Abstract
The limited understanding of the molecular mechanism underlying MYCN-amplified (MNA) neuroblastoma (NB) has hindered the identification of effective therapeutic targets for MNA NB, contributing to its higher mortality rate compared to MYCN non-amplified (non-MNA) NB. Therefore, a comprehensive analysis integrating metabolomics and transcriptomics was conducted to systematically investigate the MNA NB. Metabolomics analysis utilized plasma samples from 28 MNA NB patients and 68 non-MNA NB patients, while transcriptomics analysis employed tissue samples from 15 MNA NB patients and 37 non-MNA NB patients. Notably, joint metabolomics and transcriptomics analysis was performed. A total of 46 metabolites exhibited alterations, with 21 displaying elevated levels and 25 demonstrating reduced levels in MNA NB. In addition, 884 mRNAs in MNA NB showed significant changes, among which 766 mRNAs were higher and 118 mRNAs were lower. Joint-pathway analysis revealed three aberrant pathways involving glycerolipid metabolism, purine metabolism, and lysine degradation. This study highlights the substantial differences in metabolomics and transcriptomics between MNA NB and non-MNA NB, identifying three abnormal metabolic pathways that may serve as potential targets for understanding the molecular mechanisms underlying MNA NB.
Keywords: MYCN amplification, Neuroblastoma, Metabolomics, Transcriptomics, Therapeutic target, Molecular mechanism
Subject terms: Cancer metabolism, Cancer, Tumour biomarkers
Introduction
Neuroblastoma (NB) is a malignancy of the peripheral nervous system that arises from the embryonic neural crest, characterized by an insidious onset and rapid progression. NB constitutes 8% of all pediatric cancer cases, while accounting for 15% of childhood1–3. The clinical manifestations of NB exhibit significant heterogeneity, ranging from spontaneous regression or differentiation with an overall survival rate of 85–90%, to refractory and metastatic tumors, wherein less than 50% of patients survive even after intensive therapy4. One of the factors contributing to NB heterogeneity is MYCN amplification (MNA), which has been shown to promote NB growth and progression, and correlate with treatment resistance and unfavorable prognosis in NB5. For instance, in comparison to MNA NB, MYCN non-amplified (non-MNA) NB patients had higher event free survival (EFS) and overall survival (OS) (EFS and OS: 82.5% and 90.8% versus 36.9% and 44.8%), indicating that MNA has a tremendous impact on the prognosis of NB6. Furthermore, the prevalence of MNA in NB patients is 20–30%, whereas MNA is detected in about 50% of high risk NB (HR-NB) cases with an overall survival rate of less than 50%7,8. Studies have also shown that MYCN, as a major transcription factor, is important for normal cell proliferation and apoptosis. MNA may lead to inhibition of apoptosis signals and continuous proliferation, which may eventually lead to the development of NB9. Nevertheless, there is a dearth of studies that explicitly elucidate the molecular mechanism underlying MNA in NB or its associated therapeutic targets, thereby contributing to the elevated mortality observed in MNA NB compared to non-MNA NB10. Therefore, it is imperative to investigate the aberrant pathway of MNA NB in order to elucidate the molecular regulatory mechanism underlying MNA in NB and subsequently identify potential therapeutic targets for augmenting the survival rate of patients with MNA NB.
It is worth noting that the development of omics has been expected to make important contributions to the understanding of MYCN molecular mechanisms and the discovery of therapeutic targets of MNA NB. Metabolomics aims to characterize all small molecules in a sample to accurately reflect the biological metabolic signature of the disease, which is beneficial to understanding the pathological or physiological conditions. The MNA exerts an impact on various metabolic pathways in NB, exemplified by the ability of MYCN to bind to the E-box within the promoter region of the p53 gene and enhance p53 transcription. Notably, there exists a positive correlation between p53 expression in NB tissue and MYCN expression11. Furthermore, MYCN can bind to the promoter region of ASCT2, a glutamine carrier, promoting its transcription. Enhanced glutamine metabolism represents a crucial characteristic of malignant tumors; thus, during proliferation processes, NB cells with amplified MYCN necessitate substantial amounts of glutamine for their proliferation support12. Consequently, it is imperative to further investigate the relationship between MYCN and tumor metabolism. It has been found that MNA NB is correlated with altered expression of proteins involved in multiple metabolic processes, including enhanced glycolysis and increased oxidative phosphorylation compared with non-MNA NB13,14. Konstantinos et al.15 found that MYCN changed the sulfur transfer pathway in NB through metabolomics. Arlt et al.16 found that phosphoglycerate dehydrogenase is related to MNA through metabolomics analysis, and MNA NB has higher synthesis of serine than non-MNA NB. In addition, through metabolomics analysis, Alptekin et al.17 revealed that MNA affects purine and central carbon metabolism and reduces citrate production, leading to a decrease in the steady-state levels of cholesterol and fatty acids. Transcriptomics utilizes high-throughput sequencing techniques to investigate the comprehensive repertoire of transcribed mRNAs in specific cells, tissues, or individuals at a given temporal and physiological state. This approach enables the identification of disparities in gene expression and structure across distinct functional states, thereby facilitating the elucidation of molecular mechanisms underlying diverse pathological or physiological conditions18,19. Fan et al.20 studied differential genes between MNA NB and non-MNA NB through transcriptomics and found that FLVCR2, SCN7A, PRSS12, NTRK1 and XAGE1A could be used as biomarkers to predict the prognosis of MNA NB. Lee et al.21 conducted transcriptome analysis on non-MNA NB, providing new insights into the genomic background of non-MNA NB. In addition, research had found that EZH1 depletion in MNA NB cells resulted in significant cell death as well as xenograft inhibition22. With the rapid development of omics, multi-omics analysis has made important contributions to heart failure, colorectal cancer, bladder cancer and other diseases23–25. The integration of transcriptomics and metabolomics has been demonstrated as a robust methodology, which can enhance the comprehension of potential biological functions and molecular mechanisms underlying diseases26. Metabolomics and transcriptomics have been used to study the metabolomics and transcriptomics differences between MNA NB and non-MNA, respectively. However, a comprehensive integration of metabolomics and transcriptomics is noticeably lacking in the investigation of molecular mechanisms and therapeutic targets associated with MNA in NB. Therefore, the integration of metabolomics and transcriptomics holds immense significance in comprehending the molecular mechanisms influenced by MNA and facilitating the identification of therapeutic targets for MNA NB.
In this study, a total of 96 plasma clinical samples and 52 clinical NB tissue samples were subjected to metabolomics and transcriptomics analyses, respectively. The integration of metabolomics and transcriptomics data was employed to perform a comprehensive network analysis of MNA NB, elucidating molecular mechanisms and identifying potential therapeutic targets. The innovation of this study lies in the comprehensive analysis of the differences between MNA NB and non-MNA NBA through integrated metabolomics and transcriptomics analysis, aiming to elucidate the molecular mechanism underlying MNA and identify potential therapeutic targets. Furthermore, investigating aberrant metabolic pathways in MNA NB provides a theoretical foundation for understanding its molecular mechanisms and subsequent exploration of therapeutic targets.
Methods and materials
Sample collection
This study was granted approval by the Ethics Committee of Henan Children's Hospital (2019-H-K11), and all procedures were conducted in accordance with relevant guidelines and regulations. A total of 96 plasma samples (28 cases of MNA NB, 68 cases of non-MNA NB) and 52 NB tissue samples (15 cases of MNA NB, 37 cases of non-MNA NB) were collected and processed at the Henan Children's Hospital from October 2018 to January 2022. The plasma samples were taken from the fasting plasma of NB patients on the morning of surgery, and tissue samples were taken from tumor tissues of NB patients during surgery.
Some patients underwent tumor evaluation at our hospital but did not undergo surgery, resulting in the availability of only blood samples without corresponding NB tissue samples. Conversely, other patients were referred for surgical intervention after preoperative examinations at different facilities, yielding only tissue samples without accompanying blood samples. Consequently, a subset of patients provided both blood and tissue samples. To enhance the statistical power of our study and ensure more comprehensive results, we incorporated plasma and tissue samples from 41 children, solely tissue samples from an additional 11 children, and exclusively plasma samples from another 55 children. Patient information is illustrated using a Venn diagram in Fig. S1.
Inclusion criteria consisted of: (1) confirmed diagnosis of pathological NB; (2) the MNA status in NB tissue was clear; (3) informed consent was signed by children or their parents. Exclusion criteria consisted of: (1) complications of other diseases; (2) informed consent was not signed by children or their parents. Plasma samples were collected before the surgery and were immediately frozen at − 80 °C for metabolomics analyses. NB tissue samples were directly placed into liquid nitrogen after surgical resection until transcriptomics analysis. Tables S1 and S2 showed that there were no significant differences in the age, male to female ratio, MYCN value and gross tumor volume between MNA NB and non-MNA NB patients. On the other hand, there were statistical differences in tumor metastasis, radiological risk factors and HR-NB (based on COG classification) ratio between MNA NB and non-MNA NB patients.
Metabolomics analysis via high performance liquid chromatography-mass spectrometry (HPLC–MS)
The metabolomics approach we employ is consistent with the methodology outlined in our previous publication19. The metabolomics analysis was performed using MetaboAnalyst (https://www.metaboanalyst.ca/MetaboAnalyst/home.xhtml), which included partial least-squares discrimination analysis (PLS-DA), heatmap, volcano map, enrichment analysis, pathway analysis, and identification of biomarkers. The stability of the overall experimental results was evaluated by preparing a quality control (QC) sample, which was obtained by combining equal volumes of supernatant from all samples.
