Abstract
Human immunodeficiency virus (HIV) infection has evolved into an established global pandemic over the past four decades; however, despite massive research investment globally, the precise underlying mechanisms which are fundamental to HIV-related pathogenesis remain unclear. Single cell ribonucleic acid (RNA) sequencing methods are increasingly being used for the identification of specific cell-type transcriptional changes in HIV infection. In this scoping review, we have considered information extracted from fourteen published HIV-associated single-cell RNA sequencing-related studies, hoping to throw light on the underlying mechanisms of HIV infection and pathogenesis, and to explore potential candidate biomarkers for HIV disease progression and antiviral treatment. Generally, HIV positive individuals tend to manifest disturbances of frequency of multiple cellular types, and specifically exhibit diminished levels of CD4+ T-cells and enriched numbers of CD8+ T-cells. Cell-specific transcriptional changes tend to be linked to cell permissiveness, hyperacute or acute HIV infection, viremia, and cell productivity. The transcriptomes of CD4+ T-cell and CD8+ T-cell subpopulations are also observed to change in HIV-positive diabetic individuals, spontaneous HIV controllers, individuals with high levels of HIV viremia, and those in an acute phase of HIV infection. The transcriptional changes seen in B cells, natural killer (NK) cells, and myeloid dendritic cells (mDCs) of HIV-infected individuals demonstrate that the humoral immune response, antiviral response, and immune response regulation, respectively, are all altered following HIV infection. Antiretroviral therapy (ART) plays a crucial role in achieving immune reconstitution, in improving immunological disruption, and in mitigating immune system imbalances in HIV-infected individuals, while not fully restoring inherent cellular transcription to levels seen in HIV-negative individuals. The preceding observations not only illustrate compelling advances in the understanding of HIV-associated immunopathogenesis, but also identify specific cell-type transcriptional changes that may serve as potential biomarkers for HIV disease monitoring and therapeutic targeting.
Keywords: Single cell RNA sequencing, Human immunodeficiency virus, Acquired immunodeficiency syndrome, Frequency, Transcriptomics, Immune cells
1. Introduction
The HIV/AIDS pandemic remains an ongoing global health challenge, lacking both a viable vaccine and a cure [1]. The major barrier to HIV eradication is the enduring persistence of latent infection [2], along with the enhanced mutagenic profile of HIV [3], which contributes to the ability of HIV to evade conventional human immunological surveillance and to establish a chronic persistent infection within the host. A comprehensive understanding of key immune responses at the different HIV infection stages, the various HIV disease stages, and the diverse treatment strategies used when infected with HIV is essential for the development of effective vaccines and other curative strategies that may trigger targeted immune responses. However, individual cellular dynamics and virus-host interactions when developing and coordinating human immune responses during the HIV infective process are currently insufficiently understood, and represent a considerable knowledge gap regarding our comprehension of HIV pathology.
The emergence of single-cell RNA sequencing (scRNA-seq) technologies enables high-resolution transcriptomic analysis at single cell resolution, and allows, amongst other things, investigations into cellular heterogeneity, discovery of novel or rare cell populations, and specific cellular trajectory of developmental or differentiation processes [[4], [5], [6], [7]]. Single-cell investigations in the scientific realms of immunology, oncology, and developmental biology have significantly increased our understanding of intercellular heterogeneity and the diversity of cell subpopulations. Furthermore, scRNA-seq is increasingly being applied to investigate infectious diseases, focusing mainly on immune atlas study [e.g., the revealing of novel cell subtypes, changes in inflammatory responses, and the discovery of immune signaling pathways for differentially expressed genes (DEGs)], study related to host-pathogen interactions (such as identification of vulnerable cell types and investigation of infection dynamics), immune repertoire study, and biomarker discovery for infectious diseases [8]. Thus, using the single-cell resolution of cellular immune responses and pathogen-host interactions, we may well be able to establish a clearer immunological panorama for HIV infection, more accurately explore immune evasion and drug resistance during the process of HIV treatment, and discover specific markers that accurately predict disease prognosis and monitor the effects of treatment. Diligent focus on investigation in the preceding areas will potentially yield further evidence and insights with respect to the synoptic comprehension of the prevailing HIV immunopathophysiological milieu, and therefore materially enhance available therapeutic options for patients afflicted with HIV-related disease.
One research group has reviewed the transcriptomic signatures in host response against HIV infection [9]. The majority (55/62) of the included studies in the preceding review used bulk RNA sequencing; however, this technique only yields averaged expression data from highly diverse populations, and fails to accurately capture the heterogeneity of individual cells or enable dynamic tracking [10]. Additionally, constraints exist in the clarification of dynamic cellular transformations throughout development and differentiation processes, as well as the discovery of novel cellular states and populations (especially for rare cell subtypes) [11,12]. Another recent review [13] has summarized progress related to HIV immunopathogenesis at single cell resolution, focusing on the HIV replication cycle, new cell subsets and latently infected cells, the infected-cell exhaustion profile, and the diversity of HIV-1 reservoirs. Their observations provide crucial evidence in the quest to comprehensively understand HIV pathogenesis and pathogenicity. Nevertheless, much as yet unrevealed information remains to be learned about HIV infection, such as cell permissiveness, longitudinal analysis of HIV infection, spontaneous control of HIV, and the study of HIV-infected individuals comorbid with metabolic disease, amongst others. In the present scoping review, we have gleaned updated information from contemporary studies, and have sought to reveal and summarize some of the fundamental mechanisms which underlie immunopathogenic processes during HIV infection, and also discuss the transcriptional signatures associated with antiretroviral therapy (ART), with the intention of paving the way towards groundbreaking and novel HIV therapeutic modalities for the future.
2. An overview of scRNA-seq technology
scRNA-seq is an advanced laboratory technique that is used to characterize the transcriptomic profiles of single-cells, and has emerged as an important tool for exploring cellular heterogeneity and dynamic changes in cell populations. The complete workflow for scRNA-seq protocols shared by contemporary laboratories includes single-cell isolation, transcript capture, mRNA reverse transcription, cDNA amplification, and library construction [[14], [15], [16]]. This data analysis involves mainly two aspects at cell-type and gene level, such as differential gene expression, gene function enrichment, regulatory network reconstruction, cluster identification, differentiation trajectory inference, amongst others [[17], [18], [19], [20]].
3. Literature search
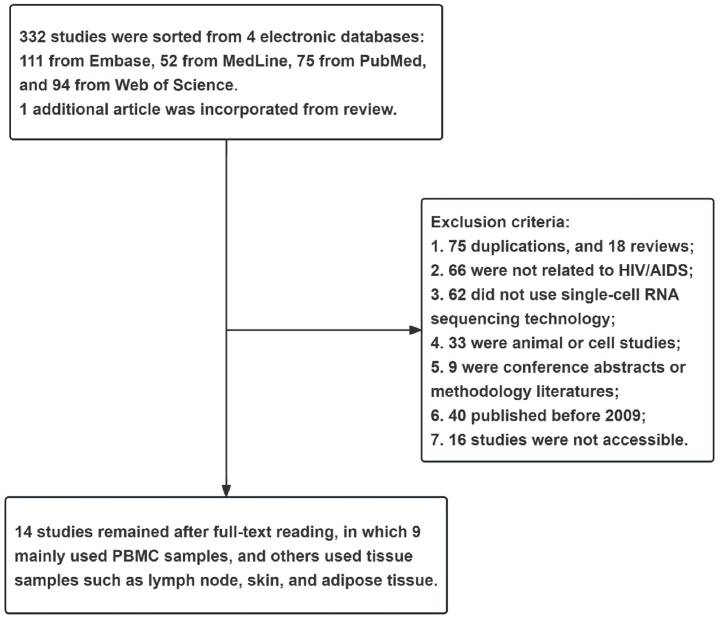
We have used key search words including "HIV" OR "AIDS" AND "single-cell RNA sequencing" OR "single-cell analysis" to identify and sort relevant published literature in PubMed, Embase, MedLine, and Web of Science from 2009 to March 15, 2022. Altogether, 332 articles were retrieved from four electronic databases. Additionally, one relevant article was retrieved from the references of a review [21] concerning HIV and single-cell analysis. Of the 333 studies that fulfilled our criteria, we examined 317 full-text documents, while sixteen studies were not accessible to our team.
After excluding duplications, reviews, conference abstracts, methodology articles, animal studies, cell studies, and studies not related to HIV/AIDS or scRNA-seq (Fig. 1), we finally included 14 studies for further analysis. These studies encompass a broad range of HIV-related clinical populations, such as hyperacute and acute HIV positive cohorts [22], individuals with high and low HIV RNA viral loads [23], ART-naïve and ART-treated HIV-infected individuals [[24], [25], [26], [27]], and HIV controllers [[28], [29], [30]], amongst others. Peripheral blood mononuclear cells (PBMCs) were the most widely used cellular sample in most included studies, followed by tissue samples, such as lymph node tissue, adipose tissue, tonsillar tissue, and skin tissue (Table 1).
