.018 for the ileS genes; Fig. 3E–F), indicating their biological importance for the NCLDVs. However, due to the limited number of sequence pairs (only 20 pairs for the ABC-F genes and six pairs for the ileS genes, respectively) meeting the criteria (70% identity over 95% sequence length as defined in ref. 29) for Ka/Ks calculation, it would be inconclusive to compare these values with those of other taxonomic groups and assess differences.
Functional validation of selected NCLDV-encoded ARGs
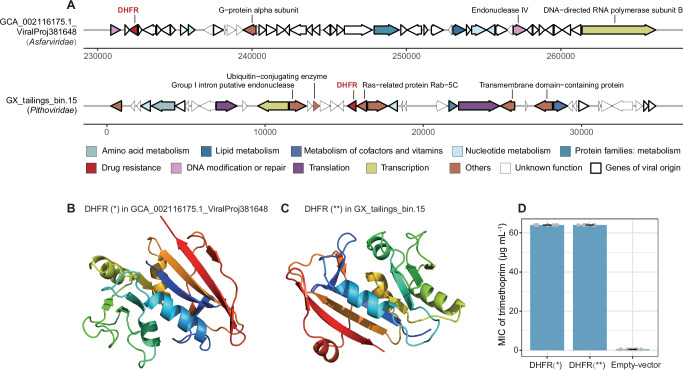
Due to the widespread presence of dfr genes in NCLDV genomes, we randomly selected two NCLDV dfr sequences (Supplementary Data 9) and validated their functionality by introducing them into a trimethoprim-sensitive E. coli strain. Among them, one is from an isolate of the family Asfarviridae, and the other from a MAG of the family Pithoviridae (Fig. 4A). The Asfarviridae isolate genome had limited annotated information in the upstream and downstream of its dfr gene, primarily featuring genes associated with DNA modification, repair, metabolism, and transcription. In contrast, the Pithoviridae MAG, assembled from mine tailings, had more annotations (15 genes) in the surrounding coding regions of its dfr gene. Notably, a few genes potentially linked to infectious diseases (including Rab-5C30 and ubiquitin-conjugating enzyme31,32), a group I intron putative endonuclease gene (potentially associated with HGT33), and two genes encoding membrane domain-containing proteins involved in membrane trafficking34 were found within 10 kb region around the dfr gene.
Fig. 4. Functional validation of two dfr sequences from NCLDVs.
A Genetic structures of the upstream and downstream coding regions of the two randomly selected NCLDV dfr genes. DHFR, dihydrofolate reductase, represents the protein product encoded by dfr gene. Function annotations follow the KEGG classification. Genes of viral origin are determined using VOG annotation. B, C Protein structures of the selected DHFRs generated by Phyre2 web platform. D Minimum inhibitory concentration (MIC) values to the antibiotic trimethoprim. DHFR (*) and DHFR (**) represent the experimental groups of Escherichia coli DH5α strains carrying recombinant plasmids with two NCLDV dfr genes, respectively (gene sequences shown in Supplementary Data 9 and protein structures shown in B and C, respectively). Control refers to E. coli DH5α strains harboring the pUC19 plasmid without any gene insert. Data are presented as mean values ± standard deviations from eight biological replicates. Source data are provided as a Source Data file on Github107.
The two NCLDV-encoded dihydrofolate reductases were predicted to show similar tertiary structures (Fig. 4B, C), along with conserved trimethoprim binding sites shared with bacterial homologs (Supplementary Fig. 4). Despite their relatively low amino acid sequence identity (33.5% and 36.3%, Supplementary Data 9) with the bacterial homologs, we speculated that these genes could function in bacteria. Because another NCLDV-encoded dihydrofolate reductase that exhibited only 22.2% amino acid sequence identity with its homolog of S. cerevisiae was reported previously to confer trimethoprim resistance when expressed in S. cerevisiae17. We introduced the two dfr genes into the E. coli 5α strain separately and tested their resistance to trimethoprim. Both dfr genes were found to be able to elevate the minimum inhibitory concentration value of the trimethoprim-sensitive E. coli strain from 0.5 to 64 µg mL−1 (Fig. 4D).
Interdependence between ARGs and MGEs in viral genomes
Besides endonucleases and insertion sequences that were previously observed in several giant virus families35, we also investigated the incidence of other various mobility-involved genes (including those encoding transposases, integrases, recombinases, resolvases, and relaxases) across the phylum Nucleocytoviricota. Insertion sequences and transposases were further integrated due to their frequent co-occurrence in annotations. For example, insertion sequences often contain one to two transposases within their structure, and both insertion sequences and transposases are typically part of transposon structures36.
Overall, 96.0% of the investigated NCLDV genomes were found to harbor MGEs (Fig. 5A). Few variations existed among different NCLDV families or among NCLDVs from different habitats in the possibility of MGE carriage (Supplementary Data 7 and Supplementary Fig. 5A). In contrast, the genomic potential of MGE carriage varied greatly among different NCLDV families or among NCLDVs from different habitats (Supplementary Data 7 and Supplementary Fig. 5B). Upon closer examination of various MGE types (Supplementary Fig. 6A), endonucleases were carried by of 87.7% of the total NCLDV genomes and ranked as the most commonly identified MGE type, followed by recombinases (69.1%), resolvases (45.8%), and insertion sequences/transposases (24.6%).
Fig. 5. Interdependency between ARGs and mobile genetic elements (MGEs) in viral genomes.
Overall incidence of MGEs in genomes of (A) NCLDVs and (E) phages. Possibility of ARG-carriage in the MGE- and MGE+ genomes in (B) NCLDVs and (F) phages. Two-sided Chi-squared test is performed to test dependency between MGE-carriage and ARG-carriage in the viral genomes. Genomic potential of ARG-carriage in MGE- and MGE+ genomes in (C) NCLDVs and (G) phages. Two-sided Wilcoxon rank-sum test is performed to evaluate the statistical significance of the differences in genomic potential of ARG carriage between MGE+ and MGE- genomes. MGE + : genomes with at least one MGE. MGE-: genomes without any MGE. Data are presented as mean values ± SEM. Each data point is one genome. n = 56 (MGE-) and 1,360 (MGE + ) for NCLDVs. n = 14,910 (MGE-) and 25,367 (MGE + ) for phages. The y-axis was truncated to zoom in on values below 0.012% in (G). Histogram of ARG-MGE distance (kb) in (D) NCLDVs and (H) phages. Source data are provided as a Source Data file on Github107.
A significant interdependence was observed between the presence of ARGs and MGEs in NCLDV genomes (Two-sided Chi-squared test: n = 1416, P = 2.6e − 6; Fig. 5B). Fifty-eight percent of MGE-positive NCLDV genomes harbored ARGs, which was higher than that (26.8%) of MGE-negative genomes. Similar patterns were observed, when different MGE types were taken into account individually (Supplementary Fig. 6B). The genomic potential of ARG carriage of NCLDVs was not significantly affected by their MGE carriage status (Two-sided Wilcoxon rank-sum test: n = 56 for MGE- and n = 1360 for MGE+ genomes respectively, P = 0.52; Fig. 5C). However, upon closer examination of different MGE types, the carriage status of endonucleases was found to significantly influence the genomic potential of ARG carriage of NCLDVs (Two-sided Wilcoxon: n = 174 for MGE- and n = 1242 for MGE+ genomes respectively, P = 1.5e-3; Supplementary Fig. 6C). The co-localization analysis showed that 37.1% of the MGEs co-occurring with ARGs located within 10 kb of their corresponding ARGs (with 72.9% within 30 kb, Fig. 5D). This kind of close association between MGEs and ARGs was relatively consistent across different MGE types (Supplementary Fig. 6D).
