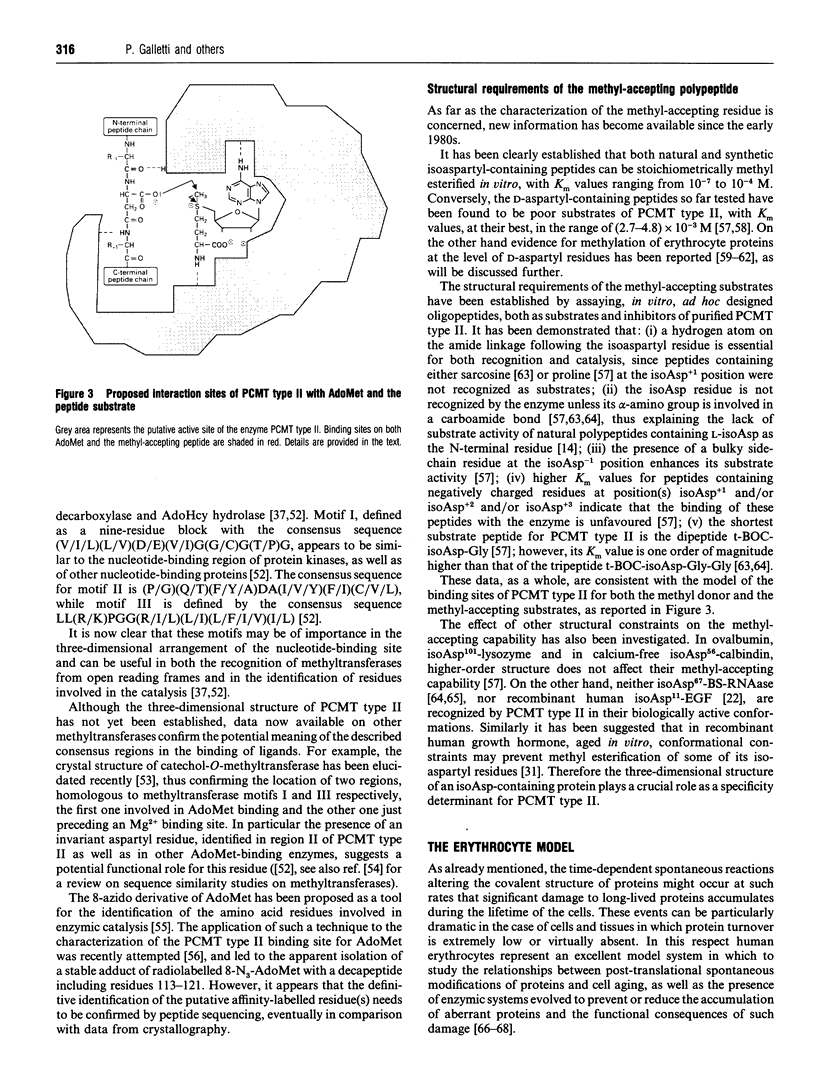
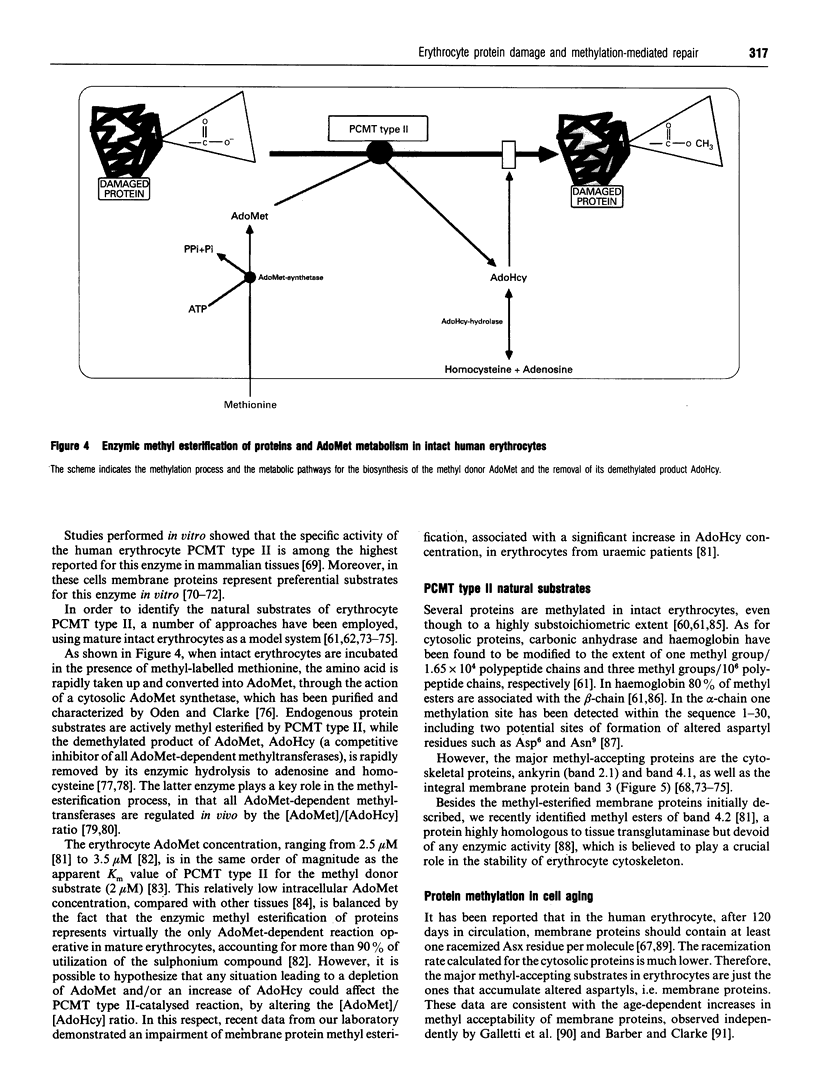
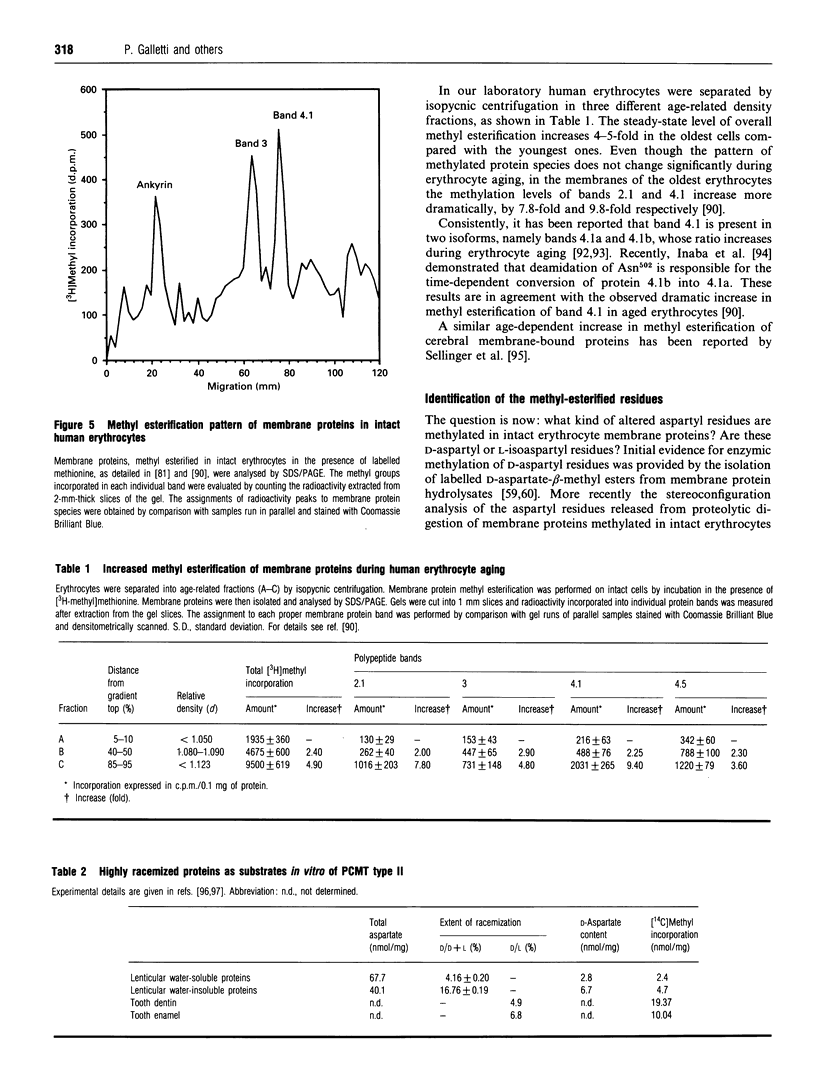
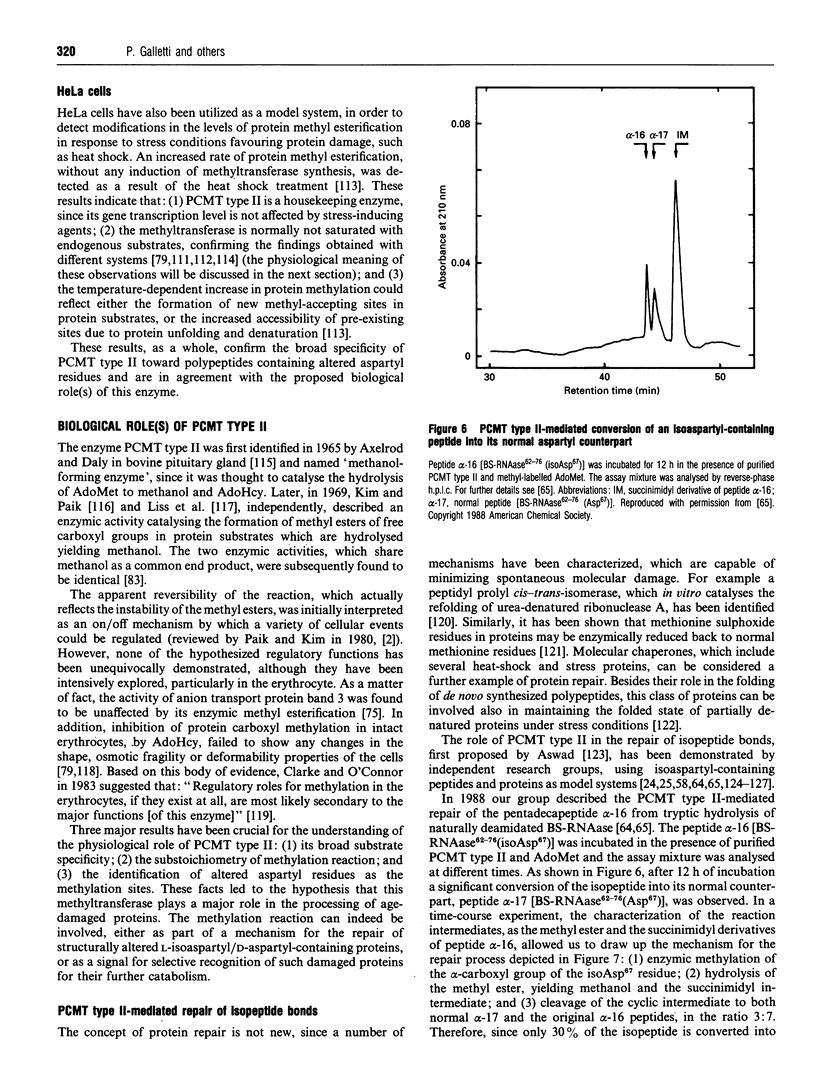
Full text
PDF
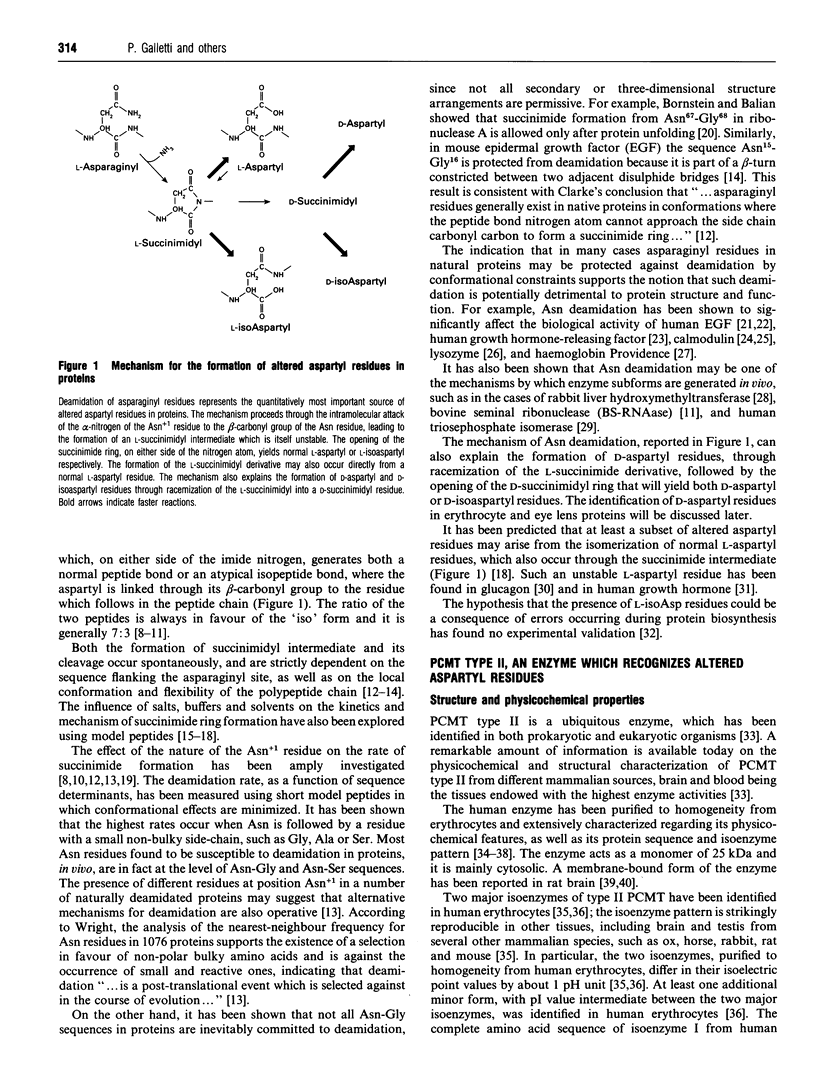


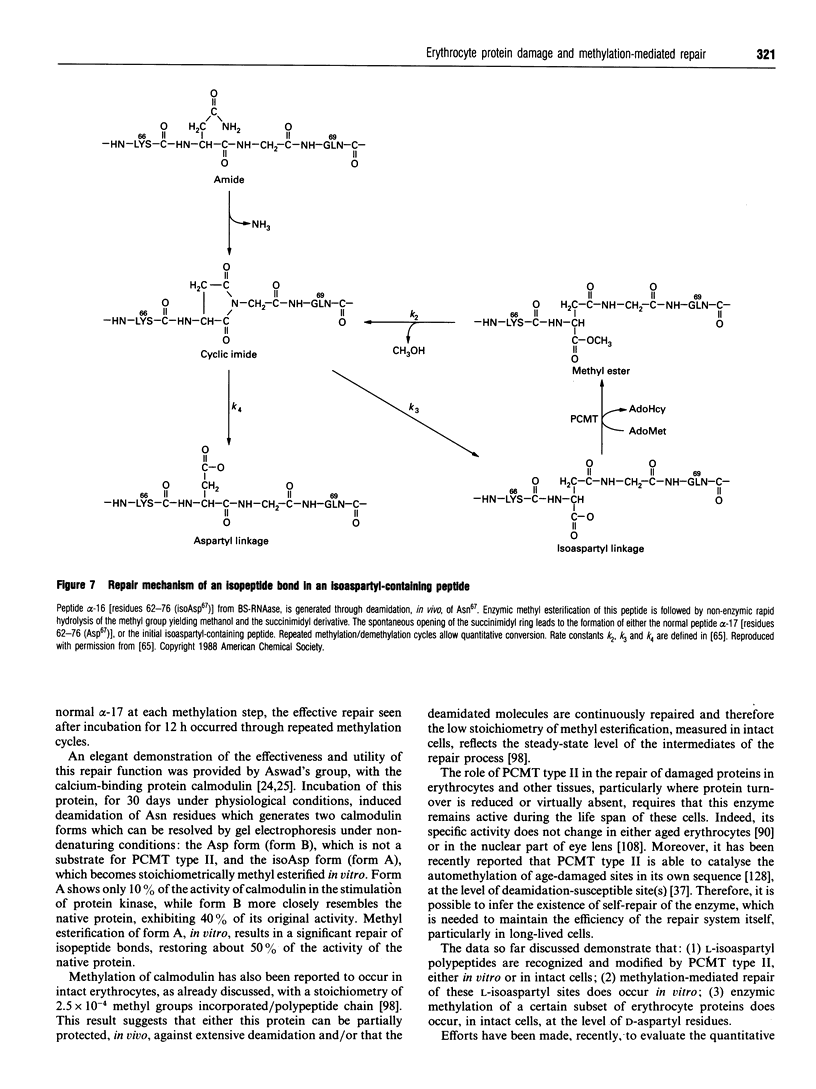
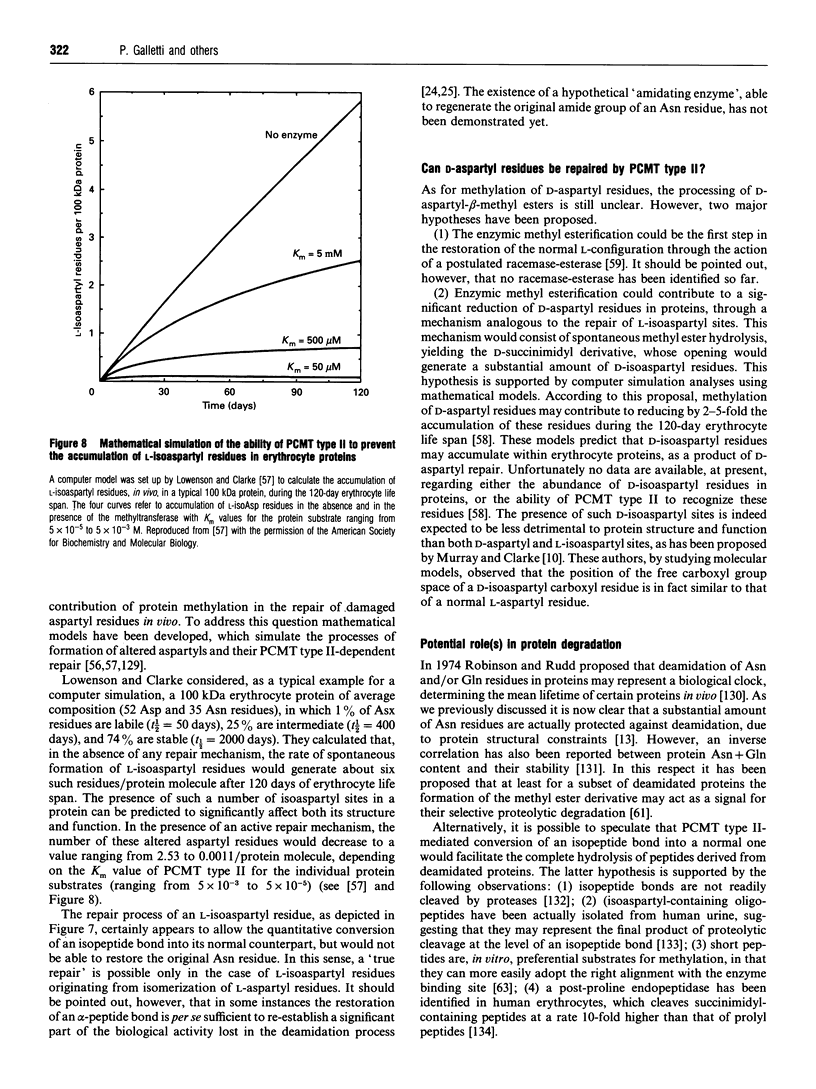
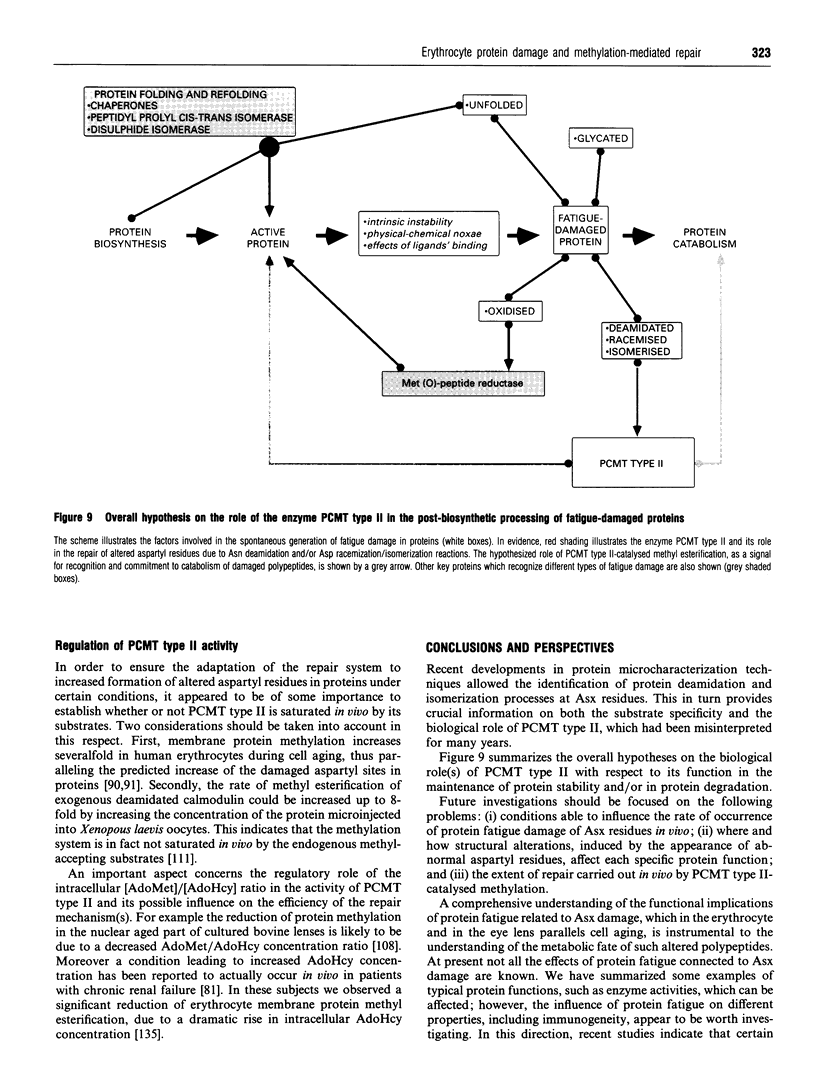


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Araki F., Nakamura H., Nojima N., Tsukumo K., Sakamoto S. Stability of recombinant human epidermal growth factor in various solutions. Chem Pharm Bull (Tokyo) 1989 Feb;37(2):404–406. doi: 10.1248/cpb.37.404. [DOI] [PubMed] [Google Scholar]
- Artigues A., Birkett A., Schirch V. Evidence for the in vivo deamidation and isomerization of an asparaginyl residue in cytosolic serine hydroxymethyltransferase. J Biol Chem. 1990 Mar 25;265(9):4853–4858. [PubMed] [Google Scholar]
- Aswad D. W. Stoichiometric methylation of porcine adrenocorticotropin by protein carboxyl methyltransferase requires deamidation of asparagine 25. Evidence for methylation at the alpha-carboxyl group of atypical L-isoaspartyl residues. J Biol Chem. 1984 Sep 10;259(17):10714–10721. [PubMed] [Google Scholar]
- Aswad D. W. Stoichiometric methylation of porcine adrenocorticotropin by protein carboxyl methyltransferase requires deamidation of asparagine 25. Evidence for methylation at the alpha-carboxyl group of atypical L-isoaspartyl residues. J Biol Chem. 1984 Sep 10;259(17):10714–10721. [PubMed] [Google Scholar]
- Axelrod J., Daly J. Pituitary gland: enzymic formation of methanol from S-adenosylmethionine. Science. 1965 Nov 12;150(3698):892–893. doi: 10.1126/science.150.3698.892. [DOI] [PubMed] [Google Scholar]
- Barber J. R., Clarke S. Inhibition of protein carboxyl methylation by S-adenosyl-L-homocysteine in intact erythrocytes. Physiological consequences. J Biol Chem. 1984 Jun 10;259(11):7115–7122. [PubMed] [Google Scholar]
- Barber J. R., Clarke S. Membrane protein carboxyl methylation does not appear to be involved in the response of erythrocytes to cytoskeletal stress. Biochem Biophys Res Commun. 1984 Aug 30;123(1):133–140. doi: 10.1016/0006-291x(84)90390-5. [DOI] [PubMed] [Google Scholar]
- Barber J. R., Clarke S. Membrane protein carboxyl methylation increases with human erythrocyte age. Evidence for an increase in the number of methylatable sites. J Biol Chem. 1983 Jan 25;258(2):1189–1196. [PubMed] [Google Scholar]
- Barber J. R., Morimoto B. H., Brunauer L. S., Clarke S. Metabolism of S-adenosyl-L-methionine in intact human erythrocytes. Biochim Biophys Acta. 1986 May 29;886(3):361–372. doi: 10.1016/0167-4889(86)90171-0. [DOI] [PubMed] [Google Scholar]
- Benkirane N., Friede M., Guichard G., Briand J. P., Van Regenmortel M. H., Muller S. Antigenicity and immunogenicity of modified synthetic peptides containing D-amino acid residues. Antibodies to a D-enantiomer do recognize the parent L-hexapeptide and reciprocally. J Biol Chem. 1993 Dec 15;268(35):26279–26285. [PubMed] [Google Scholar]
- Bonaventura J., Bonaventura C., Sullivan B., Ferruzzi G., McCurdy P. R., Fox J., Moo-Penn W. F. Hemoglobin providence. Functional consequences of two alterations of the 2,3-diphosphoglycerate binding site at position beta 82. J Biol Chem. 1976 Dec 10;251(23):7563–7571. [PubMed] [Google Scholar]
- Bornstein P., Balian G. Cleavage at Asn-Gly bonds with hydroxylamine. Methods Enzymol. 1977;47:132–145. doi: 10.1016/0076-6879(77)47016-2. [DOI] [PubMed] [Google Scholar]
