Abstract
Radiotherapy (RT) is one of major therapeutic modalities in combating breast cancer. In RT, ionizing radiation is employed to induce DNA double-strand breaks (DSBs) as a primary mechanism that causes cancer cell death. However, the induced DNA damage can also trigger the activation of DNA repair mechanisms, reducing the efficacy of RT treatment. Given the pivotal role of RAD50 protein in the radiation-responsive DNA repair pathways involving DSBs, we developed a novel polymer-lipid based nanoparticle formulation containing RAD50-silencing RNA (RAD50-siRNA-NPs) and evaluated its effect on the RAD50 downregulation as well as cellular and tumoral responses to ionizing radiation using human triple-negative breast cancer as a model. The RAD50-siRNA-NPs successfully preserved the activity of the siRNA, facilitated its internalization by cancer cells via endocytosis, and enabled its lysosomal escape. The nanoparticles significantly reduced RAD50 expression, whereas RT alone strongly increased RAD50 levels at 24 h. Pretreatment with RAD50-siRNA-NPs sensitized the cancer cells to RT with ∼2-fold higher level of initial DNA DSBs as determined by a γH2AX biomarker and a 2.5-fold lower radiation dose to achieve 50 % colony reduction. Intratumoral administration of RAD50-siRNA-NPs led to a remarkable 53 % knockdown in RAD50. The pretreatment with RAD50-siRNA-NPs followed by RT resulted in approximately a 2-fold increase in DNA DSBs, a 4.5-fold increase in cancer cell apoptosis, and 2.5-fold increase in tumor growth inhibition compared to RT alone. The results of this work demonstrate that RAD50 silencing by RAD50-siRNA-NPs can disrupt RT-induced DNA damage repair mechanisms, thereby significantly enhancing the radiation sensitivity of TNBC MDA-MB-231 cells in vitro and in orthotopic tumors as measured by colony forming and tumor regrowth assays, respectively.
Keywords: DNA damage, RAD50-siRNA-NPs, Radiotherapy, RAD50 downregulation, Triple negative breast cancer
Graphical abstract
1. Introduction
Breast cancer is a leading cause of cancer death in women worldwide [1], of which, triple negative breast cancer (TNBC) is the most aggressive subtype, with a high incidence of metastasis, resistance to existing therapies and a poor prognosis [2]. Therefore, there is an urgent need for novel and effective therapeutic strategies to combat TNBC.
Breast cancer management typically involves radiation therapy (RT) with other treatment modalities [3]. RT, via ionizing radiation, induces various forms of DNA damage, including base lesions, together with DNA single strand breaks and double-strand breaks (DSBs) [4]. Among these forms of damage, DSBs are considered the most harmful, particularly in inducing cancer cell death [4]. However, the induction of DNA DSBs also triggers DNA damage repair machinery that mitigate DNA injury in tumor cells, significantly contributing to radio-resistance of cancer, ultimately leading to treatment failure [5].
In response to genotoxic damage, cancer cells can activate their DNA damage response (DDR), involving a network of proteins capable of recognizing and repairing damaged DNA to enhance tumor cell survival [[4], [5], [6]]. Specifically, the two key intracellular mechanisms contributing to this process are homologous recombination (HR) and nonhomologous end joining (NHEJ) [[4], [5], [6]]. These responses are triggered following DSBs through the recruitment of DDR proteins, such as DNA repair protein RAD50 (RAD50) [7,8]. RAD50 can form a complex with heterotrimeric recombination 11 homolog 1 (MRE11) and Nijmegen breakage syndrome protein 1 (NBS1), named MRE11-RAD50-NBS1 (MRN) complex. This complex plays a crucial role in sensing, signaling, and repairing DSBs [9]. Upon recruitment to DSBs, the MRN complex initiates DSBs resection while maintaining DSB strand organization to facilitate repair [9]. The MRN complex is also essential for recruitment and activation of the protein kinase ataxia-telangiectasia mutated (ATM) to the site of DSBs, thus coordinating cell cycle-dependent DSB repair and the MRN signaling cascade [10]. Once activated, ATM phosphorylates several downstream substrates, including p53, checkpoint kinase 2 (CHK2), and mediator of DNA damage checkpoint 1 (MDC1). These events, subsequently recruit additional MRN complexes to sites of DNA DSBs, thereby amplifying the MRN/ATM signaling cascade [11,12].
Overexpression of RAD50 in breast tumors is recognized as a major contributor to tumor resistance to standard chemotherapy and RT in breast cancer and other types of cancer and is linked with tumor progression and lower survival rates in patients of various types of cancer [[13], [14], [15], [16]]. Therefore, silencing RAD50 in cancer cells with small interfering RNA (siRNA) has been attempted to enhance chemotherapy of breast cancer cells using lipofectamine [15,17] or RT of nasopharyngeal carcinoma using a viral vector [18]. Despite these initial studies and design of interfering RNAs for targeting various oncogenes [[19], [20], [21]], silencing RAD50 gene using safe and effective non-viral carriers to enhance RT remains to be explored [[22], [23], [24]]. Given the significant role of RAD50 in DNA DSB repair [[13], [14], [15],17,18,25] and our previous observation that RT induces RAD50 upregulation in various cancer cells, we propose to develop a new RAD50 siRNA nanoparticle system to “knockdown” RAD50 expression, thereby disrupting the function of the MRN complex to sensitize human TNBC cells and tumors to RT.
siRNAs represent a widely applicable biologic therapeutic with high specificity and capability of regulating a variety of genes that are difficult to target using conventional therapies such as antibodies and small molecule drugs in certain tissue compartments [21,26]. However, delivering siRNA to target cells and cytoplasm faces several challenges, including RNAase caused degradation, tumor accumulation and penetration, cellular uptake, and endosomal/lysosomal entrapment and degradation [20,22]. Therefore, various nanocarrier systems have been developed to obtain good in vivo stability, tissue bioavailability, and transfection efficiency of siRNAs [[27], [28], [29]]. Both viral and non-viral vectors have been employed for delivering siRNAs into the tumor site [27]. Despite high transfection efficiency, viral vectors have several disadvantages, such as high cost and high immunogenicity [30]. Thus, non-viral systems, mostly nanoparticles made of polymers, lipids, or both polymer and lipid have been studied extensively due to their low toxicity, good biocompatibility, and great versatility [27,30,31].
In past two decades, tremendous advances have been achieved in the development of non-viral nanoparticle-based RNA formulations [19,22,32]. Recently, several siRNA-NP products have been approved for human use, including ONPATTRO (patisiran), GIVLAARI (givosiran), OXLUMO (lumasiran), LEQVIO (inclisiran), AMVUTTRA (vutrisiran), and RIVFLOZATM (nedosiran), and many more formulations have entered clinical trials [19,28,33]. These products are formulated mainly based on two platform systems: lipid nanoparticles (LNP) and siRNA conjugates, such as siRNA conjugate with N-acetylgalactosamine (GalNAc) or lipid 2′-O-hexadecyl (C16) [19,28,33]. So far, LNP has been the most successful carriers for delivering nucleic acid therapeutics, such as siRNA and mRNA, propelled by the recent success of the COVID-19 vaccine [34]. In LNP formulations, PEGylation is commonly used to improve the colloidal stability of the NP formulations in solution and during blood circulation [34,35]. However, PEGylated LNP has been found to be associated with undesired side effects, including adverse immunogenic response [[34], [35], [36]]. Therefore, alternative stabilizing systems are being explored to improve the stability of LNPs and minimize side effects, such as immunogenic reactions, which will be beneficial for repeated dosing regimens. In the current work, we have replaced the PEG-lipid with our devised biocompatible terpolymer, i.e., polysorbate-80 and poly (methacrylic acid) (PMAA)-grafted starch that can cover the siRNA-NP surface with negatively charged polymer chain [31,[37], [38], [39], [40], [41]]. Our previous studies have shown that the terpolymer-based nanoparticles can recruit apolipoproteins such as apolipoprotein E, that enhances cellular uptake via low-density lipid receptor-mediated transcytosis and effectively penetrate the brain and target tumor cells [31,[37], [38], [39], [40], [41]]. Nanoparticles made of the terpolymer and various lipids can be deigned to effectively enter cancer cells via clathrin- and caveolin-dependent endocytosis [37,38]. In addition, the different water-solubility of terpolymer and lipid and pH-dependent ionization of PMAA chains can facilitate deep tumor penetration, endosomal/lysosomal escape, and intracellular transport of loaded drugs [[37], [38], [39], [40], [41], [42], [43], [44], [45], [46]].
The novel biocompatible, polymer-lipid based, RAD50 siRNA loaded nanoparticles (RAD50-siRNA-NPs) were designed and their effect on downregulating RAD50 expression was evaluated in human TNBC MDA-MB-231 cells and orthotopic xenograft tumor model. The results demonstrated that RAD50-siRNA-NPs delivered the siRNA into cytoplasm, silenced RAD50 efficiently, and enhanced radiation-induced DNA DSBs both in vitro and in vivo. Notably, the combination of RAD50-siRNA-NPs with RT significantly inhibited the clonogenicity of TNBC in vitro, when compared to RT alone. Strikingly, intratumoral injection of siRNA-NPs followed by RT at a dose of 10 Gy in the breast tumors resulted in increased DNA DSBs, cancer cell apoptosis, and profound inhibition of tumor growth compared to RT alone. These results suggest a compelling and widely applicable strategy to enhance RT response in TNBC and potentially other tumor types through disrupting the function of the DNA repair protein RAD50 using siRNA-NPs.
