Abstract
Asteraceae, the largest family of angiosperms, has attracted widespread attention for its exceptional medicinal, horticultural, and ornamental value. However, researches on Asteraceae plants face challenges due to their intricate genetic background. With the continuous advancement of sequencing technology, a vast number of genomes and genetic resources from Asteraceae species have been accumulated. This has spurred a demand for comprehensive genomic analysis within this diverse plant group. To meet this need, we developed the Asteraceae Genomics Database (AGD; http://cbcb.cdutcm.edu.cn/AGD/). The AGD serves as a centralized and systematic resource, empowering researchers in various fields such as gene annotation, gene family analysis, evolutionary biology, and genetic breeding. AGD not only encompasses high-quality genomic sequences, and organelle genome data, but also provides a wide range of analytical tools, including BLAST, JBrowse, SSR Finder, HmmSearch, Heatmap, Primer3, PlantiSMASH, and CRISPRCasFinder. These tools enable users to conveniently query, analyze, and compare genomic information across various Asteraceae species. The establishment of AGD holds great significance in advancing Asteraceae genomics, promoting genetic breeding, and safeguarding biodiversity by providing researchers with a comprehensive and user-friendly genomics resource platform.
Keywords: Asteraceae, genome, taxonomy, analysis tools, Asteraceae Genome Database (AGD)
1. Introduction
Asteraceae, recognized as the largest family of angiosperms, is globally distributed and remarkably diverse. It encompasses over 1,600 genera and approximately 25,000 species (Shen et al., 2023), including notable members such as Chrysanthemum morifolium, Artemisia caruifolia, Helianthus annuus, and Carthamus tinctorius (Zhang and Elomaa, 2024). Chrysanthemum, a prominent perennial herbaceous plant within this family, holds a revered position among China’s top ten traditional flowers and is globally considered one of the four most preeminent cut flowers. Its geometrically regular inflorescences are visually appealing, contributing to the ornamental value of Asteraceae (Elomaa, 2019). In addition, the Asteraceae family holds important medical applications, significantly contributing to human health (Rolnik and Olas, 2021). Previous research has demonstrated that sesquiterpene lactones, naturally abundant in this family, possess anticancer potential (Li et al., 2020). Furthermore, Asteraceae can be employed as an in vitro antiplatelet agent and is utilized in diverse aspects of daily life, including cosmetics and food processing (Rolnik et al., 2022).
With the remarkable advancements in genome sequencing technology, substantial progress has been made in the genome research of various species, with much attention focused on Asteraceae in recent times. Particularly, Helianthus annuus (Badouin et al., 2017), C. morifolium (Song et al., 2023a), C. nankingense (Song et al., 2018), Mikania micrantha (Liu et al., 2020), Artemisia annua (Shen et al., 2018), and Artemisia argyi have all been extensively studied (Shen et al., 2018). Despite the numerous genomic studies conducted on various Asteraceae species, the genome sequences are distributed in different databases, lacking an integrated analysis platform and comprehensive databases that consolidate the vast amount of available information. Existing databases related to Asteraceae, including the Asteraceae genome size database (GSAD) (Garnatje et al., 2011), Asteraceae sequences database (Ventimiglia et al., 2023), burdock multi-omics database (Song et al., 2023b), and HeliantHOME (Bercovich et al., 2022). These databases do not systematically capture all the findings related to the Asteraceae genome. Such as GSAD only provides the function of querying the genome sizes of most Asteraceae species. Moreover, navigating through multiple platforms to obtain the required species data can be challenging and inconvenient. Therefore, developing a unique and comprehensive database, to provide researchers with a comprehensive platform for multi-omics research is crucial to consolidate and simplify access to Asteraceae genomic information.
In this work, we established the Asteraceae Genome Database (AGD), a comprehensive repository that integrates existing genome assembly and annotation data of representative Asteraceae species. We also regularly update the AGD to include new genomic data and research findings, ensure that AGD reflects the latest scientific advancements, and provide researchers with the most current information. We anticipate AGD evolving into a preeminent platform for the in-depth analyses of genomic data related to Asteraceae plants, streamlining access and interpretation of crucial information.
2. Database construction
2.1. Data retrieval
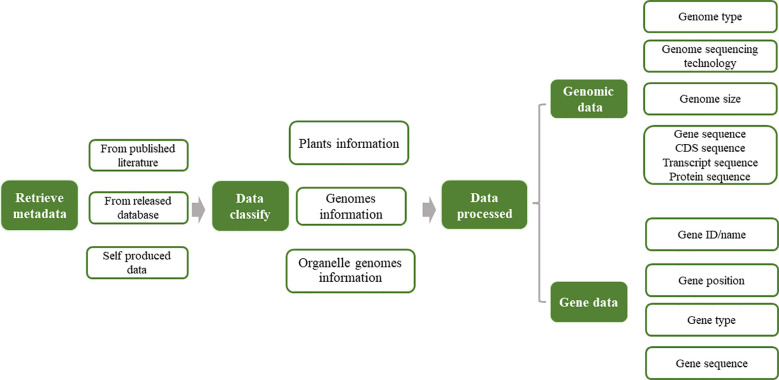
The complete omics data for Asteraceae were retrieved from various databases, including NCBI (National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/), 1 K-MPGD (1 K Medicinal Plant Genome Database, http://www.herbgenome.com/) (Su et al., 2022), GPGD (Global Pharmacopoeia Genome Database, http://www.gpgenome.com) (Liao et al., 2022a), CNCB (China National Center for Bioinformation, https://www.cncb.ac.cn/?lang=en) (CNCB-NGDC Members and Partners, 2023), GWH (Genome Warehouse, https://ngdc.cncb.ac.cn/gwh) (Chen et al., 2021), Published Plant Genomes (https://www.plabipd.de/plant_genomes_pa.ep), and GERDH (Gene Expression Regulation Database of Horticultural plants, https://dphdatabase.com) (Cheng et al., 2023). We utilized the common and scientific nomenclature for species identification, for example, ‘Sunflowers’ and ‘Helianthus annuus L’, respectively, to facilitate a comprehensive retrieval of omics data. We expanded our keyword set to include the genus name and associated taxonomic designations to ensure a comprehensive search strategy. Table 1 provides an overview of the extant genomic data available for the Asteraceae family. The AGD encompasses a diverse array of genomic data, including organelle and nuclear genomes. We employed the gffread tool (https://github.com/gpertea/gffread) to extract protein-coding, protein, and transcript sequences. These sequences were subsequently curated and integrated into our database. Figure 1 presents the analysis pipeline employed by AGD.