Transcriptomics detection through RNA-sequencing (RNA-seq) analysis
Total RNA was extracted using TRIzol reagent according to the manufacturer’s protocol. RNA purity and quantification were evaluated using the NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). RNA integrity number (RIN) was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The libraries were constructed using TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. The transcriptome sequencing and analysis were conducted by OE Biotech Co., Ltd. (Shanghai, China). The libraries were sequenced on an Illumina HiSeq X Ten platform and 150 bp paired-end reads were generated. About 48.349 M raw reads for each sample were generated. Raw data (raw reads) of fastq format were firstly processed using Trimmomatic and the low quality reads were removed to obtain clean reads27. The clean reads were mapped to the human genome (GRCh38) using HISAT228. Fragments per kilobase of exon model per million mapped fragments (FPKM) of each gene were calculated using Cufflinks and the read counts of each gene were obtained by HTSeq-count29,30. P value < 0.05 and |log2(fold change)|> 1 were set as the threshold for significantly differential expression. Hierarchical cluster analysis of differentially expressed genes was performed to demonstrate the expression pattern of genes in different groups and samples. Open database sources, including the Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG)31–33, MetaboAnalyst, Human Metabolome Database and National Center for Biotechnology Information (NCBI) were used to identify metabolic pathways.
Joint analysis of the metabolomics and transcriptomics
Finally, comprehensive transcriptomics and metabolomics analyses were conducted using MetaboAnalyst 5.0 to perform a joint-pathway analysis module for topological assessment of individual molecules (i.e., nodes) based on their position in the network. Official gene symbols and compound names, along with optional fold changes, were entered to evaluate the potential significance of each molecule within the pathway. Topological analysis encompassed assessing impact values by considering degree centrality, which measures the number of links connecting a node; betweenness centrality, which quantifies the number of shortest paths passing through a given node; and closeness centrality, which determines overall distance from a specific node to all others. Enrichment analysis employed the hypergeometric test while topology measure utilized degree centrality, and integration method involved combined queries.
Validation of representative differentially expressed genes in transcriptomics analysis
The 8 representative differential genes were selected to verify the accuracy of transcriptomics data. Corresponding primers for the differential genes were designed through the NCBI website (www.ncbi.nlm.nih.gov), and the primer design results are presented in Table S6. Reverse transcription PCR (RT-PCR) was performed using the HiScript III All-in-one RT SuperMix kit and AceQ qPCR SYBR Green Master Mix kit (Vazyme, Nanjing) based on kit instructions. The reference gene NAGK was selected as an internal control for mRNA abundance34. Fold changes in the levels of target gene mRNA were determined using the formula 2−ΔΔCt. SPSS was used for statistical analysis of sample information and analysis methods such as ANOVA and two independent samples t-test were used for difference analysis. P < 0.05 was considered as statistically significant.
Institutional review board statement
This study was granted approval by the Ethics Committee of Henan Children's Hospital (2019-H-K11), and all procedures were conducted in accordance with relevant guidelines and regulations.
Results
The research scheme
The general concept of this research is illustrated in Scheme 1. Metabolomics analysis was conducted on a total of 96 plasma samples, comprising 28 cases of MNA NB and 68 cases of non-MNA NB. Additionally, transcriptomics analysis was performed on 52 tissue samples from NB patients, including 15 cases of MNA NB and 37 cases of non-MNA NB. Ultimately, through the integration and comprehensive analysis of metabolomics and transcriptomics data, we elucidated the aberrant pathway network associated with MNA NB. This study is anticipated to provide a theoretical foundation for investigating the molecular mechanisms underlying MNA NB as well as identifying potential therapeutic targets.
Scheme 1.
Schematic diagram of the process of this study.
The Metabolome differences between MNA NB and non-MNA NB
To explore the differences in metabolites between MNA NB and non-MNA NB, plasma metabolomics analysis was first conducted using a non-targeted metabolomics-based approach.
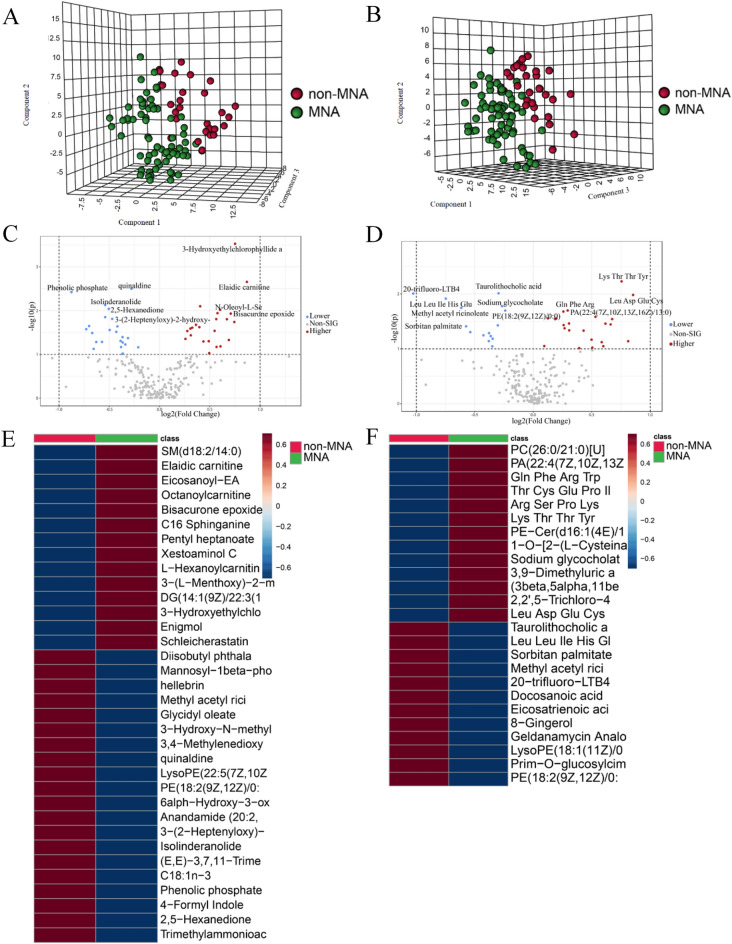
The PCA plot demonstrates the robustness of our results by revealing the close clustering of QC samples under both positive and negative modes (Fig. S2). To visually depict metabolomic differences between MNA NB and non-MNA NB, we performed cluster analysis on plasma metabolites of NB based on compound correlations, which is presented in the form of a heatmap (Fig. S3). The differences between MNA NB and non-MNA NB were visualized using PLS-DA plots in positive mode and negative mode (Fig. 1A and B), while the cross-validation scores plot is presented in Fig. S4. Additionally, volcano plots were employed to illustrate the metabolites in MNA NB and non-MNA NB under positive and negative modes (Fig. 1C and D). Metabolites exhibiting a fold change > 1.2 (fold change < 0.83) with a significance level of P < 0.05 in the volcano plot were identified as differential metabolites. In the field of metabolomics, employing positive and negative modes to characterize differentially expressed metabolites can enhance coverage: positive modes effectively capture metabolites with positive charges, such as amino acids and specific peptides; whereas negative modes are suitable for detecting metabolites with negative charges, including certain organic acids and fatty acids. Furthermore, distinct categories of metabolites exhibit varying ionization efficiencies during the process, thus utilizing positive and negative modes separately can enhance the detection sensitivity for specific groups of metabolites. Therefore, a total of 46 metabolites were identified through metabolomics analysis (Table 1). In the positive mode, a total of 28 metabolites in MNA NB exhibited significant alterations, with 13 showing higher expression and 15 showing lower expression. Similarly, in the negative mode, we identified 18 differential compounds specifically associated with MNA NB, comprising of 8 compounds with higher expression and 10 compounds with lower expression. According to the results of differential metabolite analysis, there were significant alterations in the expression of amino acids, carnitine, and esters. These findings suggest that MNA disrupts multiple metabolic classes and may impact the development of MNA NB. Additionally, considering the substantial inter-individual variability in NB, we performed an inter-group comparison heatmap alongside the differential metabolite. Heatmaps depicting the correlation among differential compounds were generated to visually represent the differential metabolites (Fig. 1E and F). In summary, the metabolomics results demonstrate substantial differences in the metabolomics between MNA NB and non-MNA samples.
Fig. 1.
The plasma metabolomics analysis between MNA NB and non-MNA NB. The PLS-DA results of MNA NB and non-MNA NB in (A) positive mode and (B) negative mode. The volcano plot of metabolite of MNA NB and non-MNA NB in (C) positive mode and (D) negative mode. The heatmap shows clear distinction of metabolites between MNA NB and non-MNA NB in (E) positive mode and (F) negative mode.
Table 1.
