Abstract
The fatal gouty disease caused by goose astrovirus genotype 2 (GAstV-2) still seriously endangers the goose industry in China, causing great economic losses. However, research on its infection mechanism has progressed relatively slowly. VP70 is the structural protein of GAstV-2 and is closely related to virus invasion and replication. To better understand the role of VP70 during GAstV-2 infection, we used immunoprecipitation and mass spectrometry to identify host proteins that interact with VP70. Here, we report that cellular vimentin (VIM) is a host binding partner of VP70. Site-directed mutagenesis showed that amino acid residues 399 to 413 of VP70 interacted with VIM. Using reverse genetics, we found that VP70 mutation disrupts the interaction of VP70 with VIM, which is essential for viral replication. Overexpression of VIM significantly promoted GAstV-2 replication, while knockdown of VIM significantly inhibited GAstV-2 replication. Laser confocal microscopy showed that VP70 protein expression induced the rearrangement of VIM, gradually aggregating from the original uniform grid to the side of the nucleus, and aggregated the originally dispersed GAstV-2 RNA in VIM. This rearrangement was associated with increased VIM phosphorylation caused by GAstV-2. Meanwhile, blocking VIM rearrangement with acrylamide substantially inhibited viral replication. These results indicate that VIM interacts with VP70 and positively regulates GAstV-2 replication, and VIM–VP70 interaction and an intact VIM network are needed for GAstV-2 replication. This study provides a theoretical basis and novel perspective for the further characterization of the pathogenic mechanism of GAstV-2-induced gouty disease in goslings.
Key words: goose astrovirus genotype 2, VP70 protein, vimentin, interaction, viral replication
INTRODUCTION
Since 2017, a highly fatal gouty disease caused by goose astrovirus genotype 2 (GAstV-2) has been reported in Shandong, Jiangsu, Anhui, and Guangdong provinces of China, with infection and mortality rates of up to 80% and 50%, respectively (Yang et al., 2018; Xu et al., 2023a; Xu et al., 2024). GAstV-2 infects geese of all breeds; however, it predominantly affects those 1–20-days-old, with the most susceptible goslings aged < 15 d (Zhang et al., 2022a; Xu et al., 2023b). GAstV-2 differs greatly from other astroviruses. In contrast to the weaker pathogenicity of other astroviruses, GAstV-2 is highly pathogenic to goslings and can directly cause gout (Liu et al., 2023; Wang et al., 2023). The main clinical symptoms are white stool, joint swelling, lying down, and an unwillingness to walk. Pathological examination reveals swollen and pale kidneys, splenomegaly, and significant urate deposits on the surfaces of the heart and liver (Li et al., 2021a). Fatal gouty diseases caused by GAstV-2 in goslings are widely prevalent in China, causing serious economic losses in the goose industry (Liu et al., 2023). Prevention and control strategies are poorly developed, and research on the pathogenic mechanisms is sparse. Although many studies have reported on GAstV-2, most have focused on epidemiology, diagnosis, and pathogenesis, lacking a preliminary discussion on the infection and pathogenic mechanism of the virus.
GAstV-2 is a single-stranded positive-sense RNA virus that lacks a cyst membrane, belongs to the genus Astrovirus, and is a member of the avian astrovirus genotype 3. The full-length genome of GAstV-2 is approximately 7.0 to 8.0 kb, including the 5′-untranslated region (5′-UTR), open reading frames ORF1a, ORF1b, and ORF2, 3′-UTR, and a poly-A tail. ORF1a and ORF1b encode viral non-structural proteins, while ORF2 encodes viral capsid proteins, involved in the recognition of related receptors on the host cell surface and the body's immune response (Chen, et al., 2020; Ren et al., 2022). The newly synthesized capsid protein is approximately 90 kDa (VP90); however, the C-terminal is cleaved by caspases to produce the VP70 protein, which is subsequently cleaved by trypsin to produce 3 structural proteins (VP34, VP27, and VP25). These 3 proteins are distributed on the viral surface and form mature virions (Cortez, et al., 2017). VP27 is the main antigenic determinant protein and is involved in the recognition of cell surface receptors and activating the host immune response. The core capsid protein VP34 also exhibits antigenicity (Bass and Upadhyayula, 1997). Therefore, VP70 plays an important role in GAstV-2 innate immunity and virus replication (Ren et al., 2022).
Vimentin (VIM) is a highly conserved intermediate fibrin protein and an important component of the cytoskeleton (Manzer et al., 2022; Martinez-Vargas et al., 2023). Owing to its prevalence within cells, its expression on the cell surface and within the extracellular space was only recently described. This underscores VIM's pivotal involvement in the invasion, infection, and replication processes of various viruses, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Li et al., 2021b; Amraei et al., 2022; Suprewicz et al., 2022), Zika virus (Zhang et al., 2022b), foot-and-mouth disease virus (Gladue et al., 2013), porcine reproductive and respiratory syndrome virus (Zheng et al., 2021), and Newcastle disease virus (Lu et al., 2023), among others (Ghosh et al., 2018). However, VIM has also been shown to elicit an inhibitory effect on the replication of certain viruses, including human parainfluenza virus (Liu et al., 2022).
In the early stage, our project team used GAstV-2 structural protein VP70 as bait to capture the host key molecules interacting with VP70 through glutathione s-transferase (GST) pull-down technology. We identified key molecules associated with GAstV-2 invasion and replication, including VIM, non-muscle myosin heavy chain 9, and fibronectin. After preliminary verification, we found that VIM had a greater effect on GAstV-2 replication than these other molecules. However, the effect of VIM on GAstV-2 replication and its regulatory mechanisms remain unclear. The current study sought to elucidate the molecular mechanisms underlying the interaction between VIM and VP70 in regulating GAstV-2 replication, providing novel insight regarding the pathogenic mechanism of GAstV-2, establishing a theoretical basis for the in-depth investigation of its pathogenesis, and providing novel perspectives and insight into the prevention and treatment of this disease.
MATERIALS AND METHODS
Cells, Viruses, and Ethics Statement
Goose embryo kidney (GEK) cells were prepared from 20-day-old embryonated goose eggs, which were purchased from Qingyuan Livestock Co. Ltd. (Guangdong, China). Briefly, aseptically removed goose embryos underwent an abdominal cavity opening, followed by removal of the kidneys. The residual blood was rinsed away using sterile phosphate buffered saline (PBS), and the kidney tissue was meticulously sectioned using scissors. The tissue pieces were washed thrice with PBS until the suspension appeared clear. The washed tissue fragments were treated with 0.25% trypsin and subjected to digestion in a 37°C water bath for 20 min; the waste solution was discarded. The cell mass was broken up using a Bartholin's duct, and the cells were obtained after filtration. These cells were cultured in Dulbecco's modified Eagle's medium (Gibco, Shanghai, China) supplemented with 10% fetal bovine serum and incubated at 37°C with 5% CO2. A strain of GAstV-2, GAstV-GD-ZJ-21-01 (GenBank: OP776629.1) (Liu et al., 2023), was isolated from a goose in Zhanjiang, Guangdong Province, and maintained in our laboratory.
All animal experiments were performed in accordance with the regulations for animal experimentation of Guangdong Province, China, and were approved by the Ethics Committee of the Institute of Animal Health, Guangdong Academy of Agricultural Sciences (No. YC-PT2023025).
Antibodies and Plasmids
The VIM primary antibody (sc-80975) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). A VIM (phospho S38) primary antibody (ab52942) was purchased from Abcam (Cambridge, MA). Mouse anti-double-stranded RNA (dsRNA, J2, 10010200) was obtained from Nordic-MuBio (Susteren, Netherlands). The GST-tagged mouse monoclonal antibody (HRP-conjugated, AF2891), Flag-tagged mouse monoclonal antibody (AF2852), Flag-tagged rabbit polyclonal antibody (AF0036), HA-tagged rabbit polyclonal antibody (AF0039), HA-tagged mouse monoclonal antibody (AF2858), Alexa Fluor 647-labeled goat anti-mouse/rabbit IgG (H+L, A0473/A0468), Alexa Fluor 488-labeled goat anti-mouse/rabbit IgG (H+L, A0428/A0423), GAPDH mouse monoclonal antibody (AF0006), and HRP-labeled goat anti-mouse IgG (H+L) antibody (A0192) were obtained from Beyotime (Shanghai, China). The mouse monoclonal antibody against GAstV-2 VP27 was prepared and provided by Professor Zongyan Chen of the Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences.