- Bornstein P., Balian G. The specific nonenzymatic cleavage of bovine ribonuclease with hydroxylamine. J Biol Chem. 1970 Sep 25;245(18):4854–4856. [PubMed] [Google Scholar]
- Brennan T. V., Clarke S. Spontaneous degradation of polypeptides at aspartyl and asparaginyl residues: effects of the solvent dielectric. Protein Sci. 1993 Mar;2(3):331–338. doi: 10.1002/pro.5560020305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brot N., Weissbach H. Biochemistry and physiological role of methionine sulfoxide residues in proteins. Arch Biochem Biophys. 1983 May;223(1):271–281. doi: 10.1016/0003-9861(83)90592-1. [DOI] [PubMed] [Google Scholar]
- Brunauer L. S., Clarke S. Age-dependent accumulation of protein residues which can be hydrolyzed to D-aspartic acid in human erythrocytes. J Biol Chem. 1986 Sep 25;261(27):12538–12543. [PubMed] [Google Scholar]
- Capasso S., Mazzarella L., Sica F., Zagari A. Deamidation via cyclic imide in asparaginyl peptides. Pept Res. 1989 Mar-Apr;2(2):195–200. [PubMed] [Google Scholar]
- Capasso S., Mazzarella L., Zagari A. Deamidation via cyclic imide of asparaginyl peptides: dependence on salts, buffers and organic solvents. Pept Res. 1991 Jul-Aug;4(4):234–238. [PubMed] [Google Scholar]
- Carter D. A., McFadden P. N. Determination of beta-isomerized aspartic acid as the corresponding alcohol. J Protein Chem. 1994 Jan;13(1):97–106. doi: 10.1007/BF01891997. [DOI] [PubMed] [Google Scholar]
- Carter D. A., McFadden P. N. Trapping succinimides in aged polypeptides by chemical reduction. J Protein Chem. 1994 Jan;13(1):89–96. doi: 10.1007/BF01891996. [DOI] [PubMed] [Google Scholar]
- Clarke S. Propensity for spontaneous succinimide formation from aspartyl and asparaginyl residues in cellular proteins. Int J Pept Protein Res. 1987 Dec;30(6):808–821. doi: 10.1111/j.1399-3011.1987.tb03390.x. [DOI] [PubMed] [Google Scholar]
- Clarke S. Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu Rev Biochem. 1992;61:355–386. doi: 10.1146/annurev.bi.61.070192.002035. [DOI] [PubMed] [Google Scholar]
- DE LA HABA G., CANTONI G. L. The enzymatic synthesis of S-adenosyl-L-homocysteine from adenosine and homocysteine. J Biol Chem. 1959 Mar;234(3):603–608. [PubMed] [Google Scholar]
- Di Donato A., Galletti P., D'Alessio G. Selective deamidation and enzymatic methylation of seminal ribonuclease. Biochemistry. 1986 Dec 30;25(26):8361–8368. doi: 10.1021/bi00374a005. [DOI] [PubMed] [Google Scholar]
- Economou-Petersen E., Henriksen K. F., Guldberg P., Güttler F. Molecular basis for nonphenylketonuria hyperphenylalaninemia. Genomics. 1992 Sep;14(1):1–5. doi: 10.1016/s0888-7543(05)80274-5. [DOI] [PubMed] [Google Scholar]
- Fischer G., Bang H. The refolding of urea-denatured ribonuclease A is catalyzed by peptidyl-prolyl cis-trans isomerase. Biochim Biophys Acta. 1985 Mar 22;828(1):39–42. doi: 10.1016/0167-4838(85)90006-8. [DOI] [PubMed] [Google Scholar]
- Freitag C., Clarke S. Reversible methylation of cytoskeletal and membrane proteins in intact human erythrocytes. J Biol Chem. 1981 Jun 25;256(12):6102–6108. [PubMed] [Google Scholar]
- Fu J. C., Ding L., Clarke S. Purification, gene cloning, and sequence analysis of an L-isoaspartyl protein carboxyl methyltransferase from Escherichia coli. J Biol Chem. 1991 Aug 5;266(22):14562–14572. [PubMed] [Google Scholar]
- Fujioka M. Mammalian small molecule methyltransferases: their structural and functional features. Int J Biochem. 1992 Dec;24(12):1917–1924. doi: 10.1016/0020-711x(92)90287-b. [DOI] [PubMed] [Google Scholar]