2. Materials and Methods
2.1. Cell culture and reagents
MDA-MB-231 cells (derived from the pleural effusion of a breast cancer patient) and MCF-10A cells (derived from the mammary gland) were obtained from ATTC (Manassas, VA, USA) and used in this study as a model of TNBC and normal breast cells, respectively. The cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10 % fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA, USA). Cell lines were grown at 37 °C in a humidified incubator (NuAire, Plymouth, MN, USA) with 5 % CO2 (Linde, Mississauga, Ontario, Canada). The RAD50 siRNA and scrambled siRNA were obtained from Thermo Fisher Scientific (Waltham, MA, USA). Antibodies specific to the phosphorylated form of H2A histone family member X (γ-H2AX) and β-actin were purchased from Abcam (Cambridge, UK), while antibodies against cleaved caspase-3 was obtained from Cell Signaling Technology (Danvers, MA, USA).
2.2. Preparation and characterization of siRNA nanoparticles
The RAD50-siRNA-NPs were prepared using oil-in-water emulsion method. Briefly, an aqueous solution of siRNA was mixed with an ethanolic solution containing cationic phospholipid 1,2-dioleoyl-3-trimethylammonium propane (DOTAP), cholesterol and helper phospholipid 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) at a molar ratio of 50:35:15. The complexation of siRNA with cationic lipid was performed using a N/P ratio of 3, corresponding to three positive DOTAP nitrogen charges for every negative siRNA phosphate. After mixing for 3 min, an aqueous solution of an amphiphilic terpolymer synthesized in our laboratory, comprised of poly(methacrylic acid) and polysorbate 80-grafted starch [[42], [43], [44], [45], [46], [47]], was added at a weight equivalent to three times the total lipid content. The emulsion was sonicated for 30 s with an ultrasonic processor probe (100 Hz, 5 mm probe depth, Heischer UP100H, Germany). Subsequently, the siRNA-NPs were collected by adding them to ice cold RNase-free water. To remove any residual free siRNA and concentrate siRNA-NPs, purification was carried out using an Amicon 100 KDa centrifuge filter (Sigma-Aldrich, St. Louis, MO, USA) at 2000 rpm. The concentrated sample was then mixed with an appropriate amount of a cryoprotectant (e.g. sucrose) at a 10 to 20 molar ratio of cryoprotectant to total lipids of cryoprotectant (e.g. sucrose), frozen at −20 °C and lyophilized over night at −80 °C using a Labconco FreeZone 6 Plus Freeze Dryer Lyophilizer freeze dryer (Labconco, USA). The lyophilized siRNA-NPs were stored at −20 °C for further use. After reconstitution in 4 % dextrose in deionized water, the particle size and zeta potential of RAD50-siRNA-NPs were determined at 37 °C using a Malvern ZetaSizer (Nano ZS) system (Malvern, UK).
2.3. Determination of encapsulation efficiency
The efficiency of siRNA encapsulation was assessed using the Quant-iT Ribogreen RNA Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) in accordance with the manufacturer's instructions. The Ribogreen indicator, a fluorescent dye, exhibits strong fluorescence signals upon binding to free nucleic acids. The fluorescence intensity is directly correlated with the amount of free nucleic acid and gradually decreases due to the increasing encapsulation of siRNA with the cationic lipid and polymer complex. The encapsulation of siRNA was validated by measuring the free siRNA in the nanoparticle formulation. Specifically, siRNA encapsulation was determined by measuring the free siRNA in the nanoparticle suspension at the final stage of synthesis and in the filtrate collected during the washing steps. The encapsulation efficiency was determined by comparing the fluorescence intensity (excitation wavelength 500 nm/emission wavelength 525 nm) of free siRNA in the nanoparticle suspension to the signal of siRNA initially introduced in the synthesis process.
2.4. In vitro transfection of RAD50 siRNA into cells
MDA-MB-231 cells were seeded in 6-well plates (100,000 cells per well), and incubated in 2 mL of DMEM containing 10 % FBS for 24 h at 37 °C. The medium was subsequently replaced by fresh medium containing different concentrations of RAD50-siRNA-NPs, free RAD50 siRNA in solution without NPs (free-siRNA) or Scrambled siRNA-loaded NPs (Scr.siRNA-NPs). Cells were incubated for additional 48 h, 96 h, or 120 h. Then, the cells were washed in isosmotic phosphate-buffered saline (PBS) buffer (100 mM, pH 7.4), trypsinized and the proteins were extracted using radioimmunoprecipitation assay (RIPA) lysis buffer supplemented with protease inhibitor cocktail. The RAD50 expression levels were subsequently assessed using Western Blotting.
2.5. Cellular uptake and intracellular trafficking of siRNA-NPs
To investigate the internalization and intracellular localization of siRNA-NPs, Cyanine3 (Cy3) labeled scrambled siRNA was incorporated into the nanoparticles. In addition, the polymer used to construct the RAD50-siRNA-NPs was conjugated with Cyanine5 (Cy5). MDA-MB-231 cells (20,000 cells/well) were seeded onto 6-well plates and treated with siRNA-NPs (10 nM siRNA). Following 2 or 6 h of incubation at 37 °C, the media was removed, and the cells were washed with PBS. Subsequently, the cells were stained with LysoTracker Green to label endosomes following the manufacturer's instructions (Thermo Fisher Scientific), along with the application of 0.5 mM Hoechst 33342 to stain cell nuclei. Cells were then imaged using confocal laser scanning microscopy (CLSM). To investigate relative levels of endocytosis, cells were incubated with Cy5 conjugated siRNA-NPs at both 37 °C and 4 °C for 2 h, and the cell uptake of RAD50-siRNA-NPs was visualized using CLSM.
2.6. Effect of RAD50-siRNA-NPs treatment and RT on DNA damage repair
The MDA-MB-231 cells were cultured on 6-well plates (100 K cells/well) in 2 mL of DMEM supplemented with 10 % FBS for 24 h. Subsequently, the culture medium was replaced with fresh medium containing 150 nM of RAD50-siRNA-NPs or free-siRNA. After 48 h of additional incubation, cells were irradiated with a single dose of 5 Gy using X-RAD 320 system (Precision X-Ray, North Branford, CT. USA; 320 kV, 2.7 Gy/min). The cells were then allowed to grow for an additional 24 h. Relative levels of γH2AX DNA DSBs marker were then assessed using Western blotting.
2.7. Clonogenic cell survival and treatment response
MDA-MB-231 cells were plated into 6-well plates at a density of 100 K cells/well in 2 mL of DMEM supplemented with 10 % FBS and allowed to adhere overnight. Subsequently, 150 nM of RAD50-siRNA-NPs or free siRNA was added, and cells were incubated for another 48 h. Following the incubation, cells were exposed to a radiation dose of 2, 4, or 6 Gy. Immediately post-irradiation, cells were trypsinized, reseeded onto 6-well plates at a density of 500 cells/well, and incubated for 7 days. Afterwards, the cells were washed with PBS and fixed with 10 % formalin for 10 min, and stained with 0.5 % crystal violet in 20 % methanol/H2O (v/v) for 10 min. The cells were then rinsed with PBS, and colonies counted. Plating efficiency (PE) was determined using the formula:
| (1) |
Survival percent was determined using the formula:
| (2) |
The experimental results were then fitted to a linear-quadratic model on GraphPad Prism 8.0.2 (San Diego, CA, USA).
| (3) |
2.8. Animals and tumor model
Female severe combined immunodeficiency (SCID) mice aged 7–8 weeks were purchased from Ontario Cancer Institute (Toronto, Ontario, Canada). An orthotopic breast tumor was established by injection of MDA-MB-231 cells (1.0 × 106) in 20 μl of growth medium into the mammary fat pad of mouse under isoflurane anesthesia. All in vivo studies were conducted in accordance with the ethical and legal requirements of the Animals for Research Act of Ontario and the Federal Canadian Council on Animal Care Guidelines for the Care and Use of Laboratory Animals. The animal protocols used in these studies were approved by the University of Toronto Animal Care Committee and/or University Health Network Animal Care Committee (Animal Use Protocol 6698).
2.9. Evaluation of in vivo RAD50 silencing and biomarkers
About 3 weeks after the tumor inoculation, when tumors reached the required size (100 mm3), the mice were randomly divided into 4 experimental groups: control (saline), RAD50-siRNA-NPs, free-RAD50-siRNA and Scr.siRNA-NPs. For the treatment involving RAD50-siRNA-NPs, free-siRNA or Scr.siRNA-NPs treatments, 0.5 nmol siRNA in 30 μl of PBS was intratumorally (IT) injected daily for 3 consecutive days. Twenty-four hours following the final injection, the mice were sacrificed, and tumor tissues were harvested for analysis of RAD50. For the biomarkers study, on day 4 following the injection of the above treatments, tumors were exposed to radiation (10 Gy) using small animal irradiator (SmART+) XRAD system (Precision X-Ray Inc., Madison, CT, USA). After 24 h, tumors were collected for analysis of cleaved caspase3 and γH2AX expression using Western blotting and/or immunohistochemistry (IHC).