The coding sequence for VP70 was obtained from the total RNA of GAstV-2 through reverse transcription polymerase chain reaction (RT-PCR), and the HA tag was ligated to the end of the coding sequence of VP70 using homologous recombination and PCR. The PCR products were cloned into pCAGGS and pGEX6P1 (GST tag) to generate pCAGGS-VP70-HA and pGEX6P1-VP70, respectively. The VIM coding sequence was amplified from the total RNA of GEK cells using RT-PCR, and the Flag tag was ligated to the end of the VIM coding sequence using homologous recombination and PCR. The PCR products were cloned into pCAGGS and pGEX6P1 (GST-tag) to generate pCAGGS-VIM-Flag and pGEX6P1-VIM, respectively. The coding region of VP70 was divided into 4 segments for truncated expression: VP70/A, VP70/B, VP70/C, and VP70/D. The HA tag was fused and cloned into the pCAGGS vector to generate pCAGGS-VP70/A-HA, pCAGGS-VP70/B-HA, pCAGGS-VP70/C-HA, and pCAGGS-VP70/D-HA vectors. The linearized vector and corresponding fragments of the PCR products were ligated using a ClonExpress II One Step Cloning kit (Vazyme, Nanjing, China). The primer sequences used for plasmid construction (Supplementary Table S1), and all plasmids were confirmed by sequencing (Sangon Biotech, Shanghai, China). To obtain mutant VP70/B proteins with the amino acid replaced by alanine, the coding region of the mutant VP70/B was synthesized by Sangon Biotech (Shanghai, China) and cloned into pCAGGS to produce plasmids (pCAGGS-VP70/B-S1 to pCAGGS-VP70/B-S10).
Immunoprecipitation and Mass Spectrometry Assay
The pGEX6P1-VP70 and pGEX6P1 plasmids were transformed into an Escherichia coli expression system to obtain large amounts of GST-VP70 and GST proteins, respectively. The total proteins of GEK cells were lysed in NP40 lysis buffer. Immunoprecipitation was performed as described below. Briefly, the GST-VP70 protein or GST protein lysate (500 μL) was mixed with 20 μL of anti-GST magnetic beads (Beyotime, Shanghai, China). Then, an appropriate amount of cell protein lysate was added and incubated for 6 h at 4 °C with gentle shaking. The supernatant was discarded, and the bead pellets were washed thrice with tris-buffered saline with tween-20 (TBST). The immunoprecipitated proteins were boiled with sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) loading buffer for 10 min and subsequently analyzed using 10% SDS–PAGE. Following electrophoresis, protein bands were stained with Coomassie Blue Fast Staining Solution (Beyotime, Shanghai, China) and manually excised from the SDS–PAGE gel using a matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI–TOF MS) assay (Sangon Biotech, Shanghai, China).
Coimmunoprecipitation of GAstV-2 VP70 and VIM
GEK cells were co-transfected with the 2 plasmids (pCAGGS-VP70-HA and pCAGGS-VIM-Flag) and cultured for 48 h. The cells were lysed using NP40 lysis buffer (Beyotime, Shanghai, China) on ice for 30 min. The lysate (500 μL) was incubated with anti-HA tag or anti-Flag magnetic beads at 4°C for 6 h with gentle agitation. The supernatant was then discarded, and the bead pellets were washed thrice with TBST buffer for the Co-IP assay. The immunoprecipitated proteins were boiled with SDS–PAGE sample loading buffer for 10 min and subsequently analyzed using western blotting. VP70 protein was truncated into 3 segments, and the key region of interaction between VP70 and VIM was identified by Co-IP. The key amino acid sites of the interaction could be identified by transfecting the cells with site-directed mutagenesis plasmids of amino acids in the key region of the interaction.
GST Pull-Down Assay
The purification of recombinant GST fusion proteins, expressed in Escherichia coli Agilent BL21, was carried out by transforming the cells with the respective plasmids: pGEX6P1, pGEX6P1-VP70, or pGEX6P1-VIM. Anti-GST magnetic beads (20 μL, Beyotime, Shanghai, China) were incubated with soluble GST fusion protein (1 mg) or GST control. They were then combined with specific amounts of cell lysates (1 mg) transfected with the pCAGGS-VIM-Flag or pCAGGS-VP70-HA plasmids. The mixture was incubated at 4°C for 6 h with gentle agitation; the supernatant was discarded and bead pellets were washed thrice with TBST buffer for the GST pull-down assay. The immunoprecipitated proteins were boiled with SDS–PAGE sample loading buffer for 10 min and subsequently analyzed by western blotting.
Western Blotting
The cells were washed thrice with PBS and lysed in NP40 lysis buffer (containing phenylmethylsulfonyl fluoride and phosphatase inhibitors) for 30 min on ice. The lysates were clarified by centrifugation at 12,000 × g and 4°C for 10 min. Equal protein amounts from each group were separated by SDS–PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were blotted and incubated with the indicated antibodies. The membranes were visualized using a TANON imaging system. Image J software was used for gray-scale quantification of protein bands to analyze the differences in protein expression levels among different groups.
Immunofluorescence and Confocal Microscopy
GEK cells grown onto 12-mm glass coverslips were co-transfected with the plasmids pCAGGS-VIM-Flag and pCAGGS-VP70-HA. At different time points after transfection, cells were fixed with 4% formaldehyde for at least 20 min and washed 3 times with PBS. Then, cells were incubated with primary antibody against HA-tag and Flag-tag and stained with fluorescent secondary antibodies of different species and colors, and the cell nuclei were stained using DAPI. After washing 3 times, the cells were observed using a laser confocal microscope (BZ-X800LE; KEYENCE, Japan). Immunofluorescence staining of GAstV-2, dsRNA, and other target molecules was performed according to the methods described above.
Construction of Mutant GAstV-2
To identify the key amino acid sites on VP70 that interact with VIM, we mutated the key site and rescued the virus to strengthen the confidence of the key site. Plasmid pACYCDuet-1 containing the full-length sequence of the GAstV-2 cDNA was constructed by Sangon Biotech Co., Ltd. (Shanghai, China). In brief, the whole GAstV-2 genome sequence was synthesized and cloned into the pACYCDuet-1 vector to obtain the reverse genetic plasmid pACYC-GAstV-2. The pACYC-GAstV-2-Mut-s3 plasmid is a derivative of pACYC-GAstV-2 with 15 alanine substitutions at positions 399 to 413 of VP70. This is the key site of VP70 binding to VIM. In brief, pACYC-GAstV-2 was linearized using the unique restriction sites Ale I and Ahd I. The sequence between restriction sites Ale I and Ahd I covered the mutation site mentioned above, and this partial gene sequence was synthesized and linked with the linearized vector by homologous recombination to obtain pACYC-GAstV-2-Mut-s3. The pattern diagram of infectious clone construction and mutation is shown in Supplementary Figure S1. The pACYC-GAstV-2-Mut-s3 plasmid was transfected into LMH cells using Lipo8000 transfection reagent (Beyotime, Shanghai, China) according to the manufacturer's instructions. The supernatants of transfected cells were collected at 72 h post-transfection, and the TCID50 of the obtained mutant GAstV-2 virus was determined. The supernatants of transfected cells were harvested at 48 h post-transfection and seeded into new GEK cells for 3 consecutive blind passages.
