Highlights
-
•
Clinical significance: high expression of GGCT and REST was associated to poor prognosis in Glioma.
-
•
Function: GGCT silencing inhibited the proliferation, migration and induced apoptosis and tumor growth of glioma cells in vitro and vivo; REST overexpression restored the repression of xenograft tumor formation induced by GGCT knockdown.
-
•
In mechanism: REST inhibited miR-34a-5p mRNA expression, and miR-34a-5p suppressed GGCT expression by targeting its 3′UTR, forming a positive regulatory loop in glioma.
-
•
GGCT/REST/miR-34a-5p axis holds promising potential as a therapeutic target, offering a potential breakthrough in the treatment of glioma.
Keywords: GGCT, REST, Glioma, miR-34a-5p
Abstract
Background
γ-Glutamylcyclotransferase (GGCT), an enzyme crucial in glutathione metabolism, has emerged as a participant in tumorigenesis. The present study is designed to elucidate the biological role and molecular mechanisms underlying GGCT in glioma.
Methods
Gene Expression Profiling Interactive Analysis (GEPIA), Chinese Glioma Genome Atlas (CGGA), and PrognoScan online databases were utilized to examine the expressions and clinical prognosis of GGCT and REST in glioma. Cell Counting Kit-8 (CCK-8), Transwell, Wound healing, and Flow cytometric assays, and RNA-sequencing analysis were employed to uncover the molecular role of GGCT and REST. Prediction of Differentially expressed microRNA (DE-miRNAs) and miRNAs targeting GGCT 3′ Untranslated Region (UTR) was performed using miRanda online datasets. Finally, Real time-quantitative Polymerase Chain Reaction (RT-qPCR), western blot and dual luciferase reporter gene activity analysis were employed to confirm a positive feedback loop involving GGCT/REST/miR-34a-5p in glioma cells.
Results
High expression of GGCT was correlated with poor prognosis in glioma. GGCT silencing demonstrated inhibitory effects on the proliferation, migration, and induction of apoptosis in T98G and U251 cells. Mechanistically, GGCT downregulated REST expression and modulated cancer-associated pathways in glioma cells. High expression of REST was associated with poor prognosis in glioma. In vitro and in vivo experiments showed that REST overexpression restored the repression of proliferation, invasion, migration, and xenograft tumor formation induced by GGCT knockdown. Furthermore, the study uncovered that REST inhibited miR-34a-5p mRNA expression, and miR-34a-5p suppressed GGCT expression by targeting its 3′UTR, forming a positive regulatory loop in glioma. Notably, the inhibitor of miR-34a-5p restored the role of REST silencing in decreasing GGCT expression in glioma cells.
Conclusions
GGCT/REST/miR-34a-5p axis holds promising potential as a therapeutic target, offering a potential breakthrough in the treatment of glioma.
Graphical abstract
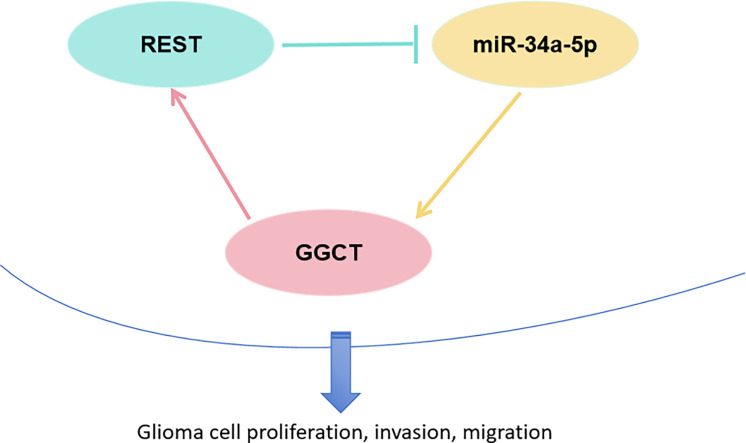
A positive feedback loop between GGCT and REST promoted the malignant progression of glioma by regulating the transcriptional inhibition of miR-34a-5p. The GGCT/REST/miR-34a-5p may be potential targets bringing a breakthrough for the treatment for glioma.
Introduction
Glioma, originating from glial cells, is the predominant malignant primary brain tumor in adults. It is categorized into localized and diffuse gliomas, with further subdivisions into grades I-IV based on malignancy [1]. Lower grade gliomas (LGG) encompass grade I and II, while higher grade gliomas (HGG) include grade III and IV [2]. Glioblastoma (GBM), a grade IV tumor, constitutes a significant proportion of all gliomas and primary central nervous system malignant tumors [3]. Current treatment modalities for GBM involve gross total resection, focal radiotherapy, chemotherapy with temozolomide, and a specific radiotherapy dose [4]. Despite these efforts, the prognosis remains unsatisfactory, with a five-year survival rate below 5 % and a median survival rarely exceeding 16 months [5]. Consequently, the identification of novel glioma biomarkers and the exploration of the roles and mechanisms of new genes are imperative for developing effective treatments.
γ-Glutamylcyclotransferase (GGCT) is an enzyme integral to glutathione metabolism, and its abnormal expression has been linked to carcinogenesis and development [6]. Recent studies have highlighted the overexpression of GGCT in various cancer tissues, positioning it as a noteworthy target for regulating tumor cell proliferation [7,8]. Research indicates that GGCT may play a role in tumor cell invasion and metastasis 8. Silencing GGCT has shown to effectively suppress proliferation, migration, and induction of apoptosis in colorectal cancer cells [9]. Notably, GGCT serves as a biomarker for diagnosing esophageal cancer from high-grade intraepithelial neoplasia to subepithelial invasion. Knockout of GGCT has shown efficacy in inhibiting ovarian cancer cell proliferation and migration, suggesting its utility as a prognostic marker and potential therapeutic target [10]. The association of GGCT with the survival rates of patients with breast cancer and osteosarcoma further underscores its significance as a diagnostic and prognostic biomarker [11]. GGCT was upregulated in glioma [12]. Knockdown of GGCT inhibits cell growth and promotes cell death in glioblastoma cells A172 [13]. GGCT knockdown decreases cell proliferation and colony formation, GGCT overexpression promotes cell proliferation and colony formation in glioma cells [14]. The mechanism of action of GGCT in gliomas is a complex and multifaceted process. Our previous research has indeed revealed its mechanism of promoting cell growth through the Notch-Akt signaling pathway [14]. However, we believe that there are still many unresolved mysteries in this field, and the reasons for the upregulation of GGCT in gliomas remain unclear.
Repressor element 1-silencing transcription factor (REST), also known as neuron-restricted silencing factor, functions as a negative regulator of neuron-specific expression with a diverse array of selective splicing phenomena [15]. REST is widely expressed in embryonic stem cells and neural stem cells, playing a crucial role in regulating cell differentiation, nerve regeneration, neuroprotection, and cognitive function [16]. Recent studies have identified REST expression in certain tumor tissues and cell lines, emphasizing its significant role in cancer progression [17]. Specifically, research has demonstrated that elevated REST expression in patients with GBM is associated with poor prognosis. This association is attributed to REST's inhibitory effect on the differentiation of neurons into glioma cells, thereby contributing to the development of GBM [18,19]. Moreover, heightened REST expression diminishes the chemosensitivity of GBM patients, leading to shortened overall survival [20].
The primary aim of this study was to assess the expression levels and functional role of GGCT in glioma through a comprehensive analysis involving bioinformatics evaluations and functional assays. Subsequently, we delved into the downstream functional mechanisms and identified potential targets of GGCT using RNA-sequencing datasets. Particularly intriguing was the examination of the in vivo and in vitro assays, which revealed a positive feedback loop involving GGCT and REST, contributing to enhanced tumor growth. Our research reveals that the GGCT/REST/miR-34a-5p positive feedback loop shows potential as a biomarker and a target for therapeutic interventions in glioma.
Methods and materials
Bioinformatic analysis
The Gene Expression Profiling Interactive Analysis (GEPIA) database, and Chinese Glioma Genome Atlas (CGGA) were explored to obtain in the expression level of GGCT and REST in LGG and GCM patient tissues. The Kaplan-Meier survival curves were analyzed concerning the different expression levels of GGCT/REST in LGG and GBM patient via the GEPIA and PrognoScan database (Gene expression Omnibus (GEO) database (GSE4271, GSE4412, MGH-glioma) was used to analysis the overall survival of glioma patients). The miRNAs of targeted GGCT 3′ Untranslated Region (3′UTR) were predicted by the miRanda database. The Cancer Genome Atlas (TCGA)-GBM were analyzed the expression of genes in tumor tissues and normal tissues.