2.10. Effect of treatment on tumor growth inhibition
Tumor-bearing mice were randomly divided into 4 groups: saline, saline + RT (10 Gy), RAD50-siRNA-NP alone, and RAD50-siRNA-NP + RT (10 Gy). Animals were treated IT with saline or RAD50-siRNA-NPs (0.5 nmol siRNA in 30 μl of PBS) daily for 3 consecutive days. On day 4, tumors were exposed to radiation (10 Gy) using SmART+ XRAD system. Subsequently, mice were monitored, and tumor size was measured using a Vernier caliper every 2 days for a duration of 20 days. Tumor volume (V) was calculated using the formula:
| V= W2 × L/2 |
where W and L are the shortest and longest diameters, respectively.
2.11. Statistical analysis
Statistical analyses of the data were performed using GraphPad one-way ANOVA followed by Tukey's post-hoc test. Error bars represent mean ± SD. Probability (P) values less than 0.05 (P < 0.05) were considered statistically significant and are designated as P < 0.05 (*), P < 0.005 (**), P < 0.0005 (***), and P < 0.0001 (****).
3. Results
3.1. Synthesis and characterization of siRNA-NPs
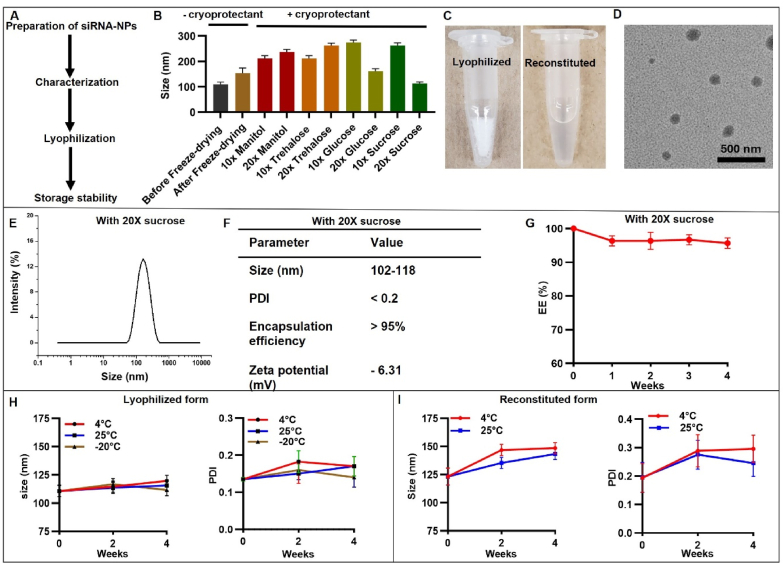
The detailed synthesis of RAD50-siRNA-NPs is described in the Materials and Methods section. Fig. 1A shows the steps we followed in the preparation of RAD50-siRNA-NP. Following the preparation, the impact of freeze-drying on stability of the RAD50-siRNA-NPs was investigated (Fig. 1B). The size of RAD50-siRNA-NPs was measured before and after freeze-drying in the presence of different cryoprotectants (mannitol, trehalose, glucose or sucrose) at different concentrations. Without cryoprotectants, the size of RAD50-siRNA-NP increased by 1.4-fold following freeze-drying. In the presence of 10 × 10 moles of cryoprotectant per mole of total lipids) or 20 × trehalose, mannitol, glucose or 10 × sucrose, the size of RAD50-siRNA-NPs was significantly increased compared to the size of nanoparticles prior to freeze-drying. However, the nanoparticle size remained comparable to the size at pre-freeze-drying stage in the presence of 20 × of sucrose (Fig. 1B). Therefore, 20 × sucrose was selected as a cryoprotective agent for the RAD50-siRNA-NP formulation in the subsequent in vitro and in vivo studies. The lyophilized RAD50-siRNA-NPs appeared as a white powder, which turned to almost transparent solution after reconstitution in water (Fig. 1C). The size and morphology of the RAD50-siRNA-NPs were examined using transmission electron microscopy (TEM). The nanoparticles exhibited a spherical shape and narrow size distribution, with no evidence of aggregation (Fig. 1D). The RAD50-siRNA-NPs exhibited an average particle size between 102 and 118 nm (Fig. 1E). The polydispersity index (PDI) of the particles was less than 0.2, aligning with the results from the TEM analysis (Fig. 1F). Additionally, the siRNA encapsulation efficiency was over 95 % quantified using Quant-iT Ribogreen assay, and the zeta potential was measured at −6.31 mV (Fig. 1F). Interestingly, the RAD50-siRNA-NPs formulation retained >95 % of siRNA, encapsulation efficiency (EE), when the formulation was kept at −20 °C for four weeks (Fig. 1G). To assess the storage stability of lyophilized RAD 50 siRNA-NPs, the particle size and PDI of siRNA-NPs were monitored up to 4 weeks, when stored at 25 °C, 4 °C or- 20 °C. The results indicated that the particle size and PDI of the lyophilized powder remained constant under all three storage conditions for the entire 4-week duration (Fig. 1H). The stability of reconstituted RAD50-siRNA-NP liquid form stored at 4 °C or 25 °C was also evaluated over a 4-week period, which showed a slight increase in the size and PDI during this storage period. (Fig. 1I).
Fig. 1.
Characterization of RAD50-siRNA-NPs. A. Flowchart of siRNA-NPs preparation. B. The size of RAD50-siRNA-NPs before and after freeze drying in the presence of different cryoprotectants. C. Appearance of lyophilized and reconstituted RAD50-siRNA-NPs. D. TEM image of RAD50-siRNA-NPs. E. Size distribution of RAD50-siRNA-NPs lyophilized using 20 × sucrose and after reconstitution in RNAs free water. F. Size, PDI, Encapsulation efficiency and zeta potential of RAD50-siRNA-NPs with 20 × sucrose. G. Encapsulation efficiency of RAD50-siRNA-NPs stored at −20 °C for four weeks. Size and PDI measurements of lyophilized RAD50-siRNA-NPs when stored at 25 °C, 4 °C or −20 °C (H), or reconstituted liquid form (I) for 4 weeks. Results presented as mean ± SD for 3 independent experiments. P < 0.05 was considered statistically significant. P < 0.05 (*), P < 0.005 (**), P < 0.0005 (***), and P < 0.0001.
3.2. Lysosomal escape and RAD50 knockdown efficiency of RAD50-siRNA-NPs in MDA-MB-231 cells in vitro
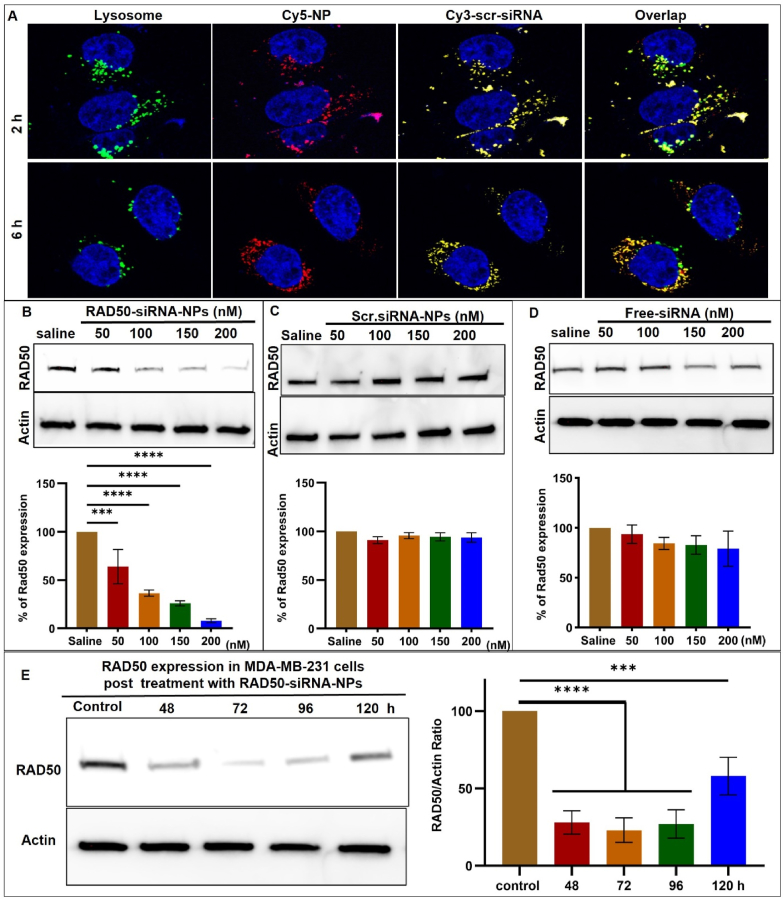
To investigate the cellular uptake, intracellular trafficking, and RAD50 silencing efficiency of the RAD50-siRNA-NPs, MDA-MB-231 cells were treated with siRNA-NPs. For cell uptake and intracellular trafficking studies, cells were treated with siRNA-NPs containing Cy5-conjugated terpolymer (red) and a Cy3-labeled scrambled siRNA (Scr.siRNA-NPs) (orange) (Fig. 2A). Following treatment with Scr.siRNA-NPs, lysosomes were labeled with LysoTracker Green probe (green), and cell nuclei stained with Hoechst 33342 (blue). CLSM images were acquired 2 h and 6 h following Scr.siRNA-NPs treatment. After 2 h of incubation with siRNA-NPs, efficient uptake into lysosomes was confirmed by co-localization of the fluorescent NPs (red) and Cy3-Scr.siRNA (yellow) signal with lysosomes (green) (Fig. 2A). Following 6 h of incubation, a decrease in co-localization of Scr.siRNA-NPs and lysosomes was observed as evidenced by the separation of Scr.siRNA signal from NPs and lysosomes [31,48]. This indicates a process of releasing siRNA from NPs which escaped from lysosomes. The continued presence of Scr.siRNA in the intracellular compartment at 6 h suggests that the siRNA-NP formulation was able to facilitate siRNA release into the cytoplasm. Additionally, the incubation of RAD50-siRNA-NPs with MDA-MB-231 cells at either 37 °C or 4 °C revealed a significantly higher uptake of RAD50-siRNA-NPs at 37 °C than at 4 °C (Fig. S1), suggesting that RAD50-siRNA-NPs internalization was mediated by energy-dependent endocytosis.
