Abstract
Cytidine deaminase (CDA) converts cytidine and deoxycytidine into uridine and deoxyuridine as part of the pyrimidine salvage pathway. Elevated levels of CDA are found in pancreatic tumors and associated with chemoresistance. Recent evidence suggests that CDA has additional functions in cancer cell biology. In this work, we uncover a novel role of CDA in pancreatic cancer cell metabolism. CDA silencing impairs mitochondrial metabolite production, respiration, and ATP production in pancreatic cancer cells, leading to a so-called Pasteur effect metabolic shift towards glycolysis. Conversely, we find that CDA expression promotes mitochondrial biogenesis and oxidative phosphorylation, independently of CDA deaminase activity. Furthermore, we observe that patient primary cells overexpressing CDA are more sensitive to mitochondria-targeting drugs. Collectively, this work shows that CDA plays a non-canonical role in pancreatic cancer biology by promoting mitochondrial function, which could be translated into novel therapeutic vulnerabilities.
Subject terms: Pancreatic cancer, Energy metabolism
Cytidine deaminase (CDA) promotes mitochondrial biogenesis independently of its deaminase activity, making CDA-overexpressing pancreatic cancer cells more sensitive to mitochondria-targeting drugs.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is projected to become the second leading cause of cancer-related death, with one in 64 individuals expected to die from this disease1. The poor prognosis is due to the late detection of PDAC, at which point patients are typically no longer candidates for surgical resection2,3. Moreover, 80% of patients relapse within five years following surgery4. PDAC is diagnosed 90% of the time at an advanced or metastatic stage, where current treatments, mainly chemotherapies, are largely ineffective, offering only minimal increases in survival and often causing detrimental side effects4. Until very recently, molecular-targeted therapies have largely failed to improve patient survival5. Pancreatic tumors quickly develop resistance or tolerance to conventional treatments, and pre-existing functional heterogeneity also contributes to PDAC chemoresistance6. Recent advances in PDAC management include the use of PARP inhibitors for patients with germline metastatic PDAC exhibiting a BRCAness genotype, who have responded to first-line platinum-based chemotherapy7. Additionally, the combination of neoadjuvant chemotherapy with surgery has significantly increased patient survival8. Despite these advances, novel therapies are urgently needed to improve the poor prognosis associated with PDAC.
Multiscale analyses have revealed significant heterogeneity in tumors among PDAC patients6. PDAC development is primarily driven by successive genetic alterations in the early stages and by epigenetic modifications in the metastatic stages2. This inter-patient tumoral heterogeneity complicates the discovery of a universal tumor marker or cure but may offer new therapeutic opportunities for some patients. Several proteins are associated with PDAC treatment resistance and have potential as therapeutic targets3. One of the most promising candidates is the cytidine deaminase (CDA), which recycles free pyrimidines, by generating uridine and deoxyuridine from cytidine and deoxycytidine, respectively, for DNA and RNA synthesis9. CDA also deaminates chemotherapeutically active deoxycytidine analogs, such as Ara-C, gemcitabine, and decitabine, transforming them into inactive molecules9,10. In PDAC patients, high CDA activity correlates with disease progression under gemcitabine-based therapy11. Furthermore, gemcitabine treatment increases CDA expression12, and commensal CDA from PDAC microbiome plays also a key role in chemoresistance13. CDA expression also drives therapeutic decision for using epigenetically modified forms of deoxycytidine to kill tumor cells14. Interestingly, we and others found that CDA is expressed in PDAC at a basal level14,15, prior to therapy, suggesting a yet unexplored role per se for this enzyme during oncogenesis. In line with this, we discovered that CDA exerts an unexpected role on DNA replication that can be exploited for therapeutic intervention15. In addition, we found that CDA targeting also results in metabolic alterations in experimental tumors15. Very recently, CDA silencing in cancer cells was associated with a perturbation of the energetic metabolism16, echoing our previous findings.
In this study, we explored the link between CDA and the mitochondrial function in cells derived from PDAC. Using metabolomic profiling, we found that the silencing of CDA in PDAC cells alters the level of several metabolites produced by the mitochondria, but also decreases mitochondrial respiration and ATP production. It rewires cancer cells metabolism toward glycolysis. Molecular explorations indicate that CDA promotes mitochondrial biogenesis independently of its known catalytic activity. Primary cultures derived from patient tumors also indicate that cells overexpressing CDA are more sensitive to drugs targeting the mitochondria. Collectively, our findings demonstrate a previously unknown role of CDA in mitochondrial function and oxydative phosphorylation (OXPHOS) regulation in PDAC cells. These findings further highlight the complexity of metabolic reprogramming in this tumor type and suggest potential therapeutic avenues for targeting mitochondrial metabolism in specific subsets of patients with high CDA expression.
Results
We generated lentiviral particles encoding for CDA small hairpins that were used to transduced Mia PaCa-2 cells as a model of PDAC. The next day, transduced cells were selected with blasticidin for three days. At this time point, CDA mRNA expression is inhibited by 95 ± 5% (p < 0.005, Fig. S1A), CDA protein is not detectable by western blot (Figs. S1B and S6E) and there is no difference in cell numbers between control-transduced, or CDA shRNA-transduced cells (Fig. S1C). Follow-up studies indicate that cell proliferation is significantly inhibited in cells expressing CDA shRNA as compared to control cells (p < 0.005, Fig. S1D).
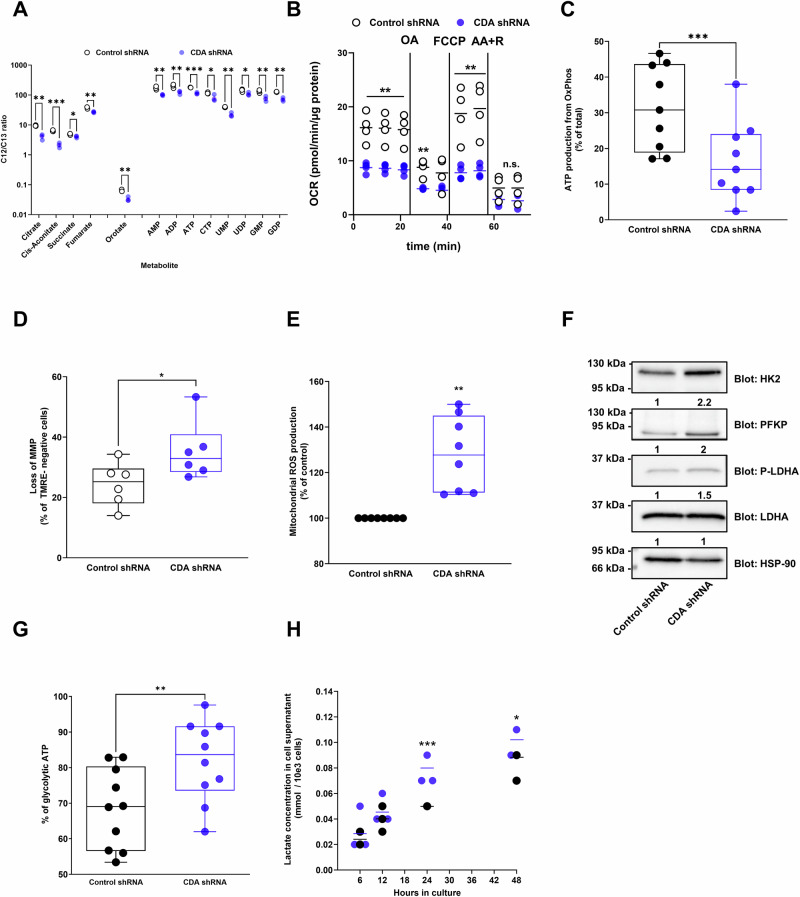
We performed metabolomics analyses following CDA silencing and before the inhibition of cell proliferation and found that the level of 13 metabolites out of 28 tested (Table 1) are altered following CDA targeting. Several metabolites of the tricarboxylic acid cycle (TCA, i.e. citrate, cis-aconitate, succinate, fumarate) were significantly decreased in cells expressing shRNA against CDA as compared to control cells (ranging from p < 0.05 to p < 0.005, Fig. 1A). In addition, the level of orotate, AMP, GMP, ADP, UDP and CTP was significantly diminished in cells depleted of CDA (ranging from p < 0.05 to p < 0.005, Fig. 1A), probably reflecting a global decrease of purine and pyrimidine synthesis. We further analyzed the mitochondrial respiration using the Seahorse analyzer (Fig. 1B) and found that the energetic balance (−32% ± 16, p < 0.05, Fig. S1E), the basal mitochondrial respiration (−47% ± 20, p < 0.05, Figs. S1F and S6F), the maximal respiratory capacity (−61% ± 9, p < 0.01, Fig. S1G), and the respiration-linked ATP production (−53% ± 15, p < 0.01, Fig. S1H) are severely compromised in PDAC cells expressing shRNA against CDA as compared to control. We confirmed that ATP production from OXPHOS is reduced when cells expressed a shRNA against CDA (−48% ± 17, p < 0.005, Fig. 1C). Last, CDA silencing resulted in a greater loss of mitochondrial membrane potential (1.5-fold ± 0.1, p < 0.05, Figs. 1D and S5A) and in the production of mitochondrial reactive oxygen species (ROS, 1.3-fold ± 0.16, p < 0.01, Figs. 1E and S5B), consistent with a massive mitochondrial defect in cells depleted for the enzyme.
