Abstract
Komagataella phaffii (Pichia pastoris) is a methylotrophic yeast that is favored by industry and academia mainly for expression of heterologous proteins. However, its full potential as a host for bioproduction of valuable compounds cannot be fully exploited as genetic tools are lagging behind those that are available for baker’s yeast. The emergence of CRISPR-Cas9 technology has significantly improved the efficiency of gene manipulations of K. phaffii, but improvements in gene-editing methods are desirable to further accelerate engineering of this yeast. In this study, we have developed a versatile vector-based CRISPR-Cas9 method and showed that it works efficiently at different genetic loci using linear DNA fragments with very short targeting sequences including single-stranded oligonucleotides. Notably, we performed site-specific point mutations and full gene deletions using short (90 nt) single-stranded oligonucleotides at very high efficiencies. Lastly, we present a strategy for transient inactivation of nonhomologous end-joining (NHEJ) pathway, where KU70 gene is disrupted by a visual marker (uidA gene). This system enables precise CRISPR-Cas9-based editing (including multiplexing) and facilitates simple reversion to NHEJ-proficient genotype. In conclusion, the tools presented in this study can be applied for easy and efficient engineering of K. phaffii strains and are compatible with high-throughput automated workflows.
Keywords: Cas9, oligonucleotide, mutagenesis, multiplexing, KU70, transient-NHEJ, uidA marker
A streamlined CRISPR-Cas9-based toolbox that improves the engineering process of Komagataella phaffii yeast by implementing short single-stranded DNA sequences for precise introduction of desired genetic modifications.
Introduction
Komagataella phaffii (aka. Pichia pastoris) is a methylotrophic yeast that has gained its popularity as a preferred host for heterologous protein expression and is being widely used for academic and industrial purposes (Yang and Zhang 2018, Werten et al. 2019). Most importantly is the use of a one carbon substrate (e.g. methanol) for bioconversion or production of high value products. However, in recent years, K. phaffii has also been increasingly applied as a cell factory for production of small value-added molecules using various carbon sources as substrates; reviewed in Peña et al. (2018) and Yang and Zhang (2018). For example, Gassler et al. (2020) has recently demonstrated that K. phaffii can be engineered to assimilate CO2. In addition, industrially relevant physiological characteristics have boosted K. phaffii application in fundamental research (Karbalaei et al. 2020, Bernauer et al. 2021). Thus, with rapidly growing interest in this organism, the development of efficient methods for gene editing that are compatible with high throughput experiments are necessary.
Gene editing in the K. phaffii is hampered by the dominance of the nonhomologous end-joining (NHEJ) DNA repair pathway, which favors random integration over specific genetic engineering that can be obtained by homologous recombination (HR)-mediated repair (Näätsaari et al. 2012). The emergence of CRISPR-Cas9 technology has revolutionized gene engineering of many nonmodel yeasts (Stovicek et al. 2017, Cai et al. 2019) and has also been successfully adapted to K. phaffii (Weninger et al. 2016), where the repertoire of CRISPR-Cas9 tools steadily increases (Gu et al. 2019, Liu et al. 2019, Dalvie et al. 2020, Cai et al. 2021, Liao et al. 2021, Gao et al. 2022, Wang et al. 2023). DNA double strand breaks (DSBs) induced by a CRISPR nuclease allow easy gene inactivation via erroneous NHEJ-based repair, as well as it increases the efficiency of template guided gene editing mediated by HR. However, recent studies have demonstrated that CRISPR nuclease induced DNA DSBs, despite the intended edits, also induce unwanted off-target mutations and/or result in gross chromosomal rearrangements in NHEJ-proficient strain backgrounds (Schusterbauer et al. 2022, Garrigues et al. 2023). Thus, to improve gene-targeting efficiency and to minimize occurrence of unwanted mutations it is desirable to inactivate the NHEJ pathway. However, the presence of the ku70 mutation may influence the final phenotype of the engineered strain. For example, deletion of YKU70 in S. cerevisiae confers a temperature-sensitive phenotype, affects telomere length, and expression of telomere-proximal genes (Fellerhoff et al. 2000). Moreover, elimination of NHEJ negatively affects transformation and growth rates and increases sensitivity to radiation and other DNA-damaging conditions (Carvalho et al. 2010, Näätsaari et al. 2012). Recently, Shao et al. (2022) have demonstrated significant improvement in production of recombinant protein when ku70 mutation was complemented. Thus, after CRISPR-based cell factory construction in an NHEJ-deficient background, it is recommended to reinstate the NHEJ pathway before conducting a comprehensive performance evaluation; and methods that allow simple ways to restore NHEJ are therefore desirable.
CRISPR sets the stage for simple multiplexing and high-throughput genetic-engineering. In K. phaffii, this potential is poorly exploited and methods that simplify delivery of multiple sgRNAs and gene targeting substrate (GTS) assembly are in demand. In this work, we have developed a highly efficient CRISPR-Cas9 method for engineering of prototrophic K. phaffii strains. Our method is based on one plasmid system that facilitates simultaneous coexpression of Cas9 and single or multiple arrays of sgRNA cassettes. We have evaluated our method by modifying multiple genetic loci with linear GTSs equipped with homology arms of varying size. Importantly, we have demonstrated that short (90 nt) single-stranded DNA oligonucleotides are sufficient to mediate repair of Cas9 induced DSBs, thus significantly reducing time and cost needed for construction of complex GTSs. Moreover, we present a method for transient disruption of the NHEJ pathway by inserting the visually selectable uidA (Jefferson et al. 1987) marker into KU70. The system enables precise Cas9-mediated gene modifications and simultaneous, easy to detect, recovery of the NHEJ pathway to the wild type state. Moreover, the color marker associated with the NHEJ pathway facilitates effortless selection of yeast colonies that are proficient for HR, and which have therefore most likely undergone the intended genetic manipulations.
Materials and methods
Strains and cultivation media
Escherichia coli DH5α strain was used for plasmid assembly and amplification. In all cases, E. coli strains were cultivated in lysogeny broth (LB) medium (Bertani 1951) supplemented with 100 mg/l of ampicillin (Sigma). Komagataella phaffii (CBS 2612) was used as the genetic background for all yeast experiments. Komagataella phaffii was cultivated in yeast extract peptone dextrose (YPD) medium containing 1% yeast extract, 2% peptone, and 2% glucose (and 2% of agar for solid media). For the selection of transformed yeast strains, YPD medium was supplemented with 100 mg/l of nourseothricin (NTC; Werner BioAgents). For blue colony screen (uidA marker), solid medium was additionally supplemented with 60 mg/l X-Gluc (Thermo Fisher Scientific).