Overexpression of VIM
The pCAGGS-VIM-Flag plasmid was transfected into GEK cells, and the pCAGGS plasmid was used as a control. At 24 h after transfection, the cells were infected with GAstV-2 at a multiplicity of infection (MOI) of 1.0 at 37°C. After 2 h of adsorption, the cells were washed 3 times with PBS and cultured with fresh medium at 37°C. At 12 and 24 hpi, the infected cells were then collected for immunofluorescence staining, reverse transcription fluorescent quantitative PCR (RT-qPCR), and western blotting analysis, and the virus titers in the supernatant were determined.
Knockdown by Small Interfering RNA
Based on the coding sequence of the VIM gene in GEK cells, siRNA targeting VIM (GenBank accession number XM_048061750.1) and negative control siRNA were designed and synthesized by RioBio (Guangzhou, China). siRNA transfection was performed using the riboFECTTM reagent (RioBio, Guangzhou, China) according to the manufacturer's instructions. At 24 h after transfection, the siRNA-transfected cells were infected with GAstV-2 at an MOI of 1.0 and cultured for 12 and 24 h. The infected cells were then collected for immunofluorescence staining, RT-qPCR, and western blotting analysis, and the virus titers in the supernatant were determined.
RT-qPCR
The total RNA of GEK cells was extracted using an RNA extraction kit (Fastagen Biotech, Shanghai, China) according to the manufacturer's recommendations. First-strand cDNA was synthesized from 500 ng of the total RNA as a template along with random primers, using the PrimeScript RT Reagent kit (TaKaRa, Japan). Then, RT-qPCR was performed employing Hieff® qPCR SYBR Green Master Mix (Yeasen, Shanghai, China) on a Roche LightCycler96 System (Roche Diagnostics, Basel, Switzerland). The gene expression levels were quantified using the 2−ΔΔCt method and normalized to GAPDH expression. The primer sequences are listed in Table 1.
Table 1.
Reverse transcription quantitative PCR primers used in this study.
| Primer1 | Sequence (5′–3′) | GenBank |
|---|---|---|
| GAstV-2 ORF2-F | GGGTGATCCGCAAGGAAATA | OP776629.1 |
| GAstV-2 ORF2-R | AAGTTTCGCCAGGGTTAGAG | |
| GAPDH-F | GAAAAGTCATCCCTGAGCTC | MG674174.1 |
| GAPDH-R | GGTCCACGACAGAGACGTTGG |
Forward and reverse primers are indicated by F and R, respectively.
Cell Viability Assay
Cytotoxicity was measured using the MTT assay with an Enhanced Cell Counting Kit-8 (Beyotime, Shanghai, China). The assay was performed according to the user manual. Briefly, GEK cells were plated in 96-well plates overnight and then treated with acrylamide or KN-93 at different concentrations. After 24 h, CCK-8 reagent was added to the cells in the plate, the cells were incubated at 37°C for 4 h, and the absorption was measured at 450 nm using a microplate reader (BioTek, VT).
Drug Treatment Assay
The GEK cells were infected with GAstV-2 at an MOI of 1.0 and 37°C. After 2 h of adsorption, the cells were washed 3 times with PBS and cultured with fresh medium at 37°C. Then, cells were treated with acrylamide (150 μM) or KN-93 (100 μM), and the corresponding blank solvent was used as a control. After 12 and 24 h, the treated cells were collected for immunofluorescence staining, RT-qPCR, and western blotting analysis, and the virus titers in the supernatant were determined.
Statistical Analysis
Data were analyzed using the Prism software (GraphPad Prism 9, La Jolla, CA). Statistical tests were performed using Student's t-test for one-group and two-group comparisons and one-way analysis of variance (ANOVA) with Bonferroni's post-tests for multiple-group comparisons. A p-value of less than 0.05 was considered significant (*p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001).
RESULTS
GAstV-2 VP70 Interacted With VIM and Induced VIM Rearrangement
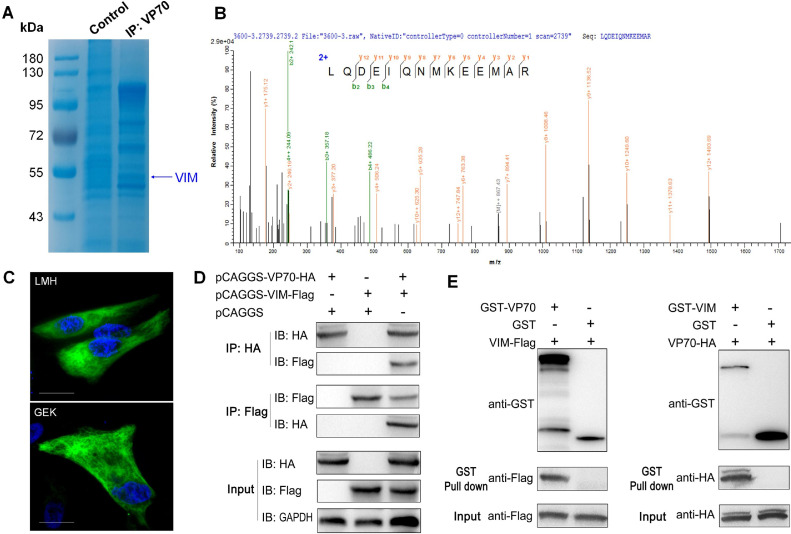
As a structural protein of GAstV-2, VP70 is closely related to the replication of the virus. However, the exact role of VP70 in GAstV-2 replication remains unclear, and the key host molecules that interact with it are still unknown. To identify the key host molecules that interact with VP70, we performed immunoprecipitation combined with mass spectrometry. In order to identify the key host molecules interacting with VP70, we used VP70 as the bait protein and GEK cell lysate as the protein library to be screened by immunoprecipitation combined with mass spectrometry analysis. We identified several host protein molecules that interacted with VP70, including VIM (Figures 1A and B), non-muscle myosin heavy chain 9, fibronectin, and so on. The preliminary experiment found that VIM had a greater effect on GAstV-2 replication, and so it was further studied. In the normal state, VIM was evenly distributed in the cytoplasm in a grid pattern, and a small amount was also distributed on the cell membrane (Figure 1C). To further verify the interaction between VP70 and VIM, Co-IP (Figure 1D) and GST pull-down (Figure 1E) assays were performed; the 2 proteins interacted in vivo and in vitro, providing a robust basis for subsequent investigations.
Figure 1.
Interaction of GAstV-2 VP70 with VIM. (A) Coomassie Brilliant Blue staining after SDS–PAGE electrophoresis. GST-VP70 fusion protein served as the bait protein, while the GEK cell lysate was used as the target protein library for GST pull-down analysis. (B) MALDI–TOF MS spectra of peptides of VIM. The distinct bands were carefully excised and sent to Sangon Biotech for MALDI-TOF MS assay, revealing VIM. (C) Immunofluorescence staining of VIM in LMH and GEK cells (scale bars: 15 μm). (D and E) Co-IP and GST pull-down assays of VP70 and VIM using both forward and reverse pull-down approaches. These results indicate a direct interaction between the VP70 protein and VIM.