Table 1.
List of the metabolites analyzed during this study
| Metabolite name | Abbreviation |
|---|---|
| 2-phosphoglycerate | 2-PG |
| 6-phosphogluconate | 6-PG |
| Alpha-ketoglutarate | a-KG |
| Adenosine diphosphate | ADP |
| Adenosine monophosphate | AMP |
| Adenosine triphosphate | ATP |
| Cytidine diphosphate | CDP |
| Cis-Aconitate | N/A |
| Citrate | N/A |
| Cytidine triphosphate | CTP |
| Fructose 1-phosphate | F1P |
| Fructose 1,6-bisphosphate | FBP |
| Fumarate | N/A |
| Guanosine diphosphate | GDP |
| Guanosine monophosphate | GMP |
| Orotate | N/A |
| Phosphoenolpyruvate | PEP |
| Ribulose-5-phosphate and ribose-5-phosphate | Ribu5P+Rib5P |
| Sedoheptulose-7-phosphate | Sed7P |
| Shikimate-3-phosphate | Shikimate-3P |
| Succinate | N/A |
| Uridine diphosphate | UDP |
| UDP-glucose | UDP-Glu |
| Uridine monophosphate | UMP |
| Uridine triphosphate | UTP |
| 5-phosphoribosyl-1-pyrophosphate | PRPP |
| Cytidine Monophosphate | CMP |
| Phosphorylated serine | P-Serine |
N/A not applicable.
Fig. 1. CDA depletion alters mitochondrial function and rewires pancreatic cancer cells toward glycolysis.
A Metabolomic analysis of Mia PaCa-2 cells expressing control shRNA or shRNA targeting CDA, 3 days following transduction. Results are mean ± s.e.m. of three experiments. AMP adenosine monophosphate, ADP adenosine diphosphate, ATP adenosine triphosphate, CTP cytosine triphosphate, UMP uridine monophosphate, UDP uridine diphosphate, GMP guanosine monophosphate, GMP guanosine monophosphate, GDP guanosine diphosphate. B Oxygen consumption rate measurement in Mia PaCa-2 cells expressing shRNA against CDA vs. control cells. Results are mean of four experiments performed in triplicate. The horizontal bar represents the mean of each biological repeat. FCCP carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone, R rotenone, OA oligomycin A, AA antimycin A. C Quantification of ATP production from OXPHOS in Mia PaCa-2 cells expressing CDA shRNA or control shRNA. Results are mean ± min to max of nine experiments performed in triplicate. D Mitochondrial membrane potential (MMP) analysis in Mia PACA-2 cells with CDA shRNA or control shRNA. Results are mean ± min to max of six experiments performed in triplicate. E Mitochondrial reactive oxygen species (ROS) production in Mia PaCa-2 cells expressing shRNA against CDA or control cells. Results are mean ± s.e.m. of eight experiments performed in duplicate. F Western blot analysis of hexokinase II (HK2), phosphofructo kinase platelet (PFKP), lactate dehydrogenase A (LDHA) and LDHA phosphorylation in Mia PaCa-2 cells expressing shRNA against CDA or control cells. Heat-shock protein 90 (HSP-90) expression was used as a control. Fold change in protein expression is indicated. Representative of four experiments. G Quantification of ATP production from glycolysis in Mia PaCa-2 cells expressing CDA shRNA or control shRNA. Results are mean ± min to max of ten experiments performed in triplicate. H Lactate content in the supernatant of Mia PaCa-2 cells expressing CDA shRNA or control shRNA. Results are mean of three experiments performed in triplicate. The horizontal bar represents the mean of each biological repeat. *p < 0.05, **p < 0.01, ***p < 0.005.
PDAC cells are well known for their ability to adapt and alter their metabolic pathways to support survival and rapid proliferation17. Facing mitochondrial dysfunction, PDAC cells can trigger metabolic reprogramming to favor aerobic glycolysis17. This can occur via altered gene expression and signaling pathways that promote glycolytic enzyme expression and activity17. We found that the expression of hexokinase II (HKII), that catalyses the phosphorylation of glucose to glucose-6-phosphate in the first step of glycolysis, and of phosphofructokinase platelet (PFKP), that catalyses the phosphorylation of fructose-6-phosphate to fructose-1,6-bisphosphate in the third step of glycolysis, are significantly increased by 2.2-fold ±0.2 and 2-fold ±0.2, respectively, in CDA depleted cells (Fig. 1F).