Compared to NCLDVs, phages had a lower possibility of MGE carriage (63.0%, Fig. 5E), with substantial variations detected among phages from different families or from different habitats (Supplementary Data 8 and Supplementary Fig. 7C). A more pronounced impact of the MGE presence on ARG carriage was observed in phages than in NCLDVs. On the one hand, 3.28% of MGE-positive phage genomes carried ARGs, a proportion higher than that (0.423%) of MGE-negative genomes (Two-sided Chi-squared test: n = 40,277, P = 4.7e−22; Fig. 5F). On the other hand, MGE-positive phage genomes were found to have an average of 0.009% of their total ORFs as ARGs, which was significantly higher than that (0.005%) of MGE-negative phage genomes (Two-sided Wilcoxon: n = 14,910 for MGE- and n = 25,367 for MGE+ genomes respectively, P = 3.7e-22; Fig. 5G). Similar trends were observed, when different MGE types of phages were considered individually (Supplementary Fig. 7). A closer association between MGEs and ARGs was recorded in phages than in NCLDVs, as evidenced by the results that 53.3% of the MGEs co-occurring with ARGs of phages were located within 10 kb of their corresponding ARGs (with 85.9% within 30 kb, Fig. 5H).
Presence of VFs in viral genomes
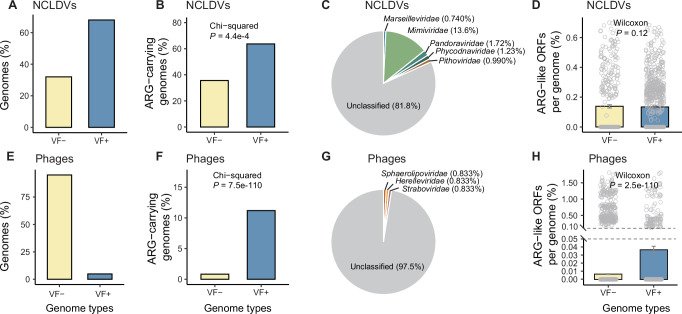
We further looked into the NCLDV genomes that carry both ARGs and VFs. A total of 2487 VF-like ORFs were annotated in the studied NCLDV genomes (Supplementary Data 10), resulting in 68.0% of the studied NCLDV genomes being identified to carry VFs (Fig. 6A). VF-positive genomes exhibited a higher possibility (63.7%) of ARG carriage compared to VF-negative genomes (35.4%; two-sided Chi-squared test: n = 1416, P = 4.4e-4; Fig. 6B). NCLDVs with both VF and ARG carriage were primarily from unknown families (81.8%, Fig. 6C) or the family Mimiviridae (13.6%). The genomic potential of ARG carriage of NCLDVs showed no correlation with their VF carriage status (Two-sided Wilcoxon rank-sum test: n = 453 and 963 for VF- and VF+ genomes respectively, P = 0.12; Fig. 6D).
Fig. 6. Carriage of virulence factors (VFs) in viral genomes and its relationship with ARG-carriage.
Overall incidence of VFs in genomes of (A) NCLDVs and (E) phages. Possibility of ARG-carriage in the VF- and VF+ genomes in (B) NCLDVs and (F) phages. Two-sided Chi-squared test is performed to test dependency between VF-carriage and ARG-carriage in the viral genomes. Family composition of the genomes carrying both ARGs and VFs in (C) NCLDVs and (G) phages. Genomic potential of ARG-carriage in VF- and VF+ genomes in (D) NCLDVs and (H) phages. Two-sided Wilcoxon rank-sum test is performed to evaluate the statistical significance of the differences in genomic potential of ARG carriage between VF+ and VF- genomes. Data are presented as mean values ± SEM. Each data point is one genome. n = 453 (VF-) and 963 (VF + ) for NCLDVs. n = 38,310 (VF-) and 1967 (VF + ) for phages. The y-axis was truncated to zoom in on values below 0.05% in (H). VF + : genomes with at least one VF. VF-: genomes without any VF. Source data are provided as a Source Data file on Github107.
Compared to NCLDVs, phages exhibited a much lower possibility of VF carriage (4.88%, Fig. 6E and Supplementary Data 11). However, the interdependence between the presence of ARGs and VFs was more pronounced in phages, as evidenced by the observation that 11.2% of VF-positive phage genomes carried ARGs, a proportion 13.5 times higher than that of VF-negative phage genomes (Two-sided Chi-squared test: n = 40,277, P = 7.5e-110; Fig. 6F). Furthermore, the genomic potential of ARG carriage was also significantly higher in VF-positive phage genomes than in VF-negative phage genomes (Two-sided Wilcoxon rank-sum test: n = 38,310 and 1967 for VF- and VF+ genomes respectively, P = 2.5e-110; Fig. 6H). Phages with both VF and ARG carriage were predominantly from unknown families (97.5%, Fig. 6G).
Discussion
As one of the most significant advances in biology over the past decades, the discovery of giant viruses whose particle size can reach up to 2.3 µm in length and genome size can be as large as 2.5 Mb37–39, has challenged the classical concept of virus40,41. Furthermore, the increasing availability of giant virus MAGs recovered from various samples has heightened our understanding of the genetic make-up, diversity, and potential ecological roles of giant viruses40,41. Despite these advances, the recovery of giant virus MAGs without contaminating DNA sequences (including those of prokaryotes rich in ARGs) still remains a challenging task40,41. Therefore, one would expect that the detection of ARGs from giant virus MAGs likely results in overestimation of the possibility and genomic potential of ARG carriage of giant viruses. Such an expectation, however, seems not to be supported by our results. The overall possibility (Fig. 1B) and genomic potential (Fig. 1C) of ARG carriage of giant virus MAGs were lower than those of giant virus isolates. Comparing isolates and MAGs from the same giant virus family (e.g., Mimiviridae or Prasinoviridae) enables a more confirmative demonstration that the ARGs carriage of MAGs had not been overestimated (Fig. 1B, C). Likewise, our results suggested that, phages showed no bias in MAGs towards overestimating ARG carriage compared to isolates (Fig. 1E, F).
A total of 35 giant virus genomes were reported previously to encode ARGs17–19. In this context, one of the most striking findings of our study was that 39.5% of the investigated giant virus genomes (Fig. 1B; i.e., 560 out of the 1416 genomes) were shown to carry ARGs. Such a proportion was approaching that (47%) for bacteria42. The detection of ARGs from viral sequences is always sensitive to the thresholds used for sequence similarity analysis43. Given that some viral ARGs were shown to exhibit low sequence identities with cellular ARGs (as low as 20.4%)7,17,18, an exploratory threshold of sequence identity (i.e., 25%) was employed in our study. Despite this, the other cutoff values used by us to annotate ARGs were conformable to or more stringent than those criteria widely used in the literature (see Methods for more details). Nonetheless, there were two lines of evidence that our selection of the exploratory threshold made a negligible contribution to the observed unexpectedly large possibility of ARG carriage of giant viruses. First, at least one previously reported NCDLV-encoded beta-lactamase gene (GenBank: AUL78925.1; whose product expressed by E. coli was able to hydrolyse a beta‑lactam and penicillin G)18 was not identified as an ARG by any of the ARG annotation methods used in this study, probably due to the higher alignment cutoffs set (e.g., the 80% target and query sequence coverage; see Methods for more details). Second, the ARG-like ORFs in the studied phage genomes, on average, accounted for 0.008% of the total predicted genes (Fig. 1F), which was lower than that (0.02%) reported by Debroas & Siguret who employed a conservative threshold to detect ARGs in virome data from public databases6.