- Galletti P., Ciardiello A., Ingrosso D., Di Donato A., D'Alessio G. Repair of isopeptide bonds by protein carboxyl O-methyltransferase: seminal ribonuclease as a model system. Biochemistry. 1988 Mar 8;27(5):1752–1757. doi: 10.1021/bi00405a055. [DOI] [PubMed] [Google Scholar]
- Galletti P., Iardino P., Ingrosso D., Manna C., Zappia V. Enzymatic methyl esterification of a deamidated form of mouse epidermal growth factor. Int J Pept Protein Res. 1989 Jun;33(6):397–402. doi: 10.1111/j.1399-3011.1989.tb00215.x. [DOI] [PubMed] [Google Scholar]
- Galletti P., Ingrosso D., Manna C., Sica F., Capasso S., Pucci P., Marino G. Enzymatic methyl esterification of synthetic tripeptides: structural requirements of the peptide substrate. Detection of the reaction products by fast-atom-bombardment mass spectrometry. Eur J Biochem. 1988 Oct 15;177(1):233–239. doi: 10.1111/j.1432-1033.1988.tb14367.x. [DOI] [PubMed] [Google Scholar]
- Galletti P., Ingrosso D., Nappi A., Gragnaniello V., Iolascon A., Pinto L. Increased methyl esterification of membrane proteins in aged red-blood cells. Preferential esterification of ankyrin and band-4.1 cytoskeletal proteins. Eur J Biochem. 1983 Sep 1;135(1):25–31. doi: 10.1111/j.1432-1033.1983.tb07613.x. [DOI] [PubMed] [Google Scholar]
- Galletti P., Ki Paik W., Kim S. Methyl acceptors for protein methylase II from human-erythrocyte membrane. Eur J Biochem. 1979 Jun;97(1):221–227. doi: 10.1111/j.1432-1033.1979.tb13106.x. [DOI] [PubMed] [Google Scholar]
- Galletti P., Manna C., Ingrosso D., Iardino P., Zappia V. Hypotheses on the physiological role of enzymatic protein methyl esterification using human erythrocytes as a model system. Adv Exp Med Biol. 1991;307:149–160. doi: 10.1007/978-1-4684-5985-2_14. [DOI] [PubMed] [Google Scholar]
- Galletti P., Paik W. K., Kim S. Selective methyl esterification of erythrocyte membrane proteins by protein methylase II. Biochemistry. 1978 Oct 3;17(20):4272–4276. doi: 10.1021/bi00613a025. [DOI] [PubMed] [Google Scholar]
- Geiger T., Clarke S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J Biol Chem. 1987 Jan 15;262(2):785–794. [PubMed] [Google Scholar]
- George-Nascimento C., Lowenson J., Borissenko M., Calderón M., Medina-Selby A., Kuo J., Clarke S., Randolph A. Replacement of a labile aspartyl residue increases the stability of human epidermal growth factor. Biochemistry. 1990 Oct 16;29(41):9584–9591. doi: 10.1021/bi00493a012. [DOI] [PubMed] [Google Scholar]
- Gilbert J. M., Fowler A., Bleibaum J., Clarke S. Purification of homologous protein carboxyl methyltransferase isozymes from human and bovine erythrocytes. Biochemistry. 1988 Jul 12;27(14):5227–5233. doi: 10.1021/bi00414a042. [DOI] [PubMed] [Google Scholar]
- Groenen P. J., van den Ijssel P. R., Voorter C. E., Bloemendal H., de Jong W. W. Site-specific racemization in aging alpha A-crystallin. FEBS Lett. 1990 Aug 20;269(1):109–112. doi: 10.1016/0014-5793(90)81131-7. [DOI] [PubMed] [Google Scholar]
- Haley E. E., Corcoran B. J., Dorer F. E., Buchanan D. L. Beta-aspartyl peptides in enzymatic hydrolysates of protein. Biochemistry. 1966 Oct;5(10):3229–3235. doi: 10.1021/bi00874a024. [DOI] [PubMed] [Google Scholar]
- Hendrick J. P., Hartl F. U. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- Inaba M., Gupta K. C., Kuwabara M., Takahashi T., Benz E. J., Jr, Maede Y. Deamidation of human erythrocyte protein 4.1: possible role in aging. Blood. 1992 Jun 15;79(12):3355–3361. [PubMed] [Google Scholar]