PCR and sequencing
All DNA fragments for plasmid or GTS construction were amplified by polymerase chain reaction (PCR) using Phusion U Master Mix (ThermoFisher) according to the manufacturer’s recommendations. All primers used in this study were obtained from Integrated DNA Technologies and are listed in Supplementary Table S1. All PCR fragments were gel-purified using NucleoSpin Gel and PCR Clean–up Kit (MACHEREY-NAGEL) by following manufacturer’s protocol. Plasmid minipreps were prepared using GeneElute Plasmid MiniPrep Kit (Sigma-Aldrich) according to the manufacturer’s protocol. New DNA constructs were validated by Sanger sequencing using the Mix2Seq service (Eurofins). All plasmids used and constructed in this work are listed in Supplementary Table S2. Synthetic DNA sequences were ordered as gBlocks from IDT (https://eu.idtdna.com).
Diagnostic PCR reactions were used to confirm mutations in yeast transformants. Routinely, eight random colonies per each biological replicate were selected for PCR analysis. All colony PCRs were done using Quick-Load® Taq 2X Master Mix (New England Biolabs) according to the manufacturer’s recommendation. Prior to PCR, samples were thermally pretreated to lyze the cells (98°C for 5 min, 4°C for 30 s, 98°C for 5 min, and 80°C for 10 min). The size of PCR fragments were determined by 1% agarose gel electrophoresis. In selected cases, PCR fragments were column-purified and Sanger sequenced using the service provided by Mix2Seq (Eurofins).
DNA assembly
Construction of sgRNA and Cas9 expressing plasmids
Plasmids used in this work were constructed by USER cloning (Nour-Eldin et al. 2006) using the uracil-specific excision reagent (USER™). The basic Cas9 expression plasmid (pDIV151) was assembled by inserting PCR fragments containing the GAP1 promoter amplified from genomic DNA (gDNA) of K. phaffii (CBS 2612) and the cas9-ScCYC1t cassette amplified from plasmid pCfB2312 (Stovicek et al. 2015) into a vector fragment obtained by linearizing pDIV019 (Strucko et al. 2021) with AsiSI/Nb.BsmI. For the relevant PCR primers, see Supplementary Table S2 and Supplementary Fig. S1. The basic sgRNA-Cas9 plasmid (pDIV153) was constructed by inserting three PCR fragments: the TEF1 promoter from K. phaffii, an sgRNA–tRNA construct from pFC902 plasmid (Nødvig et al. 2018), and the AOX terminator from K. phaffii, into AsiSI/Nb.BsmI digested pDIV151 vector. Derivatives of pDIV153 expressing different single sgRNAs were assembled as follows: for each unique sgRNA cassette two PCRs fragments amplified from the pDIV153 plasmid using specific primer sets (see Supplementary Fig. S1 for details). Next, the resulting PCR fragment pairs were individually USER-fused into the linearized pDIV151 backbone, resulting in the following plasmids: pDIV259 (sgRNA1-IS1), pDIV260 (sgRNA1-IS3), pDIV270 (sgRNA1-KU70), pDIV272 (sgRNA2-uidA), pDIV273 (sgRNA1-ADE2), and pDIV274 (sgRNA2-ADE2). A Cas9 vector expressing two sgRNAs (PEP4 and uidA) was assembled as follows: three different PCR fragments were amplified from pDIV153 by specific primer pairs (see Supplementary Fig. S1 for details) and USER-fused into pDIV151 backbone, resulting in a pDIV602 plasmid. A detailed plasmid construction procedure for all Cas9 and sgRNA expressing vectors is depicted in Supplementary Fig. S1. The list of all plasmids used in this work is in Supplementary Table S2. The relevant plasmids and their maps will be available at https://www.addgene.org/Uffe_Mortensen/upon publication of this manuscript.
Construction of plasmids harboring GTSs
The assembly of mVenus expressing cassettes flanked by long homology (LH) sequences guiding to specific loci [Integration site IS1 (GenBank: FR839628.1:1356524..1358623) or IS3 (GenBank: FR839629.1:839629..394663)] in the genome of K. phaffii was done in two steps. First, an mVenus expressing plasmid (pDIV479) was constructed by cloning TEF1 promoter amplified from K. phaffii gDNA, and an mVenus-ScCYC1t cassette amplified from a synthetic gBlock (IDT), into a PacI/Nt.BbvCI linearized vector pAC125 (Hansen et al. 2011). Second, locus specific upstream and downstream homology arms were amplified from K. phaffii gDNA, and individually USER-cloned into a PacI/Nt.BbvCI linearized pDIV479 vector. The two resulting plasmids were denoted as pDIV505 (for integration site IS1), and pDIV506 (for integration site IS3). Similarly, the assembly of the uidA expressing cassette for disruption of KU70 homolog was done in two steps. First, an uidA expressing plasmid (pDIV033) was constructed by USER-cloning TEF1 promoter amplified from S. cerevisiae gDNA, and an uidA-ScCYC1t cassette amplified from a synthetic gBlock (IDT), into AsiSI/Nb.BsmI linearized pDIV019 vector. Second, the KU70 disrupting vector (pDIV643) was constructed by cloning KU70 locus-specific upstream and downstream homology sequences amplified from K. phaffii gDNA, and ScTEF1p-uidA-CYC1t cassette, amplified from pDIV033, into PacI/Nt.BbvCI linearized pAC125 vector. A detailed cloning procedure for all GTS expressing vectors is depicted in Supplementary Fig. S2. Prior to transformation into K. phaffii, plasmids pDIV505, pDIV506, and pDIV643 were linearized with NotI enzyme in order to liberate linear DNA fragments (GTSs).
Construction of PCR-based GTSs
The mVenus expressing cassettes flanked by short homology (SH) sequences guiding to specific loci in the genome of K. phaffii were obtained by PCR using template pDIV479 as template and primers pairs (PR_DIV1509 + PR_DIV1510) and (PR_DIV1511 + PR_DIV1512). These primer sets contain 60 bp tails homologous to upstream and downstream regions of IS1 and IS3 loci, respectively. A GTS used to restore the wild-type KU70 genotype was PCR-amplified from K. phaffii gDNA with a specific primer pair (PR_DIV2660 + PR_DIV2661). A detailed procedure for construction of PCR-based GTSs is depicted in Supplementary Fig. S3.
Design of 20 nt variable parts of sgRNAs
The variable 20-nt targeting sections of each sgRNA (see Table 1) were designed using CRISPRdirect (https://crispr.dbcls.jp/); an online tool developed by Naito et al. (2015).