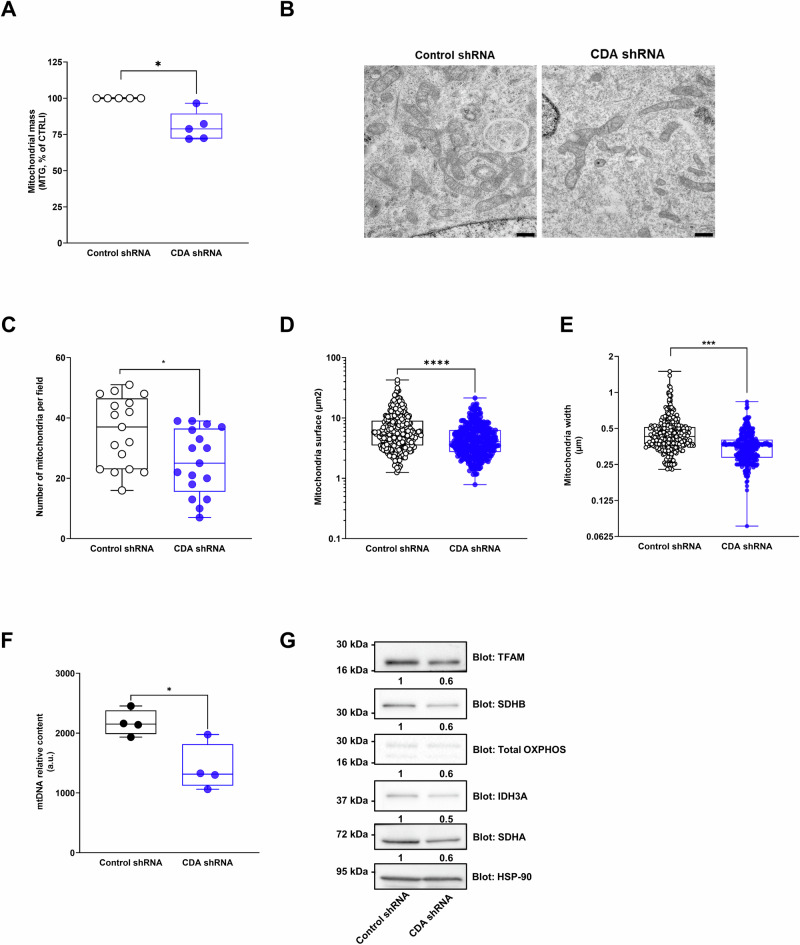
In addition, we found that Lactate dehydrogenase A (LDHA), that catalyses the conversion of pyruvate to lactate, is activated by phosphorylation in CDA depleted cells (Fig. 1F). These results correlate with an increased ATP production from glycolysis (1.35 fold ±0.07, p < 0.01, Fig. 1G) and greater levels of lactate in the cell supernatant (1.3 fold ±0.17, p < 0.005 and p < 0.05, Fig. 1H) in these cells as compared to control cells, respectively. These results are in favor of a metabolic rewiring of PDAC cells toward glycolysis provoked by a CDA depletion. Overall, our findings suggest that a CDA depletion in PDAC cells impairs mitochondrial function, alters metabolite levels and promotes metabolic plasticity. Next, we explored the molecular basis of mitochondrial dysfunction in CDA depleted cells. We found that the mitochondrial mass is significantly decreased in PDAC cells expressing CDA shRNA (−20% ± 5, p < 0.05, Fig. 2A), three days following transduction, when cell proliferation is not yet impaired. Using transmission electronic microscopy (Fig. 2B), we found that mitochondria number (−29% ± 5, p < 0.05, Fig. 2C), surface (−33% ± 3 p < 0.001, Fig. 2D) and width (−23% ± 1, p < 0.01, Fig. 2E) are decreased in PDAC cells in which CDA expression is silenced. We further analyzed a marker of mitochondrial fission (phosphorylated Dynamin-related protein 1 DRP1) and found no significant changes in response to a CDA depletion (Fig. S1I). However, silencing CDA resulted in a decreased mitochondrial DNA (mtDNA) content (−35% ± 14, p < 0.05, Fig. 2F). It also downregulated the expression of Transcription Factor A Mitochondrial (TFAM, −40% ± 12, Fig. 2G), a nuclear-encoded and master regulator of mitochondrial biogenesis that controls mtDNA transcription, replication, and packaging, and of proteins from the mitochondrial complex such as NADH:Ubiquinone Oxidoreductase Subunit B8 (NDUFB8 from complex I, −40% ± 6, Fig. 2G), succinate dehydrogenase complex flavoprotein subunit A and B (SDHA and SDHB, −40% ± 9 and −40% ± 13, respectively, Fig. 2G), cytochrome c oxidase subunit 2 (COX II from complex IV, −30% ± 9, Fig. 2G), and isocitrate dehydrogenase (IDH3A, −50% ± 9, Fig. 2G). These results point towards a role of CDA on mitochondrial biogenesis rather than on mitochondrial dynamics.
Fig. 2. CDA promotes mitochondrial biogeneis in pancreatic cancer cells.
A Quantification of mitochondrial mass using mitotracker red (MTR) in Mia PaCa-2 cells expressing CDA shRNA or control shRNA. Results are mean ± min to max of five independent experiments analyzed in duplicate. Representative captions (B, scale: 5 µm) of Mia PACA-2 cells with CDA shRNA or control shRNA analyzed by electron microscopy for mitochondria number (n = 17 fields, C), surface (n = 330 mitochondria, D) or width (n = 330 mitochondria, E). Results are mean ± min to max of two independent experiments. F Mitochondrial DNA (mtDNA) content analysis by qPCR, in Mia PACA-2 cells with CDA shRNA or control shRNA. Results are mean ± min to max of our experiments performed in triplicate. G Western blot analysis of Mitochondrial transcription factor A (TFAM), Succinate Dehydrogenase Complex Flavoprotein Subunit B (SDHB), cytochrome c oxidase subunit 2 (Cox II), NADH:Ubiquinone Oxidoreductase Subunit B8 (NDUFB8), Isocitrate Dehydrogenase (NAD(+)) 3 Catalytic Subunit Alpha (IDH3A) and Succinate Dehydrogenase Complex Flavoprotein Subunit A (SDHA) expression in Mia PaCa-2 cells expressing shRNA against CDA or control cells. Heat-shock protein 90 (HSP-90) expression was used as a control. Fold change in protein expression is indicated. Representative of four independent experiments. *p < 0.05, ***p < 0.005, ****p < 0.001.
We next investigated the mechanisms by which CDA regulates mitochondrial biogenesis in PDAC cells. By western blot, we found that CDA is not a mitochondrial protein (Figs. S2A and S6G). We employed Tetrahydrouridine (THU) and Diazepinone riboside (DR) pharmacological inhibitors that both target CDA catalytic pocket and so deaminase activity (Fig. S2B). These inhibitors did not inhibit PDAC cell proliferation (Fig. S2C). In addition, neither THU nor DR treatment showed a significant effect on the ATP production from OXPHOS, on the relative mtDNA content and on the expression of SDHA and IDH3A in PDAC cells (Figs. S2D–F and S6H). These findings suggest that the role of CDA in mitochondrial biogenesis involves mechanisms other than its catalytic activity. As a consequence, we generated a Mia PaCa-2 cell line that overexpresses a wild-type (WT) or a minimally active CDA mutant (E67Q)18. Bioluminescence resonance energy transfer (BRET2) donor saturation experiments showed that while CDA WT homodimerizes, CDA E67Q mutant does not (Fig. S3A, left panel). GFP2 and RLuc8 signals indicated that CDA E67Q is slightly less expressed than CDA WT (Fig. S3A, middle and right panel, respectively). BRET2 competition experiments validated that CDA E67Q did not dimerize (Fig. S3B). As CDA functions as a dimer of dimers, these later results suggest that E67Q mutant should show limited catalytic activity, that was confirmed in another study when expressed in cancer cells, as CDA E67Q activity is decreased by 97% as compared to the wild-type protein15.
