Abstract
Goose astrovirus (GAstV) has been widespread in China since 2016, causing significant growth inhibition and gout symptoms in goslings and leading to substantial economic losses in the goose industry. To better understand the epidemiological characteristics of GAstV in Guangdong Province, 682 samples were collected from geese with suspected GAstV infection across different regions of Guangdong Province from January 2022 to January 2024. Virus isolation, identification, and genetic evolution analysis were performed. The results showed that all samples were GAstV positive, with 52.64% co-infected with GAstV-1 and GAstV-2, and 42.38% positive for GAstV-2 alone, indicating that GAstV-2 remains the most prevalent subtype. Additionally, three GAstV isolates were identified using molecular detection, immunofluorescence, and transmission electron microscopy on LMH cells or goose embryos. Compared with GDYJ2304 and other reported GAstV-2 strains, the ORF2 region of the GDYJ2210 isolates lacked 3 bases, and the replication ability of GDYJ2210 was significantly higher than that of GDYJ2304. Whole genome sequence alignment and genetic evolution analysis revealed that the GDFS2209 isolate was located in the GAstV-1 branch, with a sequence similarity of 89.70 to 99.00% to GAstV-1 reference strains. The GDYJ2210 and GDYJ2304 isolates were located in the GAstV-2 branch, showing a sequence similarity of 96.80 to 98.90% to GAstV-2 reference strains. These results demonstrated that the GAstV isolates were highly similar to each other despite being prevalent in 5 different regions of Guangdong Province. These findings enhance the understanding of the genetic diversity and evolution of GAstV and may facilitate the development of effective preventive strategies.
Key words: goose astrovirus, isolation, molecular epidemiology, phylogenetic analysis
INTRODUCTION
Goose astrovirus (GAstV) is a single-stranded RNA virus with a total genome length of 6.1 to 7.9 kb. The genome structure comprises a 5′-untranslated region (UTR), ORF1a, ORF1b, and ORF2 open reading frames, a 3′-UTR, and a polyadenine tail (Niu et al., 2018; Yang et al., 2018; Zhang et al., 2018b). The open reading frames ORF1a and ORF1b encode the nonstructural proteins of GAstV, while ORF2 encodes the structural proteins, prompting the assembly of immature virions in the cell (Krishna, 2005; Cortez et al., 2017; Zhang et al., 2018a). The ORF2 gene shows low conservation and significant diversity. Therefore, sequence similarity of the ORF2 gene has become one of the important criteria for identifying different astrovirus species (Koci and Schultz-Cherry, 2002; Smyth et al., 2012; Ren et al., 2020) Based on this difference, GAstV can be divided into two genotypes, GAstV-1 and GAstV-2 (Zhang et al., 2022; Xu et al., 2023b).
Since 2016, new diseases have been reported in several important goose-raising provinces in China, with urate deposition in goose joints and internal organs as the main clinical symptom (Chen et al., 2020a,b), commonly known as gout pathogen detection, pathogen isolation, and gene sequencing have determined that the new disease is caused by a novel GAstV (Liu et al., 2023; Wang et al., 2023; Xu et al., 2023a). Both GAstV-1 and GAstV-2 are seriously harmful to goslings (Wei et al., 2024; Zhu et al., 2024). According to relevant research, GAstV-2 is the predominant type in clinical practice. The discovery of novel GAstV not only increases the diversity of astrovirus species but also broadens the disease patterns caused by astroviruses (Peng et al., 2023; Wang et al., 2023; Xu et al., 2023b). GAstV primarily affects goslings and shows susceptibility across different goose breeds. Currently, the infection rate among geese in clinics is increasing with a (Li et al., 2024; Xu et al., 2023a) morbidity rate of up to 80% and a mortality rate of up to 50%, causing a serious blow to China's goose industry.
Guangdong province is one of the major goose-breeding provinces in China, ranking high in both the number of geese and the yield of goose products. However, the epidemic characteristics and pathogenicity of GAstV in Guangdong province are still not fully understood. In-depth epidemiological survey data are crucial to fully understand the current epidemiological characteristics of GAstV and to formulate more effective prevention and control measures. In this study, samples of suspected GAstV infection were collected in Guangdong province over the past 2 yr. The epidemiological characteristics of GAstV were clarified through virus isolation and identification, molecular biological detection, and bioinformatics analysis. This study provides data support for the scientific prevention and control of gouty disease caused by GAstV in goslings.
MATERIALS AND METHODS
Cells, Embryos, Virus, and Antibodies
The LMH cells were cultured in DMEM/F12 (Gibco, Thermo Fisher Scientific, Inc., Shanghai, China) and supplemented with 10% fetal bovine serum at 37°C with 5% CO2. Nine-day-old Magang goose embryos were purchased from Qingyuan Livestock Co. Ltd. (Guangdong, China). The VP27 polyclonal antibody against GAstV-1 and GAstV-2 was previously prepared and stored in our laboratory.
Samples Collection and Ethics Statement
From January 2022 to January 2024, eight goose farms located in Yangjiang City, Foshan City, Zhaoqing City, Qingyuan City, Jiangmen City, Zhanjiang City, Guangzhou City, and Yunfu City in Guangdong Province were sampled. A total of 682 geese with typical gout symptoms were collected. Liver, kidney, and other tissue samples were taken from these sick geese. The affected geese showed emaciation, slow growth, reluctance to move, and even paralysis. Necropsy revealed uric acid deposits on the surface of multiple organs or in the joint cavity, kidney enlargement and bleeding, and some geese had enlarged livers with white spots. The liver and kidney tissues were collected and stored at −80°C for later use.
