Abstract
Background
Mitochondrial dysfunction and metabolic reprogramming can lead to the development and progression of hepatocellular carcinoma (HCC). Ferredoxin 1 (FDX1) is a small mitochondrial protein and recent studies have shown that FDX1 plays an important role in tumor cuproptosis, but its role in HCC is still elusive. In this study, we aim to investigate the expression and novel functions of FDX1 in HCC.
Methods
FDX1 expression was first analyzed in publicly available datasets and verified by immunohistochemistry, qRT-PCR and Western blot. In vitro and in vivo experiments were applied to explore the functions of FDX1. Non-targeted metabolomics and RNA-sequencing were used to determine molecular mechanism. mRFP-GFP-LC3 lentivirus transfection, Mito-Tracker Red and Lyso-Tracker Green staining, transmission electron microscopy, flow cytometry, JC-1 staining, etc. were used to analyze mitophagy or ROS levels. Hydrodynamic tail vein injection (HTVi) and patient-derived organoid (PDO) models were used to analyze effect of FDX1 overexpression.
Results
FDX1 expression is significantly downregulated in HCC tissues. FDX1 downregulation promotes HCC cell proliferation, invasion in vitro and growth, metastasis in vivo. In addition, FDX1 affects metabolism of HCC cells and is associated with autophagy. We then confirmed that FDX1 deficiency increases ROS levels, activates mitophagy and the PI3K/AKT signaling pathway in HCC cells. Interestingly, scavenging ROS attenuates the tumor-promoting role and mitophagy of FDX1 downregulation. The results of HTVi and PDO models both find that FDX1 elevation significantly inhibits HCC progression. Moreover, low FDX1 expression is associated with shorter survival and is an independent risk factor for prognosis in HCC patients.
Conclusions
Our research had investigated novel functions of FDX1 in HCC. Downregulation of FDX1 contributes to metabolic reprogramming and leads to ROS-mediated activation of mitophagy and the PI3K/AKT signaling pathway. FDX1 is a potential prognostic biomarker and increasing FDX1 expression may be a potential therapeutic approach to inhibit HCC progression.
Keywords: Hepatocellular carcinoma, Ferredoxin 1, Mitophagy, Reactive oxygen species, PI3K/AKT signaling pathway
1. Introduction
Liver cancer is the most common cause of cancer-related death in China, ranking fourth in terms of incidence and second in terms of mortality among men [1]. Hepatocellular carcinoma (HCC) is the most common subtype of primary liver cancer and accounts for 75–85% of all cases of liver cancer [2]. HCC is often diagnosed at an advanced stage when curative surgical treatment is no longer an option. Current molecularly targeted therapies and immunotherapies have shown limited clinical benefit due to drug resistance or recurrence, highlighting the urgent need for a deeper understanding of the molecular mechanisms underlying HCC progression and the identification of novel therapeutic targets to develop more effective treatments.
Ferredoxin 1 (FDX1) is a small iron-sulfur protein that transfers electrons from NADPH to mitochondrial cytochrome P450 via ferredoxin reductase (FDXR), and is involved in steroid, cholesterol and bile acid metabolism [3]. FDX1 plays an important role in maintaining metabolic homeostasis. A recent study has shown that FDX1 is involved in mouse embryonic development and that its deletion can lead to alterations in lipid metabolism, steatohepatitis, and adenocarcinoma [4]. In lung adenocarcinoma, FDX1 knockdown can lead to significant alterations in metabolites involved in glucose metabolism, fatty acid oxidation and amino acid metabolism [5]. Interestingly, an increasing number of studies suggest that FDX1 is associated with cuproptosis, namely copper-dependent, regulated cell death. FDX1 is an upstream regulator of protein lipoylation and can reduce Cu2+ to its more toxic form Cu1+, while deletion of FDX1 confers resistance to copper-induced cell death [6,7]. In addition to regulating cuproptosis, a recent study showed that FDX1 also regulates apoptosis and autophagy in polycystic ovary syndrome [8]. In glioma cells, C-MYC could upregulate FDX1 expression and inhibit the expression of mitochondrial autophagy protein (LC3), thereby inhibiting mitophagy [9]. Transcriptome sequencing of FDX1 overexpressing lung adenocarcinoma cells revealed that FDX1 plays a pivotal role in increasing GPRIN2 expression while concurrently suppressing PI3K signaling [10]. These studies suggest a variety of functions for FDX1.
In HCC, current studies of FDX1 also focus on cuproptosis and are predominantly bioinformatic analyses [11,12]. FDX1 is regulated by the LINC02362/hsa-miR-18a-5p axis, and overexpression of FDX1 can increase the sensitivity of HCC cells to oxaliplatin through cuproptosis [13]. Ferroptosis inducers can stabilize the FDX1 protein, which further promotes protein lipoylation and the transfer of reduced copper ions, thereby enhancing copper ionophores-induced cuproptosis in primary liver cancer [14]. In this study, we systematically investigated the expression of FDX1 in HCC, its functional roles, and potential interventions, with the aim of gaining further insight into the biological role of FDX1 and providing new targets for the treatment of HCC.
2. Materials and methods
2.1. Bioinformatics analysis
The FDX1 mRNA expression data, clinical data, and survival data in The Cancer Genome Atlas (TCGA) LIHC database were downloaded from the UCSC Xena browser (http://xena.ucsc.edu/) [15]. GSE36376 and GSE76427 in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) were used to analyze FDX1 mRNA expression. Protein expression of FDX1 in liver cancer was downloaded from UALCAN (https://ualcan.path.uab.edu/index.html) using data from the Clinical Proteomic Tumor Analysis Consortium (CPTAC) [16].
2.2. Patients and specimens
80 pairs of randomly selected HCC tissues and matched para-cancerous tissues were obtained from the Department of Hepatobiliary Surgery, Hunan Provincial People's Hospital/The First Affiliated Hospital of Hunan Normal University. The samples were fixed, embedded and then made into tissue microarrays (TMA). Clinicopathological data were collected for each patient. All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and later versions. Informed consent to be included in the study, or the equivalent, was obtained from all patients. This study was approved by the ethical committee of the Hunan Provincial People's Hospital/The First Affiliated Hospital of Hunan Normal University.
2.3. Immunohistochemical staining (IHC)
The universal two-step test kit (ZSGB-BIO, Beijing, China) was used for IHC of HCC tissues and xenografts. Briefly, sections were deparaffinized, hydrated, and blocked endogenous peroxidase, then sequentially incubated with primary antibody, reaction enhancer, and enhanced enzyme-labeled sheep anti-mouse/rabbit IgG polymer, and finally, DAB was used to develop the colors and hematoxylin to stain the nuclei. The staining intensity (SI) was multiplied by the percentage of positive cells (PP) to calculate the immunoreactive score (IRS) as previously described [17].
2.4. RNA extraction and qRT-PCR
Total RNA in HCC frozen tissues and cells was extracted using TRIzol™ reagent (Invitrogen, Carlsbad, CA). Reverse transcription was performed using BeyoRT™ II First Strand cDNA Synthesis Kit with gDNA Eraser (Beyotime, Shanghai, China). qRT-PCR was then performed using the BeyoFast™ SYBR Green qPCR Mix (Beyotime, Shanghai, China) in a QuantStudio 3 Real-Time PCR System (Applied Biosystems, Carlsbad, CA). GAPDH was used as an internal control. The 2−ΔCt or 2−ΔΔCT method was used for quantification. Sequences of primers were listed below: GAPDH-F: 5′-GAACGGGAAGCTCACTGG-3′, GAPDH-R: 5′- GCCTGCTTCACCACCTTCT-3’; FDX1-F: 5′- GACAATATGACTGTTCGAGTGCC-3′, FDX1-R: 5′- AGTTCAGGAGGTCTTGCCCA-3’.
2.5. Western blot
Total protein in HCC frozen tissues and cells was extracted using RIPA lysis buffer containing protease inhibitors (NCM, Suzhou, China). Protein concentrations were determined using a BCA protein assay kit (NCM, Suzhou, China). The equivalent amount of protein was separated by ExpressCast PAGE Gel Preparation kit (NCM, Suzhou, China) and transferred to PVDF membranes. After incubation with the appropriate antibody, the bands were developed using an ultrasensitive ECL chemiluminescence kit (NCM, Suzhou, China). Antibodies used in this study are listed in Table S1.
2.6. Transfection
FDX1 overexpression (Vector name: GV341) and knockdown (Vector name: GV112) lentiviruses (GeneChem, Shanghai, China) were transfected into corresponding HCC cells according to the manufacturer's instructions. Forty-eight hours after transfection, 2 μg/mL puromycin (Biosharp, Hefei, China) was used to select stable clones. The transfection efficiency was verified by qRT–PCR and Western blot. Sequences of shRNA were listed below: FDX1-shRNA1: 5′-TGGTGAAACATTAACAACCAA-3’; FDX1-shRNA2: 5′-GATGCCAGACAATCCATTGAT-3’; FDX1-shRNA3: 5′-GCAATCACTGATGAGGAGAAT-3’; FDX1-shCtrl: 5′-TTCTCCGAACGTGTCACGT-3’.
2.7. Cell proliferation assays
For the CCK-8 assay, approximately 1 × 103 cells were seeded into each well of a 96-well plate. 10 μl CCK-8 solution was added to each well and absorbance was measured at 450 nm after 1h incubation in a cell culture incubator. CCK-8 absorbance values were monitored for five consecutive days. For the colony formation assay, 5 × 102 cells were seeded in each well of a 6-well plate and cultured in a cell incubator for 2 weeks. The colonies were then stained with crystal violet and counted.
2.8. Flow cytometry
For cell cycle analysis, cells were harvested, washed with PBS, fixed in 70% pre-chilled ethanol, and stored overnight at −20 °C. The fixed cells were then stained with propidium iodide (PI) and DNA content was determined using a flow cytometer. The results were analyzed using ModFit software. For apoptosis analysis, the harvested HCC cells were stained with Annexin V-FITC and PI solution (Multisciences, Hangzhou, China). After incubation at room temperature for 20min in the dark, the cells were measured using a flow cytometer, and the results were analyzed using FlowJo software. To determine mitochondrial ROS levels, we gently collected HCC cells and stained with 5 μM Mitosox™ Red mitochondrial superoxide indicator (Invitrogen, Carlsbad, CA) at 37 °C for 30min in the dark. To determine intracellular ROS levels, HCC cells were collected and stained with 10 μM dihydroethidium (DHE) probe (Beyotime, Shanghai, China) at 37 °C for 30min in the dark. The cells were then washed and measured using a flow cytometer. Flow cytometry was performed using a CytoFLEX™ flow cytometer (Beckman Coulter, Brea, CA).
