Abstract
Background
Activating and inhibitory receptors of natural killer (NK) cells such as NKp, NKG2, or CLEC are highly relevant to cold tumors including glioblastoma (GBM). Here, we aimed to characterize the expression of these receptors in GBM to gain insight into their potential role as modulators of the intratumoral microenvironment.
Methods
We performed a transcriptomic analysis of several NK receptors with a focus on the activating receptor encoded by KLRC2, NKG2C, among bulk and single-cell RNA sequencing GBM data sets. We also evaluated the effects of KLRC2-overexpressing GL261 cells in mice treated with or without programmed cell death protein-1 (PD-1) monoclonal antibody (mAb). Finally, we analyzed samples from two clinical trials evaluating PD-1 mAb effects in patients with GBM to determine the potential of NKG2C to serve as a biomarker of response.
Results
We observed significant expression of several inhibitory NK receptors on GBM-infiltrating NK and T cells, which contrasts with the strong expression of KLRC2 on tumor cells, mainly at the infiltrative margin. Neoplastic KLRC2 expression was associated with a reduction in the number of myeloid-derived suppressor cells and with a higher level of tumor-resident lymphocytes. A stronger antitumor activity after PD-1 mAb treatment was observed in NKG2Chigh-expressing tumors both in mouse models and patients with GBM whereas the expression of inhibitory NK receptors showed an inverse association.
Conclusions
This study explored the role of neoplastic NKG2C/KLRC2 expression in shaping the immune profile of GBM and suggests that it is a predictive biomarker for positive responses to immune checkpoint inhibitor treatment in patients with GBM. Future studies could further validate this finding in prospective trials.
Keywords: NK Cell Lectin-Like Receptor Subfamily K, Central Nervous System Cancer, Tumor microenvironment - TME, Macrophage, Immune Checkpoint Inhibitor
WHAT IS ALREADY KNOWN ON THIS TOPIC.
WHAT THIS STUDY ADDS
We report for the first time that NKG2C, a receptor traditionally associated with immune cells, is also expressed by GBM tumor cells, especially at the invasive front. The presence of NKG2C on tumor cells correlates with an altered immune profile and increased responsiveness to immune checkpoint inhibition.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Characterizing the macrophage population and the presence of NKG2C and other NK cell receptors could help identify patients with GBM most likely to benefit from checkpoint blockade therapy. The extent of surgical resection and the spatial distribution of these biomarkers should be considered to optimize patient selection for immunotherapy.
Background
Glioma is classified based on the status of the isocitrate dehydrogenase 1/2 (IDH1/2) gene. The IDH-mutant (IDHmut) subgroup mostly includes grade 2/3 gliomas and a few grade 4 astrocytomas, whereas the term glioblastoma (GBM) refers to IDH-wild type (IDHwt) grade 4 tumors.1 The immune landscape of GBM is very different from that of other cancers, characterized by a strong immunosuppressive phenotype enriched in tumor-associated macrophages and exhausted lymphocytes.2 3 This phenotype is built on a bidirectional interplay between immune and tumor cells that release cytokines to recruit T-regulatory lymphocytes and promote the polarization of M2-like macrophages and microglia.4 These cells, in turn, shape the transcriptional profile of GBM.5
The discovery of immune checkpoint regulators has led to the generation of new therapies for a large number of otherwise therapy-resistant cancers. Drugs that restore the immune response, including cytotoxic T-lymphocyte associated protein 4 (CTLA-4) and programmed cell death protein-1 (PD-1) monoclonal antibodies (mAbs), have achieved enormous success at improving overall survival for patients with melanoma, lung cancer, and renal carcinoma, while also being investigated for other malignancies as well.6 These checkpoint inhibitors overcome the inhibition of CD8+ T and natural killer (NK) cells in the lymph node and the tumor itself. However, clinical trials using PD-1 mAbs in patients with GBM have yet to demonstrate a survival improvement7,10 even though checkpoint inhibitor antibodies appear to penetrate the brain.11 The unique microenvironment of GBM has been used as a rationale to explain why only a small percentage of glial tumors respond to this strategy. Furthermore, the absence of reliable biomarkers hampers efforts to enhance the success rate of upcoming clinical trials.
NK receptors, expressed by T and NK cells, play a key role in cancer immunosurveillance and represent a potentially important element in the response to immunotherapy with checkpoint inhibitors.12 In gliomas, NK receptor proteins, such as CD161/Clec5B, have been identified as new control points in tumor development.13 Furthermore, GBM has low major histocompatibility complex (MHC) class I expression.14 In this situation, the interaction between NKG2D and its ligands appears to be highly relevant for tumor cell killing.15
Here, we have performed a thorough analysis of NK receptor genes in GBM samples and have found a striking overexpression of KLRC2, which encodes for the activating receptor NKG2C, in GBM cells and is predominantly localized at the infiltrative margin of the tumor. We demonstrated that when NKG2C is overexpressed by GL261 cells, the immune phenotype transforms into a microenvironment that favors fewer myeloid cells with decreased immunosuppressive properties. The results presented here also show that the induction of NKG2C expression is able to enhance the antitumor effect of PD-1 mAb treatment that fuels the entrance of cytotoxic lymphocytes. The analysis of GBM after patient treatment with PD-1 mAb suggests that high levels of NKG2C and low levels of inhibitory receptors correlate with higher overall survival in response to immunotherapy and underscores their potential use as predictive biomarkers.
Methods
The workflow followed in this manuscript is graphically represented in online supplemental table 2A.
Human samples
For the flow cytometry study and the MRI-guided analysis (online supplemental table 1A) the samples were obtained after the patient’s written consent, in accordance with the Declaration of Helsinki
Sequential tumor tissue samples from a phase II clinical trial in patients with newly diagnosed or relapsed GBM that was conducted at Clinic University of Navarra were also analyzed (NCT02550249). Patients from two subgroups were recruited to the study; (1) patients who required salvage surgery to treat relapsed disease (n = 11) as well as (2) patients who required surgery for primary tumor resection (n = 2). Clinicopathological characteristics of patients are summarized in online supplemental table 1A. Treatment consisted of a 3 mg/kg dose of nivolumab given 2 weeks (±3 days) before the surgery. Patients received postsurgical doses of nivolumab every 2 weeks (±3 days) until radiologic progression or unacceptable toxicity. For the three patients who underwent primary surgical treatment, nivolumab was stopped to receive standard-of-care chemoradiotherapy (CRT). In two of these cases, nivolumab was reintroduced following completion of CRT.9
Mouse cells
The GL261 murine glioma cell line was purchased from Creative Biolabs and grown in Dulbecco's Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 0.1% penicillin (100 U/mL), and streptomycin (100 µg/mL). The cells were passaged (1:2) every 3 days to ensure logarithmic growth and tested monthly for Mycoplasma contamination by a luminescent detection kit (MycoAlert Mycoplasma Detection, Lonza). The retroviral vectors used to transduce the cells were pLV-mKLRC2 (pLV(Exp)-Bsd-mPGK>mKlrc2(NM_010653.4), Vector Builder) and pLV-GFP (pLV(Exp)-Bsd-EF1A>EGFP, Vector Builder). All cell lines were maintained in a 37°C humidified incubator with 5% CO2 and passaged to maintain 70% confluency. Infected cells were selected in the presence of blasticidin (InvivoGen). For tumor implantation into C57BL/6 mice, cells were trypsinized with 0.05% trypsin—EDTA, washed, and resuspended at a final concentration of 5×103 cells/2 µL.
Co-culture experiments GL261 and macrophages
Macrophages were obtained by intraperitoneal injection of 3% thioglycollate medium into two adult C57 mice. The inflammatory response was allowed to proceed for 3 days before the mice were euthanized and macrophages were isolated as previously described.16 GFP and KLRC2 GL261 cells were co-cultured with peritoneal macrophages in three different ratios (1:10, 1:2, and 1:1 macrophage:GL261 proportion) in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% FBS for the first 24 hours. Subsequently, the medium was changed to RPMI 1640 supplemented with 2% FBS for 24 hours, followed by 48 hours in RPMI 1640 before collecting the pellet for messenger RNA (mRNA) isolation. The detailed protocol and workflow are provided in online supplemental table 2E.
In vivo assays
The Research Ethics and Animal Welfare Committee at “Instituto de Salud Carlos III” approved and reviewed animal experiments in accordance with the European Union and national directives (PROEX 055/19). To establish orthotopic allografts, intracranial transplantation was performed on C57BL/6 mice by injecting a 2 µL volume containing GL261 cells into the striatum using a Hamilton syringe. The injection was done using a Stoelting’s Stereotaxic device with the coordinates A-P at −0.5 mm, M-L at +2 mm, and D-V at −3 mm relative to bregma.