Differential expressed metabolites between MNA NB and non-MNA NB.
| NO. | Metabolites | VIP value | Fold change | P value | Expression | Mode |
|---|---|---|---|---|---|---|
| 1 | Phenolic phosphate | 2.0419 | 0.5447 | 0.004 | Lower | Positive |
| 2 | (E,E)-3,7,11-Trimethyl-2,6,10-dodecatrienyl heptanoate | 1.3231 | 0.64625 | 0.052 | Lower | Positive |
| 3 | Anandamide (20:2, n-6) | 1.2881 | 0.67228 | 0.052 | Lower | Positive |
| 4 | Isolinderanolide | 1.9276 | 0.6873 | 0.007 | Lower | Positive |
| 5 | Methyl acetyl ricinoleate | 1.6441 | 0.70866 | 0.028 | Lower | Positive |
| 6 | Glycidyl oleate | 1.8371 | 0.72134 | 0.015 | Lower | Positive |
| 7 | 3-(2-Heptenyloxy)-2-hydroxypropyl undecanoate | 1.7345 | 0.74772 | 0.023 | Lower | Positive |
| 8 | Diisobutyl phthalate | 1.4004 | 0.76426 | 0.049 | Lower | Positive |
| 9 | LysoPE(22:5(7Z,10Z,13Z,16Z,19Z)/0:0) | 1.2718 | 0.76678 | 0.068 | Lower | Positive |
| 10 | 6α-Hydroxy-3-oxo-5β-cholan-24-oic Acid | 1.1805 | 0.77431 | 0.097 | Lower | Positive |
| 11 | Mannosyl-1beta-phosphomy-coketide C30 | 1.5977 | 0.77486 | 0.040 | Lower | Positive |
| 12 | 3,4-Methylenedioxy-benzoic acid | 1.3702 | 0.77529 | 0.053 | Lower | Positive |
| 13 | 3-Hydroxy-N-methylpyridinium | 1.3362 | 0.78963 | 0.059 | Lower | Positive |
| 14 | PE(18:2(9Z,12Z)/0:0) | 1.4403 | 0.80419 | 0.042 | Lower | Positive |
| 15 | quinaldine | 2.3218 | 0.82394 | 0.003 | Lower | Positive |
| 16 | C16 Sphinganine | 1.5118 | 1.2103 | 0.029 | Higher | Positive |
| 17 | 3-(l-Menthoxy)-2-methylpropane-1,2-diol | 1.5552 | 1.2366 | 0.026 | Higher | Positive |
| 18 | Pentyl heptanoate | 1.5625 | 1.3151 | 0.024 | Higher | Positive |
| 19 | Eicosanoyl-EA | 1.3906 | 1.4033 | 0.050 | Higher | Positive |
| 20 | SM(d18:2/14:0) | 1.0469 | 1.4078 | 0.093 | Higher | Positive |
| 21 | DG(14:1(9Z)/22:3(10Z,13Z,16Z)/0:0)[iso2] | 1.4438 | 1.4807 | 0.067 | Higher | Positive |
| 22 | l-Hexanoylcarnitinen-butyl ester | 1.7871 | 1.4904 | 0.011 | Higher | Positive |
| 23 | N-Oleoyl-l-Serine | 1.8365 | 1.5179 | 0.010 | Higher | Positive |
| 24 | Schleicherastatin 3 | 1.1984 | 1.5215 | 0.065 | Higher | Positive |
| 25 | Xestoaminol C | 1.4081 | 1.612 | 0.047 | Higher | Positive |
| 26 | Bisacurone epoxide | 1.7433 | 1.6327 | 0.012 | Higher | Positive |
| 27 | ( ±)-Octanoylcarnitine | 1.614 | 1.6736 | 0.018 | Higher | Positive |
| 28 | Elaidic carnitine | 2.1475 | 1.825 | 0.002 | Higher | Positive |
| 29 | Methyl acetyl ricinoleate | 1.9165 | 0.64537 | 0.018 | Lower | Negative |
| 30 | Leu Leu Ile His Glu | 2.0268 | 0.65018 | 0.025 | Lower | Negative |
| 31 | Sorbitan palmitate | 1.6781 | 0.6902 | 0.054 | Lower | Negative |
| 32 | Docosanoic acid | 1.5511 | 0.74246 | 0.066 | Lower | Negative |
| 33 | Prim-O-glucosylcimifugin | 1.5826 | 0.75566 | 0.044 | Lower | Negative |
| 34 | Eicosatrienoic acid | 1.5399 | 0.78633 | 0.072 | Lower | Negative |
| 35 | ( ±)8-Gingerol | 1.6914 | 0.82259 | 0.048 | Lower | Negative |
| 36 | PE(18:2(9Z,12Z)/0:0) | 1.883 | 0.82922 | 0.013 | Lower | Negative |
| 37 |
Taurolithocholic acid 3-glucuronide |
2.0878 | 0.76077 | 0.019 | Lower | Negative |
| 38 | LysoPE(18:1(11Z)/0:0) | 1.2226 | 0.79486 | 0.091 | Lower | Negative |
| 39 | Arg Ser Pro Lys | 1.6222 | 1.2435 | 0.035 | Higher | Negative |
| 40 | Gln Phe Arg Trp | 1.884 | 1.3103 | 0.008 | Higher | Negative |
| 41 | PA(22:4(7Z,10Z,13Z,16Z)/13:0) | 1.8049 | 1.4571 | 0.020 | Higher | Negative |
| 42 | (3beta,5alpha,11beta,17beta)-9-Fluoro-17-methyland-rostane-3,11,17-triol | 1.7148 | 1.5435 | 0.026 | Higher | Negative |
| 43 | 3,9-Dimethyluric acid | 1.7036 | 1.5673 | 0.030 | Higher | Negative |
| 44 | PE-Cer(d16:1(4E)/19:0) | 1.3171 | 1.5996 | 0.057 | Higher | Negative |
| 45 | Thr Cys Glu Pro Ile | 1.7754 | 1.7976 | 0.015 | Higher | Negative |
| 46 | Leu Asp Glu Cys | 2.0686 | 2.1534 | 0.004 | Higher | Negative |
The altered pathways and biomarkers between MNA NB and non-MNA NB acquired using metabolomics-based approach
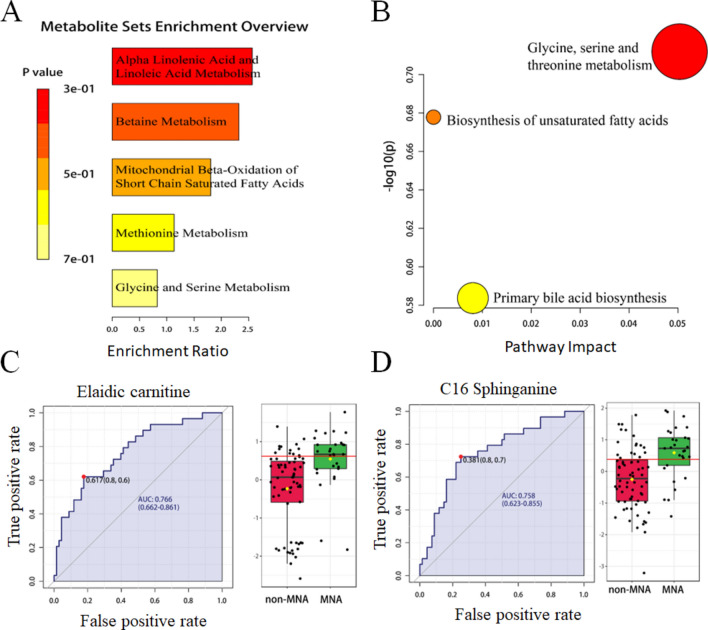
The enrichment and pathway analyses were performed on differential metabolites to identify abnormal metabolic pathways between MNA NB and non-MNA NB. Based on the 46 most altered metabolites, significant differences were observed in alpha linolenic acid and linoleic acid metabolism, betaine metabolism, mitochondrial beta-oxidation of short chain saturated fatty acids, methionine metabolism, glycine and serine metabolism in the enrichment analysis (Fig. 2A). In the pathway analysis (Fig. 2B), significant differences were found in glycine, serine and threonine metabolism, biosynthesis of unsaturated fatty acids, and primary bile acid biosynthesis. Furthermore, to investigate the impact of MNA on plasma biomarkers in metabolomics and facilitate subsequent clinical diagnosis, we conducted receiver operating characteristic (ROC) curve analysis for differential metabolites. The expression levels of elaidic carnitine and C16 sphinganine were found to be significantly different between MNA NB and non-MNA NB groups, with all ROC curves exhibiting an area under the curve (AUC) greater than 0.7 (Fig. 2C, D). Additional biomarkers are presented in Fig. S5 and Table S3.
Fig. 2.
The altered pathways and biomarkers in metabolomics. (A) The enrichment analysis of differential metabolism revealed various metabolic changes between MNA NB and non-MNA NB. (B) The pathway analysis revealed significant abnormalities in the pathways between MNA NB and non-MNA NB. The representative metabolic biomarker ROC curve and boxplot of (C) elaidic carnitine and (D) C16 Sphinganine.
Therefore, regulatory metabolic pathways of MNA in NB were identified to include alpha linolenic acid and linoleic acid metabolism, beta oxidation of short chain fatty acids harvested from mitochondria, methyl metabolism, glycine, serine and threonine metabolism, biosynthesis of unsaturated fatty acids, and primary bile acid biosynthesis. Additionally, elaidic carnitine and C16 Sphinganine were discovered as potential biomarkers for clinical treatment. In summary, metabolomics revealed multiple abnormal metabolic pathways between MNA NB and non-MNA NB that provide a theoretical basis for investigating the molecular mechanisms underlying MNA NB.