2.9. Cytoskeleton staining and immunofluorescence
HCC cells were seeded on confocal Petri dishes and cultured for 24h for attachment. Cells were fixed with 4% paraformaldehyde solution. For cytoskeleton staining, the cells were then stained with rhodamine-conjugated phalloidin (Roche, Basel, Switzerland) according to the manufacturer's protocol. For immunofluorescence, cells were incubated with primary antibodies followed by the appropriate fluorescent secondary antibody. DAPI was used for nuclear staining.
2.10. Cell migration and invasion assays
The wound healing assay was used to determine the ability of cells to migrate. HCC cells were seeded on six-well plates, grown to confluence, and then scratched with a pipette tip (10 μL). Cell invasiveness was measured using Matrigel-coated Transwell inserts (Corning Incorporated, NY). Equivalent amounts of cells were added in the upper chamber. After 24h, the cells in the upper membrane were removed and the inserts were fixed with methanol and stained with crystal violet. The scratch width and cells in the lower membrane were recorded under an inverted microscope. Before cell migration and invasion assays, cells were pre-treated with 10 μg/ml mitomycin-C to inhibit cell proliferation.
2.11. Animal studies
BALB/c nude mice (male, 4–6 weeks old) were used to establish HCC xenograft models, including subcutaneous and orthotopic models. For the subcutaneous model, approximately 5 × 106 HCC cells in 200 μL cold PBS were injected subcutaneously into the right flank (n = 5). The length (L) and width (W) of the tumors were monitored weekly using a vernier caliper. The tumor volume was calculated as follows: volume (mm3) = (W2 × L)/2. For the orthotopic model, the above subcutaneous tumors were cut into 1–2 mm3 cubes and transplanted into the mice liver (n = 5). Six weeks later, the mice were euthanized, and the livers and lungs were harvested. The final volume of the orthotopic tumors were measured. The xenograft tumors were sent for pathological examination. For NAC intervention, the mice were given drinking water supplemented with 6.5 g/L NAC. To induce HCC formation in mice, we performed hydrodynamic transfection in combination with Sleeping Beauty (SB)-mediated somatic integration as previously described [18]. Briefly, 20 μg of pT3-EF1α-c-Met (Addgene, Plasmid#31784) and 20 μg of pT3-EF1αH N90-beta-catenin (Addgene, Plasmid#86499) together with 5 μg of SB transposase (Addgene, Plasmid#34879) were added in 2 ml saline, and then injected into AAV-TBG-Ctrl or AAV-TBG-FDX1 C57BL/6 mice (6–8 weeks old, 5 mice per group) by hydrodynamic tail vein injection (HTVi). The livers of the mice were monitored using a small animal ultrasound imaging device and harvested for pathological examination after three months. All animal experiments were carried out in accordance with the relevant laws and approved by the Institutional Animal Care and Use Committee of the Hunan Provincial People's Hospital/The First Affiliated Hospital of Hunan Normal University.
2.12. TUNEL assay
Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) staining was performed in subcutaneous tumors using the Colorimetric TUNEL Apoptosis Assay Kit (Beyotime, Shanghai, China) according to the manufacturer's protocol. Briefly, sections were deparaffinized, hydrated, and then treated with proteinase K and 3% hydrogen peroxide solution. Sections were further treated with a biotin labelling solution and terminated. Streptavidin-HRP working solution and DAB were used to develop the colors and hematoxylin to stain the nuclei.
2.13. Non-targeted metabolomics and data analysis
Approximately 1 × 107 HCC cells (repeat six samples per group) were collected and metabolites were extracted with 80% methanol. All chromatographic separations were performed using an UltiMate 3000 UPLC System (Thermo Fisher Scientific, Waltham, MA). An ACQUITY UPLC T3 column (100 mm*2.1 mm, 1.8 μm, Waters, Milford, MA) was used for the reversed-phase separation. The column oven was maintained at 40 °C. Gradient elution conditions were set as follows: 0~0.8 min, 2 % B; 0.8~2.8 min, 2 %–70 % B; 2.8~5.6 min, 70 %–90 % B; 5.6–6.4 min, 90 %–100 % B; 6.4–8.0 min, 100 % B; 8.0~8.1 min, 100 %–2 % B; 8.1~10 min, 2 % B. A high-resolution tandem mass spectrometer TripleTOF 6600 (SCIEX, Framingham, MA) was used to detect metabolites eluted from the column. The Q-TOF was operated in both positive and negative ion modes. The acquired MS data pretreatments were performed using XCMS software. The online KEGG, HMDB database was used to annotate the metabolites by matching the exact molecular mass data (m/z) of samples with those from the database. The VIP value was calculated and a cut-off value of 1.0 was used to select important features.
2.14. Double-labeled lentivirus mRFP-GFP-LC3 transfection
HCC cells were seeded on confocal Petri dishes and cultured for 24h for attachment. Cells were then transfected with mRFP-GFP-LC3 lentivirus (Hanbio, Shanghai, China) according to the manufacturer's protocol. Nuclei were stained with Hoechst 33342.
2.15. Assessment of lysosome and mitochondria colocalization
HCC cells were prepared in confocal Petri dishes. Mitochondria of live cells were stained with 100 nM Mito-Tracker Red (Beyotime, Shanghai, China) working solution at 37 °C for 20min. The cells were then further stained with 50 nM Lyso-Tracker Green (Beyotime, Shanghai, China) working solution at 37 °C for 15min. Cell nuclei were stained with Hoechst 33342. The co-localization of lysosomes and mitochondria was analyzed using ImageJ software.
2.16. Transmission electron microscopy (TEM)
Adherent HCC cells were washed twice with pre-cooled PBS, and then collected and fixed with 2.5% glutaraldehyde at 4 °C for 24h. The samples were then treated with 1% osmic acid fixative solution for 2h, dehydrated through graded acetone, and further embedded in resin embedding agent. Ultrathin sections were prepared, stained with uranyl acetate and lead citrate, and examined by electron microscopy JEM-1400 (JEOL, Japan).
2.17. RNA-sequencing and data analysis
Total RNA was isolated from HCC cells using the Magzol Reagent (Magen, China) according to the manufacturer's protocol. The quantity and integrity of RNA was assessed using the K5500 ultra-micro spectrophotometer (Kaiao, Beijing, China) and the Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA) separately. The mRNA was then enriched and fragmented to approximately 200bp. The RNA fragments were then subjected to first- and second-strand cDNA synthesis followed by adaptor ligation and low-cycle enrichment according to the instructions of the NEBNext® Ultra™ RNA Library Prep Kit for Illumina. The libraries were sequenced by Illumina sequencing platform (Illumina, San Diego, CA) with 150bp paired-end reads at Ribobio Co. Ltd (Ribobio, Guangzhou, China). Raw data were processed using Trimmomatic tools (v 0.36). HTSeq (v 0.12.4) was used to count the number of reads mapped to each gene. Statistically significant differentially expressed genes (DEGs) were obtained with an adjusted P-value threshold of <0.05 and |log2(fold change)| > 1 using the DESeq2 software. The clusterProfiler package in R Bioconductor was used to identify and visualize the KEGG pathways enriched by all DEGs.
2.18. JC-1 staining
HCC cells were prepared in confocal Petri dishes, 1 ml of JC-1 staining working solution (Beyotime, Shanghai, China) was added and incubated at 37 °C for 20min in a cell incubator. The supernatant was then aspirated and washed twice with JC-1 staining buffer. JC-1 aggregates present in normal mitochondria show red fluorescence, while JC-1 monomers present in unhealthy mitochondria produce green fluorescence. Cell nuclei were stained with Hoechst 33342.
2.19. Establishment of patient-derived organoids (PDOs) and lentiviral transfection
HCC tissue was obtained from fresh surgical specimens under sterile conditions. The tissue was washed by cold DMEM containing 1% penicillin/streptomycin and cut into 1–3 mm3 pieces and further dissociated by TrypLE (Thermo Fisher Scientific, Waltham, MA) at 37 °C. The HCC cells were then filtered through a 100 μM cell strainer. Next, the HCC cells were mixed with Matrigel and placed in a cell culture incubator for 15min to allow them to solidify. The PDOs were cultured in human liver organoid culture medium (Orgen Biotech, Guangzhou, China) and passaged every 1–2 weeks. For lentiviral transfection, PDOs were dissociated, collected and suspended in culture medium. Lentivirus was added to the medium and the organoid–lentivirus mixture was transferred to a Matrigel-precoated plate and incubated at 37 °C for 12 h. The transfected PDOs were collected and cultured with complete medium. The size of the PDOs was monitored at different time intervals.
2.20. Statistical analyses
Statistical analyses were performed using SPSS 20.0 (SPSS Inc., Chicago, IL) or OriginPro 2021 (OriginLab, Northampton, MA). Data are presented as the mean ± SD of at least three independent experiments. Differences between the two groups were analyzed by Student's t-test when the variance was homogeneous or by the Mann-Whitney U test when the variance was non-homogeneous. Survival curves were plotted using the Kaplan–Meier method and analyzed using the log-rank test. Univariate and multivariate analyses were performed using the Cox proportional hazards model to identify independent risk factors. A P value (two-tailed) of less than 0.05 was considered statistically significant.