Table 1.
sgRNAs used in this study. Note, sgRNA names indicate intended gene targets.
| sgRNA name | 20-nt variable region of sgRNAs | Plasmid |
|---|---|---|
| sgRNA1-URA3 | ACGACGACTGTTTGCGCTTA | pDIV153 |
| sgRNA1-ADE2 | GCTGCGCAAGACCACATCGA | pDIV273 |
| sgRNA2-ADE2 | CAATGGAGACCGAAGTGTTG | pDIV274 |
| sgRNA1-KU70 | AAGCAATACGACATCCACGA | pDIV270 |
| sgRNA1-IS1 | GAAACAGTCCAACAGTAAGC | pDIV259, pDIV519a, pDIV520b, pDIV521c |
| sgRNA1-IS3 | TGTTGATAATGGACCGTGGG | pDIV260, pDIV519a, pDIV520b, pDIV521c |
| sgRNA1-IS5 | CTGGCCAATCTCATCTCATG | pDIV521c |
| sgRNA1-IS7 | GAAACTGGGGAGATGCAGAG | pDIV520b and pDIV521c |
| sgRNA2-uidA | TACTTCAGAAGCGTTGGACC | pDIV272 and pDIV602a |
| sgRNA1-PEP4 | TGGTTCAGGAAAGTCCATGG | pDIV581 and pDIV602a |
aPlasmid encodes two sgRNA species, bthree sgRNA species, and cfour sgRNA species.
Transformation of K. phaffii
All K. phaffii strains were transformed using an electroporation method slightly modified from Lin-Cereghino et al. (2005). In brief, a single colony was inoculated into 5 ml of YPD media and incubated O/N at 30°C with 200 rpm shaking. Next day, appropriate volume of O/N culture was transferred into a fresh 50 ml of YPD in 500 ml baffled shake flask to a starting OD of ∼0.1. Fresh cultures were incubated at 30°C with orbital shaking at 200 rpm until OD of ∼1.5 was achieved (∼5 h). Cells were collected by centrifugation at 3000 × g for 5 min, the supernatant was discarded and the pellet was resuspended in 20 ml of Transformation Buffer (100 mM LiAc, 10 mM DTT, 0.6 M sorbitol, and 6 mM Tris HCl pH 8.5) and incubated at room temperature for 30 min. Cells were centrifuged at 3000 × g for 5 min, and the supernatant was discarded. Pellets were resuspended in 1.5 ml 1 M ice-cold sorbitol and transferred to sterile 2 ml eppendorf tubes. The pellets were washed three times using 1 M ice-cold sorbitol followed by centrifugation at 10 000 × g for 30 s; and then resuspended in 2 ml of 1 M ice-cold sorbitol. The cell suspension was distributed into 10 sterile 1.5 ml eppendorf tubes (200 µl per vial) and used for transformation, or frozen at −80°C for later use. Routinely, ∼0.2 pmol of plasmid was added to the mix, and 1 pmol of linear DNA was added when required. The cell-DNA mix was incubated on ice for 5 min and transferred to an ice-cold 2 mm electroporation cuvette. Cells were electroporated by Bio-Rad MicroPulser Electroporator using predefined “Pic” settings for P. pastoris (1 pulse at 2 kV) and immediately transferred into 2 ml eppendorf tubes containing 0.8 ml of ice-cold YPD (containing 1 M Sorbitol). The cell suspension was incubated for 3 h at 30°C without shaking and plated on selective media. Transformation plates were incubated for 3–5 days at 30°C. Each transformation experiment was done in four replicates.
K. phaffii mutant strain construction
Two NHEJ-deficient K. phaffii strains were constructed as follows: sDIV0360, containing a single nucleotide deletion (A32Δ) in KU70, was made by transforming wild-type strain sDIV0084 (CBS 2612) with the pDIV270 plasmid. Transformants were selected and streak purified on solid YPD + NTC medium. The mutation was validated by Sanger sequencing (Supplementary Fig. S4).
sDIV291 containing ku70::uidA was obtained cotransforming wild-type strain sDIV0084 with the pDIV270 plasmid and linear GTS containing uidA and incubated on solid YPD + NTC + X-Gluc medium. The GTS was liberated from the plasmid pDIV643 by NotI and gel purified prior to use. Transformants containing the ku70::uidA allele were identified by colony PCR (see text below). Strains sDIV291 and sDIV360 were cured for sgRNA–Cas9 plasmids prior to storage and for use in subsequent experiments.
Image acquisitions
Komagataella phaffii colonies on solid medium were photographed using an SLR camera (Nikon D90). Settings for the white light photography were as follows: ISO Speed—ISO250, F-stop—f/7.1, exposure time—1/80 s, and focal length—60 mm. Yellow fluorescence (mVenus) was examined by using an in-house built setup employing a light and filter set (NIGHTSEA™) in combination with a digital camera, for details, see Vanegas et al. (2019). For acquisition of fluorescent images in all cases the exposure time was fixed to 1.6 s and the remaining settings were as follows: ISO Speed—ISO200, F-stop—f/7.1, and focal length—60 mm.
Results and discussion
CRISPR-Cas9 tool design and validation
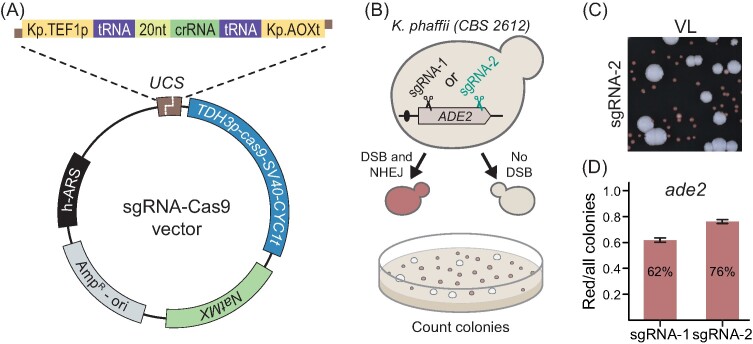
The strength of the CRISPR-Cas technology is that its sgRNA-guided CRISPR endonuclease can be effortlessly reprogrammed to cleave at almost any sequence in the genome to set the stage for targeted gene editing. However, the success and efficiency of CRISPR tools relies highly on well-balanced expression of specific sgRNA and Cas9, and often requires multiple steps of optimization as reported in the first example for K. phaffii (Weninger et al. 2016). In addition, vector features (i.e. ARS, backbone, and selection marker) influences cas9 expression and play key roles in editing (Gu et al. 2019). Based on the accumulated knowledge, we have designed a plasmid-based CRISPR-Cas9 gene-editing system for K. phaffii (Fig. 1A and the section “Materials and methods”). Specifically, we made a CRISPR vector, based on pDIV019 backbone, which is able to replicate in a selectable manner in K. phaffii as it contains a synthetic autonomously replicating sequence (h-ARS) and a dominant NatMX marker, which are both compatible with different yeast species (Strucko et al. 2021). Moreover, the vector is equipped with a cas9 gene-expression cassette composed by the K. phaffii TDH3p (GAP1p) promoter, cas9-NLS (SV40) open reading frame adopted from DiCarlo et al. (2013), and a CYC1t terminator from S. cerevisiae, and a USER cloning cassette (UCS) (Nour-Eldin et al. 2006) allowing for effortless integration of expression constructs (Fig. 1A) encoding one or more sgRNAs, see below.