Fig. 1.
Flowchart of article screening.
Table 1.
Characteristics of included studies.
| Author (publication year) | Subjects & Sample sizes | Study site | Age, median [IQR] | Gender | Race | ART initiation and ART duration | CD4+ T-cell counts (cells/μL) | HIV RNA (copies/ml) | Sample type | Platforms | Methods of sequencing | RF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kazer (2020) | From FRESH cohort; 4 patients |
NA | 22.5 (21, 24) | NA | NA | Only number 3 patient-initiated ART 2.3 years after baseline | Pre-infection: 884 (676.75, 916); baseline: 238 (212, 335.25); Week 1: 157 (140.5, 214.5); Week 2: 347 (244.5, 389.5); Week 3: 190.5 (74.5, 373.5); Week 4: 501 (322.5, 585); Month 4: 196 (140.75, 353.5); Year 1: 659 (510.5, 823.5). | NA | PBMC | BD SORP FACSAria II | Integrated single-cell analysis | 22 |
| Wang (2020) | 6 HIV-infected individuals, 4 healthy individuals | U.S.A. | 46 (30.5, 58.25); two without specific age | 1 female, 9 male | NA | Yes (except 1 HIV controller) | 451 (227, 629.25); HIV controller: unknown. |
3 donors with a high viral load and 3 with a low viral load (>100,000 and < 20) | PBMC | 10x Genomics | scRNA-seq | 23 |
| Liu (2020) | 25 HIV patients (virologically suppressed) | NA | 57 (51, 62) | Male: 17; Female: 8 | African American (17), White/Caucasian (6),Hispanic (1), Asian (1) | 181 (105, 246) months | NA | Virologically suppressed | PBMC and leukapheresis sample | NA | Sort-Seq | 24 |
| de Armas (2019) | 6 HIV patients, 6 non-HIV patients | NA | HIV: 64 (62, 66.75); non-HIV: 64 (61.25, 66). | HIV group: 5 male, 1 female; non-HIV group: 3 male, 3 female | NA | Yes | NA | Virologically suppressed on ART | PBMC | Illumina MiSeq (BD) |
Single cell gene expression analysis | 25 |
| Farhadian (2018) | Virologically suppressed HIV (3 patients) versus. healthy control (2 participants, none had active neurological disease or other infection) | U.S.A. | HIV participants: 47(40, 49); uninfected participants: 54(51.5, 56.5) | All male | NA | Yes | Nadir CD4+ T-cells [infected participants]: 237.5(157.75, 317.25); current CD4+ T-cells [infected participants]: 391(424, 967). nadir CD4+ T-cells [uninfected participants]: NA; current CD4+ T-cells [uninfected participants]:848.5(811.75, 885.25). | Infected participants: plasma HIV RNA levels <20 for more than 1 year; CSF HIV RNA: 68.5 (53.25,83.75) |
CSF and blood | NA | scRNA-seq | 26 |
| Josefsson (2013) | Chronically infected with HIV-1 (Fiebig stage VI); 5 patients |
U.S.A. | 29.9 (29.7, 33.2) | 5 male | White/Latino: 2; Black: 3 | Untreateda | 484 (220, 758); 2 patients (3, 4) had CD4+ percent <14 %. Palpable lymph nodes were identified in the axilla or inguinal regions and entire lymph nodes were excised. | 9991 (6195,35576) | Lymph node tissue; PBMC | NA | Single-cell sequencing | 27 |
| Holla (2021) | HIV:HCs = 3:3 (sc-RNA seq) | U.S.A. | NA |
HIV: 1 female, 2 male; Healthy donors: 3 female |
HIV: African (Zimbabwe),Multirace/Hispanic, African American; Healthy donor: African American |
NA | HIV1: 1366; HIV2: 458; HIV3: 677 |
HIV1: <40, HIV2: 185, HIV3: 1353 |
PBMC | Illumina HiSeq 3000 platform | Pipeliner RNA-seq | 57 |
| Angin (2019) | Non-ART and virologically suppressed HICs (n = 5) and cART individuals (n = 5). Longitudinal follow-up (4,6,8,10,12,14 days after diagnosis/infection) |
HIV controllers: ANRS CO21 Codex cohort; cART: ANRS PRIMO cohort or CHU Kremlin-Bicêtre and CHU Georges Pompidou in Paris. |
NA | NA | NA | ART group has initiated ART for at least 2 years | NA |
HIV controllers whose last five consecutive plasma HIV RNA were <400; ART group: virologically suppressed |
PBMC | Fluidigm | Single-cell gene expression | 28 |
| Nguyen (2019) | 12 HIV controllers, 14 ART, 18 CP, 7 Acute | San Francisco (USA),Mexico, Philadelphia | 52 (47–59), 40 (32–58), 31 (24–37) and 23 (20–27) | 10 male: 1 female (male transform to female); 8 male, 1 female, 5 unknown; 10 male, 1 female, 7 unknown; 7 male | NA | NA |
HIV controller group: 881 (660–1172); ART group: 544 (359–601); Chronic progressor group: 348 (175–521); Acute group: 306 (238–370) |
HIV controller group: <40; ART group: <40; Chronic progressor group: 51396 (12845–213173); Acute group: 1996027 (1297461–4421034) |
PBMC; lymph node tissue | HiSeq 4000 (Illumina) | Integrated single-cell approach (Illumina Nextera) | 29 |
| Varadarajan (2011) | HIV controllers (2 patients) versus. untreated viremic progressors (2 patients) | U.S.A. | NA | NA | NA | No | NA | HIV controllers (<50); untreated viremic progressors (>2000, median, 3692, range 189–34172) | PBMCs | SMART-RACE RT-PCR kit (BD). | TCR α and β chain sequencing | 30 |
| Saluzzo (2021) | Cohort 1 (HIV ‘late ART’, 3 patients and 3 healthy donors); cohort 2 (HIV ‘early ART’, 6 patients, 11 controls), cohort 3 (HIVSeq 5 controls) and cohort 4 (HIV HPV: HIV-infected patients [n = 11] and HIV- patients [n = 10]). | Vienna | NA | NA | NA | Yes; Cohort 1: after diagnosis (average 50 months) and was on therapy for more than 5 years at time of tissue sampling; Cohort 2: immediately after diagnosis (average 13 ± 12 days). |
Cohort 1: nadir at diagnosis (<350, average 209 ± 130). Cohort 2: average 507 ± 154. |
Cohort 1: <1000 | Skin, Blood (PBMC) | 10x Genomics, the Illumina NovaSeq platform |
scRNA-seq and scTCRαβ-seq | 34 |
| Rato (2017) | HIV: non-infected CD4+ T-cells from high (∼40 % GFP+) and low (∼10 % GFP+) permissive individuals. 1 high and 1 low permissive donor; 2 uninfected CD4+ T-cell donors (3 for validation) |
NA | NA | NA | NA | NA | NA | NA | PBMC (followed by CD4+ T-cell isolation using negative selection and magnetic separation) | Fluidigm C1 platform | scRNA-seq and Bulk RNA-seq | 33 |
| Wanjalla (2021) | HIV-positive non-diabetes (Group 1, 5 patients) versus. HIV-positive diabetes (Group 2, 6 patients) versus. HIV-negative diabetes (Group 3, 5 patients) | U.S.A. | Group 1 (41 [31,32]) versus. Group 2 (53[43, 55.5]) versus. Group 3 (49 [33,34]) | Group 1 (4M, 1F) versus. Group 2 (4M, 2F) versus. Group 3 (3M, 2F) | Group 1 (2 African American, 2 Caucasian, 1 other) versus. Group 2 (4 African American, 2 Caucasian) versus. Group 3 (5 Caucasian) | Yes, on long-term ART (1.5–20.35 years) |
Group 1: CD4+ T-cells at start: 655 [500, 661]; current CD4+ T-cells:1133 [781, 1256]; Group 2: CD4+ T-cells at start: 513.5 [304.25, 683.75]; current CD4+ T-cells: 823.5 [554.75, 950.5] |
HIV-1 RNA<50 for the 12 months prior to adipose biopsy | Adipose tissue, blood [PBMC] | Illumina MiSeq (for sc-TCR sequencing); Illumina NextSeq (for 3′ and 5′ RNaseq) | SMART seq2 and MARS-seq approach | 35 |
| Murray (2020) | 6 HIV patients | U.S.A. | 46 (31.25, 51.75) | Male: Female = 5:1 | NA | 3 start ART [6 (4, 9.5) years]; 3 ART-naïve | Normal levels of CD4+ T-cells in the blood, i.e.,∼500–1500 CD4+ T-cells |
ART-treated individuals: 2 with HIV RNA<50; 1 with HIV RNA<400; ART-naïve individuals: 5970; 304,742; 19,998 |
mDCs | Illumina | ScRNA-seq | 31 |
NA: not applicable.