- Inaba M., Maede Y. Correlation between protein 4.1a/4.1b ratio and erythrocyte life span. Biochim Biophys Acta. 1988 Oct 6;944(2):256–264. doi: 10.1016/0005-2736(88)90439-7. [DOI] [PubMed] [Google Scholar]
- Ingrosso D., Clarke S. Human erythrocyte D-aspartyl/L-isoaspartyl methyltransferases: enzymes that recognize age-damaged proteins. Adv Exp Med Biol. 1991;307:263–276. doi: 10.1007/978-1-4684-5985-2_24. [DOI] [PubMed] [Google Scholar]
- Ingrosso D., Fowler A. V., Bleibaum J., Clarke S. Sequence of the D-aspartyl/L-isoaspartyl protein methyltransferase from human erythrocytes. Common sequence motifs for protein, DNA, RNA, and small molecule S-adenosylmethionine-dependent methyltransferases. J Biol Chem. 1989 Nov 25;264(33):20131–20139. [PubMed] [Google Scholar]
- Ingrosso D., Kagan R. M., Clarke S. Distinct C-terminal sequences of isozymes I and II of the human erythrocyte L-isoaspartyl/D-aspartyl protein methyltransferase. Biochem Biophys Res Commun. 1991 Feb 28;175(1):351–358. doi: 10.1016/s0006-291x(05)81242-2. [DOI] [PubMed] [Google Scholar]
- Jamaluddin M., Kim S., Paik W. K. Studies on the kinetic mechanism of S-adenosylmethionine: protein O-methyltransferase of calf thymus. Biochemistry. 1975 Feb 25;14(4):694–698. doi: 10.1021/bi00675a008. [DOI] [PubMed] [Google Scholar]
- Johnson B. A., Aswad D. W. Fragmentation of isoaspartyl peptides and proteins by carboxypeptidase Y: release of isoaspartyl dipeptides as a result of internal and external cleavage. Biochemistry. 1990 May 8;29(18):4373–4380. doi: 10.1021/bi00470a017. [DOI] [PubMed] [Google Scholar]
- Johnson B. A., Aswad D. W. Optimal conditions for the use of protein L-isoaspartyl methyltransferase in assessing the isoaspartate content of peptides and proteins. Anal Biochem. 1991 Feb 1;192(2):384–391. doi: 10.1016/0003-2697(91)90553-6. [DOI] [PubMed] [Google Scholar]
- Johnson B. A., Murray E. D., Jr, Clarke S., Glass D. B., Aswad D. W. Protein carboxyl methyltransferase facilitates conversion of atypical L-isoaspartyl peptides to normal L-aspartyl peptides. J Biol Chem. 1987 Apr 25;262(12):5622–5629. [PubMed] [Google Scholar]
- Johnson B. A., Shirokawa J. M., Hancock W. S., Spellman M. W., Basa L. J., Aswad D. W. Formation of isoaspartate at two distinct sites during in vitro aging of human growth hormone. J Biol Chem. 1989 Aug 25;264(24):14262–14271. [PubMed] [Google Scholar]
- Kagan R. M., Clarke S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch Biochem Biophys. 1994 May 1;310(2):417–427. doi: 10.1006/abbi.1994.1187. [DOI] [PubMed] [Google Scholar]
- Kim S., Galletti P., Paik W. K. In vivo carboxyl methylation of human eruthrocyte membrane proteins. J Biol Chem. 1980 Jan 25;255(2):338–341. [PubMed] [Google Scholar]
- Kim S. Purification and properties of protein methylase II. Arch Biochem Biophys. 1973 Aug;157(2):476–484. doi: 10.1016/0003-9861(73)90665-6. [DOI] [PubMed] [Google Scholar]
- Kim S. Purification and properties of protein methylase II. Arch Biochem Biophys. 1973 Aug;157(2):476–484. doi: 10.1016/0003-9861(73)90665-6. [DOI] [PubMed] [Google Scholar]
- Korsgren C., Lawler J., Lambert S., Speicher D., Cohen C. M. Complete amino acid sequence and homologies of human erythrocyte membrane protein band 4.2. Proc Natl Acad Sci U S A. 1990 Jan;87(2):613–617. doi: 10.1073/pnas.87.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladino C. A., O'Connor C. M. Identification of a site for carboxyl methylation in human alpha-globin. Biochem Biophys Res Commun. 1991 Oct 31;180(2):742–747. doi: 10.1016/s0006-291x(05)81128-3. [DOI] [PubMed] [Google Scholar]