Figure 1.
The design of the CRISPR-Cas9 gene-editing system for K. phaffii. (A) A schematic representation of the sgRNA–cas9 expression vector. UCS—USER cloning site. (B) Screening setup to assess the functionality of our sgRNA–cas9 tool. Wild-type transformants form large white colonies while ade2 transformants form small red/pink colonies. (C) A snippet of a typical transformation plate in an NHEJ-proficient K. phaffii strain transformed with the pDIV274 plasmid (encoding sgRNA-2). VL—visible light. (D) Ratios of red/total colonies obtained from two different experiments employing plasmids coding for two different sgRNA species, sgRNA-1 and sgRNA-2, which target the ADE2 gene in different positions as indicated in panel B. Error bars represent standard deviation based on four biological replicates.
Currently, several methods for expression of sgRNA in K. phaffii are available, but the most common ones rely on RNA Pol-II-based expression cassettes equipped with self-cleaving ribozyme sequences for liberation of the functional sgRNA (Weninger et al. 2016, Liao et al. 2021) or Pol-III cassettes flanked with tRNA sequences (Dalvie et al. 2020). However, assemblies of the reported sgRNA expression cassettes are rather complicated; especially, when more than two sgRNAs need to be expressed. To allow for simple sgRNA expression cassette construction, we adopted a system we originally developed for Debaryomyces hansenii, (Strucko et al. 2021). Specifically, the cassette consists of the Pol-II TEF1p promoter from K. phaffii, an sgRNA coding sequence flanked by heterologous tRNA–Gly sequences from Aspergillus nidulans, and a AOXt terminator from K. phaffii (see Fig. 1A and the section “Materials and methods”). By using USER fusion (Nour-Eldin et al. 2006), simple and complex gRNA expression cassettes can easily be introduced into the CRISPR vector as described in Supplementary Fig. S5, Nødvig et al. (2018) and Strucko et al. (2021). In the first test of the experiment, we successfully inserted two, three, or four copies of an mVenus expression cassette into two, three, and four different loci in the genome in a multiplex CRISPR reaction (Supplementary Fig. S5).
To assess the functionality of our design, we constructed two sgRNA–Cas9 expressing CRISPR plasmids sgRNA1-ADE2 (pDIV273) and sgRNA2-ADE2 (pDIV274) targeting two different sequences in the ADE2 gene of K. phaffii (Fig. 1B and the section “Materials and methods”). In this experiment, we exploited that mutation of ADE2 can be easily assessed by visual screening as ade2 cells accumulate red pigment (Fig. 1C), (Roman 1956). Moreover, we chose this gene as a target to demonstrate robustness of our method. Specifically, ADE2 disruption in K. phaffii is accompanied by a significant fitness loss (Du et al. 2012) transformants that escape CRISPR mutagenesis will prevail in case CRISPR mutagenesis is inefficient. The two plasmids were independently transformed into K. phaffii to test whether they were able to induce sgRNA–Cas9-mediated mutations via erroneous NHEJ DNA repair. As anticipated, red and white transformants were formed in both cases (Supplementary Fig. S6). The fractions of red to total transformants (Fig. 1D) in the two experiments were similar (62%–76%, respectively), and were comparable with previously reported efficiencies (Liao et al. 2021). Sanger sequencing of the ade2 locus in three random red colonies from each experiment showed that all six mutants contained disruptive indels at the Cas9 cleavage sites in ADE2, which are expected outcomes of erroneous NHEJ DNA repair (see Supplementary Fig. S7). We conclude that our CRISPR method can be used to efficiently generate gene mutations even in a case where the mutant products grow poorly compared to the wild-type strain.
CRISPR-Cas9-induced gene targeting using linear DNA fragments
Komagataella phaffii, like many other nonconventional yeasts and higher eukaryotes, prefers to insert DNA received by transformation into the genome via the NHEJ pathway. To increase the gene-targeting efficiency, it may therefore be an advantage to eliminate the NHEJ pathway; and for K. phaffii, it has been demonstrated that gene targeting efficiencies are dramatically improved in strains containing a disrupted KU70 homolog (Näätsaari et al. 2012, Wang et al. 2023). In addition, NHEJ inactivation enables efficient use of the TAPE tool to assess whether specific sgRNA species are able to facilitate Cas9 cleavage at the target sites (Vanegas et al. 2017, Nødvig et al. 2018). For these reasons, we constructed an NHEJ-deficient ku70 strain (sDIV360) by inactivating KU70 via single nucleotide deletion (A32Δ) resulting in a frameshift mutation and a stop codon (see the section “Materials and methods” and Supplementary Fig. S4). A subsequent TAPE experiment analysing sgRNA1-ADE2 and sgRNA2-ADE2 plasmids confirmed that the NHEJ pathway was inactivated and both sgRNAs were efficiently guiding Cas9 nuclease to the intended locus (see Supplementary Fig. S8).