Table 1.
Species and genome data in Asteraceae.
| Species | Accession number | Assembly Level | Genome size | References |
|---|---|---|---|---|
| Arctium lappa | JAKOEK000000000 | Chromosome | 1.73 Gb | (Fan et al., 2022) |
| Carthamus tinctorius | GWHBJIR00000000 | Chromosome | 1.17 Gb | (Chen et al., 2023a) |
| Cynara cardunculus | SUB874020 | Chromosome | 1,084 Mb | (Scaglione et al., 2016) |
| Saussurea involucrata | SAMN36288184 | Chromosome | 2452 Mb | (Sun et al., 2023) |
| Silybum marianum | JAWIMA000000000 | Chromosome | 694.4 Mb | (Kim et al., 2024) |
| Ambrosia artemisiifolia | PRJNA967341 | Chromosome | 1.13 Gb | (Laforest et al., 2024) |
| Ambrosia trifida | PRJNA967341 | Chromosome | 2.02 Gb | (Laforest et al., 2024) |
| Artemisia argyi | PRJCA010808 | Chromosome | 3.89 Gb | (Chen et al., 2023b) |
| Helianthus annuus | MNCJ02000000 | Chromosome | 3.6 Gb | (Badouin et al., 2017) |
| Mikania micrantha | SZYD00000000 | Chromosome | 1.8 Gb | (Liu et al., 2020) |
| Lactuca sativa | PRJCA007442 | Chromosome | 2.6 Gb | (Shen et al., 2023) |
| Artemisia annua | – | Chromosome | 1.11 Gb | (Liao et al., 2022b) |
| Erigeron breviscapus | PRJNA525743 | Chromosome | 1.4 Gb | (He et al., 2021) |
| Bidens hawaiensis | SAMN18676211 | Chromosome | 6.67 Gb | (Bellinger et al., 2022) |
| Artemisia tridentata | SAMN24662005 | Chromosome | 4.2Gb | (Melton et al., 2022) |
| Chrysanthemum indicum | – | Chromosome | 3.11Gb | (Deng et al., 2024) |
| Chrysanthemum lavandulifolium | JAHFWF000000000 | Chromosome | 2.60 Gb | (Wen et al., 2022) |
| Chrysanthemum makinoi | JP131333 | Chromosome | 3.1 Gb | (Van Lieshout et al., 2022) |
| Chrysanthemum nankingense | – | Chromosome | 3.07 Gb | (Song et al., 2018) |
| Chrysanthemum seticuspe | GCA_019973895.1 | Chromosome | 3.05 Gb | (Nakano et al., 2021) |
| Chrysanthemum morifolium | PRJNA796762 PRJNA895586 |
Chromosome | 8.15 Gb | (Song et al., 2023a) |
| Conyza canadensis | SUB535309 | Chromosome | 335 Mb | (Peng et al., 2014) |
| Dittrichia graveolens | PRJNA919087-8 | Chromosome | 835 Mb | (McEvoy et al., 2023) |
| Glebionis coronaria | JANFOE000000000 | Chromosome | 6.8 Gb | (Wang et al., 2022) |
| Helianthus tuberosus | PRJNA918503 | Chromosome | 21Gb | (Wang et al., 2024) |
| Helichrysum umbraculigerum | PRJEB52026 | Chromosome | 1.3 Gb | (Berman et al., 2023) |
| Pluchea indica | PRJCA004930 | Chromosome | 495.4 Mb | (He et al., 2022) |
| Pulicaria dysenterica | PRJEB50479 | Chromosome | 833.2Mb | (Christenhusz et al., 2023) |
| Scalesia atractyloides | PRJEB52418 | Chromosome | 3.2Gb | (Cerca et al., 2022) |
| Smallanthus sonchifolius | JAKNSE000000000 | Chromosome | 2.72 Gb | (Fan et al., 2022) |
| Stevia rebaudiana | PRJNA684944 | Chromosome | 1416 Mb | (Xu et al., 2021) |
| Tagetes erecta | – | Chromosome | 707.21Mb | (Xin et al., 2023) |
| Tanacetum cinerariifolium | PRJDB8358 | Chromosome | 7.1Gb | (Yamashiro et al., 2019) |
| Tanacetum coccineum | PSUB016075 | Chromosome | 9.4 Gb | (Yamashiro et al., 2022) |
| Cichorium endivia | JAKOPN000000000 | Chromosome | 0.89Gb | (Zhang et al., 2022) |
| Cichorium intybus | JAKNSD000000000 | Chromosome | 1.28Gb | (Fan et al., 2022) |
| Lactuca saligna | PRJEB56287 | Chromosome | 2.27 Gb | (Shen et al., 2023) |
| Lactuca virosa | PRJEB50301 | Chromosome | 3.7 Gb | (Xiong et al., 2023) |
| Taraxacum mongolicum | PRJCA005187 | Chromosome | 790 Mb | (Lin et al., 2022) |
| Taraxacum kok-saghyz | PRJCA005187 | Chromosome | 1.1 Gb | (Lin et al., 2022) |
Figure 1.