- Ladino C. A., O'Connor C. M. Methylation of atypical protein aspartyl residues during the stress response of HeLa cells. J Cell Physiol. 1992 Nov;153(2):297–304. doi: 10.1002/jcp.1041530209. [DOI] [PubMed] [Google Scholar]
- Ladino C. A., O'Connor C. M. Protein carboxyl methylation and methyl ester turnover in density-fractionated human erythrocytes. Mech Ageing Dev. 1990 Aug;55(2):123–137. doi: 10.1016/0047-6374(90)90020-g. [DOI] [PubMed] [Google Scholar]
- Lauster R. Evolution of type II DNA methyltransferases. A gene duplication model. J Mol Biol. 1989 Mar 20;206(2):313–321. doi: 10.1016/0022-2836(89)90481-6. [DOI] [PubMed] [Google Scholar]
- Li C., Clarke S. Distribution of an L-isoaspartyl protein methyltransferase in eubacteria. J Bacteriol. 1992 Jan;174(2):355–361. doi: 10.1128/jb.174.2.355-361.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist J. A., McFadden P. N. Automethylation of protein (D-aspartyl/L-isoaspartyl) carboxyl methyltransferase, a response to enzyme aging. J Protein Chem. 1994 Jan;13(1):23–30. doi: 10.1007/BF01891989. [DOI] [PubMed] [Google Scholar]
- Liss M., Maxam A. M., Cuprak L. J. Methylation of protein by calf spleen methylase. A new protein methylation reaction. J Biol Chem. 1969 Mar 25;244(6):1617–1622. [PubMed] [Google Scholar]
- Lou L. L., Clarke S. Carboxyl methylation of human erythrocyte band 3 in intact cells. Relation to anion transport activity. Biochem J. 1986 Apr 1;235(1):183–187. doi: 10.1042/bj2350183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou L. L., Clarke S. Enzymatic methylation of band 3 anion transporter in intact human erythrocytes. Biochemistry. 1987 Jan 13;26(1):52–59. doi: 10.1021/bi00375a008. [DOI] [PubMed] [Google Scholar]
- Lowenson J. D., Clarke S. Recognition of D-aspartyl residues in polypeptides by the erythrocyte L-isoaspartyl/D-aspartyl protein methyltransferase. Implications for the repair hypothesis. J Biol Chem. 1992 Mar 25;267(9):5985–5995. [PubMed] [Google Scholar]
- Lowenson J. D., Clarke S. Spontaneous degradation and enzymatic repair of aspartyl and asparaginyl residues in aging red cell proteins analyzed by computer simulation. Gerontology. 1991;37(1-3):128–151. doi: 10.1159/000213255. [DOI] [PubMed] [Google Scholar]
- Lowenson J. D., Clarke S. Structural elements affecting the recognition of L-isoaspartyl residues by the L-isoaspartyl/D-aspartyl protein methyltransferase. Implications for the repair hypothesis. J Biol Chem. 1991 Oct 15;266(29):19396–19406. [PubMed] [Google Scholar]
- Lowenson J., Clarke S. Does the chemical instability of aspartyl and asparaginyl residues in proteins contribute to erythrocyte aging? The role of protein carboxyl methylation reactions. Blood Cells. 1988;14(1):103–118. [PubMed] [Google Scholar]
- MacLaren D. C., Kagan R. M., Clarke S. Alternative splicing of the human isoaspartyl protein carboxyl methyltransferase RNA leads to the generation of a C-terminal -RDEL sequence in isozyme II. Biochem Biophys Res Commun. 1992 May 29;185(1):277–283. doi: 10.1016/s0006-291x(05)80987-8. [DOI] [PubMed] [Google Scholar]
- Masters P. M., Bada J. L., Zigler J. S., Jr Aspartic acid racemisation in the human lens during ageing and in cataract formation. Nature. 1977 Jul 7;268(5615):71–73. doi: 10.1038/268071a0. [DOI] [PubMed] [Google Scholar]
