Abstract
Molecular similarities between embryonic and malignant cells can be exploited to target tumors through specific signatures absent in healthy adult tissues. One such embryonic signature tumors express is oncofetal chondroitin sulfate (ofCS), which supports disease progression and dissemination in cancer. Here, we report the identification and characterization of phage display-derived antibody fragments recognizing two distinct ofCS epitopes. These antibody fragments show binding affinity to ofCS in the low nanomolar range across a broad selection of solid tumor types in vitro and in vivo with minimal binding to normal, inflamed, or benign tumor tissues. Anti-ofCS antibody drug conjugates and bispecific immune cell engagers based on these targeting moieties disrupt tumor progression in animal models of human and murine cancers. Thus, anti-ofCS antibody fragments hold promise for the development of broadly effective therapeutic and diagnostic applications targeting human malignancies.
Subject terms: Targeted therapies, Molecular medicine, Tumour biomarkers, Cancer imaging, Cancer microenvironment
Cancer cells often acquire molecular patterns of fast-growing embryonic tissues to enable propagation and invasion, which distinguish tumour tissues from their healthy adult counterpart. Here authors develop antibody fragments which specifically target oncofetal chondroitin sulphate on the cancer cell surface and in the tumour stroma, with the antibodies achieving therapeutic effect in multiple mouse models in antibody drug conjugate and bispecific immune cell engager formats.
Introduction
Cancer is a global health problem with an estimated 19 million new cases and 10 million deaths per year1. Treatments typically involve surgery, chemotherapy, and radiation. In the past decade, there have been promising developments in systemic immunotherapies and targeted biologics, such as antibody-drug conjugates (ADC), with market approval granted for 14 ADCs, primarily for hematological cancers or HER2+ tumors2. The primary challenge in developing new targeted therapies lies in the limited number of actionable cancer targets, especially in the context of heterogeneous solid tumors.
The existence of molecular similarities between embryonic and malignant tissues has been noted since the beginning of the 20th century3,4. These similarities probably reflect shared requirements for rapid cellular proliferation, tissue invasion, angiogenesis, and immune evasion5–9. Cancer’s aberrant glycosylation patterns have emerged as promising targets, with several carbohydrate epitopes exhibiting oncofetal origins10. An example is oncofetal chondroitin sulfate (ofCS), initially identified as a distinct type of chondroitin sulfate (CS) through the selective tropism of Plasmodium falciparum malaria-infected red blood cells, adhering to the placental syncytial trophoblasts through the VAR2CSA parasite-encoded lectin11–15. This interaction allows the sequestration of malaria-infected blood cells in placental vasculature, preventing circulation and clearance through splenic filtration16–19. ofCS is a specific type of glycosaminoglycan attached to proteins on the cell surface and in the extracellular matrix (ECM). Glycosaminoglycans consist of long unbranched polysaccharide chains of alternating disaccharide units with sulfations at various positions, and can form distinct structural configurations, affecting biological functions in health and disease20. The exact ofCS sulfation pattern, defined by VAR2CSA binding, remains unclear. Structural analyses indicate that ofCS is composed of a chain longer than tetradeca-saccharides (>dp14) with islands of high 4-O sulfation of N-Acetylgalactosamine (GalNAc) residues21. Cryo-electron microscopy (cryo-EM) of VAR2CSA:ofCS complex supports that the oncofetal specificity is linked to the sulfation pattern of a long CS chain, but also the length of the chain22. Despite the obvious potential of ofCS as a tumor target for broadly effective therapy, clinical translation of recombinant VAR2CSA (rVAR2) as a targeting agent faces challenges due to rapid liver clearance, risk of immunogenicity, and complex manufacturing scale-up processes. Exploring alternative translational approaches for targeting ofCS may overcome these challenges and pave the way for the first broadly effective targeted cancer treatment.
In this work, we develop and characterize three broadly tumor-specific antibody fragments with unique binding to two distinct ofCS epitopes, and validate their potential as tumor-agnostic cancer-targeting vehicles.
Results
Phage display-derived antibody fragments are specific to cancer cells
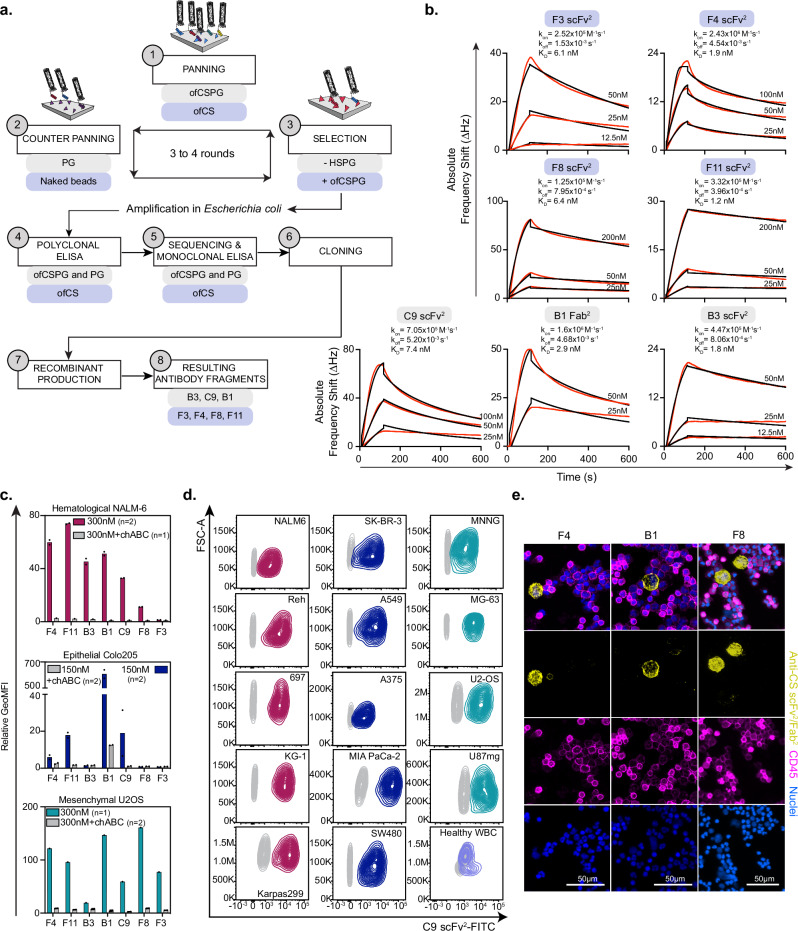
Directing high-affinity monospecific antibody responses to distinct glycosaminoglycans with specific sulfation patterns appearing on a long chain is challenging. Our attempts to seroconvert animals to make antibodies to ofCS were unsuccessful, likely due to extreme self-restriction to these fetal glycan structures. Consequently, we explored phage display as an alternative approach. To maximize the chance of identifying antibody sequences with specificity towards ofCS, we designed phage-display strategies using rVAR2-validated ofCS source material and applied it to a panel of non-overlapping phage libraries (Fig. 1a). Specifically, we purified two key reagents containing ofCS; (i) ofCS-modified proteoglycans (ofCSPG), overexpressed in immortalized Chinese Hamster Ovary (CHO) cells and purified from the cellular fraction by ion-exchange chromatography; and (ii) placental-ofCS purified from human term placenta. The presence of ofCS chains in the source material was confirmed by binding to rVAR2 (Supplementary Fig. 1a). Due to the highly negatively charged nature of CS, there was a risk of attracting unspecific charge:charge interactions during the phage-display panning, potentially complicating the selection of ofCS-specific antibody sequences. To mitigate this, counter-screens were performed on negatively charged heparan sulfate proteoglycan (HSPG). This was supplemented by an additional counter-screen using chondroitinase ABC (chABC)-treated ofCSPG (now termed PG) to eliminate protein-specific phages or phages binding degraded stumps of CS on the PG. Three phage-display libraries were investigated. The naive human LiAB-SFMAXTM (single-chain variable fragments-scFv) and synthetic human HuCAL (antibody binding fragment-Fab)23,24 libraries were panned on ofCSPG over 3–4 rounds, while the ALTHEA GoldTM human (scFv)25 library was panned on ofCS source material. Ninety-six phage clones were randomly picked from rounds 3 and 4 of LiAB-SFMAXTM library and tested in an enzyme-linked immunoassay (ELISA) against ofCSPG and PG. In round 3, 37 clones were found to be specific for ofCSPG with minimal binding to PG, revealing 7 unique sequences. This process was repeated in round 4, but here all identified sequences were identical to the ones obtained in round 3. Four out of the 7 phages coded similar VH domains, and 5 exhibited stronger binding to HSPG than to ofCSPG, suggesting unspecific charge-charge interactions (Supplementary Fig. 1b). This left two phage clones C9 and B3. The screening of these phage clones indicated higher binding to ofCSPG than to HSPG and the binding could be out-competed by rVAR2. Furthermore, they did not show binding to PG (Supplementary Figs. 1b, c) and were thus down-selected for recombinant expression.
Fig. 1. Phage-display derived antibody fragments are specific to cancer cells.
a Overview of the phage display process, illustrating key molecules involved at each step (1-panning, 2-counter panning, 3-phage selection, 4-polyclonal ELISA, 5-clone sequencing and monoclonal ELISA, 6-cloning, 7-recombinant production of positive candidates, 8-resulting antibody fragments) of the panning strategies performed on naive LiAb-SFMAXTM (scFv) and synthetic HuCAL (Fab) (gray boxes) or ALTHEA Gold semi-synthetic (scFv) library (purple boxes). b Sensograms depicting the interaction of resulting recombinant antibody fragments (F3 scFv2, F4 scFv2, F8 scFv2, F11 scFv2, C9 scFv2, B1 Fab2, and B3 scFv2) to ofCS, purified from human placenta and immobilized on a quartz crystal microbalance biosensor chip. Red curves correspond to experimental points and rate constants (association rate kon, dissociation rate koff, binding constant KD) were determined using a 1:2 fitting model with fitted curves in black. c Flow cytometry binding of V5-tagged-scFv2 (F4, F11, B3, C9, F8, and F3) and FLAG-tagged-Fab2 (B1) to cancer cell lines (hematological leukemic NALM-6 (top), epithelial colorectal Colo205 (middle) and mesenchymal glioblastoma U2OS (bottom)). The binding was measured as relative GeoMFI probed with FITC-labeled secondary antibodies without (colored) or with (gray) chABC treatment, with number of data point (n) indicated for each group. d C9 scFv2 binding to a larger panel of human cancer cell lines (hematological (red), epithelial (blue), and mesenchymal (green)) and to healthy purified white blood cells (purple), at 150 nM concentration, compared to the secondary antibody control (gray). e Immunofluorescence staining of A549 lung cancer cells spiked into healthy donor blood with PE-labeled scFv2 (F4, F8) or Fab2 (B1) (yellow), anti-CD45-Alexa647 (pink) and DAPI (blue). Top panel is a merged image of the three individuals shown below. These scans are representative of an image dimension of 2286 tiles and 41.06 mm × 14.03 mm per image (B1/F4) or 130 tiles and 5.12 mm × 2.91 mm (F8). Source data are provided as a Source data file.
Primary screening of the HuCAL library resulted in 74 phage clones binding to ofCSPG more than 5-fold over background. A single sequence (B1) was identified in 20 of the clones and a monoclonal ELISA confirmed binding to ofCSPG (and not PG) that could be competed with rVAR2 (Supplementary Fig. 1d, e).
Sequencing of 576 phage clones from the panning of the ALTHEATM library identified 17 hits with single or dual specificity for purified ofCS and ofCSPG, and no binding to HSPG. Following preliminary specificity assessment, sequence analyses, and confirmed rVAR2 competition, 4 clones were selected for further testing (F3, F4, F8, and F11) (Supplementary Fig. 1f).
In summary, only 7 out of 109 sequences, from three different libraries, were found to have potential ofCS specificity, reflecting the historical challenges of developing antibodies towards glycosaminoglycan subtypes. To address possible bias in testing the specificity of antibody fragments presented on a phage we cloned and recombinantly expressed the antibody fragments corresponding to the seven sequences. Different formats were expressed as (i) scFv; (ii) Fab; and (iii) tandem scFv (T-scFv) (Supplementary Figs. 1g and 2a). Furthermore, the monomeric proteins were dimerized through the SpyCatcher (SpyC)-SpyTag (SpyT) split-intein system to increase avidity26 creating Fab2 and scFv2. ofCS binding kinetics for each antibody fragment were assessed using a Quartz Crystal Microscale (QCM) biosensor and combined with ELISA binding experiments. The 7 recombinant antibody fragments exhibited high-affinity binding to ofCS, ranging from 1.2 to 7.4 nM, with no binding to HSPG (Fig. 1b and Supplementary Fig. 2b). When presented on the phage, the C9 clone did show some binding to HS. However, the recombinant pure antibody showed very high specificity to CS without any binding to HS (Supplementary Fig. 2b).
The 7 ofCS-targeting antibody fragments were subsequently tested for binding to a broad panel of cancer cell lines representing a range of epithelial, mesenchymal, and hematological cell lines. Initial tests on hematological NALM-6, epithelial Colo205, and mesenchymal U2OS showed different profiles among the antibody fragments (Fig. 1c). F4, F11, C9, and B1 bound all the cell lines while B3 predominantly bound to NALM-6, and F8 and F3 bound strongly to U2OS cells but weakly to NALM-6 and Colo205. chABC treatment validated CS specificity (Fig. 1c). Further analyses showed that the antibody fragments bound a broader panel of cancer cell lines with minimal binding to human white blood cells (Fig. 1c, d and Supplementary Figs. 2c, d). To validate these findings, we performed fluorescence immunocytochemical analysis of A549 lung cancer cells spiked into healthy donor blood, and stained with F4, F8, and B1. This demonstrated specific binding to A549 cells with limited binding to white blood cells, as indicated by a lack of co-localization with anti-CD45 staining (Fig. 1e). These data underscore the specific binding of the antibody fragments to tumor cells in a CS-dependent manner, with minimal binding to the normal blood cell compartment.