3. Results
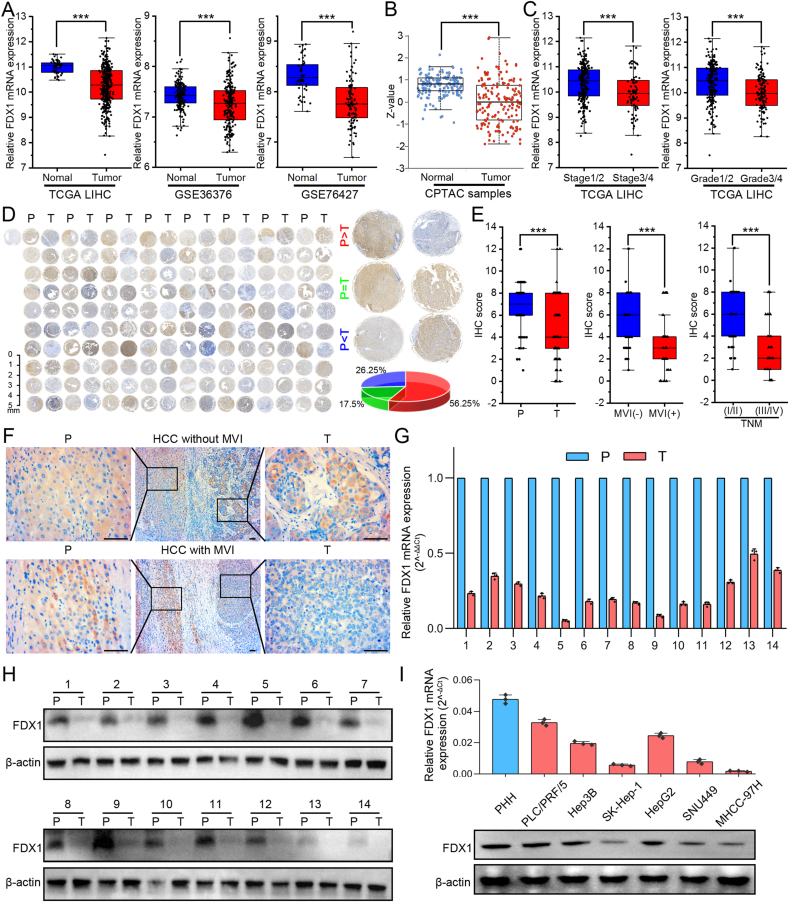
3.1. FDX1 expression is downregulated in HCC
To investigate FDX1 expression in HCC, we first analyzed FDX1 mRNA and protein levels in public databases, including GEO (GSE36376, GSE76427), TCGA and CPTAC databases. The results from these databases showed significantly downregulated FDX1 expression in HCC tissues compared with corresponding normal liver tissues (Fig. 1A and B). In addition, patients with more advanced stage and grade of HCC had significantly lower levels of FDX1 expression (Fig. 1C). To further validate the expression of FDX1 in HCC tissues, we performed IHC assay in TMA with 80 pairs of randomly selected HCC tissues and matched para-cancerous tissues. In most of the samples (56.25%), FDX1 protein expression was higher in para-cancerous tissue than in tumor tissue (Fig. 1D). IHC score of tumor tissue was significantly lower than that of para-cancerous tissue. Furthermore, HCC with microvascular invasion (MVI) and advanced TNM stage had significantly lower FDX1 expression (Fig. 1E). The presence of MVI indicated that HCC was more invasive and that FDX1 expression may be lower in these tumors (Fig. 1F). qRT-PCR (Fig. 1G) and Western blot (Fig. 1H, Fig. S1A) were then performed on 14 paired HCC and para-cancerous tissues and showed that FDX1 was downregulated in HCC tissues. Moreover, FDX1 expression was lower in human liver cancer cell lines (PLC/PRF/5, Hep3B, SK-Hep-1, HepG2, SNU449, MHCC-97H) compared with primary human hepatocytes (PHH) (Fig. 1I, Fig. S1B).
Fig. 1.
FDX1 expression is downregulated in HCC. (A) The expression of FDX1 mRNA in liver cancer was analyzed in publicly available databases, including the TCGA and GEO (GSE36376, GSE76427) databases. (B) FDX1 protein expression in liver cancer was analyzed using data from the CPTAC. (C) FDX1 mRNA expression stratified by tumor stage and grade was analyzed in the TCGA liver cancer database (LIHC). (D) FDX1 protein expression was detected by IHC assay in TMA of 80 pairs of randomly selected HCC tissues (T) and matched para-cancerous tissues (P). The pie chart on the right shows the percentages according to the expression levels in para-cancerous and tumor tissues. (E) According to IHC scores of TMA, FDX1 protein expression was analyzed in para-cancerous tissue (P)/HCC tissue (T), with/without microvascular invasion (MVI), and early TNM stage (I/II)/advanced TNM stage (III/IV). (F) Representative IHC images showed FDX1 protein expression in para-cancerous tissue and HCC tissue with or without MVI. Scale bars, 100 μm. (G–H) FDX1 mRNA expression was assessed by qRT-PCR (G) and Western blot (H) in 14 pairs of randomly selected HCC tumor and para-cancerous tissues. (I) FDX1 mRNA and protein expression in HCC cell lines were determined by qRT-PCR and Western blot. Data are mean ± SD of three independent experiments. ***P < 0.001.
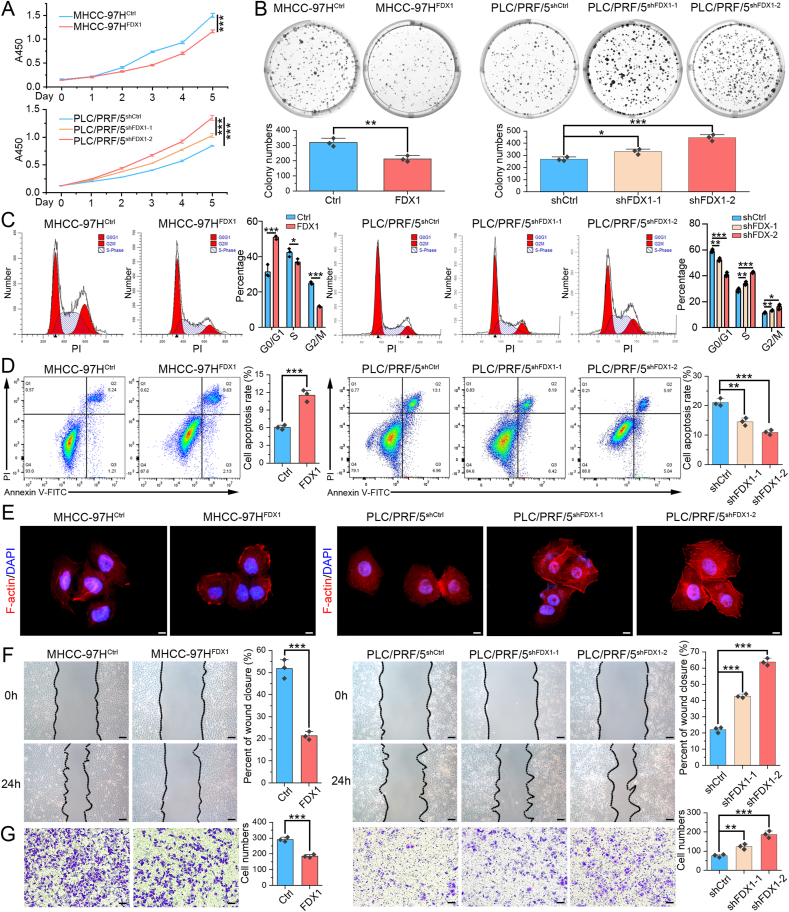
3.2. FDX1 inhibits proliferation, migration, invasion and promote apoptosis of HCC cells in vitro
According to the FDX1 expression level in HCC cell lines, we selected MHCC-97H to stably overexpress FDX1, PLC/PRF/5 to knockdown FDX1, and examined the role of FDX1 on cell behavior in vitro. The overexpression and knockdown efficiency were verified by qRT-PCR (Fig. S1C) and Western blot (Fig. S1D). Both CCK8 and colony formation assays indicated that FDX1 overexpression inhibited the proliferation of MHCC-97H cells, whereas FDX1 knockdown promoted the proliferation of PLC/PRF/5 cells (Fig. 2A and B). The results of flow cytometry for cell cycle analysis showed that overexpression of FDX1 increased cells in G1 phase and decreased cells in S and G2/M phase, whereas the knockdown of FDX1 caused opposite results (Fig. 2C). Flow cytometry for apoptosis analysis revealed that FDX1 overexpression could induce apoptosis of MHCC-97H cells, while FDX1 knockdown inhibited apoptosis of PLC/PRF/5 cells (Fig. 2D). Together, these results showed that FDX1 could inhibit cell proliferation, induce cell cycle arrest and promote apoptosis in HCC. Intriguingly, we observed that FDX1 could affect morphology of cytoskeleton, namely a looser and more spreading pattern in cells with low FDX1 expression, suggesting that they may be more aggressive (Fig. 2E). We then assessed the influence of FDX1 on the migratory and invasive phenotypes of HCC cells using wound healing and Transwell assays. Overexpression of FDX1 inhibited both migration and invasion abilities of HCC cells, whereas knockdown of FDX1 significantly promoted these phenotypes (Fig. 2F and G). To further confirmed above results, we performed the gain- and loss-of-function experiments in another two HCC cell lines (SNU449 and Hep3B). The results demonstrated that FDX1 overexpression in SNU449 could also inhibit cell proliferation (Figs. S2A–S2B), promote apoptosis (Fig. S2C), and inhibit migration, invasion (Figs. S2D–S2E). Contrary results were observed after knocking down of FDX1 in Hep3B cells (Fig. S2).
Fig. 2.
FDX1 inhibits proliferation, migration, invasion and promote apoptosis of HCC cells in vitro. (A) CCK8 assay showed the proliferative curves of FDX1-overexpressing MHCC-97H, FDX1-knockdown PLC/PRF/5 and their control cells. (B) Colony formation assays were used to evaluate the proliferation ability of HCC cell lines with overexpression or knockdown of FDX1. (C) The proportion of HCC cells with FDX1 overexpression or knockdown in G0/G1, G2/M, and S phases of cell division was analyzed by flow cytometry. (D) The rate of apoptosis in HCC cells with FDX1 overexpression or knockdown and in their control cells was analyzed by flow cytometry. (E) Cytoskeleton of indicated HCC cells visualized by staining of F-actin with rhodamine-conjugated phalloidin. DAPI was used to stain the cell nuclei. Scale bars, 5 μm. (F) Wound healing assay showed the migration ability of MHCC-97HFDX1, PLC/PRF/5shFDX1-1, PLC/PRF/5shFDX1-2 and their control cells at 0 and 24 h. Scale bars, 50 μm. (G) Transwell invasion assay showed the invasion ability of MHCC-97HFDX1, PLC/PRF/5shFDX1-1, PLC/PRF/5shFDX1-2 and their control cells at 24 h. Scale bars, 50 μm. Data shown as mean ± SD of triplicate independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
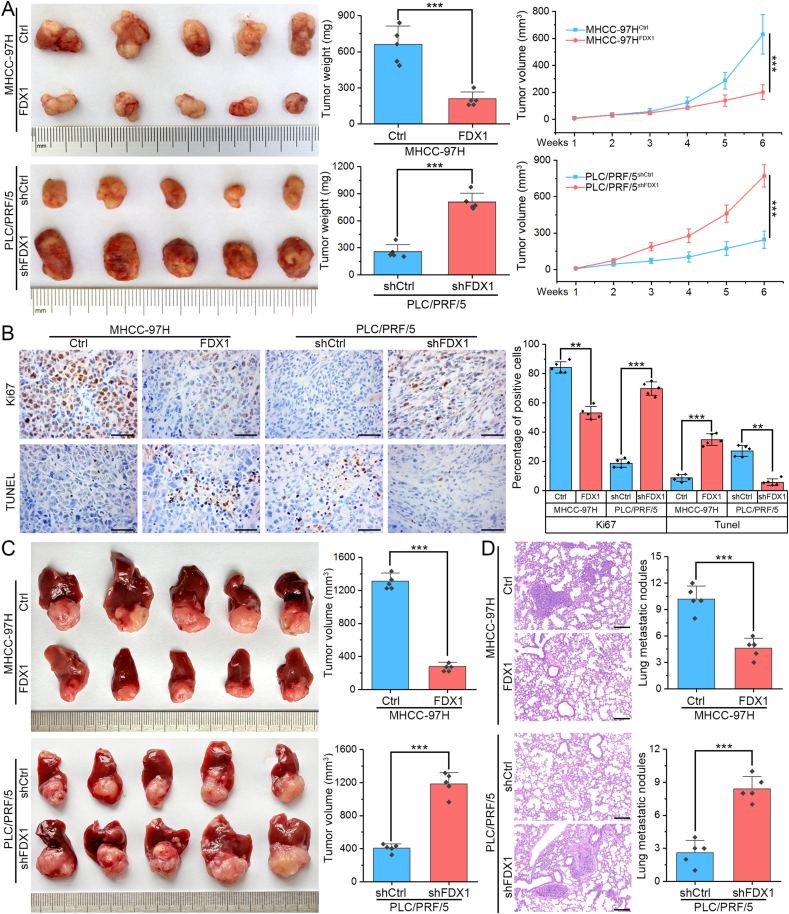
3.3. FDX1 inhibits HCC tumor growth and metastasis in vivo
To determine the biological functions of FDX1 in vivo, we established subcutaneous and orthotopic tumor models in nude mice. By plotting growth curves and measuring final tumor weights, subcutaneous implantation experiments showed that ectopic expression of FDX1 in MHCC-97H significantly impaired tumor growth. Conversely, knockdown of FDX1 in PLC/PRF/5 obviously promoted the growth of xenografted tumors (Fig. 3A). The subcutaneous tumors were then fixed, embedded and sectioned. IHC detection of Ki67 expression showed that FDX1 overexpression could inhibit HCC cell proliferation, while TUNEL assay showed that FDX1 overexpression could promote cell apoptosis, opposite effects were observed in FDX1 knockdown tumors (Fig. 3B). The orthotopic tumor model further confirmed that MHCC-97HFDX1 cell-derived tumors grew smaller than control cell-derived tumors, whereas PLC/PRF/5shFDX1 cell-derived tumors were larger than control tumors (Fig. 3C). The lung metastases were assessed by serial pathological sections and H&E staining. The number of lung metastases was significantly reduced in the FDX1 overexpression group compared with the control group. In contrast, orthotopic tumors from PLC/PRF/5shFDX1 cells had more lung metastases compared with tumors from control cells (Fig. 3D).