RNA-Sequencing and pathway enrichment analysis
Total RNA was extracted from T98G cells, with and without silenced GGCT, using the TRIzol reagent following standard procedures. The mRNA-Seq library was prepared. The library sequencing was performed, and the raw data were deposited. First, the RNA-sequencing data of the 3 negative control (NC) group and 3 GGCT shRNA groups were normalized, the Differentially Expressed Genes (DEGs) were identified by the R package DEseq2, and the threshold of value was set as P-value < 0.05 and |log2 (fold change) | ≥ 1.
The Database for Annotation, Visualization and Integrated Discovery (DAVID) database was utilized to perform Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis in order to investigate the functions and potential signaling pathways of DEGs. KEGG pathway enrichment analysis was performed [21,22]. The terms were significant enrichment with P-value < 0.05.
Cell culture
GBM cells (T98G and U251) were acquired from the American Type Culture Collection (ATCC). The cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) medium added with 10 % fetal bovine serum (FBS, Life Technologies), 100 μg/mL streptomycin, and 100 U/mL penicillin (Gibco, USA) at a humidified atmosphere containing 5 % CO2 at 37 °C.
Lentiviral vector construction
The lentiviral overexpression vector pCDH—CMV-MCS-EF1α-copGFP (System Biosciences, USA) and lentiviral interference vector: pSIH1-H1-copGFP (System Biosciences, USA), were purchased to construct lentivirus-based GGCT/REST overexpression and silencing vectors, along with a negative control. The lentiviral packaging kit (Invitrogen, USA) was used to package the carried lentiviral particles into HEK-293T cells (Saibai-Kang Biological Technology Co., Ltd., Shanghai, China). After 48 h, the cell supernatant was collected as the lentivirus, with a titer of 1 × 108 TU/ml. For lentivirus-mediated cell infection, 1 × 105 cells were seeded into a 6-well plate, and when cell confluence reaches 60–70 %, add an appropriate amount of packaged lentivirus (MOI=10, working titer approximately 1 × 106 TU/ml) and 5 μg/mL polybrene (TR-1003, Merck, USA) to the medium for transfection. Four hours post-transfection, add an equal amount of medium to dilute polybrene, replace with fresh medium 24 h post-transfection, and use 1 μg/mL puromycin (A1113803, Thermo Fisher, USA) for resistance selection 48 h post-transfection to obtain stable cell lines. GGCT shRNA-1: Sense:5′-GCUGGAGUAUCAAGAAGUUTT-3′ and antisense: 5′-AACUUCUCUUGAUACUCCAGCTT-3′; GGCT shRNA-2: sense:5′-GCAAUAGAACCAAAUGACUAUTT-3′ and antisense: 5′-AUAGUCAUUUGGUUCUAUUGCTT-3′; GGCT shRNA-3: forward, 5′-GCTCAGCGGGAGGAGGCTATATTGCAAGAGAGAACGTTT-3′ and reverse, 5′-ACTTGAACAAAGCACGCGGAGGCACAGTGTCCTCT-3′; miR-34a-5p inhibitor: forward, 5′-GTATGATTCACCACCTTCTC-3′ and reverse, 5′-CTCAACTGGTGTCGTGGAG-3′
Real time-quantitative polymerase chain reaction (RT‒qPCR) assay
Total RNA was extracted by using TRIzol (ThermoFisher, USA) from T98G and U251 cells. The concentration and purity of the extracted total RNA were assessed using the nanodrop2000 UV spectrophotometer (ThermoFisher, USA). Following the instructions of the PrimeScript RT reagent Kit (RR047A, Takara, Japan), mRNA was reverse transcribed to generate cDNA. Real-time fluorescence quantitative PCR was performed using the 7500 Fast Real-time PCR System (4,351,106, ThermoFisher, USA) under the following reaction conditions: initial denaturation at 95 °C for 10 min, denaturation at 95 °C for 10 s, annealing at 60 °C for 20 s, extension at 72 °C for 34 s for 40 cycles. GAPDH was used as a reference gene with primers synthesized by TaKaRa. The relative transcription levels of the target gene were calculated using the comparative threshold cycle method (2−ΔΔCT) method. The primer sequences were as follows: GAPDH-F: 5′-GAAGGTGAAGGTCGGAGTC-3′, GAPDH-R: 5′-GAAGATGGTGATGGGATTT-3′; GGCT-F: 5′-CTCTGGATGAGCAAGAAGGG-3′, GGCT-R: 5′- ATTTTCTTTTGCACCCATGC-3′; REST-F: 5′-GAGGCGTGGCAGACTATGC-3′, REST-R: 5′-CTTGTACTCCGTCAGCTGA-3′; U6-F: 5′-CTCGCTTCGGCAGCACA-3′, U6-R: 5′-AACGCTTCACGAATTTGCGT-3′; miR-34a-5p-F: 5′-ACACTCCAGCTGGGTGGCAGTGTCTTAGC-3′, miR-34a-5p-R: 5′-CTCAACTGGTGTCGTGGA-3′.
Cell proliferation assay
Cell counting kit-8 (CCK-8, CA1210, Solebao Technology Co., Ltd., Beijing, China) was used to detect the cell proliferation. In brief, T98G and U251 cells were seeded in 96 well plates after transfection and cultured in an incubator. At 6, 24, 48, and 72 h, the 96 well plates were removed and added with 10 μL of CCK-8 solution. After 2 h of incubation, the absorbance of OD450 nm was read using a microplate reader.
Flow cytometric assay
Cell apoptosis was measured using Annexin V-FITC/PI kit (Sigma-Aldrich, USA). Briefly, T98G and U251 cells were collected and washed with PBS after transfection. The cells were then incubated in 500 μL binding buffer for 15 min before adding 5 μL of Annexin V-FITC/PI and staining the cells for another 15 min. Finally, apoptotic cells were detected by flow cytometry analysis (BD Biosciences, USA).
Cell migration assay
T98G and U251 cells were resuspended in serum-free medium at a concentration of 1 × 105 cells/100 μL and seeded into the upper chamber of Transwell chambers placed in a 24-well plate. The lower chamber was filled with 600 µl of medium containing 10 % FBS. Transwell chambers were removed after 48 h, stained with 0.1 % crystal violet. After washing with PBS, pictures were taken under a micromicroscope and counted.
For the wound healing assay, T98G and U251 cells were seeded in 6-well cell culture plates and allowed to form a monolayer. A 200 μL pipette tip was used to create a scratch on the monolayer of cells, which were then washed with Phosphate-Buffered Saline (PBS). Cellular migration into the wounded area was observed under a microscope and photographs were taken at 0 and 36 h after the scratch was made.
Western blotting assay
Total proteins were acquired from the cells and lysed with RIPA (P0013C, Biyuntian, China). Total protein concentration was determined by BCA assay (P0012S, Biyuntian, China) 12 % Sodium Dodecyl Sulfate (SDS)-polyacrylamide gels were used to separated proteins by electrophoresis. The proteins were transferred to a Polyvinylidene Fluoride (PVDF) membrane. Sealing membranes were blocked with 5 % skim milk for 1 h at room temperature. Primary antibodies were incubated overnight at 4 °C; Membranes were washed three times with PBS for 5 min each. The secondary antibodies were incubated for 1 h at room temperature. The membrane was washed three times with PBS. The Enhanced Chemiluminescence (ECL) luminescence solution was used for visualization. Image was used to analyze the relative expression of protein bands.
