Abstract
With more than 130 clinical trials and 8 approved gene therapy products, adeno-associated virus (AAV) stands as one of the most popular vehicles to deliver therapeutic DNA in vivo. One critical quality attribute analyzed in AAV batches is the presence of residual DNA, as it could pose genotoxic risks or induce immune responses. Surprisingly, the presence of small cell-derived RNAs, such as microRNAs (miRNAs), has not been investigated previously. In this study, we examined the presence of miRNAs in purified AAV batches produced in mammalian or in insect cells. Our findings revealed that miRNAs were present in all batches, regardless of the production cell line or capsid serotype (2 and 8). Quantitative assays indicated that miRNAs were co-purified with the recombinant AAV particles in a proportion correlated with their abundance in the production cells. The level of residual miRNAs was reduced via an immunoaffinity chromatography purification process including a tangential flow filtration step or by RNase treatment, suggesting that most miRNA contaminants are likely non-encapsidated. In summary, we demonstrate, for the first time, that miRNAs are co-purified with AAV particles. Further investigations are required to determine whether these miRNAs could interfere with the safety or efficacy of AAV-mediated gene therapy.
Keywords: gene therapy, AAV vectors, bioproduction, quality control, residual nucleic acids, microRNA, HEK293 mammalian cells, Sf9 insect cells
Graphical abstract
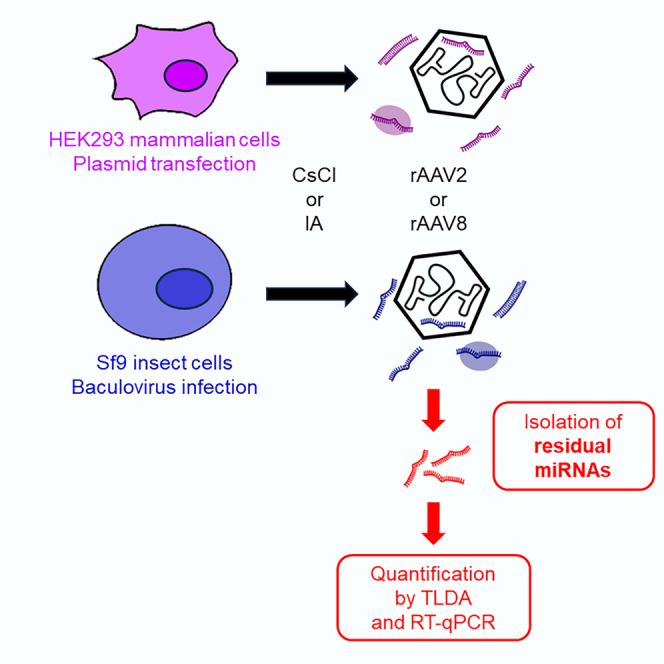
The presence of residual microRNAs in purified rAAV batches was investigated. MicroRNAs contaminate AAV vectors regardless of production cell platform or serotype. Immunoaffinity purification and RNase treatment are effective in reducing their abundance. Further research is necessary to assess the impact of miRNA contaminants on AAV vectors safety and efficacy.
Introduction
Advanced therapy medicinal products (ATMPs) have revolutionized medicine, providing curative treatments for previously incurable diseases such as genetic disorders. Based on engineering of nucleic acids, viruses, cells, or tissues, ATMPs are the fruit of decades of research and development. The corollary is that these new drugs are more complex to characterize compared with chemicals or recombinant proteins. One of the most popular viruses used for human gene therapy is the adeno-associated virus (AAV). Long regarded as poorly immunogenic, recent severe adverse events and deaths in clinical trials have raised concerns about immune response to these products.1,2,3 Despite the market authorization of eight AAV-derived drugs worldwide,4 greater efforts are needed regarding the characterization of AAV vectors. A recombinant AAV (rAAV) contains a single-stranded therapeutic DNA molecule protected by an icosahedral capsid. Either produced in mammalian or in insect cells, purified rAAV can contain various process- and product-related impurities.5 Among those, nucleic acids can come from the cell line (host cell DNA), the vector or helper plasmids, the recombinant baculovirus, or the helper viruses used for AAV production. The presence of residual DNA poses risks of oncogenicity and immunogenicity.6 Therefore, DNA impurities must be specifically identified, quantified, and minimized.7,8 While residual DNA external to the capsids can be more easily removed through DNase treatment9 or alternative purification steps, unwanted encapsidated DNA is more challenging to eliminate. Remarkably, the presence of small cell-derived RNAs, such as microRNAs (miRNAs), has never been investigated and no recommendation currently exists for controlling miRNA in rAAV products.10 miRNAs are small non-coding RNA (ncRNA) molecules of 20–25 nucleotides that trigger translational repression and mRNA degradation. Involved in various biological processes, they are transferable from cell to cell and are highly stable, especially when associated with an Argonaute (Ago) protein. miRNA expression has been found to be dysregulated in cancer cells, with certain miRNAs, known as oncomirs, linked to specific oncogenic events. The most used cell line for AAV vector production is HEK293, a human embryonic kidney cell line transformed with human adenovirus serotype 5 sheared DNA. Like other immortalized cells, HEK293 have been shown to exhibit dysregulated and specific miRNA activity profiles.11 However, whether AAV vectors produced in HEK293 cells are contaminated with cellular miRNAs remains unknown. In this study, research-grade AAV vector batches produced in mammalian cells (HEK293) or in insect cells (Spodoptera frugiperda, Sf9) were analyzed for the presence of miRNAs by reverse-transcription qPCR (RT-qPCR). Our findings indicate that miRNAs are enriched in rAAV batches compared with other small RNAs (sRNAs) and are primarily external to the AAV capsids.