Given the large genome size of NCLDVs41, one would expect that the more widespread presence of ARGs in NCLDVs than in phages (Fig. 1) could be attributed to a simple scenario that the genomic potential of ARG carriage of viruses increased with their genome size. To address this point, we examined the correlations between the genomic potential of ARG carriage of isolated viruses and their genome size. While a weak positive correlation between the genomic potential of ARG carriage and genome size was observed for overall isolated phages (Supplementary Fig. 8B), no significant relationship was recorded for overall isolated NCLDVs (Supplementary Fig. 8A). More surprisingly, a significant negative correlation between the genomic potential of ARG carriage and genome size was observed for several NCLDV families (Supplementary Fig. 8A). These results indicated that the mechanisms by which NCLDVs acquired ARGs were likely not the same as those of phages.
Among all currently known families of NCDLVs, Poxviridae exhibited not only the highest possibility but also the highest genomic potential of ARG carriage (Fig. 1B, C). A subset of poxviruses comprises the causative agents of human smallpox and cowpox44, with rifampin being utilized for treatment45. Uncoincidentally, it was observed that all the ARGs carried by Poxiviridae were rifampin resistance (rif) genes (Fig. 2C and Supplementary Data 3). Since the Poxviridae genomes analyzed in this study were all obtained from host-associated environments (Supplementary Data 1), we proposed that the presence of the rif genes within Poxiviridae likely stemmed from a direct selection under the pressure of the antiviral agent, rifampin.
There were at least two other possible reasons why giant viruses carry ARGs. First, certain ARG-encoded proteins could exert crucial functions in the reproduction of giant viruses while they were also antibiotic targets. Such proteins likely included dihydrofolate reductase encoded by dfr gene and isoleucyl-tRNA synthetase encoded by ileS gene (Table 1). Dihydrofolate reductase is involved in the production of tetrahydrofolic acid, the active form of folate that is essential for all living organisms in various biosynthetic pathways such as amino acid and nucleic acid metabolism46. Given its significance in fundamental life processes, dihydrofolate reductase has served as a drug target in both prokaryotic and eukaryotic microbial pathogens46,47. Specifically, common antibiotic combinations like sulfamethoxazole–trimethoprim, inhibiting enzymes in the folate pathway, have been applied in treating infections caused by prokaryotic pathogens such as Nocardia asteroids and an eukaryotic microbial pathogens Pneumocystis carinii48, although eukaryotic dihydrofolate reductase exhibits certain degree of inherent resistance to trimethoprim compared to its prokaryotic counterpart49. A prior study has shown that one dihydrofolate reductase gene from the giant virus family Marseilleviridae, when expressed in S. cerevisiae, conferred resistance to trimethoprim in the fungus17. In this study, we extended the previous finding by demonstrating that the two dfr genes from Asfarviridae and Pithoviridae respectively, when transferred to E. coli, were able to confer resistance to trimethoprim in the bacterium (Fig. 4). A closer look at the two functionally validated dfr genes of this study showed that they both placed within the eukaryotic clade on the gene tree (Supplementary Fig. 9). Within this context, the ability of those dfr genes falling within other clades on the gene tree (Fig. 3A) to confer trimethoprim resistance phenotypes in fungi and/or bacteria deserves further research. As to ileS gene, its product isoleucyl-tRNA synthetase is vital for protein translation across multiple kingdoms of life50. The antibiotic mupirocin effectively inhibits bacterial isoleucyl-tRNA synthetase but not its eukaryotic counterpart27. Several previous studies have revealed that the mupirocin-resistant types of isoleucyl-tRNA synthetase in bacteria exhibited a greater sequence similarity to eukaryotic sequences than to those of bacterial mupirocin-sensitive types, suggesting their potential origin from inherently resistant sequences in eukaryotes, possibly via HGT28,51,52. Similarly, we found that bacterial ileS genes conferring mupirocin-resistant phenotypes clustered separately from those with sensitive phenotypes, displaying a closer resemblance to eukaryotic ileS than to the mupirocin-sensitive bacterial clade (Fig. 3C). Moreover, the ileS genes of giant viruses were shown to occupy an intermediate position between eukaryotic ileS and bacterial resistant ileS in the gene tree (Fig. 3C), implying that the ileS genes of giant viruses likely exhibit similar inherent resistance traits as those found in eukaryotes. Note also that both dfr and ileS were revealed to evolve towards functional conservatism (especially in giant viruses), as illustrated by their Ka/Ks values < 1 (Fig. 3D, F)29.
Second, some ARG-encoded proteins could have evolved to be pleiotropic rather than mere as an agent to resist antibiotics. For example, after expression in E. coli, the beta-lactamase encoded by Tupanvirus deep ocean was able to not only hydrolyze beta-lactam but also degrade RNA from its amoebal host and a variety of bacteria18. Such an RNase activity could help the giant virus to take over its host and to interact with its sympatric bacteria. A recent study has revealed that T. deep ocean did interact with an intracellular bacterial symbiont of its host53. Although we didn’t identify any beta-lactamase genes carried by giant viruses, we found that some giant viruses carried streptogramin vat acetyltransferase genes whose resistance mechanism (i.e., antibiotic inactivation; Table 1) falls into the same category as that of beta-lactamase genes. We thus considered the possibility that streptogramin vat acetyltransferase encoded by giant viruses could have also become a pleiotropic protein. Nonetheless, further biochemical experiments are needed to test our hypothesis.
There are some eukaryotes whose cells are infected not only by giant viruses but also by a variety of microbes54. A typical example is amoeba, which are well-known “Trojan horses” for giant viruses and human pathogens55. Moreover, HGT between mimivirus and intra-amoebal bacteria has been reported56,57. As such, the widespread presence of ARGs in giant viruses (Fig. 1B, C) raises concerns about both their origin and their potential to be transferred to intracellular microbes (especially pathogens) of their hosts. That is, the implications of a strong interdependence between the possibility of ARG carriage of giant viruses and the presence of MGEs in their genomes (Fig. 5A, B) were twofold. On the one hand, it indicated that MGEs could have play an important role in the acquisition of ARGs by giant viruses. In agreement with this notion, transposable elements in Mimiviridae and Pandoravirus were proposed to have a crucial impact on their genome formation and evolution58,59. On the other hand, it hinted that a considerable proportion of ARGs of giant viruses had substantial dissemination potential. Taking endonucleases (i.e., the most dominant MGE type that co-occurred with NCLDV-encoded ARGs; Supplementary Fig. 6A) as an example, they are characterized by their ability to cleave nucleotide chains into smaller fragments60 and have been recently proposed to be able to confer mobility to their associated genes located within nearby regions of up to 10 kb in viruses61. Note that 14.2% of the total NCLDV-encoded ARGs fell within such an active range of endonucleases (Supplementary Fig. 6D), including the functional validated dfr gene from the family Pithoviridae (Fig. 4A). In this context, one may expect that, once being transferred to intracellular microbes of their hosts, certain NCLDV-encoded ARGs can help the microbes survive better under antibiotic stress.
The simultaneous carriage of ARGs and VFs by giant viruses is of particular concern. Besides human smallpox virus, cowpox virus, and African swine fever virus that have long been known as pathogens, several members of Mimiviridae, Marseilleviridae, and Phycodnaviridae were reported to be opportunistic human pathogens in some instances62–64. Despite this, the pathogenicity of other giant viruses remains poorly understood. In this study, we showed that up to 68.0% of the investigated giant virus genomes harbored VFs (Fig. 6A). Moreover, 63.7% of the VF-positive giant virus genomes also carried ARGs, encompassing almost all ARG-carrying giant virus families (Fig. 6B, C).