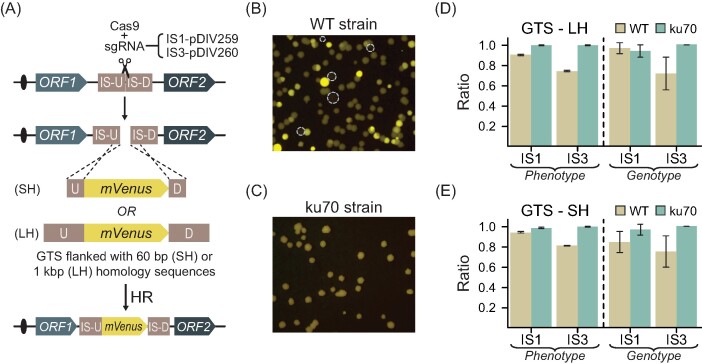
Next, we examined whether our CRISPR-based method allowed for efficient gene targeting in KU70 and ku70 cells. As a proof of concept, we decided to integrate GTSs expressing yellow fluorescent protein (mVenus), into two integration sites (ISs) located on two different chromosomes of K. phaffii. The ISs were positioned in intergenic genetic regions (Fig. 2A) between transcriptionally active genes (Liang et al. 2012) to ensure high expression and at the same time minimize accompanying fitness defects. IS1 and IS3 are positioned on chromosome I and chromosome II, respectively (see the section “Materials and methods”). For each IS, we employed two GTSs containing either long ∼1 kb (LH) or short 60 bp (SH) homology arms to evaluate the effect of the flanking homology sequence length on targeting efficiency. When we compared the transformation plates obtained with the two different strain backgrounds, we noticed that the fluorescence levels of the individual transformants obtained with wild-type strains varied considerably indicating that wild-type transformants contained variable copy numbers of GTSs (see Fig. 2B and Supplementary Fig. S9). Copy numbers higher than one is likely due to the result of NHEJ mediated polymerization of GTSs prior to incorporation into the IS and/or NHEJ-mediated integration of GTSs into other random sites in the genome. In agreement with this view, ku70 strains produced transformants that all produced yellow fluorescence at the same level (Fig. 2C and Supplementary Fig. S9). Moreover, the level of fluorescence obtained with ku70 transformants matched the lowest levels obtained with KU70 transformants indicating that the latter contained a single mVenus copy.
Figure 2.
Gene integration strategy using CRISPR-Cas9. (A) Schematic depiction of gene targeting using intergenic chromosomal regions. GTS—gene targeting substrate, SH—short homology, LH—long homology sequence, and IS-U and IS-D—sequences upstream and downstream of the IS. Snippets of typical transformation plate detecting for yellow fluorescence light emitted by (B) an NHEJ-proficient WT strain, and (C) NHEJ-deficient strains emitting yellow fluorescence light. Dashed circles show the location of nonfluorescent colonies. Gene-targeting efficiencies of GTSs with (D) Experimental results using GTSs with 1 kb LH sequences and (E) SH sequences targeting into IS1 and IS3. Phenotype—results show the ratio of fluorescent colonies versus total colonies observed on transformation plates. Genotype—results show the ratio of the number of colonies with a correctly integrated GTS versus the total number of tested colonies for the corresponding GTS. Error bars represent standard deviation based on four biological replicates.
Next, we determined the efficiency of gene targeting in KU70 cells. With the GTSs containing LH arms matching either IS1 or IS3, we found that 90% or 74% of the transformants were fluorescent, respectively. Importantly, these numbers correlate with the numbers of transformants that were correctly targeted by the GTS as judged by diagnostic PCR reactions indicating that all fluorescent colonies contain a GTS in the desired locus. Almost identical results were obtained in the targeting experiment employing the GTS containing SH arms (Fig. 2D and E). Hence, although GTSs integrate efficiently into both ISs, the efficiency was slightly higher for the setup targeting IS1. More importantly, as the same numbers were obtained by using either long or SH arms at both IS1 and IS3, we find that no advantage was gained by extending the length of the homology arm from 60 to 1000 bps.
With ku70 cells, the gene-targeting efficiency was even higher than with KU70 cells. Hence, the number of fluorescent and correctly targeted transformants exceeded 95% in all four experiments.
Similar to wild-type cells, no advantage was achieved by adding the longer homology arms. On the other hand, the suboptimal targeting of GTSs into IS3 observed with KU70 was not observed with ku70 cells (Fig. 2D and E and Supplementary Figs S10 and S11), and for that reason CRISPR mediated gene targeting appears more robust in the absence of NHEJ.
Single-stranded DNA oligonucleotides are efficient GTSs
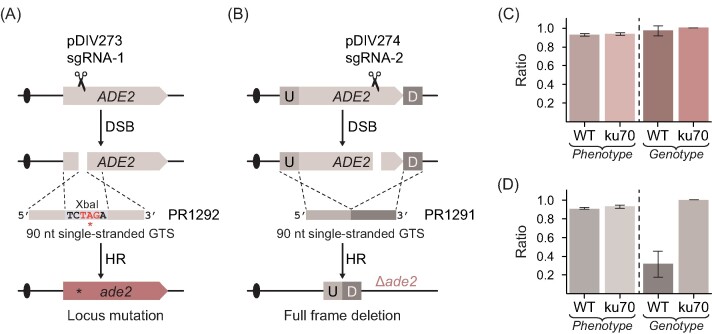
In metabolic engineering experiments, it is often necessary to rewire the metabolism of the host by multiple gene deletions and/or specific point mutations. In K. phaffii, this is typically achieved by using linear double stranded GTSs constructed by PCR (Dalvie et al. 2022), in steps, which are time consuming and costly in large scale experiments. In the yeast S. cerevisiae, short double-stranded oligonucleotides have been successfully applied for gene editing in CRISPR experiments (DiCarlo et al. 2013). Conversely, single-stranded oligonucleotides have been used for gene editing in filamentous fungi (Nødvig et al. 2018, Vanegas et al. 2019) and the nonconventional yeast D. hansenii (Strucko et al. 2021). Thus, we investigated whether short single-stranded DNA can be applied as GTS in K. phaffii. In a first pilot test, we investigated whether a single-stranded oligonucleotide based GTS could be used to delete a gene in a CRISPR experiment. For this purpose we designed a CRISPR vector (pDIV581) expressing an sgRNA targeting the PEP4 gene (coding for vacuolar aspartyl protease). Next, we cotransformed this plasmid along with a single-stranded oligonucleotide based GTS (PR2268) matching equal sizes of sequences up- (45 nt) and downstream (45 nt) of PEP4. With a 90-nt oligonucleotide, pep4 deletion strains were easily obtained with a success rate of ∼75% (Supplementary Fig. S12). With shorter oligonucleotides the efficiency rapidly decreased as GTSs composed by 60 and 40 nt produced deletions with 20% and 0% efficiencies, respectively. Encouraged by these results, we tested whether the method could be used equally well to delete another gene; and perhaps even be used to introduce a subtle mutation. For this purpose, we designed single-stranded 90-mers (ss-GTSs) that are able to either introduce a stop codon (PR1292) or completely delete the ADE2 gene (PR1291) (Fig. 3A and B). We performed experiments in both WT and ku70 strain backgrounds by cotransforming specific CRISPR plasmids encoding Cas9–sgRNA nucleases that are able to cleave ADE2 at strategic places (see Fig. 3A and B) with the relevant ss-GTSs as repair template. In all cases, ADE2 was mutated very efficiently as the ratio of red to total number of transformants varied from 92% to 95% in (Fig. 3C and D). We next genotyped randomly selected red colonies (N = 32) from each experiment to assess whether single-stranded oligonucleotide-guided genetic modifications were introduced as intended (see the section “Materials and methods” and Supplementary Fig. S13). When the mutagenic stop-codon oligonucleotide (PR1292) was used, 97% of analysed red colonies contained correct edits in WT background as compared to 100% in ku70 background (Fig. 3C). When the gene-deletion oligonucleotide (PR1291) was applied, 31% and 100% of the red colonies contained the expected gene deletion in WT and ku70 strains, respectively (Fig. 3D). These results demonstrate that short single-stranded oligonucleotides can be efficiently used as GTSs for precise CRISPR based gene editing in K. phaffii, in particular if the NHEJ pathway is inactivated.