Analysis pipeline for data retrieval in AGD.
2.2. Supplements to plant and genome information
Taxonomic resources and phenotypic images were obtained from iplant (https://www.iplant.cn/), Wikipedia (https://encyclopedia.thefreedictionary.com/), and Flora of China (http://flora.huh.harvard.edu/china/mss/intindex.htm). We documented the key details of each genomic publication, including the title, publication date, journal, and the unique PubMed identifier. We conducted a careful manual review of the associated academic articles for each genome to obtain information such as the genome size, assembly level, and the number of predicted genes. Moreover, we extracted the details of the pertinent annotation files.
2.3. Database implementation
The database is supported by Django (https://www.djangoproject.com/), uWSGI (https://uwsgi-docs-zh.readthedocs.io/zh-cn/latest/), and Nginx (https://nginx.org/en/). MySQL (https://www.mysql.com/) is used for the data management and organization of AGD. To provide a smooth and friendly user interface, bootstrap (v.4, https://v4.bootcss.com/), fontawesome (v.free-6.4.0, https://fontawesome.com/), and layUI, (https://layui.dev/docs/2/form/select.html#normal) were employed to improve the interface visual. The statistical results are displayed using bootstrap-table (https://getbootstrap.com/docs/4.0/content/tables/) and ECharts (https://echarts.apache.org/zh/index.html).
2.4. Analysis tools
Eight bioinformatics tools have been integrated into AGD, namely, BLAST (Camacho et al., 2009), JBrowse (Skinner et al., 2009), SSR Finder (Castelo et al., 2002), Heatmap (Verhaak et al., 2006), Primer3 (Rozen and Skaletsky, 2000), PlantiSMASH (Kautsar et al., 2017), CRISPRCasFIDER (Couvin et al., 2018), and HmmSearch (Rehmsmeier and Vingron, 2001). The BLAST service was constructed using the SequenceServer application, which serves as a robust front-end for BLAST. The AGD capabilities are enhanced by embedding JBrowse 2, a new version of the genome visualization tool (Diesh et al., 2023). The SSR web interface was developed to identify SSRs in user-submitted sequences, taking inspiration from the MISA page (https://webblast.ipk-gatersleben.de/misa/index.php?action=1). Protein domains are identified using the HmmSearch program within the HMMER (v.3.3.2) software suite. The Heatmap tool can provide the heat map determined from the expression profile data. Moreover, a PCR primer design tool is embedded into the system, allowing users to adopt the capabilities of Primer. PlantiSMASH is integrated to detect known secondary metabolic gene clusters present within chromosome-level genomes. The identification of CRISPR arrays and Cas proteins is facilitated by the tools provided within the AGD platform.
3. Results
3.1. Structure of AGD
AGD comprises three main parts, including modules, data, and tools ( Figure 2 ). It incorporates six primary modules: Home, Browse, Search, Tools, Visualization, and Contact&Help, each serving distinct functions to facilitate user interaction and data exploration. We have collected genomic data from 40 Asteraceae species, of which seven genomic information that can be queried and downloaded, have been uploaded to the AGD. We are committed to continually improving and expanding the AGD. Furthermore, AGD includes organellar genomic data from 15 Asteraceae species, which adds valuable genetic information to the database. The database is further enriched with large of high-quality photographs showcasing a diverse array of Asteraceae plants.
Figure 2.
Framework of three parts at AGD.
AGD also integrates eight related tools with diverse functionalities and datasets. BLAST for ortholog recognition across a spectrum of plant species, SSR Finder for simple sequence repeats detection, and JBrowse for an immersive genome exploration experience. For protein domain identification, we have integrated HmmSearch, while primer design is facilitated through our proprietary tool. Furthermore, AGD now features PlantiSMASH for secondary metabolite analysis and CRISPRCasFinder for CRISPR-associated system identification, both of which have been embedded within the AGD for user convenience ( Figure 2 ).
3.2. Browse
In the Browse module, users can browse through comprehensive list pages (plant, genome, organellar genomic); utilize interactive filters to narrow down datasets based on specific attributes, such as species hierarchy, assembly level, and herbal characteristics; and explore data subsets that possess the desired attribute. This module can also provide the detailed information, including herb names, habitats, genome version/level, data sources, characteristics, and descriptions.
3.3. Search
AGD has a separate search page where users can quickly find data of interest. The search box allows users to select a species or field and enter keywords. Recorded searches are displayed as a word cloud, and the results page provides a summary table with clickable hyperlinks for more details.