Luciferase reporter assay
The pmirGLO-GGCT plasmids and pmirGLO-GGCT Mut (mutated site) plasmid were transfected into miR-34a-5p inhibitor T98G stable cell line or T98G pLKO.1-puro (empty vector). The recombinant plasmids were transfected into T98G cells by using Lipofectamine 3000. After 48 h, cells were lysed using 1 × Passive lysis buffer. The luciferase activity was measured by Dual-Luciferase Reporter Assay System (Promega, WI, USA). The mutation was generated as following:
GGCT- Wild Type (WT): …UAGCUCACUUGAUGAUACUGCUG…
GGCT- Mutation (MUT): …UAGCUCACUUGAUCGGUGACGUG…
In vivo tumor xenograft models
BALB/c nude mice were randomly divided into three groups. T98G cells transfected with control, GGCT shRNA, GGCT shRNA + REST OV (n = 5, per groups). The stable expressed cells were resuspended in 0.25 mL PBS containing 5.0 × 105 cells, and the 0.25 mL cell suspension was subcutaneously injected into each nude mouse. The xenograft tumor size was measured and the volume was calculated. The length, width and thickness measurements were obtained using calipers, and the tumor volume was calculated. The formula for calculating tumor volume is: V (mm3) = (a × b2)/2, where “a” represents the tumor's maximum diameter and “b” represents the vertical height of the tumor.
Immunohistochemistry
Tumor tissue specimens were embedded, sectioned, deparaffinized, dehydrated in a gradient of alcohol, and rinsed twice in distilled water. They were then incubated in 3 % methanol hydrogen peroxide for 20 min, rinsed in distilled water for 2 min, and washed in 0.1 M PBS for 3 min. Antigen retrieval was performed using a water bath, followed by cooling with tap water. Normal goat serum blocking solution (Shanghai Haoran Biotechnology Co., Ltd., Shanghai, China) was applied onto the tissue sections and left at room temperature for 20 min before removing excess liquid carefully. Primary antibodies against Ki-67 (1:1000, Abcam, UK), GGCT (1:1000, Abcam, UK), and REST (1:1000, Abcam, UK) were then added to the tissue sections and incubated overnight at 4 °C. After washing three times in 0.1 M PBS for 5 min each, secondary antibodies of goat anti-rabbit IgG (ab6785, 1:500, Abcam, Cambridge, UK) were applied onto the tissue sections and left at 37 °C for 20 min. This was followed by the addition of the working solution of horseradish peroxidase-labeled streptavidin at 37 °C for 20 min. The sections were then stained with DAB (Guangzhou Weijia Technology Co., Ltd., Guangzhou, China), washed after completion of staining, counterstained with hematoxylin (Shanghai Bogu Biotechnology Co., Ltd., Shanghai, China) for 1 min, rinsed in water, differentiated in 1 % ammonia water, dehydrated in a gradient of alcohol, cleared in xylene, mounted with neutral resin, observed under a microscope and photographed.
Statistical analysis
Statistical analyses were performed using SPSS21. Data are presented as mean ± standard deviation (SD), each conducted in triplicate. Student's t-test was untiled to compare the means with multiple comparisons. A p-value <0.05 was considered statistically significant.
Results
GGCT expression was upregulated in glioma cancer and associated with a poor prognosis
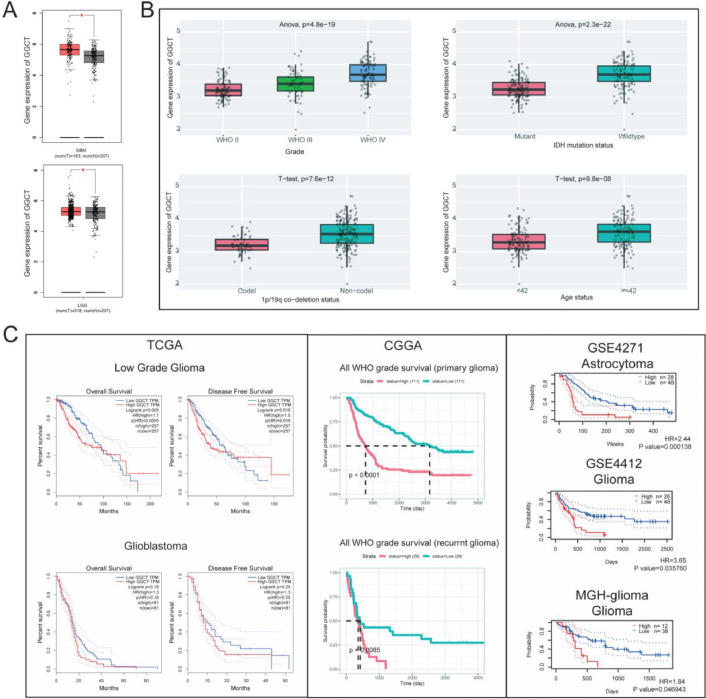
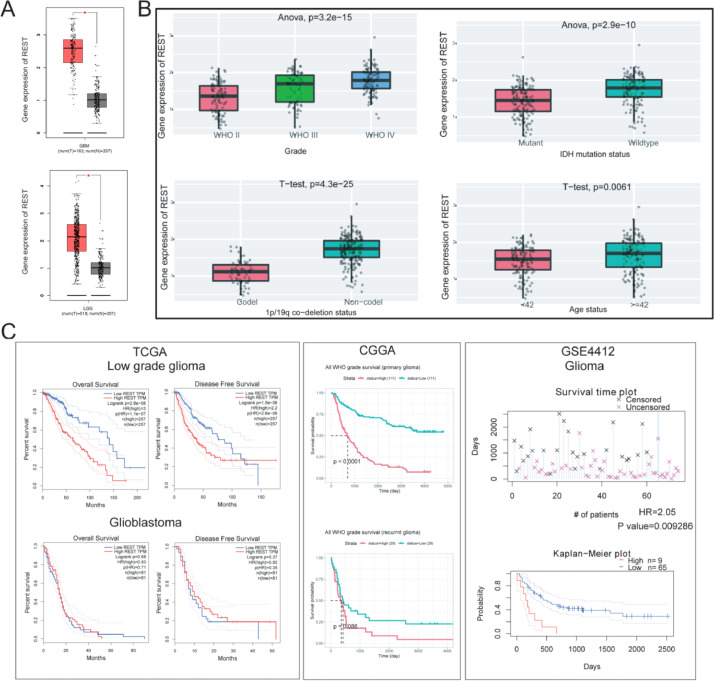
To explore the clinical value and potential role of GGCT in glioma progression, we analyzed GGCT expression in GBM and LGG cancer tissues and normal tissues. GGCT is overexpressed in GBM and LGG tumor tissues compared with normal tissues from TCGA dataset (Fig. 1A). Bioinformatic analyses from the CGGA database (Fig. 1B) showed obviously elevated GGCT expression in high grade glioma (WHO IV), IDH wildtype, 1p/19q co-deletion non-codel, age (≥42). Next, overall survival analysis in diverse datasets showed GGCT high expression was associated with poor prognosis of glioma (Fig. 1C). High expression of GGCT was also significantly associated with the shorter overall survival (OS) and Disease-Free Survival (DFS) of LGG patients in TCGA dataset. We found there was no statistically significance in GBM patient. We also performed the survival analysis of primary glioma and recurrent glioma in CGGA, and there was a tendency of high GGCT predicting a poorer prognosis in primary and recurrent glioma. Meanwhile, high expression of GGCT has a poorer prognosis in GEO dataset. Overall, GGCT was aberrantly overexpressed in glioma and correlated with poor prognosis.
Fig. 1.
Expression levels and prognosis value of GGCT in glioma. (A) The expression of GGCT was analyzed in GBM and LGG. (B) The expression of GGCT was analyzed in Grade, IDH mutation status, 1p/19q co-deletion status and age status of glioma. (C) Overall survival of disease-free survival of GGCT high expression/low expression was analyzed in Low grade glioma, Glioblastoma, CGGA datasets, GEO datasets of glioma. * P < 0.05, ** P < 0.01, *** P < 0.001.