Results
Residual miRNAs are present and enriched compared with other sRNAs in purified AAV vector batches
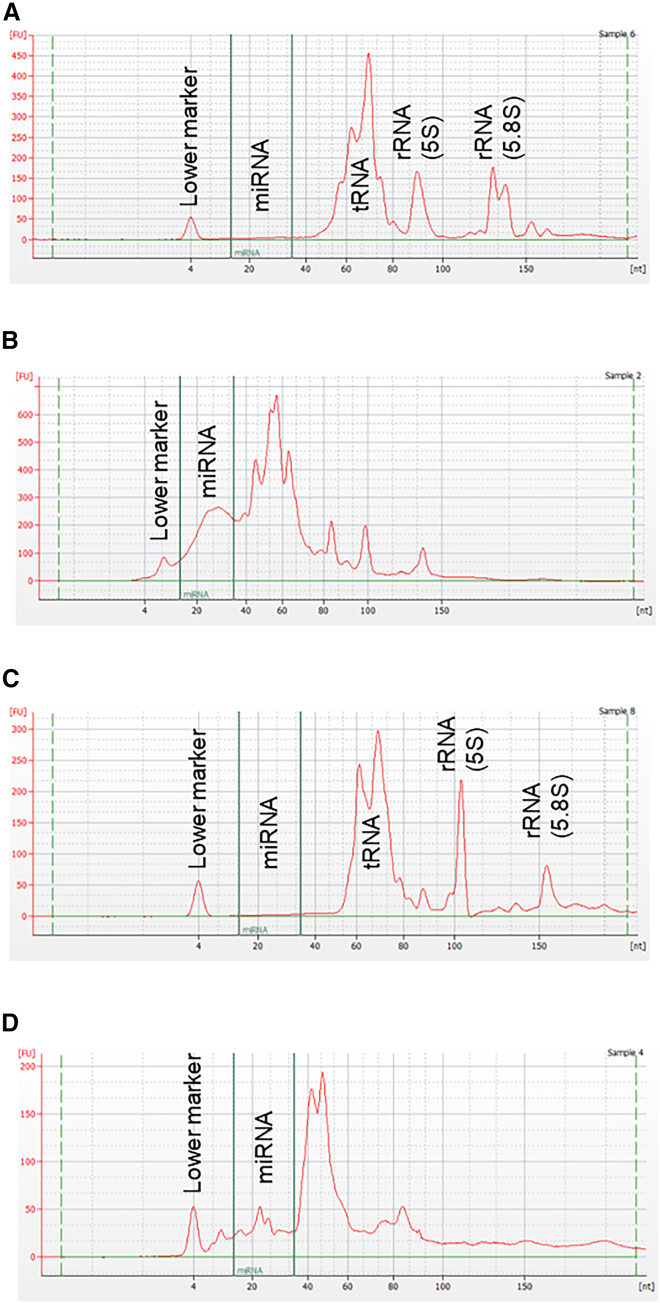
The presence of residual miRNA was investigated in AAV8 research-grade batches produced in either HEK293 mammalian cells or Sf9 insect cells. To account for variability introduced by the purification process, AAV vectors purified by CsCl-gradient ultracentrifugation (CsCl) or immunoaffinity chromatography (IA) were analyzed. After total RNA extraction, total miRNA concentration was measured by fluorescence using the Qubit microRNA assay kit. The concentration of miRNA in purified AAV8 batches ranges from 0.22 to 2.67 ng/μL (n = 4) for the HEK293 platform and from 2.26 to 5.95 ng/μL in the Sf9 platform (n = 3) (Table S1) which corresponds to 0.8 to 9.6 ng of residual miRNA per vector dose of 1 × 1010 vector genomes (vg). To further characterize the residual content of sRNA, automated electrophoresis was realized on an Agilent Small RNA chip. sRNAs include (1) the housekeeping transfer RNA (tRNA) of 76–90 nucleotides in length and the 5S and 5.8S ribosomal RNA (rRNA) of 120–121 and 156 nucleotides, respectively, as well as (2) the regulatory ncRNA, including miRNAs of 26–31 bases in size. The sRNA profile of HEK293 cells (Figure 1A) or Sf9 cells (Figure 1C) was compared with the electropherograms obtained for AAV vectors (Figures 1B and 1D). rAAV batches contain miRNA, tRNA, and rRNA. miRNAs constitute 18.0% to 59.5% of sRNA in rAAV, whereas miRNAs represent 3.6% to 16.7% of cellular sRNA content (Table 1). In conclusion, miRNAs were co-purified with AAV vectors and were found at quantifiable amount in all batches analyzed. Since miRNAs are more stable than other sRNAs, it could explain the fact that they are overrepresented in final stocks and resist better the purification process.
Figure 1.
Small RNA content of AAV vector batches
Total RNA was extracted from HEK293 mammalian cells (A), HEK293-derived rAAV (batch 17) (B), Sf9 insect cells (C), and Sf9-derived rAAV (batch 21) (D). AAV8-GFP vectors (shown in B and D) were purified by CsCl density gradient ultracentrifugation. Electropherograms were obtained after capillary electrophoresis on an Agilent Small RNA chip (6–150 nucleotides) with the miRNA region set to the range 15–35 nucleotides (area between the two green lines). miRNA, microRNA; tRNA, transfer RNA; rRNA, ribosomal RNA. The scale of the x axis is displayed in nucleotides [nt], and the y axis represents fluorescence units [FU].
Table 1.
Proportion of miRNA among sRNAs
| Cell platform | Sample | miRNA/sRNA (%) |
|---|---|---|
| HEK293 | cells (n = 5) | 4.3–16.7 |
| rAAV (n = 6) | 18.0–59.5 | |
| Sf9 | cells (n = 4) | 3.6–9.6 |
| rAAV (n = 5) | 24.5–36.8 |
AAV-CMV-eGFP batches were produced in HEK293 and Sf9 cells at small scale (400 mL to 1 L) and purified by double CsCl gradient ultracentrifugation. The percentage of miRNAs (15–35 nucleotides) to sRNAs (0–280 nucleotides) was determined after resolution by automated electrophoresis on an Agilent Small RNA chip.
Quantification of selected miRNA in AAV vectors produced in HEK293 cells using RT-qPCR
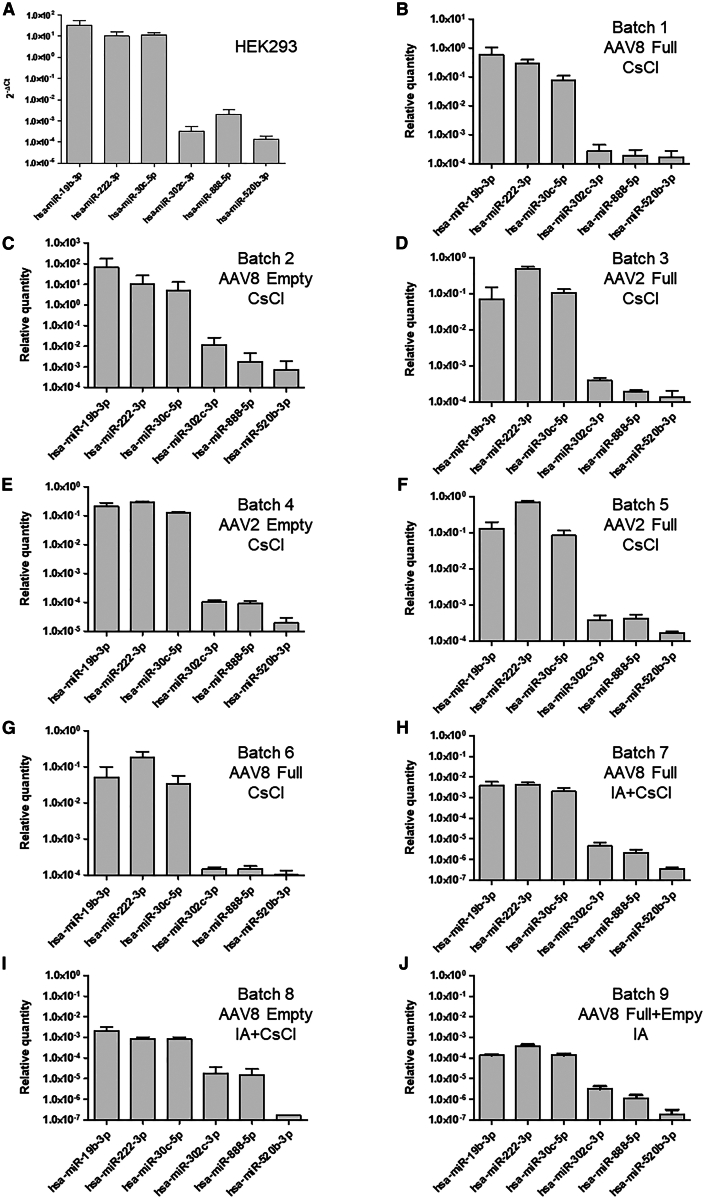
To examine the origin of contaminating miRNA and the impact of purification parameters on their residual level, we selected six miRNAs that are expressed in mammalian cells12,13 (Table S2). Three miRNAs are abundant in HEK293 producing cells: hsa-miR-19b-3p, hsa-miR-222-3p, and hsa-miR-30c-5p. Three miRNAs are expressed at low level in HEK293: hsa-miR-302c-3p, hsa-miR-888-5p, and hsa-miR-520b-3p. For more biological relevance, the selection was additionally made on the following criteria: one miRNA per cluster, the available primers are specific to the miRNA, the miRNA is expressed in a broad spectrum of tissues and involved in oncogenesis or cell proliferation. Each miRNA was quantified by RT-qPCR in HEK293 cells and in nine rAAV batches purified by CsCl (Figure 2). Titers of full AAV vectors purified by CsCl or IA range from 3.8 × 1011 to 2.6 × 1013 vg/mL (Table S3), with vector purity between 73.7% and 100% (Figure S1). The relative miRNA quantity was determined using the ΔCt method thanks to the Cel-miR-39 spike-in control (SIC). To compare batches, data were normalized to the culture volume and the final volume of the purified lot. Overall, the miRNA expression profile in rAAV correlates with those of HEK293. Noteworthy, miR-19b, a miRNA belonging to the oncomiR-1 cluster, is one of the most represented miRNAs found in HEK293-derived vectors.