All animal experiments were performed in accordance with the regulation for animal experimentation of Guangdong Province, China, and were permitted by the Ethics Committee of Institute of Animal Health, Guangdong Academy of Agricultural Sciences (No. YC-PT2022040).
Pathological Autopsy and HE Staining
The diseased goose was aseptically dissected, and the heart, liver, spleen, lung, kidney, gallbladder, and other organs were observed for urate deposition or other lesions after exposing the chest and abdomen. After recording the clinical sample information, significant lesions in the liver, kidney, and other tissues and organs were collected and immersed in formalin solution. Once the tissues were completely immersed, they were sent to Servicebio (Wuhan, China) for paraffin section preparation and HE staining. CaseViewer software was used to view the scanning results of tissue sections, perform histopathological examinations, and take timely records.
RNA Extraction and GAstV Detection
After mixing the tissue samples with sterile phosphate buffer (PBS, pH 7.4) to form a tissue suspension, the samples were homogenized by grinding using a frozen tissue grinder. The supernatant was collected by centrifugation at 10,000 r/min for 10 minutes at 4°C. The tissue-grinding fluid supernatant was used for nucleic acid extraction and virus isolation. Viral nucleic acid was extracted from the tissue supernatant using an automatic nucleic acid extraction instrument (TIANLONG, Xi'an, China). Different genotypes of GAstV were detected by reverse transcription polymerase chain reaction with specific primers (Table 1).
Table 1.
Specific PCR primers for GAstV detection of different genotypes.
| Primer name | Accession number | Primer sequences (5′-3′) | Product (bp) |
|---|---|---|---|
| GAstV-1-F | MH410610.1 | CCGACAAGGTCACTGTCTCA | 436 |
| GAstV-1-R | CCTGCAAGTGGTGTAAATCG | ||
| GAstV-2-F | MG934571.1 | ACCTTCAATGTTGGCTATCGT | 280 |
| GAstV-2-R | ATTGAAATCACATAGTCCCCAT |
Isolation and Identification of GAstV
The tissue grinding supernatant was filtered through a 0.22 μm filter, and the filtrate was inoculated through the chorioallantoic membrane of 10 to 12-day-old goose embryos, 200 μL per embryo, and sealed with medical sterile tape. The eggs were incubated in a 37°C incubator. After 5 d of inoculation, the eggs were placed in a 4°C refrigerator to contract blood vessels and euthanize the embryos. Goose embryos were inoculated and passaged 5 times. Nucleic acid was extracted from the allantoic fluid of each generation for GAstV detection by PCR.
In addition, the filtrate was inoculated into LMH cells. When the monolayer of LMH cells reached a density of about 80%, the supernatant was discarded, 500 μL of filtrate was added, and the cells were incubated for 2 hours. After rinsing and washing, fresh medium was added for culture. The cytopathic effect of GAstV on LMH cells was observed under a microscope every 24 h. After maintaining the cell culture for 5 d, the culture flask was placed in a −80°C refrigerator and subjected to three freeze-thaw cycles. The culture was then centrifuged at 12,000 r/min for 5 min, and the supernatant was collected. The next generation of LMH cells was inoculated as described above and passed for 3 generations. Nucleic acid was extracted from the cell culture supernatant of each generation for GAstV detection by PCR.
Indirect Immunofluorescence Analysis
When LMH cells reached approximately 80% density in a 6-well plate monolayer, they were washed three times with PBS and incubated with the virus solution for 2 h, while the negative control was incubated with an equal volume of PBS. The cells were then cultured in a fresh medium for 48 h. The supernatant was discarded, 4% paraformaldehyde was added, and the cells were fixed at room temperature for 20 min. After washing three times with PBS, GAstV-1 or GAstV-2 VP27 polyclonal antibody was added, and the cells were incubated at 4°C overnight. After washing three times, Alexa Fluor 488 fluorescent secondary antibody was added and incubated for 1 h at 37°C in the dark. After washing 3 times, DAPI was added for nuclear staining. After a final wash with PBS, the cells were observed and photographed under a fluorescence-inverted microscope.
Transmission Electron Microscopy
The GAstV-infected LMH cells were fixed with glutaraldehyde. The cell layer was gently scraped off the bottom of the culture flask using a cell scraper. The cell precipitates were transferred into a centrifuge tube using a Pasteur pipette. The centrifuge speed did not exceed 3,000 r/min for 2 min, and the cell precipitates were approximately the size of mung beans. After discarding the supernatant, a new fixative was added, and the cells were left in suspension. They were fixed at room temperature in the dark for 30 min and then transferred to a 4°C refrigerator for storage. Subsequently, they were sent to Servicebio (Wuhan, China) for transmission electron microscope observation.
Whole Genome Cloning and Sequencing of GAstV
The RNA extracted above was reverse transcribed according to the instructions provided with the PrimeScript RT Master Mix (Takara, Dalian, China) reagent. Subsequently, the entire GAstV genome was amplified in segments using the primers listed in (Supplementary Table S1). The PCR products were recovered and purified by agarose gel electrophoresis, then ligated into the pMD18-T vector (Takara, Dalian, China) and transformed into E. coli DH5α competent cells (Takara, Dalian, China). The strains were expanded and cultured, and the plasmid DNA was extracted and sent to Sangon Biotech (Shanghai, China) for sequencing.