Fig. 2.
Lysosomal escape and RAD50 silencing ability of the RAD50-siRNA-NPs. A) CLSM images of MDA-MB-231 acquired at 2 and 6 h after incubation with 10 nM of labeled Scr.siRNA-NPs demonstrating the relative intracellular distribution of lysosomes (green), NPs (red), siRNA (orange) and nuclei (blue). B, C, D) Western blot and quantification for RAD50 knockdown efficiency at various concentrations of RAD50-siRNA-NPs, Scr.siRNA-NPs and free-siRNA. E) Western blot and quantification for RAD50 expression for up to 120 h after cells were incubated with 150 nM of RAD50-siRNA-NPs for 48 h, which was reconstituted from the lyophilized powder and stored at 4 °C for 4 weeks. Results presented as mean ± SD for 3 independent experiments. P < 0.05 was considered statistically significant. P < 0.05 (*), P < 0.005 (**), P < 0.0005 (***), and P < 0.0001.
To evaluate the effectiveness of RAD50-siRNA-NPs in silencing RAD50 in vitro, MDA-MB-231 cells were exposed to different concentrations (50, 100, 150, 200 nM) of either RAD50-siRNA-NPs, scr-siRNA-NPs or free siRNA for 48 h. The results demonstrated that treatment with RAD50-siRNA-NPs resulted in a significant knockdown of RAD50 expression, with a concentration of 200 nM exhibiting an approximately 90 % knockdown relative to saline controls (Fig. 2B). In contrast, neither free siRNA nor Scr.siRNA-NPs led to reductions in RAD50 protein levels, when compared to saline controls (Fig. 2C and D). Taken together, these results demonstrate that that RAD50-siRNA-NPs successfully delivered, protected, released bioactive siRNA, and effectively silenced RAD50 protein expression in TNBC MDA-MB-231 cells.
To investigate how long the RAD50 knockdown can last, MDA-MB-231 cells were treated with 150 nM of RAD50-siRNA-NPs for 48 h, which was reconstituted from the lyophilized powder and stored at 4 °C for 4 weeks. Then the expression of RAD50 protein measured by Western blotting at varying times. Fig. 2E shows that RAD50 expression is significantly reduced by >75 % for 96 h (4 days). At 120 h (5 days) post RAD50-siRNA-NPs treatment, the expression of RAD50 increased but was still ∼40 % lower than the untreated cells (Fig. 2E). These results confirm that our lyophilized RAD50-siRNA-NP formulation can maintain the bioactivity of the siRNA after 4 weeks storage at 4 °C and downregulate the RAD50 for at least 5 days.
3.3. RAD50 silencing by RAD50-siRNA-NPs overcomes RT-induced DNA damage response and increases DNA DSBs
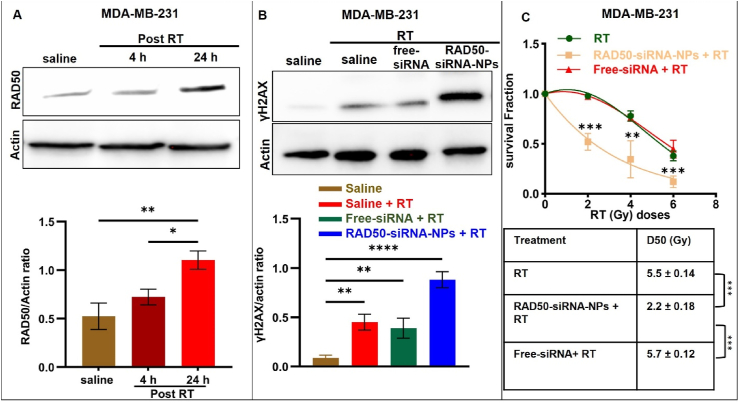
The expression of RAD50 in MDA-MB-231 cells in vitro 4 and 24 h following a single dose of 5 Gy radiation was investigated. As shown in Fig. 3A, there was no significant increase in RAD50 expression at 4 h post-radiation with a 1.5-fold increase, when compared to saline. However, at 24 h after RT, RAD50 expression significantly increased, reaching a 2.1-fold elevation compared to the saline control. Since the treatment with Scr.siRNA-NPs did not reduce RAD50 expression, we only compared free-siRNA and RAD50-siRNA-NPs in all subsequent in vitro experiments. To assess the impact of RAD50-siRNA-NPs on RAD50 knockdown with respect to RT-induced DNA damage, MDA-MB-231 cells were treated with free-siRNA or RAD50-siRNA-NPs (150 nM) for 48 h followed by irradiation with 5 Gy. At 24 h post irradiation, γ-H2AX expression, serving as a maker of DNA damage repair, was evaluated by Western analysis. As shown in Fig. 3B, the combination of free siRNA with RT did not increase the level of γH2AX, when compared to RT alone. In contrast, pretreatment with RAD50-siRNA-NPs resulted in a 53 % increase in γH2AX expression compared to radiation alone or radiation plus free-siRNA. Collectively, these findings demonstrate that the NPs delivered RAD50 siRNA dramatically enhancing the efficacy of RAD50 knockdown in a functionally meaningful way.
Fig. 3.
Mode and effect of RAD50 silencing on an ionizing radiation induced DNA damage response. A) Western analysis and RAD50/actin ratio of RAD50 expression in MDA-MB-231 cells 4 and 24 h following treatment with 5 Gy in vitro. B) Western analysis of DNA damage marker γ-H2AX in MDA-MB-231 and gamma H2AX/actin ratio for cells treated with 150 nM free-siRNA or RAD50-siRNA-NPs for 48 h followed by 5 Gy radiation in vitro and collected 24 h post treatment. C) Clonogenic assay of MDA-MB-231 cells treated with free-siRNA or RAD50-siRNA-NPs (150 nM) for 48 h followed by various doses of radiation (2, 4, 6 Gy) in vitro. Results presented as mean ± SD for 3 independent experiments. P < 0.05 was considered statistically significant. P < 0.05 (*), P < 0.005 (**), P < 0.0005 (***), and P < 0.0001 (****).
3.4. RAD50-siRNA-NPs enhance the radiation efficacy on cancer cell clonogenicity in vitro
The radio-sensitization effect of RAD50 knockdown using RAD50-siRNA-NPs on MDA-MB-231 cells was assessed using clonogenic assay. Cells were plated onto 6-well plates and incubated at 37 °C for 24 h prior to treatment with saline, free-siRNA or RAD50-siRNA-NPs (150 nM siRNA) for 48 h and then irradiated with 2, 4, 6 or 8 Gy. As shown in Fig. 3C and Fig. S2, free-siRNA plus RT had no effect on clonogenicity compared to RT alone. In contrast, the combination of RAD50-siRNA-NPs and RT significantly inhibited clonogenicity compared to RT alone, as evidenced by a 2.75-fold decrease in the D50 value from 5.5 Gy to 2 Gy (Fig. 3C). These results indicate that NP-mediated delivery of RAD50 siRNA significantly enhances the radiosensitivity of MDA-MB-231 cells compared to the treatment with free RAD50 siRNA.