3.4. Tools
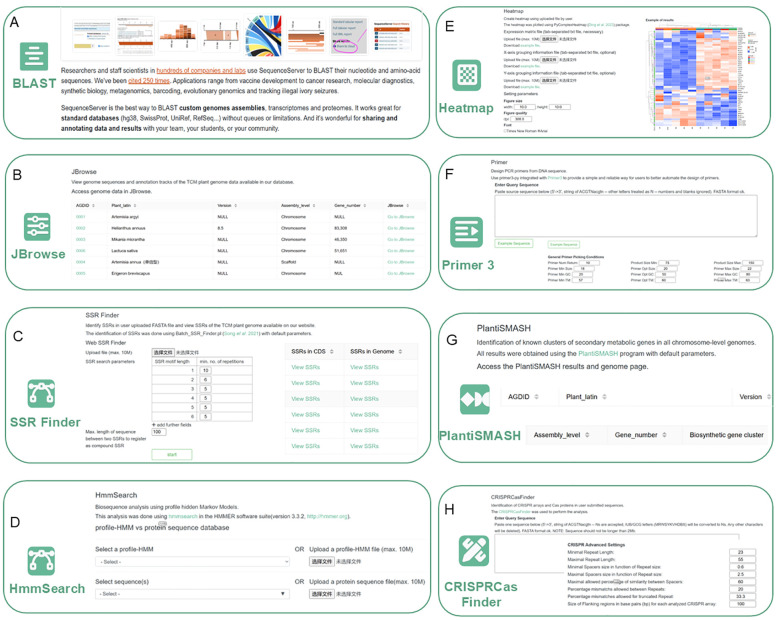
AGD has embedded several online analysis tools to facilitate the systematic analysis of Asteraceae plant genomes. For example, homology searches and the visualization of results can be performed by SequenceServer in BLAST. Users can input query sequences or upload a file in FASTA format, and select a database for the search. The available BLAST options are automatically set based on the query sequence type and selected database ( Figure 3A ). JBrowse can display the integrated data of three genomes and annotated genomic datasets. Users can upload their data for visualization and comparison with AGD datasets. JBrowse enables genome sequence browsing, viewing gene information, and data comparison ( Figure 3B ). In addition, the SSR Finder module identifies SSRs in uploaded sequences and displays SSRs found in AGD coding sequences ( Figure 3C ). HmmSearch analyzes gene families using profile-HMMs ( Figure 3D ) and Heatmap generates visual representations of data matrices ( Figure 3E ). Primer3 can be adopted to design primers for PCR experiments ( Figure 3F ), while PlantiSMASH predicts biosynthetic gene clusters in plants ( Figure 3G ) and CRISPRCasFinder identifies CRISPR-Cas systems in genomes ( Figure 3H ).
Figure 3.
Eight tools at AGD. (A) Blast. (B) JBrowse. (C) SSR Finder. (D) HmmSearch. (E) Heatmap. (F) Primer3. (G) PlantiSMASH. (H) CRISPRCasFinder.
3.5. Visualization
We implement ECharts to display the data contained in AGD. Users can access this tool through the visualization buttons on the navigation bar, which serves as the starting point for exploring the database. The AGD visualization interface offers simple statistics, including the number of plants in the Asteraceae family and the number of Asteraceae and organellar genomes. Users can also examine detailed charts for specific taxonomic subsets by engaging with the corresponding category tabs. The taxonomic hierarchy of the flora is represented with a Sunburst diagram, which allows for the expansion of any segment upon user interaction, and is accompanied by a set of controls below the diagram to facilitate the retrieval of pertinent records. In the genomic data representation block, we include a donut chart featuring smoothed edges to delineate the distribution of genomes across various size spectra. Users can extract corresponding data entries by interacting with any segment of the chart.
3.6. Contact and help
We have included a feedback form within the contact module, tailored for users to conveniently submit their inquiries, concerns, and suggestions regarding various issues. Our email address is displayed on the contact page, ensuring swift and straightforward communication with our team. To strengthen the accessibility of the user interface, we present detailed step-by-step instructions on the help page on how to utilize the primary modules.
4. Discussion
From 2000 to 2020, 1,144 genomes of 782 plant species were sequenced (Xie et al., 2024). Compared to ~10 years ago, high-quality genome assembly has become relatively easier, and there has been a tremendous leap in genome assembly. Due to the remarkable advancements in sequencing technology, a vast array of species has been sequenced (Yang et al., 2024a), and a total of 2,836 genomes from 1,410 plant species was available by 2023 (Xie et al., 2024). Of course, the genome assembly quality has also improved rapidly (Yang et al., 2024b). These afforded the emergence of several databases dedicated to housing their genomes, such as the 1 K medicinal plant genome database (Su et al., 2022), the Rosaceae genome database (Jung et al., 2019), the cucurbit genomics database (Zheng et al., 2019), and the Portal of Juglandaceae (Guo et al., 2020), Traditional Chinese Medicine Plant Genome database Traditional Chinese Medicine Plant Genome database (TCMPG; http://cbcb.cdutcm.edu.cn/TCMPG/) (Meng et al., 2022), and so on ( Supplementary Table S1 ). Asteraceae, the largest family of flowering plants, is renowned for its medicinal, horticultural, and ornamental value. However, research on these plants faces several challenges. The diverse habitats of the Asteraceae family have led to the widespread dispersion of its resources. Additionally, many Asteraceae species are polyploids with large and diverse genomes, posing significant challenges for scientific research due to their genetic complexity. Meanwhile, the continuous advancement of sequencing technologies has facilitated the extensive publication of genomic and genetic resources for various Asteraceae species.