ofCS-scFvs exhibit cancer-selective reactivity in tissues
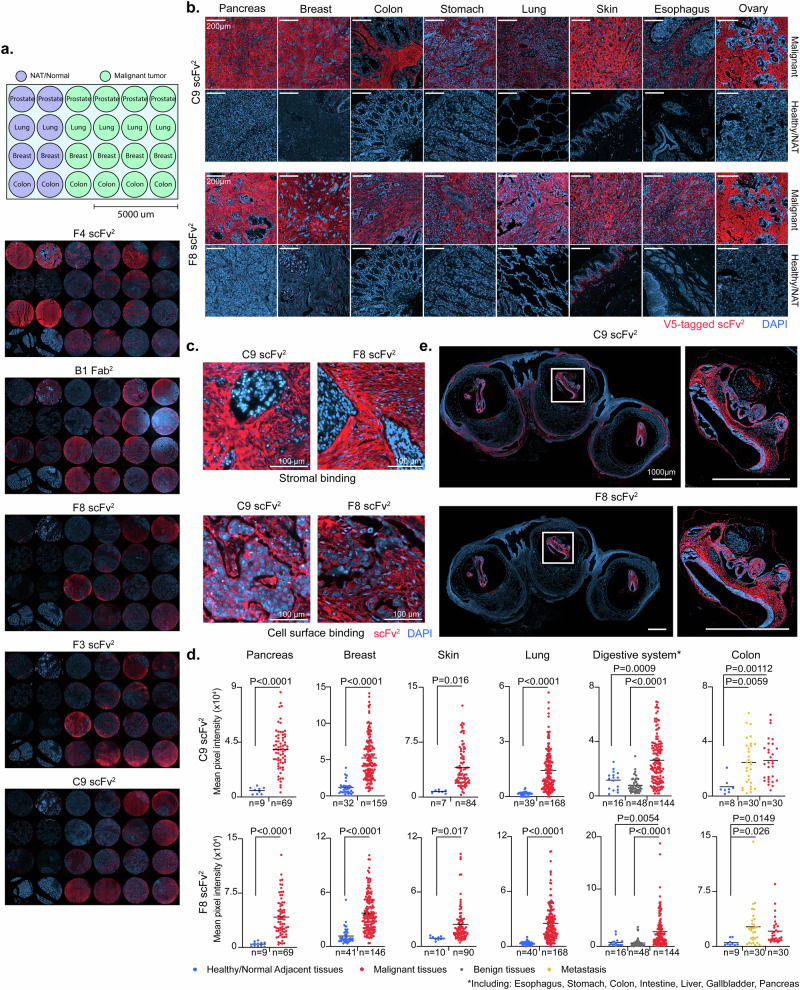
CS is prevalent in the ECM and pericellular glycocalyx of normal tissues, while ofCS is exclusive to the placenta and tumors27. We next evaluated the ability of the antibody fragments to discriminate between ofCS in malignant/fetal tissues and CS in normal tissue. We tested the binding of F8, F3, B1, C9, and F4 to prostate, lung, breast, and colon carcinomas versus the binding to normal or normal adjacent tissues (Fig. 2a). Results indicated that F4 recognized a non-cancer-specific CS epitope abundant in normal prostate and breast tissues. B1 displayed strong staining for malignant tissues as compared to most healthy tissues, but reacted with healthy prostate and breast tissues. Remarkably, F8, F3, and C9 exclusively stained tumor tissues with minimal binding to CS structures in healthy tissues (Fig. 2a). Considering these data, and the similar fine-specificity of F8 and F3—as demonstrated later—F8 and C9 were selected for further analysis in expanded patient cohorts across various cancer types (Supplementary Figs. 3–5). The two antibody fragments stained the vast majority of tumor sections with some variations in the staining patterns, binding partly cancer cell surfaces and partly the tumor stroma (Fig. 2b, c). Supportive to the stromal binding, we show by flow cytometry that both antibody fragments bind to human cancer-associated fibroblasts (CAF). Interestingly, unlike C9, the F8 scFv2 did not bind murine CAFs (Supplementary Fig. 6). Generally, minimal or absent staining to normal tissues was observed, except for some staining in testis tissue (F8 and C9), the basal membrane of the skin (F8), and weak staining in the brain (C9) (Fig. 2b and Supplementary Figs. 7a, b). C9 and F8 were applied to six human tissue microarrays (TMA) with malignant and non-malignant tissues from the pancreas, breast, skin, lung, and the digestive system (including esophagus, stomach, colon, intestine, liver, gallbladder, and pancreas). Quantitative analysis, conducted in standardized settings, showed that F8 and C9 ofCS-specific antibody fragments discriminated between normal and malignant tissues from the pancreas (P < 0.0001), breast (P < 0.0001), skin (C9: P < 0.016; F8: P < 0.017), lung (P < 0.0001), the digestive system (C9: P < 0.0009; F8: P = 0.0054) and colon (C9: P = 0.00112; F8: P = 0.0149) (Fig. 2d). C9 and F8 also distinguished benign from malignant tissues in a large TMA with various tumors from the digestive system (P < 0.0001) and bound similarly to primary tumors and metastatic tissues in colon cancer arrays (Fig. 2d and Supplementary Fig. 7c). Robust and specific staining of human placenta tissues confirmed the oncofetal nature of F8 and C9 epitopes (Supplementary Fig. 7d). C9 staining predicted poor survival of patients after cisplatin chemotherapy, highlighting ofCS as an attractive target, also in this patient group. We did not find a difference in ofCS expression between early and late stages of bladder cancer, colon adenocarcinoma, and breast ductal carcinoma, supporting that ofCS is a tumor agnostic target presents from early tumor onset throughout disease progression (Supplementary Fig. 8a, b).
Fig. 2. ofCS-scFvs exhibit cancer-selective reactivity in tissues.
a Immunofluorescence (IF) analysis of paraffin-embedded fixed (FFPE) human tissue TP241b micro-array of healthy/normal adjacent (NAT) and malignant tissues from prostate, lung, breast and colon using 25 nM V5-tagged scFv2 (F4, F8, F3, C9) or V5-tagged Fab2 (B1) detected with Anti-V5-Alexa647 (red). DAPI was used for nuclear staining (blue). b Panel of malignant (top) and healthy (bottom) FFPE sections of different tissue types stained with C9 or F8 scFv2 as described in (a). c IF staining of FFPE lung adenocarcinoma (top) and carcinoma (bottom) sections from LC2085 TMA with C9 or F8 scFv2 as described in (a). d Mean pixel intensities of tissue micro-array (TMA) from different cancer types with number of sections (n) indicated for each group, stained as described in (a) comparing healthy/normal to malignant tissues from the pancreas (P < 0.0001), breast (P < 0.0001), skin (C9: P < 0.016; F8: P < 0.017), lung (P < 0.0001) the digestive system (C9: P < 0.0009; F8: P = 0.0054) and colon (C9: P = 0.00112; F8: P = 0.0149); healthy/normal to benign from the digestive system (P < 0.0001); and healthy/normal to metastatic colorectal tissues (C9: P = 0.0059; F8: P = 0.026). Intensities were measured on Fiji software and two-tailed unpaired parametric t-test was applied. e IF staining of mouse embryos stained with scFv2 as described in (a). To the right, a zoom-in of the dashed box is shown. Source data are provided as a Source data file.
A major challenge for the preclinical development of human antibody-based therapies is the limited cross-reactivity to rodent proteins, making preclinical testing in animal models less informative in identifying risks and benefits before human testing. Previous findings demonstrated that ofCS, as defined by the rVAR2 protein, was detectable in both human and murine tumor tissues27. Consistent with that observation, C9 and F8 stained mouse embryos in uterine pockets (Fig. 2e). To confirm the induction of the ofCS epitopes during endogenous murine tumor development, pancreatic tumors spontaneously developed in genetically engineered mice (KpC model)28 were stained. C9 stained early-onset lesions of malignancy originating from the pancreatic epithelium in the KpC model, highlighting distinct ofCS-positive regions across the pancreas (Supplementary Fig. 7e).
In summary, these data demonstrate that the F8 and C9 antibody fragments detect ofCS modifications in humans and rodents that are presented on proteoglycans in the fetal and malignant tissue compartments.
F8 and C9 antibody fragments localize to solid tumors in vivo
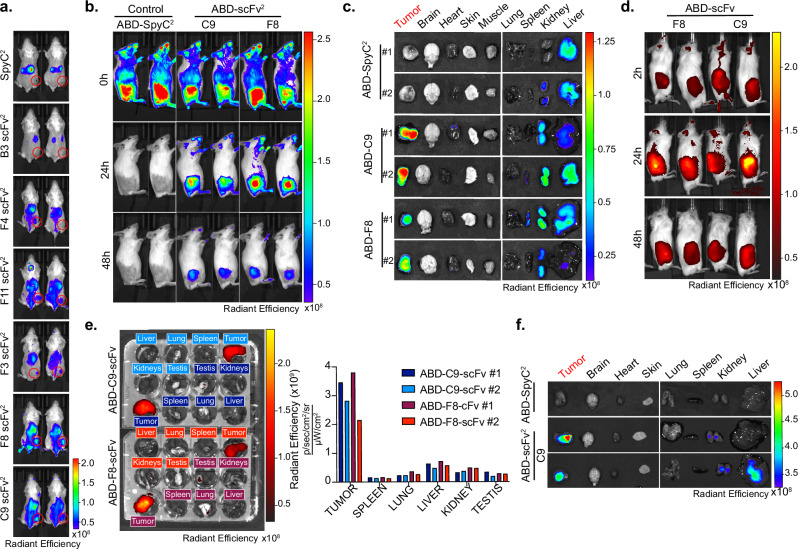
We next evaluated the antibody fragments’ ability to localize to ofCS-expressing tumors in various murine and human cancer models in mice. Tissues from these tumors reacted with F8 and C9 and the reactivity was inhibited by chABC treatment. The reactivity was outcompeted by chondroitin sulfate A (CSA) but not by HS (Supplementary Fig. 9a, c, and e), again highlighting the CS specificity of the antibody fragments. We employed the Alexa750-modified Spycatcher dimer (SpyC2) to enable the dimerization and homogeneous fluorescent labeling of scFvs containing a SpyT. The naked Alexa750-SpyC2 was used as a control (Supplementary Fig. 9f). We first evaluated tumor localization of the 3 tumor-tissue-specific antibody fragments (F3, F8, and C9) and included a broader selection of scFv2s (B3, F4, and F11) in the 4T1 breast cancer allograft model (Fig. 3a). In vivo imaging System (IVIS) scans performed 24 h post-injection showed SpyC2 control predominantly in the kidneys and liver, indicating clearance. Likewise, all scFv2 constructs accumulated in the kidneys, suggesting a similar clearance pathway as SpyC2. Regarding tumor specificity, B3 and F4 exhibited minimal-to-absent tumor localization. F3 and F11 showed intermediate tumor localization while F8 and C9 demonstrated enhanced tumor localization with most of the signal detected in the tumor area. Moreover, F8 and C9 displayed a stronger overall signal in mice after 24 h, indicating longer circulation time (Fig. 3a). In summary, the in vitro tumor specificity of C9 and F8 was confirmed in vivo.
Fig. 3. F8 and C9 antibody fragments localize to solid tumors in vivo.
a In Vivo Imaging system (IVIS) localization of 50 μg Alexa750-labeled antibody (SpyC2, B3, F4, F11, F3, F8, and C9) injected in 4T1 allografted mice with two mice per construct. Tumor area is encircled in red. b IVIS localization of C9 and F8 scFv2 fused to albumin binding domain (ABD) in Karpas299 lymphoma xenografted mice after injection (0 h), 24 h, and 48 h after injection. c Ex vivo IVIS signals from the mice in panel b. 48 h after injection of ABD-control, ABD-C9, ABD-F8. d IVIS localization in patient-derived xenograft (PDX) model of neuroendocrine prostate cancer in male mice as described in b in vivo, and e ex vivo of mice from (d). 48 h, with radiant efficiency measured for each organ. f Ex vivo scanning in MiaPaca2 pancreatic xenograft model 24 h after injection of indicated proteins. Source data are provided as a Source data file.
To extend the antibody fragments’ plasma half-life, we genetically fused an albumin binding domain (ABD) to F8, C9, and SpyC2 control, which similar to previous studies29, dramatically increased plasma half-life as demonstrated with the C9 construct (Supplementary Fig. 9g). Alexa750-labeled ABD-SpyC2, ABD-F8, and ABD-C9 (scFv2 formats) were tested for tumor localization in a Karpas299 cell line-derived xenograft (CDX) lymphoma mouse model (Fig. 3b and Supplementary Fig. 9h). In vivo scans performed within 5 min (0 h) confirmed successful intravenous (i.v.) injections with a well-distributed whole-body signal. At 24 h post-injection, ABD-F8 and ABD-C9 accumulated in the tumor while the ABD control did now show tumor localization. The signal remained in the tumor area after 48 h demonstrating high tumor specificity of both antibody fragments, supporting the potential use of these antibody fragments for cancer imaging and therapeutic delivery. Residual signals in the extremities of the animals suggested continued protein circulation after 24 and 48 h (Fig. 3b). Ex vivo scans of organs collected 48 h post-protein injection confirmed tumor accumulation for both antibody fragments (Fig. 3c). ABD-C9 exhibited sporadic low signals in the heart, likely due to the circulating labeled antibody in the blood, as C9 did not react with normal mouse heart tissue (Supplementary Fig. 9i). Importantly, despite F8 reaction to skin tissue in vitro (Fig. 2b), the ABD-F8 antibody did not localize to the skin in vivo, and similarly, neither antibody fragment was observed in the brain (Fig. 3c). Next, we established a patient-derived xenograft (PDX) model of neuroendocrine prostate cancer in male mice that would also allow us to assess potential testis localization of F8 and C9, as observed in the in vitro analysis (Supplementary Fig. 7b). ABD-F8 and ABD-C9 rapidly localized to the tumor tissues, persisting for at least 48 h (Fig. 3d). Ex vivo analyses showed specific tumor accumulation with minimal signal in the testis (Fig. 3e). Similar specific tumor accumulation for ABD-C9 was obtained in the MiaPaca2 pancreatic CDX model (Fig. 3f) and in the A549 lung cancer CDX model (Supplementary Fig. 9j), highlighting the pan-cancer targeting of the antibodies
In conclusion, F8 and C9 antibody fragments specifically localize to murine and human tumors in vivo with limited off-target accumulation.
Anti-ofCS antibody therapies elicit anti-tumor activity in vivo
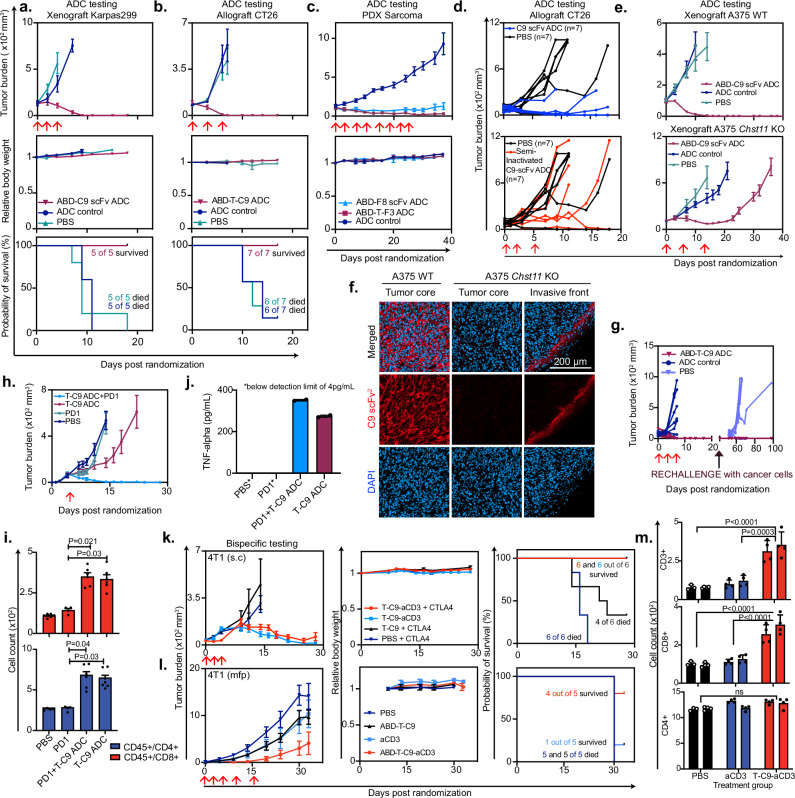
We next evaluated the therapeutic potential of the ofCS-specific antibody fragments as ADCs, in ofCS-expressing tumor models (Supplementary Figs. 9a and 10a–e). ADCs were produced using different antibody fragment formats (monomeric scFv, dimeric T-scFv, or scFv2) derived from C9, F8, and F3. These reagents were coupled to the commonly used cytotoxic payload, monomethyl auristatin E (MMAE) with a cleavable valine-citrulline linker30. The F8-ADC, F3-ADC, and C9-ADC conferred strong cytotoxicity towards human melanoma A375 wild type (WT), murine colorectal CT26, human lymphoma Karpas299, human lung A549, human glioblastoma U87mg, human osteosarcoma MG63, and human pancreas MiaPaca2 cancer cells in vitro with half-maximal inhibitory concentration (IC50) in the nanomolar ranges. Monomeric antibody fragments had higher IC50, likely reflecting a higher off rate in vitro. Importantly, the cytotoxicity of C9-ADC was nullified when C9 binding to A375 was outcompeted by soluble CSA, or when ofCS or glycosaminoglycan expression was abolished through gene knockout (KO) in A375 Chst11 KO and A375 B4GALT7 KO, respectively, verifying that the cytotoxicity is determined by the ofCS specificity and internalization of the antibody (Supplementary Fig. 10f, g).