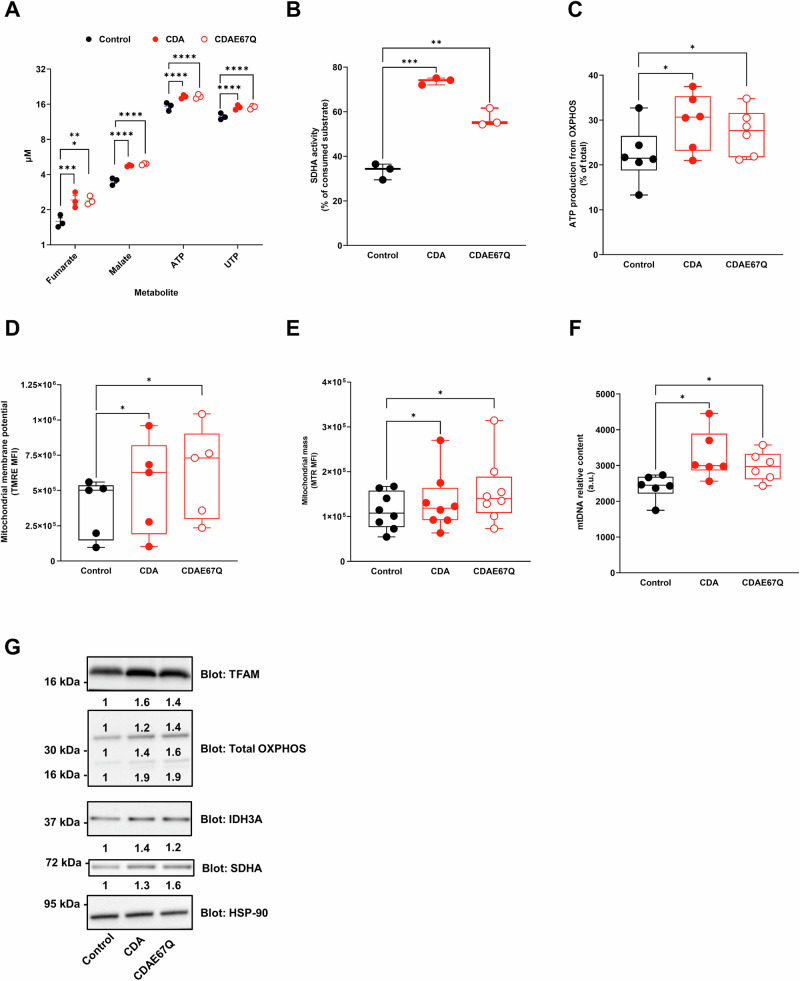
Remarkably, CDA WT and CDA E67Q expression significantly increased the production of fumarate, malate, ATP and UTP in PDAC cells (ranging from p < 0.005 to p < 0.001, Fig. 3A). We confirmed that SDHA activity is significantly increased in cells expressing either WT CDA (2.2 fold ±0.3, p < 0.005, Fig. 3B) or E67Q CDA (1.7 fold ±0.1, p < 0.01, Fig. 3B). We also found that ATP production from OXPHOS is increased by either WT CDA (1.3 fold ±0.09, p < 0.05, Fig. 3C) or E67Q CDA expression (2.2 fold ±0.09, p < 0.05, Fig. 3C). However, CDA overexpression had no effect on lactate production and glucose consumption. We then performed functional studies and identified that WT CDA and E67Q CDA expression increases mitochondrial membrane potential (1.4 fold ±0.2, p < 0.05 for WT CDA and 1.7 fold ±0.2, p < 0.05 for E67Q CDA, respectively, Figs. 3D and S5C), mitochondrial mass (1.8 fold ±0.08, p < 0.05 for WT CDA and 1.4 fold ±0.1, p < 0.05 for E67Q CDA, respectively, Figs. 3E and S5D), and relative mtDNA content (1.35 fold ±0.08, p < 0.05 for WT CDA and 1.25 fold ±0.05, p < 0.05 for E67Q CDA, respectively, Fig. 3F) as compared to control cells. We identified a significant increase in DNA polymerase gamma (POLG) mRNA levels, that is responsible for synthesizing new mtDNA strands during replication, and could be involved in the increased mtDNA content in cells overexpressing WT CDA and E67Q CDA (Fig. S4A). However, modulating CDA expression did not result in changes of the expression of proteins involved in the regulation of mitochondrial fusion (Mitofusin-1 Mfn1; optic atrophy-1 Opa1) or fission (phosphorylated Dynamin-related protein 1 DRP1, Figs. S4B and S6I). On the contrary, WT CDA and E67Q CDA expression positively correlated with markers of mitochondrial biogenesis (TFAM), structure and function (mitochondrial complex proteins, IDH3A, SDHA, Figs. 3G and S6C). Collectively, these findings suggest that CDA is involved in the regulation of mitochondrial biogenesis in PDAC cells, but its impact may occur through mechanisms independent of its known catalytic activity.
Fig. 3. CDA regulates mitochondrial biogenesis in pancreatic cancer cells through mechanisms independent of its known catalytic activity.
A Metabolomic analysis of Mia PaCa-2 cells expressing WT CDA or E67Q CDA mutant. Cells expressing luciferase were used as control. Results are mean ± s.e.m. of three experiments. ATP adenosine triphosphate, UMP uridine triphosphate. B Succinate Dehydrogenase Complex Flavoprotein Subunit B (SDHB) activity in Mia PaCa-2 cells expressing WT CDA or E67Q CDA mutant, with cells expressing luciferase used as control. Results are mean ± s.e.m. of three experiments performed in triplicate. C Quantification of ATP production from OXPHOS in control Mia PaCa-2 cells and cells expressing WT CDA or E67Q CDA. Results are mean ± min to max of six experiments performed in triplicate. D Mitochondrial membrane potential analysis in control Mia PaCa-2 cells and cells expressing WT CDA or E67Q CDA. Results are mean ± min to max of five experiments performed in triplicate. E Quantification of mitochondrial mass using mitotracker red (MTR) in control Mia PACA-2 cells and cells expressing WT CDA or E67Q CDA. Results are mean ± min to max of eight independent experiments performed in duplicate. F Mitochondrial DNA (mtDNA) content analysis by qPCR, in control Mia PaCa-2 cells and cells expressing WT CDA or E67Q CDA. Results are mean ± min to max of six experiments performed in triplicate. G Western blot analysis of Mitochondrial transcription factor A (TFAM), Succinate Dehydrogenase Complex Flavoprotein Subunit B (SDHB), cytochrome c oxidase subunit 2 (Cox II), NADH:Ubiquinone Oxidoreductase Subunit B8 (NDUFB8), Isocitrate Dehydrogenase (NAD(+)) 3 Catalytic Subunit Alpha (IDH3A) and Succinate Dehydrogenase Complex Flavoprotein Subunit A (SDHA) expression in control Mia PaCa-2 cells and cells expressing WT CDA or E67Q CDA. Heat-shock protein 90 (HSP-90) expression was used as a control. Fold change in protein expression is indicated. Representative of three experiments. *p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.001.
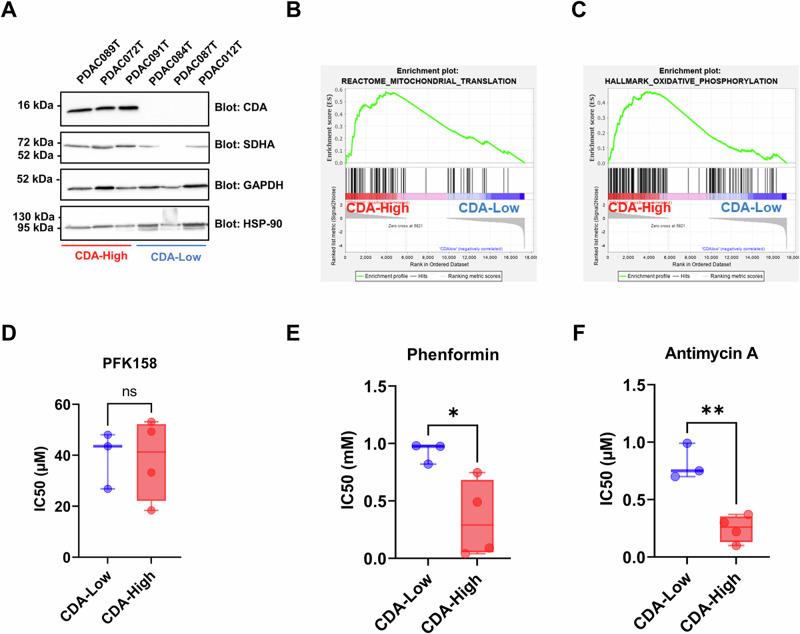
Our findings point towards an unexpected role of CDA in PDAC as it promotes mitochondrial biogenesis that correlates with increased OXPHOS capacity. We then asked whether this phenotype would create a metabolic vulnerability in primary cultures derived from patient tumors with a known level of CDA. Two distinct groups composed of three primary PDAC cells were established based on their CDA protein expression level. These groups were referred to as “CDA-High” and “CDA-Low” (Figs. 4A and S6D). Remarkably, we observed that CDA expression positively correlates with the expression of SDHA in these cells (Figs. 4A and S6D). We next performed gene signature enrichment analysis (GSEA) to assess the differences in gene expression patterns between “CDA-High” and “CDA-Low” groups. The analysis revealed that gene signatures related to mitochondrial translation (normalized enrichment score NES = 2.3, p < 0.01, Fig. 4B) and OXPHOS (NES = 2.1, p < 0.01, Fig. 4C) are significantly enriched in primary PDAC cells with a high CDA expression.