Transcriptomics analysis uncovers the abnormal expression gene between MNA NB and non-MNA NB
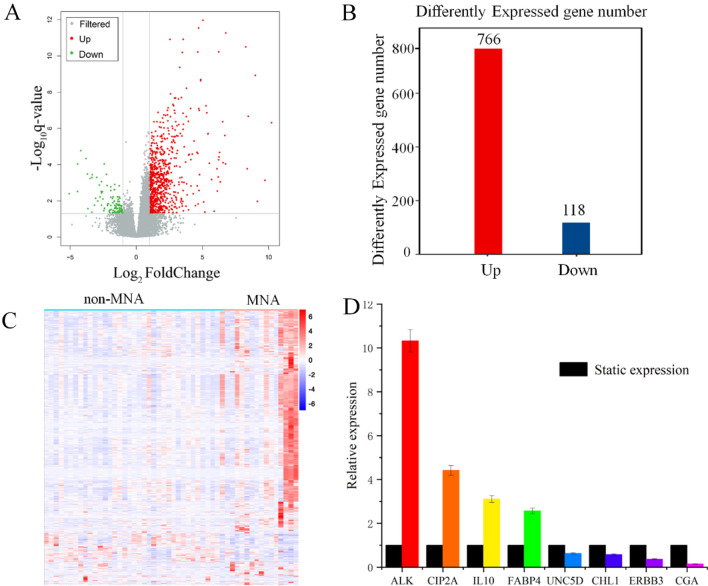
To further investigate the transcriptome differences between MNA NB and non-MNA NB, we conducted a comparative analysis of the transcriptomics from 15 MNA NB tissues and 37 non-MNA NB tissues. The detailed results of RNA quality assessment, including total RNA concentration, A260/A280, A260/A230, 28S/18S ratio, and RIN values for the extracted samples are presented in Table S4. These results confirm that the RNA quality of our NB samples meets the required standards for subsequent analyses. Additionally, preprocessing of sequencing data revealed high-quality raw bases ranging from 6.49G to 7.76G per sample, clean bases ranging from 6.00G to 7.22G per sample, and Q30 base percentages ranging from 92.59 to 95.18% across all samples. Furthermore, the GC content ranged from 44.29 to 50.53%, indicating consistent high data quality among different samples (Table S5). The FPKM values (Fig. S6) and the total number of detected mRNAs in each sample (Fig. S7) reflect the expression abundance of genes across different samples, thereby highlighting inter-sample variations. Moreover, to visually represent the transcriptomics disparities between MNA NB and non-MNA NB, alterations in genes affected by MNA were initially depicted using volcano plots. Differential genes were identified among NB tissue RNAs with P < 0.05 and |log2(fold change)|> 1 in the volcano plot (Fig. 3A). A total of 884 differential genes were discovered, comprising of 766 higher expression genes and 118 lower expression genes (Fig. 3B). The top 20 higher expression and lower expression genes in both MNA and non-MNA NB are presented in Tables 2 and 3 respectively. Furthermore, a cluster analysis heatmap was employed to present the differential gene expression patterns more intuitively (Fig. 3C), demonstrating significant distinctions between MNA NB and non-MNA NB. Based on the transcriptomics results and NB-related literature35–42, 8 reported differential genes related to NB were selected for RT-PCR validation. These genes included ALK, CIP2A, IL10, FABP4, UNC5D, CHL1, ERBB3 and CGA. RT-PCR results showed that the relative expression levels of ALK, CIP2A, IL10 and FABP4 were higher in MNA NB than in non-MNA NB, while the relative expression levels of UNC5D, CHL1, ERBB3 and CGA were lower in non-MNA NB. In particular, the transcriptomics results were consistent with the RT-PCR results, showing the reliability of the transcriptomics results (Fig. 3D, Table S6). In summary, the transcriptomics results showed that MNA NB and non-MNA NB metabolomes were significantly different.
Fig. 3.
The NB tissue transcriptomics and validation. (A) The volcano plot shows differentially expressed genes between MNA NB and non-MNA NB. (B) Number of differentially expressed genes of MNA NB compared to non-MNA NB. (C) The heatmap shows segregation of MNA NB and non-MNA NB based on transcriptomics analysis. (D) Expression trends of genes in RT-PCR were consistent with transcriptomics results.
Table 2.
The top 20 genes significantly higher expression between MNA NB and non-MNA NB.
| NO. | Gene | Description | Fold change | P value |
|---|---|---|---|---|
| 1 | H4C8 | H4 clustered histone 8 | 32.563 | 6.31E−17 |
| 2 | H2AC11 | H2A clustered histone 11 | 28.999 | 1.43E−12 |
| 3 | H3C4 | H3 clustered histone 4 | 28.915 | 1.79E−12 |
| 4 | H2BC9 | H2B clustered histone 9 | 26.890 | 1.67E−10 |
| 5 | H4C5 | H4 clustered histone 5 | 26.245 | 1.18E−10 |
| 6 | H2AC8 | H2A clustered histone 8 | 26.071 | 3.39E−16 |
| 7 | H2BC18 | H2B clustered histone 18 | 24.947 | 2.87E−14 |
| 8 | WDR72 | WD repeat domain 72 | 24.864 | 2.12E−07 |
| 9 | ZNF732 | zinc finger protein 732 | 20.561 | 2.69E−08 |
| 10 | ABCA12 | ATP binding cassette subfamily A member 12 | 18.707 | 2.41E−07 |
| 11 | H3C1 | H3 clustered histone 1 | 17.349 | 5.77E−07 |
| 12 | H4C9 | H4 clustered histone 9 | 17.067 | 1.17E−10 |
| 13 | H2AC13 | H2A clustered histone 13 | 15.954 | 2.54E−07 |
| 14 | KCNH5 | potassium voltage-gated channel subfamily H member 5 | 15.419 | 2.85E−07 |
| 15 | TEX15 | testis expressed 15, meiosis and synapsis associated | 15.180 | 9.92E−10 |
| 16 | H2BC11 | H2B clustered histone 11 | 13.812 | 5.03E−12 |
| 17 | TRIM71 | tripartite motif containing 71 | 13.666 | 4.92E−10 |
| 18 | PLIN4 | perilipin 4 | 32.563 | 8.61E−07 |
| 19 | PLIN1 | perilipin 1 | 28.999 | 6.80E−08 |
| 20 | H3C10 | H3 clustered histone 10 | 28.915 | 1.70E−09 |
Table 3.
The top 20 genes significantly lower expression between MNA NB and non-MNA NB.
| NO. | Gene | Description | Fold Change | P value |
|---|---|---|---|---|
| 1 | CYP11B1 |
cytochrome P450 family 11 subfamily B member 1 |
0.030 | < 0.001 |
| 2 | HSD3B2 | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 | 0.046 | < 0.001 |
| 3 | GRP | gastrin releasing peptide | 0.046 | 1.16E−06 |
| 4 | NCMAP | non-compact myelin associated protein | 0.054 | 1.28E−07 |
| 5 | CRH | corticotropin releasing hormone | 0.071 | 4.85E−07 |
| 6 | CGA | glycoprotein hormones, alpha polypeptide | 0.077 | 6.26E−06 |
| 7 | GSTA1 | glutathione S-transferase alpha 1 | 0.079 | < 0.001 |
| 8 | CYP21A2 |
cytochrome P450 family 21 subfamily A member 2 |
0.088 | 1.36E−05 |
| 9 | CYP17A1 |
cytochrome P450 family 17 subfamily A member 1 |
0.097 | 0.0047 |
| 10 | UCN3 | urocortin 3 | 0.097 | 1.15E−05 |
| 11 | ADCYAP1 | adenylate cyclase activating polypeptide 1 | 0.099 | 7.18E−06 |
| 12 | PNPLA5 |
patatin like phospholipase domain containing 5 |
0.111 | 0.0014 |
| 13 | FIBCD1 | fibrinogen C domain containing 1 | 0.118 | < 0.001 |
| 14 | SST | somatostatin | 0.125 | 1.14E−05 |
| 15 | SBK2 | SH3 domain binding kinase family member 2 | 0.132 | < 0.001 |
| 16 | PROK1 | prokineticin 1 | 0.143 | 4.31E−05 |
| 17 | SLC25A48 | solute carrier family 25 member 48 | 0.149 | < 0.001 |
| 18 | FGF16 | fibroblast growth factor 16 | 0.157 | < 0.001 |
| 19 | SFRP5 | secreted frizzled related protein 5 | 0.160 | 2.44E−05 |
| 20 | LRRC38 | leucine rich repeat containing 38 | 0.160 | 0.0013 |
KEGG and GO analysis between MNA NB and non-MNA NB using transcriptomics-based approach
The differential genes between MNA NB and non-MNA NB were utilized for GO analysis and KEGG analysis to identify aberrant pathways. Metabolic pathways associated with the obtained differential genes were analyzed through GO analysis, aiming to elucidate their potential biological functions (Figs. 4A and S8). Regarding biological processes, the top 3 aberrant expressions consisted of cellular process, single-organism process and biological regulation. Regarding cellular component, the top 3 significantly aberrant expressions were cell, cell part and organelle. Regarding molecular function, the top 3 significantly aberrant expressions were binding, catalytic activity and molecular transducer activity. We further conducted KEGG prediction analysis and observed that the signaling molecules and interaction pathway exhibited the most significant alterations, suggesting an alternative perspective on the biological functions of MNA NB (Figs. 4B, S9). Relationships between differential genes were visualized by protein–protein interaction (PPI) circle diagrams (Fig. S10), illustrating the close connection between genes such as CDC6, CDC45, MCM2, CCNB1 etc. The identified genes were found to be associated with the processes of mitosis and DNA replication43, as well as being implicated in abnormalities related to these processes according to the results of GO analysis (Fig. S11). This suggests that MNA may play a pivotal role in the intricate mechanisms governing mitosis and DNA replication. Consequently, cellular process, single-organism process, biological regulation, cell, cell part, organelle, binding, catalytic activity, molecular transducer activity, signaling molecules and interaction were molecular mechanisms of MNA that affected the prognosis of NB. Therefore, based on the transcriptomics method, multiple biological functional differences between MNA NB and non-MNA NB have been discovered, which is expected to provide a theoretical basis for exploring the molecular mechanism of MNA NB.