Mice were treated with anti-mouse PD-1 mAb (purified anti-mouse CD279 Antibody, BioLegend) starting on postoperative day 8, with three doses administered every other day at 10 mg/kg (250 µg/dose) by intraperitoneal injection. Control mice received equivalent doses of isotype murine IgG (Purified Rat IgG2a, κ Isotype Ctrl, BioLegend) with the same dosing schedule mice. Daily animal monitoring was carried out to ensure that humane endpoints were reached. Euthanasia was carried out if animals showed signs such as a protruded skull, hunched posture, extreme lethargy, or weight loss. Characteristics of experimental mouse model samples are summarized in online supplemental table 1B.
RNA extraction and quantitative PCR
Total RNA from the human and mouse tumors was extracted using a commercial RNA extraction kit (ReliaPrep RNA Miniprep System, Promega) and equivalent amounts (1 µg) of purified RNA were reverse transcribed using PrimeScript RT Reagent Kit (Takara). Quantitative real-time PCR was performed using the LightCycler 480 (Roche Diagnostics) with the SYBR Premix (TB Green Premix Ex Taq II, Takara) and gene-specific primers (online supplemental table 2B). Relative gene expression was calculated using the ΔΔCt method and normalized to GAPDH.
Western blot
Proteins were extracted from the cells and tissues through mechanical disaggregation using lysis buffer (50 mM Tris-pH 7.5, 300 mM NaCl, 0.5% SDS, and 1% Triton X-100) (15 min with agitation at 95°C). Protein content was determined using the BCA Protein Assay Kit (Thermo Scientific) and 30 µg of proteins were resolved by 12% SDS-PAGE and then transferred to a nitrocellulose membrane (Amersham Biosciences). The membranes were blocked and then incubated with the primary (overnight at 4°C) and the respective secondary antibody (2 hours at room temperature (RT)) (online supplemental table 2B). Protein bands were visualized with the Imager 800 (Amersham Biosciences) and quantitatively analyzed using ImageQuant TL software (Cytiva).
Immunohistochemical staining and quantification
Immunohistochemical (IHC) staining with different antibodies (online supplemental table 2B) was performed on brain tissue sections from the murine model or from patient’s tumors that were fixed in 4% formalin, paraffin-embedded, and sectioned onto slides of 4 µm thickness. Before the 3,3'-diaminobenzidine (DAB) or red staining, slides were dewaxed and stained with H&E to verify tumor presence. For the chromogenic and fluorescent detection, the slides were prior preheated for 30 min at 65°C and the BOND RXm automated advanced staining system (Leica Biosystems) programmed with the specific protocol according to the staining type was used (online supplemental table 2C).
The chromogenic slides were scanned using the Hamamatsu NanoZoomer-SQ microscope slide digitizer (C13140-01). QuPath software was used for the quantification by setting two different methods depending on the specific antibody stain. For CD3 and CD8 stains, the positive-cell detection method was used by calculating the percentage of positive cells in comparison to negative cells within a consistently defined area for each stain. In contrast, for CD68, CD206, and NKG2C stains, the pixel classification method was applied. This involved setting an intensity threshold in each region of interest corresponding to the specific markers. A signal exceeding this threshold was considered positive and the percentage of such staining in the total area was determined. The specific codes and parameters used for quantification of each stain are detailed in online supplemental table 2D.
The immunofluorescence images were acquired with a STELLARIS 8 Confocal Microscope (Leica), using HCPL APO CS2 20×/0.75NA dry objective in low-magnification images and HCPL APO CS2 63×/1.40NA oil-immersion objective in high-magnification images.
Flow cytometry
Tumor suspensions were obtained after mechanical and enzymatic disaggregation (Accumax (Merck Millipore)) (15 min at RT). Erythrocytes were lysed with Quicklysis buffer (Cytognos) and cells were incubated with hFcR Blocking (Miltenyi) prior to antibody (online supplemental table 2B) incubation (20 min at 4°C in phosphate-buffered saline 1% FBS). Viable cells were labeled with a Fixable Viability Stain (Becton Dickinson) (20 min at RT). The analysis was conducted in a MACSQuant V.10 flow cytometry (Miltenyi). Subset definition was: CD45+, lymphoid (CD45+CD11b−SSClo), myeloid (CD45+CD11b+SSClo or SSChi), microglia (CD45lowCD11b+SSClo or SSChi), neutrophils (CD45+CD11b+CD16+CD15+CD14−CD33−); MDSCs (CD45+CD11b+CD16+CD15 CD14±CD33+); macrophages (CD45+CD11b+CD16 CD15−CD14±CD33−93 MHC-II+) as previously used17 (online supplemental table 2B).
In silico studies
Single-cell RNA sequencing data sets analysis
To generate the integrated single-cell transcriptomic atlas, we downloaded different single-cell RNA sequencing (scRNA-seq) databases available in public repositories of normal brain,18 IDHmut gliomas, and GBMs.19 Subsequent data processing and analysis were performed using the Seurat package V.4.3.0. For the analysis, the two IDHmut samples were selected, and in the case of GBMs, the three cases of newly diagnosed and recurrent GBM with the highest number of counts were chosen (online supplemental table 1C). Integration and batch-corrected dimensionality reduction were performed using Harmony20 correcting for sample type. Cell types were identified using scCATCH (Single-Cell Cluster-Assisted Annotation Toolkit for Cellular Heterogeneity)21 and confirmed by canonical markers described in a previous study22 (online supplemental figure 1A).
For scRNA-seq analyses of the tumor periphery23 and PD-1 mAb therapy response,22 the already processed RDS files were downloaded from the respective public repositories, selecting only the IDHwt patients with GBM (online supplemental table 1C). Data processing and analysis that followed were also conducted using the Seurat package V.4.3.0.
RNA sequencing data set analyses
Kaplan-Meier survival analysis was conducted on GBM samples from the Cancer Genome Atlas (TCGA) and the Chinese Glioma Genome Atlas (CGGA) data sets based on KLRC2 expression using the GlioVis web platform (http://gliovis.bioinfo.cnio.es/).
Functional enrichment analyses
Gene ontology (GO) biological process (GO:BP) was performed using g:Profiler (v e109_eg56_p17_1d3191d) with g:SCS multiple testing correction method applying a significance threshold of 0.05.24
Immune landscape heatmap
Quantitative real time-polymerase chain reaction (qRT-PCR) data were normalized and the results of correlation analysis (unsupervised clustering) were visualized using the pheatmap package (Kolde R pheatmap: Pretty Heatmaps. R package V.1.0.12. https://CRAN.R-project.org/package=pheatmap. 2019) in R software.
Statistics
GraphPad Prism V.9.5.0 and R V.4.1.2 were used for statistical analysis. The normality of the variables was previously verified using the Kolmogorov-Smirnov test and the differences between pairs of experimental groups were analyzed by the Student’s t-test. To assess survival differences observed between mice injected with glioma cells, distributions of overall survival were estimated using the Kaplan-Meier method, and the p values were calculated using the log-rank (Mantel-Cox) test. A threshold of p<0.05 was used to determine the statistical significance of each experiment.
Results
Characterization of NK receptor expression in glioma
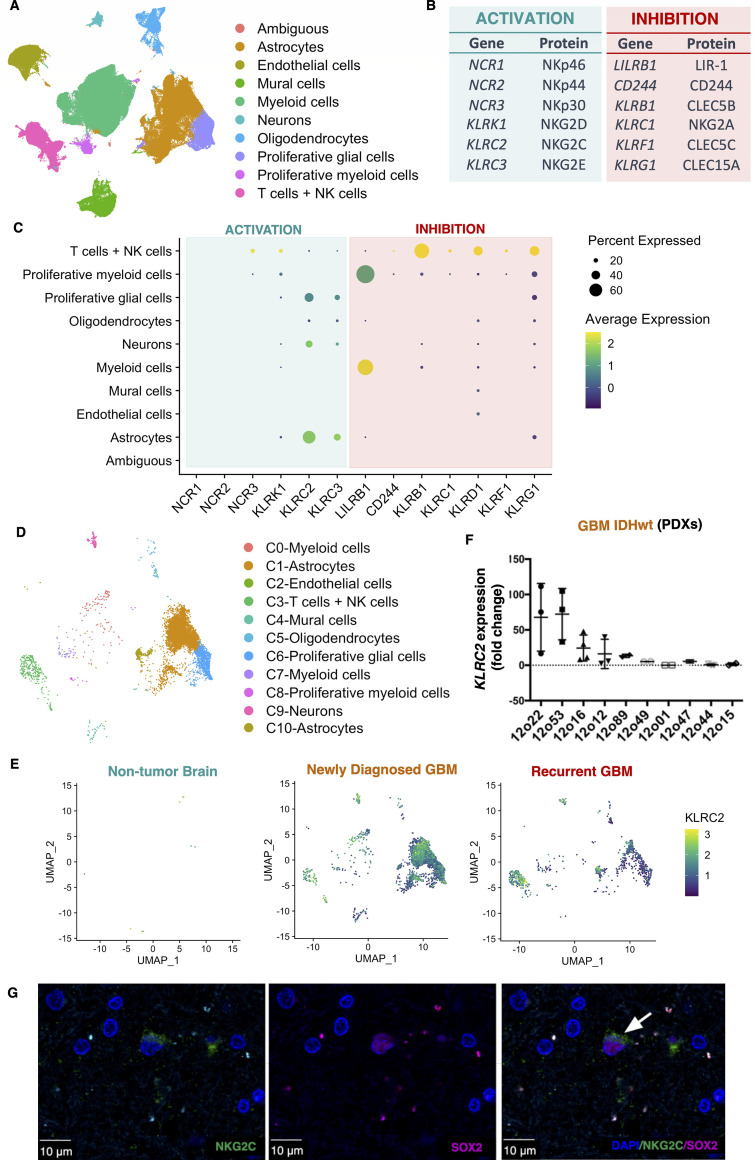
We integrated publicly available data from scRNA-seq studies of non-tumoral brain18 and glioma.19 This analysis identified different clusters of immune cell populations (macrophages, microglia and T cells), astrocytic cells and vascular cells (endothelial cells and pericytes) (figure 1A; online supplemental figure 1A). We then determined the expression of genes encoding NK receptors (figure 1B) in the different cell types of the integrated object. In agreement with a recent study,13 we found high levels of expression for the inhibitory receptors, CLEC5B (also named CD161) (encoded by KLRB1), CLEC15A (encoded by KLRG1) and NKG2A/CD94 (encoded by KLRC1/KLRD1) in the cluster of T cells and NK cells, although few of them also express the activating receptor NKG2D (encoded by KLRK1) and NKp30 (encoded by NCR3) (figure 1C). Monocyte/macrophage Ig-like receptor (MIR) 7, LIR-1, was mostly expressed by myeloid cells while NKG2E (encoded by KLRC3) and NKG2C (encoded by KLRC2) were mostly expressed by glial cells including the proliferative cluster (figure 1C). KLRC3 was previously characterized as a gene differentially expressed in GBM cancer stem cells,25 but there were no previous reports of the glial expression of KLRC2. To further explore which cells are expressing NKG2C, we selected KLRC2+cells from our integrated database (figure 1D) and compared the expression level of the KLRC2 gene in the different sample types. Our analysis revealed that KLRC2+cells are almost absent in normal brain tissue while they are abundant in GBM tumors (figure 1E). Furthermore, the analysis revealed a significant decrease in KLRC2 expression in glial cells in recurrent GBM compared with newly diagnosed tumors (figure 1E) (online supplemental figure 1B,C, online supplemental table 3A). By contrast, inhibition-related genes such as LILRB1, KLRB1 and KLRG1 show a higher expression after tumor recurrence (online supplemental figure 1C). To obtain an independent confirmation of the neoplastic expression of KLRC2, we conducted a qRT-PCR analysis on a collection of patient-derived GBM xenografts (PDX)26 using human-specific primers to exclude mRNAs from host cells. The results confirmed the expression of human KLRC2 expression in several PDX models (figure 1F). Moreover, immunofluorescence (IF) staining of human GBM samples showed a clear NKG2C expression in SOX2+cells (figure 1G), SOX2 being a pan-glioma marker that is widely used to detect GBM cells.27
Figure 1. NKG2C is expressed on neoplastic cells in gliomas. (A) UMAP plot annotated according to each cell type of the integrated object (control and glioma scRNA-seq databases) (online supplemental table 1C). (B) List of the main genes and their corresponding proteins that are involved in the activation or inhibition of immune cell function; particularly regarding natural killer (NK) cells. (C) Dot plot showing the percentage and average expression of activation and inhibition markers in the different cell types of the integrated object (control and glioma scRNA-seq databases). The dot.min value was set to 0.01, meaning only genes with expression levels above 1% are represented by a dot. (D) UMAP plot with the cells expressing KLRC2 detailing the clusters annotated according to each cell type of the integrated object. (E) UMAP plot of cells with positive KLRC2 expression in non-tumor brain, newly diagnosed glioblastoma (GBM) and recurrent GBM. (E) UMAP plot of cells with positive KLRC2 expression in non-tumor brain, newly diagnosed GBM and recurrent GBM. (F) Human KLRC2 relative expression in human GBM patient-derived xenografts (PDXs) (n=3). GAPDH expression was used for normalization. (G) Representative image of the immunofluorescence analysis of SOX2 (magenta) and NKG2C (green) in patients with GBM. The white arrow points to a double-positive cell. Nuclei were counterstained with DAPI. Scale: 10 µm. DAPI, 4′,6-diamidino-2-phenylindole; IDHwt, isocitrate dehydrogenase-wild type; scRNA-seq, single-cell RNA sequencing; UMAP, uniform manifold approximation and projection.
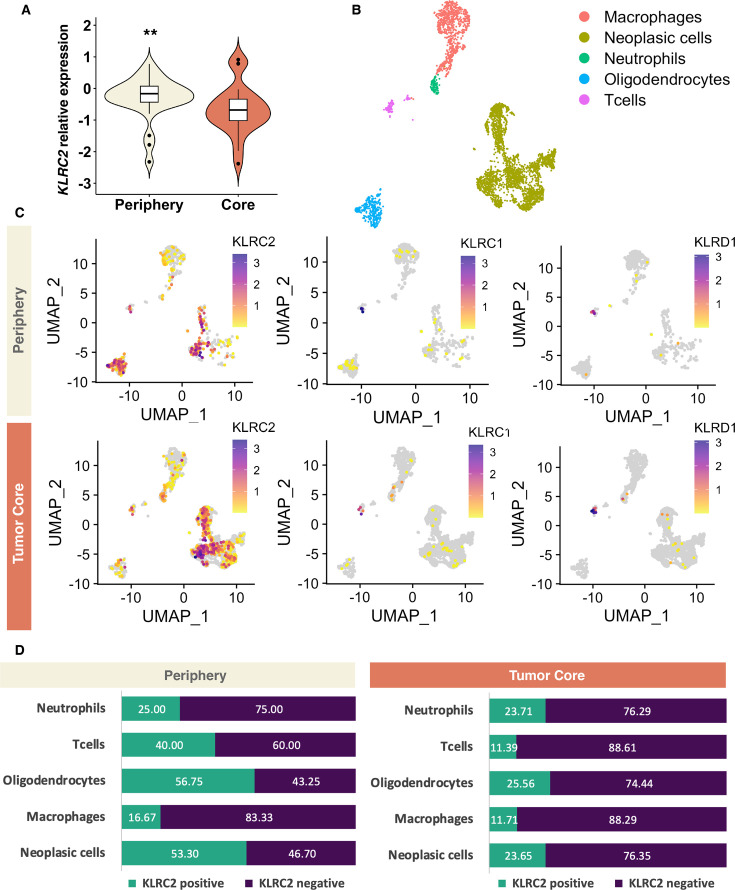
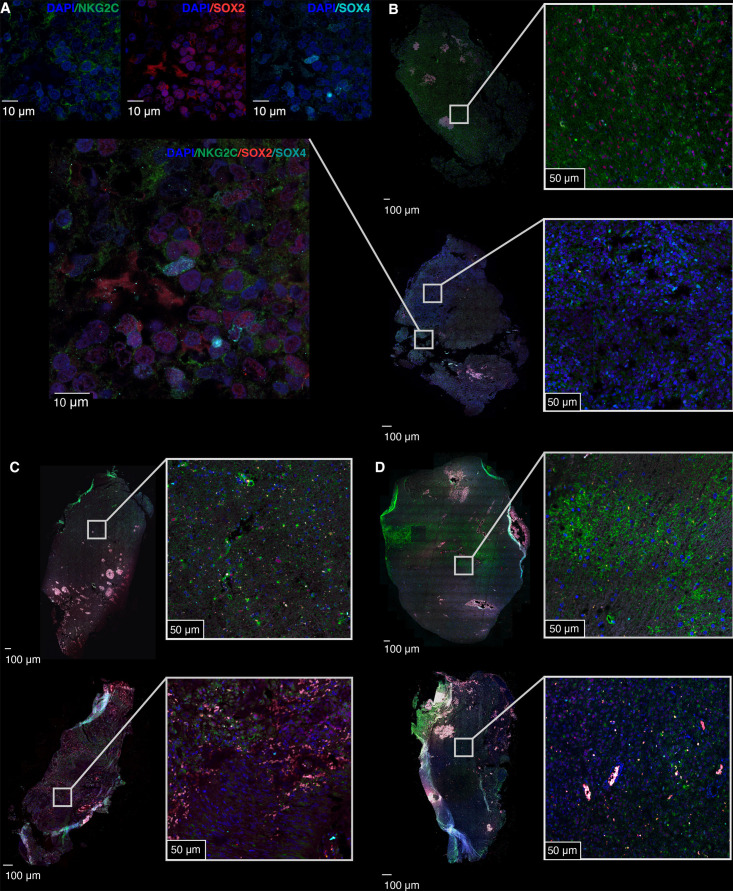
For a more comprehensive characterization of KLRC2 expression in glioma, we employed a new cohort of regionally annotated biopsies of the tumor core and periphery. The qRT-PCR analysis showed that KLRC2 expression is more pronounced in the peritumoral zone compared with the tumor mass, finding differences in paired (online supplemental figure 2A,B) and unpaired analyses (figure 2A). An in-silico analysis of published scRNA-seq data (figure 2B)23 corroborated the spatial distribution of KLRC2 expression and indicated a higher percentage of cells expressing this gene in the periphery in the clusters annotated as neoplastic cells, oligodendrocytes and T cells (figure 2C,D; online supplemental table 3B). To corroborate these results, we performed an IF analysis on regionally segregated sections from patients with GBM. In agreement with the previous results (figure 1G), NKG2C staining was found in SOX2+cells, but also in SOX4+cells (figure 3A). Notably, we observed stronger NKG2C expression in the peripheral region compared with the tumor core in different GBM cases (figure 3B–D). In summary, our results indicate that KLRC2 is expressed by glioma cells with a particularly high expression in the peripheral zone of GBM, a region containing highly invasive tumor cells that express oligodendrocyte-lineage markers (online supplemental figure 2C).28
Figure 2. Intertumoral and intratumoral heterogeneity of NKG2C expression in IDHwt GBM. (A) KLRC2 relative expression (qRT-PCR) in the periphery (n=30) versus tumor core (n=18) of IDHwt GBM. P values using unpaired t-test: **p≤0.01, ***p≤0.001. (B) UMAP plot annotated according to each cell type of the integrated GBM scRNA-seq database (GSE117891). A detailed description of the samples and counts used for the study can be found in online supplemental table 1C). (C) UMAP plot of cells with positive KLRC2, KLRC1 and KLRD1 expression in the periphery and tumor core of the GBM scRNA-seq database (GSE117891). (D) Bar chart of the percentage of cells expressing KLRC2 in each cell type in both the periphery and tumor core of the GBM scRNA-seq database (GSE117891). Significant differences were observed in the proportions of KLRC2-positive cells, especially in neoplastic cells (p=5.2e-22), oligodendrocytes (p=7.1e-7), and T cells (p=7.0e-3). The statistical analysis was conducted using the χ2 test. The statistical analysis was conducted using the χ2 test and can be found in more detail in online supplemental table 3B. GBM, glioblastoma; IDHwt, isocitrate dehydrogenase-wild type; qRT-PCR, quantitative real time-polymerase chain reaction; scRNA-seq, single-cell RNA sequencing; UMAP, uniform manifold approximation and projection.
Figure 3. Intertumoral and intratumoral heterogeneity of NKG2C expression in three patients with GBM. (A) Representative high-magnification image showing the co-expression of SOX2 (magenta), SOX4 (cyan), and NKG2C (green) in tumor cells. The upper panels display individual DAPI/NKG2C, DAPI/SOX2, and DAPI/SOX4 staining. (B–D) Representative images of the immunofluorescence analysis of the expression of SOX2 (magenta), SOX4 (cyan), and NKG2C (green) in the periphery (top images) and tumor core (bottom images) of three different patients with GBM. Nuclei were counterstained with DAPI (blue). The left images show the overall tissue structure with boxes indicating regions of interest magnified on the right. Scale: 10 µm (A) 100 µm for the main images and 50 µm for the insets (B–D). DAPI, 4',6-diamidino-2-phenylindole; GBM, glioblastoma; qRT-PCR, quantitative real time-polymerase chain reaction; UMAP, uniform manifold approximation and projection.
Glioma cell KLRC2 expression reshapes the immune microenvironment of mouse and human glioblastoma
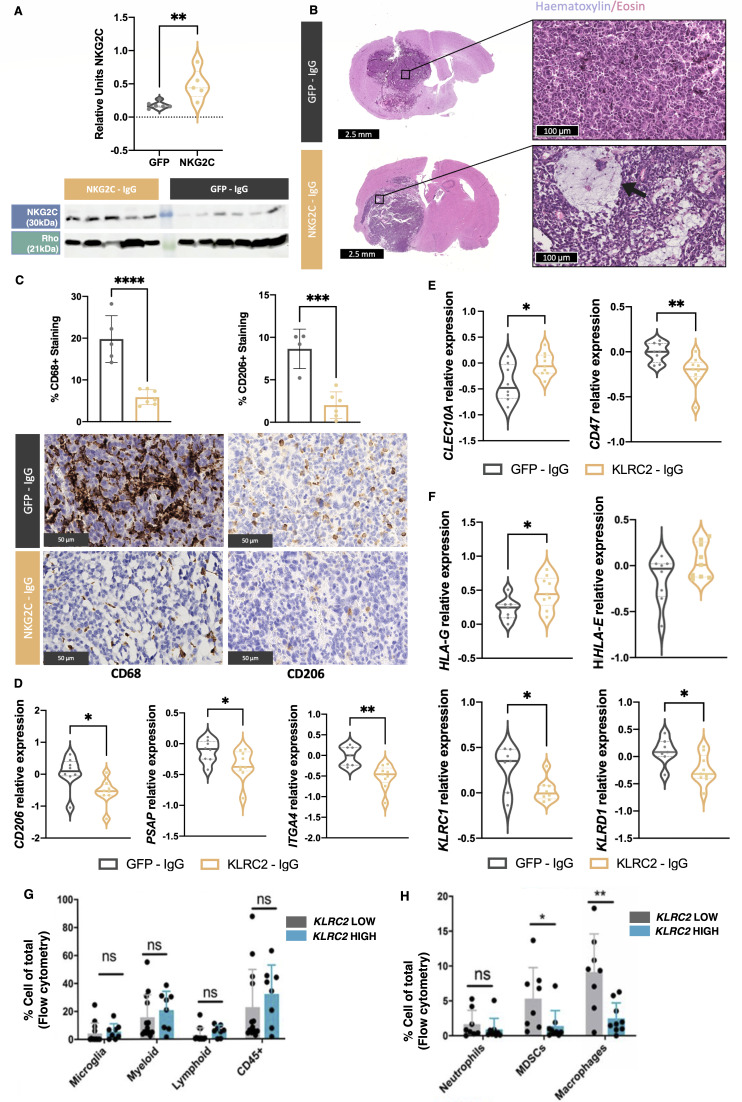
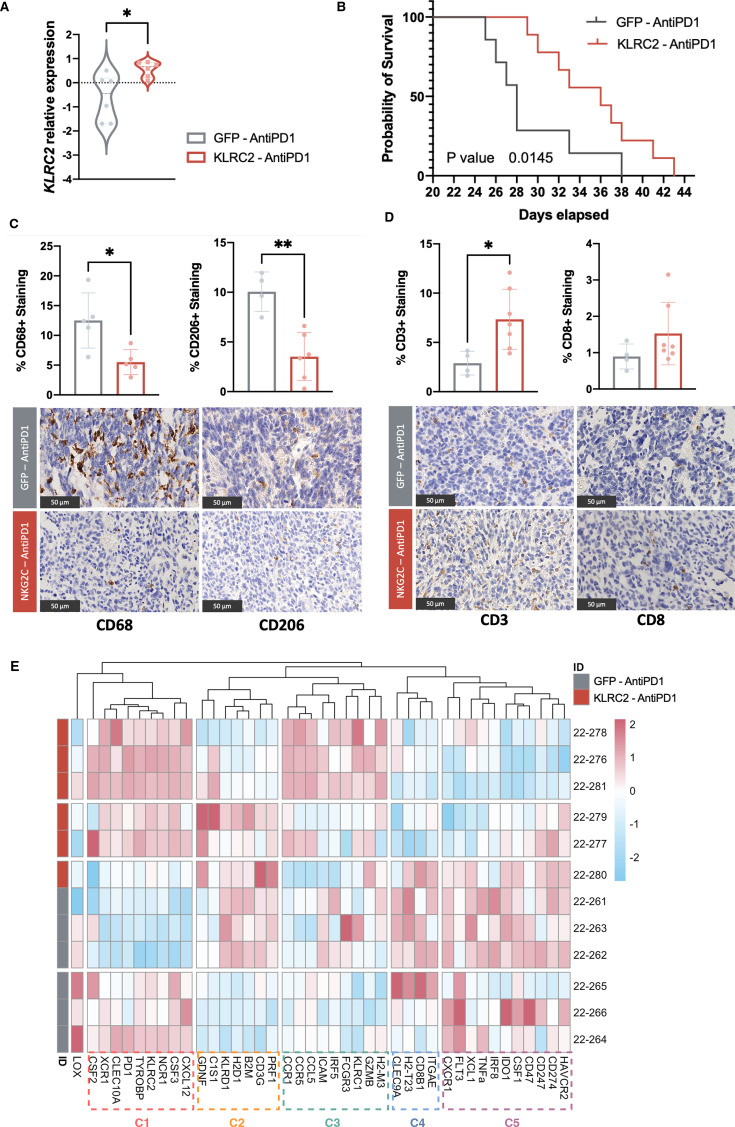
To study the role of KLRC2 expression in glioma cells, we used a lentiviral construct to overexpress the gene in GL261 cells. The efficacy of this overexpression in vitro was validated at mRNA (online supplemental figure 3A) and protein level (online supplemental figure 3B). These cells were injected into the brain of C57BL/6 mice and we observed a similar overexpression pattern at mRNA (online supplemental figure 3A) and protein level (figure 4A) in the tumors. While there was no significant change in tumor burden (online supplemental figure 3C), histological analysis of tumors with KLRC2 overexpression revealed a fibrillar matrix with microcystic formations (figure 4B) that are characteristic of astrocytic histology and a better prognosis.29 30 Additionally, IHC analysis of the tumor tissue showed a robust decrease in infiltrating myeloid cells and CD206 staining (figure 4C), which is a typical marker of immunosuppressive macrophages. We also noted the downregulation of M2-related gene expression (figure 4D), a significant increase in the dendritic cell marker CLEC10A, and a decrease in the classical “don’t-eat-me signal” CD47, (figure 4E) in KLRC2-GL261 compared with GFP-GL261 tumors (figure 4E). Conversely, we did not detect significant changes in the number of CD3+ or CD8+ cells during KLRC2 overexpression (online supplemental figure 3D). Notably, there was an upregulation of several MHC molecules including HLA-G (encoded by H2-M3) and a downregulation of NKG2A/CD94 (encoded by KLRD1/KLRC1) in NKG2C overexpressing tumors (figure 4F). Similar changes in macrophage polarization were observed when we co-cultured GL261 cells with peritoneal macrophages. KLRC2 overexpression induced the downregulation of M2 markers (CD206 and ARG) and the upregulation of iNOS, an M1-related gene (online supplemental figure 3E), suggesting a direct effect of tumor cells expressing NKG2C on the profile of the associated macrophages.
Figure 4. Effect of KLRC2 overexpression in murine and human glioma. GFP cDNA-expressing GL261 or KLRC2 cDNA-expressing GL261 cells were injected into the striatum of C57BL/6 mice. (A) Western blot analysis quantified NKG2C expression in GL261 brain tumors with Rho as the normalization control (GFP, n=6; NKG2C, n=5). (B) Representative H&E staining of tumor sections. KLRC2 cDNA-expressing GL261 brain tumors show microcystic formations (arrow). (C) Representative immunohistochemical staining of tumor sections using a CD68 or a CD206-specific antibody. Quantification of each staining is shown above (n=5). (D–F) qRT-PCR analysis of the expression for several macrophage-related (D) immune-related (E) or KLRC2- associated (F) gene markers in GFP versus KLRC2 cDNA-expressing GL261 brain tumors (n=7). (G–H) Flow cytometry assessment of immune cell population percentages in human glioma categorized by KLRC2 expression levels (n=8). *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001. Scale: 2.5 mm and 100 µm (B) 50 µm (C). cDNA, complementary DNA; GFP, green fluorescent protein; qRT-PCR, quantitative real time-polymerase chain reaction.
To validate the findings from the mouse model, we examined a cohort of patient tumors that were previously characterized for tumor microenvironment composition via flow cytometry analysis.17 We categorized the tumors based on high or low KLRC2 expression as measured by qRT-PCR. No differences were observed in the number of leukocytes, lymphocytes, microglia, or total myeloid cells in the groups of tumors (figure 4G). However, a detailed analysis of myeloid populations revealed a significant decrease in the number of myeloid-derived suppressor cells and macrophages in glioma expressing high KLRC2 levels (figure 4H).
Glioma cell KLRC2 overexpression enhances the antitumor effect of PD-1 blockade in mice
Overall, our results suggest that KLRC2 expression in neoplastic cells changes the immune profile of gliomas, reduces the number of immunosuppressive macrophages, and alters the expression of molecules involved in antigen presentation and antitumor activity. To analyze if these changes could affect glioma response to checkpoint inhibitors, two groups of animals injected with green fluorescent protein (GFP)-expressing control or KLRC2-overexpressing GL261 cells (figure 5A) were treated with a low dose of PD-1 mAb in parallel to the animals of figure 4 that received control IgG. The treatment induced a decrease in tumor burden in the KLRC2 group compared with the GFP group (figure 5B). Moreover, anti-PD-1 treatment only induced a significant delay in tumor growth after the injection of KLRC2-GL261 cells and not after the injection of control GFP cells (online supplemental figure 4A). The IHC analysis of PD-1 mAb treated tumors revealed a decreased number of myeloid cells, particularly those with a protumoral activity (figure 4C). Moreover, we observed again the presence of microcysts (online supplemental figure 4B) and the upregulation of HLA-G (encoded by H2-M3) (online supplemental figure 4C) after KLRC2 overexpression.
Figure 5. KLRC2 cDNA expression enhances the antitumor immune response against a brain tumor after treatment with PD-1 mAb. (A) qRT-PCR-based KLRC2 expression in tumor tissues comparing GFP control with KLRC2 in the PD-1 mAb-treated group (n=6). (B) Mouse survival rates post-injection of GFP (n=7) or KLRC2 (n=9) GL261 cells and treatment that is depicted by Kaplan-Meier curves (p value=0.0145). (C–D) Representative immunohistochemical staining of tumor sections using CD68 and CD206 (n=5) (C) or CD3 and CD8 (GFP, n=4; NKG2C, n=7) (D) antibodies. Quantification of each staining is shown on the top. (E) Heatmap of normalized expression levels of genes associated with the immune environment. The heatmap’s color gradation corresponds to the relative expression levels of targeted markers. *p<0.05, **p<0.01. Scale: 50 µm (C–D). GFP, Green Fluorescent Protein; mAb, monoclonal antibodies; PD-1, programmed cell death protein-1; qRT-PCR, quantitative real time-polymerase chain reaction.
In contrast to what we had observed in IgG-treated tumors, there was a clear upregulation of the number of CD3+ cells and a tendency for more CD8+ cells in KLRC2 overexpressing gliomas after PD-1 mAb treatment (figure 5D). Moreover, there was a strong activation of the expression for several genes expressed by NK cells and cytotoxic lymphocytes, including GZMB, NCR1, CCR1, TYROBP, XCR1, and CSF3 (online supplemental figure 4D) with a reduction in the transcription for several immunosuppressive markers including IDO1, CXCR1, and CD47 (online supplemental figure 4E).
We performed unsupervised clustering of the PD-1 mAb-treated samples according to the expression profile of immune environment-related genes. We were able to differentiate two large tumor clusters based on KLRC2 overexpression and confirmed the changes at the immune environment level (figure 5E). The heatmap showed that cluster 3, whose genes are directly related to the immune response mediated by lymphocytes or cytotoxicity driven by NK cells (online supplemental figure 4F) (online supplemental table 3C), is induced in KLRC2-overexpressing GL261 tumors. By contrast, the expression of cluster 5, that is, enriched in immunosuppressive genes (online supplemental table 3C), is higher in control tumors (figure 5E).
Tumor cell NKG2C overexpression as a biomarker for PD-1 mAb treatment response in patients with GBM
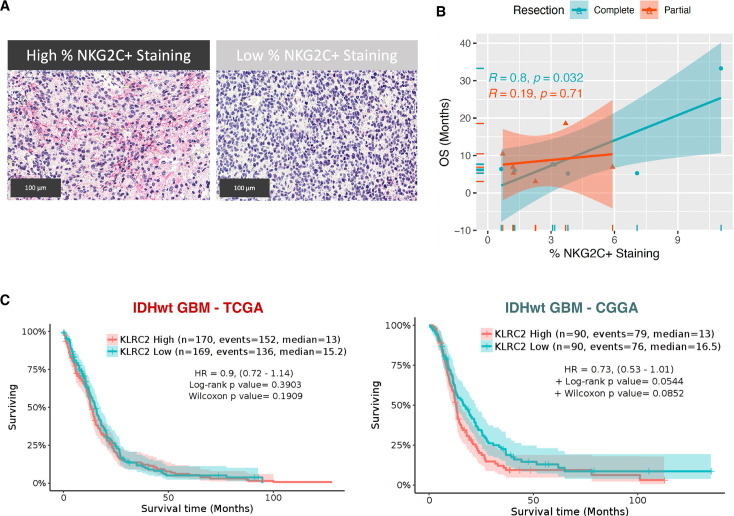
The results presented so far led us to hypothesize that NKG2C expression in glioma cells could induce a distinct immune pattern that favors the antitumor response in the presence of PD-1 mAb treatment. To test this hypothesis in patients, we performed an IHC analysis using an NKG2C-specific antibody in a cohort of baseline samples from patients who had been treated with neoadjuvant nivolumab.9 We categorized patients based on the extent of surgical resection, distinguishing those receiving complete resection (>95% of tumor removal) from those with partial or non-complete resections (between 80% and 95% of tumor removal). Interestingly, the amount of NKG2C staining (figure 6A) in samples before nivolumab treatment correlated positively with overall survival of the patients, but only for those receiving complete surgical resections (figure 6B). Notably, KLRC2 expression in primary or recurrent tumors does not have a prognostic value in the absence of immunotherapy (figure 6C).
Figure 6. Relationship between NKG2C Expression and PD-1 mAb treatment efficacy in IDHwt patients with GBM. (A) Representative image of the immunohistochemical analysis (red) of NKG2C in patient tumor sections. (B) Correlation analysis between NKG2C+staining and overall survival (OS), stratified by the extent of tumor resection (complete (n=7) or partial (n=6)). The correlation coefficient (R) and p values are provided for each resection group. (C) Kaplan-Meier survival curves of IDHwt GBM divided according to the KLRC2 expression in the TCGA and CGGA studies. Scale: 100 µm. CGGA, Chinese Glioma Genome Atlas; GBM, glioblastoma; IDHwt, isocitrate dehydrogenase-wild type; mAb, monoclonal antibodies; PD-1, programmed cell death protein-1; TCGA, the Cancer Genome Atlas.
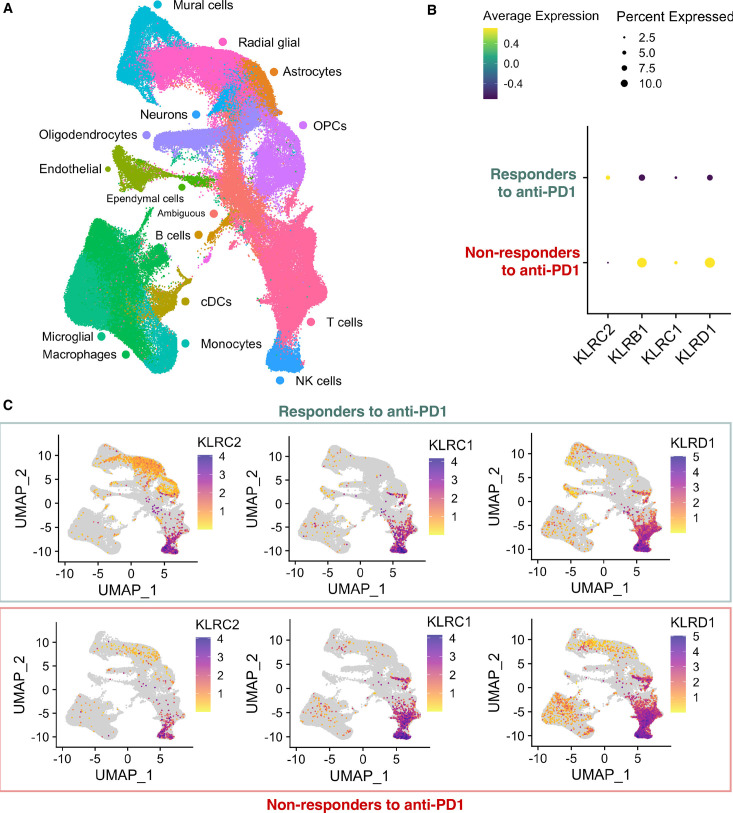
To further validate the value of NKG2C expression as a biomarker to predict the response to PD-1 mAb treatment, we conducted an in-silico analysis of a recently published scRNA-seq data set from patients receiving pembrolizumab and anlotinib (a tyrosine kinase inhibitor) neoadjuvant combination therapy (figure 7A). The authors classified the group of patients as responders if their tumor volume either remained stable or continuously shrunk over at least 3 months or non-responders.22 Remarkably, the expression of KLRC2 was higher in tumors from patients who responded to immunotherapy, with a particular enrichment in glial cell clusters, including radial glial cells, astrocytes and oligodendrocyte progenitor cells (OPCs) between responders and non-responders (figure 7B,C) (online supplemental figure 5A). Furthermore, our analysis revealed that other NK activating receptors (NKCR1, NCR2, NCR3, KLRK1, or KLRC3) did not exhibit significant overexpression in the responders’ group when assessed using the predefined criteria (adjusted p value≤1.0E-04 and average log2 fold change ≥1.0) (online supplemental table 3D). Notably, the expression of NKG2A/CD94 (encoded by KLRC1/KLRD1) (figure 7B–C), as well as other inhibitory receptors (online supplemental figure 5B,C), showed the opposite trend with higher expression in the non-responding tumors.
Figure 7. Association of the neoplastic expression of KLRC2 and other NK receptors with the response to anti-PD-1 in patients. (A) UMAP plot annotated by cell type within the isocitrate dehydrogenase-wild type glioblastoma single cell RNA sequencing database (CRA011176) (online supplemental table 1C). (B) Dot plot showing the percentage and average expression of KLRC2, KLRB1, KLRC1, and KLRD1 markers among responders and non-responders to anti-PD-1. The dot.min value was set to 0.01, meaning only genes with expression levels above 1% are represented by a dot. (C) UMAP plot of cells with positive KLRC2, KLRC1, and KLRD1 expression among responder and non-responder patients. cDCs, conventional dendritic cells; NK, natural killer; OPCs, oligodendrocyte progenitor cells; PD-1, programmed cell death protein-1; UMAP, uniform manifold approximation and projection.
These results reinforce the idea that the immune remodeling induced by NKG2C expression in glioma cells does not have a strong impact on tumor burden without immunotherapy. In contrast, NKG2C expression facilitates GBM to become more sensitive to immune checkpoint inhibitor treatment and the quantification of this receptor becomes useful as a biomarker of response to PD-1 mAb-based immunotherapies.
Discussion/conclusion
NK cell receptors have emerged as possible regulators of the tumor immune microenvironment. However, a detailed characterization of NK-related genes in glioma has been missing until now. Our comprehensive analysis of the expression of these genes confirms previous reports showing high expression of NK receptors in NK and T cells in GBM.13 However, we described here for the first time the expression of NKG2C in neoplastic glial cells. This adds to the well-known plasticity of GBM cells that have been shown to acquire myeloid markers to attract additional immunosuppressive macrophages into the tumor milieu and further promote immune evasion.31 In fact, GBM cells can transition between different glioma phenotypes, and even transform into endothelial cells or pericytes to fuel tumor growth and to escape from therapies.32 KLRC3, encoding NKG2E and representing another NK activating receptor, was previously characterized as a gene that is differentially expressed in GBM cancer stem cells and promotes cell invasion and radioresistance through TYROBP (encoded by DAP12) and GSK3β.25 In our analysis, KLRC2 that encodes for NKG2C has an even stronger expression in glial cells and particularly at the GBM infiltrating margin, suggesting that this receptor could also induce a more invasive profile to the cells. Although we did not describe the mechanism of action NKG2C in glioma cells, we found an increase in TYROBP expression in KLRC2-overexpressing tumors suggesting a similar function to that of NKG2E, possibly through autocrine and paracrine interactions with other cells. However, we cannot discard the induction of cell-intrinsic pathways in the presence of NKG2C.
Using both a mouse model and samples from patients, our study reveals a novel function for KLRC2 expression by GBM cells, inducing a change in the immune profile with a clear reduction in the number of protumoral macrophages. While these changes do not impact tumor burden, they significantly enhance the response of tumors to immune checkpoint inhibition. This is in accordance with recent findings indicating that the depletion of immunosuppressive macrophages expressing CD7333 or SIGLEC922 synergizes with PD-1 or CTLA-4 mAbs. Additionally, the activation of MAPK, either through mutations in the BRAF/PTPB11 pathway present in 2–3% of GBM or through other mechanisms, has been consistently associated with longer overall survival in response to immunotherapy.34,36 Notably, the presence of phospho-ERK-positive cells in tumors was linked to the abundance of microglia with an inflammatory profile.34 These results underscore the critical role of macrophages in the response to immunotherapy and the opportunities for better treatment through combination therapies.37 Moreover, our results point to the fact that OPCs should also be considered as possible targets to modulate the immune profile of glioma. Notably, the highest accumulation of KLRC2 expression in patients responding to nivolumab was found in the oligodendrocyte cluster (figure 7), underscoring the relevance of oligodendrocyte lineage cells in the control of the immune microenvironment of glioma, as happens in other diseases where they can serve as antigen-presenting cells.38
Our results suggest that NKG2C accumulates in the peritumoral zone of GBM, a region that expands beyond the contrast-enhancing tumor core and contains infiltrating tumor cells that are supposed to be responsible for the recurrence of GBM.39,41 Although the tumor microenvironment of the extratumoral zone of GBM is still poorly characterized, previous investigations have suggested that is enriched in microglial cells, while the intratumoral region possesses a higher proportion of blood-derived macrophages42 43 and immunosuppressive markers.44 45 These findings collectively suggest that the microenvironment at the invasive front of glioma is more favorable to the activation of an antitumor immune response, at least in part due to NKG2C expression in glioma cells. Consequently, immunotherapies may exhibit greater efficacy against tumor cells residing in non-contrast-enhancing areas. In contrast, patients who have undergone non-complete resections may retain a more immunosuppressed residual disease. This could explain why NKG2C expression did not have a predictive value in patients who did not undergo complete resections and contained larger amounts of immunosuppressive tumor when they received PD-1 inhibitor treatment. It is surprizing that none of the published clinical trials have incorporated the extent of resection as a variable to study the response to checkpoint inhibitors despite its clear impact on patient survival and even in the absence of immunotherapies.46 47
Initial studies determined that while PD-1/PD-L1 expression can serve as a biomarker for response, as observed in other types of cancer, although the data in patients with GBM are controversial.36 48 Similarly, a high mutational burden does not appear to be a robust predictor of response in GBM.49 Our results highlight the relevance of characterizing the macrophage population and the repertoire of antigen-presenting molecules50 and NK receptors (NKG2C in particular) to identify gliomas with a higher probability of responding to immune checkpoint regulators. Additionally, it will be highly interesting to determine whether the predictive value of the proposed biomarkers could be enhanced when assessed in the peripheral zone rather than in the tumor core. In any case, this combinatorial approach could refine the categorical decision-making for future trials with checkpoint inhibitors or other immunotherapies in patients with GBM.
Another important corollary of our study is the relevance of NK receptors as checkpoint modulators of the immune profile for GBM that reinforces the potential of NK cell-based immunotherapy as a novel GBM treatment strategy.51 52 In GBM, there is a high expression of the non-classical HLA-E and HLA-G, which contribute to the expression of peptides that interact with NK cells expressing NKG2A/CD94 (the inhibitory receptor) or those expressing NKG2C/CD94, which could have an important anti-glioma effect.53 Based on our results, we can propose that NKG2C-positive cells or specific peptides to activate this receptor54 should be tested in patients with GBM in synergism with PD-1 inhibitor treatments or other immunotherapies.
supplementary material
Footnotes
Funding: This work was supported in part by Ministerio de Ciencia, Innovación y Universidades and FEDER funds: grants PI20/00434 (MER-R), RTC2019- 006860-1 (MER-R), PI21/01406 (JMS-S), PI22/01171 (RG), PI21/01168 (AP-N), PI21CIII/00002 (PS-G), TED2021-132318B-I00 (PS-G), P2022/BMD-7344 (PS-G), Sara Borrell Contracts CD21/00080 (BS-C), CD22CIII/00001(OdD), Miguel Servet Contract CP21/00116 (RG); by National Institutes of Health (NIH) grants: R01NS097851 (DAW), R01NS129835 (DAW), K02AG068617 (DAW); by Cancer Research UK (C18915/A29362) (MER-R); by American Cancer Society grant RSG-21-058-01-CCE (DAW); by the American Brain Tumor Association Research Collaboration Grant ARC2300007 (PS-G, DAW); and by Fundación Científica Asociación Española Contra el Cáncer (FC-AECC) Ideas Semilla grant IDEAS20095SÁNC (PS-G).
Provenance and peer review: Not commissioned; externally peer reviewed.
Patient consent for publication: Not applicable.
Ethics approval: This study involves human participants and was approved by Comité de Ética de la Investigación del Hospital 12 de Octubre (CEI_21/480 and CEI_18/02). Participants gave informed consent to participate in the study before taking part.
Data availability free text: The data that support the findings of this study and the materials used are available on request from the corresponding authors.
Contributor Information
Olaya de Dios, Email: olaya.dedios@isciii.es.
M Angeles Ramírez-González, Email: ma.ramirez@externos.isciii.es.
Irene Gómez-Soria, Email: i.gomez@externos.isciii.es.
Berta Segura-Collar, Email: bsegura.imas12@salud.madrid.org.
Juliana Manosalva, Email: j.manosalva@isciii.es.
Diego Megías, Email: diego.megias@isciii.es.
Carlos E De Andrea, Email: ceandrea@unav.es.
Leticia Fernández-Rubio, Email: lfernandezru@unav.es.
Aurelio Hernández-Laín, Email: aurelio.hlain@salud.madrid.org.
Juan M Sepúlveda-Sánchez, Email: juanmanuel.sepulveda@salud.madrid.org.
Maria E Rodriguez-Ruiz, Email: mrruiz@unav.es.
Ángel Pérez-Núñez, Email: apnunez@salud.madrid.org.
Derek A Wainwright, Email: dwainwr@luc.edu.
Ricardo Gargini, Email: ricgargini.imas12@h12o.es.
Pilar Sánchez-Gómez, Email: psanchezg@isciii.es.
Data availability statement
Data are available upon reasonable request.
References
- 1.Louis DN, Perry A, Wesseling P, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol . 2021;23:1231–51. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity. 2018;48:812–30. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gieryng A, Pszczolkowska D, Walentynowicz KA, et al. Immune microenvironment of gliomas. Lab Invest. 2017;97:498–518. doi: 10.1038/labinvest.2017.19. [DOI] [PubMed] [Google Scholar]
- 4.Friedmann-Morvinski D, Hambardzumyan D. Monocyte-neutrophil entanglement in glioblastoma. J Clin Invest. 2023;133:e163451. doi: 10.1172/JCI163451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitt MJ, Company C, Dramaretska Y, et al. Phenotypic Mapping of Pathologic Cross-Talk between Glioblastoma and Innate Immune Cells by Synthetic Genetic Tracing. Cancer Discov. 2021;11:754–77. doi: 10.1158/2159-8290.CD-20-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma P, Allison JP. Dissecting the mechanisms of immune checkpoint therapy. Nat Rev Immunol. 2020;20:75–6. doi: 10.1038/s41577-020-0275-8. [DOI] [PubMed] [Google Scholar]
- 7.Omuro A, Reardon DA, Sampson JH, et al. Nivolumab plus radiotherapy with or without temozolomide in newly diagnosed glioblastoma: Results from exploratory phase I cohorts of CheckMate 143. Neurooncol Adv. 2022;4:vdac025. doi: 10.1093/noajnl/vdac025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim M, Weller M, Idbaih A, et al. Phase III trial of chemoradiotherapy with temozolomide plus nivolumab or placebo for newly diagnosed glioblastoma with methylatedMGMTpromoter. Neuro-oncology. 2022;24:1935–49. doi: 10.1093/neuonc/noac116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schalper KA, Rodriguez-Ruiz ME, Diez-Valle R, et al. Neoadjuvant nivolumab modifies the tumor immune microenvironment in resectable glioblastoma. Nat Med. 2019;25:470–6. doi: 10.1038/s41591-018-0339-5. [DOI] [PubMed] [Google Scholar]
- 10.Reardon DA, Brandes AA, Omuro A, et al. Effect of Nivolumab vs Bevacizumab in Patients With Recurrent Glioblastoma: The CheckMate 143 Phase 3 Randomized Clinical Trial. JAMA Oncol. 2020;6:1003–10. doi: 10.1001/jamaoncol.2020.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tawbi HA, Forsyth PA, Algazi A, et al. Combined Nivolumab and Ipilimumab in Melanoma Metastatic to the Brain. N Engl J Med. 2018;379:722–30. doi: 10.1056/NEJMoa1805453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frazao A, Rethacker L, Messaoudene M, et al. NKG2D/NKG2-Ligand Pathway Offers New Opportunities in Cancer Treatment. Front Immunol. 2019;10:661. doi: 10.3389/fimmu.2019.00661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathewson ND, Ashenberg O, Tirosh I, et al. Inhibitory CD161 receptor identified in glioma-infiltrating Tcells by single-cell analysis. Cell. 2021;184:1281–98. doi: 10.1016/j.cell.2021.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeung JT, Hamilton RL, Ohnishi K, et al. LOH in the HLA class I region at 6p21 is associated with shorter survival in newly diagnosed adult glioblastoma. Clin Cancer Res. 2013;19:1816–26. doi: 10.1158/1078-0432.CCR-12-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lerner EC, Woroniecka KI, D’Anniballe VM, et al. CD8+ T cells maintain killing of MHC-I-negative tumor cells through the NKG2D-NKG2DL axis. Nat Cancer . 2023;4:1258–72. doi: 10.1038/s43018-023-00600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernández-SanMiguel E, Gargini R, Cejalvo T, et al. Ocoxin Modulates Cancer Stem Cells and M2 Macrophage Polarization in Glioblastoma. Oxid Med Cell Longev. 2019;2019:9719730. doi: 10.1155/2019/9719730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cejalvo T, Gargini R, Segura-Collar B, et al. Immune Profiling of Gliomas Reveals a Connection with IDH1/2 Mutations, Tau Function and the Vascular Phenotype. Cancers (Basel) 12:3230. doi: 10.3390/cancers12113230. n.d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang AC, Vest RT, Kern F, et al. A human brain vascular atlas reveals diverse mediators of Alzheimer’s risk. Nature New Biol. 2022;603:885–92. doi: 10.1038/s41586-021-04369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdelfattah N, Kumar P, Wang C, et al. Single-cell analysis of human glioma and immune cells identifies S100A4 as an immunotherapy target. Nat Commun. 2022;13:767. doi: 10.1038/s41467-022-28372-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–96. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao X, Liao J, Lu X, et al. scCATCH: Automatic Annotation on Cell Types of Clusters from Single-Cell RNA Sequencing Data. i Sci. 2020;23:100882. doi: 10.1016/j.isci.2020.100882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mei Y, Wang X, Zhang J, et al. Siglec-9 acts as an immune-checkpoint molecule on macrophages in glioblastoma, restricting T-cell priming and immunotherapy response. Nat Cancer . 2023;4:1273–91. doi: 10.1038/s43018-023-00598-9. [DOI] [PubMed] [Google Scholar]
- 23.Yu K, Hu Y, Wu F, et al. Surveying brain tumor heterogeneity by single-cell RNA-sequencing of multi-sector biopsies. Natl Sci Rev. 2020;7:1306–18. doi: 10.1093/nsr/nwaa099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raudvere U, Kolberg L, Kuzmin I, et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–8. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheray M, Bessette B, Lacroix A, et al. KLRC3, a Natural Killer receptor gene, is a key factor involved in glioblastoma tumourigenesis and aggressiveness. J Cell Mol Med. 2017;21:244–53. doi: 10.1111/jcmm.12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segura-Collar B, Garranzo-Asensio M, Herranz B, et al. Tumor-Derived Pericytes Driven by EGFR Mutations Govern the Vascular and Immune Microenvironment of Gliomas. Cancer Res. 2021;81:2142–56. doi: 10.1158/0008-5472.CAN-20-3558. [DOI] [PubMed] [Google Scholar]
- 27.Al-Dalahmah O, Argenziano MG, Kannan A, et al. Re-convolving the compositional landscape of primary and recurrent glioblastoma reveals prognostic and targetable tissue states. Nat Commun. 2023;14:2586. doi: 10.1038/s41467-023-38186-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brooks LJ, Clements MP, Burden JJ, et al. The white matter is a pro-differentiative niche for glioblastoma. Nat Commun. 2021;12:2184. doi: 10.1038/s41467-021-22225-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komori T. Grading of adult diffuse gliomas according to the 2021 WHO Classification of Tumors of the Central Nervous System. Lab Invest. 2022;102:126–33. doi: 10.1038/s41374-021-00667-6. [DOI] [PubMed] [Google Scholar]
- 30.Burger PC, Green SB. Patient age, histologic features, and length of survival in patients with glioblastoma multiforme. Cancer. 1987;59:1617–25. doi: 10.1002/1097-0142(19870501)59:9<1617::aid-cncr2820590916>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 31.Gangoso E, Southgate B, Bradley L, et al. Glioblastomas acquire myeloid-affiliated transcriptional programs via epigenetic immunoediting to elicit immune evasion. Cell. 2021;184:2454–70. doi: 10.1016/j.cell.2021.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segura-Collar B, Mata-Martínez P, Hernández-Laín A, et al. Blood-Brain Barrier Disruption: A Common Driver of Central Nervous System Diseases. Neuroscientist. 2022;28:222–37. doi: 10.1177/1073858420985838. [DOI] [PubMed] [Google Scholar]
- 33.Goswami S, Walle T, Cornish AE, et al. Immune profiling of human tumors identifies CD73 as a combinatorial target in glioblastoma. Nat Med. 2020;26:39–46. doi: 10.1038/s41591-019-0694-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arrieta VA, Chen AX, Kane JR, et al. ERK1/2 phosphorylation predicts survival following anti-PD-1 immunotherapy in recurrent glioblastoma. Nat Cancer . 2021;2:1372–86. doi: 10.1038/s43018-021-00260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arrieta VA, Duerinck J, Burdett KB, et al. ERK1/2 Phosphorylation Predicts Survival in Recurrent Glioblastoma Following Intracerebral and Adjuvant PD-1/CTLA-4 Immunotherapy: A REMARK-guided Analysis. Clin Cancer Res. 2024;30:379–88. doi: 10.1158/1078-0432.CCR-23-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao J, Chen AX, Gartrell RD, et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med. 2019;25:462–9. doi: 10.1038/s41591-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sampson JH, Gunn MD, Fecci PE, et al. Brain immunology and immunotherapy in brain tumours. Nat Rev Cancer. 2020;20:12–25. doi: 10.1038/s41568-019-0224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrington EP, Bergles DE, Calabresi PA. Immune cell modulation of oligodendrocyte lineage cells. Neurosci Lett. 2020;715:134601. doi: 10.1016/j.neulet.2019.134601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemée J-M, Clavreul A, Menei P. Intratumoral heterogeneity in glioblastoma: don’t forget the peritumoral brain zone. Neuro Oncol . 2015;17:1322–32. doi: 10.1093/neuonc/nov119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giambra M, Di Cristofori A, Valtorta S, et al. The peritumoral brain zone in glioblastoma: where we are and where we are going. J of Neuroscience Research . 2023;101:199–216. doi: 10.1002/jnr.25134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Diaz C, Pöysti A, Mereu E, et al. Glioblastoma cell fate is differentially regulated by the microenvironments of the tumor bulk and infiltrative margin. Cell Rep. 2023;42:112472. doi: 10.1016/j.celrep.2023.112472. [DOI] [PubMed] [Google Scholar]
- 42.Darmanis S, Sloan SA, Croote D, et al. Single-Cell RNA-Seq Analysis of Infiltrating Neoplastic Cells at the Migrating Front of Human Glioblastoma. Cell Rep. 2017;21:1399–410. doi: 10.1016/j.celrep.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Z, Feng X, Herting CJ, et al. Cellular and Molecular Identity of Tumor-Associated Macrophages in Glioblastoma. Cancer Res. 2017;77:2266–78. doi: 10.1158/0008-5472.CAN-16-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura R, Ohara K, Sasaki H, et al. Difference in Immunosuppressive Cells Between Peritumoral Area and Tumor Core in Glioblastoma. World Neurosurg. 2018;120:e601–10. doi: 10.1016/j.wneu.2018.08.133. [DOI] [PubMed] [Google Scholar]
- 45.Rahimi Koshkaki H, Minasi S, Ugolini A, et al. Immunohistochemical Characterization of Immune Infiltrate in Tumor Microenvironment of Glioblastoma. J Pers Med. 2020;10:112. doi: 10.3390/jpm10030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karschnia P, Young JS, Dono A, et al. Prognostic validation of A new classification system for extent of resection in glioblastoma: Areport of the RANO resect group. Neuro Oncol. 2023;25:940–54. doi: 10.1093/neuonc/noac193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cahill DP. Extent of Resection of Glioblastoma: A Critical Evaluation in the Molecular Era. Neurosurg Clin N Am. 2021;32:23–9. doi: 10.1016/j.nec.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khasraw M, Reardon DA, Weller M, et al. PD-1 Inhibitors: Do they have a Future in the Treatment of Glioblastoma? Clin Cancer Res. 2020;26:5287–96. doi: 10.1158/1078-0432.CCR-20-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGrail DJ, Pilié PG, Rashid NU, et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol. 2021;32:661–72. doi: 10.1016/j.annonc.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simonds EF, Lu ED, Badillo O, et al. Deep immune profiling reveals targetable mechanisms of immune evasion in immune checkpoint inhibitor-refractory glioblastoma. J Immunother Cancer . 2021;9:e002181. doi: 10.1136/jitc-2020-002181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morimoto T, Nakazawa T, Maeoka R, et al. Natural Killer Cell-Based Immunotherapy against Glioblastoma. Int J Mol Sci . 2023;24:2111. doi: 10.3390/ijms24032111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Golán I, Rodríguez de la Fuente L, Costoya JA. NK Cell-Based Glioblastoma Immunotherapy. Cancers (Basel) 2018;10:10.:522. doi: 10.3390/cancers10120522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murad S, Michen S, Becker A, et al. NKG2C+ NK Cells for Immunotherapy of Glioblastoma Multiforme. Int J Mol Sci . 2022;23:10.:5857. doi: 10.3390/ijms23105857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huisman BD, Guan N, Rückert T, et al. High-throughput characterization of HLA-E-presented CD94/NKG2x ligands reveals peptides which modulate NK cell activation. Nat Commun . 2023;14:4809. doi: 10.1038/s41467-023-40220-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon reasonable request.