Phylogenetic Analysis and Sequence Comparison
The SeqMan software was used to assemble the sequencing data. The GAstV sequence and the astrovirus reference sequence from the GenBank database were saved in a FASTA format file. MegAlign software was utilized for similarity comparison and nucleotide similarity analysis. Simultaneously, MEGA7 software was employed to compare the nucleotide sequence data. The Neighbor-Joining method was selected to analyze the nucleotide differences between the sequences and to construct the genetic evolution tree.
RESULTS
Pathological Autopsy
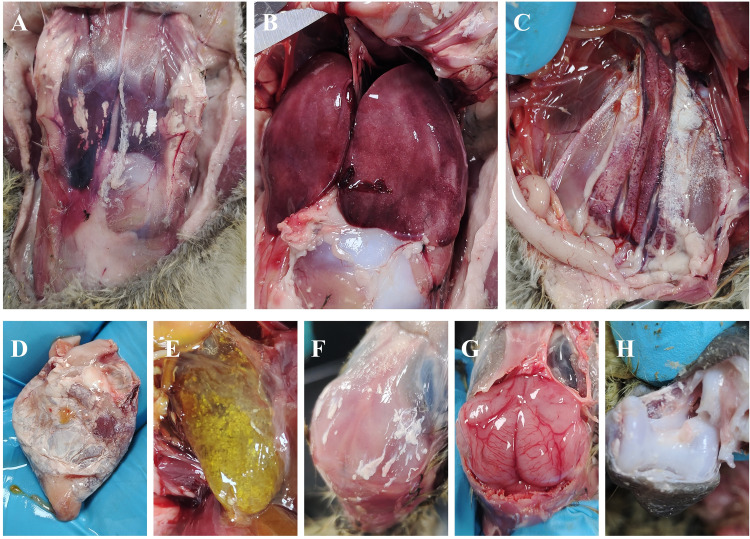
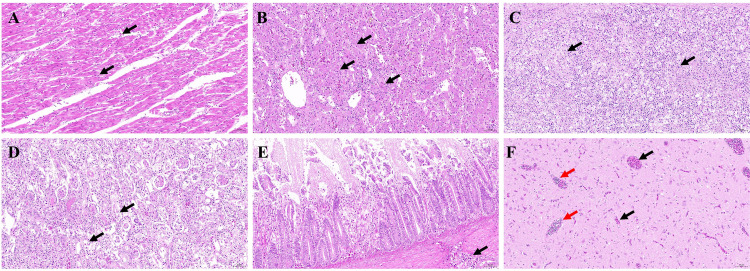
Necropsy of the goose with gout (Figure 1) revealed extensive deposits of urate in subcutaneous hemorrhages, kidneys, ureters, heart, and joints. Additionally, the liver of the sick goose was enlarged, bleeding, and congested. The gallbladder was filled with white urine crystals, and the brain exhibited signs of congestion. Pathological tissues were collected for HE staining, and the results (Figure 2) indicated cardiac hemorrhage, loose arrangement of myocardial cells, and uneven staining of myocardial fibers. The arrangement of liver cells was disordered, with multiple foci showing necrotic cells accompanied by a small amount of lymphocyte infiltration. The spleen cells were arranged disorderly, with numerous irregular nuclei and a small amount of necrotic cell debris observed. Necrosis and exfoliation of a large number of renal tubular epithelial cells were observed in the kidney, with renal cells exhibiting disorganization and pyknotic and fragmented nuclei. Extensive shedding of the intestinal villous epithelium and scattered infiltration of vascular lymphocytes in the muscular layer were noted. Brain tissue showed significant vascular congestion, with white blood cells observed in the lumen of many vessels. These findings provide valuable insights into the pathological effects of GAstV infection on goslings.
Figure 1.
Autopsy lesions of geese with gout disease. (A) Subcutaneous urate deposition; (B) Hepatomegaly and congestion; (C) Kidney and ureter enlargement, urate deposition; (D) The heart is covered with urate deposit membrane; (E) The gallbladder is full with urate crystals; (F) Subcutaneous urate deposition in brain; (G) cerebral congestion and hemorrhage; (H) Urate deposits in the joint cavity.
Figure 2.
Histopathological examination of geese with naturally infected GAstV (×200). (A) Heart bleeding; (B) Multifocal necrosis of hepatocytes; (C) A large number of cells with large volume and irregular nucleolar shape can be seen in the spleen; (D) Renal tubular epithelial cells necrosis and exfoliation; (E) There is scattered infiltration of lymphocytes in the muscular vessels of intestinal tissue; (F) Blood vessel congestion in brain tissue (black arrow), white blood cells can be seen in the vascular lumen (red arrow).
Molecular Diagnosis of GAstV
A total of 682 samples were analyzed by RT-PCR (Supplementary Figure S1), all of which tested positive for GAstV. Among them, co-infection of GAstV-1 and GAstV-2 accounted for 52.64% (359/682), while 42.38% (289/682) tested positive for GAstV-2 and negative for GAstV-1. Additionally, 4.99% (34/682) were positive for GAstV-1 and negative for GAstV-2. These data indicate that goose farms in Guangdong province continue to be affected by GAstV.
According to the date of sample collection, the detection results of GAstV in different seasons were pooled and analyzed (Table 2). The percentage of GAstV-positive samples in spring was 16.72% (114/682), in summer it was 26.39% (180/682), in autumn it was 29.18% (199/682), and in winter it was 27.71% (189/682). These findings suggest that the positive rate of GAstV is higher in autumn and winter.
Table 2.