Figure 2.
Quantification of miRNAs in HEK293-derived AAV vector batches
MiRNAs were quantified by RT-qPCR in HEK293 cell extract (A) and in AAV vector batches produced in HEK293 mammalian cells (B–J). AAV8 full (B) and empty (C) particles were purified by CsCl, as well as AAV2 full (D) and empty (E) particles. Quantification was performed from duplicate lots of full AAV2 (F) and full AAV8 (G) purified by CsCl. AAV8 full (H) and empty (I) particles were purified from the same bulk by IA followed by CsCl gradient ultracentrifugation. Batch 9 corresponds to an AAV8 purified by IA (J). The same cassette (cytomegalovirus [CMV] promoter and enhanced green fluorescent protein [eGFP] transgene) was used for all AAV vectors. MicroRNAs were extracted in triplicate and quantified in duplicate by SYBR Green-based RT-qPCR. The relative quantity of each miRNA was determined using the ΔCt method. For AAV, data were normalized to account for culture and batch volumes.
To determine if the vector genome had an impact on the degree of residual miRNA, empty and full fractions of a same bulk were purified by CsCl density gradient ultracentrifugation. The overall profiles of full and empty capsids are similar (Figure 2), except for empty AAV8 (batch 2), which presents a higher level of residual miRNA. Moreover, we analyzed the impact of the AAV serotype in the quantification of residual miRNAs and found that the relative quantity of miRNA was equivalent for AAV2 (batches 3 and 5) and AAV8 (batches 1 and 6), showing a low lot-to-lot variability. Indeed, the profiles of the six selected miRNAs were well correlated (Table S4), irrespective of the vector type. In conclusion, the amount of miRNA is independent of the particle type (full vs. empty) and capsid serotype (2 vs. 8).
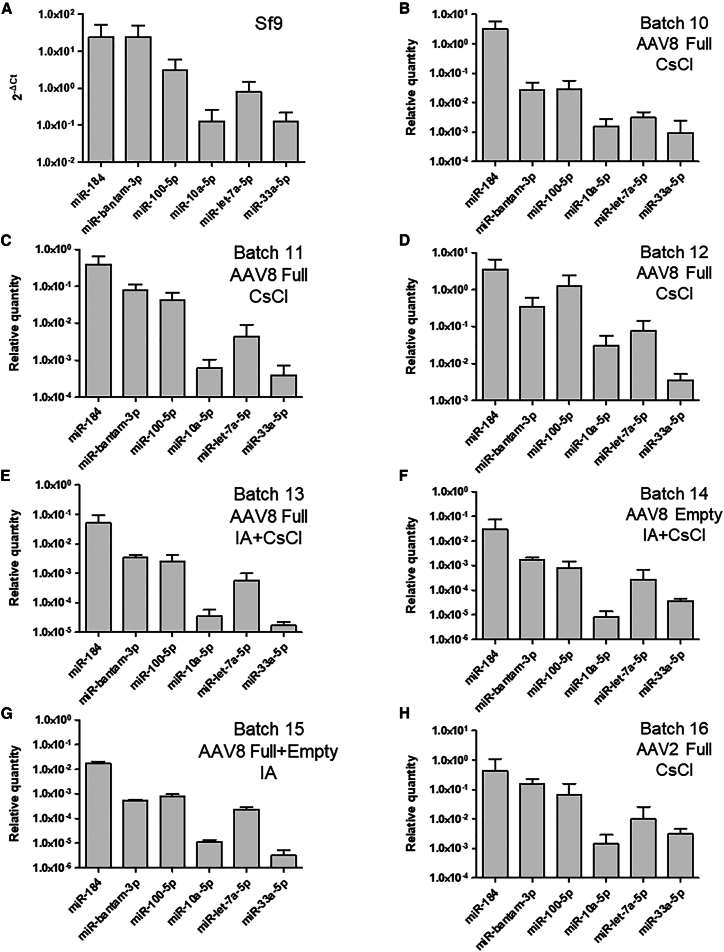
Quantification of selected miRNA in AAV vectors batches produced in Sf9 cells
Akin to the study performed with vectors derived from HEK293 cells, for Sf9-derived vectors, six miRNAs expressed in Bombyx mori caterpillar were selected for RT-qPCR. Based on Mehrabadi et al.’s publication about the microRNAome of Spodoptera frugiperda cells,14 we selected three highly expressed and three weakly expressed miRNAs in Sf insect cells. Referring to miRBase, these miRNAs show a perfect homology to human miRNAs with a conserved preferred strand, except for miR-10a with a 1 base shift compared with human miR and miR-bantam, which lacks a human counterpart. Of note, miR-bantam and miR-184 represent around 65% of all the miRNAs found in Sf9 cells.14 The different batches analyzed by RT-qPCR are listed in Table S3. Titers of full AAV vectors purified by CsCl or IA range from 1.8 × 1012 to 3.2 × 1013 vg/mL (Table S3), with vector purity between 56.1% and 84.7% (Figure S2). Again, miRNA abundance in rAAV batches correlates with the proportion of each miRNA in Sf9 cells (Figure 3A). The capsid serotype, purification process, and particle type do not modify the general miRNA profile as shown by the Pearson correlation coefficient R2 ≥ 0.93 (Table S5).
Figure 3.