Figure 3.
Single-stranded oligonucleotide-based gene editing in K. phaffii. (A) Schematic depiction of site-specific mutagenesis using a short single-stranded GTS. DSB—double strand break, HR—homologous recombination. The oligonucleotide PR1292 contains an XbaI recognition site designed to introduce an in-frame stop codon. (B) Schematic depiction of specific gene-deletion mutagenesis using a short single-stranded GTS (PR1291) for full deletion. U—upstream and D—downstream. (C) Results of ADE2 locus mutation efficiencies using a short oligonucleotide as GTS. The bars show the ratio of red colonies with the expected GTS directed mutation versus total number of tested red colonies for the corresponding GTS in WT—(NHEJ proficient) and in ku70 (NHEJ deficient) strains. Error bars represent standard deviation based on four biological replicates. (D) Results of ADE2 deletion efficiencies using a short oligonucleotide as GTS. The bars show the ratio of red colonies with correctly integrated GTS versus total number of tested red colonies for the corresponding GTS in WT—(NHEJ proficient) and in ku70 (NHEJ deficient) strains. Error bars represent standard deviation based on four biological replicates.
Visual marker system for easy distinction of NHEJ-deficient/proficient strains
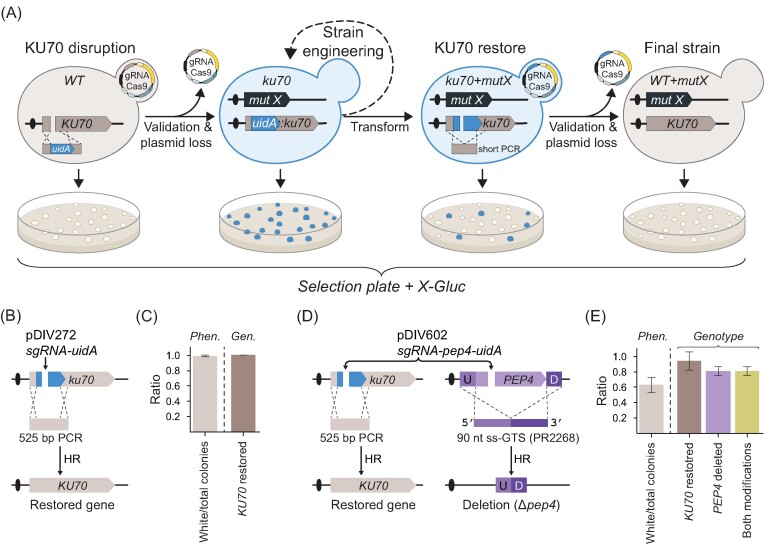
Restoration of the NHEJ pathway in CRISPR-engineered strains prior to functional characterization and data interpretation is advisable. To facilitate this routine, we have developed a transient KU70 gene disruption system that allows visual distinction between NHEJ-deficient and -proficient cells. Specifically, NHEJ was eliminated by inserting the common color-marker gene uidA gene (Jefferson et al. 1987) into the ORF of KU70 to disrupt its gene function. As a result, ku70::uidA strains can therefore easily be identified on media supplemented with X-Gluc as they turn blue. Once desirable modification(s) have been introduced into the ku70::uidA strain, the NHEJ pathway can be easily restored in a CRISPR event by using a Cas9–sgRNA nuclease targeting the uidA cassette and a short PCR repair fragment that allows the KU70 gene to be restored. Subsequent plating on X-Gluc media readily identifies KU70 strains as they turn white (Fig. 4A).
Figure 4.
Color-based sensor to assess KU70 functionality in K. phaffii. (A) A schematic illustration of how the uidA marker is used as a sensor during CRISPR gene editing. Firstly, the KU70 gene is disrupted by the uidA cassette to produce blue NHEJ-deficient ku70::uidA strain colonies. Secondly, the ku70::uidA strain is used for (iterative) gene-editing (necessary validation for desired mutations not shown). Thirdly, in the last round of gene editing, ku70::uidA is restored to WT in a CRISPR event that excises uidA to produce white colonies. Fourthly, strains forming white colonies are streak-purified and validated for desired mutations and loss of the CRISPR plasmid. (B) Detailed experimental setup to restore disrupted KU70 gene using a short PCR-based GTS. (C) KU70 gene restoration efficiency. Phen.—the ratio of white colonies versus total colonies observed on transformation plates; Gen.—the ratio of colonies with correctly restored KU70 versus total number of tested white colonies. Error bars represent standard deviation based on four biological replicates. (D) Multiplex experimental setup for simultaneous restoration of KU70 and deletion of the PEP4 gene. U—upstream and D—downstream sequence of PEP4. (E) Results of an experiment performed using the scheme presented in panel (D), see text for details. Left of dashed line: Phen.—phenotypical analysis of the transformants. The ratio of white colonies versus total colonies observed on transformation plates is shown. Right of dashed line: genotypic analysis of white colonies by diagnostic PCR reactions. The ratios of analysed transformants containing correctly restored KU70, correct PEP4 deletions, and both alterations are shown as indicated. Error bars represent standard deviation based on three biological replicates.