Fig. 4.
The GO and KEGG analysis between MNA NB and non-MNA NB in transcriptomics. (A) GO analysis of biological processes, molecular functions and cellular components organization between MNA NB and non-MNA NB. (B) KEGG analyzes from six aspects of cellular processes, environmental information, genetic information processing, human diseases, metabolism, organismal systems between MNA NB and non-MNA NB.
Integrated transcriptomics and metabolomics analyses between MNA NB and non-MNA NB
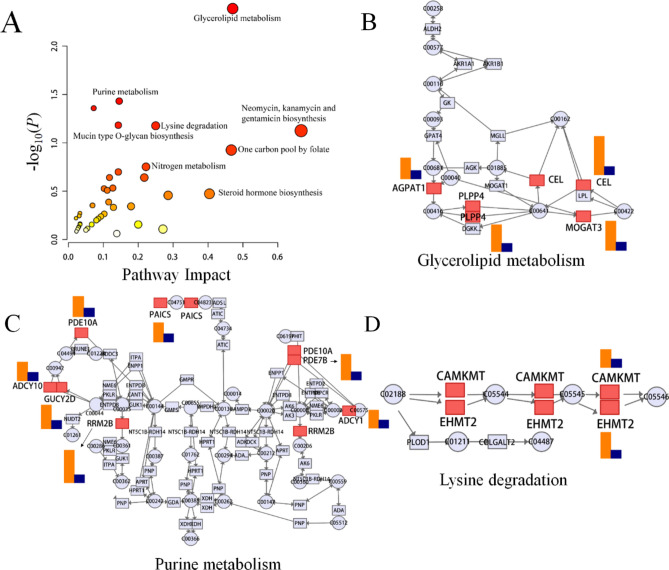
Multi-omics research employs omics integration, which facilitates the integration of data and regulatory relationships across various levels. This approach enables a comprehensive exploration of the mechanistic actions of specific genes in diseases from diverse perspectives and facets. In order to establish connections between crucial metabolites and genes via shared metabolic pathways, we systematically investigated the altered pathways of MNA in NB and conducted joint-pathway analysis using MetaboAnalyst 5.0. The modified pathways are depicted in Fig. 5A and summarized in Table 4, encompassing glycerolipid metabolism, purine metabolism, lysine degradation, as well as other metabolic pathways. These findings suggest that MNA impacts NB through these specific metabolic routes. Glycerolipid metabolism, shown in Fig. 5B, had P values < 0.05 and an impact coefficient of 0.470, and included significantly changed genes consisting of AGPAT1, CEL, PLPP4 and MOGAT3. As shown in Fig. 5C, in the purine metabolism, the expression levels of PDE10A, PAICS, PDE7B, ADCY10, GUCY2D, RRM2B and ADCY1 were altered in the MNA NB compared to non-MNA NB. Figure 5D shows that in lysine degradation, CAMKMT and EHMT2 were altered in MNA NB. Abnormal pathways suggest that the molecular mechanism of MNA changes in NB. Related differentially expressed genes by joint-pathway analysis are shown in Table 5 and other altered pathways are shown in Fig. S11, including mucin type O-glycan biosynthesis, nitrogen metabolism, one carbon pool by folate pathway, starch and sucrose metabolism. Hence, glycerolipid metabolism, purine metabolism, and lysine degradation were found to be altered in MNA NB through joint metabolomics and transcriptomics analysis, which could provide a theoretical basis for future treatment of MNA NB.
Fig. 5.
Integrated transcriptomics and metabolomics analysis of NB metabolic pathways. (A) Joint-pathway analysis of differential pathway between MNA NB and non-MNA NB. (B) The glycerolipid metabolism pathway, (C) the purine metabolism pathway and (D) the lysine degradation pathway with altered significantly genes in MNA NB compared to non-MNA NB. (Significant overexpression in red, and no significant changes in grey. The size of the bubbles represents the number of genes enriched, with larger bubbles indicating a higher enrichment level while the color indicates the significance of enrichment, the redder the color, the higher the significance. In (B–D), the orange color represents the corresponding gene's fold change value, while blue indicates the baseline value.)
Table 4.
Differential metabolic pathways based on joint-pathway analysis.
| NO. | Pathway name | Match status | P value | Impact | ||
|---|---|---|---|---|---|---|
| 1 | Glycerolipid metabolism | 4/35 | 0.004 | 0.47059 | ||
| 2 | Purine metabolism | 7/166 | 0.037 | 0.14545 | ||
| 3 | Drug metabolism—other enzymes | 4/70 | 0.044 | 0.072464 | ||
| 4 | Mucin type O-glycan biosynthesis | 2/22 | 0.066 | 0.14286 | ||
| 5 | Lysine degradation | 3/49 | 0.067 | 0.25 | ||
| 6 | Neomycin, kanamycin and gentamicin biosynthesis | 1/4 | 0.075 | 0.66667 | ||
| 7 | One carbon pool by folate | 2/31 | 0.119 | 0.46667 | ||
| 8 | Nitrogen metabolism | 1/10 | 0.177 | 0.22222 | ||
| 9 | Starch and sucrose metabolism | 2/43 | 0.200 | 0.14286 | ||
| 10 | Retinol metabolism | 2/47 | 0.229 | 0.21739 | ||
Table 5.
Related differentially expressed genes by joint-pathway analysis.
| Gene | Enriched pathway | Function |
|---|---|---|
| AGPAT1 | Glycerolipid metabolism | 1-acylglycerol-3-phosphate O-acyltransferase 1 |
| PLPP4 | Glycerolipid metabolism | Phospholipid phosphatase 4 |
| CEL | Glycerolipid metabolism | Carboxyl ester lipase |
| MOGAT3 | Glycerolipid metabolism | Monoacylglycerol O-acyltransferase 3 |
| PAICS | Purine metabolism | Phosphoribosylaminoimidazole carboxylase and Phosphoribosylaminoimidazo-lesuccinocarboxamide synthase |
| PDE10A | Purine metabolism | Phosphodiesterase 10A |
| ADCY10 | Purine metabolism | Adenylate cyclase 10 |
| GUCY2D | Purine metabolism | Guanylate cyclase 2D, retinal |
| PDE7B | Purine metabolism | Phosphodiesterase 7B |
| RRM2B | Purine metabolism |
Ribonucleotide reductase regulatory TP53 inducible subunit M2B |
| CAMKMT | Lysine degradation | Calmodulin-lysine N-methyltransferase |
| EHMT2 | Lysine degradation | Euchromatic histone lysine methyltransferase 2 |
| GALNT8 |
Mucin type O-glycan biosynthesis |
Polypeptide N-acetylgalactosaminyltransferase 8 |
| CA1 | Nitrogen metabolism | Carbonic anhydrase 1 |
| DHFR | One carbon pool by folate | Dihydrofolate reductase |
| TYMS | One carbon pool by folate | Thymidylate synthetase |
| HK1 | Starch and sucrose metabolism | Hexokinase 1 |
| GYG1 | Starch and sucrose metabolism | Glycogenin 1 |
Discussion
It is well known that the MNA is closely associated with the late stages of NB and poor prognostic outcomes. It has been found that MNA is a potential carcinogenic driver in the developing nervous system and leads to the development of NB10,44. However, MNA is not always positively correlated with mRNA or protein expression, suggesting a more complex interaction of MNA with NB45. In this study, 96 clinical plasma samples and 52 clinical NB tissue samples were analyzed and 884 differential genes and 46 differential metabolites were identified. Metabolites elaidic carnitine and C16 sphinganine were found to be key metabolites in MNA NB. The metabolomics and transcriptomics characteristics of MNA NB were demonstrated and a comprehensive network analysis was carried out. Compared with non-MNA NB, MNA NB showed significant differences in glycerolipid metabolism, purine metabolism, amino acid metabolism. The genes GPAT1, CEL, PLPP4, MOGAT3, PDE10A, PAICS, PDE7B, ADCY10, GUCY2D, RRM2B, ADCY1, CAMKMT and EHMT2 involved in glycerolipid metabolism, purine metabolism, and amino acid metabolism were regulated by MNA. Our study provides deep insight into the molecular mechanism of MNA on NB, and identified key metabolites as potential targets of MNA, providing a basis for future therapeutic research on NB.