Quantification of miRNAs in Sf9-derived AAV vector batches
MiRNAs were quantified by RT-qPCR from Sf9 cell extract (A) and in rAAV produced in Sf9 insect cells (B–H). Three batches of AAV8-GFP (B–D) and one batch of AAV2-GFP (H) purified by CsCl were tested. AAV8 full (E) and empty (F) particles were purified from the same bulk by IA followed by CsCl gradient ultracentrifugation. Batch 15 corresponds to an AAV8 purified by IA (G). The same cassette (CMV-eGFP) was used for all AAV vectors. MicroRNAs were extracted in triplicate and quantified in duplicate by SYBR Green-based RT-qPCR. The relative quantity of each miRNA was calculated using the ΔCt method. For AAV, data were normalized to account for culture and batch volumes.
Residual miRNAs are representative of the cellular miRNA content
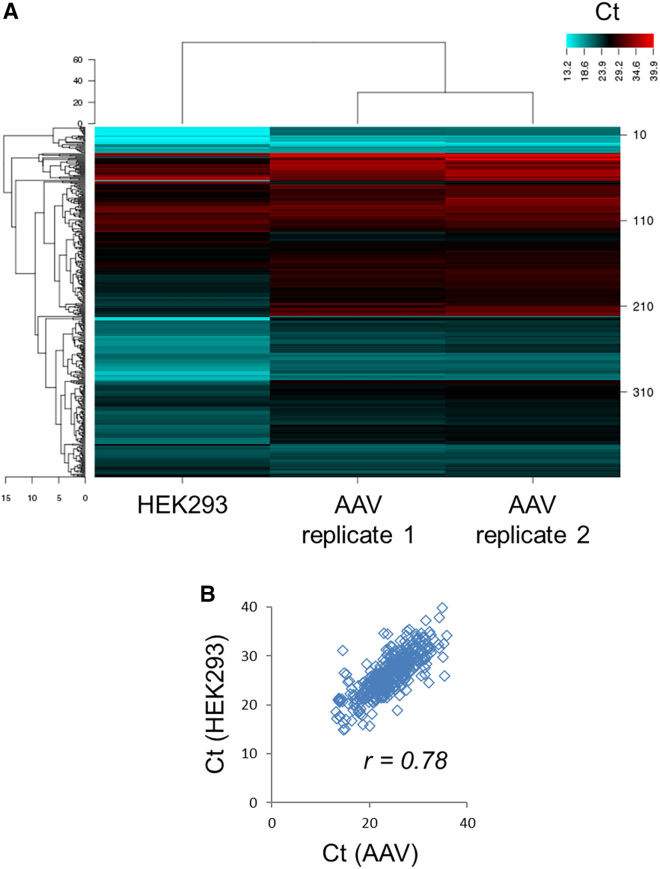
To confirm the correlation between miRNA profiles in rAAV and in producing cells, a TaqMan low-density array (TLDA) analysis was realized for AAV8 vectors, allowing a precise quantification of the 754 human miRNAs (Figure 4A). The results are in accordance with RT-qPCR data. Among the top ten more expressed miRNAs in HEK293 cells, four were previously selected for RT-qPCR analysis, which is in good agreement with our selection criteria (Table 2). With an R2 of 0.78 (Figure 4B), it demonstrates that the AAV miRNA profile correlates with miRNA abundance in the producing cell line.
Figure 4.
Comparison of miRNA content in HEK293-producing cells and AAV vector by TLDA
Human miRNAs were quantified in HEK293 cells and an AAV8 vector batch (n = 2). AAV particles were purified by CsCl gradient ultracentrifugation. Results are represented as a heatmap (A) and a Ct correlation graph (B).
Table 2.
List of the most abundant miRNAs expressed in HEK293 cells and quantification in derived AAV batch as determined by TLDA
| Name | HEK293 Ct |
AAV8 (n = 2) Ct |
|---|---|---|
| hsa-miR-17 | 12.3 | 15.9 |
| hsa-miR-106a | 12.8 | 16.4 |
| hsa-miR-19ba | 12.8 | 16.7 |
| hsa-miR-20a | 13.7 | 17.3 |
| hsa-miR-520ba | 14.9 | 15.1 |
| hsa-miR-222a | 15.1 | 17.1 |
| hsa-miR-302ca | 15.1 | 15.1 |
| hsa-miR-221 | 16.4 | 18.6 |
| hsa-miR-34a | 18.1 | 18.7 |
| hsa-miR-520f | 18.3 | 15.6 |
miRNA selected for RT-qPCR.
In cells, miRNAs are often associated with Ago proteins. Ago-2 was detected by western blot in a unique AAV batch that corresponds to AAV8 empty capsids (Figure S3, lane 4). The presence of Ago-2 proteins is related to a poor purity of the empty CsCl fraction. Indeed, miRNA quantity was approximatively 2 logs higher in the empty fraction than in the full fraction purified from the same bulk (Figures 2B and 2C, batch 1 compared with batch 2). Furthermore, the percentage of purity of batch 2 is low (76%) with a band above VP1 on the SDS-PAGE gel (Figure S1, lane 5).
MicroRNAs are also known to be selectively packaged, secreted, and transferred between cells in exosomes. No exosome was detected by an anti-CD63 western blot in the purified AAV batches (data not shown), suggesting that miRNAs are not embedded into exosomes.
A significant proportion of miRNAs is external to AAV capsids
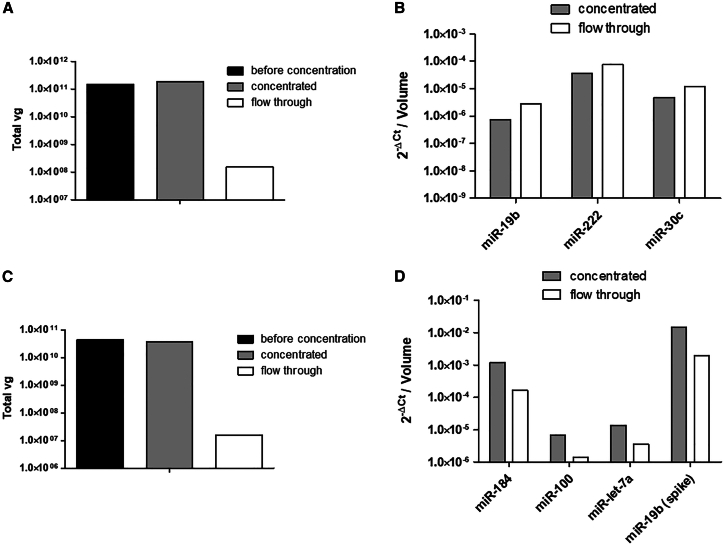
A filtration assay was developed to determine if residual RNAs are encapsidated into the AAV particles or are external to the capsids (Figure 5). AAV samples were loaded onto a centrifugal filter of 100 kDa MWCO usually used to concentrate AAV vectors. The rationale for the assay is the assumption that miRNA encapsidated into the capsids will be retained in the concentrated (C) fraction with AAV particles, while those external to the capsid will go in the flowthrough (FT) fraction. The filter capacity to retain AAV particles was confirmed by qPCR showing the same total vector genome copy number before and after concentration (Figures 5A and 5C). The three most abundant miRNAs were then quantified by RT-qPCR in the C and the FT fractions. For the AAV8 batch produced in HEK293, miRNA concentrations in FT represent 68%–80% of total residual miRNA (Figure 5B). Taking into consideration that FT corresponds to 61% of the total volume (FT + C) after centrifugation, these data are consistent with a passive filtration of free miRNA. For Sf9-derived AAV8, miRNA concentrations in FT range between 12% and 21% of the total residual miRNA (Figure 5D). The FT fraction volume corresponding to 51% of the total volume, a substantial proportion of miRNA was retained in the C fraction, either as free or encapsidated miRNA. To better study miRNA distribution, a synthetic hsa-miR-19b mimic was spiked in the Sf9-derived AAV sample before concentration. Considering that miR-19b is not expressed in Sf9 cells, it was used as a control of free miRNA (external to the capsids). Like Sf9 endogenous miRNAs, a percentage of 11% of the spiked miR-19b was distributed in the FT, suggesting that a significant proportion of residual miRNAs is outside AAV capsids.