Knockdown of GGCT inhibited proliferation, invasion, migration and induced apoptosis of glioma cells in vitro
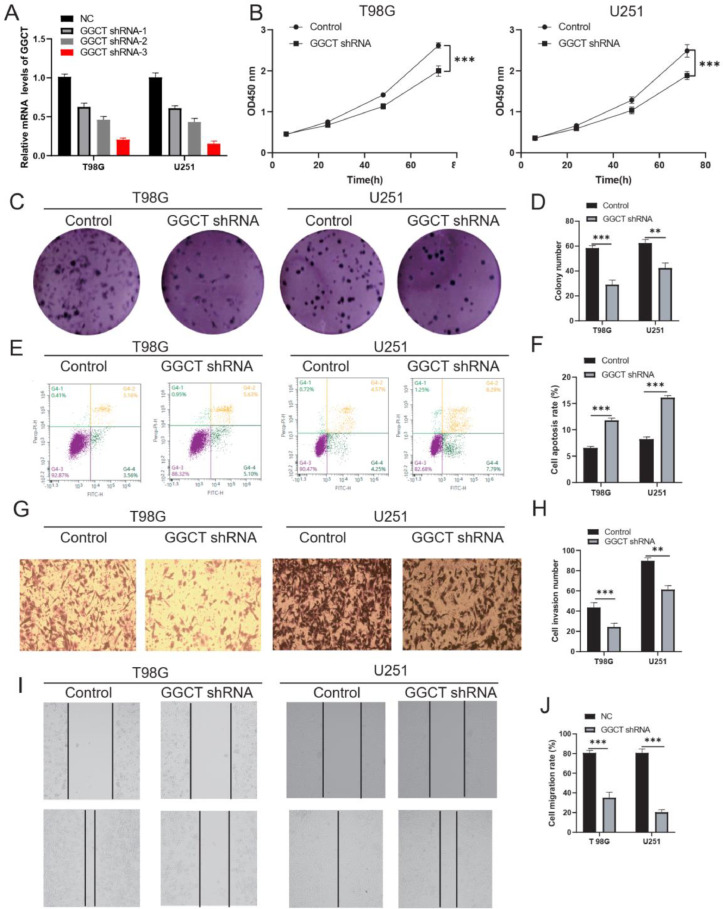
The function of GGCT in glioma cancer remains unclear and requires further investigation. First, we transfected three shRNAs (shRNA1, shRNA2, and shRNA3) into glioma cell T98G and U251. As a result, RT-qPCR assay indicated that shRNA3 displayed the most significant efficiency for GGCT knockdown among T98G and U251 cells (Fig. 2A). GGCT shRNA3 was selected for functional study of GGCT in T98G and U251 cells.CCK-8 assay shown that knockdown of GGCT significantly reduced the proliferation of T98G and U251 cells (Fig. 2B). Silencing of GGCT reduced colony formation of T98G and U251 cells (Fig. 2C and 2D). Next, knockdown of GGCT enhanced the apoptotic rates of T98G and U251 cell by flow cytometry assay (Fig. 2E and 2F). Next, transwell assay verified that cell invasion was inhibited in the GGCT shRNA group compared with NC group (Fig. 2G,2H). Wound healing assay revealed that GGCT silencing decreased cell migration ability of T98G and U251 cell (Fig. 2I-J). The results indicated that knockdown of GGCT repressed cell malignant phenotype in T98G and U251 cells.
Fig. 2.
GGCT silencing inhibited cell malignant phenotype of T98G and U251 in vitro. (A) RT-qPCR analysis of the knockdown efficiency of GGCT lentiviruses. (B) CCK-8 assay detected the cell proliferation. (C) Colon formation was used to explore the effect of GGCT on the proliferation of T98G and U251 cells. (D) Cell clone formation number was calculated. (E) Flow cytometry assay was used to verify the effect of GGCT on the apoptosis of T98G and U251 cells. (F) Cell apoptosis rate was calculated. (G) Transwell assay was used to detect the cell invasion. (H) Cell migration number was calculated. (I) Wound healing assay was performed to explore the cell migration. (J) Area wound healed (%) was analyzed. * P < 0.05, ** P < 0.01, *** P < 0.001.
RNA-sequencing revealed that GGCT silencing regulated cancer related pathways and inhibited rest in glioma
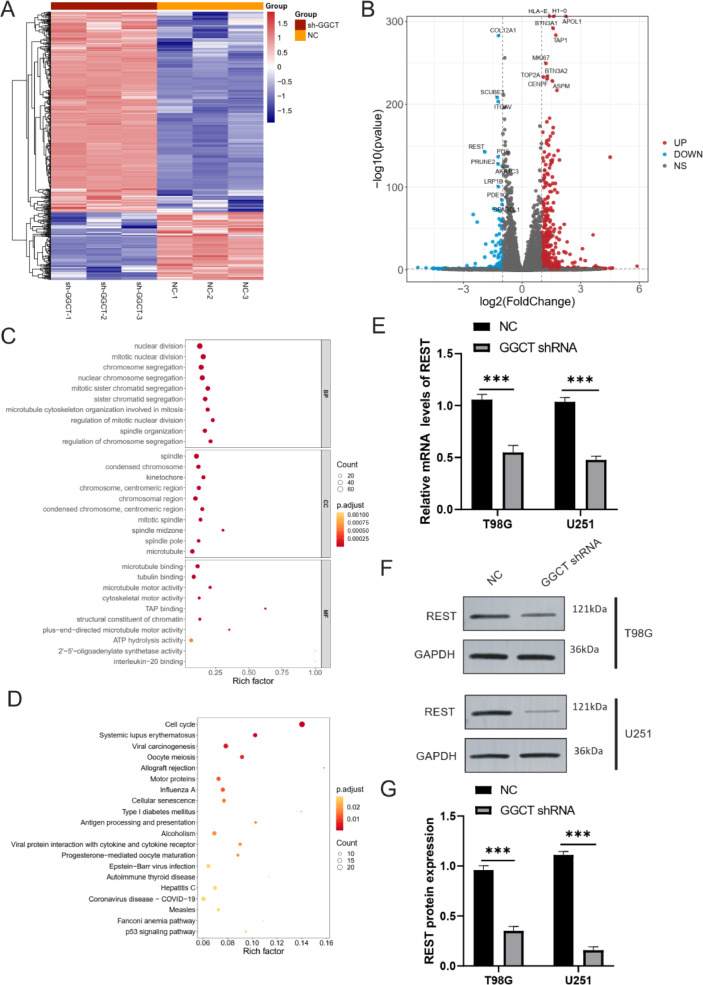
The above functional and phenotype of GGCT paves us to explore the underlying mechanism of GGCT in glioma cells. In order to explore the impact of GGCT silencing on tumor related pathways and genes, we knocked down GGCT in T98G cells and performed RNA sequencing to detect changes in downstream gene expression, followed by biological pathway and functional enrichment analysis. The heatmap of 457 differentially expressed genes in between NC groups and GGCT shRNA groups, including 342 significantly upregulated genes and 113 significantly downregulated gens in GGCT shRNA groups (Fig. 3A). The volcano map of top 10 upregulated genes (APOL1, HLA-E, H1–0, BTN3A1, MKI67, TOP2A, CENPF, ASPM, BTN3A2, TAP1) and downregulated genes (COL12A1, SCUBE3, ITGAV, REST, PI15, PRUNE2, AKR1C3, LRP1B, PDE1C)in GGCT shRNA groups (Fig. 3B). GO function enrichment analysis of the differentially expressed genes showed significantly enrichment of mitotic nuclear division, microtubule binding, mitotic spindle, mitosis and etc. (Fig. 3C). KEGG pathway enrichment analysis of the differentially expressed genes showed significantly enrichment of Cell cycle, Cellular senescence, P53 signaling pathway and etc. (Fig. 3D). Further, we focus on the expression of upregulated and downregulated genes in the top 10 in glioma to identify key molecules regulated by GGCT. The mRNA expression of ITGAV, REST, HLA_E, APOL1, TAP1, BTN3A1, MKI67, TOP2A, CENPF, ASPM, and BTN3A2 were significantly upregulated, and the mRNA expression of PRUNE2, LRP1B, PDE1C were significantly downregulated in glioma tissues compared normal tissues form TCGA-GBM dataset (Supplementary Figure 1A). Moreover, top10 upregulated/downregulated genes in GGCT shRNA expression were verified by RT-qPCR assay. Knockdown of GGCT reduced the oncogenes (ITGAV and REST) mRNA levels in T98G and U251 cells (Supplementary Figure 1B and 1C). Therefore, REST is considered a key downstream molecule of GGCT.
Fig. 3.
Functional enrichment analysis of DEGs in NC and GGCT shRNA T98G cells. (A) Heatmap of DEGs in the NC group compared with the GGCT shRNA group of T98G cell. (B) Volcano plot showed that upregulated genes (Red) and downregulated genes (Blue) in GGCT shRNA group compared with NC group. (C) The top10 enriched biological process (BP), CC and MF of the DEGs. (D) The top20 enriched KEGG pathway of DEGs. (E) RT-qPCR assay was used to verify the GGCT mRNA expression in T98G and U251 cells. (F) Western blot assay was used to detect the REST protein expression in T98G and U251 cells. (G) REST protein expression was analyzed in T98G and U251 cells of NC and GGCT shRNA. * P < 0.05, ** P < 0.01, *** P < 0.001.