- McFadden P. N., Clarke S. Chemical conversion of aspartyl peptides to isoaspartyl peptides. A method for generating new methyl-accepting substrates for the erythrocyte D-aspartyl/L-isoaspartyl protein methyltransferase. J Biol Chem. 1986 Sep 5;261(25):11503–11511. [PubMed] [Google Scholar]
- McFadden P. N., Clarke S. Conversion of isoaspartyl peptides to normal peptides: implications for the cellular repair of damaged proteins. Proc Natl Acad Sci U S A. 1987 May;84(9):2595–2599. doi: 10.1073/pnas.84.9.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden P. N., Clarke S. Methylation at D-aspartyl residues in erythrocytes: possible step in the repair of aged membrane proteins. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2460–2464. doi: 10.1073/pnas.79.8.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden P. N., Clarke S. Protein carboxyl methyltransferase and methyl acceptor proteins in aging and cataractous tissue of the human eye lens. Mech Ageing Dev. 1986 Mar;34(1):91–105. doi: 10.1016/0047-6374(86)90107-7. [DOI] [PubMed] [Google Scholar]
- McFadden P. N., Horwitz J., Clarke S. Protein carboxyl methyltransferase from cow eye lens. Biochem Biophys Res Commun. 1983 Jun 15;113(2):418–424. doi: 10.1016/0006-291x(83)91742-4. [DOI] [PubMed] [Google Scholar]
- Momand J. A., Clarke S. The fidelity of protein synthesis: can mischarging by aspartyl-tRNA(Asp) synthetase lead to the formation of isoaspartyl residues in proteins? Biochim Biophys Acta. 1990 Sep 3;1040(2):153–158. doi: 10.1016/0167-4838(90)90070-v. [DOI] [PubMed] [Google Scholar]
- Momand J., Clarke S. Rapid degradation of D- and L-succinimide-containing peptides by a post-proline endopeptidase from human erythrocytes. Biochemistry. 1987 Dec 1;26(24):7798–7805. doi: 10.1021/bi00398a040. [DOI] [PubMed] [Google Scholar]
- Mueller T. J., Jackson C. W., Dockter M. E., Morrison M. Membrane skeletal alterations during in vivo mouse red cell aging. Increase in the band 4.1a:4.1b ratio. J Clin Invest. 1987 Feb;79(2):492–499. doi: 10.1172/JCI112839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray E. D., Jr, Clarke S. Metabolism of a synthetic L-isoaspartyl-containing hexapeptide in erythrocyte extracts. Enzymatic methyl esterification is followed by nonenzymatic succinimide formation. J Biol Chem. 1986 Jan 5;261(1):306–312. [PubMed] [Google Scholar]
- Murray E. D., Jr, Clarke S. Synthetic peptide substrates for the erythrocyte protein carboxyl methyltransferase. Detection of a new site of methylation at isomerized L-aspartyl residues. J Biol Chem. 1984 Sep 10;259(17):10722–10732. [PubMed] [Google Scholar]
- O'Connor C. M., Clarke S. Carboxyl methylation of cytosolic proteins in intact human erythrocytes. Identification of numerous methyl-accepting proteins including hemoglobin and carbonic anhydrase. J Biol Chem. 1984 Feb 25;259(4):2570–2578. [PubMed] [Google Scholar]
- O'Connor C. M., Clarke S. Methylation of erythrocyte membrane proteins at extracellular and intracellular D-aspartyl sites in vitro. Saturation of intracellular sites in vivo. J Biol Chem. 1983 Jul 10;258(13):8485–8492. [PubMed] [Google Scholar]
- O'Connor C. M., Germain B. J. Kinetic and electrophoretic analysis of transmethylation reactions in intact Xenopus laevis oocytes. J Biol Chem. 1987 Jul 25;262(21):10404–10411. [PubMed] [Google Scholar]
- O'Connor C. M. Regulation and subcellular distribution of a protein methyltransferase and its damaged aspartyl substrate sites in developing Xenopus oocytes. J Biol Chem. 1987 Jul 25;262(21):10398–10403. [PubMed] [Google Scholar]