Figure 5.
Filtration assay on Amicon centrifugal filters
AAV8-GFP were produced in HEK293 (A and B) or in Sf9 cells (C and D) and purified by the CsCl gradient method. AAV vectors were loaded on a centrifugal filter of 100 kDa MWCO that does not allow the AAV particles to pass through. A synthetic miR-19 was spiked before filtration in the Sf9-derived AAV sample, as control of free miRNA. Vector genomes (A and C) were quantified by qPCR before concentration, and in the concentrated (C) and flowthrough (FT) fractions. The quantity of residual miRNAs (B and D) was quantified by RT-qPCR in the C and FT fractions after centrifugation.
Immunoaffinity chromatography process and RNase treatment reduce residual miRNA level
A critical issue of vector safety is determining whether miRNAs can be effectively removed from AAV batches. For both production platforms, vectors purified by IA seems to contain less miRNAs than CsCl-purified rAAV (batches 8 and 9 on Figure 2, batch 13 and 15 on Figure 3A). For a side-by-side comparison, AAV8 particles were purified from the same biomass either by IA or CsCl, and miRNAs were quantified by RT-qPCR. Using a process including an IA column followed by a tangential flow filtration (TFF), the amount of three miRNAs was reduced by approximatively 100-fold, and to a lower extend for miR222 and miR-30c (Figure S4). We concluded that the chromatography process (IA + TFF) allows a better clearance of residual miRNAs than ultracentrifugation on CsCl gradients, which is also in agreement with previous experiments showing that residual miRNAs are mainly non-encapsidated.
A complementary option to reduce residual miRNA level could be to add an RNase digestion step as part of the purification process. Widely used for the removal of RNA from proteins and DNA preparations, RNase A and RNase T1 were tested on an AAV8-GFP lot produced in Sf9 cells and purified on density gradients. These two endonucleases hydrolyze single-stranded RNA with different sequence specificity. RNase A cleaves RNA after pyrimidine bases, whereas RNase T1 specifically degrades RNA at G residues. The RNase treatment was performed before capsid denaturation and miRNA extraction. The quantity of miR-184, abundant in Sf9, and of miR-19b mimic spike was checked by RT-qPCR before and after RNase digestion. Both miRNAs harbor several C, U, and G nucleotides (Table S2). A combined effect was observed for the control miR-19b level with a drastic drop of more than 5 logs (Figure 6). A treatment with RNase cocktail reduced the level of residual miRNA-184 by 2 logs. In conclusion, we established that IA purification and RNase treatment can be efficient in limiting residual miRNA levels in AAV batches.
Figure 6.
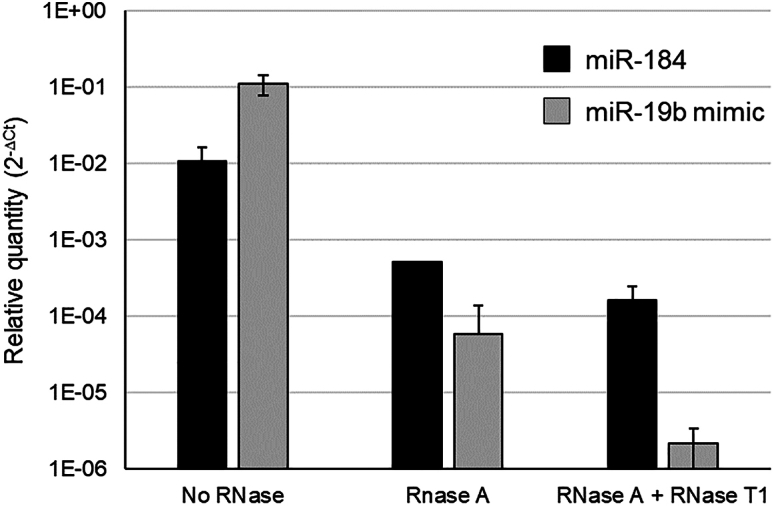
Removal of miRNA by RNase pretreatment of rAAV batches
RNase digestion was realized in duplicate on a CsCl-purified AAV8-GFP batch produced in Sf9 cells. RT-qPCR was performed targeting miR-184 (black) and miR-19b mimic (gray) that was spiked before RNase treatment. The miRNA relative quantity was calculated using the SIC by the ΔCt method for 200 μL of AAV.
Discussion
The purity of AAV vectors is paramount for both gene transfer efficiency and patient safety. While monitoring residual host cell DNA is mandatory, investigation into residual RNA has been lacking. However, RNA could serve as an immunostimulatory molecule within the innate immune system. In this study, we aimed to investigate the presence of miRNAs, known to be more stable than long mRNA molecules. Our data revealed the presence of residual miRNAs in purified AAV preparations at concentrations below 6 ng/μL, and the proportion of each miRNA correlated with the expression level of the production cells. These findings were confirmed in vector batches produced in human cells (HEK293) and in insect cells (Sf9), indicating a general observation rather than a phenomenon exclusive to one system. Since miRNAs act as post-transcriptional regulators of mRNA targets, the primary risk associated with the inadvertent transfer of such sRNAs alongside AAV vectors is gene dysregulation. Among the miRNAs detected in AAV vector produced in the HEK293 cells, miR-19b15 and miR-22216 are two oncogenic miRNAs implicated in cell proliferation, miR-30c is involved in inflammation17 and miR-888 is upregulated in hepatocellular carcinoma.18,19 As miRNA expression profile vary depending on the production cell line,11 it may be worthwhile to extend our investigation to other AAV production platforms utilizing HEK293T or HeLa cells.
In our study, miRNA concentration was slightly higher in Sf9-derived AAV vectors than in those produced with the HEK293 platform (Table S1). Considering the species specificity of miRNAs, it can be speculated that miRNAs originating from Sf9 insect cells might have less homology to human transcripts, potentially resulting in lower activity in humans. In addition, baculovirus infection of insect cells leads to suppression of cellular miRNAs, except for miR-184 and miR-10,14 which could aid in reducing residual miRNA levels in the final AAV stocks. While baculoviruses derived from Autographa californica multiple nucleopolyhedrovirus (AcMNPV) are widely used for biomolecule production, the identification of the first baculovirus miRNA occurred only a decade ago.20 Five miRNAs are encoded by AcMNPV, playing multiple roles in virus infection.21 Based on the assumption that these miRNAs will not bind to human target transcripts, the presence of baculovirus miRNAs was not explored.