REST is negative regulator of neuron specific expression. Previous study has reported that high expression of REST reduced the chemosensitivity of patients with GBM and shortens the overall survival. Next, RT-qPCR assay confirmed that GGCT silencing inhibited the mRNA expression of REST in T98G and U251 cells (Fig. 3E). Additionally, western blot assay indicated that knockdown of GGCT reduced the REST protein expression in T98G and U251 cells (Fig. 3F and 3G). Overall, RNA-sequencing revealed that GGCT silencing regulated cancer related pathways and downregulated REST in Glioma cells.
REST expression was upregulated in glioma cancer and associated with a poor prognosis
Moreover, we analysis the expression and clinical significance of REST in glioma. REST was overexpressed in GBM and LGG tumor tissues compared with normal tissues from TCGA dataset (Fig. 4A). Bioinformatic analyses from the CGGA database (Fig. 4B) showed obviously elevated REST expression in high grade glioma (WHO IV), IDH wildtype, 1p/19q co-deletion non-codel, age (≥42). Next, overall survival analysis in diverse datasets showed GGCT high expression was associated with poor prognosis of glioma (Fig. 4C). High expression of REST was also significantly associated with the shorter overall survival and disease-specific survival of LGG patients in TCGA dataset. We found there was no statistically significance in GBM patient. We also performed the survival analysis of REST in primary glioma and recurrent glioma, and there was a tendency of high REST predicting a poorer prognosis in primary and recurrent glioma. Meanwhile, high expression of REST has a poorer prognosis in GEO dataset. Overall, REST was aberrantly overexpressed in glioma and correlated with poor prognosis.
Fig. 4.
Expression and prognosis value of REST in glioma. (A) The expression of REST was analyzed in GBM and LGG. (B) The expression of REST was analyzed in Grade, IDH mutation status, 1p/19q co-deletion status and age status of glioma. (C) Overall survival of disease-free survival of REST high expression/low expression was analyzed in Low grade glioma, Glioblastoma, CGGA datasets, GSE4412 dataset. * P < 0.05, ** P < 0.01, *** P < 0.001.
REST promoted proliferation, invasion, migration and inhibited apoptosis of glioma cells in vitro
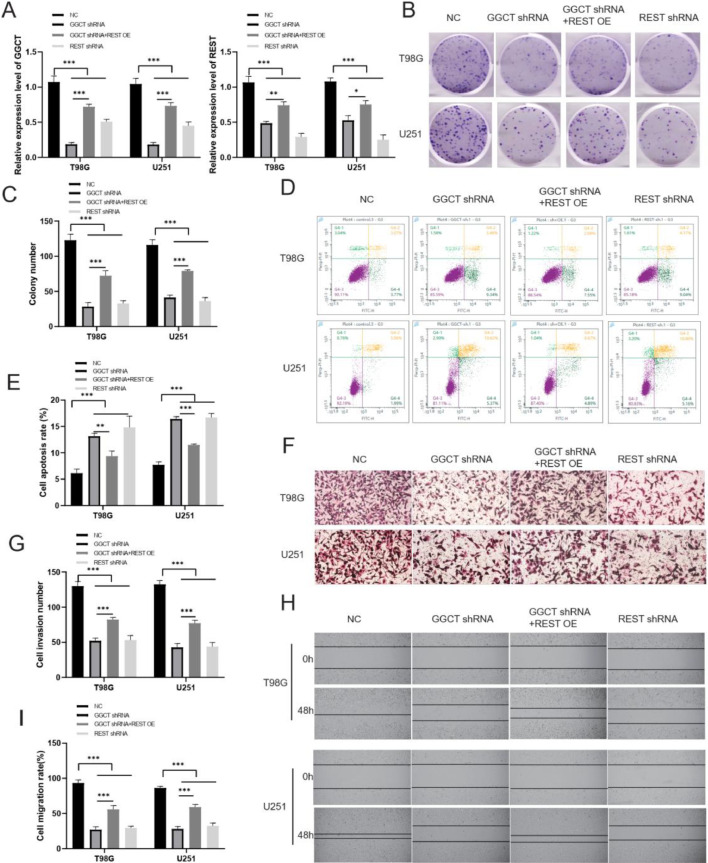
The function of REST in glioma cancer remains unclear and requires further investigation. Cell functional experiments were divided into four groups: NC, GGCT shRNA, GGCT shRNA + REST OE, REST shRNA group. Firstly, RT-qPCR assay indicated that GGCT and REST expression were reduced in GGCT shRNA group and REST shRNA group compared with control, and REST overexpression restored the GGCT and REST expression (Fig. 5A). Silencing of GGCT/REST reduced colony formation of T98G and U251 cells, REST overexpression restored the effect of GGCT shRNA on colony formation of T98G and U251 cells (Fig. 5B and 5C). Next, knockdown of REST enhanced the apoptotic rates of T98G and U251 cell, and REST overexpression restored the effect of GGCT shRNA on T98G and U251 cell apoptosis by flow cytometry assay (Fig. 5D and 5E). Next, transwell assay verified that cell invasion was inhibited in the REST shRNA group compared with NC group (Fig. 5F,5G). Wound healing assay revealed that REST silencing decreased cell migration ability of T98G and U251 cell, and REST overexpression restored the effect of GGCT shRNA on T98G and U251 cell migration (Fig. 5H,5I). The results indicated that knockdown of REST repressed cell malignant phenotype in T98G and U251 cells.
Fig. 5.
REST acts as an oncogene in vitro. (A) The mRNA level of GGCT and REST were validated through RT-qPCR assay. (B) Colon formation was used to explore the effect of REST on the proliferation of T98G and U251 cells. (C) Cell clone formation number was calculated. (D) Flow cytometry assay was used to verify the effect of REST on the apoptosis of T98G and U251 cells. (E) Cell apoptosis rate was calculated. (F) Transwell assay was used to detect the cell invasion. (G) Cell migration number was calculated. (H) Wound healing assay was performed to explore the cell migration. (I) Area wound healed (%) was analyzed. * P < 0.05, ** P < 0.01, *** P < 0.001.
Knockdown of GGCT suppressed the tumor growth via downregulated REST in vivo
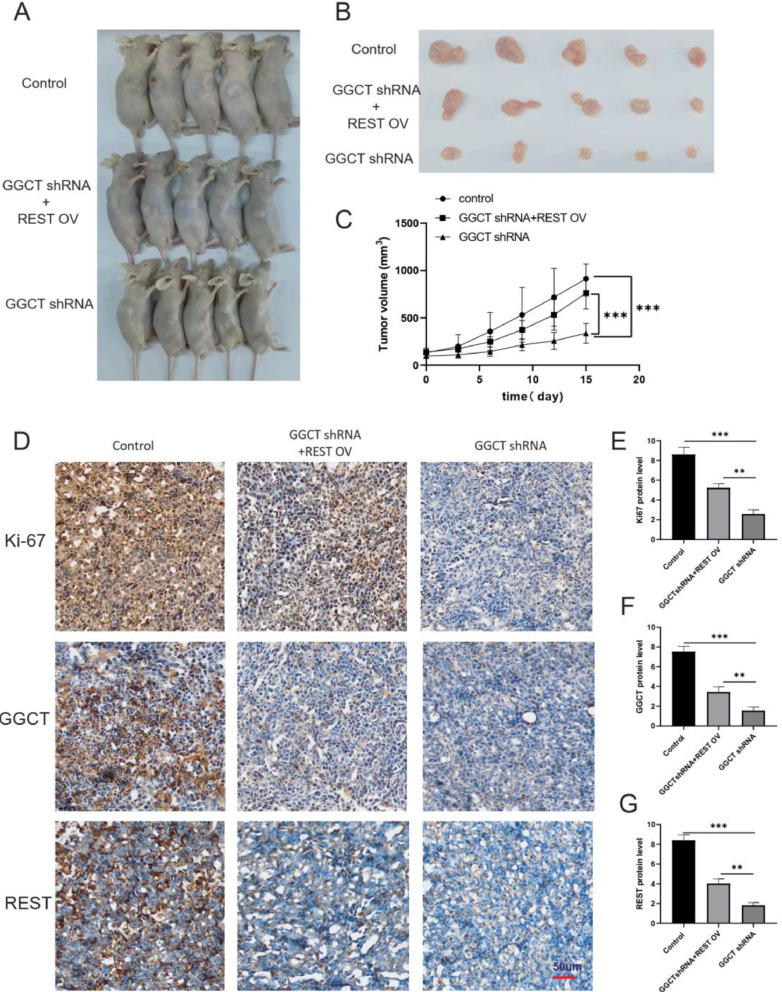
To assess the role of GGCT via REST in glioma tumorigenesis in vivo, we studies effect of GGCT silencing and REST overexpression on tumor growth by inducing tumor formation in nude BALB/c mice through subcutaneous injection of tumor cells. The mice were randomly divided into three groups: the control group (T98G cells), T98G cells with stably GGCT shRNA expression (GGCT shRNA), and T98 cells with stably GGCT shRNA + REST overexpression (REST OV). All group T98 cells were injected subcutaneously into 5 male BALB/c nude mice (Fig. 6A). Tumor size, volume, and weight in each group were measured to assess tumor growth. As shown in Fig. 6B and 6C, GGCT silencing significantly reduced tumor growth and volume, REST overexpression significantly restored the GGCT silencing reduced tumor growth and volume in vivo. Next, Ki-67 protein (a cell proliferation marker[17])expression was detected by IHC staining, and GGCT silencing significantly inhibited Ki-67 expression and REST overexpression restored the Ki-67 expression (Fig. 6D and 6E). Next, IHC staining results showed that GGCT and REST protein expression were reduced in GGCT shRNA group compared with control, and REST overexpression restored the GGCT and REST protein expression (Fig. 6D-6G). Therefore, GGCT silencing suppressed the tumor formation, growth of glioma cell via inhibited REST pathway in vivo.