3.5. Effect of RAD50-siRNA-NPs on in vivo RAD 50 silencing and radiation response in an orthotopic breast tumor model
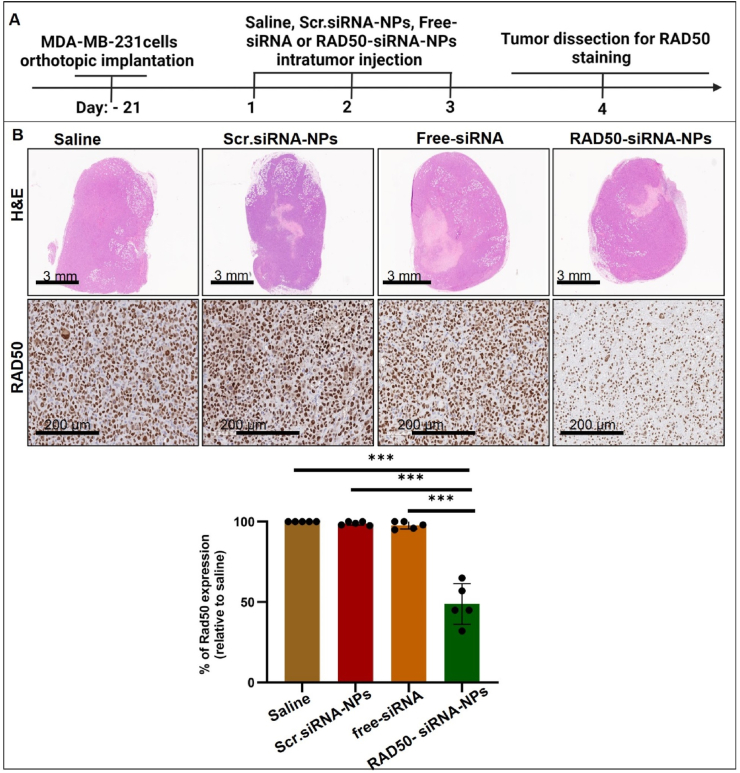
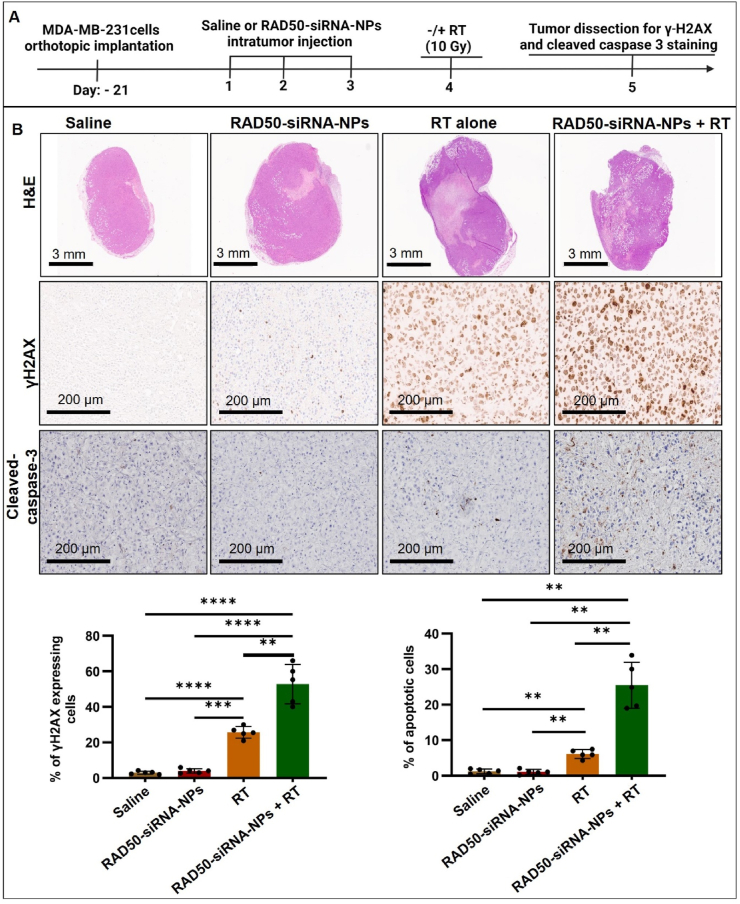
To examine the in vivo effects of RAD50-siRNA-NPs, MDA-MB-231 xenograft tumor bearing mice were injected IT with either saline, Scr.siRNA-NPs, free-siRNA or RAD50-siRNA-NPs (0.5 nmol siRNA per dose) for three consecutive days (Fig. 4A). On day 4, mice were sacrificed, and tumors were collected and subjected to IHC staining for RAD50. As shown in Fig. 4B, the results demonstrated that neither Scr.siRNA nor free-siRNA led to significant downregulation of RAD50 expression. In contrast, injection of RAD50-siRNA-NPs resulted in an ∼50 % knockdown in RAD50 expression (Fig. 4B). To assess the functional consequences of this reduction, the effect of RAD50-siRNA-NPs treatment on radiation-induced DNA damage and cell death was further examined. Given that both Scr.siRNA-NPs and free-siRNA treatments did not result in significant RAD50 knockdown in vivo, this study was focused solely on examining the effect of RAD50-siRNA-NPs with or without RT. The MDA-MB-231 tumor-bearing mice were randomly divided into 4 groups: saline, RAD50-siRNA-NPs, RT alone (10 Gy) or RAD50-siRNA-NPs + RT (10 Gy). The mice were injected IT for three consecutive days at a dose of 0.5 nmole siRNA or saline followed by 10 Gy irradiation on the following day of the last injection. Tumors were collected at 24 h post RT and subjected to IHC staining for γ-H2AX and activated caspase-3. As shown in Fig. 5, RT alone induced the accumulation of γ-H2AX and caspase-3 cleavage compared to saline or RAD50-siRNA-NPs alone groups which showed no effect. However, the RAD50-siRNA-NPs plus radiation treatment group demonstrated a significant enhancement of γ-H2AX expression and caspase-3 cleavage, by 2-fold and 4.5-fold, respectively, compared to the RT alone group.
Fig. 4.
The RAD50-siRNA-NPs significantly downregulates the expression of RAD50 in MDA-MB-231 tumor cells in vivo. A) Experimental treatment schedule of orthotopic MDA- MB-231 tumors grown in SCID mice. Mice were treated with saline control, Scr.siRNA-NPs, free-siRNA, or RAD50-siRNA-NPs (5 mice in each group). B) Representative images of H&E and IHC stained tumors for hematoxylin/eosin and RAD50 (brown) at 24 h post the last IT injection of saline, Scr.siRNA or RAD50-siRNA-NPs (0.5 nmol siRNA). The quantification of RAD50-positive area relative to saline control, where each data point represents average value in two tumor slices. Data are presented as mean ± SD. P < 0.05 (*), P < 0.005 (**), P < 0.0005 (***), and P < 0.0001 (****).
Fig. 5.
RAD50 silencing using RAD50-siRNA-NPs plus RT results in enhanced DNA damage and induced apoptosis in MDA-MB-231 tumors in vivo. A) Schematic treatment schedule of orthotopic MDA-MB-231 tumors grown in SCID mice. B) Representative images of tumor tissue stained by H&E and IHC for γ-H2AX and cleaved caspase-3 after the treatment with saline control, Scr.siRNA-NPs, free-siRNA or RAD50-siRNA-NPs (5 mice in each group). Quantification of image data for the indicated markers compared to saline control. Two slices of each tumor were analyzed and their average value was used for the plot. Data are presented as mean ± SD. P < 0.05 (*), P < 0.005 (**), P < 0.0005 (***), and P < 0.0001 (****).
3.6. RAD50-siRNA-NPs treatment significantly enhances antitumor efficacy of RT
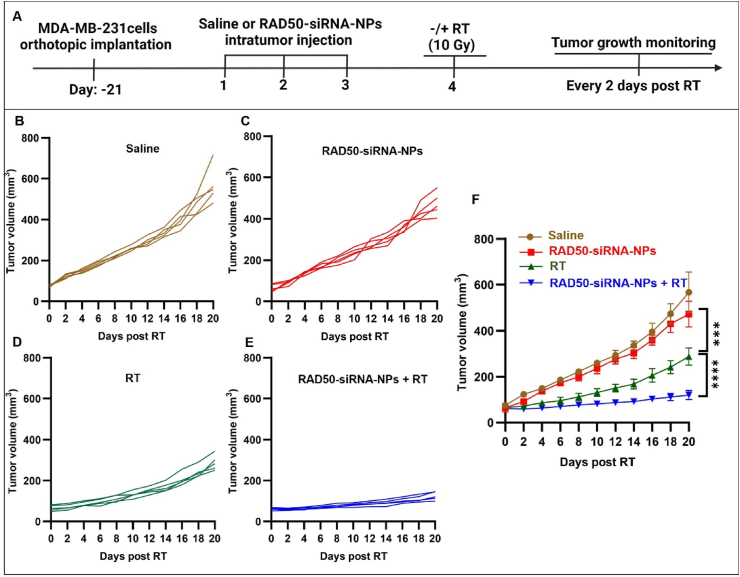
The effect of RAD50 silencing on enhancing RT efficacy was investigated further in orthotopic MDA-MB-231 tumor-bearing SCID mice. The treatment schedule is depicted in Fig. 6A. Three weeks after tumor inoculation (day-21), each mouse bearing the MDA-MB-231 xenograft received one IT treatment per day for three consecutive days of either saline or RAD50-siRNA-NPs (0.5 nmol siRNA). Twenty-four hours after the last treatment, the tumors were exposed to radiation (10 Gy), and the tumor volumes were measured every two days over a period of 3 weeks. As shown in Fig. 6B, radiation alone significantly inhibited tumor growth, when compared to saline or RAD50-siRNA-NPs groups, whereas the combination of RAD50-siRNA-NPs and radiation resulted in substantial increase in the inhibition of tumor growth, achieving a 2.5-fold smaller in tumor volume, when compared to the RT alone group (Fig. 6B). These findings suggest that RAD50 silencing using RAD50-siRNA-NPs significantly enhanced RT efficacy in delaying tumor growth.
Fig. 6.
RAD50 silencing by RAD50-siRNA-NPs plus RT inhibited tumor growth of MDA-MB-231 tumor-bearing mice. (A) Schematic of treatments of RAD50-siRNA-NPs and RT of orthotopic MDA-MB-231 tumors grown in SCID mice. (B) Individual mouse tumor growth curves for saline group, (C) RAD50-siRNA-NPs group, (D) saline + RT (10 Gy) and (E) RAD50-siRNA-NPs + RT treated animals. (F) Comparison of tumor growth curves for the four treatment groups, saline, RAD50-siRNA-NPs (0.5 nmol siRNA), saline + RT alone (10 Gy), and RAD50-siRNA-NPs (0.5 nmol siRNA) + RT (10 Gy) treatment, 5 mice in each group. RT was locally delivered on day 4 after daily IT injection of saline or RAD50-siRNA-NPs for 3 consecutive days. Data are expressed as mean ± SD. P < 0.05 (*), P < 0.005 (**), P < 0.0005 (***), and P < 0.0001 (****).