Fig. 3.
FDX1 inhibits HCC tumor growth and metastasis in vivo. (A) Subcutaneous tumor models were established using MHCC-97HFDX1, PLC/PRF/5shFDX1 and their control cells (n = 5). Subcutaneous tumors were harvested and photographed (left panel). Tumor weight (middle panel) and growth curves (right panel) of subcutaneous tumors were determined and compared. (B) Ki67 expression levels and apoptosis-positive cells in subcutaneous tumor models derived from MHCC-97HFDX1, PLC/PRF/5shFDX1 and their control cells were detected by IHC and TUNEL assays, respectively. Scale bars, 100 μm. (C) Orthotopic tumor models were established using MHCC-97HFDX1, PLC/PRF/5shFDX1 and their control cells (n = 5). Orthotopic tumors were harvested and photographed (left panel). Tumor volume (right panel) of orthotopic tumors were determined and compared. (D) The lungs of the mice were harvested and examined by serial pathological sections using H&E staining. Representative images of lung metastases were shown (left panel) and the number of nodules in each group was compared (right panel). Scale bars, 200 μm **P < 0.01; ***P < 0.001.
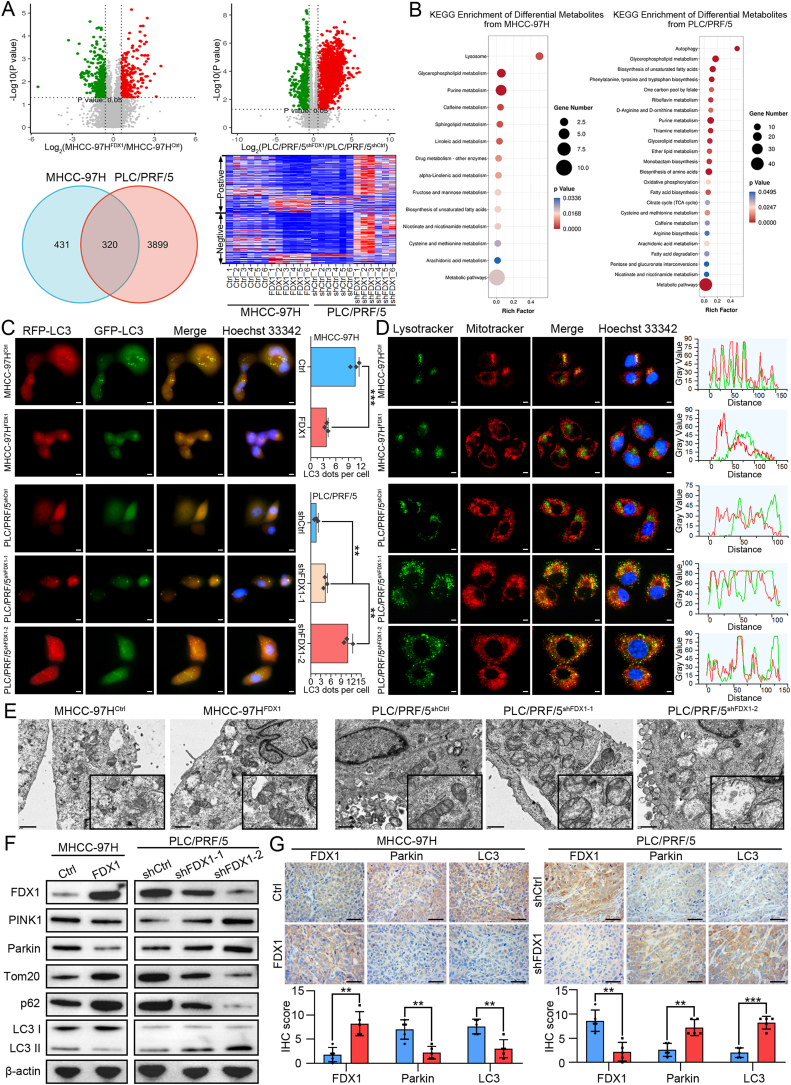
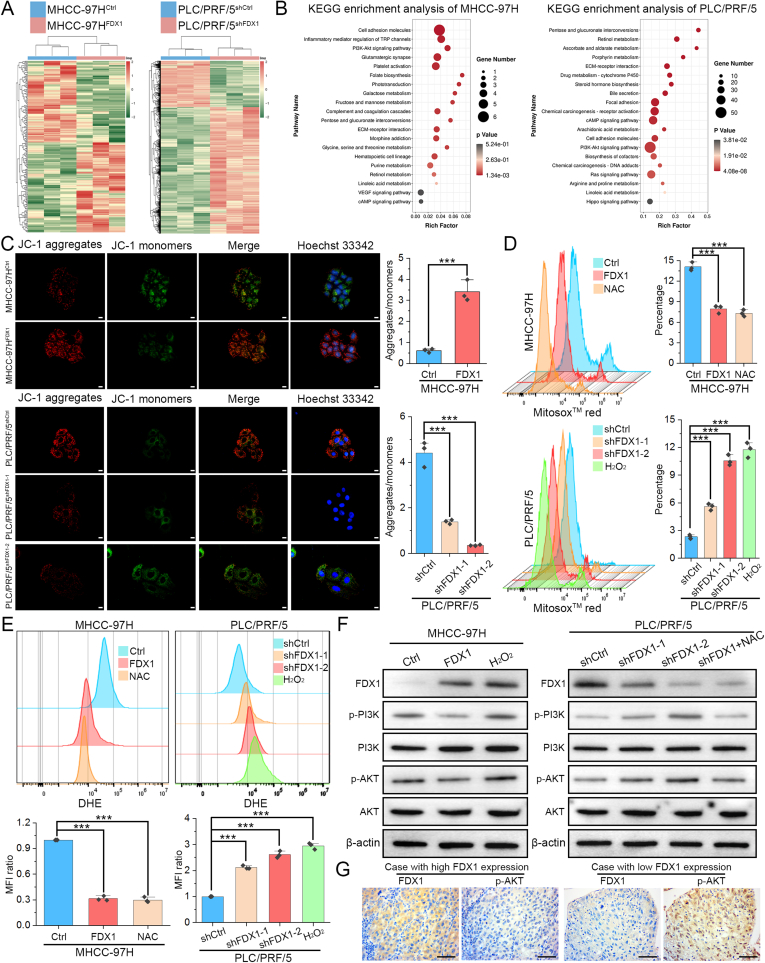
3.4. FDX1 regulates metabolism and its downregulation facilitates mitophagy of HCC cells
Previous study showed that FDX1 protein is located in the mitochondrion [19]. By performing double immunofluorescence staining of FDX1 and mitochondrial marker LRPPRC, we found that FDX1 is indeed mainly co-expressed with mitochondria in HCC cells (Fig. S3A). To understand whether FDX1 is involved in the metabolism of HCC cells, we performed non-targeted metabolomics on FDX1 overexpressing and knockdown HCC cells. Indeed, volcano diagram showed that overexpression or knockdown of FDX1 resulted in significant changes in the levels of many metabolites (Fig. 4A). The Venn diagram showed that there were 320 kinds of common differential metabolites in FDX1 overexpressing and knockdown HCC cells. The heatmap of these metabolites showed a clear difference between MHCC-97HFDX1, PLC/PRF/5shFDX1 and their control cells in either positive ion mode or negative ion mode (Fig. 4A). KEGG enrichment analysis for metabolism by differential metabolites showed that FDX1 could regulate many aspects of metabolism in HCC, mainly including lipid metabolism and amino acid metabolism (Fig. 4B). Intriguingly, KEGG enrichment analysis for cellular processes by differential metabolites revealed that FDX1 is associated with lysosome and autophagy (Fig. 4B). Metabolic reprogramming of tumor cells can support activation of autophagy or mitophagy [20,21]. Considering that FDX1 functions mainly in mitochondria, we speculated that FDX1 may be involved in autophagy of HCC, especially in mitophagy. We first monitored autophagy by transfecting mRFP-GFP-LC3 lentivirus in HCC cells. FDX1 overexpression resulted in a significant decrease in mRFP-GFP-LC3 dot accumulation, whereas FDX1 knockdown could lead to opposite results, indicating that FDX1 downregulation may promote formation of autophagosomes (Fig. 4C). Furthermore, overexpression of FDX1 in MHCC-97H decreased the co-localization of mitochondria and lysosomes, while knockdown of FDX1 in PLC/PRF/5 obviously enhanced overlapping signals of MitoTracker and LysoTracker, indicating that FDX1 may regulate mitophagy of HCC cells (Fig. 4D). Consistently, mitochondrial damage and autophagosome formation were more frequently observed in HCC cells with low FDX1 expression by TEM (Fig. 4E). We performed Western blot to detect autophagy/mitophagy-related proteins in HCC cells. Overexpression of FDX1 downregulated PINK1 and Parkin, upregulated Tom20 and p62, and decreased the LC3-II/LC3-I ratio, whereas knockdown of FDX1 caused opposite effects (Fig. 4F, Fig. S3B). Finally, we performed IHC in the above subcutaneous tumors to detect the expression of FDX1, Parkin and LC3, and the results showed that these protein expression trends were consistent with Western blot, further confirming that FDX1 could regulate PINK1/Parkin-dependent mitophagy (Fig. 4G).