Fig. 4. CDA expression correlates with mitochondrial transcriptomic signature enrichment and sensitivity to mitochondrial-targeting drugs in primary cultures from pancreatic cancer patients.
A Western blot analysis of CDA and SDHA expression in PDAC primary cells. Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) and heat-shock protein 90 (HSP90) expression were used as control. Representative of three independent experiments. Gene set enrichment analysis for mitochondrial translation (B, reactome) and oxidative phosphorylation (C, hallmarks) signature enrichment in CDA-High and CDA-Low expressing primary PDAC cells. Primary cells with high and low CDA expression were treated in dose response and cell confluence was monitored using the Incucyte Zoom to calculate the half maximal inhibitory concentration (IC50) of PFK-158 (D), phenformin (E) and antimycin A (F). Results are mean ± s.e.m. of two independent experiment with six repetitions per cell line. *p < 0.05, **p < 0.01.
Finally, we calculated the half maximal inhibitory concentration (IC50) of PFK-158, a phosphofructokinase inhibitor that impairs glycolysis, of Phenformin and Antimycin A that target the mitochondrial electron transport chain (ETC) complexes I and III, respectively, on primary PDAC cell confluence. We found that PFK-158 strongly inhibited cell confluence, regardless of CDA expression levels (39.4 µM ± 6.5 vs. 38.5 µM ± 7.9, respectively, Fig. 4D). In contrast, PDAC patients’ cells with a high CDA expression were significantly more sensitive to treatment by Phenformin than cells with low level of the protein (0.3 mM ± 0.1 vs. 0.92 mM ± 0.05, p < 0.05, respectively, Fig. 4E) or by Antimycin A (0.2 µM ± 0.05 vs. 0.8 µM ± 0.09, p < 0.01, respectively, Fig. 4F). Collectively, our findings established a first link between CDA expression, increased mitochondrial function and a reliance on OXPHOS for PDAC cell proliferation.
Discussion
The importance of mitochondria for the initiation and the progression of pancreatic tumorigenesis is now well accepted19. However, PDAC tumors show heterogeneity in energetic metabolism20 and the underlying molecular mechanisms that regulate mitochondrial function and OXPHOS in PDAC cells remain largely unknown. Here, we show that CDA has a new and unexpected role on mitochondria biology and OXPHOS in PDAC cells. This study establishes another connection between enzymes involved in pyrimidine production and mitochondrial function. Indeed, dihydroorotate dehydrogenase (DHODH) from the de novo pyrimidine synthesis localizes into the mitochondrial inner membrane21. DHODH depends on respiration to catalyze the fourth and last step in pyrimidine metabolism, and in turn, participates to OXPHOS with its electron acceptor Coenzyme Q21. Here, we found that CDA is not detected in PDAC cells mitochondria. In addition, our results strongly suggest that CDA exerts its new role independently of the pyrimidine metabolism, as pharmacological inhibitors of the enzyme are ineffective in altering mitochondrial energy production, that was both stimulated to a similar level by wild-type and catalytically inactive CDA.
We found that targeting CDA shifts Mia PaCa-2 cells metabolism from OXPHOS to glycolysis which is highly evocative of the well described Warburg effect, and characterized in this study by the increase of the expression of HKII, the activation of LDHA, the increase of ATP and lactate production from glycolysis. While these results provide another demonstration of PDAC cells metabolic plasticity, increased glycolysis could also account for the deterioration of mitochondrial function that was measured following CDA silencing. In addition, primary PDAC cells are equally sensitive to the glycolytic inhibitor PFK158, regardless of the expression of CDA. This strongly suggest that an increased glycolysis consecutive to a CDA targeting is a compensatory mechanism rather than a causative factor. We next explored whether CDA participates in mitochondrial dynamics and/or biogenesis in PDAC cells. Indeed, PDAC cells commonly present with highly fragmented mitochondria22, probably resulting from mitochondrial fission, that correlate with higher OXPHOS activity and tumor aggressiveness. Interestingly, a recent study demonstrated that targeting DHODH promotes mitofusin-2 (Mfn2) expression, mitochondrial fusion, mitophagy and inhibits OXPHOS to eventually improve the survival of mice with spontaneous PDAC23. Our results show no significant changes in the expression of proteins involved in the regulation of mitochondrial fusion (MFN1, MFN2, Opa1) or fission (phosphorylated DRP1) in response to CDA silencing or overexpression. Interestingly, we found that CDA expression positively correlates with mtDNA content, mitochondrial biogenesis, structural and functional proteins expression, and overall, with mitochondrial mass. mtDNA replication is regulated by nuclear-encoded factors such as the mitochondrial-specific DNA polymerase, known as POLG, that is responsible for synthesizing new mtDNA strands during replication. Preliminary finding indicates that WT CDA and E67Q CDA expression results in modest, yet significant, induction of POLG expression in PDAC cells. On the other hand, we found during this study that the expression of TFAM, that plays a crucial role in initiating transcription and packaging mtDNA into nucleoids, is correlated to CDA expression in PDAC cells. While the results obtained are of interest, this study comes with limitations, particularly considering the reliance on a single cell line model for nearly all experiments, highlighting the need for further validation across diverse cellular contexts to ensure broader applicability to determine how CDA regulates the expression of mitochondrial biogenesis-promoting genes in PDAC cells. As we found that this effect is independent of CDA activity, we speculate that CDA may act as a co-regulator of the transcription factors that regulates the aforementioned genes expression such as nuclear respiratory factor 1 (NRF-1), peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1α), or estrogen-related receptor alpha (ERRα), or may favor the communication between the nuclear and the mitochondrial compartments that is crucial for maintaining mtDNA integrity and function. Nevertheless, one has to consider that the nucleotide imbalance that we demonstrate in this study following CDA knock-down could also impact the metabolic profile in these cells, as nucleotide shortage can hinder the proper replication and repair of mtDNA and compromise the synthesis of essential mitochondrial proteins, leading to defects in the respiratory chain and oxidative phosphorylation. In addition, nucleotide imbalance may also contribute to oxidative stress, leading to the production of reactive oxygen species (ROS), that may damage mitochondrial proteins, lipids, and DNA, further impairing mitochondrial function.
Last, we show here that the increase of OXPHOS activity in cells expressing a high level of CDA uncloaks novel therapeutic vulnerabilities in primary PDAC cultures for mitochondria targeting-drugs, such as Phenformin. Unfortunately, Phenformin, that shows antitumor activity in patient-derived xenografts24, has been so far disappointing in clinical trials, including in PDAC, due to a general lack of efficacy and safety issues25. As Phenformin side effects can now be clinically managed26, our results suggest that CDA expression levels may help identify PDAC patients who could potentially benefit from mitochondria-targeting therapies.
Methods
Cell lines and culture conditions
Mia PaCa-2, Capan-2 and Hela cell lines were obtained from ATCC, validated by STR analysis and cultured in Dulbecco Modified Eagle Medium (DMEM, Thermo Fisher Scientific) 4.5 g/L glucose containing 10% fetal bovine serum (FBS, ThermoFisher Scientific), 100 IU/mL penicillin (Thermo Fisher Scientific), 100 µg/mL streptomycin (Thermo Fisher Scientific) and 250 ng/mL fungizone (Thermo Fisher Scientific), 2 mM glutamine (Sigma). Cultures were monthly verified for mycoplasma contamination and maintained in culture media containing 100 IU/mL anti mycoplasma plasmocin (Invivogen). Cells were incubated at 37 °C with 5% CO2. Patient-derived primary cells from the PacaOMICS cohort were generated from PDAC tumors and a kind gift of Dr N. Dusetti (Centre de Recherche en Cancérologie de Marseille, Team « pancreatic cancer) and cultured as previously described27.