1 patient had received transient ART but had not undergone any consistent therapy for over 2 years prior to biopsy.
4. Frequencies and transcriptional signatures of immune cell clusters in HIV positive individuals
The impairment of immune function caused by HIV infection is not limited only to CD4+ T-cell depletion and exhaustion, but also affects other immune cell populations to varying degrees, including B-cells, NK cells, CD8+ T-cells, and myeloid dendritic cells (mDCs), amongst others. The effect of these depleted cell populations may likely be to further exacerbate dysfunction of the immune system in the host, which would thus permit uncontrolled viral replication and rapid progression of HIV disease. scRNA-seq has been applied to analyze the manner in which HIV infection may perturb immune cell population transcriptomes, and has revealed marked changes in the immune response to viral infection, and is distinguished by the production of antiviral cytokines (e.g., type I IFNs)/chemokines), direct cytotoxic responses, and antiviral antibody production.
4.1. CD4+ T-cells
The primary target of HIV infection are CD4+ T-cells, and these cells are considered to be the predominant cell type responsible for harboring latent virus. Significantly reduced CD4+ T-cell pool and impaired CD4+ T-cell functionality are hallmarks of HIV infection [35,36]. CD4+ T-cells may be stratified into multiple subtypes with specific molecular characteristics, with the subtypes each having distinct biological functions. This heterogeneous population, with diverse phenotypic characteristics and regulatory mechanisms, intimately reflects the overall immunological functionality of the body, and closely correlates with the onset, progression, and prognosis of different diseases. However, it remains incompletely understood as to which precise CD4+ T-cell subpopulations may be selectively depleted or expanded among HIV-positive individuals, and also how HIV infection influences each of their respective transcriptomes. Exploration of the cellular heterogeneity seen in HIV infection, as revealed by scRNA-seq, may help researchers gain additional insights into individual cellular dynamics, cell-cell cooperativity, virus-host interactions, and biomarker discovery.
Profiling of the single-cell landscape characterizes the overall CD4+ T-cell population composition during both infectious and physiological settings, which offers crucial insights into understanding the immunopathogenic mechanisms which are fundamental to HIV infection. In total, five studies reported on the changes of frequency of CD4+ T-cells using PBMCs, skin, or adipose tissue samples extracted from various HIV-infected individuals. Their observations reveal that different populations of CD4+ T-cells, including exhausted memory CD4+ T-cells (CD4-Tex), effector memory CD4+ T-cells (CD4-Tem), and skin CD4+ tissue-resident memory T-cells (CD4-Trm) consistently exhibit exhaustion signatures during HIV infection. In contrast, some specific subclusters of CD4+ T-cells, including hyper-permissive cells (viz., CD25highCD298highCD63highCD317high CD4+ T-cells), and CX3CR1+GPR56+CD57+ (C-G-C+) CD4+ T-cells are substantially enriched in highly permissive HIV-infected individuals, and also in adipose tissue of HIV-infected individuals comorbid with diabetes. Thus, the inherent composition and relative proportions for different CD4+ T-cells found in samples from different anatomical sites are markedly altered in response to HIV infection.
Specifically, Wang et al., performed a comparative analysis of CD4+ T-cell counts among people with varying HIV viral loads and healthy controls (HCs) by using PBMC sampling on the 10X Genomics platform [23]. Their observations revealed that individuals with a high HIV viral load exhibited significantly lower populations of CD4+ T-naïve and CD4-Tem cells compared to their healthy counterparts, while also presenting a distinct cluster of cells referred to as CD4-Tex. Conversely, individuals with a low viral load demonstrate a reduction in the CD4-Tem cluster and an absence of a CD4-Tex cluster [23]. These observations indicate that variations to the absolute number of and percentage variation with respect to different CD4+ T-cell subclusters are proportional to the prevailing viral load, and also that a reduction of CD4-Tex cell numbers may be considered to be an indicator of aggressive invasion by HIV. Similarly, Rato et al., conducted scRNA-seq and bulk RNA-seq on the Fluidigm C1 platform, and compared the specific PBMC single cell transcriptional differences between one highly permissive and one lowly permissive HIV individual. They observed an up to 28-fold enrichment of the hyper-permissive cell subpopulation, i.e., the CD25highCD298highCD63highCD317high CD4+ T-cell population, in comparison to the population of cells that are unsorted [37]. Hence, the utilization of scRNA-seq facilitates the quantification of HIV-susceptible cell proportions that can be effectively defined by the combined biomarkers.
The precise mechanisms responsible for chronic tissue-related immunodeficiency observed in individuals with HIV infection remain poorly understood. Two recent studies have applied several different approaches in an attempt to characterize the CD4+ T-cell population in specific tissues. In the first study, Saluzzo et al. [38], observed a depletion of skin-derived pro-inflammatory CD4-Trm cells following HIV infection, with replenishment of these cells occurring only on early initiation of ART. The authors further revealed that reduced in situ expression of CXCR3 is linked to dysregulation of skin CD4-Trm cells, which exhibit a propensity towards a tolerogenic, Th2-like phenotype. Mirroring observations in the skin, perilesional CXCR3+ CD4-Trm cells are also depleted in the mucosa of HIV-infected patients with HPV-related anal intraepithelial neoplasia (AIN) in contrast to non-HIV-infected individuals. In summary, the loss of CXCR3+ CD4-Trm cells is linked to severe and irreversible tissue-confined immunodeficiency, and maintenance of a healthy population of CXCR3+ Trm cells may prevent long-term complications. In the second study, Wanjalla's study group studied adipose tissue from six patients with comorbid HIV and diabetes and observed a high proportion of cytomegalovirus-specific C-G-C+ CD4+ T-cells in these patients, while HIV-negative diabetic patients predominantly exhibited CD69+ CD4+ T-cells [39]. Additionally, the preceding group believe that adipose tissue-derived C-G-C+ CD4+ T-cell subsets may exert a substantial influence on adipocyte function, which in turn increases the risk of metabolic complications among individuals with HIV infection [39]. These observations therefore may have significant implications for HIV-positive people who are at high risk of developing the metabolic syndrome.
Now recognized to be an indispensable laboratory tool for research scientists, scRNA-seq allows investigators to discern alterations in the immune cellular landscape during HIV infection, and to uncover the distinct roles played by the various cellular populations, thus shedding light on the underlying and hitherto enigmatic mechanisms known to be inherent to HIV immunopathogenesis.
Four of our included studies revealed the specific transcriptional signatures of CD4+ T-cells among HIV-positive people using cells isolated from PBMCs, tonsillar tissue, or adipose tissue (Supplementary Table 1). These transcriptional functional alterations in CD4+ T-cells mainly relate to HIV disease progression and immunosuppression. One subcluster of CD4+ T-cells, i.e., the CD4+ T-helper (Th) 17 cell subcluster, is particularly susceptible to HIV infection. Depleted expression of CD4+ Th17 cells at gut mucosal sites in HIV positive individuals causes disruption of gut barrier integrity, and subsequently results in microbial translocation-induced chronic systemic inflammation, which is contemporarily recognized as a hallmark of HIV disease progression [40]. In studies included in our analysis, genes related to the Th17 transcription factor, retinoic acid-related orphan receptor C (RORC), were increased in productive cells, and the highly expressed genes in CD4+ memory T-cells from HIV-infected individuals with diabetes were also involved in Th17 cell differentiation pathways. Sustained interferon (IFN) signaling is recognized as a key factor in driving immunosuppression [31,41]. IFN-stimulated genes are markedly up-regulated in CD4+ T-cells in individuals with a high viral load. Conversely, a down-regulation in the expression of IFN-induced genes is observed in the purified hyper-permissive cell subset.