Subsequently, we evaluated the in vivo efficacy of various ADC formats, maintaining equimolar MMAE amounts between the drug and the control. In the Karpas299 CDX lymphoma xenograft model, all mice in the C9-ADC group were tumor-free after 3 treatments, while aggressive tumor growth was observed in the ABD-SpyC2 MMAE controls and the PBS-treated animals (Fig. 4a). No weight loss was observed in the C9-ADC group indicated limited off-target toxicity (Fig. 4a and Supplementary Fig. 10h). In the CT26 CDX colon allograft model, the C9-ADC had tumor-free outcomes after 3 treatments, while 6 out of 7 mice in the control group reached humane endpoint 12 days after the final injection (Fig. 4b). As in the Karpas299 model, multiple treatments were well tolerated (Fig. 4b and Supplementary Fig. 10h).
Fig. 4. Anti-ofCS antibody therapies elicit anti-tumor activity in vivo.
Tumor burden (mean +/− SEM mm3), body weight and survival curves of mice. a Xenografted with Karpas299 and treated with ABD-C9-scFv ADC, ADC control, or PBS (n = 5). b Allografted with colorectal cancer cells CT26 and treated with ABD-tandem-C9 (ABD-T-C9) or ADC control (n = 7). c Xenografted with patient-derived-xenograft (PDX) sarcoma cells and received F3 or F8-based ADCs (n = 6). d Individual tumor burden (mm3) of mice allografted with CT26 cells and treated with C9-scFv ADC, a lysine-conjugated semi-inactivated C9-ADC, or PBS (n = 7). e Tumor burden (mean +/− SEM mm3) of mice xenografted with melanoma A375 wild type (WT) or Chst11 knock-out (KO), treated with ABD-C9-scFv ADC, ADC control, or PBS (n = 5). f Analysis of tissues from (e), stained with C9-scFv2 (red) and DAPI for nuclei (blue). g Tumor burden (mean +/− SEM mm3) of cured mice from (b), re-challenged with CT26 cells, compared to naive mice injected with cells for the first time (PBS). h Same as in (e), but mice were allografted with CT26 cells and treated with T-C9 ADC alone, in combination with PD1, PD1 alone, or PBS (n = 7). i Immunoprofiling of tumor suspensions from mice treated as in (h), counting CD45+/CD4+ and CD45+/CD8+ populations (PDL-1, n = 2), PDL-1 + T-C9 ADC, T-C9, ADC, and PBS, n = 3)). with triplicate samples. Two-tailed nested t-test compared means and non-significant results were labeled “ns”. j TNFalpha levels from pooled plasma of mice in (i) measured in ELISA in duplicates. Levels in PBS and PD1 groups were below detection limit and marked with an asterisk. k Similar to (a), but mice received 4T1 cells subcutaneously (s.c), and were treated with T-C9-aCD3 with or without anti-CTLA4 checkpoint inhibitor and T-C9 as control (n = 6). l Similar to (e), but mice received 4T1 cells in the mammary fat pad (mfp) and were treated with ABD-T-C9-aCD3 and ABD-T-C9 as control (n = 5). m Immune cell counts (CD3+, CD8+, and CD4+) of 4T1 mfp tumors (n = 2) after three treatments with PBS, aCD3, or T-C9-aCD3. Replicate staining was performed (n = 4) and two-tailed nested t-test was applied to compare groups. Red arrows indicate days of treatment. Source data are provided as a Source data file.
We next tested the efficacy of the C9-ADC in a low-passage hard-to-treat PDX model of human pancreatic ductal adenocarcinoma (PDAC). Dosing was done with a pre-fixed regimen, at the contract research organization (CRO) EPOBerlin, and the mice were dosed multiple times until the control groups reached humane endpoints. Remarkably, the C9-ADC effectively restrained PDAC PDX tumor growth, while the control group rapidly progressed to a humane endpoint (Supplementary Fig. 10h). Notably, multiple treatments were well tolerated despite the higher dosage used in the study (Supplementary Fig. 10i). We next tested ADC formulations of F8 and F3 in a PDX model of undifferentiated pleomorphic sarcoma (UPS), a mesenchymal type of cancer with very limited clinical treatment options. We first validated that the UPS PDX tissue expressed the ofCS epitope (Supplementary Fig. 10e). Similarly to the PDAC PDX model, this was performed at EPOBerlin as a blinded pre-fixed study with rigorous dosing. The F8-ADC and F3-ADC inhibited tumor growth after 1–2 treatments, while the control group progressed towards a humane endpoint (Fig. 4c). Similar to C9-ADC, multiple treatments with F8-ADC and F3-ADC were well tolerated by the mice (Fig. 4c and Supplementary Fig. 10i).
Next, we verified that the efficacy observed in vivo was attributed to the ofCS specificity. First, we semi-inactivated the ofCS binding of C9-ADC through blockage of free amines and thiols to obstruct its binding region and tested its efficacy in a CT26 model. The correlation between in vivo efficacy and in vitro binding was evident: the semi-inactivated ADC showed a moderate to null effect compared to the untreated (PBS) mice, in comparison with the non-inactivated ADC showing a strong therapeutic effect (Fig. 4d and Supplementary Fig. 10j). Secondly, we tested the C9-ADC in melanoma A375 models, genetically knocked out for CHST11 4-O sulfotransferase and thus not fully capable of making the ofCS C9 epitope (Chst11 KO). The C9 ADC completely abolished tumor growth in the WT A375 CDX abundantly expressing the target, with a clear differentiation in efficacy compared to the KO where all mice were euthanized with large tumors at day 35. Interestingly, we did observe a moderate, but only initial, effect of the C9-ADC on the Chst11 KO tumors (Fig. 4e). To address this, we stained the tumor tissues with the C9 antibody and found that the Chst11 KO tumor core was ofCS negative, as we expected. However, the boundaries of the solid tumor stained positive, as a result of ofCS-expressing infiltrating murine stromal cells. These results were in line with previous flow cytometry data on C9 binding to murine CAFs (Fig. 4f and Supplementary Fig. 6). These findings, using the semi-inactivated scFv and the KO cells, indicate that the efficacy of the anti-ofCS ADC depends on ofCS binding.
ADCs can trigger immunogenic cell death resulting in immunological memory against the primary tumor hindering recurrence31,32. To assess whether mice treated with the C9-ADC displayed immunological memory towards the treated tumor type, we re-challenged cured mice from Fig. 4b with inoculation of new tumor cells 35 days after the last ADC treatment. None of the mice previously cured by C9-ADC treatment were able to develop new tumors, while a new group of naive control mice developed tumors at the expected rate (Fig. 4g). These findings suggest an active involvement of the immune system and the induction of immunological memory after C9-ADC treatment.
To further study this, we repeated the CT26 study and administered a single dose (sub-optimal for complete regression) of T-C9-ADC alone or in combination with anti-PDL-1 antibodies, compared to single-agent anti-PDL-1 antibody control arm (Fig. 4h). T-C9-ADC treatment fully eradicated tumors in a durable manner when treated in combination with anti-PDL-1 checkpoint inhibitors—an effect not observed with anti-PDL-1 alone nor with a single dose of the T-C9-ADC. Immune-profiling of tumors from each treatment group revealed a significant increase in CD8+ and CD4+ cells in the two ADC treatment groups (with and without anti-PDL-1) (Fig. 4i and Supplementary Fig. 11e), indicating that the immune cell compartment contributes to the anti-tumor effects of the ADC. Furthermore, analysis of pooled plasma samples from all mice in each treatment arm revealed elevated levels of tumor-necrotic-factor alpha (TNF-alpha) in ADC-treated groups compared to controls (Fig. 4j). TNF-alpha is known to be secreted by various innate immune cells in the TME33,34 and is recognized for its tumor-suppressive properties such as cancer cell killing through apoptosis or T-cell effector activation35. Altogether, the data suggest that the anti-ofCS ADC induces immunological cell death and highlights the capacity of the ADC to potentiate checkpoint inhibitors for added synergistic efficacy.
Targeting ofCS could form the basis of other therapeutic strategies besides ADCs. To investigate additional treatment platforms, we produced a bispecific immune cell engager comprised of C9 T-scFv (T-C9) fused to murine anti-CD3 scFv (T-C9-aCD3). The dual binding specificity of T-C9-aCD3 to ofCS and CD3 receptors was validated in ELISA and by flow cytometry using murine T-cells and murine 4T1 breast cancer cells (Supplementary Fig. 11a–c). Murine 4T1 (s.c.) breast cancer tumors were treated peritumorally with T-C9-aCD3 with and without adjuvant treatment with anti-CTLA4 checkpoint inhibitors. T-C9-aCD3 treatment led to complete tumor regression in all mice independently of co-administration of anti-CTLA4 (Fig. 4k). These results were confirmed in a syngeneic 4T1 model with tumor cells injected into the mammary fat pads (mfp) treated intravenously. In this model, administration of the anti-ofCS-aCD3 bispecific molecule stalled tumor growth (Fig. 4l). Moreover, as expected, we observed a higher prevalence of CD3+ and CD8 + T cells in the tumors of mice treated with the T-C9-aCD3 as compared to the control groups, correlating the immunological response with efficacy (Fig. 4m and Supplementary Fig. 11d).
Combined, these data indicate that our anti-ofCS antibodies can be functionalized as ADCs, and have the potential to serve as bispecific immune cell engagers for diverse cancer-targeting therapies.
Distinct CS sulfation patterns define antibody binding specificities
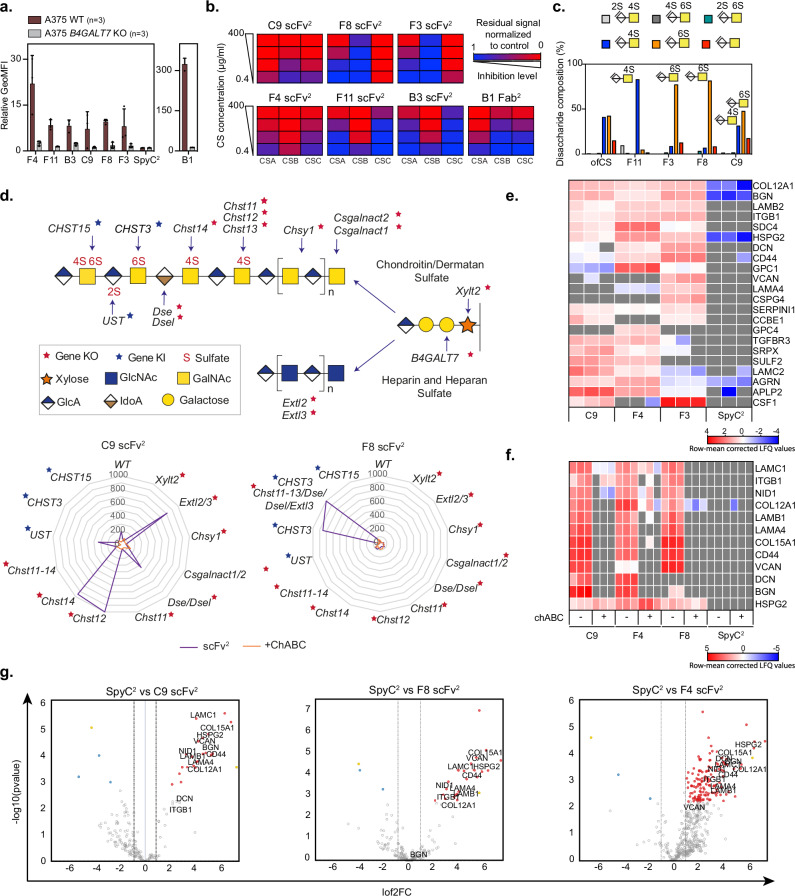
After defining antibody fragments targeting ofCS modifications with limited binding to CS in normal tissues, we attempted to determine the fine structure of the binding epitopes. First, we abrogated glycosaminoglycan biosynthesis in human A375 melanoma cells by CRISPR-cas9 KO of B4GALT7, encoding an enzyme involved in the formation of the tetra-saccharide linker for glycosaminoglycans on proteins36,37. KO of B4GALT7 resulted in complete loss of binding, indicating strict specificity for glycosaminoglycans (Fig. 5a). To confirm specificity to CS, we performed a glycosaminoglycan inhibition ELISA where binding to recombinant ofCSPG or purified ofCS was challenged by pre-incubation with commercial chondroitin sulfate A (CSA, Sigma), chondroitin sulfate B (CSB, Sigma), or chondroitin sulfate C (CSC, VWR) (Fig. 5b and Supplementary Fig. 12a). The ofCS binding of F8 was outcompeted by CSC at 0.4 μg/ml, indicating strong preference for 6-O sulfation (CSC). In contrast, C9 binding to ofCSPG was inhibited by all three reagents, suggesting specificity towards a more complex epitope. However, this pattern was not markedly different from the non-tumor-specific F4 antibody, even though F4, F11, and B3 were mostly inhibited by CSB (also named dermatan sulfate, DS) (Fig. 5b).
Fig. 5. Distinct CS sulfation patterns define the antibody binding specificities.
a Flow cytometry measured binding (relative GeoMFI +/− SEM) of 150 nM F4, F11, B3, C9, F8 and F3 scFv2, SpyC2 and B1 Fab2 to A375 WT melanoma cells or with B4GALT7 knocked out (KO) (n = 3). b Inhibition of the different antibody fragments’ ability to bind ofCS with soluble CSA, CSB, or CSC (0.4–400 μg/ml) measured in ELISA competition assay. c Composition of intact ofCS and the disaccharides binding the different antibody fragments (F11, F3, F8, C9 scFv2) by footprint analysis. d C9 and F8 scFv2 binding to genetically engineered CHO cells. The graphic overview (top) of the GAG biosynthesis pathways highlights the key genes studied and their assigned functions. Stars indicate genes investigated by knock-out (red) or knock-in (blue). Radar plot axis depict normalized mean fluorescence intensity of 200 nM C9 and F8 scFv2 binding to cells (n = 1–3) without (purple lines) or with chABC treatment (orange lines). e Heatmap of statistically significant hits obtained by Mass Spectrometry analysis of protein pull-downs from cell supernatants with C9, F4 and F3 scFv2 and SpyC2 as a control run in triplicates (n = 3). Two-sided unpaired t-test with Benjamin–Hochberg correction for multiple hypotheses testing was applied to determine differentially expressed proteins (significance threshold set to 1% and fold change to 2). Missing values are colored in gray, and lo2 transformed label-free quantification (LFQ) values are row mean normalized. f Heatmap representing row mean corrected LFQ values of hits obtained by MS analysis of CSPG pull-downs from human colon biopsy (H352) with C9, F4, and F8 scFv2 and SpyC2 in combination with chABC treatment as indicated, prior to enrichment analyzed as in (e). g Volcano plot of human colon biopsy (H352) processed and analyzed as in (e). with selected CSPG hits highlighted in black for each protein. Source data are provided as a Source data file.
We next examined the biochemical composition of the epitopes bound by the three ofCS specific antibody fragments, F3, F8, and C9, and F11 as a control (Fig. 5c). Immobilized scFv2 were incubated with ofCS, treated with chABC to remove unprotected saccharides and salt-eluted protected moieties interacting with the antibody fragments. Analysis showed that (i) F11 predominantly enriched 4-O sulfated CS (CSA and/or CSB); (ii) F3 and F8 predominantly enriched 6-O sulfated CS (CSC); and (iii) C9 enriched a heterogeneous and likely long CS chain containing domains with 4-O, 6-O, and non-sulfated disaccharides, suggesting a complex epitope (Fig. 5c). Interestingly, pull-downs without chABC treatment showed that F8 captured CS chains containing high amounts of 4-O and non-sulfated groups, suggesting that its epitope is a 6-O sulfated domain within a long CS chain with various CS motifs (Supplementary Figs. 12b, 13, and 14). In summary, we identified two distinct ofCS epitopes: one was defined by C9 and another by F3 and F8.