MNA affects glycerolipid metabolism in NB
In the joint-pathway analysis, glycerolipid metabolism was found to be significantly changed in MNA NB where MYCN can directly regulate glycerolipid synthesis, degradation and accumulation46. Additionally, secondary messengers of glycerolipids activate downstream oncogenic signaling and serve as a fatty acid reservoir for energy storage, preventing the accumulation of toxic fatty acids that support tumor growth, corroborating the positive effect of MNA on tumors47. Glycerolipid metabolism had been proven to be a potential independent prognostic factor in colon cancer and positively correlated with cancer hallmark pathways including bile acid metabolism, xenobiotic metabolism, and peroxisome and negatively correlated with pathways such as interferon gamma response, allograft rejection, apoptosis, and inflammatory response (P < 0.05)48. Moreover, the abnormal expression of glycerolipid metabolism in gastric cancer and bladder cancer shows that glycerolipid metabolism plays a key role in tumors49;50. Nevertheless, few studies have reported the changes of glycerolipid metabolism in NB and no research has found the relationship between MNA and glycerolipid metabolism. Therefore, this study raises the question of whether it is possible to improve the prognosis of MNA NB by studying the relationship between MNA and glycerolipid metabolism.
According to the results, there were significant differences in the expression of AGPAT1, CEL, PLPP4, and MOGAT3 genes. AGPAT1 has been used as a novel colonic biomarker for discriminating between ulcerative colitis with and without primary sclerosing cholangitis, and abnormalities of AGPAT1 have also been found between MNA NB and non-MNA NB51. Therefore, it is necessary to further study whether AGPAT1 can also be a therapeutic target of MNA NB. In addition, MYCN is also believed to play a key role in promoting fatty acid metabolism for sustainable tumor cell growth, and the cell survival of MYCN is also highly dependent on fatty acid uptake, which is consistent with the significant differences in fatty acid metabolism in our metabolomics results, suggesting that fatty acid metabolism may be a promising strategy for HR-NB patients46. In conclusion, the discovery of the potential link between glycerolipid metabolism and MNA provides a new perspective for understanding the molecular mechanism of NB.
MNA influenced purine metabolism in NB
The joint-pathway analysis found that purine metabolism was significantly altered in the MNA NB. Purine, as a rich substrate in organism, which includes DNA, RNA, nucleosides and nucleotides, AMP, ADP, ATP, GMP, GDP, GTP, and cyclic forms of cAMP and cGMP, is an important raw material for cell proliferation and an important factor for immune regulation. Purine is involved in the stabilization of immune regulation and the formation of energy carriers and functions, thereby influencing the growth of both cancer and non-cancer cells52–54. Purine metabolism and purine biosynthesis pathway activities were significantly activated in patients with a poor prognosis of hepatocellular carcinoma, and purine metabolism has also been confirmed to change in ovarian cancer, gastric cancer, breast cancer and other cancers53,55,56. However, the relationship between MNA and purine metabolism has not yet been reported. This study found that purine metabolism has changed significantly due to MNA. Therefore, we demonstrated that there is a complex relationship between MNA and purine metabolism. Exploring the relationship between MNA and purine, and using purine metabolism to inhibit tumor development is a plausible future research direction. Furthermore, it has been found that PAICS knockout in MNA cells significantly reduces cell proliferation, colony formation, migration capacity and DNA synthesis57. Chakravarthi et al.58 found that PAICS plays an important role in the proliferation and invasion of prostate cancer cells, identifying it as an effective therapeutic target. Therefore, we also believe that PAICS may be an effective therapeutic target for MNA NB. Overall, the change of purine metabolism is of great significance in MNA NB, which lays a foundation for clarifying the molecular mechanism of MNA NB and finding effective therapeutic targets.
MNA affect amino acid metabolism in NB
Through metabolomics and joint-pathway analysis, our results showed that amino acid metabolism changed dramatically in MNA NB. In metabolomics, we found significant differences in glycine, serine and threonine metabolism. Serine-glycine metabolism has been found to be critical for tumorigenesis59,60. On the other hand, serine and glycine are synthesized from glycolysis through oxidation of the intermediate 3-PGA, which consists of two processes: de novo serine synthesis from glucose and reversible interconversion of serine into glycine61. These products fuel one-carbon metabolism. In addition, Xia et al.62 found that NB cells with MNA show increased transcriptional activation of the serine-glycine-one-carbon biosynthetic pathway and an increased dependence on this pathway for supplying glucose-derived carbon for serine and glycine synthesis. Metabolic abnormalities of serine and glycine were also observed in other cancers, such as glioblastoma and non-small cell lung cancer63,64. In the joint-pathway analysis, we found that lysine degradation was a significantly changed pathway. Lysine is an essential amino acid for the human body, and it must be taken in sufficient amount to maintain protein synthesis. It has been found that the lysine degradation pathway is clinically relevant due to the occurrence of two severe neurometabolic disorders (pyridoxine-dependent epilepsy and glutaric aciduria type 1)65. In addition, lysine degradation may also be related to the development of early myocardial hypertrophy66. Moreover, lysine degradation was found to be significantly different between MNA NB and non-MNA NB, and the potential mechanism of lysine degradation in MNA NB requires further study. Overall, this study showed that amino acid metabolism plays a significant role in the initiation and progression of MNA NB, and the disclosure of amino acid metabolism is expected to offer novel opportunities for advancing the treatment and prognostic evaluation of NB.
Potential therapeutic targets of MNA NB
The discovery of effective therapeutic targets of MNA NB is very important for improving the therapeutic effect of MNA NB. The study conducted by Schonheer et al.67 demonstrated that both wild-type and acquired ALK mutants can induce the transcription of the MYCN promoter through the activation of the downstream molecule ERK, thereby activating MYCN mRNA transcription in neurons and NB. The ALK gene is a direct transcriptional target of MYCN, and MYCN-induced ALK activation contributed to the occurrence and development of NB68–70. Hasan et al., discovered that MYCN has a positive feedback loop which directly regulates ALK expression, thereby enhancing MYCN's oncogenic activity and promoting rapid malignant transformation. Additionally, overexpression of MYCN induces promoter activity of the ALK gene, resulting in elevated levels of ALK expression in NB cells71. The present study further substantiates the significant upregulation of ALK in MNA NB tissues, thereby reinforcing the strong correlation between ALK and MNA. In human cancer cells, CDK2 is an essential component of the cell cycle with key function in tumorigenesis72. Our findings also confirmed the relationship between CDK2 and MNA, thus further investigation into the underlying mechanism between CDK2 and MNA may help promote a new treatment strategy for MNA NB73. Nozato et al.74 found that the expression level of ERBB3 was significantly reduced in MNA NB, confirming that the low expression of ERBB3 affected the progression of NB by affecting the mechanism of epithelial-mesenchymal transition. The decreased expression of ERBB3 was also associated with MNA NB and poor survival rate. The transcriptomics and RT-PCR findings in our study further support the notion of diminished ERBB3 gene expression in MNA NB, thus reinforcing the established association between ERBB3 and MNA. Therefore, these genes should be further studied as potential therapeutic targets to improve the prognosis of MNA NB.
According to the results presented in Table 2, our findings indicate that the significantly altered differentially expressed genes primarily pertain to histones, which are fundamental components of chromatin organization. Histones form a protein complex around which DNA is wrapped, forming nucleosomes. These histone proteins can undergo specific amino acid residue modifications such as acetylation or methylation, and these modifications play a crucial role in regulating gene expression75. The regulation of gene expression through the remodeling of tryptophan structure is governed by histone deacetylases, a class of enzymes. However, dysregulation in the expression and activity of these enzymes leads to an imbalance in histone acetylation, thereby promoting the progression of NB76.
Based on our KEGG results, we have identified the significant involvement of the immune system in NB. The immune evasion of NB is facilitated by various mechanisms, including low immunogenicity, upregulation of immune checkpoint molecules, and secretion of immunomodulatory mediators. These characteristics make it a unique model for studying tumor immunity. Currently, it is widely accepted that the diminished immunogenicity in NB can be attributed to MNA, which leads to reduced expression of MHC-I in HR-NB patients compared to those with LR-NB. This poses a significant challenge for T cell-mediated immunotherapy77. On the contrary, NB cells secrete transforming growth factor β1 and galectin-1, which induce functional impairment in cytotoxic T lymphocytes and NK cells, consequently leading to immunosuppression78.
Based on joint analysis, we have identified a correlation between drug metabolism enzymes and aberrant biosynthesis of neomycin, kanamycin, and gentamicin. However, it is important to note that these antibiotics are not utilized in the clinical management of NB; therefore, further investigation is required to establish any specific potential association.
Conclusion
In summary, a metabolomic-based approach identified 46 differential metabolites between MNA NB and non-MNA NB. Enrichment analysis revealed significant differences in the metabolism of alpha linolenic acid, linoleic acid, and betaine. Transcriptomics analysis further identified 884 differential genes between MNA NB and non-MNA NB. GO analysis demonstrated significant alterations in biological functions such as cellular processes, single organizational processes, and biological regulations when comparing MNA NB to non-MNA NB. In addition, the KEGG analysis revealed significant alterations in signaling molecules and interactions between MNA NB and non-MNA NB. Specifically, the joint-pathway analysis demonstrated notable disparities in the expression of glycerolipid metabolism, purine metabolism, and lysine degradation pathways between MNA-NB and non-MNA NB, suggesting that MNA impacts glycerolipid metabolism, purine metabolism, and amino acid metabolism in NB. Furthermore, ALK and CDK2 were identified as potential therapeutic targets for MNA NB. In conclusion, this study provides a theoretical foundation for investigating the molecular mechanisms underlying MNA NB and identifying potential therapeutic targets.