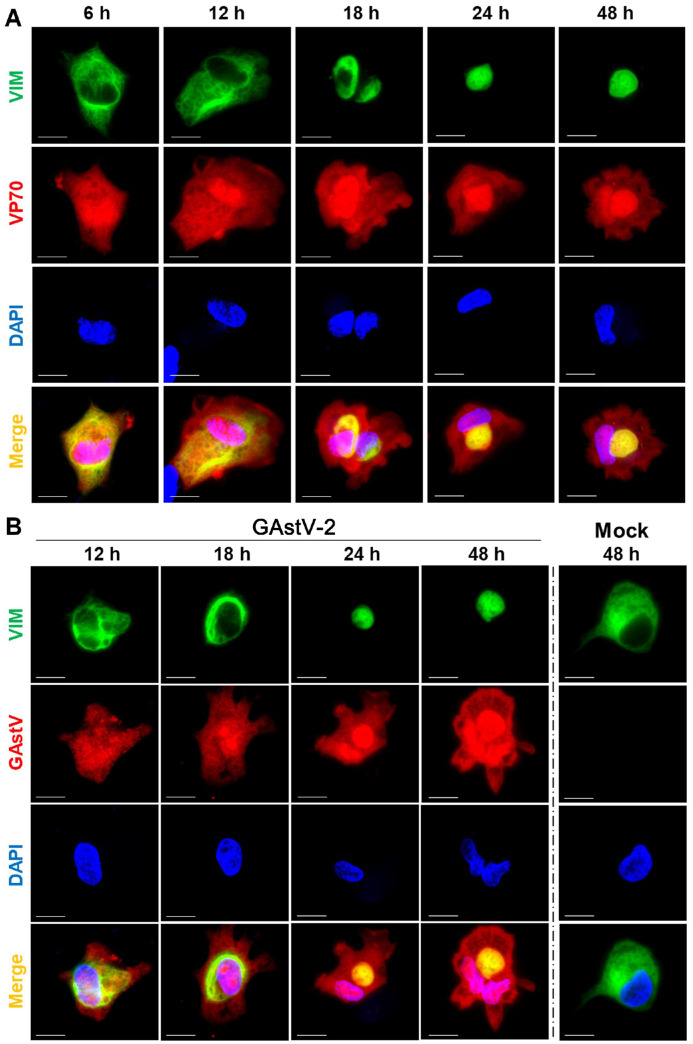
Upon further investigation of the interaction between VIM and VP70 using confocal analysis, we found that, aside from their consistent co-localization with the expression of VP70 protein, VIM's structure was rearranged. Specifically, its scattered distribution across the cell membrane and cytoplasm gradually aggregated, ultimately localizing alongside the nucleus (Figure 2A). We further used live GAstV-2 (1.0 MOI) to infect GEK cells, revealing that the structure of VIM was rearranged upon GAstV-2 infection as well (Figure 2B). VIM rearrangement was completed as early as 18 h after VP70 expression or GAstV-2 infection. Therefore, we hypothesized that VIM rearrangement plays an important role in GAstV-2 replication.
Figure 2.
VP70 co-localizes with VIM and drives GAstV-2 to induce VIM rearrangement. (A) Immunofluorescence colocalization analysis of VIM and VP70. The plasmids pCAGGS-VIM-Flag and pCAGGS-VP70-HA were co-transfected into GEK cells and stained with HA-tag, Flag-tag antibodies, and DAPI at different time points. Laser confocal microscopy reveals that VIM gradually shifts from its distribution in the cell membrane and cytoplasm to the nucleus with extended VP70 expression, ultimately accumulating alongside the nucleus (scale bars: 15 μm). (B) GAstV-2 infection induces VIM rearrangement in GEK cells. The cells were transfected with pCAGGS-VIM-Flag plasmid and subsequently infected with GAstV-2; immunofluorescence staining was performed at various times after viral infection. Changes in VIM morphology and structure were observed using laser confocal microscopy at different time points (scale bars: 15 μm).
Identification of the VIM Binding Site on GAstV-2 VP70 and its Role in Viral Replication
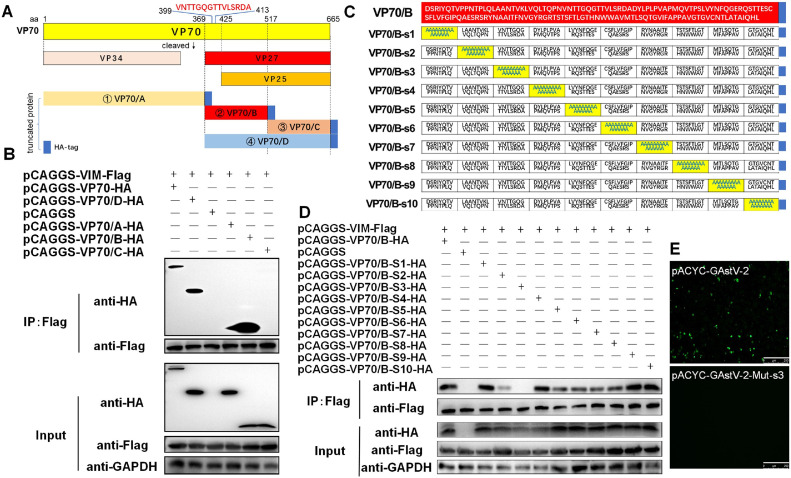
In order to identify the critical amino acid sites governing the interaction between GAstV-2 VP70 and VIM, the VP70 protein was divided into 4 fragments (VP70/A, VP70/B, VP70/C, VP70/D) to achieve truncated expression (Figure 3A). The key regions that participate in its interaction with VIM were identified using Co-IP assays. The C-terminus of VP70 (VP70/B) was the key binding domain of VP70 for VIM (Figure 3B). Subsequently, alanine-scanning mutagenesis of VP70/B was performed to identify the key amino acid sites involved in these interactions. We constructed 10 mutant VP70/B proteins, containing 15 alanine residues substituting the native amino acid residues (Figure 3C). The ability of these VP70/B mutants to bind VIM was analyzed by Co-IP. The VP70/B protein containing mutations in area 3 (VP70/B-s3) was unable to bind VIM (Figure 3D), demonstrating that the amino acid residues (aa 399–413) in area 3 of VP70 are critical for the interaction between VP70 and VIM.
Figure 3.
Identification of the VIM binding site on GAstV-2 VP70, essential for viral replication. (A) Schematic representation of the truncated expression of VP70, which was sequentially truncated to 4 segments, VP70/A, VP70/B, VP70/C, and VP70/D. (B) Co-IP analysis of VP70 truncates and VIM. Each truncated VP70 variant was separately co-expressed with VIM in GEK cells, and subsequently subjected to Co-IP assay. The VP70/B region is crucial for the interaction between VP70 and VIM. (C) Schematic representation of site-directed mutagenesis in the VP70/B region. It was mutated to alanine via segment-by-segment mutation in groups of 15 amino acids. (D) Co-IP analysis of the 10 VP70/B mutants (S1–S10) co-expressed with VIM. The s3 mutant is incapable of interacting with VIM. (E) GEK cells were transfected with the plasmids pACYC- GAstV-2-Mut-S3 and pACYC- GAstV-2, respectively. After 48 h, the virus in the supernatant was harvested to infect new GEK cells, and immunofluorescence staining of GAstV-2 was performed (scale bars: 250 μm). Transfection with pACYC- GAstV-2-Mut-s3 was not able to produce viable progeny.
To further investigate the significance of the interaction between GAstV-2 VP70 and VIM, reverse genetics was used to mutate the aa 399 to 413 of VP70 to alanine to obtain GAstV-2 mutants (pACYC-GAstV-2-Mut-s3); the unmutated wild strain was used as a control (pACYC- GAstV-2). These plasmids were transfected into GEK cells, and supernatants of transfected cells were then used to infect new GEK cells and passaged 3 times. Immunofluorescence results of GAstV-2 showed that transfection with pACYC-GAstV-2 produced live virus; however, transfection with pACYC-GAstV-2-Mut-s3 was not able to produce virus (Figure 3E). These data demonstrate that the interaction between VP70 and VIM is critical for GAstV-2 replication. However, we cannot rule out other possibilities at this point, that is, these mutations in VP70 may have other effects by altering the structure of VP70 or by inhibiting the interaction of VP70 with other proteins.