We tested the functionality of our system by cotransforming the ku70::uidA (sDIV291) strain with sgRNA-uidA (pDIV272) plasmid and a PCR fragment harboring the restoring sequence of KU70 gene (Fig. 4B). In this experiment, virtually all (∼98%) of transformed cells formed white colonies on X-Gluc media (Fig. 4C and Supplementary Fig. S14). The genotyping of 32 randomly selected white colonies revealed that in 100% of cases the KU70 gene was restored to the wild-type sequence (Fig. 4C and Supplementary Fig. S14). Based on the latter results, we conclude that our system offers quick and efficient reversion of ku70 to wild-type. Next, we asked whether our ku70::uidA system can be applied in multiplex experiments involving gene editing of a desired locus and simultaneous reversion of the edited strain to NHEJ-proficient state.
Exploiting multiplexing, we decided to explore whether it could be possible to delete the PEP4 gene and restore ku70::uidA locus to wild-type KU70 in a single round of transformation (see Fig. 4D). We performed this experiment by cotransforming the NHEJ-deficient (sDIV291) strain with a plasmid pDIV602 (expressing two sgRNAs targeting Cas9 to PEP4 and uidA) and two repair templates, a PCR fragment to restore KU70 and an ss-GTS (PR2268) to delete the PEP4 gene. Transformants were successfully obtained, but the number of colonies per transformation were noticeably lower as compared to the previous experiments where only ku70 locus was targeted (Supplementary Fig. S14). Moreover, only ∼60% of the transformants displayed the white phenotype on X-Gluc plates (Fig. 4E and Supplementary Fig. S14). Based on this result, we note that the efficiency of a specific gene-editing event appears reduced when it is part of a multiplexing experiment as compared to the efficiency obtained in an experiment where it is introduced as the only modification. Next, to examine for the overall multiplexing efficiency, we genotyped 20 randomly selected white colonies on X-Gluc media (see the section “Materials and methods” and Supplementary Fig. S14). This analysis showed that ∼92% of the transformants contained a restored KU70 gene and 80% contained the pep4 genotype (Fig. 4E). However, when we examined the transformants that were KU70 they were all pep4 showing that cells that are proficient for CRISPR gene editing are capable of introducing two modifications at high efficiency. The latter suggests that the colony color itself can therefore serve as an indicator for selection of cells that most likely underwent the intended genetic modification(s), in this case a PEP4 deletion and ku70 reversion.
Concluding remarks
In this study, we have developed a CRISPR-Cas9-based method that can serve as an improved addition to the currently available gene engineering toolbox for K. phaffii. We have demonstrated that our CRISPR tools can be applied for editing in either NHEJ-proficient or -deficient strain backgrounds. Additionally, our results strongly indicate and support multiple previous studies showing that NHEJ-deficient strain backgrounds are beneficial for gene-editing experiments. Importantly, we have demonstrated that short single-stranded oligonucleotides facilitate DNA DSB repair induced by CRISPR nuclease, and thus, can be used to introduce specific gene deletions or mutations with very high efficiency. Successful application of short single-stranded oligonucleotides will dramatically reduce the cost and time associated with construction of GTSs in large scale experiments. Lastly, we address the widely ignored fact that nonfunctional KU70 gene is associated with unwanted effects that can potentially compromise the performance of K. phaffii in physiological characterizations as well as when it is applied as a cell factory. Thus, we have devised a color-based method that allows for transient color marker-based inactivation of KU70 and successfully demonstrated its application in genetic engineering. Lastly, we envision that our color marker-based system could serve as a highly useful add-on that can be easily implemented into automated strain construction workflows.
Supplementary Material
Acknowledgement
We thank Irina Borodina for kindly providing the pCfB2312 plasmid.
Contributor Information
Tomas Strucko, Section for Synthetic Biology, Department of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads 223, 2800 Kongens Lyngby, Denmark.
Adrian-E Gadar-Lopez, Section for Synthetic Biology, Department of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads 223, 2800 Kongens Lyngby, Denmark.
Frederik B Frøhling, Section for Synthetic Biology, Department of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads 223, 2800 Kongens Lyngby, Denmark.
Emma T Frost, Section for Synthetic Biology, Department of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads 223, 2800 Kongens Lyngby, Denmark.
Esther F Iversen, Section for Synthetic Biology, Department of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads 223, 2800 Kongens Lyngby, Denmark.
Helen Olsson, Section for Synthetic Biology, Department of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads 223, 2800 Kongens Lyngby, Denmark.
Zofia D Jarczynska, Section for Synthetic Biology, Department of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads 223, 2800 Kongens Lyngby, Denmark.
Uffe H Mortensen, Section for Synthetic Biology, Department of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads 223, 2800 Kongens Lyngby, Denmark.
Author contributions
T.S., Z.D.J., and U.H.M. designed the research. T.S., A.E.G.L., F.B.F., E.T.F., E.F.I., and H.O. performed the research. T.S., A.E.G.L., F.B.F., and E.T.F., analyzed the data. T.S. and U.H.M. wrote the manuscript.
Conflict of interest
No conflict of interest to declare.
Funding
This research was supported by the Innovation Fund Denmark (grant numbers 6150–00031B and 0224–00121B).