4. Discussion
Cancer cells activate a variety of cellular pathways to overcome DNA damage through the activation of DNA-repair mechanisms to promote their survival and reducing the effectiveness of RT [49]. The targeting of DNA damage repair proteins has gained increasing attention as a way to improve cancer treatments [50]. Among these proteins, RAD50 has been identified as playing a pivotal role in the DNA repair process [51]. Elevated tumor expression of RAD50 is associated with radioresistance and poor survival in cancer patients undergoing RT [13,25,52]. These findings emphasize the importance of strategies to overcome DDR-mediated cell survival to improve the clinical outcome of RT.
With this in mind, we have developed a siRNA-NP formulation (RAD50-siRNA-NPs) for IT delivery of an siRNA for targeting RAD50. Utilizing the oil-in-water self-assembly nanoparticle approach, we have successfully designed and developed a biocompatible polymer-lipid complex for the efficient encapsulation of siRNA. Ensuring the stability of RAD50-siRNA-NPs is crucial, particularly in the context of utilizing them as therapeutic agents [53]. The lyophilization of siRNA-NPs not only prolongs their shelf life, but also amplifies the stability and effectiveness of the encapsulated payloads [54]. Our investigation revealed that sucrose surpassed other cryoprotectants in preserving the properties of RAD50-siRNA-NPs during the lyophilization process. The lyophilized RAD50-siRNA-NPs exhibited stability under both ambient and low-temperature conditions, positioning them as an improved carrier system for siRNA (Fig. 1).
Lysosomal entrapment poses a significant challenge in the field of gene therapy [55]. Typically, NPs are internalized by cancer cells through endocytosis, subsequently entering the acidic endosomes and enzyme-rich lysosomes where they face the risk of enzymatic degradation of the therapeutic payload, particularly RNAs with respect to gene therapy [55]. We demonstrate here that our novel polymer-lipid siRNA-NP formulation is able to both effectively penetrate cancer cells to aid egress of siRNAs and to escape from the lysosomes reaching to the cytoplasm. It appears that this process of lysosomal escape involves surface charge alterations. The relatively negative surface charge of siRNA-NPs is very important, as it not only plays a role in the kinetics of siRNA-NPs, but also enhances the stability of NPs [56]. By introducing a negative surface charge, siRNA-NPs can minimize aggregation through the electrostatic repulsion between particles during storage, in solution, and even after administration in the bloodstream [57]. An example of this is seen in lipid nanoparticles (LNPs), which are widely used for siRNA delivery, with most clinical lipid NPs (LNPs) utilizing ionizable cationic phospholipids [34].
One drawback of such lipid formulations is their overall neutral surface charge under physiologic pH [34]. To address this, a PEG-lipid is commonly used in LNP formulations due to its hydrophilicity and bulkiness, facilitating surface coverage and reducing serum protein adsorption, prolonging circulation and minimizing kidney and mononuclear phagocyte system (MPS) clearance [58]. However recent studies demonstrate that COVID-19 vaccines utilizing LNPs can induce anti-PEG antibody production, potentially contributing to hypersensitivity reactions [35,36,59]. Additionally, such effects may promote immune-mediated clearance of systemically administered PEGylated nanomedicines, limiting their efficacy [35].
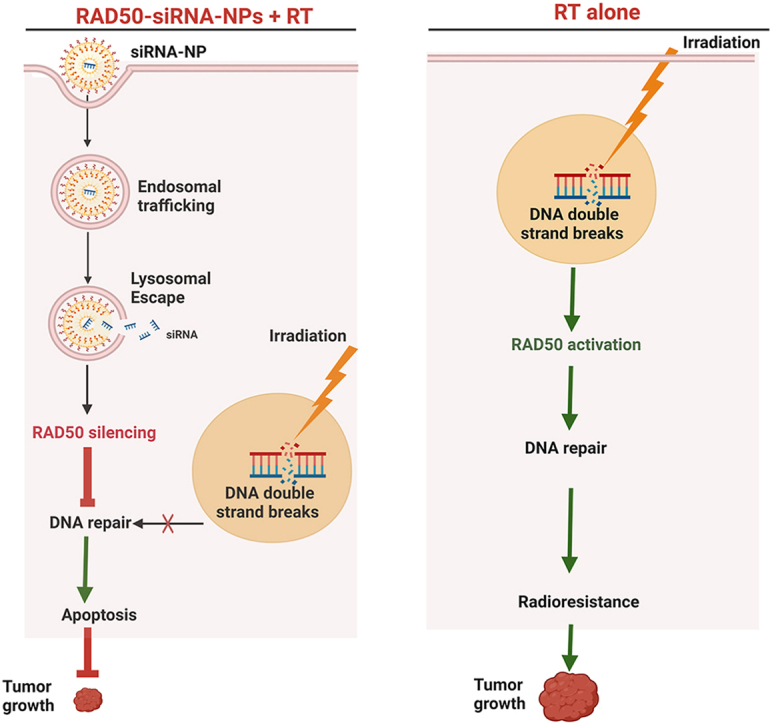
To overcome such limitations, the current polymer-lipid based siRNA carrier system is proposed. In this system, the surface of RAD50-siRNA-NPs is covered with a biocompatible terpolymer shell that is introduced during the final step of the synthesis process, eliminating the need for PEG and covering the lipid-siRNA complex. The terpolymer side chain contains abundant carboxylic acid groups resulting in a net negative surface charge of NPs under physiological conditions. The negative surface charge such RAD50-siRNA-NPs enhances their stability without the need of the PEG due to electrostatic repulsion between particles [60]. Upon cellular uptake and transport to early endosomes (pH < 6.0), the polymer carboxylic acid group start to protonate, neutralizing the polymer charge [61]. As a result, cationic DOTAP becomes the dominant surface charge, enabling stronger binding of RAD50-siRNA-NPs with the endosomal lipid membrane. This interaction aids in the disruption of the membrane layer, facilitating NP escape from endosomes to the cytoplasm. The relative lower pH in the lysosomes reduces interaction of RNA and lipid-polymer complex, facilitating the release of RNA from RAD50-siRNA-NPs to the cytoplasm (Fig. 2A).
Our in vitro investigations conducted on MDA-MB-231 TNBC cells clearly indicate effective delivery of siRNA into the cytoplasm using the polymer-lipid nanocomplex as a carrier system. The results have not only demonstrated the steps required for the successful delivery of the particles but have also revealed that these polymer-lipid NPs servs as a robust protective shield, ensuring the preservation of siRNA integrity. Consequently, this protective mechanism enabled significant reduction of RAD50 protein levels in cancer cells in vitro by 90 % and in tumor tissue by 53 % as evidenced by Western blot and IHC analysis (Fig. 2, Fig. 4B).
The RAD50 protein plays an important role in radiation resistance The impact of downregulating RAD50 protein using RAD50-siRNA-NPs on radiation sensitization is evident in both in vitro and in vivo settings. Interestingly, the knockdown of RAD50 markedly enhanced the radiation effect by reducing the D50 radiation dose to achieve 50 % survival of TNBC cells by 2.5-fold, when compared to RT alone or free siRNA plus RT (Fig. 3C). The in vivo treatment with RAD50-siRNA-NPs for three consecutive days followed by RT resulted in a notable 2-fold increase in DNA DSBs and a 2.4-fold increase in tumor cell apoptosis, when comparison to RT alone (Fig. 5). These results led to a substantial inhibition of tumor growth by 2.5-fold by the combination therapy (RAD50-siRNA-NPs + RT), when compared to RT alone at day 20 post-treatment (Fig. 6f). To the best of our knowledge, such a profound enhancement of RT by non-viral RAD50-siRNA-NPs has not been reported before and this is the first formulation of RAD50-siRNA-NPs to target RAD50 and the translation of this approach to enhance the antitumor efficacy of RT in vitro and in vivo.
The radiosensitizing effect of RAD50 silencing of RAD50-siRNA-NPs is attributable to the disruption of the MRN complex and inhibition of DNA damage response [62]. The noteworthy suppression of tumor growth in in mice treated with RAD50-siRNA-NPs followed by RT, reinforces the argument that targeting RAD50 before exposure to radiation is crucial for improving the antitumor efficacy of RT.
5. Conclusion
We have successfully developed a new RAD50-siRNA-NP system using a rationally designed polymer-lipid hybrid composition with clinical translatability for silencing the DNA damage repair protein RAD50 to enhance the efficacy of RT in TNBC. The RAD50-siRNA-NPs exhibited very high encapsulation efficiency, excellent storage stability, successful intracellular delivery, and effective capability of silencing RAD50 in vitro and in vivo. We demonstrated that IT delivery of siRNA-NPs prior to RT profoundly increased the levels of DNA DSBs and tumor cell apoptosis induced by standard RT, leading to 2.5-fold enhancement of RT efficacy in vitro and in vivo. The results suggest that targeting RAD50 using siRNA-NPs to enhance antitumor effect of RT in TNBC tumors could be a novel strategy to treat this aggressive form of breast cancer. The findings of this work will pave the way for further advancement of this strategy, facilitating the translation of this technology from the laboratory to clinical application.