- O'Connor C. M., Yutzey K. E. Enhanced carboxyl methylation of membrane-associated hemoglobin in human erythrocytes. J Biol Chem. 1988 Jan 25;263(3):1386–1390. [PubMed] [Google Scholar]
- Oden K. L., Clarke S. S-adenosyl-L-methionine synthetase from human erythrocytes: role in the regulation of cellular S-adenosylmethionine levels. Biochemistry. 1983 Jun 7;22(12):2978–2986. doi: 10.1021/bi00281a030. [DOI] [PubMed] [Google Scholar]
- Oliva A., Galletti P., Zappia V., Paik W. K., Kim S. Studies on substrate specificity of S-adenosylmethionine: protein-carboxyl methyltransferase from calf brain. Eur J Biochem. 1980 Mar;104(2):595–602. doi: 10.1111/j.1432-1033.1980.tb04463.x. [DOI] [PubMed] [Google Scholar]
- Ota I. M., Clarke S. Calcium affects the spontaneous degradation of aspartyl/asparaginyl residues in calmodulin. Biochemistry. 1989 May 2;28(9):4020–4027. doi: 10.1021/bi00435a058. [DOI] [PubMed] [Google Scholar]
- Ota I. M., Clarke S. Enzymatic methylation of L-isoaspartyl residues derived from aspartyl residues in affinity-purified calmodulin. The role of conformational flexibility in spontaneous isoaspartyl formation. J Biol Chem. 1989 Jan 5;264(1):54–60. [PubMed] [Google Scholar]
- Ota I. M., Clarke S. Multiple sites of methyl esterification of calmodulin in intact human erythrocytes. Arch Biochem Biophys. 1990 Jun;279(2):320–327. doi: 10.1016/0003-9861(90)90498-n. [DOI] [PubMed] [Google Scholar]
- Ota I. M., Ding L., Clarke S. Methylation at specific altered aspartyl and asparaginyl residues in glucagon by the erythrocyte protein carboxyl methyltransferase. J Biol Chem. 1987 Jun 25;262(18):8522–8531. [PubMed] [Google Scholar]
- Ota I. M., Gilbert J. M., Clarke S. Two major isozymes of the protein D-aspartyl/L-isoaspartyl methyltransferase from human erythrocytes. Biochem Biophys Res Commun. 1988 Mar 30;151(3):1136–1143. doi: 10.1016/s0006-291x(88)80484-4. [DOI] [PubMed] [Google Scholar]
- Perna A. F., Ingrosso D., De Santo N. G., Galletti P., Zappia V. Mechanism of erythrocyte accumulation of methylation inhibitor S-adenosylhomocysteine in uremia. Kidney Int. 1995 Jan;47(1):247–253. doi: 10.1038/ki.1995.31. [DOI] [PubMed] [Google Scholar]
- Perna A. F., Ingrosso D., Zappia V., Galletti P., Capasso G., De Santo N. G. Enzymatic methyl esterification of erythrocyte membrane proteins is impaired in chronic renal failure. Evidence for high levels of the natural inhibitor S-adenosylhomocysteine. J Clin Invest. 1993 Jun;91(6):2497–2503. doi: 10.1172/JCI116485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai J., Bhagwat A. S., Pósfai G., Roberts R. J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989 Apr 11;17(7):2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. B., Rudd C. J. Deamidation of glutaminyl and asparaginyl residues in peptides and proteins. Curr Top Cell Regul. 1974;8(0):247–295. doi: 10.1016/b978-0-12-152808-9.50013-4. [DOI] [PubMed] [Google Scholar]
- Rogers S. W., Rechsteiner M. Degradation of structurally characterized proteins injected into HeLa cells. Tests of hypotheses. J Biol Chem. 1988 Dec 25;263(36):19850–19862. [PubMed] [Google Scholar]
- Romanik E. A., O'Connor C. M. Methylation of microinjected isoaspartyl peptides in Xenopus oocytes. Competition with protein carboxyl methylation reactions. J Biol Chem. 1989 Aug 25;264(24):14050–14056. [PubMed] [Google Scholar]