Proliferation assays
Lentiviral particles expressing CDA-directed small hairpins (sh) RNA were generated and used to transduce Mia PaCa-2 cells for 24 h, as previously described15. Control cells were transduced with lentiviral vectors encoding for control shRNAs. The next day, transduced cells were selected with 5 µl/mL of blasticidin (Invivogen) for 3 days and counted using a ZI Coulter (Beckman). For longitudinal studies, six thousand blasticidin-selected cells were seeded in 96-well plates in 100 µl of complete medium.cell confluence was monitored non-invasively using the IncuCyte Zoom, up to 10 days following transduction (Sartorius, Essen BioScience).
Metabolic analysis
Metabolomic analysis: 5.105 cells were seeded in 6-well plates with 2 mL medium for 24 h. Metabolites were quenched and extracted in a one-step process as described previously28. C13 IDMS standards added into the quenching solution were used to quantify metabolites. Samples were analyzed by Metatoul platform (Toulouse). Exometabolome analysis: 3.104 cells were seeded in 48-well plates with 500 µL medium. Medium was changed with DMEM A14430 (no glucose, no glutamine, no phenol red, Thermo Fisher Scientific) containing 1% FBS, 4.5 g/L glucose, 5 mM L-Glutamine, 1 mM pyruvate. Medium was collected, filtered on 0.2 µm filters, and quenched in liquid nitrogen after 3, 6, 12, 24 and 48 h following medium change. Cells were counted at each time for data normalization. Glucose, glutamine, pyruvate and lactate content in the supernatant were measured by Nuclear magnetic resonance (NMR) (Metatoul platform, Toulouse). Succinate Dehydrogenase activity was measured using the Succinate Dehydrogenase Activity Colorimetric Assay Kit from Biovision following the provider’s recommendation.
DNA extraction for mitochondrial DNA relative quantification
105 cells were seeded in 6-well plates with 2 mL medium for 24 h. Total cellular DNA was extracted with QIAamp DNA mini kit (Qiagen) according to the manufacturer’s instructions. DNA quality and quantity were measured using Nanodrop (Thermo Scientific). mtDNA relative content was quantified by qPCR as described previously29, using the following oligos to amplify H. sapiens mitochondrial tRNA-Leu(UUR) DNA with forward primer 5′- CAC CCA AGA ACA GGG TTT GT-3′ and reverse primer 5′-TGG CCA TGG GTA TGT TGT TA-3′. Amplification of H. sapiens nuclear B2-microglobulin DNA amplification with forward primer 5′- TGC TGT CTC CAT GTT TGA TGT ATC T-3′ and reverse primer 5′- TCT CTG CTC CCC ACC TCT AAG T-3′ was used as an internal control.
Protein extraction and immunoblot analysis
5.105 cells were seeded in 100-mm culture dishes with 10 mL medium for 24 h. For soluble protein detection, cell pellets were incubated in RIPA buffer supplemented with 10 µL/mL protease inhibitor (Sigma-Aldrich). After 15 min on ice, samples were centrifuged (15 min at 12,000 rpm and 4 °C) and supernatants containing soluble proteins were collected. For cytoplasmic/nuclear extract isolation, cell pellets were resuspended in 10 mM TrisHCl, pH 7.4 containing 1.5 mM MgCl2, 5 mM KCl, 0.5 mM dithiothreitol, 0.5%NP40® and 0.5 mM PMSF complemented with 10 µL/mL protease inhibitor (Sigma-Aldrich) and incubated on ice for 10 min. Following centrifugation for 15 min at 2000 rpm and 4 °C, the supernatant corresponding to the cytoplasmic fraction was collected and the pellet was washed and centrifuged twice with the same buffer as previously described. The pellet was then resuspended in 20 mM TrisHCl, 0.025% glycerol, 1.5 mM MgCl, 0.5 mM PMSF, 0.2 mM EDTA, 0.5 mM DTT and 0. 4 M NaCl complemented with 10 µL/mL protease inhibitor (Sigma-Aldrich) and incubated on ice for 15 min. After centrifugation at 12,000 rpm for 20 min and 4 °C, the supernatant corresponding to the nuclear fraction was collected. Mitochondrial isolation was performed as described previously30. The pellet containing the mitochondrial fraction was solubilized in RIPA buffer as described for total cell lysate. Extracts were separated by SDS-PAGE under reducing conditions, transferred to a nitrocellulose membrane and analyzed by immunoblotting using the indicated antibodies from Cell Signaling (LDHA clone C4B5, # 3582, HSP-90 (#4874), P-LDHA Tyr 10, #8176, HK II C6465, #2867, TFAM D5C8, #8076, SDHA D6J9M, #11998, SDHB E3H92, #92648, P-DRP1 S616 #3455, P-DRP1 S637 #4867, OPA-1 D6U6N, # 80471, Mfn1 D6E25, #14739, VDAC #4866, cytochrome-c D18C7, #11940, beta tubulin 6F3, #2146, Cox IV 3E11, #4850, PDI C81H6, #3501), Abclonal (CDA, A13959) and Abcam (Total OXPHOS ab110411) following the manufacturer recommended protocol. Corresponding HRP conjugate secondary antibodies were purchased from Promega. Signal was detected using the ECL system (BioRAD) according to the manufacturer’s instructions. Densitometry of the bands was done using Chemidoc system (BioRAD) and image Lab software.
CDA enzymatic activity assay
5.105 cells were seeded in 100-mm culture dishes with 10 mL medium for 24 h. The cytidine deaminase activity assay kit was performed following the manufacturer’s instruction with 20 µg of total proteins (BioVision, #K451-100).
Flow cytometry
5.105 cells were seeded in 100-mm culture dishes with 10 mL medium for 24 h. Cells were collected by trypsinisation and resuspended in PBS for the following treatments: 200 µM cyanide 3-chlorophenylhydrazone (CCCP, Sigma) for 15 min as TMRE negative control, 10 µM 2′,7′ – dichlorofluorescin diacetate (DCF-DA, Invitrogen), 20 µM MitoSox, 200 nM TMRE (Sigma), 100 nM MitoTracker Green (MTG, Sigma) for 20 min. After wash, cells were resuspended in Annexin V Binding Buffer (BD Pharmingen) and 1 µL/mL anti-annexin V antibody (BD Pharmingen) was added to measure cell viability. Intracellular ROS levels were measured by flow cytometry using the DCF-DA dye (total ROS) and MitoSox (mitochondrial ROS), mitochondrial membrane potential and mitochondrial mass were measured with TMRE and MTG respectively in live cells (AnV negative cells). Data collection was made with a MACSQUANT 10 (Miltenyi Biotec). Gating strategy was performed on size exclusion and/or viability markers (Fig. S5), and data analysis was performed using FlowJo (FlowJo). Cell population of interest abundance ranged from 80 to 90% during the final step of gating/analysis.
Measurement of ATP production
ATP was measured using the Cell Titer Glo® kit (Promega). 8.105 cells were plated into a white 96-well plate (Corning) for 24 h. Medium was changed and cells were treated with 100 µL of water/PBS (control) or 100 µM sodium iodoacetate (IA) (to block glycolytic ATP production) both alone or in combination with 30 µM FCCP (to block mitochondrial ATP production). Following 1 h incubation, 100 µL of Cell Titer Glo® reaction mix solution was added to each well. Plates were then assayed for luminescence with a Clariostar (BMG LABTECH). Percentage of glycolytic ATP = 100 × (ATPPBS/water − ATPIA)/(ATPPBS/water − ATPIA+FCCP). Percentage of mitochondrial ATP = 100-% Glycolytic ATP.