The positive rate of GAstV in different seasons.
| Types of GAstV infection | The number of samples and its proportion |
||||
|---|---|---|---|---|---|
| Spring (n = 114) |
Summer (n = 180) |
Autumn (n = 199) |
Winter (n = 189) |
Total (n = 682) |
|
| GAstV-1 a | 0 | 2 | 7 | 25 | 34 |
| 0.00% | 1.11% | 3.52% | 13.23% | 4.99% | |
| GAstV-2 b | 50 | 76 | 96 | 67 | 289 |
| 43.86% | 42.22% | 48.24% | 35.45% | 42.38% | |
| GAstV-1+2 c | 64 | 102 | 96 | 97 | 359 |
| 56.14% | 56.67% | 48.24% | 51.32% | 52.64% | |
| Total | 114 | 180 | 199 | 189 | 682 |
| 16.72% | 26.39% | 29.18% | 27.71% | 100% | |
positive for GAstV-1 and negative for GAstV-2.
positive for GAstV-2 and negative for GAstV-1.
co-infected with GAstV-1 and GAstV-2.
The association between GAstV and different growth stages of geese was explored based on the age information of geese with clinical onset (Table 3). Geese aged 1-15 days accounted for 40.18% (274/682) of GAstV PCR-positive samples, while those aged 16 to 30 d accounted for 39% (266/682). Geese aged 31 to 50 d represented 15.10% (103/682) of the total, those aged 51 to 80 d accounted for 5.13% (35/682), and geese aged over 80 d represented 0.59% (4/682) of the total. This distribution aligns with the observation that GAstV mainly affects goslings.
Table 3.
The positive rate of GAstV in geese at different ages.
| The number of samples and its proportion |
||||||
|---|---|---|---|---|---|---|
| Types of GAstV infection | 1-15 d (n = 274) |
16-30 d (n = 266) |
31-50 d (n = 103) |
51-80 d (n = 35) |
>80 d (n = 4) |
Total (n = 682) |
| GAstV-1 a | 14 | 11 | 7 | 0 | 0 | 32 |
| 5.11% | 4.14% | 6.80% | 0.00% | 0.00% | 4.69% | |
| GAstV-2 b | 55 | 129 | 80 | 23 | 2 | 289 |
| 20.07% | 48.50% | 77.67% | 65.71% | 50.00% | 42.38% | |
| GAstV-1+2 c | 205 | 126 | 16 | 12 | 2 | 361 |
| 74.82% | 47.37% | 15.53% | 34.29% | 50.00% | 52.93% | |
| Total | 274 | 266 | 103 | 35 | 4 | 682 |
| 40.18% | 39.00% | 15.10% | 5.13% | 0.59% | 100% | |
positive for GAstV-1 and negative for GAstV-2.
positive for GAstV-2 and negative for GAstV-1.
co-infected with GAstV-1 and GAstV-2.
Virus Isolation and Identification
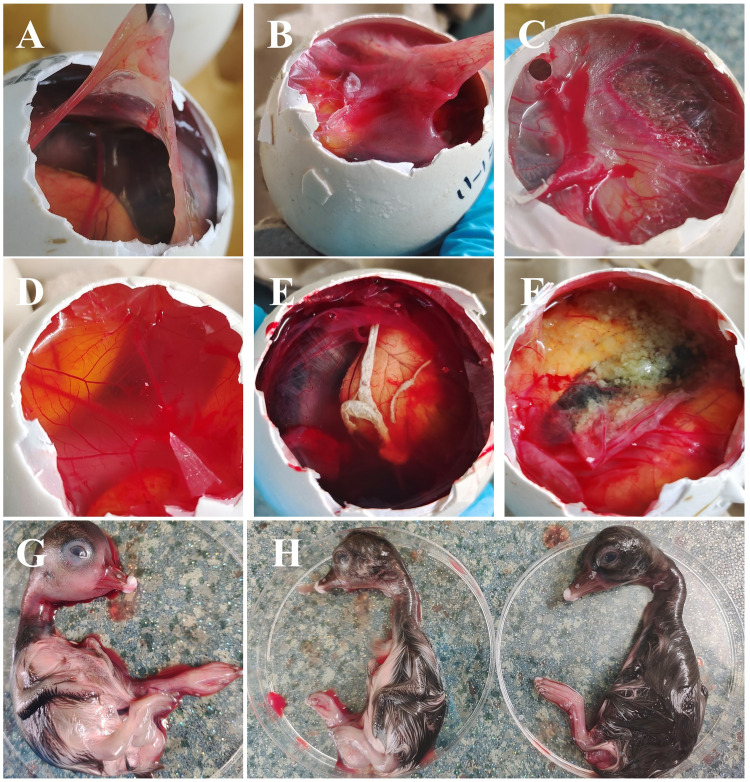
Filter the grinding solution of the diseased material and inoculate it into the goose embryos. After 5 days, the air chamber was opened, and the chorioallantoic membrane significantly thickened, with scattered yellow-white or lump-white urate deposits in the allantoic fluid. After collecting the allantoic fluid, the goose embryos were removed for observation. The head and neck of the goose embryos were swollen, the embryo body was underdeveloped, obviously atrophic or swollen, and the coat was sparse (Figure 3).
Figure 3.
Goose embryo infected with GAstV. (A) Normal goose embryo chorioallantoic membrane; (B-C) Thickening of chorioallantoic membrane in geese infected with GAstV; (D) normal goose embryo allantoic fluid; (E) There were lumps of urate deposits in allantoic fluid of goose embryo infected with GAstV; (F) There were scattered yellow and white urate crystals in allantoic fluid of goose embryo infected with GAstV; (G) Swelling of goose embryo infected with GAstV; (H) Goose embryos infected with GAstV were small in size and sparsely coated (left), while no significant changes were observed in control geese embryos (right).