Fig. 4.
FDX1 regulates metabolism and its downregulation facilitates mitophagy of HCC cells. (A) Non-targeted metabolomics analysis of metabolites in indicated HCC cells. The volcano diagram shows the metabolites that were significantly altered after overexpression or knockdown of FDX1 (top panel). Venn diagram shows the common altered metabolites in HCC cells after overexpression or knockdown of FDX1 (lower left panel). Heatmap shows these common altered metabolites in positive ion mode and negative ion mode (lower right panel). (B) KEGG enrichment analysis for metabolism by differential metabolites from non-targeted metabolomics analysis. (C) Representative images of MHCC-97HFDX1, PLC/PRF/5shFDX1 and their control cells after transfecting with mRFP-GFP-LC3 lentivirus. Hoechst 33342 was used for nuclear staining. Scale bars, 5 μm. (D) Mitochondria and lysosomes of MHCC-97HFDX1, PLC/PRF/5shFDX1 and their control cells were stained by MitoTracker-Red and LysoTracker-Green, respectively. Hoechst 33342 was used for nuclear staining. Scale bars, 5 μm. (E) Representative transmission electron microscopy images of MHCC-97HFDX1, PLC/PRF/5shFDX1 and their control cells. Scale bars, 1 μm. (F) Western blot analysis of PINK1, Parkin, Tom20, p62, and LC3 I/II in HCC cells with FDX1 overexpression or knockdown. (G) Protein expression of FDX1, Parkin and LC3 was examined by IHC in subcutaneous tumors derived from the indicated cells. Scale bars, 100 μm. Data presented as mean ± SD of triplicate independent experiments. **P < 0.01; ***P < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Downregulation of FDX1 activates PI3K/AKT signaling pathway by increasing reactive oxygen species (ROS) levels in HCC
We then carried out RNA sequencing to investigate the potential molecular mechanisms that may be affected by FDX1 in HCC. Overexpressing or knocking down FDX1 in HCC clearly altered the expression of many genes (Fig. 5A). KEGG enrichment analysis of DEGs in MHCC-97H and PLC/PRF/5 cells indicated that FDX1 not only affects metabolism of HCC cells, but also regulates the PI3K/AKT signaling pathway (Fig. 5B). Meanwhile, KEGG enrichment analysis for environmental information processing by the above differential metabolites also showed that FDX1 could regulate PI3K/AKT signaling pathway (Figs. S4A and B). Considering the important role of FDX1 in mitochondria, we then evaluated mitochondrial membrane potential (MMP), mitochondrial ROS levels and intracellular ROS levels in HCC cells using JC-1, MitoSox™ red and DHE staining, respectively. The results revealed that overexpression of FDX1 increased JC-1 aggregate formation but reduced JC-1 monomer levels in MHCC-97H cells, whereas knockdown of FDX1 in PLC/PRF/5 cells induced contrary effects (Fig. 5C). MitoSox™ red and DHE staining both showed that FDX1 could reduce ROS levels in HCC cells and its downregulation increased both mitochondrial and intracellular ROS levels (Fig. 5D and E). These results confirmed that FDX1 plays important role in maintaining mitochondrial stability and its downregulation may cause mitochondrial depolarization and oxidative stress. Excessive ROS can lead to mitochondrial damage, depolarization and mitophagy [22]. ROS could also activate PI3K/AKT signaling pathway through various mechanisms [23]. Much evidence has shown that the interplay between ROS and the PI3K/AKT signaling pathway coordinates the occurrence of autophagy [24]. Therefore, ROS may play a central role in mediating the functions of FDX1 in HCC. To investigate whether FDX1 regulates the PI3K/AKT signaling pathway by affecting ROS production, we treated HCC cells with H2O2 (10 μM) or ROS scavenger N-acetyl-l-cysteine (NAC) and the mitochondrial ROS scavenger MitoTEMPO (MT). Appropriate concentrations of NAC (5 mM) or MT (100 μM) could significantly reduce ROS levels in MHCC-97H (Fig. S4C). We found that FDX1 inhibits the PI3K/AKT signaling pathway while H2O2 treatment reactivates the PI3K/AKT signaling pathway in MHCC-97HFDX1 cells. In PLC/PRF/5 cells, knockdown of FDX1 activated the PI3K/AKT signaling pathway. However, after treatment with NAC or MT, the PI3K/AKT pathway was again inhibited (Fig. 5F, Figs. S4D–S4E). To further illustrate the relationship between FDX1 and PI3K/AKT signaling pathway, we treated MHCC-97HFDX1 cells with the PI3K-specific agonist 740 Y–P and found that 740 Y–P could partially restore cell proliferation (Figs. S4F–S4G), migration (Fig. S4H), and invasion (Fig. S4I). While in PLC/PRF/5shFDX1 cells, the PI3K inhibitor LY294002 could partly inhibit cell proliferation, migration, and invasion (Figs. S4F–S4I). IHC analysis of clinical HCC samples revealed that FDX1 expression was negatively correlated with p-AKT expression, further suggesting that FDX1 could regulate the PI3K/AKT signaling pathway in HCC (Fig. 5G).
Fig. 5.
Downregulation of FDX1 activates PI3K/AKT signaling pathway by increasing reactive oxygen species (ROS) levels in HCC. (A) RNA sequencing was performed in MHCC-97HFDX1, PLC/PRF/5shFDX1 and their control cells. Heatmap shows the differentially expressed genes between the FDX1 overexpression cells, the FDX1 knockdown cells and their corresponding control cells. (B) KEGG enrichment analysis of differentially expressed genes in MHCC-97H and PLC/PRF/5 cells with FDX1 overexpression and knockdown, respectively. (C) Mitochondrial membrane potential (MMP) was determined by JC-1 staining. Hoechst 33342 was used for nuclear staining. The ratio of aggregates/monomers was compared in the right bar graph. Scale bars, 10 μm. (D–E) Mitochondrial and intracellular ROS in MHCC-97HFDX1, PLC/PRF/5shFDX1 and their control cells were detected by staining with MitoSox™ red (D) and DHE (E), respectively. ROS scavenger NAC (5 mM) and inducer H2O2 (10 μM) were used as controls. (F) Western blot analysis showed the protein expression of p-PI3K, p-AKT, and their total protein expression levels in MHCC-97HFDX1, PLC/PRF/5shFDX1 and their control cells treated with either NAC (5 mM) or H2O2 (10 μM). (G) Protein expression of FDX1 and p-AKT was examined by IHC in HCC tissues with high and low FDX1 expression. Scale bars, 100 μm. The data are presented as mean ± SD of three independent experiments. ***P < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
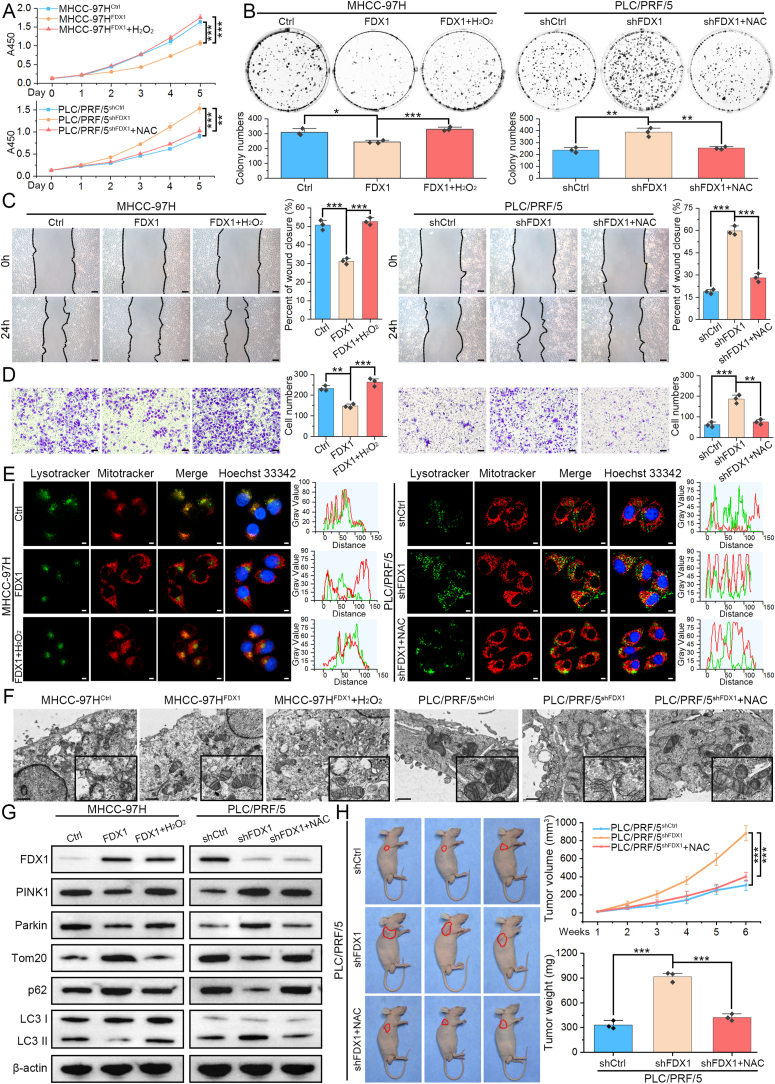
3.6. FDX1 inhibits HCC progression and mitophagy by suppressing ROS production
To determine whether FDX1 function is mediated by dysregulation of ROS levels, we performed several functional rescue assays by interfering with ROS levels. As shown by CCK8 and colony formation assays, overexpression of FDX1 inhibited the proliferation of MHCC-97H, which was restored after treatment with H2O2; whereas knockdown of FDX1 in PLC/PRF/5 significantly promoted its proliferation, which was further reversed by ROS scavenging with NAC (Fig. 6A and B). Wound healing and Transwell assays also showed that the inhibited migratory and invasive capacities of MHCC-97HFDX1 cells were restored by the addition of H2O2, while the addition of NAC in PLC/PRF/5shFDX1 further inhibited its migration and invasion ability (Fig. 6C and D). Since the source of ROS is mitochondria, we also used the mitochondrial ROS scavenger MT to validate these findings. Similarly, MT could also inhibit cell proliferation (Figs. S5A–S5B), migration, and invasion (Fig. S5C) of PLC/PRF/5 cells after FDX1 knockdown. Furthermore, MitoTracker and LysoTracker staining demonstrated that H2O2 could promote the colocalization of mitochondria and lysosomes in MHCC-97HFDX1 cells, whereas NAC or MT treatment obviously disassociated mitochondria and lysosomes in PLC/PRF/5shFDX1, suggesting that the regulation of mitophagy by FDX1 may be also mediated by ROS (Fig. 6E, Fig. S5D). To further investigate and verify the role of FDX1 in mitophagy regulation, we treated PLC/PRF/5shFDX1 cells with selective vacuolar (V)-ATPase inhibitor Bafilomycin A1, the lysosomal inhibitor Chloroquine, and the mitophagy inhibitor Mdivi-1. The results showed that these autophagy inhibitors could inhibit the co-localization of mitochondria and lysosomes, namely the occurrence of mitophagy (Fig. S5D). The effects of ROS on mitophagy of HCC cells with FDX1 overexpression and knockdown were verified by TEM (Fig. 6F). To further confirm these results, Western blot to detect autophagy/mitophagy-related proteins showed that H2O2 increased the expression of PINK1, Parkin and the LC3-II/LC3-I ratio, and decreased Tom20 and p62 expression in MHCC-97HFDX1 cells, whereas NAC or MT treatment led to opposite results in PLC/PRF/5shFDX1 cells (Fig. 6G, Figs. S5E–S5F). Finally, we again established subcutaneous tumor model using PLC/PRF/5shCtrl and PLC/PRF/5shFDX1 cells. Knockdown of FDX1 promoted subcutaneous tumor growth, which was further inhibited by treatment of mice with NAC (Fig. 6H). IHC and TUNEL assays also confirmed that NAC could decrease the expression of Ki67, inhibit the PI3K/AKT signaling pathway and mitophagy, and promote apoptosis in PLC/PRF/5shFDX1 cell-derived tumors (Fig. S5G).