The Global Compositae Database (https://www.compositae.org/gcd/index.php) boasts an extensive collection of approximately 33,057 recognized species. A large number of databases provide partial information on Asteraceae data, yet the data available is quite restricted, such as the GERDH databases, while offering valuable resources for horticultural crops, are limited in scope as they only cover a small number of closely related Asteraceae species (Cheng et al., 2023). According to the published plant genome website, 40 Asteraceae species have had their genomes sequenced, each with varying degrees of assembly completeness and distributed in different databases. Currently, genomes, organelle genomes, and some genetic resources of Asteraceae are distributed in different databases, resulting in the need to spend a lot of time collecting this information before many bioinformatics analyses, lacking a unique and comprehensive database that integrates a large amount of available information on Asteraceae genomics and genetic resources. We recognized that constructing an Asteraceae genome database provides researchers with a comprehensive and user-friendly genomics resource platform, which is very important for advancing Asteraceae genomics and promoting genetic breeding.
Based on this, the Asteraceae Genome Database (AGD) introduces 15 organelle genomes and 7 genomic information of Asteraceae that can be queried and downloaded, along with related genetic information, it provides a data update mechanism, improved user interface design, and advanced data analysis tools (including BLAST, JBrowse, SSR Finder, HmmSearch, Heatmap, Primer3, PlantiSMASH, and CRISPRCasFinder). As an integrated repository for genomic, genotypic, and taxonomic data, it is committed to promoting research on Asteraceae species.
In this work, we developed AGD to manage this wealth of data on the Asteraceae species effectively. It integrates genomic data from multiple species, offering a platform for comparative and functional genomics analysis. This integration is pivotal as it uncovers conserved and variable regions within the genomes, shedding light on gene functions and evolutionary patterns across the family. This strengthens phylogenetic studies, genetic breeding, and drug development specifically for Asteraceae plants. Moreover, we provide robust data analysis and visualization tools, as well as comprehensive and insightful data support for Asteraceae plant research, thereby propelling scientific advancements in related fields.
5. Conclusion
The AGD was established as an integrated database resource dedicated to collecting the genomic-related data of the Asteraceae family, including genomic datasets, organellar genomes, and phenotypic information. Equipped with a suite of useful tools, including BLAST, JBrowse, SSR Finder, HmmSearch, Heatmap, Primer3, PlantiSMASH, and CRISPRCasFinder, the AGD offers researchers valuable resources for genomic analysis. The database is freely accessible online at http://cbcb.cdutcm.edu.cn/AGD/. The AGD serves as a comprehensive repository of genome, genotype, and taxonomy data, and stands as a valuable resource for the entire research community of Asteraceae.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the talented person scientific research start funds subsidization project of Chengdu University of Traditional Chinese Medicine (project code: 030040015). The key special project of the National Key Research and Development Program of the Ministry of Science and Technology in 2023, “Modernization of Traditional Chinese Medicine”: Spatio-temporal Analysis of Quality Formation of Chinese Herbal Medicines and Demonstration of Pseudo-cultivation Research (SQ2023YFC3500127).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .
Author contributions
LW: Supervision, Writing – original draft. HY: Supervision, Writing – original draft. GX: Methodology, Software, Visualization, Writing – review & editing. ZL: Supervision, Validation, Writing – review & editing. FM: Data curation, Methodology, Writing – review & editing. LS: Data curation, Writing – review & editing. XL: Formal analysis, Writing – review & editing. YZ: Visualization, Writing – review & editing. GZ: Data curation, Writing – review & editing. XY: Data curation, Writing – review & editing. WC: Supervision, Writing – review & editing. CS: Supervision, Writing – review & editing. BZ: Supervision, Writing – review & editing.
Conflict of interest
Author LW was employed by the company China Resources Sanjiu Medical & Pharmaceutical Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1445365/full#supplementary-material
References
- Badouin H., Gouzy J., Grassa C. J., Murat F., Staton S. E., Cottret L., et al. (2017). The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 546, 148–152. doi: 10.1038/nature22380 [DOI] [PubMed] [Google Scholar]
- Bellinger M. R., Datlof E. M., Selph K. E., Gallaher T. J., Knope M. L. (2022). A genome for bidens hawaiensis: A member of a hexaploid hawaiian plant adaptive radiation. J. Hered 113, 205–214. doi: 10.1093/jhered/esab077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercovich N., Genze N., Todesco M., Owens G. L., Légaré J.-S., Huang K., et al. (2022). HeliantHOME, a public and centralized database of phenotypic sunflower data. Sci. Data 9, 735. doi: 10.1038/s41597-022-01842-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman P., de Haro L. A., Jozwiak A., Panda S., Pinkas Z., Dong Y., et al. (2023). Parallel evolution of cannabinoid biosynthesis. Nat. Plants 9, 817–831. doi: 10.1038/s41477-023-01402-3 [DOI] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: architecture and applications. BMC Bioinf. 10, 421. doi: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo A. T., Martins W., Gao G. R. (2002). TROLL–tandem repeat occurrence locator. Bioinformatics 18, 634–636. doi: 10.1093/bioinformatics/18.4.634 [DOI] [PubMed] [Google Scholar]