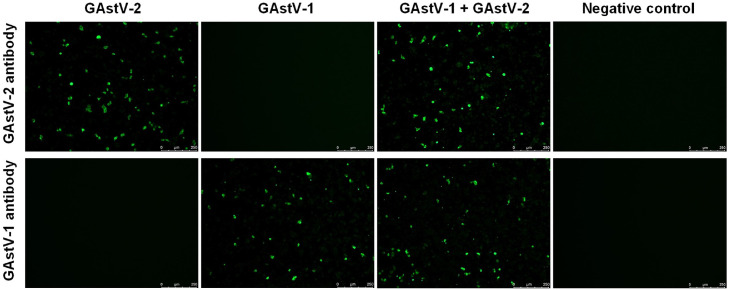
To further validate the isolated virus, IFA experiments were conducted using VP27 antibodies capable of specifically recognizing different genotypes of GAstV. The results (Figure 4) showed that LMH cells inoculated with the virus exhibited green fluorescence specific to the GAstV VP27 antibody, while the control group cells did not show green fluorescence. Finally, three GAstV strains were obtained in this study: GAstV-1 strains GDFS2209, GAstV-2 strains GDYJ2304 and GDYJ2210, respectively.
Figure 4.
Indirect immunofluorescence was used to analyze GAstV infection in LMH cells.
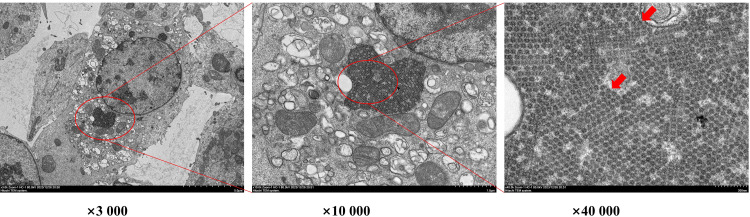
The nucleic acid of the virus fluid was extracted and detected by RT-PCR. The results showed that it does not contain other common goose-derived viruses, including ambush virus, reovirus, calicivirus, parvovirus, circovirus, fowl adenovirus, and avian influenza virus (Supplementary Figure S2). Meanwhile, the results of the hemagglutination test showed that these isolates could not agglutinate red blood cells (Supplementary Figure S3), consistent with GAstV lacking hemagglutination, indicating that the virus obtained was relatively pure. The GAstV virus particles in LMH cells were observed by transmission electron microscopy. The virus particles were located in LMH cells and were spherical in shape with a diameter of about 25 nm (Figure 5).
Figure 5.
Observation of GAstV virions in LMH cells by transmission electron microscopy. Red arrows indicate GAstV virion.
Whole Genome Sequencing and Characterization of GAstV
The whole genome sequences of the GAstV isolates mentioned above were divided into multiple segments for PCR amplification and finally spliced together (Supplementary Figure S4). In this study, a total of three GAstV strains were isolated. The full length of the GAstV-1 GDFS2209 gene was 7288 bp, while the full-length gene of GAstV-2 strain GDYJ2210 was 7180 bp, and that of GAstV-2 strain GDYJ2304 was 7183 bp. All three GAstV isolates exhibited the typical GAstV gene structure, comprising 5′ UTR-ORF1a-ORF1b-ORF2-3′ UTR-poly (A) tail.
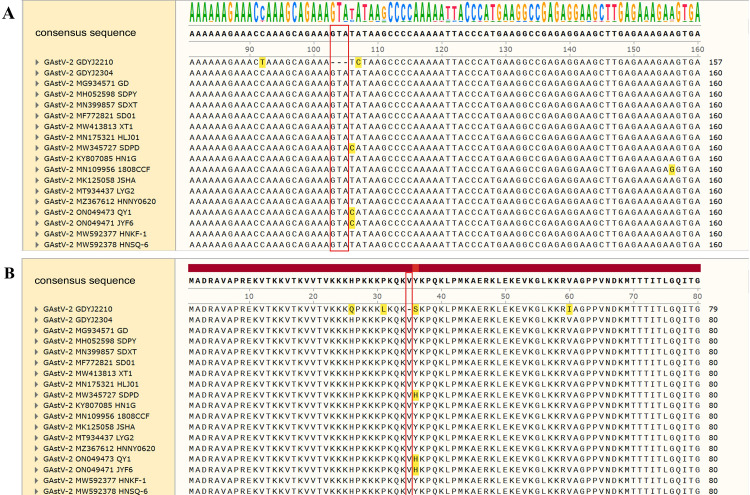
The GAstV-2 GDYJ2210 strain lacked three consecutive bases (GTA) at nucleotides 103-105 of the ORF2 region compared with the GAstV-2 GDYJ2304 strain and the GAstV-2 strain recorded in GenBank (Figure 6). Furthermore, the GAstV-2 GDYJ2210 strain exhibited a deletion of the amino acid valine at position 35 of ORF2. Additionally, the proliferation ability of the mutant isolate strain in LMH cells was significantly higher than that of the non-mutant strain (Figure 7). The ORF2 gene is the main structural protein of GAstV, closely associated with the infectivity and antigenicity of the virus. The deletion mutation in the ORF2 gene may have an impact on the biological characteristics of the virus. Therefore, the influence and significance of these mutations need to be further investigated. It should be noted that the GAstV-1 GDFS2209 isolated in this study did not have regular mutation characteristics in the gene sequence with other GAstV-1 strains.