Fig. 6.
FDX1 inhibits HCC cell progression and mitophagy by suppressing ROS production. (A–B) CCK8 (A) and colony formation (B) assays showed the proliferation ability of the indicated HCC cells after further treatment with H2O2 (10 μM) or NAC (5 mM). (C–D) After treatment with H2O2 (10 μM) or NAC (5 mM), the migration and invasion ability of HCC cells with overexpression or knockdown of FDX1 was determined by wound healing (C) and Transwell (D) assays, respectively. Scale bars, 50 μm. (E) In FDX1 overexpressing or knockdown cells, the colocalization of mitochondria and lysosomes was determined by staining with MitoTracker-Red and LysoTracker-Green after treatment with H2O2 (10 μM) or NAC (5 mM). Hoechst 33342 was used for nuclear staining. Scale bars, 5 μm. (F) Representative transmission electron microscopy images of MHCC-97HFDX1, PLC/PRF/5shFDX1 and their control cells after further treatment with H2O2 (10 μM) or NAC (5 mM). Scale bars, 1 μm. (G) Western blot analysis of PINK1, Parkin, Tom20, p62, and LC3 I/II in HCC cells with FDX1 overexpression or knockdown after further treatment with H2O2 (10 μM) or NAC (5 mM). (H) Subcutaneous tumor model established using PLC/PRF/5shFDX1 and its control cells were further treated by NAC. Tumor weight and growth curves of subcutaneous tumors were determined and compared. The data are presented as mean ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
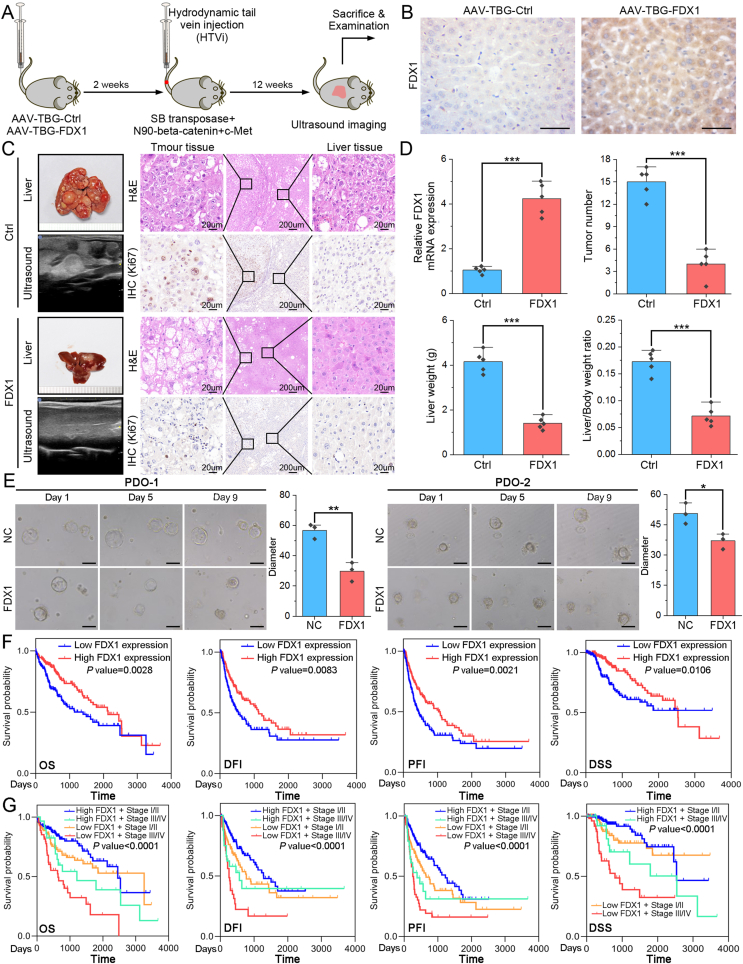
3.7. FDX1 elevation inhibits HCC progression and its downregulation predicts poor prognosis of HCC patients
To further confirm the tumor suppressor role of FDX1 in HCC and explore potential interventions, we overexpressed FDX1 in mouse liver by adeno-associated virus (AAV) with a liver-specific thyroid hormone-binding globulin (TBG) promoter. The FDX1-overexpressing or control mice were then co-injected with SB-mediated oncogenes using HTVi method (Fig. 7A). HCC, but not cholangiocarcinoma, models were successfully constructed using the HTVi method with SB-mediated c-Met and beta-catenin integration (Fig. S6A). FDX1 expression in AAV-TBG-Ctrl/AAV-TBG-FDX1 mice was confirmed by IHC (Fig. 7B). Through small animal ultrasound imaging or ex vivo liver, we found that liver-specific overexpression of FDX1 significantly inhibits tumor progression. Cell proliferation marker, Ki67, was also decreased in AAV-TBG-FDX1 mice compared with control mice (Fig. 7C). FDX1 overexpression in HCC cells was confirmed by qRT-PCR. Besides, FDX1-overexpressing mice had fewer tumor number in the liver, a lower liver weight and a lower liver-to-body weight ratio than control mice (Fig. 7D). We then established HCC PDO model and found that overexpression of FDX1 resulted in organoids with a smaller diameter than those in the control group, suggesting that FDX1 may compromise the ability of organoids to proliferate (Fig. 7E). Finally, we analyzed the clinical significance of FDX1 in HCC using data from the TCGA LIHC database. FDX1 expression is significantly associated with T stage, pathologic stage, histologic grade, vascular invasion, gender, adjacent hepatic tissue inflammation, and AFP level (all with P < 0.05, Table S2). Kaplan–Meier survival analysis showed that HCC patients with low FDX1 expression had significantly shorter overall survival (OS), disease-free interval (DFI), progression-free interval (PFI) and disease specific survival (DSS) than those with high FDX1 expression (Fig. 7F). Moreover, HCC patients with low FDX1 expression and advanced pathologic stage had the worst prognosis, while those with high FDX1 expression and early pathologic stage showed the best prognosis (Fig. 7G). Univariate and multivariate analysis also showed that low FDX1 expression is an independent risk factor for OS, DFI, PFI and DSS (Fig. S6B).
Fig. 7.
FDX1 elevation inhibits HCC progression and its downregulation predicts poor prognosis of HCC patients. (A) Schematic diagram of liver-specific overexpression of FDX1 in mice and establishment of HTVi model using sleeping beauty (SB) transposase-mediated beta-catenin/c-Met delivery. (B) IHC assay showed the FDX1 protein expression in AAV-TBG-Ctrl and AAV-TBG-FDX1 mice. Scale bars, 100 μm. (C) Representative small animal ultrasound images, ex-vivo mouse liver, H&E staining and IHC for Ki67 of liver tissue from the HTVi model. (D) Bar graphs showed FDX1 mRNA expression, tumor number, liver weight and liver/body weight ratio in the HTVi models with or without FDX1 overexpression. (E) Representative images of two HCC PDOs at day 1, day 5 and day 9 with or without FDX1 overexpression. The size of the organoids was calculated and compared. Scale bars, 50 μm. (F) Kaplan–Meier analysis of overall survival (OS), disease-free interval (DFI), progression-free interval (PFI) and disease specific survival (DSS) in HCC patients from the TCGA LIHC database with high or low FDX1 expression. (G) Survival curves for OS, DFI, PFI and DSS after stratification based on FDX1 expression and pathologic stage. *P < 0.05; **P < 0.01; ***P < 0.001.
4. Discussion
FDX1 plays a key role in cuproptosis by acting as a reductase to reduce Cu(II) to Cu(I) [25]. In HCC, many bioinformatic analyses have found that cuproptosis-related genes, including FDX1, are associated with tumor progression, prognosis, immune infiltration, and therapeutic responses [[26], [27], [28], [29]]. High FDX1 expression could decrease HCC cell viability, lead HCC cell sensitive to Cu2+, and knockdown of FDX1 promotes HCC cell proliferation and migration, indicating that FDX1 is also correlated with HCC cuproptosis [12]. Nevertheless, systematic experimental studies in HCC are still scarce, its potential molecular mechanisms of regulating HCC progression are still unclear and require further in-depth investigation.