- Cerca J., Petersen B., Lazaro-Guevara J. M., Rivera-Colón A., Birkeland S., Vizueta J., et al. (2022). The genomic basis of the plant island syndrome in Darwin’s giant daisies. Nat. Commun. 13, 3729. doi: 10.1038/s41467-022-31280-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Guo M., Dong S., Wu X., Zhang G., He L., et al. (2023. a). A chromosome-scale genome assembly of Artemisia argyi reveals unbiased subgenome evolution and key contributions of gene duplication to volatile terpenoid diversity. Plant Commun. 4, 100516. doi: 10.1016/j.xplc.2023.100516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Guo S., Hu X., Wang R., Jia D., Li Q., et al. (2023. b). Whole-genome and genome-wide association studies improve key agricultural traits of safflower for industrial and medicinal use. Hortic. Res. 10, uhad197. doi: 10.1093/hr/uhad197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Ma Y., Wu S., Zheng X., Kang H., Sang J., et al. (2021). Genome warehouse: A public repository housing genome-scale data. Genomics Proteomics Bioinf. 19, 584–589. doi: 10.1016/j.gpb.2021.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Zhang H., Song J., Jiang J., Chen S., Chen F., et al. (2023). GERDH: an interactive multi-omics database for cross-species data mining in horticultural crops. Plant J. 116, 1018–1029. doi: 10.1111/tpj.16350 [DOI] [PubMed] [Google Scholar]
- Christenhusz M. J. M., Fay M. F., Royal Botanic Gardens Kew Genome Acquisition Lab. Darwin Tree of Life Barcoding collective. Plant Genome Sizing collective. Wellcome Sanger Institute Tree of Life programme et al. (2023). The genome sequence of common fleabane, Pulicaria dysenterica (L.) Bernh. (Asteraceae). Wellcome Open Res. 8, 447. doi: 10.12688/wellcomeopenres.20003.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CNCB-NGDC Members and Partners (2023). Database resources of the national genomics data center, China national center for bioinformation in 2023. Nucleic Acids Res. 51, D18–D28. doi: 10.1093/nar/gkac1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvin D., Bernheim A., Toffano-Nioche C., Touchon M., Michalik J., Néron B., et al. (2018). CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 46, W246–W251. doi: 10.1093/nar/gky425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Yang P., Zhang Q., Wu Q., Feng L., Shi W., et al. (2024). Genomic insights into the evolution of flavonoid biosynthesis and O-methyltransferase and glucosyltransferase in Chrysanthemum indicum. Cell Rep. 43, 113725. doi: 10.1016/j.celrep.2024.113725 [DOI] [PubMed] [Google Scholar]
- Diesh C., Stevens G. J., Xie P., De Jesus Martinez T., Hershberg E. A., Leung A., et al. (2023). JBrowse 2: a modular genome browser with views of synteny and structural variation. Genome Biol. 24, 74. doi: 10.1186/s13059-023-02914-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elomaa P. (2019). My favourite flowering image: a capitulum of Asteraceae. J. Exp. Bot. 70, e6496–e6498. doi: 10.1093/jxb/erw489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W., Wang S., Wang H., Wang A., Jiang F., Liu H., et al. (2022). The genomes of chicory, endive, great burdock and yacon provide insights into Asteraceae palaeo-polyploidization history and plant inulin production. Mol. Ecol. Resour 22, 3124–3140. doi: 10.1111/1755-0998.13675 [DOI] [PubMed] [Google Scholar]
- Garnatje T., Canela M.ÁCheckt. a., Garcia S., Hidalgo O., Pellicer J., Sánchez-Jiménez I., et al. (2011). GSAD: a genome size in the Asteraceae database. Cytometry A 79, 401–404. doi: 10.1002/cyto.a.21056 [DOI] [PubMed] [Google Scholar]
- Guo W., Chen J., Li J., Huang J., Wang Z., Lim K.-J. (2020). Portal of Juglandaceae: A comprehensive platform for Juglandaceae study. Hortic. Res. 7, 35. doi: 10.1038/s41438-020-0256-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Dong X., Zhang G., Fan W., Duan S., Shi H., et al. (2021). High quality genome of Erigeron breviscapus provides a reference for herbal plants in Asteraceae. Mol. Ecol. Resour 21, 153–169. doi: 10.1111/1755-0998.13257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Feng X., Chen Q., Li L., Li S., Han K., et al. (2022). Evolution of coastal forests based on a full set of mangrove genomes. Nat. Ecol. Evol. 6, 738–749. doi: 10.1038/s41559-022-01744-9 [DOI] [PubMed] [Google Scholar]
- Jung S., Lee T., Cheng C.-H., Buble K., Zheng P., Yu J., et al. (2019). 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 47, D1137–D1145. doi: 10.1093/nar/gky1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautsar S. A., Suarez Duran H. G., Blin K., Osbourn A., Medema M. H. (2017). plantiSMASH: automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Res. 45, W55–W63. doi: 10.1093/nar/gkx305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. D., Shim J., Hwang J. H., Kim D., El Baidouri M., Park S., et al. (2024). Chromosome-level genome assembly of milk thistle (Silybum marianum (L.) Gaertn.). Sci. Data 11, 342. doi: 10.1038/s41597-024-03178-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforest M., Martin S. L., Bisaillon K., Soufiane B., Meloche S., Tardif F. J., et al. (2024). The ancestral karyotype of the Heliantheae Alliance, herbicide resistance, and human allergens: Insights from the genomes of common and giant ragweed. Plant Genome 17, e20442. doi: 10.1002/tpg2.20442 [DOI] [PubMed] [Google Scholar]