Fig. 6.
GGCT knockdown repressed the formation of xenograft tumors via REST. (A) Representative images of each group in vivo xenograft tumors (n = 5). (B) Representative images of tumor harvested from nude mice inoculated with T98G cells of Control group, GGCT shRNA + REST OV group, GGCT shRNA group. (C) Tumor volume was measured every 3 days (n = 5). (D) Ki-67, GGCT and REST protein expression was detected by IHC staining in tumor tissues of Control group, GGCT shRNA + REST OV group, GGCT shRNA group. (E) Ki-67, (F) GGCT, (G) REST protein expression was statistical analyzed from IHC staining results (n = 5). * P < 0.05, ** P < 0.01, *** P < 0.001.
REST increased GGCT expression via inhibiting miR-34a-5p expression in glioma cells
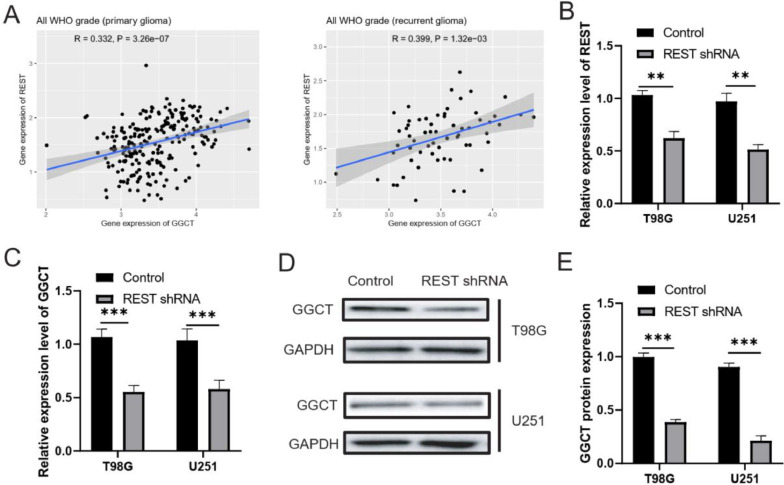
It is widely acknowledged that REST plays a vital role in cancer progression, by transcription regulation of its target genes. Therefore, we explored that the regulate correlation GGCT with REST in glioma cells. Firstly, we found that GGCT expression was positively correlated with REST expression in All WHO grade (primary glioma and recurrent glioma) (Fig. 7A). Next, REST knockdown significantly reduced REST and GGCT mRNA level in T98G and U251 cells (Fig. 7B and 7C). Further, Western blotting assay revealed that REST silencing significantly recued GGCT protein expression in T98G and U251 cells (Fig. 7D and 7E). Together, these results suggested that REST silencing decreased GGCT expression in glioma cells.
Fig. 7.
REST increased GGCT expression in TG98G and U251 cells. (A) The GGCT expression was positively correlated with REST expression. (B) RT-qPCR assay was used to verify the transfected efficacy of REST. (C) RT-qPCR assay was used to detect the mRNA expression of GGCT. (D) Western blotting assay was used to detect GGCT protein level. (E) The GGCT protein level was measured. * P < 0.05, ** P < 0.01, *** P < 0.001.
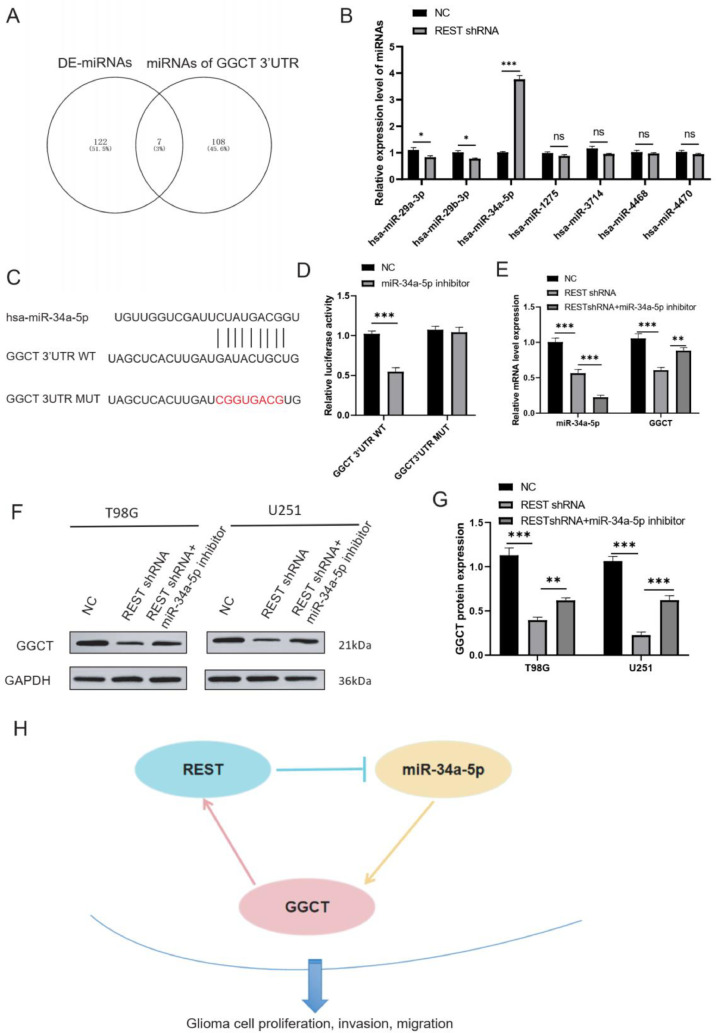
REST is an inhibitory transcription factor that acts by binding to miRNAs to regulate target genes expression. Here, we hypothesized that REST increased GGCT expression by inhibiting miRNA transcriptional expression in glioma. To identify the key miRNA in between REST and GGCT in glioma cell, we analyzed the differentially expressed miRNAs in glioma tumor compared with normal groups based on online website (https://www.biosino.org/dbDEMC/index). We found that total of 129 differentially expressed miRNAs in between glioma tissues and normal tissues (Supplementary Table 1). Further, the total of 115 miRNAs of targeted GGCT 3′UTR was predicted by using miRanda dataset (Supplementary Table 2). Venn diagram showed the common 7 miRNAs in between DE-miRNAs and miRNAs of GGCT 3′UTR (Fig. 8A). RT-qPCR assay confirmed that REST silencing significantly upregulated hsa-miR-34a-5p, and downregulated hsa-miR-29a-3p, hsa-miR-29b-3p in T98G cell (Fig. 8B). has-miR-34a-5p was chosen to further study. As shown in Fig. 8C, hsa-miR-34a-5p targeted GGCT 3′UTR WT and MUT sites. Relative luciferase activity analysis confirmed that hsa-miR-34a-5p targeted GGCT 3′UTR in T98G cells (Fig. 8D). RT-qPCR analysis revealed that has-miR-34a-5p inhibitor restored the role of REST decreased GGCT mRNA expression in T98G cells. Next, western blotting assay confirmed that REST silencing inhibited GGCT protein expression, and miR-34a-5p inhibitor restored the effect of REST shRNA on the GGCT protein expression in T98G and U251 cells (Fig. 8F and 8G). Therefore, REST increased GGCT expression via inhibiting miR-34a-5p expression in glioma cells (Fig. 8H).