Supplementary Information
Acknowledgements
The authors sincerely thank all participants for participating in this study, as this study would not be possible without their valuable contributions.
Author contributions
The manuscript was written by B. D.; Y. Z. and P. Z. collected the data; M. Z., Z. Y., L. L. and L. H. analyzed the data; Q. W. organized the charts. X. Z. were responsible for the experimental design, while W. Z. oversaw the overall study. All authors critically reviewed the manuscript.
Funding
This work were funded by National Natural Science Foundation of China (32201237), Scientific and technological projects of Henan province (222102310270, 222102310109, 232102311135), Henan medical science and technology program (LHGJ20220767), Henan International Joint Laboratory for Prevention and Treatment of Pediatric Disease foundation (EKB202204).
Data availability
We have submitted the raw RNA-seq data to NCBI (https://www.ncbi.nlm.nih.gov/sra) under the accession number PRJNA884866. Besides, we have uploaded mass spectrometry data to the MetaboLights (https://www.ebi.ac.uk/metabolights/) with the number of MTBLS6352.
Competing interests
The authors declare no competing interests.
Informed consent
Informed consent was signed by children or their parents.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Qionglin Wang, Email: wangqionglin2020@163.com.
Xianwei Zhang, Email: zhangxw956658@126.com.
Wancun Zhang, Email: zhangwancun@126.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-71211-x.
References
- 1.Beaudry, P. et al. A pilot study on the utility of serum metabolomics in neuroblastoma patients and xenograft models. Pediatr. Blood Cancer63, 214–220 (2016). 10.1002/pbc.25784 [DOI] [PubMed] [Google Scholar]
- 2.Zhou, X. et al. Therapy resistance in neuroblastoma: Mechanisms and reversal strategies. Front. Pharmacol.14, 1114295 (2023). 10.3389/fphar.2023.1114295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu, J. et al. Perioperative hypertension and anesthetic management in patients undergoing resection of neuroblastoma. Paediatr. Anaesth.33, 577–582 (2023). 10.1111/pan.14673 [DOI] [PubMed] [Google Scholar]
- 4.Lundberg, K. I., Treis, D. & Johnsen, J. I. Neuroblastoma heterogeneity, plasticity, and emerging therapies. Curr. Oncol. Rep.24, 1053–1062 (2022). 10.1007/s11912-022-01270-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang, H. et al. A Mycn-independent mechanism mediating secretome reprogramming and metastasis in Mycn-amplified neuroblastoma. Sci. Adv.9, eadg6693 (2023). 10.1126/sciadv.adg6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Irwin, M. S. et al. Revised neuroblastoma risk classification system: A report from the children’s oncology group. J. Clin. Oncol.39, 3229–3241 (2021). 10.1200/JCO.21.00278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee, J. W. et al. Clinical significance of Mycn amplification in patients with high-risk neuroblastoma. Pediatr. Blood Cancer.65, e27257 (2018). 10.1002/pbc.27257 [DOI] [PubMed] [Google Scholar]
- 8.Chui, C. Effects of preoperative chemotherapy on neuroblastoma with Mycn amplification: A surgeon’s perspective. World J. Pediatr. Surg.3, e000129 (2020). 10.1136/wjps-2020-000129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rickman, D. S., Schulte, J. H. & Eilers, M. The expanding world of N-Myc–driven tumors. Cancer Discov.8, 150–163 (2018). 10.1158/2159-8290.CD-17-0273 [DOI] [PubMed] [Google Scholar]
- 10.Otte, J., Dyberg, C., Pepich, A. & Johnsen, J. I. Mycn function in neuroblastoma development. Front. Oncol.10, 624079 (2021). 10.3389/fonc.2020.624079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, L. et al. P53 is a direct transcriptional target of Mycn in neuroblastoma. Cancer Res.70, 1377–1388 (2010). 10.1158/0008-5472.CAN-09-2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren, P. et al. Atf4 and N-Myc coordinate glutamine metabolism in Mycn-amplified neuroblastoma cells through Asct2 activation. J. Pathol.235, 90–100 (2015). 10.1002/path.4429 [DOI] [PubMed] [Google Scholar]
- 13.Oliynyk, G. et al. Mycn-enhanced oxidative and glycolytic metabolism reveals vulnerabilities for targeting neuroblastoma. Iscience21, 188–204 (2019). 10.1016/j.isci.2019.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tjaden, B. et al. N-Myc-induced metabolic rewiring creates novel therapeutic vulnerabilities in neuroblastoma. Sci. Rep.10, 7157 (2020). 10.1038/s41598-020-64040-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Floros, K. V. et al. Mycn upregulates the transsulfuration pathway to suppress the ferroptotic vulnerability in Mycn-amplified neuroblastoma. Cell Stress6, 21–29 (2022). 10.15698/cst2022.02.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arlt, B. et al. Inhibiting phosphoglycerate dehydrogenase counteracts chemotherapeutic efficacy against Mycn-amplified neuroblastoma. Int. J. Cancer148, 1219–1232 (2021). 10.1002/ijc.33423 [DOI] [PubMed] [Google Scholar]
- 17.Alptekin, A. et al. Glycine decarboxylase is a transcriptional target of Mycn required for neuroblastoma cell proliferation and tumorigenicity. Oncogene38, 7504–7520 (2019). 10.1038/s41388-019-0967-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa, V., Angelini, C., De Feis, I. & Ciccodicola, A. Uncovering the complexity of transcriptomes with Rna-Seq. J. Biomed. Biotechnol.2010, 1–19 (2010). 10.1155/2010/853916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du, B. et al. Joint analysis of the metabolomics and transcriptomics uncovers the dysregulated network and develops the diagnostic model of high-risk neuroblastoma. Sci. Rep.13, 16991 (2023). 10.1038/s41598-023-43988-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan, X. et al. A comprehensive analysis of potential prognostic biomarkers for Mycn-amplified neuroblastoma. Zhongguo Dang Dai Er Ke Za Zhi.22, 262–268 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, E. et al. Genomic profile of Mycn non-amplified neuroblastoma and potential for immunotherapeutic strategies in neuroblastoma. BMC Med. Genom.13, 171 (2020). 10.1186/s12920-020-00819-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinno, Y. et al. Polycomb Ezh1 regulates cell Cycle/5-fluorouracil sensitivity of neuroblastoma cells in concert with Mycn. Cancer Sci.113, 4193–4206 (2022). 10.1111/cas.15555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spyropoulos, F. et al. Metabolomic and transcriptomic signatures of chemogenetic heart failure. Am. J. Physiol. Heart Circul. Physiol.322, H451–H465 (2022). 10.1152/ajpheart.00628.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao, P. et al. Integration of transcriptomics and metabolomics reveals the antitumor mechanism underlying tadalafil in colorectal cancer. Front. Pharmacol.13, 793499 (2022). 10.3389/fphar.2022.793499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loras, A. et al. Integrative metabolomic and transcriptomic analysis for the study of bladder cancer. Cancers11, 686 (2019). 10.3390/cancers11050686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavill, R., Jennen, D., Kleinjans, J. & Briedé, J. J. Transcriptomic and metabolomic data integration. Brief. Bioinform.17, 891–901 (2016). 10.1093/bib/bbv090 [DOI] [PubMed] [Google Scholar]
- 27.Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics30, 2114–2120 (2014). 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, D., Langmead, B. & Salzberg, S. L. Hisat: A fast spliced aligner with low memory requirements. Nat. Methods12, 357–360 (2015). 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts, A., Trapnell, C., Donaghey, J., Rinn, J. L. & Pachter, L. Improving Rna-Seq expression estimates by correcting for fragment bias. Genome Biol.12, R22 (2011). 10.1186/gb-2011-12-3-r22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell, C. et al. Transcript assembly and quantification by Rna-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol.28, 511–515 (2010). 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa, M. Kegg: Kyoto encyclopedia of genes and genomes. Nucleic. Acids. Res.28, 27–30 (2000). 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci.28, 1947–1951 (2019). 10.1002/pro.3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res.51, D587–D592 (2023). 10.1093/nar/gkac963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang, Y. et al. Dual isothermal amplification all-in-one approach for rapid and highly sensitive quantification of plasma circulating Mycn gene of neuroblastoma. Anal. Biochem.658, 114922 (2022). 10.1016/j.ab.2022.114922 [DOI] [PubMed] [Google Scholar]
- 35.O’Donohue, T. et al. Differential impact of Alk mutations in neuroblastoma. Jco Precis. Oncol.5, 492–500 (2021). 10.1200/PO.20.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bo, L. et al. Bioinformatics analysis of the Cdk2 functions in neuroblastoma. Mol. Med. Rep.17, 3951–3959 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Urso, C. J. & Zhou, H. Role of Cd36 in palmitic acid lipotoxicity in Neuro-2a neuroblastoma cells. Biomolecules11, 1567 (2021). 10.3390/biom11111567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhen, Z. et al. Involvement of Il-10 and Tgf-Β in Hla-E-mediated neuroblastoma migration and invasion. Oncotarget7, 44340–44349 (2016). 10.18632/oncotarget.10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miao, L. et al. Fabp4 deactivates Nf-Κb-Il1Α pathway by ubiquitinating atpb in tumor-associated macrophages and promotes neuroblastoma progression. Clin. Transl. Med.11, e395 (2021). 10.1002/ctm2.395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, H. et al. Unc5D regulates P53-dependent apoptosis in neuroblastoma cells. Mol. Med. Rep.9, 2411–2416 (2014). 10.3892/mmr.2014.2100 [DOI] [PubMed] [Google Scholar]