In summary, we reveal that the diversity and incidence of ARGs in NCLDVs are much higher than previously recognized. We also obtain evidence that some NCLDV-encoded ARGs have the potential to confer resistance phenotypes and exhibit close associations with MGEs and VFs. Our results highlight that the functions and associated potential health risks of NCLDV-encoded ARGs deserve much more attention.
Methods
Public viral genome collection
In this study, the terms “NCLDVs” and “giant viruses” both specifically refer to members within the phylum Nucleocytoviricota. Public NCLDV genomes were obtained from a seminal paper by Aylward and colleagues in 2021 (archived in the Giant Virus Database, https://faylward.github.io/GVDB/, accessed on 2022/10/19)65. Through conducting a comprehensive screening of the NCBI RefSeq database and related references, the authors selected a set of 1383 high-quality genomes of the phylum Nucleocytoviricota to identify phylogenetic marker genes. This set encompassed the genomes of almost all currently available cultured isolates and representative MAGs from diverse habitats across the globe65. All of the 1383 genomes were used directly in this study (Supplementary Data 1).
Public phage genomes were obtained from the CheckV (v1.5) complete viral genomes database, which initially contained 62,895 phage genomes identified through a systematic search of the NCBI GenBank database, publicly available metagenomes, metatranscriptomes and metaviromes66. We excluded those phage genomes that did not have habitat information or could not be classified at any taxonomic level of phages from our further analysis, resulting in a total of 39,689 public phage genomes being used in this study (Supplementary Data 2).
Obtaining viral sequences from mine tailings metagenomes
A country-scale sampling of mine tailings (an under-sampled habitat type both in the literature and in the public datasets) from 39 mine sites across China was conducted by ourselves in July and August 2018. Three mine tailings samples at a depth of 0–20 cm were taken at each site. 10–30 g of tailings per sample were extracted for total genomic DNA (using FastDNA Spin kit, MP Biomedicals, Santa Ana, CA), which were subsequently used for library construction (with NEBNext Ultra II DNA PCR-free Library Prep Kit, New England Biolabs, Ipswich, MA, USA), and shotgun-sequencing on the MiSeq platform with PE150 mode (Illumina, San Diego, CA, USA). A total of 115 metagenomes with 69.6 ± 14.0 Gb clean reads per sample (more information can be found in a previous study67) were generated, and assembled into contigs using MEGAHIT (v1.2.9).
Binning was performed to generate NCLDV MAGs from the tailings metagenomes according to a previously published protocol68. Briefly, contigs were screened with a minimum length of 5 kb, and putative NCLDV contigs were identified by either of the following criteria: (1) classified as “NCLDV” by the random forest classifier published by Schulz et al.68; (2) contained at least two nucleo-cytoplasmic virus orthologous groups (NCVOGs) based on HMMs of 20 ancestral NCVOGs employed by Yutin et al.69; and (3) contained the NCLDV polB gene (NCVOG0038)70. Putative NCLDV contigs were first assessed for coverage information using Bowtie (v2.4.5)71, Samtools (v1.15.1)72, and the jgi_summarize_bam_contig_depths script. Subsequently, contigs were pooled and binned using MetaBAT273 with default parameters, with the contig coverage serving as input for the binning process. Bins were then de-duplicated using dRep (v3.3.0)74, de-contaminated and quality-checked following Schulz’s protocol68. Thirty-three NCLDV MAGs were generated in this study, and their taxonomy was inferred by constructing a phylogenetic tree using them and the known NCLDV genomes (Supplementary Fig. 10) published by Aylward et al.65. IQ-TREE (v.1.6.12) was utilized to construct the tree, using concatenated protein alignments of seven marker genes including SFII (DEAD/SNF2-like helicase), RNAPL (DNA-directed RNA polymerase alpha subunit), PolB (DNA polymerase family B), TFIIB (transcription initiation factor IIB), TopoII (DNA topoisomerase II), A32 (Packaging ATPase), and VLTF3 (Poxvirus late transcription factor VLTF3), employing the LG + I + F + G4 model, 1000 ultrafast bootstrap replicates. Poxviridae was used as an outgroup for the tree construction. Meta information of the NCLDV sequences used in this study were presented in Supplementary Data 1.
Phage sequences from our own mine tailings metagenomes were annotated by VirSorter2 (v2.2.3) with a minimum length of 10 kb75. CheckV software66 was further employed to remove potential host regions at the end of prophages and to evaluate genome quality. Only sequences with CheckV quality tiers of “complete” (n = 9) or “high-quality” (n = 579) were retained for further analysis, resulting in a total of 588 phage genomes from the mine tailings being used in this study. Taxonomy of phages were annotated using geNomad software with the “end-to-end” command and default settings and genomad_db_v1.776. Meta information of the phage sequences used in this study were presented in Supplementary Data 2.
Annotation of ARGs and characterization of ARG-carriage
ARGs were annotated using multiple methods as follows. First, DeepARG software (v1.0.2), which employs a deep learning algorithm specifically designed to enhance annotation accuracy particularly for novel ARG sequences, was applied on the viral protein sequences using the “LS” model77. Sequences were further filtered to retain those with alignment identity >25%, both target and query coverage >80%, and e < 1e-10 in the alignment step, and a final prediction probability over 80% in the machine learning prediction step. Second, sequence alignment was conducted using diamond (v2.1.8.162) blastp command to align viral protein sequences against the CARD (https://card.mcmaster.ca/, accessed on 2024.03.10)20, SARG (v3.2.1-S, https://smile.hku.hk/ARGs/Indexing, accessed on 2024.03.14)78, and NCBI NDARO (https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/, accessed on 2024.03.19)79 databases, respectively. For each viral sequence, top alignment with e < 1e-10, percent of identity >25%, and both target and query coverage >80% were kept for final integration. Third, viral protein sequences were also aligned against profile HMMs from the SARGfam database (https://smile.hku.hk/SARGs, accessed on 2024.03.21) and Reference HMM Catalog of the NCBI NDARO platform (https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/, accessed on 2024. 03.19) using HMMER (v3.1b2). Alignments with both domain and sequence scores > 40, domain e < 1e-15 and sequence e < 1e-10 were retained, and top hit with the lowest domain e-value was designated as the annotation for each viral protein. Among the above cutoffs, a percent identity cutoff of 25% was adopted considering that some viral ARGs were reported to exhibit considerable novelty, with sequence identities ranging from 20.4% to 67.3% compared to cellular ARGs7,17. The other cutoffs were conformable to or more stringent than those employed in recent publications7,80–85. Sequences annotated by DeepARG or those by at least two of the abovementioned four databases (i.e., CARD, SARG, SARGfam, and NCBI NDARO) were finally retained as ARGs.
Two quantitative parameters were used to describe the ARG carriage of a given viral taxonomic group. The first one was “Possibility of ARG carriage”, which refers to the proportion of genomes within a given group that carry ARGs, expressed as a percentage of the total number of genomes in that group (Eq. 1). The second one was “Genomic potential of ARG carriage”, which is defined as the percentage of ARG-like ORFs relative to the total number of ORFs in a given genome (Eq. 2)86. The genomic potential of ARG carriage of a given group can be then calculated as the average value of the genomic potential of all genomes within that group.
| 1 |
| 2 |
Annotation and characterization of MGEs and VFs
Insertion sequences were annotated with ISEScan (v1.7.2.3)87 with default settings. Other types of mobility-involved genes including integrases, transposase, resolvases, recombinases, relaxases, and endonucleases were annotated using the following methods: (1) aligning protein sequences against a self-compiled mobility gene database referenced in a previous publication67 using Diamond blastp; (2) aligning protein sequences against profile HMMs collected from CONJscan (v2.0.1)88, Phage_Finder (v2.1)89, ICEberg (v2.0)90, and Jiang et al.91, as described in our previous publication92; and (3) using DRAM (v1.3.5)93 implemented with KEGG and PFAM databases and the default settings. The cutoffs for diamond blastp and hmmsearch for MGEs were the same as those in the annotation of ARGs. Virulence factors were annotated by aligning the viral protein sequences against the VFDB (http://www.mgc.ac.cn/VFs, accessed on 2022.06.07) using diamond blastp with the same cutoffs as those used in the ARG annotation. Four parameters, including “Possibility of MGE carriage”, “Genomic potential of MGE carriage”, “Possibility of VF carriage”, and “Genomic potential of VF carriage”, were calculated in a way similar to that for ARGs.