Quantification of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) by Seahorse
Cells were seeded at 4.104 cells per well in Seahorse XF24 tissue culture plates (Seahorse Bioscience Europe). The sensor cartridge was placed into the calibration buffer medium supplied by Seahorse Biosciences to hydrate overnight. Twenty-four hours later, medium was changed with 500 µL of XF base minimal DMEM medium containing 10 mM glucose, 1 mM pyruvate and 2 mM glutamine at pH 7.4. After one-hour incubation at 37 °C in CO2 free-atmosphere, basal oxygen consumption rate (OCR, as a mitochondrial respiration indicator) and extracellular acidification rate (ECAR, as a glycolysis indicator) were performed using the XF analyzer, according to the manufacturer’s instructions. The data were normalized to protein content, determined by Bradford assay upon lysis of the cells (using 0.1 N NaOH) immediately after the assay.
Transmission electronic microscopy
2.105 cells were seeded in 6-well plates with 2 mL medium for 24 h. Cells were fixed with 2% glutaraldehyde in 0.1 M Sorensen phosphate buffer (pH = 7.4), washed with the Sorensen phosphate buffer (0.1 M) for 12 h, then were post-fixed with 1% OsO4 in Sorensen phosphate buffer (Sorensen phosphate 0.05 M, glucose 0.25 M, 1%OsO4) for 1 h, washed twice with distilled water and pre-stained with 2% uranyl acetate aqueous solution for 12 h. Samples were dehydrated in ascending ethanol solutions and embedded in epoxy resin (Epon 812, Electron Microscopy Sciences). After 24 h of polymerization at 60 °C, ultrathin sections (70 nm thick) were mounted on 150 mesh collodion-coated copper grids and post-stained with 3% uranyl acetate in 50% ethanol and with 8.5% lead citrate before being examined on a HT 7700 Hitachi electron microscope at an accelerating voltage 80 KV. Number of mitochondria per cell and number of cristae per mitochondria were counted by blinded users without indication of experimental groups. Length and width of mitochondria was measured by ImageJ.
Functional enrichment analysis
Enrichment analyses were performed using GSEA Hallmarks and Reactome31 and most other pathways were extracted from MSigDB. No genes were removed during the analysis.
Molecular cloning
CDA WT and E67Q were cloned between NcoI/XhoI sites of the pEF-MCS-GFP2, pEF-MCS-RLuc8 vectors32 and pEF-MCS-mCherry vector. pEF-MCS-mCherry vector was generated by digesting pEF-MCS-GFP2 plasmid by NotI/XbaI sites and the mCherry was then cloned into this digested plasmid by PCR using NotI/XbaI sites.
BRET2 saturation and competition assays
For all BRET experiments (titration curves and competition assays), 6.5.105 HEK293T were seeded in each well of six-well plates. After 24 h at 37 °C, cells were transfected with a total of 1.6 µg of DNA mix, containing the donor + acceptor ± competitor plasmids, using JetPrime transfection reagent (Polyplus Transfection). For the BRET donor saturation assays, cells were transfected with 0.05 µg of donor and with an increased amount of acceptor plasmid (0.0125, 0.025, 0.05, 0.1, 0.25, 0.5, 0.75, and 1 µg of DNA) equalized to a total amount of 1.6 µg of DNA with an empty vector pEF-cyto-myc. In dose-response competition experiments, competitors were transfected with the following amount of DNA: 0.1, 0.5, and 1 µg with a 1:1 donor:acceptor DNA plasmid ratio (i.e., 0.05 µg : 0.05 µg of DNA plasmid). Cells were detached 24 h later, washed with phosphate-buffered saline (PBS), and seeded in a white 96-well plate (clear bottom, #6005181; PerkinElmer) in Opti-MEM without phenol red medium complemented with 4% FBS, and cells were incubated for an additional 20 to 24 h at 37 °C before the BRET assay reading. A step-by-step BRET protocol is provided elsewhere33.
BRET2 measurements
BRET2 signal was determined immediately after injection of coelenterazine 400a substrate (10 µM final concentration) to cells (Cayman Chemicals) using a CLARIOstar plate reader (BMG Labtech) with a luminescence module. Total GFP2 fluorescence was detected with excitation and emission peaks set at 400 ± 15 and 510 nm ± 20, respectively. The total mCherry fluorescence was detected with excitation and emission peaks set at 570 ± 15 and 620 ± 20 nm, respectively. Total RLuc8 luminescence was measured with an emission peak set at 480 nm ± 80. The BRET signal or BRET ratio corresponds to the light emitted by the GFP2 acceptor constructs (515 nm ± 30) upon addition of coelenterazine 400a divided by the light emitted by the RLuc8-CDA (mutant) donor constructs (410 nm ± 80). The background signal is subtracted from that BRET ratio using the donor-only negative control where only the RLuc8-CDA (mutant) fusion plasmid is transfected into the cells. The normalized BRET ratio is the BRET ratio normalized to the negative control (mCherry alone) during a competition assay. The total GFP2, mCherry and RLuc8 signals were used to control the protein expression level from each plasmid.
Statistics and reproducibility
No sample size calculation was performed, and we performed at least three biological independent experiments. Experiments were performed in at least three technical replicates. No data were excluded from the analysis. Unpaired Student’s t or Wilcoxon–Mann–Whitney tests were used to determine the statistical significance of differences between two groups using GraphPad Prism 9 software with the default settings. Methods of statistical analysis are indicated in the figure captions. Values are presented as *p < 0.05, **p < 0.01 and ***p < 0.005 and ****p < 0.001. Error bars are s.e.m. unless otherwise stated.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
The authors thank the financial support of the “Fondation Toulouse Cancer Santé” (A.F., P.C.), “Inserm” (A.L.), “Région Occitanie” (A.L.), “Université Paul Sabatier UT3” (M.G.), “Fondation de France” (N.B.), “Ligue Nationale Contre le Cancer” (A.L.) and “Inserm Transfert” (CDA inhibitors). The authors thank the MetaToul platform for its experimental expertise. The authors thank Ms Emilie Martin and Ms Catherine Zanibellato for technical assistance.
Author contributions
Conceptualization: Pierre Cordelier, Jean-Emmanuel Sarry, and Anthony Lemarié. Investigations: Audrey Lumeau, Audrey Frances, Marie Sorbara, Marion Gayral, Estelle Saland, Lucille Stuani, Delphine Pagan, and Naima Hanoun. Supervision: Pierre Cordelier, Jean-Emmanuel Sarry, Jean-Charles Portais, Nicolas Bery, Louis Buscail, Jérôme Torrisani, Nelson Dusetti, and Anthony Lemarié. Writing—original draft: Pierre Cordelier. Writing—review & editing: Jean-Emmanuel Sarry, Nelson Dusetti, Nicolas Bery, Anthony Lemarié, Jérôme Torrisani, Audrey Lumeau, and Delphine Pagan. Funding acquisition: Pierre Cordelier (PI), Jean-Emmanuel Sarry (co-PI), and Nicolas Bery. Data analysis: Jean-Emmanuel Sarry, Audrey Lumeau, Pierre Cordelier, Nelson Dusetti, Anthony Lemarié, Audrey Frances, Marion Gayral, Nicolas Bery, and Jean-Charles Portais.
Peer review
Peer review information
Communications Biology thanks Yogen Saunthararajah, Sandra Pérez-Torras and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Giulia Bertolin and Johannes Stortz.