As a general principle, reducing the amount of residual miRNA in purified batches can mitigate the risk of inadvertent transfer and subsequent modification of gene expression in patients. Our results demonstrate that most residual miRNAs are external to AAV capsids and susceptible to RNase digestion and/or removal by purification methods such as chromatography and TFF. This study has examined research-grade vector stocks with varying degrees of purity. In contrast, for clinical studies, vector manufacturing protocols include more stringent purification steps that might contribute to reduce residual miRNAs. Indeed, purification of AAV vectors according to current good manufacturing practices involves treating the bulk harvest with nucleases capable of degrading DNA and RNA, as well as multiple steps of depth filtration, TFF, and sterile filtration. In addition, immunoaffinity columns are commonly used as a primary capture step, typically followed by ion-exchange chromatography as a polishing strategy.
In summary, our data demonstrate, for the first time, the presence of residual miRNAs in purified AAV batches. Although further investigations are necessary to determine whether these sRNAs can be transferred to targeted tissues and taken up by cells in vivo, it is prudent to explore this phenomenon on a broader scale and including the analysis of clinically relevant AAV batches.
Materials and Methods
Plasmid constructs and recombinant baculoviruses
AAV and helper plasmids used for the production of recombinant AAV2/2 and 2/8 in mammalian and insect cells are described by Penaud-Budloo et al.22 In brief, the donor plasmid pFB-GFP contains a reporter cassette of 3.3 kbp composed of the human cytomegalovirus promoter, the enhanced green fluorescent protein (eGFP) reporter gene, and the 3′ untranslated region of the human hemoglobin b gene, flanked by pSub201-derived flop ITRs. The plasmid pFB-GFP was verified by Sanger sequencing and subsequently used either for transfection of HEK293 cells or for the generation of the baculovirus expression vector BEV-GFP as described in Penaud-Budloo et al.22 The stability of the BEV stocks was validated trough ten serial passages by real-time PCR and the identity of the P2 stocks was verified by Sanger sequencing.23 The infectious unit (IU) titer or the recombinant baculovirus batches was determined by cell size assay.23
AAV vector production, purification, and quality control
To produce rAAV2/2 and AAV2/8 in mammalian cells, HEK293 cells were cotransfected with the pDP2 or pDP8 helper plasmid, respectively, and the pFB-GFP vector plasmid. More precisely, HEK293 cells at 70%–80% confluence were cotransfected with 550 μg of helper plasmid and 275 μg of vector plasmid per CellSTACK 5 culture chambers (Corning Life Sciences) following the calcium phosphate precipitation method. Six hours post-transfection, the culture medium was replaced by DMEM with 1% penicillin/streptomycin without FBS. For AAV2 vectors, cells and culture supernatant were recovered 72 h after transfection and treated with 1% Triton X-100 (Sigma-Aldrich) for 1 h at 37°C and 1 U/mL of Benzonase (Merck) for an additional 1 h at 37°C. For AAV8 vectors, the supernatant was recovered 96 h after transfection. AAV2 and AAV8 particles were polyethylene glycol precipitated overnight. After centrifugation, the pellet was resuspended in TBS and treated with 25 U/mL of Benzonase for 1 h at 37°C.
For AAV vector production in insect cells, Spodoptera frugiperda Sf9 cells from Thermo Fisher Scientific were grown at 27°C in Sf-900 III SFM in spinner flasks or 2-L bioreactors (Sartorius Biostat B). Sf9 cells were infected at a density of 1 × 106 cells/mL with two baculoviruses: the recombinant BEV-GFP harboring the AAV vector genome and the BEV-rep2cap2 or BEV-rep2Cap8 for serotype 2 or 8 production, respectively. Each BEV was added to the cells at a multiplicity of infection of 1 IU per baculovirus. 96 h after infection, cells were lysed by the addition of 0.5% Triton X-100 (Sigma-Aldrich) for 2.5 h at 27°C. The crude bulk was clarified by centrifugation for 5 min at 1,000 × g or by depth filtration using Opticap disposable capsule filters (Merck Millipore) if the bulk volume was ≥ 2 L.
After clarification, recombinant AAV particles were purified either by (1) double CsCl density gradient ultracentrifugation as described in Ayuso et al.24 (CsCl), (2) IA, or (3) IA followed by one round of CsCl gradient ultracentrifugation (IA + CsCl). Full and empty particles were recovered separately from the first CsCl gradient. The IA process consists of a purification step on a POROS CaptureSelect AAV8 high-performance affinity resin (Thermo Fisher Scientific) followed by TFF using a MidiKros 100-kDa mPES hollow fiber filter (Repligen). AAV vectors were finally formulated in Dulbecco’s phosphate-buffered saline (Lonza) containing Ca2+ and Mg2+, and 0.001% Pluronic F-68 (Sigma-Aldrich) for IA-purified batches.
The vg titer of the purified rAAV batches was determined by real-time PCR targeting the GFP transgene. To remove DNA contaminants external to the capsids, 3 μL of rAAV stock was pretreated with 20 U of DNase I (Roche) in a total volume of 200 μL of DNase reaction buffer (13 mM Tris [pH 7.5], 0.12 mM CaCl2, 5 mM MgCl2) for 45 min at 37°C. DNA was then extracted using the High Pure Viral Nucleic Acid kit (Roche) and the vg copy number was determined using the following primers and probe: GFP For 5′-AGTCCGCCCTGAGCAAAGA-3′, GFP Rev 5′-GCGGTCACGAACTCCAGC-3′, and GFP probe 5′-FAM-CAACGAGAAGCGCGATCACATGGTC-TAMRA-3′. Total particle titers were determined by ELISA using the AAV8 titration enzyme-linked immunosorbent assay kit (Progen Biotechnik) following the manufacturer’s instructions.
Vector purity was evaluated by SDS-PAGE followed by silver staining using the PlusOne silver staining kit (GE Healthcare Life Sciences). The percentage of AAV VP proteins to total proteins was determined with GeneTools software (Syngene).
Finally, the proportion of empty and full particles was calculated from analytical ultracentrifugation data. Sedimentation velocity experiments were performed as described by Saleun et al.25
miRNA isolation
Total RNA, including miRNAs, was extracted from HEK293 and Sf9 cells in triplicate using the miRNeasy mini kit (QIAGEN). In brief, a pellet of 1 × 106 cells was resuspended in 20 μL of phosphate-buffered saline 1× and in 700 μL of QIAzol by vortexing for 1 min. After 5 min incubation at room temperature, 3.5 μL of miRNeasy serum/plasma SIC (QIAGEN) that was previously diluted at 1.6 × 108 copies/μL in RNase-free water was added to the mix. The synthetic SIC corresponds to a C. elegans miR-39 miRNA mimic and was used for RT-qPCR normalization. After homogenization, 140 μL of chloroform was added and extraction steps were realized following the manual instructions. Elution from an RNeasy Mini Spin column was performed with 14 μL of RNase-free water and extracted RNA was stored at −80°C before analysis.