CRediT authorship contribution statement
Abdulmottaleb E. Zetrini: Writing – original draft, Visualization, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Azhar Z. Abbasi: Writing – review & editing, Writing – original draft, Project administration, Investigation, Formal analysis, Data curation, Conceptualization. Chunsheng He: Writing – review & editing, Project administration, Methodology, Formal analysis, Data curation, Conceptualization. HoYin Lip: Writing – review & editing, Methodology, Formal analysis. Ibrahim Alradwan: Software, Investigation, Formal analysis. Andrew M. Rauth: Writing – review & editing. Jeffrey T. Henderson: Writing – review & editing. Xiao Yu Wu: Writing – review & editing, Supervision, Resources, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported in part by grants from the Canadian Institutes of Health Research, Natural Sciences and Engineering Council of Canada, and Cancer Research Society to X.Y. Wu. We also thank the Canada Council for the Arts for the Killam Research Fellowship to X.Y. Wu, the Ministry of Education of Libya scholarship to A. Zetrini, the King Abdulaziz City for Science and Technology (KACST) for the scholarship to I. Alradwan, and the funding to the STTARR facilities.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2024.101206.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics. CA A Cancer J. Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Xiong N., Wu H., Yu Z. Advancements and challenges in triple-negative breast cancer: a comprehensive review of therapeutic and diagnostic strategies. Front. Oncol. 2024;14 doi: 10.3389/fonc.2024.1405491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haussmann J., Corradini S., Nestle-Kraemling C., Bölke E., Njanang F.J.D., Tamaskovics B., et al. Recent advances in radiotherapy of breast cancer. Radiat. Oncol. 2020;15:1–10. doi: 10.1186/s13014-020-01501-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan M.A., Lawrence T.S. Molecular pathways: overcoming radiation resistance by targeting DNA damage response pathways. Clin. Cancer Res. 2015;21:2898–2904. doi: 10.1158/1078-0432.CCR-13-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Y., Song Y., Wang R., Wang T. Molecular mechanisms of tumor resistance to radiotherapy. Mol. Cancer. 2023;22:96. doi: 10.1186/s12943-023-01801-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groelly F.J., Fawkes M., Dagg R.A., Blackford A.N., Tarsounas M. Targeting DNA damage response pathways in cancer. Nat. Rev. Cancer. 2023;23:78–94. doi: 10.1038/s41568-022-00535-5. [DOI] [PubMed] [Google Scholar]
- 7.Ranjha L., Howard S.M., Cejka P. Main steps in DNA double-strand break repair: an introduction to homologous recombination and related processes. Chromosoma. 2018;127:187–214. doi: 10.1007/s00412-017-0658-1. [DOI] [PubMed] [Google Scholar]
- 8.Nickoloff J.A., Boss M., Allen C.P., LaRue S.M. Translational research in radiation-induced DNA damage signaling and repair. Transl. Cancer Res. 2017;6:S875. doi: 10.21037/tcr.2017.06.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Syed A., Tainer J.A. The MRE11–RAD50–NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu. Rev. Biochem. 2018;87:263–294. doi: 10.1146/annurev-biochem-062917-012415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villa M., Cassani C., Gobbini E., Bonetti D., Longhese M.P. Coupling end resection with the checkpoint response at DNA double-strand breaks. Cell. Mol. Life Sci. 2016;73:3655–3663. doi: 10.1007/s00018-016-2262-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu L., Luo K., Lou Z., Chen J. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc. Natl. Acad. Sci. USA. 2008;105:11200–11205. doi: 10.1073/pnas.0802885105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rt Abraham. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y., Gudikote J., Giri U., Yan J., Deng W., Ye R., et al. RAD50 expression is associated with poor clinical outcomes after radiotherapy for resected non–small cell lung CancerPrognostic role of RAD50 in resected NSCLC. Clin. Cancer Res. 2018;24:341–350. doi: 10.1158/1078-0432.CCR-17-1455. [DOI] [PubMed] [Google Scholar]
- 14.Karamat U., Ejaz S. Overexpression of RAD50 is the marker of poor prognosis and drug resistance in breast cancer patients. Curr. Cancer Drug Targets. 2021;21:163–176. doi: 10.2174/1568009620666201009125507. [DOI] [PubMed] [Google Scholar]
- 15.Abad E., Civit L., Potesil D., Zdrahal Z., Lyakhovich A. Enhanced DNA damage response through RAD50 in triple negative breast cancer resistant and cancer stem‐like cells contributes to chemoresistance. FEBS J. 2021;288:2184–2202. doi: 10.1111/febs.15588. [DOI] [PubMed] [Google Scholar]
- 16.Zhang M., Liu G., Xue F., Edwards R., Sood A.K., Zhang W., et al. Copy number deletion of RAD50 as predictive marker of BRCAness and PARP inhibitor response in BRCA wild type ovarian cancer. Gynecol. Oncol. 2016;141:57–64. doi: 10.1016/j.ygyno.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flores-Pérez A., Rafaelli L.E., Ramírez-Torres N., Aréchaga-Ocampo E., Frías S., Sánchez S., et al. RAD50 targeting impairs DNA damage response and sensitizes human breast cancer cells to cisplatin therapy. Cancer Biol. Ther. 2014;15:777–788. doi: 10.4161/cbt.28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang L., Huang J., Wang K., Li J., Yan R., Zhu L., et al. Targeting Rad50 sensitizes human nasopharyngeal carcinoma cells to radiotherapy. BMC Cancer. 2016;16:1–12. doi: 10.1186/s12885-016-2190-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Androsavich J.R. Frameworks for transformational breakthroughs in RNA-based medicines. Nat. Rev. Drug Discov. 2024:1–24. doi: 10.1038/s41573-024-00943-2. [DOI] [PubMed] [Google Scholar]
- 20.Ratnayake G., Bain A.L., Fletcher N., Howard C.B., Khanna K.K., Thurecht K.J. RNA interference to enhance radiation therapy: targeting the DNA damage response. Cancer Lett. 2018;439:14–23. doi: 10.1016/j.canlet.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 21.Barata P., Sood A.K., Hong D.S. RNA-targeted therapeutics in cancer clinical trials: current status and future directions. Cancer Treat Rev. 2016;50:35–47. doi: 10.1016/j.ctrv.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 22.Akinc A., Maier M.A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019;14:1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 23.Liu L., Zhou Y., Xu A., Wang C., Li N., Huang H., et al. Non‐viral nucleic acid delivery system for RNA therapeutics. Advanced Therapeutics. 2023;6 [Google Scholar]
- 24.Sahu K.K., Pradhan M., Singh D., Singh M.R., Yadav K. Non-viral nucleic acid delivery approach: a boon for state-of-the-art gene delivery. J. Drug Deliv. Sci. Technol. 2023;80 [Google Scholar]
- 25.Xu W., Wang J., Sheng H., Qu Y., Wang H., Zhu Y., et al. Prognostic implication and functional annotations of Rad50 expression in patients with prostate cancer. J. Cell. Biochem. 2020;121:3124–3134. doi: 10.1002/jcb.29580. [DOI] [PubMed] [Google Scholar]
- 26.Kim H.J., Yi Y., Kim A., Miyata K. Small delivery vehicles of siRNA for enhanced cancer targeting. Biomacromolecules. 2018;19:2377–2390. doi: 10.1021/acs.biomac.8b00546. [DOI] [PubMed] [Google Scholar]
- 27.Swaminathan G., Shigna A., Kumar A., Byroju V.V., Durgempudi V.R., Kumar L Dinesh. RNA interference and nanotechnology: a promising alliance for next generation cancer therapeutics. Frontiers in Nanotechnology. 2021;3 [Google Scholar]
- 28.Yan Y., Liu X., Lu A., Wang X., Jiang L., Wang J. Non-viral vectors for RNA delivery. J.Controlled Release. 2022;342:241–279. doi: 10.1016/j.jconrel.2022.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu B., Zhong L., Weng Y., Peng L., Huang Y., Zhao Y., et al. Therapeutic siRNA: state of the art. Signal Transduct. Targeted Ther. 2020;5:101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian Z., Liang G., Cui K., Liang Y., Wang Q., Lv S., et al. Insight into the prospects for RNAi therapy of cancer. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.644718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmed T., Liu F.F., Wu X.Y. An update on strategies for optimizing polymer-lipid hybrid nanoparticle-mediated drug delivery: exploiting transformability and bioactivity of PLN and harnessing intracellular lipid transport mechanism. Expet Opin. Drug Deliv. 2024;21:245–278. doi: 10.1080/17425247.2024.2318459. [DOI] [PubMed] [Google Scholar]
- 32.Cullis P.R., Felgner P.L. The 60-year evolution of lipid nanoparticles for nucleic acid delivery. Nat. Rev. Drug Discov. 2024:1–14. doi: 10.1038/s41573-024-00977-6. [DOI] [PubMed] [Google Scholar]