Gene tree construction
To construct gene trees for representative ARGs from NCLDVs, phages, eukaryotes, and prokaryotes, the protein sequences of ARG-like ORFs in NCLDVs and phages annotated in this study were extracted and their orthologs in prokaryotes and eukaryotes were collected either from the literature or from the public databases. Specifically, representative protein sequences of the dfr gene encoding dihydrofolate reductase in prokaryotes were downloaded from NCBI using the accession list provided in a previous study which systematically evaluated their phylogeny94. Orthologs of dihydrofolate reductase from eukaryotes were downloaded from eggNOG (v5.0.0)95 under the ID KOG1324. All dihydrofolate reductase sequences were examined with the PF00186 HMM model downloaded from the Pfam database (https://www.ebi.ac.uk/interpro/) and only those with sequence e-value below 1e-5 were retained for gene tree construction. Sequences from prokaryotes and eukaryotes were clustered at 95% identity over 90% of the shorter ORF length to eliminate redundancy. Additionally, taxonomy-based filtering was applied by randomly selecting one sequence from each family or order depending on the available sequence count, aiming to minimize the number of sequences while preserving optimal sequence diversity.
Homologs of F-subtype ATP-binding cassette protein in prokaryotes and eukaryotes were directly extracted from a previous literature that evaluated their structural and functional diversification across the tree of life96. The original publication collected 16,848 F-subtype ATP-binding cassette protein sequences, and five were randomly selected from each subfamily for gene tree construction to simplify visualization while retaining sequence diversity.
To download sequences of isoleucyl-tRNA synthetase encoded by the ileS gene, we searched NCBI with “isoleucyl-tRNA synthetase” as the keyword (with source database set as “UniprotKB” to minimize redundancy, accessed on 2023.08.23), downloaded 112 protein sequences from the search results, and manually checked the annotation and attached references of each record for those conferring the mupirocin resistance phenotypes. Additionally, some ileS sequences with either confirmed mupirocin resistant or sensitive phenotypes were collected from a previous publication52. Eukaryotic ileS-encoded proteins were downloaded from eggNOG database under the ID KOG0434. Sequences were dereplicated at 95% identity over 90% of the shorter protein length before tree construction.
To construct the gene trees, protein sequences were aligned using MAFFT (v7.490)97 and trimmed using trimAI (v1.4, parameter -gt 0.1)98. IQ-TREE (v2.1.2)99 was used to build maximum likelihood phylogenetic trees with 1000 ultrafast bootstrap replicates (parameter --alrt 1000 --bnni). Gene trees were visualized using ‘ggtree’ package (v3.0.4) in R software (v4.1.0, R Foundation for Statistical Computing).
Ka/Ks ratio
The Ka/Ks ratios of selected ARGs were calculated for NCLDVs, phages, bacteria, archaea and eukaryotes, respectively. On the one hand, the ARG protein sequences for different taxonomic groups were clustered individually at 75% identity over at least 95% of their lengths using CD-hit29. Sequences in each cluster were separated into all possible pairs, and each pair was subjected to the following Ka/Ks analysis. On the other hand, for ARGs with only protein sequences available, the matched nucleotide sequences were linked and fetched from the NCBI Nucleotide database using the ‘rentrez’ package (v1.2.3) in R software (v4.1.0). Subsequently, the ARG protein sequences, as well as the nucleotide sequences aligned with MAFFT97 were used as input for the ParaAT (v2.0) software100 to generate alignment files in the axt format, which were then processed using the KaKs (v3.0) software101 to calculate the Ka/Ks ratios.
Functional characterization of NCLDV dfr genes
Two dfr genes from NCLDVs (Supplementary Data 3) were synthesized by General Bioscience Co., Ltd (Anhui, China) and subcloned into the expression vector pUC19 plasmid, respectively102. The two recombinant plasmids were then transformed separately into E. coli DH5α strain. Another E. coli DH5α strain harboring pUC19 plasmid without any gene insert was used as the negative control. The positive clones were screened by Mueller–Hinton Broth (MHB) containing 100 μg mL−1 ampicillin102.
The E. coli DH5α strains carrying different pUC19 plasmids were cultured overnight in MHB and subjected to trimethoprim susceptibility test using the broth-dilution method103, with concentrations of trimethoprim tested at 0, 0.5, 1, 2, 4, 8, 16, 32, 64, and 128 μg mL−1. The minimum inhibitory concentration of trimethoprim against a given strain was defined as the lowest concentration that inhibited ≥80% growth of that strain compared to the growth control104.
Statistics and reproducibility
In this study, we utilized a comprehensive public dataset of virus genomes as the main data for all analyses. Consequently, no statistical methods were employed to predetermine the sample size, as the sample size was restrained by the available public dataset. No data were excluded from the analyses. For the antimicrobial susceptibility test, individuals involved in the experiments were blinded to the experiment groups to ensure unbiased measurements.
Two-group comparisons were performed with two-sided Wilcoxon rank-sum test. Multiple-group comparisons were performed with Kruskal–Wallis test. Dependency between two binary variables were tested with two-sided Chi-squared test. The multiple sequence alignments plots were generated using ESPript 3 web platform105. Gene structure plots were generated using gggenes package. Protein structures were generated using Phyre2 web platform106. R software (v4.1.0) was used for the statistics analysis and plotting.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank Professor AJM Baker (Universities of Melbourne and Queensland, Australia, and Sheffield, UK) for his help in English language editing. This work was supported financially by the National Natural Science Foundation of China (42077117 and 32470001 to J.T.L., 32470094 to X.Z.Y., 42177009 to J.-L.L., & 41830318 to W.S.S.) and the National Key R&D Program of China (2023YFC3207300 to J.T.L.).
Author contributions
J.T.L., X.Z.Y., J.-.L.L. and W.S.S. conceived and designed the experiments; J.L.L., S.W.F., P.J., S.J.Z., S.Y.L., Y.Y.Z., Y.Q.G. and B.L. performed the experiments; X.Z.Y., J.-L.L., P.W., S.W.F. and Z.W. analyzed the data; X.Z.Y., J.T.L. and J.-.L.L. wrote the first draft of the manuscript; all authors revised the manuscript.
Peer review
Peer review information
Nature Communications thanks Jeffrey Blanchard, Maite Muniesa, Sofia Rigou and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Data availability
Metagenomic sequencing data for the tailings samples used in this study have been deposited in the NCBI BioProject database under the accession number PRJNA1085405. The giant virus sequences and phage contigs generated from tailings metagenomes in this study have been deposited in the ENA Sequence Read Archive database under the accession number PRJEB74361 and PRJEB78842, respectively. Source data of this paper are provided on Github107.
Code availability
The codes used in this study are available on GitHub107.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xinzhu Yi, Jie-Liang Liang.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51936-z.
References
- 1.WHO. “Thirteenth general programme of work, 2019–2023”. https://apps.who.int/iris/bitstream/handle/10665/324775/WHO-PRP-18.1-eng.pdf (2019).
- 2.De la Cruz, F. & Davies, J. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol.8, 128–133 (2000). 10.1016/S0966-842X(00)01703-0 [DOI] [PubMed] [Google Scholar]
- 3.Watson, B. N. J. et al. CRISPR-Cas-mediated phage resistance enhances horizontal gene transfer by transduction. mBio9, e02406–e02417 (2018). 10.1128/mBio.02406-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lekunberri, I., Villagrasa, M., Balcázar, J. L. & Borrego, C. M. Contribution of bacteriophage and plasmid DNA to the mobilization of antibiotic resistance genes in a river receiving treated wastewater discharges. Sci. Total Environ.601-602, 206–209 (2017). 10.1016/j.scitotenv.2017.05.174 [DOI] [PubMed] [Google Scholar]
- 5.Jankowski, P. et al. Metagenomic community composition and resistome analysis in a full-scale cold climate wastewater treatment plant. Environ. Microbiome17, 3 (2022). 10.1186/s40793-022-00398-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Debroas, D. & Siguret, C. Viruses as key reservoirs of antibiotic resistance genes in the environment. ISME J.13, 2856–2867 (2019). 10.1038/s41396-019-0478-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moon, K. et al. Freshwater viral metagenome reveals novel and functional phage-borne antibiotic resistance genes. Microbiome8, 75 (2020). 10.1186/s40168-020-00863-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, M.-L. et al. Viral community and virus-associated antibiotic resistance genes in soils amended with organic fertilizers. Environ. Sci. Technol.55, 13881–13890 (2021). 10.1021/acs.est.1c03847 [DOI] [PubMed] [Google Scholar]
- 9.Colomer-Lluch, M., Jofre, J. & Muniesa, M. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS One6, e17549 (2011). 10.1371/journal.pone.0017549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlson, T. A. & Chelm, B. K. Apparent eukaryotic origin of glutamine synthetase II from the bacterium Bradyrhizobium japonicum. Nature322, 568–570 (1986). 10.1038/322568a0 [DOI] [Google Scholar]
- 11.Brown, J. R., Zhang, J. & Hodgson, J. E. A bacterial antibiotic resistance gene with eukaryotic origins. Curr. Biol.8, R365–R367 (1998). 10.1016/S0960-9822(98)70238-6 [DOI] [PubMed] [Google Scholar]
- 12.Li, Z. & Bock, R. Rapid functional activation of a horizontally transferred eukaryotic gene in a bacterial genome in the absence of selection. Nucleic Acids Res.47, 6351–6359 (2019). 10.1093/nar/gkz370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank, J. A. & Feschotte, C. Co-option of endogenous viral sequences for host cell function. Curr. Opin. Virol.25, 81–89 (2017). 10.1016/j.coviro.2017.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koonin, E. V. & Krupovic, M. The depths of virus exaptation. Curr. Opin. Virol.31, 1–8 (2018). 10.1016/j.coviro.2018.07.011 [DOI] [PubMed] [Google Scholar]
- 15.Irwin, N. A. T., Pittis, A. A., Richards, T. A. & Keeling, P. J. Systematic evaluation of horizontal gene transfer between eukaryotes and viruses. Nat. Microbiol.7, 327–336 (2022). 10.1038/s41564-021-01026-3 [DOI] [PubMed] [Google Scholar]
- 16.Iyer, L. M., Balaji, S., Koonin, E. V. & Aravind, L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res.117, 156–184 (2006). 10.1016/j.virusres.2006.01.009 [DOI] [PubMed] [Google Scholar]
- 17.Mueller, L., Hauser, P. M., Gauye, F. & Greub, G. Lausannevirus encodes a functional dihydrofolate reductase susceptible to proguanil. Antimicrob. Agents Chemother.61, e02573–16 (2017). 10.1128/AAC.02573-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colson, P. et al. A protein of the metallo-hydrolase/oxidoreductase superfamily with both beta-lactamase and ribonuclease activity is linked with translation in giant viruses. Sci. Rep.10, 21685 (2020). 10.1038/s41598-020-78658-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rigou, S., Santini, S., Abergel, C., Claverie, J.-M. & Legendre, M. Past and present giant viruses diversity explored through permafrost metagenomics. Nat. Commun.13, 5853 (2022). 10.1038/s41467-022-33633-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alcock, B. P. et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res.51, D690–D699 (2023). 10.1093/nar/gkac920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sköld, O. Resistance to trimethoprim and sulfonamides. Vet. Res.32, 261–273 (2001). 10.1051/vetres:2001123 [DOI] [PubMed] [Google Scholar]
- 22.Coque, T. M., Singh, K. V., Weinstock, G. M. & Murray, B. E. Characterization of dihydrofolate reductase genes from trimethoprim-susceptible and trimethoprim-resistant strains of Enterococcus faecalis. Antimicrob. Agents Chemother.43, 141–147 (1999). 10.1128/AAC.43.1.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su, W. et al. Ribosome protection by antibiotic resistance ATP-binding cassette protein. Proc. Natl. Acad. Sci. USA115, 5157–5162 (2018). 10.1073/pnas.1803313115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hetem, D. J. & Bonten, M. J. Clinical relevance of mupirocin resistance in Staphylococcus aureus. J. Hosp. Infect.85, 249–256 (2013). 10.1016/j.jhin.2013.09.006 [DOI] [PubMed] [Google Scholar]
- 25.Bugg, T. D. et al. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry30, 10408–10415 (1991). 10.1021/bi00107a007 [DOI] [PubMed] [Google Scholar]
- 26.Jung, Y. H. et al. Characterization of two newly identified genes, vgaD and vatH, conferring resistance to streptogramin A in Enterococcus faecium. Antimicrob. Agents Chemother.54, 4744–4749 (2010). 10.1128/AAC.00798-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakama, T., Nureki, O. & Yokoyama, S. Structural basis for the recognition of isoleucyl-adenylate and an antibiotic, mupirocin, by isoleucyl-tRNA synthetase. J. Biol. Chem.276, 47387–47393 (2001). 10.1074/jbc.M109089200 [DOI] [PubMed] [Google Scholar]
- 28.Sassanfar, M., Kranz, J. E., Gallant, P., Schimmel, P. & Shiba, K. A eubacterial Mycobacterium tuberculosis tRNA synthetase is eukaryote-like and resistant to a eubacterial-specific antisynthetase drug. Biochemistry35, 9995–10003 (1996). 10.1021/bi9603027 [DOI] [PubMed] [Google Scholar]
- 29.Kondrashov, F. A., Rogozin, I. B., Wolf, Y. I. & Koonin, E. V. Selection in the evolution of gene duplications. Genome Biol.3, research0008.0001 (2002). 10.1186/gb-2002-3-2-research0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mire, C. E., White, J. M. & Whitt, M. A. A spatio-temporal analysis of matrix protein and nucleocapsid trafficking during vesicular stomatitis virus uncoating. PLoS Pathog.6, e1000994 (2010). 10.1371/journal.ppat.1000994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo, J., Chen, Y., Guo, Y., Li, H. & Zhang, S. The E2 ubiquitin-conjugating enzyme CfRad6 regulates the autophagy and pathogenicity of Colletotrichum fructicola on Camellia oleifera. Phytopathol. Res.5, 39 (2023). 10.1186/s42483-023-00191-z [DOI] [Google Scholar]
- 32.Eini, O. et al. Interaction with a host ubiquitin-conjugating enzyme is required for the pathogenicity of a geminiviral DNA β satellite. Mol. Plant Microbe Interact.22, 737–746 (2009). 10.1094/MPMI-22-6-0737 [DOI] [PubMed] [Google Scholar]
- 33.Loizos, N., Tillier, E. R. & Belfort, M. Evolution of mobile group I introns: recognition of intron sequences by an intron-encoded endonuclease. Proc. Natl. Acad. Sci. USA91, 11983–11987 (1994). 10.1073/pnas.91.25.11983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho, W. & Stahelin, R. V. Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Bioph. Biom.34, 119–151 (2005). 10.1146/annurev.biophys.33.110502.133337 [DOI] [PubMed] [Google Scholar]
- 35.Filée, J. Giant viruses and their mobile genetic elements: the molecular symbiosis hypothesis. Curr. Opin. Virol.33, 81–88 (2018). 10.1016/j.coviro.2018.07.013 [DOI] [PubMed] [Google Scholar]
- 36.Partridge, S. R., Kwong, S. M., Firth, N. & Jensen, S. O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev.31, e00088–00017 (2018). 10.1128/CMR.00088-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.La Scola, B. et al. A giant virus in amoebae. Science299, 2033 (2003). 10.1126/science.1081867 [DOI] [PubMed] [Google Scholar]
- 38.Philippe, N. et al. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science341, 281–286 (2013). 10.1126/science.1239181 [DOI] [PubMed] [Google Scholar]
- 39.Abrahão, J. et al. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun.9, 749 (2018). 10.1038/s41467-018-03168-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abergel, C., Legendre, M. & Claverie, J.-M. The rapidly expanding universe of giant viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus. FEMS Microbiol. Rev.39, 779–796 (2015). 10.1093/femsre/fuv037 [DOI] [PubMed] [Google Scholar]
- 41.Schulz, F., Abergel, C. & Woyke, T. Giant virus biology and diversity in the era of genome-resolved metagenomics. Nat. Rev. Microbiol.20, 721–736 (2022). 10.1038/s41579-022-00754-5 [DOI] [PubMed] [Google Scholar]
- 42.Li, L.-G., Xia, Y. & Zhang, T. Co-occurrence of antibiotic and metal resistance genes revealed in complete genome collection. ISME J.11, 651–662 (2017). 10.1038/ismej.2016.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Enault, F. et al. Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J.11, 237–247 (2017). 10.1038/ismej.2016.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliveira, G. P., Rodrigues, R. A., Lima, M. T., Drumond, B. P. & Abrahão, J. S. Poxvirus host range genes and virus–host spectrum: a critical review. Viruses9, 331 (2017). 10.3390/v9110331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ben-Ishai, Z. V. I., Heller, E., Goldblum, N. & Becker, Y. Rifampicin poxvirus and trachoma agent: rifampicin and poxvirus replication. Nature224, 29–32 (1969). 10.1038/224029a0 [DOI] [PubMed] [Google Scholar]
- 46.Bertacine Dias, M. V., Santos, J. C., Libreros-Zúñiga, G. A., Ribeiro, J. A. & Chavez-Pacheco, S. M. Folate biosynthesis pathway: mechanisms and insights into drug design for infectious diseases. Future Med. Chem.10, 935–959 (2018). 10.4155/fmc-2017-0168 [DOI] [PubMed] [Google Scholar]
- 47.Fairlamb, A. H., Gow, N. A. R., Matthews, K. R. & Waters, A. P. Drug resistance in eukaryotic microorganisms. Nat. Microbiol.1, 16092 (2016). 10.1038/nmicrobiol.2016.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cockerill, F. R. & Edson, R. S. Trimethoprim-sulfamethoxazole. Mayo Clin. Proc.66, 1260–1269 (1991). 10.1016/S0025-6196(12)62478-1 [DOI] [PubMed] [Google Scholar]
- 49.Liu, J., Bolstad, D. B., Bolstad, E. S., Wright, D. L. & Anderson, A. C. Towards new antifolates targeting eukaryotic opportunistic infections. Eukaryot. Cell8, 483–486 (2009). 10.1128/EC.00298-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yarus, M. & Berg, P. Recognition of tRNA by isoleucyl-tRNA synthetase: effect of substrates on the dynamics of tRNA-enzyme interaction. J. Mol. Biol.42, 171–189 (1969). 10.1016/0022-2836(69)90037-0 [DOI] [PubMed] [Google Scholar]
- 51.Hodgson, J. E. et al. Molecular characterization of the gene encoding high-level mupirocin resistance in Staphylococcus aureus J2870. Antimicrob. Agents Chemother.38, 1205–1208 (1994). 10.1128/AAC.38.5.1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yanagisawa, T. & Kawakami, M. How does Pseudomonas fluorescens avoid suicide from its antibiotic pseudomonic acid?: Evidence for two evolutionarily distinct isoleucyl-tRNA synthetases conferring self-defense. J. Biol. Chem.278, 25887–25894 (2003). 10.1074/jbc.M302633200 [DOI] [PubMed] [Google Scholar]
- 53.Arthofer, P., Delafont, V., Willemsen, A., Panhölzl, F. & Horn, M. Defensive symbiosis against giant viruses in amoebae. Proc. Natl. Acad. Sci. USA119, e2205856119 (2022). 10.1073/pnas.2205856119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Casadevall, A. Evolution of intracellular pathogens. Annu. Rev. Microbiol.62, 19–33 (2008). 10.1146/annurev.micro.61.080706.093305 [DOI] [PubMed] [Google Scholar]
- 55.Greub, G. & Raoult, D. Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev.17, 413–433 (2004). 10.1128/CMR.17.2.413-433.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boyer, M. et al. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl. Acad. Sci. USA106, 21848–21853 (2009). 10.1073/pnas.0911354106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moliner, C., Fournier, P.-E. & Raoult, D. Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol. Rev.34, 281–294 (2010). 10.1111/j.1574-6976.2009.00209.x [DOI] [PubMed] [Google Scholar]
- 58.Desnues, C. et al. Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc. Natl. Acad. Sci. USA109, 18078–18083 (2012). 10.1073/pnas.1208835109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun, C., Feschotte, C., Wu, Z. & Mueller, R. L. DNA transposons have colonized the genome of the giant virus Pandoravirus salinus. BMC Biol.13, 38 (2015). 10.1186/s12915-015-0145-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chevalier, B. S. & Stoddard, B. L. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res.29, 3757–3774 (2001). 10.1093/nar/29.18.3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barth, Z. K., Dunham, D. T. & Seed, K. D. Nuclease genes occupy boundaries of genetic exchange between bacteriophages. NAR Genom. Bioinform.5, lqad076 (2023). 10.1093/nargab/lqad076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yolken, R. H. et al. Chlorovirus ATCV-1 is part of the human oropharyngeal virome and is associated with changes in cognitive functions in humans and mice. Proc. Natl. Acad. Sci. USA111, 16106–16111 (2014). 10.1073/pnas.1418895111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aherfi, S., La Scola, B., Pagnier, I., Raoult, D. & Colson, P. The expanding family Marseilleviridae. Virology466-467, 27–37 (2014). 10.1016/j.virol.2014.07.014 [DOI] [PubMed] [Google Scholar]
- 64.Saadi, H. et al. First isolation of Mimivirus in a patient with pneumonia. Clin. Infect. Dis.57, e127–e134 (2013). 10.1093/cid/cit354 [DOI] [PubMed] [Google Scholar]
- 65.Aylward, F. O., Moniruzzaman, M., Ha, A. D. & Koonin, E. V. A phylogenomic framework for charting the diversity and evolution of giant viruses. PLoS Biol.19, e3001430 (2021). 10.1371/journal.pbio.3001430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nayfach, S. et al. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol.39, 578–585 (2021). 10.1038/s41587-020-00774-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yi, X. et al. Globally distributed mining-impacted environments are underexplored hotspots of multidrug resistance genes. ISME J.16, 2099–2113 (2022). 10.1038/s41396-022-01258-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schulz, F. et al. Giant virus diversity and host interactions through global metagenomics. Nature578, 432–436 (2020). 10.1038/s41586-020-1957-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yutin, N., Wolf, Y. I., Raoult, D. & Koonin, E. V. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol. J.6, 223 (2009). 10.1186/1743-422X-6-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Endo, H. et al. Biogeography of marine giant viruses reveals their interplay with eukaryotes and ecological functions. Nat. Ecol. Evol.4, 1639–1649 (2020). 10.1038/s41559-020-01288-w [DOI] [PubMed] [Google Scholar]