For AAV vectors, miRNAs were extracted in triplicate using the miRNeasy Serum/Plasma kit (QIAGEN) following the manufacturer’s instructions. In brief, 1 mL of QIAzol were added to 200 μL of purified AAV. After vortexing and 5 min incubation at room temperature, 3.5 μL of miRNeasy serum/plasma SIC (QIAGEN) diluted at 1.6 × 108 copies/μL were added to the mix. After homogenization by inversion, 200 μL of chloroform was added and extraction steps were realized using the RNeasy MinElute Spin columns following the manual instructions. The elution step was realized in 14 μL of RNase-free water. Extracted miRNAs were stored at −80°C before subsequent steps.
Quantification and characterization of total residual miRNAs
After miRNA extraction, miRNA concentration of each AAV sample was determined from three biological replicates and two measurements per replicate with the Qubit 4 Fluorometer (Thermo Fisher Scientific) using the Qubit miRNA assay kit (Thermo Fisher Scientific). In addition, the sRNA profile was visualized by automated electrophoresis on the 2100 Bioanalyzer instrument (Agilent) using the Small RNA chip (Agilent). Cellular extracts were diluted at 40 ng/μL before analysis. Electropherograms were analyzed with the 2100 Expert software (Agilent). The percentage of miRNAs (15–35 nucleotides) to sRNAs (0–280 nucleotides) was automatically calculated from concentrations in pg/μL.
Quantification of selected miRNAs by RT-qPCR
Selected miRNAs listed in Table S2 were quantified by RT-qPCR. Reverse transcription of mature miRNAs was realized from 10 μL of RNA extract using the miScript II RT kit with HiSpec buffer (QIAGEN). Complementary DNA was generated following the manual instructions after 1 h incubation at 37°C and 5 min inactivation at 95°C in the GeneAmp PCR System 9700 thermocycler (Thermo Fisher Scientific). Negative controls were included by replacing the MiScript RT mix by RNase-free water. Complementary DNAs were finally diluted 1:10 in RNase-free H2O and stored at −20°C.
Each miRNA was then quantified from the cDNA, in duplicate, using the miScript SYBR Green PCR kit (QIAGEN), the miRNA-specific forward primer (miScript Primer Assay, QIAGEN) and the reverse primer (miScript Universal Primer, QIAGEN) targeting the universal tag of the oligo-dT primer. Cycling conditions used for real-time PCR on the StepOne Plus real-time PCR system (Thermo Fisher Scientific, Applied Biosystems) were in accordance with the kit technical specifications. Before analysis, the ΔRn threshold was established at 0.2 for all qPCR assays. Data were normalized to the Cel-miR-39 SIC and the relative quantity of each miRNA was determined using the ΔCt method with the following formulae:
For cells:
For AAV:
TLDA analysis
Total RNA was reverse transcribed with the Megaplex Primer Pools A and B, or A only (human version 3), and miRNAs were quantified with the TaqMan Array Human MicroRNA Cards A and B set v.3.0, or Card A only (Thermo Fisher Scientific) on the 7900HT Real-Time PCR System (Thermo Fisher Scientific, Applied Biosystems) according to the manufacturer’s guidelines. Quantification cycle (Cq) values were calculated with the Applied Biosystems SDS software v.2.3 by using automatic baseline with a threshold set at 0.1.
Filtration assays
Filtration assays were performed from CsCl-purified AAV8 produced either in HEK293 cells or Sf9 cells. Prior to filtration, 2 × 109 copies of a miR-19b mimic were spiked in 200 μL of Sf9-derived sample. An Amicon Ultra-4 centrifugal filter of 100 kDa MWCO (Millipore, Merck) was pre-wet with 2 mL dPBS and centrifuged for 2 min at 3,000 rpm at 20°C. The FT was discarded, 500 μL of AAV vectors was loaded onto the filter, and a short centrifugation was performed 30 s at 3,000 rpm at 20°C for particle concentration. The concentrated sample and the FT were recovered from the top and bottom of the tube, respectively. The three more abundant miRNAs were quantified by RT-qPCR from 200 μL of each fraction (before concentration, concentrated, and FT). Data were normalized by the volume of each fraction. In parallel, the copy number of vector genomes was determined by qPCR from 10 μL of each fraction similarly to the quality control assay.
RNase digestion assay
For the RNase treatment of rAAV, the following mixture was prepared: 150 μL of AAV vector, 50 μL of RNase, 75 μL of RNase buffer (10 mM Tris-HCl, 5 mM MgCl2 [pH 7.5]) and 4.2 × 108 copies of a miR-19b mimic. For the “no RNase” condition, RNase was replaced with an equivalent volume of RNase buffer. For the “RNase A” condition, RNase A at 7,000 U/mL (QIAGEN) was used. For the “RNase A + RNase L” condition, Ambion RNase cocktail enzyme mix (Thermo Fisher Scientific) was added, comprising RNase A (500 U/mL) and RNase T1 (20,000 U/mL). After incubation 2 h at 37°C, QIAzol was added to the mixture, along with the Cel-miR-39 SIC (QIAGEN) for normalization. miRNAs were then isolated and quantified by RT-qPCR.
Statistical analysis
The Pearson correlation coefficient and Mann-Whitney U-test were carried out using GraphPad Prism 5 (GraphPad Software). The mean and the standard deviation of the biological and technical replicates were calculated and represented on the figures.
Data and code availability
RT-qPCR raw data of TLDA analyses were deposited in the Gene Expression Omnibus,26 an international public repository, under accession number GSE270691.
Acknowledgments
We would like to warmly thank the Vector Core of TaRGeT laboratory (Nantes, France) for AAV vector production, purification, and quality controls. We are also grateful to Adrien Léger for giving his scientific input during the initiation of the study. Automated electrophoresis was performed at the Genomics and Core Facility of Nantes (GenoA, Nantes, France). This research was supported by the Région Pays de la Loire, the Centre hospitalier universitaire (CHU) de Nantes, Nantes Université, and the Institut National de la Santé et de la Recherche Médicale (INSERM).
Author contributions
M.P.-B. designed experiments, supervised the study, and wrote the manuscript. E.A. designed experiments, participated to the scientific supervision, reviewed the manuscript, and raised funds. O.A. raised funds and provided resources required for the study. Q.L. developed the RT-qPCR assays and realized experiments. L.J.-L. realized the TLDA analysis. E.L. analyzed the TLDA data, realized experiments, and performed bioinformatics analysis. S.P. performed miRNA quantification and designed the filtration experiment. F.B. designed and realized RNase tests. C.R. and V.B. supervised the purification process of AAV vectors. A.G.-D. performed experiments and bioinformatics analyses. J.-B.D. provided valuable scientific discussion.
Declaration of interests
M.P.-B. and E.A. are inventors of patents related to AAV gene therapy licensed to biopharma companies.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2024.101305.