Figure 6.
Location of gene deletion in GAstV-2 GDYJ2210. (A) Nucleotide deletion site; (B) Amino acid deletion site.
Figure 7.
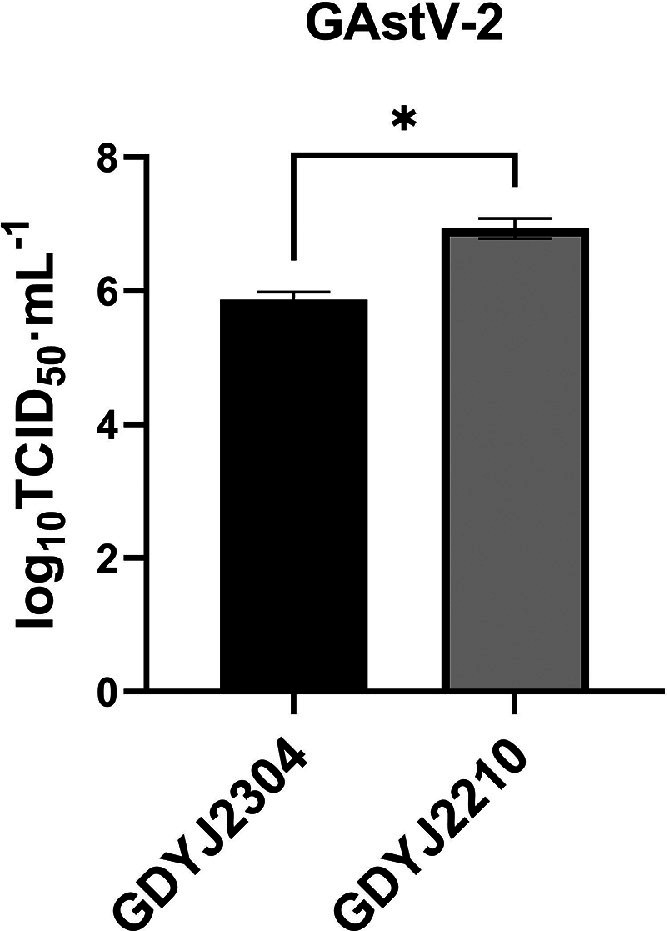
Proliferation ability of GAstV-2 with gene deletion was stronger than that without gene deletion.
Phylogenetic Analysis and Classification of GAstV
The GAstV strain isolated in this study was aligned with other avian astroviruses. The results showed that the GAstV-1 GDFS2209 strain isolated in this study was located in the GAstV-1 branch (Figure 8), with a sequence similarity ranging from 89.70 to 99.00% with the GAstV-1 reference strain. The sequence similarity with the GAstV-2 reference strain was 54.80 to 55.40%. The GAstV-2 GDYJ2210 and GAstV-2 GDYJ2304 strains isolated in this study were located in the GAstV-2 branch, exhibiting a sequence similarity of 54.50 to 55.00% with the GAstV-1 reference strain and 96.80 to 98.90% with the GAstV-2 reference strain. The sequence similarity between GAstV-1 GDFS2209, GAstV-2 GDYJ2210, and GAstV-2 GDYJ2304 strains and other avian astroviruses ranged from 45.90% to 61.60% (Supplementary Table S2).
Figure 8.
Genetic evolution analysis of GAstV complete genome sequence. Black triangles represent GAstV isolated in this study.
DISCUSSION
Since 2016, a highly fatal gouty disease caused by GAstV has been reported in Jiangsu, Anhui, Shandong, and Guangdong provinces of China, with an infection rate and mortality rate of up to 80% and about 50%, respectively (Li et al., 2024; Wei et al., 2024) Up to now, fatal gouty disease caused by GAstV is still widely prevalent in China, which has caused serious economic losses to the goose industry. In order to better understand the epidemic situation of GAstV in Guangdong province, we collected goose samples suspected of GAstV infection from January 2022 to January 2024 for molecular diagnosis, virus isolation and identification, and genetic analysis of GAstV, providing epidemiological data support for strengthening GAstV prevention and control.
Among the 682 samples suspected of being infected with GAstV, all samples tested positive for GAstV by PCR. Co-infection with GAstV-1 and GAstV-2 accounted for the highest proportion and was the main form of infection in clinical practice. Moreover, co-infected tissue samples exhibited more significant urate deposition, indicating more severe goose gout symptoms (Zhou et al., 2024). Additionally, we found that the positive rate of GAstV in autumn and winter was relatively higher than that in other seasons, which may be related to public health measures such as reduced goose resistance due to cold climate conditions or poor ventilation caused by indoor insulation in winter. This parallels the epidemic characteristics of other viral diseases such as influenza (Xu et al., 2023c).
The detection results of GAstV in geese at different ages indicated a gradual decrease in the number of infected geese with age. The highest proportion of GAstV infection was observed in goslings aged 1-30 days, accounting for 79.18% (540/682), suggesting their susceptibility to GAstV. Geese aged 51-80 days represented 5.13% (35/682) of GAstV-positive samples. Additionally, cases of GAstV infection were found in adult geese over 80-days-old, with the oldest being 119 days old. A previous study reported GAstV infection and gout in 70-day-old geese (Zhu et al., 2022b), consistent with our findings, further supporting that geese of different ages can be susceptible to GAstV.