References
- Bernauer L, Radkohl A, Lehmayer LGK et al. Komagataella phaffii as emerging model organism in fundamental research. Front Microbiol. 2021;11:1–16. 10.3389/fmicb.2020.607028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. 10.1128/JB.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai P, Duan X, Wu X et al. Recombination machinery engineering facilitates metabolic engineering of the industrial yeast Pichia pastoris. Nucleic Acids Res. 2021;49:7791–805. 10.1093/nar/gkab535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai P, Gao J, Zhou Y. CRISPR-mediated genome editing in non-conventional yeasts for biotechnological applications. Microb Cell Fact. 2019;18:1–12. 10.1186/s12934-019-1112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho NDSP, Arentshorst M, Jin Kwon M et al. Expanding the ku70 toolbox for filamentous fungi: establishment of complementation vectors and recipient strains for advanced gene analyses. Appl Microbiol Biotechnol. 2010;87:1463–73. 10.1007/s00253-010-2588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalvie NC, Leal J, Whittaker CA et al. Host-informed expression of CRISPR guide RNA for genomic engineering in Komagataella phaffii. ACS Synth Biol. 2020;9:26–35. 10.1021/acssynbio.9b00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalvie NC, Lorgeree T, Biedermann AM et al. Simplified gene knockout by CRISPR-Cas9-induced homologous recombination. ACS Synth Biol. 2022;11:497–501. 10.1021/acssynbio.1c00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Norville JE, Mali P et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–43. 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Battles MB, Nett JH. A color-based stable multi-copy integrant selection system for Pichia pastoris using the attenuated ADE1 and ADE2 genes as auxotrophic markers. Bioengineered. 2012;3:32–37. 10.4161/bbug.3.1.17936. [DOI] [PubMed] [Google Scholar]
- Fellerhoff B, Eckardt-Schupp F, Friedl AA. Subtelomeric repeat amplification is associated with growth at elevated temperature in yku70 mutants of Saccharomyces cerevisiae. Genetics. 2000;154:1039–51. 10.1093/genetics/154.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Ye C, Cheng J et al. Enhancing homologous recombination efficiency in Pichia pastoris for multiplex genome integration using short homology arms. ACS Synth Biol. 2022;11:547–53. 10.1021/acssynbio.1c00366. [DOI] [PubMed] [Google Scholar]
- Garrigues S, Peng M, Kun RS et al. Non-homologous end-joining-deficient filamentous fungal strains mitigate the impact of off-target mutations during the application of CRISPR/Cas9. mBio. 2023;0:e00668–23. 10.1128/mbio.00668-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassler T, Sauer M, Gasser B et al. The industrial yeast Pichia pastoris is converted from a heterotroph into an autotroph capable of growth on CO2. Nat Biotechnol. 2020;38:210–6. 10.1038/s41587-019-0363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Gao J, Cao M et al. Construction of a series of episomal plasmids and their application in the development of an efficient CRISPR/Cas9 system in Pichia pastoris. World J Microbiol Biotechnol. 2019;35:79. 10.1007/s11274-019-2654-5. [DOI] [PubMed] [Google Scholar]
- Hansen BG, Salomonsen B, Nielsen MT et al. Versatile enzyme expression and characterization system for Aspergillus nidulans, with the Penicillium brevicompactum polyketide synthase gene from the mycophenolic acid gene cluster as a test case. Appl Environ Microbiol. 2011;77:3044–51. 10.1128/AEM.01768-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–7. 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbalaei M, Rezaee SA, Farsiani H. Pichia pastoris: a highly successful expression system for optimal synthesis of heterologous proteins. J Cell Physiol. 2020;235:5867–81. 10.1002/jcp.29583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Wang B, Pan L et al. Comprehensive structural annotation of Pichia pastoris transcriptome and the response to various carbon sources using deep paired-end RNA sequencing. BMC Genomics. 2012;13:738. 10.1186/1471-2164-13-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X, Li L, Jameel A et al. A versatile toolbox for CRISPR-based genome engineering in Pichia pastoris. Appl Microbiol Biotechnol. 2021;105:9211–8. 10.1007/S00253-021-11688-Y. [DOI] [PubMed] [Google Scholar]
- Lin-Cereghino J, Wong WW, Xiong S et al. Condensed protocol for competent cell preparation and transformation of the methylotrophic yeast Pichia pastoris. BioTechniques. 2005;38:44–48. 10.2144/05381BM04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Shi X, Song L et al. CRISPR–Cas9-mediated genomic multiloci integration in Pichia pastoris. Microb Cell Fact. 2019;18:144. 10.1186/s12934-019-1194-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näätsaari L, Mistlberger B, Ruth C et al. Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS One. 2012;7:e39720. 10.1371/JOURNAL.PONE.0039720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y, Hino K, Bono H et al. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31:1120–3. 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nødvig CS, Hoof JB, Kogle ME et al. Efficient oligo nucleotide mediated CRISPR-Cas9 gene editing in Aspergilli. Fungal Genet Biol. 2018;115:78–89. 10.1016/j.fgb.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Nour-Eldin HH, Hansen BG, Nørholm MHH et al. Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 2006;34:e122. 10.1093/nar/gkl635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña DA, Gasser B, Zanghellini J et al. Metabolic engineering of Pichia pastoris. Metab Eng. 2018;50:2–15. 10.1016/j.ymben.2018.04.017. [DOI] [PubMed] [Google Scholar]
- Roman H. Studies of gene mutation in Saccharomyces. Cold Spring Harbor Symp Quant Biol. 1956;21:175–85. 10.1101/SQB.1956.021.01.015. [DOI] [PubMed] [Google Scholar]
- Schusterbauer V, Fischer JE, Gangl S et al. Whole genome sequencing analysis of effects of CRISPR/Cas9 in Komagataella phaffii: a budding yeast in distress. J Fungi. 2022;8:992. 10.3390/jof8100992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Xue C, Liu W et al. High-level secretory production of leghemoglobin in Pichia pastoris through enhanced globin expression and heme biosynthesis. Bioresour Technol. 2022;363:127884. 10.1016/j.biortech.2022.127884. [DOI] [PubMed] [Google Scholar]
- Stovicek V, Borodina I, Forster J. CRISPR–Cas system enables fast and simple genome editing of industrial Saccharomyces cerevisiae strains. Metab Eng Commun. 2015;2:13–22. 10.1016/j.meteno.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stovicek V, Holkenbrink C, Borodina I. CRISPR/Cas system for yeast genome engineering: advances and applications. FEMS Yeast Res. 2017;17. 10.1093/femsyr/fox030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strucko T, Andersen NL, Mahler MR et al. A CRISPR/Cas9 method facilitates efficient oligo-mediated gene editing in Debaryomyces hansenii. Synth Biol. 2021;6:ysab031. 10.1093/synbio/ysab031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanegas KG, Jarczynska ZD, Strucko T et al. Cpf1 enables fast and efficient genome editing in Aspergilli. Fungal Biol Biotechnol. 2019;6:6. 10.1186/s40694-019-0069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanegas KG, Lehka BJ, Mortensen UH. SWITCH: a dynamic CRISPR tool for genome engineering and metabolic pathway control for cell factory construction in Saccharomyces cerevisiae. Microb Cell Fact. 2017;16:25. 10.1186/s12934-017-0632-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li Y, Jin Z et al. A novel CRISPR/Cas9 system with high genomic editing efficiency and recyclable auxotrophic selective marker for multiple-step metabolic rewriting in Pichia pastoris. Synth Syst Biotechnol. 2023;8:445–51. 10.1016/j.synbio.2023.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weninger A, Hatzl A-M, Schmid C et al. Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris. J Biotechnol. 2016;235:139–49. 10.1016/j.jbiotec.2016.03.027. [DOI] [PubMed] [Google Scholar]
- Werten MWT, Eggink G, Cohen Stuart MA et al. Production of protein-based polymers in Pichia pastoris. Biotechnol Adv. 2019;37:642–66. 10.1016/j.biotechadv.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhang Z. Engineering strategies for enhanced production of protein and bio-products in Pichia pastoris: a review. Biotechnol Adv. 2018;36:182–95. 10.1016/j.biotechadv.2017.11.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.