Data availability
With the exception of the transcriptomic analysis preformed on CDA expressing cell lines (GSE253662), all data supporting the findings of this study are available within the paper. Supplementary Data 1 contains the underlying data source data for the graphs shown in this study. Supplementary Information includes Fluorescence-activated cell sorting (FACS) gating strategies (Fig. S5) and uncropped western blots (Fig. S6).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06760-y.
References
- 1.Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin.73, 17–48 (2023). 10.3322/caac.21763 [DOI] [PubMed] [Google Scholar]
- 2.Kleeff, J. et al. Pancreatic cancer. Nat. Rev. Dis. Prim.2, 16022 (2016). 10.1038/nrdp.2016.22 [DOI] [PubMed] [Google Scholar]
- 3.Buscail, L., Bournet, B. & Cordelier, P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol.10.1038/s41575-019-0245-4 (2020). [DOI] [PubMed]
- 4.Neoptolemos, J. P. et al. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat. Rev. Gastroenterol. Hepatol.15, 333–348 (2018). 10.1038/s41575-018-0005-x [DOI] [PubMed] [Google Scholar]
- 5.Hosein, A. N., Dougan, S. K., Aguirre, A. J. & Maitra, A. Translational advances in pancreatic ductal adenocarcinoma therapy. Nat. Cancer3, 272–286 (2022). 10.1038/s43018-022-00349-2 [DOI] [PubMed] [Google Scholar]
- 6.Connor, A. A. & Gallinger, S. Pancreatic cancer evolution and heterogeneity: integrating omics and clinical data. Nat. Rev. Cancer22, 131–142 (2022). 10.1038/s41568-021-00418-1 [DOI] [PubMed] [Google Scholar]
- 7.Golan, T. et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med.381, 317–327 (2019). 10.1056/NEJMoa1903387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conroy, T. et al. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N. Engl. J. Med.379, 2395–2406 (2018). 10.1056/NEJMoa1809775 [DOI] [PubMed] [Google Scholar]
- 9.Frances, A. & Cordelier, P. The emerging role of cytidine deaminase in human diseases: a new opportunity for therapy? Mol. Ther.28, 357–366 (2020). 10.1016/j.ymthe.2019.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neff, T. & Blau, C. A. Forced expression of cytidine deaminase confers resistance to cytosine arabinoside and gemcitabine. Exp. Hematol.24, 1340–1346 (1996). [PubMed] [Google Scholar]
- 11.Serdjebi, C. et al. Rapid deaminator status is associated with poor clinical outcome in pancreatic cancer patients treated with a gemcitabine-based regimen. Pharmacogenomics14, 1047–1051 (2013). 10.2217/pgs.13.93 [DOI] [PubMed] [Google Scholar]
- 12.Amit, M. & Gil, Z. Macrophages increase the resistance of pancreatic adenocarcinoma cells to gemcitabine by upregulating cytidine deaminase. Oncoimmunology2, e27231 (2013). 10.4161/onci.27231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geller, L. T. et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science357, 1156–1160 (2017). 10.1126/science.aah5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zauri, M. et al. CDA directs metabolism of epigenetic nucleosides revealing a therapeutic window in cancer. Nature524, 114–118 (2015). 10.1038/nature14948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lumeau, A. et al. Cytidine deaminase resolves replicative stress and protects pancreatic cancer from DNA-targeting drugs. Cancer Res.84, 1013–1028 (2024). 10.1158/0008-5472.CAN-22-3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mameri, H. et al. Cytidine deaminase deficiency in tumor cells is associated with sensitivity to a naphthol derivative and a decrease in oncometabolite levels. Cell Mol. Life Sci.79, 465 (2022). 10.1007/s00018-022-04487-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biancur, D. E. & Kimmelman, A. C. The plasticity of pancreatic cancer metabolism in tumor progression and therapeutic resistance. Biochim. Biophys. Acta Rev. Cancer1870, 67–75 (2018). 10.1016/j.bbcan.2018.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cambi, A. et al. Identification of four amino acid residues essential for catalysis in human cytidine deaminase by site-directed mutagenesis and chemical modifications. Protein Eng.11, 59–63 (1998). 10.1093/protein/11.1.59 [DOI] [PubMed] [Google Scholar]
- 19.Reyes-Castellanos, G., Masoud, R. & Carrier, A. Mitochondrial metabolism in PDAC: from better knowledge to new targeting strategies. Biomedicines8, E270 (2020). 10.3390/biomedicines8080270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masoud, R. et al. Targeting mitochondrial complex I overcomes chemoresistance in high OXPHOS pancreatic cancer. Cell Rep. Med.1, 100143 (2020). [DOI] [PMC free article] [PubMed]
- 21.Boukalova, S. et al. Dihydroorotate dehydrogenase in oxidative phosphorylation and cancer. Biochim. Biophys. Acta Mol. Basis Dis.1866, 165759 (2020). 10.1016/j.bbadis.2020.165759 [DOI] [PubMed] [Google Scholar]
- 22.Anderson, G. R. et al. Dysregulation of mitochondrial dynamics proteins are a targetable feature of human tumors. Nat. Commun.9, 1677 (2018). 10.1038/s41467-018-04033-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu, M. et al. Mitochondrial fusion exploits a therapeutic vulnerability of pancreatic cancer. JCI Insight5, e126915 (2019). 10.1172/jci.insight.126915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajeshkumar, N. V. et al. Treatment of pancreatic cancer patient-derived xenograft panel with metabolic inhibitors reveals efficacy of phenformin. Clin. Cancer Res.23, 5639–5647 (2017). 10.1158/1078-0432.CCR-17-1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey, C. J. & Turner, R. C. Metformin. N. Engl. J. Med.334, 574–579 (1996). 10.1056/NEJM199602293340906 [DOI] [PubMed] [Google Scholar]
- 26.Pollak, M. Potential applications for biguanides in oncology. J. Clin. Invest.123, 3693–3700 (2013). 10.1172/JCI67232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraunhoffer, N. A. et al. Evidencing a pancreatic ductal adenocarcinoma subpopulation sensitive to the proteasome inhibitor carfilzomib. Clin. Cancer Res.26, 5506–5519 (2020). 10.1158/1078-0432.CCR-20-1232 [DOI] [PubMed] [Google Scholar]
- 28.Martano, G. et al. Fast sampling method for mammalian cell metabolic analyses using liquid chromatography-mass spectrometry. Nat. Protoc.10, 1–11 (2015). 10.1038/nprot.2014.198 [DOI] [PubMed] [Google Scholar]
- 29.Rooney, J. P. et al. PCR based determination of mitochondrial DNA copy number in multiple species. Methods Mol. Biol.1241, 23–38 (2015). 10.1007/978-1-4939-1875-1_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kappler, L. et al. Purity matters: a workflow for the valid high-resolution lipid profiling of mitochondria from cell culture samples. Sci. Rep.6, 21107 (2016). 10.1038/srep21107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillespie, M. et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res.50, D687–D692 (2022). 10.1093/nar/gkab1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bery, N. et al. BRET-based RAS biosensors that show a novel small molecule is an inhibitor of RAS-effector protein-protein interactions. Elife7, e37122 (2018). [DOI] [PMC free article] [PubMed]
- 33.Bery, N. et al. Bioluminescence Resonance Energy Transfer 2 (BRET2)-Based RAS Biosensors to Characterize RAS Inhibitors. Curr Protoc Cell Biol.83, e83 (2019). 10.1002/cpcb.83 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
With the exception of the transcriptomic analysis preformed on CDA expressing cell lines (GSE253662), all data supporting the findings of this study are available within the paper. Supplementary Data 1 contains the underlying data source data for the graphs shown in this study. Supplementary Information includes Fluorescence-activated cell sorting (FACS) gating strategies (Fig. S5) and uncropped western blots (Fig. S6).