Current studies reveal that the expression of FDX1 is regulated by miRNA [13], transcription factor [30], or mRNA m6A-modification [31]. FDX1 is often down-regulated in tumor tissues, but can be up-regulated in some tumors, e.g. in gastric cancer [31,32], implying that the function of FDX1 is tissue-specific. Therefore, clarifying the expression profile of FDX1 in HCC is a prerequisite for understanding the role of FDX1 in HCC. In addition to the bioinformatic analysis using public databases that revealed FDX1 mRNA downregulation in HCC, we further confirmed that FDX1 protein expression was also significantly downregulated using IHC analysis of HCC tissue microarray (TMA). Intriguingly, HCC with MVI or in advanced stage showed more lower FDX1 expression compared with HCC without MVI or in early stage, respectively, indicating that FDX1 may be associated with HCC progression. By upregulating or knocking down FDX1 expression in HCC cells, we found that FDX1 significantly prevented cell cycle progression, promoted apoptosis, and inhibited cell migration and invasion. In vivo subcutaneous and orthotopic models further confirmed that FDX1 could suppress HCC tumor growth and metastasis. FDX1 regulated by METTL3/miR-21-5p axis in non-small-cell lung cancer could inhibit proliferation, invasion, migration, apoptosis in vitro and tumorigenesis in vivo [33]. Although FDX1 is known to be an essential mediator of cuproptosis, these results also reveal that FDX1 may regulate other forms of cell death, such as apoptosis, and further influence tumor cell viability and dissemination.
FDX1 is localized in the mitochondrial matrix and involved in steroidogenesis, Fe–S cluster biosynthesis, and lipoylation [34]. Mitochondria are the central site of oxidative metabolism, where carbohydrates, lipids and amino acids are finally oxidized to release energy, and liver is the major metabolic organ in the body. We therefore speculated that aberrant expression of FDX1 in mitochondria may lead to metabolic reprogramming of HCC cells, thereby affecting the status of tumor cells. In this context, we performed non-targeted metabolomics analysis and identified many differential metabolites associated with lipid metabolism and amino acid metabolism and regulated by FDX1. Aberrant metabolism meets the rapidly increasing energy demands of tumor cells but can lead to mitochondrial damage [35]. Mitophagy is a special type of autophagy that could effectively remove damaged or depolarized mitochondria by combining with autophagosomes and autolysosomes through the PINK1/Parkin pathway, the main regulatory mechanism of mitophagy [36]. Our results showed that FDX1 maintained mitochondrial homeostasis and reduced mitophagy levels in HCC cells, whereas FDX1 deficiency caused metabolic reprogramming and activation of PINK1/Parkin-mediated mitophagy. Activation of mitophagy in FDX1 downregulated HCC cells could remove or recycle damaged mitochondria, which may facilitate tumor metabolism and progression. The specific molecular mechanisms by which FDX1 regulates the metabolism of HCC require further investigation.
Although FDX1 has been shown to be a regulator of mitophagy in HCC, the underlying molecular mechanisms are still unclear. We further performed RNA sequencing and KEGG enrichment analysis, which showed that FDX1 was associated with the PI3K/AKT signaling pathway. PI3K/AKT pathway could promotes autophagy in malignant tumors [37,38]. PI3K/AKT pathway is also a mitophagy-related signaling pathway in which AKT, a serine/threonine kinase, is involved in regulating PINK1 accumulation and Parkin recruitment to the damaged mitochondria [[39], [40], [41]]. We also found that FDX1 could stabilize MMP and reduce mitochondrial and intracellular ROS levels, while its downregulation led to a decrease of MMP and an elevation of ROS levels in HCC. Aberrant metabolism in mitochondria produces a large amount of ROS, including superoxide anion (O2−), hydrogen peroxide (H2O2), hydroxyl radical (·OH), and singlet oxygen (1O2) [42]. ROS can not only directly activate mitophagy, but also activate the PI3K/AKT signaling pathway to indirectly regulate mitophagy [24]. Therefore, loss of FDX1 in HCC may contribute to abnormal metabolism and subsequent elevation of ROS, which mediates the role of FDX1 in regulating HCC proliferation, invasion and mitophagy.
We further used HTVi and PDO models to investigate the role of FDX1 in HCC. Upregulation of FDX1 could inhibit tumor progression and PDO growth, which confirmed that FDX1 is a tumor suppressor gene in HCC. Direct activation of FDX1 function using small molecule drugs may contribute to the treatment of HCC. However, no relevant studies of agonists directly targeting FDX1 have been reported to date. As FDX1 expression is downregulated in HCC and its protein expression level is relatively low, the anti-cancer effect of direct activation of FDX1 may be limited. Some solutions have been proposed to deal with the problem of clinical treatment. One approach is to take advantage of the involvement of FDX1 in cuproptosis, and the use of the copper ionophore elesclomol can enhance the anti-cancer effect of FDX1 [14]. Another approach is to induce FDX1 expression with drugs. For example, in ovarian tissue, Chia-Chun Wu et al. found through pharmacogenetic screening that drugs such as CX-5461, Temozolomide, FTY-720, Erlotinib and Sapitinib may have the potential to target and regulate FDX1 expression [43]. The third approach is to use ROS scavengers or mitophagy inhibitors in HCC patients with low FDX1 expression. Although these approaches need to be further demonstrated, they may provide new clues for the treatment of HCC. Intriguingly, it has been found that NK cells, macrophages and B-cell are infiltrated in high FDX1-expressing HCC tissues, while PD-1 expression was higher in low FDX1-expressing HCC tissues [29]. Targeting FDX1 in combination with the latest promising immunotherapy regimens may help to prolong survival and improve quality of life for HCC patients.
We also found that low FDX1 expression is associated with poor prognosis in HCC patients, and its combination with tumor stage could identify patients with worse prognosis. Some cuproptosis-related gene prognostic models involving FDX1 have been constructed [27,28], finding more FDX1-related prognostic biomarkers and conducting multi-center validation are required to build a more optimized prognostic model. Some studies have identified serum metabolites with good diagnostic performance, such as phenylalanyl-tryptophan and glycocholate [44], urea [45], retinol and retinal [46], etc. As FDX1 is not a secreted protein, direct analysis of FDX1 protein expression may not be meaningful for clinical diagnosis. Our results showed that FDX1 affects the metabolism of HCC cells, and retrospective analysis of differential metabolites levels in the serum of HCC patients with FDX1 downregulation and combination of the non-targeted metabolomics results may help to identify new diagnostic markers.
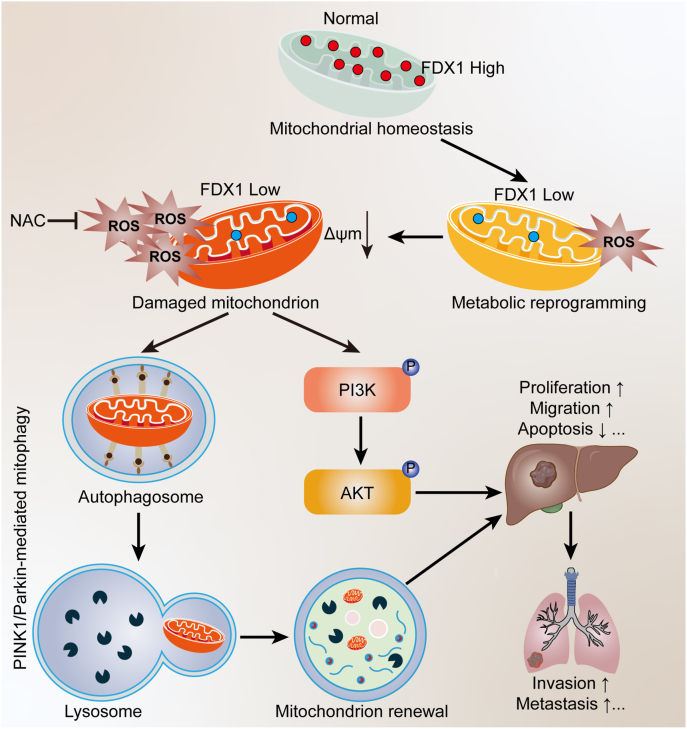
In conclusion, we have identified a novel role for FDX1 in regulating HCC progression. We elucidated that mitochondrial protein FDX1 deficiency could lead to metabolic reprogramming and oxidative stress, which activates PINK1/Parkin-mediated mitophagy and PI3K/AKT pathway to facilitate HCC proliferation, invasion and metastasis (Fig. 8). Thus, FDX1 is a tumor suppressor in HCC that may serve as a novel prognostic predictor and therapeutic target for HCC patients.
Fig. 8.
Schematic diagram of the mechanism of FDX1 downregulation to facilitate HCC progression.
Ethics declarations
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and later versions. Informed consent to be included in the study, or the equivalent was obtained from all patients. This study was approved by the ethical committee of the Hunan Provincial People's Hospital/The First Affiliated Hospital of Hunan Normal University. All institutional and national guidelines for the care and use of laboratory animals were followed.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
CRediT authorship contribution statement
Bo Sun: Writing – review & editing, Writing – original draft, Software, Methodology, Funding acquisition, Data curation, Conceptualization. Peng Ding: Methodology, Investigation. Yinghui Song: Methodology, Formal analysis, Data curation. Jia Zhou: Visualization, Validation, Investigation. Xu Chen: Validation, Resources, Data curation. Chuang Peng: Writing – review & editing, Supervision, Resources, Project administration, Data curation. Sulai Liu: Writing – review & editing, Supervision, Project administration, Funding acquisition, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 82303186, 22374045); the Natural Science Foundation of Hunan Province (Grant No. 2023JJ40370); the China Postdoctoral Science Foundation (Grant No. 2023M731068); Huxiang Youth Talent of Innovative Talent Programme of Hunan Province (2024RC3232).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2024.103302.
Contributor Information
Chuang Peng, Email: pengchuangcn@163.com.
Sulai Liu, Email: liusulai@hunnu.edu.cn.