- Li Q., Wang Z., Xie Y., Hu H. (2020). Antitumor activity and mechanism of costunolide and dehydrocostus lactone: Two natural sesquiterpene lactones from the Asteraceae family. BioMed. Pharmacother. 125, 109955. doi: 10.1016/j.biopha.2020.109955 [DOI] [PubMed] [Google Scholar]
- Liao B., Hu H., Xiao S., Zhou G., Sun W., Chu Y., et al. (2022. a). Global Pharmacopoeia Genome Database is an integrated and mineable genomic database for traditional medicines derived from eight international pharmacopoeias. Sci. China Life Sci. 65, 809–817. doi: 10.1007/s11427-021-1968-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B., Shen X., Xiang L., Guo S., Chen S., Meng Y., et al. (2022. b). Allele-aware chromosome-level genome assembly of Artemisia annua reveals the correlation between ADS expansion and artemisinin yield. Mol. Plant 15, 1310–1328. doi: 10.1016/j.molp.2022.05.013 [DOI] [PubMed] [Google Scholar]
- Lin T., Xu X., Du H., Fan X., Chen Q., Hai C., et al. (2022). Extensive sequence divergence between the reference genomes of Taraxacum kok-saghyz and Taraxacum mongolicum. Sci. China Life Sci. 65, 515–528. doi: 10.1007/s11427-021-2033-2 [DOI] [PubMed] [Google Scholar]
- Liu B., Yan J., Li W., Yin L., Li P., Yu H., et al. (2020). Mikania micrantha genome provides insights into the molecular mechanism of rapid growth. Nat. Commun. 11, 340. doi: 10.1038/s41467-019-13926-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy S. L., Lustenhouwer N., Melen M. K., Nguyen O., Marimuthu M. P. A., Chumchim N., et al. (2023). Chromosome-level reference genome of stinkwort, Dittrichia graveolens (L.) Greuter: A resource for studies on invasion, range expansion, and evolutionary adaptation under global change. J. Hered 114, 561–569. doi: 10.1093/jhered/esad033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton A. E., Child A. W., Beard R. S., Dumaguit C. D. C., Forbey J. S., Germino M., et al. (2022). A haploid pseudo-chromosome genome assembly for a keystone sagebrush species of western North American rangelands. G3 12, jkac122. doi: 10.1093/g3journal/jkac122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F., Tang Q., Chu T., Li X., Lin Y., Song X., et al. (2022). TCMPG: an integrative database for traditional Chinese medicine plant genomes. Hortic. Res. 9, uhac060. doi: 10.1093/hr/uhac060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M., Hirakawa H., Fukai E., Toyoda A., Kajitani R., Minakuchi Y., et al. (2021). A chromosome-level genome sequence of Chrysanthemum seticuspe, a model species for hexaploid cultivated chrysanthemum. Commun. Biol. 4, 1167. doi: 10.1038/s42003-021-02704-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Lai Z., Lane T., Nageswara-Rao M., Okada M., Jasieniuk M., et al. (2014). De novo genome assembly of the economically important weed horseweed using integrated data from multiple sequencing platforms. Plant Physiol. 166, 1241–1254. doi: 10.1104/pp.114.247668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M., Vingron M. (2001). Phylogenetic information improves homology detection. Proteins 45, 360–371. doi: 10.1002/prot.1156 [DOI] [PubMed] [Google Scholar]
- Rolnik A., Olas B. (2021). The plants of the asteraceae family as agents in the protection of human health. Int. J. Mol. Sci. 22, 3009. doi: 10.3390/ijms22063009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolnik A., Stochmal A., Olas B. (2022). The in vitro anti-platelet activities of plant extracts from the Asteraceae family. BioMed. Pharmacother. 149, 112809. doi: 10.1016/j.biopha.2022.112809 [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386. doi: 10.1385/1-59259-192-2:365 [DOI] [PubMed] [Google Scholar]
- Scaglione D., Reyes-Chin-Wo S., Acquadro A., Froenicke L., Portis E., Beitel C., et al. (2016). The genome sequence of the outbreeding globe artichoke constructed de novo incorporating a phase-aware low-pass sequencing strategy of F1 progeny. Sci. Rep. 6, 19427. doi: 10.1038/srep19427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F., Qin Y., Wang R., Huang X., Wang Y., Gao T., et al. (2023). Comparative genomics reveals a unique nitrogen-carbon balance system in Asteraceae. Nat. Commun. 14, 4334. doi: 10.1038/s41467-023-40002-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Zhang L., Liao Z., Wang S., Yan T., Shi P., et al. (2018). The Genome of Artemisia annua Provides Insight into the Evolution of Asteraceae Family and Artemisinin Biosynthesis. Mol. Plant 11, 776–788. doi: 10.1016/j.molp.2018.03.015 [DOI] [PubMed] [Google Scholar]
- Skinner M. E., Uzilov A. V., Stein L. D., Mungall C. J., Holmes I. H. (2009). JBrowse: a next-generation genome browser. Genome Res. 19, 1630–1638. doi: 10.1101/gr.094607.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Liu Y., Song A., Dong G., Zhao H., Sun W., et al. (2018). The Chrysanthemum nankingense Genome Provides Insights into the Evolution and Diversification of Chrysanthemum Flowers and Medicinal Traits. Mol. Plant 11, 1482–1491. doi: 10.1016/j.molp.2018.10.003 [DOI] [PubMed] [Google Scholar]