Fig. 8.
REST increased GGCT expression via inhibiting miR-34a-5p expression in glioma cells. (A) Veen diagram was used to visualize the common miRNAs in DE-miRNAs and miRNAs of target GGCT 3′UTR. (B) RT-qPCR assay was used to confirm the miRNAs of REST negatively regulate in T98G cells. (C) has-miR-34a-5p binds to the site of GGCT 3′UTR. (D) Dual-luciferase reporter genes activity assay in T98G cells. (E) RT-qPCR assay was used to detect the GGCT and miR-34a-5p mRNA expression in T98G cells. (F) Western blot assay was used to detect the GGCT protein expression. (G) GGCT protein expression level was statistic in T98 an dU251 cells. (H) Schematics diagram depicting the key findings of this study. * P < 0.05, ** P < 0.01, *** P < 0.001.
Discussion
GGCT, initially recognized for its involvement in γ-glutathione synthesis and degradation [23], has recently garnered attention for its heightened expression in various tumor tissues [24,25]. GGCT influence cell morphological variations, senescence, autophagy, epithelial-mesenchymal transition, and intracellular signaling pathways in diseases [7,24]. GGCT assumes diverse roles in cancer onset and progression, emerging as a potential molecular target for treating gastrointestinal cancers [26]. Our study revealed a significant overexpression of GGCT in glioma, which correlated with unfavorable prognosis. Notably, GGCT knockdown exhibited in a significant inhibition of cell proliferation, migration, invasion, and induction of apoptosis in T98G and U251 cells. Mechanistically, RNA-sequencing analysis revealed that GGCT silencing regulated cancer related pathways and inhibited REST in glioma. Elevated REST expression has been associated with heightened invasiveness in glioma patients [27]. Furthermore, REST, found to be upregulated in glioma tissues and U87 and U251 cells, demonstrated a significant impact on proliferation and apoptosis when its expression was inhibited [28]. In vivo and in vitro experiments solidified the observation that GGCT knockdown suppressed tumor growth, cell proliferation, invasion and migration by downregulating REST. These findings underscore the intricate interplay between GGCT and REST in glioma progression and offer insights into potential therapeutic avenues.
REST, recognized as an inhibitory transcription factor, exerts its function by binding to neuron-restricted silencing factors found in numerous neuronal genes [10,29]. The C-terminal domain of REST interacts with its cofactor coREST, which, in turn, recruits various epigenetic regulators [30]. REST exhibits versatility in chromatin remodeling, involving both local and distal modifications such as alterations in nucleosome positioning and histone modifications [31]. Additionally, REST plays a pivotal role in miRNA regulation alongside CREB by governing genes involved in miRNA biosynthesis [32]. Notably, REST inhibits miR-124 and miR-203, thereby promoting oncogenic properties in GBM stem cells [32]. Several of the miRNAs regulated by REST participate in feedback loops crucial for the self-renewal and maintenance of neural stem cells [33].
In our investigation, we discerned that REST, upon transfection, exerts transcriptional repression on miR-34a-5p expression in glioma cells. A prior study has documented that circITGA7 enhances glioma cell proliferation by modulating the miR-34a-5p/VEGFA axis [34]. The activation of positive feedback loops stands out as a crucial mechanism driving the progression of glioma [35,36]. Intriguingly, our findings indicate that GGCT is regulated by miR-34a-5p, serving as a direct target of miR-34a-5p in glioma cells. Notably, we have unveiled an unprecedented positive feedback loop involving GGCT and REST, contributing to the advancement of glioma and tumorigenesis. REST, functioning as an inhibitory transcription factor, suppresses the transcription of miR-34a-5p in glioma cells. The diminished expression of miR-34a-5p fosters the upregulation of GGCT in glioma cells, subsequently elevating REST expression. The development of new inhibitors targeting GGCT specifically holds promise as a novel therapeutic avenue to impede glioma growth and recurrence.
The interaction between GGCT and REST is a complex process, and our research mainly focuses on the regulation of GGCT by REST through miR-34a-5p. We recognize that studying the regulation of REST by GGCT will provide additional depth and dimension to our understanding. The regulation of REST by GGCT is an important research area, and we plan to explore this pathway in future studies. We believe that this will offer us a more comprehensive perspective to comprehend the intricate interactions between these two proteins.
Conclusions
In summary, our study has unveiled a positive feedback loop involving GGCT and REST that propels the malignant progression of glioma through the regulatory transcriptional inhibition of miR-34a-5p. The GGCT/REST/miR-34a-5p axis emerges as a potential therapeutic target, offering a promising avenue for advancing glioma treatment.
Declarations
Ethics approval and consent to participate
The Animal experiment protocol listed below has been reviewed and approved by Laboratory animal management ethics committee of Xiamen University (Approval No 2,023,051).
Consent for publication
No conflict of interest exits in the submission of this manuscript, and manuscript is approved by all authors for publication.
Availability of data and materials
All data can be accessed by contacting the corresponding author.
CRediT authorship contribution statement
Shang-Hang Shen: Writing – original draft, Conceptualization. Si-Fang Chen: Methodology, Formal analysis. Jian-Feng Guo: Validation, Software. Zhan-Xiang Wang: Writing – review & editing, Visualization.
Declaration of competing interest
The authors have no conflict of interest.
Acknowledgement
None.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2024.102083.