Host Cell VIM Positively Regulates GAstV-2 Replication
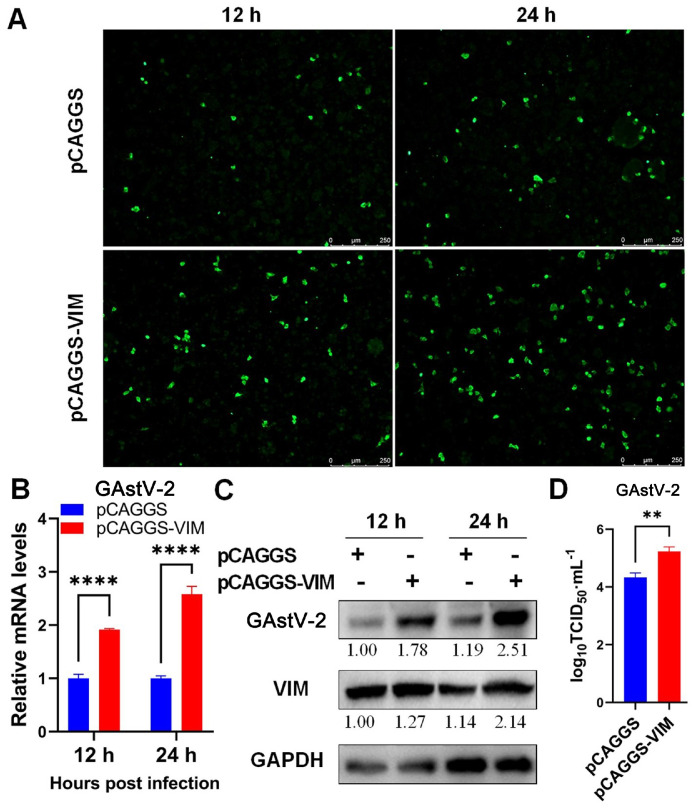
To explore the role of VIM in GAstV-2 replication, the pCAGGS-VIM-Flag plasmid was transfected into GEK cells, GAstV-2 was infected 24 h later at 1.0 MOI, and the viral load of GAstV-2 in cells was analyzed 12 and 24 h after infection. The results showed that overexpression of VIM significantly increased the proportion of GAstV-2 positive cells (Figure 4A), as well as mRNA and protein expression levels of GAstV-2 (Figures 4B and 4C) in the cells. Additionally, the virus titer in the supernatants of VIM overexpressing cells also increased compared with that of the control group (Figure 4D). This implied that elevated VIM levels significantly promoted the replication of GAstV-2.
Figure 4.
Overexpression of vimentin promotes GAstV-2 replication. (A) Immunofluorescence analysis of GAstV-2 following VIM overexpression (scale bars: 250 μm). (B and C) GAstV-2 mRNA and protein expression levels were significantly increased following VIM overexpression. (D) The virus titer in the supernatant of VIM overexpressing cells significantly increased compared with that of the control group. ** p < 0.01; **** p < 0.0001.
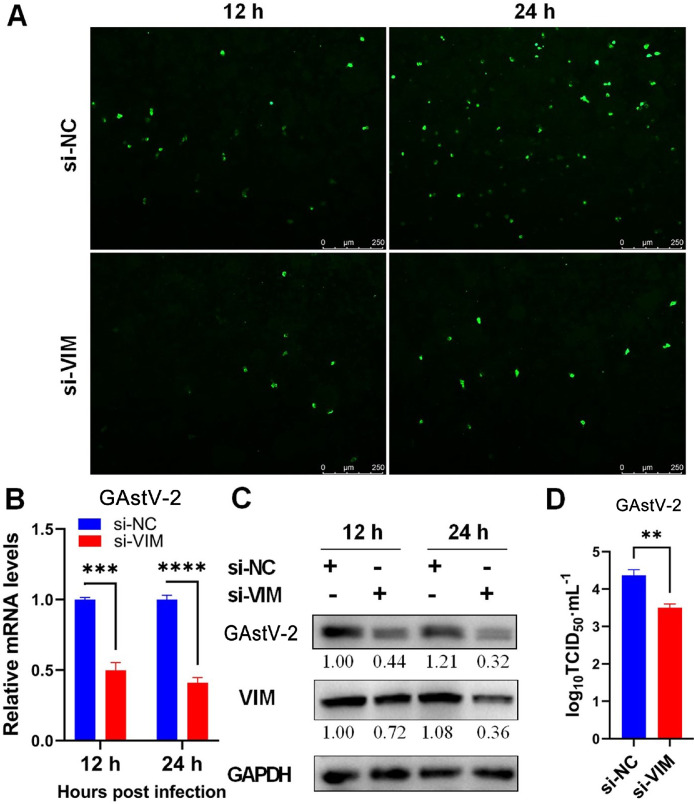
To further verify the effect of VIM in GAstV-2 replication, siRNA knockdown assays were conducted. GEK cells were transfected with either siRNA against VIM or a non-targeting negative-control siRNA. GAstV-2 was infected 24 h later at 1.0 MOI, and the viral load of GAstV-2 in cells was analyzed 12 and 24 h after infection. A significant reduction was observed in the proportion of GAstV-2-positive cells (Figure 5A) as well as GAstV-2 mRNA and protein expression levels (Figures 5B and 5C) in the cells upon VIM knockdown. Meanwhile, the viral titer in the supernatant was significantly reduced when VIM was knocked down (Figure 5D), indicating that VIM downregulation significantly inhibits GAstV-2 replication.
Figure 5.
Knockdown of VIM impairs GAstV-2 replication. (A) Immunofluorescence analysis of GAstV-2 following VIM knockdown (scale bars: 250 μm). (B and C) GAstV-2 mRNA and protein expression levels were significantly decreased following VIM knockdown. (D) The virus titer in the supernatant of VIM knockdown cells significantly decreased compared with that of the control group. ** p < 0.01, *** p < 0.001, **** p < 0.0001.
In addition, we performed an antibody blocking assay using a VIM monoclonal antibody to analyze the effect of cell surface VIM on GAstV-2 invasion. The results showed that blocking VIM on the cell surface also significantly inhibited virus entry (Supplementary Figure S2), suggesting that VIM may play a role as a coreceptor; however, further research is still needed.
VIM Rearrangement Acts as an Organizer for the Integrity of the Viral Replication Complex and is Required for High-Efficiency GAstV-2 Replication
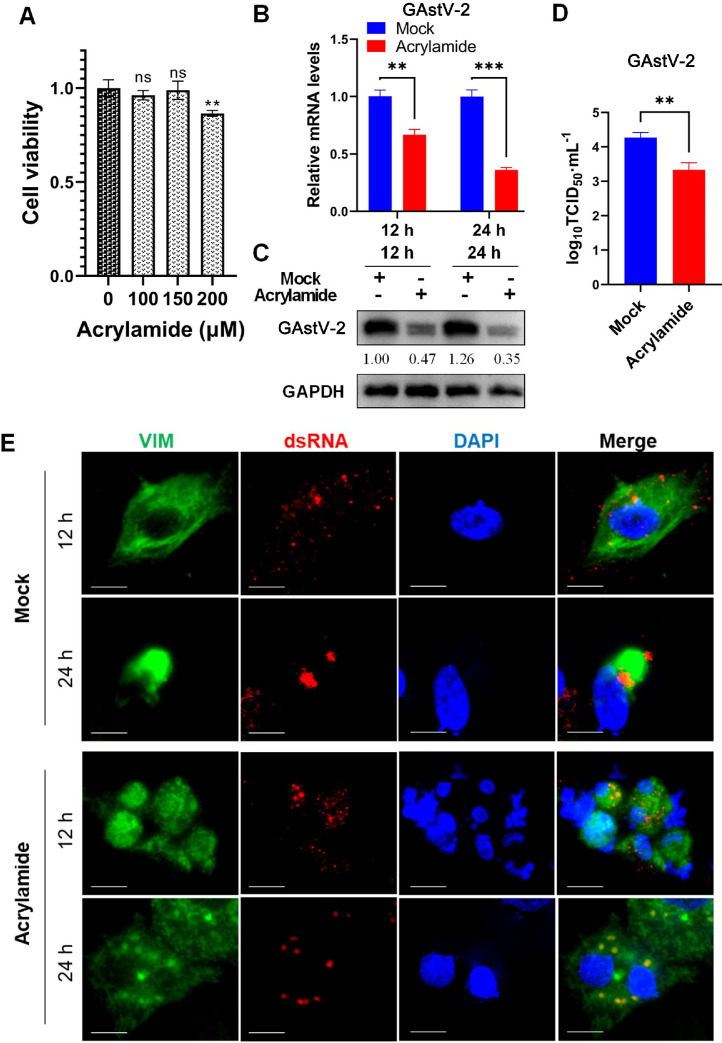
In this study, we used acrylamide (ACR) (Ma et al., 2020), a relevant inhibitor that disrupts the rearranged VIM structure, to investigate its impact on viral replication. There was no significant toxic effect of ACR on cells at working concentrations of 150 μM or below (Figure 6A). Therefore, 150 μM was used as the ACR working concentration for subsequent studies. The results showed that ACR significantly reduced the mRNA and protein expression levels of GAstV-2 in the cells (Figures 6B and 6C), as well as the viral titer in the supernatant (Figure 6D). These data suggested that VIM rearrangement plays a positive role in GAstV-2 replication.