Another research group performed scRNA-seq by utilizing the Illumina MiSeq sequencer, and used tonsillar tissue to evaluate the transcriptional signatures among productive HIV-infected cells, latent HIV-infected cells, and HIV uninfected cells [42]. Their study observed that: ⅰ) genes related to the Th17 transcription factor RORC are upregulated in productive cells, in comparison with HIV uninfected cells; ⅱ) productive cells express the highest number of HIV transcripts (env, tat, and gag genes, among others) per cell (median 38.5), while latent HIV-infected cells had very few HIV transcripts per cell (median 1.5); ⅲ) a small set of genes, such as those related to histone cluster 1 (HIST1) and pro-inflammatory cytokine IL32, exhibited upregulation in latent cells with respect to uninfected and productively infected cells, indicating the involvement of these molecular pathways in HIV persistence.; ⅳ) twenty-four DEGs were identified in latent infected cells versus uninfected cells. These DEGs are associated with protein targeting and viral gene expression, the respiratory electron transport chain and protein folding [42]. Wanjalla et al., using adipose tissue samples, identified the top highly expressed genes in CD4+ memory T-cells from people with HIV infection comorbid with diabetes, in contrast to people with diabetes but without HIV infection. Their observations revealed that the identified genes were significantly enriched in multiple pathways, such as the Th1 and Th2 cell differentiation pathways, and the T-cell receptor signaling pathway, to name just a few [39].
Wang's research team has documented immune cell exhaustion in PBMCs sampled from six individuals who were HIV-positive (three with high viral loads vs. three with low viral loads). Wang and colleagues observed that in individuals with high viral loads, the expression of IFN-stimulated genes (including IFIH1, IFIT2, and IFI6) was remarkably upregulated in CD4+ T-cells with respect to HCs [23]. Rato's research group [37] noted the PBMC-specific transcriptional function in individuals with high and low permissiveness to HIV infection, and revealed conflicting results with respect to IFN-stimulated genes. They observed that the purified hyper-permissive cell subset is distinguished by a downregulation of IFN-induced genes, i.e., 95 genes exhibited a distinct downregulation in CD25highMRK3high (more permissive) cells, when compared against CD25high (less permissive) cells, and these genes were associated with the type I IFN pathway [37].
Overall, scRNA-seq enables a comprehensive exploration of the profound alterations to the composition and functionality of the CD4+ T-cell population throughout HIV infection by identifying distinct surface markers, cytokines, or transcription factors associated with specific T-cell subpopulations. This provides valuable insights into the functional attributes of immune cells, the regulatory mechanisms governing immune responses, disease progression and pathogenic mechanisms and pathways, as well as educated speculation with respect to the potential for personalized therapeutic strategies for HIV-related disease.
4.2. CD8+ T-cells
CD8+ T-cells are of critical importance to adaptive immune response against viral infections. Although not directly targeted by HIV, CD8+ T-cells are capable of mounting anti-HIV responses through effector mechanisms such as the direct lysis of infected cells and the secretion of cytokines and chemokines during both the early and chronic phases of HIV infection [13]. scRNA-seq has enabled the transcriptional profiling of individual immune cells, revealing cellular heterogeneity and the immunomodulatory molecular mechanisms of the CD8+ T-cell sub-population during HIV-1 infection.
Five studies have reported on the alterations in the frequency of CD8+ T-cells or their subtypes obtained from PBMCs or lymphoid tissues [22,23,[28], [29], [30]], and their observations have revealed that although CD4+ T-cells are eventually depleted during HIV infection, CD8+ T-cell numbers continue to increase until the very late phase of HIV infection [43]. In contrast, the variations in different CD8+ T-cell subset frequency are not consistent during HIV infection. Using scRNA-seq on PBMC samples from four ART-naïve individuals before and longitudinally during acute HIV infection, Kazer et al., observed a rapid rise in CD8+ T-cell frequency within the first month of infection, when compared to pre-infection [22]. In the absence of ART, a higher frequency of proliferating CD8+ T-cells in spontaneous HIV controllers (also referred to as HIV controllers, and generally defined as individuals who maintain HIV viral loads below detection levels in the absence of ART [44]) compared to non-HIV controllers was also observed in the preceding study [22]. Interestingly, Nguyen et al., conducted an integrated single-cell analysis using the FACSAria II flow cytometer for single-cell sorting and the HiSeq 4000 sequencing system. In comparison to HIV progressors, the preceding authors also noticed that spontaneous HIV controllers had larger frequencies of HIV-specific CD8+ T-cells in their lymph nodes [29]. Notably however, they observed no statistically significant difference in HIV-specific CD8+ T-cell frequency between ART-naïve individuals and ART-treated individuals [29]. The preceding results indicate that rapid expansion of CD8+ T-cell numbers contributes to active control of viral replication in the early stages of infection. Overall, the composition and proportions of individual CD8+ T-cell subsets are variably abnormal at distinct stages of the HIV infection cycle.
scRNA-seq analysis continues to shed light on immune cell complexity and diversity of immune cell populations in the context of HIV infection. This technique has been reported to be capable of characterizing the heterogeneity of the immune cell transcriptome induced by HIV infection. Wang et al. [23], for instance, demonstrated that CD8-Tex cells exhibit distinct epigenetic and transcriptional characteristics compared to CD8-Tem cells and terminally differentiated cells, secrete fewer cytokines, and are characterized by up-regulated cell-surface expression of a panel of inhibitory receptors, including KLRG1, CD160, and TIGIT. The expression of these inhibitory receptors has also been recognized as a hallmark of exhausted T-cell populations in pathogen-infected and tumor-bearing animals [[33], [45], [46]]. More importantly, the blocking of KLRG1 has been shown to significantly rejuvenate HIV-specific CD8+ T-cell function [23], which may potentially be a immunotherapeutic target against HIV infection. In individuals infected with HIV, CD8-Tex cells specifically upregulate expression of some genes, including CMC1, TRAPPC1, COTL-1, and KLRG-1, and downregulate expression of other genes, viz., ITGB1, GZMB, and PRF1 [23]. These altered genes are mainly involved in cytoskeletal function, energy metabolism, and vesicle transportation. The preceding observations further suggest that CD8-Tex cells exhibit a phenotype characterized by lower effector function compared to normal CD8-Tem cells. Additionally, two differentially expressed immune response genes, i.e., NF-kB inhibitor COMMD6 and IFN-responsive transcription factor IRF1 are upregulated and downregulated, respectively, in CD8-Tex cells, which is consistent with the HIV-induced suppression of immune signaling. Additionally, cells within the CD8-Tem-IFNhi cluster (a cell cluster which undergoes a significant upregulation of IFN-response genes) exhibit elevated IFN stimulated gene expression, including that of IFIT3, ISG15, and OASL, which is consistent with a trend towards an enhanced host antiviral immune response in the host. Similarly, Buggert et al., have also reported on interferon signatures in HIV-specific CD8+ T-cells resident within lymphoid tissues [47].
The functional characteristics of CD8+ T-cells have been reported to correlate with effective immune control of HIV infection. In analyzing PBMCs collected from four participants testing positive for HIV (treatment naïve) during the study, Kazer et al., noted that cytotoxic CD8+ T-cells highly expressed GZMB and PRF1 during acute infection, potentially contributing to control of initial-stage HIV infection [22]. Growing evidence suggests that CD8+ T-cells play a pivotal role in the attainment of a functional cure for HIV disease. We thus further outline the single-cell transcriptional profile of CD8+ T-cells of HIV controllers in Supplementary Table 1. Angin et al., analyzed 96 subset-specific genes in 1440 individual central memory CD8+ T-cells (CD8+ TCMs) from five HIV controllers and five ART-treated individuals on the Fluidigm platform, and revealed contrasting profiles of gene expression in these cells [28]. A comparative analysis of CD8+ TCMs from individuals undergoing ART and those with controlled HIV infection revealed a significant upregulation of a diverse range of genes associated with effector functions, including genes encoding (i) β-chemokines (XCL1, CCL3, and CCL3L1), which have been demonstrated to impede progression of HIV-1 infection [48]; (ii) granzymes (GZMK, GZMB), which are crucial for the exertion of antiviral and cytotoxicity by CD8+ T-cells [49]; and (iii) the tumor necrosis factor (TNF) family of receptors associated with CD8+ T-cell-induced apoptosis, such as TNF-related apoptosis-inducing ligand receptor (TRAIL) and Fas ligand (FASL). The above results indicate that HIV-specific CD8+ TCMs have enormous antiviral potential, and this is consistent with research findings demonstrating HIV-control of CD8+ TCMs which effectively inhibits HIV infection in vitro [28]. Notably, despite the general consensus that CD8+ T-cells are not a major source of type I interferon (IFN) production, cells from HIV controllers exhibit higher levels of IFNB transcript expression than individuals treated with ART. Additionally, several key genes upregulated in the mTORC2 pathway associated with survival were seen in CD8+ TCMs from HIV controllers, while in ART-exposed individuals, these cells predominantly upregulate genes related to the mTORC1/activation/glycolysis pathway, and the inhibitory receptor LAG3 [28]. Collectively, these points underscore the molecular uniqueness of HIV-specific CD8+ T-cell specimens taken from HIV controllers, and point to distinct gene expression profiles matched with improved cell survival, and the sustained potential to generate more effective anti-HIV responses under challenging circumstances.