Appendix. Supplementary materials
References
- 1.Chen R., Smith-Cohn M., Cohen A.L., Colman H. Glioma subclassifications and their clinical significance. Neurotherapeutics. 2017;14(2):284–297. doi: 10.1007/s13311-017-0519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sturm D., Pfister S.M., Jones D.T.W. Pediatric gliomas: current concepts on diagnosis, biology, and clinical management. J. Clin. Oncol. 2017;35(21):2370–2377. doi: 10.1200/JCO.2017.73.0242. [DOI] [PubMed] [Google Scholar]
- 3.Aldape K., Zadeh G., Mansouri S., Reifenberger G., von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129(6):829–848. doi: 10.1007/s00401-015-1432-1. [DOI] [PubMed] [Google Scholar]
- 4.Ostrom Q.T., Bauchet L., Davis F.G., Deltour I., Fisher J.L., Langer C.E., et al. The epidemiology of glioma in adults: a "state of the science" review. Neuro Oncol. 2014;16(7):896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexander B.M., Cloughesy T.F. Adult glioblastoma. J. Clin. Oncol. 2017;35(21):2402–2409. doi: 10.1200/JCO.2017.73.0119. [DOI] [PubMed] [Google Scholar]
- 6.Li H.N., Zhang H.M., Li X.R., Wang J., Xu T., Li S.Y., et al. MiR-205-5p/GGCT attenuates growth and metastasis of papillary thyroid cancer by regulating CD44. Endocrinology. 2022;163(4) doi: 10.1210/endocr/bqac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H.M., Li Z.Y., Dai Z.T., Wang J., Li L.W., Zong Q.B., et al. Interaction of MRPL9 and GGCT promotes cell proliferation and migration by activating the MAPK/ERK pathway in papillary thyroid cancer. Int. J. Mol. Sci. 2022;23(19) doi: 10.3390/ijms231911989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W., Chen L., Xiang H., Hu C., Shi W., Dong P., et al. Knockdown of GGCT inhibits cell proliferation and induces late apoptosis in human gastric cancer. BMC. Biochem. 2016;17(1):19. doi: 10.1186/s12858-016-0075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Q., Zhou Y., Li Y., Liao Z. GGCT promotes colorectal cancer migration and invasion via epithelial-mesenchymal transition. Oncol. Lett. 2020;20(2):1063–1070. doi: 10.3892/ol.2020.11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y., Wu T., Wang Y., Yang L., Hu C., Chen L., et al. gamma-Glutamyl cyclotransferase contributes to tumor progression in high grade serous ovarian cancer by regulating epithelial-mesenchymal transition via activating PI3K/AKT/mTOR pathway. Gynecol. Oncol. 2018;149(1):163–172. doi: 10.1016/j.ygyno.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 11.Nohara Y., Taniguchi K., Ii H., Masuda S., Kawakami H., Matsumoto M., et al. Development of an activity-based chemiluminogenic probe for gamma-glutamylcyclotransferase. Org. Biomol. Chem. 2023;21(29):5977–5984. doi: 10.1039/d3ob00655g. [DOI] [PubMed] [Google Scholar]
- 12.Kageyama S., Ii H., Taniguchi K., Kubota S., Yoshida T., Isono T., et al. Mechanisms of tumor growth inhibition by depletion of gamma-glutamylcyclotransferase (GGCT): a novel molecular target for anticancer therapy. Int. J. Mol. Sci. 2018;19(7) doi: 10.3390/ijms19072054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taniguchi K., Ii H., Kageyama S., Takagi H., Chano T., Kawauchi A., et al. Depletion of gamma-glutamylcyclotransferase inhibits cancer cell growth by activating the AMPK-FOXO3a-p21 axis. Biochem. Biophys. Res. Commun. 2019;517(2):238–243. doi: 10.1016/j.bbrc.2019.07.049. [DOI] [PubMed] [Google Scholar]
- 14.Shen S.H., Yu N., Liu X.Y., Tan G.W., Wang Z.X. Gamma-glutamylcyclotransferase promotes the growth of human glioma cells by activating Notch-Akt signaling. Biochem. Biophys. Res. Commun. 2016;471(4):616–620. doi: 10.1016/j.bbrc.2016.01.165. [DOI] [PubMed] [Google Scholar]
- 15.Singh S., Ramamoorthy M., Vaughan C., Yeudall W.A., Deb S., Palit Deb S. Human oncoprotein MDM2 activates the Akt signaling pathway through an interaction with the repressor element-1 silencing transcription factor conferring a survival advantage to cancer cells. Cell Death. Differ. 2013;20(4):558–566. doi: 10.1038/cdd.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J.H., Chai Y.G., Hersh L.B. Expression patterns of mouse repressor element-1 silencing transcription factor 4 (REST4) and its possible function in neuroblastoma. J. Mol. Neurosci. 2000;15(3):205–214. doi: 10.1385/JMN:15:3:205. [DOI] [PubMed] [Google Scholar]
- 17.Wong T.L., Ng K.Y., Tan K.V., Chan L.H., Zhou L., Che N., et al. CRAF methylation by PRMT6 regulates aerobic glycolysis-driven hepatocarcinogenesis via ERK-Dependent PKM2 nuclear relocalization and activation. Hepatology. 2020;71(4):1279–1296. doi: 10.1002/hep.30923. [DOI] [PubMed] [Google Scholar]
- 18.Yucebas M., Yilmaz Susluer S., Onur Caglar H., Balci T., Dogan Sigva Z.O., Akalin T., et al. Expression profiling of RE1-silencing transcription factor (REST), REST corepressor 1 (RCOR1), and Synapsin 1 (SYN1) genes in human gliomas. J. BUON. 2016;21(4):964–972. [PubMed] [Google Scholar]
- 19.Marisetty A.L., Lu L., Veo B.L., Liu B., Coarfa C., Kamal M.M., et al. REST-DRD2 mechanism impacts glioblastoma stem cell-mediated tumorigenesis. Neuro Oncol. 2019;21(6):775–785. doi: 10.1093/neuonc/noz030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang J., Meng Q., Zhao W., Tong P., Li P., Zhao Y., et al. An expression based REST signature predicts patient survival and therapeutic response for glioblastoma multiforme. Sci. Rep. 2016;6:34556. doi: 10.1038/srep34556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 2000e-pub ahead of print (1):28. [DOI] [PMC free article] [PubMed]
- 22.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28(11) doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito Y., Taniguchi K., Ii H., Horinaka M., Kageyama S., Nakata S., et al. Identification of c-Met as a novel target of gamma-glutamylcyclotransferase. Sci. Rep. 2023;13(1):11922. doi: 10.1038/s41598-023-39093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia C., Gao J., Wang L., Li Z., Dong Z., Yao L., et al. miR-877 inhibits the proliferation, migration, and invasion of osteosarcoma cells by targeting gamma-glutamylcyclotransferase. Endocr. J. 2021;68(9):1109–1116. doi: 10.1507/endocrj.EJ20-0752. [DOI] [PubMed] [Google Scholar]
- 25.Sun W., Wang X., Li G., Ding C., Wang Y., Su Z., et al. Development of a thyroid cancer prognostic model based on the mitophagy-associated differentially expressed genes. Discov. Oncol. 2023;14(1):173. doi: 10.1007/s12672-023-00772-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kageyama S., Hanada E., Ii H., Tomita K., Yoshiki T., Kawauchi A. Gamma-Glutamylcyclotransferase: a novel target molecule for cancer diagnosis and treatment. Biomed. Res. Int. 2015;2015 doi: 10.1155/2015/345219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G., Yang X., Qi M., Li M., Dong M., Xu R., et al. Systematic analysis identifies REST as an oncogenic and immunological biomarker in glioma. Sci. Rep. 2023;13(1):3023. doi: 10.1038/s41598-023-30248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang D., Li Y., Wang R., Li Y., Shi P., Kan Z., et al. Inhibition of REST suppresses proliferation and migration in glioblastoma cells. Int. J. Mol. Sci. 2016;17(5) doi: 10.3390/ijms17050664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majumder S. REST in good times and bad: roles in tumor suppressor and oncogenic activities. Cell Cycle. 2006;5(17):1929–1935. doi: 10.4161/cc.5.17.2982. [DOI] [PubMed] [Google Scholar]
- 30.Alsaqati M., Davis B.A., Wood J., Jones M.M., Jones L., Westwood A., et al. NRSF/REST lies at the intersection between epigenetic regulation, miRNA-mediated gene control and neurodevelopmental pathways associated with Intellectual disability (ID) and Schizophrenia. Transl. Psychiatry. 2022;12(1):438. doi: 10.1038/s41398-022-02199-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng D., Zhao K., Mehler M.F. Profiling RE1/REST-mediated histone modifications in the human genome. Genome Biol. 2009;10(1):R9. doi: 10.1186/gb-2009-10-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marisetty A.L., Singh S.K., Nguyen T.N., Coarfa C., Liu B., Majumder S. REST represses miR-124 and miR-203 to regulate distinct oncogenic properties of glioblastoma stem cells. Neuro Oncol. 2017;19(4):514–523. doi: 10.1093/neuonc/now232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nassar A., Kodi T., Satarker S., Gurram P.C., Fayaz S.M., Nampoothiri M. Astrocytic transcription factors REST, YY1, and putative microRNAs in Parkinson's disease and advanced therapeutic strategies. Gene. 2024;892 doi: 10.1016/j.gene.2023.147898. [DOI] [PubMed] [Google Scholar]
- 34.Qi L., Wang W., Zhao G., Jiang H., Zhang Y., Zhao D., et al. Circular RNA circitga7 accelerates glioma progression via miR-34a-5p/VEGFA axis. Aging. 2021;13(9):13138–13152. doi: 10.18632/aging.202996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu T., Hu J., Han B., Tan S., Jia W., Xin Y. A positive feedback loop of lncRNA-RMRP/ZNRF3 axis and Wnt/beta-catenin signaling regulates the progression and temozolomide resistance in glioma. Cell Death Dis. 2021;12(11):952. doi: 10.1038/s41419-021-04245-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang Y., Zhao J., Liu Y., Hu J., Gao L., Wang H., et al. CircKPNB1 mediates a positive feedback loop and promotes the malignant phenotypes of GSCs via TNF-alpha/NF-kappaB signaling. Cell Death Dis. 2022;13(8):697. doi: 10.1038/s41419-022-05149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data can be accessed by contacting the corresponding author.