Next, we employed a genetically engineered CHO cell-based glycosaminoglycan array platform (GAGOme) to dissect the biosynthetic and genetic regulation of the antibody epitopes (Fig. 5d and Supplementary Fig. 12c). This array employs libraries of isogenic cells with knock-out (KO) and knock-in (KI) of key genes involved in glycosaminoglycan biosynthesis. The cell lines display distinct glycosaminoglycan structures on the surface allowing for flow cytometry-based binding analysis38–40. We then tested the ability of C9 and F8 to bind to these cell lines. While C9 scFv2 bound WT cells, the binding was abrogated by complete loss of glycosaminoglycan (Xylt2 KO) or selective CS biosynthesis (Chsy1 KO). Selective loss of HS biosynthesis (Extl2/3 KO) enhanced binding, suggesting a compensatory CS increase. Altogether, the data highlight a high C9 specificity to CS and no binding to HS. Abrogation of DS epimerization (Dse/Dsel KO) slightly enhanced C9 binding indicating a preference for CS. We then analyzed the requirement for 4-O-sulfation of CS, which is a differentially regulated biosynthetic step involving multiple isoenzymes41. C9 binding was lost with complete 4-O-sulfation abrogation (Chst11-14 KO) and loss of CHST11 isoenzyme (Chst11 KO). The loss of other 4-O-sulfotransferases (Chst12 KO or Chst14 KO) markedly enhanced C9 binding, confirming a preference for 4-O-sulfated CS and indicating that the binding epitope involves a more complex sulfation pattern. CHST12 4-O-sulfotransferase differs from the CHST11 isoenzyme by its capacity to introduce 4-O-sulfation at the non-reducing end of the CS chain42, leading to the production of shorter CS chains with end-terminal sulfation. Consequently, CHST12 depletion might lead to CS elongation. Surprisingly, introducing 6-O-sulfation into WT CHO cells by either of the two 6-O-sulfotransferases (CHST3 KI or CHST15 KI) completely abolished C9 binding. Notably, the disaccharide analysis suggests that these cells may have relatively short chains. In contrast to C9, the F8 scFv2 did not bind WT CHO cells. However, CHO cells lack endogenous 6-O-sulfation capacities, and introducing 6-O-sulfation (CHST3 KI) induced strong binding while introducing capacity for synthesis of 4,6-disulfated CS (CHST15 KI) did not. It should be noted that only small quantities of the 4,6-O-sulfated disaccharide were introduced. Moreover, the abrogation of 4-O-sulfation combined with the introduction of 6-O-sulfation (CHST3 KI in Chst11-13/Dse/Dsel/Extl3 KO) did not significantly affect binding (Fig. 5d and Supplementary Fig. 12c).
As CS is found on many proteoglycans (CSPG)43, we next identified the repertoire of CSPGs in tumor cells carrying ofCS chains. For that, we coated beads with C9, F4, and F3 scFv2, purified CSPGs from a pool of supernatant from four cancer cell line cultures (Fig. 5e), and analyzed them by mass spectrometry44. The generated heat maps, based on proteins that have previously been described to be modified with CS or ofCS44–47, revealed that the CS antibody fragments bound to a wide range of CS-modified proteins, both membrane-bound and secreted CSPGs (Fig. 5e and Supplementary Table 1). Interestingly, the proteins pulled down by C9 and F3 were not identical, confirming that the two antibody fragments target different epitopes. Similar experiments with patient-derived colon cancer biopsy homogenates confirmed the ofCS signature on a broad selection of CSPGs (Fig. 5f and Supplementary Data 1), including biglycan, syndecan-4, CD44, and versican, all previously described as overexpressed in multiple cancer types and/or playing roles in metastasis formation48–51. F4, recognizing a non-cancer specific glycan motif, pulled down the broadest range of CSPGs (Fig. 5g) indicating shared glycan motifs between ofCS and normal CS (Fig. 5g, Supplementary Fig. 12d, e and Supplementary Data 1). From the patient-derived colon cancer biopsies, we predominantly isolated ofCS-modified CSPGs predicted to be in the ECM, such as versican, collagens, and laminins (Fig. 5g, Supplementary Fig. 12d, e and Supplementary Data 1). Combined, these data show the existence of two distinct ofCS epitopes that can be found as modifications on a wider range of cell surface expressed proteins and secreted proteoglycans.
Antibody-CS complexing depends on oligomerization on a long CS chain
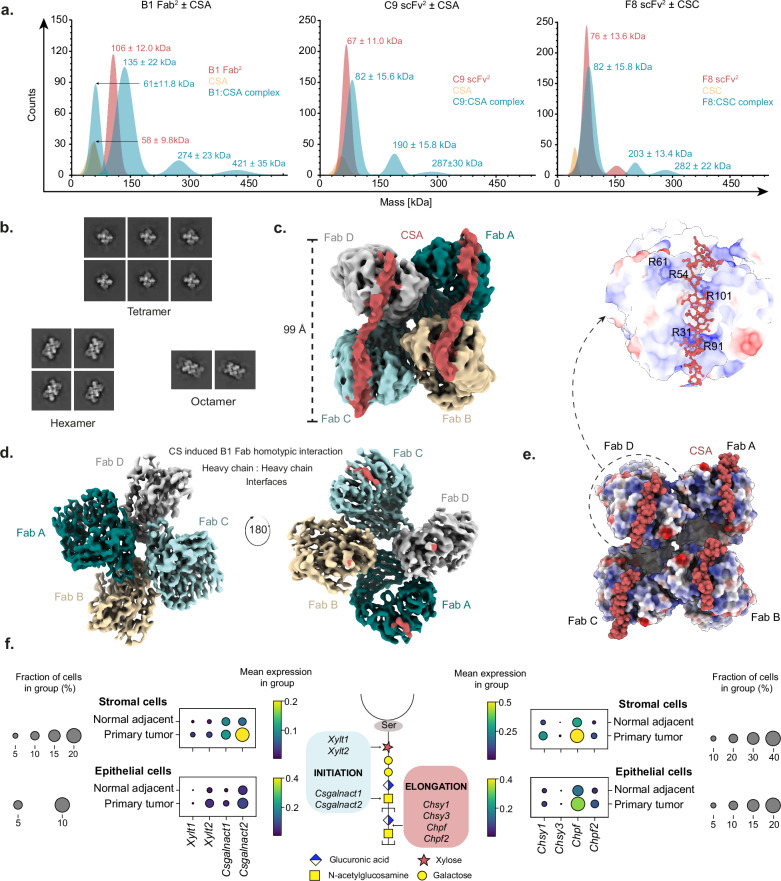
The specificity of VAR2CSA to ofCS is partially dependent on the glycosaminoglycan chain length21 with structural analysis identifying an interaction interface stretching over a CS 20-mer52, which we note is longer than the hypothetical interaction with CS and larger than common epitope domains bound by variable loops of an antibody fragment. To analyze the antibody fragments’ interaction with CS, mass photometry experiments were performed on different ratios of antibody-CS mixtures (Fig. 6a and Supplementary Fig. 15). The resulting mass photometry contrasts revealed complexes of greater mass than the antibody-CS complex alone and identified scFv2/Fab2:CS complexes as dimeric (200–300 kDa, i.e., a total of four monomeric scFv/Fab) and trimeric (300–400 kDa, i.e., a total of six monomeric scFv/Fab) structures. Such oligomers could arise from monovalent antibody binding on a long CS chain, like pearls on a string, or could alternatively be higher-order oligomers in a single interaction. To define this better, we performed cryo-EM analysis of antibody fragments in complex with CS (Supplementary Fig. 16). A cryo-EM structure of the monomeric B1 Fab complex was obtained whereas the C9 and F8 structures did not resolve. Despite sharing some ofCS-specific features, B1 showed residual binding to CS in normal tissues (Fig. 2a). Initial 2D classification of the B1 Fab:CS complex revealed multimerization, suggesting Fab-to-Fab homotypic interaction upon CS ligand binding (Fig. 6b and Table 1). High-pressure liquid chromatography (HPLC) analysis on apo B1 Fab and B1 Fab complexed with CS at varying molar concentrations confirmed the B1 Fab multimerization as CS-ligand induced. Further data processing on tetrameric Fabs revealed a Fab-Fab homotypic interaction (Fig. 6c and Table 1). Outputs from 3D flex train followed by 3D flex generator promulgated high inter-Fabs conformational movements, with restricted flexibility at the Fab-Fab homotypic interfaces (Supplementary Movie 1). Interestingly, two CS cryo-EM densities traversing across the variable regions of the Fabs were observed. One density corresponded to a CS glycan chain traversing across Fab A to Fab B and the other traversing across Fab C to Fab D (Fig. 6d). Molecular Dynamics flexible fitting (MDFF) to model two CS glycan chains onto tetrameric Fab and a subsequent 1000 ns explicit solvent atomistic MD simulation of the Fab tetramer bound to two CS glycan chains demonstrated that the CS chains remained attached to the Fab surface (Supplementary Movie 2). The observed high-avidity binding to CS is likely facilitated by these occurring homotypic interactions, contributing to the antibody fragment’s ability to target different sulfation patterns along a CS chain. While the bound CS chain was poorly resolved, the analysis of the electrostatic surface potential revealed a positively charged surface on the variable chain fragment, suggesting charge-dependent interactions with the highly negatively charged CS chain52 (Fig. 6e). The length of the CS glycan chain traversing the Fabs was 99 Å, indicating that a long CS chain is necessary for the formation of high-avidity binding. Previous studies demonstrated that VAR2CSA specificity and affinity depended on a long CS structure21. Moreover, our findings indicated that the B1 homotypic interaction covers a remarkably long CS chain, and observation of C9 and F8 forming oligomers when complexed with CS further implies the presence of a recurring epitope along a long chain (Fig. 6a).
Fig. 6. Antibody-CS complexing depends on oligomerization on a long CS chain.
a Mass photometry contrasts of dimerized antibody fragments (B1, C9, and F8) analyzed alone (red) or in combination with soluble CSA or CSC (green). Soluble uncomplexed CS (yellow) was analyzed as a control. b 2D classification of B1 Fab:CSA complex from cryo-EM. c Cryo-EM density map depicting CS glycan chains (red) traversing across B1 Fabs in a tetramer complex. d Heavy chain:heavy chain homotypic interactions of B1 Fab. e Structure of B1 Fab interaction with CSA showing electrostatic surface potential of the complex. Above, a zoom-in of the dashed box is shown. f Single-cell RNAseq analysis of CS-related genes in a non-small lung carcinoma (NSCLC) transcriptome atlas (left and right) and a schematic overview of studied CS synthesis pathways (middle).
Table 1.
Cryo-EM collection, refinement, and validation statistics
| Homotypic interacting B1 Fabs bound to chondroitin sulfate A (EMDB-17362) (PDB-8P2E) | |
|---|---|
| Data collection and processing | |
| Microscope | Titan Krios |
| Camera | Falcon III |
| Magnification | 130,000 |
| Voltage (kV) | 300 |
| Electron exposure (e–/Å2) | 40 |
| Defocus range (μm) | −1.2 to −2.8 |
| Pixel size (Å) | 0.832 |
| Symmetry imposed | C1 |
| Initial particle images (no.) | 1,257,314 |
| Final particle images (no.) | 235,115 |
|
Map resolution (Å) FSC threshold (0.143) |
3.3 (based on 3DFSC) |
| Map sharpening B factor (Å2) | 107.8 |
| FSC model (0/0.143/0.5) | 2.8/3.2/3.8 (Masked) |
| Refinement | |
| Initial model used (PDB code) | N/A (Alphafold) |
| Model composition | |
| Non-hydrogen atoms | 12,786 |
| Ligands | N/A |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.018 |
| Bond angles (°) | 1.693 |
| Validation | |
| MolProbity score | 1.33 |
| Clashscore | 5.1 |
| Poor rotamers (%) | 0.14 |
| Ramachandran plot | |
| Favored (%) | 97.73 |
| Allowed (%) | 1.92 |
| Disallowed (%) | 0.36 |
| Cβ outliers (%) | 0 |
|
Model vs data CC mask/box/peaks/volume |
0.72/0.82/0.64/0.7 |
To determine what cells in the tumor environment can produce longer CS chains, we re-analyzed previous data from a high-resolution single-cell atlas of 309 non-small cell lung cancer patients (NSCLC)53. Interestingly, we found that primary tumor cells had increased expression of the CS polymerization factor Chpf, and the CS synthesis initiation gene Csgalnact2. Both initiation and elongation factors were also upregulated in tumor-infiltrating stromal cells compared to normal adjacent stromal cells (Fig. 6f and Supplementary Figs. 17a–c). Moreover, a subset of plasma cells also upregulated some of these factors with no difference between normal and tumor compartments (Supplementary Fig. 17c), highlighting that CS polymerization is just one of several requirements for the generation of the ofCS epitope.
In summary, these data suggest that the scFvs, similarly to rVAR2, bind an extended CS chain enabled through oligomerization on the epitope21,52. Our results further indicate that the oncofetal epitopes arise as distinct sulfation domains in an extended CS chain.
Discussion
ofCS is present in embryonic and malignant tissues but absent from normal or inflamed tissues27. These specific ofCS modifications, attached to different membrane-bound or ECM CSPGs44,54,55, could play crucial roles in cancer cell motility, proliferation, and immune evasion47,56. Until now, the malaria-derived rVAR2 protein has been the only ofCS-targeting reagent, widely used for ofCS characterization in human tumors27,46,47,54,57–62. Its interaction with ofCS stretches over 100 Å and the specificity relies on distinct sulfation patterns along the CS chain22,63. Notably, antibody epitopes typically cover 3–5 monosaccharides64 (10–15 Å) making it challenging to generate antibodies binding multiple disaccharides. Despite extensive efforts to immunize mice, rats, and rabbits with purified ofCS, no detectable humoral immune responses were elicited, possibly due to mammalian restrictions in developing antibodies against crucial fetal epitopes and the low immunogenicity of carbohydrates65. To address these challenges, we employed phage display technology, utilizing different commercial antibody libraries. Our screening involved panning on ofCS-modified molecules, counter panning on similarly charged HSPG, and validation through rVAR2 binding competition. From all libraries, we identified a few CS-specific antibody fragments, among which only three (C9, F3, and F8) exhibited high tumor specificity. These antibodies target two distinct ofCS features, with C9 preferring 4-O sulfation, and F3 and F8 preferring 6-O sulfation.
The distinct CS features recognized by C9, F3, and F8 were shown abundantly expressed in various human cancers, both in primary tumors and metastatic lesions. Notably, their expression in benign tumors, inflamed tissues, and healthy specimens was minimal, despite the wide abundance of CS in these tissues. We employed multiple strategies to characterize the molecular epitopes recognized by C9 and F8/F3, which consistently indicated that the antibodies targeted intricate sulfation patterns present over long stretches of CS. The complex biosynthesis of CS chains, involving numerous isoenzymes with distinct properties, allows great diversity in CS modifications and results in varied molecular epitopes for glycosaminoglycan-binding proteins. We defined 4-O and 6-O sulfation as critical events for C9 and F8/F3 binding, respectively. Gene KI and KO studies demonstrated complex regulation of the glycosaminoglycan biosynthesis machinery for ofCS expression including coordinated regulation of multiple sulfotransferase isoenzymes that either promote or inhibit the ofCS formation. Furthermore, the data point to changes in the CS biosynthesis that favor longer CS chains. Combined, we found that ofCS encompasses at least two distinct molecular epitopes with domains of 4-O- or 6-O-sulfated motifs over longer CS stretches.
Pull-down experiments on patient-derived colorectal biopsy homogenates validated the ofCS modification of a wide range of proteins, such as VCAN and distinct collagens. Although abundantly expressed in normal tissues, VCAN’s role in cancer has been well described, encompassing growth factors tethering, tumor immune evasion, and angiogenesis66. Thus, our findings suggest that the unique ofCS modifications could distinguish these proteins in malignancy. Furthermore, a contrasting expression of the two ofCS epitopes on various protein cores was observed, demonstrated by the selection of proteoglycans enriched by C9 and F8/F3. However, it should be noted that these results might be confounded by the generally lower extraction of membrane-bound proteins with poor solubility during the procedure.
Various cell types contribute to the presentation of ofCS within the tumor micro-environment, as supported by our RNAseq data on NSCLC, and tissue staining revealing expression in both the stromal compartment and on the surface of tumor cells. Consequently, the composition of the tumor microenvironment, varying across tissue types and disease progression, may influence the exact expression pattern of ofCS.
We demonstrate that the antibody fragments’ specificity to distinct CS types arises from an interaction along a long CS chain. Using mass photometry, we demonstrated that antibody binding to CS resulted in macromolecular assemblies indicative of self-organization into multimeric structures. Cryo-EM analysis of CS:B1 Fab fragment complex revealed that the B1 variable regions forming oligomers were primarily composed of tetramers binding to two CS chains of ~100 Å (or one chain of at least 200 Å). Only a few examples of this type of homotypic antibody fragment oligomerization upon ligand binding have been described67 and is likely a very rare mechanism. This could account for the low number of specific hits obtained from our extensive screening efforts and may explain how the selected antibody fragments achieve their high affinity for ofCS.
The ofCS-specific antibody fragments displayed effective tumor targeting across various animal models, leading to the development of a preclinical data package for future clinical development. F8 and C9 demonstrated specific tumor localization, illustrating potential applications in tumor imaging and targeted therapeutic delivery. Remarkably, C9 and F8/F3, whether formulated as ADCs or anti-CD3 bispecific molecules, exhibited broad and robust therapeutic efficacy. In immunocompetent tumor models, the ADC efficacy was linked to the immune system’s involvement, which could be further boosted by the co-administration of a checkpoint inhibitor. Importantly, mice, like humans, were fully capable of producing the two ofCS epitopes during fetal development, making the murine system suitable for evaluating potential off-tumor binding and associated. Encouragingly, repeated treatments of the ADCs in four different murine tumor models showed no signs of distress nor weight loss, suggesting that the ofCS epitopes are primarily restricted to the malignant tissue compartments. Interestingly, the ofCS expression level in murine tumor models (CDX and PDX tumors) was lower compared to the levels found in most human tissue biopsies. Particularly, the F8/F3 ofCS epitope was primarily present in selected PDX models and only expressed at lower levels in CDX models. This stands in contrast to the high ofCS density in human malignant tissues that is probably due to low amount of ofCS expressing CAFs in CDX models, as demonstrated by lack of F8 binding to murine CAFs. This implies a potential underestimation of the therapeutic potential of the antibody fragments in mice. The lower expression of ofCS in murine CDX/PDX systems can likely be ascribed to the inability of these models to fully recreate the tumor stroma and ECM found in primary tumors. This was supported by our analyses in NSCL cancers showing that infiltrating stromal cells overexpress CS elongation factors compared to stromal cells outside the tumor. Overall, the antibody fragments reported in this study could provide an effective route for targeting the vast majority of cancers. They have the potential to deliver cytokines, immune cells, radiotherapies, immunotherapies, or ADC payloads, to cancer cells and the tumor microenvironment.
In conclusion, we developed antibody fragments that can access ofCS in a broad range of solid tumors. Specificity to the ofCS epitopes depends on oligomerization of the antibody fragments on the CS chains, allowing the detection of different tissue-specific sulfation patterns along an elongated CS chain. We defined two distinct ofCS epitopes and the antibody fragments exhibited high cancer-specific reactivity in most tumors, holding the potential for precision targeting of tumors. Preclinical studies in rodent models demonstrated the safety and efficacy of these antibody fragments for targeted cancer therapy formulated as ADCs and as bispecific immune engagers. These results augment further clinical development and translation of ofCS biologics into broadly effective anti-cancer treatments, imaging, and tissue diagnostics.
Methods
Ethical approval
Animal studies in Copenhagen were approved by the Animal Experiments Inspectorate (P23-118, P23-103, P23-071, and P21-P19). Ethical approvals were obtained from Dyreforsøgstilsynet Danmark.
Experiments on LTL370 Prostate Patient-Derived-Xenograft (PDX) models were approved by the Animal Care Committee at the University of British Columbia (A22-0206, A19-0324).
Experiments on PDAC and Sarcoma PDX models were done at a contract research organization (CRO EPO Berlin), in accordance with the United Kingdom Coordinating Committee on Cancer Research regulations for the Welfare of Animals and of the German Animal Protection Law and approved by the local responsible authorities. The tissues for immunofluorescence staining were obtained from commercialized tissue micro-array (US BioMax, Inc). Approval of the local ethical committees was given, and informed consent was obtained from all patients prior to sample acquisition and experimentation. Their tissue samples were excised by licensed medical doctors, received from certified hospitals, collected with informed consent from the donors and relatives, and diagnosed and identified by at least two evaluators. All patient data were used in an anonymized fashion according to the ethical guidelines.
The in vitro use of colorectal tumor biopsies obtained from Saelland University Hospital (Køge) was approved by the Danish Regional Ethical Committee (De Videnskabsetiske Komiteers) (SJ-826).
Phage display generation of antibody fragments binding ofCS
The phage display technology was utilized to generate human antibody fragments. Phage libraries were bio-panned over 3 to 4 rounds for successful enrichment of ofCS binders. Each round consisted of three main steps. Firstly, the panning step selects the positive binders to our antigen (ofCSG or ofCS) by incubating phages with ofCS-modified molecules. Next, the counter-panning step was employed to deplete low affinity and unspecific binders on a counter-antigen (PG or naked beads). Lastly, the selection step was performed either positively on an ofCS-carrying molecule or negatively on an ofCS-depleted molecule. At each round, potential binding candidates were eluted, amplified in Escherichia coli (E. coli), and tested over a succession of Polyclonal phage ELISA against ofCSPG and PG. The clones displaying the most promising binding properties were sequenced and further tested on Monoclonal phage ELISA to assess their binding to ofCSPG and PG.
The first strategy involved two libraries, namely the naive LiAb-SFMAXTM (scFv) and the synthetic HuCAL (Fab)23,24 where ofCSPG and were used as antigens and counter-antigen, respectively. The counter-selection was carried out against HSPG (Sigma, #H4777) to deplete binders to negative charges. In this method, an additional competition ELISA was performed to further select clones sharing some epitope with VAR2CSA protein. The second strategy was run on the ALTHEA Gold semi-synthetic (scFv) library25, where purified ofCS was used for panning, and ofCSPG was used as the positive selection target.
Attana QCM biosensor binding kinetic measurements
Two LNB carboxyl chips were equilibrated at a flow rate of 10 μL/min in coupling buffer composed of 10 nM HEPES, 150 nM NaCl, 0.005% Triton-x100 (pH 7.4) on an Attana A200 quartz crystal microbalance (QCM) biosensor (Attana AB). Both chip surfaces were activated using S-NHS and EDC and conjugated through primary amines with 50 μg/ml Streptavidin, following the manufacturer’s instructions. Blocking of the chip surface was done using 1 M ethanolamine. Next, 100 μg/ml placental-purified biotinylated ofCS diluted into 1xPBS (pH 7.4) was injected over the A-channel chip. The B-channel reference chip was kept uncoated to assess the background binding of the proteins. The success of each coating step was verified by a frequency shift, indicating a mass change after coupling. After equilibration in running buffer (1xPBS, pH 7.4), a 2-fold dilution series of each analyte from 200 nM to 25 nM was injected at a flow rate of 20 μl/min to assess the protein binding to ofCS. Between each injection, the chip was regenerated using 0.01 M NaOH. The binding kinetics (kon, koff) were fitted using a 1:2 binding model in TraceDrawer software (Ridgeview Instruments AB).
Cell culture
Adherent cells were washed 3× in sterile 1xPBS depleted in Calcium and Magnesium to remove residual fetal bovine serum (FBS) and detached with 0.05% trypsin solution containing EDTA (GibcoTM, #25300-054). Pre-warmed fresh media was added to inhibit the trypsin and cells were transferred to a tube before centrifugation for 5 min at 350 × g. Suspension cells were directly transferred to a tube and centrifuged the same way. Supernatants were discarded and cells were resuspended in fresh media before being seeded in a new flask containing warm culture media. Cancer cells were maintained in culture at 70–80% confluency in a 5% CO2 atmosphere at 37 °C and split 2 to 3 times per week. Culture media and growth conditions (adherent or in suspension) suggested by the vendor were used for each cell line. The list of cancer cell lines used for flow cytometry experiments is presented in Supplementary Table 2 including cell type (hematological, epithelial, mesenchymal), origin (human, mouse), disease, and accession number (Cellosaurus ID).
Genetic engineering and handling of CHO cells were performed as previously described38.
Flow cytometry binding experiments on cancer cells
Flow cytometry was employed to evaluate the binding of antibody fragments to cancer cell lines, human white blood cells (WBC), and engineered CHO cells.
Adherent cancer cells were cultured until they reached 80% confluency and then detached with a non-enzymatic CellStripper solution (Corning #25-056-Cl). Following dissociation, cells were counted on a Nucleocounter® (NC-200, ChemoMetec), spun down at 400 × g for 5 min, and reconstituted in 1xPBS with 2% Fetal Bovine Serum (PBS2). Suspension cells were directly counted, spun down, and resuspended in PBS2. Binding assays were performed with 100,000 cells incubated 30 min at 37 °C in PBS2, or with 0.2 mg/mL chABC as control. 300 nM V5-tagged scFv2 (F3, F4, F8, F11, B3, C9), FLAG-tagged Fab2 (B1) or His-tagged SpyC2 diluted in PBS2 was added and incubated for 30 min at 4 °C. After a couple of washes in PBS2, 3 μg/ml mouse anti-FLAG antibody (Sigma, #F3165) was added to the B1 wells and incubated 30 min at 4 °C. Following a second round of washes, secondary antibodies were added to the wells: 1:500 Anti-V5-FITC (Invitrogen, #R963-25), 1:100 Anti-mouse-IgG-FITC (Vector Laboratories, #FI-2000) or 1:200 Anti-His-Alexa488 (Qiagen, #1019199). After three final washes, cells were analyzed by flow cytometry on a BD Fortessa 3-laser analyzer (Becton Dickinson).
Binding assays to CHO cells by flow cytometry were performed with ~100,000 cells incubated for 30 min at 37 °C with PBS or as control 0.2 mg/mL chABC. After washing with PBS with 1% Bovine Serum Albumin (BSA), cells were incubated with 200 nM C9 or F8 scFv2 for 30 min at 4 °C. Cells were washed with PBS with 1% BSA and incubated with 1:500 anti-V5-FITC antibody (Invitrogen, #R963-25) for 30 min at 4 °C. After three final washes, cells were analyzed by flow cytometry on a BD Fortessa 3-laser analyzer (Becton Dickinson).
All acquired data were analyzed on FlowJoTM (Becton Dickinson). Cell population was selected on the Side-Forward scatter plot (SSC/FSC) by discarding dead cells, and debris based on to their size. Single cells were picked on the FSC-H/FSC-A plot for further analysis. Geometric Mean intensity or Mean Intensity were calculated. Signals were min–max normalized to the antibody control.
Binding assays to WBC were performed on fresh blood drawn from a healthy donor, collected in K2EDTA tubes, and processed within 1 h of collection. Red blood cells were lysed in a lysis buffer containing NH4Cl, KHCO3, and EDTA. WBCs were recovered, counted, and resuspended in 1xDulbecco’ Phosphate buffer (DPBS) with 2% Fetal Bovine Serum (GibcoTM, #A4766801) (DPBS2). The assay was performed as described above on ~200,000 cells where live cells were stained with Zombie aqua viability marker (BioLegend) for 15 min at room temperature. Subsequently, 300 nM V5-tagged scFv2 (F3, F4, F8, F11, B3, C9) or His-tagged SpyC2 was added in combination with 1:200 Anti-CD45-Alexa647 (Invitrogen, clone HI30). As a compensation control, 1:200 Anti-CD45-FITC (Miltenyi Biotec, clone 5BS1) was used. After a last round of washes, cells were analyzed on a CytoFLEX S instrument (Beckman Coulter). Acquired data were analyzed as described above where single cells were picked on SSC-H/SSC-A, gated on live cells as defined by a dead cell marker, and subsequently on CD45+ cells, as defined by the unstained cell marker. Geometric Mean intensity was calculated and normalized to the antibody control.
Anti-ofCS staining of cancer cells spiked into blood
Blood from healthy individuals was collected in K2-EDTA-tubes and processed within 1 h of collection. F8, F4, and B1 constructs were biotinylated as described above and subsequently, coupled to a PE fluorophore, for 30 min at room temperature. A549 cancer cells were grown to ~80% confluence and detached using StemPro® Accutase (Gibco, #A11105-01). The cells were resuspended in a 2 ml conditioned medium and the cell concentration was counted manually using a hemacytometer. Right before spike-in, the cells were pre-diluted in Dulbecco’s PBS without Calcium and Magnesium (Sigma-Aldrich, #D8537). 10 µl of the cell suspension (10,000 cells/ml) was added to 1 ml of healthy donor blood for spike-in of 1000 or 100 cells.
The red blood cells (RBC) were lysed by diluting the spiked blood samples in RBC lysis buffer reaching a final concentration of 0.01 M Potassium hydrogen carbonate, 0.155 M Ammonium chloride, and 0.1 mM EDTA. Following RBC lysis, the samples were centrifuged at 400 × g at RT for 8 min and washed with 6 ml Dulbecco’s PBS. The cell pellet was gently resuspended in Dulbecco’s PBS supplemented with 2% FBS and kept on ice. The samples were stained for 30 min at 4 °C with 4 nM of PE-labeled F8, B1 or F4 in combination with anti-CD45-Alexa647 (1:10, Invitrogen, clone: HI30, #51-0459-42). The samples were washed twice in 0.5 ml of Dulbecco’s PBS + 2% FBS following fixation by 4% paraformaldehyde. The cell nuclei were labeled with 5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI, Fischer Scientific, #D1306), and the samples were mounted on microscopy slides and scanned on a fluorescent scanning microscope. For the F4 and B1 samples, the slides were scanned on a Zeiss Axio Z1 automated slide scanner (×20 magnification, 0.8 NA objective). The F8 samples were scanned on CytationTM 5 Cell Imaging Multi-Mode Reader (×20 magnification, 0.8 NA). The images were analyzed by the Gen5 software (BioTeK, Version 3.10) or the Zeiss Zen blue software.
Immunofluorescence staining of paraffin-embedded fixed tissues
Murine tissues were harvested from our mice model, fixed in 4% paraformaldehyde overnight, and transferred to 70% ethanol before paraffin embedding. Sections slides were prepared by the Histolab Core Facility at the University of Copenhagen at 3 µm thickness using a microtome. Human tissue-micro-arrays (TMA) were purchased from Biomax. The list of arrays used in this manuscript is presented in Supplementary Table 3.
Glass slides were baked for 30 min at 60 °C to melt paraffin and allow adhesion of tissue to the glass. Deparaffinization and rehydration were executed in a succession of treatments in Xylene Substitute (Tissue-Tek®, Sakura, #94-1466), absolute ethanol (Honeywell, #02860), 96% Ethanol (VWR, #20824.385) 70% Ethanol (VWR, #83801.360), tap water and distilled ultra-pure water. Immunofluorescence staining was carried out in a humidified chamber to avoid drying of the tissues. In some experiments, in-house produced chABC enzyme was applied to the control sections at 0.2 mg/ml for 1 h at 37 °C. The untreated tissues were incubated in the same conditions in 1xPBS. The tissues were blocked for 1 h with 1xPBS, 5% Fetal Bovine Serum (FBS) (GibcoTM, #A4766801), 1% BSA (Sigma, #A3059) and washed 2 times with 1xPBS. Tissues were then incubated for 1 h at room temperature with 25 nM of V5 tagged-scFv2 or FLAG-tagged-B1 Fab2 diluted in assay buffer (1xPBS, 0.25% BSA). Staining was followed by a 3 × 5 min wash with 1xPBS before pursuing with 1 h incubation of Anti-V5-Alexa647 (Invitrogen, #451098) or Anti-Flag-Alexa647 (Abcam, #ab245893) antibodies in the dark. A final 3 × 5 min wash was completed before cell nuclei staining with DAPI (Life Technologies, #D1306) in 1xPBS, and a last wash in ultra-pure water. The tissue slides were dried for 30 min and mounted with an aqueous mounting media (Dako, #33025). Imaging was done on a Zeiss Axio Z1 automated slide scanner (×20 magnification, 0.8 NA objective), and image analysis on the ZEN lite software. The mean signal intensity in the Alexa647 channel was quantified for each section on Fiji software and plotted. Cores without DAPI were excluded from the analysis. Data were plotted on GraphPad (Prism), and a parametric two-sided unpaired t-test was applied for comparing groups.
Establishment of cell-derived xenograft models (CDX)
Female BALB/cAnNRJ (#000651) (CT26, 4T1), female CB-17/lcr-Prkdc scid/scid/Rj (#001803) (Karpas299), and female BALB/cAnNRJ-Foxn1nu/nu (# mice (A549, MiaPaca2, A375 WT and A375 Chst11 KO) were purchased from Janvier 000711) age of 6–8 weeks. The mice were kept in a 12-light/12-dark cycle period at 40–60% humidity with an ambient temperature of 20 °C at the animal facility in the Department for Experimental Medicine at the University of Copenhagen in accordance with the FELASA Rodent Health Surveillance program.
All cells were thawed and cultured up to at least three passages and maintained at 50–80% confluency before injections in mice. Cell suspension of Karpas injected subcutaneously (s.c.) (1 million cells/mouse), A549 s.c. (1 million cells/mouse), 4T1 s.c. (75,000 cells/mouse), 4T1 mfp (10,000 cells into the mfp), CT26 s.c. (250,000 cells/mouse), A375 WT and Chst11KO s.c. (1 million cells/mouse) prepared in PBS were injected subcutaneously or to the mammary fat pad (mfp) using a 25 G needle in the left flank of the mice to establish the xenograft models. Tumors were measured using a caliper tool and volume was calculated according to the formula 0.5236* length*(width)2.
Establishment of patient-derived xenografts (PDX)
PDX models of PDAC (Panc12536) and sarcoma (Sarc9593) were established from fresh patient tumor fragments at EPO Berlin CRO. Tumor fragments of 3 × 3 mm were subcutaneously transplanted to female Rj:NMRI-Foxn1nu/nu nude (#007850) recipient mice. into the left flank. Mice were maintained under sterile and controlled conditions (22 °C, 50% relative humidity, 12 h light–dark cycle, autoclaved food and bedding, and acidified drinking water). Tumor growth was measured twice weekly, and tumors were routinely passaged at tumor volume (TV) = 1 cm³.
The LTL370 prostate tumor model was established in male NRG mice (#007799) following the method described previously68 where tumor volumes (mm3) were calculated according to the formula: (π*length*width*depth)/6.
IVIS localization study in mice models
Mice with a tumor size of 150–250 mm3 were included in the study and their tail vein was dilated by heat to facilitate protein injection. Each study (4T1, Karpas, A549, MiaPaca2, LTL370) involved the injection of 50 µg of Alexa-750 labeled scFv2 fragment per mice in the tail vein. Equimolar amount of control protein was injected. The sex of the mice was taken into account for assessing antibody localization in reproductive organs (testis or ovary). Following injection, mice were immediately scanned using an In Vivo Imaging System (IVIS) to confirm appropriate injection and circulation of proteins. Subsequent scanning was conducted 24 h and 48 h post-injection to monitor tumor localization while the mice were anesthetized. Ex vivo scanning was performed after 24 h or 48 h to examine organ distribution. Mice were euthanized by cervical dislocation (or isoflurane then CO2 for the LTL370 model) and whole body was perfused by injecting 1xPBS through the lower left ventricle of the heart while it was still beating. Tumor, brain, heart, skin, muscle, lung, spleen, kidney, and liver were harvested and washed in 1xPBS before display on the mat for scanning. Manual exposure at an excitation wavelength of 752 nm and an emission of 776 nm, medium binning, F-stop 1 was used for each scan to have the same exposure time and ensure comparability of different conditions. Ex vivo surface fluorescence of the organs was quantified as average radiance efficiency ([p/s/cm2/sr]/[µW/cm2]) by using Living Image software (Perkin Elmer).
Antibody-drug-conjugate drug response testing on CDX models
Mice were randomly grouped based on an average tumor size of 100–150 mm3. All treatments were administered i.v. In the Karpas299 efficacy study, mice were administered 3 treatments based upon 2.14 nmol of MMAE for both treatment groups ABD-C9 scFv ADC (Drug:Antibody ratio, DAR = 0.6) and ADC control (ABD-SpyC, DAR = 2). In the CT26 study (Fig. 4b), mice were treated 3 times based upon 0.6 nmol of MMAE with ABD-T-C9 ADC (DAR = 2), and ADC control (ABD-SpyC-DAR = 2). In the second CT26 study (Fig. 4d), mice were treated 3 times based upon 1.08 nmol of MMAE, with ABD-C9-scFv ADC (DAR = 2.1) and semi-inactivated ABD-C9-scFv ADC (DAR = 2.1). In the A375 WT and Chst11 KO studies, mice were treated 3 times based upon 2.14 nmol of MMAE, with ABD-C9-scFv-ADC (DAR = 0.9) and ADC control (ABS-SpyC, DAR = 2). In the CT26 study combining ADC with checkpoint inhibitors (Fig. 4h), mice were treated 1 time with 2 mg/kg of ABD-T-C9 ADC (DAR = 2) intravenously, 10 mg/kg of anti-PDL-1 (PD1) antibody (CD279, BioXCell) intraperitoneally, or a combination of both. All treatments and measurements were single-blinded. Using the principles of the three Rs (Replacement, Reduction, and Refinement) and previous results, we chose the smallest possible sample sizes. Mice were euthanized when either the experimental or humane endpoint was reached. Humane endpoints were set at: tumor volume exceeding 1000 mm3, weight loss <20%, ulcerations exceeding a number of 3, 5 mm in length or 3 mm in depth, and/or when the general well-being of the animal was impaired e.g., normal body functions compromised, lack of food and liquid intake, defecation, urination or impaired motor skills, and/or lack of activity and inability to stand on the hind legs, and/or signs of respiratory distress. The experimental endpoint was aimed at achieving tumor regression or better survival in the treatment groups. When sub-cutaneous tumors were palpable, a digital caliper was used to measure tumor dimensions in width [mm] and length [mm] always reporting the larger diameter as width and the perpendicular diameter as length. Individual tumor volumes were calculated by the equation for all of our CDX model: TV[cm3] = lenght × width2 × 0.5. When relevant, following anesthesia and cervical dislocation, blood samples from mice were collected in EDTA tubes from the eye and put on mice. Plasma samples were prepared within 1 h after collection by centrifuging the blood at 1500 × g for 15 min at 4 °C. TNF-alpha contents in plasma were measured using a kit (BioLegend).
Antibody-drug-conjugate drug response testing of PDX models
PDX models of PDAC (Panc12536) and sarcoma (Sarc9593) were analyzed for their response against different ADCs. Tumors were measured and volumes calculated as described in the section above. Once the tumors reached the predefined mean starting volume of about 0.1 cm³ mice were randomly assigned to one control and treatment groups. PDAC PDX mice were treated biweekly (nine times in total), with ABD-C9-T ADC (DAR = 2) and control ADC (ABD-SpyC; DAR = 3.7) using a dosage of 8 and 2 mg/kg respectively, corresponding to equimolar amounts of MMAE. Sarcoma PDX mice were treated biweekly (eight times in total), with ABD-T-F3 ADC (DAR = 2.4), ABD-F8-scFv ADC (DAR = 1.1), and control ADC (ABD-SpyC; DAR = 3) using a dosage of 8, 7.2, and 2 mg/kg, respectively, corresponding to equimolar amounts of MMAE. For evaluation of therapeutic response, tumor burden was monitored. Health status and body weight (BW) for all mice were recorded at least twice weekly to control for toxic adverse effects. Mice were euthanized when either the experimental or humane endpoint was reached. Humane endpoints were set at: tumor volume exceeding 1.5 cm3, body weight loss exceeding 20% of initial weight, or severe clinical observations as described in the section above. All data were plotted on GraphPad (Prism) software and edited on Illustrator (Adobe).
Anti-CD3 bispecific drug response testing in a 4T1 xenograft model
4T1 mice models (s.c. and mfp) were tested for their response to bispecific treatment. 4T1 s.c. tumors were peritumorally treated with 20 μg T-C9 coupled to 14.5 μg aCD3 (equal molar amounts) in PBS (10 μL in total) on days 10, 12, and 14 post-tumor inoculation. 100 μg murine anti-mCTLA-4 (Invivofit) was intraperitoneally administered on days 10, 12, and 14 after tumor inoculation. 4T1 mfp tumors were given intravenously 5 treatments on days 14, 17, 20, 24, and 29 after tumor inoculation with 40 μg T-C9, 100 μg T-C9-aCD3 or 60 μg aCD3. Experimental and humane endpoints were defined as in the previous section “Antibody-drug-conjugate drug response testing on CDX models”. Tumors were measured and volumes were calculated as described in the same section.
Flow binding on tumor cell suspensions
Balb/c-mice were injected with either 4T1+ luc cells (100,000 cells/mouse) into the mammary fat pad (mfp) or CT26 cells (250,000 cells/mouse) subcutaneously. When the tumors reached 100 mm3 in size the first treatment injections were performed, followed by 2 subsequent injections every 3 days. The treatments used were (i) PBS, aCD3 (i.v., 53 μg/mouse), or T-C9-aCD3 (i.v., 120 μg/mouse) in the 4T1 study, (ii) PBS, anti-PDL-1 (i.p., bioXCell 10 mg/kg), anti-PDL-1 (i.p.)+T-C9-ADC (i.v., DAR = 2, 2 mg/kg), or T-C9-ADC (i.v., DAR = 2, 2 mg/kg) alone in the CT26 study. Mice were euthanized and the tumors were harvested, disrupted through a cell strainer, and washed in PBS2. CD16/CD32 mouse BD Fc block (clone 2.4G2, BD Biosciences) was added, and the cells were washed and stained with antibodies binding to CD3 (clone 17A2, PerCP/Cy5.5, BioLegend, Cat#100218), CD4 (clone GK1.5, APC/Cy7, BioLegend, Cat#100414), CD8 (clone 53-6.7, APC, Invitrogen, Cat #48-0441-82), or CD45 (clone 30-F11, PE, Biolegend, Cat#103106). After a last wash with PBS2, cells were resuspended in PBS2, and read on on BD Fortessa 5-laser analyzer (Becton Dickinson). Data were analyzed on FlowJoTM (Becton Dickinson) and plotted on GraphPad (Prism).
CS competition on ofCSPG/ofCS coated ELISA plate
To understand the specificity of each antibody fragment towards different CS subtypes, we performed competition assays. The ELISA plate were coated with 3 μg/ml of ofCS (F3, F4, F8, F11) or ofCSPG (B3, C9, B1) overnight at 4 °C, washed in 1xPBS, 0.5%Tween20 and blocked using 5% skim milk. Before adding samples to the plate, 100 nM of V5-tagged scFv2 (F3, F4, F8, F11, B3, C9) or FLAG-tagged Fab2 (B1) were pre-incubated with [0–400] μg/ml of soluble chondroitin sulfate (CSA (Sigma-Aldrich #C9819), CSB (Sigma-Aldrich #C3788), CSC (MP Biochemicals, #12678-07-8) in 1xPBS.1:5000 anti-V5-HRP (Invitrogen #R961-25) or 3 μg/ml anti-FLAG (Sigma, #A8592) antibodies were used for detection and plate were developed with TMB Plus2 (Kementec, #4395). The reaction was stopped with 0.2 M H2SO4 (Honeywell Fluka #35348) and Optical densities (OD) were read on an ELISA reader at 450 nm. The inhibition level was assessed by normalizing OD450 to the control well depleted in glycosaminoglycan. Data were plotted in GraphPad (Prism) and edited in Illustrator (Adobe).
Full chain and footprint analysis of CS binding to phage-derived scFv2
SpyC2 was coupled to HiTrap NHS-activated HP column (Cytiva, #17071601) following the manufacturer’s guidelines. To remove any excess active groups, a buffer containing 0.5 M Ethanolamine and 0.5 M NaCl, pH 8.3, was employed. Non-specifically bound proteins were washed out with 0.1 M Sodium Acetate, 0.5 M NaCl, and pH4 buffer. Anti-CS scFv monomers were then immobilized onto the column using the SpyT-SpyC interaction and incubated for 1 h, RT. The column was then washed with PBS before being loaded with 200 μg of ofCS purified from the placenta, and washed with digestion buffer containing 50 nM Tris, 50 nM NH4OAc, pH 7.9. For the full chain analysis, the bound ofCS was eluted with 2 M NH4OAc and subsequently lyophilized. In the footprint analysis, 50 mU/ml of chABC (Sigma-Aldrich, #C3667) diluted in digestion buffer was loaded and incubated for 1.5 h at room temperature. Following several washes of 250 nM NH4OAc, the bound chABC treated ofCS was eluted as described above.
Lyophilized samples were then resuspended in 40 mM sodium acetate and digested to disaccharide products by the addition of 0.5 mU/μl chABC and incubation overnight at 37 °C. Disaccharides were then labeled with 2-aminoacridone (AMAC) and subjected to Ultra Performance Liquide Chromatography (UPLC) C18 chromatography as previously described38. In detail, samples were lyophilized and labeled through resuspending of samples in 5 μl 0.1 M AMAC in acetic acid/dimethyl sulfoxide (DMSO) (vol/vol 3:17) followed by 15 min incubation at room temperature. 5 μL 1 M NaCNBH3 was subsequently added, and samples were incubated for 3 h at 45 °C. Labeled CS was purified from free AMAC through the addition of 500 µl acetone followed by centrifugation at 20,000 × g for 20 min and removal of acetone was repeated twice. Dried CS disaccharides were dissolved in 2% acetonitrile and analyzed on a Waters Acquity UPLC system using a BEH C18 column (2.1 × 150 mm, 1.7 μm, Waters), measuring fluorescence at 525 nm. 80 mM ammonium acetate (pH 5.5) was used as mobile phase A and acetonitrile as mobile phase B at a flow rate of 0.2 mL/min where mobile phase B increases from 3 to 13% over 30 min. 20 pmol AMAC-labeled CS disaccharides standard (Iduron) was analyzed prior to samples for quantification and identification of CS in samples.
CSPG pull-downs from cell supernatant and human cancer biopsies
Sera-Mag SpeedBeads Blocked Streptavidin particles (Cytiva, #21152104010150) were washed twice in Pierce Protein-Free (PBS) Blocking Buffer (ThermoScientific, #37572) and coated with 30 ng biotinylated rVAR2 or biotinylated anti-CS scFv2 per µg beads. Control beads were conjugated with SpyC2 protein alone. Excess protein was removed by three washes of Protein-Free buffer.
50 µg coated beads were incubated for 60 min either with 500 µL mix containing equal volumes of cell supernatants (PC-3M-luc-C6, MG63, SW480, and BEWO), or with 50 μg of human colon biopsy homogenate (H352 or H386). As a control, homogenates were incubated with 1 μg chABC for 1 h at 37 °C before bead isolation. Finally, the beads were washed three times in Dulbecco’s PBS without Calcium and Magnesium (Sigma-Aldrich, #D8537) prior to a final wash step in ultrapure water. All liquid was removed before storing the beads at −80 °C until mass spectrometry processing.
The cell supernatants were generated by culturing confluent cells for two days in serum-free media followed by 1 h treatment with 25 µM phorbol 12-myristate 13-acetate (Sigma-Aldrich, #P8139).
Mass spectrometry analysis of CSPG pull-downs
All samples were prepared in triplicates (n = 3) to allow statistical analysis. Washed beads were incubated for 30 min with elution buffer 1 (2 M Urea, 50 mM Tris-HCl pH 7.5, 2 mM DTT, 20 µg/ml trypsin) followed by a second elution for 5 min with elution buffer 2 (2 M Urea, 50 mM Tris-HCl pH 7.5, 10 mM Chloroacetamide). Both eluates were combined and further incubated at room temperature overnight. Tryptic peptide mixtures were acidified to 1% TFA and loaded on Evotips (Evosep). Peptides were separated on 15 cm, 150 μM ID columns packed with C18 beads (1.9 μm) (Pepsep) on an Evosep ONE HPLC applying the ‘30 samples per day’ method and injected via a CaptiveSpray source and 10 μm emitter into a timsTOF pro mass spectrometer (Bruker) operated in PASEF mode69. Raw mass spectrometry data were analyzed with MaxQuant (v1.6.15.0). Peak lists were searched against the human Uniprot FASTA database combined with the sequences of the human antibody fragments as well as 262 common contaminants by the integrated Andromeda search engine. False discovery rate was set to 1% for both peptides (minimum length of 7 amino acids) and proteins. “Match between runs” (MBR) was enabled with a Match time window of 0.7, and a Match ion mobility window of 0.05 min. Relative protein amounts were determined by the MaxLFQ algorithm with a minimum ratio count of two.
Processing and statistical analysis of label-free quantification (LFQ) protein expression data was performed using python3.9.0 Proteins tagged as potential contaminants, identified by matches to the decoy database, and identified only by modified sites, were excluded from the dataset. LFQ intensity values were log2 transformed, and uninformative proteins were removed by filtering out the ones with less than 2 valid values, in at least one group. The remaining missing values were imputed sample-wise using the MinProb method, where values are randomly drawn from a Gaussian distribution (downshift = 1.8, width = 0.2). Differentially expressed proteins were determined by performing two-sided unpaired t-tests with python package scipy (v1.11.2), and correcting for multiple hypothesis testing with statsmodels (v0.14.0) Benjamin–Hochberg correction. The significance threshold was set to 1% and the fold change to 2. For the heat map visualization (matplotlib package v3.7.1) of selected hits, protein LFQ values were row mean normalized and clustered by row. Imputed values were removed after clustering shown in gray. We omitted the non-significant hits (−1<log2FC < 1 and −log10pvalue < 2) in the Supplementary Data 1.
Mass photometry analysis of antibody:CS complexes
The mass photometry instrument70 was started, and emersion oil (Thorlabs #MOIL-30) was placed on the mass photometry objective. A glass slide (GLW storing systems #ZK25) and a gasket (Re-useable Culture well gasket 3 mm diameter × 1 mm depth, Sigma #GBL103250-10EA) were placed on top of the emersion oil. Next, 18 µl of PBS was placed into the gasket, and the mass photometry objective was adjusted and focused. The Mass Photometry instrument was then calibrated by the addition of 2 µl of protein standards of molecular weights 66 kDa, 146 kDa, 480 kDa, and 1048 kDa from Native Mark Unstained Protein Standard (ThermoFisher, #LC0725) in PBS. Immediately after the addition of the sample the mass photometry recording was started. Analysis was performed with the mass photometry software, and a calibration was created on the standard protein markers. The instrument for B1 samples focused on 13.5 µl PBS and 6,5 µl of sample was added. Samples recorded were B1 Fab2 alone (58 nM) and B1 Fab2 (375 nM):CSA (3.9 μg/ml). The instrument for C9 samples focused on 16 µl PBS and 4 µl of sample was added. Samples were CSA alone (7.8 μg/ml), C9 scFv2 alone (25 nM), C9 scFv2 (375 nM):CSA (7.8 μg/ml). The instrument for F8 samples focused on 15 µl PBS and 5 µl of sample was added. Samples were CSC alone (15.6 μg/ml), F8 scFv2 alone (25 nM), F8 scFv2 (375 nM):CSC (15.6 μg/ml). The Mass photometry mass distributions were plotted with an 8.3 kDa bin size and a best-fit Gaussian distribution was added.
Cryo-EM sample preparation
The initial step involved complexing B1 Fab with CSA (Sigma-Aldrich #C9819) in TBS buffer, pH 7.5, at a final concentration of 0.15 mg/ml of B1 Fab after complexing with 2 mg/ml of CSA. The mixture was then subjected to quantifoil 1.2/1.3 300 C mesh, which was plasma washed by glow discharging with Leica Coater ACE200 for 30 s at 10 mA. After incubating for 5 seconds, the grids were blotted for 3 s at 4 ˚C with 100% humidity. The complex was plunge-frozen in liquid ethane using Vitrobot Mark IV (Thermofisher) and stored in liquid nitrogen for subsequent data collection.
Cryo-EM data collection and processing
Data collection of the B1 Fab:CSA complex was obtained on a TITAN Krios Electron Microscope (FEI), 300 kV. 5819 micrographs (2135: un-tilt, 3684: 30˚ tilt) were collected using a Falcon-III direct electron detector operating in counting mode at a pixel size of 0.832 Å and a total dose of 44 e/Å2 over 40 frames with defocus range of −1 to −2.6 µm using Thermofisher EPU data collection software. The movies were patch motion-corrected for beam-induced motion and patch CTF estimation was done using cryoSPARC v4.1.271. Blob particle picking was performed on all micrographs with a minimum particle diameter of 100 Å and a maximum of 200 Å. Particles extracted at 360 pixels box size were used to perform 2D classification. Template-based particle selection was done to re-extract particles from micrographs by Local Motion Correction with dose-weighting at a box size of 360 pixels. An initial 1,257,314 particle stack was extracted and subjected to iterative rounds of reference-free 2D classifications to identify class averages corresponding to tetrameric complex. 3D ab initio classification was performed, followed by heterogeneous refinement to obtain one good class that was further non-uniform (NU) heterogeneous refined (Table 1). Gold-Standard Fourier Shell Correlation (GSFSC) resolution was calculated to be 3.3 Å at FSC 0.143 by 3dFSC job and local resolution was estimated. The resolution values are likely to be overestimated due to some orientation bias and particle heterogeneity. To visualize any conformational changes on the tetrameric cryo-EM map, the 200,000 particles and the map from NU refine job were used as input for the 3D flex data preparation job. A base number tetra cell of 20 and rigidity weight of 0.5 were used for 3D flex Mesh Prep job. The 3D flex mesh and the prepared particles with map were used to run 3D flex train with the number of latent dimensions 2. Flex generator was used to produce a volume series of 100 frames depicting the flexibility conformational movements and the movie (Movie 1) was made in ChimeraX72.
Cryo-EM structure modeling and refinement
Initial coordinates of B1 Fab were generated by Alphafold273 and docked into the cryoEM density map in UCSF Chimera74. The structure was constructed iteratively with COOT75 followed by real-space refinement in PHENIX package76 and Rosetta relax77. Data collection parameters and model refinement statistics are reported in Table 1. The CS density was modeled as a 12mer and subsequently fitted manually to the low-resolution cryo-EM density region on the highly positively charged surfaces of the Fab complex. The docked structures were further refined via MDFF with an implicit solvent78. A three-step protocol with an increasing map potential scaling factor from 0.3 to 0.7 was used for MDFF. The MDFF model of B1 Fab tetramer complexed with CS was subsequently solvated in a periodic cubic box (with a side of 13.8 nm) with TIP3P water molecules containing Na+ and Cl− ions at 0.15 M, resulting in approximately 267,000 atoms in total. The CHARMM36m force field was used for the protein79. A weak position restraint potential (with a force constant of 10 kJ/mol/nm2) was added to the protein backbone to maintain its cryo-EM conformation. Force field parameters for CSA were generated using the Glycan Modeler module in the CHARMM-GUI web interface80. Neighbor searching was performed every 20 steps. The PME algorithm was used for electrostatic interactions with a cut-off of 1.2 nm. A reciprocal grid of 120 × 120 × 120 cells was used with 4th-order B-spline interpolation. A single cut-off of 1.2 nm was used for Van der Waals interactions. Temperature coupling was done with the v-rescale algorithm. Pressure coupling was done with the Parrinello-Rahman algorithm. To validate the MDFF model, a 1000 ns MD simulation was performed using Gromacs 2022.581.
Single-cell RNAseq analysis
The extended version of integrated non-small cell lung carcinoma (NSCLC) transcriptome atlas consisting of 1.2 M cells from 309 lung cancer patients from previously published studies53 in AnnData object format (h5ad) was downloaded [Dataset (https://cellxgene.cziscience.com/collections/edb893ee-4066-4128-9aec-5eb2b03f8287)] and imported in Scanpy version 1.9.1.
All cell-type annotations were used from the original study53. For the further analysis ‘primary tumor’ and ‘normal adjacent’ samples from 10X Genomics and BD-Rhapsody assay platforms were extracted from the dataset and merged into a separate AnnData object. The analysis was performed on approximately 87,000 epithelial and 10,000 stromal cells derived from 108 patients and the expression of CS-related genes was visualized with dotplot or Uniform Manifold Approximation and Projection (UMAP).
Data analysis and graphical visualization was performed with scanpy v.1.9.1, anndata v.0.8.0, umap v.0.5.3, numpy v.1.23.5, scipy v.1.10.1, pandas v.2.0.0, scikit-learn v.1.2.2, statsmodels v.0.13.2, pynndescent v.0.5.8, and python-igraph v.0.10.2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
The authors would like to acknowledge Chumpol Hueyploo, Benjamin Jacobsen, Muhammad Tauseef Mukhtar, Sabrina Damiri, Julie Zeberg Salwin, and Raminta Jonauskaite for excellent technical support. We would like to thank Nanna Skeltved for supplying us with some reagents and sharing her expertise on VAR2-based bispecific proteins. At University of Copenhagen, Faculty of Healthy and Medical Sciences, we would like to thank the Core Facility for Integrated Microscopy for support and assistance in cryo-EM data collection and tissue scanning; the Histolab for embedding and cutting murine tissue specimens, the Core Facility for Flow cytometry and Single Cell Analysis for the assistance in flow cytometry experiments. Mass spectrometry analysis were performed by the Proteomics Research Infrastructure (PRI) at the University of Copenhagen (UCPH), supported by the Novo Nordisk Foundation (NNF) (grant agreement number NNF19SA0059305). At Vancouver Prostate Centre, we would like to thank the Molecular Pathology & Imaging Core for processing human and murine tissue specimens. The mice embryo tissue slides were kindly shared by Dr. Pamela Hoodless from British Columbia Research Center (BCRC), Vancouver, British Columbia, Canada. The pancreas tissue slides from KPC mice were kindly shared by Lesley P. Ferguson and Tannishtha Reya from Sanford Consortium for Regenerative Medicine, La Jolla, CA, USA. We would like to thank Chris D. Madsen, Lund University, Sweden, for sharing the CAFs cell lines with us, and Line Mathiesen, University of Copenhagen, Denmark, for providing us fresh human placenta tissues. This work was funded by Innovation Fund Denmark (to E.V.C.; 9065-00216A), Lundbeck Foundation (to S.S.R.R. and T.L., R344-2020-934; to K.W., R324-2019-1855); the National Key Research and Development Program of China (to Yong W., 2021YFF1200404); the Fundamental Research Funds for the Central Universities of China (to Yong W., K20220228); NIH Prostate Cancer PNW-SPORE (to M.D., 1016339, 223493; 5P50 CA097186-17); the Canadian Institutes of Health Research (CIHR) (to M.D., PJT-153092 and PJT- 470421); The Novo Nordisk Foundation (to R.L.M., NNF22OC0073736; to A.S., NNF19OC0058387); NNF Tandem grant (to A.S., NNF21OC0068192); NNF Distinguished Innovator grant (to A.S., NNF22OC0076055); the Carlsberg Foundation (to R.L.M., CF20-0412; to R.K., CF22-0354); the Danish National Research Foundation (to H.C., DNRF107); EMBO Postdoctoral Fellowship (to F.G.; ALTF 105-2023).
Author contributions
E.V.C., S.C., R.D., T.G., M.D., and A.S. designed the research; E.V.C., T.G., A.S., and M.D. wrote the main manuscript text; E.V.C. prepared the main figures and Supplementary Figs. files; F.G. and R.M. prepared Fig. 5d; S.S.R.R. prepared the Fig. 6b–e; S.S.R.R., Yo.W., and K.W. performed the structural analyses and prepared the Supplementary Movies 1 and 2; E.V.C., A.M.S., S.C., R.D., S.S.R.R., L.D., M.A.N., T.M.C., A.S., J.O., C.L., A.M.J., D.R., J.M., S.W., A.G.L., R.T.G.K., F.G., Yo.W., I.N., M.M., H.Z.O., and M.S.Ø. performed the experiments; K.W., M.Ø.A., R.L.M., T.L., M.A.N., H.C., M.W., D.W., T.G., M.D., and A.S. supervised; A.R.C., A.M., M.W., and D.W. performed the bioinformatics analyses of large datasets; Y.H.C., Yu.W., and I.G. provided useful reagents; All authors contributed to discussing the data and reviewing the manuscript.
Peer review
Peer review information
Nature Communications thanks Haotian Lei, James McCarthy and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data relating to the findings in the article are contained in the manuscript and Supporting information. The atomic coordinates and electron-microscopy data have been deposited in the RCSB Protein Data Bank (https://www.rcsb.org) and in Electron Microscopy Data Bank (https://www.ebi.ac.uk/emdb/) under the entry PDB: 8P2E and EMDB: 17362. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE82 partner repository with the dataset identifier PXD044455 (https://www.ebi.ac.uk/pride/archive/projects/PXD34723319). Source data are provided with this paper.
Competing interests
A.S., M.D., M.Ø.A., T.M.C., M.A.N., and T.G.T. are shareholders of VAR2 Pharmaceuticals. The antibodies are subject to a patenting process owned by VAR2 Pharmaceuticals. H.C. is a shareholder of GlycoDisplay ApS, GlycoZym ApS, GlycoZym Inc, now GO Therapeutics Inc. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Elena Ethel Vidal-Calvo, Email: elena@sund.ku.dk.
Mads Daugaard, Email: mads.daugaard@ubc.ca.
Ali Salanti, Email: salanti@sund.ku.dk.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51781-0.