Three GAstV strains were isolated and identified from tissue samples using LMH cells and goose embryos. The GDFS2209 strain was identified as GAstV-1 through phylogenetic tree and genetic similarity analysis, while GDYJ2210 and GDYJ2304 belonged to GAstV-2. The genetic similarity between GAstV-1 and GAstV-2 was only 54.50%-55.50%. Currently, there remains a lack of an efficient and standardized in vitro culture system for GAstV, significantly hindering research progress. Previous studies reported that GAstV-1 strains ZJC14 (MZ819185) and TZ03 (MW353015) could propagate in goose embryos but not in LMH cells (Wang et al., 2021). The GAstV-2 ZJLD20 strain (MZ819184) could proliferate in LMH cells but not in embryos. Similarly, the GAstV-1 GDFS2209 strain in this study could adapt to goose embryos but not to LMH cells. Moreover, the GAstV-2 GDYJ2210 and GDYJ2304 strains could proliferate stably in LMH cells but not effectively in goose embryos, suggesting differences in the adaptability of different GAstV genotypes to various culture systems. Additionally, spherical virions with a diameter of 25 nm were observed in LMH cells in this study, consistent with the size and morphology of GAstV virions previously reported in LMH cells (Zhu et al., 2022a).
Interestingly, we found that the GAstV-2 GDYJ2210 strain lacks 3 consecutive bases in the gene sequence compared with the GAstV-2 GDYJ2304 strain and other GAstV-2 reference strains. These three bases were located in the N-terminal region of ORF2, which did not cause a frameshift mutation and deleted exactly one amino acid. Moreover, the proliferation ability of the mutant strain was significantly higher than that of the wild-type strain in LMH cells. ORF2 encodes the viral capsid protein, which exhibits the largest diversity in the whole genome(Smyth et al, 2012). It is involved in the recognition of related receptors on the cell surface and the body's immune response and is closely related to the invasion and replication of viruses (Ren et al., 2020). The virulence and pathogenicity of the deletion of these three clips are currently unknown, but further studies are planned. However, recent studies have reported a significant increase in the morbidity and mortality of gouty disease caused by GAstV in goslings (Xu et al., 2024), which may be related to the gene sequence deletion at this position.
In conclusion, this study collected cases of suspected GAstV infection over the past 2 yr, all of which tested positive for the GAstV molecule. Among them, co-infection of GAstV-1 and GAstV-2 was predominant, with a higher positive rate observed in autumn and winter. Furthermore, a gene deletion was identified in the GAstV-2 gene sequence, which was associated with enhanced replication ability. These findings underscore the importance of long-term monitoring of GAstV's molecular epidemiological characteristics and the need for intensified research into its pathogenesis. This study contributes to a better understanding of the epidemiological characteristics and genetic heterogeneity of GAstV in Guangdong province, laying a foundation for the development of effective measures to protect waterfowl from GAstV infection.
DISCLOSURES
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
This study was supported by the National Key R&D Program of China (2022YFD1801000), Special Fund for Scientific Innovation Strategy–Construction of High Level Academy of Agriculture Science (202110TD, R2020PY-JX014, R2020QD-049, R2020PY-JC001), open competition program of the top ten critical priorities of Agricultural Science and Technology Innovation for the 14th Five-Year Plan of Guangdong Province (2022SDZG02), Talent Introduction Project for Excellent PhD of Guangdong Academy of Agricultural Sciences (R2023YJ-YB2001).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2024.104143.
Appendix. Supplementary materials
REFERENCES
- Chen H., Zhang B., Yan M., Diao Y., Tang Y. First report of a novel goose astrovirus outbreak in Cherry Valley ducklings in China. Transbound. Emerg. Dis. 2020;67:1019–1024. doi: 10.1111/tbed.13418. [DOI] [PubMed] [Google Scholar]
- Chen Q., Xu X., Yu Z., Sui C., Zuo K., Zhi G., Ji J., Yao L., Kan Y., Bi Y., Xie Q. Characterization and genomic analysis of emerging astroviruses causing fatal gout in goslings. Transbound. Emerg. Dis. 2020;67:865–876. doi: 10.1111/tbed.13410. [DOI] [PubMed] [Google Scholar]
- Cortez V., Meliopoulos V.A., Karlsson E.A., Hargest V., Johnson C., Schultz-Cherry S. Astrovirus Biology and Pathogenesis. Annu. Rev. Virol. 2017;4:327–348. doi: 10.1146/annurev-virology-101416-041742. [DOI] [PubMed] [Google Scholar]
- Koci M.D., Schultz-Cherry S. Avian astroviruses. Avian Pathol. 2002;31:213–227. doi: 10.1080/03079450220136521. [DOI] [PubMed] [Google Scholar]
- Krishna N.K. Identification of structural domains involved in astrovirus capsid biology. Viral Immunol. 2005;18:17–26. doi: 10.1089/vim.2005.18.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Luo J., Shang J., Zhang F., Deng C., Feng Y., Meng G., Jiang W., Yu X., Liu H. Epidemiological investigation and pathogenicity analysis of waterfowl astroviruses in some areas of China. Front. Microbiol. 2024;15 doi: 10.3389/fmicb.2024.1375826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Li L., Dong J., Zhang J., Huang Y., Zhai Q., Xiang Y., Jin J., Huang X., Wang G., Sun M., Liao M. Global analysis of gene expression profiles and gout symptoms in goslings infected with goose astrovirus. Vet. Microbiol. 2023;279 doi: 10.1016/j.vetmic.2023.109677. [DOI] [PubMed] [Google Scholar]