- Sato M., Yoshida T., Tuboi S. Primary structure of rat brain protein carboxyl methyltransferase deduced from cDNA sequence. Biochem Biophys Res Commun. 1989 May 30;161(1):342–347. doi: 10.1016/0006-291x(89)91602-1. [DOI] [PubMed] [Google Scholar]
- Sellinger O. Z., Kramer C. M., Conger A., Duboff G. S. The carboxylmethylation of cerebral membrane-bound proteins increases with age. Mech Ageing Dev. 1988 May;43(2):161–173. doi: 10.1016/0047-6374(88)90044-9. [DOI] [PubMed] [Google Scholar]
- Sellinger O. Z., Kramer C. M. The carboxylmethylation of membrane-bound proteins in the aging rat brain. Adv Exp Med Biol. 1988;231:269–280. doi: 10.1007/978-1-4684-9042-8_21. [DOI] [PubMed] [Google Scholar]
- Sumner J., Jencks D. A., Khani S., Matthews R. G. Photoaffinity labeling of methylenetetrahydrofolate reductase with 8-azido-S-adenosylmethionine. J Biol Chem. 1986 Jun 15;261(17):7697–7700. [PubMed] [Google Scholar]
- Syed S. K., Kim S., Paik W. K. Identification of the S-adenosyl-L-methionine binding site of protein-carboxyl O-methyltransferase using 8-azido-S-adenosyl-L-methionine. Biochemistry. 1993 Mar 9;32(9):2242–2247. doi: 10.1021/bi00060a016. [DOI] [PubMed] [Google Scholar]
- Terwilliger T. C., Clarke S. Methylation of membrane proteins in human erythrocytes. Identification and characterization of polypeptides methylated in lysed cells. J Biol Chem. 1981 Mar 25;256(6):3067–3076. [PubMed] [Google Scholar]
- Tuong A., Maftouh M., Ponthus C., Whitechurch O., Roitsch C., Picard C. Characterization of the deamidated forms of recombinant hirudin. Biochemistry. 1992 Sep 8;31(35):8291–8299. doi: 10.1021/bi00150a024. [DOI] [PubMed] [Google Scholar]
- Vidgren J., Svensson L. A., Liljas A. Crystal structure of catechol O-methyltransferase. Nature. 1994 Mar 24;368(6469):354–358. doi: 10.1038/368354a0. [DOI] [PubMed] [Google Scholar]
- Voorter C. E., Roersma E. S., Bloemendal H., de Jong W. W. Age-dependent deamidation of chicken alpha A-crystallin. FEBS Lett. 1987 Sep 14;221(2):249–252. doi: 10.1016/0014-5793(87)80935-3. [DOI] [PubMed] [Google Scholar]
- Voorter C. E., de Haard-Hoekman W. A., van den Oetelaar P. J., Bloemendal H., de Jong W. W. Spontaneous peptide bond cleavage in aging alpha-crystallin through a succinimide intermediate. J Biol Chem. 1988 Dec 15;263(35):19020–19023. [PubMed] [Google Scholar]
- Wright H. T. Sequence and structure determinants of the nonenzymatic deamidation of asparagine and glutamine residues in proteins. Protein Eng. 1991 Feb;4(3):283–294. doi: 10.1093/protein/4.3.283. [DOI] [PubMed] [Google Scholar]
- Xie H., Clarke S. Methyl esterification of C-terminal leucine residues in cytosolic 36-kDa polypeptides of bovine brain. A novel eucaryotic protein carboxyl methylation reaction. J Biol Chem. 1993 Jun 25;268(18):13364–13371. [PubMed] [Google Scholar]
- Yamada H., Ueda T., Kuroki R., Fukumura T., Yasukochi T., Hirabayashi T., Fujita K., Imoto T. Isolation and characterization of 101-beta-lysozyme that possesses the beta-aspartyl sequence at aspartic acid-101. Biochemistry. 1985 Dec 31;24(27):7953–7959. doi: 10.1021/bi00348a017. [DOI] [PubMed] [Google Scholar]
- Yuan P. M., Talent J. M., Gracy R. W. Molecular basis for the accumulation of acidic isozymes of triosephosphate isomerase on aging. Mech Ageing Dev. 1981 Oct;17(2):151–162. doi: 10.1016/0047-6374(81)90081-6. [DOI] [PubMed] [Google Scholar]
- Zappia V., Zydek-Cwick R., Schlenk F. The specificity of S-adenosylmethionine derivatives in methyl transfer reactions. J Biol Chem. 1969 Aug 25;244(16):4499–4509. [PubMed] [Google Scholar]