Supplemental information
References
- 1.Muhuri M., Maeda Y., Ma H., Ram S., Fitzgerald K.A., Tai P.W., Gao G. Overcoming innate immune barriers that impede AAV gene therapy vectors. J. Clin. Invest. 2021;131 doi: 10.1172/JCI143780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa-Verdera H., Unzu C., Valeri E., Adriouch S., González Aseguinolaza G., Mingozzi F., Kajaste-Rudnitski A. Understanding and Tackling Immune Responses to Adeno-Associated Viral Vectors. Hum. Gene Ther. 2023;34:836–852. doi: 10.1089/hum.2023.119. [DOI] [PubMed] [Google Scholar]
- 3.Duan D. Lethal immunotoxicity in high-dose systemic AAV therapy. Mol. Ther. 2023;31:3123–3126. doi: 10.1016/j.ymthe.2023.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghauri M.S., Ou L. AAV Engineering for Improving Tropism to the Central Nervous System. Biology. 2023;12:186. doi: 10.3390/biology12020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Penaud-Budloo M., François A., Clément N., Ayuso E. Pharmacology of Recombinant Adeno-associated Virus Production. Mol. Ther. Methods Clin. Dev. 2018;8:166–180. doi: 10.1016/j.omtm.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brimble M.A., Winston S.M., Davidoff A.M. Stowaways in the cargo: Contaminating nucleic acids in rAAV preparations for gene therapy. Mol. Ther. 2023;31:2826–2838. doi: 10.1016/j.ymthe.2023.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gimpel A.L., Katsikis G., Sha S., Maloney A.J., Hong M.S., Nguyen T.N.T., Wolfrum J., Springs S.L., Sinskey A.J., Manalis S.R., et al. Analytical methods for process and product characterization of recombinant adeno-associated virus-based gene therapies. Mol. Ther. Methods Clin. Dev. 2021;20:740–754. doi: 10.1016/j.omtm.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright J.F. Product-Related Impurities in Clinical-Grade Recombinant AAV Vectors: Characterization and Risk Assessment. Biomedicines. 2014;2:80–97. doi: 10.3390/biomedicines2010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bucher K., Rodríguez-Bocanegra E., Wissinger B., Strasser T., Clark S.J., Birkenfeld A.L., Siegel-Axel D., Fischer M.D. Extra-viral DNA in adeno-associated viral vector preparations induces TLR9-dependent innate immune responses in human plasmacytoid dendritic cells. Sci. Rep. 2023;13:1890. doi: 10.1038/s41598-023-28830-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Recommendations for the evaluation of animal cell cultures as substrates for the manufacture of biological medicinal products and for the characterization of cell banks, Annex, 3, World Health Organization Technical Report Series No 978. https://www.who.int/publications/m/item/animal-cell-culture-trs-no-978-annex3
- 11.Tian W., Dong X., Liu X., Wang G., Dong Z., Shen W., Zheng G., Lu J., Chen J., Wang Y., et al. High-throughput functional microRNAs profiling by recombinant AAV-based microRNA sensor arrays. PLoS One. 2012;7 doi: 10.1371/journal.pone.0029551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panwar B., Omenn G.S., Guan Y. miRmine: a database of human miRNA expression profiles. Bioinforma. Oxf. Engl. 2017;33:1554–1560. doi: 10.1093/bioinformatics/btx019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehrabadi M., Hussain M., Asgari S. MicroRNAome of Spodoptera frugiperda cells (Sf9) and its alteration following baculovirus infection. J. Gen. Virol. 2013;94:1385–1397. doi: 10.1099/vir.0.051060-0. [DOI] [PubMed] [Google Scholar]
- 15.Olive V., Bennett M.J., Walker J.C., Ma C., Jiang I., Cordon-Cardo C., Li Q.-J., Lowe S.W., Hannon G.J., He L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang D., Sang Y., Sun T., Kong P., Zhang L., Dai Y., Cao Y., Tao Z., Liu W. Emerging roles and mechanisms of microRNA-222-3p in human cancer (Review) Int. J. Oncol. 2021;58:20. doi: 10.3892/ijo.2021.5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ceolotto G., Giannella A., Albiero M., Kuppusamy M., Radu C., Simioni P., Garlaschelli K., Baragetti A., Catapano A.L., Iori E., et al. miR-30c-5p regulates macrophage-mediated inflammation and pro-atherosclerosis pathways. Cardiovasc. Res. 2017;113:1627–1638. doi: 10.1093/cvr/cvx157. [DOI] [PubMed] [Google Scholar]
- 18.Li Y.-B., Sun F.-N., Ma X.-Y., Qu H., Yu Y. MiR-888 promotes cell migration and invasion of hepatocellular carcinoma by targeting SMAD4. Eur. Rev. Med. Pharmacol. Sci. 2019;23:2020–2027. doi: 10.26355/eurrev_201903_17241. [DOI] [PubMed] [Google Scholar]
- 19.Jiang Q., Wang Y., Hao Y., Juan L., Teng M., Zhang X., Li M., Wang G., Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J., Xing K., Xiong P., Liang H., Zhu M., Zhao J., Yu X., Ning X., Li R., Wang X. Identification of miRNAs encoded by Autographa californica nucleopolyhedrovirus. J. Gen. Virol. 2021;102:XX. doi: 10.1099/jgv.0.001510. [DOI] [PubMed] [Google Scholar]
- 21.Motta L.F., Cerrudo C.S., Belaich M.N. A Comprehensive Study of MicroRNA in Baculoviruses. Int. J. Mol. Sci. 2024;25:603. doi: 10.3390/ijms25010603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penaud-Budloo M., Lecomte E., Guy-Duché A., Saleun S., Roulet A., Lopez-Roques C., Tournaire B., Cogné B., Léger A., Blouin V., et al. Accurate Identification and Quantification of DNA Species by Next-Generation Sequencing in Adeno-Associated Viral Vectors Produced in Insect Cells. Hum. Gene Ther. Methods. 2017;28:148–162. doi: 10.1089/hgtb.2016.185. [DOI] [PubMed] [Google Scholar]
- 23.Jacob A., Brun L., Jiménez Gil P., Ménard L., Bouzelha M., Broucque F., Roblin A., Vandenberghe L.H., Adjali O., Robin C., et al. Homologous Recombination Offers Advantages over Transposition-Based Systems to Generate Recombinant Baculovirus for Adeno-Associated Viral Vector Production. Biotechnol. J. 2021;16 doi: 10.1002/biot.202000014. [DOI] [PubMed] [Google Scholar]
- 24.Ayuso E., Blouin V., Lock M., McGorray S., Leon X., Alvira M.R., Auricchio A., Bucher S., Chtarto A., Clark K.R., et al. Manufacturing and characterization of a recombinant adeno-associated virus type 8 reference standard material. Hum. Gene Ther. 2014;25:977–987. doi: 10.1089/hum.2014.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saleun S., Mas C., Le Roy A., Penaud-Budloo M., Adjali O., Blouin V., Ebel C. Analytical ultracentrifugation sedimentation velocity for the characterization of recombinant adeno-associated virus vectors sub-populations. Eur. Biophys. J. 2023;52:367–377. doi: 10.1007/s00249-023-01650-3. [DOI] [PubMed] [Google Scholar]
- 26.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RT-qPCR raw data of TLDA analyses were deposited in the Gene Expression Omnibus,26 an international public repository, under accession number GSE270691.