- 33.Brown K.M., Nair J.K., Janas M.M., Anglero-Rodriguez Y.I., Dang L.T., Peng H., et al. Expanding RNAi therapeutics to extrahepatic tissues with lipophilic conjugates. Nat. Biotechnol. 2022;40:1500–1508. doi: 10.1038/s41587-022-01334-x. [DOI] [PubMed] [Google Scholar]
- 34.Kulkarni J.A., Witzigmann D., Chen S., Cullis P.R., van der Meel R. Lipid nanoparticle technology for clinical translation of siRNA therapeutics. Acc. Chem. Res. 2019;52:2435–2444. doi: 10.1021/acs.accounts.9b00368. [DOI] [PubMed] [Google Scholar]
- 35.Tenchov R., Sasso J.M., Zhou Q.A. PEGylated lipid nanoparticle formulations: immunological safety and efficiency perspective. Bioconjug.Chem. 2023;34:941–960. doi: 10.1021/acs.bioconjchem.3c00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guerrini G., Gioria S., Sauer A.V., Lucchesi S., Montagnani F., Pastore G., et al. Monitoring anti-PEG antibodies level upon repeated lipid nanoparticle-based COVID-19 vaccine administration. Int. J. Mol. Sci. 2022;23:8838. doi: 10.3390/ijms23168838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amini M.A., Ahmed T., Liu F.F., Abbasi A.Z., Soeandy C.D., Zhang R.X., et al. Exploring the transformability of polymer-lipid hybrid nanoparticles and nanomaterial-biology interplay to facilitate tumor penetration, cellular uptake and intracellular targeting of anticancer drugs. Expet Opin. Drug Deliv. 2021;18:991–1004. doi: 10.1080/17425247.2021.1902984. [DOI] [PubMed] [Google Scholar]
- 38.Ahmed T., Liu F.F., He C., Abbasi A.Z., Cai P., Rauth A.M., et al. Optimizing the design of blood–brain barrier-penetrating polymer-lipid-hybrid nanoparticles for delivering anticancer drugs to glioblastoma. Pharm. Res. (N. Y.) 2021;38:1897–1914. doi: 10.1007/s11095-021-03122-9. [DOI] [PubMed] [Google Scholar]
- 39.Li J., Cai P., Shalviri A., Henderson J.T., He C., Foltz W.D., et al. A multifunctional polymeric nanotheranostic system delivers doxorubicin and imaging agents across the blood–brain barrier targeting brain metastases of breast cancer. ACS Nano. 2014;8:9925–9940. doi: 10.1021/nn501069c. [DOI] [PubMed] [Google Scholar]
- 40.Zhang T., Lip H., He C., Cai P., Wang Z., Henderson J.T., et al. Multitargeted nanoparticles deliver synergistic drugs across the blood–brain barrier to brain metastases of triple negative breast cancer cells and tumor‐associated macrophages. Adv. Healthcare Mater. 2019;8 doi: 10.1002/adhm.201900543. [DOI] [PubMed] [Google Scholar]
- 41.Yen T.C., Abbasi A.Z., He C., Lip H., Park E., Amini M.A., et al. Biocompatible and bioactivable terpolymer-lipid-MnO2 Nanoparticle-based MRI contrast agent for improving tumor detection and delineation. Materials Today Bio. 2024;25 doi: 10.1016/j.mtbio.2024.100954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordijo C.R., Abbasi A.Z., Amini M.A., Lip H.Y., Maeda A., Cai P., et al. Design of hybrid MnO2‐polymer‐lipid nanoparticles with tunable oxygen generation rates and tumor accumulation for cancer treatment. Adv. Funct. Mater. 2015;25:1858–1872. [Google Scholar]
- 43.He C., Li J., Cai P., Ahmed T., Henderson J.T., Foltz W.D., et al. Two‐step targeted hybrid nanoconstructs increase brain penetration and efficacy of the therapeutic antibody trastuzumab against brain metastasis of HER2‐positive breast cancer. Adv. Funct. Mater. 2018;28 [Google Scholar]
- 44.Park E., Li L.Y., He C., Abbasi A.Z., Ahmed T., Foltz W.D., et al. Brain‐Penetrating and disease site‐targeting manganese dioxide‐polymer‐lipid hybrid nanoparticles remodel microenvironment of Alzheimer's disease by regulating multiple pathological pathways. Adv. Sci. 2023 doi: 10.1002/advs.202207238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zetrini A.E., Lip H., Abbasi A.Z., Alradwan I., Ahmed T., He C., et al. Remodeling tumor immune microenvironment by using polymer-lipid-manganese dioxide nanoparticles with radiation therapy to boost immune response of castration-resistant prostate cancer. Research. 2023;6:247. doi: 10.34133/research.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shalviri A., Chan H.K., Raval G., Abdekhodaie M.J., Liu Q., Heerklotz H., et al. Design of pH-responsive nanoparticles of terpolymer of poly (methacrylic acid), polysorbate 80 and starch for delivery of doxorubicin. Colloids Surf. B Biointerfaces. 2013;101:405–413. doi: 10.1016/j.colsurfb.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 47.Lip H., Amini M.A., Zetrini A., Cai P., Abbasi A.Z., Bristow R.G., et al. Redox-responsive nanoparticles enhance radiation therapy by altering multifaceted radio-resistance mechanisms in human castration-resistant prostate cancer cells and xenografts. Radiother. Oncol.170. 2022:213–223. doi: 10.1016/j.radonc.2022.02.026. [DOI] [PubMed] [Google Scholar]
- 48.Wang Z., Zhang R.X., Zhang T., He C., He R., Ju X., et al. In situ proapoptotic peptide-generating rapeseed protein-based nanocomplexes synergize chemotherapy for cathepsin-B overexpressing breast cancer. ACS Appl. Mater. Interfaces. 2018;10:41056–41069. doi: 10.1021/acsami.8b14001. [DOI] [PubMed] [Google Scholar]
- 49.Deng R., Tang J., Ma J.G., Chen S.P., Xia L.P., Zhou W.J., et al. PKB/Akt promotes DSB repair in cancer cells through upregulating Mre11 expression following ionizing radiation. Oncogene. 2011;30:944–955. doi: 10.1038/onc.2010.467. [DOI] [PubMed] [Google Scholar]
- 50.Raimundo L., Calheiros J., Saraiva L. Exploiting DNA damage repair in precision cancer therapy: BRCA1 as a prime therapeutic target. Cancers. 2021;13:3438. doi: 10.3390/cancers13143438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang R., Zhou P. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Targeted Ther. 2020;5:60. doi: 10.1038/s41392-020-0150-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams D.L., Adams D.K., He J., Kalhor N., Zhang M., Xu T., et al. Sequential tracking of PD-L1 expression and RAD50 induction in circulating tumor and stromal cells of lung cancer patients undergoing RadiotherapyTracking PD-L1 and RAD50 in tumor-derived blood cells in NSCLC. Clin. Cancer Res. 2017;23:5948–5958. doi: 10.1158/1078-0432.CCR-17-0802. [DOI] [PubMed] [Google Scholar]
- 53.Sajid M.I., Moazzam M., Kato S., Cho K Yeseom, Tiwari R.K. Overcoming barriers for siRNA therapeutics: from bench to bedside. Pharmaceuticals. 2020;13:294. doi: 10.3390/ph13100294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi M., McHugh K.J. Strategies for overcoming protein and peptide instability in biodegradable drug delivery systems. Adv. Drug Deliv. Rev. 2023 doi: 10.1016/j.addr.2023.114904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pittella F., Zhang M., Lee Y., Kim H.J., Tockary T., Osada K., et al. Enhanced endosomal escape of siRNA-incorporating hybrid nanoparticles from calcium phosphate and PEG-block charge-conversional polymer for efficient gene knockdown with negligible cytotoxicity. Biomaterials. 2011;32:3106–3114. doi: 10.1016/j.biomaterials.2010.12.057. [DOI] [PubMed] [Google Scholar]
- 56.Honary S., Zahir F. Effect of zeta potential on the properties of nano-drug delivery systems-a review (Part 2) Trop. J. Pharmaceut. Res. 2013;12:265–273. [Google Scholar]
- 57.Mendes B.B., Conniot J., Avital A., Yao D., Jiang X., Zhou X., et al. Nanodelivery of nucleic acids. Nature Reviews Methods Primers. 2022;2:24. doi: 10.1038/s43586-022-00104-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malcolm D.W., Wang Y., Overby C., Newman M., Benoit D.S. Delivery of RNAi-based therapeutics for bone regeneration. Curr. Osteoporos. Rep. 2020;18:312–324. doi: 10.1007/s11914-020-00587-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moghimi S.M. Allergic reactions and anaphylaxis to LNP-based COVID-19 vaccines. Mol. Ther. 2021;29:898–900. doi: 10.1016/j.ymthe.2021.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Naeye B., Deschout H., Caveliers V., Descamps B., Braeckmans K., Vanhove C., et al. In vivo disassembly of IV administered siRNA matrix nanoparticles at the renal filtration barrier. Biomaterials. 2013;34:2350–2358. doi: 10.1016/j.biomaterials.2012.11.058. [DOI] [PubMed] [Google Scholar]
- 61.Bazban-Shotorbani S., Hasani-Sadrabadi M.M., Karkhaneh A., Serpooshan V., Jacob K.I., Moshaverinia A., et al. Revisiting structure-property relationship of pH-responsive polymers for drug delivery applications. J.Controlled Release. 2017;253:46–63. doi: 10.1016/j.jconrel.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 62.Bian L., Meng Y., Zhang M., Li D. MRE11-RAD50-NBS1 complex alterations and DNA damage response: implications for cancer treatment. Mol. Cancer. 2019;18:1–14. doi: 10.1186/s12943-019-1100-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.