References
- 1.Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Fu, Z., Li, S., Han, S., Shi, C. & Zhang, Y. Antibody drug conjugate: the “biological missile” for targeted cancer therapy. Sig Transduct. Target Ther.7, 1–25 (2022). 10.1038/s41392-022-00947-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watanabe, K. & Hakomori, S. I. Status of blood group carbohydrate chains in ontogenesis and in oncogenesis. J. Exp. Med.144, 644–653 (1976). 10.1084/jem.144.3.644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banwo, O., Versey, J. & Hobbs, J. R. New oncofetal antigen for human pancreas. Lancet1, 643–645 (1974). 10.1016/S0140-6736(74)93197-3 [DOI] [PubMed] [Google Scholar]
- 5.Baston-Büst, D. M., Götte, M., Janni, W., Krüssel, J.-S. & Hess, A. P. Syndecan-1 knock-down in decidualized human endometrial stromal cells leads to significant changes in cytokine and angiogenic factor expression patterns. Reprod. Biol. Endocrinol.8, 133 (2010). 10.1186/1477-7827-8-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang, Y. & Zhao, S. Vascular Biology of the Placenta (Morgan & Claypool Life Sciences, 2010). [PubMed]
- 7.Xu, G., Guimond, M.-J., Chakraborty, C. & Lala, P. K. Control of proliferation, migration, and invasiveness of human extravillous trophoblast by decorin, a decidual product. Biol. Reprod.67, 681–689 (2002). 10.1095/biolreprod67.2.681 [DOI] [PubMed] [Google Scholar]
- 8.Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell100, 57–70 (2000). 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- 9.Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell144, 646–674 (2011). 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 10.Freire-de-Lima, L. Sweet and sour: the impact of differential glycosylation in cancer cells undergoing epithelial–mesenchymal transition. Front. Oncol.4, 59 (2014). [DOI] [PMC free article] [PubMed]
- 11.Fried, M. & Duffy, P. E. Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science272, 1502–1504 (1996). 10.1126/science.272.5267.1502 [DOI] [PubMed] [Google Scholar]
- 12.Tuikue Ndam, N. G. et al. High level of var2csa transcription by Plasmodium falciparum isolated from the placenta. J. Infect. Dis.192, 331–335 (2005). 10.1086/430933 [DOI] [PubMed] [Google Scholar]
- 13.Salanti, A. et al. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J. Exp. Med.200, 1197–1203 (2004). 10.1084/jem.20041579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ayres Pereira, M. et al. Placental sequestration of Plasmodium falciparum malaria parasites is mediated by the interaction between VAR2CSA and chondroitin sulfate A on syndecan-1. PLoS Pathog.12, e1005831 (2016). 10.1371/journal.ppat.1005831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salanti, A. et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol. Microbiol.49, 179–191 (2003). 10.1046/j.1365-2958.2003.03570.x [DOI] [PubMed] [Google Scholar]
- 16.Smith, J. D. et al. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell82, 101–110 (1995). 10.1016/0092-8674(95)90056-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamain, B., Gratepanche, S., Miller, L. H. & Baruch, D. I. Molecular basis for the dichotomy in Plasmodium falciparum adhesion to CD36 and chondroitin sulfate A. Proc. Natl Acad. Sci. USA99, 10020–10024 (2002). 10.1073/pnas.152321599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyes, S., Horrocks, P. & Newbold, C. Antigenic variation at the infected red cell surface in malaria. Annu. Rev. Microbiol.55, 673–707 (2001). 10.1146/annurev.micro.55.1.673 [DOI] [PubMed] [Google Scholar]
- 19.Su, X. Z. et al. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell82, 89–100 (1995). 10.1016/0092-8674(95)90055-1 [DOI] [PubMed] [Google Scholar]
- 20.Soares da Costa, D., Reis, R. L. & Pashkuleva, I. Sulfation of glycosaminoglycans and its implications in human health and disorders. Annu. Rev. Biomed. Eng.19, 1–26 (2017). 10.1146/annurev-bioeng-071516-044610 [DOI] [PubMed] [Google Scholar]
- 21.Spliid, C. B. et al. The specificity of the malarial VAR2CSA protein for chondroitin sulfate depends on 4-O-sulfation and ligand accessibility. J. Biol. Chem.297, 101391 (2021). 10.1016/j.jbc.2021.101391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma, R. et al. Structural basis for placental malaria mediated by Plasmodium falciparum VAR2CSA. Nat. Microbiol6, 380–391 (2021). 10.1038/s41564-020-00858-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knappik, A. et al. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J. Mol. Biol.296, 57–86 (2000). [DOI] [PubMed]
- 24.Prassler, J. et al. HuCAL PLATINUM, a synthetic fab library optimized for sequence diversity and superior performance in mammalian expression systems. J. Mol. Biol.413, 261–278 (2011). 10.1016/j.jmb.2011.08.012 [DOI] [PubMed] [Google Scholar]
- 25.Valadon, P. et al. ALTHEA Gold LibrariesTM: antibody libraries for therapeutic antibody discovery. mAbs11, 516–531 (2019). 10.1080/19420862.2019.1571879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zakeri, B. et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA109, E690–E697 (2012). [DOI] [PMC free article] [PubMed]
- 27.Salanti, A. et al. Targeting human cancer by a glycosaminoglycan binding malaria protein. Cancer Cell28, 500–514 (2015). 10.1016/j.ccell.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hingorani, S. R. et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell4, 437–450 (2003). 10.1016/S1535-6108(03)00309-X [DOI] [PubMed] [Google Scholar]
- 29.Zorzi, A., Linciano, S. & Angelini, A. Non-covalent albumin-binding ligands for extending the circulating half-life of small biotherapeutics. Medchemcomm10, 1068–1081 (2019). 10.1039/C9MD00018F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, C. et al. Clinical pharmacology of vc-MMAE antibody–drug conjugates in cancer patients: learning from eight first-in-human Phase 1 studies. MAbs12, 1699768 (2019). 10.1080/19420862.2019.1699768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hingorani, D. V. et al. Monomethyl auristatin antibody and peptide drug conjugates for trimodal cancer chemo-radio-immunotherapy. Nat. Commun.13, 3869 (2022). 10.1038/s41467-022-31601-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heiser, R. A. et al. Brentuximab Vedotin–Driven Microtubule Disruption Results in Endoplasmic Reticulum Stress Leading to Immunogenic Cell Death and Antitumor Immunity. Mol. Cancer Ther.23, 68–83 (2024). 10.1158/1535-7163.MCT-23-0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Josephs, S. F. et al. Unleashing endogenous TNF-alpha as a cancer immunotherapeutic. J. Transl. Med.16, 242 (2018). 10.1186/s12967-018-1611-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laha, D., Grant, R., Mishra, P. & Nilubol, N. The role of tumor necrosis factor in manipulating the immunological response of tumor microenvironment. Front Immunol.12, 656908 (2021). 10.3389/fimmu.2021.656908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nie, H. et al. Phosphorylation of FOXP3 controls regulatory T cell function and is inhibited by TNF-α in rheumatoid arthritis. Nat. Med.19, 322–328 (2013). 10.1038/nm.3085 [DOI] [PubMed] [Google Scholar]
- 36.Okajima, T., Yoshida, K., Kondo, T. & Furukawa, K. Human homolog of Caenorhabditis elegans sqv-3 gene is galactosyltransferase I involved in the biosynthesis of the glycosaminoglycan-protein linkage region of proteoglycans. J. Biol. Chem.274, 22915–22918 (1999). 10.1074/jbc.274.33.22915 [DOI] [PubMed] [Google Scholar]
- 37.Almeida, R. et al. Cloning and expression of a proteoglycan UDP-galactose:β-xylose β1,4-galactosyltransferase I. J. Biol. Chem.274, 26165–26171 (1999). 10.1074/jbc.274.37.26165 [DOI] [PubMed] [Google Scholar]
- 38.Chen, Y.-H. et al. The GAGOme: a cell-based library of displayed glycosaminoglycans. Nat. Methods15, 881–888 (2018). 10.1038/s41592-018-0086-z [DOI] [PubMed] [Google Scholar]
- 39.Narimatsu, Y. et al. Genetic glycoengineering in mammalian cells. J. Biol. Chem.296, 100448 (2021). 10.1016/j.jbc.2021.100448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Büll, C., Joshi, H. J., Clausen, H. & Narimatsu, Y. Cell-based glycan arrays—a practical guide to dissect the human glycome. STAR Protoc.1, 100017 (2020). 10.1016/j.xpro.2020.100017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mikami, T. & Kitagawa, H. Biosynthesis and function of chondroitin sulfate. Biochim. Biophys. Acta1830, 4719–4733 (2013). 10.1016/j.bbagen.2013.06.006 [DOI] [PubMed] [Google Scholar]
- 42.Izumikawa, T., Koike, T. & Kitagawa, H. Chondroitin 4-O-sulfotransferase-2 regulates the number of chondroitin sulfate chains initiated by chondroitin N-acetylgalactosaminyltransferase-1. Biochem. J.441, 697–705 (2012). 10.1042/BJ20111472 [DOI] [PubMed] [Google Scholar]
- 43.Noborn, F., Nikpour, M., Persson, A., Nilsson, J. & Larson, G. Expanding the Chondroitin Sulfate Glycoproteome - But How Far? Front Cell Dev. Biol.9, 695970 (2021). 10.3389/fcell.2021.695970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toledo, A. G. et al. An affinity chromatography and glycoproteomics workflow to profile the chondroitin sulfate proteoglycans that interact with malarial VAR2CSA in the placenta and in cancer. Glycobiology30, 989–1002 (2020). 10.1093/glycob/cwaa039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agerbaek, M. Ø. et al. Burkitt lymphoma expresses oncofetal chondroitin sulfate without being a reservoir for placental malaria sequestration. Int J. Cancer140, 1597–1608 (2017). 10.1002/ijc.30575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bang-Christensen, S. R. et al. Capture and detection of circulating glioma cells using the recombinant VAR2CSA malaria protein. Cells8, 998 (2019). 10.3390/cells8090998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clausen, T. M. et al. Oncofetal chondroitin sulfate glycosaminoglycans are key players in integrin signaling and tumor cell motility. Mol. Cancer Res.14, 1288–1299 (2016). 10.1158/1541-7786.MCR-16-0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacobsen, F. et al. Up-regulation of biglycan is associated with poor prognosis and PTEN deletion in patients with prostate cancer. Neoplasia19, 707–715 (2017). 10.1016/j.neo.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pinto, F., Santos-Ferreira, L., Pinto, M., Gomes, C. & Reis, C. The extracellular small leucine-rich proteoglycan biglycan is a key player in gastric cancer aggressiveness. Cancers13, 1330 (2021). 10.3390/cancers13061330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu, Y. et al. High expression of syndecan-4 is related to clinicopathological features and poor prognosis of pancreatic adenocarcinoma. BMC Cancer22, 1042 (2022). 10.1186/s12885-022-10128-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, Q. et al. Upregulation of versican associated with tumor progression, metastasis, and poor prognosis in bladder carcinoma. BioMed. Res. Int.2021, 1–11 (2021). 10.1155/2021/5047501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, K. et al. Cryo-EM reveals the architecture of placental malaria VAR2CSA and provides molecular insight into chondroitin sulfate binding. Nat. Commun.12, 2956 (2021). 10.1038/s41467-021-23254-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salcher, S. et al. High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. Cancer Cell40, 1503–1520.e8 (2022). 10.1016/j.ccell.2022.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oo, H. Z. et al. Oncofetal chondroitin sulfate is a highly expressed therapeutic target in non-small cell lung cancer. Cancers13, 4489 (2021). 10.3390/cancers13174489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khazamipour, N. et al. Oncofetal chondroitin sulfate: a putative therapeutic target in adult and pediatric solid tumors. Cells9, 818 (2020). 10.3390/cells9040818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Al-Nakouzi, N. et al. Reformation of the chondroitin sulfate glycocalyx enables progression of AR-independent prostate cancer. Nat. Commun.13, 4760 (2022). 10.1038/s41467-022-32530-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clausen, T. M. et al. Real-time and label free determination of ligand binding-kinetics to primary cancer tissue specimens; a novel tool for the assessment of biomarker targeting. Sens Biosens. Res.9, 23–30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seiler, R. et al. An oncofetal glycosaminoglycan modification provides therapeutic access to cisplatin-resistant bladder cancer. Eur. Urol.72, 142–150 (2017). 10.1016/j.eururo.2017.03.021 [DOI] [PubMed] [Google Scholar]
- 59.Agerbæk, M. Ø. et al. The VAR2CSA malaria protein efficiently retrieves circulating tumor cells in an EpCAM-independent manner. Nat. Commun.9, 3279 (2018). 10.1038/s41467-018-05793-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clausen, T. M. et al. A simple method for detecting oncofetal chondroitin sulfate glycosaminoglycans in bladder cancer urine. Cell Death Discov.6, 65 (2020). 10.1038/s41420-020-00304-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nordmaj, M. A. et al. Development of a bispecific immune engager using a recombinant malaria protein. Cell Death Dis.12, 353 (2021). 10.1038/s41419-021-03611-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skeltved, N. et al. Bispecific T cell-engager targeting oncofetal chondroitin sulfate induces complete tumor regression and protective immune memory in mice. J. Exp. Clin. Cancer Res.42, 106 (2023). 10.1186/s13046-023-02655-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gowda, A. S. P. et al. Structural basis for the adherence of Plasmodium falciparum-infected erythrocytes to chondroitin 4-sulfate and design of novel photoactivable reagents for the identification of parasite adhesive proteins. J. Biol. Chem.282, 916–928 (2007). 10.1074/jbc.M604741200 [DOI] [PubMed] [Google Scholar]
- 64.Ramsland, P. A., Farrugia, W., Bradford, T. M., Mark Hogarth, P. & Scott, A. M. Structural convergence of antibody binding of carbohydrate determinants in Lewis Y tumor antigens. J. Mol. Biol.340, 809–818 (2004). 10.1016/j.jmb.2004.05.037 [DOI] [PubMed] [Google Scholar]
- 65.Tommasone, S. et al. The challenges of glycan recognition with natural and artificial receptors. Chem. Soc. Rev.48, 5488–5505 (2019). 10.1039/C8CS00768C [DOI] [PubMed] [Google Scholar]
- 66.Du, W. W., Yang, W. & Yee, A. J. Roles of versican in cancer biology-tumorigenesis, progression and metastasis. Histol. Histopathol.28, 701–713 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Martin, G. M. et al. Affinity-matured homotypic interactions induce spectrum of PfCSP structures that influence protection from malaria infection. Nat. Commun.14, 4546 (2023). 10.1038/s41467-023-40151-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin, D. et al. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res.74, 1272–1283 (2014). 10.1158/0008-5472.CAN-13-2921-T [DOI] [PubMed] [Google Scholar]
- 69.Meier, F. et al. Online parallel accumulation–serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteom.17, 2534–2545 (2018). 10.1074/mcp.TIR118.000900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sonn-Segev, A. et al. Quantifying the heterogeneity of macromolecular machines by mass photometry. Nat. Commun.11, 1772 (2020). 10.1038/s41467-020-15642-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017). 10.1038/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- 72.Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci.30, 70–82 (2021). 10.1002/pro.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goddard, T. D., Huang, C. C. & Ferrin, T. E. Visualizing density maps with UCSF Chimera. J. Struct. Biol.157, 281–287 (2007). 10.1016/j.jsb.2006.06.010 [DOI] [PubMed] [Google Scholar]
- 75.Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr66, 486–501 (2010). 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D. Struct. Biol.74, 531–544 (2018). 10.1107/S2059798318006551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang, R. Y.-R. et al. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. eLife5, e17219 (2016). 10.7554/eLife.17219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trabuco, L. G., Villa, E., Mitra, K. & Frank, J. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure16, 673–683 (2008). [DOI] [PMC free article] [PubMed]
- 79.Huang, J. et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods14, 71–73 (2017). 10.1038/nmeth.4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee, J. et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput.12, 405–413 (2016). 10.1021/acs.jctc.5b00935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Páll, S. et al. Heterogeneous parallelization and acceleration of molecular dynamics simulations in GROMACS. J. Chem. Phys.153, 134110 (2020). 10.1063/5.0018516 [DOI] [PubMed] [Google Scholar]
- 82.Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res.50, D543–D552 (2021). 10.1093/nar/gkab1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data relating to the findings in the article are contained in the manuscript and Supporting information. The atomic coordinates and electron-microscopy data have been deposited in the RCSB Protein Data Bank (https://www.rcsb.org) and in Electron Microscopy Data Bank (https://www.ebi.ac.uk/emdb/) under the entry PDB: 8P2E and EMDB: 17362. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE82 partner repository with the dataset identifier PXD044455 (https://www.ebi.ac.uk/pride/archive/projects/PXD34723319). Source data are provided with this paper.