- 41.Ognibene, M. et al. Chl1 gene acts as a tumor suppressor in human neuroblastoma. Oncotarget9, 25903–25921 (2018). 10.18632/oncotarget.25403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilzen, A. et al. Erbb3 is a marker of a ganglioneuroblastoma/ganglioneuroma-like expression profile in neuroblastic tumours. Mol. Cancer.12, 70 (2013). 10.1186/1476-4598-12-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He, Y. et al. Integrated transcriptome analysis reveals the impact of photodynamic therapy on cerebrovascular endothelial cells. Front. Oncol.11, 731414 (2021). 10.3389/fonc.2021.731414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swartling, F. J. et al. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-Myc. Cancer Cell.21, 601–613 (2012). 10.1016/j.ccr.2012.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eberherr, C. et al. Targeting excessive Mycn expression using Mln8237 and Jq1 impairs the growth of hepatoblastoma cells. Int. J. Oncol.54, 1853–1863 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Tao, L. et al. Mycn-driven fatty acid uptake is a metabolic vulnerability in neuroblastoma. Nat. Commun.13, 3728 (2022). 10.1038/s41467-022-31331-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ackerman, D. et al. Triglycerides promote lipid homeostasis during hypoxic stress by balancing fatty acid saturation. Cell Rep.24, 2596–2605 (2018). 10.1016/j.celrep.2018.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, Z. et al. Multi-omics characterization of a glycerolipid metabolism-related gene enrichment score in colon cancer. Front. Oncol.12, 881953 (2022). 10.3389/fonc.2022.881953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiong, Z. et al. Exploration of lipid metabolism in gastric cancer: A novel prognostic genes expression profile. Front. Oncol.11, 712746 (2021). 10.3389/fonc.2021.712746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen, P., Chen, J., He, L., Du, C. & Wang, X. Identification of Circrna-Mirna-Mrna regulatory network in bladder cancer by integrated analysis. Urol. Int.105, 705–715 (2021). 10.1159/000512066 [DOI] [PubMed] [Google Scholar]
- 51.Vessby, J. et al. Agpat1 as a novel colonic biomarker for discriminating between ulcerative colitis with and without primary sclerosing cholangitis. Clin. Transl. Gastroenterol.13, e00486 (2022). 10.14309/ctg.0000000000000486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.La Grotta, R. et al. Anti-inflammatory effect of Sglt-2 inhibitors via uric acid and insulin. Cell. Mol. Life Sci.79, 273 (2022). 10.1007/s00018-022-04289-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu, J. et al. Targeting purine metabolism in ovarian cancer. J. Ovarian Res.15, 1–93 (2022). 10.1186/s13048-022-01022-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Vitto, H., Arachchige, D., Richardson, B. & French, J. The intersection of purine and mitochondrial metabolism in cancer. Cells10, 2603 (2021). 10.3390/cells10102603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Syniachenko, O. V., Aliiev, R. F., Iermolaieva, M. V. & Bondar, V. G. The changes in purine metabolism in gastric cancer. Gastroenterology53, 223–229 (2019). 10.22141/2308-2097.53.4.2019.182401 [DOI] [Google Scholar]
- 56.Chen, X. & Chen, J. Mir-10B-5P-mediated upregulation of Piezo1 predicts poor prognosis and links to purine metabolism in breast cancer. Genomics.114, 110351 (2022). 10.1016/j.ygeno.2022.110351 [DOI] [PubMed] [Google Scholar]
- 57.Cheung, C. H. Y. et al. “Abstracts of the 77Th Annual Meeting of the Japanese Cancer Association; 2018 Sept 27–29; Osaka, Japan” as Cancer Science, Supplement 2, Vol 109 (2018). Cancer Sci.109(Suppl 2), 1–1444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chakravarthi, B. V. S. K. et al. Expression and role of Paics, a De Novo purine biosynthetic gene in prostate cancer. The Prostate77, 10–21 (2017). 10.1002/pros.23243 [DOI] [PubMed] [Google Scholar]
- 59.Ding, J. et al. The histone H3 methyltransferase G9a epigenetically activates the serine-glycine synthesis pathway to sustain cancer cell survival and proliferation. Cell Metab.18, 896–907 (2013). 10.1016/j.cmet.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao, E. et al. Kdm4C and Atf4 cooperate in transcriptional control of amino acid metabolism. Cell Rep.14, 506–519 (2016). 10.1016/j.celrep.2015.12.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao, E., Hou, J. & Cui, H. Serine-glycine-one-carbon metabolism: Vulnerabilities in Mycn-amplified neuroblastoma. Oncogenesis N. Y. N. Y.9, 14 (2020). 10.1038/s41389-020-0200-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia, Y. et al. Metabolic reprogramming by Mycn confers dependence on the serine-glycine-one-carbon biosynthetic pathway. Cancer Res.79, 3837–3850 (2019). 10.1158/0008-5472.CAN-18-3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liao, L. et al. Upregulation of phosphoserine phosphatase contributes to tumor progression and predicts poor prognosis in non-small cell lung cancer patients. Thorac. Cancer10, 1203–1212 (2019). 10.1111/1759-7714.13064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim, D. et al. Shmt2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature520, 363–367 (2015). 10.1038/nature14363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leandro, J. & Houten, S. M. The lysine degradation pathway: Subcellular compartmentalization and enzyme deficiencies. Mol. Genet. Metab.131, 14–22 (2020). 10.1016/j.ymgme.2020.07.010 [DOI] [PubMed] [Google Scholar]
- 66.Liu, J., Hu, J., Tan, L., Zhou, Q. & Wu, X. Abnormalities in lysine degradation are involved in early cardiomyocyte hypertrophy development in pressure-overloaded rats. BMC Cardiovasc. Disord.21, 403 (2021). 10.1186/s12872-021-02209-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schönherr, C. et al. Anaplastic lymphoma kinase (Alk) regulates initiation of transcription of Mycn in neuroblastoma cells. Oncogene31, 5193–5200 (2012). 10.1038/onc.2012.12 [DOI] [PubMed] [Google Scholar]
- 68.Htike, W., Islam, M. A., Hasan, M. T., Ferdous, S. & Rifat, M. Factors associated with treatment delay among tuberculosis patients referred from a tertiary hospital in Dhaka City: A cross-sectional study. Public Health Action3, 317–322 (2013). 10.5588/pha.13.0067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kramer, M., Ribeiro, D., Arsenian-Henriksson, M., Deller, T. & Rohrer, H. Proliferation and survival of embryonic sympathetic neuroblasts by Mycn and activated Alk signaling. J. Neurosci.36, 10425–10439 (2016). 10.1523/JNEUROSCI.0183-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schönherr, C. et al. Anaplastic lymphoma kinase (Alk) regulates initiation of transcription of Mycn in neuroblastoma cells. Oncogene.31, 5193–5200 (2012). 10.1038/onc.2012.12 [DOI] [PubMed] [Google Scholar]
- 71.Hasan, M. K. et al. Alk is a Mycn target gene and regulates cell migration and invasion in neuroblastoma. Sci. Rep.3, 3450 (2013). 10.1038/srep03450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liang, X. H. et al. Mapre1 promotes cell cycle progression of hepatocellular carcinoma cells by interacting with Cdk2. Cell Biol. Int.44, 2326–2333 (2020). 10.1002/cbin.11442 [DOI] [PubMed] [Google Scholar]
- 73.Gogolin, S. et al. Cdk4 inhibition restores G1-S arrest in Mycn-amplified neuroblastoma cells in the context of doxorubicin-induced Dna damage. Cell Cycle12, 1091–1104 (2014). 10.4161/cc.24091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nozato, M., Kaneko, S., Nakagawara, A. & Komuro, H. Epithelial-mesenchymal transition-related gene expression as a new prognostic marker for neuroblastoma. Int. J. Oncol.42, 134–140 (2013). 10.3892/ijo.2012.1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DuBois, S. G. & Park, J. R. Neuroblastoma and histone demethylation. N. Engl. J. Med.379, 1476–1477 (2018). 10.1056/NEJMcibr1806782 [DOI] [PubMed] [Google Scholar]
- 76.Phimmachanh, M., Han, J. Z. R., Donnell, Y. E. I., Latham, S. L. & Croucher, D. R. Histone deacetylases and histone deacetylase inhibitors in neuroblastoma. Front. Cell. Dev. Biol.8, 578770 (2020). 10.3389/fcell.2020.578770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aiken, T. J. et al. Mechanism of effective combination radio-immunotherapy against 9464D-Gd2, an immunologically cold murine neuroblastoma. J. Immunother. Cancer10, e004834 (2022). 10.1136/jitc-2022-004834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wienke, J. et al. The immune landscape of neuroblastoma: Challenges and opportunities for novel therapeutic strategies in pediatric oncology. Eur. J. Cancer144, 123–150 (2021). 10.1016/j.ejca.2020.11.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have submitted the raw RNA-seq data to NCBI (https://www.ncbi.nlm.nih.gov/sra) under the accession number PRJNA884866. Besides, we have uploaded mass spectrometry data to the MetaboLights (https://www.ebi.ac.uk/metabolights/) with the number of MTBLS6352.