Figure 6.
Acrylamide disrupts the network of VIM and prevents its rearrangement to inhibit GAstV-2 replication. (A) Toxicity analysis of different concentrations of acrylamide on GEK cells. (B and C) GAstV-2 mRNA and protein expression levels following acrylamide treatment. (D) The virus titer in the supernatant of acrylamide-treated cells significantly decreased compared with that of the control group. (E) Immunofluorescence colocalization analysis of VIM and GAstV-2 dsRNA. Effect of acrylamide on GAstV-2 induced VIM rearrangement. Immunofluorescence staining for dsRNA, VIM, and DAPI was performed 12 and 24 h after virus infection (scale bars: 15 μm). ns p >0.05, ** p < 0.01, *** p < 0.001.
To elucidate the associated mechanism, we used laser confocal microscopy, revealing that VIM in ACR-treated cells did not aggregate alongside the nucleus; instead, it was scattered throughout the cytoplasm. In the control group, GAstV-2 dsRNA was clustered and surrounded by VIM, while after ACR treatment, viral dsRNA was dispersed in the cytoplasm (Figure 6E). It has been speculated that the viral replication complex (RC) is unable to form a centralized and stable structure, which is crucial for efficient replication. These results suggest that VIM rearrangement is important for GAstV-2 replication and an intact VIM network is required.
GAstV-2 Infection Increases VIM Phosphorylation and Induces Its Rearrangement
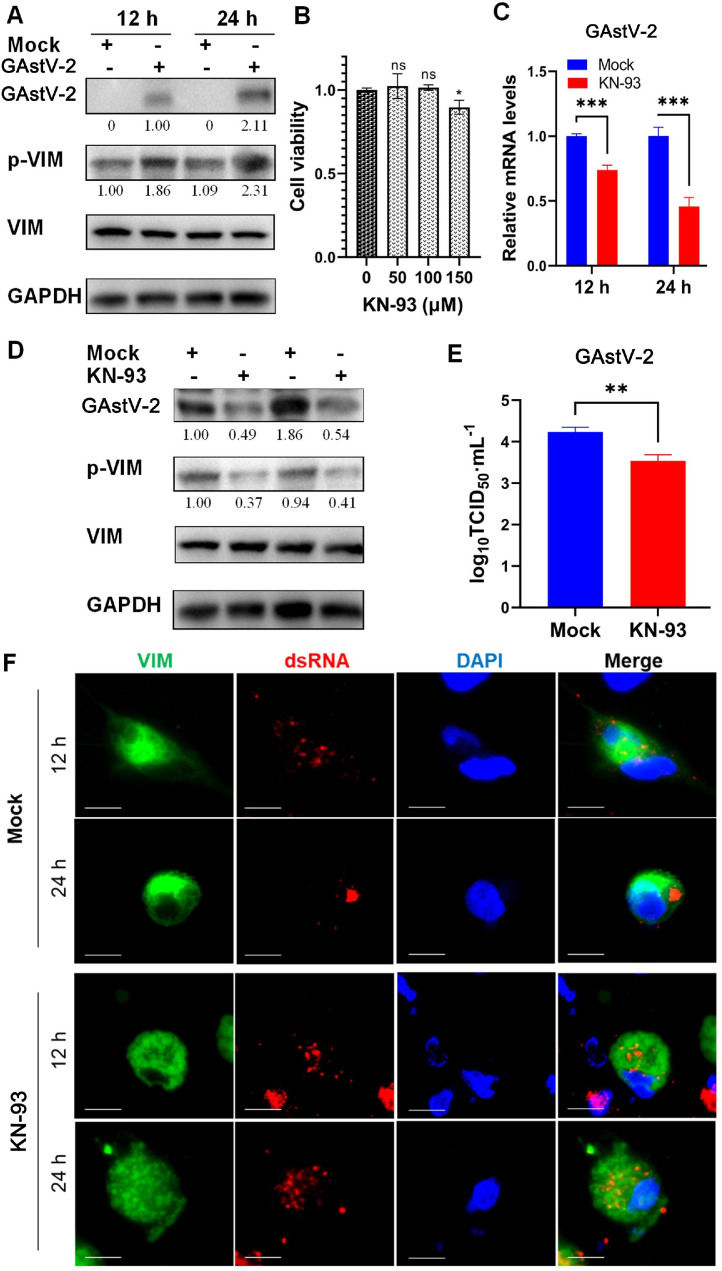
To explore the mechanism underlying the VIM rearrangement triggered by GAstV-2, we investigated changes in VIM phosphorylation before and after GAstV-2 infection. A significant increase was observed in VIM phosphorylation within GAstV-2-infected cells compared with the control group 12 h after infection. This increase occurred prior to the onset of VIM rearrangement. After 24 h of infection with GAstV-2, VIM rearrangement was triggered by the virus, and its phosphorylation level was further increased compared with the control group (Figure 7A). Moreover, it showed that GAstV-2 had no significant effect on VIM expression. These results indicate that the rearrangement of VIM is related to the increase in its phosphorylation induced by GAstV-2.
Figure 7.
GAstV-2 infection increases VIM phosphorylation and induces its rearrangement. (A) Phosphorylation level of VIM after GAstV-2 infection. VIM expression did not change significantly in GAstV-2 infected cells. (B) Toxicity analysis of different concentrations of KN-93 on GEK cells. (C and D) Effect of KN-93 on GAstV-2 mRNA and protein expression levels and VIM phosphorylation in GEK cells. (E) The virus titer in the supernatant of KN-93 treated cells significantly decreased compared with that of the control group. (F) Immunofluorescence colocalization analysis of VIM and dsRNA showing the effect of inhibiting VIM phosphorylation on its rearrangement. Cells transfected with pCAGGS-VIM-Flag plasmid were infected with GAstV-2 and treated with KN-93 to inhibit VIM phosphorylation. Immunofluorescence staining for dsRNA, VIM, and DAPI was performed at 12 and 24 h after virus infection (scale bars: 15 μm). ns p>0.05, * p < 0.05, ** p < 0.01, *** p < 0.001.
To further confirm that VIM rearrangement is closely related to its phosphorylation, we used KN-93 inhibitor to inhibit VIM phosphorylation (Zheng et al., 2021). The cell viability analysis showed that KN-93 had no significant cytotoxicity to cells at concentrations below or equal to 100 μM (Figure 7B). Therefore, cells were treated at a working concentration of 100 μM and subjected to subsequent analysis. Treatment of cells with the VIM phosphorylation inhibitor KN-93 significantly reduced the mRNA and protein expression of GAstV-2 (Figures 7C and 7D), as well as the viral titer in the supernatant (Figure 7E). Furthermore, confocal microscopy observations revealed that VIM in KN-93-treated cells did not aggregate alongside the nucleus; instead, it remained scattered in the cytoplasm. In the control group, GAstV-2 dsRNA was clustered and surrounded by VIM; however, after treatment with KN-93, the dsRNA of the virus was dispersed in the cytoplasm (Figure 7F). These data suggest that VIM rearrangement is closely related to an increase in its phosphorylation and positively affects GAstV-2 replication.
DISCUSSION
In this study, we report that VIM interacts with GAstV-2′s VP70 protein and that this interaction plays an important role in promoting the replication of GAstV-2. Furthermore, this is the first study to report an interaction between VIM- and GAstV-2-associated proteins, establishing a strong molecular and biological basis for the development of drugs aimed at preventing and treating gout caused by GAstV-2.