- Niu X., Tian J., Yang J., Jiang X., Wang H., Chen H., Yi T., Diao Y. Novel goose astrovirus associated gout in Gosling, China. Vet. Microbiol. 2018;220:53–56. doi: 10.1016/j.vetmic.2018.05.006. [DOI] [PubMed] [Google Scholar]
- Peng Z., Gao D., Song X., Huang H., Zhang X., Jiang Z., Qiao H., Bian C. Isolation and genomic characterization of one novel goose astrovirus causing acute gosling gout in China. Sci. Rep. 2023;13:10565. doi: 10.1038/s41598-023-37784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Li T., Zhang X., Yao X., Gao W., Xie Q., Zhang J., Shao H., Wan Z., Qin A., Ye J. OASL triggered by novel goose astrovirus via ORF2 restricts its replication. J. Virol. 2020;94 doi: 10.1128/JVI.01767-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth V.J., Todd D., Trudgett J., Lee A., Welsh M.D. Capsid protein sequence diversity of chicken astrovirus. Avian Pathol. 2012;41:151–159. doi: 10.1080/03079457.2011.652938. [DOI] [PubMed] [Google Scholar]
- Wang A., Xie J., Wu Z., Liu L., Wu S., Feng Q., Dong H., Zhu S. Pathogenicity of a goose astrovirus 2 strain causing fatal gout in goslings. Microb. Pathog. 2023;184 doi: 10.1016/j.micpath.2023.106341. [DOI] [PubMed] [Google Scholar]
- Wang A.P., Zhang S., Xie J., Gu L.L., Wu S., Wu Z., Liu L., Feng Q., Dong H.Y., Zhu S.Y. Isolation and characterization of a goose astrovirus 1 strain causing fatal gout in goslings. China. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F., He D., Wu B., Diao Y., Tang Y. Isolation, Identification, and Pathogenicity of a goose astrovirus genotype 1 strain in goslings in China. Viruses. 2024;16:541. doi: 10.3390/v16040541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Gao L., Zhu P., Chen S., Chen Z., Yan Z., Lin W., Yin L., Javed M.T., Tang Z., Chen F. Isolation, identification, and pathogenicity analysis of newly emerging gosling astrovirus in South China. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1112245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Jiang B., Cheng Y., Gao Z., He Y., Wu Z., Wang M., Jia R., Zhu D., Liu M., Zhao X., Yang Q., Wu Y., Zhang S., Huang J., Ou X., Gao Q., Sun D., Cheng A., Chen S. Molecular epidemiology and virulence of goose astroviruses genotype-2 with different internal gene sequences. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1301861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Wu Z., He Y., Jiang B., Cheng Y., Wang M., Jia R., Zhu D., Liu M., Zhao X., Yang Q., Wu Y., Zhang S., Huang J., Ou X., Sun D., Cheng A., Chen S. Molecular characterization of a virulent goose astrovirus genotype-2 with high mortality in vitro and in vivo. Poult. Sci. 2024;103 doi: 10.1016/j.psj.2024.103585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Tang L., Gu X., Bo S., Ming L., Ma M., Zhao C., Sun K., Liu Y., He G. Characterization of avian influenza A (H4N2) viruses isolated from wild birds in Shanghai during 2019 to 2021. Poult. Sci. 2023;102 doi: 10.1016/j.psj.2023.102948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Tian J., Tang Y., Diao Y. Isolation and genomic characterization of gosling gout caused by a novel goose astrovirus. Transbound. Emerg. Dis. 2018;65:1689–1696. doi: 10.1111/tbed.12928. [DOI] [PubMed] [Google Scholar]
- Zhang F., Li H., Wei Q., Xie Q., Zeng Y., Wu C., Yang Q., Tan J., Tan M., Kang Z. Isolation and phylogenetic analysis of goose astrovirus type 1 from goslings with gout in Jiangxi province. China. Poult. Sci. 2022;101 doi: 10.1016/j.psj.2022.101800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Cao Y., Wang J., Fu G., Sun M., Zhang L., Meng L., Cui G., Huang Y., Hu X., Su J. Isolation and characterization of an astrovirus causing fatal visceral gout in domestic goslings. Emerg. Microbes Infect. 2018;7:71. doi: 10.1038/s41426-018-0074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ren D., Li T., Zhou H., Liu X., Wang X., Lu H., Gao W., Wang Y., Zou X., Sun H., Ye J. An emerging novel goose astrovirus associated with gosling gout disease. China. Emerg Microbes Infect. 2018;7:152. doi: 10.1038/s41426-018-0153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Cui Y., Wang C., Wu H., Xiong H., Qi K., Liu H. Characterization of natural co-infection with goose astrovirus genotypes I and II in gout affected goslings. Avian Pathol. 2024;53:146–153. doi: 10.1080/03079457.2023.2295341. [DOI] [PubMed] [Google Scholar]
- Zhu M., Guo Z., Xu H., Li X., Chen H., Cao R., Lv Y. Aminoguanidine alleviates gout in goslings experimentally infected with goose astrovirus-2 by reducing kidney lesions. Poult Sci. 2024;103 doi: 10.1016/j.psj.2024.103484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Miao Y., Wang J., Bai W., Yang X., Yu S., Guo D., Sun D. Isolation, identification, and pathogenicity of a goose astrovirus causing fatal gout in goslings. Vet. Microbiol. 2022;274 doi: 10.1016/j.vetmic.2022.109570. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Wang H., Hua J., Ye W., Chen L., Ni Z., Yun T., Ma J., Yao H., Bao E., Zhang C. Isolation and pathogenicity of a novel goose astrovirus from overfed adult landaise geese in China. Viruses. 2022;14:2806. doi: 10.3390/v14122806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.