In addition to PBMC samples, Nguyen et al., used an integrated single-cell approach to document the transcriptional profiling of lymphoid tissue CD8+ T-cells of HIV controllers and chronic progressors who were not receiving ART [29]. According to their investigational outcomes, the lymph node-derived CD8+ T-cells from HIV controllers were distinguished by lower expression of cytotoxic granule-associated genes (PRF1 and GZMB), while GZMB expression was indeed upregulated in the circulating CD8+ T-cells of HIV controllers [28]. Transcript expression for genes encoding cytolytic molecules, such as TNFSF10, FASL, GZMA, and GZMH [50,51] was either comparable between HIV controllers and chronic progressors, or enriched in HIV-specific CD8+ T-cells in the lymph nodes of chronic progressors, as compared to HIV controllers. Similar to observations reported by Wang et al. [23], researchers further demonstrated that cells extracted from the lymph nodes of chronic progressors upregulate inhibitory receptor genes e.g., CD244, TIGIT, and LAG3, as well as markers associated with an exhausted phenotype (KLRG1 and EOMES) [[32], [34], [52]]. In contrast, HIV-specific CD8+ T-cells from HIV controller lymph nodes predominantly upregulate IL7R expression, which is essential for maintenance of homeostasis [53], and several chemokines/cytokines like CCL5 and IL32, together suggesting a highly functional memory phenotype. Furthermore, Nguyen et al., analyzed 2264 DEGs, and observed that these genes were enriched in immune-related pathways, including “defense response”, “immune system process”, and “immune response” [29]. Most importantly, the preceding group have identified eleven transcripts that code for predictive secretory factors, which are specifically elevated in HIV-specific CD8+ T-cells originating from the lymph nodes of HIV controllers. The eleven transcripts in question have been found to contain genes that code for TNF, CCL5, RNASE1, and IL32, all of which have been demonstrated to suppress virus replication [[54], [55], [56]]. Thus, the preceding observations provide a feasible explanation for the effectiveness of CD8+ T-cells in spontaneously controlling virus replication and preserving inherent immune functions.
Together, the preceding observations suggest that individual CD8+ T-cells isolated from disparate HIV-positive patients display distinct single-cell transcriptional characteristics due to specific host factors, and thus do not necessarily respond uniformly or analogously to HIV infection. scRNA-seq analysis on PBMCs and lymph nodes has confirmed that CD8+ T-cells from HIV controllers are significantly enriched in effector functional genes and survival genes, whereas CD8+ T-cells in chronic progressors are clearly enriched in genes associated with activation, exhaustion, and IFN-stimulation.
4.3. B-cells
The human B-cell compartment is significantly affected by HIV infection, with a notable expansion of B-cell numbers [57]. Wang et al., have reported on numerical changes to the B-cell population after HIV infection, and have observed that HIV positive individuals have an elevated fraction of B-cells in the PBMC population, when compared to HCs [23]. Moreover, three studies have reported on the specific altered transcriptional genes and the corresponding functional changes seen in B-cells following HIV infection [23,25,57], as shown in Supplementary Table 1. After HIV infection, B-cells are hyper-activated; however, the antibody response is suppressed [31]. In concordance with this, Wang et al., recognized that expression of immunoglobulin genes (IGLC2, IGLC3, and IGKC, among others), the naïve B-cell marker gene (TCL1A), and the activated B-cell marker gene (CD27) are either completely absent or dramatically downregulated in HIV positive individuals. Conversely, the B-cell inhibitory receptor gene (LILRB1) is found to be upregulated [23]. The B-cell receptor (BCR) possesses an inherent ability to discriminate antigen affinity. Upon encountering antigens, B-cells generate antibodies with diverse specificities in the BCR, triggering signals that lead to B-cell proliferation and antibody production. Holla et al., observed that three naïve B-cell clusters differentially express BCR-driven activation associated genes, including IER2, NFKBIA, and JunB, indicating that the heterogeneity of naïve B-cell clusters could well be driven by levels of BCR activation [57]. Atypical B-cells (ABCs) (which are a subpopulation of B-cells that are CD21− CD27− and was originally observed in tonsillar tissue, and later in peripheral blood [58]) isolated from HIV positive individuals highly express ITGB2, CD69, SELL, and CD62L, indicating a more-activated B-cell state [57]. Interestingly, Armas et al., observed that transcriptional genes such as PTEN, PPP3CA, and LILRB1 are overexpressed, while DUSP4, CD86, SYK, and STAT5A are downregulated in H1N1-specific PBMC memory B-cells in HIV-positive individuals. These genes play major roles in regulating immune functions in virologically suppressed HIV positive individuals [25].
The preceding scRNA-seq findings thus demonstrate that even whilst not being infected by HIV, the numbers and functionality of the B-cell compartment may be altered in the setting of HIV-infected cells secondary to hyper-activation and elevated apoptosis, resulting in suboptimal antibody responses to common infectious agents. Consequently, restoration of B-cell functionality may potentially contribute in part to the antibody-mediated control against HIV infection.
4.4. NK cells
Increasing research evidence points towards a role that NK cells may play in mediating immune response associated with HIV infection. Two recent studies have reported on the specific transcriptional changes in NK cells under different HIV infection scenarios (Supplementary Table 1) [22,23]. Data gleaned from these studies have shown that a subpopulation of proliferative and cytotoxic NK cells are enriched in the initial phases of acute HIV infection, and normal NK cell function is impaired by chronic HIV infection. Additionally, the secretion of NK cell cytokine (an antiviral response) and their cytotoxic functioning are regulated by a variety of activatory and inhibitory receptors. However, no consistent up- or down-regulatory trend has been observed in either activatory receptor genes or inhibitory receptor genes. The reason for these divergent findings may be related to the following factors: ⅰ) there is a paucity of published studies concerning this specific realm of scientific endeavor; ⅱ) the presence of limited sample sizes in those studies that have been published (up to six enrolled HIV positive individuals); ⅲ) cognizance of the specific individual HIV infection state or disease stage of subjects enrolled in the studies (for example, subjects with high viral load, HIV RNA >100,000 copies/mL, high or low CD4+ T-cell counts, on ART or not, amongst other factors)
Kazer et al., assessed dynamic transcriptional changes in NK cells among individuals having both hyperacute and acute HIV infection using PBMC samples [22]. They observed that the NK cluster comprised the largest proportion of proliferating cells during the hyperacute phase of HIV infection. Participants who maintained viral loads of <1000 viral copies/mL in the absence of ART at 2.75 years post-infection showed an enrichment of a specific subpopulation of proliferative cytotoxic NK cells in the initial phase of acute infection prior to the emergence of CD8+ T-cells [22]. This suggests potentially novel roles for NK cells in the context of acute HIV infection, which may potentially be relevant for future viral control. Thus, the numerical change in proliferative cytotoxic NK cell abundance may be seen to function as a marker for potential HIV disease progression in ART-naïve HIV positive individuals with low viral loads.
Although NK cells play an essential role in the antiviral response, chronic HIV infection is known to correlate with impaired NK function. In concordance with this, Wang et al., observed a decreased expression of genes involved in the production of cytokine signalling proteins (e.g., IL12RB and IL18RAP) [23]. Costanzo et al., reported a downregulation of IL18RAP, which is an accessory protein that promotes IL-18 signalling in NK cells from HIV-positive individuals [59]. Vaccination with an HIV vaccine appeared to mitigate this reduction, suggesting a potential normalization of IL18RAP expression levels post-vaccination [59]. The preceding group also observed that HIV infection significantly alters expression of several NK receptor genes in HIV positive individuals with high viral loads, when compared with HCs [23]. Of the activating receptor genes, CD84, CD2, SLAMF7, and CD69 exhibit increased expression levels, whereas CD160 and NCR3 expression were observed to be at decreased levels. On the other hand, there is a decrease in the expression of inhibitory receptor genes SIGLEC7, LAIR2, and KLRB1, while TIGIT, LAG3, and LILRB1 show an increase in their expression levels. Also, IFNG, TNF and BIRC3, which are considered to be indicators of NK cell activation, were reported to be highly expressed [23].
Results revealed by scRNA-seq analysis suggests that HIV infection may also induce NK cell dysfunction by disruption of its effector functions, which include cellular cytotoxicity and the capacity to secrete cytokines, which together with T-cell exhaustion may be involved in HIV-associated immunodeficiency. The fundamental mechanisms which underlie aberrant NK cell-related function warrants further in-depth studies, thus offering the potential to discover novel approaches that may enhance immunological protection or achieve an HIV cure.
4.5. mDCs
As the most potent activators of CD4+ T-cells, mDCs play a pivotal role in regulating immunological responses. PBMC single-cell transcriptional analysis has shown that mDCs from HIV controllers exhibit unique antiviral immune responses and enhanced immune and metabolic states, suggesting that mDCs from HIV controllers display traits of innate immune cells. Martin-Gayo's research group have documented the antiviral immune response of a subset of mDCs isolated from PBMCs of three HIV controllers [60]. The CD64 and PD-L1 surface molecules are upregulated on these antiviral mDCs, and TLR3 along with poly:IC may induce their activation. One recent study identified a long-noncoding RNA, named MIR4435-2HG, that is present at a higher concentration in the mDCs of HIV controllers. This long-noncoding RNA was observed to be associated with enhanced immune and metabolic states in HIV controllers, suggesting that these mDCs express traits of a trained innate immune cell [61].
The preceding findings suggest that mDCs from HIV controllers may exhibit similar antiviral functionality as innate immune cells, and may be distinguished from HIV non-controllers based on their specific markers via scRNA-seq technology.
5. Dynamic transcriptional analysis in HIV positive individuals
5.1. Longitudinal transcriptomic analysis before and after HIV infection
Until recently, most HIV-related scRNA-seq studies have been cross-sectional; however, one recent study sought to explore the manner in which frequency of immune cells change before and after HIV infection (Supplementary Table 1) [22]. Kazer et al., examined changes in the frequency of immune cells at various timepoints ranging from pre-infection through one year post viral detection, and observed that the proportion of CD4+ T-cell cluster was diminished, while the monocyte cluster, the cytotoxic T-cell cluster, and the NK cell cluster were enriched after HIV infection [22]. Importantly, they observed that individuals who maintained low viremia following infection without ART showed higher frequencies of proliferating cytotoxic T-cells and a premature subpopulation of NK cells (i.e., proliferative, cytotoxic NK cells) one week after the detection of HIV viremia. These proliferating NK cells may function alongside proliferating cytotoxic T-cells early in infection, thus mitigating the proliferating cytotoxic T-cell antigenic load and subsequent exhaustion, which may be viewed as potential therapeutic targets for control of HIV [62]. Additionally, the observed DEGs in immune cells (such as those for IRF7, IFN-γ, IL8, and IL-6) are involved in the IFN pathway, suggesting a sustained role of innate immunity during acute infection [22]. Therefore, performing scRNA-seq analysis on PBMCs from HIV-positive individuals prior to ART initiation may provide valuable insights into the early immune response, potentially influencing subsequent clinical outcomes.
With the novel approach of scRNA-seq, Kazer et al., have successfully characterized the broad dynamics of peripheral immune cell frequencies, and have identified specific cell subpopulations and DEGs that may be associated with potentially therapeutic and preventive applications. Ethical and practical challenges exist in obtaining further samples from untreated individuals infected with HIV, and thus further investigations in nonhuman primates may provide valuable insights into the crucial role that early immunological responses play in controlling HIV in the long term. Moreover, these investigational observations have significant implications for future research endeavors that aim to examine the impact of early ART initiation on the longitudinal dynamics of the HIV response.
5.2. Transcriptomic signatures in ART-naïve and ART-treated HIV individuals
Recently, one research team reported a significant decrease in the frequency of naïve CD8+ T-cells in ART-naïve individuals, compared to that in HCs [63]. After ART initiation, naïve and memory CD8+ T-cell frequency increase marginally, but remain at comparatively lower levels, compared to HCs. Consistent with these findings, Wang et al., also observed that the frequency of naïve CD8+ T-cells in ART-treated HIV patients was significantly lower than that of HCs [23]. Additionally, effector CD8+ T-cell subpopulations, namely CD8+ Effector Memory-GZMK, CD8+ Effector-GZMH, CD8+ Effector-GNLY, CD8+ CD38, and CD8+ CD16 cells are dramatically increased in numbers in ART-naïve individuals, compared to those in HCs [63]. The five effector CD8+ T-cell subpopulations each respond differently to the initiation of ART. ART use induces a significant reduction of CD8+ Effector Memory-GZMK cell and CD8+ CD38 cell frequency, but only partially influences CD8+ Effector-GZMH cell and CD8+ CD16 cell frequency. Interestingly, the enhanced CD8+ Effector-GNLY cell frequency is further expanded during treatment with ART, as is the CXCR5+CD8+ T-cell subpopulations, which undergoes continuous proliferation after ART is initiated. Analysis of samples from ART-treated individuals with high viral loads has revealed the presence of a CD8-Tex cluster, which is not observed in samples obtained from individuals with low viral loads [23]. Consequently, immunological limitations and abnormalities tend to persist long-term in individuals with HIV infection, despite treatment with ART.
Murray's research group, using isolated mDCs, documented transcriptional signatures among three ART-naïve HIV positive individuals, three ART-treated HIV positive individuals, and four HIV negative individuals (Supplementary Table 1) [35]. Their research team observed that: ⅰ) 120 DEGs were identifiable among ART-treated HIV positive individuals when compared against HIV negative individuals. Seventy one of the 120 DEGs were specific to use of ART, while 49 of the DEGs were common to mDCs sampled from ART-naïve individuals; ⅱ) The antigen processing and presentation pathway and Fc receptor-mediated endocytosis pathway are examples of pathway alterations shared between mDCs from ART-naïve and ART-treated HIV positive individuals. Overall, the preceding findings suggest that ART does not fully restore transcription to levels seen in HIV negative individuals when mDCs sampled from treated HIV positive individuals are assessed.
Additionally, only one published study [38] has investigated the association between the timing of ART initiation and PBMC transcriptomic changes in HIV positive individuals, and the study observed that CD4+ T-cell abundance and the CD4:CD8 ratio observed in PBMCs is related to whether ART is initiated or not, and also to the relative timing of ART initiation. Saluzzo et al. [38], quantified CD4+ T-cell changes, and observed that CD4+ T-cell depletion is ameliorated among ART-treated HIV positive individuals (after ART initiation, there was a cessation of CD4+ T-cell depletion, and CD4+ T-cell counts subsequently increased). An increase in blood CD4:CD8 ratio was also observed in the preceding study. Furthermore, patients who initiated ART early manifested a significant increase of the CD4:CD8 ratio within one year, compared to that seen when ART was deferred.
The preceding findings reinforce the fact that ART plays a pivotal role in achieving immune reconstitution, in improving immune disorders, and in mitigating imbalances within the immune systems of individuals infected with HIV. However, even with successful ART treatment, restoration of immune cell population transcription to levels observed in HIV-uninfected individuals remains unattainable in contemporary times. This may likely be attributable to the persistently abnormal immunological status seen in the HIV-infected human body, and includes contributions made by other influencing factors such as abnormal immune activation, the effect of the viral reservoir, non-AIDS-related complications, and pathological alterations to the gut microbiome and the systemic immunopathological implications thereof, amongst other factors.
6. Concluding remarks and future perspectives
Immune dysregulation is a hallmark of HIV pathogenesis [64]. The preceding single-cell-level revelations related to cellular frequencies and transcriptional changes in immune cells of HIV positive individuals indicate that CD4+ T-cells exhibit exhaustion signatures during HIV infection, which is consistent with transcriptional functional alterations which are mainly related to the IFN, immune modulation, and inflammatory pathways. In contrast to CD4+ T-cells, a higher frequency of traditional and novel subclusters of CD8+ T-cells are detected in HIV positive individuals, and these are consistent with DEG enrichment observed in glycolysis, senescence, and exhaustion. The cellular frequencies and transcriptional changes seen in CD4+ T-cells, CD8+ T-cells, B-cells, NK cells, and mDCs are all altered following HIV progression. Additionally, ART plays a crucial role in achieving immune reconstitution, in improving immunological disorders, and in mitigating immune system imbalances in HIV-positive individuals; however, ART does not comprehensively restore transcription to levels reported in HIV-negative individuals. These observations significantly advance our understanding of the fundamental mechanisms involved in HIV disease progression, and also of the implicit potential for targeted treatment of HIV-related disease.
We are cognizant of the fact that the sample sizes of enrolled subjects in the contemporarily published HIV-related single cell transcriptomic studies are relatively small, and that the HIV positive individuals participating in these studies are characterized by an elevated level of diversity. Larger future scRNA-seq studies, therefore, are warranted to formulate standardized inclusion and exclusion criteria, to establish multi-center clinical cohorts, and to dynamically collect and collate samples in tandem with HIV disease evolution, with the objective of determining dynamic transcriptomic changes in PBMCs, lymphoid immune cells, and cerebrospinal fluid (CSF) immune cells. Furthermore, it is critically important to formulate accurate and reliable methods to definitively identify crucially important immune cell types and their subclusters, and to precisely track the molecular targets of these cells at distinct stages of the HIV infection continuum. This will pave the way for the development of novel targets for disease monitoring, diagnosis, and treatment. With respect to the observed critical cell types, flow cytometry technology should further be utilized to enrich specific cellular types, which will contribute to the mapping of the specific changes to those cell subclusters that are observed during HIV infection. As far as ART-naïve and ART-treated HIV positive patients are concerned, current transcriptomics technology is capable of accurately distinguishing transcriptional differences between ART-responders and ART-nonresponders, of identifying transcriptomic biomarkers associated with the efficacy of ART, and of determining novel therapeutic targets for HIV positive individuals.
In addition to transcriptomics, combined multi-omics, which incorporates the interrelated realms of epigenomics, translatomics, lipidomics, metabolomics, and proteomics, will undoubtedly be consequential in future medical research in order to comprehensively understand and appropriately respond to the ongoing threat to human health represented by the enigmatic underlying mechanisms which are integral to the immunopathogenesis of HIV infection and disease.
Funding
The work was supported by the Joint Medical Research Projects of Chongqing Municipal Health Committee and Chongqing Municipal Bureau of Science and Technology (NO.2024MSXM088).
CRediT authorship contribution statement
Ting Zhao: Writing – original draft. Yixian Jing: Writing – original draft, Visualization, Conceptualization. Yao Li: Writing – original draft, Visualization, Conceptualization. Yinqiu Huang: Writing – original draft. Yanqiu Lu: Writing – review & editing, Conceptualization. Yaokai Chen: Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We extend our gratitude to Dr. Vijay Harypursat for his valuable contributions in revising, copy-editing, proofreading, and enhancement of the quality of the English language used this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e35856.
Contributor Information
Ting Zhao, Email: zhaoting9277@163.com.
Yixian Jing, Email: 15023160120@163.com.
Yao Li, Email: yaoli0823@hotmail.com.
Yinqiu Huang, Email: huangyinqiu917@163.com.
Yanqiu Lu, Email: doctorlu828@163.com.
Yaokai Chen, Email: yaokaichen@hotmail.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Mcbrien J.B., Mavigner M., Franchitti L., et al. Robust and persistent reactivation of SIV and HIV by N-803 and depletion of CD8+ cells. Nature. 2020;578(7793):154–159. doi: 10.1038/s41586-020-1946-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fromentin R., Dafonseca S., Costiniuk C.T., et al. PD-1 blockade potentiates HIV latency reversal ex vivo in CD4 + T cells from ART-suppressed individuals. Nat. Commun. 2019;10(1):814. doi: 10.1038/s41467-019-08798-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almodovar S., Cicalini S., Petrosillo N., Flores S.C. Pulmonary hypertension associated with HIV infection: pulmonary vascular disease: the global perspective. Chest. 2010;137(6 Suppl):6S–12S. doi: 10.1378/chest.09-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lonnberg T., Svensson V., James K.R., et al. Single-cell RNA-seq and computational analysis using temporal mixture modelling resolves Th1/Tfh fate bifurcation in malaria. Sci. Immunol. 2017;2(9) doi: 10.1126/sciimmunol.aal2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buettner F., Natarajan K.N., Casale F.P., et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat. Biotechnol. 2015;33(2):155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 6.Patel A.P., Tirosh I., Trombetta J.J., et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L., Dong J., Yan L., et al. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell. 2017;20(6):858–873. doi: 10.1016/j.stem.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 8.G. Luo, Q. Gao, S. Zhang, B. Yan, Probing infectious disease by single-cell RNA sequencing: Progresses and perspectives, Comp. Struct. Biotechnol. J. 182020) 2962-2971 10.1016/j.csbj.2020.10.016. [DOI] [PMC free article] [PubMed]
- 9.Judge M., Parker E., Naniche D., Le Souef P. Gene expression: the key to understanding HIV-1 infection? Microbiol. Mol. Biol. Rev. 2020;84(2) doi: 10.1128/MMBR.00080-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papalexi E., Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018;18(1):35–45. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- 11.Paik D.T., Cho S., Tian L., Chang H.Y., Wu J.C. Single-cell RNA sequencing in cardiovascular development, disease and medicine. Nat. Rev. Cardiol. 2020;17(8):457–473. doi: 10.1038/s41569-020-0359-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villani A.C., Satija R., Reynolds G., et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356(6335) doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.S.D. Zaongo, V. Harypursat, Y. Chen, Single-cell sequencing facilitates elucidation of HIV immunopathogenesis: a review of current literature, Front. Immunol. 132022) 828860 10.3389/fimmu.2022.828860. [DOI] [PMC free article] [PubMed]
- 14.Stahl P.L., Salmen F., Vickovic S., et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353(6294):78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 15.Haque A., Engel J., Teichmann S.A., Lonnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9(1):75. doi: 10.1186/s13073-017-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bock C., Boutros M., Camp J.G., et al. The organoid cell atlas. Nat. Biotechnol. 2021;39(1):13–17. doi: 10.1038/s41587-020-00762-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ke M., Elshenawy B., Sheldon H., Arora A., Buffa F.M. Single cell RNA-sequencing: a powerful yet still challenging technology to study cellular heterogeneity. Bioessays. 2022;44(11) doi: 10.1002/bies.202200084. [DOI] [PubMed] [Google Scholar]
- 18.Li B., Gould J., Yang Y., et al. Cumulus provides cloud-based data analysis for large-scale single-cell and single-nucleus RNA-seq. Nat. Methods. 2020;17(8):793–798. doi: 10.1038/s41592-020-0905-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mikolajewicz N., Gacesa R., Aguilera-Uribe M., Brown K.R., Moffat J., Han H. Multi-level cellular and functional annotation of single-cell transcriptomes using scpipeline. Commun. Biol. 2022;5(1):1142. doi: 10.1038/s42003-022-04093-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.P. Ma, P. Zhang, S. Chen, et al., Immune cell landscape of patients with diabetic macular edema by single-cellRNA analysis, Front. Pharmacol. 122021) 754933 10.3389/fphar.2021.754933. [DOI] [PMC free article] [PubMed]
- 21.Brandt L., Cristinelli S., Ciuffi A. Single-cell analysis reveals heterogeneity of virus infection, pathogenicity, and host responses: HIV as a pioneering example. Annu. Rev. Virol. 2020;7(1):333–350. doi: 10.1146/annurev-virology-021820-102458. [DOI] [PubMed] [Google Scholar]
- 22.Kazer S.W., Aicher T.P., Muema D.M., et al. Integrated single-cell analysis of multicellular immune dynamics during hyperacute HIV-1 infection. Nat. Med. 2020;26(4):511–518. doi: 10.1038/s41591-020-0799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S., Zhang Q., Hui H., Agrawal K., Karris M., Rana T.M. An atlas of immune cell exhaustion in HIV-infected individuals revealed by single-cell transcriptomics. Emerg. Microbes Infect. 2020;9(1):2333–2347. doi: 10.1080/22221751.2020.1826361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu R., Yeh Y.J., Varabyou A., et al. Single-cell transcriptional landscapes reveal HIV-1-driven aberrant host gene transcription as a potential therapeutic target. Sci. Transl. Med. 2020;12(543) doi: 10.1126/scitranslmed.aaz0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Armas L.R., Pallikkuth S., Pan L., et al. Single cell profiling reveals PTEN overexpression in influenza-specific B cells in aging HIV-infected individuals on anti-retroviral therapy. Sci. Rep. 2019;9(1):2482. doi: 10.1038/s41598-019-38906-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farhadian S.F., Mehta S.S., Zografou C., et al. Single-cell RNA sequencing reveals microglia-like cells in cerebrospinal fluid during virologically suppressed HIV. JCI Insight. 2018;3(18) doi: 10.1172/jci.insight.121718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Josefsson L., Palmer S., Faria N.R., et al. Single cell analysis of lymph node tissue from HIV-1 infected patients reveals that the majority of CD4+ T-cells contain one HIV-1 DNA molecule. PLoS Pathog. 2013;9(6) doi: 10.1371/journal.ppat.1003432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Angin M., Volant S., Passaes C., et al. Metabolic plasticity of HIV-specific CD8+ T cells is associated with enhanced antiviral potential and natural control of HIV-1 infection. Nat. Metab. 2019;1(7):704–716. doi: 10.1038/s42255-019-0081-4. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen S., Deleage C., Darko S., et al. Elite control of HIV is associated with distinct functional and transcriptional signatures in lymphoid tissue CD8+ T cells. Sci. Transl. Med. 2019;11(523) doi: 10.1126/scitranslmed.aax4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varadarajan N., Julg B., Yamanaka Y.J., et al. A high-throughput single-cell analysis of human CD8+ T cell functions reveals discordance for cytokine secretion and cytolysis. J. Clin. Invest. 2011;121(11):4322–4331. doi: 10.1172/JCI58653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Saout C., Hasley R.B., Imamichi H., et al. Chronic exposure to type-I IFN under lymphopenic conditions alters CD4 T cell homeostasis. PLoS Pathog. 2014;10(3) doi: 10.1371/journal.ppat.1003976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buggert M., Tauriainen J., Yamamoto T., et al. T-bet and Eomes are differentially linked to the exhausted phenotype of CD8+ T cells in HIV infection. PLoS Pathog. 2014;10(7) doi: 10.1371/journal.ppat.1004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chew G.M., Fujita T., Webb G.M., et al. TIGIT marks exhausted T cells, correlates with disease progression, and serves as a target for immune restoration in HIV and SIV infection. PLoS Pathog. 2016;12(1) doi: 10.1371/journal.ppat.1005349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.J. Tauriainen, L. Scharf, J. Frederiksen, et al., Perturbed CD8+ T cell TIGIT/CD226/PVR axis despite early initiation of antiretroviral treatment in HIV infected individuals, Sci. Rep. 72017) 40354, 10.1038/srep40354. [DOI] [PMC free article] [PubMed]