Initially, we aimed to identify the key host molecules that interact with GAstV-2 VP70. During this process, VIM gradually appeared. After reviewing the literature and preliminary experiments, we became more interested in VIM which is an important component of the cytoskeleton and a highly conserved intermediate fiber protein. VIM can affect cell adhesion and migration, participate in cell signal transduction, regulate cell apoptosis, proliferation, and chemotactic movement, and function as an alarmin (Zhao et al., 2018). In this study, we first demonstrated the direct interaction between VIM and the GAstV-2 structural protein, VP70; the key amino acid site was pinpointed to area 3 of VP70/B, encompassing residues 399–413 of VP70. Additionally, this interaction is essential for the replication of GAstV-2. While studying the interaction of VIM with VP70 using laser confocal microscopy, we found that VP70 induced VIM rearrangement. We then infected the cells with live virus, and similar observations were made for intracellular VIM. VP70 expressed during viral replication emerged as the major factor responsible for VIM rearrangement induced by GAstV-2, providing the basis for further studies. We speculated that this might be a key factor affecting GAstV-2 replication.
To further analyze the effect of VIM on GAstV-2 replication, we altered the expression level of VIM in the cells using various methods. The results showed that VIM significantly promoted GAstV-2 replication. This conclusion is consistent with studies on various other viruses (Song et al., 2016; Zheng et al., 2021; Zhang et al., 2022b). However, there are also exceptions, as reported for foot-and-mouth disease, in which VIM overexpression in PK-15 cells inhibits viral replication (Ma et al., 2020) and the replication of human parainfluenza virus type 3 (Liu et al., 2022). Hence, although VIM is widely distributed among cells, its effects on the invasion and replication of different viruses are dynamic. On the other hand, we found that blocking VIM on the surface of GEK cells significantly inhibited their infection by GAstV-2. Therefore, we speculate that VIM may function as a co-receptor for GAstV-2; nevertheless, this can only be verified after the identification of its main receptor, which we are pursuing. Recently, VIM has also been found distributed in small amounts on the cell surface and in the extracellular environment. Moreover, it is reportedly important in the invasion and infection of various microorganisms (Lalioti et al., 2022; Martinez-Vargas et al., 2023). For example, VIM is an attachment factor that promotes the entry of SARS-CoV-2 into human endothelial cells by binding to the viral S protein and promoting SARS-CoV-2 infection. Furthermore, VIM acts as a co-receptor during SARS-CoV-2 infection and can synergistically interact with the ACE2 receptor (Amraei et al., 2022; Lalioti et al., 2022).
As mentioned above, VP70 induces the rearrangement of VIM. To analyze the role of VIM rearrangement in GAstV-2 replication, we used ACR to disrupt the grid structure of VIM, preventing its rearrangement. The results showed that the replication of GAstV-2 was significantly reduced after blocking VIM rearrangement, indicating that VIM rearrangement plays an important role in the replication of GAstV-2. Furthermore, confocal microscopy revealed that VIM rearrangement could encapsulate and cluster viral dsRNAs. We speculate that this allows the viral nucleic acid to form an efficient replication complex. However, when VIM rearrangement was prevented, dsRNA remained scattered and did not aggregate to form efficient replication complexes, which might inhibit GAstV-2 replication. This phenomenon has also been observed in the interaction between other viruses and VIM (Teo and Chu, 2014; Yu et al., 2020; Zhang et al., 2022b); however, this study is the first to report the interaction between GAstV-2 and VIM, laying a foundation for more in-depth research on GAstV-2 pathogenesis. Having answered this question, a new question arises as to why GAstV-2 and VP70 induce VIM rearrangement. The occurrence of VIM rearrangement has been suggested to be related to the level of its phosphorylation (Sihag et al., 2007; Zheng et al., 2021). Owing to the limited availability of commercial VIM phosphoantibodies limited to specific species sources, there is a lack of phosphoantibodies targeting goose VIM in the market. Consequently, our approach involved human VIM phosphoantibodies. After testing several antibodies, we successfully identified a monoclonal antibody targeting phosphorylation site 38 of human VIM that could be effectively applied to goose samples. Although imperfect, its ability to detect changes in the phosphorylation levels of VIM in geese renders it a valuable tool. The results showed that GAstV-2 infection increased VIM phosphorylation in GEK cells. When VIM was rearranged, its phosphorylation level increased further. After inhibiting the phosphorylation of VIM using a KN-93 inhibitor, VIM rearrangement was not induced by GAstV-2. Viral dsRNA was also unable to aggregate or form replication complexes. Therefore, GAstV-2 infection promotes VIM phosphorylation, leading to VIM rearrangement and active regulation of viral replication.
In conclusion, our results show for the first time that VIM interacts with GAstV-2 VP70 and positively modulates GAstV-2 replication in GEK cells. The interaction between VP70 and VIM is crucial for virus survival, as the GAstV-2 genome containing VP70 mutations disrupts this interaction, resulting in the failure of virus replication in cell cultures. Additionally, an intact VIM network is required for efficient replication of GAstV-2. Therefore, this study not only helps us better understand the mechanism of GAstV-2 replication in GEK cells but may also contribute to the development of new antiviral drugs in the future.
DISCLOSURES
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
This work was supported by the National Key R&D Program of China (2022YFD1801000), Open competition program of the top ten critical priorities of Agricultural Science and Technology Innovation for the 14th Five-Year Plan of Guangdong Province (2022SDZG02), Project of Collaborative Innovation Center of GDAAS (XTXM202202, XT202207), Talent Introduction Project for Excellent PhD of Guangdong Academy of Agricultural Sciences (R2023YJ-YB2001), and Opening Project of State Key Laboratory of Swine and Poultry Breeding Industry (2023QZ-NK05).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.psj.2024.104146.