- Song A., Su J., Wang H., Zhang Z., Zhang X., Van de Peer Y., et al. (2023. a). Analyses of a chromosome-scale genome assembly reveal the origin and evolution of cultivated chrysanthemum. Nat. Commun. 14, 2021. doi: 10.1038/s41467-023-37730-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Yang Y., Xu L., Bian C., Xing Y., Xue H., et al. (2023. b). The burdock database: a multi-omic database for Arctium lappa, a food and medicinal plant. BMC Plant Biol. 23, 86. doi: 10.1186/s12870-023-04092-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Yang L., Wang D., Shu Z., Yang Y., Chen S., et al. (2022). 1 K Medicinal Plant Genome Database: an integrated database combining genomes and metabolites of medicinal plants. Hortic. Res. 9, uhac075. doi: 10.1093/hr/uhac075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Zhang A., Landis J. B., Shi W., Zhang X., Sun H., et al. (2023). Genome assembly of the snow lotus species Saussurea involucrata provides insights into acacetin and rutin biosynthesis and tolerance to an alpine environment. Hortic. Res. 10, uhad180. doi: 10.1093/hr/uhad180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lieshout N., Van Kaauwen M., Kodde L., Arens P., Smulders M. J. M., Visser R. G. F., et al. (2022). De novo whole-genome assembly of Chrysanthemum makinoi, a key wild chrysanthemum. G3 12, jkab358. doi: 10.1093/g3journal/jkab358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventimiglia M., Bosi E., Vasarelli L., Cavallini A., Mascagni F. (2023). Letter to the editor: ASTER-REP, a database of asteraceae sequences for structural and functional studies of transposable elements. Plant Cell Physiol. 64, 365–367. doi: 10.1093/pcp/pcad008 [DOI] [PubMed] [Google Scholar]
- Verhaak R. G. W., Sanders M. A., Bijl M. A., Delwel R., Horsman S., Moorhouse M. J., et al. (2006). HeatMapper: powerful combined visualization of gene expression profile correlations, genotypes, phenotypes and sample characteristics. BMC Bioinf. 7, 337. doi: 10.1186/1471-2105-7-337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wang A., Chen R., Xu D., Wang H., Jiang F., et al. (2024). Haplotype-resolved chromosome-level genome of hexaploid Jerusalem artichoke provides insights into its origin, evolution, and inulin metabolism. Plant Commun. 5, 100767. doi: 10.1016/j.xplc.2023.100767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wang A., Wang H., Jiang F., Xu D., Fan W. (2022). Chromosome-level genome of a leaf vegetable Glebionis coronaria provides insights into the biosynthesis of monoterpenoids contributing to its special aroma. DNA Res. 29, dsac036. doi: 10.1093/dnares/dsac036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., Li J., Wang L., Lu C., Gao Q., Xu P., et al. (2022). The chrysanthemum lavandulifolium genome and the molecular mechanism underlying diverse capitulum types. Hortic. Res. 9, uhab022. doi: 10.1093/hr/uhab022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Gong X., Yang K., Huang Y., Zhang S., Shen L., et al. (2024). Technology-enabled great leap in deciphering plant genomes. Nat. Plants 10, 551–566. doi: 10.1038/s41477-024-01655-6 [DOI] [PubMed] [Google Scholar]
- Xin H., Ji F., Wu J., Zhang S., Yi C., Zhao S., et al. (2023). Chromosome-scale genome assembly of marigold (Tagetes erecta L.): An ornamental plant and feedstock for industrial lutein production. Hortic. Plant J. 9, 1119–1130. doi: 10.1016/j.hpj.2023.04.001 [DOI] [Google Scholar]
- Xiong W., van Workum D. M., Berke L., Bakker L. V., Schijlen E., Becker F. F. M., et al. (2023). Genome assembly and analysis of Lactuca virosa: implications for lettuce breeding. G3 13, 11. doi: 10.1093/g3journal/jkad204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Yuan H., Yu X., Huang S., Sun Y., Zhang T., et al. (2021). The chromosome-level Stevia genome provides insights into steviol glycoside biosynthesis. Hortic. Res. 8, 129. doi: 10.1038/s41438-021-00565-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro T., Shiraishi A., Nakayama K., Satake H. (2022). Draft genome of tanacetum coccineum: genomic comparison of closely related tanacetum-family plants. Int. J. Mol. Sci. 23, 7039. doi: 10.3390/ijms23137039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro T., Shiraishi A., Satake H., Nakayama K. (2019). Draft genome of Tanacetum cinerariifolium, the natural source of mosquito coil. Sci. Rep. 9, 18249. doi: 10.1038/s41598-019-54815-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wang Y., Liu W., He T., Liao J., Qian Z., et al. (2024. a). Genome-wide pan-GPCR cell libraries accelerate drug discovery. Acta Pharm. Sin. B. doi: 10.1016/j.apsb.2024.06.023 [DOI] [Google Scholar]
- Yang H., Wang C., Zhou G., Zhang Y., He T., Yang L., et al. (2024. b). A haplotype-resolved gap-free genome assembly provides novel insight into monoterpenoid diversification in Mentha suaveolens “Variegata. Hortic. Res. 11, uhae022. doi: 10.1093/hr/uhae022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Elomaa P. (2024). Development and evolution of the Asteraceae capitulum. New Phytol. 242, 33–48. doi: 10.1111/nph.19590 [DOI] [PubMed] [Google Scholar]
- Zhang B., Wang Z., Han X., Liu X., Wang Q., Zhang J., et al. (2022). The chromosome-scale assembly of endive (Cichorium endivia) genome provides insights into the sesquiterpenoid biosynthesis. Genomics 114, 110400. doi: 10.1016/j.ygeno.2022.110400 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Wu S., Bai Y., Sun H., Jiao C., Guo S., et al. (2019). Cucurbit Genomics Database (CuGenDB): a central portal for comparative and functional genomics of cucurbit crops. Nucleic Acids Res. 47, D1128–D1136. doi: 10.1093/nar/gky944 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .