Abstract
Background
Triple-negative breast cancer (TNBC) is an aggressive subtype of breast cancer with a poor prognosis particularly in the metastatic setting. Treatments with anti-programmed cell death protein-1/programmed death-ligand 1 (PD-L1) immune checkpoint inhibitors (ICI) in combination with chemotherapies have demonstrated promising clinical benefit in metastatic TNBC (mTNBC) but there is still an unmet need, particularly for patients with PD-L1 negative tumors. Mechanisms of resistance to ICIs in mTNBC include the presence of immunosuppressive tumor-associated macrophages (TAMs) in the tumor microenvironment (TME). Eganelisib is a potent and selective, small molecule PI3K-γ inhibitor that was shown in preclinical studies to reshape the TME by reducing myeloid cell recruitment to tumors and reprogramming TAMs from an immune-suppressive to an immune-activating phenotype and enhancing activity of ICIs. These studies provided rationale for the clinical evaluation of eganelisib in combination with the anti-PD-L1 atezolizumab and nab-paclitaxel in firstline mTNBC in the phase 2 clinical trial MAcrophage Reprogramming in Immuno-Oncology-3 (MARIO-3, NCT03961698). We present here for the first time, in-depth translational analyses from the MARIO-3 study and supplemental data from eganelisib monotherapy Ph1/b study in solid tumors (MARIO-1, NCT02637531).
Methods
Paired pre-treatment and post-treatment tumor biopsies were analyzed for immunophenotyping by multiplex immunofluorescence (n=11), spatial transcriptomics using GeoMx digital spatial profiling (n=12), and PD-L1 immunohistochemistry, (n=18). Peripheral blood samples were analyzed using flow cytometry and multiplex cytokine analysis.
Results
Results from paired tumor biopsies from MARIO-3 revealed gene signatures of TAM reprogramming, immune activation and extracellular matrix (ECM) reorganization. Analysis of PD-L1 negative tumors revealed elevated ECM gene signatures at baseline that decreased after treatment. Gene signatures of immune activation were observed regardless of baseline PD-L1 status and occurred in patients having longer progression-free survival. Peripheral blood analyses revealed systemic immune activation.
Conclusions
This is the first report of translational analyses including paired tumor biopsies from a phase 2 clinical study of the first-in-class PI3K-γ inhibitor eganelisib in combination with atezolizumab and nab-paclitaxel in frontline mTNBC. These results support the mechanism of action of eganelisib as a TAM-reprogramming immunotherapy and support the rationale for combining eganelisib with ICI and chemotherapy in indications with TAM-driven resistance to ICI.
Keywords: Immune Checkpoint Inhibitors, Macrophage, Breast Cancer, Immunotherapy, Tumor microenvironment - TME
WHAT IS ALREADY KNOWN ON THIS TOPIC
Reprogramming tumor-associated macrophages (TAMs) through inhibition of PI3K-γ is a promising therapeutic approach that has been extensively studied and validated in the preclinical setting including syngeneic models of myeloid-rich breast cancer where the loss of PI3K-γ enhances the effects of chemotherapy and overcomes resistant to immune checkpoint therapy.
WHAT THIS STUDY ADDS
This study is the first report of translational analyses from a phase 2 clinical study of the first-in-class PI3K-γ inhibitor eganelisib, in combination with immune checkpoint therapy (atezolizumab) and chemotherapy (nab-paclitaxel) in 1 L metastatic triple-negative breast cancer (mTNBC). Translational analyses revealed evidence of TAM reprogramming and immune activation consistent with that previously characterized in a preclinical setting and further demonstrated immune activation regardless of programmed death-ligand 1 (PD-L1) tumor status.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
The immune activation observed in a subset of PD-L1 negative tumors is intriguing given the unmet need in this patient population. These data support the rationale for further evaluation of PI3K-γ inhibition in combination with immune checkpoint therapy and chemotherapy in patients with mTNBC regardless of PD-L1 status and potentially more broadly in oncology indications where TAMs are a factor of resistance to therapy.
Background
Triple-negative breast cancer (TNBC), characterized by negative expression of estrogen, progesterone and human epidermal growth factor receptor-2, represents 15–20% of breast cancers and is the most aggressive subtype.1 Approximately 46% of patients with TNBC develop distant metastasis where the median survival is only 13.3 months.2 Treatment options for frontline metastatic TNBC (mTNBC) historically included only chemotherapy, with recent advances including immunotherapy, PARP inhibitors and antibody–drug conjugates. Anti-programmed cell death protein-1 (PD-1)/programmed death-ligand 1 (PD-L1) immune checkpoint inhibitors (ICIs) have shown promising activity when combined with chemotherapy in patients with mTNBC whose tumors are PD-L1 positive, with improved progression-free and overall survival compared with chemotherapy alone.3 4 Despite this progress, there is still a need for improvement in frontline mTNBC treatment, particularly in patients with PD-L1 negative tumors, which represent the majority of patients with mTNBC.
One important mechanism of resistance to ICIs is the presence of immunosuppressive cells in the tumor microenvironment (TME), including T regulatory cells, myeloid-derived suppressor cells (MDSCs) and tumor-associated macrophages (TAMs).5 Among these, TAMs are the most abundant and correlate with poor prognosis across many cancer types, including TNBC.6 As a result, several therapeutic approaches targeting TAMs have been developed and evaluated in the clinic, including approaches to suppress, block trafficking or, more recently, reprogram TAMs.7 8 TAM reprogramming seeks to leverage macrophages plasticity to turn immunosuppressive, tumor-supporting M2-like TAMs into immune-activating, tumor-suppressing M1-like TAMs.9 Suppressing the trafficking of MDSCs to the TME and reprogramming TAMs through inhibition of PI3K-γ is a promising therapeutic approach that has been extensively studied and validated in the preclinical setting including models of TNBC.10,12 PI3K-γ, selectively expressed in leukocytes and preferentially within myeloid cells, promotes mammalian target of rapamycin (mTOR)-dependent C/EBPb activation while blocking NFkB signaling resulting in gene transcription of immunosuppressive factors such as TGF-β1, Arg1 and IL-10.1013,16 PI3K-γ blockade reverses these effects, reshaping the TME toward an immune-activated, anti-tumor state characterized by macrophage reprogramming, T-cell activation and infiltration and enhanced gene signatures of IFN-γ signaling and antigen presentation.10,1214
Eganelisib (IPI-549) is a first-in-class, oral, potent and highly selective PI3K-γ inhibitor developed as a macrophage reprogramming cancer immunotherapy with the potential to treat a broad range of cancers.17 The addition of eganelisib enhanced responses to ICIs (eg, anti−PD-1, anti−PD-L1, anti−cytotoxic T-lymphocytes-associated protein 4 (CTLA-4) antibodies) in multiple syngeneic tumor models and overcame resistance to ICIs in myeloid-rich ICI-refractory tumor models including a syngeneic TNBC model.10 11 Further, a combination of eganelisib to chemotherapy (eg, nanoparticle albumin-bound paclitaxel) and anti−PD-1 antibody led to improved survival compared with either chemotherapy alone or chemotherapy plus anti−PD-1 in a spontaneous syngeneic TNBC model.18 Together these data support the rationale for exploring the combination of eganelisib and ICI therapy with or without chemotherapy in the clinic.
Cohort A of MAcrophage Reprogramming in Immuno-Oncology (MARIO)-3 is a single-arm phase 2 clinical study designed after IMpassion130 to evaluate the activity of eganelisib in combination with atezolizumab/nab-paclitaxel in both PD-L1 positive and PD-L1 negative subgroups in front line mTNBC patients (NCT03961698).4 19 Preliminary efficacy data at a median duration of follow-up of 9.9 months revealed median progression-free survival (PFS) of 11.0 months in the PD-L1 positive subgroup (n=14) and 7.3 months in the PD-L1 negative subgroup (n=27).19 Although cross-trial comparisons are only hypothesis-generating, this data is encouraging compared with the historical IMpassion130 benchmark with median progression free survival (mPFS) of 7.5 months and 5.6 months in PD-L1 positive (n=185) and PD-L1 negative (n=266) subgroups, respectively.4 Future analyses with longer follow-up will be required to further characterize the triplet combination of eganelisib and atezolizumab/nab-paclitaxel in PD-L1 positive and PD-L1 negative mTNBC.
Here, we report translational findings from MARIO-3 including paired pre-treatment and post-treatment tumor biopsy analyses demonstrating immune activation in both PD-L1 negative and PD-L1 positive tumor types and supporting a role of eganelisib as a TAM reprogramming immunotherapy within the triple regimen. Peripheral blood analyses from MARIO-1 eganelisib monotherapy provide further support for the mechanism of drug action.
Materials and methods
MARIO-3 (NCT03961698)
MARIO-3 is a single-arm phase 2 study of eganelisib (30 mg once daily) in combination with standard dosing and regimen of atezolizumab and nab-paclitaxel in 62 patients with inoperable locally advanced or mTNBC. Radiographic evaluations and Response Evaluation Criteria in Solid Tumors (RECIST) V.1.1 assessments were conducted every 8 weeks for 12 months and then at least every 12 weeks until confirmed progression of disease, alternate anticancer therapy, or withdrawal of consent. Median PFS was determined in months, censoring on the last tumor assessment (April 14, 2023 datacut). Peripheral blood, and serum were collected pre-dose at cycle 1 day 1 (C1D1), cycle 1 day 15±2 days (C1D15) and day 1 of each 28-day cycle≥C2D1±3 days. Biopsies were collected at day 0 and 2 months post-treatment.
MARIO-1 (NCT02637531)
MARIO-1 is a phase 1/1b first-in-human study evaluating the safety and tolerability of once-daily eganelisib as monotherapy and in combination with nivolumab in patients with advanced solid tumors.20 A total of 39 patients were treated with eganelisib monotherapy in study MARIO-1 which consisted of a 3+3 dose escalation ranging from 10 mg to 60 mg (n=19) and a 60 mg monotherapy dose expansion phase (n=20).20 Peripheral blood, and serum were collected pre-dose at C1D1, C1D8, and day 1 of each 28-day cycle≥C2D1.
Digital spatial profiling of tumor biopsy FFPE tissues
Formalin-fixed paraffin-embedded (FFPE) tissue from 12 paired biopsies (pre-treatment and 2 months on-treatment) was analyzed for spatially resolved profiling of 1800 RNA targets from the cancer transcriptome atlas panel using NanoString’s GeoMx digital spatial profiling (DSP) platform performed at Canopy Biosciences, Hayward, California, USA (Bruker Nano, Billerica, Massachusetts, USA). Fluorescent labeled antibodies targeting CD68 (Santa Cruz catalog#SC-20060 AF594), CD45 and PanCK (NanoString morphology kit catalog#GMX-RNA-MORPH-HST-12) were used to identify the spatial location of macrophages, leukocytes, and tumor cells, respectively. Photocleavable RNA probes were released from each cellular segment by ultraviolet (UV) light and sequenced using next-generation sequencing (NGS) readout with an Illumina NovaSeq 6000. For each 5-micron slide, 4 regions of interest (∼450,000 mm2 diameter) were selected and segmented into CD45+, CD68+and PanCK+areas for collection. For analysis, NGS counts were normalized as recommended by NanoString using Q3 normalization (third quartile normalization). Linear mixed effects models were used to characterize changes in gene expression between pre-treatment and on-treatment samples. Gene set enrichment analysis using the Reactome pathway database identified changes in pathways related to treatment. The R package visNetwork was used to visualize the relationship between differentially expressed genes and pathways.
Multiplex immunofluorescence
FFPE samples were processed and analyzed with the MultiOmyx platform at NeoGenomics (Fort Meyers, Florida, USA) using sequential rounds of staining with Cy3-conjugated antibodies and Cy5-conjugated antibodies targeting human leukocyte antigen DR (HLA-DR) (Novus catalog#NB100-64358), CD68 (BioLegend catalog#916104), CD8 (Dako catalog#M7103), Granzyme B (Dako catalog#M7235) and PanCK (Sigma-Aldrich catalog#C5992, Abcam Catalog#ab213135). Cell densities were measured as mean cell counts per area across∼20 regions of interest (0.832 mm × 0.702 mm). The ratio of HLA-DR+CD68+ to HLA-DR-CD68+macrophages was calculated using mean cell counts for each measure taken from the same region of interest. Images were collected using IN Cell Analyzer 2200 Bioimager (GE Healthcare).
PD-L1 immunohistochemistry
FFPE samples were processed and analyzed by CellCarta (Naperville, Illinois, USA) for PD-L1 immunohistochemistry analysis using Ventana SP142 antibody. Determination of PD-L1 status was evaluated based on immune cell score (IC) defined as the proportion of tumor area occupied by PD-L1 tumor-infiltrating immune cells of any intensity, with a cut-off for PD-L1 positive≥1% IC.
Peripheral blood analyses
Results
Study design
Translational analyses were performed on peripheral blood and paired tumor biopsy samples collected from a single-arm phase 2 study of eganelisib in combination with atezolizumab and nab-paclitaxel (MARIO-3). Peripheral blood analyses from MARIO-3 are reported for patients having a full set of day 0, day 15, and day 28 post-treatment samples and included multiplex cytokine analysis (n=45) and flow cytometry immunophenotyping (n=43). A total of 29 paired biopsies (within 3 months prior to the first dose and week 8±7 days) were collected from MARIO-3, 21 of which were evaluable by one or more of the translational analyses reported here. To support the role of eganelisib within the triplet regimen, additional peripheral blood analyses were performed with samples collected from patients treated with eganelisib monotherapy in a phase 1/1b dose escalation/expansion study (MARIO-1). The 30 mg, 40 mg and 60 mg dose groups were combined for monotherapy analyses based on pharmacokinetic (PK)/pharmacodynamic (PD) data showing potent PI3K-γ inhibition throughout the dosing intervals.20
Peripheral blood analyses reveal systemic immune activation on treatment
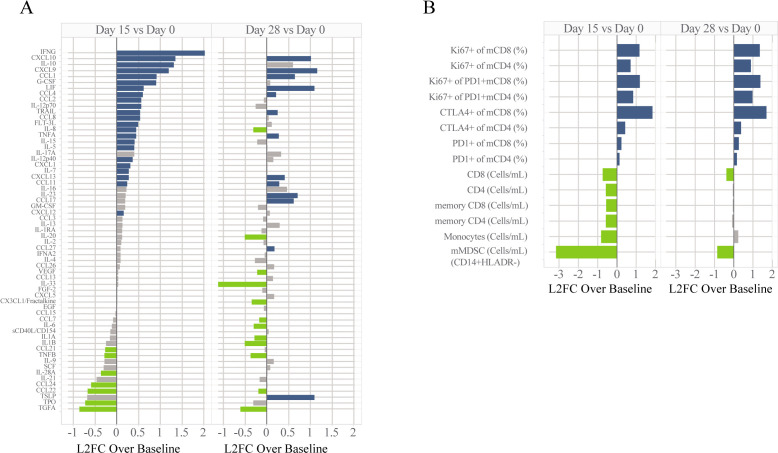
Multiplex cytokine and chemokine quantitative analysis performed on serum collected from MARIO-3 patients revealed an increase in immune-activating chemokines including IFNG, CXCL9, CXCL10, IL-12p70 and IL-15, and a decrease in immune suppressive chemokines including CCL22 and TGFA at day 15 post-treatment with the triplet regimen (figure 1A).21,23 The anti-inflammatory factor IL-10 was also upregulated post-treatment, potentially as an immune dampening mechanism. Some effects were transient in nature with the peak activity observed at the early timepoints including upregulation of IFNG and IL12p70 whereas others were sustained through day 28 including upregulation of CXCL9 and CXCL10 among others (figure 1A).
Figure 1. Peripheral blood analyses for MARIO-3. Analytes for cytokine (A) and immune cell subsets (B) determined by Luminex and flow cytometry, respectively are expressed as log2FC over baseline for samples collected at day 15 and day 28 after treatment. Absolute cell counts for monocytic MDSCs (HLAR-CD14+CD11b+CD33+), monocytes (HLADR+CD14+CD11b+CD33b+), CD8+T cells, CD4+T cells, memory CD8 T-cells (mCD8; CD45RA-CD8+CD3+), memory CD4 T cells (mCD4; CD45RA-CD4+CD3+) and subpopulations including per cent of memory CD8/CD4 T-cells expressing exhaustion markers PD-1 and CTLA-4, proliferation (Ki67+) of memory CD8/CD4 T-Cells, and proliferation (Ki67+) of exhausted (PD1+) memory CD8/CD4 T-Cells are shown (B). One-way repeated measures ANOVA was performed with colors representing significance and direction of change; blue: significant upregulation, green: significant downregulation, gray: not significant. For cytokine analysis, p-values were adjusted using Bonferroni corrections for multiple comparisons. ANOVA, analysis of variance; CTLA-4, cytotoxic T-lymphocytes-associated protein 4; MARIO-3, MAcrophage Reprogramming in Immuno-Oncology-3; MDSCs, myeloid-derived suppressor cells; PD-1, programmed cell death protein-1.
Flow cytometry analyses of peripheral blood samples collected from MARIO-3 showed systemic immune activation characterized by a decrease in immunosuppressive monocytic MDSCs (mMDSC) and an increase in activated (Ki67+) memory T-cells at day 15 and day 28 post-treatment compared with pre-treatment baseline (figure 1B). While a transient reduction of monocytes (CD14+HLADR+) was observed, the reduction of mMDSCs (CD14+HLADR−) was of greater magnitude and duration compared with monocytes (figure 1B). A modest transient reduction in CD4, CD8, memory CD4 and memory CD8 cells was observed, likely a consequence of chemotherapy-mediated lymphodepletion (figure 1B). However, the remaining T-cells showed evidence of activation with an upregulation of proliferating (Ki67+) memory CD4/CD8 cells (figure 1B). Importantly, increases in the percentage of proliferating PD1+memory CD8/CD4 cells at day 15 and day 28 post-treatment were observed, consistent with the reinvigoration of exhausted memory T-cells (figure 1B). The percentage of memory CD8 and memory CD4 T-cells expressing CTLA-4 was increased on day 15 and day 28 after treatment with the largest effect on memory CD8 cells (figure 1B). A modest increase in the percentage of mCD4 and mCD8 cells expressing PD-1 was also observed.
Importantly, peripheral blood analyses from eganelisib monotherapy dose escalation/expansion in patients with advanced solid tumors (MARIO-1)20 showed similar upregulation of CXCL9, downregulation of CCL22, proliferation of memory CD8+T cells and reinvigoration of PD-1+memory CD8+ T cells and a trend toward increased CXCL10 and reduced mMDSCs at day 28 post-treatment (online supplemental figure 1).
Immunofluorescence analysis of paired biopsies reveals macrophage activation and T-cell infiltration in PD-L1 positive and PD-L1 negative tumors
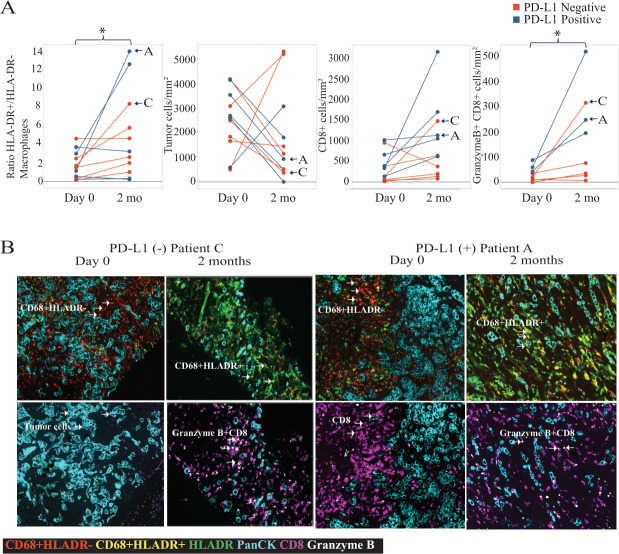
To investigate treatment-related changes to the TME, MultiOmyx immunofluorescence analysis was performed on pre-treatment and post-treatment paired tumor biopsies collected from MARIO-3. Cell densities for distinct immune populations were quantified from 11 paired biopsies (figure 2A). An increase in the ratio of HLA-DR+activated macrophages to HLA-DR− immunosuppressive macrophages as well as an increase in the density of CD8+T cells and activated (Granzyme B+) CD8+T cells was observed in paired biopsies including both PD-L1 negative and PD-L1 positive tumor types after 2-month treatment with eganelisib+atezolizumab + nab-paclitaxel (figure 2A). Representative images from a PD-L1 negative (patient C) and PD-L1 positive (patient A) tumor biopsy pair are shown (figure 2B). Micrographs from the PD-L1 negative tumor (patient C) demonstrate an increase in the ratio of HLA-DR+to HLA-DR− macrophages (yellow), and density of CD8+T cells (purple) and activated Granzyme B+CD8+ T cells (white) after treatment. Micrographs from the PD-L1 positive tumor (patient A) show high baseline CD8+T cells (purple) and an increase in the density of activated (Granzyme B+) CD8+T cells (white) and the ratio of HLA-DR+to HLA-DR− macrophages (yellow) after treatment (figure 2B).
Figure 2. Immunofluorescence analysis of immune cell subsets from MARIO-3 paired tumor biopsies. FFPE tissue collected from pre-treatment (day 0) and on-treatment (2 months) biopsies (n=11) stained using MultiOmyx are quantified using the mean of∼20 ROIs and color coded according to baseline PD-L1 status (PD-L1 negative (red); PD-L1 positive (blue)) (A). Representative images from a PD-L1 negative (patient C) and PD-L1 positive (patient A) tumor biopsy pair are shown to visualize TAM reprogramming (CD68+HLA-DR− (red), CD68+HLA-DR+ (yellow), and density of CD8 T-Cells (purple), Granzyme B+CD8+ T Cells (white) and tumor PanCK+ (cyan) cells. Post treatment changes were characterized using one-way ANOVA *p<0.05. ANOVA, analysis of variance; FFPE, formalin-fixed paraffin-embedded; ROI, region of interest; PD-L1, programmed death-ligand 1; TAM, tumor-associated macrophage.
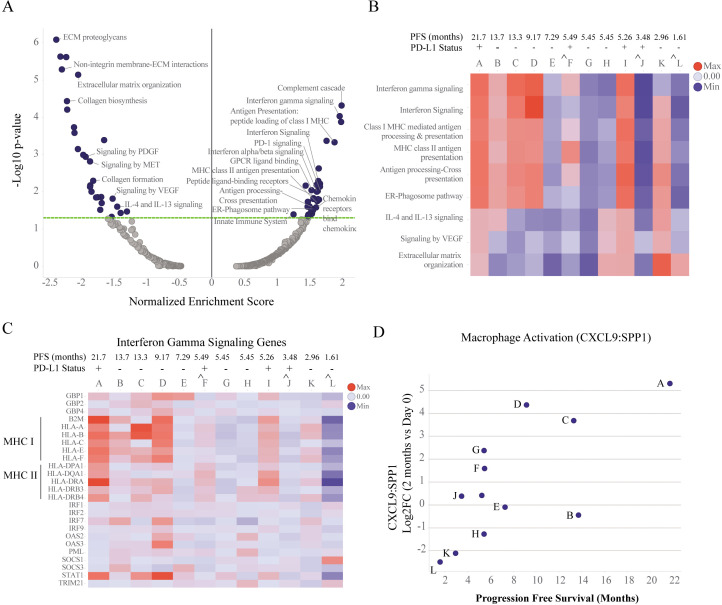
Digital spatial profiling of paired biopsies reveals gene signatures of immune activation and ECM reorganization in leukocytes-enriched regions of the TME
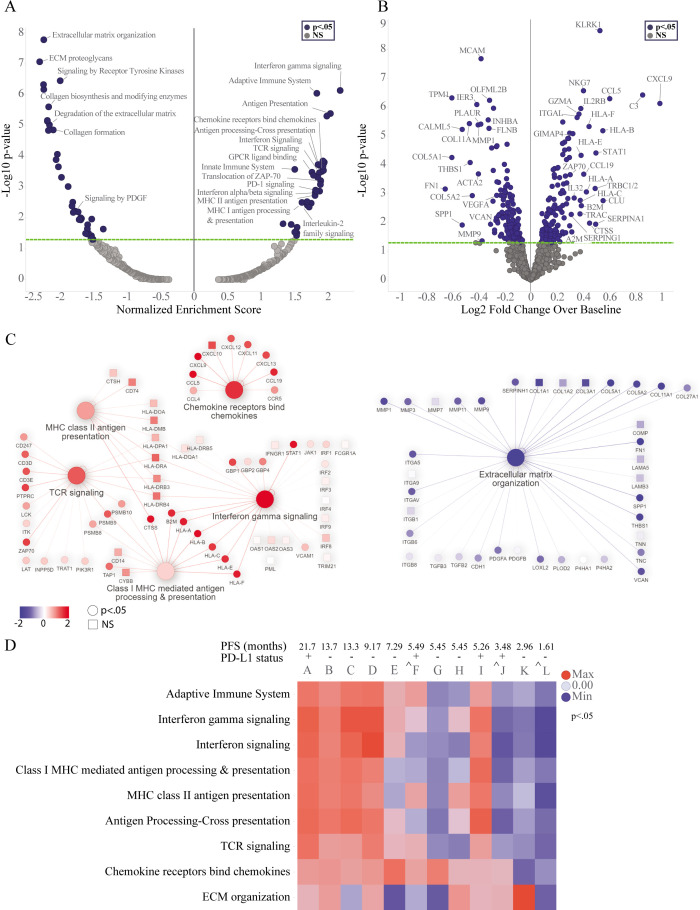
To identify gene expression changes within specific cell types of the TME including leukocytes (CD45+) and macrophages (CD68+), DSP was performed on paired biopsies collected at baseline and 2 months after treatment with eganelisib + atezolizumab + nab-paclitaxel from MARIO-3. DSP was performed using the Cancer Transcriptome Atlas panel representing 1800 RNA targets and 55 pathways critical to immune response, tumor biology and the microenvironment. Reactome pathway analysis (figure 3A) and differential gene expression (DEG) (figure 3B, online supplemental table 2) were performed on the collective group of paired biopsies (n=12). Enrichment of pathways associated with immune activation including the adaptive immune system, interferon-γ signaling, antigen presentation, chemokines and TCR signaling, and suppression of pathways associated with extracellular matrix organization (ECM) were observed after treatment (figure 3A). The strongest upregulated genes, based on log2Fold change and significance included CXCL9 and CCL5 involved in T-cell chemotaxis, complement component C3, and KLRK1, a gene expressed on activated immune cells including natural killer (NK)-cells, T-cells and macrophages (figure 3B, online supplemental table 2). The strongest downregulated genes included ECM proteins SPP1, FN1, COL5A1, COL11A1 and TPM1 (figure 3B, online supplemental table 2). Gene expression for gene sets contributing to the core enrichment of select pathways from the group analysis are represented by network plots (figure 3C). Immune-related pathways were highly interconnected. Upregulated genes that contributed to the core enrichment of key pathways included interferon regulated genes (STAT1, GBP1, GBP2, GPB4), major histocompatibility complex (MHC) I antigen presentation (B2M, HLA-A, HLA-B, HLA-C, HLA-E, HLA-F, TAP1), MHC II antigen presentation genes (CTSS, CD74, HLA-DRA, HLA-DMB), chemokines (CXCL9, CXCL10, CXCL11, CXCL12, CXCL13, CCL4, CCL5, CCL19, CCR5), and T-cell receptor signaling (CD3D, CD3E, CD247, PSMB8, PSMB9, PSMB10, ZAP70) (figure 3C, online supplemental table 2) among others. Genes contributing to the core enrichment of the ECM organization pathway included fibrogenic cytokines (PDGFA, PDGFB), integrins (ITGA5, ITGAV, ITGB6, ITGB8), matrix metalloproteinases and other matrix remodeling enzyme systems (MMP1, MMP3, MMP9, MMP11, PLAU, PLAT, PLAUR), collagens (COL5A1, COL5A2, COL11A1, COL27A1), and other ECM proteins (SPP1, FN1, VCAN) (figure 3C, online supplemental table 2). In addition, downregulation of genes associated with angiogenesis (VEGF) and immune suppression including ITGAV, ITGB8, TGFB2, and TGFB3 were also observed (figure 3C, online supplemental table 2).
Figure 3. Digital spatial profiling (DSP) analysis of 12 paired biopsies collected pre-treatment (day 0) and on-treatment (2 months) from MARIO-3 using anti-CD45+antibody guide to mark leukocytes. GSEA with Reactome pathways was performed to characterize changes in pathway expression within leukocyte enriched regions and visualized with volcano plots (A, left panel). Treatment related changes in gene expression were determined using linear mixed effects model and visualized by volcano plots (B, right panel). Network plots were used to visualize connectivity among pathways, direction of change, and significance of genes contributing to the core enrichment of pathways p<0.05 (circles), p≥0.05 (squares). GSEA was performed on paired tumor biopsies from individual patients, and normalized enrichment scores from representative pathways are shown using heat maps, *p<0.05 (D). ˆeganelisib hold with continued atezolizumab/nab-paclitaxel for 2 weeks prior to sample collection. mPFS (RECIST V.1.1) from April 14, 2023 datacut is indicated for each patient (D). ECM, extracellular matrix; GSEA, gene set enrichment analysis; MARIO-3, MAcrophage Reprogramming in Immuno-Oncology-3; PD-1, programmed cell death protein-1; PD-L1, programmed death-ligand 1; PFS, progression-free survival; RECIST, response evaluation criteria in solid tumor; TCR, T-cell receptor.
Individual patient digital spatial profiling analysis reveals immune activation in a subset of patients
To investigate potential relationships between TME changes and clinical outcomes, pathway analyses were performed on individual paired biopsy samples and annotated with PFS (online supplemental table 1). Individual patient analyses revealed that the immune activation signature including the adaptive immune system, interferon-γ signaling and antigen presentation pathways observed in the group analysis (n=12) was primarily driven by patients A–D and I (figure 3D, online supplemental table 3). PFS (RECIST V.1.1) for each patient (A–L) show that patients A–D have the longest PFS of the group. An exploratory analysis of samples collected from patients experiencing transient eganelisib-only dose hold with uninterrupted atezolizumab/nab-paclitaxel for 2 weeks immediately prior to biopsy collection and PFS≤5.49 months (patients F, J, L) showed a lack of enrichment of many immune-related pathways including interferon-gamma and antigen presentation pathways (figure 3D, online supplemental table 1). The reduction in ECM organization was variable across samples (figure 3D). Changes in gene expression for genes contributing to the core enrichment of the adaptive immune system pathway from the group analysis were visualized for individual patient samples using heat maps. Antigen presentation genes including MHC Class I (HLA-A, HLA-B, HLA-C, HLA-E, HLA-F, TAP1) and MHC II (HLA-DMB, HLA_DOA, HLA-DPA1, HLA-DQA1, HLA-DRA, HLA-DRB3, HLA-DRB4, HLA-DRB4, CD74) were among the genes strongly upregulated in patients A–D with longer PFS (online supplemental figure 2).
Increased PD-L1 expression observed by IHC in a subset of paired tumor biopsies
To evaluate PD-L1 dynamics between pre-treatment and post-treatment paired biopsies, PD-L1 status was measured using the SP142 Ventana assay (IC≥1% for PD-L1 positive) in 18 paired biopsies collected from MARIO-3 (online supplemental figure 3). Upregulation of PD-L1 was observed in 67% (12 out of 18) of paired biopsies including 80% of baseline PD-L1 positive (4 out of 5) and 62% of baseline PD-L1 negative (8 out of 13) tumors. Of interest, 6 paired samples changed from PD-L1 negative to PD-L1 positive status post-treatment (IC≥1% cut-off).
DSP analysis reveals immune activation regardless of PD-L1 status and ECM reorganization in PD-L1-negative tumors
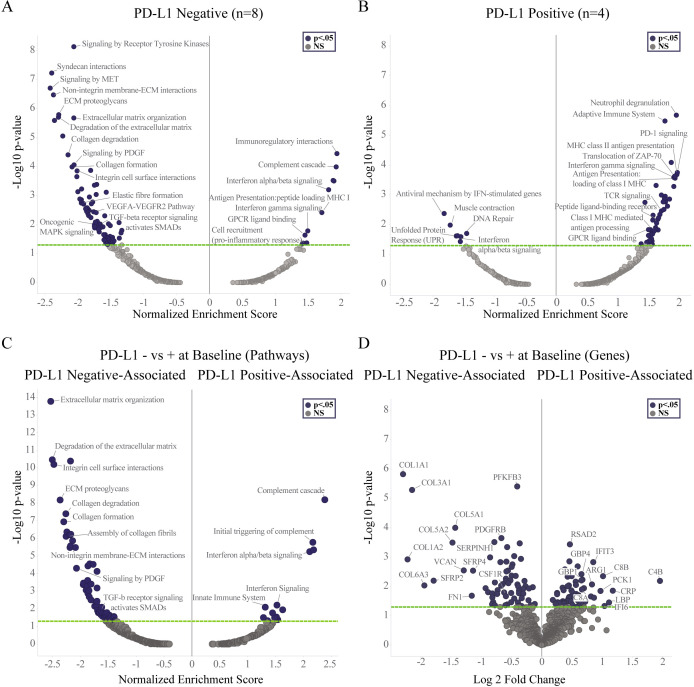
Pathway and DEG analyses from DSP of leukocytes were performed on samples grouped according to baseline PD-L1 status. Upregulation of genes and pathways consistent with immune activation including interferon gamma signaling, antigen presentation, and GPCR ligand binding pathways were observed in both PD-L1 negative and PD-L1 positive tumors (figure 4A,B, online supplemental table 4). Genes and pathways involved in ECM organization and matrix remodeling desmoplasia and metastasis,24,28 including COL1A1, COL3A1, COL5A1, COL5A2, COL11A1, ITGB1, ITGAV, ITGA5, MMP1, MMP3, MMP9, PLAU, PLAUR, MFGE8, FN1, PDGFA, and PDGFRB, were specifically downregulated in the PD-L1 negative, but not PD-L1 positive subgroup (figure 4A,B, online supplemental table 4). To investigate whether differences existed between the TME of PD-L1 negative versus PD-L1 positive subgroups at baseline, DEG and pathway analysis were performed comparing the two subgroups against each other using pre-treatment samples. Pathways and genes associated with the baseline PD-L1 negative subgroup are represented on the left side of the volcano plots with negative normalized enrichment score (NES) and Log2FC values and those associated with baseline PD-L1 positive subgroup are shown on the right side of the volcano plots with positive NES and Log2FC values (figure 4C,D). Genes and pathways related to ECM organization were strongly associated with baseline PD-L1 negative relative to baseline PD-L1 positive subgroups and included genes associated with collagen COL1A1, COL1A2, COL3A1, COL5A1, COL5A2, COL11A1, and matrix remodeling desmoplasia and metastasis (PLAU, PLAUR, ITGAV, ITGA5, FN1) (figure 4C,D, online supplemental table 5). In contrast, genes and pathways associated with baseline PD-L1 positive relative to baseline PD-L1 negative subgroup were primarily involved in complement cascade, interferon and T-cell signaling including C4B, IFIT2, IFIT3, IFI6, GBP1, GBP4, CD8A, and CD8B among others (figure 4C,D, online supplemental table 5). Genes associated with immunosuppressive macrophages were observed in pre-treatment samples from both PD-L1 subgroups with CSF1R and Arg1 associated with PD-L1 negative and PD-L1 positive subgroups, respectively (figure 4D, online supplemental table 5).
Figure 4. Digital spatial profiling (DSP) analysis within CD45+leukocytes in paired biopsies from MARIO-3 grouped according to baseline PD-L1 status. GSEA was performed within leukocyte enriched regions. Treatment related changes are represented with volcano plots for PD-L1 negative subgroup (n=8) (A, left panel) and PD-L1 positive subgroup (n=4) (A, right panel). GSEA (left panel) and DEG (right panel) showing differences between PD -L1 negative and PD-L1 positive subgroups at baseline (Day 0) are shown with volcano plots (B). DEG, differential gene expression; ECM, extracellular matrix; GSEA, gene set enrichment analysis; MARIO-3, MAcrophage Reprogramming in Immuno-Oncology-3; PD-L1, programmed death-ligand 1; PFS, progression-free survival.
Digital spatial profiling within macrophages-enriched regions reveals gene signatures consistent with TAM reprogramming in a subset of paired biopsies
To investigate the effect of treatment on TAM reprogramming, DEG and pathway analysis were performed within macrophage (CD68+) enriched regions using DSP. Like that observed in total leukocytes (figure 3A), upregulation of interferon-gamma signaling, chemokines and antigen presentation and suppression of ECM genes and pathways were observed in macrophage-enriched regions (figure 5A, online supplemental table 6). Unlike that observed in total leukocytes, pathways linked to immunosuppressive macrophages including IL-4/IL-13 and VEGF were observed only in macrophage-enriched regions (figure 5A). Individual patient sample analysis within macrophage-enriched regions revealed that enrichment of pathways associated with TAM reprogramming including interferon-gamma signaling, and MHC Class I and Class II antigen presentation pathways were primarily driven by patients A, C, D and I (figure 5B, online supplemental table 7). Reduction in pathways associated with immune suppression (IL-4/IL-13 and VEGF) and/or ECM was observed in a subset of patients (figure 5B, online supplemental table 7). Changes in gene expression for genes contributing to the core enrichment of the interferon-gamma pathway in the group analysis were visualized on an individual sample basis with heatmaps (figure 5C, online supplemental table 8). Consistent with the pathway analysis, interferon-gamma pathway related genes were upregulated in a subset of patient samples including A, C, D and I and dampened in eganelisib-only dose hold patients F, J and L (figure 5C).
Figure 5. Digital spatial profiling (DSP) analysis of 12 paired biopsies collected pre-treatment (day 0) and on-treatment (2 months) from MARIO-3 using anti-CD68+antibody guide to mark macrophages. GSEA with Reactome pathways was performed and normalized enrichment scores were calculated for macrophage enriched regions. Treatment related changes are represented with volcano plots (A). GSEA was performed on individual paired tumor biopsies, and normalized enrichment scores from representative pathways are shown using heat maps, *p<0.05 (B). Gene expression of genes contributing to the core enrichment of the interferon-gamma signaling pathway identified in the group analysis (n=12) were plotted as log2FC over baseline on an individual sample basis using heat maps (C). Log2FC changes in CXCL9:SPP1 ratio for on-treatment relative to baseline were plotted for individual paired biopsies against PFS (D). ˆeganelisib hold with continued atezolizumab/nab-paclitaxel for 2 weeks prior to sample collection. mPFS (RECIST V.1.1) from April 14, 2023 datacut is indicated for each patient (B,C). ECM, extracellular matrix; GSEA, gene set enrichment analysis; MARIO-3, MAcrophage Reprogramming in Immuno-Oncology-3; PD-1, programmed cell death protein-1; PD-L1, programmed death-ligand 1; PFS, progression-free survival; RECIST, response evaluation criteria in solid tumors.
It was recently proposed that the ratio of the expression of CXCL9 and SPP1 (CXCL9:SPP1) can be used to define TAM polarity in several cancers and has a stronger prognostic association than conventional M1-like and M2-like macrophage markers.29 We hypothesized that changes in the CXCL9:SPP1 ratio in macrophage-enriched regions could be used as a measure of TAM reprogramming on treatment with eganelisib combination. Interestingly, the strongest increase in CXCL9:SPP1 ratio was observed in patients A, C and D with prolonged PFS, consistent with the enrichment of TAM reprogramming pathways observed in these samples (figure 5D).
Discussion
While the combination of ICI and chemotherapy has shown encouraging survival benefit over chemotherapy alone for patients with PD-L1 positive tumors, patients do eventually progress so more effective treatment strategies are needed in mTNBC.3 4 In addition, a combination of ICI with chemotherapy does not improve survival for patients with PD-L1 negative tumors, leaving these patients with high unmet needs. Strategies to target resistance mechanisms including immunosuppressive TAMs are explored to realize the full potential of immune therapies, particularly in the PD-L1 negative setting. The TNBC portion of the single-arm, phase 2 MARIO-3 study (NCT03961698) replicated key inclusion/exclusion criteria from the benchmark IMpassion130 study and sought to evaluate the activity of the first-in-class PI3K-γ inhibitor and TAM reprogramming agent eganelisib in combination with atezolizumab/nab-paclitaxel in PD-L1 positive and PD-L1 negative subgroups. Preliminary results from MARIO-3 revealed encouraging mPFS (months) in patients with both PD-L1 positive and PD-L1 negative tumors suggesting a potentially positive, contributory role of eganelisib within the triplet regimen19 (online supplemental table 1).
This is the first report to describe translational findings from the evaluation of eganelisib in combination with atezolizumab and nab-paclitaxel in frontline mTNBC, including analysis from peripheral blood and paired tumor biopsies. In this study, DSP revealed transcriptional changes within macrophage-enriched regions consistent with TAM reprograming to a pro-inflammatory state including enrichment of interferon-gamma signaling, and MHC I and MHC II antigen presentation pathways as well as increased CXCL9:SPP1 ratio in a subset of paired biopsies (figure 5A–D, online supplemental table 6). In an orthogonal approach, TAM reprogramming, defined by an increase in the ratio of HLA-DR+activated macrophages to HLA-DR− suppressive macrophages was also observed (figure 2). In summary, DSP and immunofluorescence analyses both reveal evidence of TAM reprogramming in a subset of paired tumor biopsies from MARIO-3, consistent with eganelisib mechanism of action and preclinical findings.
Immune activation observed in post-treatment biopsies, including enrichment of interferon-gamma signaling, MHC-I and MHC-II antigen presentation and T-cell activation pathways within leukocyte enriched regions (figure 3A) as well as increased CD8+T cell density and Granzyme B+activated CD8+ T cells within the TME (figure 2) are in line with preclinical studies showing immune activation with similar characteristics on PI3K-γ blockade11 13 suggesting eganelisib is contributing to the overall effect. Importantly, our data show a shift to an immune-activated state in tumor biopsies after treatment with the triplet not only in subsets of PD-L1 positive tumors, but also in subsets of PD-L1 negative tumor types using orthogonal approaches (figures2 4). Interestingly, the TME of PD-L1 negative tumors differed from PD-L1 positive tumors and were dominated by genes associated with ECM organization, matrix remodeling, desmoplasia and metastasis that were downregulated in post-treatment biopsies (figure 4). The dysregulation of ECM genes by TGF-β signaling in cancer-associated fibroblasts have been linked to immune evasion and immunotherapy failure.30 The enrichment of ECM genes observed at baseline in PD-L1 negative tumors from MARIO-3 suggest a more fibrotic, potentially ICI-resistant state compared with PD-L1 positive tumors which may further benefit from a combination of TME modulators such as eganelisib.
Peripheral blood analyses from MARIO-3 demonstrated systemic immune activation including proliferation and reinvigoration of memory CD8+T cells, upregulation of IFN-γ, CXCL9, CXCL10, and IL-15 and reduced immunosuppressive mMDSC cells and CCL22, a cytokine expressed by immunosuppressive macrophages (figure 1A,B). IFN-γ, CXCL9 and CXCL10 promote T-cell recruitment and activation, while IL-15 promotes NK cell activation and T-cell memory; each has been associated with stimulation of immune responses and positive outcomes in cancer therapy.21,23 Importantly, peripheral blood analyses from MARIO-1 eganelisib monotherapy revealed similar results to MARIO-3 for key cytokines and immune cell populations, supporting a role for eganelisib within the triplet regimen (online supplemental figure 1).
It is noteworthy that similar immune and ECM changes observed in eganelisib-treated patients were observed on PI3K-γ inhibition in in vitro cultured monocyte/macrophages and/or in vivo preclinical models of cancer.1011 14,16 31 Upregulation of MHC II members (HLA-DR, HLA-DQ, CIITA, CD74) and T-cell activating cytokines (CXCL9, CXCL10, CCL5, IL12) in MDSCs, TAMs and dendritic cells (DCs), upregulation of MHC I on all tumor-associated cells; upregulation of T and NK cell activation markers IFNG, ITGAL, GZMK, GZMA, IL2RG,CD3D, ZAP70, RORA, ITGA4, ITK, CD8B, and LCK and others; and downregulation of desmoplasia and metastasis-associated genes PDGFA, PDGFB, TGF, VEGFA, PLAU, PLAT, MFGE8, VCAN, COL5A1, MMP1 and MMP9, were also observed on PI3K-γ inhibition in animal models of cancer.10 31 In particular, inhibition of PI3K-γ reduced ECM deposition, enhanced T-cell recruitment to tumors, prevented metastasis, and promoted survival in genetically engineered models of pancreatic cancer and TNBC.10 31
An intriguing finding from our DSP analysis of paired tumor biopsies was the potential association between gene signatures of TAM reprogramming and immune activation with clinical outcomes based on PFS. Patients whose tumors showed evidence of TAM reprogramming defined by enrichment of interferon-gamma, and MHC I and MHC II pathways within macrophages and/or increased ratio of CXCL9:SPP1 were among those with the longest PFS of the group (figure 5B,D). Consistent with a contributory role of eganelisib, samples collected from patients that experienced eganelisib-only dose holds prior to biopsy collection showed reduced evidence of TAM reprogramming and shorter PFS relative to the group (figure 5B,D, patients F, J, L). While a controlled study with larger sample numbers would be required to determine if there is a link between eganelisib TAM reprogramming and clinical outcome, results from this small sub-analysis support a potential contribution of eganelisib within the triplet regimen and warrants further study. DSP analysis of leukocyte-enriched regions showed similar results with 4 out of the 5 patients with gene signatures of immune activation, defined by significant enrichment of IFNG, adaptive immune system, MHC I, and MHC II pathways, having the longest PFS of the group (figure 3D, patients A–D) with dampened effects in patients experiencing eganelisib-only dose hold and shorter PFS.
The immune response is a highly regulated biological process involving many factors including checkpoint proteins that maintain homeostasis. The goal of immune checkpoint therapy in the context of cancer is to bypass checkpoints to achieve a sustained anti-tumor immune response. Compensatory upregulation of checkpoint proteins including CTLA-4 and immune tolerance factors including IL-10 can limit the full potential of immunotherapies.32 We observed evidence of these compensatory changes after treatment with the triplet in MARIO-3 including an increase in PD-L1 expressing immune cells within the tumor (online supplemental figure 3) and an increase in PD-1 and CTLA-4 expressing memory CD4/CD8 T-Cells and IL-10 in peripheral blood (figure 1). The upregulation of PD-L1 observed in PD-L1 negative tumors supports a rationale for further testing of eganelisib and PD-L1/PD-1 inhibitors in the treatment of PD-L1 negative tumors. In addition, improved tumor growth inhibition has been observed on the addition of anti-CTLA-4 to eganelisib+anti-PD1 therapy in TNBC preclinical models, providing rationale for exploration of this chemotherapy-free triple immunotherapy in the clinic.11
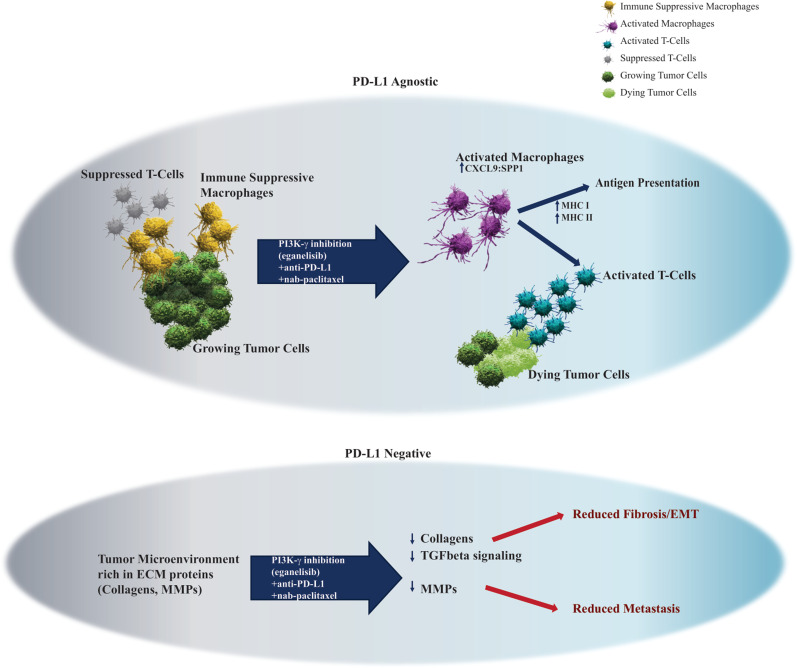
In summary, MARIO-3 TNBC tumor biopsy analyses revealed, in a subset of patients, evidence of TAM reprogramming and immune activation regardless of PD-L1 status as well as ECM reorganization within PD-L1 negative tumor types. Based on these findings, we propose a model for the mechanism of action of the triple therapy shown in figure 6, whereby eganelisib triggers TAM reprogramming, relieving T-cell suppression and together with atezolizumab mediated checkpoint blockade and nab-paclitaxel mediated tumor cell killing results in a sustained anti-tumor immune response. In the context of PD-L1 negative tumors with elevated ECM proteins at baseline, treatment with the triplet results in gene signatures associated with reduced desmoplasia and metastasis. Further studies will be required to determine if these changes translate to reduced metastasis in the clinic as has been observed in PI3K-γ -/- mice.10
Figure 6. Model for mechanism of action of the triple regimen in both PD-L1 positive and PD-L1 negative tumors. In the absence of treatment, the presence of TAMs contributes to an immune suppressed TME that prevents T-cell activation and anti-tumor immunity. Treatment with eganelisib triggers TAM reprogramming to an activated state marked by high CXCL9:SPP1 ratio and elevated MHC I and MHC II expression, resulting in enhanced antigen presentation and relief of T-cell suppression. Combination with nab-paclitaxel mediated tumor cell killing and tumor antigen release and atezolizumab mediated checkpoint blockade, the triple therapy results in sustained T-cell activity and anti-tumor immunity. For PD-L1 negative tumors with elevated ECM proteins in the TME relative to PD-L1 positive tumors, the triple therapy results in reduced gene signatures of EMT and fibrosis. ECM, extracellular matrix organization; EMT, epithelial-mesenchymal transition; PD-L1, programmed death-ligand 1; TAM, tumor-associated macrophages; TME, tumor microenvironment.
We realize the limitations of interpreting translational data from a single-arm study with small sample numbers which include an inability to decipher the contribution of components. While trends between immune activation/TAM reprogramming and clinical activity were observed, the sample size is too small to assign statistical significance. Caution must be taken when interpreting results when combining with drugs which may elicit similar pharmacodynamic changes. Immune activation and PD-L1 upregulation have been observed with atezolizumab/nab-paclitaxel combination33 and preclinical studies suggest a role for checkpoint blockade and paclitaxel in macrophage polarization,34 35 therefore it is not possible to decipher which drug is causing these pharmacodynamic changes in a single-arm study. However, a recent report describing albumin nanoparticle containing eganelisib and paclitaxel demonstrated improved macrophage reprogramming with the combination compared with either drug alone in preclinical breast cancer models.18 Despite the limitations of this study, evidence of systemic immune activation in peripheral blood from MARIO-1 with eganelisib monotherapy (online supplemental figure 1) and evidence of TAM reprogramming in post-treatment biopsies, including within PD-L1 negative tumors from MARIO-3 (figures2 5) are consistent with a role for eganelisib within the triplet. Importantly, upregulation of MCH I, MHC II, CXCL9 and downregulation of SPP1, MMPs, PLAU, PDGF observed in tumors biopsies from MARIO-3 were also observed in PI3K-γ -/- macrophages in preclinical models.10 31 Future randomized controlled studies with more patients will be required to further refine the mechanistic contribution of components and establish the clinical efficacy of the triple regimen.
In conclusion, the translational data from MARIO-3, presented here for the first time, are consistent with the TAM reprogramming mechanism of action previously characterized in preclinical studies. The immune activation observed in a subset of PD-L1 negative tumors is intriguing given the unmet need in this patient population. These data support the rationale for further evaluation of the first-in-class PI3K-γ inhibitor eganelisib in combination with ICI and chemotherapy in patients with mTNBC regardless of PD-L1 status and potentially more broadly to other PI3K pathway inhibitors or TAM-targeting agents in oncology indications where TAMs are a factor of resistance to therapy.
supplementary material
Acknowledgements
We thank the clinicians, patients and their families for participating in the MAcrophage Reprogramming in Immuno-Oncology (MARIO)-1 and MARIO-3 clinical trials. We acknowledge Roche Genentech for supplying atezolizumab for MARIO-3 and for valuable input on the manuscript. We would like to acknowledge Canopy Biosciences/Bruker Nano (Hayward, California, USA; Billerica, Massachusetts, USA) for performing the digital spatial profiling, NeoGenomics (Fort Meyers, Florida, USA) for the MultiOmyx multiplex immunofluorescence, HistogeneX/CellCarta (Naperville, Illinois, USA) for the programmed death-ligand 1 immunohistochemistry, Primity Bio/Cell Carta (Fremont, California, USA) for the flow cytometry and the Forsyth Institute for the cytokine analysis.
Footnotes
Funding: This work was funded by Infinity Pharmaceuticals.
Provenance and peer review: Not commissioned; externally peer reviewed.
Patient consent for publication: Not applicable.
Ethics approval: NCT03961698 (MARIO-3) study was approved by the institutional review board (WIRB 20191478) on April 7, 2019. NCT02637531 (MARIO-1) study was approved by the institutional review board IntegReview START2015.31 on December 15, 2015. Participants gave informed consent to participate in the study before taking part.
Contributor Information
Brenda C O'Connell, Email: brendaoconnell5@gmail.com.
Charley Hubbard, Email: charleyjhubbard@gmail.com.
Nora Zizlsperger, Email: noraziz@gmail.com.
Donna Fitzgerald, Email: dfitzgerpa@gmail.com.
Jeffrey L Kutok, Email: jlkutok@gmail.com.
Judith Varner, Email: jvarner@health.ucsd.edu.
Robert Ilaria, Jr, Email: cancerdoc@mac.com.
Melody A Cobleigh, Email: melody_cobleigh@rush.edu.
Dejan Juric, Email: juric.dejan@mgh.harvard.edu.
Kate H R Tkaczuk, Email: ktkaczuk@umm.edu.
Anthony Elias, Email: anthony.elias@ucdenver.edu.
Arielle Lee, Email: arielle.lee@uttyler.edu.
Shaker Dakhil, Email: shaker.dakhil@cancercenterofkansas.com.
Erika Hamilton, Email: Erika.hamilton@scri.com.
Hatem Soliman, Email: Hatem.Soliman@moffitt.org.
Stephane Peluso, Email: stefpeluso@mac.com.
Data availability statement
Data are available upon reasonable request.
References
- 1.Almansour NM. Triple-Negative Breast Cancer: A Brief Review About Epidemiology, Risk Factors, Signaling Pathways, Treatment and Role of Artificial Intelligence. Front Mol Biosci. 2022;9:836417. doi: 10.3389/fmolb.2022.836417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yin L, Duan JJ, Bian XW, et al. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020;22:61. doi: 10.1186/s13058-020-01296-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortes J, Rugo HS, Cescon DW, et al. Pembrolizumab plus Chemotherapy in Advanced Triple-Negative Breast Cancer. N Engl J Med. 2022;387:217–26. doi: 10.1056/NEJMoa2202809. [DOI] [PubMed] [Google Scholar]
- 4.Schmid P, Rugo HS, Adams S, et al. Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2020;21:44–59. doi: 10.1016/S1470-2045(19)30689-8. [DOI] [PubMed] [Google Scholar]
- 5.Sharma P, Hu-Lieskovan S, Wargo JA, et al. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell. 2017;168:707–23. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan Z-Y, Luo R-Z, Peng R-J, et al. High infiltration of tumor-associated macrophages in triple-negative breast cancer is associated with a higher risk of distant metastasis. Onco Targets Ther. 2014;7:1475–80. doi: 10.2147/OTT.S61838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mantovani A, Marchesi F, Malesci A, et al. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14:399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pittet MJ, Michielin O, Migliorini D. Clinical relevance of tumour-associated macrophages. Nat Rev Clin Oncol. 2022;19:402–21. doi: 10.1038/s41571-022-00620-6. [DOI] [PubMed] [Google Scholar]
- 9.Molgora M, Colonna M. Turning enemies into allies-reprogramming tumor-associated macrophages for cancer therapy. Med . 2021;2:666–81. doi: 10.1016/j.medj.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaneda MM, Messer KS, Ralainirina N, et al. PI3Kγ is a molecular switch that controls immune suppression. Nature New Biol. 2016;539:437–42. doi: 10.1038/nature19834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Henau O, Rausch M, Winkler D, et al. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kγ in myeloid cells. Nature New Biol. 2016;539:443–7. doi: 10.1038/nature20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carnevalli LS, Taylor MA, King M, et al. Macrophage Activation Status Rather than Repolarization Is Associated with Enhanced Checkpoint Activity in Combination with PI3Kγ Inhibition. Mol Cancer Ther. 2021;20:1080–91. doi: 10.1158/1535-7163.MCT-20-0961. [DOI] [PubMed] [Google Scholar]
- 13.Lanahan SM, Wymann MP, Lucas CL. The role of PI3Kγ in the immune system: new insights and translational implications. Nat Rev Immunol. 2022;22:687–700. doi: 10.1038/s41577-022-00701-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmid MC, Avraamides CJ, Dippold HC, et al. Receptor tyrosine kinases and TLR/IL1Rs unexpectedly activate myeloid cell PI3kγ, a single convergent point promoting tumor inflammation and progression. Cancer Cell. 2011;19:715–27. doi: 10.1016/j.ccr.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmid MC, Franco I, Kang SW, et al. PI3-kinase γ promotes Rap1a-mediated activation of myeloid cell integrin α4β1, leading to tumor inflammation and growth. PLoS One. 2013;8:e60226. doi: 10.1371/journal.pone.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmid MC, Kang SW, Chen H, et al. PI3Kγ stimulates a high molecular weight form of myosin light chain kinase to promote myeloid cell adhesion and tumor inflammation. Nat Commun. 2022;13:1768. doi: 10.1038/s41467-022-29471-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans CA, Liu T, Lescarbeau A, et al. Discovery of a Selective Phosphoinositide-3-Kinase (PI3K)-γ Inhibitor (IPI-549) as an Immuno-Oncology Clinical Candidate. ACS Med Chem Lett. 2016;7:862–7. doi: 10.1021/acsmedchemlett.6b00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song Y, Bugada L, Li R, et al. Albumin nanoparticle containing a PI3Kγ inhibitor and paclitaxel in combination with α-PD1 induces tumor remission of breast cancer in mice. Sci Transl Med. 2022;14:eabl3649. doi: 10.1126/scitranslmed.abl3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatem S, Hargis J, Elias A, et al. Abstract P5-16-02: Updated efficacy, safety and translational data from MARIO-3, a phase II open-label study evaluating a novel triplet combination of eganelisib (IPI-549), atezolizumab (atezo), and nab-paclitaxel (nab-pac) as first-line (1L) therapy for locally advanced or metastatic triple-negative breast cancer (TNBC) Cancer Res. 2022;82:5–16. doi: 10.1158/1538-7445.SABCS21-P5-16-02. [DOI] [Google Scholar]
- 20.Hong DS, Postow M, Chmielowski B, et al. Eganelisib, a First-in-Class PI3Kγ Inhibitor, in Patients with Advanced Solid Tumors: Results of the Phase 1/1b MARIO-1 Trial. Clin Cancer Res. 2023;29:2210–9. doi: 10.1158/1078-0432.CCR-22-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tokunaga R, Zhang W, Naseem M, et al. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation - A target for novel cancer therapy. Cancer Treat Rev. 2018;63:40–7. doi: 10.1016/j.ctrv.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoenborn JR, Wilson CB. Regulation of interferon-gamma during innate and adaptive immune responses. Adv Immunol. 2007;96:41–101. doi: 10.1016/S0065-2776(07)96002-2. [DOI] [PubMed] [Google Scholar]
- 23.Steel JC, Waldmann TA, Morris JC. Interleukin-15 biology and its therapeutic implications in cancer. Trends Pharmacol Sci. 2012;33:35–41. doi: 10.1016/j.tips.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox TR. The matrix in cancer. Nat Rev Cancer. 2021;21:217–38. doi: 10.1038/s41568-020-00329-7. [DOI] [PubMed] [Google Scholar]
- 25.Patras L, Paul D, Matei IR. Weaving the nest: extracellular matrix roles in pre-metastatic niche formation. Front Oncol. 2023;13:1163786. doi: 10.3389/fonc.2023.1163786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kothari AN, Arffa ML, Chang V, et al. Osteopontin-A Master Regulator of Epithelial-Mesenchymal Transition. J Clin Med. 2016;5:39. doi: 10.3390/jcm5040039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Lin X, Sun P. uPAR, beyond regulating physiological functions, has orchestrated roles in cancer (Review) Int J Oncol. 2022;61:151. doi: 10.3892/ijo.2022.5441. [DOI] [PubMed] [Google Scholar]
- 28.Tian J, Wang V, Wang N, et al. Identification of MFGE8 and KLK5/7 as mediators of breast tumorigenesis and resistance to COX-2 inhibition. Breast Cancer Res. 2021;23:23. doi: 10.1186/s13058-021-01401-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bill R, Wirapati P, Messemaker M, et al. CXCL9:SPP1 macrophage polarity identifies a network of cellular programs that control human cancers. Science. 2023;381:515–24. doi: 10.1126/science.ade2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakravarthy A, Khan L, Bensler NP, et al. TGF-β-associated extracellular matrix genes link cancer-associated fibroblasts to immune evasion and immunotherapy failure. Nat Commun. 2018;9:4692. doi: 10.1038/s41467-018-06654-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaneda MM, Cappello P, Nguyen AV, et al. Macrophage PI3Kγ Drives Pancreatic Ductal Adenocarcinoma Progression. Cancer Discov. 2016;6:870–85. doi: 10.1158/2159-8290.CD-15-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–5. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emens LA, Cruz C, Eder JP, et al. Long-term Clinical Outcomes and Biomarker Analyses of Atezolizumab Therapy for Patients With Metastatic Triple-Negative Breast Cancer: A Phase 1 Study. JAMA Oncol. 2019;5:74–82. doi: 10.1001/jamaoncol.2018.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gubin MM, Esaulova E, Ward JP, et al. High-Dimensional Analysis Delineates Myeloid and Lymphoid Compartment Remodeling during Successful Immune-Checkpoint Cancer Therapy. Cell. 2018;175:1014–30. doi: 10.1016/j.cell.2018.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wanderley CW, Colón DF, Luiz JPM, et al. Paclitaxel Reduces Tumor Growth by Reprogramming Tumor-Associated Macrophages to an M1 Profile in a TLR4-Dependent Manner. Cancer Res. 2018;78:5891–900. doi: 10.1158/0008-5472.CAN-17-3480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon reasonable request.