Abbreviations
- HCC
Hepatocellular carcinoma
- FDX1
Ferredoxin 1
- TCGA
The Cancer Genome Atlas
- GEO
the Gene Expression Omnibus
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- TMA
Tissue microarray
- IHC
Immunohistochemistry
- ROS
Reactive oxygen species
- DHE
Dihydroethidium
- NAC
N-acetyl-l-cysteine
- HTVi
Hydrodynamic tail vein injection
- SB
Sleeping Beauty
- TUNEL
Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling
- TEM
Transmission electron microscopy
- PDO
Patient-derived organoid
- DEG
Differentially expressed genes
- MVI
Microvascular invasion
- PHH
Primary human hepatocytes
- MMP
Mitochondrial membrane potential
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Xia C., Dong X., Li H., Cao M., Sun D., He S., et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin. Med. J. 2022;135:584–590. doi: 10.1097/CM9.0000000000002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Sheftel A.D., Stehling O., Pierik A.J., Elsasser H.P., Muhlenhoff U., Webert H., et al. Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11775–11780. doi: 10.1073/pnas.1004250107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohibi S., Zhang Y., Perng V., Chen M., Zhang J., Chen X. Ferredoxin 1 is essential for embryonic development and lipid homeostasis. Elife. 2024;13 doi: 10.7554/eLife.91656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Z., Ma Y., Guo X., Du Y., Zhu Q., Wang X., et al. FDX1 can impact the prognosis and mediate the metabolism of lung adenocarcinoma. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.749134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsvetkov P., Coy S., Petrova B., Dreishpoon M., Verma A., Abdusamad M., et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375:1254–1261. doi: 10.1126/science.abf0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsvetkov P., Detappe A., Cai K., Keys H.R., Brune Z., Ying W., et al. Mitochondrial metabolism promotes adaptation to proteotoxic stress. Nat. Chem. Biol. 2019;15:681–689. doi: 10.1038/s41589-019-0291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xing J., Qiao G., Luo X., Liu S., Chen S., Ye G., et al. Ferredoxin 1 regulates granulosa cell apoptosis and autophagy in polycystic ovary syndrome. Clin. Sci. 2023;137:453–468. doi: 10.1042/CS20220408. [DOI] [PubMed] [Google Scholar]
- 9.Guowei L., Xiufang L., Qianqian X., Yanping J. The FDX1 methylation regulatory mechanism in the malignant phenotype of glioma. Genomics. 2023;115 doi: 10.1016/j.ygeno.2023.110601. [DOI] [PubMed] [Google Scholar]
- 10.Liu M., Wu S., Wu H., Zhou Y., Zhang X., Zhu D., et al. Ferredoxin 1: a gatekeeper in halting lung adenocarcinoma progression through activation of the GPRIN2 signaling pathway. J. Transl. Med. 2024;22:510. doi: 10.1186/s12967-024-05277-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang C., Zeng Y., Guo X., Shen H., Zhang J., Wang K., et al. Pan-cancer analyses confirmed the cuproptosis-related gene FDX1 as an immunotherapy predictor and prognostic biomarker. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.923737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quan Y., Li W., Yan R., Cheng J., Xu H., Chen L. Tumor cuproptosis and immune infiltration improve survival of patients with hepatocellular carcinoma with a high expression of ferredoxin 1. Front. Oncol. 2023;13 doi: 10.3389/fonc.2023.1168769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quan B., Liu W., Yao F., Li M., Tang B., Li J., et al. LINC02362/hsa-miR-18a-5p/FDX1 axis suppresses proliferation and drives cuproptosis and oxaliplatin sensitivity of hepatocellular carcinoma. Am. J. Cancer Res. 2023;13:5590–5609. [PMC free article] [PubMed] [Google Scholar]
- 14.Wang W., Lu K., Jiang X., Wei Q., Zhu L., Wang X., et al. Ferroptosis inducers enhanced cuproptosis induced by copper ionophores in primary liver cancer. J. Exp. Clin. Cancer Res. 2023;42:142. doi: 10.1186/s13046-023-02720-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldman M.J., Craft B., Hastie M., Repecka K., McDade F., Kamath A., et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020;38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandrashekar D.S., Karthikeyan S.K., Korla P.K., Patel H., Shovon A.R., Athar M., et al. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27. doi: 10.1016/j.neo.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun B., Zhong F.J., Xu C., Li Y.M., Zhao Y.R., Cao M.M., et al. Programmed cell death 10 promotes metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma via PP2Ac-mediated YAP activation. Cell Death Dis. 2021;12:849. doi: 10.1038/s41419-021-04139-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X., Calvisi D.F. Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am. J. Pathol. 2014;184:912–923. doi: 10.1016/j.ajpath.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joshi P.R., Sadre S., Guo X.A., McCoy J.G., Mootha V.K. Lipoylation is dependent on the ferredoxin FDX1 and dispensable under hypoxia in human cells. J. Biol. Chem. 2023;299 doi: 10.1016/j.jbc.2023.105075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferro F., Servais S., Besson P., Roger S., Dumas J.F., Brisson L. Autophagy and mitophagy in cancer metabolic remodelling. Semin. Cell Dev. Biol. 2020;98:129–138. doi: 10.1016/j.semcdb.2019.05.029. [DOI] [PubMed] [Google Scholar]
- 21.Zhang T., Liu Q., Gao W., Sehgal S.A., Wu H. The multifaceted regulation of mitophagy by endogenous metabolites. Autophagy. 2022;18:1216–1239. doi: 10.1080/15548627.2021.1975914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Y., Li Z., Zhang S., Zhang T., Liu Y., Zhang L. Cellular mitophagy: mechanism, roles in diseases and small molecule pharmacological regulation. Theranostics. 2023;13:736–766. doi: 10.7150/thno.79876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoxhaj G., Manning B.D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer. 2020;20:74–88. doi: 10.1038/s41568-019-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kma L., Baruah T.J. The interplay of ROS and the PI3K/Akt pathway in autophagy regulation. Biotechnol. Appl. Biochem. 2022;69:248–264. doi: 10.1002/bab.2104. [DOI] [PubMed] [Google Scholar]
- 25.Xue Q., Kang R., Klionsky D.J., Tang D., Liu J., Chen X. Copper metabolism in cell death and autophagy. Autophagy. 2023;19:2175–2195. doi: 10.1080/15548627.2023.2200554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao X., Chen J., Yin S., Shi J., Zheng M., He C., et al. The expression of cuproptosis-related genes in hepatocellular carcinoma and their relationships with prognosis. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.992468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z., Zeng X., Wu Y., Liu Y., Zhang X., Song Z. Cuproptosis-related risk score predicts prognosis and characterizes the tumor microenvironment in hepatocellular carcinoma. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.925618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding L., Li W., Tu J., Cao Z., Li J., Cao H., et al. Identification of cuproptosis-related subtypes, cuproptosis-related gene prognostic index in hepatocellular carcinoma. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.989156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ke C., Dai S., Xu F., Yuan J., Fan S., Chen Y., et al. Cuproptosis regulatory genes greatly contribute to clinical assessments of hepatocellular carcinoma. BMC Cancer. 2023;23:25. doi: 10.1186/s12885-022-10461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X., Jia J.H., Zhang M., Meng Q.S., Yan B.W., Ma Z.Y., et al. Adrenomedullin/FOXO3 enhances sunitinib resistance in clear cell renal cell carcinoma by inhibiting FDX1 expression and cuproptosis. Faseb. J. 2023;37 doi: 10.1096/fj.202300474R. [DOI] [PubMed] [Google Scholar]
- 31.Sun L., Zhang Y., Yang B., Sun S., Zhang P., Luo Z., et al. Lactylation of METTL16 promotes cuproptosis via m(6)A-modification on FDX1 mRNA in gastric cancer. Nat. Commun. 2023;14:6523. doi: 10.1038/s41467-023-42025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Q., Yu M., Du X., Li Y., Lv J., Jiang X., et al. The role of cuproptosis key factor FDX1 in gastric cancer. Curr. Pharmaceut. Biotechnol. 2024 doi: 10.2174/0113892010301997240527162423. [DOI] [PubMed] [Google Scholar]
- 33.Qian S., Liu J., Liao W., Wang F. METTL3 promotes non-small-cell lung cancer growth and metastasis by inhibiting FDX1 through copper death-associated pri-miR-21-5p maturation. Epigenomics. 2023;15:1237–1255. doi: 10.2217/epi-2023-0230. [DOI] [PubMed] [Google Scholar]
- 34.Zulkifli M., Spelbring A.N., Zhang Y., Soma S., Chen S., Li L., et al. FDX1-dependent and independent mechanisms of elesclomol-mediated intracellular copper delivery. Proc. Natl. Acad. Sci. U. S. A. 2023;120 doi: 10.1073/pnas.2216722120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song C., Pan S., Zhang J., Li N., Geng Q. Mitophagy: a novel perspective for insighting into cancer and cancer treatment. Cell Prolif. 2022;55 doi: 10.1111/cpr.13327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choubey V., Zeb A., Kaasik A. Molecular mechanisms and regulation of mammalian mitophagy. Cells. 2021;11 doi: 10.3390/cells11010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao X., Wang W., Li Y., Yang D., Li X., Shen C., et al. HSP90AA1-mediated autophagy promotes drug resistance in osteosarcoma. J. Exp. Clin. Cancer Res. 2018;37:201. doi: 10.1186/s13046-018-0880-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H., Liu Q., Hu Y., Yin C., Zhang Y., Gao P. Linc00707 regulates autophagy and promotes the progression of triple negative breast cancer by activation of PI3K/AKT/mTOR pathway. Cell Death Dis. 2024;10:138. doi: 10.1038/s41420-024-01906-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mundekkad D., Cho W.C. Mitophagy induced by metal nanoparticles for cancer treatment. Pharmaceutics. 2022;14 doi: 10.3390/pharmaceutics14112275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soutar M., Kempthorne L., Miyakawa S., Annuario E., Melandri D., Harley J., et al. AKT signalling selectively regulates PINK1 mitophagy in SHSY5Y cells and human iPSC-derived neurons. Sci. Rep. 2018;8:8855. doi: 10.1038/s41598-018-26949-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo X., Zhang J., Guo C., Jiang N., Zhang F., Jiao Q., et al. Solute carrier family 35 member A2 regulates mitophagy through the PI3K/AKT/mTOR axis, promoting the proliferation, migration, and invasion of osteosarcoma cells. Gene. 2024;898 doi: 10.1016/j.gene.2023.148110. [DOI] [PubMed] [Google Scholar]
- 42.Chen T.H., Wang H.C., Chang C.J., Lee S.Y. Mitochondrial glutathione in cellular redox homeostasis and disease manifestation. Int. J. Mol. Sci. 2024;25 doi: 10.3390/ijms25021314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu C.C., Li C.J., Lin L.T., Lin P.H., Wen Z.H., Cheng J.T., et al. Cuproptosis-related gene FDX1 identified as a potential target for human ovarian aging. Reprod. Sci. 2024 doi: 10.1007/s43032-024-01573-0. [DOI] [PubMed] [Google Scholar]
- 44.Luo P., Yin P., Hua R., Tan Y., Li Z., Qiu G., et al. A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma. Hepatology. 2018;67:662–675. doi: 10.1002/hep.29561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bai C., Wang H., Dong D., Li T., Yu Z., Guo J., et al. Urea as a by-product of ammonia metabolism can Be a potential serum biomarker of hepatocellular carcinoma. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.650748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han J., Han M.L., Xing H., Li Z.L., Yuan D.Y., Wu H., et al. Tissue and serum metabolomic phenotyping for diagnosis and prognosis of hepatocellular carcinoma. Int. J. Cancer. 2020;146:1741–1753. doi: 10.1002/ijc.32599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Data will be made available on request.