- 71.Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods9, 357–359 (2012). 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics25, 2078–2079 (2009). 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kang, D. D. et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ7, e7359 (2019). 10.7717/peerj.7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olm, M. R., Brown, C. T., Brooks, B. & Banfield, J. F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J.11, 2864–2868 (2017). 10.1038/ismej.2017.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roux, S., Enault, F., Hurwitz, B. L. & Sullivan, M. B. VirSorter: mining viral signal from microbial genomic data. PeerJ3, e985 (2015). 10.7717/peerj.985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Camargo, A. P. et al. Identification of mobile genetic elements with geNomad. Nat. Biotechnol. 42, 1303–1312 (2023). [DOI] [PMC free article] [PubMed]
- 77.Arango-Argoty, G. et al. DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome6, 23 23 (2018). 10.1186/s40168-018-0401-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yin, X. et al. ARGs-OAP v2.0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes. Bioinformatics34, 2263–2270 (2018). 10.1093/bioinformatics/bty053 [DOI] [PubMed] [Google Scholar]
- 79.Feldgarden, M. et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep.11, 12728 (2021). 10.1038/s41598-021-91456-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Che, Y. et al. High-resolution genomic surveillance elucidates a multilayered hierarchical transfer of resistance between WWTP- and human/animal-associated bacteria. Microbiome10, 16 (2022). 10.1186/s40168-021-01192-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang, B. et al. Tackling soil ARG-carrying pathogens with global-scale metagenomics. Adv. Sci.10, 2301980 (2023). 10.1002/advs.202301980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Madrigal, P. et al. Machine learning algorithm to characterize antimicrobial resistance associated with the International Space Station surface microbiome. Microbiome10, 134 (2022). 10.1186/s40168-022-01332-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang, J. et al. Metagenomics insights into the profiles of antibiotic resistome in combined sewage overflows from reads to metagenome assembly genomes. J. Hazard. Mater.429, 128277 (2022). 10.1016/j.jhazmat.2022.128277 [DOI] [PubMed] [Google Scholar]
- 84.Sivalingam, P. et al. Extracellular DNA includes an important fraction of high-risk antibiotic resistance genes in treated wastewaters. Environ. Pollut.323, 121325 (2023). 10.1016/j.envpol.2023.121325 [DOI] [PubMed] [Google Scholar]
- 85.Liu, W. et al. Temporal dynamics and contribution of phage community to the prevalence of antibiotic resistance genes in a full-scale sludge anaerobic digestion plant. Environ. Sci. Technol.58, 6296–6304 (2024). 10.1021/acs.est.4c00712 [DOI] [PubMed] [Google Scholar]
- 86.Eichorst, S. A. et al. Genomic insights into the Acidobacteria reveal strategies for their success in terrestrial environments. Environ. Microbiol.20, 1041–1063 (2018). 10.1111/1462-2920.14043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xie, Z. & Tang, H. ISEScan: automated identification of insertion sequence elements in prokaryotic genomes. Bioinformatics33, 3340–3347 (2017). 10.1093/bioinformatics/btx433 [DOI] [PubMed] [Google Scholar]
- 88.Abby, S. S., Néron, B., Ménager, H., Touchon, M. & Rocha, E. P. C. MacSyFinder: a program to mine genomes for molecular systems with an application to CRISPR-Cas systems. PLoS One9, e110726 (2014). 10.1371/journal.pone.0110726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fouts, D. E. Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res.34, 5839–5851 (2006). 10.1093/nar/gkl732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu, M. et al. ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res.47, D660–D665 (2019). 10.1093/nar/gky1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jiang, X., Hall, A. B., Xavier, R. J. & Alm, E. J. Comprehensive analysis of chromosomal mobile genetic elements in the gut microbiome reveals phylum-level niche-adaptive gene pools. PLoS One14, e0223680 (2019). 10.1371/journal.pone.0223680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yi, X. et al. Phytostabilization mitigates antibiotic resistance gene enrichment in a copper mine tailings pond. J. Hazard. Mater.443, 130255 (2023). 10.1016/j.jhazmat.2022.130255 [DOI] [PubMed] [Google Scholar]
- 93.Shaffer, M. et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res.48, 8883–8900 (2020). 10.1093/nar/gkaa621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sánchez-Osuna, M., Cortés, P., Llagostera, M., Barbé, J. & Erill, I. Exploration into the origins and mobilization of di-hydrofolate reductase genes and the emergence of clinical resistance to trimethoprim. Microb. Genom.6, mgen000440 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huerta-Cepas, J. et al. EggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res.47, D309–D314 (2019). 10.1093/nar/gky1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Murina, V. et al. ABCF ATPases involved in protein synthesis, ribosome assembly and antibiotic resistance: structural and functional diversification across the tree of life. J. Mol. Biol.431, 3568–3590 (2019). 10.1016/j.jmb.2018.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol.30, 772–780 (2013). 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics25, 1972–1973 (2009). 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol.37, 1530–1534 (2020). 10.1093/molbev/msaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang, Z. et al. ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem. Biophys. Res. Commun.419, 779–781 (2012). 10.1016/j.bbrc.2012.02.101 [DOI] [PubMed] [Google Scholar]
- 101.Zhang, Z. KaKs_calculator 3.0: calculating selective pressure on coding and non-coding sequences. Genom. Proteom. Bioinf.20, 536–540 (2022). 10.1016/j.gpb.2021.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ambrose, S. J. & Hall, R. M. Novel trimethoprim resistance gene, dfrA35, in IncC plasmids from Australia. J. Antimicrob. Chemother.74, 1863–1866 (2019). 10.1093/jac/dkz148 [DOI] [PubMed] [Google Scholar]
- 103.Reller, L. B., Weinstein, M., Jorgensen, J. H. & Ferraro, M. J. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin. Infect. Dis.49, 1749–1755 (2009). 10.1086/647952 [DOI] [PubMed] [Google Scholar]
- 104.EUCAST. “Broth microdilution - EUCAST reading guide v 4.0” EUCAST. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/2022_manuals/Reading_guide_BMD_v_4.0_2022.pdf (2022).
- 105.Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res.42, W320–W324 (2014). 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc.10, 845–858 (2015). 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yi, X. et al. Giant viruses as reservoirs of antibiotic resistance genes. Github. 10.5281/zenodo.13234118 (2024). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Metagenomic sequencing data for the tailings samples used in this study have been deposited in the NCBI BioProject database under the accession number PRJNA1085405. The giant virus sequences and phage contigs generated from tailings metagenomes in this study have been deposited in the ENA Sequence Read Archive database under the accession number PRJEB74361 and PRJEB78842, respectively. Source data of this paper are provided on Github107.
The codes used in this study are available on GitHub107.