- 35.S.M. Murray, Y. Zhang, D.C. Douek, R.P. Sekaly, Myeloid cells enriched for a dendritic cell population from people living with HIV have altered gene expression not restored by antiretroviral therapy, Front. Immunol. 112020) 261. 10.3389/fimmu.2020.00261. [DOI] [PMC free article] [PubMed]
- 36.Kim Y., Anderson J.L., Lewin S.R. Getting the "kill" into "shock and kill": strategies to eliminate latent HIV. Cell Host Microbe. 2018;23(1):14–26. doi: 10.1016/j.chom.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rato S., Rausell A., Munoz M., Telenti A., Ciuffi A. Single-cell analysis identifies cellular markers of the HIV permissive cell. PLoS Pathog. 2017;13(10) doi: 10.1371/journal.ppat.1006678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saluzzo S., Pandey R.V., Gail L.M., et al. Delayed antiretroviral therapy in HIV-infected individuals leads to irreversible depletion of skin- and mucosa-resident memory T cells. Immunity. 2021;54(12):2842–2858. doi: 10.1016/j.immuni.2021.10.021. [DOI] [PubMed] [Google Scholar]
- 39.Wanjalla C.N., Mcdonnell W.J., Ram R., et al. Single-cell analysis shows that adipose tissue of persons with both HIV and diabetes is enriched for clonal, cytotoxic, and CMV-specific CD4+T cells. Cell Rep. Med. 2021;2(2) doi: 10.1016/j.xcrm.2021.100205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiche S.T., Zhang Y., Sarnello D., et al. Th17 cell master transcription factor RORC2 regulates HIV-1 gene expression and viral outgrowth. Proc. Natl. Acad. Sci. U. S. A. 2021;118(48) doi: 10.1073/pnas.2105927118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Catalfamo M., Di Mascio M., Hu Z., et al. HIV infection-associated immune activation occurs by two distinct pathways that differentially affect CD4 and CD8 T cells. Proc. Natl. Acad. Sci. U. S. A. 2008;105(50):19851–19856. doi: 10.1073/pnas.0810032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.L.R. de Armas, C. Gavegnano, S. Pallikkuth, et al., The effect of JAK1/2 inhibitors on HIV reservoir using primary lymphoid cell model of HIV latency, Front. Immunol. 122021) 720697, 10.3389/fimmu.2021.720697. [DOI] [PMC free article] [PubMed]
- 43.Cao W., Mehraj V., Trottier B., et al. Early initiation rather than prolonged duration of antiretroviral therapy in HIV infection contributes to the normalization of CD8 T-cell counts. Clin. Infect. Dis. 2016;62(2):250–257. doi: 10.1093/cid/civ809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saez-Cirion A., Pancino G. HIV controllers: a genetically determined or inducible phenotype? Immunol. Rev. 2013;254(1):281–294. doi: 10.1111/imr.12076. [DOI] [PubMed] [Google Scholar]
- 45.Peretz Y., He Z., Shi Y., et al. CD160 and PD-1 co-expression on HIV-specific CD8 T cells defines a subset with advanced dysfunction. PLoS Pathog. 2012;8(8) doi: 10.1371/journal.ppat.1002840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Czerwinska P., Rucinski M., Wlodarczyk N., et al. Therapeutic melanoma vaccine with cancer stem cell phenotype represses exhaustion and maintains antigen-specific T cell stemness by up-regulating BCL6. OncoImmunology. 2020;9(1) doi: 10.1080/2162402X.2019.1710063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buggert M., Nguyen S., Salgado-Montes De Oca G., et al. Identification and characterization of HIV-specific resident memory CD8+ T cells in human lymphoid tissue. Sci. Immunol. 2018;3(24) doi: 10.1126/sciimmunol.aar4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cocchi F., Devico A.L., Garzino-Demo A., Arya S.K., Gallo R.C., Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270(5243):1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 49.Migueles S.A., Osborne C.M., Royce C., et al. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity. 2008;29(6):1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.J.H. Russell, T.J. Ley, Lymphocyte-mediated cytotoxicity, Annu. Rev. Immunol. 202002) 323-370, 10.1146/annurev.immunol.20.100201.131730. [DOI] [PubMed]
- 51.Voskoboinik I., Whisstock J.C., Trapani J.A. Perforin and granzymes: function, dysfunction and human pathology. Nat. Rev. Immunol. 2015;15(6):388–400. doi: 10.1038/nri3839. [DOI] [PubMed] [Google Scholar]
- 52.Wherry E.J., Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 2015;15(8):486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carrette F., Surh C.D. IL-7 signaling and CD127 receptor regulation in the control of T cell homeostasis. Semin. Immunol. 2012;24(3):209–217. doi: 10.1016/j.smim.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zapata W., Aguilar-Jimenez W., Feng Z., et al. Identification of innate immune antiretroviral factors during in vivo and in vitro exposure to HIV-1. Microbes Infect. 2016;18(3):211–219. doi: 10.1016/j.micinf.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 55.Monteleone K., Di Maio P., Cacciotti G., et al. Interleukin-32 isoforms: expression, interaction with interferon-regulated genes and clinical significance in chronically HIV-1-infected patients. Med. Microbiol. Immunol. 2014;203(3):207–216. doi: 10.1007/s00430-014-0329-2. [DOI] [PubMed] [Google Scholar]
- 56.Ribeiro-Dias F., Saar G.R., de Lima S.L., Dos S.J., Joosten L.A. Interleukin 32: a novel player in the control of infectious diseases. J. Leukoc. Biol. 2017;101(1):39–52. doi: 10.1189/jlb.4RU0416-175RR. [DOI] [PubMed] [Google Scholar]
- 57.Holla P., Dizon B., Ambegaonkar A.A., et al. Shared transcriptional profiles of atypical B cells suggest common drivers of expansion and function in malaria, HIV, and autoimmunity. Sci. Adv. 2021;7(22) doi: 10.1126/sciadv.abg8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ehrhardt G.R., Hsu J.T., Gartland L., et al. Expression of the immunoregulatory molecule FcRH4 defines a distinctive tissue-based population of memory B cells. J. Exp. Med. 2005;202(6):783–791. doi: 10.1084/jem.20050879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Costanzo M.C., Kim D., Creegan M., et al. Transcriptomic signatures of NK cells suggest impaired responsiveness in HIV-1 infection and increased activity post-vaccination. Nat. Commun. 2018;9(1):1212. doi: 10.1038/s41467-018-03618-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin-Gayo E., Cole M.B., Kolb K.E., et al. A reproducibility-based computational framework identifies an inducible, enhanced antiviral state in dendritic cells from HIV-1 elite controllers. Genome Biol. 2018;19(1):10. doi: 10.1186/s13059-017-1385-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hartana C.A., Rassadkina Y., Gao C., et al. Long noncoding RNA MIR4435-2HG enhances metabolic function of myeloid dendritic cells from HIV-1 elite controllers. J. Clin. Invest. 2021;131(9) doi: 10.1172/JCI146136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoffmann M., Pantazis N., Martin G.E., et al. Exhaustion of activated CD8 T cells predicts disease progression in primary HIV-1 infection. PLoS Pathog. 2016;12(7) doi: 10.1371/journal.ppat.1005661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X.M., Zhang J.Y., Xing X., et al. Global transcriptomic characterization of T cells in individuals with chronic HIV-1 infection. Cell Discov. 2022;8(1):29. doi: 10.1038/s41421-021-00367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Margolis D.M., Archin N.M., Cohen M.S., et al. Curing HIV: seeking to target and clear persistent infection. Cell. 2020;181(1):189–206. doi: 10.1016/j.cell.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.