Appendix. Supplementary materials
REFERENCES
- Amraei R., Xia C., Olejnik J., White M.R., Napoleon M.A., Lotfollahzadeh S., Hauser B.M., Schmidt A.G., Chitalia V., Muhlberger E., Costello C.E., Rahimi N. Extracellular vimentin is an attachment factor that facilitates SARS-CoV-2 entry into human endothelial cells. Proc Natl Acad Sci U S A. 2022;119 doi: 10.1073/pnas.2113874119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass D.M., Upadhyayula U. Characterization of human serotype 1 astrovirus-neutralizing epitopes. J. Virol. 1997;71:8666–8671. doi: 10.1128/jvi.71.11.8666-8671.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Xu X., Yu Z., Sui C., Zuo K., Zhi G., Ji J., Yao L., Kan Y., Bi Y., Xie Q. Characterization and genomic analysis of emerging astroviruses causing fatal gout in goslings. Transbound. Emerg. Dis. 2020;67:865–876. doi: 10.1111/tbed.13410. [DOI] [PubMed] [Google Scholar]
- Cortez V., Meliopoulos V.A., Karlsson E.A., Hargest V., Johnson C., Schultz-Cherry S. Astrovirus biology and pathogenesis. Annu. Rev. Virol. 2017;4:327–348. doi: 10.1146/annurev-virology-101416-041742. [DOI] [PubMed] [Google Scholar]
- Ghosh P., Halvorsen E.M., Ammendolia D.A., Mor-Vaknin N., O'Riordan M.X.D., Brumell J.H., Markovitz D.M., Higgins D.E. Invasion of the brain by listeria monocytogenes is mediated by InlF and host cell vimentin. mBio. 2018;9 doi: 10.1128/mBio.00160-18. e00160-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladue D.P., O'Donnell V., Baker-Branstetter R., Holinka L.G., Pacheco J.M., Fernandez Sainz I., Lu Z., Ambroggio X., Rodriguez L., Borca M.V. Foot-and-mouth disease virus modulates cellular vimentin for virus survival. J. Virol. 2013;87:6794–6803. doi: 10.1128/JVI.00448-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalioti V., Gonzalez-Sanz S., Lois-Bermejo I., Gonzalez-Jimenez P., Viedma-Poyatos A., Merino A., Pajares M.A., Perez-Sala D. Cell surface detection of vimentin, ACE2 and SARS-CoV-2 Spike proteins reveals selective colocalization at primary cilia. Sci. Rep. 2022;12:7063. doi: 10.1038/s41598-022-11248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Y., Hu W.Q., Liu T.N., Zhang H.H., Opriessnig T., Xiao C.T. Isolation and evolutionary analyses of gout-associated goose astrovirus causing disease in experimentally infected chickens. Poult. Sci. 2021;100:543–552. doi: 10.1016/j.psj.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wu J., Zhou J., Yuan B., Chen J., Wu W., Mo L., Qu Z., Zhou F., Dong Y., Huang K., Liu Z., Wang T., Symmes D., Gu J., Sho E., Zhang J., Chen R., Xu Y. A vimentin-targeting oral compound with host-directed antiviral and anti-inflammatory actions addresses multiple features of COVID-19 and related diseases. mBio. 2021;12 doi: 10.1128/mBio.02542-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Li L., Dong J., Zhang J., Huang Y., Zhai Q., Xiang Y., Jin J., Huang X., Wang G., Sun M., Liao M. Global analysis of gene expression profiles and gout symptoms in goslings infected with goose astrovirus. Vet. Microbiol. 2023;279 doi: 10.1016/j.vetmic.2023.109677. [DOI] [PubMed] [Google Scholar]
- Liu P., Zhang S., Ma J., Jin D., Qin Y., Chen M. Vimentin inhibits alpha-tubulin acetylation via enhancing alpha-TAT1 degradation to suppress the replication of human parainfluenza virus type 3. PLoS Pathog. 2022;18 doi: 10.1371/journal.ppat.1010856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Liu K., Chen Y., Gao R., Hu Z., Hu J., Gu M., Hu S., Ding C., Jiao X., Wang X., Liu X., Liu X. Cellular vimentin regulates the infectivity of Newcastle disease virus through targeting of the HN protein. Vet. Res. 2023;54:92. doi: 10.1186/s13567-023-01230-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Ling Y., Li P., Sun P., Cao Y., Bai X., Li K., Fu Y., Zhang J., Li D., Bao H., Chen Y., Li Z., Wang Y., Lu Z., Liu Z. Cellular vimentin interacts with foot-and-mouth disease virus nonstructural protein 3A and negatively modulates viral replication. J. Virol. 2020;94 doi: 10.1128/JVI.00273-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzer H.S., Villarreal R.I., Doran K.S. Targeting the BspC-vimentin interaction to develop anti-virulence therapies during Group B streptococcal meningitis. PLoS Pathog. 2022;18 doi: 10.1371/journal.ppat.1010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vargas M., Cebula A., Brubaker L.S., Seshadri N., Lam F.W., Loor M., Rosengart T.K., Yee A., Rumbaut R.E., Cruz M.A. A novel interaction between extracellular vimentin and fibrinogen in fibrin formation. Thromb. Res. 2023;221:97–104. doi: 10.1016/j.thromres.2022.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Li T., Zhang W., Zhang X., Zhang X., Xie Q., Zhang J., Shao H., Wan Z., Qin A., Ye J., Gao W. Identification of three novel B cell epitopes in ORF2 protein of the emerging goose astrovirus and their application. Appl. Microbiol. Biotechnol. 2022;106:855–863. doi: 10.1007/s00253-021-11711-2. [DOI] [PubMed] [Google Scholar]
- Sihag R.K., Inagaki M., Yamaguchi T., Shea T.B., Pant H.C. Role of phosphorylation on the structural dynamics and function of types III and IV intermediate filaments. Exp Cell Res. 2007;313:2098–2109. doi: 10.1016/j.yexcr.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T., Fang L., Wang D., Zhang R., Zeng S., An K., Chen H., Xiao S. Quantitative interactome reveals that porcine reproductive and respiratory syndrome virus nonstructural protein 2 forms a complex with viral nucleocapsid protein and cellular vimentin. J. Proteomics. 2016;142:70–81. doi: 10.1016/j.jprot.2016.05.009. [DOI] [PubMed] [Google Scholar]
- Suprewicz L., Swoger M., Gupta S., Piktel E., Byfield F.J., Iwamoto D.V., Germann D., Reszec J., Marcinczyk N., Carroll R.J., Janmey P.A., Schwarz J.M., Bucki R., Patteson A.E. Extracellular vimentin as a target against SARS-CoV-2 host cell invasion. Small. 2022;18 doi: 10.1002/smll.202105640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo C.S., Chu J.J. Cellular vimentin regulates construction of dengue virus replication complexes through interaction with NS4A protein. J. Virol. 2014;88:1897–1913. doi: 10.1128/JVI.01249-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Xie J., Wu Z., Liu L., Wu S., Feng Q., Dong H., Zhu S. Pathogenicity of a goose astrovirus 2 strain causing fatal gout in goslings. Microb. Pathog. 2023;184 doi: 10.1016/j.micpath.2023.106341. [DOI] [PubMed] [Google Scholar]
- Xu J., Gao L., Zhu P., Chen S., Chen Z., Yan Z., Lin W., Yin L., Javed M.T., Tang Z., Chen F. Isolation, identification, and pathogenicity analysis of newly emerging gosling astrovirus in South China. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1112245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Jiang B., Cheng Y., Gao Z., He Y., Wu Z., Wang M., Jia R., Zhu D., Liu M., Zhao X., Yang Q., Wu Y., Zhang S., Huang J., Ou X., Gao Q., Sun D., Cheng A., Chen S. Molecular epidemiology and virulence of goose astroviruses genotype-2 with different internal gene sequences. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1301861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Wu Z., He Y., Jiang B., Cheng Y., Wang M., Jia R., Zhu D., Liu M., Zhao X., Yang Q., Wu Y., Zhang S., Huang J., Ou X., Sun D., Cheng A., Chen S. Molecular characterization of a virulent goose astrovirus genotype-2 with high mortality in vitro and in vivo. Poult Sci. 2024;103 doi: 10.1016/j.psj.2024.103585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Tian J., Tang Y., Diao Y. Isolation and genomic characterization of gosling gout caused by a novel goose astrovirus. Transbound. Emerg. Dis. 2018;65:1689–1696. doi: 10.1111/tbed.12928. [DOI] [PubMed] [Google Scholar]
- Yu Y.N., Zheng Y., Hao S.S., Zhang Z., Cai J.X., Zong M.M., Feng X.L., Liu Q.T. The molecular evolutionary characteristics of new isolated H9N2 AIV from East China and the function of vimentin on virus replication in MDCK cells. Virol. J. 2020;17:78. doi: 10.1186/s12985-020-01351-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Deng T., Song Y., Liu J., Jiang Z., Peng Z., Guo Y., Yang L., Qiao H., Xia Y., Li X., Wang Z., Bian C. Identification and genomic characterization of emerging goose astrovirus in central China, 2020. Transbound. Emerg. Dis. 2022;69:1046–1055. doi: 10.1111/tbed.14060. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhao S., Li Y., Feng F., Li M., Xue Y., Cui J., Xu T., Jin X., Jiu Y. Host cytoskeletal vimentin serves as a structural organizer and an RNA-binding protein regulator to facilitate Zika viral replication. Proc. Natl. Acad. Sci. U S A. 2022;119 doi: 10.1073/pnas.2113909119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Liu J., Sun W., Zeng R., Li T., Ma R. Long non-coding RNA-ENST00000434223 suppresses tumor progression in gastric cancer cells through the Wnt/β-catenin signaling pathway. Int. J. Biol. Macromol. 2018;120:491–501. doi: 10.1016/j.ijbiomac.2018.08.079. [DOI] [PubMed] [Google Scholar]
- Zheng X.X., Li R., Qiao S., Chen X.X., Zhang L., Lu Q., Xing G., Zhou E.M., Zhang G. Vimentin rearrangement by phosphorylation is beneficial for porcine reproductive and respiratory syndrome virus replication in vitro. Vet. Microbiol. 2021;259 doi: 10.1016/j.vetmic.2021.109133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.